Abstract
Sphingosine 1-phosphate lyase (SGPL1) insufficiency (SPLIS) is a syndrome which presents with adrenal insufficiency, steroid-resistant nephrotic syndrome, hypothyroidism, neurological disease, and ichthyosis. Where a skin phenotype is reported, 94% had abnormalities such as ichthyosis, acanthosis, and hyperpigmentation. To elucidate the disease mechanism and the role SGPL1 plays in the skin barrier we established clustered regularly interspaced short palindromic repeats-Cas9 SGPL1 KO and a lentiviral-induced SGPL1 overexpression (OE) in telomerase reverse-transcriptase immortalised human keratinocytes (N/TERT-1) and thereafter organotypic skin equivalents. Loss of SGPL1 caused an accumulation of S1P, sphingosine, and ceramides, while its overexpression caused a reduction of these species. RNAseq analysis showed perturbations in sphingolipid pathway genes, particularly in SGPL1_KO, and our gene set enrichment analysis revealed polar opposite differential gene expression between SGPL1_KO and _OE in keratinocyte differentiation and Ca2+ signaling genesets. SGPL1_KO upregulated differentiation markers, while SGPL1_OE upregulated basal and proliferative markers. The advanced differentiation of SGPL1_KO was confirmed by 3D organotypic models that also presented with a thickened and retained stratum corneum and a breakdown of E-cadherin junctions. We conclude that SPLIS associated ichthyosis is a multifaceted disease caused possibly by sphingolipid imbalance and excessive S1P signaling, leading to increased differentiation and an imbalance of the lipid lamellae throughout the epidermis.
Supplementary key words: lipid lamellae imbalance, sphingolipid accumulation, organotypics, pyridoxal 5′-phosphate-dependent aldehyde lyase, differentiation, calcium signaling, ceramides, gene set enrichment analysis, keratinocytes, epidermis
Sphingosine-1-phosphate lyase insufficiency (SPLIS) is a multisystemic disorder caused by bi-allelic loss-of-function mutations in sphingosine 1-phosphate lyase (SGPL1). The syndrome can present with multiple comorbidities including ichthyosis, primary adrenal insufficiency, steroid resistant nephrotic syndrome, hypothyroidism, neurological disease, and hypogonadism (1, 2, 3, 4). Mutations in SGPL1 are rare, morbidity significant, and 50% mortality in utero or early childhood (1, 2, 3, 4, 5). Where investigated, 94% had skin abnormalities such as ichthyosis, acanthosis, and hyperpigmentation (1, 2, 5). SPLIS-associated ichthyosis presents with orthokeratotic hyperkeratosis and a thickened stratum corneum (1, 2). Similarly, rodent KO of Sgpl1 or S1P-lyase inhibition, both result in mild epidermal hyperplasia and orthokeratotic hyperkeratosis (6).
SGPL1, an endoplasmic reticulum (ER) pyridoxal 5′-phosphate-dependent aldehyde lyase, performs the final degradative step in the sphingolipid pathway, irreversible cleavage of sphingosine-1-phosphate (S1P) (Fig. 1A). Patient fibroblasts and KO HeLa cells with no SGPL1 activity show increased levels of S1P and upstream accumulation of sphingolipid intermediates including ceramides and sphingosine, which are integral to the formation of the lipid lamellae, the protective barrier of the epidermis (7). Imbalance of the equimolar ratio of ceramides, cholesterol, and free fatty acids, either from defects in lipid trafficking (Harlequin ichthyosis) or mutations in the sphingolipid metabolic pathway (autosomal recessive congenital ichthyosis 9), has been shown to disrupt the barrier and may render it non-functional (8, 9, 10).
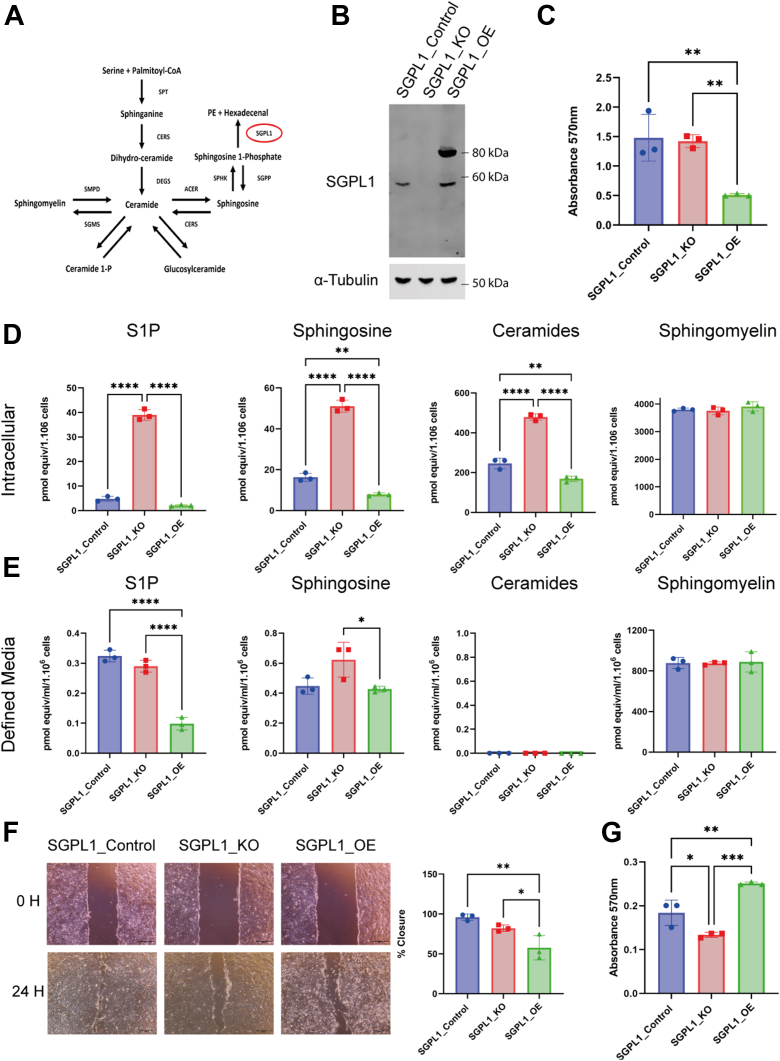
Fig. 1.
Loss and overexpression of SGPL1 results in a perturbance of sphingolipid species levels and significant changes in growth and migration. A: Schematic of the sphingolipid pathway where SGPL1 performs the final degradative process. B: SGPL1_Control shows a detectable band at 60 KDa, SGPL1_KO show no detectable band and SGPL1_OE show endogenous SGPL1 at 60 KDa and a band at 90 KDa representing SGPL1 linked to GFP. C: Proliferation rate measured of cells grown in serum-free media by MTT assay after 96 h. D, E: Levels of intracellular and extracellular sphingolipid intermediates in SGPL1_Control, SGPL1_KO and SGPL1_OE grown in FBS-containing media. F: Migration assay and quantification of wound gap closure (%) of cell grown in FBS media. (All statistical analyses were conducted using one-way ANOVAs with Tukey’s multiple comparison test, n = 3. Scale bar = 500 μm). G: Proliferation rates measured by MTT assay of cells grown in FBS after 96 h. SGPL1, Sphingosine 1-phosphate lyase.
S1P itself is a bioactive metabolite that promotes differentiation and inhibits proliferation of keratinocytes, through Ca2+ release and upregulation of p21 and p27 (11, 12, 13). S1P can function both intracellularly and extracellularly to increase [Ca2+]i, by signaling through one of five S1P receptors (receptor operated calcium entry) or potentially by acting as a secondary regulator of calcium mobilisation through store-operated calcium entry (SOCE) (12, 14, 15, 16, 17). In contrast, upstream sphingosine attenuates SOCE, as well as external calcium entry by blocking voltage operated calcium channels (18, 19) and can trigger calcium release from acidic stores such as lysosomes to increase [Ca2+]i (20). Ceramides can also lessen SOCE, elicit calcium release, and promote differentiation depending on their chain length, the number, and location of their hydroxyl groups (21). It is evident, therefore, that sphingolipid species have pleiotropic effects on [Ca2+]i.
In this study, using cells and 3D organotypic keratinocyte cultures, we show that SPLIS-associated ichthyosis is due to a multifaceted process. Loss of SGPL1 causes accumulation of S1P and upstream sphingolipids, including ceramides. High levels of S1P result in increased differentiation due to depletion of ER Ca2+ stores and concurrently, this heightened differentiation causes early release of lipid species throughout the epidermis and an imbalance of the lipid lamellae. We also show a dichotomy between SGPL1 loss and overexpression with cells displaying a polarity in differentiation and proliferation.
Materials and methods
Cell culture
The N/TERT-1 cell line, telomerase reverse-transcriptase immortalised human keratinocytes, was derived from foreskin tissue and supplied by Professor Rheinwald (Harvard Medical School) (22). Cells were grown in FAD media ((3:1 (v/v) DMEM:F12) 10% FBS, 5 μg/ml insulin, 0.4 μg/ml hydrocortisone, 10−10 M cholera enterotoxin, 10 ng/ml EGF, and 1% penicillin/streptomycin). FBS was substituted with charcoal stripped FBS and cells were grown for one passage before being plated for experiments as indicated. Human dermal fibroblasts were isolated from the dermis of donor abdominal skin from an adult female (a gift from the Philpott group, Blizard Institute, QMUL) and grown in DMEM, containing 10% FBS and 1% penicillin/streptomycin. To investigate monolayer differentiation, cells were grown in EpiLife low-calcium media (Gibco MEPI500CA) supplemented with Human Keratinocyte Growth Supplement (Gibco S0015).
Clustered regularly interspaced short palindromic repeats KO of SGPL1
SGPL1 KO nTERT cells were produced using second generation lentiviral transduction using lenti clustered regularly interspaced short palindromic repeats (CRISPR) v2 (Addgene #52961), pCMVΔR8.2 (Addgene #12263) and pDM2.G (Addgene #12259). CRISPR guides published by Dr Brügger’s group, designed to be cloned in using BBS1, were as follows FWD; 5′-CACCGTAATTGCATGGAGTGTCGTG-3′, REV; 5′-AAACCACGACACTCCATGCAATTAC-3′ (23). HEK293T cells were transfected using PEI with 800 ng transfer plasmid and 600 ng of both packaging and envelope plasmids. After 48 h nTERTs were transduced with virus containing media at a ratio of 3:1 (normal media: virus containing media). Media was changed to normal growth media after 24 h and cells were left to grow for 72 h. Positively transduced cells were selected with puromycin (2 mg/ml), single cell sorted and propagated from a 96-well plate. Clones were initially tested for loss of SGPL1 expression through Western blot. Genomic DNA was extracted from potential KO clones (Nucleon BACC3 gDNA Extraction kit) and SGPL1 exon 3 was amplified using the following primers FWD; 5′-GGAATGACCTTGCCCTTG-3′ and REV; 5′-TCAGCAATTAGTTTGAGGCTAGT-3′. DNA was sequenced by Sanger Sequencing (Eurofins Genomics) and mutations were identified and translated using BioEdit Sequence Alignment Editor. Control cells were produced using the same method but with no guide RNA cloned into the LentiCRISPR plasmid.
Creation of overexpression cell line
nTERTs overexpressing SGPL1 were created by lentiviral transduction of WT nTERTS using pCMV-SGPL1-C-GFPspark (Origene), containing SGPL1 linked to GFP, in the same method as above. After seven days, cells were selected for GFP and propagated. SGPL1 overexpression was validated by Western blot.
Western blot analysis
Protein lysates were prepared using radioimmunoprecipitation assay buffer (Thermo Fisher Scientific) and protease inhibitors (Roche) and quantified using a bicinchoninic acid assay (ThermoFisher Scientific). Twenty micrograms of protein was resolved on 4–12% SDS-PAGE gradient gels and transferred to a nitrocellulose membrane. The following primary antibodies were used: SGPL1 (1:500, R & D systems, AF5535), Loricrin (1:500, Biolegend, #905103), Alpha-Tubulin (1:10,000, Abcam, ab4074), and GAPDH (1:10,000, Abcam, ab8245). Bands were visualised using IRDye secondary antibodies and an Odyssey machine (LI-COR). Band intensities were measured using ImageJ (NIH, Bethesda, MD, https://imagej.net/software/fiji/) and normalised against corresponding loading controls. SGPL1 expression in SGPL1_OE was quantified with endogenous and lentiviral expression collectively.
Lipidomics
Cells were grown in 10 cm2 dishes to ∼70% confluency in triplicate. Media was centrifuged to remove cells and frozen at 80°C. Cells were trypsinised, counted, and 2 × 106 cells were pelleted and frozen at −80°C. Lipidomic analysis for components of the sphingolipid pathway were at the Research Unit on BioActive Molecules, Department of Biomedicinal Chemistry IQAC-CSIC, Barcelona (Spain).
Seven hundred fifty microliters of a methanol-chloroform (2:1, vol/vol) solution containing internal standards (N-dodecanoylsphingosine, N-dodecanoylglucosylsphingosine, N-dodecanoylsphingosylphosphorylcholine, C17-sphinganine, and C17-sphinganine-1-phosphate, 0.2 nmol each, from Avanti Polar Lipids) were added to samples. Samples were extracted at 48°C overnight and cooled, 75 μl of 1 M KOH in methanol was added, and the mixture was incubated for 2 h at 37°C. Following addition of 75 μl of 1 M acetic acid, samples were evaporated to dryness, and stored at −20°C until the analysis of sphingolipids. Before the analysis 150 μl of methanol were add to the samples, centrifuged at 13,000 g for 5 min, and 130 μl of the supernatant were transferred to a new vial and injected.
Lipids were measured with an Acquity ultraperformance liquid chromatography (UPLC) system (Waters) connected to a TOF (LCT Premier XE) detector controlled with Waters/Micromass MassLynx software (https://www.waters.com/waters/en_US/MassLynx-Mass-Spectrometry-Software-/nav.htm?cid=513164&locale=en_US). The acquisition range of the TOF detector was m/z 50–1,500, the capillary voltage was set to 3.0 kV, the desolvation temperature was 350°C, and the desolvation gas flow rate was 600 l/h. For long chain base-phosphate quantification, analysis of the extracts was performed by UPLC-MS/MS with a system consisting of a Acquity UPLC system (Waters) connected to a triple-quadrupole mass spectrometer (Xevo TQ-S, Waters) and controlled with Waters/Micromass MassLynx software. Positive identification of compounds was based on the accurate mass measurement with an error <5 ppm and its LC retention time, compared with that of standards (Cer and SM with the following acyl chain C16:0, C18:0, C20:0; C24:0, C24:1, So, dhSo, S1P, and C36:1-PE). Quantification with UPLC-TOF instrument was carried out using the extracted ion chromatogram of each compound, using 50 mDa windows. Values for the supernatant were normalised by ml and total cell number. Experiment consisted of three biological repeats per cell line.
Proliferation
Proliferation was measured using an MTT assay. Cells were seeded at 5,000/well in multiple 96-well plates. After 96 h, supernatant was removed and 50 μl fresh media and 50 μl 12 mM MTT solution in PBS was added. After 4 h, all media was removed and 100 μl DMSO was added to solubilise the formazan product. After mixing the plate was read at 570 nm. Experiment consisted of three biological repeats each containing five technical repeats/plate.
Migration
In a 12 well plate, 200,000 cells were seeded and propagated to confluency. Cells were treated with 5 μg/ml of Mitomycin C for 2 h, scratched with a p20 tip, and imaged at 0 and 24 h. Experiment consisted of three biological repeats each containing three technical repeats/plate. Wound gap at 0 and 24 h was measured using ImageJ and the reduction in size was calculated as a percentage of the initial wound gap.
RNAseq
RNA was extracted from keratinocytes grown in FBS media when 70% confluent using Rneasy spin columns according to the manufacturer’s instructions (Qiagen). RNA was assessed for quantity and quality using the Nanodrop 8000 spectrophotometer and 15 μl of RNA at 10 ng/μl was sent to Eurofins Genomics (Germany) for library preparation and sequencing. All fastq files have been deposited in the NCBI Gene Expression Omnibus (GEO) repository (GSE207499).
Primary bioinformatic analysis was conducted using the DNA Nexus platform. Reads data underwent quality control using FastQC Reads Quality Control version 3.0.1 (28/10/2020). Read alignment was performed by HISAT2 version 1.03 (1/6/2017) (24) and differential expression analysis was done by subread_featureCounts version 0.1.0 (21/3/2019) (25) and DESEQ2 (R.3.2-packages-quantification modified 11/12/2017) (26). Secondary analysis was performed using the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) (27, 28) and the gene set enrichment analysis (GSEA) software (27) (https://www.gsea-msigdb.org/gsea/index.jsp). Differentially expressed gene (DEGs) were classified as those with an adjusted P < 0.05. Analysis of DEGs and creation of heatmaps and dotplots was performed using R Studio software (https://posit.co/downloads/).
3D model generation
3D organotypic models were created as previously described (29). Briefly, a Type 1 collagen matrix, populated with primary human dermal fibroblasts was seeded into a transwell insert and left to polymerise for 1 h. SGPL1_Control or SGPL1_KO keratinocytes were subsequently seeded atop the matrix. Models were lifted to the air-liquid interface and grown for 2 weeks. After two weeks models were cut from the insert with the membrane, fixed with 4% paraformaldehyde, and embedded in paraffin. Models were sectioned at 5 μm.
Immunofluorescence and histological analysis
Paraffin sections were hydrated using decreasing concentrations of ethanol and either stained with H&E or underwent antigen retrieval (heat induced epitope retrieval) prior to immunofluorescent staining. Primary antibodies used were Keratin 14 (1:100, CRUK), Keratin 10 (1:150, Abcam, ab76318), Involucrin (1:100, Thermo Fisher Scientific, #MA5-11803), Loricrin (1:200, Biolegend, #905103), Filaggrin (1:500, SCBT, sc66192), and E-Cadherin (1:50, Abcam, ab1416). Images were captured using an Epifluorescent Leica Microscope.
Statistical analysis
All statistical analysis was conducted using Graphpad Prism 9 (Graphpad software Inc, CA) (https://www.graphpad.com/features). All test were either one-way ANOVAs or t tests and are detailed in figure legends. For all Figures; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 ∗∗∗∗P < 0.0001.
Some figures generated in this paper were created using Servier free medical art (https://smart.servier.com/).
Results
Proliferation and migration of keratinocytes is affected by loss or overexpression of SGPL1
We developed a CRISPR-Cas9 SGPL1_KO keratinocyte cell line using nTERTs, as a biologically relevant model of human skin previously utilised to study other forms of ichthyosis (Fig. 1B) (30, 31). SGPL1_KO cells had compound heterozygous mutations in exon 3 of SGPL1, [c.142_143insT] + [c.141_145delCGTGT], predicted to cause frameshifts and early protein truncations ([p.V48fs∗10] + [p.V48fs∗8], respectively). We also created a stable cell line overexpressing SGPL1 linked to GFP (SGPL1_OE) to further explore the effect of an imbalance in the sphingolipid rheostat and compared both to an isogenic control cell line (SGPL1_Control). Western blotting revealed the expected 60 kDa protein in SGPL1_Control, no protein in the SGPL1_KO, presumably because of nonsense-mediated RNA decay and 8-fold overexpression in SGPL1_OE (Fig. 1B).
We performed LC-MS of sphingolipid species to determine the effect of loss or overexpression of SGPL1 on upstream sphingolipids. Analysis of cell pellets to examine intracellular species indicated that loss of SGPL1 resulted, as expected, in accumulation of intracellular S1P, as well as upstream sphingolipids; sphingosine and ceramide species (Fig. 1D). Overexpression of SGPL1 reduced S1P levels somewhat (P = 0.13), and significantly decreased sphingosine and ceramide species, indicating full functionality of the GFP-tagged enzyme (Fig. 1D). No change in sphingomyelin was detected in either case.
LC-MS analysis of the media the cells were grown in showed SGPL1 loss had no effect on extracellular S1P and possibly increased sphingosine (P = 0.06) but overexpression resulted in a significant decrease in S1P compared to SGPL1_Control and SGPL1_KO (Fig. 1E), while it had no effect on sphingosine. No ceramide species were detected and neither loss nor overexpression of SGPL1 had any effect on sphingomyelin levels. Levels of extracellular S1P and sphingosine were markedly lower than intracellular (S1P levels were 0.32 pmol/ml/1 × 106 cells compared to 4.7 pmol/1 × 106 cells respectively for SGPL1_Control).
SGPL1_OE grew at a significantly slower rate than SGPL1_Control or SGPL1_KO with no significant difference between the latter two (Fig. 1C). The migratory capacity of cells showed a similar pattern to proliferation. There was no difference between the migratory speed of SGPL1_Control and SGPL1_KO but SGPL1_OE closed the gap significantly slower (Fig. 1F). Taken together, these data suggest that depletion of extracellular sphingolipid species, impacts the rate of cell proliferation and migration.
However, when cells were grown in serum-free media, the loss of SGPL1 caused a reduction in growth rate while its overexpression resulted in an increased growth rate (Fig. 1G) in keeping with previous reports (11, 32) and here reflecting intracellular sphingolipid levels. This reversal in growth rate of SGPL1_OE highlights the complex interplay of intracellular and extracellular S1P signaling. To allow for extracellular S1P signalling, as well as the creation of 3D organotypic models, the cells were grown in FBS-containing media for all further experiments.
SGPL1 disruption highlights its importance to oxidative phosphorylation/CYP450 action
Principal component analysis of RNAseq data demonstrated good separation between the three cell types (Fig. 2A). Using a threshold of adjusted P < 0.05, analysis showed 6,405 genes were differentially expressed between SGPL1_Control and SGPL1_KO, 2,151 between SGPL1_Control and SGPL1_OE, and 6,837 between SGPL1_KO and SGPL1_OE (Fig. 2B). 833 DEGs were common between all three comparisons and heatmap visualisation displays the contrasting effects of KO and OE of SGPL1 on these genes (Fig. 2C and supplemental Table S1).
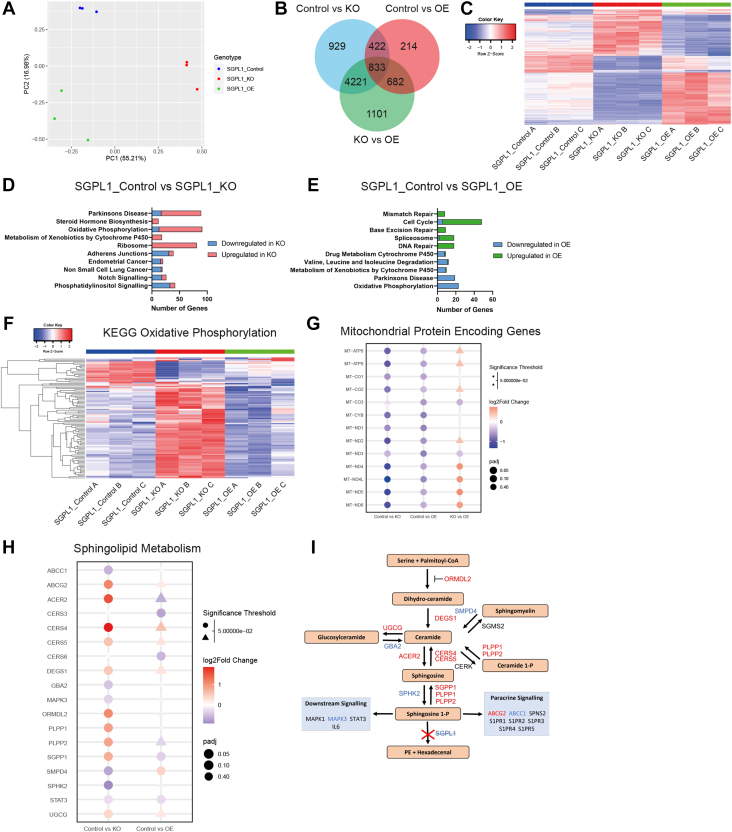
Fig. 2.
RNAseq analysis shows loss and overexpression of SGPL1 causes a dichotomy of gene expression. A: Principal component analysis (PCA plot) displaying the nine samples used for RNAseq analysis with samples clustered by genotype. B: Venn diagram of differentially expressed genes displaying the number of genes present in each comparison SGPL1_Control versus SGPL1_KO, SGPL1_Control versus SGPL1_OE, and SGPL1_KO versus SGPL1_OE. C: Heatmap showing the z-scores of all genes common between all three above comparisons. D, E: The top five enriched pathways in each direction for SGPL1_KO and SGPL1_OE compared to SGPL1_Control. The number of DEGs in each gene set is demonstrated on the x-axis. F: Heatmap showing the z scores of all genes in the KEGG oxidative phosphorylation. G: Dotplot showing the P values and log2Fold change of mitochondrial encoded genes in all comparisons. H: Dotplot showing the P values and log2Fold change of SGPL1_Control versus SGPL1_KO and SGPL1_Control versus SGPL1_OE of genes encoding enzymes involved in the sphingolipid metabolism pathway. I: Sphingolipid metabolism pathway with enzymes colour coded according to log2fold change of SGPL1_Control versus SGPL1_KO. Genes upregulated in SGPL1_KO are red and genes downregulated are blue. SGPL1, Sphingosine 1-phosphate lyase.
Reflecting this, GSEA revealed a dichotomy between the effect of loss and overexpression of SGPL1 (Fig. 2D, E). Mitochondrial-associated gene sets “Oxidative Phosphorylation” and “Parkinson’s Disease” were enriched in both SGPL1_Control versus SGPL1_KO and SGPL1_Control versus SGPL1_OE comparisons, with the majority of genes upregulated in SGPL1_KO and downregulated in SGPL1_OE. A further enriched gene set in SGPL1_KO was KEGG_Ribosome (KEGG is the Kyoto Encyclopedia of Genes and Genomes) (https://www.genome.jp/kegg/pathway/hsa/hsa03010.html, accessed 09.08.22), 81 of 88 genes were upregulated, implying an increase in translation within the cell. In SGPL1_OE the top five most enriched gene sets are all involved in cell proliferation, including cell cycle, mismatch repair and DNA repair (Fig. 2E, supplemental Tables S2 and S3).
Further analysis of the oxidative phosphorylation KEGG gene set (https://www.genome.jp/kegg/pathway/hsa/hsa00190.html, accessed 09.08.22) demonstrated that of a total of 132 genes, 91 were differentially expressed in SGPL1_KO (13 downregulated and 78 upregulated), and 23 (all downregulated) in SGPL1_OE (P < 0.05) (Fig. 2F). Interestingly, all 13 mitochondrial protein encoding genes were downregulated in SGPL1_OE and 12 of the 13 were downregulated in SGPL1_KO, with only MT_CO3 not significantly differentially expressed in SGPL1_KO (Fig. 2G). Comparison of mitochondrial protein encoding genes between SGPL1_KO and SGPL1_OE revealed that loss of SGPL1 possibly causes greater mitochondrial perturbation with four genes significantly lower in SGPL1_KO and a further four displaying a decreasing trend. Of the remaining 10 downregulated DEGs in SGPL1_OE, eight were upregulated in SGPL1_KO displaying an opposite expression in nuclear encoded oxidative phosphorylation genes.
Sphingolipid pathway enzyme regulation is affected by loss and overexpression of SGPL1
Differential expression of genes encoding sphingolipid enzymes was investigated, Fig. 2H shows a dot plot of all DEGs in the pathway for either SGPL1_Control versus SGPL1_KO or SGPL1_Control versus SGPL1_OE analysis and Fig. 2I shows their location in the pathway (colour coded for SGPL1_Control vs. SGPL1_KO analysis). Strikingly, SGPP1, PLPP1, and PLPP2 were all upregulated in SGPL1_KO, and SPHK2 downregulated, indicating that S1P is being dephosphorylated to sphingosine. Concurrently, CERS4 and CERS5, which convert sphingosine to ceramide are upregulated, SMPD4, which converts sphingomyelin to ceramide is downregulated, and ORMDL2, which inhibits the production of dihydro-ceramide, the start of the de novo synthesis pathway, is upregulated. Furthermore, UGCG (conversion of ceramide to glucosylceramide) is upregulated and its converse, GBA2, is downregulated. The ATP-binding cassette transporter ABCG2, which transports S1P out of cells (33), is upregulated indicating that SGPL1_KO are attempting to export excess S1P. There is however no difference in S1P levels in the media between SGPL1_Control and SGPL1_KO cells (Fig. 1E). Taken together this indicated SGPL1_KO respond to high levels of S1P, sphingosine and ceramide by inhibiting de novo synthesis, downregulating the salvage pathway, exporting S1P, and promoting conversion back to ceramide.
Despite a reduction in sphingosine and ceramide in the SGPL1_OE, we see fewer DEGs in the pathway. Only two genes were upregulated SGPL1, as expected, and SMPD4, possibly to increase ceramide levels and three downregulated genes: SGPP1, CERS3, and CERS6. OE cells may therefore upregulate the salvage pathway and downregulate specific enzymes to replenish sphingosine and S1P levels.
Analysis of other lipid metabolic pathways shows the disruption that loss or overexpression of SGPL1 has on wider lipid metabolism. Of the 42 genes in the “KEGG fatty acid metabolism” geneset (https://www.gsea-msigdb.org/gsea/msigdb/human/geneset/KEGG_FATTY_ACID_METABOLISM.html, accessed 11.11.22) 17 were differentially expressed in SGPL1_KO (14 upregulated, 3 downregulated) and 12 in SGPL1_OE (2 upregulated, 10 downregulated) (supplemental Fig. S1 and supplemental Table S4). In particular this highlighted upregulation of enzymes involved in fatty acid degradation (KEGG pathway hsa00071) in SGPL1_KO compared to SGPL1_Control (ACAA1, ACAA2, ACADM, ACADSB, ADH5, ALDH3A2, ALDH7A1, ALDH9A1, ECHS1, ECI1, ECI2, HADH, HADHA, HADHB) with 9/10 downregulated genes in SGPL1_OE also in this pathway (ACADS, ACOX1, ACOX3, ACSL1, ALDH7A1, CPT1A, CPT2, HADHA, HADHB) with four common to both comparisons (ACADM, ALDH7A1, HADHA, and HADHB). Taken together this suggests that the loss or gain of SGPL1 affects the wider lipidome.
Genes involved in keratinocyte differentiation and calcium regulation are differentially expressed
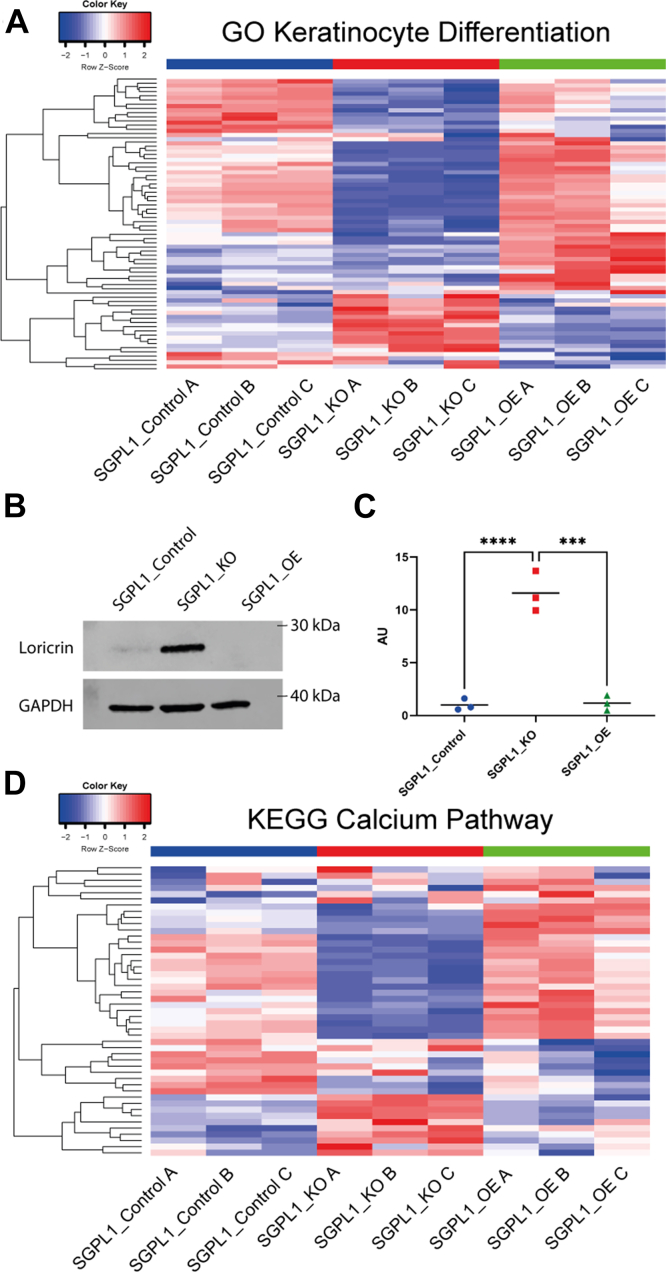
We investigated the transcriptomes for specific gene sets involved in keratinocyte differentiation. Analysis of the 142 genes in the “GOBP Keratinocyte Differentiation” gene set (http://www.gsea-msigdb.org/gsea/msigdb/cards/GOBP_KERATINOCYTE_DIFFERENTIATION.html, accessed 09.08.22) revealed that in SGPL1_KO, when compared to SGPL1_Control there were 60 DEGs with 38 genes downregulated and 24 upregulated (P < 0.05) (Fig. 3A and supplemental Table S5). The gene with the highest fold change (log2fold change; 2.09) was RARRES3 (PLAAT4/TIG3), a retinoid response protein that promotes keratinocyte terminal differentiation and inhibits proliferation (34, 35). The following three most upregulated genes were CASP14, which is confined to differentiating keratinocytes and plays a role in barrier formation, giving rise to autosomal recessive congenital ichthyosis 12 when mutated (36, 37), ALOX15B, needed for barrier function (38), and AKR1C3, required for differentiation-associated gene regulation and colocalizes with KRT10, which is also upregulated (39). In SGPL1_OE there were fewer DEGs (32, 17 downregulated and 15 upregulated) (Fig. 3A and supplemental Table S6) with 13 genes displaying a mirrored effect to SGPL1_KO (Fig. 3A) (AQP3, AKR1C3, SGPP1, KLK5, KRT10, ASAH1, CLIC4, SFN, TGM1, CDH3, FLNB, KRT17, ZBED2). Furthermore, differentiation marker involucrin is downregulated, as is ABCA12, a protein localised to the granular layer in vivo (31). Increased protein expression of the terminal differentiation marker loricrin confirmed that in monolayer culture SGPL1_KO cells were at an advanced differentiated state (Fig. 3B, C).
Fig. 3.
Loss and overexpression of SGPL1 causes a dichotomy in growth rate and overexpression results in a lower differentiated state. A: Heatmap showing the z scores of all genes in the “GO Keratinocyte Differentiation” gene set. B: Western blot of terminal differentiation marker loricrin of cell lysates extracted from cells grown in monolayer in FBS media. C: Densitometry analysis of loricrin Western blot. (One-way ANOVA with Tukey’s multiple comparison test, n = 3). D: Heatmap showing the z scores of all genes in the “KEGG Calcium Pathway” gene set. SGPL1, Sphingosine 1-phosphate lyase.
With calcium the main driver of keratinocyte differentiation, we analyzed the expression of genes in the KEGG calcium signaling pathway (178 genes) (https://www.genome.jp/kegg/pathway/hsa/hsa04020.html, accessed 09.08.22). In SGPL1_KO there were 39 DEGs with 18 genes upregulated and 21 downregulated, a larger response than SGPL1_OE which had 15 DEGs, seven upregulated and eight downregulated (Fig. 3B, Supplemental Tables S5 and S6) The three genes with the largest negative fold change in SGPL1_KO were PDE1C, ITPR3, and PLCG1, all of which are involved in the S1P-mediated ER calcium release through S1PR3, suggesting SGPL1_KO downregulated the S1PR3 pathway, possibly due to high intracellular S1P levels. Interestingly all calmodulin genes were upregulated, further suggesting a response to high [Ca2+]i. In SGPL1_OE, PDE1C, EGFR, and ADCY3 were upregulated, all of which were downregulated in SGPL1_KO. This demonstrates further the dichotomy between the effects of loss and overexpression of SGPL1.
SGPL1 KO models demonstrate heightened differentiation, a thickened corneum, and aberrant intercellular junctions
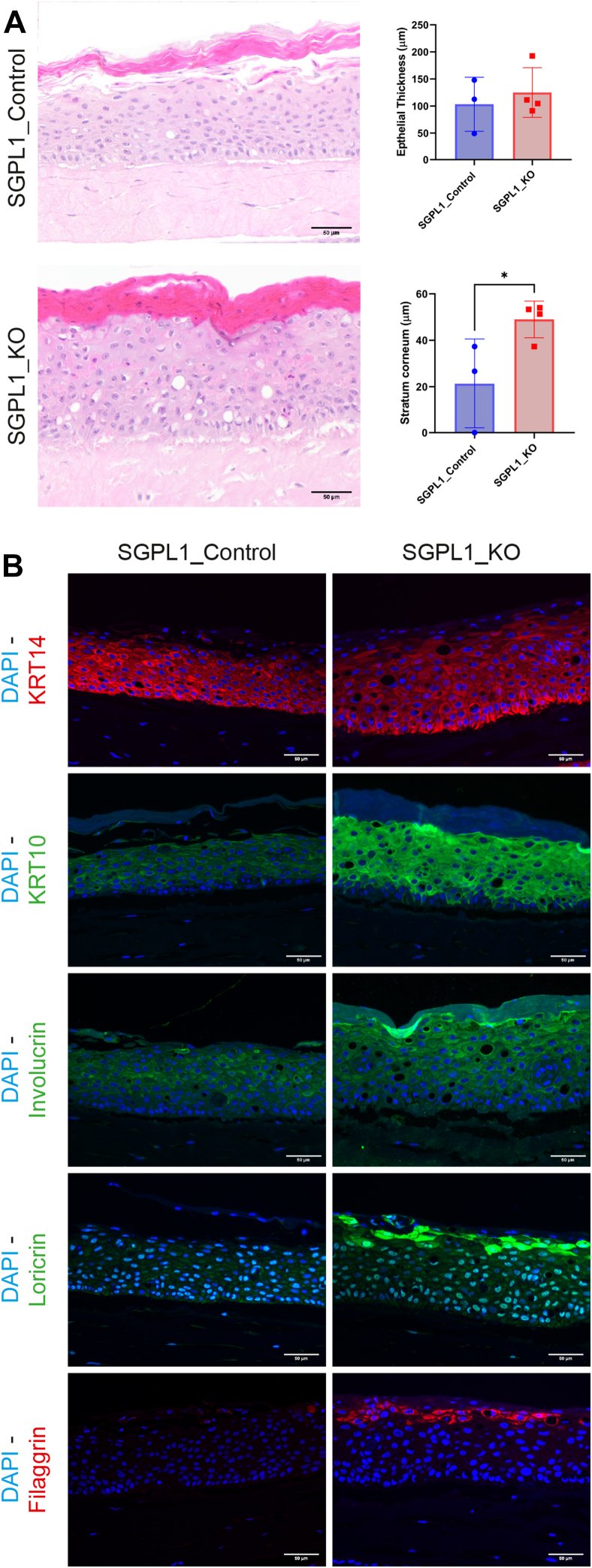
To further analyze the contrasting differentiation of the cell lines in a more in vivo like environment and to investigate whether SGPL1_KO recreated the SPLIS-associated ichthyosis phenotype in vitro we constructed 3D organotypic models of both SGPL1_Control and SGPL1_KO. Both models displayed stratification and differentiation. SGPL1_KO models produced a significantly thicker stratum corneum, with no difference in overall epidermal thickness (Fig. 4A). We saw little difference in basal marker keratin 14 but differentiation markers keratin 10, involucrin, loricrin, and filaggrin all displayed markedly higher expression in SGPL1_KO models (Fig. 4B). Images were taken at the same exposure for comparative analysis. Normal differentiation marker expression is present in SGPL1_Control models (supplemental Fig. S2). This confirmed that when SGPL1_KO were grown in the presence of extracellular calcium and in 3D, an environment closely mimicking in vivo, they displayed markedly more differentiation. Further differences between the models were found at cell-cell junctions; E-cadherin staining demonstrated a breakdown of intercellular junctions in SGPL1_KO models that was not observed in SGPL1_Control models (Fig. 5). Intracellular staining was also observed in SGPL1_KO models, indicating aberrant E-Cadherin processing.
Fig. 4.
SGPL1_KO models present with a thickened stratum corneum and increased differentiation. A: Representative H&E-stained images of SGPL1_Control and SGPL1_KO 3D organotypic models. Total epidermal and stratum corneum thickness were measured in μm using ImageJ. Scale bar = 50 μm. B: Immunofluorescent staining of organotypic models for differentiation markers KRT14, KRT10, involucrin, loricrin, and filaggrin. Images were captured at the same exposure and gain for comparison. Scale bar = 50 μm. (Statistical analysis was conducted using unpaired t test, n = three-fourths, ∗P < 0.05.). SGPL1, Sphingosine 1-phosphate lyase.
Fig. 5.
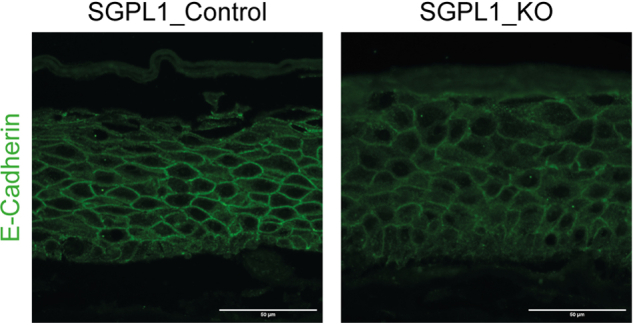
SGPL1_KO organotypic models display abnormal intercellular junctions. Immunofluorescent staining of SGPL1_Control and SGPL1_KO 3D organotypic models for junctional marker E-cadherin. Scale bar = 50 μm. SGPL1, Sphingosine 1-phosphate lyase.
Discussion
Little work has previously concentrated on SPLIS-associated ichthyosis. The effects of S1P on keratinocytes and its links to psoriasis and atopic dermatitis have been studied through extracellular S1P treatment and transient knockdown of SGPL1 but these methods fail to account for prolonged S1P exposure or upstream sphingolipid species accumulation over a significant time period (11, 32, 40). Here we have created SGPL1 KO and overexpression cell lines to account for this and investigated the complexity of the sphingolipid pathway and its role in the disease mechanism of SPLIS-associated ichthyosis.
It has previously been shown that accumulation of S1P in keratinocytes, grown in serum-free media, induces growth arrest and differentiation (11). Our results in similar serum-free conditions correlate with this as SGPL1_KO, which contains high levels of intracellular S1P, grow at a reduced rate. Concurrently, SGPL1_OE cells, which have lowered S1P and significantly less sphingosine and ceramides, grow at an increased rate. Previous research demonstrates that S1P upregulates the cell cycle inhibitors p21 and p27 and therefore reduced levels of S1P would result in less growth inhibition (11).
Interestingly however, this phenomenon was eradicated when grown in FBS-containing media where proliferation and migration of the cells followed a strikingly similar pattern to the S1P concentration in the media, with SGPL1_Control & SGPL1_KO>SGPL1_OE. In Sgpp1−/− mice, which also have increased S1P, overexpression of Ki67 and epidermal hyperplasia were observed and S1P has been shown to be proproliferative in other cell types in vitro (41, 42, 43). This implies that extracellular S1P could have a proproliferative capacity in keratinocytes, in contrast to its intracellular form. It is important to remember, however, that the reduction of sphingosine and ceramide species as well as media components could influence cell proliferation.
The upregulation of enzymes in the sphingolipid pathway indicates that SGPL1_KO cells actively respond to accumulation of S1P to promote conversion of downstream lipids to higher order species and inhibit de novo synthesis. Importantly the cells also downregulated the salvage pathway, responsible for 50–90% of sphingolipid biosynthesis while de novo synthesis contributes only 10% (44, 45). This active process highlights the cells attempt to maintain the sphingolipid rheostat by altering gene expression. No differences were observed in sphingomyelin concentrations in the cells or media. This is possibly due to the much higher abundance of sphingomyelin. Essential for cell membranes, intracellular levels were 794 times higher than that of S1P and therefore loss of SGPL1 may have little effect on sphingomyelin concentrations.
We also show that, at the RNA level, the loss or gain of SGPL1 expression affects more than just sphingolipid pathway enzymes, with differential expression of fatty acid degradation genes, upregulated in SGPL1_KO and downregulated in SGPL1_OEs. The fatty acid β-oxidation pathway is the major pathway for the degradation of fatty acids and its upregulation could be due to the increased accumulation of sphingolipids within the SGPL1_KO cells. What effect this has on fatty acid species in this model is currently unknown but altered levels could contribute to the phenotype here since it has also recently been shown that increased levels of short chain fatty acids promote skin barrier function and keratinocyte differentiation, potentially to protect against atopic dermatitis-like skin inflammation (46). GSEA analysis highlighted the enrichment of genes involved in Parkinson’s disease and oxidative phosphorylation in both SGPL1_KO and SGPL1_OE cells. Many genes in these gene sets are associated with the mitochondria and this supports previous findings that SGPL1 deficiency is associated with mitochondrial perturbation (7). The common downregulation of mitochondrial protein encoding genes in both SGPL_KO and _OE cell lines indicates that imbalance of the sphingolipid pathway in either direction could lead to a perturbation of mitochondrial function.
Interestingly, the top five enriched gene sets/pathways in SGPL1_OE are all involved in cell proliferation with the majority of DEGs being upregulated in SGPL1_OE, indicative of an increase in proliferation, which we observe in cells grown in serum-free conditions. A recent study of five different ichthyoses indicated that of the top 20 enriched pathways across all types of the disease, 16 were directly related to the cell cycle and replication (47). This identifies increased proliferation as a trait of ichthyosis and is of particular importance for the treatment of SPLIS-associated ichthyosis. Gene therapy rescuing the expression of SGPL1 would have to be carefully titred to ensure that reduced S1P levels did not have further detrimental effect.
The use of organotypic models of human skin allows for the investigation of keratinocyte differentiation in 3D, at the air-liquid interface, and allows paracrine signaling from fibroblasts, providing a superior model to monolayer culture. SPLIS-associated ichthyosis presents with an orthokeratotic hyperkeratosis and a thickened stratum corneum and this was observed in our organotypic models (1), which could therefore serve as a disease model for therapeutic interventions. It is important to note, however, that although organotypic models of skin have been beneficial for the study of the disease mechanism behind SPLIS-associated ichthyosis, they do have limitations, such as the lack of an immune component or other cells present in in vivo skin. Further lipidomic analysis could be conducted on such models to understand the wider lipidome required for skin barrier function within these tissue substitutes.
A breakdown of intercellular junctions could also contribute to the skin defect. E-Cadherin staining in SGPL1_KO models showed malformed junctions and intracellular retention. Darier Disease is caused by a mutation in ATP2A2, a sarco/ER Ca2+-ATPase pump , which when defective abolishes Ca2+ transport into the ER, causing high cytosolic Ca2+, low ER Ca2+, and abnormal keratinisation (46, 48, 49). Depletion of ER Ca2+ leads to protein misfolding and accumulation with consequent ER stress (49, 50). In Darier Disease, or cells treated with thapsigargin, an inhibitor of sarco/ER Ca2+, the processing of E-Cadherin, along with other junctional markers, is disturbed resulting in punctate staining and abnormal organisation, akin to our SGPL1_KO models (48, 49). Accumulation of S1P may therefore cause depletion of ER calcium and consequently impair processing of E-cadherin and intercellular junctions.
To conclude, we show that SPLIS-associated ichthyosis is a result of a multifaceted process. Our results reveal opposite effects of a loss or gain of SGPL1 function in 2D culture. While its loss leads to an increase in expression of differentiation genes and reduced growth, its overexpression leads to decreased differentiation and increased growth rate, an effect that is media-specific. This increased differentiation may be due to aberrant calcium handling and abnormal E-Cadherin localization. These are present in our 3D human skin equivalents and therefore highlights them as possible disease models for therapeutic interventions.
Data availability
The RNA-seq data sets were deposited in GEO (NCBI) public repository, GEO accession number: GSE207499.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Acknowledgments
The authors would like to thank Dr Jordan Lee for the dermal fibroblasts he provided for the 3D organotypic cultures. We are grateful to the Pathology Facility and the Light Microscopy Facility at Charterhouse Square for their help capturing and analysing data (Core Award C16420/A18066).
Author contributions
C. J. S., J. L. W., C. H., and J. C. investigation; C. J. S., J. L. W., C. H., and J. C. methodology; C. J. S., J. L. W., C. H., and J. C. formal analysis; C. J. S. and C. H. visualization; E. O. T. resources; C. J. S. writing-original draft; J. L. W., M. P. C., E. O. T., R. P., and L. A. M. writing-review and editing; R. P. and L. A. M. conceptualization; R. P. and L. A. M. supervision; R. P. and L. A. M funding acquisition.
Funding and additional information
This work was supported by Barts Charity grants MGU0361 to L. A. M. and R. P.; MGU0438 to L. A. M. and Medical Research Council (MRC) UK Clinical Academic Research Partner Grant (MR/T02402X/1) to R. P.
Supplemental data
Supplemental Figure S2.
References
- 1.Prasad R., Hadjidemetriou I., Maharaj A., Meimaridou E., Buonocore F., Saleem M., et al. Sphingosine-1-phosphate lyase mutations cause primary adrenal insufficiency and steroid-resistant nephrotic syndrome. J. Clin. Invest. 2017;127:942–953. doi: 10.1172/JCI90171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lovric S., Goncalves S., Gee H.Y., Oskouian B., Srinivas H., Choi W.I., et al. Mutations in sphingosine-1-phosphate lyase cause nephrosis with ichthyosis and adrenal insufficiency. J. Clin. Invest. 2017;127:912–928. doi: 10.1172/JCI89626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janecke A.R., Xu R., Steichen-Gersdorf E., Waldegger S., Entenmann A., Giner T., et al. Deficiency of the sphingosine-1-phosphate lyase SGPL1 is associated with congenital nephrotic syndrome and congenital adrenal calcifications. Hum. Mutat. 2017;38:365–372. doi: 10.1002/humu.23192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi Y.J., Saba J.D. Sphingosine phosphate lyase insufficiency syndrome (SPLIS): a novel inborn error of sphingolipid metabolism. Adv. Biol. Regul. 2019;71:128–140. doi: 10.1016/j.jbior.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linhares N.D., Arantes R.R., Araujo S.A., Pena S.D.J. Nephrotic syndrome and adrenal insufficiency caused by a variant in SGPL1. Clin. Kidney J. 2018;11:462–467. doi: 10.1093/ckj/sfx130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumann J., Grevot A., Ledieu D., Wolf A., Schubart A., Piaia A., et al. Reduced activity of sphingosine-1-phosphate lyase induces podocyte-related glomerular proteinuria, skin irritation, and platelet activation. Toxicol. Pathol. 2015;43:694–703. doi: 10.1177/0192623314565650. [DOI] [PubMed] [Google Scholar]
- 7.Maharaj A., Williams J., Bradshaw T., Güran T., Braslavsky D., Casas J., et al. Sphingosine-1-phosphate lyase (SGPL1) deficiency is associated with mitochondrial dysfunction. J. Steroid Biochem. Mol. Biol. 2020;202:105730. doi: 10.1016/j.jsbmb.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akiyama M., Sugiyama-Nakagiri Y., Sakai K., McMillan J.R., Goto M., Arita K., et al. Mutations in lipid transporter ABCA12 in harlequin ichthyosis and functional recovery by corrective gene transfer. J. Clin. Invest. 2005;115:1777–1784. doi: 10.1172/JCI24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelsell D.P., Norgett E.E., Unsworth H., Teh M.T., Cullup T., Mein C.A., et al. Mutations in ABCA12 underlie the severe congenital skin disease harlequin ichthyosis. Am. J. Hum. Genet. 2005;76:794–803. doi: 10.1086/429844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckl K.M., Tidhar R., Thiele H., Oji V., Hausser I., Brodesser S., et al. Impaired epidermal ceramide synthesis causes autosomal recessive congenital ichthyosis and reveals the importance of ceramide acyl chain length. J. Invest. Dermatol. 2013;133:2202–2211. doi: 10.1038/jid.2013.153. [DOI] [PubMed] [Google Scholar]
- 11.Jeon S., Song J., Lee D., Kim G.T., Park S.H., Shin D.Y., et al. Inhibition of sphingosine 1-phosphate lyase activates human keratinocyte differentiation and attenuates psoriasis in mice. J. Lipid Res. 2020;61:20–32. doi: 10.1194/jlr.RA119000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lichte K., Rossi R., Danneberg K., ter Braak M., Kurschner U., Jakobs K.H., et al. Lysophospholipid receptor-mediated calcium signaling in human keratinocytes. J. Invest. Dermatol. 2008;128:1487–1498. doi: 10.1038/sj.jid.5701207. [DOI] [PubMed] [Google Scholar]
- 13.Hong J.H., Youm J.K., Kwon M.J., Park B.D., Lee Y.M., Lee S.I., et al. K6PC-5, a direct activator of sphingosine kinase 1, promotes epidermal differentiation through intracellular Ca2+ signaling. J. Invest. Dermatol. 2008;128:2166–2178. doi: 10.1038/jid.2008.66. [DOI] [PubMed] [Google Scholar]
- 14.Blom T., Slotte J.P., Pitson S.M., Törnquist K. Enhancement of intracellular sphingosine-1-phosphate production by inositol 1,4,5-trisphosphate-evoked calcium mobilisation in HEK-293 cells: endogenous sphingosine-1-phosphate as a modulator of the calcium response. Cell. Signal. 2005;17:827–836. doi: 10.1016/j.cellsig.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Pulli I., Asghar M.Y., Kemppainen K., Törnquist K. Sphingolipid-mediated calcium signaling and its pathological effects. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:1668–1677. doi: 10.1016/j.bbamcr.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Hopson K.P., Truelove J., Chun J., Wang Y., Waeber C. S1P activates store-operated calcium entry via receptor- and non-receptor-mediated pathways in vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 2011;300:C919–C926. doi: 10.1152/ajpcell.00350.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujii K., Machida T., Iizuka K., Hirafuji M. Sphingosine 1-phosphate increases an intracellular Ca2+ concentration via S1P3 receptor in cultured vascular smooth muscle cells. J. Pharm. Pharmacol. 2014;66:802–810. doi: 10.1111/jphp.12214. [DOI] [PubMed] [Google Scholar]
- 18.Calloway N., Vig M., Kinet J.P., Holowka D., Baird B. Molecular clustering of STIM1 with Orai1/CRACM1 at the plasma membrane depends dynamically on depletion of Ca2+ stores and on electrostatic interactions. Mol. Biol. Cell. 2009;20:389–399. doi: 10.1091/mbc.E07-11-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Titievsky A., Titievskaya I., Pasternack M., Kaila K., Törnquist K. Sphingosine inhibits voltage-operated calcium channels in GH4C1 cells. J. Biol. Chem. 1998;273:242–247. doi: 10.1074/jbc.273.1.242. [DOI] [PubMed] [Google Scholar]
- 20.Höglinger D., Haberkant P., Aguilera-Romero A., Riezman H., Porter F.D., Platt F.M., et al. Intracellular sphingosine releases calcium from lysosomes. Elife. 2015;4 doi: 10.7554/eLife.10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon Y.B., Kim C.D., Youm J.-K., Gwak H.S., Park B.D., Lee S.H., et al. Novel synthetic ceramide derivatives increase intracellular calcium levels and promote epidermal keratinocyte differentiation. J. Lipid Res. 2007;48:1936–1943. doi: 10.1194/jlr.M700185-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Dickson M.A., Hahn W.C., Ino Y., Ronfard V., Wu J.Y., Weinberg R.A., et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerl M.J., Bittl V., Kirchner S., Sachsenheimer T., Brunner H.L., Luchtenborg C., et al. Sphingosine-1-phosphate lyase deficient cells as a tool to study protein lipid interactions. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 26.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith C.J., Parkinson E.K., Yang J., Pratten J., O'Toole E.A., Caley M.P., et al. Investigating wound healing characteristics of gingival and skin keratinocytes in organotypic cultures. J. Dent. 2022;125:104251. doi: 10.1016/j.jdent.2022.104251. [DOI] [PubMed] [Google Scholar]
- 30.Smits J.P.H., Niehues H., Rikken G., van Vlijmen-Willems I., van de Zande G., Zeeuwen P., et al. Immortalized N/TERT keratinocytes as an alternative cell source in 3D human epidermal models. Sci. Rep. 2017;7:11838. doi: 10.1038/s41598-017-12041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enjalbert F., Dewan P., Caley M.P., Jones E.M., Morse M.A., Kelsell D.P., et al. 3D model of harlequin ichthyosis reveals inflammatory therapeutic targets. J. Clin. Invest. 2020;130:4798–4810. doi: 10.1172/JCI132987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogler R., Sauer B., Kim D.S., Schafer-Korting M., Kleuser B. Sphingosine-1-phosphate and its potentially paradoxical effects on critical parameters of cutaneous wound healing. J. Invest. Dermatol. 2003;120:693–700. doi: 10.1046/j.1523-1747.2003.12096.x. [DOI] [PubMed] [Google Scholar]
- 33.Takabe K., Kim R.H., Allegood J.C., Mitra P., Ramachandran S., Nagahashi M., et al. Estradiol induces export of sphingosine 1-phosphate from breast cancer cells via ABCC1 and ABCG2. J. Biol. Chem. 2010;285:10477–10486. doi: 10.1074/jbc.M109.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scharadin T.M., Eckert R.L. TIG3: an important regulator of keratinocyte proliferation and survival. J. Invest. Dermatol. 2014;134:1811–1816. doi: 10.1038/jid.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jans R., Sturniolo M.T., Eckert R.L. Localization of the TIG3 transglutaminase interaction domain and demonstration that the amino-terminal region is required for TIG3 function as a keratinocyte differentiation regulator. J. Invest. Dermatol. 2008;128:517–529. doi: 10.1038/sj.jid.5701035. [DOI] [PubMed] [Google Scholar]
- 36.Lippens S., Kockx M., Knaapen M., Mortier L., Polakowska R., Verheyen A., et al. Epidermal differentiation does not involve the pro-apoptotic executioner caspases, but is associated with caspase-14 induction and processing. Cell Death Differ. 2000;7:1218–1224. doi: 10.1038/sj.cdd.4400785. [DOI] [PubMed] [Google Scholar]
- 37.Rendl M., Ban J., Mrass P., Mayer C., Lengauer B., Eckhart L., et al. Caspase-14 expression by epidermal keratinocytes is regulated by retinoids in a differentiation-associated manner. J. Invest. Dermatol. 2002;119:1150–1155. doi: 10.1046/j.1523-1747.2002.19532.x. [DOI] [PubMed] [Google Scholar]
- 38.Kim S.N., Akindehin S., Kwon H.J., Son Y.H., Saha A., Jung Y.S., et al. Anti-inflammatory role of 15-lipoxygenase contributes to the maintenance of skin integrity in mice. Sci. Rep. 2018;8:8856. doi: 10.1038/s41598-018-27221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantel A., Carpenter-Mendini A.B., VanBuskirk J.B., De Benedetto A., Beck L.A., Pentland A.P. Aldo-keto reductase 1C3 is expressed in differentiated human epidermis, affects keratinocyte differentiation, and is upregulated in atopic dermatitis. J. Invest. Dermatol. 2012;132:1103–1110. doi: 10.1038/jid.2011.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Japtok L., Baumer W., Kleuser B. Sphingosine-1-phosphate as signaling molecule in the skin: relevance in atopic dermatitis. Allergo J. Int. 2014;23:54–59. doi: 10.1007/s40629-014-0008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young N., Van Brocklyn J.R. Roles of sphingosine-1-phosphate (S1P) receptors in malignant behavior of glioma cells. Differential effects of S1P2 on cell migration and invasiveness. Exp. Cell Res. 2007;313:1615–1627. doi: 10.1016/j.yexcr.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshino O., Yamada-Nomoto K., Kano K., Ono Y., Kobayashi M., Ito M., et al. Sphingosine 1 phosphate (S1P) increased IL-6 expression and cell growth in endometriotic cells. Reprod. Sci. 2019;26:1460–1467. doi: 10.1177/1933719119828112. [DOI] [PubMed] [Google Scholar]
- 43.Allende M.L., Sipe L.M., Tuymetova G., Wilson-Henjum K.L., Chen W., Proia R.L. Sphingosine-1-phosphate phosphatase 1 regulates keratinocyte differentiation and epidermal homeostasis. J. Biol. Chem. 2013;288:18381–18391. doi: 10.1074/jbc.M113.478420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillard B.K., Clement R.G., Marcus D.M. Variations among cell lines in the synthesis of sphingolipids in de novo and recycling pathways. Glycobiology. 1998;8:885–890. doi: 10.1093/glycob/8.9.885. [DOI] [PubMed] [Google Scholar]
- 45.Tettamanti G., Bassi R., Viani P., Riboni L. Salvage pathways in glycosphingolipid metabolism. Biochimie. 2003;85:423–437. doi: 10.1016/s0300-9084(03)00047-6. [DOI] [PubMed] [Google Scholar]
- 46.Sakuntabhai A., Ruiz-Perez V., Carter S., Jacobsen N., Burge S., Monk S., et al. Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nat. Genet. 1999;21:271–277. doi: 10.1038/6784. [DOI] [PubMed] [Google Scholar]
- 47.Kim M., Mikhaylov D., Rangel S.M., Pavel A.B., He H., Renert-Yuval Y., et al. Transcriptomic analysis of the major orphan ichthyosis subtypes reveals shared immune and barrier signatures. J. Invest. Dermatol. 2022;142:2363–2374. doi: 10.1016/j.jid.2022.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savignac M., Simon M., Edir A., Guibbal L., Hovnanian A. SERCA2 dysfunction in Darier disease causes endoplasmic reticulum stress and impaired cell-to-cell adhesion strength: rescue by Miglustat. J. Invest. Dermatol. 2014;134:1961–1970. doi: 10.1038/jid.2014.8. [DOI] [PubMed] [Google Scholar]
- 49.Celli A., Mackenzie D.S., Zhai Y., Tu C.L., Bikle D.D., Holleran W.M., et al. SERCA2-controlled Ca2+-dependent keratinocyte adhesion and differentiation is mediated via the sphingolipid pathway: a therapeutic target for Darier's disease. J. Invest. Dermatol. 2012;132:1188–1195. doi: 10.1038/jid.2011.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data sets were deposited in GEO (NCBI) public repository, GEO accession number: GSE207499.