Abstract
Prostate cancer remains one of the most fatal malignancies in men in the United States. Predicting the course of prostate cancer is challenging given that only a fraction of prostate cancer patients experience cancer recurrence after radical prostatectomy or radiation therapy. This study examined the expressions of 14 fusion genes in 607 prostate cancer samples from the University of Pittsburgh, Stanford University, and the University of Wisconsin–Madison. The profiling of 14 fusion genes was integrated with Gleason score of the primary prostate cancer and serum prostate-specific antigen level to develop machine-learning models to predict the recurrence of prostate cancer after radical prostatectomy. Machine-learning algorithms were developed by analysis of the data from the University of Pittsburgh cohort as a training set using the leave-one-out cross-validation method. These algorithms were then applied to the data set from the combined Stanford/Wisconsin cohort (testing set). The results showed that the addition of fusion gene profiling consistently improved the prediction accuracy rate of prostate cancer recurrence by Gleason score, serum prostate-specific antigen level, or a combination of both. These improvements occurred in both the training and testing cohorts and were corroborated by multiple models.
Prostate cancer remains a leading cause of cancer-related death in men in the United States. In 2021, 34,500 US men died from prostate cancer, while 268,490 new cases were diagnosed.1 Most prostate cancers develop slowly. Surgical treatments such as radical prostatectomy are effective in curing cancer. However, patients present with distal metastasis or recurrence after surgical resection.
Some analyses of data from the Surveillance, Epidemiology, and End Results database, maintained by the National Cancer Institute, have shown that patients having prostate cancer with distal metastasis had a high risk for prostate cancer–related death.2,3 Thus, patients at a high risk for prostate cancer recurrence at the time of diagnosis may benefit from early radiotherapy or anti-androgen or other adjunctive chemotherapy and thereby have a reduced risk for mortality.
Currently, the Gleason score of the primary prostate cancer at the time of diagnosis is the main criterion used for predicting the outcomes of patients with prostate cancer. A high Gleason score (eg, 8 to 10) has been associated with an increased risk for prostate cancer recurrence after radical prostatectomy, while a Gleason score of 6 has been associated with a low risk for recurrence.4 The contemporary initial management of patients with a Gleason score of 6 is observation (active surveillance and watchful waiting). Using a combination of Gleason score, prostate-specific antigen (PSA) level, age, and other clinical factors, several nomograms have been developed to gauge the risk for prostate cancer recurrence. These tools have been used with variable success in the predicting clinical outcomes in patients with prostate cancer.5, 6, 7 However, these tools provide little insight into the mechanisms of the disease.
Numerous mutations,8 gene fusions,9, 10, 11, 12 chromosome alterations,13,14 and epigenetic abnormalities15, 16, 17, 18 have been discovered in patients with prostate cancer. In particular, gene fusion events appear widespread and frequent in patients with prostate cancer. Even though some fusion genes such as TMPRSS2-ETS/ERG have been extensively studied, the relationship between gene fusion events and clinical outcomes in patients with prostate cancer remains unclear. In previous studies, 14 fusion genes were detected in prostate cancer samples, with various frequencies ranging from 6% to 80%.10,11,19, 20, 21 Many of these fusion transcripts were shed into the bloodstream and were readily detectable in the blood or serum samples from patients.20,22 Among these fusion genes, MAN2A1-FER, PTEN-NOLC1, and SLC45A2-AMACR induce spontaneous liver cancer in a short period of time when coupled with somatic Pten knockout in mice.10,19,21 Yet their potential in predicting the course of prostate cancer is not known. This study determined whether the presence of these fusion genes in prostate cancer samples can be used for predicting the recurrence of prostate cancer.
Materials and Methods
Tissue Samples
There was a total of 607 prostate cancer tissue specimens from the University of Pittsburgh Medical Center (UPMC; Pittsburgh, PA), Stanford University Medical Center (Stanford, CA), and the University of Wisconsin–Madison Medical Center (Madison, WI). The sample size was estimated by power analysis (293 on 80% versus 70% comparison) and the availability of clinical specimens. Samples from patients who received radiation or hormone therapy prior to radical prostatectomy were excluded. The samples from UPMC were obtained from the University of Pittsburgh Tissue Bank in compliance with institutional regulatory guidelines and comprised 301 prostate cancer samples, including 271 prostate cancer samples with annotated clinical information available (Supplemental Table S1). The recurrence status of prostate cancer was defined as a serum PSA level of >0.2 ng/mL on at least two consecutive tests obtained after radical prostatectomy. All of the samples were obtained in accordance with the guidelines approved by the Institutional Review Board of University of Pittsburgh. All methods were performed in accordance with relevant guidelines and regulations. Informed-consent exemptions were obtained from the University of Pittsburgh Institutional Review Board. All cancer samples were macrodissected. Samples with at least 50% cancer cells were included in the study. Samples of prostate cancer tissues obtained from other institutions included 112 from Stanford University (Supplemental Table S2) and 194 from the University of Wisconsin–Madison (191 samples annotated with clinical information) (Supplemental Table S3). The procedure of obtaining the tissue samples was in full compliance with the guidelines of those institutions.
Quantitative Real-Time RT-PCR Methods
Total RNA was extracted from the cells using TRIzol (InvitroGen, Carlsbad, CA). The quality of the extracted RNA was assessed through 260:280 and 260:230 ratio analyses by NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The samples that passed quality control were accepted for further analysis. The first stranded cDNA was synthesized from approximately 2 μg of the total RNA template from each sample. Random hexamers and Superscript II (InvitroGen) were incubated with the RNA at 42°C for 2 hours. One microliter of each cDNA sample was used for the TaqMan PCR reactions, with 50 heat cycles for standard formalin-fixed paraffin-embedded samples, as follows: 94°C for 30 seconds, 61°C for 30 seconds, and 72°C for 30 seconds, using the primers and probes listed in Table 1. The PCR reactions were performed in the QuantStudio 3 real-time PCR thermocycler system (Thermo Fisher Scientific) or the Mastercycler RealPlex2 system (Eppendorf, Inc., Framingham, MA). A negative control with no DNA template and a synthetic positive control were included in each batch of reactions. Samples with a cycle threshold (CT) of ≤45 were considered positive for fusion gene detection, while those with a CT of >45 were considered negative. If a negative control showed any CT, the results of the entire batch were discarded. If a positive control failed, the results of the batch of samples were abandoned. The results from TaqMan quantitative real-time RT-PCRs were shown in Supplemental Tables S1–S3. The PCR products from 18% to 100% of the positive samples were sequenced to verify the fusion genes using the Sanger sequencing method.
Table 1.
Primers and Probes
| Fusion gene | Primers | Probe |
|---|---|---|
| MAN2A1-FER | F:5′-AGCGCAGTTTGGGATACAGCA-3′ R:5′-CTTTAATGTGCCCTTATATACTTCACC-3′ |
5′-/56-FAM/TCAGAAACA/ZEN/GCCTATGAGGGAAATT/3IABkFQ/-3′ |
| SLC45A2-AMACR | F:5′-TTGATGTCTGCTCCCATCAGG-3′ R:5′-CAGCTGGAGTTTCTCCATGAC-3′ |
5'-/56-FAM/AAGAGGGCA/ZEN/TGGTAGTGGAGGC/3IABkFQ/-3′ |
| CCNH-C5orf30 | F:5′-AAAGTTATTTATCAGAGAGTCTGATGCTG-3′ R:5′-CTGTTCTACTCCAGGTATTTTCATTATATC-3′ |
5'-/56-FAM/ACAGGCAAG/ZEN/TTCTGTTCTCTTTCAGCA/3IABkFQ/-3′ |
| MTOR-TP53BP1 | F:5′-TGATAGACCAGTCCCGGGATG-3′ R:5′-CCACTGACATTCCCAGAACAAG-3′ |
5'-/56-FAM/TGTCAGCCT/ZEN/GTCAGAATCCAAGTCAAG/3IABkFQ/-3′ |
| TRMT11-GRIK2 | F:5′-CCCTTAACAGGTATCTGCTCC-3′ R:5′-CCCATTGGGCCAGATTCCACA-3′ |
5'-/56-FAM/CGGAACTCC/ZEN/AGATGCTCCTGCG/3IABkFQ/-3′ |
| LRRC59-FLJ60017 | F:5′-GTGACTGCTTGGATGAGAAGC-3′ R:5′-GTTGATGAGCAGCCATTGAGC-3′ |
5'-/56-FAM/CAGTGTGCA/ZEN/AACAAGGTGACTGGAAG/3IABkFQ/-3′ |
| TMEM135-CCDC67 | F:5′-GAGACCATCTTACTGGAAGTTCC-3′ R:5′-TGGTACTCTTCCACCTGTTGG-3′ |
5'-/56-FAM/TTTGCCCTT/ZEN/GGTGAGTCTTAAAAGGAAC/3IABkF/-3′ |
| KDM4B-AC011523.2 | F:5′-CACACCGAGGACATGGACCT-3′ R:5′-CTCAGATCCAGGCTTGCTTAC-3′ |
5'-/56-FAM/ACAGCATCA/ZEN/ACTACCTGCACTTTGGG/3IABkFQ/-3′ |
| CLTC-ETV1 | F:5′-CCTTCCTCCTACATGGAAGTTG-3′ R:5′-CTTGATTTTCAGTGGCAGGCC-3′ |
5′-/56-FAM/CTGCCAATA/ZEN/CTAGTGTGGCTTTT/3IABkFQ/-3′ |
| PCMTD1-SNTG1 | F:5′-CTGGAGAGCTTCATCAAAAATAG-3′ R:5′-CACTTCTCGGGCAATCTCAACA-3′ |
5′-/56-FAM/AGCTTTGAT/ZEN/AAACTGCTCTCCAGAATGTTG/3IABkFQ/-3′ |
| ACPP-SEC13 | F:5′-CCTCATGGCCACAAGGATTTG-3′ R:5′-TGAGGCTTCCAGGTACAACAG-3′ |
5′-/56-FAM/CCAGATTGG/ZEN/CTGCAATGCCGTC/3IABkFQ/3IABkFQ/-3′ |
| DOCK7-OLR1 | F:5′-TAAAACAAGGGTCAATGTCACTCAT-3′ R:5′-CAGTCTGGATCTTTAGGTCATCA-3′ |
5′-/56-FAM/AGACACAGC/ZEN/AGGATGCCAATG/3IABkFQ/-3′ |
| ZMPSTE24-ZMYM4 | F:5′-GAGGAAGAAGGGAACAGTGAAGA-3′ R:5′-CTGGAATAGGGCTCAGTAAAAATGTTATC-3′ |
5′-/56-FAM/AGACACAGC/ZEN/AGGATGCCAATG/3IABkFQ/-3′ |
| PTEN-NOLC1 | F:5′-AAGCCAACCGATACTTTTCTCCA-3′ R:5′-ATAGATGTCTAAGAGGGAAGAGG-3′ |
5'-/56-FAM/AGACACAGC/ZEN/AGGATGCCAATG/3IABkFQ/-3′ |
| ACTB | F:5′-ACCCCACTTCTCTCTAAGGAG-3′ R:5′-GCAATGCTATCACCTCCCCTG-3′ |
5'-/56-FAM/CCAGTCCTC/ZEN/TCCCAAGTCCACAC/3IABkFQ/-3′ |
F, forward; R, reverse.
Prediction Model on Fusion Gene Profile
Fusion gene machine-learning methods were introduced to predict the recurrence status of prostate cancer. These machine-learning algorithms generally take in the fusion gene status and generate a prediction probability per sample. For fusion profiling, the semi-quantitative status of each fusion gene based on CT cycles was tabulated across all of the tumor samples. The optimal CT cycle of each fusion gene was obtained based on the differentiation between the recurrent and nonrecurrent status of the samples from the UPMC cohort. Several machine-learning algorithms were applied to the fusion gene profiling data, specifically: support vector machine,23 random forest (RF) modeling,24,25 linear discriminant analysis (LDA),26 and logistic regression.27 For all of these methods, leave-one-out cross-validation (LOOCV) was performed on the training cohort to evaluate the prediction algorithms and select the best parameters of 14 fusion gene combinations. The best algorithms were then applied to the whole training cohort to train a model and to the testing cohort. Eventually, the training and testing cohorts were pooled together to generate the model most accurate in predicting recurrence based on LOOCV. All biostatistical analyses were performed using R programming and available R software packages (randomForest, MASS, and e1071; R Foundation, https://www.r-project.org).
Prediction Model Integrating Fusion Genes, Gleason Score, and Serum PSA
Clinical features such as Gleason score and serum PSA were also available for the prediction of cancer recurrence. The machine-learning algorithm was first applied to these clinical features individually. With regard to Gleason score, the combined Gleason score optimal for use in predicting recurrence was selected. For serum PSA, the cutoff value that best differentiated recurrence from nonrecurrence was chosen. In order to integrate fusion gene profiling, Gleason score, and serum PSA, the machine learning models described in the Materials and Methods were applied to all three of the features together to train the optimal model and generate the prediction probability for the fusion + Gleason + PSA model. If the probability was ≤0.5, it was predicted as nonrecurrent. If the probability was >0.5, it was predicted as recurrent. Similarly, fusion gene status combined with Gleason score generated probability for fusion + Gleason models, while fusion combined with serum PSA prediction generated probability for fusion + PSA models. Similar to models involving only fusion gene data, the models integrating fusion gene profiling, Gleason score, and serum PSA were applied to the training cohort. The best parameters selected by LOOCV were used as the final model for the training cohort and were then applied to the validation cohort for evaluation. Eventually, both cohorts were pooled together to provide a final prediction model for recurrent cases. All of the biostatistics analyses were performed using R programming.
Results
Fusion Genes Are Frequently Present in Prostate Cancer Samples
The role of fusion genes in promoting the metastasis/recurrence of prostate cancer is still poorly understood. Previous studies have shown that the fusion genes MAN2A1-FER, TRMT11- GRIK2, MTOR-TP53BP1, CCNH-C5orf30, KDM4B-AC011523.2, SLC45A2-AMACR, TMEM135-CCDC67, LRRC59-FLJ60017, CLTC-ETV1, PCMTD1-SNTG1, ACPP-SEC13, DOCK7-OLR1, ZMPSTE24-ZMYM4, and PTEN-NOLC1 are present in prostate cancers, with various frequencies.10,11 Herein, data from a multi-institutional cohort that included 271 samples of radical prostatectomy with adequate clinical information from UPMC, 191 from University of Wisconsin–Madison, and 112 from Stanford Medical Center were analyzed to determine whether these fusion genes were accurate in predicting the clinical outcomes in patients with prostate cancers. Eligible patients with nonrecurrent samples had clinical follow-ups at least 5 years after surgical treatment.
As shown in Supplemental Table S4, all 14 fusion genes were detected in the prostate cancer samples from the combined cohorts. SLC45A2-AMACR had the highest detection rate (86.8%) of all fusion genes in the combined cohorts, ranging from 80.1% in the UPMC cohort to 93.2% in the Wisconsin cohort. This was followed by MAN2A1-FER (76.5%), ZMPSTE24-ZMYM4 (70.7%), and PTEN-NOLC1 (66.4%), while TMEM135-CCDC67 had the lowest frequency, only 1.2% of the samples were positive for this fusion gene. In general, the frequencies of the fusion genes were comparable among the three cohorts, except CCNH-C5orf30, which was detected with a significantly higher frequency in the Wisconsin cohort (78% versus 29.5% and 33.9% in the UPMC and Stanford cohorts, respectively).
Fusion Gene Expressions Associated with Clinical and Pathologic Features of Prostate Cancer
Association analysis in the UPMC cohort showed that the presence of MTOR-TP53BP1 (P = 0.0028), KDM4B-AC011523.2 (P = 0.02), ACPP-SEC13 (P = 0.007), and DOCK7-OLR1 (P = 0.03) in the prostate cancer samples was associated with an increased risk for a high combined Gleason score (8 to 10), while CCNH-C5orf30 (P = 0.01) was associated with a low combined Gleason score (6 or 7). In addition, the presence of MAN2A1-FER (P = 0.046), MTOR-TP53BP1 (P = 0.0018), KDM4B-AC011523.2 (P = 0.025), and PCMTD1-SNTG1 (P = 0.021) was associated with a high Gleason score, while CCNH-C5orf30 was associated with a low Gleason score (P = 0.0027). The presence of MAN2A1-FER (P = 0.01) and MTOR-TP53BP1 (P = 0.007) in a prostate cancer sample was also associated with a more advanced pathologic cancer stage (T3/4), while the presence of CCNH-C5orf30 was associated with cancers of a less invasive stage (T2) (P = 0.027). Strong expression of MAN2A1-FER (CT ≤ 35, P = 0.0008) and the presence of MTOR-TP53BP1 (P = 0.0007) were associated with a higher preoperative serum PSA level. Six fusion genes were associated lymph node involvement: MAN2A1-FER (P = 0.0036), TRMT11-GRIK2 (P = 0.025), MTOR-TP53BP1 (P = 0.0088), SLC45A2-AMACR (P = 0.028), PCMTD1-SNTG1 (P = 0.033), and DOCK7-OLR1 (P = 0.0031). Similar to lymph node involvement, six fusion genes were associated with an increased risk for biochemical recurrence of prostate cancer: MAN2A1-FER (P = 9.4 × 10−6), TRMT11-GRIK2 (P = 0.007), MTOR-TP53BP1 (P = 4.97 × 10−6), PCMTD1-SNTG1 (P = 0.00018), ACPP-SEC13 (P = 0.0019), and DOCK7-OLR1 (P = 0.0017).
Interestingly, the presence of CCNH-C5orf30 was associated with a decreased risk for the recurrence of prostate cancer (P = 0.00026).
To investigate whether fusion genes were also associated with similar clinical characteristics of prostate cancer samples in independent cohorts, association analyses were performed on the Stanford and Wisconsin cohorts. In the Wisconsin cohort, 17.3% of prostate cancer cases were recurrent, while in the Stanford cohort, 62.5% were recurrent. To make the analyses balanced and comparable, the Wisconsin and Stanford cohorts were combined into one external cohort, with sample number and clinical characteristics similar to those from UPMC (39.5% cases from 271 samples were recurrent). The combined cohort had a total of 303 prostate cancer samples, including 297 samples with available clinical follow-up information. Thirty-four percent of the samples (102/297) from the combined cohort had known prostate cancer recurrence. Association analyses of the combined external cohort showed that the presence of MTOR-TP53BP1 (P = 0.03), LRRC59-FLJ60017 (P = 0.02), and CLTC-ETV1 (P = 0.006) was associated with a higher Gleason score. Strong expressions of MAN2A1-FER (CT ≤ 34, P = 0.006) and PTEN-NOLC1 (CT ≤ 33, P = 0.04) were also associated with a higher Gleason score. The presence of PTEN-NOLC1 was associated with a higher preoperative serum PSA level (P = 0.03). The expressions of DOCK7-OLR1 and ZMPSTE24-ZMYM4 were associated lower PSA-free survival (both, P = 0.04). In contrast, good expression of CCNH-C5orf30 (CT ≤ 37) was associated with a lower Gleason score (P = 0.005), lower PSA level (P = 4.1 × 10−5), a lower recurrence rate (P = 0.0006), and better PSA-free survival (P = 0.0002).
Fusion Gene–Based Machine-Learning Models to Predict Prostate Cancer Recurrence in the UPMC Cohort
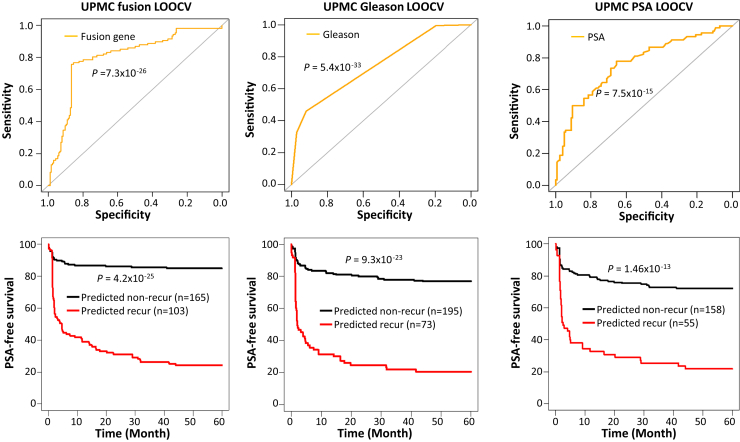
To investigate whether individual fusion genes or combinations of fusion genes were predictive of outcomes in patients with prostate cancer recurrence, multiple machine-learning models utilizing various combinations of fusions with optimal intensity cutoffs were employed to analyze the UPMC prostate cancer cohort based on the LOOCV method. A total of 764 models were constructed, of which 457 had prediction accuracy rates above 70% (Supplemental Table S5). The support vector machine model, which combined the detection of six fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), PCMTD1-SNTG1 (CT ≤ 38), and ACPP-SEC13 (CT ≤ 40)], produced an accuracy of 81.9%, with a sensitivity of 76.6% and a specificity of 85.4%. The model also generated a Youden index of 0.62 (Figure 1 and Supplemental Table S5). The PSA-free survival analysis of the six-fusion support vector machine model showed that 24.3% of patients survived 5 years PSA-free if the cancer was predicted as recurrent, while 85% of patients had no recurrence for at least 5 years if the cancer was predicted as nonrecurrent (P = 4.2 × 10−25) (Figure 1).
Figure 1.
Prediction of prostate cancer recurrence by fusion gene profiling, Gleason score, and serum PSA level in the UPMC cohort. Top panels: Receiver operating characteristic curves from the support vector machine model, which combines the detection of six fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), PCMTD1-SNTG1 (CT ≤ 38), and ACPP-SEC13 (CT ≤ 40)] (left), Gleason score (middle), or serum PSA levels (right). Bottom panels: Kaplan-Meier analyses of PSA-free survival of prostate cancer patients predicted by a support vector machine model that detects six fusion gene (left), Gleason score (middle), and serum PSA (right).
Incorporation of Fusion Gene Detection Enhances Gleason Score Prediction of Prostate Cancer Recurrence in the UPMC Cohort
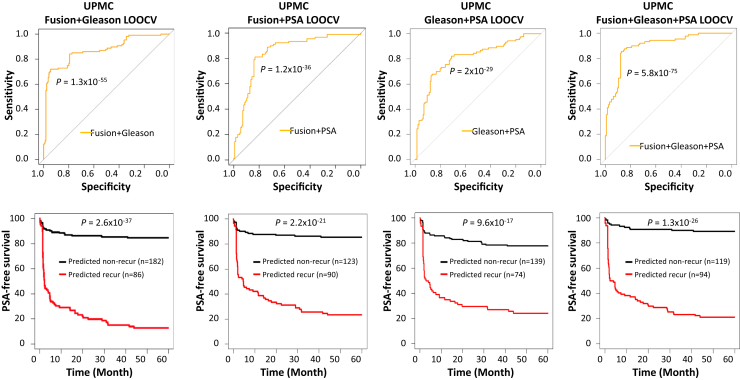
The prediction analysis based on Gleason scores showed that a cutoff of the Gleason score at 8 in the UPMC cohort had the best prediction: 77.9% accuracy, with a sensitivity of 57% and a specificity of 91.5% (Figure 1 and Supplemental Table S6). To investigate whether the combination of fusion gene profiling and Gleason score enhanced the prediction of prostate cancer recurrence, Gleason score was incorporated into the machine-learning LOOCV analysis. A total of 442 models of different combinations showed an accuracy above 80% when fusion gene profiling was combined with Gleason score (Supplemental Table S7). As shown in Figure 2 and Supplemental Table S7, a support vector machine model using the detection of six fusions [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR- TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), PCMTD1-SNTG1 (CT ≤ 38), and ACPP-SEC13 (CT ≤ 40)] + Gleason score accurately predicted prostate cancer recurrence in 85.2% of cases, with a sensitivity of 72% and a specificity of 94%. The survival analysis showed that only 12.8% of patients had recurrence-free survival of 5 years after surgery if the cancer was predicted as recurrent. In contrast, 84.6% of patients had recurrence-free survival of 5 years after surgery if the cancer was predicted as nonrecurrent. These results represented an improvement over the use of Gleason score alone, with 20.5% having recurrence-free survival for 5 years after surgery if Gleason score was 8 or above, and 76.9% having no recurrence if Gleason score was 7 or less (Figures 1 and 2).
Figure 2.
Fusion genes enhance predictions by Gleason score, serum PSA level, and the combination of both in the UPMC cohort. Top panels: Receiver operating characteristic curves from a support vector machine model that detects six fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), PCMTD1-SNTG1 (CT ≤ 38), and ACPP-SEC13 (CT ≤ 40)] + Gleason score (left), a support vector machine model that detects five fusion genes [MAN2A1-FER (CT ≤ 34), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), PCMTD1-SNTG1 (CT ≤ 38), and ACPP-SEC13 (CT ≤ 40)] + PSA (second from the left), Gleason score + PSA logistic model (third from the left), and a random forest model that uses the detection of three fusion genes [MAN2A1-FER (CT ≤ 34), CCNH-C5orf30 (negative), DOCK7-OLR1 (CT ≤ 41)] + Gleason score + PSA (right). Bottom panels: Kaplan-Meier analyses of PSA-free survival in prostate cancer patients predicted by a support vector machine model that uses the detection of six fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), PCMTD1-SNTG1 (CT ≤ 38), and ACPP-SEC13 (CT ≤ 40)] + Gleason score (left), a support vector machine model that uses the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), PCMTD1-SNTG1 (CT ≤ 38), and ACPP-SEC13 (CT ≤ 40)] + PSA (second from the left), Gleason score + PSA logistic model (third from the left), and a random forest model that uses the detection of three fusion genes [MAN2A1-FER (CT ≤ 34), CCNH-C5orf30 (negative), DOCK7-OLR1 (CT ≤ 41)] + Gleason + PSA (right).
Fusion Gene Detection Improves PSA Prediction of Prostate Cancer Recurrence in the UPMC Cohort
The use of serum PSA alone was moderately effective in predicting the recurrence of prostate cancer. A high serum PSA level was correlated with the risk for prostate cancer recurrence. Indeed, a PSA of >9.77 ng/mL correctly predicted 73.5% of cases of prostate cancer recurrence in the UPMC cohort, with a sensitivity of 50% and a specificity of 90.4% (Figure 1 and Supplemental Table S8). When fusion gene profiling was combined with the PSA prediction analysis, 265 models of different combinations showed prediction accuracy rates above 75%. The top predictor was a support vector machine model that incorporated a serum PSA level cutoff of 9.77 ng/mL and the presence of five fusion genes, MAN2A1-FER (CT ≤ 34), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), PCMTD1-SNTG1 (CT ≤ 38), and ACPP-SEC13 (CT ≤ 40) (Figure 2 and Supplemental Table S9), which produced 82.3% accuracy, with 80% sensitivity and 84% specificity. Survival analyses showed that 23.3% of patients survived 5 years PSA-free if the cancer was predicted as recurrent, while 85.4% of patients survived 5 years PSA-free if the cancer was predicted as nonrecurrent (P = 2.2 × 10−21) (Figure 2). This finding represented a moderate improvement over the use of PSA used alone: 21.8% PSA-free survival for 5 years if PSA was above 9.77 ng/mL, and 72.2% PSA-free survival if PSA was below 9.77 ng/mL (P = 1.46 × 10−13) (Figure 1).
Combination of Fusion Gene Profiling, Serum PSA, and Gleason Score in Predicting the Recurrence of Prostate Cancer in the UPMC Cohort
To investigate whether a combination of serum PSA, Gleason score, and fusion gene profiling improved the prediction of prostate cancer recurrence further, 385 models with various combinations based on the best intensity cutoffs using LOOCV were constructed. A total of 317 models yielded prediction accuracy rates of 80% or better (Supplemental Table S10). The RF model, which combined Gleason score, serum PSA, and the detection of three fusion genes [MAN2A1-FER (CT ≤ 34), CCNH-C5orf30 (negative), and DOCK7-OLR1 (CT ≤ 41)], produced the highest Youden index, with 84.7% accuracy, 84.4% sensitivity, and 84.8% specificity (Figure 2 and Supplemental Table S10). These results represented an improvement over the use of Gleason score + serum PSA: 78.6% accuracy, with 64.4% sensitivity and 88.8% specificity (Figure 2 and Supplemental Table S11). Survival analyses showed that 21.3% of prostate cancer patients survived 5 years PSA-free after surgery if the cancer was predicted as recurrent by the RF model, while 89.1% of patients experienced no recurrence for 5 years after surgery if the cancer was predicted as nonrecurrent (P = 1.3 × 10−26) (Figure 2). On the other hand, the best Gleason score + serum PSA model (logistic) generated a 21.1% PSA-free survival for 5 years if the cancer was predicted as recurrent, and 78.2% PSA-free survival for 5 years if the cancer was predicted as nonrecurrent (P = 9.6 × 10−17) (Figure 2).
Stanford/Wisconsin Cohort Validation of Fusion Gene Profiling Enhances the Prediction of Prostate Cancer Recurrence
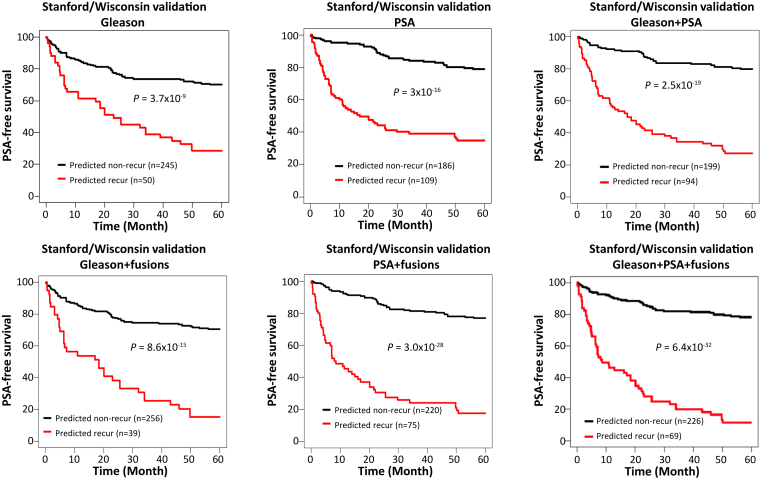
Next, 764 machine-learning models trained using data from the UPMC cohort were applied to the Stanford/Wisconsin cohort. However, none of the models had a prediction accuracy rate reaching 70% (Supplemental Table S12). The optimized cutoff of Gleason score based on data from the UPMC cohort was then applied to predict the outcomes of prostate cancer patients from the Stanford/Wisconsin cohort (combined Gleason score of >8 as recurrent). The results produced a Youden index of 0.27 and yielded 72.4% accuracy, with 34.3% sensitivity and 92.3% specificity (P = 4.4 × 10−17) (Supplemental Table S13 and Supplemental Figure S1). To investigate whether fusion gene detection enhanced the prediction of prostate cancer recurrence by Gleason score, 764 model algorithms developed from the UPMC cohort were applied to the Stanford/Wisconsin cohort for cross-validation, with 52 models yielding prediction accuracy rates exceeding 72.5% (Supplemental Table S14). One was an LDA model that integrated two fusion genes [TRMT11-GRIK2 (CT ≤ 43) and CCNH-C5orf30 (negative)] with Gleason score, which yielded the highest Youden index, 0.3, and a prediction accuracy of 75%, with 32.3% sensitivity and 96.9% specificity (Supplemental Table S14 and Supplemental Figure S1). The same model also accurately predicted 79% of cases in the UPMC cohort (Supplemental Table S7). Survival analysis showed that 70.6% of patients survived 5 years without the recurrence of prostate cancer when the cancer was predicted as nonrecurrent, while only 15.4% of patients survived 5 years when the cancer was predicted as recurrent by the model (P = 8.6 × 10−15) (Figure 3). These findings represented a moderate improvement over Gleason score alone: 70.2% survived 5 years without recurrence if Gleason score was 7 or lower, while 28.7% survived a similar period without recurrence if Gleason score was 8 or above (P = 3.7 × 10−9) (Figure 3).
Figure 3.
Fusion gene algorithms from UPMC cohort improve PSA-free survival predictions by Gleason score, serum PSA, or the combination of both in the Stanford/Wisconsin cohort. Top panels: Kaplan-Meier analyses of PSA-free survival in prostate cancer patients in the Stanford/Wisconsin cohort predicted by Gleason (cutoff = 8, left), PSA (cutoff = 9.77 ng/mL, middle), or Gleason score + PSA (logistic, right). Bottom panels: Kaplan-Meier analyses of PSA-free survival in prostate cancer patients in the Stanford/Wisconsin cohort, predicted by linear discriminant analysis (LDA) model using the detection of four fusion genes [TRMT11-GRIK2 (CT ≤ 43), CCNH-C5orf30 (negative), CLTC-ETV1 (CT ≤ 37), and ACPP-SEC13 (CT ≤ 40)] + Gleason score (left), a logistic model using the detection of three fusion genes [TRMT11-GRIK2 (CT ≤ 43), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)] + PSA (middle), and an LDA model using the detection of four fusion genes [TRMT11-GRIK2 (CT ≤ 43), CCNH-C5orf30 (negative), ACPP-SEC13 (CT ≤ 40), and DOCK7-OLR1 (CT ≤ 41)] + Gleason score + PSA (right).
PSA used as the sole criterion for predicting prostate cancer recurrence in the Stanford/Wisconsin cohort based on the training data from the UPMC cohort yielded 74.7% accuracy, with 67.6% sensitivity and 78.5% specificity (Supplemental Table S15 and Supplemental Figure S1). Among 56 models of fusion gene profiling + serum PSA, the prediction accuracy rate exceeded 75% (Supplemental Table S16). A logistic model using the detection of three fusion genes [TRMT11-GRIK2 (CT ≤ 43), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)] integrated with PSA yielded 78.9% accuracy, with 56% sensitivity and 90.8% specificity (Supplemental Table S16 and Supplemental Figure S1). The same model predicted 80% of recurrence correctly in the UPMC cohort (Supplemental Table S9). Survival analysis showed that 77% of patients survived 5 years without cancer recurrence when the cancer was predicted as nonrecurrent, while only 17.8% of patients had no recurrence in 5 years if the cancer was predicted as recurrent (P = 3.0 × 10−28) (Figure 3). These findings represented a moderate improvement over survival prediction by PSA alone: 78.8% of patients survived 5 years without recurrence if PSA was <9.77 ng/mL, while 38.8% of patients survived 5 years without recurrence if PSA was >9.77 ng/mL (P = 3 × 10−16) (Figure 3).
Combining PSA and Gleason score improved the prediction of prostate cancer recurrence to 76.8%, with a Youden index 0.45 (Supplemental Table S17). This model was more accurate than the use of either PSA or Gleason score alone. To investigate whether the integration of fusion gene profiling, PSA level, and Gleason score further improved the prediction accuracy rate, 764 algorithms developed from the UPMC training cohort were applied to the Stanford/Wisconsin cohort for validation analysis. Seventy-three algorithms produced an accuracy exceeding 77% (Supplemental Table S18). Among them, an LDA model that integrated the detection of four fusion genes [TRMT11-GRIK2 (CT ≤ 43), CCNH-C5orf30 (negative), ACPP-SEC13 (CT ≤ 40), and DOCK7-OLR1 (CT ≤ 41)] with Gleason score and serum PSA level generated a prediction accuracy of 79.5%, with 53.9% sensitivity and 92.8% specificity and a Youden index of 0.47 (Supplemental Figure S1 and Supplemental Table S18). The same model had 82.3% prediction accuracy in the UPMC cohort (Supplemental Table S10). Survival analyses showed that 78% of patients survived 5 years without recurrence if the cancer was predicted as nonrecurrent by the model using four fusion genes + Gleason + PSA LDA, while only 11.6% of patients experienced no recurrence in the same period if the cancer was predicted as recurrent (P = 6.4 × 10−32) (Figure 3). These results represented an improvement on the optimal Gleason + PSA model: 78% of patients survived 5 years without recurrence if the cancer was predicted as nonrecurrent, while 26.7% of patients experienced no recurrence if the cancer was predicted as recurrent (P = 2.5 × 10−19) (Figure 3). In general, these results indicated that the addition of the fusion gene algorithm improved the prediction accuracy rate of PSA and/or Gleason score on prostate cancer recurrence in two independent cohorts.
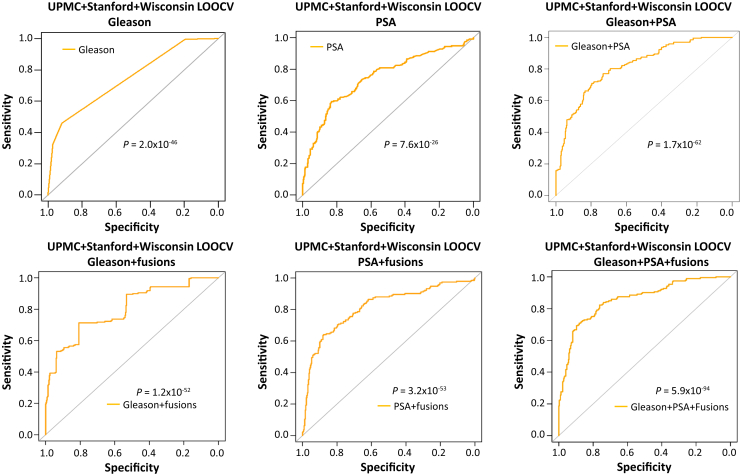
Combining UPMC, Stanford, and University of Wisconsin Cohorts for Cross-Validation Prediction
With all of the cohorts combined (574 cases), Gleason score alone (cutoff = 8, optimal) yielded 75% accuracy (Supplemental Table S19). Most (440) of the fusion gene–containing algorithms combined with Gleason score exceeded an accuracy of 76% based on LOOCV (Supplemental Table S20). One of the models called five fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)].
The RF model improved the Gleason prediction accuracy from 75% to 78% (Figure 4 and Supplemental Table S20). Serum PSA also yielded a prediction accuracy of 74.2% when using the optimal cutoff (9.77 ng/mL) (Figure 4 and Supplemental Table S21). When serum PSA was incorporated into an RF model that used the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), CLTC-ETV1 (CT ≤ 37), and ACPP-SEC13 (CT ≤ 40)], the accuracy of predicting prostate cancer recurrence was improved to 78.7% (Figure 4 and Supplemental Table S22). With an RF model of Gleason score, serum PSA, and the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)], the prediction accuracy improved to 82.4%, with 68.8% sensitivity and 90.6% specificity, and a Youden index of 0.59 (Figure 4 and Supplemental Table S23). These results represented an improvement in prediction accuracy over the use of combined serum PSA + Gleason score: 77.1% accuracy, 55.8 sensitivity, 90% specificity, and a Youden index of 0.46 (Figure 4 and Supplemental Table S24).
Figure 4.
Fusion gene algorithm improves prediction of prostate cancer recurrence in combined cohorts of UPMC, Stanford, and Wisconsin, by Gleason score, serum PSA level, or the combination of both. Top panels: Receiver operating characteristic curves from Gleason (left), PSA (middle), or Gleason + PSA (right) logistic models. Bottom panels: Receiver operating characteristic curves from a random forest model using the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)] + Gleason score (left), a random forest model using the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), CLTC-ETV1 (CT ≤ 37), and ACPP-SEC13 (CT ≤ 40)] + PSA (middle), and a random forest model using the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)] + Gleason + PSA (right).
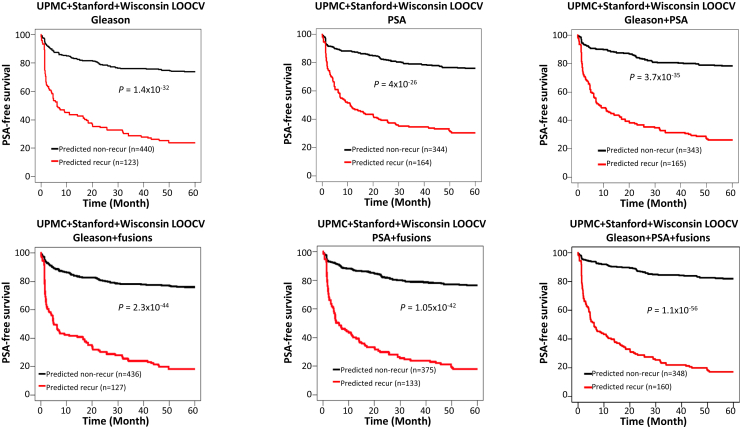
The survival analysis showed that 76% of patients survived 5 years recurrence-free if the cancer was predicted as nonrecurrent by the fusion + Gleason RF model, while 18.4% of patients were prostate cancer-free if the cancer was predicted as recurrent by the same model (P = 2.3 × 10−44) (Figure 5). This combination yielded an improvement over the use of Gleason score alone: 73.8% PSA-free survival if the Gleason score was 7 or lower, and 23.9% PSA-free survival if the Gleason score was 8 or above (P = 1.4 × 10−32). When PSA and fusion algorithms were combined, 76.5% of patients were prostate cancer free for 5 years if the cancer was predicted as negative for recurrence, and 17.9% of patients were prostate cancer free for 5 years if the cancer was predicted as positive for recurrence (P = 1.05 × 10−42) (Figure 5). These results compared favorably against prediction by PSA alone: 76% of patients survived 5 years recurrence-free if serum PSA was <9.77 ng/mL, and 30.5% of patients had cancer recurrence in 5 years if serum PSA was above 9.77 ng/mL. When fusion gene profiling, Gleason score, and PSA algorithm were combined, the prediction results were further improved: 81.9% of patients were prostate cancer recurrence free for 5 years after surgery if the cancer was predicted as nonrecurrent by the fusion + Gleason + PSA RF model, while only 17.2% patients were cancer recurrence free if the cancer was predicted as recurrent by the same model (P = 1.1 × 10−56) (Figure 5). On the other hand, with the Gleason + PSA logistic model, 78.3% of patients had no cancer recurrence for 5 years if the cancer was predicted as nonrecurrent by the model, and 26.2% of patients had no cancer recurrence for 5 years if the cancer was predicted as recurrent (P = 3.7 × 10−35).
Figure 5.
Fusion gene–containing algorithms enhance PSA-free survival prediction by Gleason score, serum PSA level, or the combination of both, in the combined cohorts of UPMC, Stanford, and Wisconsin. Top panels: Kaplan-Meier analyses of PSA-free survival of prostate cancer patients in the combined cohorts by Gleason (cutoff = 8, left), PSA (cutoff = 9.77 ng/mL, middle), Gleason + PSA (logistic model, right). Bottom panels: Kaplan-Meier analyses of PSA-free survival of prostate cancer patients in the combined cohorts by random forest model using the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)] + Gleason score (left), random forest model using the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), CLTC-ETV1 (CT ≤ 37), and ACPP-SEC13. (CT ≤ 40)] + PSA (middle), and a random forest model using the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)] + Gleason score + PSA (right).
Learning Models Most Consistent Among Cohorts
The models that worked well in the Stanford/Wisconsin validation appeared the most consistent models for clinical application: the LDA model integrating Gleason score, serum PSA level, and the detection of four fusion genes [TRMT11-GRIK2 (CT ≤ 43), CCNH-C5orf30 (negative), ACPP-SEC13 (CT ≤ 40), and DOCK7-OLR1 (CT ≤ 41)] yielded 79.5% accuracy in the Stanford/Wisconsin cohort, 82.3% in the UPMC cohort, and 81.8% in the combined UPMC/Stanford/Wisconsin cohorts. Similarly, the LDA model that integrated Gleason score with the detection of two fusion genes [TRMT11-GRIK2 (CT ≤ 43) and CCNH-C5orf30 (negative)] yielded 75% accuracy in the Stanford/Wisconsin cohort, 79% in the UPMC cohort, and 77.8% in the combined UPMC/Stanford/Wisconsin cohort. When only serum PSA was available, the logistic model using PSA integrated with the detection of three fusion genes [TRMT11-GRIK2 (CT ≤ 43), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)] yielded 78.9% accuracy in the Stanford/Wisconsin cohort, 80% in the UPMC cohort, and 76% in the RF combined UPMC/Stanford/Wisconsin cohort.
Discussion
Prediction of the clinical course of prostate cancer remains challenging. Most cases of organ-confined prostate cancer are curable by radical prostatectomy or radiation therapy. A fraction of prostate cancer patients experience recurrent cancer and die from the disease. Gleason score and serum PSA level have been widely used as the basis for predicting clinical outcomes in prostate cancer patients. The results from this study showed that fusion gene models were important contributing factors in the prediction of the recurrence of prostate cancer. The enhancement of PSA and/or Gleason grading by fusion gene status was quite robust. The use of detecting several hundred combinations of fusion genes in different algorithmic models improved the accuracy over predicting prostate cancer recurrence by Gleason score, serum PSA, or the combination of both.
This enhancement appeared in different cohorts with highly variable clinical characteristics. The wide variety of models that improved prediction may also be useful in overcoming the heterogeneity issue of the cancer samples in which different fusion gene patterns may appear in different loci. These machine-learning models can be utilized in several scenarios: in a patient with prostate cancer diagnosed using a Gleason score and a recent serum PSA level, the fusion gene + Gleason + PSA model may be useful in predicting the risk for prostate cancer recurrence, with an accuracy ranging from 79.5% to 84.7%. If serum PSA is not available, the fusion gene profiling + Gleason model can be useful in predicting the recurrence of prostate cancer, with an accuracy of 74% to 85.2%. In the absence of a Gleason score, the fusion gene profiling + PSA model yielded a prediction accuracy from 78.9% to 82.3%. In a patient with radical prostatectomy, these models may be useful in determining whether additional adjuvant therapy is needed. It is also possible to combine these fusion gene–prediction models with other methods, such as prostate imaging reporting and data system28 or prostate genome decipher classifier,29 to improve the prediction further.
Overfitting is a potential problem associated with machine-learning methods. Indeed, significant variations in both clinical features and fusion gene detection were present among cases from the UPMC, Stanford, and Wisconsin cohorts. Despite these variations, the addition of fusion gene profiling to models using Gleason score and/or serum PSA consistently improved the accuracy of predicting prostate cancer recurrence in all of the cohorts. Some fusion genes were consistently associated with clinical features in both the UPMC and Stanford/Wisconsin cohorts: The presence of MTOR-TP53BP1 and strong expression of MAN2A1-FER (CT ≤ 34) were associated with a higher Gleason score in both cohorts. The expression of DOCK7-OLR1 was associated with prostate cancer recurrence. However, the presence of CCNH-C5orf30 signaled a lower Gleason score, lower cancer recurrence, and better PSA-free survival in all of the cohorts. CCNH-C5orf30 fusion features a truncated cyclin H protein and an intact independent C5orf30. Cyclin H protein (CCNH) is an important regulator of cell cycle progression to mitosis30,31 and basal RNA transcription.32 The truncated CCNH from the gene fusion lacks H5′ and HC domain, and is defective in binding cyclin-dependent kinase (CDK)-7 protein.33 Such defects may prevent CCNH from promoting cell mitosis and RNA transcription. The truncated CCNH protein due to the gene fusion may have a negative impact on prostate cancer progression.
This study reported a new tool for predicting clinical outcomes in patients with prostate cancer. In comparison with Gleason score and PSA, fusion gene profiling has added value for clinical patient management because some gene fusions are important molecular processes in generating prostate cancer. These fusion genes are readily detectable in blood samples from prostate cancer patients. Thus, it is possible to build similar prediction models based on the fusion gene status of blood/serum samples from prostate cancer patients. Some of these fusion genes are proven cancer drivers,10,19,21 while some others are functional knockouts of tumor suppressors.34 Thus, the detection of fusion genes provides new mechanistic insight into prostate cancer progression. In patients whose samples are positive for MAN2A1-FER, the fusion gene sensitizes the cancer cells to crizotinib and canertinib because of the ectopic tyrosine kinase activity of the fusion protein.21 Cancer cells positive for PTEN-NOLC1 are sensitive to cyclopropanecarboxylic acid-(3-[6-(3-trifluoromethyl-phenylamino)-pyrimidin-4-ylamino]-phenyl)-amide, a potent epidermal growth factor receptor (EGFR) inhibitor, because PTEN-NOLC1 promotes the expression of EGFR and its downstream signaling molecules,10 while cancer cells positive for SLC45A2-AMACR are sensitive to SCH772984, an inhibitor of ERKs, due to the direct activation of ERK2 by the translocated α-methylacyl–coenzyme A racemase (AMACR) protein.19 Cancer cells harboring any of these gene fusions can be targetable by gene-editing technology through the insertion of a suicide gene into the breakpoints of their recombinant genome.35 Thus, the incorporation of fusion gene detection into the prostate cancer–diagnostic scheme benefits patients with regard to diagnosis, prognosis, cancer progression surveillance, and treatment.
Acknowledgments
We thank Songyang Zheng and Yan Luo for their technical support.
Footnotes
Supported partly by NIH National Cancer Institute grant 1R56CA229262-01 (J.-H.L.), US Department of Defense grant W81XWH-16-1-0541 (J.-H.L., J.D.B., and D.J.), and NIH National Institute of Diabetes and Digestive and Kidney Diseases grant P30- DK120531-01.
Y.-P.Y. and S.L. contributed equally to this article.
Disclosures: A provisional US patent application has been filed (number 63/393,030, "Systems and Methods for Predicting Prostate Cancer Recurrence"; J.-H.L., Y.-P.Y., S.L., J.N., and G.M., University of Pittsburgh).
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2022.12.013.
Supplemental Data
Fusion gene profiling enhances predictions by Gleason score, serum PSA level, or the combination of both in the Stanford/Wisconsin cohort. Top panel: Receiver operating characteristic curves from Gleason score (left), serum PSA level (middle), and a combination of Gleason score and serum PSA level (logistic model). Bottom panel: Receiver operating characteristic curves from a linear discriminant analysis (LDA) model of the detection of two fusion genes [TRMT11-GRIK2 (Ct ≤ 43) and CCNH-C5orf30 (negative)] + Gleason (left), a logistic model of the detection of three fusion genes [TRMT11-GRIK2 (Ct ≤ 43), CCNH-C5orf30 (negative), and ACPP-SEC13 (Ct ≤ 40)] + PSA (middle), and an LDA model of the detection of four fusion genes [TRMT11-GRIK2 (Ct ≤ 43), CCNH-C5orf30 (negative), ACPP-SEC13 (Ct ≤ 40), and DOCK7-OLR1 Ct ≤ 41)] + Gleason + PSA (right).
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Zhao F., Wang J., Chen M., Chen D., Ye S., Li X., Chen X., Ren G., Yan S. Sites of synchronous distant metastases and prognosis in prostate cancer patients with bone metastases at initial diagnosis: a population-based study of 16,643 patients. Clin Transl Med. 2019;8:30. doi: 10.1186/s40169-019-0247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elmehrath A.O., Afifi A.M., Al-Husseini M.J., Saad A.M., Wilson N., Shohdy K.S., Pilie P., Sonbol M.B., Alhalabi O. Causes of death among patients with metastatic prostate cancer in the US from 2000 to 2016. JAMA Netw Open. 2021;4:e2119568. doi: 10.1001/jamanetworkopen.2021.19568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gleason D.F., Mellinger G.T. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol. 1974;111:58–64. doi: 10.1016/s0022-5347(17)59889-4. [DOI] [PubMed] [Google Scholar]
- 5.Hou G.D., Zheng Y., Zheng W.X., Gao M., Zhang L., Hou N.N., Yuan J.R., Wei D., Ju D.E., Dun X.L., Wang F.L., Yuan J.L. A novel nomogram predicting the risk of positive biopsy for patients in the diagnostic gray area of prostate cancer. Sci Rep. 2020;10:17675. doi: 10.1038/s41598-020-74703-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephenson A.J., Scardino P.T., Eastham J.A., Bianco F.J., Jr., Dotan Z.A., Fearn P.A., Kattan M.W. Preoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Natl Cancer Inst. 2006;98:715–717. doi: 10.1093/jnci/djj190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X., Ning Q., Jin K., Zhang T., Ma X. Development and validation of a preoperative nomogram for predicting survival of patients with locally advanced prostate cancer after radical prostatectomy. BMC Cancer. 2020;20:97. doi: 10.1186/s12885-020-6565-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C., Asangani I.A., Ateeq B., Chun S.Y., Siddiqui J., Sam L., Anstett M., Mehra R., Prensner J.R., Palanisamy N., Ryslik G.A., Vandin F., Raphael B.J., Kunju L.P., Rhodes D.R., Pienta K.J., Chinnaiyan A.M., Tomlins S.A. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J.E., Shah R.B., Pienta K.J., Rubin M.A., Chinnaiyan A.M. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science (New York, NY) 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 10.Luo J.H., Liu S., Tao J., Ren B.G., Luo K., Chen Z.H., Nalesnik M., Cieply K., Ma T., Cheng S.Y., Chen Q., Michalopoulos G.K., Nelson J.B., Bhargava R., Zhang J., Ma D., Jarrard D., Pennathur A., Luketich J.D., DeFranco D.B., Monga S.P., Tseng G., Yu Y.P. Pten-NOLC1 fusion promotes cancers involving MET and EGFR signalings. Oncogene. 2021;40:1064–1076. doi: 10.1038/s41388-020-01582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Y.P., Ding Y., Chen Z., Liu S., Michalopoulos A., Chen R., Gulzar Z.G., Yang B., Cieply K.M., Luvison A., Ren B.G., Brooks J.D., Jarrard D., Nelson J.B., Michalopoulos G.K., Tseng G.C., Luo J.H. Novel fusion transcripts associate with progressive prostate cancer. Am J Pathol. 2014;184:2840–2849. doi: 10.1016/j.ajpath.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo J.H., Liu S., Zuo Z.H., Chen R., Tseng G.C., Yu Y.P. Discovery and classification of fusion transcripts in prostate cancer and normal prostate tissue. Am J Pathol. 2015;185:1834–1845. doi: 10.1016/j.ajpath.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y.P., Liu S., Huo Z., Martin A., Nelson J.B., Tseng G.C., Luo J.H. Genomic copy number variations in the genomes of leukocytes predict prostate cancer clinical outcomes. PLoS One. 2015;10:e0135982. doi: 10.1371/journal.pone.0135982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y.P., Song C., Tseng G., Ren B.G., Laframboise W., Michalopoulos G., Nelson J., Luo J.H. Genome abnormalities precede prostate cancer and predict clinical relapse. Am J Pathol. 2012;180:2240–2248. doi: 10.1016/j.ajpath.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao S.G., Chen W.S., Li H., Foye A., Zhang M., Sjostrom M., et al. The DNA methylation landscape of advanced prostate cancer. Nat Genet. 2020;52:778–789. doi: 10.1038/s41588-020-0648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y.P., Ding Y., Chen R., Liao S.G., Ren B.G., Michalopoulos A., Michalopoulos G., Nelson J., Tseng G.C., Luo J.H. Whole-genome methylation sequencing reveals distinct impact of differential methylations on gene transcription in prostate cancer. Am J Pathol. 2013;183:1960–1970. doi: 10.1016/j.ajpath.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo J.H., Ding Y., Chen R., Michalopoulos G., Nelson J., Tseng G., Yu Y.P. Genome-wide methylation analysis of prostate tissues reveals global methylation patterns of prostate cancer. Am J Pathol. 2013;182:2028–2036. doi: 10.1016/j.ajpath.2013.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damaschke N.A., Gawdzik J., Avilla M., Yang B., Svaren J., Roopra A., Luo J.H., Yu Y.P., Keles S., Jarrard D.F. CTCF loss mediates unique DNA hypermethylation landscapes in human cancers. Clin Epigenetics. 2020;12:80. doi: 10.1186/s13148-020-00869-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuo Z.H., Yu Y.P., Ren B.G., Liu S., Nelson J., Wang Z., Tao J., Pradhan-Sundd T., Bhargava R., Michalopoulos G., Chen Q., Zhang J., Ma D., Pennathur A., Luketich J., Satdarshan Monga P., Nalesnik M., Luo J.H. Oncogenic activity of solute carrier family 45 member 2 and alpha-methylacyl-coenzyme a racemase gene fusion is mediated by mitogen-activated protein kinase. Hepatol Commun. 2022;6:209–222. doi: 10.1002/hep4.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y.P., Liu S., Nelson J., Luo J.H. Detection of fusion gene transcripts in the blood samples of prostate cancer patients. Sci Rep. 2021;11:16995. doi: 10.1038/s41598-021-96528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z.H., Yu Y.P., Tao J., Liu S., Tseng G., Nalesnik M., Hamilton R., Bhargava R., Nelson J.B., Pennathur A., Monga S.P., Luketich J.D., Michalopoulos G.K., Luo J.H. Man2a1-fer fusion gene is expressed by human liver and other tumor types and has oncogenic activity in mice. Gastroenterology. 2017;153:1120–1132. doi: 10.1053/j.gastro.2016.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y.P., Tsung A., Liu S., Nalesnick M., Geller D., Michalopoulos G., Luo J.H. Detection of fusion transcripts in the serum samples of patients with hepatocellular carcinoma. Oncotarget. 2019;10:3352–3360. [PMC free article] [PubMed] [Google Scholar]
- 23.Cortes C., Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 24.Amit Y., Geman D. Shape quantization and recognition with randomized trees. Neural Comput. 1997;9:1545–1588. [Google Scholar]
- 25.Bauer E., Kohavi R. An empirical comparison of voting classification algorithms: bagging, boosting, and variants. Mach Learn. 1999;36:105–139. [Google Scholar]
- 26.McLachlan G.J. Wiley; New York, NY: 2004. Discriminant Analysis and Statistical Pattern Recognition. [Google Scholar]
- 27.Tolles J., Meurer W.J. Logistic regression: relating patient characteristics to outcomes. JAMA. 2016;316:533–534. doi: 10.1001/jama.2016.7653. [DOI] [PubMed] [Google Scholar]
- 28.Rosenkrantz A.B., Oto A., Turkbey B., Westphalen A.C. Prostate Imaging Reporting and Data System (PI-RADS), version 2: a critical look. AJR Am J Roentgenol. 2016;206:1179–1183. doi: 10.2214/AJR.15.15765. [DOI] [PubMed] [Google Scholar]
- 29.Den R.B., Santiago-Jimenez M., Alter J., Schliekelman M., Wagner J.R., Renzulli Ii J.F., Lee D.I., Brito C.G., Monahan K., Gburek B., Kella N., Vallabhan G., Abdollah F., Trabulsi E.J., Lallas C.D., Gomella L.G., Woodlief T.L., Haddad Z., Lam L.L., Deheshi S., Wang Q., Choeurng V., du Plessis M., Jordan J., Parks B., Shin H., Buerki C., Yousefi K., Davicioni E., Patel V.R., Shah N.L. Decipher correlation patterns post prostatectomy: initial experience from 2 342 prospective patients. Prostate Cancer Prostatic Dis. 2016;19:374–379. doi: 10.1038/pcan.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makela T.P., Parvin J.D., Kim J., Huber L.J., Sharp P.A., Weinberg R.A. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:5174–5178. doi: 10.1073/pnas.92.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher R.P., Morgan D.O. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 32.Shiekhattar R., Mermelstein F., Fisher R.P., Drapkin R., Dynlacht B., Wessling H.C., Morgan D.O., Reinberg D. CDK-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 33.Andersen G., Busso D., Poterszman A., Hwang J.R., Wurtz J.M., Ripp R., Thierry J.C., Egly J.M., Moras D. The structure of cyclin H: common mode of kinase activation and specific features. EMBO J. 1997;16:958–967. doi: 10.1093/emboj/16.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Y.P., Liu P., Nelson J., Hamilton R.L., Bhargava R., Michalopoulos G., Chen Q., Zhang J., Ma D., Pennathur A., Luketich J., Nalesnik M., Tseng G., Luo J.H. Identification of recurrent fusion genes across multiple cancer types. Sci Rep. 2019;9:1074. doi: 10.1038/s41598-019-38550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z.H., Yu Y.P., Zuo Z.H., Nelson J.B., Michalopoulos G.K., Monga S., Liu S., Tseng G., Luo J.H. Targeting genomic rearrangements in tumor cells through Cas9-mediated insertion of a suicide gene. Nat Biotechnol. 2017;35:543–550. doi: 10.1038/nbt.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fusion gene profiling enhances predictions by Gleason score, serum PSA level, or the combination of both in the Stanford/Wisconsin cohort. Top panel: Receiver operating characteristic curves from Gleason score (left), serum PSA level (middle), and a combination of Gleason score and serum PSA level (logistic model). Bottom panel: Receiver operating characteristic curves from a linear discriminant analysis (LDA) model of the detection of two fusion genes [TRMT11-GRIK2 (Ct ≤ 43) and CCNH-C5orf30 (negative)] + Gleason (left), a logistic model of the detection of three fusion genes [TRMT11-GRIK2 (Ct ≤ 43), CCNH-C5orf30 (negative), and ACPP-SEC13 (Ct ≤ 40)] + PSA (middle), and an LDA model of the detection of four fusion genes [TRMT11-GRIK2 (Ct ≤ 43), CCNH-C5orf30 (negative), ACPP-SEC13 (Ct ≤ 40), and DOCK7-OLR1 Ct ≤ 41)] + Gleason + PSA (right).