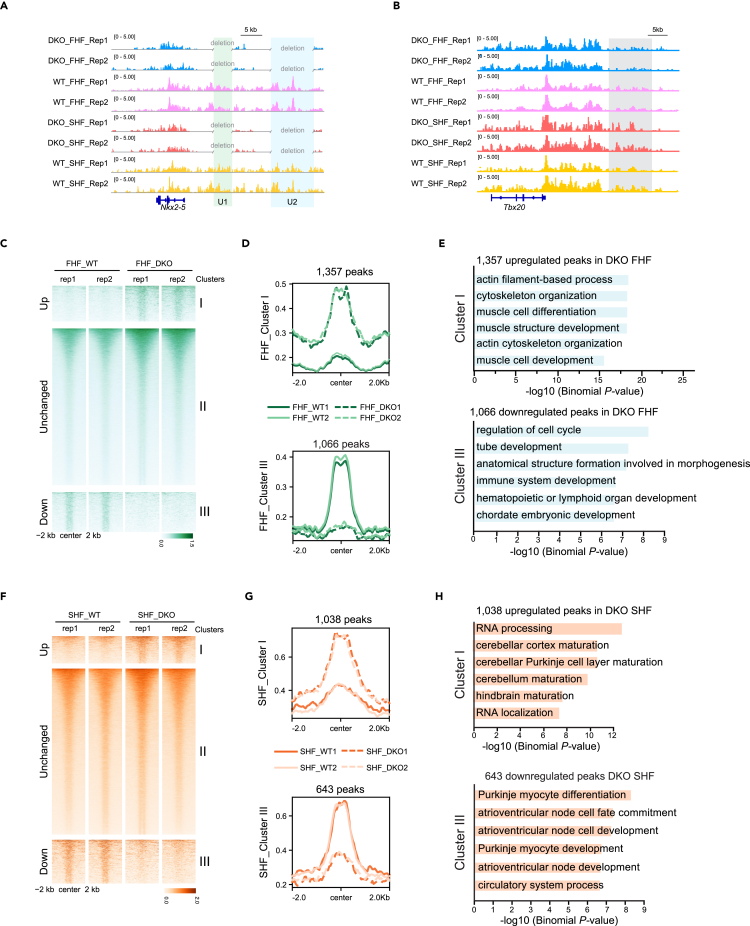
Figure 4.
Differential enhancer dynamics between FHF and SHF of E8.25 DKO and WT hearts
(A) Track view showing H3K27ac signals in FHF and SHF of E8.25 WT/DKO hearts around Nkx2-5 locus. U1 and U2 enhancers were highlighted by green and blue shading, respectively.
(B) Track view showing the H3K27ac in situ ChIP-seq signals in FHF and SHF of E8.25 WT/DKO hearts around the Tbx20 locus. Stronger H3K27ac signals around the Tbx20 enhancer in SHF of DKO hearts were highlighted by gray shading.
(C) Heatmaps showing H3K27ac signals at 60,836 peaks in E8.25 FHF of WT and DKO hearts. Groups consist of K-means clusters: cluster I (increased; n = 1,357), cluster II (unchanged; n = 58,413), and cluster III (decreased; n = 1,066). The heatmaps and aggregate plots were both generated with DeepTools.
(D) Aggregate plots of H3K27ac signals at ± 2 kb of peak centers of cluster I and cluster III in (C).
(E) Top 6 enriched GO terms of genes nearby upregulated and downregulated peaks in (C). p-value was calculated by Binomial test.
(F) Heatmaps showing H3K27ac signals at 44,375 peaks in E8.25 SHF of WT and DKO hearts. Groups consist of K-means clusters: cluster I (increased; n = 1,038), cluster II (unchanged; n = 42,694), and cluster III (decreased; n = 643). Heatmaps and aggregate plots were generated with DeepTools.
(G) Aggregate plots of H3K27ac signals at ± 2 kb of peak centers of cluster I and cluster III in (F).
(H) Top 6 enriched GO terms of genes nearby upregulated and downregulated peaks in (F). p-value was calculated by Binomial test. See also Figure S3.