Abstract
Background
Differentiating among cardiac diseases associated with left ventricular hypertrophy (LVH) informs diagnosis and clinical care.
Objective
To evaluate if artificial intelligence–enabled analysis of the 12-lead electrocardiogram (ECG) facilitates automated detection and classification of LVH.
Methods
We used a pretrained convolutional neural network to derive numerical representations of 12-lead ECG waveforms from patients in a multi-institutional healthcare system who had cardiac diseases associated with LVH (n = 50,709), including cardiac amyloidosis (n = 304), hypertrophic cardiomyopathy (n = 1056), hypertension (n = 20,802), aortic stenosis (n = 446), and other causes (n = 4766). We then regressed LVH etiologies relative to no LVH on age, sex, and the numerical 12-lead representations using logistic regression (“LVH-Net”). To assess deep learning model performance on single-lead data analogous to mobile ECGs, we also developed 2 single-lead deep learning models by training models on lead I (“LVH-Net Lead I”) or lead II (“LVH-Net Lead II”) from the 12-lead ECG. We compared the performance of the LVH-Net models to alternative models fit on (1) age, sex, and standard ECG measures, and (2) clinical ECG-based rules for diagnosing LVH.
Results
The areas under the receiver operator characteristic curve of LVH-Net by specific LVH etiology were cardiac amyloidosis 0.95 [95% CI, 0.93–0.97], hypertrophic cardiomyopathy 0.92 [95% CI, 0.90–0.94], aortic stenosis LVH 0.90 [95% CI, 0.88-0.92], hypertensive LVH 0.76 [95% CI, 0.76-0.77], and other LVH 0.69 [95% CI 0.68-0.71]. The single-lead models also discriminated LVH etiologies well.
Conclusion
An artificial intelligence–enabled ECG model is favorable for detection and classification of LVH and outperforms clinical ECG-based rules.
Keywords: Artificial intelligence, Electrocardiography, Hypertrophic heart disease, Hypertrophic cardiomyopathy, Cardiac amyloidosis
Key Findings.
-
•
A 12-lead electrocardiogram (ECG)-based artificial intelligence algorithm (“LVH-Net”) classified cardiac diseases associated with left ventricular hypertrophy (LVH), including cardiac amyloidosis, hypertrophic cardiomyopathy, aortic stenosis, hypertensive heart disease, and other causes of LVH, with favorable performance. LVH-Net outperformed comparison models based on quantitative electrocardiogram (ECG) measures, including heart rate and standard ECG measures, as well as traditional ECG-based rules for diagnosing LVH used in clinical practice.
-
•
Single-lead versions of LVH-Net trained on 12-lead ECG data from lead I (“LVH-Net Lead I”) and lead II (“LVH-Net lead II”) also outperformed the comparison models for classification of cardiac diseases associated with LVH, suggesting that these models may be applicable to mobile ECG data.
-
•
LVH-Net may have clinical utility as a fully automated ECG-based model for detection and classification of LVH.
Introduction
Left ventricular hypertrophy (LVH) is defined as increased left ventricular mass and is associated with cardiovascular morbidity and mortality.1,2 LVH is caused by a spectrum of cardiovascular diseases, including hypertension, aortic stenosis, hypertrophic cardiomyopathy, and cardiac amyloidosis, among others. Discriminating among etiologies of increased left ventricular mass has important implications for treatment and prognosis.3, 4, 5
Deep learning models have shown promise in classifying etiologies of ventricular hypertrophy, including cardiac amyloidosis and hypertrophic cardiomyopathy, using 12-lead electrocardiograms (ECGs).6,7 Developing models that can differentiate cardiac diseases associated with LVH may facilitate efficient diagnosis and treatment, and potentially prevent morbidity. Given the increased availability of mobile devices capable of recording single-lead ECGs, models that accurately discriminate conditions associated with LVH on single-lead ECGs may enable scalable detection of disease in large populations. We sought to develop deep learning models to detect LVH and classify cardiac diseases that cause LVH using both 12-lead and single-lead ECG data.
Using a large retrospective sample of individuals receiving longitudinal cardiology care, we identified cardiovascular diseases that cause LVH, including cardiac amyloidosis, hypertrophic cardiomyopathy, LVH owing to hypertension, LVH owing to aortic stenosis, and LVH owing to other causes. We assessed the extent to which a pretrained deep learning model applied to both 12-lead and single-lead ECG waveform data could detect LVH and discriminate among etiologies of LVH. We compared the performance of the deep learning models against alternative models using structured ECG data, and to traditional ECG-based rules for diagnosing LVH. We also evaluated potential associations between deep learning model predictions and clinical outcomes.
Methods
Study sample
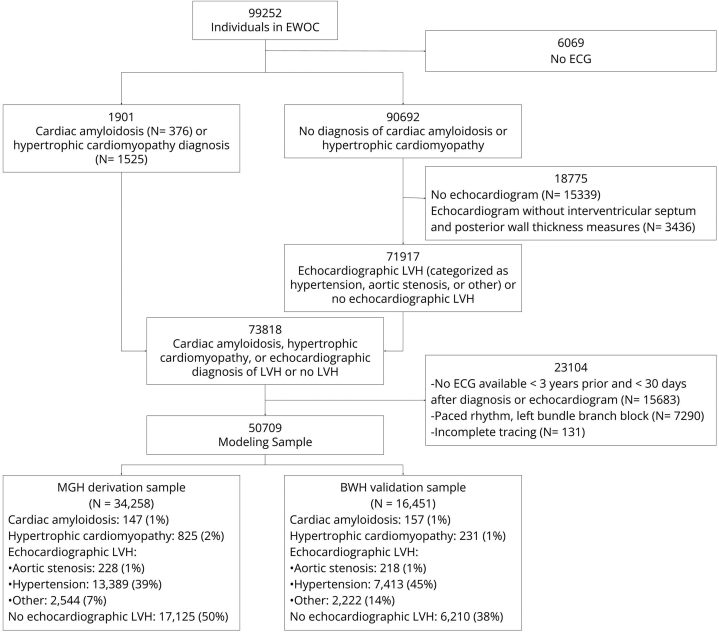
We identified adult patients aged 18 years or older who received longitudinal ambulatory cardiology care at a multi-institutional academic healthcare system (Mass General Brigham). We identified 99,252 patients with at least 1 pair of cardiology clinic visits 1–3 years apart occurring between 2000 and 2019. We collected a broad range of electronic health record (EHR) data, including demographics, anthropometrics, vital signs, narrative notes, laboratory results, medication lists, radiology and cardiology diagnostic test results, pathology reports, and procedural and diagnostic administrative billing codes, as previously described.8 We refer to the retrospective dataset as the Enterprise Warehouse of Cardiology (EWOC). For the present analysis, patients without an ECG (n = 6069) were excluded, resulting in 93,138 patients eligible for inclusion (Figure 1).
Figure 1.
CONSORT diagram of Massachusetts General Hospital (MGH) derivation and Brigham and Women’s Hospital (BWH) validation samples. ECG = electrocardiogram; EWOC = Enterprise Warehouse of Cardiology; LVH = left ventricular hypertrophy.
LVH ascertainment and classification
We identified potential patients with cardiac diseases associated with LVH based on clinical diagnoses, cardiac imaging, and pathology data. Ascertainment and adjudication of cardiac amyloidosis and hypertrophic cardiomyopathy diagnoses were performed by a clinical reviewer (Supplemental Methods). Patients without a diagnosis of cardiac amyloidosis or hypertrophic cardiomyopathy after adjudication were still considered for inclusion in the echocardiographic LVH categories below. We established mutually exclusive categories of LVH-related diseases for all analyses (Supplemental Table 1). Study availability and diagnostic modality sources for cardiac amyloidosis and hypertrophic cardiomyopathy are presented in Supplemental Tables 2–5.
We included all individuals with cardiac amyloidosis and hypertrophic cardiomyopathy identified by comprehensive review of cardiac imaging and pathology data (Supplemental Methods) in our definition of LVH regardless of echocardiographic criteria for LVH (see below), given the potential imperfect sensitivity of echocardiography for LVH in these diseases.9,10 Although echocardiographic LVH was not required for a cardiac amyloidosis or hypertrophic cardiomyopathy diagnosis, 273 of 304 (89%) individuals with cardiac amyloidosis and 874 of 1056 (83%) individuals with hypertrophic cardiomyopathy met echocardiographic LVH criteria prior to diagnosis.
After identification of individuals with cardiac amyloidosis or hypertrophic cardiomyopathy, we evaluated the remaining individuals with an echocardiogram to identify those with echocardiographic LVH (Figure 1). We defined echocardiographic LVH by a string-matching algorithm applied to echocardiogram reports (Supplemental Table 6) and an alternative definition of an average combined interventricular septal and posterior wall thickness >11 mm.11 We further delineated individuals with echocardiographic LVH by likely etiology hierarchically, resulting in mutually exclusive categories: LVH due to aortic stenosis (“aortic stenosis LVH”), LVH due to hypertension (“hypertensive LVH”), and LVH due to another cause (“other LVH”). We first assessed for aortic stenosis LVH, defined as a concurrent diagnosis of moderate or greater aortic stenosis on an echocardiogram with LVH. Following identification of individuals with aortic stenosis LVH, we identified individuals with hypertensive LVH, defined as echocardiographic LVH with a prior diagnosis of hypertension (see clinical feature ascertainment). Individuals with echocardiographic LVH that did not meet criteria for an aortic stenosis or hypertension etiology were grouped into the “LVH other” category. Finally, remaining individuals were categorized as “No LVH” if they had at least 1 echocardiogram report available that reported both posterior wall thickness and interventricular septal thickness but did not meet echocardiographic LVH criteria.
ECG acquisition
Study sample ECGs were performed during routine clinical care by GE Healthcare machines (models MAC5500 and MAC5000) and accessed via the MUSE Cardiology Information System database of Mass General Brigham (GE Healthcare; “MUSE” software versions 8.0 and 9.0). ECG waveforms contain voltage measurements for each of the 12 leads sampled at 240 (8%), 250 (41%), or 500 Hz (51%) for a 10-second duration. ECGs were not excluded based on sampling frequency. ECGs missing a complete 10-second tracing for each lead were excluded (n = 131, Figure 1). The MUSE database includes automated and physician-entered diagnostic statements; tabular ECG measures including axes, intervals, and lead-specific voltage amplitudes; and raw ECG waveforms. Similar to prior studies, and to facilitate comparison to clinical ECG LVH rules (see below), we additionally excluded ECGs with physician- or computer-entered diagnoses of electronically paced rhythms and left bundle branch block (n = 7290, Figure 1).7,12
We selected an index ECG from each individual closest to the date of hypertrophic cardiomyopathy or cardiac amyloidosis diagnosis, or closest to the date of the earliest echocardiogram demonstrating LVH. In the case of the “No LVH” category, the date of the earliest echocardiogram was used. ECGs performed more than 3 years before or 30 days after the date of diagnosis or echocardiogram were excluded (n = 15,688, Figure 1). The mean time from the index ECG to the date of diagnosis was 75 ± 191 days.
Clinical feature ascertainment
Age, sex, race, height, weight, and systolic blood pressure were ascertained from the EHR. Patient age was defined at the time of the index ECG. Vital sign data including height, weight, and systolic blood pressure were ascertained from the closest EHR entry prior to the date of the index ECG. Antihypertensive use was determined via a medication list (Supplemental Table 7).13 Additional patient characteristics, also ascertained prior to the date of the index ECG, were defined using previously published groupings of International Classification of Diseases, 9th and 10th revision diagnosis codes.8 Definitions for clinical features used in the analysis are provided in Supplemental Table 8.
Statistical analysis
Model derivation was performed among individuals for whom an index ECG was performed at the Massachusetts General Hospital (MGH), and model validation was performed among individuals for whom the index ECG was performed at the Brigham and Women’s Hospital (BWH) (Figure 1). Individuals who had ECGs at both sites were prioritized for inclusion in the MGH derivation sample. The partitioned datasets comprised independent ECGs and patients.
For the deep learning model, we first transformed each index ECG into a 320-dimensional vector representation using a previously described deep learning approach known as Patient Contrastive Learning of Representations (PCLR), which is optimized for training efficiency (Figure 2).14 We used a publicly available PCLR convolutional neural network trained on 3 million inpatient and outpatient ECGs from MGH that creates numerical representations of ECG waveforms that emphasize differences between ECGs from different individuals and similarities between ECGs from the same individual. Unlike other ECG deep learning models, PCLR is designed not to predict a specific clinical outcome, but rather to produce 320-dimensional numerical representations of ECG waveforms that can be used as input into linear models such as logistic regression. Linear interpolation is applied before neural network preprocessing so that each input is 2500 voltage measurements regardless, allowing for variability in sample frequency. The PCLR numerical representations, along with patient age and sex, were then included in a multinomial logistic regression model with Ridge regularization (“LVH-Net”), trained to simultaneously classify cardiac amyloidosis, hypertrophic cardiomyopathy, aortic stenosis LVH, hypertensive LVH, other LVH, and no LVH, where the “No LVH” group served as the baseline comparator. Ridge regularization parameters were tuned by 5-fold cross-validation. We also developed 2 analogous single-lead deep learning models, “LVH-Net Lead I” and “LVH-Net Lead II,” using 320-dimensional PCLR representations derived from ECG waveform data from only the respective leads (Figure 2). We selected leads I and II for single-lead modeling based on their frequent availability among mobile ECG technology.15 The single-lead model deep learning representations used as input into LVH-Net Lead I and II were derived from separate PCLR convolutional neural network models trained on waveform data from only those leads.
Figure 2.
Modeling overview. Modeling schema of comparison models (A) and deep learning models (B). A: The Age and Sex Model includes terms for patient age and sex in a logistic regression model. The ECG Measures Model includes quantitative electrocardiogram (ECG) measures along with patient age and sex in a logistic regression model. B: In LVH-Net, the 12-lead ECG waveform is first converted into a 320-dimensional representation using PCLR (Patient Contrastive Learning of Representations; see text). The representation, along with patient age and sex, are input into a logistic regression model that is trained to classify left ventricular hypertrophy (LVH) etiology, including cardiac amyloidosis, hypertrophic cardiomyopathy, aortic stenosis LVH, hypertensive LVH, other LVH, and no LVH. LVH-Net Lead I and LVH-Net Lead II models use PCLR representations derived only from ECG waveform data of ECG leads I and II, respectively.
We also trained comparator models, including an age and sex model and an “ECG Measures” model including age, sex, heart rate, PR, QRS, and QT intervals, and voltage amplitudes from the lead V1 S wave, aVL R wave, V3 S wave, V5 R wave, and V6 R wave (Figure 2). The ECG measures were selected based on previously described relations with LVH-related diagnoses including hypertrophic cardiomyopathy and cardiac amyloidosis,16,17 and inclusion in traditional ECG LVH criteria (see below).18,19 Where possible, we extracted maximum lead voltage from automated measurements in MUSE and inferred missing maximum voltage amplitudes where these values were missing (Supplemental Methods). Automated voltage measurements were present for 78% of ECGs in the derivation set and 88% of ECGs in the validation set. The voltage inference model performed with a mean absolute error of 0.05 mV in a separate validation set comprised of MGH ECGs.
We assessed model performance in the validation sample using area under the receiver operating characteristic curve (AUROC), and area under the precision-recall curve (AUPRC) for classification of each LVH etiology vs no LVH. We generated distributions of AUROC and AUPRC from 1000-iteration bootstrapping with replacement. We estimated 95% confidence intervals for AUROC and AUPRC from these distributions. We also measured P values for metric differences between pairs of models by applying the 2-sided Wilcoxon signed rank test to the bootstrapped distributions. We compared test characteristics including positive predictive value (PPV), negative predictive value (NPV), positive likelihood ratio (LR+), and negative likelihood ratio (LR-), and the number needed to screen (NNS) at probability thresholds corresponding to model sensitivities and specificities of 99%, 90%, and 80%.20 The number needed to screen estimates the number of individuals a model must classify as positive to detect 1 true positive. Confidence intervals for test characteristics were calculated using an exact binomial method. In sensitivity analyses, we used the last echocardiogram as the reference date of the “No LVH” category in the validation sample, which resulted in an increase in the median year of the No LVH index ECGs from 2012 to 2015 with equivalent year ranges (2000–2019) compared to the primary analysis. We also performed sensitivity analyses in which we assessed deep learning model performance for 1 vs rest classification of each LVH etiology (ie, data from the LVH etiology of interest is treated as positive, and all other classes including no LVH are treated as negative). As part of the 1 vs rest sensitivity analyses, we evaluated model performance in the subgroup of individuals in the validation set demonstrating echocardiographic LVH.
To gain clinical insight into how the deep learning model classifies individuals, we evaluated differences in ECG waveforms corresponding to individuals predicted to be at the highest (³95th) and lowest risk (£5th) percentiles of LVH-Net predicted risk of cardiac amyloidosis and hypertrophic cardiomyopathy. We selected median waveforms from leads I, II, V1, and V5 for evaluation, and aligned the median waveforms at the onset of the P wave to facilitate comparison.
We also assessed whether commonly used clinical ECG LVH rules can discriminate among cardiac diseases causing LVH from those without LVH and compared performance of clinical ECG LVH rules to the deep learning models. We applied 3 commonly used ECG-based rules for diagnosing LVH: Cornell voltage criteria (R wave in aVL + S wave in V3 > 28 mm for males or > 20 mm for females), Modified Cornell criteria (R wave in aVL > 11 mm), and Sokolow-Lyon criteria (S wave in V1 + tallest R wave in V5–V6 > 35 mm).18,19 An ECG was considered to demonstrate rule-based LVH if any of the 3 criteria were met. The number of patients that met clinical ECG LVH criteria by LVH etiology is shown in Supplemental Table 9.
We compared the deep learning models to the clinical ECG LVH rules by first calculating the specificity, sensitivity, PPV, and NPV of the clinical ECG LVH rules for detection of echocardiographic LVH and each LVH etiology vs no LVH. We then found the threshold probability at which the deep learning models yielded an equivalent specificity or sensitivity to the clinical rules, and calculated the corresponding sensitivity (or specificity) and PPV of the deep learning models using this threshold. We then visualized the test characteristics and associated 95% confidence intervals for LVH-Net and the clinical ECG rules for comparison.
We evaluated potential associations between LVH-Net predicted probability of cardiac amyloidosis and hypertrophic cardiomyopathy with incident heart failure, incident atrial fibrillation, and mortality, after excluding individuals in the validation sample with known cardiac amyloidosis or hypertrophic cardiomyopathy. We defined incident heart failure and atrial fibrillation using previously described groupings of diagnostic codes (Supplemental Table 8).21,22 For time-to-event analyses, person-time began at time of the index ECG and ended at the primary outcome, death, or last clinical encounter. For each analysis, we omitted individuals with the primary outcome occurring prior to, or on the same day as, the index ECG. Incidence rates were calculated by dividing the number of incident events by 100 person-years of follow-up. We fit multivariable Cox proportional hazards models adjusted for age and sex to assess the relations between LVH-Net probabilities of cardiac amyloidosis and hypertrophic cardiomyopathy with heart failure, atrial fibrillation, or mortality as the outcomes. Prior to regression, LVH-Net probabilities were first transformed to the logit scale using the logit transformation logit(x) = log (x/[1 − x]) to achieve an approximately normal distribution of probabilities.23 Hazard ratios for LVH-Net probabilities are reported per standard deviation on the logit scale. We also calculated age- and sex-stratified cumulative incidence rates for each clinical outcome.
All analyses were performed in Python v3.8 using packages “pandas,” “sklearn,” “tensorflow,” and “lifelines.”24, 25, 26, 27, 28
Results
Study sample
We included 50,709 individuals with a diagnosis of cardiac amyloidosis, hypertrophic cardiomyopathy, echocardiographic LVH, or no echocardiographic LVH in the modeling sample (Figure 1). The MGH derivation sample included 34,258 individuals (68%), among whom 40% were female and mean age was 63.1 ± 15.3 years. The BWH validation sample included 16,451 individuals (32%), among whom 44% were female and mean age was 62.7 ± 15.0 years. LVH etiologies in the derivation and validation samples included the following: cardiac amyloidosis (n = 147 vs 157), hypertrophic cardiomyopathy (n = 825 vs 231), aortic stenosis LVH (n = 228 vs 218), hypertensive LVH (n = 13,389 vs 7413), other LVH (n = 2544 vs 2222), and no echocardiographic LVH (n = 17, 125 vs 6210). Characteristics of the derivation and validation samples are presented in Table 1.
Table 1.
Characteristics of derivation (Massachusetts General Hospital) and validation (Brigham and Women’s Hospital) samples
| Derivation sample (MGH N = 34,258) | Validation sample (BWH N = 16,451) | |
|---|---|---|
| Age, y | 63.2 ± 15.3 | 62.7 ± 15.0 |
| Female sex | 13,622 (39.8) | 7309 (44.4) |
| Height, in | 66.8 ± 7.8 | 66.8 ± 11.1 |
| Weight, lb | 183.4 ± 50.1 | 184.3 ± 48.6 |
| Systolic blood pressure, mm Hg | 128.7 ± 18.5 | 132.0 ± 19.8 |
| Diastolic blood pressure, mm Hg | 74.4 ± 10.9 | 75.7 ± 12.5 |
| Diabetes | 6512 (19.0) | 3665 (22.3) |
| Coronary artery disease | 17,212 (50.2) | 8052 (48.9) |
| Myocardial infarction | 7444 (21.7) | 5198 (31.6) |
| Valvular disease | 5791 (16.9) | 3053 (18.6) |
| Atrial fibrillation | 10,615 (31.0) | 5132 (31.2) |
| Obesity | 2415 (7.0) | 1361 (8.3) |
| Chronic kidney disease | 5594 (16.3) | 2959 (18.0) |
| Hypertension | 24,926 (72.8) | 11,342 (68.9) |
| Antihypertensive medication | 26,641 (77.8) | 13,871 (84.3) |
| Ethnicity | ||
| White | 29,888 (87.2) | 13,315 (80.9) |
| Black | 1125 (3.3) | 1239 (7.5) |
| Asian | 920 (2.7) | 348 (2.1) |
| Hispanic | 612 (1.8) | 595 (3.6) |
| Other | 971 (2.8) | 482 (2.9) |
| Mixed | 1 (0.0) | 1 (0.0) |
| Unknown | 745 (2.2) | 472 (2.9) |
| Clinical ECG LVH rules | ||
| aVL | 3154 (9.2) | 1397 (8.5) |
| Sokolow-Lyon | 1179 (3.4) | 562 (3.4) |
| Cornell | 2763 (8.1) | 1278 (7.8) |
| Any† | 5463 (15.9) | 2459 (14.9) |
Data are displayed as mean ± standard deviation or n (%) unless otherwise noted.
BWH = Brigham and Women’s Hospital; ECG = electrocardiogram; LVH = left ventricular hypertrophy; MGH = Massachusetts General Hospital.
Clinical ECG LVH criteria “Any” category aggregates aVL, Sokolow-Lyon, and Cornell criteria.
Model performance
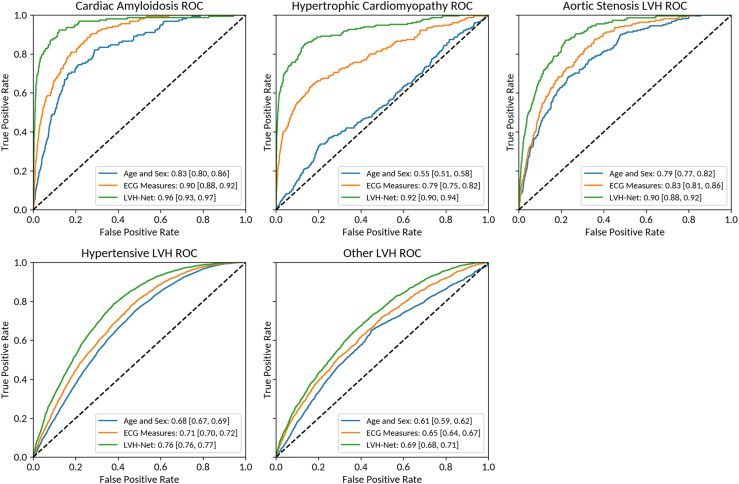
When applied to the validation sample, LVH-Net significantly outperformed the comparison models for classification of each LVH etiology vs no LVH as judged by the AUROC and AUPRC (Figure 3 and Supplemental Figure 1). LVH-Net discrimination point estimates for classification of LVH etiologies were as follows: cardiac amyloidosis (AUROC 0.96 [95% CI, 0.94–0.97]), hypertrophic cardiomyopathy (AUROC 0.92 [95% CI, 0.90–0.94]), aortic stenosis LVH (AUROC 0.90 [95% CI, 0.88–0.92]), hypertensive LVH (AUROC 0.76 [95% CI, 0.76–0.77]), and other LVH (AUROC 0.69 [95% CI, 0.68–0.70]) (Figure 3). The AUPRCs for classification of LVH etiologies were as follows: cardiac amyloidosis 0.55 [95% CI, 0.48–0.63], hypertrophic cardiomyopathy 0.59 [95% CI, 0.53–0.66], aortic stenosis LVH 0.29 [95% CI, 0.24–0.35], hypertensive LVH 0.76 [95% CI, 0.75–0.77], and other LVH 0.44 [95% CI, 0.42–0.45] (Supplemental Figure 1). LVH-Net performance remained consistent in sex-stratified analyses (Supplemental Table 10), as well as in additional analyses stratified by age and clinical ECG rule positivity (Supplemental Table 11). Sensitivity analyses in which the No LVH group was defined by the last available echocardiogram included an additional 1523 individuals in the No LVH group compared to the primary analysis owing to increased availability of ECGs meeting inclusion criteria. In these analyses, LVH-Net model performance remained significantly higher than the comparison models (Supplemental Table 12).
Figure 3.
Receiver operating characteristic curves (ROC) of LVH-Net and comparison models by left ventricular hypertrophy (LVH) etiology vs no LVH. ROC curves of comparison models (age and sex, electrocardiogram [ECG] measures) and LVH-Net by LVH etiology vs no LVH comparator with corresponding area under the ROC and 95% confidence interval shown in the legend. Dashed line shows expected performance of “no skill” random classifier at area under the curve of 0.5.
When differentiating a specific cause of LVH (eg, cardiac amyloidosis) from all other causes of LVH (including no LVH), we found that LVH-Net also outperformed the other comparison models, classifying cardiac amyloidosis and hypertrophic cardiomyopathy with AUROCs of 0.93 [95% CI, 0.90–0.95] and 0.88 [95% CI, 0.85–0.91], respectively (Supplemental Figures 2 and 3, Supplemental Table 13). One vs rest performance remained consistent in age- and sex-stratified analyses (Supplemental Table 14). We found that in the subgroup of 10,174 (62%) individuals in the validation set demonstrating echocardiographic LVH, discriminatory performance remained high for both cardiac amyloidosis (AUROC 0.92 [95% CI, 0.90--0.95]) and hypertrophic cardiomyopathy (AUROC 0.86 [95% CI, 0.83–0.90]) (Supplemental Table 14).
Deep learning models trained on single-lead waveforms (LVH-Net Leads I and II) also accurately classified LVH etiology vs no LVH (Supplemental Figures 4 and 5). The AUROCs for LVH-Net Lead I classification of cardiac amyloidosis and hypertrophic cardiomyopathy were 0.90 [95% CI, 0.88–0.92] (P < .05 for comparison to ECG Measures AUROC), and 0.90 [95% CI, 0.88–0.92] (P < .05), respectively, while the AUROCs for LVH-Net Lead II classification of cardiac amyloidosis and hypertrophic cardiomyopathy were 0.93 [95% CI, 0.91–0.95] (P < .05) and 0.85 [95% CI, 0.82–0.88] (P < .05), respectively. Performance of the single-lead models was maintained in age- and sex-stratified analyses (Supplemental Tables 15 and 16). Performance of the single-lead models for 1 vs rest classification of LVH etiology is shown in Supplemental Figures 2 and 3. Calibration curves for the deep learning and comparison models are presented in Supplemental Figures 6 and 7.
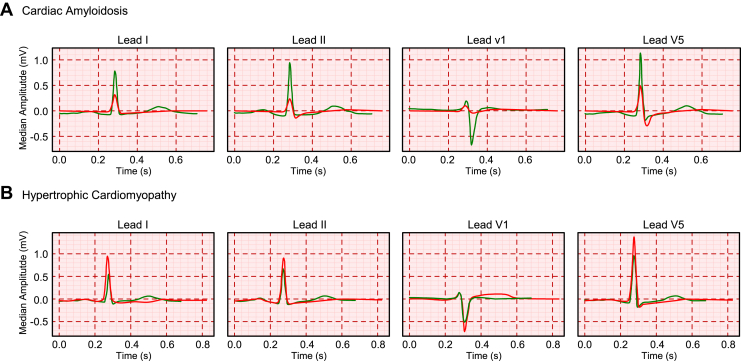
ECG waveforms from individuals at highest predicted risk (³95th percentile) of cardiac amyloidosis had lower voltages and wider QRS complexes compared to waveforms from individuals at lower risk (£5th percentile), whereas waveforms from individuals at higher predicted risk of hypertrophic cardiomyopathy had higher voltages and wider QRS complexes compared to waveforms from lower-risk individuals (Figure 4).
Figure 4.
Median waveforms of electrocardiograms (ECGs) predicted to be at low risk (5th percentile) (green) and high risk (95th percentile) (red) of LVH-Net risk of A: cardiac amyloidosis and B: hypertrophic cardiomyopathy. Grid measures correspond to standard ECG scaling (1 small box per 0.1 mm on y-axis, and 1 small box per 0.2 seconds on x-axis). Median waveforms are shown for leads I, II, V1, and V5.
We compared test characteristics among the models across a range of sensitivity and specificity thresholds for LVH etiologies vs no LVH in Supplemental Tables 17–21. At a set specificity of 99%, LVH-Net demonstrated a corresponding PPV of 57.8% [95% CI, 50.1–65.5], positive likelihood ratio of 53.8, and number need to screen of 2 for detection of cardiac amyloidosis and a PPV of 65.8 [95% CI, 50.6–71.9], positive likelihood ratio of 51.5, and number need to screen of 2 for detection of hypertrophic cardiomyopathy (Supplemental Table 22). In 1 vs rest analyses, LVH-Net had a corresponding PPV of 26.6% [95% CI, 19.7–33.5] and 32.8% [95% CI, 26.8–38.9] for detection of cardiac amyloidosis and hypertrophic cardiomyopathy, respectively, at a set specificity of 99% (Supplemental Table 23).
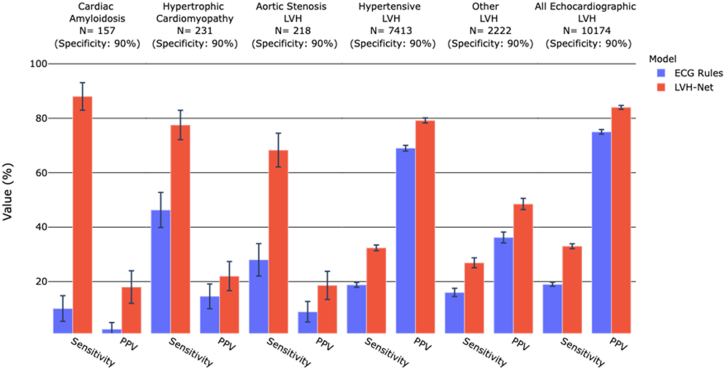
LVH-Net outperformed the clinical ECG rules for classification of LVH etiology (Figure 5 and Supplemental Figure 8). LVH-Net was also more sensitive than clinical ECG-based rules for detecting echocardiographic LVH. Of 10,174 individuals with echocardiographic LVH in the validation sample, LVH-Net detected 3411 cases (LVH-Net sensitivity = 33.8%) compared to 1918 by the clinical ECG rules (ECG rule sensitivity = 18.9%) at the observed clinical rule specificity of 90% (P < .05 for comparison).
Figure 5.
Test characteristics of clinical electrocardiogram (ECG) rules vs deep learning models for classification of left ventricular hypertrophy (LVH) etiology vs no LVH at equivalent specificity. Grouped bar plot of sensitivity and positive predictive value (PPV) for classification of LVH etiology and echocardiographic LVH vs no LVH by clinical ECG LVH rules (blue) and LVH-Net (red). LVH-Net test characteristics were calculated at the probability cutoff that yielded a specificity equal to the specificity of the clinical ECG LVH rules. The specificity of the clinical ECG rules is shown above each facet, along with the number of cases in the validation sample. Error bars represent 95% confidence interval.
Association between predicted LVH-Net risk and clinical outcomes
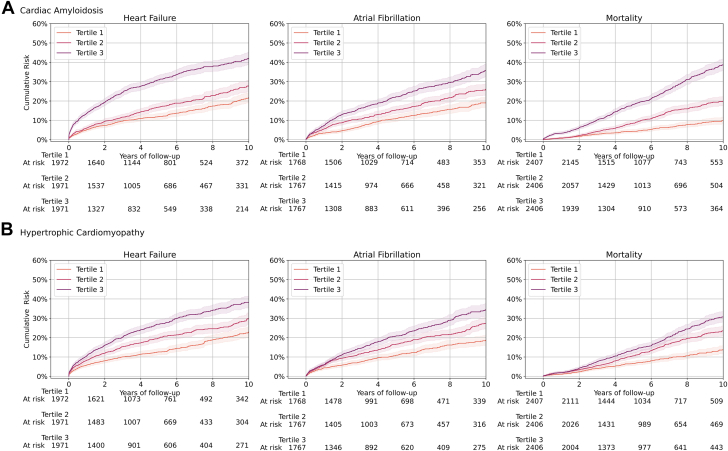
Greater LVH-Net predicted risk of cardiac amyloidosis and hypertrophic cardiomyopathy was associated with incident heart failure, atrial fibrillation, and mortality (Figure 6 and Supplemental Figure 9). For example, the cumulative incidence of mortality at 10 years was 39% in the highest LVH-Net predicted cardiac amyloidosis tertile vs 10% in the lowest tertile, and 31% in the highest predicted cardiomyopathy tertile vs 13% in the lowest. In age- and sex-adjusted proportional hazards models, LVH-Net predicted cardiac amyloidosis was associated with a 1.65-fold [95% CI, 1.58–1.72] increased hazard (per 1-unit standard deviation increase on the logit scale) for mortality, whereas LVH-Net predicted hypertrophic cardiomyopathy was associated with a 1.12-fold [95% CI, 1.08–1.17] increased hazard. Age- and sex-stratified cumulative incidence curves for each outcome are presented in Supplemental Figures 10–13, and age- and sex-adjusted proportional hazards model results are presented in Supplemental Table 24.
Figure 6.
Cumulative incidence of clinical outcomes by predicted LVH-Net risk of cardiac amyloidosis and hypertrophic cardiomyopathy. Cumulative incidence of heart failure, atrial fibrillation, and mortality by tertiles of increasing predicted LVH-Net risk of A: cardiac amyloidosis and B: hypertrophic cardiomyopathy in the validation sample, excluding individuals with known cardiac amyloid and hypertrophic cardiomyopathy. At-risk counts for each tertile are shown below each plot. Error bars represent 95% confidence intervals.
Discussion
We report the results of LVH-Net and LVH-Net Leads I and II, a collection of deep learning models for classification of cardiac diseases associated with LVH using 12-lead and single-lead ECG waveforms. We found that LVH-Net accurately classifies diagnoses across a range of cardiac diseases associated with LVH, including cardiac amyloidosis, hypertrophic cardiomyopathy, and aortic stenosis, with AUROCs of 0.95, 0.92, and 0.90 respectively. LVH-Net, and single-lead versions of LVH-Net, outperformed comparison models fit on quantitative ECG measures, as well as commonly used clinical ECG LVH rules, in the classification of LVH and LVH etiologies. We also demonstrate that predicted LVH-Net risk of cardiac amyloidosis and hypertrophic cardiomyopathy were associated with clinical outcomes including heart failure, atrial fibrillation, and mortality, indicating that model predictions are associated with expected cardiovascular outcomes.3,29 With strong performance across LVH and LVH etiologies, LVH-Net may have potential clinical utility as a fully automated point-of-care screening tool for LVH and rare LVH etiologies with application to both 12-lead ECGs and mobile single-lead ECG devices.
Prior work has demonstrated the capability of ECG-based deep learning models for classification of hypertrophic cardiomyopathy, cardiac amyloidosis, and echocardiographic LVH.6,7,30 LVH-Net expands on this work by enabling simultaneous discrimination of echocardiographic LVH and multiple cardiac diseases associated with LVH, including hypertrophic cardiomyopathy and cardiac amyloidosis. We demonstrate that the use of a pretrained, publicly available contrastive learning model produces meaningful model inputs that can be applied to other classification problems. Although deep learning models have not yet been widely implemented in clinical practice, expanding the range of diseases classified by a single model such as LVH-Net could streamline implementation and, importantly, reduce the computational complexity of applying individual deep learning models in parallel. Our work also demonstrates the potential application of LVH-Net to single-lead ECG data equivalent to what is currently available from mobile ECG devices.
Left ventricular hypertrophy is a common clinical entity found in about 5%–15% of the general population, and is associated with significant cardiovascular morbidity.1,2 Treatment and regression of LVH is associated with improved cardiovascular outcomes, and therefore identification of untreated individuals with LVH presents a public health opportunity.31 Though the 12-lead ECG has been used to screen for LVH, clinical ECG rules employed in routine clinical practice are cumbersome and insensitive.32,33 LVH-Net is fully automated and also demonstrated significantly improved sensitivity for classification of echocardiographic LVH and LVH etiologies when compared to the clinical ECG LVH rules, and may therefore offer greater utility in identifying individuals in the community at increased cardiovascular risk. Individuals identified as having a high probability of LVH may benefit from clinical evaluation and echocardiogram. In addition, LVH-Net may have application in the longitudinal management of patients such as those with hypertensive LVH by supplying additional information on treatment effect marked by LVH regression.
In addition to classifying individuals with LVH, LVH-Net is also able to accurately classify LVH etiology. Cardiac amyloidosis and hypertrophic cardiomyopathy are important diseases associated with significant cardiovascular morbidity, and expedient diagnosis of these diseases is essential for improving patient outcomes and identifying at-risk relatives.3,5,34 Currently, there are no standardized screening approaches for cardiac amyloidosis and hypertrophic cardiomyopathy, and traditional ECG diagnosis of these diseases relies on insensitive or nonspecific findings.35,36 At a specificity threshold of 99%, individuals classified as having cardiac amyloidosis or hypertrophic cardiomyopathy by LVH-Net had a greater than 1 in 2 chance of carrying either of these critical diagnoses, while at the same time, over 99% of individuals classified as negative did not have either of these diagnoses. Furthermore, both models yielded a low number needed to screen of 2 at a specificity of 99%, suggesting that for every 2 individuals flagged as positive by LVH-Net, 1 new disease diagnosis would be expected (assuming full availability and 100% accuracy of confirmatory diagnostic testing for each positive LVH-Net prediction). Although PPV, NPV, and NNS are affected by the disease prevalence in our sample, our results nevertheless suggest that LVH-Net could increase the yield of advanced diagnostic testing for these diseases.
Subgroup analyses restricted to individuals with echocardiographic LVH demonstrate that LVH-Net retains performance for hypertrophic cardiomyopathy and cardiac amyloidosis even among individuals already known to have imaging evidence of LVH. In addition to screening healthy individuals, LVH-Net may therefore hold additional utility in facilitating discrimination of the etiology of echocardiographic LVH and indicating the need for further testing to confirm the diagnosis.
Results of LVH-Net models trained using single-lead data show the potential application of our model to single-lead ECG data. Prior work has shown that deep learning models trained on 12-lead and single-lead ECG waveforms, which leverage the relative ubiquity of ECG data compared to mobile data, can be subsequently applied to mobile ECG device data without retraining.37,38 Here, the single-lead deep learning models demonstrated favorable performance, highlighting the ability of deep learning tools to extract meaningful representations from limited ECG data in discriminating LVH. Our findings may be applicable to mobile ECG devices, which present an emerging opportunity for scalable population-based cardiovascular screening. However, more work must be done to evaluate the generalizability of the LVH-Net models to wearable device data and the population of device users.39,40
Our study must be interpreted in the context of its limitations. Use of mutually exclusive LVH cardiomyopathy definitions may misclassify individuals with multiple LVH etiologies. In addition, given our reliance on gold-standard diagnostic data for identification of individuals with hypertrophic cardiomyopathy and cardiac amyloidosis, individuals in our dataset may have a more pronounced phenotype of these diseases, making classification an easier task. Furthermore, transthyretin amyloidosis is under-recognized, and so undiagnosed individuals in our sample may be misclassified.41 Notably, owing to limitations in available data we were unable to classify the athletic heart, as well as other rare causes of LVH such as glycogen storage diseases. We were also unable to subtype cardiac amyloid (light chain vs transthyretin) present in our dataset. The study population was largely composed of older white individuals (87% white), and though age-stratified analysis showed consistent performance, our results may not generalize to younger individuals or those with varying racial or ethnic compositions. ECGs were performed during routine clinical care and individuals without an ECG (though few) were excluded, limiting the study sample. Furthermore, LVH is associated with sex-specific differences in cardiovascular risk and, although our results were consistent across sex strata, further study may be necessary to extend these results to larger samples.42,43 In addition, although the model was derived and validated within 2 separate hospitals, these sites are mainly referral centers and exist within the same healthcare system, which may introduce selection bias and increases the prevalence of rare diseases such as cardiac amyloidosis compared to the general population. Further work is required to show that our results are generalizable to other healthcare-related or community-based settings and to prospectively validate our model and the observed associations with clinical outcomes.
In conclusion, we demonstrate that deep learning models using ECG-based representations offer favorable discrimination of LVH diagnoses, including cardiac amyloidosis and hypertrophic cardiomyopathy. LVH-Net outperforms both clinical ECG rules and more complex models that include ECG-based measurements. Predicted LVH-Net risk of cardiovascular disease is strongly associated with observed clinical outcomes. LVH-Net and single-lead versions of LVH-Net may have clinical utility in point-of-care screening for left ventricular hypertrophy, as well as rare diseases such as cardiac amyloidosis and hypertrophic cardiomyopathy.
Acknowledgments
Funding Sources
Investigators were supported by National Institutes of Health grants R38HL150212 (Dr Haimovich); R01HL139731 (Dr Lubitz), R01HL157635 (Dr Lubitz), R01HL134893 (Dr Ho), R01HL140224 (Dr Ho), K24HL153669 (Dr Ho), 2R01HL092577 (Dr Ellinor), and T32HL007208 (Dr Khurshid); American Heart Association grants 18SFRN34250007 (Dr Lubitz) and 18SFRN34110082 (Dr Ellinor); and by a grant from the European Union, MAESTRIA 965286 (Dr Ellinor).
Disclosures
Dr Lubitz is a full-time employee of Novartis Institutes for Biomedical Research as of July 18, 2022. Dr Lubitz has received sponsored research support from Bristol Myers Squibb, Pfizer, Boehringer Ingelheim, Fitbit, Medtronic, Premier, and IBM, and has consulted for Bristol Myers Squibb, Pfizer, Blackstone Life Sciences, and Invitae. Dr Ellinor receives sponsored research support from Bayer AG and IBM and has consulted for Novartis, MyoKardia, and Bayer AG. Dr Ho has received sponsored research support from Bayer AG. Dr Batra receives sponsored research support from Bayer AG and IBM and has consulted for Novartis and Prometheus Biosciences. Dr Philippakis is a Venture Partners and employee at GV, has consulted for Novartis and Rakuten, and receives sponsored research support from Verily, Microsoft, IBM, Intel, Pfizer, Abbvie, Biogen, Ionnis, and Bayer. Dr Batra receives sponsored research support from IBM, and consults for Novartis and Prometheus Biosciences. The other authors report no conflicts.
Authorship
All authors attest they meet the current ICMJE criteria for authorship
Patient Consent
All patients provided written informed consent.
Ethics Statement
The authors designed the study and gathered and analyzed the data according to the Helsinki Declaration guidelines on human research. The research protocol used in this study was reviewed and approved by the institutional review board.
Disclaimer
Given his role as Editor-in-Chief, David McManus had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to Dr Hamid Ghanbari.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.cvdhj.2023.03.001.
Appendix. Supplementary data
References
- 1.Vakili B.A., Okin P.M., Devereux R.B. Prognostic implications of left ventricular hypertrophy. Am Heart J. 2001;141:334–341. doi: 10.1067/mhj.2001.113218. [DOI] [PubMed] [Google Scholar]
- 2.Savage D.D., Garrison R.J., Kannel W.B., et al. The spectrum of left ventricular hypertrophy in a general population sample: the Framingham Study. Circulation. 1987;75:I26–I33. [PubMed] [Google Scholar]
- 3.Gertz M.A., Dispenzieri A. Systemic amyloidosis recognition, prognosis, and therapy: a systematic review. JAMA. 2020;324:79–89. doi: 10.1001/jama.2020.5493. [DOI] [PubMed] [Google Scholar]
- 4.Gradman A.H., Alfayoumi F. From left ventricular hypertrophy to congestive heart failure: management of hypertensive heart disease. Prog Cardiovasc Dis. 2006;48:326–341. doi: 10.1016/j.pcad.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Makavos G., Kappaairis C., Tselegkidi M.E., et al. Hypertrophic cardiomyopathy: an updated review on diagnosis, prognosis, and treatment. Heart Fail Rev. 2019;24:439–459. doi: 10.1007/s10741-019-09775-4. [DOI] [PubMed] [Google Scholar]
- 6.Goto S., Mahara K., Beussink-Nelson L., et al. Artificial intelligence-enabled fully automated detection of cardiac amyloidosis using electrocardiograms and echocardiograms. Nat Commun. 2021;12:2726. doi: 10.1038/s41467-021-22877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko W.Y., Siontis K.C., Attia Z.I., et al. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J Am Coll Cardiol. 2020;75:722–733. doi: 10.1016/j.jacc.2019.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Khurshid S., Reeder C., Harrington L., et al. Cohort design and natural language processing to reduce bias in electronic health records research: the Community Care Cohort Project. medRxiv. 2021;26 doi: 10.1038/s41746-022-00590-0. 05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boldrini M., Cappelli F., Chacko L., et al. Multiparametric echocardiography scores for the diagnosis of cardiac amyloidosis. JACC Cardiovasc Imaging. 2020;13:909–920. doi: 10.1016/j.jcmg.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Rickers C., Wilke N.M., Jerosch-Herold M., et al. Utility of cardiac magnetic resonance imaging in the diagnosis of hypertrophic cardiomyopathy. Circulation. 2005;112:855–861. doi: 10.1161/CIRCULATIONAHA.104.507723. [DOI] [PubMed] [Google Scholar]
- 11.Rawlins J., Bhan A., Sharma S. Left ventricular hypertrophy in athletes. Eur J Echocardiogr. 2009;10:350–356. doi: 10.1093/ejechocard/jep017. [DOI] [PubMed] [Google Scholar]
- 12.Hancock E.W., Deal B.J., Mirvis D.M., et al. AHA/ACCF/HRS recommendations for the standardization and interpretation of the electrocardiogram: part V: electrocardiogram changes associated with cardiac chamber hypertrophy: a scientific statement from the American Heart Association Electrocardiography and Arrhythmias Committee, Council on Clinical Cardiology; the American College of Cardiology Foundation; and the Heart Rhythm Society. Endorsed by the International Society for Computerized Electrocardiology. J Am Coll Cardiol. 2009;53:992–1002. doi: 10.1016/j.jacc.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 13.Hulme O.L., Khurshid S., Weng L.C., et al. Development and validation of a prediction model for atrial fibrillation using electronic health records. JACC Clin Electrophysiol. 2019;5:1331–1341. doi: 10.1016/j.jacep.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamant N, Reinertsen E, Song S, Aguirre A, Stultz C, Batra P. Patient contrastive learning: a performant, expressive, and practical approach to ECG modeling. arXiv 2021:2104.04569. [DOI] [PMC free article] [PubMed]
- 15.Al-Alusi M.A., Ding E., McManus D.D., Lubitz S.A. Wearing your heart on your sleeve: the future of cardiac rhythm monitoring. Curr Cardiol Rep. 2019;21:158. doi: 10.1007/s11886-019-1223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson J.N., Grifoni C., Bos J.M., et al. Prevalence and clinical correlates of QT prolongation in patients with hypertrophic cardiomyopathy. Eur Heart J. 2011;32:1114–1120. doi: 10.1093/eurheartj/ehr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubrey S.W., Cha K., Anderson J., et al. The clinical features of immunoglobulin light-chain (AL) amyloidosis with heart involvement. QJM. 1998;91:141–157. doi: 10.1093/qjmed/91.2.141. [DOI] [PubMed] [Google Scholar]
- 18.Peguero J.G., Lo Presti S., Perez J., Issa O., Brenes J.C., Tolentino A. Electrocardiographic criteria for the diagnosis of left ventricular hypertrophy. J Am Coll Cardiol. 2017;69:1694–1703. doi: 10.1016/j.jacc.2017.01.037. [DOI] [PubMed] [Google Scholar]
- 19.Schillaci G., Verdecchia P., Borgioni C., et al. Improved electrocardiographic diagnosis of left ventricular hypertrophy. Am J Cardiol. 1994;74:714–719. doi: 10.1016/0002-9149(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 20.Liu V.X., Bates D.W., Wiens J., Shah N.H. The number needed to benefit: estimating the value of predictive analytics in healthcare. J Am Med Inform Assoc. 2019;26:1655–1659. doi: 10.1093/jamia/ocz088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khurshid S., Keaney J., Ellinor P.T., Lubitz S.A. A simple and portable algorithm for identifying atrial fibrillation in the electronic medical record. Am J Cardiol. 2016;117:221–225. doi: 10.1016/j.amjcard.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goff D.C., Jr., Pandey D.K., Chan F.A., Ortiz C., Nichaman M.Z. Congestive heart failure in the United States: is there more than meets the I(CD code)? The Corpus Christi Heart Project. Arch Intern Med. 2000;160:197–202. doi: 10.1001/archinte.160.2.197. [DOI] [PubMed] [Google Scholar]
- 23.Christopoulos G., Graff-Radford J., Lopez C.L., et al. Artificial intelligence-electrocardiography to predict incident atrial fibrillation: a population-based study. Circ Arrhythm Electrophysiol. 2020;13 doi: 10.1161/CIRCEP.120.009355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinney W. :pandas-dev/pandas Pandas 1.0.3 (Version v1.0.3). Zenodo. 2020.
- 25.Pedregosa F., Varoquaux G., Gramfort A., et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 26.Van Rossum G., Drake F.L. 2009. Python 3 Reference Manual. [Google Scholar]
- 27.Abadi M, Agarwal A, Barham P, et al. TensorFlow: large-scale machine learning on heterogeneous systems. arXiv:1603.04467.
- 28.Davidson-Pilon C. lifelines: survival analysis in Python. Journal of Open Source Software. 2019;4:1317. [Google Scholar]
- 29.Maron B.J. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 30.Kwon J.M., Jeon K.H., Kim H.M., et al. Comparing the performance of artificial intelligence and conventional diagnosis criteria for detecting left ventricular hypertrophy using electrocardiography. Europace. 2020;22:412–419. doi: 10.1093/europace/euz324. [DOI] [PubMed] [Google Scholar]
- 31.Devereux R.B., Wachtell K., Gerdts E., et al. Prognostic significance of left ventricular mass change during treatment of hypertension. JAMA. 2004;292:2350–2356. doi: 10.1001/jama.292.19.2350. [DOI] [PubMed] [Google Scholar]
- 32.Cyrille N.B., Goldsmith J., Alvarez J., Maurer M.S. Prevalence and prognostic significance of low QRS voltage among the three main types of cardiac amyloidosis. Am J Cardiol. 2014;114:1089–1093. doi: 10.1016/j.amjcard.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 33.Lakdawala N.K., Thune J.J., Maron B.J., et al. Electrocardiographic features of sarcomere mutation carriers with and without clinically overt hypertrophic cardiomyopathy. Am J Cardiol. 2011;108:1606–1613. doi: 10.1016/j.amjcard.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rapezzi C., Merlini G., Quarta C.C., et al. Systemic cardiac amyloidoses: disease profiles and clinical courses of the 3 main types. Circulation. 2009;120:1203–1212. doi: 10.1161/CIRCULATIONAHA.108.843334. [DOI] [PubMed] [Google Scholar]
- 35.Savage D.D., Seides S.F., Clark C.E., et al. Electrocardiographic findings in patients with obstructive and nonobstructive hypertrophic cardiomyopathy. Circulation. 1978;58:402–408. doi: 10.1161/01.cir.58.3.402. [DOI] [PubMed] [Google Scholar]
- 36.Murtagh B., Hammill S.C., Gertz M.A., Kyle R.A., Tajik A.J., Grogan M. Electrocardiographic findings in primary systemic amyloidosis and biopsy-proven cardiac involvement. Am J Cardiol. 2005;95:535–537. doi: 10.1016/j.amjcard.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 37.Bayoumy K., Gaber M., Elshafeey A., et al. Smart wearable devices in cardiovascular care: where we are and how to move forward. Nat Rev Cardiol. 2021;18:581–599. doi: 10.1038/s41569-021-00522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giudicessi J.R., Schram M., Bos J.M., et al. Artificial intelligence-enabled assessment of the heart rate corrected QT interval using a mobile electrocardiogram device. Circulation. 2021;143:1274–1286. doi: 10.1161/CIRCULATIONAHA.120.050231. [DOI] [PubMed] [Google Scholar]
- 39.Halcox J.P.J., Wareham K., Cardew A., et al. Assessment of remote heart rhythm sampling using the AliveCor heart monitor to screen for atrial fibrillation: the REHEARSE-AF study. Circulation. 2017;136:1784–1794. doi: 10.1161/CIRCULATIONAHA.117.030583. [DOI] [PubMed] [Google Scholar]
- 40.Chung E.H., Guise K.D. QTC intervals can be assessed with the AliveCor heart monitor in patients on dofetilide for atrial fibrillation. J Electrocardiol. 2015;48:8–9. doi: 10.1016/j.jelectrocard.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto H., Yokochi T. Transthyretin cardiac amyloidosis: an update on diagnosis and treatment. ESC Heart Fail. 2019;6:1128–1139. doi: 10.1002/ehf2.12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerdts E., Izzo R., Mancusi C., et al. Left ventricular hypertrophy offsets the sex difference in cardiovascular risk (the Campania Salute Network) Int J Cardiol. 2018;258:257–261. doi: 10.1016/j.ijcard.2017.12.086. [DOI] [PubMed] [Google Scholar]
- 43.Liao Y., Cooper R.S., Mensah G.A., McGee D.L. Left ventricular hypertrophy has a greater impact on survival in women than in men. Circulation. 1995;92:805–810. doi: 10.1161/01.cir.92.4.805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.