Abstract
Objective:
To catalogue and evaluate response biomarkers correlated with autism spectrum disorder (ASD) symptoms to improve clinical trials.
Methods:
A systematic review of Medline, Embase, and Scopus was conducted in April 2020. Seven criteria were applied to focus on original research that includes quantifiable response biomarkers measured alongside ASD symptoms. Interventional studies or human studies that assessed the correlation between biomarkers and ASD-related behavioral measures were included.
Results:
5,799 independent records yielded 280 manuscripts for review that reported on 940 biomarkers – 755 of which were unique to a single publication. Molecular biomarkers were the most frequently assayed, including cytokines, growth factors, measures of oxidative stress, neurotransmitters, and hormones, followed by neurophysiology (e.g., electroencephalogram and eye-tracking), neuroimaging (e.g., fMRI), and other physiological measures. Studies were highly heterogeneous, including phenotype, demographics, tissues assayed, and methods for biomarker detection. With a median total sample size of 64, almost all of the reviewed studies were only powered to identify biomarkers with large effect sizes. Reporting of individual-level values and summary statistics was inconsistent, hampering mega- and meta-analysis. Biomarkers assayed in multiple studies yielded mostly inconsistent results, revealing a “replication crisis”.
Conclusions:
There is currently no response biomarker with sufficient evidence to inform ASD clinical trials. This review highlights methodological imperatives for ASD biomarker research necessary to make definitive progress: consistent experimental design, correction for multiple comparisons, formal replication, sharing of sample-level data, and preregistration of study designs. Systematic ‘big data’ analyses of multiple potential biomarkers could accelerate discovery.
Keywords: Autism Spectrum Disorders, pharmacodynamic biomarkers, response biomarkers, endophenotypes, symptom severity, clinical trials, intervention
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental syndrome defined in DSM-5 by a combination of symptoms across two clinical domains: “Persistent deficits in social communication and social interaction” and “Restricted, repetitive patterns of behavior, interests, or activities”, which can include sensory processing difficulties (1). It is a common disorder, currently estimated to affect almost 2% of eight-year-old children leading to substantial morbidity (2).
Biomarkers are characteristics that can be measured accurately and reproducibly in individual patients to provide objective and quantifiable metrics of clinically relevant processes (3). They can reflect typical biological processes, pathogenic processes, or responses to an exposure or intervention (4) and include molecular, physiological, or anatomical measures (5).
To date, no biomarkers have passed the Food and Drug Administration (FDA)’s Center for Drug Evaluation and Research Biomarker Qualification Program (CDER-BQP) multi-step approval process to be “qualified” for use in ASD. The FDA and National Institute of Health (NIH) Biomarker Working Group generated the “Biomarkers, EndpointS, and other Tools” (BEST) resource to harmonize biomarker terminology. BEST defines several biomarker categories based on use case, such as diagnostic and safety biomarkers. Here, we focus on response biomarkers, defined by BEST as “A biomarker used to show that a biological response, potentially beneficial or harmful, has occurred in an individual who has been exposed to a medical product or an environmental agent” (4). Given the strong evidence for pathological vulnerability during fetal and perinatal development (6, 7), the challenges of early detection of social deficits, and the current paucity of somatic treatments that target ASD-defining deficits (8), more reliable, biologically-based assays would be transformative. In light of the substantial recent progress in the genetics and biology of ASD and the associated promise of identifying novel molecular treatment targets (9), identifying reliable response biomarkers in ASD could revolutionize the field, providing a standardized metric to assess and refine therapeutic strategies (5).
There is ample reason to be optimistic that response biomarkers can be found. Over the past decade substantial progress has been made in identifying specific genes that dramatically increase the risk for ASD (6). Moreover, the study of these definitive molecular risk factors, both individually and collective, have identified a wide range of potential biological mechanisms (6, 10) and also provided evidence that these genes converge to disrupt a smaller number of molecular pathways, cell types, and circuits in particular brain regions at specific points in development, resulting in the clinical phenotype (6, 11-15). Indeed, while the genetic contribution to ASD has been defined only in a minority of affected individuals, findings to date strongly suggest that markers of altered biological processes are likely identifiable, whether there is contribution from rare large effect mutations, common polygenic inheritance, or environmental factors – all of which play a role in ASD pathogenesis (16).
The heterogeneity of ASD etiology is mirrored in the clinic in the heterogeneity of symptoms and co-morbid disorders. The diagnosis itself is defined by a constellation of behaviors in multiple combinations (17) and while all cases must exhibit impairments in the two DSM-5 defined clinical domains, other important features, such as sensory processing impairment, cognitive impairment, language delay, or stereotypic behavior are present only in subsets of patients. This extensive heterogeneity and the absence of a clearly defined proxy for ASD symptoms in experimental model systems complicate the search for biomarkers, the pursuit of causal relationships, and the development of therapeutics.
To catalogue and evaluate progress in identifying a response biomarker in ASD, we performed a systematic literature review with the objective of documenting which biomarkers have been tested and whether any biomarkers showed evidence of replication. We identified 1,057 original research articles that include a biomarker in individuals with ASD or ASD-related animal models (Fig. 1, Table S1). To focus on response biomarkers, an in-depth review led us to a subset of 280 articles that included both a biomarker and a measurement of ASD symptoms, with either a therapeutic intervention, or showing evidence of the relationship between the biomarker and ASD behaviors. This analysis demonstrated that, to date, no ASD response biomarkers meet the exacting standards necessary to inform clinical trials and highlighted key methodological imperatives necessary for the field to achieve success.
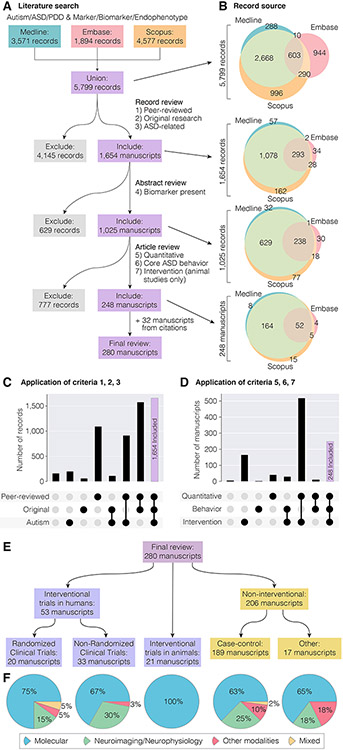
Figure 1. Overview of database search.
A) 5,799 independent records were identified from searches of Medline, Embase, and Scopus. Applying seven inclusion criteria in three steps yielded 248 articles, with an additional 32 included from citations giving a final review set of 280 articles. B) At each step of the application of inclusion criteria the overlap between the three databases is shown by Venn diagrams. C-D) The number of records that met or did not meet combinations of the first three criteria (C) or fifth to seventh criteria (D) is shown in these upset plots. A complete record of this process is recorded in Table S1. E) The 280 articles are subdivided by experimental design. F) Types of biomarker assessed in each of the five types of experimental design.
Methods
A systematic search of the literature was performed following the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (18), though we note our study was not registered in advance and includes an additional 32 articles identified from citations of the reviewed papers that were not captured by our initial database search (Table S2). In April 2020, three databases (Medline, Embase, and Scopus) were searched for relevant articles from January 1st 1900 to February 29th 2020 with the terms: (autism, ASD, pervasive developmental disorder, or PDD) and (biomarker, marker, or endophenotype). Exact search terms, methodologies, and the results of these searches are detailed in the supplementary materials (Supplemental Methods and Table S1). Our initial search identified 3,571 Medline records, 1,894 Embase records, and 4,577 Scopus records (Fig. 1A). Duplicate records within and across these databases were identified to yield a union of 5,799 independent records (Fig. 1, Table S1).
AA and ASJ independently applied a first round of filtering based on the title, article type, and language of the record based on three inclusion criteria:
Criterion 1: The article must be peer-reviewed and published in English.
Criterion 2: The article must describe original research.
Criterion 3: The article must focus on non-syndromic ASD, though it may use a different term such as pervasive developmental disorders (PDD) or Asperger’s.
Applying these criteria, we retained 1,654 records (28.5%, 1,654/5,799) and excluded 4,145 records (Fig. 1A-C). The majority of exclusions (70.2%, 2,908/4,145) were due to not meeting criterion 3 (focus on ASD), followed by criterion 2 (original research) in 1,121 of the remaining 1,237 (90.6%), see Fig. 1C. For the 1,654 records retained, we assessed our fourth inclusion criteria by reading the abstract and, if necessary, the full article:
Criterion 4: The article must describe new data assessing at least one biomarker.
Applying this fourth criteria identified 1,025 records of biomarkers in ASD (62.0%, 1,025/1,654) and excluded 629 (Fig. 1A, 1B). For all 1,025 records, we assessed the full article to apply three further criteria:
Criterion 5: The biomarker assessed must have been both quantifiable and potentially variable (i.e., not fixed/structural)
Criterion 6: The research must have included a measure of ASD severity using behavioral measures or scales to assess social-communication and/or repetitive/restrictive behaviors. Non-interventional studies must also assess the association between these measures and the biomarker.
Criterion 7: If the research was based on animal models, there must have been an intervention.
To assess these criteria, each article was assessed independently by at least two of the authors (AA, ASJ, MP, MB, EU) and discrepancies were reviewed by an additional author. Applying the remaining criteria yielded a final sample of 248 articles (24.2%, 248/1,025) and excluded 777 (Fig. 1A-B, D). The majority of exclusions (94.3%, 733 of 777) were due to criterion 6 (requirement for a measure of ASD severity), with 567 of the 733 being case-control studies for which the study needed to assess the relationship between the biomarker and ASD severity for inclusion (Fig. 1D). We note that these 777 excluded articles may nevertheless provide insights into potential ASD diagnostic biomarkers for future study, and we include a complete list of these articles in Table S1. From the citations of these 248 articles, we identified a further 32 articles that met our seven inclusion criteria, but that had not been identified by the initial search, due to the absence of the words “biomarker”, “marker”, or “endophenotype” in the title or abstract, to yield 280 articles for final review. Table S1 details all 5,799 articles and the outcome as these criteria were applied sequentially, plus 32 articles identified from references cited by the articles we reviewed. We developed a standardized data extraction form, which five review authors (AA, ASJ, MP, MB and EU) used to extract data manually from all 280 eligible studies. If data were unclear or ambiguous a consensus decision was taken by all five authors. The following metrics were extracted: biomarkers, study design, sample size, trial registration number if applicable (i.e., interventional study), ASD diagnostic criteria, participant age, sex, and cognitive ability, inclusion/exclusion criteria, intervention including dose and duration, primary outcomes, and behavioral measures (Tables S3 and S4). For the most frequently analyzed biomarkers we also extracted the following outcomes: statistical association for the biomarker, direction of effect for the biomarker, whether the biomarker correlated with behavioral symptoms and, for interventional studies, whether the intervention led to behavioral improvement. Where multiple outcomes were stated, the result based on the largest sample size was recorded. Missing data were recorded as ‘Not stated’. For response biomarkers with consistent directions of effect across multiple studies (Tables 1 and 2), the following outcomes were also extracted: t statistic or mean and standard deviation in cases and controls; studies in which these metrics were not reported were excluded from this step.
Table 1.
Most cited biomarkers: non-interventional studies
| BM | Reference | Tissue/Measurement | Sample size [ASD/CT] | Age range (years) or ASD mean (SD) |
Biomarker outcome |
ASD Behavior measure | Biomarker x Behavior Correlation |
PMID |
|---|---|---|---|---|---|---|---|---|
| Glutathione (GSH) | Geier et al., 2009 | Plasma | 148 [28/120] | 2–13 | CT > ASD | CARS | No | 18817931 |
| Adams et al., 2011 | Plasma | 99 [55/44] | 5-16 | CT > ASD | ATEC, PDDBI, SAS | Yes | 21651783 | |
| Lakshmi et al., 2011 | Plasma | 90 [45/45] | 4-12 | CT > ASD | CARS | No | 21338594 | |
| Frye et al., 2013 | Plasma | 72 [18/54] | 8.2 (3.1) | CT > ASD | SRS, VABS | No | 23778583 | |
| Vijayashankar et al., 2014 | Saliva | 60 [40/20] | 4-12 | CT > ASD | CARS | Yes | None | |
| Durieux et al., 2016 | Brain (MRS) | 50 [21/29] | 18-50 | Not different | ADI-R, ADOS | No | 26290215 | |
| Meguid et al., 2018 | Plasma | 120 [90/30] | 2-7 | CT > ASD | CARS | Yes | None | |
| IL-6 | Emanuele et al., 2010 | Serum | 50 [22/28] | 19-42 | ASD > CT | ADI-R-soc, VABS | Not reported | 20097267 |
| Ashwood et al., 2011a | PBMC | 72 [37/32] | 2-5 | Not stated | AbBC, ADOS, regression, VABS | No | 20705131 | |
| Ashwood et al., 2011b | Plasma | 223 [97/87/39DD] | 2-5 | ASD > DD, CT | ADI-R, regression | Yes | 20833247 | |
| Napolioni et al., 2013 | Plasma | 50 [25/25] | 0-25 | Not different | SRS | No | 23497090 | |
| Ricci et al., 2013 | Serum | 58 [29/29] | 2-21 | ASD > CT | CARS | No | 23604965 | |
| Yang et al., 2015 | Plasma | 64 [33/31] | 4.5 (1.3) | ASD > CT | CARS | Yes | 25453766 | |
| Ferguson et al., 2016 | Serum | 120 [120/0]1 | 6-18 | NA | VABS | Yes | 27181180 | |
| Jácome et al., 2016 | Plasma | 32 [17/15] | 3-9 | ASD > CT | CARS | Yes | 27983615 | |
| Careaga et al., 2017 | PBMC, LPS stimulated | 50 [50/16] | 2-4 | Not stated | ADOS | Yes | 26493496 | |
| Guloksuz et al., 2017 | Plasma | 75 [40/35] | 7.0 (4.0) | Not different | CARS, regression | Not reported | 28099628 | |
| Krakowiak et al., 2017 | Blood spot | 303 [214/62/27DD] | Neonatal | Not different | VABS | No | 26392128 | |
| Masi et al., 2017 | Plasma | 144 [113/31]2 | 2-18 | Not stated | SRS | No | 29214007 | |
| Ning et al., 2019 | Serum | 204 [102/102] | 4.5 (1.3) | ASD > CT | CARS | Yes | 30503813 | |
| Prosperi et al., 2019 | Plasma | 85 [85/0]1 | 2-6 | NA | Regression | No | 31835709 | |
| BDNF | Correia et al., 2010 | Platelet-rich plasma | 196 [146/50] | 2-18 | ASD > CT | CARS | Yes | 20662941 |
| Mansour et al., 2010 | Serum | 40 [20/20] | 3-12 | CT > ASD | CARS | No | None | |
| Ray et al., 2011 | Plasma | 39 [21/18] | 2-18 | CT > ASD | CARS | No | 21731612 | |
| Ricci et al., 2013 | Serum | 58 [29/29] | 2-21 | ASD > CT | CARS | No | 23604965 | |
| Zhang et al., 2014 | Serum | 120 [60/60] | 3.8 (1.2) | ASD > CT | CARS | Yes | 24984148 | |
| Segura et al., 2015 | Whole blood, RNA | 31 [21/10] | 12-44 | ASD > CT | ToM | No | 25535174 | |
| Wang et al., 2015 | Serum | 150 [75/75] | 4.0 (1.3) | ASD > CT | CARS | Yes | 26103118 | |
| Meng et al., 2017 | Serum | 164 [82/82] | 4.0 (1.3) | ASD > CT | CARS | Yes | 26820673 | |
| Francis et al., 2018 | Serum | 71 [45/26] | 2.5-3.5 | CT > ASD | ADOS, CARS, VABS | No | 29532458 | |
| Serotonin | Alabdali et al., 2014 | Platelet-free plasma | 82 [52/30] | 3-12 | CT > ASD | CARS, SRS | Yes | 24400970 |
| Yang et al., 2015 | Whole blood | 64 [33/31] | 12.4 (2.4) | ASD > CT | CARS | Yes | 25453766 | |
| Russo et al., 2013 | Plasma | 77 [48/29] | 8.0 (ND) | ASD > CT | Pfeiffer Q | Yes | 23825437 | |
| Alabdali et al., 2014 | Plasma | 82 [52/30] | 3-12 | ASD > CT | CARS, SRS | Yes | 24400970 | |
| GABA | Brix et al., 2015 | Brain, anterior cingulate | 38 [14/24] | 10.2 (1.9) | Not different | ASSQ | Yes | 26157380 |
| Carvalho Pereira et al., 2018 | Brain, mOFC | 34 [20/14] | 10-18 | Not different | ADI-R | Yes | 29177616 | |
| Meguid et al., 2018 | Plasma | 120 [90/30] | 2-7 | ASD > CT | CARS | Yes | None | |
| Saleem et al., 2020 | Plasma | 118 [54/64] | 5.6 (2.1) | Not different | CARS | No | 32021195 | |
| Oxytocin | Alabdali et al., 2014 | Platelet-free plasma | 82 [52/30] | 3-12 | CT > ASD | CARS, SRS | Yes | 24400970 |
| Oztan et al., 2018 | CSF | 72 [36/36] | 1.5-9 | Not stated | ADOS | No | 30152888 | |
| Aita et al., 2019 | Serum | 79 [54/25] | 16-60 | Not stated | CARS, RBS-R | Yes | 30640053 | |
| Alpha power | Van Hecke et al., 2009 | EEG, right temporal-parietal | 33 [19/14] | 8-12 | Not different | SRS, SSRS | No | 19630897 |
| Cornew et al., 2012 | EEG, absolute and relative power | 50 [27/23] | 6-15 | ASD > CT | SRS | Yes (relative power) | 22207057 | |
| Kozhushko et al., 2018 | EEG, group independent components | 112 [42/70] | 4-9 | Not different | Self-devised scale | No | 29577946 | |
| Keehn et al., 2017 | EEG, posterior left, central, right | 40 [19/21] | 12-17 | CT > ASD | ADOS, SRS | No | 29170759 | |
| Sutton at al., 2005 | EEG, central and parietal | 43 [23/20] | 9-14 | ASD > CT | ASSQ, ASAS, BASC | Not reported | 15679529 | |
| Fixation time (proportional looking times) | Bacon et al., 2019 | GeoPref, SocPref, JA | 49 [49/0]3 | 1.9 (ND)* | NA | ADOS, VABS, SRS | Yes (GeoPref) | 31647314 |
| Shic et al., 2019 | Direct gaze, speech | 97 [50/47] | 1.96 (0.26) | CT > ASD | ADOS | Yes | 31471912 | |
| Åsberg et al., 2017 | Dynamic emotional faces | 115 [57/58] | 10-44 | CT > ASD | ADOS, ADI-R, AQ | No | 27891819 | |
| Swanson et al., 2013 | In/congruent gaze | 45 [21/24] | 3.58-9.41 | Not different | ADOS, SRS | Yes (high SRS) | None | |
| Wang et al., 2018 | Dyadic bid and sandwich | 301 [112/36/163DD] | 1.86 (0.25) | CT > ASD | ADOS, ADI-R | Yes | 29651331 | |
| Klin et al., 2002 | Eyes in dynamic social scene | 30 [15/15] | 15.1 (7.2) | CT > ASD | ADOS-Soc, VABS | Yes | 12215080 | |
| Frazier et al., 2016 | ARI | 79 [40/39] | 3-8 | Not different | ADOS, SRS, CBCL | Yes | 27015721 | |
| Murias et al., 2018 | Social vs. non-social | 25 [25/0]1,† | 2-6 | NA | ADOS, VABS, BASC, PDDBI | Yes | 29193826 | |
| Laidi et al., 2017 | Eyes avatars | 59 [33/26] | 18-64 | Not stated | ADOS | No | 28931838 | |
| Fractional Anisotropy | Lin et al., 2019 | DTI | 24 [13/11] | 8.9 (1.9) | ASD > CT | ADI-R, ADOS | No | 30755585 |
| Gibbard et al., 2013 | DTI | 50 [25/25] | 18.8-33.3 | CT > ASD | AQ | Yes | 24179854 | |
| Barnea-Goraly et al., 2010 | DTI | 37 [13/13/11] | 10.5(2) | CT > ASD | ADI-R, ADOS | No | 20921121 | |
| Lange et al., 2010 | DTI | 60 [30/30] | 7-28 | Not different | SRS | No | 21182212 | |
| Functional activation (after biological motion) | Moessnang et al., 2020 | Whole-brain/fMRI | 394 [205/189] | 6-30 | Not different | SRS | Yes | 32087753 |
| Kaiser et al., 2010 | Whole-brain/fMRI | 62 [25/20/17] | 4-17 | CT > ASD | SRS | Yes | 21078973 | |
| Björnsdotter et al., 2016 | Whole-brain/fMRI | 275 [22/215/38] | 4.0-17.7 | CT > ASD | SRS, VABS, ADOS | Yes | 27096285 | |
| Functional connectivity (resting state) | Jung et al., 2014 | DMN/fMRI | 40 [19/21] | 16-40 | CT > ASD | AQ | Yes | 24955232 |
| Neufeld J., 2018 | DMN/fMRI | 150 [29/121]# | 8-23 | Not different | SRS-2 | Yes | 28761079 | |
| Yerys et al., 2015 | DMN/fMRI | 44 [22/22] | 8-13 | CT > ASD (post DMN) | ADOS | Yes | 26484047 | |
| Jann et al., 2015 | DMN/fMRI | 39 [17/22] | 13.8(2.0) | CT > ASD (post DMN) | ADOS, SRS | Yes | 26445698 | |
| Respiratory sinus arrhythmia | Van Hecke et al., 2009 | Cardiorespiratory | 33 [19/14] | 8-12 | CT > ASD | ADOS, SRS | Yes | 19630897 |
| Patriquin et al., 2011 | Cardiorespiratory | 23 [23/0]1 | 4-7 | NA | SICS, SRS, SSP | Yes | 22212893 | |
| Klusek et al., 2013 | Cardiorespiratory | 107 [40/28/39FXS] | 4-15 | Not different | ADOS, CASL-PJ, PRS-SA | No | 24432860 | |
| Guy et al., 2014 | Cardiorespiratory | 36 [14/22] | 14.2 (3.7) | CT > ASD | ADOS, BRIEF, SRS, VABS | Yes | 24752681 |
monozygotic and dizygotic twins.
cross sectional
sex differences
longitudinal
child/father dyads
age of entry (followed up at 8 years of age)
part of an open-label trial; AbBC = Aberrant Behavior Checklist; AD = Axial diffusivitv; ADI-R = Autism Diagnostic Interview-Revised; ADI-Rsoc = ADI-R social domain; ADOS = Autism Diagnostic Observation Scale; ARI = Autism Risk Index (average of dwelling times to a priori social and non-social target regions of interest); ASAS = Australian Scale for Asperger Syndrome; ASD = Autism spectrum disorder; AQ = Autism Spectrum Quotient; ASSQ = Autism Spectrum Screening Questionnaire; ATEC = Autism Treatment Evaluation Checklist; BASC = Behavioral Assessment System for Children; BDNF = Brain-derived neurotrophic factor; BM = Biomarker; BRIEF = Behavior Rating Inventory of Executive Function; CARS = Childhood Autism Rating Scale; CASL-P:=Pragmatic Judgment subtest of the Comprehensive Assessment of Spoken Language; CBCL = Child Behavior Checklist; CBF = cerebral blood flow; CSF = cerebrospinal fluid; CT = Control; DD = Developmental Delay; diffs = differences; DMN= Default Mode Network; EEG = Electroencephalogram; FA = fractional anisotropy; FXS = Fragile X Syndrome; GABA = Gamma aminobutyric acid; GeoPref = Geometrical Preference; gIC = group Independent Components; GSH = Glutathione; IL-6 = Interleukin-6; JA = Joint Attention; LPS = Lipopolysaccharide; MD = mean diffusivity; mOFC = Medial orbitofrontal cortex; ND = Not Defined; PBMC = Peripheral blood mononuclear cell; PDD-BI = Pervasive Developmental Disorder Behavior Inventory; PMID = PubMed reference number; PRS-SA: Pragmatic Rating Scale-School Age; RBS-R = Repetitive Behavior Scale-Revised; RD = radial diffusivity; RNA = Ribonucleic acid; RS Resting State; RSA = Respiratory sinus arrhythmia; SAS = Severity of Autism Scale; sex diffs = sex differences; SICS = Social Interaction Coding Scale; Soc = Socialization; SocPref = Social Preference; SRS = Social Responsiveness Scale; SSP = Short Sensory Profile; SSRS = Social Skills Rating System; TD = Typical Development; TSwek = tensor skewness; ToM = Theory of Mind; VABS = Vineland Adaptive Behavior Scales, Social domain; XSkew = hemispheric asymmetry tensor skewness
Table 2.
Most cited biomarkers: interventional studies
| BM | Reference | Modality | Study Design | Intervention | Sample size [drug/placebo] |
Age (years) |
Biomarker outcome |
ASD Behavior measures |
Behavioral improvement |
Biomarker x Behavior Correlation* |
PMID |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutathione (GSH) | Bertoglio et al., 2010 | Plasma | RDBPC, crossover | Methyl B12 | 30/0 | 3-8 | Not different | AbBC, CARS, CBCL, PIA-CV | No | Not tested | 20804367 |
| Al-Ayadhi et al., 2013 | Plasma | RDBPC | Camel milk | 49/11 | 2-12 | Post > Pre | CARS, WSQ | Yes | Not tested | 24069051 | |
| Faber et al., 2015 | Plasma | Open-label | Cleanroom | 10/0 | 3-12 | Not stated | ATEC, CARS, GARS-2, PDDBI | No | Not tested | 25887094 | |
| Wink et al., 2016 | Plasma | RDBPC | N-acetylcysteine | 16/15 | 4-12 | Post > Pre | SRS, VABS | No | Not tested | 27103982 | |
| IL-6 | Tsilioni et al., 2015 | Serum | Open-label | Diet | 40/0 | 4-10 | Pre > Post | VABS | Yes | Yes | 26418275 |
| Melamed et al., 2018 | Blood | Open-label | Immunoglobulin, intravenous | 17/0 | 4-14 | Not different | ADOS, SRS | Yes | Not tested | 29427532 | |
| BDNF | Makkonen et al., 2011 | CSF | Open-label | Fluoxetine | 13/0 | 5-16 | Pre > Post | ATEC | Yes | Not tested | 29056860 |
| Pardo et al., 2013 | CSF, plasma | Open-label | Minocycline | 11/0 | 3-5 | Pre > Post | CGI-anchored, VABS | No | Not tested | 23566357 | |
| Hellings et al., 2015 | Serum | Open-label | Loxapine | 16/0 | 13-65 | Post > Pre | RBS-R | Yes | Not tested | 25782098 | |
| Serotonin | Bent et al., 2018 | Urine | Open-label | Sulforaphane | 21/0 | 5-22 | Not stated | AbBC, SRS | Yes | Yes | 29854372 |
| GABA | Connery et al., 2018 | Plasma | Open-label | Immunoglobulin, intravenous | 31/0 | 9.7 (4.4) | Not different | AbBC, SRS | Yes | Not tested | 30097568 |
| Oxytocin | Andari et al., 2010 | Plasma | RDBPC, cross-over | Oxytocin, intranasal | 33/0 | 17-39 | Post > Pre | Social behavior | Yes | Not tested | 20160081 |
| Parker et al., 2017 | Plasma | RDBPC | Oxytocin, intranasal | 32/0 | 6-12 | Not stated | SRS | Yes | Yes | 28696286 | |
| Alpha power | Kang et al., 2019 | EEG (resting state) | Open-label | rTMS treatment | 16/16 | 7.8 (2.1) | Post > Pre | AuBC | Yes | Not tested | 31228356 |
| Murias et al., 2018 | EEG (social vs non-social) | Open-label | Autologous umbilical cord blood | 25/0 | 2-6 | Post > Pre | VABS | Yes | Yes | 30070044 | |
| Fixation time (proportional looking times) | Bradshaw et al., 2019 | Eyes, mouth, face; social vs nonsocial | Pilot, randomized | PRISM | 12/11 | 1.5-4 | Not different | ADOS, VABS | No | No | 30891960 |
| Umbricht et al., 2017 | Social vs non-social | RDBPC, cross-over | V1a receptor antagonist, intravenous | 19/0 | 18-45 | Post > Pre (social) | ASR, RMET, SCIT, AbBC reduced | Yes | Not tested | 27711048 | |
| Andari et al., 2010 | Mouth, eyes, face | RDBPC, cross-over | Oxytocin, intranasal | 20/0 | 17-39 | Post > Pre | Social behavior | Yes | Yes | 20160081 | |
| Fractional Anisotropy | Carpenter et al., 2019 | DTI | Open-label | Autologous umbilical cord blood | 19/0 | 2-6 | Post > Pre | VABS | Yes | Yes | 30620122 |
| Functional activation (after biological motion) | Ventola et al., 2015 | Whole-brain | Other CT | PRT | 15/0 | 4.5-7.7 | Post > Pre (in one of 2 groups) | ADOS, SRS-2 | Yes | Yes | 25370452 |
| Yang et al., 2016 | Whole-brain | Open-label | PRT | 20/0 | 5.90 (1.07) | Not stated | SRS | Yes | Yes | 27845779 | |
| Yang et al., 2017 | Whole-brain | Pilot | VR-SCT | 17/0 | 18-31 | Not stated | ACS-SP, SAT | Yes | Yes | 28384509 |
correlation was not tested, the authors looked for the association of severity of symptoms in relation to the biomarker under study
proportional looking times; AbBC = Aberrant Behavior Checklist; ADOS = Autism Diagnostic Observation Scale; ARS = Affective Speech Recognition; ASD = Autism Spectrum Disorder; ATEC = Autism Treatment Evaluation Checklist; ACS= WMS-IV Social Perception Subtest; AuBC = Autism Behavior Checklist; BASC = Behavior Assessment System for Children; BDNF = Brain-derived neurotrophic factor; BM = Biomarker; CARS = Childhood Autism Rating Scale; CBCL = Child Behavior Checklist; CGI = Clinical Global Impression; CSF = cerebrospinal fluid; DTI = Diffusion Tensor Imaging; EEG = Electroencephalogram; FU = Follow-up after treatment; GABA = Gamma aminobutyric acid; GARS-2 = Gilliam Autism Rating Scale-2; GSH = Glutathione; IL-6 = Interleukin-6; Other CT = Other Clinical Trial; PDDBI = Pervasive Developmental Disorder Behavior Inventory; PIA-CV = Parent Interview for Autism-Clinical Version; PMID = PubMed reference number; PRISM = Pivotal Response Intervention for Social Motivation; PRT= Pivotal Response Treatment; RBS-R = Repetitive Behavior Scale-Revised; RDBPC = Randomized double-blind placebo-controlled; RMET = Reading the Mind in the Eyes Test; rTMS = Repetitive transcranial magnetic stimulation; SAT= Social Attribution Task; SCIT = Scripted Communication and Interaction Test; SRS = Social Responsiveness Scale; VABS = Vineland Adaptive Behavior Scales; VR-SCT= Virtual reality Social Cognition treatment; WSQ = Wing Subgroups Questionnaire.
Data analysis and statistical methods
Power calculations (Fig. 2) were performed using the ‘TTestIndPower’ function in the Python ‘statsmodels’ library to perform a two-sided t test with α values of 0.05 (nominal) and (5.3 x 10−5, Bonferroni correction for 940 biomarkers). Biomarkers reported in more than one study are displayed as co-publication networks using Cytoscape with the default ‘ForceDirected’ layout. For response biomarkers with consistent directions of effect across multiple studies (i.e., gluthathione) Cohen’s d was estimated from the t statistic (‘t2d’ function in the Python ‘psych’ library) or mean and standard deviations (Supplemental methods) and converted to Hedges’ g* with 95% confidence intervals (Table S7 and Supplemental methods). Hedges’ g* values were represented alongside 95% confidence intervals and sample size to provide insight into potential sample size biases (Fig. 3D, 3E). Due to the small number of studies and their heterogeneous designs, the data were not subjected to meta-analysis, or statistical assessment of heterogeneity, robustness, or bias.
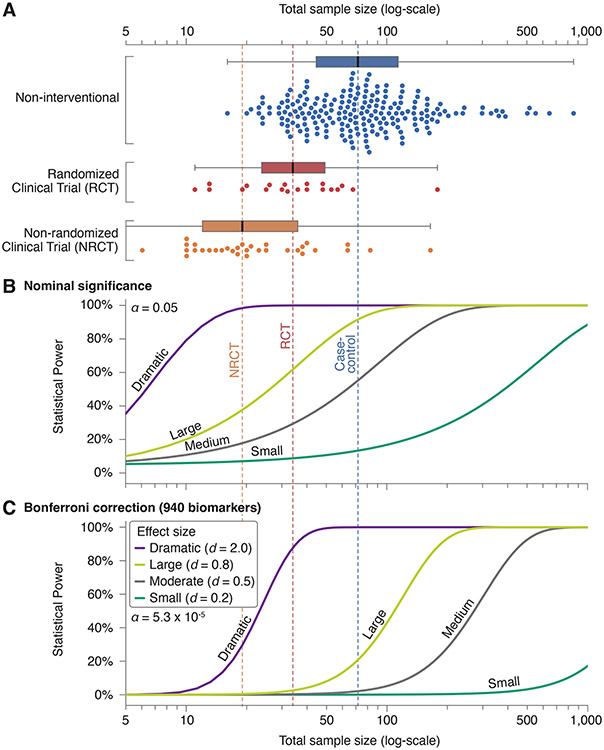
Figure 2. Sample sizes of biomarker studies alongside statistical power calculations.
A) Total sample size (x-axis, logarithmic scale) was calculated by adding all cases and controls in each study. Studies are divided by design (Fig. 1), with a boxplot showing the median sample size (thick black line and vertical dashed lines), the interquartile range (filled box), and data points within 1.5 interquartile ranges (whiskers). Each point of the swarmplot below the corresponding boxplot shows the total sample size of one of the 259 papers reviewed that included human subjects. B) Curves showing the statistical power against total sample size using a two-sided t-test with equal numbers of cases and controls (ratio = 1) at nominal significance (α = 0.05) for four effect sizes: Dramatic (Cohen’s d = 2), Large (Cohen’s d = 0.8), Moderate (Cohen’s d = 0.5), and Small (Cohen’s d = 0.2). C) The analysis in ‘B’ is repeated correcting for all 940 biomarkers with the Bonferroni method (α = 5.3 x 10−5).
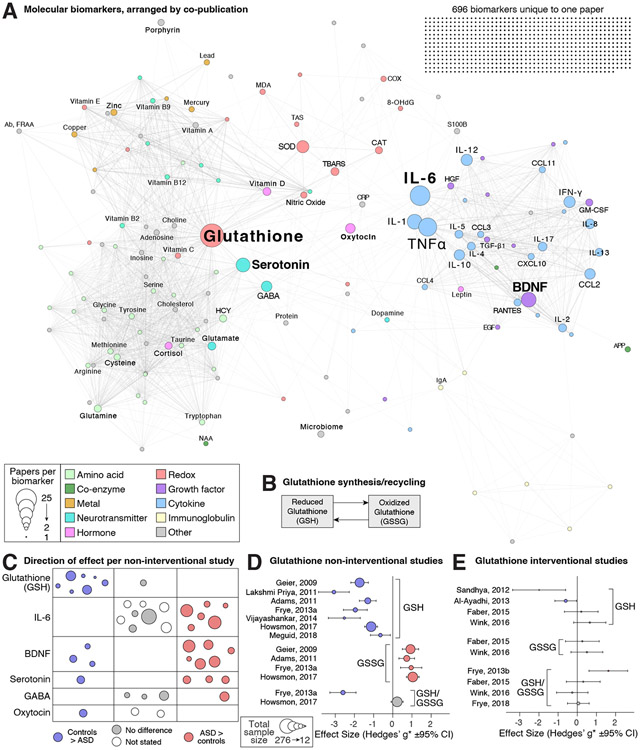
Figure 3. Molecular biomarkers.
A) The 846 unique molecular biomarkers are represented as nodes (circles) with sizes corresponding to the number of articles in which they are assayed and color representing functional category. Edges (lines) represent co-analysis of the two biomarkers in the same article and the network is displayed with a force-directed layout. B) When reduced glutathione (GSH) is oxidized it forms a dimer called oxidized glutathione (GSSG). C) The direction of effect (color and location) and total sample size (circle size) is shown for six frequently assayed biomarkers in non-interventional studies; each circle represents one study (Table 1). D) The effect size and 95% confidence interval for the difference in glutathione between ASD cases and controls. A positive effect size represents a higher concentration in cases. E) The effect size and 95% confidence interval for the difference in glutathione between treated ASD cases and untreated ASD cases. A positive effect size represents a higher concentration in those receiving the treatment. Hedges’ g* is similar to Cohen’s d, using the same scale but with a correction for small sample sizes (Supplementary methods). Abbreviations: HCY: Homocysteine, GABA: Gamma-aminobutyric acid, MDA: Malondialdehyde, SOD: Superoxide dismutase, TBARS: Thiobarbituric acid-reactive substances, CAT: Catalase, BDNF: Brain-Derived Neurotrophic Factor, TNFα: Tumor Necrosis Factor, GM-CSF: Granulocyte-Macrophage Colony Stimulating Factor (CSF2), CCL2: C-C motif chemokine ligand 2 (MCP-1), CCL3: C-C motif chemokine ligand 3 (MIP-1α), CCL4: C-C motif chemokine ligand 4 (MIP-1-beta), CCL5: C-C motif chemokine ligand 5 (RANTES), IFN-γ: Interferon gamma (IFNG), CRP: C-reactive protein, IL: interleukin, CXCL10: C-X-C motif chemokine ligand 10 (IP-10).
Results
Our final review included 280 articles (Table S1) of which 53 were human studies that included an intervention, 206 were human studies without an intervention, and 21 were interventional animal studies (Fig. 1E-F). The 53 human interventional studies included 20 randomized clinical trials (RCTs) and 33 non-randomized clinical trials (NRCTs), most of which were open-label trials. The non-interventional studies were mostly case-control design (189/206), with the remaining 17 including longitudinal and cross-sectional cohorts. Across all 280 studies, we divided the biomarkers into three groups (Fig. 1E-F): molecular (Table S5, e.g., glutathione, serotonin), neuroimaging and neurophysiological (Table S5, e.g., functional MRI, electroencephalography, eye-tracking), and other modalities (Table S5, e.g., heart rate). In total, we identified 940 unique biomarkers across all included studies of which 846 were molecular, 82 were neuroimaging/neurophysiological, and 12 were other modalities (Tables S5, S6).
Sample sizes and multiple comparisons
To provide insight into the level of evidence provided by the biomarker literature reviewed, we considered the statistical power at varying levels of biomarker effect size alongside the total sample size in the human studies reviewed (Fig. 2). The power calculation was initially performed with a two-sided t-test at nominal significance, i.e., α=0.05 (Fig. 2B). Candidate gene studies have highlighted the perils of relying on nominal significance as an appropriate threshold when numerous groups are engaging in parallel discovery efforts (19, 20). Appropriate correction for multiple comparisons has led to replicable findings in genomics, therefore we repeated the power calculation using the Bonferroni method to correct for all 940 biomarkers assessed, α=5.3 x 10−5 (Tables S5, S6). Non-interventional studies were generally larger than clinical trials (Fig. 2A). Most non-interventional and randomized clinical trials were adequately powered to identify a biomarker with a large effect size (Cohen’s d of ≥0.8) at nominal significance (Fig. 2B), however, few were capable of identifying a moderate effect size (Cohen’s d of ≥0.5) or a biomarker with large effect size after correction for multiple biomarkers (Fig. 2C). Non-randomized clinical trials included the fewest samples and were only powered to identify biomarkers with dramatic effects (Cohen’s d of ≥2.0) at nominal significance (Fig. 2B).
Molecular biomarkers
The most frequently assayed group of biomarkers was molecular (Table S5, Fig. 3), the great majority of which were measured peripherally via blood, though there was considerable variation both between and within individual biomarkers. We identified 846 unique molecules across 189 manuscripts, the majority of which (696 molecules, 82.2%) were unique to one manuscript (Fig. 3A). By considering the frequency and overlap of the remaining 150 molecules across the manuscripts, we generated a network of molecule co-publication. Two major groups were apparent, one made up of cytokines and growth factors (e.g., Interleukin-6 [IL-6], Brain-Derived Neurotrophic Factor [BDNF]) and the other a combination of amino acids, neurotransmitters (e.g., Cysteine, Serotonin, GABA), and hormones (e.g., Vitamin D). Between these two groups were molecules relating to reduction/oxidation (redox), including glutathione, the most frequently assayed molecule. Most papers report an association between a molecular marker and ASD diagnostic status. True biomarkers should have a consistent association with ASD symptoms, including magnitude and direction of effect, across multiple analyses, therefore we collated the reported outcomes of the most frequently analyzed molecules across the main molecular classes (Fig. 3C, Tables 1 and 2). While the direction of effect was usually reported, the magnitude of effect, summary statistics that enabled the magnitude to be calculated (e.g., mean and standard deviation), or individual-level values were often not, preventing meta- or mega-analysis.
Redox metabolism:
Glutathione is assessed in 25 of the reviewed papers (Fig. 3). All measures were peripheral and 14 included sufficient details to quantify the magnitude of observed effect (Fig. 3D-E, Table S7). A decrease in reduced glutathione (GSH), generally studied in plasma (Table 1), is reported in children diagnosed with ASD in seven out of eight case-control cohorts (Fig. 3C) with a median Hedges’ g*, equivalent to Cohen’s d with a sample size correction, of −1.77 (Fig. 3D, Table 1, Table S7). Four of these papers also report a corresponding increase in oxidized glutathione (GSSG) in ASD cases (Fig. 3D). The glutathione results were the most consistent in direction and magnitude of effect size of any of the biomarkers examined in depth. Interventional studies, mostly aimed at reducing oxidative stress, showed inconsistent results (Fig. 3E, Table 2 and S8).
Cytokines:
Thirty-three papers described assays of cytokine immune signaling molecules in ASD, with Interleukin 6 (IL-6) being the most frequently assayed (21 papers). All papers used peripheral measures, except one that assayed cerebrospinal fluid (CSF). Fourteen papers compare IL-6 levels between ASD cases and controls (Table 1). Eight did not report any IL-6 association, while the remaining six report an IL-6 increase (pro-inflammatory state), which was correlated with the severity of ASD symptoms (stereotypies or social impairment), especially in those with regressive ASD (21) or with gastrointestinal disorders (22). Two interventional studies, aimed at reducing systemic inflammation, report improvements in ASD-related behaviors, one of which is accompanied by an IL-6 reduction (Table 2 and S8).
Growth factors:
Thirty-three papers reported on growth factors, often in tandem with cytokines; all were assayed peripherally except one assayed in CSF. Brain-derived neurotrophic factor (BDNF) was the most frequently assayed growth factor, with 14 papers reporting levels in serum/plasma and one in CSF. Nine case-control studies all reported association with ASD, however the direction of effect varied – an increase was associated with ASD in six and a decrease in three. Those reporting an increase tended to be larger (120 vs. 50 mean total samples) and include more severe cases (Table 1). Three interventional studies also showed inconsistent direction of effect for BDNF (Table 2 and S8).
Neurotransmitters:
Neurotransmitters were measured in 25 papers (two in brain via spectroscopy, the remainder were all peripheral), of which 14 included serotonin and ten included GABA (Fig. 3). Three of four case-control analyses reported higher serotonin levels in ASD cases, while one showed the opposite (Table 1, Fig. 3C); one interventional study in humans reported correlation with behavior (Table 2). The remaining studies, many of which were interventional studies in animal models, showed inconsistent effects on serotonin levels (Table S8). For GABA, one case-control study of plasma GABA levels reported higher levels in children with ASD while another did not; two brain spectroscopy analyses showed no differences. A single interventional trial reported no associated between GABA levels and ASD.
Hormones:
Twenty-eight of the reviewed papers included hormonal assays, with oxytocin being the most frequently assayed. Most assays were peripheral, with the exception of two analyses of CSF (oxytocin/vasopressin and leptin). One of three case-control papers reported an association, with oxytocin lower in those with ASD (Table 1, Fig. 3C). The two intervention studies conducted in humans assessed intranasal oxytocin, and while both reported some degree of improvement in social behavior, only one reported increased levels of oxytocin (Table 2) (23, 24). Two more interventional studies were conducted in animal models. Again, both showed improvement in the behavioral measures assessed while only one reported an increase in the biomarker (Table S8).
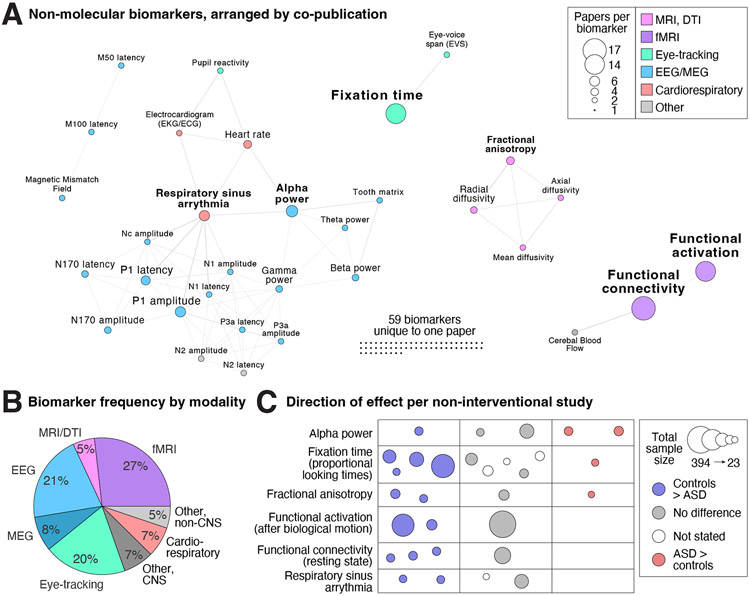
Neurophysiological biomarkers
The second most frequently assayed category of putative biomarkers was neurophysiological (46 manuscripts, Figs. 1F, 4B). Within this category, the two main approaches were electroencephalography (EEG)/magnetoencephalography (MEG), with 28 papers describing 50 unique biomarkers, and eye-tracking, with 19 papers describing 21 unique biomarkers. Across EEG/MEG analyses, the most frequently assessed biomarker was alpha power (Tables 1 and 2, Fig. 4), which showed inconsistent results across studies with variable directions of effect if an association was detected. N170 has been proposed as a diagnostic biomarker in ASD (25); our review included three case-control analyses that assess correlation with severity (Table S6). Within the eye-tracking manuscripts, fixation time was the most frequently assayed outcome. Seven case-control studies assessed the proportion of looking time, however the visual stimulus varied widely. Four of the seven reported controls looking for longer at social behaviors, speech, or eyes while three did not (Table 1, Fig. 4); one of three interventional studies showed correlation with severity.
Figure 4. Non-molecular biomarkers.
A) The 94 unique non-molecular biomarkers are represented as nodes (circles) with size corresponding to the number of articles in which they are assayed and color representing biomarker category. Edges (lines) represent co-analysis of the two biomarkers in the same article and the network is displayed with a force-directed layout. B) Distribution of the total number of papers split by modality. C) The direction of effect (location and color) and total sample size (circle size) is shown for six most frequently assayed biomarkers; each circle represents one study (Table 1). Abbreviations: CNS: Central nervous system; DTI: Diffusion tensor imaging; EEG: electroencephalography; MEG: magnetoencephalography.
Neuroimaging biomarkers
The third most frequently assayed category was neuroimaging with 35 manuscripts (Figs. 1F, 4B), including 25 manuscripts detailing functional MRI (fMRI) analyses, most of which assessed resting state functional connectivity or task-related functional activation (Fig. 4, Table S6). Of the remaining neuroimaging manuscripts, six described diffusion tensor imaging (DTI) with fractional anisotropy being the most common metric (Fig. 4) and the direction of effect varying across the four case-control studies and one open-label trial (Tables 1 and 2, Fig. 4). The remainder of neuroimaging studies applied diverse neuroimaging techniques (Table S6).
Seventeen functional connectivity studies assessed brain networks in the resting state or, occasionally, during a task. Five studies assayed whole-brain network connectivity, with four different analytical approaches and no replicated findings. Of these, two used Independent Component Analysis but the connectivity patterns detected did not overlap (26, 27). Another study was a meta-analysis across two multisite repositories to assess 654 individuals (28). This meta-analysis employed a Bayesian model to identify three ASD-associated “factors”, all within the default network. Only the first factor correlated with ASD symptoms and was defined by hypoconnectivity within and between perceptual/motor networks and hyperconnectivity between perceptual/motor and association networks and between somatomotor and subcortical regions. The other eleven studies assessed specific networks; only the default mode network (DMN) and the salience network were studied in more than one. All four default mode network studies (Table 1 and 2, Fig. 4) reported decreased connectivity between the medial prefrontal cortex (mPFC) and the posterior cingulate cortex (PCC), which was negatively correlated with social impairment in only two. The salience network studies did not show replicated patterns or symptomatic correlation.
Across the 13 functional activation studies, the tasks most frequently assessed were attention to biological motion versus scrambled motion and social psychological tasks, such as nonverbal judgments after viewing a situation or game. Whole-brain analyses were generally used for initial discovery with specific region-task combinations followed up in subsequent targeted analyses. Task activation correlated with ASD severity most frequently in the superior temporal sulcus, parieto-temporal junction, and medial and inferior prefrontal cortices (Table 1, Table S6). Regional activation was included in five clinical trials, conducted by two research groups, focusing on oxytocin and pivotal response treatment respectively. Promising results from an RCT of single intranasal dose of oxytocin under lab conditions (29) were followed by improvement in ADOS reciprocity scores following six-weeks of treatment (30) that correlated with greater task-independent resting state functional connectivity between the anterior cingulate cortex and dorsomedial prefrontal cortex. However, larger trials have failed to replicate improvements in ADOS score with intranasal oxytocin (31).
Physiological biomarkers
The final category of biomarkers we identified were physiological measures outside the central nervous system (e.g., movement, heart rate). Eleven papers included such measures with respiratory sinus arrhythmia (RSA), the physiological change in heart rate with breathing, being the most frequently assayed. Four papers assess RSA, of which two reported reduced RSA in individuals with ASD (Table 1).
Discussion
Through a systematic review of quantitative biomarkers in ASD, we identified 280 papers that detailed analyses of 940 potential response biomarkers. The majority of papers reported an association between a biomarker and ASD, yet no biomarkers have been qualified by the Food and Drug Administration’s Center for Drug Evaluation and Research Biomarker Qualification Program (CDER-BQP). Furthermore, biomarkers assessed multiple times mostly reveal both inconsistent evidence of ASD association and variable direction of effect (Figs. 3 and 4, Tables 1 and 2). These discrepancies suggest a reproducibility crisis, as observed in other fields of biomedical research (32, 33). Our review identifies small sample sizes (Fig. 2), inadequate correction for multiple comparisons (Fig. 2), and the absence of replication cohorts as contributory factors. Given the high degree of positive findings despite minimal replication, it is likely that there is also a substantial publication bias, though this was not assessed statistically due to the limited reporting of quantitative outcomes in many analyses.
Against this background, distinguishing biomarkers that show true association with ASD symptoms is challenging. Mega-analysis of individual-level biomarker values would facilitate clear comparisons across biomarkers and assessment of biomarker-wide significant results; however, few studies include individual-level data. Similarly, meta-analysis, based on summary statistics, would also allow consistent comparisons across the field; surprisingly, many studies did not include these metrics, furthermore the heterogeneity of study design (e.g., phenotypic, demographics, methods) complicates comparison across multiple biomarkers. Well-known developmental changes are rarely taken into account in study designs (30). Replication should distinguish true positive associations; focusing on the most frequently assayed biomarkers in each class; the majority do not show consistent results (Figs. 3, 4).
The most consistent results were lower levels of reduced glutathione (GSH) in ASD, observed in seven out of eight case-control cohorts, with corresponding changes in oxidized glutathione (GSSG) (34). The median effect size of −1.77 (Hedges’ g/Cohen’s d, Fig. 3D) is substantial. This pattern could be consistent with true ASD association, or it could arise by chance from the 940 biomarkers assayed and/or publication bias. A rigorous, large-scale, pre-registered analysis is required to resolve these two possibilities; with this effect size, the results should be definitive. Glutathione is considered to be a marker of oxidative stress in ASD, a hypothesis based on early findings of increased lactic acid in some children with ASD and the high frequency of ASD in children with genetic defects in mitochondrial enzymes (35). To date, no gene related to oxidative stress has been associated with ASD through common or rare variation, suggesting a causal role is unlikely, though it is possible that oxidative stress reflects nonspecific systemic dysfunction.
Many of the biomarkers assessed were cytokines, aiming to detect a pro-inflammatory state. Inflammation is a component of the maternal immune activation (MIA) and lipopolysaccharide (LPS) animal models that induce social impairments, furthermore gene expression analyses of the postmortem human cortex show a consistent up-regulation of co-expressed modules enriched for immune-related genes in ASD (14, 36, 37), however, neither common nor rare variation associated with ASD implicate immune processes. Pro-inflammatory IL-6 was the most frequently studied cytokine and most studies failed to identify association with ASD (Fig. 3), but those that did consistently reported higher IL-6 in ASD cases, often correlated with severity. In a meta-analysis of pro-inflammatory cytokines in ASD, IFN-γ, IL-1β, IL-6, and TNF-α reached nominal significance (38), however, these results do not survive correction for the 21 cytokines assayed, let alone the 940 identified in this review.
The growth factor BDNF and neurotransmitter serotonin both play critical roles in neurodevelopment and neurophysiology and highlight the importance of developmental age. Both BDNF and serotonin have some evidence to support a role as diagnostic biomarkers (39-42), however, physiological levels and correlation with ASD vary across development (41, 43). Hyperserotonemia was one of the first biomarkers implicated in ASD, with early studies showing that a third of the subjects with ASD showed increased blood levels (44), a finding that has been validated in a recent meta-analysis (45). Subsequent human and animal studies showed a physiological reduction in serotonin with increasing age. This age-related reduction is attenuated in autistic individuals (43, 46), so that ASD-related hyperserotonemia is most apparent in late childhood (47). While the papers we reviewed all report an association of BDNF and serotonin levels with ASD, the direction of effect varies (Table 1, Fig. 3C). Prior evidence of ASD association may have been incorrect, alternatively heterogeneous ages of individuals in these cohorts may have masked or augmented the ASD-related differences.
The extensive variability of biomarkers, both within and between individuals, presents one of the biggest challenges to biomarker research. Likely sources of variability include symptom severity, co-morbidities, developmental age, sex, ancestry, genetic variation, environmental conditions (e.g., diet, medications, infections), time-of-day, sample processing methods, tissue assayed, and experimental assay. The blood-brain barrier is expected to be a major source of variability, leading to different results if molecular biomarkers are assayed centrally or peripherally. This will vary between molecules, for example, plasma-brain correlation has been demonstrated for BDNF in rodents, but not in humans (48), while such correspondence has not been established for GABA (49) or most other biomarkers. Differences between central and peripheral assays may also occur across development as the permeability of the blood-brain barrier varies with maturity. Within the central or peripheral compartments, the tissue assayed is also critical. For example, platelets store serotonin, so platelet-rich blood provides more accurate assays. Where possible, recording likely sources of variability enables their inclusion as covariates in statistical models, for example correcting for population stratification based on genotypic data. For unrecognized variables, careful experimental design, including selection of cases and controls, is critical. In discovery cohorts, large sample sizes are essential to overcome this variability, while longitudinal analyses in the same individuals can help delineate the major sources of intra- and inter-individual variability in validation studies. Identifying these key covariates will be critical to define the homogenous cohorts in which a specific biomarker may augment a clinical trial.
Neuroimaging and neurophysiological studies have clear potential for detecting biomarkers, however, sample sizes are modest (median total sample size = 44, Table S6) and a myriad of techniques, instruments, tasks, brain regions, and data processing methods pose additional challenges to distinguishing true biomarkers (Fig. 4). Both alpha power and fixation time, the most frequently assayed biomarkers in neurophysiological studies, showed inconsistent direction of effect across studies (Table 1, Fig. 4C). As for neuroimaging, both task activation and functional connectivity studies implicated brain regions that are generally considered part of the “social brain” (50), including the medial prefrontal and temporal cortices. Activation of regions in the medial prefrontal cortex and the parieto-temporal junction replicate across different study designs and show changes that correlate with ASD symptoms in clinical trials (29, 30, 51). Functional connectivity analyses show consistent ASD-association for the default mode network with decreased connectivity between the medial prefrontal cortex and posterior cingulate cortex with some correlation with ASD symptoms (52-55).
The variability issues that embroil neuroimaging markers are compounded by variability in data acquisition methods, such as differences in hardware, image acquisition sequence parameters, tasks, as well as differences in data analytic approaches including data post-processing and quality control as well as focus on various brain regions. While there is some replication in studies implicating social brain regions and the default mode network, the heterogeneity between studies prevents clear conclusions at this time. Data repositories, such as the Autism Brain Imaging Data Exchange (ABIDE), and open-source data analysis pipelines (e.g., ABIDE imaging masks and analysis pipelines) are enabling a new generation of larger-scale neuroimaging analyses with clear correction for multiple comparisons and open datasets for replication (56). It remains to be seen whether these initial findings prove to be robustly replicated in subsequent studies and whether some of the methodological difficulties are overcome best by pooling multiple studies or designing large-scale studies with consistent methods.
Animal models enable direct measurement of brain tissue and greater control over experimental conditions. All biomarkers assessed in animals were molecular and many overlapped with the molecules and classes most frequently assayed in human studies (Fig. 3, Table S8). The model most frequently used in the reviewed studies was the BTBR mouse, which is defined solely on ASD-like behaviors (57). The reliance on purely behavioral phenotypes, particularly in rodents, has generally not been productive for illuminating the biology of psychiatric phenotypes, with rare exceptions, e.g., brexanolone (58). In ASD, particularly given the discovery of dozens of large-effect mutations that can subserve the creation of “construct-valid” animal models, reliance on the BTBR system has come under increasing scrutiny and at present has questionable relevance to the human syndrome. In addition, several environmentally induced models including exposure to MIA and VPA have been studied, but it is unclear to what extent these model common etiological mechanisms of ASD in humans. After applying our review criteria (Fig. 1), none of the animal models included were based on ASD-associated genes. Relaxing our exclusion criteria by discounting criterion 6 and 7 (Fig. 1) identifies a further 53 animal studies, but even here only a few genetic models are used (e.g., 16p11.2, MECP2, FMR1, Table S1). With CRISPR-Cas9 gene editing and biorepositories, genetic model systems are an underutilized resource in biomarker discovery, including genetic animal models, and isogenic and patient-derived human cell culture models.
Limitations of the review
While we identified over one thousand original research articles that included putative biomarkers in ASD, we focused on about a quarter of these that were most relevant to the BEST-defined response biomarkers (see Methods and Table S1) (4), by selecting manuscripts that assessed whether biomarkers correlate with ASD symptom intensity. We note that biomarkers can overlap between classes, for example a diagnostic biomarker that distinguished individuals with high or low ASD liability might also change in relation to a behavior-of-interest. However, since no ASD biomarker in any class has been qualified by CDER-BQP and since assessing correlation to symptom severity is a logical follow-up analysis for promising diagnostic biomarkers, it is unlikely that in depth analysis of the other 750 manuscripts would change our main conclusions. We note that we did not systematically assess whether these conclusions generalize to other biomarker classes.
Our search is also limited by the accuracy and sensitivity of ASD severity measures, especially in the older studies. We cast a wide net, including instruments the authors of each study utilized as a measure of autistic severity, unless the measure was solely assessing global functioning or disability. Consequently, some of the measures included are confounded by behavioral or intellectual impairment. Severity measures and validation efforts continue to improve (59); it remains to be seen whether currently available metrics can detect the modest short-term changes that are likely to be necessary for evaluating biomarkers or therapeutic response.
Our most in-depth analysis focused on twelve biomarkers assayed across multiple papers (Table 1), allowing us to assess replication to distinguish true biomarkers. An alternative strategy would be to rank all biomarkers assayed by effect size or p-value to find the most promising. However, we found the analytic methods and summary statistics reported to be too heterogeneous and incomplete for this approach; it is possible that true biomarkers are included in those studied but do not currently stand out from the crowd (Table S4, S5).
Finally, we note several items missing from the PRISMA checklist, specifically that the review and protocol were not registered in advance, that we included 32 articles identified from citations but not the initial search, and that formal analysis of risk of bias, robustness, and heterogeneity were not performed due to the heterogeneity of the studies reviewed.
Future studies
The ASD biomarker field is reminiscent of the era of candidate gene discovery, in which technological and biobank limitations necessitated a focus on small numbers of loci in small cohorts, which in turn led to a replication crisis (19, 20). Many of the lessons learned from candidate gene approaches are transferable to biomarkers, including the need for larger sample sizes, appropriate multiple comparisons (Fig. 2), and replication cohorts. Correcting statistical significance thresholds to reflect all biomarkers assayed in a study is a bare minimum. A higher statistical threshold is required to overcome publication bias. Ideally, this threshold would be based on the total number of effective tests across all biomarkers, estimated from the degree of interdependence between biomarkers (60). Until such estimates can be made, family-wise error correction (e.g., Bonferroni) for all biomarkers tested to date, currently about 1,000, is a simple and conservative approach. Alternatively, widespread sharing of individual-level data and key metrics (e.g., tissue, collection conditions, assay, demographics, deep phenotyping) would allow false-discovery rates to be estimated in multi-biomarker mega-analyses. The widespread data sharing required would be simplified by the adoption of community-wide standards, for example using standardized ontologies and machine-readable file formats to share biomarker results and key metrics. Once true biomarkers are identified for specific subgroups of ASD individuals, specific developmental stages or specific symptoms, their usefulness for monitoring change due to specific interventions could be tested in clinical trials. Only when the evidence for the biomarker is comparable to the confidence in existing behavioral measures of ASD severity will biomarkers become a useful outcome measure for interventional studies.
Based on our review, we provide a list of recommendations to help identify and distinguish true ASD biomarkers (Box 1). Many of these recommendations are already being applied in some recent neuroimaging studies and through the Food and Drug Administration’s Center for Drug Evaluation and Research Biomarker Qualification Program process initiated by Autism Biomarkers Consortium for Clinical Trials (ABC-CT). Advances in genetics, such Mendelian randomization (61) and CRISPR-derived model systems, and technology, such as proteomics and metabolomics, have the potential to greatly accelerate the hunt for biomarkers. Though ASD biomarkers remain elusive, there is immense potential if community-wide efforts can be paired with rigorous scientific methodology.
Box 1: Recommendations for ASD Biomarker Research.
Individual biomarker values should be shared for each sample to enable mega-analyses, ideally in standardized machine-readable formats.
Key demographic, deep phenotypic, and experimental data should be shared for each sample, including age, sex, self-reported and/or genotypic ancestry, medications, and comorbid conditions including intellectual disability, seizures, and motor delay, tissue assayed, time-of-day, and experimental assay. Sharing these details would enable them to be included as covariates and to identify more homogenous subsets.
Biomarkers and meta-data should be registered against standard schema or anatomical atlases (e.g., Chemical Entities of Biological Interest, PRotein Ontology) to facilitate comparisons and mega-/meta-analyses.
Consortia should promote consistent methodologies and larger cohorts with consistent phenotypic and genotypic data.
Case-control biomarker discovery cohorts should be adequately powered and should include an independent replication cohort.
A clear statement should be included about the approach to multiple comparisons, both in the paper and across the biomarker field (equivalent to genome-wide significance).
As is standard practice in human interventional trials, non-interventional biomarker studies should be preregistered to define the research plan prior to data collection.
Once suitable biomarkers are identified, cohorts selected for clinical trials should be tailored to both the biomarker and the intervention.
Supplementary Material
Acknowledgements:
This work was supported by the Spanish Ministry of Science and Innovation, the National Institute for Mental Health (NIMH, R01 MH129751, S.J.S.), the Simons Foundation Autism Research Initiative (SFARI, #736613, S.J.S.), Instituto de Salud Carlos III, co-financed by ERDF Funds from the European Commission, “A way of making Europe”, CIBERSAM. Madrid Regional Government (B2017/BMD-3740 AGES-CM-2), European Union Structural Funds and European Union Seventh Framework Program and H2020 Program; Fundación Familia Alonso, Fundación Alicia Koplowitz and Fundación Mutua Madrileña. These funding sources had no role or influence on the systematic review or manuscript. We thank Kenia Martinez, PhD, CIBERSAM, University Autonoma, Madrid, for her insights into the neuroimaging section of the paper.
Footnotes
Conflicts of Interest
MP has served in advisory or consultancy role for Exeltis and Servier and she is involved in clinical trials promoted by Servier and for F. Hoffmann La Roche Ltd. ASJC has served in an advisory or consultancy role for F. Hoffmann La Roche Ltd and she is involved as consultant in clinical trials conducted by Servier. AA, EU and MB have nothing to declare. LLC has received consulting income from Neuronetics, AffectNeuro, Neurolief, Janssen, Sage Therapeutics, Sunovion, and Otsuka. She has research support from Neuronetics, AffectNeuro, Janssen, and Nexstim. NVK has served as a consultant for Neurocrine Biosicences, Inc. WMM is a member of the American Psychiatric Association (APA) Council on Research representing ECT and Neuromodulation Therapies. He is compensated as the chair of the DSMB for the NIA multicenter study. He is on the Board of Skyland Trail and 3Keys. He is a paid consultant for Signant Health and Sage Therapeutics. He has received past funding from the Stanley Foundation, Soterix, Neuronetics, NeoSync and Cervel Neurotherapeutics. He has endowed chair funded by the JB Fuqua Foundation. He is an employee of Emory University School of Medicine. CBN has served as a consultant for ANeuroTech (division of Anima BV), Signant Health, Sunovion Pharmaceuticals, Inc., Janssen Research & Development LLC, Magstim, Inc., Navitor Pharmaceuticals, Inc., Intra-Cellular Therapies, Inc., EMA Wellness, Acadia Pharmaceuticals, Axsome, Sage, BioXcel Therapeutics, Silo Pharma, XW Pharma, Neuritek, Engrail Therapeutics, Corcept Therapeutics Pharmaceuticals Company. He is a stockholder of Xhale, Seattle Genetics, Antares, BI Gen Holdings, Inc., Corcept Therapeutics Pharmaceuticals Company, EMA Wellness. He serves on the Scientific Advisory Boards of ANeuroTech (division of Anima BV), Brain and Behavior Research Foundation (BBRF), Anxiety and Depression Association of America (ADAA), Skyland Trail, Signant Health, Laureate Institute for Brain Research (LIBR), Inc., Magnolia CNS, Heading Health. He serves on the Board of Directors for Gratitude America, ADAA, Xhale Smart, Inc. He holds patents for “Method and devices for transdermal delivery of lithium” (US 6,375,990B1) and “Method of assessing antidepressant drug therapy via transport inhibition of monoamine neurotransmitters by ex vivo assay” (US 7,148,027B2). CIR has served as a consultant for Epiodyne, received research grant support from Biohaven Pharmaceuticals, and a stipend from APA Publishing for her role as Deputy Editor at The American Journal of Psychiatry. ASW has served as a consultant for Livanova and Dandelion Science. He receives device donations for Medtronic. He has both granted and pending patents related to subject matter discussed in the article. SJS receives research funding from BioMarin Pharmaceutical Inc.
References
- 1.Association AP. Diagnostic and statistical manual of mental disorders, 5th Edition (DSM-V): American Psychiatric Publishing, Washington, DC; 2013. [Google Scholar]
- 2.Maenner MJ, Shaw KA, Baio J. Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2016. MMWR Surveillance Summaries. 2020;69(4):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5(6):463–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Group F-NBW. BEST (Biomarkers, EndpointS, and other Tools) resource. 2016. [PubMed] [Google Scholar]
- 5.Califf RM. Biomarker definitions and their applications. Exp Biol Med (Maywood). 2018;243(3):213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180(3):568–84.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155(5):997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lord C, Brugha TS, Charman T, Cusack J, Dumas G, Frazier T, et al. Autism spectrum disorder. Nat Rev Dis Primers. 2020;6(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sestan N, State MW. Lost in Translation: Traversing the Complex Path from Genomics to Therapeutics in Autism Spectrum Disorder. Neuron. 2018;100(2):406–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51(3):431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willsey AJ, Morris MT, Wang S, Willsey HR, Sun N, Teerikorpi N, et al. The Psychiatric Cell Map Initiative: A Convergent Systems Biological Approach to Illuminating Key Molecular Pathways in Neuropsychiatric Disorders. Cell. 2018;174(3):505–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramaswami G, Won H, Gandal MJ, Haney J, Wang JC, Wong CCY, et al. Integrative genomics identifies a convergent molecular subtype that links epigenomic with transcriptomic differences in autism. Nature communications. 2020;11(1):4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Forn M, Boitnott A, Akpinar Z, De Rubeis S. Linking Autism Risk Genes to Disruption of Cortical Development. Cells. 2020;9(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46(8):881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones RM, Lord C. Diagnosing autism in neurobiological research studies. Behavioural brain research. 2013;251:113–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS med. 2009;6(7):e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson EC, Border R, Melroy-Greif WE, de Leeuw CA, Ehringer MA, Keller MC. No Evidence That Schizophrenia Candidate Genes Are More Associated With Schizophrenia Than Noncandidate Genes. Biological psychiatry. 2017;82(10):702–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan LE, Ostacher M, Ballon J. How genome-wide association studies (GWAS) made traditional candidate gene studies obsolete. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2019;44(9):1518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashwood P, Krakowiak P, Hertz-Picciotto I, Hansen R, Pessah IN, Van de Water J. Altered T cell responses in children with autism. Brain, behavior, and immunity. 2011;25(5):840–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Napolioni V, Ober-Reynolds B, Szelinger S, Corneveaux JJ, Pawlowski T, Ober-Reynolds S, et al. Plasma cytokine profiling in sibling pairs discordant for autism spectrum disorder. Journal of neuroinflammation. 2013;10(1):813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andari E, Duhamel J-R, Zalla T, Herbrecht E, Leboyer M, Sirigu A. Promoting social behavior with oxytocin in high-functioning autism spectrum disorders. Proceedings of the National Academy of Sciences. 2010;107(9):4389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker KJ, Oztan O, Libove RA, Sumiyoshi RD, Jackson LP, Karhson DS, et al. Intranasal oxytocin treatment for social deficits and biomarkers of response in children with autism. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(30):8119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Food And Drug Administration U. LOI Decission Letter: N170 to upright human faces 2019. [Available from: https://www.fda.gov/media/127494/download. [Google Scholar]
- 26.Xu S, Li M, Yang C, Fang X, Ye M, Wei L, et al. Altered Functional Connectivity in Children With Low-Function Autism Spectrum Disorders. Frontiers in neuroscience. 2019;13:806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uddin LQ, Supekar K, Lynch CJ, Khouzam A, Phillips J, Feinstein C, et al. Salience network-based classification and prediction of symptom severity in children with autism. JAMA psychiatry. 2013;70(8):869–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang S, Sun N, Floris DL, Zhang X, Di Martino A, Yeo BTT. Reconciling Dimensional and Categorical Models of Autism Heterogeneity: A Brain Connectomics and Behavioral Study. Biological psychiatry. 2020;87(12):1071–82. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe T, Abe O, Kuwabara H, Yahata N, Takano Y, Iwashiro N, et al. Mitigation of Sociocommunicational Deficits of Autism Through Oxytocin-Induced Recovery of Medial Prefrontal Activity: A Randomized Trial. JAMA psychiatry. 2014;71(2):166–75. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe T, Kuroda M, Kuwabara H, Aoki Y, Iwashiro N, Tatsunobu N, et al. Clinical and neural effects of six-week administration of oxytocin on core symptoms of autism. Brain : a journal of neurology. 2015;138(11):3400–12. [DOI] [PubMed] [Google Scholar]
- 31.Yamasue H, Okada T, Munesue T, Kuroda M, Fujioka T, Uno Y, et al. Effect of intranasal oxytocin on the core social symptoms of autism spectrum disorder: a randomized clinical trial. Molecular psychiatry. 2020;25(8):1849–58. [DOI] [PubMed] [Google Scholar]
- 32.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2(8):e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pashler H, Wagenmakers EJ. Editors' Introduction to the Special Section on Replicability in Psychological Science: A Crisis of Confidence? Perspect Psychol Sci. 2012;7(6):528–30. [DOI] [PubMed] [Google Scholar]
- 34.Bjørklund G, Tinkov AA, Hosnedlová B, Kizek R, Ajsuvakova OP, Chirumbolo S, et al. The role of glutathione redox imbalance in autism spectrum disorder: A review. Free radical biology & medicine. 2020;160:149–62. [DOI] [PubMed] [Google Scholar]
- 35.Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Molecular psychiatry. 2012;17(3):290–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science. 2019;364(6441):685–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J, et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nature communications. 2014;5:5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saghazadeh A, Ataeinia B, Keynejad K, Abdolalizadeh A, Hirbod-Mobarakeh A, Rezaei N. A meta-analysis of pro-inflammatory cytokines in autism spectrum disorders: Effects of age, gender, and latitude. Journal of psychiatric research. 2019;115:90–102. [DOI] [PubMed] [Google Scholar]
- 39.Armeanu R, Mokkonen M, Crespi B. Meta-analysis of BDNF levels in autism. Cellular and molecular neurobiology. 2017;37(5):949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saghazadeh A, Rezaei N. Brain-Derived Neurotrophic Factor Levels in Autism: A Systematic Review and Meta-Analysis. Journal of autism and developmental disorders. 2017;47(4):1018–29. [DOI] [PubMed] [Google Scholar]
- 41.Qin X-Y, Feng J-C, Cao C, Wu H-T, Loh YP, Cheng Y. Association of peripheral blood levels of brain-derived neurotrophic factor with autism spectrum disorder in children: a systematic review and meta-analysis. JAMA pediatrics. 2016;170(11):1079–86. [DOI] [PubMed] [Google Scholar]
- 42.Zheng Z, Zhang L, Zhu T, Huang J, Qu Y, Mu D. Peripheral brain-derived neurotrophic factor in autism spectrum disorder: a systematic review and meta-analysis. Scientific reports. 2016;6:31241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandana SR, Behen ME, Juhász C, Muzik O, Rothermel RD, Mangner TJ, et al. Significance of abnormalities in developmental trajectory and asymmetry of cortical serotonin synthesis in autism. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 2005;23(2–3):171–82. [DOI] [PubMed] [Google Scholar]
- 44.Cook EH Jr., Leventhal BL, Freedman DX. Free serotonin in plasma: autistic children and their first-degree relatives. Biological psychiatry. 1988;24(4):488–91. [DOI] [PubMed] [Google Scholar]
- 45.Gabriele S, Sacco R, Persico AM. Blood serotonin levels in autism spectrum disorder: a systematic review and meta-analysis. European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology. 2014;24(6):919–29. [DOI] [PubMed] [Google Scholar]
- 46.Chugani DC, Muzik O, Behen M, Rothermel R, Janisse JJ, Lee J, et al. Developmental changes in brain serotonin synthesis capacity in autistic and nonautistic children. Ann Neurol. 1999;45(3):287–95. [DOI] [PubMed] [Google Scholar]
- 47.Yang CJ, Tan HP, Du YJ. The developmental disruptions of serotonin signaling may involved in autism during early brain development. Neuroscience. 2014;267:1–10. [DOI] [PubMed] [Google Scholar]
- 48.Klein AB, Williamson R, Santini MA, Clemmensen C, Ettrup A, Rios M, et al. Blood BDNF concentrations reflect brain-tissue BDNF levels across species. The international journal of neuropsychopharmacology. 2011;14(3):347–53. [DOI] [PubMed] [Google Scholar]
- 49.Schür RR, Draisma LW, Wijnen JP, Boks MP, Koevoets MG, Joëls M, et al. Brain GABA levels across psychiatric disorders: A systematic literature review and meta-analysis of (1) H-MRS studies. Human brain mapping. 2016;37(9):3337–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schurz M, Radua J, Aichhorn M, Richlan F, Perner J. Fractionating theory of mind: a meta-analysis of functional brain imaging studies. Neuroscience and biobehavioral reviews. 2014;42:9–34. [DOI] [PubMed] [Google Scholar]
- 51.Moessnang C, Baumeister S, Tillmann J, Goyard D, Charman T, Ambrosino S, et al. Social brain activation during mentalizing in a large autism cohort: the Longitudinal European Autism Project. Molecular autism. 2020;11(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung M, Kosaka H, Saito DN, Ishitobi M, Morita T, Inohara K, et al. Default mode network in young male adults with autism spectrum disorder: relationship with autism spectrum traits. Molecular autism. 2014;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jann K, Hernandez LM, Beck-Pancer D, McCarron R, Smith RX, Dapretto M, et al. Altered resting perfusion and functional connectivity of default mode network in youth with autism spectrum disorder. Brain Behav. 2015;5(9):e00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neufeld J, Kuja-Halkola R, Mevel K, Cauvet É, Fransson P, Bölte S. Alterations in resting state connectivity along the autism trait continuum: a twin study. Molecular psychiatry. 2018;23(7):1659–65. [DOI] [PubMed] [Google Scholar]
- 55.Yerys BE, Gordon EM, Abrams DN, Satterthwaite TD, Weinblatt R, Jankowski KF, et al. Default mode network segregation and social deficits in autism spectrum disorder: Evidence from non-medicated children. NeuroImage Clinical. 2015;9:223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Q, Becker B, Jiang X, Zhao Z, Zhang Q, Yao S, et al. Decreased interhemispheric functional connectivity rather than corpus callosum volume as a potential biomarker for autism spectrum disorder. Cortex; a journal devoted to the study of the nervous system and behavior. 2019;119:258–66. [DOI] [PubMed] [Google Scholar]
- 57.Ellegood J, Crawley JN. Behavioral and Neuroanatomical Phenotypes in Mouse Models of Autism. Neurotherapeutics. 2015;12(3):521–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maguire J, Mody I. Steroid hormone fluctuations and GABA(A)R plasticity. Psychoneuroendocrinology. 2009;34 Suppl 1(Suppl 1):S84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiggins LD, Barger B, Moody E, Soke G, Pandey J, Levy S. Brief report: the ADOS calibrated severity score best measures autism diagnostic symptom severity in pre-school children. Journal of autism and developmental disorders. 2019;49(7):2999–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32(3):227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt AF, Finan C, Gordillo-Marañón M, Asselbergs FW, Freitag DF, Patel RS, et al. Genetic drug target validation using Mendelian randomisation. Nature communications. 2020;11(1):3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.