Abstract
Msx2 is a homeobox gene expressed in multiple embryonic tissues which functions as a key mediator of numerous developmental processes. YY1 is a bi-functional zinc finger protein that serves as a repressor or activator to a variety of promoters. The role of YY1 during embryogenesis remains unknown. In this study, we report that Msx2 is regulated by YY1 through protein–DNA interactions. During embryogenesis, the expression pattern of YY1 was observed to overlap in part with that of Msx2. Most notably, during first branchial arch and limb development, both YY1 and Msx2 were highly expressed, and their patterns were complementary. To test the hypothesis that YY1 regulates Msx2 gene expression, P19 embryonal cells were used in a number of expression and binding assays. We discovered that, in these cells, YY1 activated endogenous Msx2 gene expression as well as Msx2 promoter–luciferase fusion gene activity. These biological activities were dependent on both the DNA binding and activation domains of YY1. In addition, YY1 bound specifically to three YY1 binding sites on the proximal promoter of Msx2 that accounted for this transactivation. Mutations introduced to these sites reduced the level of YY1 transactivation. As bone morphogenetic protein type 4 (BMP4) regulates Msx2 expression in embryonic tissues and in P19 cells, we further tested whether YY1 is the mediator of this BMP4 activity. BMP4 did not induce the expression of YY1 in early mouse mandibular explants, nor in P19 cells, suggesting that YY1 is not a required mediator of the BMP4 pathway in these tissues at this developmental stage. Taken together, these findings suggest that YY1 functions as an activator for the Msx2 gene, and that this regulation, which is independent of the BMP4 pathway, may be required during early mouse craniofacial and limb morphogenesis.
INTRODUCTION
During vertebrate embryogenesis, a group of transcription factors that contains a conserved DNA binding homeodomain controls pattern formation and organogenesis (1,2). One such factor is Msx2 and, despite extensive studies of its involvement in developmental outcomes, the upstream molecular regulation of Msx2 remains to be fully understood. Msx2 is a member of the Msx family of homeobox genes expressed in a variety of embryonic tissues involved in epithelial–mesenchymal interactions, pattern formation and apoptosis (3–5). Mutations in the human MSX2 gene, along with corroborating genetically engineered mouse models, demonstrated that Msx2 determines sutural patency in the developing cranium (6–11). Msx2 is also involved in craniofacial morphogenesis including cranial neural crest cell apoptosis (12–14), and the formation of cartilage (15–17), tooth (18,19) and eye (20,21). During limb development, Msx2 is involved in interdigital apoptosis, limb outgrowth and the maintenance of the activity of the apical ectodermal ridge (AER) (22–24).
In order to identify regulators of Msx2 expression, the mouse and chick promoters of the gene have been molecularly dissected. Proximal promoter regions conferring specific expression to the AER of developing limbs have been identified (25,26). Another fragment directing expression to a subset of cells within the suture sites during embryonic, fetal and neonatal stages has also been described (9). The regulation of Msx2 in limb and calvarial tissues was thought to be disparate due to differences in the pattern of DNase I hypersensitive sites present (27). During development, although the Msx2 gene is often placed as a downstream target of the BMP4 signaling pathway, it remains unclear through which transcription factor(s) the promoter of Msx2 gene is regulated.
YY1 is a transcription factor with four zinc finger motifs. Three of the four finger motifs are members of the GLI family of zinc fingers (28). YY1 functions as a repressor in some promoters and an activator in other promoters depending on the context of the promoters (reviewed in 29–31). Apart from its zinc finger domains, YY1 protein contains both activation and repression domains (32,33). YY1 has also been found as an initiator sequence-binding protein that directs and activates transcription in vitro (34). Furthermore, YY1 influences transcription by protein–protein interaction with other transcription factors such as Sp1, c-Myc and p300 (35–39). Despite biochemical characterizations of transcriptional regulation by YY1, the role of YY1 during mammalian embryogenesis is unknown. Targeted disruption of YY1 in mice resulted in peri-implantation lethality and complete resorption by E8.5, due to failure in the formation of the egg cylinder (40). However, a subset of heterozygotes survived to mid-gestation, displayed growth retardation and neurulation defects suggesting that YY1 functions during neural development. The localization of YY1 also suggests that YY1 has additional roles during embryogenesis (40). As YY1 is present in many cell types, and its binding sites are distributed in a variety of cellular gene promoters, we propose that YY1 is expressed during mouse embryogenesis and regulates developmental specific genes such as Msx2.
In this report, we tested the hypothesis that YY1 regulates Msx2 transcription. We identified that YY1 and Msx2 expression patterns overlapped, especially in regions with elevated expression levels. We discovered that YY1 induced Msx2 expression, and activated the Msx2 promoter. We demonstrated that the transactivation of Msx2 by YY1 was mediated by protein–DNA interactions employing three YY1 binding sites on the Msx2 promoter. However, we also discovered that BMP4 did not induce the expression of YY1. We conclude that YY1 functions as an activator for the expression of the Msx2 gene, along a pathway that is independent of BMP4.
MATERIALS AND METHODS
Whole mount in situ hybridization
Whole mount in situ hybridization was performed as described previously (41). Briefly, mouse embryos of embryonic day E8, E10 and E12 were isolated and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS). The specimens were permeabilized with radioimmunoprecipitation buffer. After pre-hybridization, the specimens were hybridized with 1 mg/ml dideoxygenin (DIG)-labeled riboprobes at 70°C. The specimens were then washed, blocked and further incubated with alkaline phosphatase-conjugated anti-DIG antibody (Boehringer Mannheim, Indianapolis, IN) at a 1:2000 dilution at 4°C overnight. The bound alkaline phosphatase was colorized when incubated in nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrates (Sigma, St Louis, MO).
The 390 bp probe specific for mouse YY1 was subcloned as an XhoI/XbaI fragment generated by PCR from a mouse YY1 cDNA clone (gift from Dr Ozato) into pBluescript II KS+ (Stratagene, La Jolla, CA). The sequence was confirmed by DNA sequencing. The antisense and sense RNA probes were transcribed using T7 and T3 promoters accordingly. The 458 bp probe for mouse Msx2 was used as described previously (41).
Msx2 promoter–luciferase reporter gene construction and mutagenesis
The 5′ upstream fragments of the mouse Msx2 promoter were isolated from various restriction enzyme digestions of the Msx2-5.2K-LacZ (25) and gel purified. The Msx2–luciferase fusion reporter genes were constructed by inserting the Msx2 promoter fragments into the polylinker of the luciferase reporter vector pGL3 basic (Promega Corp., Madison, WI). Msx2SS-Luc contains an insert of 550 bp (–369 to +181 bp) of the Msx2 promoter. Msx2KS-Luc contains –1451 to +181 bp of the Msx2 promoter. Msx2NS-Luc has the same insert as in Msx2SS-Luc and an additional 250 bp fragment from the 5′ end of Msx2-5.2K-LacZ. Msx2NH-Luc was created by deleting the –238 to +181 bp fragment from Msx2NS-Luc. Msx2SA-Luc has the insert of –369 to –91 bp of the Msx2 gene.
Mutagenesis in the YY1 recognition sites was performed using three sets of oligonucleotides (+ strand shown only): M1 5′-GCTCATAGTGGGAGCTTTATAAACCTTCCATGCCCTCCGCAGATTTC-3′; M2 5′-GAGGGGTTAAAAGACAAGAAACCAGACTCGGCAAGCTTCCTC-3′; and M3 5′-CTCCGCAGATTTCCAAGGTTCTCAGGCGGGAGCGTG-3′. These were synthesized and then purified by HPLC. Within these oligonucleotides, the core binding site of YY1, 5′-CAT-3′, was mutated to 5′-GGT-3′. For mutating the initiator of the Msx2 promoter, a set of oligonucleotides (+ strand) 5′-GGTTGAGCCGAGTCTCCGGCTTCCCCTCGGAG-3′ was used. Point mutations were introduced by using QuickChange Site-Directed Mutagenesis Kit (Stratagene) following specifications from the manufacturer. The annealing temperature and amplification cycles for each mutation construct were determined empirically. After mutagenesis, the DNA products were treated with DPN I and transformed into Escherichia coli XL1-Blue supercompetent cells (Stratagene). All introduced mutations were confirmed by DNA sequencing.
P19 cell culture, transient transfection, reporter gene analysis and western blotting
P19 mouse embryonal carcinoma cell line was purchased from American Type Culture Center (Rockville, MD) and maintained in 90% a-MEM supplemented with 7% fetal bovine serum and 3% calf serum. The expression vector of wild-type YY1 was a gift from Dr Ozato (42), and the mutant YY1 expression constructs YY1-Δ2–150, YY1-Δ334–414, YY1-Δ399–414 and YY1-S339/S342 were gifts from Dr Luscher (43). For transfections, P19 cells were seeded 20–24 h before transfection at a density of 3 × 104 cells per well onto a 24-well plate. When the cells reached 35–50% confluency, the medium was changed to Opti-MEM (Life Technologies, Gaithersburg, MD) and the cells transfected using LipofectAMINE Plus reagent (Life Technologies). After 24 h the cells were harvested and lysed in 200 µl of cell lysis buffer. The luciferase activity was measured using a Monolight 2010 luminometer (Analytical Luminescence Laboratories, Ann Arbor, MI) with 50 µl of cell lysate and 300 µl of assay buffer. The CMV-β-gal construct was co-transfected with the reporter for normalization of transfection efficiency. The activity of β-galactosidase was measured using an o-nitrophenyl-β-d-galactopyranoside colorimetric assay (Promega, Madison, WI). All experiments were performed in triplicate. Data were presented as mean ± standard deviation and analyzed using one-way analysis of variance or Student’s t-test. A confidence level of P < 0.05 was considered to be statistically significant. To confirm expression of the YY1 mutant constructs, western blotting was performed. Cells were lysed 24 h post-transfection in 50 mM Tris buffer pH 7.4, 150 mM sodium chloride, 0.5% Triton-X 100, 0.5% sodium deoxycholate and protease inhibitors. Total protein was measured using the BCA Protein Assay (Pierce, Rockford, IL) for each sample, and equal amounts of protein were loaded for each construct onto a 10% SDS–PAGE gel, separated and transferred onto nitrocellulose. A rabbit polyclonal antibody directed against YY1, H414 (Santa Cruz Biotechnologies, Santa Cruz, CA), was used at 1:1000 dilution as primary antibody, and a horseradish peroxidase-conjugated goat anti-rabbit (Jackson Immunoresearch Labs, West Grove, PA) secondary antibody was used. The blot was developed using ECL Kit (Amersham Pharmacia Biotech, UK).
Semi-quantitative reverse transcription–polymerase chain reaction
P19 cells were plated onto 10-cm tissue culture plates and grown for 1 day. Cells were transfected using LipofectAMINE method. Cells were lysed and homogenized by QIAshredder cell lysate homogenizer (Qiagen, Valencia, CA) 24 h after transfection. Total RNA was isolated using RNeasy Mini kit (Qiagen) and first strand cDNA was reverse transcribed using Superscript Preamplification System kit (Life Technologies) according to specifications from the manufacturers. Semi-quantitative reverse transcription–polymerase chain reaction (RT–PCR) was performed to evaluate Msx2 expression levels relative to that of β-actin. Amplimers specific for the Msx2 gene were 5′-TGGATACAGGAGCCCGGCAGATAC-3′ from sense strand of exon 1 and 5′-CTGGAGTCTGGTCCATCTGGTCTTC-3′ from antisense strand of exon 2 of the mouse Msx2 cDNA sequence (44,45), which yielded an amplified fragment of 488 bp as confirmed by DNA sequencing. Primer set for mouse β-actin for RT–PCR was purchased from Stratagene. The PCR was carried out using Ready-To-Go PCR beads (Amersham Pharmacia Biotech, Piscataway, NJ). Thirty cycles of amplification for Msx2 annealing at 62°C for 30 s, and 27 cycles for β-actin annealing at 57°C for 30 s, were determined empirically to optimize for signal and amplification linearity. After amplification, the PCR products were electrophoretically separated in agarose gel, stained with ethidium bromide, imaged and quantified by using NIH Image Version 1.6.1 (NIH, Bethesda, MD). Msx2 expression was determined by normalizing its value against that of β-actin. These experiments were conducted in triplicate. The values among groups were subject to Student’s t-test and P < 0.05 was considered as statistically significant.
P19 cell nuclear extract preparation, glutathione S-transferase–YY1 protein preparation and electrophoretic mobility shift assay
P19 cell nuclear extracts were prepared using the method of Dignam (46). The expression construct for glutathione S-transferase–YY1 fusion protein (GST–YY1) and a YY1 antibody were gifts from Dr Shi (34). The isolation of GST–YY1 was performed using GST Module kit following the protocol supplied by the manufacturer (Amersham Pharmacia Biotech). P19 nuclear extracts and GST–YY1 protein were dialysed using Slide-A-Lyzer cassette (Pierce) and the concentration was measured.
Three sets of oligonucleotides corresponding to three YY1 candidate sites on the Msx2 promoter were synthesized and then purified using polyacrylamide gel. The 5′ end of probe was labeled with [γ-32P]ATP (3000 Ci/mmol) using T4 polynucleotide kinase. The labeled probes were purified using G-25 spin columns. The YY1 binding consensus oligonucleotide and its mutant used in this assay were from Geneka Biotechnology Inc. (Montréal, Quebéc, Canada). Electrophoretic mobility shift assay (EMSA) was carried out using a YY1 NUSHIFT kit (Geneka Biotechnology, Inc.) according to specifications from the manufacturer. Twenty thousand counts of probe were mixed with 3 µg of P19 nuclear extracts or 260 ng of the GST–YY1 for 20 min at 5°C. Cold competitors were pre-incubated with P19 nuclear extract or GST–YY1 for 15 min at 5°C before the labeled probe was added. YY1 antibody used for supershifting was pre-incubated with GST–YY1 for 15 min at 5°C. The DNA–protein complexes were loaded onto 4.5% non-denaturing polyacrylamide gel in 28 mM Tris–acetate and 0.7 mM EDTA, and electrophoresis was performed at 10 V/cm at 4°C. After electrophoresis, the gel was dried and autoradiography was performed at –80°C with intensifying screens.
Mandibular process explant culture and bead implantation
Timed pregnant Swiss Webster mice were purchased (Harlan Bioproducts for Science, Indianapolis, IN) and used according to Animal Study Protocols approved by the National Institutes of Health (NIH). Embryonic day E10 embryos were microdissected and mandibular processes were collected. Organ culture of mandibular processes was performed according to methods described previously (47). Mandibular processes were cultured using serum-free, chemically defined BGJb medium (Life Technologies) supplemented with 100 µg/ml ascorbic acid. Affi-gel blue agarose beads (Bio-Rad, Hercules, CA) with a diameter of 100–120 µm were selected and soaked in 100 ng/µl human recombinant BMP4 (Genetics Institute Inc., Cambridge, MA) or PBS at room temperature for 6 h. Control PBS- or BMP4-soaked beads were implanted into the mandibular processes using a mouth-controlled micropipette under the stereomicroscope (47).
RESULTS
YY1 and Msx2 genes co-expressed in mouse limb and branchial arches
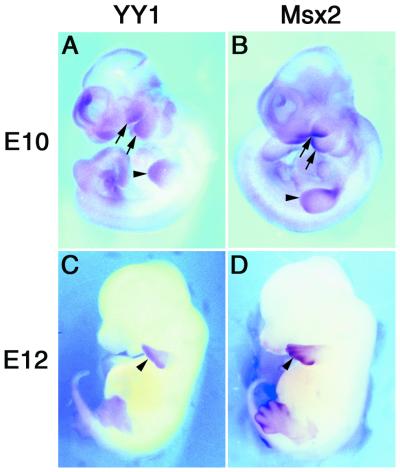
Temporal and spatial relationships between YY1 and Msx2 gene expression patterns during critical developmental stages were examined by whole mount in situ hybridization during E10 and E12 stages of development. In the E10 embryos, YY1 and Msx2 exhibited restrictive patterns of expression, including in the forebrain, midbrain, hindbrain and tail bud (Fig. 1A and B) although a low level of YY1 expression was detected throughout the embryo. However, most notably, YY1 and Msx2 were detected at high levels in the developing first and second branchial arches and limb buds. YY1 and Msx2 co-expression was limited to the distal portion of the organs. These regions were undergoing tissue interactions and outgrowth to generate the morphogenetic template for the future organs. In E12 embryos, the expression and co-expression patterns were entirely limited to the distal tips of the limbs, albeit a greater extension of Msx2 expression extending proximally into the interdigital zones (Fig. 1C and D). Previous studies demonstrated that Msx2 is a morphoregulatory molecule in signaling networks during embryogenesis (3,6). Our co-localization data suggests that YY1 and Msx2 may be involved in the same morphoregulatory events, and that the two genes may be interacting at a molecular level.
Figure 1.
Co-localization of YY1 and Msx2 in E10 and E12 mouse embryos. Whole mount in situ hybridization detected gene expression for YY1 and Msx2 in E10 (A and B) and E12 (C and D) mouse embryos, respectively. Expression of both genes was observed in the developing first and second branchial arches (arrows) and limb buds (arrowheads) at the E10 stage, and persisted in the distal limb bud at the E12 stage (arrows). Low level of YY1 expression was noted throughout the E10 embryo.
YY1 induced Msx2 gene expression and transactivated the Msx2 promoter
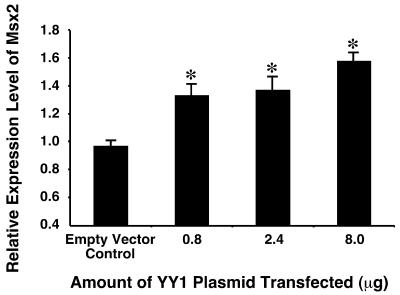
As YY1 has been shown to regulate the expression of a large number of genes by directly regulating the promoters, and that YY1 and Msx2 co-localized in several embryonic sites, we hypothesized that YY1 regulates the expression of Msx2. In order to test this hypothesis, we transfected different amounts of YY1 into P19 embryonal cells, and determined the expression level of Msx2 by semi-quantitative RT–PCR. P19 cells were chosen for the experiments because the cells present endogenous Msx2 expression, and that this expression is subject to genetic regulation (48). We observed that over-expression of YY1 induced significant increases (P < 0.01) in the expression level of Msx2 in a dose-dependent manner (Fig. 2). The increase was modest which was probably due to limited transfection efficiency and the presence of an already elevated level of endogenous Msx2.
Figure 2.
YY1 induced endogenous Msx2 expression in P19 embryonal cells. Different amounts of YY1 (x-axis) were transfected into P19 cells cultured on 10-cm plates, and Msx2 expression level was assayed by semi-quantitative RT–PCR 24 h post-transfection. Increasing amounts of YY1 resulted in significantly elevated levels of Msx2 when compared with control in which an expression vector with no YY1 insert (empty vector) was used. *P < 0.01.
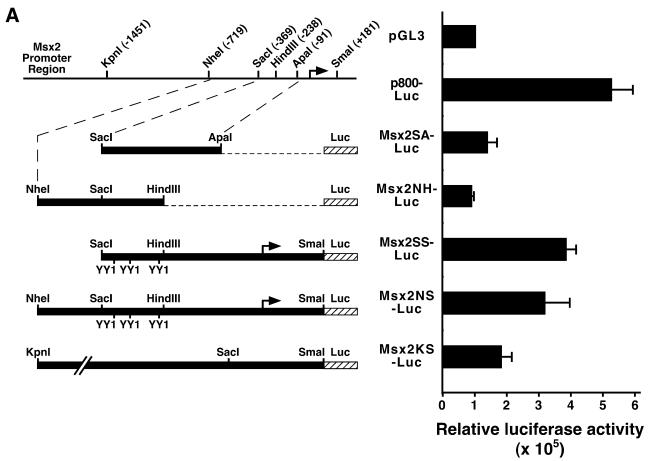
To determine whether YY1 regulates Msx2 expression by regulating the Msx2 promoter, we first subcloned various DNA fragments of the Msx2 promoter region into the luciferase reporter vector pGL3. These constructs were transfected into P19 cells and the basal promoter activities were assayed by the luciferase reporter (Fig. 3A). Upon comparison, we determined that the Msx2SS-Luc construct that contained 550 bp of the Msx2 upstream region exhibited maximum basal promoter activity. Msx2SA-Luc and Msx2NH-Luc, both of which lacked the proximal region of the Msx2 gene, had no activity above background, suggesting that the proximal region is essential for Msx2 native promoter activity. Two constructs, Msx2KS-Luc and Msx2NS-Luc, which contained a more extended upstream region, showed promoter activities that were lower than that of Msx2SS-Luc, suggesting that cis-acting repressive elements may be present in these upstream regions. Subsequently, the Msx2SS-Luc construct was selected for further experiments because it presented high promoter activity in P19 cells. Furthermore, a similar construct using β-galactosidase instead of luciferase as the reporter retained the native expression pattern when expressed in transgenic mice (25). This suggests that the promoter fragment encompasses elements that mimic endogenous Msx2 behavior and may be regulated similar to in vivo conditions.
Figure 3.
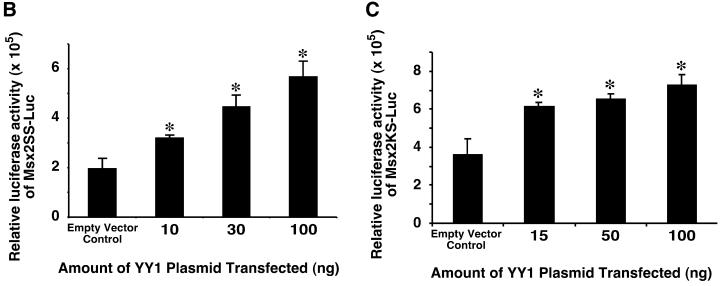
Msx2 proximal promoter bearing 550 bp of the upstream sequences exhibited maximum activity and was transactivated by YY1. Five constructs of the Msx2 promoter were generated and cloned into luciferase reporter vector pGL3 (A). These are represented schematically on the left. A schematic representation of the genomic promoter region of the Msx2 gene with restriction enzyme sites and their respective positions is shown for referencing the various constructs. The promoter activities of these constructs were assayed by the luciferase reporter and compared with the negative control using the basal vector pGL3, and the positive control using p800-Luc, which contains the PAI-1 gene promoter (74). In this assay, the Msx2SS-Luc construct displayed greater activity than the other Msx2 promoter constructs. Subsequently, different amounts of YY1 (x-axis) were co-transfected with Msx2 promoter reporter constructs into P19 cells cultured on 10-cm plates, and luciferase reporter activities were assayed from Msx2SS-Luc (B) or Msx2KS-Luc (C) constructs, 24 h post-transfection. Increasing amounts of YY1 resulted in significantly elevated levels of Msx2 promoter activities when compared with control in which an expression vector with no YY1 insert (empty vector) was used. *P < 0.01.
Since YY1 increased Msx2 expression levels, we next designed experiments to determine whether YY1 regulates the Msx2 promoter. Over-expression of YY1 in P19 cells resulted in significant increases (P < 0.01) in Msx2SS-Luc activities in a dose-dependent manner (Fig. 3B). Similarly, YY1 also transactivated the Msx2KS-Luc reporter in a dose-dependent fashion (Fig. 3C). The data indicate that the 550 bp and 1.6 kb Msx2 promoter constructs may contain YY1 responsive elements. Since over-expression of YY1 did not alter the total cell number (data not shown), we concluded that the elevated Msx2 promoter activity was a direct consequence of YY1 transactivation, and not mediated by effects on cell proliferation.
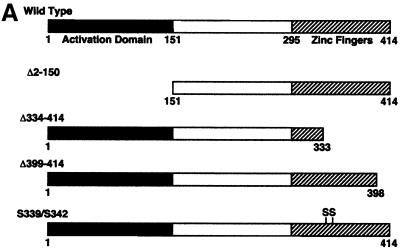
The N-terminal activation domain and the C-terminal zinc finger domains of YY1 were both required for the activity of YY1 on the Msx2 promoter
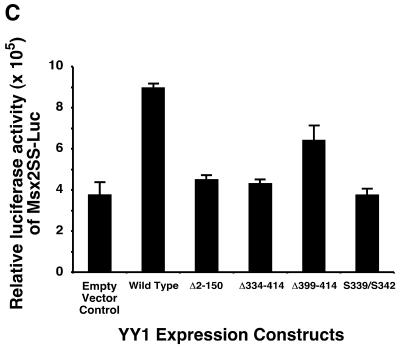
Since YY1 transactivated the Msx2 promoter, we endeavored to determine which domains on YY1 were necessary for this activity. A series of YY1 mutant constructs were obtained, which included an N-terminal truncation (Δ2–150), C-terminal truncations (Δ334–414 and Δ399–414) and amino acid substitutions (S339/S342) (Fig. 4A). The Δ334–414 and Δ399–414 YY1 mutants encoded proteins with the terminal three and one zinc finger domain deleted, respectively. The expression of these constructs was confirmed by western blotting using an antibody directed against the full length YY1 (Fig. 4B). We detected the endogenous YY1 protein at 68 kDa. The three truncation mutants yielded smaller proteins consistent with the length of the deletion. As all the sample lanes have equal amounts of protein, exogenous expression of wild-type YY1 and the point mutant form were identified as an increased band intensity of the 68 kDa protein. Each construct was co-transfected with the Msx2SS-Luc reporter and reporter activity was assayed (Fig. 4C). Consistent with our previous observations, wild-type YY1 transactivated the Msx2 promoter. The same amount of YY1 of the Δ2–150, Δ334–414 or S339/S342 mutants failed to transactivate the Msx2 promoter, whereas the Δ399–414 mutant form showed activation at a reduced level when compared with the full length wild-type construct. Taken together, our data suggested that both the N- and C-terminal domains of YY1 were required for transactivation function on the Msx2 promoter. Amino acid residues 339 and 342 in zinc finger 2 were also critical for the activity of YY1.
Figure 4.
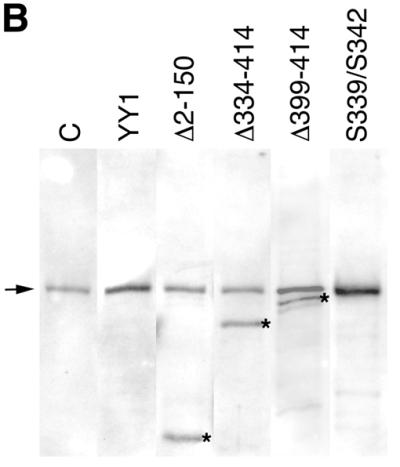
Both the N- and C-terminal domains of YY1 were required for YY1 transactivation of Msx2. Five expression constructs of wild-type and mutant YY1 were used (A). Wild-type YY1 contains an N-terminal activation domain, and C-terminal DNA binding zinc finger domain, which has four zinc finger motifs. The Δ2–150 construct is an N-terminal truncation mutant, whereas the Δ334–414 and Δ399–414 are C-terminal truncation mutants of different lengths. The S339/S342 mutant has two amino acid substitutions within the second zinc finger motif. The expression of these constructs was confirmed by western blot analysis (B). Endogenous YY1 was detected at 68 kDa (arrow) in the untransfected control (C) and in all transfected samples. The three YY1 truncation mutants were detected as additional smaller size bands (asterisks) consistent with the length of deletion in the construct. Since all lanes were loaded with equal amounts of protein, wild-type YY1 and the point mutant form were identified as increased intensity of the 68 kDa band. These YY1 constructs (x-axis) were co-transfected with Msx2SS-Luc reporter constructs into P19 cells cultured on 10-cm plates, and luciferase activities were assayed 24 h post-transfection (C). Wild-type YY1 expression resulted in significantly elevated level of Msx2 promoter activities when compared with control in which an expression vector with no YY1 insert (empty vector) was used. *P < 0.01. Δ399–414 exhibited significantly reduced level of promoter activation, whereas Δ2–150, Δ334–414 and S339/S342 had no activities at all.
Three YY1 binding sites on the Msx2 promoter mediated the function of YY1 by protein–DNA interactions
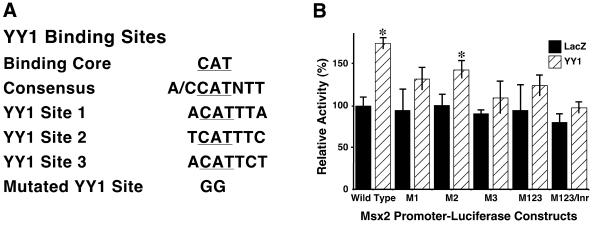
Within the 550 bp fragment of the Msx2 promoter contained in the Msx2SS-Luc construct, sequence analysis revealed three candidate YY1 core binding sites, with sequence CATNTT. Therefore, in order to determine whether these motifs mediate the transactivation function of YY1 on the Msx2 promoter, the three candidate sites were mutated individually to generate constructs M1, M2 and M3. Additionally, all three sites were mutated to produce constructs M123 and M123/Inr, in which the latter had the initiator sequence mutated (Fig. 5A). The effects of YY1 on these mutant forms of the Msx2 promoter were monitored and compared with their basal activities in the presence of LacZ control (Fig. 5B). The basal activity of wild-type Msx2SS-Luc was designated as 100%. The basal activities of the various mutant promoters were comparable with wild-type. We confirmed that YY1 induced a significant increase in wild-type Msx2 promoter activity. However, all mutant constructs displayed reduced activity when compared with the wild-type control, although that of M2 was least severe. M1, M2 and M3 had 75, 82 and 62% of full activity, respectively. When all three candidate YY1 binding sites were mutated, the activity was reduced to 71% of wild-type. Taken together, we concluded that all three candidate YY1 binding sites on the Msx2 promoter contributed to promoter activity induced by YY1. Disruption to any of these sites markedly reduced YY1 transactivation of Msx2. Since YY1 has been shown previously to bind to initiator sequences to activate transcription (39), we also mutated the initiator sequence on the Msx2 promoter to test its potential role. However, this M123/Inr construct did not result in a further significant reduction in promoter activity beyond that of the mutant constructs, suggesting that the initiator sequence in the Msx2 promoter was probably not responsible for YY1 transactivation.
Figure 5.
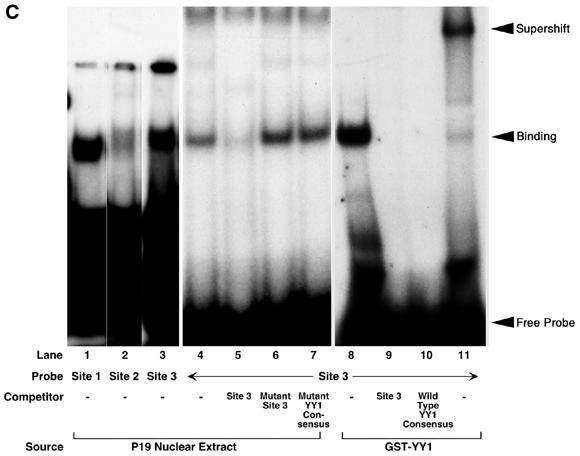
YY1 transactivated Msx2 promoter by protein–DNA binding to three YY1 binding sites on the Msx2 proximal promoter. Three candidate YY1 binding sites encompassing the core binding motif 5′-CAT-3′ were identified on the proximal Msx2 promoter and designated as YY1 sites 1, 2 and 3 (A). Using site directed mutagenesis these sites were mutated in the core sequence from 5′-CAT-3′ to 5′-GGT-3′. The three sites were mutated individually and resulted in constructs designated M1, M2 and M3, or in combinations to yield M123. Additionally, the initiator on the Msx2 promoter was also mutated from 5′-CAC-3′ to 5′-GGC-3′ to generate M123/Inr construct. These Msx2 promoter constructs (x-axis) were co-transfected with LacZ as control or YY1 expression vector into P19 cells cultured on 10-cm plates. Luciferase activities were assayed 24 h post-transfection (B). Basal wild-type Msx2 promoter activity in the presence of LacZ control was designated as 100%. YY1 induced a significant increase in this Msx2 promoter activity. However, all mutant Msx2 promoters, except M2, exhibited reduced level of activities in response to YY1 when compared with wild-type and their respective LacZ controls. *P < 0.01. M2 showed slightly decreased activity. Three sets of oligonucleotides, corresponding to the three YY1 binding sites on the Msx2 promoter, were radiolabeled and tested in EMSA for their binding to P19 nuclear extract or to a GST–YY1 fusion protein. All three probes bound, albeit at different intensities (sites 1–3, lanes 1–3, respectively). The results for all three sets of oligonucleotides were similar for additional assays and those for site 3 are shown in lanes 4–11. The site 3 probe bound to the P19 nuclear extract (lane 4), and this binding was competed off by 100-fold molar excess of cold site 3 probe (lane 5), but not by excess mutant site 3 probe (lane 6) or mutant YY1 consensus oligonucleotides (lane 7). Similarly, the site 3 probe bound to the GST–YY1 fusion protein (lane 8), and this binding was competed off by 100-fold molar excess of cold site 3 probe (lane 9) or YY1 consensus oligonucleotides (lane 10). The band was super-shifted when YY1 antibody was also added to the site 3 probe and GST–YY1 complex (lane 11).
As all three candidate YY1 binding sites contributed to the response of the Msx2 promoter upon YY1 activation, we next tested the hypothesis that YY1 binds to these motifs. Indeed, in EMSA, the nuclear extract of P19 cells bound to all three probes representing the YY1 binding sites. Site 3 probe appeared to bind most strongly, and mutations within the site (M3 construct) resulted in the most severe reduction in YY1 transactivation of the Msx2 promoter (Fig. 5B). Further analysis of site 3 demonstrated that binding of the radioactive probe (Fig. 5C, lane 4) was competed successfully by 100-fold molar excess of cold site 3 probe (Fig. 5C, lane 5). However, binding was not compromised by 100-fold molar excess of mutated site 3 oligonucleotide (Fig. 5C, lane 6), nor with 100-fold molar excess of mutated YY1 consensus sequence (Fig. 5C, lane 7). Furthermore, we tested the specificity of this binding by first expressing and purifying a GST–YY1 fusion protein, which was subsequently used in EMSA. We showed that the site 3 radioactive probe bound to the GST–YY1 fusion protein (Fig. 5C, lane 8) that was competed successfully by excess amount of cold site 3 probe (Fig. 5C, lane 9) or cold YY1 consensus oligonucleotides (Fig. 5C, lane 10). When a YY1 antibody was pre-incubated with GST–YY1 prior to adding the labeled site 3 probe, the specific band was super-shifted (Fig. 5C, lane 11). The experiments were repeated for analyzing sites 1 and 2, with similar results (data not shown). However, it should be noted that the excess amount of cold mutated site 2 probe did reduce specific binding of radioactive site 2 probe to YY1, suggesting that the binding of YY1 to site 2 was weak. Taken together, we concluded that YY1 binds to three recognition sites in the upstream region of the Msx2 promoter. The bindings were necessary for YY1 to transactivate the Msx2 promoter.
BMP4 did not induce YY1 expression in developing mouse mandibular processes
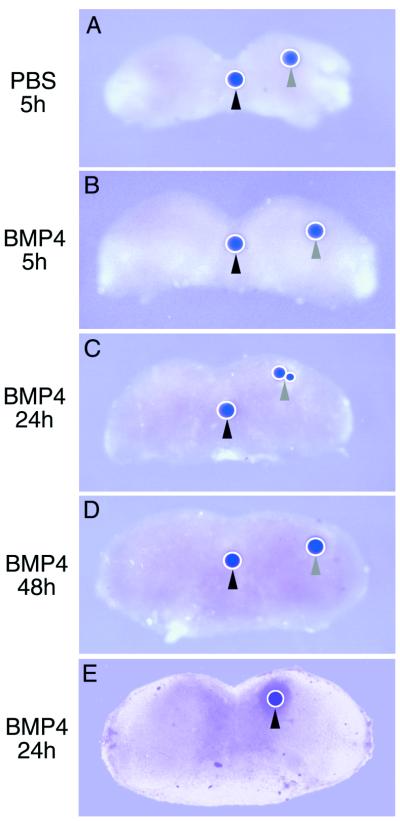
In many developmental systems, including the embryonic first branchial arch and limb, where YY1 and Msx2 were co-localized, Msx2 is induced by BMP4. Therefore, we tested the hypothesis that YY1 is the mediator of BMP4 induction of Msx2 gene expression during mandibular morphogenesis. Microbeads that were soaked in 100 ng/µl of BMP4 were implanted into E10 mouse mandibular processes, and cultured for 5, 24 and 48 h. Beads were implanted into the medial as well as the lateral portions of the explant. YY1 expression was detected by whole mount in situ hybridization (Fig. 6A–D). None of the time points and neither of the two sites of implantation exhibited an elevation of YY1 expression although endogenous YY1 expression was detected. However, BMP4-soaked beads did induce a ring of ectopic Msx2 expression surrounding the beads in the explants at 24 h (Fig. 6E). Control PBS-soaked beads had no effect. Similarly, exogenous addition of BMP4 to P19 cells did not increase the expression level of YY1 assayed by semi-quantitative RT–PCR (data not shown), although BMP4 has been demonstrated to induce ectopic Msx2 expression in P19 cells within 24 h (49). The data indicated that whereas BMP4 induced the expression of Msx2 in mandibular processes and P19 cells, YY1 was not the mediator in this pathway. YY1 activation and induction of Msx2 were independent of BMP4, and these two signaling pathways may act in parallel.
Figure 6.
BMP4-soaked beads induced Msx2 but not YY1 expression. Mandibular processes were isolated from E10 mouse embryos and explanted in organ culture. Affi-gel microbeads with a diameter of 100–150 µm were soaked in PBS as control (A), or 100 ng/µl BMP4 (B–E) and implanted into mandibular explants in the medial (black arrowheads) and lateral (gray arrowheads) positions. At different time points indicated on the left, explants were probed by whole mount in situ hybridization for YY1 (A–D) or Msx2 (E). Endogenous expression of these genes was detected in all explants. Neither PBS- nor BMP4-soaked beads were able to induce ectopic expression of YY1 around the site of implantation (A–D). However, BMP4-soaked beads induced a ring of ectopic Msx2 expression around the implanted bead after 24 h (E).
DISCUSSION
Determining the specificity for transcriptional controls during exquisite developmental process is a major problem area. In this investigation, we showed that although YY1 expression is ubiquitous, areas of elevated YY1 expression during embryogenesis overlap with Msx2 expression patterns. The new and most significant finding in pursuit of this initial observation is that YY1 binds to and activates Msx2 transcription that presents a regulatory pathway for Msx2 that is independent of BMP4 signaling.
The significance in the regulation of Msx2 transactivation by factors other than BMP4 is in the maintenance of an optimum level of Msx2 expression. The homeodomain transcription factor Msx2 has been demonstrated to be pivotal to many developmental processes including epithelial–mesenchymal interactions and programmed cell death (3), and that a precise level of expression is necessary for normal function. The significance of an exact Msx2 expression level was exemplified by the discoveries of mutations in the human gene. A proline to histidine mutation resulted in an activated gene and was associated with Boston-type craniosynostosis (7). The genotype-to-phenotype link of this mutation was confirmed by genetically engineered mouse models (8,9). Recently, loss-of-function mutations in MSX2 have been shown to be causal to the human genetic disease enlarged parietal foramina (11), which were also corroborated by animals with targeted disruption to Msx2 showing similar phenotype (10). Therefore, taken together, excess Msx2 accelerates fusion of sutures, whereas Msx2 deficiency causes persistent sutural patency, suggesting that an optimal level of Msx2 must be achieved and maintained in order for developmental programs to proceed normally. Stringent regulation of Msx2 dosage is critical for normal development and this balance, when tipped either way is deleterious. One strategy to maintain rigorous control of expression is to employ multiple signaling pathways acting in combination and channeling towards the target gene.
Despite the importance of Msx2 in developmentally regulated events, regulation of Msx2 expression has largely been attributed to that from BMP4. The expression patterns of BMP4 and Msx2 overlap greatly in time and space. Furthermore, BMP4 induced ectopic expression of Msx2 in a number of in vitro and in vivo experiments (12,41,48,50–54). Recently, studies of Smad4 functions revealed that Smad4 directly influenced the Msx2 promoter, in support of BMP signaling in the regulation of Msx2 transactivation (55). However, although BMP4 and Msx2 are intimately related, differences in expression patterns exist between the two molecules in the developing mandibular processes (41), sutures (56) and rhombomeres (57) beyond that which can be accounted for by tissue interactions. Further, the causal relationship between BMP4 and Msx2 was not sustained during positional apoptosis within the neuroepithelium (58) or in the mammary glands following ovariectomy (59). Even during calvarial development, although BMP4-soaked beads induced Msx2 expression, they did not induce premature closure of the sutures, as occurred in the Msx2 transgenic animals (8,60).
These observations, in addition to the fact that Msx2 gene dosage is critical for normal suture development, suggest that the regulation of Msx2 may be dependent on factors other than BMP4. Indeed, studies in the developing limb and rhombomeres have demonstrated that FGF, IGF and Wnt signaling may regulate the expression and functions of Msx2 (61–64). In this investigation we demonstrated that Msx2 expression is independently regulated by another transcription factor, YY1. We showed that YY1 induced the ectopic expression of Msx2. We showed that YY1 transactivated the Msx2 promoter by binding to three YY1 binding sites on the Msx2 promoter. It is possible that YY1 directly binds to and regulates Msx2. However, as YY1 interacts with a large number of proteins of the transcriptional machinery, YY1 may also participate in a regulatory transcription complex to modulate Msx2. Furthermore, YY1 may mediate the functions of additional growth and differentiation factors that regulate Msx2. Taken together, we conclude that YY1 regulates Msx2 expression, and suggest that deregulation of this interaction may account for the degree of severity of abnormalities related to abnormal level of expression of the Msx2 gene. A similar scenario was observed in some cases of cystic fibrosis in which mutation in the CFTR gene altered a YY1 binding element and resulted in increased CFTR expression (65). Moreover, failure in the interaction between YY1 and Msx2 may also cause disruption in the maintenance of the feedback loop between BMP4 and Msx2, perceivable as in the case of adontia in the diastema (66,67).
YY1 is a multifunctional transcription factor that binds to a large number of both cellular and viral promoters (29–31). Binding of YY1 to these promoters can either activate or repress transcription of target genes. Moreover, YY1 can bind to the initiator signals on promoters and initiate transcription (68). Therefore, YY1 regulates the expression of many genes, thus accounting for a widely distributed pattern of the molecule. The complexity of the activity of YY1 is further complicated by the discovery that the viral protein E1A may alter the repressive activity of YY1, and switch it into an activator (28). In our study, it is unclear how the three YY1 binding sites on the Msx2 promoter function in relationship to one another and to the activities of Msx2. We speculate that site 2 may have lower affinity for YY1 than either sites 1 or 3, since sites 1 and 3 have the invariant consensus YY1 binding core sequence of 5′-ACAT-3′, which predicts high affinity binding, whereas site 2 has a core sequence of 5′-TCAT-3′, which may result in lower affinity (69,70). However, it has been shown that YY1 participates in transcriptional complex formation and function. Therefore, the binding affinity of YY1 to the Msx2 promoter is likely to be further modulated by other transcription factors within the complex. Sites 1 and 3 are separated by only 20 bp and such physical proximity may implicate functional cooperativity. Mutations in the core sequences of each these sites resulted in the loss of YY1 response, and mutations in both sites did not attenuate the outcome. Mutations in site 2 resulted in a smaller reduction in YY1 response suggesting that site 2 may be less critical in mediating the YY1 response on the Msx2 promoter. Therefore, we suggest that YY1 acts as an enhancer binding protein through binding to upstream sites, but not through the initiator, in the Msx2 promoter.
The functions of the various domains of the YY1 molecule have been extensively explored but remain unresolved. In general, there is ample evidence that the N-terminal domain of the molecule, which has large stretches of acidic regions, is responsible for the activation function of YY1. The C-terminal domain, consisting of four zinc fingers, is necessary for the repression function of YY1 (30). The C-terminal of YY1 also has DNA binding and nuclear matrix association properties (71,72). Our findings revealed that both the N- and C-terminals of YY1 are essential for the transactivation of the Msx2 promoter. The N-terminal deletion mutant represents an abrogation of the activation domain of YY1. The C-terminal deletion mutants also display loss of activities, but can be attributed to the loss of DNA binding or nuclear localization. Complete removal of three zinc fingers (2, 3 and 4) results in the loss of activation. Removal of zinc finger 4 reduces but does not eliminate the activation, suggesting that zinc fingers 2 and 3 are more important. Since zinc finger 2 has been proposed to be critical for DNA recognition (73), we tested the activity of YY1 bearing point mutations in Lys339 and Arg342 in zinc finger 2 accordingly. The YY1-S339/S342 mutant presents no activity indicating that YY1 transactivation of the Msx2 promoter is dependent on DNA binding.
YY1 functions as a repressor to many developmentally regulated genes. Such function is necessary to ensure that these genes are temporally and spatially restricted, and that a developmental event progresses from one stage to another by regulation of stage-specific signals. However, equally important is the maintenance of morphogenetic signals and feedback loops to ensure the completion of a particular stage of differentiation. Regulation of expression level by activators and repressors is a common theme in development. We suggest that YY1 regulation of Msx2 serves to govern the crucial level of Msx2 needed for function. We further suggest that, since YY1 binding elements are widespread in many genes, the regulation by YY1 of these genes could also serve as the developmental basis of differential levels of expression and the generation of morphogenetic gradients.
Acknowledgments
ACKNOWLEDGEMENTS
We are deeply grateful for gifts from Dr Ozato at the Laboratory of Molecular Growth Regulation, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, Dr Luscher at Institut fur Molekularbiologie, Medizinische Hochschule Hannover, Germany and Dr Shi at Department of Pathology, Harvard Medical School, Boston, MA, such that we could complete our studies.
REFERENCES
- 1.Lumsden A. and Krumlauf,R. (1996) Patterning the vertebrate neuraxis. Science, 274, 1109–1115. [DOI] [PubMed] [Google Scholar]
- 2.Chariot A., Gielen,J., Merville,M.P. and Bours,V. (1999) The homeodomain-containing proteins: an update on their interacting partners. Biochem. Pharmacol., 58, 1851–1857. [DOI] [PubMed] [Google Scholar]
- 3.Bendall A.J. and Abate-Shen,C. (2000) Roles for Msx and Dlx homeoproteins in vertebrate development. Gene, 247, 17–31. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y.H., Ma,L., Kundu,R., Ignelzi,M., Sangiorgi,F., Wu,L., Luo,W., Snead,M.L. and Maxson,R. (1996) Function of the Msx2 gene in the morphogenesis of the skull. Ann. N.Y. Acad. Sci., 785, 48–58. [DOI] [PubMed] [Google Scholar]
- 5.Davidson D. (1995) The function and evolution of Msx genes: pointers and paradoxes. Trends Genet., 11, 405–411. [DOI] [PubMed] [Google Scholar]
- 6.Cohen M.M. Jr (2000) Craniofacial disorders caused by mutations in homeobox genes MSX1 and MSX2. J. Craniofac. Genet Dev. Biol., 20, 19–25. [PubMed] [Google Scholar]
- 7.Jabs E.W., Muller,U., Li,X., Ma,L., Luo,W., Haworth,I.S., Klisak,I., Sparkes,R., Warman,M.L., Mulliken,J.B. et al. (1993) A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell, 75, 443–450. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y.H., Kundu,R., Wu,L., Luo,W., Ignelzi,M.A.,Jr, Snead,M.L. and Maxson,R.E.,Jr (1995) Premature suture closure and ectopic cranial bone in mice expressing Msx2 transgenes in the developing skull. Proc. Natl Acad. Sci. USA, 92, 6137–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y.H., Tang,Z., Kundu,R.K., Wu,L., Luo,W., Zhu,D., Sangiorgi,F., Snead,M.L. and Maxson,R.E. (1999) Msx2 gene dosage influences the number of proliferative osteogenic cells in growth centers of the developing murine skull: a possible mechanism for MSX2-mediated craniosynostosis in humans. Dev. Biol., 205, 260–274. [DOI] [PubMed] [Google Scholar]
- 10.Satokata I., Ma,L., Ohshima,H., Bei,M., Woo,I., Nishizawa,K., Maeda,T., Takano,Y., Uchiyama,M., Heaney,S. et al. (2000) Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nature Genet., 24, 391–395. [DOI] [PubMed] [Google Scholar]
- 11.Wilkie A.O., Tang,Z., Elanko,N., Walsh,S., Twigg,S.R., Hurst,J.A., Wall,S.A., Chrzanowska,K.H. and Maxson,R.E.,Jr (2000) Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nature Genet., 24, 387–390. [DOI] [PubMed] [Google Scholar]
- 12.Graham A., Francis-West,P., Brickell,P. and Lumsden,A. (1994) The signalling molecule BMP4 mediates apoptosis in the rhombencephalic neural crest. Nature, 372, 684–686. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi K., Nuckolls,G.H., Tanaka,O., Semba,I., Takahashi,I., Dashner,R., Shum,L. and Slavkin,H.C. (1998) Adenovirus-mediated ectopic expression of Msx2 in even-numbered rhombomeres induces apoptotic elimination of cranial neural crest cells in ovo. Development, 125, 1627–1635. [DOI] [PubMed] [Google Scholar]
- 14.Winograd J., Reilly,M.P., Roe,R., Lutz,J., Laughner,E., Xu,X., Hu,L., Asakura,T., vander Kolk,C., Strandberg,J.D. et al. (1997) Perinatal lethality and multiple craniofacial malformations in MSX2 transgenic mice. Hum. Mol. Genet., 6, 369–379. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K., Nuckolls,G.H., Takahashi,I., Nonaka,K., Nagata,M., Ikura,T., Slavkin,H.C. and Shum,L. (2001) Msx2 is a repressor of chondrogenic differentiation in migratory cranial neural crest cells. Dev. Dyn., 222, 252–262. [DOI] [PubMed] [Google Scholar]
- 16.Mina M., Gluhak,J., Upholt,W.B., Kollar,E.J. and Rogers,B. (1995) Experimental analysis of Msx-1 and Msx-2 gene expression during chick mandibular morphogenesis. Dev. Dyn., 202, 195–214. [DOI] [PubMed] [Google Scholar]
- 17.Mina M., Gluhak,J. and Rodgers,B. (1996) Downregulation of Msx-2 expression results in chondrogenesis in the medial region of the avian mandible. Connect Tissue Res., 35, 79–84. [DOI] [PubMed] [Google Scholar]
- 18.Maas R. and Bei,M. (1997) The genetic control of early tooth development. Crit. Rev. Oral Biol. Med., 8, 4–39. [DOI] [PubMed] [Google Scholar]
- 19.Peters H. and Balling,R. (1999) Teeth. Where and how to make them. Trends Genet., 15, 59–65. [DOI] [PubMed] [Google Scholar]
- 20.Holme R.H., Thomson,S.J. and Davidson,D.R. (2000) Ectopic expression of Msx2 in chick retinal pigmented epithelium cultures suggests a role in patterning the optic vesicle. Mech. Dev., 91, 175–187. [DOI] [PubMed] [Google Scholar]
- 21.Foerst-Potts L. and Sadler,T.W. (1997) Disruption of Msx-1 and Msx-2 reveals roles for these genes in craniofacial, eye and axial development. Dev. Dyn., 209, 70–84. [DOI] [PubMed] [Google Scholar]
- 22.Ganan Y., Macias,D., Duterque-Coquillaud,M., Ros,M.A. and Hurle,J.M. (1996) Role of TGF beta and BMPs as signals controlling the position of the digits and the areas of interdigital cell death in the developing chick limb autopod. Development, 122, 2349–2357. [DOI] [PubMed] [Google Scholar]
- 23.Macias D., Ganan,Y., Ros,M.A. and Hurle,J.M. (1996) In vivo inhibition of programmed cell death by local administration of FGF-2 and FGF-4 in the interdigital areas of the embryonic chick leg bud. Anat. Embryol. (Berl.), 193, 533–541. [DOI] [PubMed] [Google Scholar]
- 24.Ferrari D., Lichtler,A.C., Pan,Z.Z., Dealy,C.N., Upholt,W.B. and Kosher,R.A. (1998) Ectopic expression of Msx-2 in posterior limb bud mesoderm impairs limb morphogenesis while inducing BMP-4 expression, inhibiting cell proliferation and promoting apoptosis. Dev. Biol., 197, 12–24. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y.H., Ma,L., Wu,L.Y., Luo,W., Kundu,R., Sangiorgi,F., Snead,M.L. and Maxson,R. (1994) Regulation of the Msx2 homeobox gene during mouse embryogenesis: a transgene with 439 bp of 5′ flanking sequence is expressed exclusively in the apical ectodermal ridge of the developing limb. Mech. Dev., 48, 187–197. [DOI] [PubMed] [Google Scholar]
- 26.Sumoy L., Wang,C.K., Lichtler,A.C., Pierro,L.J., Kosher,R.A. and Upholt,W.B. (1995) Identification of a spatially specific enhancer element in the chicken Msx-2 gene that regulates its expression in the apical ectodermal ridge of the developing limb buds of transgenic mice. Dev. Biol., 170, 230–242. [DOI] [PubMed] [Google Scholar]
- 27.Pan Z., Lichtler,A.C. and Upholt,W.B. (1998) DNase I hypersensitive sites in the chromatin of the chicken Msx2 gene differ in anterior and posterior limb mesenchyme, calvarial osteoblasts and embryonic fibroblasts. Biochem. Mol. Biol. Int., 46, 549–557. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y., Seto,E., Chang,L.S. and Shenk,T. (1991) Transcriptional repression by YY1, a human GLI-Kruppel-related protein and relief of repression by adenovirus E1A protein. Cell, 67, 377–388. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y., Lee,J.S. and Galvin,K.M. (1997) Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta, 1332, F49–F66. [DOI] [PubMed] [Google Scholar]
- 30.Thomas M.J. and Seto,E. (1999) Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene, 236, 197–208. [DOI] [PubMed] [Google Scholar]
- 31.Shrivastava A. and Calame,K. (1994) An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res., 22, 5151–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bushmeyer S., Park,K. and Atchison,M.L. (1995) Characterization of functional domains within the multifunctional transcription factor, YY1. J. Biol. Chem., 270, 30213–30220. [DOI] [PubMed] [Google Scholar]
- 33.Austen M., Luscher,B. and Luscher-Firzlaff,J.M. (1997) Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem., 272, 1709–1717. [DOI] [PubMed] [Google Scholar]
- 34.Seto E., Shi,Y. and Shenk,T. (1991) YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature, 354, 241–245. [DOI] [PubMed] [Google Scholar]
- 35.Seto E., Lewis,B. and Shenk,T. (1993) Interaction between transcription factors Sp1 and YY1. Nature, 365, 462–464. [DOI] [PubMed] [Google Scholar]
- 36.Shrivastava A., Saleque,S., Kalpana,G.V., Artandi,S., Goff,S.P. and Calame,K. (1993) Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science, 262, 1889–1892. [DOI] [PubMed] [Google Scholar]
- 37.Lee J.S., Galvin,K.M. and Shi,Y. (1993) Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc. Natl Acad. Sci. USA, 90, 6145–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J.S., See,R.H., Galvin,K.M., Wang,J. and Shi,Y. (1995) Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res., 23, 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Usheva A. and Shenk,T. (1996) YY1 transcriptional initiator: protein interactions and association with a DNA site containing unpaired strands. Proc. Natl Acad. Sci. USA, 93, 13571–13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donohoe M.E., Zhang,X., McGinnis,L., Biggers,J., Li,E. and Shi,Y. (1999) Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol., 19, 7237–7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semba I., Nonaka,K., Takahashi,I., Takahashi,K., Dashner,R., Shum,L., Nuckolls,G.H. and Slavkin,H.C. (2000) Positionally-dependent chondrogenesis induced by BMP4 is co-regulated by Sox9 and Msx2. Dev. Dyn., 217, 401–414. [DOI] [PubMed] [Google Scholar]
- 42.Flanagan J.R. (1995) Autologous stimulation of YY1 transcription factor expression: role of an insulin-like growth factor. Cell Growth Differ., 6, 185–190. [PubMed] [Google Scholar]
- 43.Austen M., Cerni,C., Luscher-Firzlaff,J.M. and Luscher,B. (1998) YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene, 17, 511–520. [DOI] [PubMed] [Google Scholar]
- 44.Bell J.R., Noveen,A., Liu,Y.H., Ma,L., Dobias,S., Kundu,R., Luo,W., Xia,Y., Lusis,A.J., Snead,M.L. et al. (1993) Genomic structure, chromosomal location and evolution of the mouse Hox 8 gene. Genomics, 16, 123–131. [DOI] [PubMed] [Google Scholar]
- 45.Monaghan A.P., Davidson,D.R., Sime,C., Graham,E., Baldock,R., Bhattacharya,S.S. and Hill,R.E. (1991) The Msh-like homeobox genes define domains in the developing vertebrate eye. Development, 112, 1053–1061. [DOI] [PubMed] [Google Scholar]
- 46.Dignam J.D. (1990) Preparation of extracts from higher eukaryotes. Methods Enzymol., 182, 194–203. [DOI] [PubMed] [Google Scholar]
- 47.Slavkin H., Nuckolls,G. and Shum,L. (2000) Craniofacial development and patterning. Methods Mol. Biol., 136, 45–54. [DOI] [PubMed] [Google Scholar]
- 48.Marazzi G., Wang,Y. and Sassoon,D. (1997) Msx2 is a transcriptional regulator in the BMP4-mediated programmed cell death pathway. Dev. Biol., 186, 127–138. [DOI] [PubMed] [Google Scholar]
- 49.Marazzi G., Wang,Y. and Sassoon,D. (1997) Msx2 is a transcriptional regulator in the BMP4-mediated programmed cell death pathway. Dev. Biol., 186, 127–138. [DOI] [PubMed] [Google Scholar]
- 50.Hollnagel A., Oehlmann,V., Heymer,J., Ruther,U. and Nordheim,A. (1999) Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J. Biol. Chem., 274, 19838–19845. [DOI] [PubMed] [Google Scholar]
- 51.Barlow A.J. and Francis-West,P.H. (1997) Ectopic application of recombinant BMP-2 and BMP-4 can change patterning of developing chick facial primordia. Development, 124, 391–398. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe Y. and Le Douarin,N.M. (1996) A role for BMP-4 in the development of subcutaneous cartilage. Mech. Dev., 57, 69–78. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi Y., Tonegawa,A., Matsumoto,K., Ueno,N., Kuroiwa,A., Noda,M. and Nifuji,A. (1996) BMP-4 mediates interacting signals between the neural tube and skin along the dorsal midline. Genes Cells, 1, 775–783. [DOI] [PubMed] [Google Scholar]
- 54.Monsoro-Burq A.H., Duprez,D., Watanabe,Y., Bontoux,M., Vincent,C., Brickell,P. and Le Douarin,N. (1996) The role of bone morphogenetic proteins in vertebral development. Development, 122, 3607–3616. [DOI] [PubMed] [Google Scholar]
- 55.Sirard C., Kim,S., Mirtsos,C., Tadich,P., Hoodless,P.A., Itie,A., Maxson,R., Wrana,J.L. and Mak,T.W. (2000) Targeted disruption in murine cells reveals variable requirement for Smad4 in transforming growth factor beta-related signaling. J. Biol. Chem., 275, 2063–2070. [DOI] [PubMed] [Google Scholar]
- 56.Rice D.P., Kim,H.J. and Thesleff,I. (1999) Apoptosis in murine calvarial bone and suture development. Eur. J. Oral Sci., 107, 265–275. [DOI] [PubMed] [Google Scholar]
- 57.Farlie P.G., Kerr,R., Thomas,P., Symes,T., Minichiello,J., Hearn,C.J. and Newgreen,D. (1999) A paraxial exclusion zone creates patterned cranial neural crest cell outgrowth adjacent to rhombomeres 3 and 5. Dev. Biol., 213, 70–84. [DOI] [PubMed] [Google Scholar]
- 58.Maden M., Graham,A., Gale,E., Rollinson,C. and Zile,M. (1997) Positional apoptosis during vertebrate CNS development in the absence of endogenous retinoids. Development, 124, 2799–2805. [DOI] [PubMed] [Google Scholar]
- 59.Phippard D.J., Weber-Hall,S.J., Sharpe,P.T., Naylor,M.S., Jayatalake,H., Maas,R., Woo,I., Roberts-Clark,D., Francis-West,P.H., Liu,Y.H. et al. (1996) Regulation of Msx-1, Msx-2, Bmp-2 and Bmp-4 during fetal and postnatal mammary gland development. Development, 122, 2729–2737. [DOI] [PubMed] [Google Scholar]
- 60.Kim H.J., Rice,D.P., Kettunen,P.J. and Thesleff,I. (1998) FGF-, BMP- and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development, 125, 1241–1251. [DOI] [PubMed] [Google Scholar]
- 61.Dealy C.N. and Kosher,R.A. (1995) Studies on insulin-like growth factor-I and insulin in chick limb morphogenesis. Dev. Dyn., 202, 67–79. [DOI] [PubMed] [Google Scholar]
- 62.Dealy C.N. and Kosher,R.A. (1996) IGF-I, insulin and FGFs induce outgrowth of the limb buds of amelic mutant chick embryos. Development, 122, 1323–1330. [DOI] [PubMed] [Google Scholar]
- 63.Ganan Y., Macias,D., Basco,R.D., Merino,R. and Hurle,J.M. (1998) Morphological diversity of the avian foot is related with the pattern of msx gene expression in the developing autopod. Dev. Biol., 196, 33–41. [DOI] [PubMed] [Google Scholar]
- 64.Ellies D.L., Church,V., Francis-West,P. and Lumsden,A. (2000) The WNT antagonist cSFRP2 modulates programmed cell death in the developing hindbrain. Development, 127, 5285–5295. [DOI] [PubMed] [Google Scholar]
- 65.Romey M.C., Pallares-Ruiz,N., Mange,A., Mettling,C., Peytavi,R., Demaille,J. and Claustres,M. (2000) A naturally occurring sequence variation that creates a YY1 element is associated with increased cystic fibrosis transmembrane conductance regulator gene expression. J. Biol. Chem., 275, 3561–3567. [DOI] [PubMed] [Google Scholar]
- 66.Chen Y., Zhang,Y., Jiang,T.X., Barlow,A.J., St Amand,T.R., Hu,Y., Heaney,S., Francis-West,P., Chuong,C.M. and Maas,R. (2000) Conservation of early odontogenic signaling pathways in Aves. Proc. Natl Acad. Sci. USA, 97, 10044–10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tureckova J., Sahlberg,C., Aberg,T., Ruch,J.V., Thesleff,I. and Peterkova,R. (1995) Comparison of expression of the msx-1, msx-2, BMP-2 and BMP-4 genes in the mouse upper diastemal and molar tooth primordia. Int. J. Dev. Biol., 39, 459–468. [PubMed] [Google Scholar]
- 68.Usheva A. and Shenk,T. (1994) TATA-binding protein-independent initiation: YY1, TFIIB and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell, 76, 1115–1121. [DOI] [PubMed] [Google Scholar]
- 69.Hyde-DeRuyscher R.P., Jennings,E. and Shenk,T. (1995) DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res., 23, 4457–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yant S.R., Zhu,W., Millinoff,D., Slightom,J.L., Goodman,M. and Gumucio,D.L. (1995) High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res., 23, 4353–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McNeil S., Guo,B., Stein,J.L., Lian,J.B., Bushmeyer,S., Seto,E., Atchison,M.L., Penman,S., van Wijnen,A.J. and Stein,G.S. (1998) Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situ: the C-terminus is a principal determinant for nuclear trafficking. J. Cell. Biochem., 68, 500–510. [PubMed] [Google Scholar]
- 72.Bushmeyer S.M. and Atchison,M.L. (1998) Identification of YY1 sequences necessary for association with the nuclear matrix and for transcriptional repression functions. J. Cell. Biochem., 68, 484–499. [PubMed] [Google Scholar]
- 73.Galvin K.M. and Shi,Y. (1997) Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol., 17, 3723–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keeton M.R., Curriden,S.A., van Zonneveld,A.J. and Loskutoff,D.J. (1991) Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor beta. J. Biol. Chem., 266, 23048–23052. [PubMed] [Google Scholar]