Abstract
We discuss the problems in defining the extent of the proteomes for completely sequenced eukaryotic organisms (i.e. the total number of protein-coding sequences), focusing on yeast, worm, fly and human. (i) Six years after completion of its genome sequence, the true size of the yeast proteome is still not defined. New small genes are still being discovered, and a large number of existing annotations are being called into question, with these questionable ORFs (qORFs) comprising up to one-fifth of the ‘current’ proteome. We discuss these in the context of an ideal genome-annotation strategy that considers the proteome as a rigorously defined subset of all possible coding sequences (‘the orfome’). (ii) Despite the greater apparent complexity of the fly (more cells, more complex physiology, longer lifespan), the nematode worm appears to have more genes. To explain this, we compare the annotated proteomes of worm and fly, relating to both genome-annotation and genome evolution issues. (iii) The unexpectedly small size of the gene complement estimated for the complete human genome provoked much public debate about the nature of biological complexity. However, in the first instance, for the human genome, the relationship between gene number and proteome size is far from simple. We survey the current estimates for the numbers of human genes and, from this, we estimate a range for the size of the human proteome. The determination of this is substantially hampered by the unknown extent of the cohort of pseudogenes (‘dead’ genes), in combination with the prevalence of alternative splicing. (Further information relating to yeast is available at http://genecensus.org/yeast/orfome)
INTRODUCTION
The total amount of DNA in a genome has little correlation with the apparent complexity of the organism that it encodes, with some amoebae carrying more than 200 times the DNA in the human genome (1; Database of Genome Sizes http://www.cbs.dtu.dk/databases/DOGS). This has been dubbed the ‘C-value’ paradox (C-value is the total haploid DNA content of an organism). The sequencing of the genomes of six eukaryotes has provided us with a related quandary: namely, how is the number of genes related to the biological complexity of an organism (termed an ‘N-value’ paradox by Claverie) (2)? How can our own supremely sophisticated species be governed by just 50–100% more genes than the nematode worm? Here, we review work on a directly related property, the size of the proteome for the sequenced eukaryotes, where the ‘proteome’ can be defined as the total number of protein-coding sequences (or ‘CDS’) used by an organism. We discuss issues arising in defining the extent of the proteomes required by yeast and the metazoan eukaryotes, and how proteome size relates to gene number, touching upon some evolutionary issues relating to proteome size.
Refining the yeast proteome
Since the yeast genome was sequenced (3), the true size of its proteome has been a point of considerable confusion. Initially, 6275 open reading frames (ORFs) of length greater than or equal to 100 codons were identified in the genome (3). Only 3.5% of these identified ORFs were spliced and there is very little alternative splicing in Saccharomyces cerevisiae to complicate definition of the proteome (4). About one-third of the initially annotated proteome had no assignable function or known protein homolog and were thus designated ‘orphans’ (5). A sizeable minority of these (390) were heuristically labeled as ‘questionable’, i.e. unlikely to encode proteins due to having bad codon usage [with a codon adaptation index (CAI; a measure of codon usage) <0.11 (6) and being short (less than 150 codons) (7)]. Smith and co-workers (8) noted that the sequence length distribution for the initial ORFs set that have no clear known protein homolog, peaks anomalously at 100–110 codons, which is close to the arbitrary minimum length cut-off point of 100 codons used in the original ORF definition. The notion of ‘questionable ORF’ (qORF) was further refined in the MIPS yeast genome database as an ORF having two or three of the following attributes: (i) a CAI value <0.11, (ii) overlap with a longer ORF and (iii) no similarity to other ORFs (http://mips.gsf.de). (Upon writing this, the current total for such MIPS qORFs is 471.)
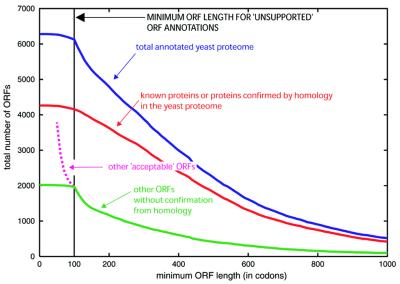
The total number of possible ORFs in the yeast genome could be described as an ‘ORFome’, which contains the true proteome as a subset. As noted above, an arbitrary minimum of 100 codons length has been used previously in the determination of yeast ORFs that are otherwise unsupported by homology or evidence of expression. For ORF lengths decreasing below 100 codons, the number of ‘acceptable’ ORFs (which have good CAI >0.11 and do not overlap a longer ORF) becomes substantially larger (Fig. 1). During annotation, any ORFs of a size less than 100 codons have generally only been kept if there is additional evidence, e.g. from previous functional characterization, protein homology or serial analysis of gene expression (SAGE) (9). (SAGE is a method that uses short sequence tags of 9–11 bp that are sufficiently informative to identify a transcript uniquely.) For ORFs greater than or equal to 100 codons, the problem is largely one of deciding on the exclusion of qORFs.
Figure 1.
Number of yeast ORFs as a function of the minimum allowed ORF length. The total number of annotated ORFs in the yeast proteome is plotted against minimum ORF length (continuous blue line). A curve for known proteins or proteins confirmed by homology to a known protein is shown (red line), along with a green curve for the remaining ORFs that have no homology to a known protein (or are not otherwise characterized). Also displayed (dotted pink line) is the total number of additional ‘acceptable’ ORFs from the yeast genome that have good codon adaptation (CAI ≥0.11) that do not overlap an annotated gene or other genomic feature. The plots are cumulative backwardly at intervals of 10 residues.
A number of studies have attempted to separate real ORFs from qORFs computationally. It is a natural property of the genetic code that alternative ORFs are generated inside of, or overlapping, a coding sequence, either in the sense or antisense strands (10,11). It is unclear to what extent these ‘generated’ ORFs can encode real proteins. Cebrat and co-workers (11–13) analyzed yeast ‘orphans’ and concluded that many of them have properties of alternative ORFs generated by the genetic code. Using a measure of codon and nucleotide composition bias (particularly at the first and second positions of codons), they calculated that the yeast proteome is much smaller than was originally proposed, comprising only 4800 ORFs. A more recent gene prediction algorithm based on nucleotide composition and tailored for S.cerevisiae yielded an estimate of less than 5645 true ORFs (14). The ‘Genolevures’ initiative to partially sequence the genomes of 13 S.cerevisiae relatives has indicated that the latter number might be nearer the true value (15). Homologs for S.cerevisiae proteins from other hemiascomycetes were detected for many orphan sequences, appearing to bring the total number of real ORFs to at least 5651. However, some of these may still be qORFs, as they may be conserved ‘generated’ ORFs like those described by Cebrat et al. (11).
If the homologs detected in the Genolevures project are real, then what of the remaining approximately 600 ORFs greater than 100 codons? These may still encode genuine proteins. First, the proteins could be rapidly evolving, making it more difficult to find homology with other organisms, and so be naturally biased against with current techniques for homolog searching and assignment. Such rapid divergence has been observed for the fly Drosophila melanogaster, for which about one-third of randomly picked cDNAs were found to be sufficiently divergent that they do not cross-hydridize with Drosophila virilis DNA, a species from which D.melanogaster diverged 40–60 million years ago (16). Secondly, they may have a marginal effect on yeast strain fitness and so be difficult to study by conventional experiments to ascertain function (17). For example, in a study of 34 S.cerevisiae genes that were judged non-essential by gene disruption (18), 70% of these genes were found to affect strain fitness marginally (19). This implies that the effective size of the yeast proteome can only be determined in a ‘selectomic’ way, i.e. from study of its behavior from generation to generation for the reproducing organism.
A number of genome-scale transcription experiments that verify yeast ORFs have been performed, using SAGE or DNA microarrays (9,20–25). When data from genome-wide cDNA microarray analysis (23), SAGE (9) and transposon tagging (26) are combined, we note that there are more than 400 annotated ORFs, that do not appear at all in these experiments (P.Harrison et al., unpublished data). On the other hand, a small number of essential genes have consistently low expression; for example, YGR113W (or DAM1), a protein that localizes to intranuclear spindles and spindle pole bodies, is expressed at consistently low levels, but is essential according to the Winzeler et al. (18) ORF disruption data. Also, it is possible that qORFs that are near a genuine expressed ORF may be spuriously determined as expressed, purely because of this proximity.
It is unclear to what extent the number of short proteins (less than 100 codons) in the yeast proteome has been underestimated. When one plots the total number of annotated yeast ORFs versus minimum ORF length, there is an obvious discontinuity at the 100-codon mark (Fig. 1). As one expands the possible ‘ORFome’ to include shorter minimum ORF size, one still finds a number of acceptable ORFs that do not overlap a previously annotated gene or other feature and have a good CAI value (CAI greater than or equal to 0.11; Fig. 1). For example, for ORF length of greater than or equal to 80 and less than 100 codons there are 198 such ORFs that have good codon usage (P.Harrison et al., unpublished data). In an early study, more than 140 potential protein-coding ORFs of between 36 and 100 codons were found, using a discriminant function based on in-phase hexamer frequencies in known and simulated ORFs (27), and later also using protein homology (28). The MIPS and SGD databases, in combination, list up to 217 short ORFs with protein homology or SAGE tag support (9). A further 48 short ORFs were determined as a result of partial genome sequencing of hemiascomycetes (15). Indeed, an experiment to identify genes in the yeast genome using a combination of transposon tagging, microarray-based expression analysis and exhaustive homology searching indicated up to 137 novel ORFs with 104 of them less than 100 codons in length and about one-third overlapping previously annotated genes (26,29,30). Further material relating to this is available at http://genecensus.org/yeast/orfome.
An additional complication relating to the size of the yeast proteome is the number of ORFs that have simple disablements (termed ‘dORFs’) and which could potentially form complete ORFs in other yeast strains. We recently surveyed the yeast genome for ‘dORFs’ and found over 100 that do not entail an existing ORF annotation (31). Further details about dORFs are described in http://genecensus.org/pseudogene/yeast.
Thus, the yeast proteome may yet vary in size over a range of more than 1700 ORFs—refinement and reannotation of the proteome will take longer for the remaining problematic ORFs, some of which appear to be refractive to conventional techniques.
Worm versus fly—why more worm proteins?
For the worm and fly, splicing is much more extensive than in yeast, and there is a minor degree of alternative splicing (currently ∼2% of the documented worm proteome arises from alternative splicing, ∼7% for fly) (32–35). Both have similar overall genome size (100 Mb for worm, 120 Mb for the euchromatic portion of the fly genome), and similar distributions of exon size, with small average numbers of exons per gene (about six exons in worm and about four exons in fly). In contrast, however, the total apparent proteome sizes of these organisms differ markedly: the original estimates were 19 099 worm and 13 601 fly coding sequences, although the proteomes comprise comparable numbers of protein families (32–35). (At the time of writing, the annotated proteome sizes are 20 009 for the worm and 14 332 for the fly.)
Notably, however, the worm has considerably more organism-specific genes (∼50%) than the fly (∼30%) (35). To investigate homology trends further, we scanned the raw genomic sequences of the worm and the euchromatic portions of the fly genome (and also yeast and human chromosomes 21 and 22 for comparison) for homologies to ‘known’ proteins in the SWISS-PROT database (36) (Table 1). An intriguing contrast arises between the profiles of homology found for the worm and fly genomes. Although the worm has substantially more annotated proteins (approximately 6000) than the fly, the amount of protein homology in the fly is actually greater, regardless of the subset of SWISS-PROT concerned. The tendency for a stable ratio of homology across different levels for worm and fly could be termed a ‘H-value’ paradox (similar to the ‘C-value’ paradox for overall genome size) (1). This relationship may result for evolutionary reasons and/or differences in genome annotation. For example, it may imply that the worm genome has undergone a contraction in its number of protein-coding genes (which included the deletion of many bacterial and metazoan homologs), followed by a late, organism-specific expansion. Alternatively, this observation may imply a small number of worm gene over-predictions.
Table 1. Calculated homology coverage or ‘H-value’ data and other characteristics for the genomes of worm, fly and yeast and the combined human chromosomes 21 and 22.
| Genome or chromosomes | Number of annotated genes | Genome size (C) (Mb) | Genomic DNA coverage by annotated genes (G) (Mb) | Total homology (H) to just bacterial proteins (Mb) | H for non-phylum homology (Mb) | H for non-organism homology (Mb) |
|---|---|---|---|---|---|---|
| Yeast | 6280 | 12 | 8.92 | 1.26 | 1.80 | 2.85 |
| Worm | 18 576 | 99 | 24.35 | 0.98 | 2.11 | 4.37 |
| Fly | 13 601 | 112 | 14.83 | 1.40 | 2.80 | 6.15 |
| Human chromosomes 21 and 22 | 924 | 69 | 1.34 | 0.06 | 0.18 | 0.57 |
NB: Mb denotes megabases.
We call the total amount of protein homology detected for each genome (in bases) the ‘H-value’. This simple, direct examination of protein homology content bypasses some of the vagaries of gene prediction algorithms. For human, data for chromosomes 21 and 22 are combined. The values for human gene annotations for human are taken from predicted genes by the program GenomeScan (60). The term ‘bacterial’ denotes homology to bacterial species, ‘non-phylum’ denotes homology to all proteins from phyla other than those represented by the organisms examined here, and ‘all’ indicates homology to proteins not from the specific organism in question. We used BLASTX (80) (with an expectation value <0.0001 and six-frame translation) to compare genomic sequence against the SWISS-PROT database (36). All other annotated features, including repeats and transposable elements, were masked for and deleted from the total protein homology coverage. The homology trends shown do not differ when we account for any possible pseudogenic homology match (data not shown), and are unlikely to be explained by an elaborate configuration of database biases. Gene exon size is also not a factor in comparing worm and fly, as their exon sizes have similar distributions (56). The value for C for the fly only comprises the euchromatic portion.
With regard to differences in genome annotation, the numbers of genes for both organisms may yet converge somewhat. During the original fly genome annotation, a total of 17 464 genes were predicted by the program GENSCAN (37), but these were believed to be about 4000 too many, and to be largely artefactual because of the lack of parameterization in GENSCAN for fly (34). However, a study on the fly genome that used GENSCAN has yielded 1042 additional candidate genes, potentially increasing the Drosophila proteome size to greater than 15 400 (38). A large initial list of 19 410 potential genes in the whole genome was predicted with GENSCAN, regardless of matches to proteins, cDNAs or ESTs, and subsequently compared in translation with ESTs, cDNAs and other proteins, with additional support from model-building of distant sequence homologs (38).
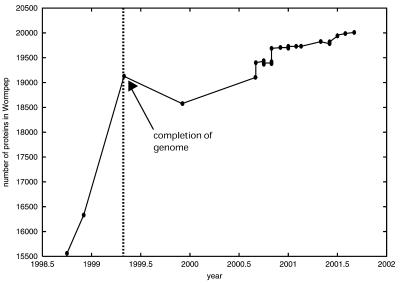
Since its publication (33), the size of the worm proteome has varied over a range of 1433 proteins (Fig. 2). This is due partly to updates and corrections in sequencing and partly to refinement of gene predictions using verifying protein and EST/cDNA homology. Projects to collate libraries of cDNAs and ESTs for the fly and worm appear to be at similar stages of ‘completeness’: for the fly, ∼42% (at the time of writing) of predicted genes have a verifying EST/cDNA (39), compared with >50% for the worm (40). Interestingly, an experiment to study genome-wide expression of 98% of predicted worm ORFs only detected expression that is significant on a ‘worm-wide’ scale for a proportion of predicted worm transcripts (∼56% detected) similar to that detected by EST/cDNA matching (40). This may imply the approach of an expression ‘detection plateau’ in the worm and a limit to the utility of methods that rely on relatively higher expression for gene detection for this organism. Similar microarray experiments have been performed for the fly; White et al. (41) studied more than 4500 unique EST clones to ascertain expression variation over the course of Drosophila metamorphosis. Andrews et al. (42) studied EST frequency and microarray expression in Drosophila testis and noted that coverage of the fly gene complement with ESTs/cDNAs is still far from complete, as only 44% of their derived cDNAs corresponded to known or predicted genes—indeed, 22% of the most highly over-expressed genes aligned with genomic sequence, but not with the original set of fly gene annotations.
Figure 2.
The variation in the size of the WormPep database over time. The size of the WormPep database is plotted against time for the period after and just prior to publication of the genome sequence. The dotted line indicates the approximate time of genome sequence completion.
For the worm, work using ORF sequence tags (OSTs) that are directly generated from predicted ORFs has by-passed reliance on higher relative expression to detect genes (43). OSTs that were made from a sample of one-eighth of nearly 10 000 genes (that had been unconfirmed by EST/cDNA) were used to obtain an estimate of about 17 300 genes in the worm genome.
So, why might the worm need more proteins than the fly, yet have comparable numbers of protein families in its proteome? Aside from questions of genome annotation, the worm may have more proteins than the fly from evolutionary considerations. The larger worm proteome may simply arise because factors such as genomic DNA deletion rate and chromosomal rearrangement have allowed it. The fly genomic DNA deletion rate (which is known to be very high from the apparent rarity of true fly pseudogenes) (44–46) may hamper the maintenance of recent gene duplications, so that they have less time to become of use. Experiments with transposable elements in D.melanogaster and the cricket species Laupala indicate a very rapid loss of genomic DNA in Drosophila (1,47,48). Drosophila also has an extremely high rate of chromosomal rearrangement (49). However, studies on families of worm chemoreceptor genes and pseudogenes also suggest that the worm has a rather high genomic DNA deletion rate (50–52). Nonetheless, the number of pseudogenes for the worm seems to be about a scale of magnitude larger than that for the fly. A preliminary survey suggests there are about 100 pseudogenes in the fly genome (P.Harrison et al., unpublished data). In comparison, the worm genome appears to have at least approximately 1100 pseudogenes, with the largest numbers associated with families of seven-transmembrane receptors (53). Indeed, the population of olfactory receptors/chemoreceptors, and other seven-transmembrane receptors in the worm (about 1100) is almost a scale of magnitude larger than in the fly (about 160 such receptors: InterPro Database http://www.ebi.ac.uk/interpro) and is >80% organism-specific (54,55).
The out-of-focus human proteome
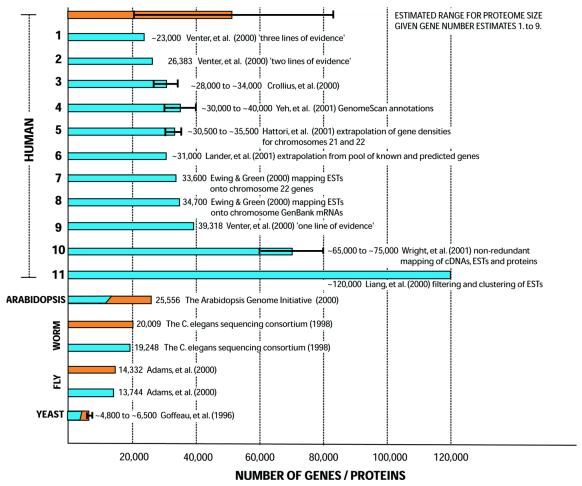
The near-complete sequencing of the human genome has yielded gene total estimates that, at first glance, seem surprisingly low; of the order of 23 000–40 000 genes (56,57). This finding has triggered much debate in the public press (58). Gene numbers arising from the two human genome sequencing projects are compared in Figure 3 to gene number estimates published just prior to the genome publications, as well as gene numbers and proteome sizes for the other eukaryotes. Venter et al. (57) identified approximately 6500 human genes previously discovered, and then annotated genes using a novel gene prediction procedure called ‘Otto’ and three other prediction algorithms that used conservation between human and mouse genomic DNA, and support from human and rodent ESTs and from protein homology. Depending on the number of ‘lines of evidence’ (e.g. a protein homology and a rodent EST provides two lines of evidence), the total predicted gene number varies between approximately 23 000 (three lines of evidence) and approximately 40 000 (one line). The International Human Genome Sequencing Consortium (IHGS) combined predicted genes from two procedures [one based on the Ensembl system that uses the ab initio gene predictor GENSCAN (37), and the other from the program Genie (59)] with the approximately 10 000 known genes in the RefSeq set of mRNAs from the NCBI, to compile a list of 31 778 predicted transcripts, arising from an estimated greater than 24 500 true genes (56). Both procedures used supporting evidence from ESTs, mRNAs and protein homology. They estimated that the predicted genes comprise ∼60% of unknown human genes, thereby arriving at a total of approximately 31 000. The program GenomeScan, a development of GENSCAN that incorporates scoring for protein homology, predicted a total of 20 000–25 000 predicted genes out of an estimated total of 30 000–40 000, including a further approximately 6500 distinct whole or partial genes relative to the IHGS gene set (60). Estimations of the number of human genes just prior to the publication of the genome, with one notable exception (which estimated about 120 000 genes) (61), yielded largely similar numbers, in the range of approximately 28 000–35 000 (Fig. 3) (62–65). Recently, Wright et al. (66) non-redundantly mapped all available cDNA, EST and protein sequence data from public databases and arrived at a considerably higher estimate of 65 000–75 000 genes or ‘transcriptional units’. An algorithm to predict the first exons of human genes (‘FirstEF’) identified about 69 000 such exons, also suggesting a much higher number of human genes (67). Hogenesch et al. (68) compared the predicted gene sets from Celera and from the IHGS and found that, collectively, 80% of novel genes were predicted by only one of the groups. Also, they performed RNA expression analysis to characterize a pool of novel genes from both sets of annotations, and found that a similar proportion of these novels genes (>80%) was found to be expressed as for a set of known human genes. This rather puzzlingly suggests that the substantial majority of the novel transcripts arising from either of the Celera or IHGS annotations are real genes. We expect that, in the future, a variety of approaches, such as the probing of arrays containing segments covering entire human chromosomes, will be a valuable tool in discovering novel gene exons.
Figure 3.
Human gene numbers and proteome size. The figure depicts, in bar chart form, the number of human genes (blue bar) from various estimates and a corresponding estimate for proteome size (orange bar). Gene numbers and proteome sizes are shown for the other sequenced eukaryotes, with the same coloring. The size of the human proteome (NCDS) can be estimated as follows: NCDS = f1.f2.Ngenes, where f1 is the proportion of gene structures that are not pseudogenic, and f2 is the ratio of the total number of distinct protein-coding transcripts to the total number of genes (arising from alternative splicing). Assuming 0.76 ≤ f1 ≤ 0.91 (see text), a minimum value of f2 = 1.16 can be derived from the alternative splicing survey of Mironov et al. (72), and a maximum value f2 = 2.22 is calculable from the alternative splicing analysis in Lander et al. (56). Using these, and the wider range for Ngenes given by Venter et al. (57) a range of approximately 20 300 to approximately 83 800 is yielded for NCDS. This range is clearly rather large, and is reminiscent of the range of values arising for estimates of Ngenes that arose in the months and years prior to publications of the human genome.
How do these estimated gene numbers relate to the size of the human proteome? Two main issues complicate the extrapolation of human proteome size from the corresponding gene numbers.
First, the prevalence of pseudogenes in the human genome is still unclear (69). Pseudogenes are either ‘processed’, i.e. resulting from reverse transcription from messenger RNA and re-integration into the genomic DNA, or ‘duplicated’, i.e. arising from duplication in the genomic DNA and subsequent disablement, most commonly through frameshift or premature stop codon formation. Processed pseudogenes will be less likely to interfere with the accuracy of gene predictions; they will, on average, tend to be longer than the average human exon size, and comprise characteristic signals, including a C-terminal poly(A) tail (70,71). Duplicated pseudogenes are more problematic for gene annotation. An exon with a disablement that is in the region of a gene may have been recently discarded evolutionarily (perhaps as part of an alternative splicing) and so may not be a part of the extant gene; also, gene prediction algorithms may shorten an exon to avoid inclusion of a disabled extension to it. In the completed chromosome 22 sequence, the annotators initially predicted at least 545 genes and 134 pseudogenes (one for every approximately 4.1 genes) (62). They surmised that 82% of these pseudogenes were processed, since they contained single spans of homology and lacked the characteristic exon structure of the closest matching gene. This implies only a small proportion of duplicated pseudogenes relative to the gene total (about one for every 25 genes). For the initial publication of chromosome 21, there was a total of 225 known and predicted genes and a corresponding total of 59 pseudogenes (one for every approximately 3.8 genes), but no assessment of the number of processed and duplicated pseudogenes was presented (63). The IHGS project estimated that ∼9% of their predicted genes may be pseudogenes, from comparison with chromosome 22 sequence data (56). Yeh et al. (60) used their program GenomeScan, to estimate that between 11 and 22% of predicted genes in a set of 20 000–25 000 were either false positives or pseudogenes. A survey on pseudogenes in chromosomes 21 and 22 yielded an estimate of one duplicated pseudogene for approximately four genes, with up to 6% of predicted gene exons being potentially pseudogenic, and up to 14% of predicted genes (69). So, in summary, an estimated range for the proportion of duplicated pseudogenes is 4–22% of predicted genes.
Secondly, alternative splicing is much more prevalent in the human than in the worm or fly. The IHGS project noted from analysis of chromosomes 19 and 22 that there may be up to about 3.2 mRNA transcripts per gene, with ∼70% involving alternatives within the coding region, and thus producing distinct proteins (56). Mironov et al. (72) performed an analysis of human alternative splicing based on alignment of EST data to genomic DNA. They observed that ∼40% of genes undergo alternative splicing. A lower bound of about 1.8 mRNA transcripts per gene can be deduced from their data (M.Gelfand, personal communication). Contrary to the survey noted above, they found that only ∼20% of alternative splicing occurred in coding regions of transcripts. Two other EST-based approaches found a similar proportion of alternatively spliced genes [∼38% (73) and >42% (74)]. Using the data from Brett et al. (73), we can deduce an overall ratio of about 2.1 mRNA transcripts per gene. Evidently, also, pseudogene assignment is complicated by alternative splicing, as it may be unclear whether a disabled exon is actually required in the gene structure or not.
The estimated proportions of duplicated pseudogenes and alternative splicing can be used to speculate about the total human proteome size (Fig. 3). The true value is more likely to be closer to approximately 84 000, as the alternative splicing surveys described above err on the conservative side (72,73). Indeed, all of the current data may under-estimate the rate of alternative splicing because it is based on transcripts observed to date, which are likely to be only a fraction of the total expressed at all times in all tissues, so the human proteome size is likely to be significantly larger than approximately 90 000.
Concluding remarks
We have examined how proteome definition for different eukaryotic organisms with (near-) complete genome sequences is progressing. But is the size of the proteome of an organism any more an indicator of biological complexity than the number of genes, or the total amount of genomic DNA? For the higher eukaryotes, alternative splicing in non-coding segments of mRNA transcripts, alternative polyadenylation and differential binding to promoter elements, and networks of interaction in genetic control all engender biological complexity in ways that are independent of the number of genes or protein-coding sequences in an organism. Claverie (2) noted that biological complexity is perhaps better understood in terms of distinct ‘transcriptome’ states, i.e. there is a combinatorial explosion in the number of possibilities as the number of genes under controlled expression gets larger. Nonetheless, knowing the manner and extent of proteome size variation between the vertebrate genomes—puffer fish (Fugu rubripes) (75), mouse (76), rat (77) and, perhaps, chimpanzee (78,79)—will yield insight into how the apparently greater biological complexity of the human species arises.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Chris Burge (MIT) for comments on the manuscript. M.G. and M.S. acknowledge support from NIH grants HG02357-01 and CA77808 Tx.
REFERENCES
- 1.Petrov D.A. (2001) Evolution of genome size: new approaches to an old problem. Trends Genet., 17, 23–28. [DOI] [PubMed] [Google Scholar]
- 2.Claverie J.M. (2001) What if there are only 30,000 human genes? Science, 291, 1255–1257. [DOI] [PubMed] [Google Scholar]
- 3.Goffeau A., Barrell,B.G, Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D, Jacq,C., Johnston,M. et al. (1996) Life with 6000 genes. Science, 274, 546, 563-567. [DOI] [PubMed] [Google Scholar]
- 4.Davis C.A., Grate,L., Spingola,M. and Ares,M.,Jr (2000) Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res., 28, 1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dujon B. (1996) The yeast genome project: what did we learn? Trends Genet., 12, 263–270. [DOI] [PubMed] [Google Scholar]
- 6.Sharp P.M. and Li,W.H. (1987) The codon adaptation index: a measure of directional synonymous codon usage bias and its potential applications. Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dujon B., Alexandraki,D., Andre,B., Ansorge,W., Baladron,V., Ballesta,J.P., Banrevi,A., Bolle,P.A., Bolotin-Fukuhara,M., Bossier,P. et al. (1994) Complete DNA sequence of yeast chromosome XI. Nature, 369, 371–378. [DOI] [PubMed] [Google Scholar]
- 8.Das S., Yu,L., Gaitatzes,C., Rogers,R., Freeman,J., Bienkowska,J., Adams,R.M., Smith,T.F. and Lindelien,J. (1997) Biology’s new Rosetta stone. Nature, 385, 29–30. [DOI] [PubMed] [Google Scholar]
- 9.Velculescu V.E., Zhang,L., Zhou,W., Vogelstein,J., Basrai,M.A., Bassett,D.E.,Jr, Hieter,P., Vogelstein,B. and Kinzler,K.W. (1997) Characterization of the yeast transcriptome. Cell, 88, 243–251. [DOI] [PubMed] [Google Scholar]
- 10.Merino E., Balbas,P., Puente,J.L. and Bolivar,F. (1994) Antisense overlapping open reading frames in genes from bacteria to humans. Nucleic Acids Res., 22, 1903–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cebrat S., Mackiewicz,P. and Dudek,M.R. (1998) The role of the genetic code in generating new codings sequences inside existing genes. Biosystems, 45, 165–176. [DOI] [PubMed] [Google Scholar]
- 12.Kowalczuk M., Mackiewicz,P., Gierlik,A., Dudek,M.R. and Cebrat,S. (1999) Total number of coding open reading frames in the yeast genome. Yeast, 15, 1031–1034. [DOI] [PubMed] [Google Scholar]
- 13.Mackiewicz P., Kowalczuk,M., Gierlik,A., Dudek,M.R. and Cebrat,S. (1999) Origin and properties of non-coding ORFs in the yeast genome. Nucleic Acids Res., 27, 3503–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C.-T. and Wang,J. (2000) Recognition of protein coding genes in the yeast genome at better than 95% accuracy based on the Z curve. Nucleic Acids Res., 28, 2804–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blandin G., Durrens,P., Tekaia,F., Aigle,M., Bolotin-Fukuhara,M., Bon,E., Casaregola,S., de Montigny,J., Gaillardin,C., Lepingle,A. et al. (2000) Genomic exploration of the hemiascomycetous yeasts: 4. The genome of Saccharomyces cerevisiae revisited. FEBS Lett., 487, 31–36. [DOI] [PubMed] [Google Scholar]
- 16.Schmid K.J. and Tautz,D. (1997) A screen for fast evolving genes from Drosophila. Proc. Natl Acad. Sci. USA, 94, 9746–9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tautz D. (2000) A genetic uncertainty problem. Trends Genet., 16, 475–477. [DOI] [PubMed] [Google Scholar]
- 18.Winzeler E.A., Shoemaker,D,D., Astromoff,A., Liang,H., Anderson,K., Andre,B., Bangham,R., Benito,R., Boeke,J.D., Bussey,H. et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 19.Thatcher J.W., Shaw,J.M. and Dickinson,W.J. (1998) Marginal fitness contributions of nonessential genes in yeast. Proc. Natl Acad. Sci. USA, 95, 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 21.Chu S., DeRisi,J., Eisen,M., Mulholland,J., Botstein,D., Brown,P.O. and Herskowitz,I. (1998) The transcriptional program of sporulation in budding yeast. Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- 22.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 24.Roth F.P., Hughes,J.D., Estep,P.W. and Church,G.M. (1998) Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nature Biotechnol., 16, 939–945. [DOI] [PubMed] [Google Scholar]
- 25.Jelinsky S.A. and Samson,L.D. (1999) Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl Acad. Sci. USA, 96, 1486–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross-Macdonald P., Coelho,P.S., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K.H., Sheehan,A., Symoniatis,D., Umansky,L. et al. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 27.Barry C., Fichant,G., Kalogeropoulos,A. and Quentin,Y. (1996) A computer filtering method to drive out tiny genes from the yeast genome. Yeast, 12, 1163–1178. [DOI] [PubMed] [Google Scholar]
- 28.Andrade M.A., Daruvar,A., Casari,G., Schneider,R., Termier,M. and Sander,C. (1997) Characterization of new proteins found by analysis of short open reading frames from the full yeast genome. Yeast, 13, 1363–1374. [DOI] [PubMed] [Google Scholar]
- 29.Vidan S. and Snyder,M. (2001) Large-scale mutagenesis: yeast genetics in the genome era. Curr. Opin. Biotech., 12, 28–34. [DOI] [PubMed] [Google Scholar]
- 30.Kumar A., Harrison,P.M., Cheung,K.-H., Lan,N., Echols,N., Bertone,P., Miller,P., Gerstein,M.B. and Snyder,M. (2002). An integrated approach for finding overlooked genes in yeast. Nat. Biotechnol., 20, 58–63. [DOI] [PubMed] [Google Scholar]
- 31.Harrison P.M., Kumar,A., Lan,N., Echols,N., Snyder,M. and Gerstein,M.B. (2002). A small reservoir of disabled ORFs in the sequenced yeast genome and its implications for the dynamics of proteome evolution. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 32.Chervitz S.A., Aravind,L., Sherlock,G., Ball,C.A., Koonin,E.V., Dwight,S.S., Harris,M.A., Dolinski,K., Mohr,S., Smith,T. et al. (1998) Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science, 282, 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 34.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 35.Rubin G.M., Yandell,M.D., Wortman,J.R., Gabor Miklos,G.L., Nelson,C.R., Hariharan,I.K., Fortini,M.E., Li,P.W., Apweiler,R., Fleischmann,W. et al. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 38.Gopal S., Schroeder,M., Pieper,U., Sczyrba,A., Aytekin-Kurban,G., Bekiranov,S., Fajardo,J.E., Eswar,N., Sanchez,R., Sali,A. and Gaasterland,T. (2001) Homology-based annotation yields 1,042 new candidate genes in the Drosophila melanogaster genome. Nature Genet., 27, 337–340. [DOI] [PubMed] [Google Scholar]
- 39.Rubin G.M., Hong,L., Brokstein,P., Evans-Holm,M., Frise,E., Stapleton,M. and Harvey,D.A. (2000) A Drosophila complementary DNA resource. Science, 287, 2222–2224. [DOI] [PubMed] [Google Scholar]
- 40.Hill A.A., Hunter,C.P., Tsung,B.T., Tucker-Kellogg,G. and Brown,E.L. (2000) Genomic analysis of gene expression in C. elegans. Science, 290, 809–812. [DOI] [PubMed] [Google Scholar]
- 41.White K.P., Rifkin,S.A., Hurban,P. and Hogness,D.S. (1999) Microarray analysis of Drosophila development during metamorphosis. Science, 286, 2179–2184. [DOI] [PubMed] [Google Scholar]
- 42.Andrews J., Bouffard,G.G., Cheadle,C., Lu,J., Becker,K.G. and Oliver,B. (2000) Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res., 10, 2030–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reboul J., Vaglio,P., Tzellas,N., Thierry-Mieg,N., Moore,T., Jackson,C., Shin-i,T., Kohara,Y., Thierry-Mieg,D., Thierry-Mieg,J. et al. (2001) Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nature Genet., 27, 332–336. [DOI] [PubMed] [Google Scholar]
- 44.Robin G.C., Russell,R.J., Cutler,D.J. and Oakeshott,J.G. (2000) The evolution of an alpha-esterase pseudogene inactivated in the Drosophila melanogaster lineage. Mol. Biol. Evol., 17, 563–575. [DOI] [PubMed] [Google Scholar]
- 45.Currie P.D. and Sullivan,D.T. (1994) Structure, expression and duplication of genes which encode phosphoglyceromutase of Drosophila melanogaster. Genetics, 138, 353–363. [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan D.T., Starmer,W.T., Curtiss,S.W., Menotti-Raymond,M. and Yum,J. (1994) Unusual molecular evolution of an Adh pseudogene in Drosophila. Mol. Biol. Evol., 11, 443–458. [DOI] [PubMed] [Google Scholar]
- 47.Petrov D.A., Lozovskaya,E.R. and Hartl,D.L. (1996) High intrinsic rate of DNA loss in Drosophila. Nature, 384, 346–349. [DOI] [PubMed] [Google Scholar]
- 48.Petrov D.A. and Hartl,D.L. (2000) Pseudogene evolution and natural selection for a compact genome. J. Heredit., 91, 221–227. [DOI] [PubMed] [Google Scholar]
- 49.Ranz J.M., Casals,F. and Ruiz,A. (2001) How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila. Genome Res., 11, 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robertson H.M. (1998) Two large families of chemoreceptor genes in the nematodes Caenorhabditis elegans and Caenorhabditis briggsae reveal extensive gene duplication, diversification, movement and intron loss. Genome Res., 8, 449–463. [DOI] [PubMed] [Google Scholar]
- 51.Robertson H.M. (2000) The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res., 10, 192–203. [DOI] [PubMed] [Google Scholar]
- 52.Robertson H.M. (2001) Updating the str and srj (stl) families of chemoreceptors in Caenorhabditis nematodes reveals frequent gene movement within and between chromosomes. Chem. Senses, 26, 151–159. [DOI] [PubMed] [Google Scholar]
- 53.Harrison P.M., Echols,N. and Gerstein,M.B. (2001) Digging for dead genes: an analysis of the characteristics and distribution of the pseudogene population in the C. elegans genome. Nucleic Acids Res., 29, 818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Remm M. and Sonnhammer,E. (2000) Classification of transmembrane protein families in the Caenorhabditis elegans genome and identification of human orthologs. Genome Res., 10, 1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bargmann C.I. (1998) Neurobiology of the Caenorhabditis elegans genome. Science, 282, 2028–2033. [DOI] [PubMed] [Google Scholar]
- 56.Lander E.S., Linton,L,M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. International Human Genome Sequencing Consortium. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 57.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 58.Wade N. (2001) Long-Held Beliefs Are Challenged By New Human Genome Analysis. In New York Times.
- 59.Kulp D., Haussler,D., Reese,M.G. and Eeckman,F.H. (1996) A generalized hidden Markov model for the recognition of human genes in DNA. Proc. Int. Conf. ISMB, 4, 134–142. [PubMed] [Google Scholar]
- 60.Yeh R.F., Lim,L.P. and Burge,C.B. (2001) Computational inference of homologous gene structures in the human genome. Genome Res., 11, 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang F., Holt,I., Pertea,G., Karamycheva,S., Salzberg,S.L. and Quackenbush,J. (2000) Gene index analysis of the human genome estimates approximately 120,000 genes. Nature Genet., 24, 239–240. [DOI] [PubMed] [Google Scholar]
- 62.Dunham I., Shimizu,N., Roe,B.A., Chissoe,S., Hunt,A.R., Collins,J.E., Bruskiewich,R., Beare,D.M., Clamp,M., Smink,L.J., Ainscough,R. et al. (1999) The DNA sequence of human chromosome 22. Nature, 402, 489–495. [DOI] [PubMed] [Google Scholar]
- 63.Hattori M., Fujiyama,A., Taylor,T.D., Watanabe,H., Yada,T., Park,H.S., Toyoda,A., Ishii,K., Totoki,Y., Choi,D.K. et al. (2000) The DNA sequence of human chromosome 21. The chromosome 21 mapping and sequencing consortium. Nature, 405, 311–319. [DOI] [PubMed] [Google Scholar]
- 64.Crollius H.R., Jaillon,O., Bernot,A., Dasilva,C., Bouneau,L., Fischer,C., Fizames,C., Wincker,P., Brottier,P., Quetier,F. et al. (2000) Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nature Genet., 25, 235–238. [DOI] [PubMed] [Google Scholar]
- 65.Ewing B. and Green,P. (2000) Analysis of expressed sequence tags indicates 35,000 human genes. Nature Genet., 232, 232–233. [DOI] [PubMed] [Google Scholar]
- 66.Wright F.A., Lemon,W.J., Zhao,W.D., Sears,R., Zhuo,D., Wang,J.P., Yang,H.Y., Baer,T., Stredney,D., Spitzner,J. et al. (2001) A draft annotation of the human genome. Genome Biol., 2, 0025.0021–0025.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davuluri R.V., Grosse,I. and Zhang,M.Q. (2001) Computational identification of promoters and first exons in the human genome. Nature Genet., 29, 412–417. [DOI] [PubMed] [Google Scholar]
- 68.Hogenesch J.B., Ching,K.A., Batalov,S., Su,A.I., Walker,J.R., Zhou,Y., Kay,S.A., Schultz,P.G. and Cooke,M.P. (2001). A comparison of the Celera and Ensembl predicted gene sets reveals little overlap in novel genes. Cell, 106, 413–415. [DOI] [PubMed] [Google Scholar]
- 69.Harrison P.M., Hegyi,H., Balasubramanian,S., Luscombe,N.M., Bertone,P., Echols,N., Johnson,T. and Gerstein,M.B. (2002). Molecular fossils in the human genome: identification and analysis of the pseudogenes on chromosomes 21 and 22. Genome Res., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vanin E.F. (1985) Processed pseudogenes: characteristics and evolution. Annu. Rev. Genet., 19, 253–272. [DOI] [PubMed] [Google Scholar]
- 71.Mighell A.J., Smith,N.R., Robinson,P.A. and Markham,A.F. (2000) Vertebrate pseudogenes. FEBS Lett., 468, 109–114. [DOI] [PubMed] [Google Scholar]
- 72.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 74.Modrek B., Resch,A., Grasso,C. and Lee,C. (2001) Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res., 29, 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Venkatesh B., Gilligan,P. and Brenner,S. (2000) Fugu: a compact vertebrate reference genome. FEBS Lett., 476, 3–7. [DOI] [PubMed] [Google Scholar]
- 76.Marra M., Hillier,L., Kucaba,T., Allen,M., Barstead,R., Beck,C., Blistain,A., Bonaldo,M., Bowers,Y., Bowles,L. et al. (1999) An encyclopedia of mouse genes. Nature Genet., 21, 191–194. [DOI] [PubMed] [Google Scholar]
- 77.Watanabe T.K., Bihoreau,M.T., McCarthy,L.C., Kiguwa,S.L., Hishigaki,H., Tsuji,A., Browne,J., Yamasaki,Y., Mizoguchi-Miyakita,A., Oga,K. et al. (1999) A radiation hybrid map of the rat genome containing 5,255 markers. Nature Genet., 22, 27–36. [DOI] [PubMed] [Google Scholar]
- 78.Varki A. (2000) A chimpanzee genome project is a biomedical imperative. Genome Res., 10, 1065–1070. [DOI] [PubMed] [Google Scholar]
- 79.McConkey E.H. and Varki,A. (2000) A primate genome project deserves high priority. Science, 289, 1295–1296. [DOI] [PubMed] [Google Scholar]
- 80.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]