Abstract
We aimed to explore the relationships between specific viral mutations/mutational patterns and ventilator-associated pneumonia (VAP) occurrence in COVID-19 patients admitted in intensive care units between October 1, 2020, and May 30, 2021. Full-length SARS-CoV-2 genomes were sequenced by means of next-generation sequencing. In this prospective multicentre cohort study, 259 patients were included. 222 patients (47%) had been infected with pre-existing ancestral variants, 116 (45%) with variant α, and 21 (8%) with other variants. 153 patients (59%) developed at least one VAP. There was no significant relationship between VAP occurrence and a specific SARS CoV-2 lineage/sublineage or mutational pattern.
Subject terms: Microbiology, Medical research
Introduction
Current data on ventilator-associated pneumonia (VAP) complicating COVID-19-associated acute respiratory distress syndrome (ARDS) have been mainly obtained during the first wave of COVID-191. Yet, bacterial superinfection may be affected by the nature of the infecting viral variants and the widespread use of corticosteroids2. There is scarce information in the literature on the rate of superinfections in COVID-19 patients, especially of VAP, according to variant types in the dexamethasone era. Potential explanations for the high incidence of VAP in COVID-19 patients include the long duration of invasive mechanical ventilation support, the high incidence of ARDS, and the administration of immune-suppressive treatment. Specific risk factors for VAP, including SARS-CoV-2-related pulmonary lesions and bacteria-virus interaction in lung microbiota, might also play a role in VAP pathogenesis. As the upper respiratory tract microbiome composition significantly changed as COVID-19 severity increased3, we hypothesize that specific SARS-CoV-2 mutations, either in the spike protein or in another viral protein, could have an impact on VAP prevalence.
In this study, we aimed to explore the relationships between SARS-CoV-2 variant lineage/sublineage and specific viral mutations/mutational patterns with VAP occurrence in COVID-19 patients in the dexamethasone era.
Patients and methods
This is a prospective multicentre observational cohort study. Patients admitted between October 1, 2020 (week 40/20) and May 30, 2021 (week 21/21) in one of the 11 participating ICUs from Greater Paris area hospitals included in the ANTICOV study (NCT04733105) were eligible for study inclusion. Inclusion criteria were as follows: age ≥ 18 years, SARS-CoV-2 infection confirmed by a positive reverse transcriptase-polymerase chain reaction (RT-PCR), patient admitted in the ICU who required mechanical ventilation more than 48 h for acute respiratory failure. Patients with a low viral load (PCR cycle threshold (Ct) > 32) in nasopharyngeal samples were not included in the study.
The primary endpoint was the difference in prevalence of the first VAP according to SARS-CoV-2 variants.
Ventilator associated pneumonia definition
All VAP episodes prospectively diagnosed by attending physicians were evaluated and retained for analyses, provided that they fulfilled the following criteria (the definition of VAP was based on current guidelines4): (1) new or progressive persistent pulmonary infiltrates on chest X-ray; combined with (2) purulent tracheal secretions; (3) fever or hypothermia (body temperature ≥ 38.5 °C or ≤ 36.5 °C, respectively) and/or leukocytosis or leukopenia (white blood cells count ≥ 10.4/mL or ≤ 4 × 10.3/mL, respectively); and (4) a positive quantitative lower respiratory tract sample (endotracheal aspirate [ETA] ≥ 10.5 colony-forming unit [CFU]/mL, bronchoalveolar lavage [BAL] fluid ≥ 10.4 CFU/mL or plugged telescopic catheter [PTC] ≥ 10.3 CFU/mL). Respiratory tract secretions were cultured for VAP diagnosis and susceptibility profiles of recovered microorganisms were recorded. VAP onset was defined as the day on which the lung sample tested positive.
SARS-CoV-2 genome sequence analysis
Full-length SARS-CoV-2 genomes from all included patients were sequenced by means of next-generation sequencing. Viral RNA was extracted from nasopharyngeal swabs in viral transport medium using NucliSENS® easyMAG kit on EMAG device (bioMérieux, Marcy-l’Étoile, France). Sequencing was performed with the Illumina COVIDSeq Test (Illumina, San Diego, California), that uses 98-target multiplex amplifications along the full SARS-CoV-2 genome. The libraries were sequenced with NextSeq 500/550 High Output Kit v2.5 (75 Cycles) on a NextSeq 500 device (Illumina). The sequences were demultiplexed and assembled as full-length genomes by means of the DRAGEN COVIDSeq Test Pipeline on a local DRAGEN server (Illumina). Lineages and clades were interpreted using Pangolin and NextClade, before being submitted to the GISAID database (https://www.gisaid.org). Phylogeny was performed after full-length genome alignment with Muscle v3.8.31 (maximum-likelihood model GTR + I; 1000 bootstrap replicates), by means of IQ-Tree v1.3.11.1 and iTOL.
Statistical analysis
We used a competing risk model (cumulative incidence function of the Gray model)5,6 to properly estimate the effect of COVID-19 on VAP development, while considering death and ventilator weaning as competing events using cmprsk package developed by Gray in R software ([http://biowww.dfci.harvard.edu/~gray/cmprsk_2.1-4.tar.gz]). Two-sided p values < 0.05 were considered significant.
Ethical approval and consent to participate
The study was approved by the Comité de Protection des Personnes Nord-Ouest IV (N° EudraCT/ID-RCB: 2020-A03009-30). Informed consent was obtained from all patients or their relatives. All study procedures and processes were conducted in accordance with the Declaration of Helsinki and with the ethical standards of the institutional and/or research committee.
Results
Between October 1, 2020, and May 30, 2021, 845 patients were admitted in one of the 11 participating ICUs. Among them, 737 had at least one positive SARS-CoV-2 RNA RT-PCR performed in a nasopharyngeal swab sample in the hospital; 413 patients with a cycle threshold (Ct) ≤ 32 and a nasopharyngeal sample that could be used for full-length viral genome sequence analysis were included in this study. Among these 413 patients, 267 were mechanically ventilated more than 48 h. Patients’ characteristics are shown in Table 1. Most patients had ARDS criteria, including 240 (93%) patients who needed prone positioning, and 52 (20%) requiring veno-venous ECMO support. Among these 259 patients, 122 (47%) had been infected with so-called ancestral “pre-existing” variants, i.e., variants circulating before the emergence of variant α, 116 (45%) with variant α (B.1.1.7), and 21 (8%) with other variants, including β (B.1.351) (n = 19, 4.6%), γ (P.1) (n = 2, 0.5%) and other variants of interest (n = 12, 2.9%). One hundred and fifty-three patients (59%) developed at least one VAP episode.
Table 1.
Characteristics of patients with Coronavirus disease 19 (COVID-19) (n = 259).
| Variables | Total (n = 259) |
|---|---|
| Age—median [IQR] | 65 [58–71] |
| Male gender | 185 (71.4%) |
| Medical history | |
| Diabetes mellitus | 80 (30.9%) |
| Congestive heart failure | 25 (9.7%) |
| Hypertension | 138 (53.3%) |
| COPD | 20 (7.7%) |
| Chronic renal failure | 36 (13.9%) |
| Dialysis | 13 (5.0%) |
| Liver cirrhosis | 7 (2.7%) |
| Current smoking | 31 (12.0%) |
| Solid cancer | 15 (5.8%) |
| Blood cancer | 13 (5.0%) |
| Organ transplant | 15 (5.8%) |
| HIV infection | 4 (1.5%) |
| Frailty scale—median [IQR] | 3 [2–4] |
| Clinical characteristics upon ICU admission | |
| SAPS II- median [IQR] | 37 [30–48] |
| Baseline SOFA- median [IQR] | 4 [3–8] |
| ARDS (Berlin definition) | 230 (88.8%) |
| Norepinephrine | 67 (25.9%) |
| Antibiotics | 186 (71.8%) |
| Treatment during stay | |
| Corticosteroids | 242 (93.4%) |
| Tocilizumab | 31 (12.0%) |
| Neuromuscular blockade | 254 (98.1%) |
| Prone position | 240 (92.7%) |
| Inhaled nitric oxide | 26 (10.0%) |
| Extra-corporeal membrane oxygenation | 52 (20.1%) |
| Death in ICU | 115 (44%) |
COPD chronic obstructive pulmonary disease, HIV human immunodeficiency virus, SAPS II Simplified Acute Physiology Score II, SOFA sequential organ failure assessment, ARDS acute respiratory distress syndrome, ICU intensive care unit; variables are shown as median [quartile 1-quartile 3] and number (percentage).
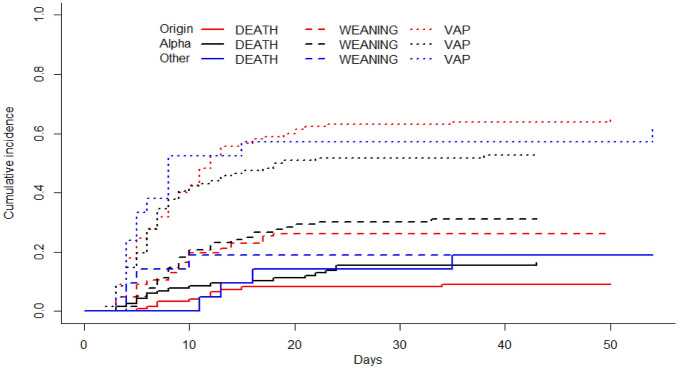
During the study period, variant α was the dominant VOC in the Greater Paris area. Thus, we first assessed whether patients infected with variant α had a higher risk of developing VAP than others (merging patients infected with preexisting variants and other variants) during the ICU stay. The Fine and Gray model showed that the probability of VAP was not significantly different in the two groups [sub-hazard ratio = 1.26 (0.62–2.54), p = 0.53, Fig. 1] after adjusting for weaning and death as competing events.
Figure1.
Day-60 cumulative probability of ventilator-associated pneumonia (VAP) in patients infected with the variant of origin (red lines), variant α (black lines) or other variants (blue lines). Cumulative incidence estimated using the Kalbfleish and Prentice method considering time from intubation to VAP (dotted line), to death (continuous line) and to weaning (dashed line).
Pre-existing and emerging variants (VOC or VOI) are characterized by multiple lineage-specific deletions and amino acid substitutions in their viral genomes. The Spike glycoprotein undergoes evolutive convergence at several amino-acid signature sites found in VOC and VOI circulating at the time of the study period. These sites were selected a priori for in-depth analysis. We found no significant relationship between VAP occurrence and any specific mutation at these sites (Table 2). These results persisted after adjusting for age, sex, antibiotic at admission and SAPS II (Table 3). Although one mutation (A67V) was found to be significantly associated with the risk of VAP (p < 0.05), the number of patients harboring this mutation was considered too small (n = 1) to draw clinically relevant conclusions.
Table 2.
Association between relevant mutations [(substitutions, deletions (Del)] in spike selected a priori and risk of ventilator-acquired pneumonia by univariable Fine and Gray model analysis.
| Mutations | Missing data | VAP | Total | SHR (CI 95%) | |
|---|---|---|---|---|---|
| Yes | No | ||||
| N = 153 | N = 106 | N = 259 | |||
| del6970 | 0 | 61 (39.9%) | 55 (51.9%) | 116 (44.8%) | 0.77 [0.56; 1.06] |
| del140145 | 0 | 61 (39.9%) | 55 (51.9%) | 116 (44.8%) | 0.77 [0.56; 1.06] |
| del242244 | 0 | 8 (5.2%) | 5 (4.7%) | 13 (5.0%) | 1.12 [0.54; 2.30] |
| L5F | 0 | 4 (2.6%) | 3 (2.8%) | 7 (2.7%) | 0.89 [0.36; 2.20] |
| L18F | 0 | 4 (2.6%) | 3 (2.8%) | 7 (2.7%) | 1.04 [0.37; 2.94] |
| A67V | 0 | 1 (0.7%) | 0 (0.0%) | 1 (0.4%) | 5.70 [4.27; 7.60] |
| D80AG | 0 | 9 (5.9%) | 5 (4.7%) | 14 (5.4%) | 1.26 [0.62; 2.57] |
| S98F | 0 | 4 (2.6%) | 3 (2.8%) | 7 (2.7%) | 1.04 [0.36; 3.05] |
| K417NT | 0 | 10 (6.5%) | 5 (4.7%) | 15 (5.8%) | 1.33 [0.69; 2.58] |
| N439K | 0 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| N450K | 0 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| L452R | 0 | 3 (2.0%) | 2 (1.9%) | 5 (1.9%) | 1.08 [0.34; 3.43] |
| S477N | 0 | 38 (24.8%) | 21 (19.8%) | 59 (22.8%) | 1.12 [0.79; 1.57] |
| T478K | 0 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| E484K | 0 | 11 (7.2%) | 5 (4.7%) | 16 (6.2%) | 1.49 [0.77; 2.88] |
| N501Y | 0 | 71 (46.4%) | 61 (57.5%) | 132 (51.0%) | 0.80 [0.59; 1.09] |
| A570D | 0 | 60 (39.2%) | 55 (51.9%) | 115 (44.4%) | 0.76 [0.55; 1.04] |
| D614G | 0 | 149 (97.4%) | 104 (98.1%) | 253 (97.7%) | 0.85 [0.35; 2.06] |
| H655Y | 0 | 2 (1.3%) | 1 (0.9%) | 3 (1.2%) | 1.32 [0.32; 5.37] |
| P681H | 0 | 61 (39.9%) | 53 (50.0%) | 114 (44.0%) | 0.81 [0.59; 1.11] |
| P681R | 0 | 2 (1.3%) | 3 (2.8%) | 5 (1.9%) | 0.57 [0.15; 2.24] |
SHR sub-hazard ratio, CI 95% 95% confidence interval; variables are shown as median [quartile 1-quartile 3] and number (percentage).
Significant values are in [bold].
Table 3.
Multivariable Fine and Gray model analysis on risk of ventilator-acquired pneumonia.
| Mutations | Missing data | SHR | CI 95% |
|---|---|---|---|
| Variant α | 0 | 0.76 | [ 0.54; 1.07 ] |
| Other SARS-CoV-2 variants a | 0 | 1.07 | [ 0.62; 1.87 ] |
| Age | 0 | 1.02 | [ 1.01; 1.04 ] |
| Male sex | 0 | 1.39 | [ 0.98; 1.98 ] |
| Antibiotic at admission | 0 | 0.84 | [ 0.59; 1.18 ] |
| SAPS II | 0 | 1.00 | [ 0.99; 1.01 ] |
SHR sub-hazard ratio, CI 95% 95% confidence interval; apre-existing variant or other variants.
Discussion
We previously failed to find a significant relationship between variant lineages, including variant α, pre-existing and other variants, and mortality7. The main results of this study show the lack of statistically significant relationship between variant lineages or the presence of any of 17 relevant spike substitutions and/or deletions selected a priori and VAP occurrence. These findings suggest that the occurrence of VAP is not related to the virological nature of the infecting agent, but to non-virological causes.
The full-length SARS-CoV-2 genomes of 413 critically ill COVID-19 patients from 11 ICUs, who were predominantly infected with pre-existing and α (B.1.1.7) variants, were sequenced and the relationship between viral sequences and VAP occurrence was studied. The main results of this study show the lack of statistically significant relationship between variant lineages or the presence of any of 17 relevant spike substitutions and/or deletions selected a priori and VAP occurrence. Previous large-scale data have suggested that patients infected with variant α (B.1.1.7), identified using the SGTF proxy, had a higher risk of dying8–10. Yet, no specific comorbidity or clinical conditions was shown to interact with variant types in the previous studies. Interestingly, following SARS-CoV-2-infected patients with different disease severities through the pathway of disease management, Grint et al. reported a weakening association between variant α (B.1.1.7) infection and mortality, highest in the primary care population, lower in the hospitalized population, not significant in patients admitted in the ICU10. Moreover, in another cohort of 496 hospitalized patients in the United Kingdom, 341 samples positive for SARS-CoV-2 on PCR were sequenced and analyzed, including 198 with B.1.1.7 variant (α) and 143 with non-B.1.1.7 variant infection. No association with variant and death was found with or without adjustment for confounding variables11.
Together, our results indicate that, in patients with the most severe forms of COVID-19 who required ICU admission, there was no particular mutational pattern associated with VAP. Therefore, the occurrence of VAP is not related to the virological nature of the infecting agent, but to other causes.
Limitations and strengths of the study
We acknowledge that our study has some limitations. In this cohort, included patients were predominantly infected with pre-existing and α (B.1.1.7) variants, while the inclusion period ended before the emergence of the δ and ο variants, precluding the generalizability of our findings to these more recent variants. The number of patients included was limited and thus the statistical power may have been too weak to show between-group differences. Yet, there was no clear trend regarding associations between variant groups/mutations and VAP, suggesting that increasing the number of patients in the cohort would not have changed the results.
Our study also has strengths, including the constitution of a prospective multicenter cohort of well-phenotyped critically ill patients, and the fact that we performed full-length SARS-CoV-2 genome sequencing in a large number of patients. The start of inclusion of the present study was October 1, 2020, which corresponded in France to the beginning of the second COVID-19 wave, a period when ICU admission policies, ICU patient load and demand, and COVID-19 management strategies were more homogeneous among centres than during the first COVID-19 wave. The high incidence of VAP described in our study is in line with that reported in previous studies1,12.
Conclusions
We found no association between the variant status or any mutational pattern in SARS CoV-2 viral genes and VAP, indicating that the occurrence of VAP is related to non-virological causes.
Acknowledgements
The authors wish to thank the patients involved in the study and all the nurses and physicians involved in patient care in the intensive care units;
Abbreviations
- ARDS
Acute respiratory distress syndrome
- ICU
Intensive care unit
- ECMO
Extra-corporeal membrane oxygenation
- VOC
Variant of concern
- VOI
Variant of interest
- VAP
Ventilator associated pneumonia
- COVID 19
Coronavirus disease 2019
Author contributions
K.R., A.M.B. and N.P. had full access to all of the study data and is deemed responsible for data integrity and accuracy of their analysis. K.R., A.M.B. and N.P. contributed to the study initial design, analysis, interpretation of data, drafting of the initial manuscript, critical revision of intellectual content, and approval of the article final version. R.A., B.P., G.V., C.E.L., T.U., J.M., T.P., D.R., R.B., Z.A.H., S.G., M.S., E.A., A.M.D., C.R., J.M.P., S.F. contributed to the study design and analysis, interpretation of data, drafting of initial manuscript, critical revision of intellectual content, and approval of the submitted version of the article.
Funding
This research received funding from the Agence Nationale de la Recherche (ANR-21-COVR-0022).
Data availability
The datasets supporting the conclusions are included within the article. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
C.-E. L. has served as consultant for Bayer Healthcare, Carmat and Thermo Fisher Brahms, and received lecture fees from MSD, Aerogen, Advanzpharma and BioMérieux, outside the submitted work. J.-M.P. has served as an advisor or speaker for Gilead, Abbvie, Merck, Assembly Biosciences and Arbutus. Other authors have no conflict of interest to disclose.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Keyvan Razazi and Anissa Martins Bexiga.
These authors jointly supervised this work: Slim Fourati and Nicolas de Prost.
References
- 1.Rouzé A, Martin-Loeches I, Povoa P, Makris D, Artigas A, Bouchereau M, Lambiotte F, Metzelard M, Cuchet P, Boulle Geronimi C, et al. Relationship between SARS-CoV-2 infection and the incidence of ventilator-associated lower respiratory tract infections: A European multicenter cohort study. Intensive Care Med. 2021;47:188–198. doi: 10.1007/s00134-020-06323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.RECOVERY Collaborative Group. Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, et al. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shilts MH, Rosas-Salazar C, Strickland BA, Kimura KS, Asad M, Sehanobish E, Freeman MH, Wessinger BC, Gupta V, Brown HM, et al. Severe COVID-19 is associated with an altered upper respiratory tract microbiome. Front. Cell. Infect. Microbiol. 2021;11:781968. doi: 10.3389/fcimb.2021.781968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leone M, Bouadma L, Bouhemad B, Brissaud O, Dauger S, Gibot S, Hraiech S, Jung B, Kipnis E, Launey Y, et al. Brief summary of French guidelines for the prevention, diagnosis and treatment of hospital-acquired pneumonia in ICU. Ann. Intensive Care. 2018;8:104. doi: 10.1186/s13613-018-0444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J. Am. Stat. Assoc. 1999;94:496–509. doi: 10.1080/01621459.1999.10474144. [DOI] [Google Scholar]
- 6.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann. Stat. 1988;16:1141–1154. doi: 10.1214/aos/1176350951. [DOI] [Google Scholar]
- 7.Fourati S, Audureau E, Arrestier R, Marot S, Dubois C, Voiriot G, Luyt C-E, Urbina T, Mayaux J, Roque-Afonso A-M, et al. SARS-CoV-2 genomic characteristics and clinical impact of SARS-CoV-2 viral diversity in critically ill COVID-19 patients: A prospective multicenter cohort study. Viruses. 2022;14:1529. doi: 10.3390/v14071529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies NG, Jarvis CI, CMMID COVID-19 Working Group. Edmunds WJ, Jewell NP, Diaz-Ordaz K, Keogh RH. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patone M, Thomas K, Hatch R, Tan PS, Coupland C, Liao W, Mouncey P, Harrison D, Rowan K, Horby P, et al. Mortality and critical care unit admission associated with the SARS-CoV-2 lineage B.1.1.7 in England: An observational cohort study. Lancet Infect. Dis. 2021;21:1518–1528. doi: 10.1016/S1473-3099(21)00318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grint DJ, Wing K, Houlihan C, Gibbs HP, Evans SJW, Williamson E, McDonald HI, Bhaskaran K, Evans D, Walker AJ, et al. Severity of SARS-CoV-2 alpha variant (B.1.1.7) in England. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021;20:754. doi: 10.1093/cid/ciab754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frampton D, Rampling T, Cross A, Bailey H, Heaney J, Byott M, Scott R, Sconza R, Price J, Margaritis M, et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: A whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021;21:1246–1256. doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Razazi K, Arrestier R, Haudebourg AF, Benelli B, Carteaux G, Decousser J-W, Fourati S, Woerther PL, Schlemmer F, Charles-Nelson A, et al. Risks of ventilator-associated pneumonia and invasive pulmonary aspergillosis in patients with viral acute respiratory distress syndrome related or not to Coronavirus 19 disease. Crit. Care Lond. Engl. 2020;24:699. doi: 10.1186/s13054-020-03417-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions are included within the article. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.