Abstract
Ensuring food safety is paramount worldwide. Developing effective detection methods to ensure food safety can be challenging owing to trace hazards, long detection time, and resource-poor sites, in addition to the matrix effects of food. Personal glucose meter (PGM), a classic point-of-care testing device, possesses unique application advantages, demonstrating promise in food safety. Currently, many studies have used PGM-based biosensors and signal amplification technologies to achieve sensitive and specific detection of food hazards. Signal amplification technologies have the potential to greatly improve the analytical performance and integration of PGMs with biosensors, which is crucial for solving the challenges associated with the use of PGMs for food safety analysis. This review introduces the basic detection principle of a PGM-based sensing strategy, which consists of three key factors: target recognition, signal transduction, and signal output. Representative studies of existing PGM-based sensing strategies combined with various signal amplification technologies (nanomaterial-loaded multienzyme labeling, nucleic acid reaction, DNAzyme catalysis, responsive nanomaterial encapsulation, and others) in the field of food safety detection are reviewed. Future perspectives and potential opportunities and challenges associated with PGMs in the field of food safety are discussed. Despite the need for complex sample preparation and the lack of standardization in the field, using PGMs in combination with signal amplification technology shows promise as a rapid and cost-effective method for food safety hazard analysis.
Keywords: Food safety, Personal glucose meter, Signal amplification, Point-of-care testing, Detection principle
Graphical abstract
Highlights
-
•
PGM is an effective tool for detecting food hazards.
-
•
Integration of various signal amplification technologies with PGMs.
-
•
The principles of PGM-based biosensing strategies are described.
-
•
The advantages and disadvantages of PGMs for biosensing applications are discussed.
-
•
Future challenges and perspectives of PGMs in the field of food safety are addressed.
1. Introduction
Food safety incidents have been frequent in recent years, affecting public health and the well-being of society and causing severe economic losses in the food industry [1]. Food safety has been recognized by the World Health Organization and its member states as an essential public health function [2] and is one of the most significant public issues worldwide [3]. Modern analytical techniques have been widely used in the food industry to ensure the quality and safety of all kinds of food, from farmland to dining tables [4].
Currently, food safety testing techniques generally include confirmatory quantitative testing and rapid screening testing techniques. Chromatographic techniques (such as gas chromatography [5] and high-performance liquid chromatography (HPLC) [6]), coupled with mass spectrometric techniques (such as gas chromatography-tandem mass spectrometry [7], liquid chromatography-tandem mass spectrometry [8], and inductively coupled plasma-mass spectrometry [9]) and spectrographic techniques (such as surface-enhanced Raman spectroscopy [10], infrared spectroscopy [11], and ultraviolet-visible spectroscopy [12]), enable highly accurate and sensitive assessments. However, they require either expensive equipment or complex operational procedures and must be performed in a laboratory by specialized technicians, which limits their widespread application. In contrast, most commercially available rapid detection techniques are immunoassay methods based on antigen-antibody specific interactions, including the classical enzyme-linked immunosorbent assay (ELISA) and lateral-flow immunochromatographic strip (LFICS). Both ELISA and LFICS analyses have the advantages of rapid operation and low cost, but ELISA requires conventional instruments such as microplate readers. Moreover, the LFICS method only allows semi-quantitative analysis, and false-negative and false-positive results are frequently observed. Therefore, the development of highly sensitive and low-cost rapid assays is of great practical importance.
Point-of-care testing (POCT) is a modern prompt detection method that meets people's demand for time, realizes rapid analysis and accurate diagnosis through portable devices [13], and develops rapidly in the scope of analysis and clinical application [14]. Compared to traditional testing methods that rely on large equipment, POCT delivers quick on-site results without needing to send samples for inspection, thereby greatly saving sample turnaround time. Concerning medical diagnosis, POCT devices, such as personal glucose meters (PGMs) and pregnancy test strips, offer advantages such as miniaturization, ease of operation, nonrequirement of professional technicians, and lack of spatial or temporal restriction [15]. Thus, POCT devices are becoming more popular among users for self-examination to track their health in real time, allowing for the early diagnosis of diseases and timely prevention and treatment [16].
In recent years, many food contamination incidents have been reported, including dioxin-contaminated eggs, milk powder contaminated with melamine, and food products that carry Salmonella bacteria, which undoubtedly alarm the public [1]. Food contamination is a problem that affects everyone; therefore, it is imperative to ensure food safety before products reach consumers. However, one challenge encountered is that most food safety incidents are characterized by abruptness, rapid spread, and a large scale [4]. These factors require the present system to have efficient on-site detection capabilities to prevent the threat of food hazards, which is impacted by practical issues, such as the cost of materials and personnel [13]. More importantly, another challenge to consider is a low concentration (trace or ultratrace amounts) of contaminants in the food matrix for sensitive detection of food hazards. In practical detection applications, researchers often face the problem that signal transduction and output cannot be triggered due to the low concentration of the target [15]. In summary, although traditional detection methods have their advantages, they still cannot function to meet all the given requirements, and a solution is urgently needed.
A series of POCT tools have been developed to detect pesticide residues [17], harmful trace elements (such as mercury [18], silver [19], and other heavy metals [20]), and mycotoxins [21] in food. These POCT devices provide effective technical support to monitor and ensure food safety in the supply chain. Commercialized PGMs are among the most popular POCT devices because of their small size, easy operation, and reliable quantification. Compared to the traditional large biochemical instruments on the market, the advantages of PGMs include low cost and quick readings [22]. Nevertheless, the detection of only glucose has restricted PGMs’ wide application. To overcome this problem, Xiang and Lu [23] introduced invertase as a signal transduction molecule for application in PGMs, successfully detecting non-glucose targets using a PGM for the first time. Furthermore, in contrast to developing a new portable device or associated electrodes with different selectivities (which is costly to develop and unfamiliar to users), this research team has demonstrated the potential for people to use a standard PGM system to detect non-glucose targets; this can greatly reduce the cost, time, and risk of bringing a new product to the market and help the transition from lab-based assays to commercial POCT.
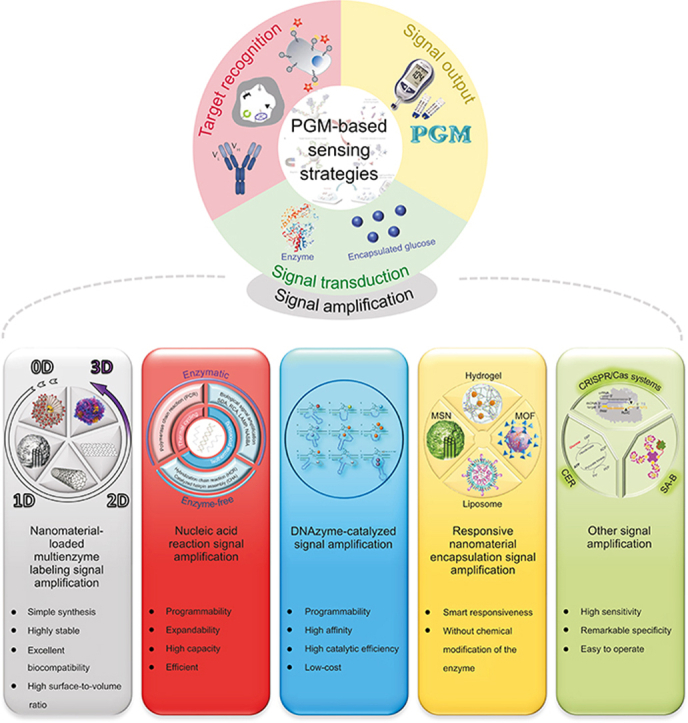
Signal amplification technology to improve the detection sensitivity and range of biosensors provides a very effective way to solve another challenge. In addition to the advantages of signal amplification technology with a strong output signal and high sensitivity, the increased diversity also provides more options for the application of different food matrices [10]. Therefore, exploring the sensing strategy of signal amplification technology combined with PGM is a developmental direction that cannot be ignored in food safety detection. To date, dozens of studies have been published on using PGMs to screen for food hazards (Fig. 1). Biological and chemical hazards are the primary food safety hazard factors, and may occur at all stages, including production, processing, packaging, distribution, retailing, consumption, and disposal [1]. Biological hazards in food generally include pathogenic bacteria and mycotoxins, while chemical hazards generally include illegal additives, drug residues, and heavy metals. In Fig. 2, we summarize the sensing targets (food hazards) of PGM-based biosensors at all stages of the food chain. Food safety is a top priority at every stage before the product reaches the consumer.
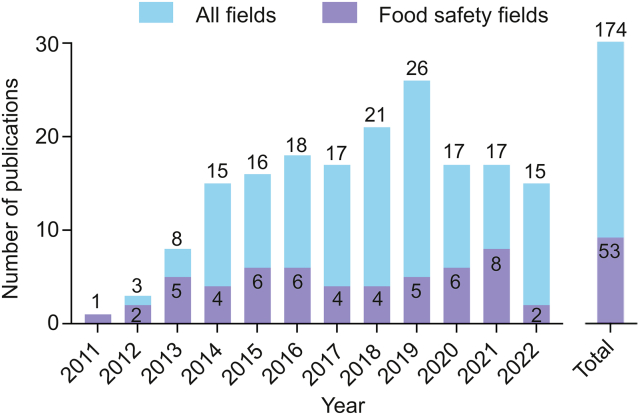
Fig. 1.
Evolution of the publication of articles on PGM-based sensing strategies in all fields and food safety fields from 2011 to 2022. Data obtained from the Web of Science. Total: 174 publications up to Nov. 2022 (Web of Science Core Collection). PGM: personal glucose meters.
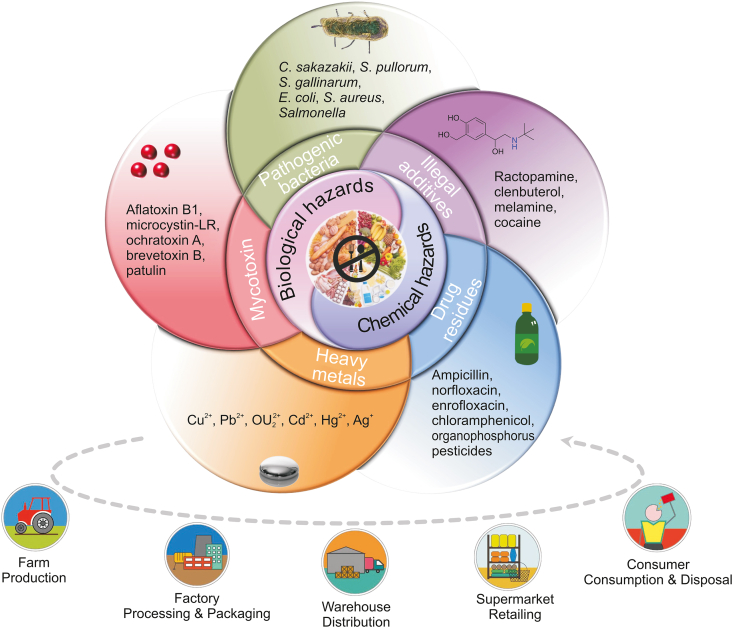
Fig. 2.
Schematic diagram of the category of sensing targets (food hazards) for PGM-based biosensors in food safety. PGM: personal glucose meters; C. sakazakii: Cronobacter sakazakii; S. pullorum: Salmonella pullorum; S. Gallinarum: Salmonella gallinarum; E. coli: Escherichia coli O157:H7; S. aureus: Staphylococcus aureus.
2. Basic principle of PGM-based sensing
PGMs provide diabetic patients with the convenience of self-measurement of blood glucose levels and have improved the life quality of millions of patients. Moreover, research has been conducted on assays targeting glucose as a biomarker in diseases other than diabetes [17]. For example, acute bacterial meningitis is diagnosed by detecting glucose concentrations in abnormal cerebrospinal fluid [24], and pathogens are detected by monitoring the bacterial metabolic consumption of glucose [25,26]. However, PGMs can only detect glucose, limiting their application against non-glucose targets. To overcome this limitation, researchers have converted non-glucose target signals to glucose signals through a signal recognition and transduction system, which dramatically improves the applicability of PGMs in the POCT of non-glucose targets [19]. PGM-based sensing of non-glucose targets generally comprises three critical factors: target recognition, signal transduction, and signal output (Fig. 3).
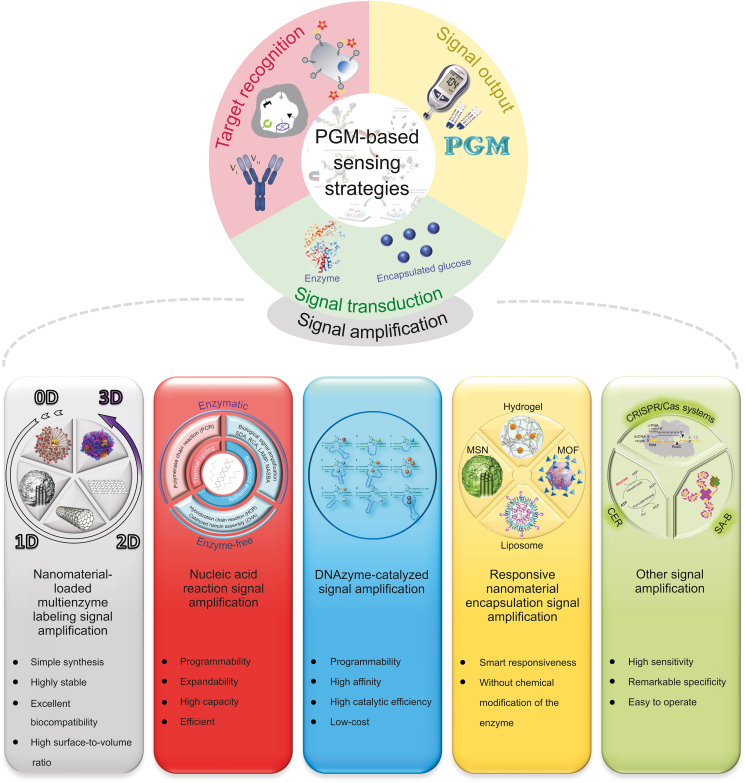
Fig. 3.
Visual overview of the review on PGM-based sensing strategies. PGM: personal glucose meters; SDA: strand displace amplification; RCA: rolling circle amplification; LAMP: loop-mediated isothermal amplification; NASBA: nucleic acid sequence-based amplification; MIP: molecularly imprinted polymer; MSN: mesoporous silica nanoparticles; MOF: metal organic framework; CRISPR/Cas systems: clustered regularly interspaced short palindromic repeats and associated proteins systems; CER: cascade enzymatic reaction; SA-B: streptavidin-biotin.
Factor 1: Recognition of a non-glucose target is realized by converting its signal to a glucose signal using biomolecules that can specifically interact with the non-glucose target, such as antibodies, aptamers, and DNAzymes [23].
Factor 2: Signal transduction involves transducing the non-glucose target signal into a signal detected by a PGM and establishing a quantitative relationship between the target and glucose concentrations [21].
Factor 3: The output of the signal. The PGM detection architecture comprises the PGM and the test strip. The former measures glucose concentration based on the changes in the current or voltage, while the latter works by immobilizing glucose oxidase (GOx) or glucose dehydrogenase (GDH) on the surface of an enzyme electrode, allowing GOx or GDH to undergo an electrochemical redox reaction with glucose in a conductive medium (ferrocene and its derivatives) to induce changes in the current or voltage [27].
3. Food safety hazard analysis based on PGM-based sensing combined with signal amplification technology
Improving sensitivity in detecting food hazards (mycotoxins, illegal additives, foodborne pathogens, heavy metal ions, pesticides, and veterinary drugs) has been a research focus [28,29]. Signal amplification technology is a commonly used approach in the biosensor-based quantitative determination of trace substances. The response signal of a biosensor consists of two components: the detection signal and the noise signal. Most of the reported studies integrate PGM-based sensing strategies with multiple signal amplification techniques to improve detection sensitivity. The most commonly used signal amplification technologies are nanomaterial-loaded multienzyme labeling, nucleic acid reaction, DNAzyme-catalysis, responsive nanomaterial encapsulation, and other technologies (Fig. 3).
This paper reviews the existing representative work on PGM-based sensing strategies combined with signal amplification technologies in food safety. The analytical principles of each study are summarized, the challenges of PGM-based sensing strategies for detecting trace hazards in food are considered, and the prospects available through the combination of signal amplification technologies are made.
3.1. Nanomaterial-loaded multienzyme labeling signal amplification technology
Nanomaterials are materials with at least one dimension in the nanoscale range (1–100 nm) in three-dimensional space or those composed of structural units in the nanoscale range [30]. Compared to macroscopic materials, nanomaterials have superior electrical conductivity, playing the role of “electron conducting wires”, which significantly increases the electron transport rate [[31], [32], [33]]. The surface and small-size effects of nanomaterials make them highly advantageous in immobilizing and modifying biomolecules by significantly increasing the immobilization load of biomolecules and improving sensor sensitivity [[34], [35], [36]]. Additionally, the superior biocompatibility of nanomaterials ensures a more stable microenvironment for biomolecules, thereby protecting biomolecular activity to the maximum extent possible [37]. Therefore, numerous studies have been conducted to combine or functionalize two or more nanomaterials with different properties by physical or chemical methods to prepare functionalized nanocomposites [38,39]. Based on their structure, nanomaterials can be classified as zero-dimensional (0D) spherical nanomaterials (e.g., silica nanoparticles, gold nanoparticles, and magnetic nanoparticles), one-dimensional (1D) nanotubes or nanowires (e.g., single-walled carbon nanotubes), two-dimensional (2D) nanosheets (e.g., graphene), or three-dimensional (3D) nanoprisms and nanoflowers [32]. Most of the existing PGM-based sensors with higher sensing efficiency rely on these nanomaterials to enhance signal amplification.
Silica nanoparticles (SiNPs) with revolutionary physicochemical properties and specific applicability have been widely used in various fields, especially in nano-vectorization, to load small molecules onto the surface or voids of microspheres [40]. Ye et al. [41] introduced antibody and GOx biofunctionalized spherical SiNPs (SiNP-GOx-IgG) and antibody monofunctionalized silica-coated magnetic nanoparticles (MNP-IgG) for target recognition in the form of sandwich structures. A large amount of GOx loaded on the surface of SiNPs catalyzes the hydrolysis of the substrate and leads to the change of glucose in the system to achieve the transduction and amplification of the signal, which successfully detected Cronobacter sakazakii in powdered infant formula. In the same year, they also used this SiNP-loaded multi-GOx labeling signal amplification method to rapidly quantify Salmonella pullorum and Salmonella gallinarum with a detection limit of 7.2 × 101 CFU/mL [42].
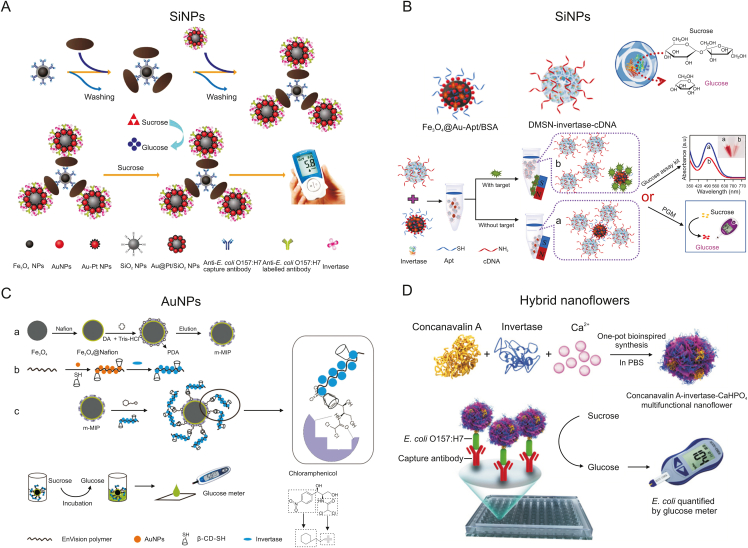
After that, this research group used poly-(4-styrenesulfonic acid-co-maleic acid) (PSSMA) as a “bridge” for the first time to functionalize SiNPs with Au–Pt bimetallic nanoparticles (Au–Pt NPs) and modify antibodies and invertase on the surface (Fig. 4A) [43]. The negative charge carried by PSSMA can act as a spacer to prevent the aggregation of SiNPs and improve the stability of the system. In addition, the loading rate of invertase was greatly improved due to the good biocompatibility of Au–Pt NPs and the enhanced surface-to-volume ratio of SiNPs. The system successfully detected Escherichia coli O157:H7 in milk samples and was comparable to the standard culture method. This concept of immobilizing enzymes on the surface of SiNPs has proven to be an effective means of increasing the amount of enzyme per unit and signal amplification. However, enzyme immobilization methods can affect the protein structure and catalytic activity due to factors such as the surface properties and geometric structure of SiNPs [44,45].
Fig. 4.
Schematic diagram of the nanomaterial-loaded multienzyme labeling signal amplification technology for food hazard detection. (A) Immunosandwich POCT with Au@Pt/SiO2 NPs and magnetic nanoparticles for detecting E. coli using a PGM. Reprinted from Ref. [43] with permission. (B) Sensing strategy based on DMSN-invertase nanoreactors and Fe3O4@Au nanocomposites for detecting AFB1. Reprinted from Ref. [46] with permission. (C) Antibody-free sandwich assay for detecting CAP in food by m-MIP cooperative EV-Au-β-CD/invertase. Reprinted from Ref. [51] with permission. (D) A PGM-based sandwich assay of concanavalin A-invertase-CaHPO4 nanoflowers and antibodies was used to detect E. coli in milk. Reprinted from Ref. [53] with permission. SiNPs: silica nanoparticles; Apt: aptamer; BSA: bovine serum albumin; DMSN: dendritic mesoporous silica nanoparticles; AuNPs: gold nanoparticles; DA: dopamine; PDA: polydopamine; Tris: tris(hydroxymethyl)aminomethane; cDNA: the complementary DNA strand to aptamer; PBS: phosphate buffered solution; POCT: point-of-care testing; Au@Pt/SiO2 NPs: Au-Pt bimetallic nanoparticles functionalized silica nanoparticles; E. coli: Escherichia coli O157:H7; PGM: personal glucose meters; AFB1: aflatoxin B1; CAP: chloramphenicol; m-MIP: magnetic molecularly imprinted polymer; EV-Au-β-CD/invertase: En Vision reagent loaded gold nanoparticles co-immobilized invertase and mercaptol-β-cyclodextrin.
To increase the relative number of SiNPs loaded with enzymes while maintaining the catalytic activity of the enzymes, dendritic mesoporous silica nanoparticles (DMSNs) with large surface curvature were introduced by Yang et al. (Fig. 4B) [46]. The DMSNs have a larger pore volume and a highly accessible internal surface area than the nonporous SiNPs. In this sensing strategy, signal amplification elements are designed with DMSNs immobilizing invertase and modifying aptamer complementary DNA strands (cDNA) on the surface. DMSNs and aflatoxin B1 (AFB1) compete for aptamers on Fe3O4@Au nanocomposites. When the target concentration of AFB1 changes, it causes a change in the number of captured DMSN-invertase-cDNA molecules. After the substrate reaction, the PGM can be applied to detect AFB1 in corn oil and wheat powder. Moreover, they also compared the strategy with other reported single-enzyme labeled aptamer sensors, and the DMSN-invertase-cDNA-based sensing strategy had a wider linear range and high sensitivity.
In addition, gold nanoparticles (AuNPs), another representative spherical nanomaterial, have become popular among researchers due to their easy preparation and unique physicochemical properties [47]. Li et al. [48] reported a strategy for the detection of clenbuterol (CLB) in pork by a competitive ELISA combined with a PGM. This strategy is that the introduced exogenous small molecule target can compete with the immunomagnetic beads for CLB-antibody (Ab1) to further prevent the immunomagnetic beads from attaching the AuNP-invertase-Ab2 conjugation. The signal value of the PGM is inversely proportional to the concentration of CLB. It is worth noting that when using nanomaterials as carriers to couple invertase and antibodies, the ratio of invertase and antibodies and the pH of the coupling buffer need to be optimized and discussed. Gao et al. [49] also developed a PGM-based competitive immunosensor for the simple and rapid detection of norfloxacin in animal-derived foods using AuNPs as a carrier for immobilized invertase.
Chen et al. [50] used an immunohistochemical reagent called envision reagent (EV) with anti-IgG antibodies attached to a poly-dextrin amine skeleton chain and AuNPs immobilized on the EV matrix as a connecting carrier to load invertase to complete the signal amplification. The method has a good linear response in the range of 1–40 ng/mL, and the detection limit is as low as 0.34 ng/mL for detecting ractopamine in pork. Furthermore, another unique aspect of this team's study [51] was antibody-free sandwich recognition based on the molecular structure of chloramphenicol (CAP) and signal amplification in combination with the EV complex (Fig. 4C). A magnetic molecularly imprinted polymer was used to selectively recognize the 2,2-dichloroacetamide segment in CAP. The residual nitrophenol part was exposed outside the imprinted site for recognition by the EV complex β-cyclodextrin (β-CD)/invertase functionalized signal tag to form the host-guest complex (EV-Au-β-CD/invertase). The magnetically separated sandwich-type complex carried invertase that catalyzed sucrose substrate hydrolysis to glucose. Under optimal conditions, this strategy showed good selectivity and sensitivity with a detection limit of 0.16 ng/mL. It is worth mentioning that this aggressive antibody-free sandwich assay can substantially reduce costs and overcome the limitations of the tendency to deactivate. Zhao et al. [52] used 2D graphene oxide as a substrate to immobilize AuNPs to load a large amount of invertase applied to the detection of microcystin-LR in drinking water.
In contrast, a novel 3D flower-like organic‒inorganic nanocomposite has attracted researchers' attention due to its mild preparation conditions and simple process, which can maintain better catalytic activity and durability of the enzyme. Ye et al. [53] successfully prepared concanavalin A-invertase-CaHPO4 hybrid nanoflowers by a one-pot bioinspired synthesis method and constructed a PGM-based sandwich immunosensor to analyze E. coli in milk (Fig. 4D). After that, Bai et al. [54] used the antimicrobial peptides cecropin P1 and magainin I as recognition elements for E. coli to prepare another invertase-embedded cecropin P1–Ca3(PO4)2 nanoflower and magnetic magainin I-phosphate nanocomposite pair for sandwich analysis of E. coli. The detection sensitivity was 10 CFU/mL, and the linear range was 10–107 CFU/mL. In addition, protein-inorganic hybrid nanoflowers can simply confer recognition ability or biological activity to other molecules by changing the protein species.
This strategy uses nanomaterials as carriers, increasing the number of loaded enzymes and directly utilizing the enzyme turnover amplification signal to achieve lower detection limits and wider linear detection ranges. However, the disadvantages cannot be ignored, such as the probe modification and immobilization of the enzyme, which often requires various modifications that will affect the structure and function of the enzymes. Moreover, the short service life of a probe is susceptible to inactivation and degradation caused by external environmental disturbances [[55], [56], [57]]. In addition, the established method involves multiple steps and is procedurally complex.
3.2. Nucleic acid reaction signal amplification technology
Nucleic acids store and transmit genetic information in biological systems and are basic materials used in nanotechnology due to their programmability and expandability [58]. For nucleic acid analysis, in vitro nucleic acid amplification techniques have been used to improve signal response and detection sensitivity, which is a strategy inspired by intracellular DNA replication and RNA transcription [59]. Thus, chemical cross-linking of nucleic acids and functional enzymes offers the possibility of integrating in vitro nucleic acid amplification techniques.
In vitro nucleic acid amplification techniques are classified into two types based on their thermal cycling requirements, with one type being thermal cycling amplification, such as polymerase chain reaction (PCR) [60]. The other is isothermal amplification, such as rolling circle amplification (RCA) [61], loop-mediated isothermal amplification (LAMP) [62], strand displacement amplification [63], catalytic hairpin assembly (CHA) [64], and hybridization chain reaction (HCR) [65].
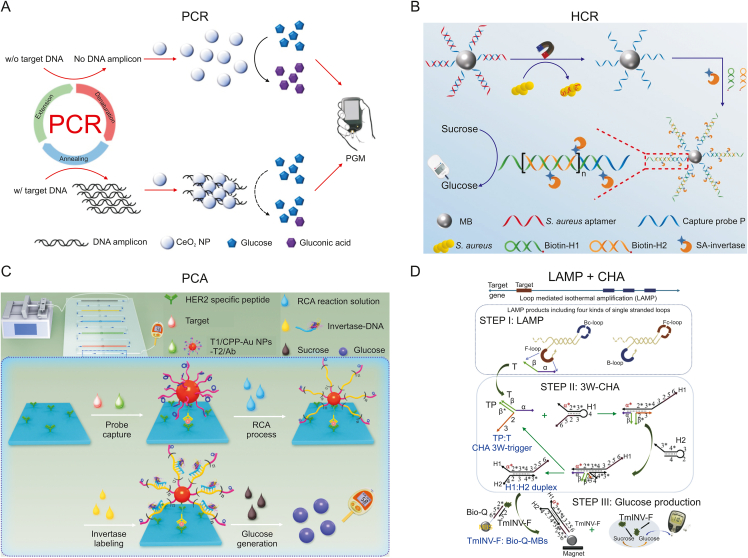
Based on the novel finding that cerium oxide nanoparticles (CeO2 NPs) have GOx activity similar to that of natural GOx, Kim et al. [66] created a method for label-free detection of target genomic DNA from E. coli by PCR combined with mimetic enzymes. Fig. 5A demonstrates in principle that when the target DNA is present, large amounts of target DNA products amplified by PCR bind to CeO2 NPs through electrostatic interactions, resulting in aggregation and reduced catalytic activity. As a result, glucose is hardly oxidized to gluconic acid, thus maintaining the system at the initial high glucose concentration. In contrast, in the absence of target DNA or the presence of nontarget DNA, no target DNA amplification product is produced, resulting in a significant decrease in the final glucose level. In addition, the researchers explored the GOx activity and mechanism of action of CeO2 NPs, which also provided a new signal transduction method for PGM-based sensing strategies.
Fig. 5.
Schematic diagram of the nucleic acid reaction signal amplification technology for food hazard detection. (A) Label-free readout based on PGM using the GOx-like activity of CeO2 NPs combined with PCR amplification. Reprinted from Ref. [66] with permission. (B) PGM combined with the HCR amplification sensing strategy for detecting S. aureus in food samples. Reprinted from Ref. [69] with permission. (C) Schematic representation of the PGM-based microfluidic immunoassay system. Reprinted from Ref. [70] with permission. (D) Working process of LAMP-three-way (3W)-CHA based on PGM for detecting two foodborne bacterial genes. Reprinted from Ref. [71] with permission. PGM: personal glucose meters; GOx: glucose oxidase; CeO2 NP: cerium oxide nanoparticle; PCR: polymerase chain reaction; HCR: hybridization chain reaction; MB: magnetic bead; SA: Streptavidin; RCA: rolling circle amplification; S. aureus: Staphylococcus aureus; HER2: human epidermal growth factor receptor 2; LAMP: loop-mediated isothermal amplification; CHA: catalytic hairpin assembly. F-loop: ssDNA loop sequence; TP: transducer probe; TmINV: thermostable invertase; Bio: biotin.
Since isothermal amplification is a simple and rapid process allowing the efficient accumulation of nucleic acid sequences at a constant temperature, it can be easily integrated into biosensors [67,68]. Unlike PCR amplification, which is intended to amplify a target gene to improve detection sensitivity, isothermal amplification usually involves modifying primers on a solid carrier and loading functional invertase. Yang et al. [69] modified the aptamer and complementary strand of Staphylococcus aureus to the surface of magnetic beads. The introduction of the target allowed the aptamer to bind specifically to the target and expose the complementary strand. The complementary strand was designed as an initiator to successfully activate the exogenous HCR system, thereby immobilizing invertase on the magnetic bead surface through streptavidin-biotin interactions (Fig. 5B). After optimization of various conditions, the sensor had a high sensitivity with a detection limit of 2 CFU/mL. This method successfully evaluated the concentration of S. aureus in real food samples, and the results were consistent with those obtained using plate counting methods. Gu et al. [70] used the immunosandwich method to identify the target and immobilize AuNPs functionalized with primers. By exploiting the characteristics of RCA, the primers produced repetitive single-strand length products, which were combined with DNA-functionalized invertases for signal amplification (Fig. 5C). Recently, Guo et al. [71] proposed a strategy to couple LAMP and three-way amplifiable CHA to the PGM (Fig. 5D). The amplification product replaced the DNA-functionalized invertase modified on the surface of magnetic beads. Glucose was generated by simple magnetic field separation and substrate catalysis to meet the requirements for sensitive detection of foodborne pathogenic bacteria by commercial PGMs. An innovative one-tube smart strategy (logical or multiplex analysis) based on PGM was developed to achieve genetic detection of C. sakazakii (ompA) and E. coli (malB). Compared with previously reported signal-free amplification strategies, the introduction of CHA successfully increased the signal amplitude by at least 12.5-fold with a detection limit sensitivity of 6.6 × 10−18 M.
Signal amplification of sensing strategies through nucleic acid cycling amplification technology is relatively simple and efficient, benefitting from the programmability and expandability of nucleic acids. Compared to PCR, isothermal amplification technology, without precisely controlled temperature cycling, makes it easier to integrate with biosensors [68]. Nevertheless, the amplification bias and inherent nonspecific amplification in isothermal amplification techniques may affect the accuracy of signal output results [72]. The complex design, as well as specific systems and enzymes, can also lead to increased assay processes and costs.
3.3. DNAzyme-catalyzed signal amplification technology
Apart from aptamers (ligand-binding nucleic acids), functional nucleic acids isolated using systematic evolution of ligands by exponential enrichment also include DNAzymes, which are widely applied in the field of biosensing [73]. DNAzymes have been shown to catalyze various chemical reactions, such as DNA ligation [74], DNA cleavage [75], DNA phosphorylation [76], and ester and amide bond hydrolysis [77]. Therefore, the successful application of DNA-functionalized invertase not only proves the feasibility of integrating invertase into in vitro nucleic acid amplification techniques but also indicates that innovative DNAzyme-based assays have been proposed for signal amplification.
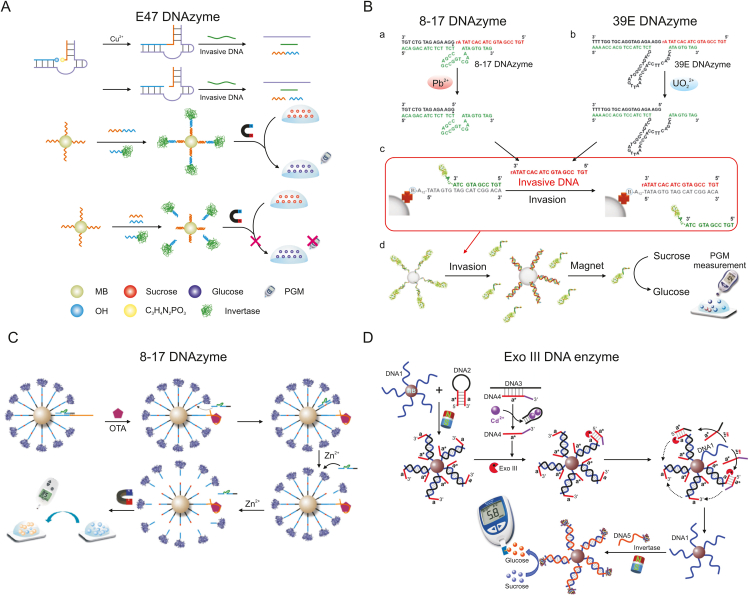
Fig. 6A illustrates a DNAzyme-assisted signal amplification strategy utilizing Cu2+-dependent DNA ligation reported by Ming et al. [78] The presence of a Cu2+ target in the system allows E47 DNAzyme to catalyze the phosphorus center nucleophilic of DNA1 and the linking of the hydroxyl group on DNA2 to form the ligation product of a phosphodiester bond, which is called the linker strand. The introduced invasive DNA forces the linker strand to release. In a system of DNA-functionalized invertase and magnetic beads, a large amount of invertase can be immobilized on the surface of magnetic beads by the linker strand for signal amplification purposes. It then reacts with the sucrose substrate to produce glucose, which the PGM reads. This strategy has a detection limit of up to 1 nM, good metal ion selectivity, and little influence from biological complexes.
Fig. 6.
Schematic diagram of the DNAzyme-catalyzed signal amplification technology for food hazard detection. (A) Detection of Cu2+ based on PGMs and a ligation DNAzyme release strategy. Reprinted from Ref. [78] with permission. (B) The detection process of hazardous metal ions based on DNAzyme and invasive DNA using PGMs. Reprinted from Ref. [79] with permission. (C) Schematic illustration of a signal amplification biosensor based on a series of cyclic hybridization and cutting reactions triggered by DNAzymes. Reprinted from Ref. [81] with permission. (D) Detection of Cd2+ in real water samples using PGM as a point-of-use device combined with Exo III and invertase amplification strategies. Reprinted from Ref. [82] with permission. PGMs: personal glucose meters; MB: magnetic bead; OH: hydroxyl; OTA: ochratoxin A; Exo III: exonuclease III.
In addition to relying on DNA ligation catalyzed by DNAzyme, the ability of DNAzyme to catalyze DNA cleavage was also successfully exploited by Xiang and Lu [79], who reported a method for the detection of hazardous metal ions in drinking water where DNAzyme binds invasive DNA (Fig. 6B). In this method, Pb2+-specific and UO22+-specific DNAzymes were selected, both of which catalyzed the cleavage of DNA substrate strands at the 3’ phosphoester bond of ribonucleotide A (rA). Longer DNA substrate strands were released as invasive DNA, forcing the release of DNA-functionalized invertase immobilized on magnetic beads. Finally, the DNA-functionalized invertase conjugate was collected by magnetic field separation, and the sucrose substrate was converted to glucose for quantitative determination using a PGM. Subsequently, Yang et al. [80] proposed a DNA walking machine concept for detecting AFB1 in bread using a similar approach combined with the catalytic cleavage ability of Pb2+-specific DNAzyme.
To detect ochratoxin A (OTA), a common small-molecule toxin in red wine, Zhang et al. [81] developed a biosensing platform for signal amplification using DNAzyme and magnetic beads (Fig. 6C). The magnetic bead-labeled substrate strand and the aptamer-capture strand hybridized with the DNAzyme strand to block substrate cleavage activity. In the presence of the OTA target, the target specifically binds to the OTA aptamer to replace the DNAzyme and induce its hybridization with the substrate strand. DNA functionalized-invertase is released by the amplification reaction that catalyzes the cyclic cleavage of the substrate strand by introducing the cofactor Zn2+. The platform successfully converted the detection of small-molecule toxins into a glucose assay using DNA-functionalized invertase and 8–17 DNAzyme with a linear detection range of 5 orders of magnitude and a detection limit as low as 0.88 pg/mL.
Unlike the DNAzyme mentioned above with catalytically active artificial DNA sequences, DNA enzymes refer to protein enzymes that act on DNA substrates and were applied by Zeng and his colleagues [82] to the detection of Cd2+ in real water samples (including lake water, river water, and pond water) (Fig. 6D). The specific binding of the aptamer to Cd2+ led to the release of DNA4, which bound to the previously immobilized DNA2 on magnetic beads to form a blunt 3ʹ-terminus of DNA2, thereby triggering the ability of exonuclease III (Exo III) to mediate 3ʹ blunt-end cleavage of double-stranded DNA (Exo III could selectively digest DNA2 from the 3ʹ to 5ʹ direction). Exo III stimulated a recycling signal amplification process for the hybridization and cleavage reactions of the system, exposing a large amount of single-stranded DNA1 on the surface of the magnetic beads. Then, DNA-functionalized invertase was introduced, thereby immobilizing invertase on the surface of the magnetic beads. This synergistic signal amplification mode by Exo III and invertase greatly improved the analytical sensitivity of Cd2+ with good selectivity for other competing metals, and the detection limit was 5 pM.
The signal amplification strategy based on DNAzyme-catalyzed cleavage demonstrates unique advantages, such as economic benefits without the need for additional proteases. The initiation of metal ion cofactors also provides more possibilities for detection methods. However, an obvious challenge for these strategies is their instability in the presence of nucleases, and they only work in relatively pure solutions [73]. As a result, a slight change in reaction conditions can result in great changes in activity.
3.4. Responsive nanomaterial encapsulation signal amplification technology
Most of the signals mentioned in the above amplification strategies involve the functional modification of enzymes to attach them to the surface of nanomaterials. Xiang and Lu [23] first realized the target's connection to glucose by DNA chemical cross-linking functionalization of invertase, making it an ideal mediator candidate associated with glucose. In addition, glucoamylase can also be used as an alternative, playing a similar role to that of invertase [[83], [84], [85], [86]]. However, the active site of invertase is mainly composed of aspartate and glutamate [87]. Although the reactive amines not involved in the active site of invertase are selected as the coupling site to preserve the enzyme's catalytic activity, it will be weakened to a certain extent during the actual chemical cross-linking functionalization process, affecting the structure and function of the enzyme [88]. The enzyme's catalytic efficiency is also susceptible to interference and analytical conditions (e.g., pH, temperature, and instability caused by structural unfolding) during the signal generation phase. Moreover, these methods require hydrolases such as convertase or glucoamylase to produce glucose for PGM detection, which inevitably leads to increased washing or separation steps. To overcome this drawback, researchers have explored a novel signal amplification strategy based on responsive nanomaterials that directly encapsulate and release enzymes or glucose to avoid the chemical crosslinking functionalization of the enzyme.
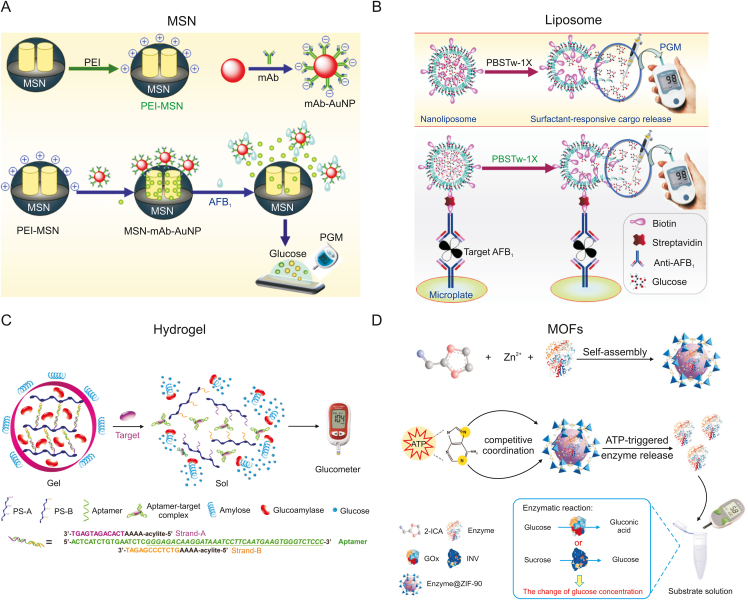
Tang's team [89] designed a simple, low-cost gating system-like immunosensing platform using AFB1 as a target model and successfully applied it to peanut samples (Fig. 7A). First, negatively charged monoclonal anti-AFB1 antibody (mAb)-labeled AuNPs (mAb-AuNPs) and positively charged polyethylenimine (PEI)-coated mesoporous silica nanocontainers (PEI-MSNs) were synthesized. The electrostatic interaction between them gated a large number of glucose molecules into the pores of the MSN. In the presence of the target AFB1, the mAb-AuNPs and PEI-MSN underwent a competitive-type displacement reaction via antigen-antibody specific binding, which resulted in the opening of the pore gate to release the loaded glucose. Notably, the strategy was validated on naturally contaminated and spiked blank peanut samples and yielded results consistent with those of the reference ELISA. In the same year, Cao et al. [90] also successfully detected brevetoxin B (PbTx-2) in seafood samples by replacing the gating system with a positively charged PEI-coated polystyrene microsphere and encapsulating glucose into a negatively charged magnetic mesoporous NiCo2O4 nanostructure (MMB) by electrostatic action. Another novel idea is to activate Pb2+-specific DNAzymes by introducing targets, which catalyze the cleavage of the DNA double-strand “gate” to release glucose in MSNs [91]. Using DNA as a functional unit to close and open micropores is also a preferable option [92]. However, the above gating system cannot be used to detect samples containing endogenous glucose, limiting its application in food safety testing.
Fig. 7.
Schematic diagram of the responsive nanomaterial encapsulation signal amplification technology for food hazard detection. (A) Schematic representation of the PGM-based immunosensing platform combined with a one-step competitive displacement reaction mode for detecting AFB1 in peanut samples. Reprinted from Ref. [89] with permission. (B) Immunosandwich sensing strategy based on nanoliposome-encapsulated glucose combined with PGM to detect AFB1. Reprinted from Ref. [93] with permission. (C) Scheme of the aptamer sensing platform for non-glucose target detection based on a target-responsive “sweet” hydrogel encapsulating glucoamylase. Reprinted from Ref. [97] with permission. (D) Schematic illustration of the PGM-based enzyme@ZIF-90 sensing platform for non-glucose target detection. Reprinted from Ref. [98] with permission. PEI-MSN: polyethylenimine-coated mesoporous silica nanocontainer; mAb: monoclonal antibody; AuNP: gold nanoparticle; PGM: personal glucose meters; PBSTw-1×: 1× phosphate buffered solution-Tween 20 buffer; PS: polymer strands; AFB1: aflatoxin B1; MOFs: metal-organic frameworks; enzyme@ZIF-90: enzyme-encapsulated zeolitic imidazole framework-90; 2-ICA: imidazole-2-carboxaldehyde; GOx: glucose oxidase; INV: sucrose invertase; ATP: adenosine-5’-triphosphate.
To address this problem, Tang's team [93] synthesized a spherical lipid bilayer nanomaterial (liposome) with glucose embedded in the lumen (Fig. 7B). This strategy was to use an immunosandwich method to capture AFB1 and connect the nanoliposomes via a streptavidin-biotin “bridge”. The nanoliposome hydrolyzes with the surfactant PBS-Tween 20 buffer, which releases glucose and combines it with a PGM to measure AFB1. This strategy uses glucose-encapsulated nanoliposomes as a signal amplification label, which is not only much lower than the detection limit of commercial ELISA kits but also has good reproducibility and specificity for complex systems. In addition, most methods based on surfactant-responsive nanodrug delivery systems are applied in biomedicine. The successful application of this strategy provides new ideas and directional development for future food safety detection. Nie et al. [94] first reported a novel chemically coupled strategy for detecting patulin in apple juice and grape juice based on nanoliposome encapsulation of glucose as an effective method for signal amplification. The team first grafted sulfhydryl (−SH) as a unique functional group for efficiently recognizing patulin on the surface of nanoliposomes encapsulated with glucose. This functionalized modification also expands the scope of nanoliposome applications.
Recently, two classes of materials, namely, 3D hydrogels composed of hydrophilic crosslinked polymer networks [95] and metal-organic frameworks (MOFs)—a new generation of crystalline porous materials [96]—have offered greater possibilities in the development of nanomaterial encapsulation and release for signal amplification due to their impressive properties, such as flexibility, biocompatibility, deformability, and high porosity, thereby receiving wide research attention. The former is shown in Fig. 7C, where Yan et al. [97] described a novel design of a target-responsive “sweet” hydrogel encapsulating glucoamylase. The aptamer-assisted cross-linked polymer strands A and B form glucoamylase-encapsulated hydrogels that are physically isolated from the substrate amylose of the external system. When a target is introduced, specific binding of the target to the aptamer causes the hydrogel to disintegrate and release glucoamylase, which catalyzes the hydrolysis of the substrate to produce glucose for quantitative measurement by the PGM. The aptamer-assisted encapsulation of glucoamylase gives the method good specificity and sensitivity. However, even the best PGM can give a reading 10% different from the reference value when measuring low glucose, an important aspect that cannot be ignored in practical applications. Alternatively, Cao et al. [98], shown in Fig. 7D, synthesized enzyme-encapsulated zeolitic imidazole framework-90 (enzyme@ZIF-90) self-assembled by combining imidazole-2-carboxaldehyde and Zn2+ with GOx or invertase. Due to the competitive coordination between the enzyme@ZIF-90 microparticles and the target, the microparticles decompose and release GOx or invertase to catalyze the substrate and produce glucose for indirect quantification of the target by PGM. This method is the first to combine a PGM with a MOF-encapsulated enzyme to achieve immediate quantification of a non-glucose target, demonstrating its potential practical application and providing a new option for detecting food hazards in the future.
It is worth mentioning that responsive nanomaterials have good biocompatibility. The signal amplification strategy constructed on this basis retains the enzyme activity to the greatest extent and avoids affecting the enzyme in the actual chemical cross-linking functionalization process. In addition, a simple trigger mechanism also reduces the washing and separation steps. When detecting low concentrations of targets in a complex food matrix, interfering contaminants are usually removed by pretreatment procedures using organic solvents. Organic solvents react with nanomaterials encapsulating glucose or enzymes, leading to their structural degradation [99]. Further, nanomaterial synthesis and modification methods may not accurately control their pore size and volume, resulting in batch differences.
3.5. Other signal amplification technologies
The main trends of PGM-based signal amplification techniques are presented in this paper, revealing technological advances in establishing POCT methods for detecting trace food hazards. In addition, several studies providing solutions with unique advantages have been reviewed.
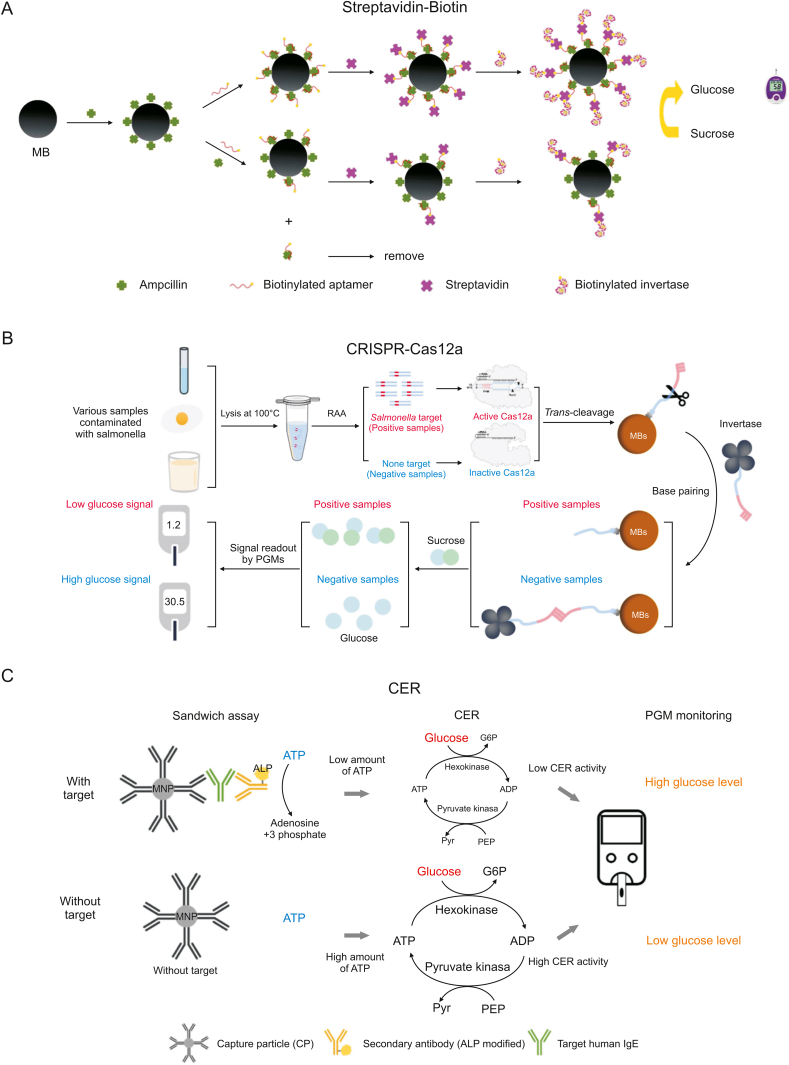
Streptavidin-biotin (SA-B) is a well-established signal amplification system for biological responses [100,101]. SA is a small-molecule glycoprotein (homotetramer) consisting of four subunits, and under certain conditions, it can form stable complexes with four B molecules [102]. Moreover, SA-B binding is one of the most vital noncovalent interactions to date [103,104]. This multilevel signal amplification system has been used widely in research, as its advantages include binding firmly to biomarker molecules and increasing the number of biomarkers significantly. As shown in Fig. 8A, Li et al. [105] used ampicillin-coated magnetic beads as a platform, with ampicillin as the target of interest, allowing the beads to compete with a sample containing free ampicillin for biotinylated aptamers and further connecting biotinylated aptamers to biotinylated invertase via highly specific SA-B interactions. This method achieved signal amplification and was successfully applied to the detection of ampicillin residues in milk.
Fig. 8.
Schematic diagram of the other signal amplification technologies for food hazard detection. (A) Schematic diagram of an aptasensor based on SA-B for detecting ampicillin in milk. Reprinted from Ref. [105] with permission. (B) Working process of the PGM-CRISPR-based quantitative assay to detect Salmonella in milk samples. Reprinted from Ref. [107] with permission. (C) Principle of CER combined with PGM for target human IgE detection. Reprinted from Ref. [114] with permission. SA-B: streptavidin-biotin; PGM: personal glucose meters; CRISPR-Cas12a: clustered regularly interspaced short palindromic repeats and associated proteins; RAA: recombinase aided amplification; MBs: magnetic beads; MNP: magnetic nanoparticles; CER: cascade enzymatic reaction; ATP: adenosine 5’-triphosphate; ALP: alkaline phosphatase; ADP: adenosine diphosphate; G6P: glucose-6-phosphate; PEP: phosphoenolpyruvic acid; Pyr: pyruvate; MNP: carboxyl-modified magnetic nanoparticles; IgE: immunoglobulin E.
In addition, the clustered regularly interspaced short palindromic repeats and associated protein system (CRISPR‒Cas system) has become a revolutionary tool for genome editing due to its unique nucleic acid cleavage capabilities. Moreover, it has also promoted innovation in signal amplification technologies for in vitro molecular diagnostics [106]. Zhou et al. [107] developed a highly sensitive method for detecting Salmonella in milk using two consecutive signal amplification steps in combination with the CRISPR‒Cas12a system (Fig. 8B). The first step was using recombinase-aided amplification (RAA) of a double-stranded target gene fragment in Salmonella. The second step was the specific recognition of the RAA product by Cas12a mediated by RNA and activation of its trans-cleavage activity that nonspecifically cleaves nearby DNA single strands. In this way, a large amount of single-stranded DNA modified on the surface of magnetic beads was cleaved, resulting in no complementary DNA binding to the DNA-functionalized invertase and a low PGM signal after treatment with sucrose substrate. Conversely, the CRISPR-Cas12a system was not activated without the target gene. A large amount of intact complementary DNA on the surface of the magnetic beads hybridized with the DNA-functionalized invertase, thereby generating glucose concentrations for high signal values from the PGM via sucrose substrate. This PGM-CRISPR method had a detection sensitivity of 5 CFU/reaction and good specificity for Salmonella and non-Salmonella isolates. Notably, the recently reported coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SRAR-CoV-2) has a high potential for transmission to humans from contaminated cold-chain food samples [108], posing a serious challenge to the food industry and food safety testing. However, SARS-CoV-2 is not considered as a foodborne pathogen [109]. Therefore, Fang et al. [110] used a similar method of PGM combined with the CRISPR‒Cas12a system to successfully detect SRAR-CoV-2 pseudovirus with a limit of detection of 50 copies/μL.
Food allergies are also important concerns, as specific allergens in food can cause hypersensitivity in the body, generating large amounts of immunoglobulin E (IgE) that harm the body [111,112]. Ahn et al. [113] introduced a novel cascade enzymatic reaction (CER, which consists of at least two enzymes and corresponding substrates) as a strategy to achieve cyclic catalytic reactions of the substrate for signal amplification. This method was integrated with PGM measurement to successfully achieve target detection with high sensitivity (Fig. 8C) [114]. This strategy identifies the target using an immunosandwich complex. It establishes a relationship between the target and alkaline phosphatase (ALP) concentrations by assessing ALP activity in regulating adenosine triphosphate (ATP). The lower the concentration of the target, the lower the amount of phosphate of ATP cleaved by ALP, and thus, the higher the initial ATP concentration that allows the hexokinase to catalyze the conversion of ATP to adenosine diphosphate (ADP) and glucose to glucose-6-phosphate (G6P). The resultant ADP was further converted into ATP via pyruvate kinase and phosphoenolpyruvic acid (PEP), thereby continuously reducing the amount of glucose in a cyclic manner to maintain signal amplification. The combination of hexokinase and pyruvate kinase promoted multiple rounds of ATP generation and consumption, leading to highly reduced glucose levels. Such cyclic catalysis of the substrate offers a new direction for developing signal amplification techniques [88].
This novel method has injected new vitality into the application of PGM in food safety detection. Although it meets the demands for high sensitivity and rapid detection for real samples, the complexity of the process and the participation of a variety of enzymes make the detection method expensive. Further studies are necessary to address these concerns.
4. Challenges and limitations
Currently, absolute quantification can be achieved with PGMs compared to lateral flow devices, such as immunochromatographic test strips. This means that PGM-based biosensing strategies should be able to provide people with more information regarding the content of the target in food safety detection, thereby helping the public ensure food safety. Although great progress has been achieved in this field, several challenges and limitations exist in using PGM-based biosensors and signal amplification technologies for food safety hazard analysis. Some of the main challenges are as follows:
-
(1)
Sample preparation: Accurate sample pretreatment is crucial for successfully detecting food safety hazards. However, sample pretreatment is usually the bottleneck of the entire testing process. It can be time-consuming and complex, requires specialized equipment and expertise, and can introduce sources of error. Currently, the main obstacle to commercializing PGMs to detect non-glucose targets is that they rely on laboratory pretreatment. To solve this problem, many researchers have tried to optimize the extraction solvent and adopt magnetic separation technology so that the detection system can complete the detection of hazardous substance samples without relying on the laboratory environment. Simplifying the sample pretreatment processing will be one of the key development directions of PGM for the quantitative detection of non-glucose targets in the future.
-
(2)
Standardization: The lack of standardization in the field can make comparing results from different studies difficult. Standardization is vital for developing and validating PGM-based biosensors for food safety hazard analysis.
-
(3)
Sensitivity and specificity: While signal amplification technologies can enhance detection sensitivity, they can also increase the risk of false positive results. Therefore, it is crucial to optimize the sensitivity and specificity of detection to minimize the risk of false positive results. Emerging signal amplification techniques (e.g., CRISPR-Cas and CER), combined with PGMs to improve sensitivity, are attracting increasing attention. Given the wide variety of food hazards, it is necessary to explore more detection methods and develop recognition elements with more functional groups, such as nanobodies, phages, and modified nucleic acids, to detect new targets with higher affinity and specificity.
-
(4)
Cost and scalability: PGM-based biosensors are generally less expensive than traditional laboratory-based methods. However, they are still in the laboratory stage, require a certain level of investment, and may not be suitable for large-scale or high-throughput analysis. Recent research advances with other related efforts, such as combining immunochromatographic test strips with PGMs, can be considered as a more effective solution [115,116]. Because test strips have a relatively long shelf life and the PGM has quantitative signal output, both have the advantages of low cost and a wide range of user groups to power the market for PGM-based biosensors.
-
(5)
Validation and regulatory approval: PGM-based biosensors for food safety hazard analysis need to be validated and receive regulatory approval before being used in the field. This process can be time-consuming and may require additional resources.
-
(6)
Complexity: On the one hand, integrating different technologies, such as signal amplification, into a PGM can make biosensors more complex and challenging to use. Although various signal amplification technologies are integrated with PGMs to ensure sensitivity, signal conversion and amplification require linking target analytes to functional intermediary enzymes (e.g., invertase and GOx), often at the expense of cost-effectiveness and simplicity. It is desirable to simplify the signal conversion and amplification processes or find easier-to-use media (e.g., nanocarriers, hydrogels, liposomes, and MOFs) in the future. In addition, given that food can be contaminated by multiple agents, it is imperative to expand the range of PGM detection targets from single analytes to complex multicomponent systems, despite the challenges involved. In contrast, the complexity of food types (sample matrices and endogenous sugars) may interfere with the detection of PGMs. Therefore, it is necessary to exclude foods containing endogenous sucrose and glucose or to develop reasonable methods to eliminate their interference in real food samples, such as treating it as a background signal [23,97], diluting it to below the detection threshold (washing step) [39,40], or removing it enzymatically [97,117].
Despite these challenges, the use of PGM-based biosensors and signal amplification technologies for food safety hazard analysis shows great potential and is an active field of research. The development of more accurate, sensitive, and user-friendly PGM-based biosensors can help to improve food safety and reduce the risk of foodborne illnesses. The relevant reports on PGM-based detection of food hazards in recent years are summarized in Table 1 [[41], [42], [43],46,[48], [49], [50], [51], [52], [53], [54],66,69,71,[72], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82],[89], [90], [91], [92], [93], [94],97,105,107,114].
Table 1.
A comparison of PGM-based sensing strategies for food hazard detection.
| Signal amplification technologies | Signal amplification element | Signal recognition element | Assay format | Signal transduction element | Food hazards | Real samples | Limit of detection (LOD) | Dynamic range | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Nanomaterial-loaded multienzyme labeling signal amplification | SiNPs | Antibody | Sandwich | GOx | Cronobacter sakazakii | Milk | 4.2 × 101 CFU/mL | 9.0 × 102–9.0 × 107 CFU/mL | [41] |
| SiNPs | Antibody | Sandwich | GOx | Salmonella pullorum and Salmonella gallinarum | N | 7.2 × 101 CFU/mL | 1.27 × 102–1.27 × 105 CFU/mL | [42] | |
| Au@Pt/SiO2 NPs | Antibody | Sandwich | Invertase | E. coli | Milk | 1.83 × 102 CFU/mL | 3.5 × 102–3.5 × 108 CFU/mL | [43] | |
| DMSNs | Aptamer | Competitive | Invertase | AFB1 | Corn oil, wheat powder |
0.74 pg/mL | 0.001–100 ng/mL | [46] | |
| AuNPs | Antibody | Competitive | Invertase | CLB | Pork, liver | 0.1 ng/mL | 0.1–100 ng/mL | [48] | |
| AuNPs | Antibody | Sandwich | Invertase | Norfloxacin | Milk, chicken, pork, shrimp |
0.5 ng/mL | 0.5–500 ng/mL | [49] | |
| EV/AuNPs | Antibody and β-CD | Sandwich | Invertase | Ractopamine | Pork | 0.34 ng/mL | 1–40 ng/mL | [50] | |
| EV/AuNPs | Magnetic molecularly imprinted polymer and β-CD | Sandwich | Invertase | CAP | Pork, fish |
0.16 ng/mL | 0.5–50 ng/mL | [51] | |
| Graphene oxide/AuNPs | Antibody | Sandwich | Invertase | Microcystin-LR | Drinking water | 0.1 ng/mL | 0.6–100 ng/mL | [52] | |
| Hybrid protein nanoflowers | Antibody and concanavalin A | Sandwich | Invertase | E. coli | Milk | 10 CFU/mL | 10–105 CFU/mL | [53] | |
| Nanocomposite pair | Peptide | Sandwich | Invertase | E. coli | Milk | 10 CFU/mL | 10–107 CFU/mL | [54] | |
| Nucleic acid reaction signal amplification | PCR | DNA | Label-free | CeO2 NP | E. coli target genomic DNA | N | 10 copies | N | [66] |
| Hybridization chain reaction | Aptamer | Competitive | Invertase | Staphylococcus aureus | Milk, water, peach juice |
2 CFU/mL | 3–3 × 103 CFU/mL | [69] | |
| LAMP-CHA | DNA and RNA | Label-free | Invertase | ompA and malB | N | 6.6 × 10−18 M | N | [71] | |
| DNAzyme-catalyzed signal amplification | E47 DNAzyme | DNAzyme | Competitive | Invertase | Cu2+ | Serum | 1 nM | 10 nM−10 μM | [78] |
| 8-17 DNAzyme and 39 E DNAzyme | DNAzyme | Competitive | Invertase | Pb2+ and UO22+ | Drinking water | 16 nM; 5 nM | 16–1000 nM; 5–1000 nM | [79] | |
| Pb2+-specific DNAzyme | DNAzyme | Competitive | Invertase | AFB1 | Bread | 10 pM | 0.02–10 nM | [80] | |
| 8-17 DNAzyme | Aptamer | Competitive | Invertase | OTA | Red wine | 0.88 pg/mL | 1 pg/mL - 300 ng/mL | [81] | |
| Exo III DNA enzyme | Aptamer | Competitive | Invertase | Cd2+ | Lake water, river water, pond water |
5 pM | 20 pM - 200 nM | [82] | |
| Responsive nanomaterial encapsulation signal amplification | PEI-MSN | Antibody | Competitive | Glucose | AFB1 | Peanut | 5 ng/kg | 0.01–15 μg/kg | [89] |
| MMB | Antibody | Competitive | Glucose | PbTx-2 | Seafood | 0.01 ng/mL | 0.01–20 ng/mL | [90] | |
| MSN | DNAzyme | Label-free | Glucose | Pb2+ | Water | 1 pM | 1 pM - 0.7 nM | [91] | |
| MSN | T-Hg2+-T | Competitive | Glucose | Hg2+ | Tap water, lake water |
0.1 nM | 0.1–80 nM | [92] | |
| Liposome | Antibody | Sandwich | Glucose | AFB1 | Peanut | 0.6 pg/mL | 0.001–10 ng/mL | [93] | |
| Liposome | Sulfhydryl (−SH) functional group | Competitive | Glucose | Patulin | Apple juice, grape juice |
0.05 ng/mL | 0.1–50 ng/mL | [94] | |
| Hydrogel | Aptamer | Competitive | Glucoamylase | Cocaine | Urine | 3.8 μM | 0–750 μM | [97] | |
| Other signal amplification | Streptavidin-Biotin | Aptamer | Competitive | Invertase | Ampicillin | Milk | 2.5 × 10−10 M | 2.5 × 10−10 − 2.5 × 10−7 M | [105] |
| CRISPR-Cas12a | RNA | Label-free | Invertase | Salmonella target gene | Milk | 5 CFU/reaction | 1 × 103 CFU/reaction | [107] | |
| CER | Antibody | Sandwich | Hexokinase | IgE | Serum | 29.59 ng/mL | 10–400 ng/mL | [114] |
“N” means not specified in the reference.
SiNPs: silica nanoparticles; GOx: glucose oxidase; Au@Pt/SiO2 NPs: Au-Pt bimetallic nanoparticles functionalized silica nanoparticles; E. coli: Escherichia coli O157:H7; DMSNs: dendritic mesoporous silica nanoparticles; AFB1: aflatoxin B1; AuNPs: gold nanoparticles; CLB: clenbuterol; EV: En Vision reagent; β-CD: β-cyclodextrin; CAP: chloramphenicol; PCR: polymerase chain reaction; CeO2 NPs: cerium oxide nanoparticles; LAMP: loop-mediated isothermal amplification; CHA: catalytic hairpin assembly; ompA and malB: genes of Cronobacter sakazakii and Escherichia coli; OTA: ochratoxin A; Exo III: exonuclease III; PEI-MSN: polyethylenimine-coated mesoporous silica nanocontainers; MMB: magnetic mesoporous NiCo2O4 nanostructure; PbTx-2: brevetoxin B; MSN: mesoporous silica nanoparticles; CRISPR-Cas12a: clustered regularly interspaced short palindromic repeats and associated proteins; CER: cascade enzymatic reaction; IgE: immunoglobulin E.
5. Conclusions and perspectives
In conclusion, PGMs are being developed as effective tools to address food safety issues. Various signal amplification technologies, combined with PGM-based biosensing strategies, remain an emerging field in food safety detection and are undergoing substantial development. This review presents typical examples and recent advances in using PGM-based assays combined with signal amplification techniques as an accurate and promising alternative to conventional analytical processes (e.g., HPLC, electrochemical workstations, and ELISA) for rapidly screening food hazards to ensure food safety.
There are several future perspectives for using PGM-based biosensors and signal amplification technologies for food safety hazard analysis. Some of the potential developments include:
-
(1)
Advances in signal amplification technologies: The use of new signal amplification technologies such as CRISPR-based detection and biosensors based on machine learning are expected to further enhance the sensitivity and specificity of detection.
-
(2)
Integration with other technologies: The integration of PGMs with other technologies, such as blockchain and the Internet of Things (IoT), can help improve traceability and food safety management by providing real-time monitoring and data analysis.
-
(3)
Point-of-care and on-site testing: The development of PGM-based biosensors that can be used for on-site or point-of-care testing will help reduce the time between sample collection and analysis and minimize the risk of food contamination.
-
(4)
Miniaturization: The miniaturization of PGM-based biosensors can further improve their portability and ease of use, making them suitable for use in remote or resource-limited settings.
-
(5)
Development of multiplexed biosensors: Multiplexing, or the ability to detect multiple targets in a single assay, can increase the efficiency and cost-effectiveness of food safety analysis.
-
(6)
Development of affordable and user-friendly biosensors: The development of low-cost and user-friendly biosensors can make them more accessible for small and medium-sized enterprises and farmers.
-
(7)
Development of biosensors with AI and ML abilities: With the integration of artificial intelligence (AI) and machine learning (ML) in biosensors, they can be trained to detect new pathogens and adapt to changing conditions.
CRediT author statement
Feng He: Conceptualization, Investigation, Visualization, Writing - Original draft preparation, and Reviewing and Editing; Haijie Wang: Investigation, Visualization, Formal analysis; Pengfei Du: Conceptualization, Funding acquisition, Project administration, Writing - Reviewing and Editing; Tengfei Li: Writing - Reviewing and Editing, Supervision; Weiting Wang: Funding acquisition, Project administration; Tianyu Tan: Investigation, Supervision, Software; Yaobo Liu: Funding acquisition, Resources, Supervision, Project administration; Yanli Ma: Investigation, Conceptualization; Yuanshang Wang: Investigation, Conceptualization; A. M. Abd El-Aty: Supervision, Writing - Reviewing and Editing.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of Shandong Province (Grant No.: ZR2020QC250), China Agriculture Research System (Grant No.: CARS-38), Modern Agricultural Technology Industry System of Shandong Province (Grant No.: SDAIT-10-10), and Key Technology Research and Development Program of Shandong (Grant Nos.: 2021CXGC010809 and 2021TZXD012).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Contributor Information
Pengfei Du, Email: dupengfei2011@163.com.
Tengfei Li, Email: litengfei@hebeu.edu.cn.
References
- 1.Aung M.M., Chang Y.S. Traceability in a food supply chain: Safety and quality perspectives. Food Control. 2014;39:172–184. [Google Scholar]
- 2.Kirk M.D., Pires S.M., Black R.E., et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng W., Tang X., Zhang Y., et al. Applications of metal-organic framework (MOF)-based sensors for food safety: Enhancing mechanisms and recent advances. Trends Food Sci. Technol. 2021;112:268–282. [Google Scholar]
- 4.Gallo M., Ferranti P. The evolution of analytical chemistry methods in foodomics. J. Chromatogr. A. 2016;1428:3–15. doi: 10.1016/j.chroma.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Miao Q., Kong W., Yang S., et al. Rapid analysis of multi-pesticide residues in lotus seeds by a modified QuEChERS-based extraction and GC-ECD. Chemosphere. 2013;91:955–962. doi: 10.1016/j.chemosphere.2013.01.104. [DOI] [PubMed] [Google Scholar]
- 6.Lu S., Wu D., Li G., et al. Facile and sensitive determination of N-nitrosamines in food samples by high-performance liquid chromatography via combining fluorescent labeling with dispersive liquid-liquid microextraction. Food Chem. 2017;234:408–415. doi: 10.1016/j.foodchem.2017.05.032. [DOI] [PubMed] [Google Scholar]
- 7.Jung M.Y., Kang J.H., Jung H.J., et al. Inorganic arsenic contents in ready-to-eat rice products and various Korean rice determined by a highly sensitive gas chromatography-tandem mass spectrometry. Food Chem. 2018;240:1179–1183. doi: 10.1016/j.foodchem.2017.08.053. [DOI] [PubMed] [Google Scholar]
- 8.Malik A.K., Blasco C., Picó Y. Liquid chromatography-mass spectrometry in food safety. J. Chromatogr. A. 2010;1217:4018–4040. doi: 10.1016/j.chroma.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 9.Djedjibegovic J., Larssen T., Skrbo A., et al. Contents of cadmium, copper, mercury and lead in fish from the Neretva river (Bosnia and Herzegovina) determined by inductively coupled plasma mass spectrometry (ICP-MS) Food Chem. 2012;131:469–476. [Google Scholar]
- 10.Chen X., Lin M., Sun L., et al. Detection and quantification of carbendazim in Oolong tea by surface-enhanced Raman spectroscopy and gold nanoparticle substrates. Food Chem. 2019;293:271–277. doi: 10.1016/j.foodchem.2019.04.085. [DOI] [PubMed] [Google Scholar]
- 11.Miaw C.S.W., Sena M.M., de Souza S.V.C., et al. Detection of adulterants in grape nectars by attenuated total reflectance Fourier-transform mid-infrared spectroscopy and multivariate classification strategies. Food Chem. 2018;266:254–261. doi: 10.1016/j.foodchem.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Cheng X., Vella A., Stasiewicz M.J. Classification of aflatoxin contaminated single corn kernels by ultraviolet to near infrared spectroscopy. Food Control. 2019;98:253–261. [Google Scholar]
- 13.Dincer C., Bruch R., Kling A., et al. Multiplexed point-of-care testing - xPOCT. Trends Biotechnol. 2017;35:728–742. doi: 10.1016/j.tibtech.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luppa P.B., Müller C., Schlichtiger A., et al. Point-of-care testing (POCT): Current techniques and future perspectives. Trends Analyt. Chem. 2011;30:887–898. doi: 10.1016/j.trac.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Geng Z., Fan Z., et al. Point-of-care testing based on smartphone: The current state-of-the-art (2017-2018) Biosens. Bioelectron. 2019;132:17–37. doi: 10.1016/j.bios.2019.01.068. [DOI] [PubMed] [Google Scholar]
- 16.Bhavadharini B., Mahalakshmi M.M., Maheswari K., et al. Use of capillary blood glucose for screening for gestational diabetes mellitus in resource-constrained settings. Acta Diabetol. 2016;53:91–97. doi: 10.1007/s00592-015-0761-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang W., Yang J., Wang F., et al. Thiocholine-triggered reaction in personal glucose meters for portable quantitative detection of organophosphorus pesticide. Anal. Chim. Acta. 2019;1060:97–102. doi: 10.1016/j.aca.2019.01.051. [DOI] [PubMed] [Google Scholar]
- 18.Xu X., Liang K., Zeng J. Highly sensitive and portable mercury(ii) ion sensor using personal glucose meter. Anal. Methods. 2015;7:81–85. [Google Scholar]
- 19.Xiao W., Gao Y., Zhang Y., et al. Enhanced 3D paper-based devices with a personal glucose meter for highly sensitive and portable biosensing of silver ion. Biosens. Bioelectron. 2019;137:154–160. doi: 10.1016/j.bios.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Xu Z., Liu Z., Xiao M., et al. A smartphone-based quantitative point-of-care testing (POCT) system for simultaneous detection of multiple heavy metal ions. Chem. Eng. J. 2020;394 [Google Scholar]
- 21.Qiu S., Yuan L., Wei Y., et al. DNA template-mediated click chemistry-based portable signal-on sensor for ochratoxin A detection. Food Chem. 2019;297 doi: 10.1016/j.foodchem.2019.05.203. [DOI] [PubMed] [Google Scholar]
- 22.Aggidis A.G.A., Newman J.D., Aggidis G.A. Investigating pipeline and state of the art blood glucose biosensors to formulate next steps. Biosens. Bioelectron. 2015;74:243–262. doi: 10.1016/j.bios.2015.05.071. [DOI] [PubMed] [Google Scholar]
- 23.Xiang Y., Lu Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rousseau G., Asmolov R., Grammatico-Guillon L., et al. Rapid detection of bacterial meningitis using a point-of-care glucometer. Eur. J. Emerg. Med. 2019;26:41–46. doi: 10.1097/MEJ.0000000000000495. [DOI] [PubMed] [Google Scholar]
- 25.Chavali R., Kumar Gunda N.S., Naicker S., et al. Detection of Escherichia coli in potable water using personal glucose meters. Anal. Methods. 2014;6:6223–6227. [Google Scholar]
- 26.Kwon D., Lee H., Yoo H., et al. Facile method for enrofloxacin detection in milk using a personal glucose meter. Sens Actuators B Chem. 2018;254:935–939. [Google Scholar]
- 27.Newman J.D., Turner A.P.F. Home blood glucose biosensors: A commercial perspective. Biosens. Bioelectron. 2005;20:2435–2453. doi: 10.1016/j.bios.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Wu L., Li G., Xu X., et al. Application of nano-ELISA in food analysis: Recent advances and challenges. Trends Analyt. Chem. 2019;113:140–156. [Google Scholar]
- 29.Li G., Zhang X., Zheng F., et al. Emerging nanosensing technologies for the detection of beta-agonists. Food Chem. 2020;332 doi: 10.1016/j.foodchem.2020.127431. [DOI] [PubMed] [Google Scholar]
- 30.Quesada-González D., Merkoçi A. Nanomaterial-based devices for point-of-care diagnostic applications. Chem. Soc. Rev. 2018;47:4697–4709. doi: 10.1039/c7cs00837f. [DOI] [PubMed] [Google Scholar]
- 31.Ratautas D., Ramonas E., Marcinkevičienė L., et al. Wiring gold nanoparticles and redox enzymes: A self-sufficient nanocatalyst for the direct oxidation of carbohydrates with molecular oxygen. ChemCatChem. 2018;10:971–974. [Google Scholar]
- 32.Tiwari J.N., Tiwari R.N., Kim K.S. Zero-dimensional, one-dimensional, two-dimensional and three-dimensional nanostructured materials for advanced electrochemical energy devices. Prog. Mater. Sci. 2012;57:724–803. [Google Scholar]
- 33.Chen Y., Zhou S., Li L., et al. Nanomaterials-based sensitive electrochemiluminescence biosensing. Nano Today. 2017;12:98–115. [Google Scholar]
- 34.Zhou Y.L., Yang M., Sun K., et al. Similar topological origin of chiral centers in organic and nanoscale inorganic structures: Effect of stabilizer chirality on optical isomerism and growth of CdTe nanocrystals. J. Am. Chem. Soc. 2010;132:6006–6013. doi: 10.1021/ja906894r. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Y., Zhu Z., Huang W., et al. Optical coupling between chiral biomolecules and semiconductor nanoparticles: Size-dependent circular dichroism absorption. Angew. Chem. Int. Ed. Engl. 2011;50:11456–11459. doi: 10.1002/anie.201103762. [DOI] [PubMed] [Google Scholar]
- 36.Li Z., Zhu Z., Liu W., et al. Reversible plasmonic circular dichroism of Au nanorod and DNA assemblies. J. Am. Chem. Soc. 2012;134:3322–3325. doi: 10.1021/ja209981n. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y., Asiri A.M., Tang Z., et al. Graphene based materials for biomedical applications. Mater. Today. 2013;16:365–373. [Google Scholar]
- 38.Zhou Y., Tian X.L., Li Y.S., et al. An enhanced ELISA based on modified colloidal gold nanoparticles for the detection of Pb(II) Biosens. Bioelectron. 2011;26:3700–3704. doi: 10.1016/j.bios.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Sun F., Sun X., Jia Y., et al. Ultrasensitive detection of prostate specific antigen using a personal glucose meter based on DNA-mediated immunoreaction. Analyst. 2019;144:6019–6024. doi: 10.1039/c9an01558b. [DOI] [PubMed] [Google Scholar]
- 40.Croissant J.G., Fatieiev Y., Almalik A., et al. Mesoporous silica and organosilica nanoparticles: Physical chemistry, biosafety, delivery strategies, and biomedical applications. Adv. Healthc. Mater. 2018;7 doi: 10.1002/adhm.201700831. [DOI] [PubMed] [Google Scholar]
- 41.Ye L., Zhao G., Dou W. An ultrasensitive sandwich immunoassay with a glucometer readout for portable and quantitative detection of Cronobacter sakazakii. Anal. Methods. 2017;9:6286–6292. [Google Scholar]
- 42.Luo Y., Dou W., Zhao G. Rapid electrochemical quantification of Salmonella Pullorum and Salmonella Gallinarum based on glucose oxidase and antibody-modified silica nanoparticles. Anal. Bioanal. Chem. 2017;409:4139–4147. doi: 10.1007/s00216-017-0361-3. [DOI] [PubMed] [Google Scholar]
- 43.Ye L., Zhao G., Dou W. An electrochemical immunoassay for Escherichia coli O157:H7 using double functionalized Au@Pt/ SiO(2) nanocomposites and immune magnetic nanoparticles. Talanta. 2018;182:354–362. doi: 10.1016/j.talanta.2018.01.095. [DOI] [PubMed] [Google Scholar]
- 44.Bai G., Xu X., Dai Q., et al. An electrochemical enzymatic nanoreactor based on dendritic mesoporous silica nanoparticles for living cell H2O2 detection. Analyst. 2019;144:481–487. doi: 10.1039/c8an01712c. [DOI] [PubMed] [Google Scholar]
- 45.Y. Wang, Y. Yang, T. Wu, et al., Dendritic porous silica nanoparticles with high-curvature structures for a dual-mode DNA sensor based on fluorometer and person glucose meter, Microchim. Acta 188 (2021), 407. [DOI] [PubMed]
- 46.L. Yang, Y. Wang, C. Yao, et al., Highly sensitive and portable aptasensor by using enzymatic nanoreactors as labels, Microchem. J. 168 (2021), 106407.
- 47.Chen Y., Xianyu Y., Jiang X. Surface modification of gold nanoparticles with small molecules for biochemical analysis. Acc. Chem. Res. 2017;50:310–319. doi: 10.1021/acs.accounts.6b00506. [DOI] [PubMed] [Google Scholar]
- 48.Li F., Zhang R., Kang H., et al. Facile and sensitive detection of clenbuterol in pork using a personal glucose meter. Anal. Methods. 2017;9:6507–6512. [Google Scholar]
- 49.Gao S., Hao J., Su D., et al. Facile and sensitive detection of norfloxacin in animal-derived foods using immuno-personal glucose meter. Eur. Food Res. Technol. 2021;247:2635–2644. [Google Scholar]
- 50.Chen S., Zhang J., Gan N., et al. An on-site immunosensor for ractopamine based on a personal glucose meter and using magnetic β-cyclodextrin-coated nanoparticles for enrichment, and an invertase-labeled nanogold probe for signal amplification. Microchim. Acta. 2014;182:815–822. [Google Scholar]
- 51.Chen S., Gan N., Zhang H., et al. A portable and antibody-free sandwich assay for determination of chloramphenicol in food based on a personal glucose meter. Anal. Bioanal. Chem. 2015;407:2499–2507. doi: 10.1007/s00216-015-8478-8. [DOI] [PubMed] [Google Scholar]
- 52.Zhao L., Teng L., Zhang J., et al. Point-of-care detection of microcystin-LR with a personal glucose meter in drinking water source. Chin. Chem. Lett. 2019;30:1035–1037. [Google Scholar]
- 53.Ye R., Zhu C., Song Y., et al. One-pot bioinspired synthesis of all-inclusive protein–protein nanoflowers for point-of-care bioassay: Detection of E. coli O157:H7 from milk. Nanoscale. 2016;8:18980–18986. doi: 10.1039/c6nr06870g. [DOI] [PubMed] [Google Scholar]
- 54.Bai H., Bu S., Wang C., et al. Sandwich immunoassay based on antimicrobial peptide-mediated nanocomposite pair for determination of Escherichia coli O157:H7 using personal glucose meter as readout. Mikrochim. Acta. 2020;187:220. doi: 10.1007/s00604-020-4200-4. [DOI] [PubMed] [Google Scholar]
- 55.Vidal B.C., Jr., Deivaraj T.C., Yang J., et al. Stability and hybridization-driven aggregation of silver nanoparticle–oligonucleotide conjugates. New J. Chem. 2005;29:812–816. [Google Scholar]
- 56.Herdt A.R., Drawz S.M., Kang Y., et al. DNA dissociation and degradation at gold nanoparticle surfaces. Colloids Surf. B Biointerfaces. 2006;51:130–139. doi: 10.1016/j.colsurfb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Qiao F., Liu J., Li F., et al. Antibody and DNA dual-labeled gold nanoparticles: Stability and reactivity. Appl. Surf. Sci. 2008;254:2941–2946. [Google Scholar]
- 58.Seeman N.C., Sleiman H.F. DNA nanotechnology. Nat. Rev. Mater. 2017;3 [Google Scholar]
- 59.Zhao Y., Zuo X., Li Q., et al. Nucleic acids analysis. Sci. China Chem. 2021;64:171–203. doi: 10.1007/s11426-020-9864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du P., Jin M., Chen G., et al. A competitive bio-barcode amplification immunoassay for small molecules based on nanoparticles. Sci. Rep. 2016;6 doi: 10.1038/srep38114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du Y., Zhou Y., Wen Y., et al. Multiplexed aptasensing of food contaminants by using terminal deoxynucleotidyl transferase-produced primer-triggered rolling circle amplification: Application to the colorimetric determination of enrofloxacin, lead (II), Escherichia coli O157:H7 and tropomyosin. Mikrochim. Acta. 2019;186:840. doi: 10.1007/s00604-019-3935-2. [DOI] [PubMed] [Google Scholar]
- 62.X. Xiang, Y. Shang, F. Li, et al., A microfluidic genoserotyping strategy for fast and objective identification of common Salmonella serotypes isolated from retail food samples in China, Anal. Chim. Acta 1201 (2022), 339657. [DOI] [PubMed]
- 63.Yin J., Liu Y., Wang S., et al. Engineering a universal and label-free evaluation method for mycotoxins detection based on strand displacement amplification and G-quadruplex signal amplification. Sens. Actuators B Chem. 2018;256:573–579. [Google Scholar]
- 64.Y. Wang, A.M. Abd El-Aty, G. Chen, et al., A competitive immunoassay for detecting triazophos based on fluorescent catalytic hairpin self-assembly, Microchim. Acta 189 (2022), 114. [DOI] [PMC free article] [PubMed]
- 65.Guo Q., Han J., Shan S., et al. DNA-based hybridization chain reaction and biotin-streptavidin signal amplification for sensitive detection of Escherichia coli O157:H7 through ELISA. Biosens. Bioelectron. 2016;86:990–995. doi: 10.1016/j.bios.2016.07.049. [DOI] [PubMed] [Google Scholar]
- 66.Kim H.Y., Park K.S., Park H.G. Glucose oxidase-like activity of cerium oxide nanoparticles: Use for personal glucose meter-based label-free target DNA detection. Theranostics. 2020;10:4507–4514. doi: 10.7150/thno.41484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qi H., Yue S., Bi S., et al. Isothermal exponential amplification techniques: From basic principles to applications in electrochemical biosensors. Biosens. Bioelectron. 2018;110:207–217. doi: 10.1016/j.bios.2018.03.065. [DOI] [PubMed] [Google Scholar]
- 68.X. Xia, H. Yang, J. Cao, et al., Isothermal nucleic acid amplification for food safety analysis, Trends Analyt. Chem. 153 (2022), 116641.
- 69.Y. Yang, T. Wu, L.P. Xu, et al., Portable detection of Staphylococcus aureus using personal glucose meter based on hybridization chain reaction strategy, Talanta 226 (2021), 122132. [DOI] [PubMed]
- 70.Gu Y., Yang X., Hu S., et al. Sensitive glucometer-based microfluidic immune-sensing platform via DNA signal amplification coupled with enzymatic reaction. Sens. Actuators B Chem. 2021;329 [Google Scholar]
- 71.Guo L., Lu B., Dong Q., et al. One-tube smart genetic testing via coupling isothermal amplification and three-way nucleic acid circuit to glucometers. Anal. Chim. Acta. 2020;1106:191–198. doi: 10.1016/j.aca.2020.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao Y., Chen F., Li Q., et al. Isothermal amplification of nucleic acids. Chem. Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 73.McConnell E.M., Cozma I., Mou Q., et al. Biosensing with DNAzymes. Chem. Soc. Rev. 2021;50:8954–8994. doi: 10.1039/d1cs00240f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yin H., Kuang H., Liu L., et al. A ligation DNAzyme-induced magnetic nanoparticles assembly for ultrasensitive detection of copper ions. ACS Appl. Mater. Interfaces. 2014;6:4752–4757. doi: 10.1021/am405482a. [DOI] [PubMed] [Google Scholar]
- 75.Liu C., Hu Y., Pan Q., et al. A microRNA-triggered self-powered DNAzyme walker operating in living cells. Biosens. Bioelectron. 2019;136:31–37. doi: 10.1016/j.bios.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 76.Li Y.F., Breaker R.R. Phosphorylating DNA with DNA. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2746–2751. doi: 10.1073/pnas.96.6.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brandsen B.M., Hesser A.R., Castner M.A., et al. DNA-catalyzed hydrolysis of esters and aromatic amides. J. Am. Chem. Soc. 2013;135:16014–16017. doi: 10.1021/ja4077233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ming J., Fan W., Jiang T.-F., et al. Portable and sensitive detection of copper(II) ion based on personal glucose meters and a ligation DNAzyme releasing strategy. Sens. Actuators B Chem. 2017;240:1091–1098. [Google Scholar]
- 79.Xiang Y., Lu Y. An invasive DNA approach toward a general method for portable quantification of metal ions using a personal glucose meter. Chem. Commun. 2013;49:585–587. doi: 10.1039/c2cc37156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang X., Shi D., Zhu S., et al. Portable aptasensor of aflatoxin B1 in bread based on a personal glucose meter and DNA walking machine. ACS Sens. 2018;3:1368–1375. doi: 10.1021/acssensors.8b00304. [DOI] [PubMed] [Google Scholar]
- 81.Zhang S., Luan Y., Xiong M., et al. DNAzyme amplified aptasensing platform for ochratoxin A detection using a personal glucose meter. ACS Appl. Mater. Interfaces. 2021;13:9472–9481. doi: 10.1021/acsami.0c20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeng L., Gong J., Rong P., et al. A portable and quantitative biosensor for cadmium detection using glucometer as the point-of-use device. Talanta. 2019;198:412–416. doi: 10.1016/j.talanta.2019.02.045. [DOI] [PubMed] [Google Scholar]
- 83.Fu X., Xu K., Ye J., et al. Glucoamylase-labeled nanogold flowers for in situ enhanced sensitivity of a glucometer-based enzyme immunoassay. Anal. Methods. 2015;7:507–512. [Google Scholar]
- 84.Taebi S., Keyhanfar M., Noorbakhsh A. A novel method for sensitive, low-cost and portable detection of hepatitis B surface antigen using a personal glucose meter. J. Immunol. Methods. 2018;458:26–32. doi: 10.1016/j.jim.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 85.Lin B., Liu D., Yan J., et al. Enzyme-encapsulated liposome-linked immunosorbent assay enabling sensitive personal glucose meter readout for portable detection of disease biomarkers. ACS Appl. Mater. Interfaces. 2016;8:6890–6897. doi: 10.1021/acsami.6b00777. [DOI] [PubMed] [Google Scholar]
- 86.Wang Z., Chen Z., Gao N., et al. Transmutation of personal glucose meters into portable and highly sensitive microbial pathogen detection platform. Small. 2015;11:4970–4975. doi: 10.1002/smll.201500944. [DOI] [PubMed] [Google Scholar]
- 87.Reddy A., Maley F. Studies on identifying the catalytic role of Glu-204 in the active site of yeast invertase. J. Biol. Chem. 1996;271:13953–13957. doi: 10.1074/jbc.271.24.13953. [DOI] [PubMed] [Google Scholar]
- 88.F. Yin, R. Cai, S. Gui, et al., A portable and quantitative detection of microRNA-21 based on cascade enzymatic reactions with dual signal outputs, Talanta 235 (2021), 122802. [DOI] [PubMed]
- 89.Tang D., Lin Y., Zhou Q., et al. Low-cost and highly sensitive immunosensing platform for aflatoxins using one-step competitive displacement reaction mode and portable glucometer-based detection. Anal. Chem. 2014;86:11451–11458. doi: 10.1021/ac503616d. [DOI] [PubMed] [Google Scholar]
- 90.Gao Z., Tang D., Xu M., et al. Nanoparticle-based pseudo hapten for target-responsive cargo release from a magnetic mesoporous silica nanocontainer. Chem. Commun. 2014;50:6256–6258. doi: 10.1039/c4cc01511h. [DOI] [PubMed] [Google Scholar]
- 91.Fu L., Zhuang J., Lai W., et al. Portable and quantitative monitoring of heavy metal ions using DNAzyme-capped mesoporous silica nanoparticles with a glucometer readout. J. Mater. Chem. B. 2013;1:6123–6128. doi: 10.1039/c3tb21155j. [DOI] [PubMed] [Google Scholar]
- 92.Liang X., Wang L., Wang D., et al. Portable and quantitative monitoring of mercury ions using DNA-gated mesoporous silica nanoparticles using a glucometer readout. Chem. Commun. 2016;52:2192–2194. doi: 10.1039/c5cc08611f. [DOI] [PubMed] [Google Scholar]
- 93.Tang J., Huang Y., Liu H., et al. Novel glucometer-based immunosensing strategy suitable for complex systems with signal amplification using surfactant-responsive cargo release from glucose-encapsulated liposome nanocarriers. Biosens. Bioelectron. 2016;79:508–514. doi: 10.1016/j.bios.2015.12.097. [DOI] [PubMed] [Google Scholar]
- 94.Nie D., Zhang Z., Guo D., et al. A flexible assay strategy for non-glucose targets based on sulfhydryl-terminated liposomes combined with personal glucometer. Biosens. Bioelectron. 2021;175 doi: 10.1016/j.bios.2020.112884. [DOI] [PubMed] [Google Scholar]
- 95.Shao Y., Jia H., Cao T., et al. Supramolecular hydrogels based on DNA self-assembly. Acc. Chem. Res. 2017;50:659–668. doi: 10.1021/acs.accounts.6b00524. [DOI] [PubMed] [Google Scholar]
- 96.Wang P.L., Xie L.H., Joseph E.A., et al. Metal–organic frameworks for food safety. Chem. Rev. 2019;119:10638–10690. doi: 10.1021/acs.chemrev.9b00257. [DOI] [PubMed] [Google Scholar]
- 97.Yan L., Zhu Z., Zou Y., et al. Target-responsive "sweet" hydrogel with glucometer readout for portable and quantitative detection of non-glucose targets. J. Am. Chem. Soc. 2013;135:3748–3751. doi: 10.1021/ja3114714. [DOI] [PubMed] [Google Scholar]
- 98.Y. Cao, F. Mo, Y. Liu, et al., Portable and sensitive detection of non-glucose target by enzyme-encapsulated metal-organic-framework using personal glucose meter, Biosens. Bioelectron. 198 (2022), 113819. [DOI] [PubMed]
- 99.Burtch N.C., Jasuja H., Walton K.S. Water stability and adsorption in metal-organic frameworks. Chem. Rev. 2014;114:10575–10612. doi: 10.1021/cr5002589. [DOI] [PubMed] [Google Scholar]
- 100.Liu N., Nie D., Zhao Z., et al. Ultrasensitive immunoassays based on biotin–streptavidin amplified system for quantitative determination of family zearalenones. Food Control. 2015;57:202–209. [Google Scholar]
- 101.Shao Y., Duan H., Zhou S., et al. Biotin-streptavidin system-mediated ratiometric multiplex immunochromatographic assay for simultaneous and accurate quantification of three mycotoxins. J. Agric. Food Chem. 2019;67:9022–9031. doi: 10.1021/acs.jafc.9b03222. [DOI] [PubMed] [Google Scholar]
- 102.Dundas C.M., Demonte D., Park S. Streptavidin-biotin technology: Improvements and innovations in chemical and biological applications. Appl. Microbiol. Biotechnol. 2013;97:9343–9353. doi: 10.1007/s00253-013-5232-z. [DOI] [PubMed] [Google Scholar]
- 103.Weber P.C., Ohlendorf D.H., Wendoloski J.J., et al. Structural origins of high-affinity biotin binding to streptavidin. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 104.Tiwary P. Molecular determinants and bottlenecks in the dissociation dynamics of biotin–streptavidin. J. Phys. Chem. B. 2017;121:10841–10849. doi: 10.1021/acs.jpcb.7b09510. [DOI] [PubMed] [Google Scholar]
- 105.Li F., Li X., Zhu N., et al. An aptasensor for the detection of ampicillin in milk using a personal glucose meter. Anal. Methods. 2020;12:3376–3381. doi: 10.1039/d0ay00256a. [DOI] [PubMed] [Google Scholar]
- 106.Huang D., Shi Z., Qian J., et al. A CRISPR-Cas12a-derived biosensor enabling portable personal glucose meter readout for quantitative detection of SARS-CoV-2. Biotechnol. Bioeng. 2021;118:1587–1596. doi: 10.1002/bit.27673. [DOI] [PubMed] [Google Scholar]
- 107.Zhou C., Huang D., Wang Z., et al. CRISPR Cas12a-based "sweet" biosensor coupled with personal glucose meter readout for the point-of-care testing of Salmonella. J. Food Sci. 2022;87:4137–4147. doi: 10.1111/1750-3841.16287. [DOI] [PubMed] [Google Scholar]
- 108.Martínez-Periñán E., García-Mendiola T., Enebral-Romero E., et al. A MoS(2) platform and thionine-carbon nanodots for sensitive and selective detection of pathogens. Biosens. Bioelectron. 2021;189 doi: 10.1016/j.bios.2021.113375. [DOI] [PubMed] [Google Scholar]
- 109.Yin L., Man S., Ye S., et al. CRISPR-Cas based virus detection: Recent Advances and perspectives. Biosens. Bioelectron. 2021;193 doi: 10.1016/j.bios.2021.113541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.B. Fang, Z. Jia, C. Liu, et al., A versatile CRISPR Cas12a-based point-of-care biosensor enabling convenient glucometer readout for ultrasensitive detection of pathogen nucleic acids, Talanta 249 (2022), 123657. [DOI] [PubMed]
- 111.Peavy R.D., Metcalfe D.D. Understanding the mechanisms of anaphylaxis. Curr. Opin. Allergy Clin. Immunol. 2008;8:310–315. doi: 10.1097/ACI.0b013e3283036a90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morais S., Tortajada-Genaro L.A., Maquieira Á., et al. Biosensors for food allergy detection according to specific IgE levels in serum. Trends Analyt. Chem. 2020;127 [Google Scholar]
- 113.Ahn J.K., Kim H.Y., Lee C.Y., et al. Label-free and washing-free alkaline phosphatase assay using a personal glucose meter. J. Biol. Eng. 2019;13 doi: 10.1186/s13036-019-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.H. Han, J. Park, J.K. Ahn, Immunoglobulin E detection method based on cascade enzymatic reaction utilizing portable personal glucose meter, Sensors (Basel) 21 (2021), 6396. [DOI] [PMC free article] [PubMed]
- 115.Zhao Y., Chen X., Lin S., et al. Integrated immunochromatographic strip with glucometer readout for rapid quantification of phosphorylated proteins. Anal. Chim. Acta. 2017;964:1–6. doi: 10.1016/j.aca.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 116.Huang H., Zhao G., Dou W. Portable and quantitative point-of-care monitoring of Escherichia coli O157:H7 using a personal glucose meter based on immunochromatographic assay. Biosens. Bioelectron. 2018;107:266–271. doi: 10.1016/j.bios.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 117.Zhang J., Xiang Y., Wang M., et al. Dose-dependent response of personal glucose meters to nicotinamide coenzymes: Applications to point-of-care diagnostics of many non-glucose targets in a single step. Angew. Chem. Int. Ed. Engl. 2016;55:732–736. doi: 10.1002/anie.201507563. [DOI] [PMC free article] [PubMed] [Google Scholar]