Abstract
Plant growth‐promoting bacteria (PGPB) may be of use for increasing crop yield and plant resilience to biotic and abiotic stressors. Using hyperspectral reflectance data to assess growth‐related traits may shed light on the underlying genetics as such data can help assess biochemical and physiological traits. This study aimed to integrate hyperspectral reflectance data with genome‐wide association analyses to examine maize growth‐related traits under PGPB inoculation. A total of 360 inbred maize lines with 13,826 single nucleotide polymorphisms (SNPs) were evaluated with and without PGPB inoculation; 150 hyperspectral wavelength reflectances at 386–1021 nm and 131 hyperspectral indices were used in the analysis. Plant height, stalk diameter, and shoot dry mass were measured manually. Overall, hyperspectral signatures produced similar or higher genomic heritability estimates than those of manually measured phenotypes, and they were genetically correlated with manually measured phenotypes. Furthermore, several hyperspectral reflectance values and spectral indices were identified by genome‐wide association analysis as potential markers for growth‐related traits under PGPB inoculation. Eight SNPs were detected, which were commonly associated with manually measured and hyperspectral phenotypes. Different genomic regions were found for plant growth and hyperspectral phenotypes between with and without PGPB inoculation. Moreover, the hyperspectral phenotypes were associated with genes previously reported as candidates for nitrogen uptake efficiency, tolerance to abiotic stressors, and kernel size. In addition, a Shiny web application was developed to explore multiphenotype genome‐wide association results interactively. Taken together, our results demonstrate the usefulness of hyperspectral‐based phenotyping for studying maize growth‐related traits in response to PGPB inoculation.
Keywords: genome‐wide association analysis, growth trait, hyperspectral wavelength, multiphenotype
Abbreviations
- B+
with plant growth‐promoting bacterial inoculation
- B−
without plant growth‐promoting bacterial inoculation
- GWA
genome‐wide association
- PGPB
plant growth‐promoting bacteria
- PH
plant height
- SD
stalk diameter
- SDM
shoot dry mass
- SNPs
single nucleotide polymorphisms
1. INTRODUCTION
Sustainably increasing food production is required considering growing demands, especially in developing nations (Laurance et al., 2014). Recent studies found that plant growth‐promoting bacteria (PGPB) are a viable option for increasing plant resilience to biotic and abiotic stressors, with the potential to increase food production (Batista et al., 2018; Compant et al., 2005; Yassue et al., 2021). PGPB can promote morphological (Mantelin, 2003) and functional (Di Benedetto et al., 2017) changes in plants. Reported effects include increased uptakes of nutrients, such as nitrogen, phosphate, potassium, and iron (Egamberdiyeva, 2007; Pii et al., 2015), and activation of responses to pathogens and abiotic stressors (Olanrewaju et al., 2017; Singh et al., 2018).
One of the challenges associated with assessing the possible benefits of PGPB is identifying plant response. Hyperspectral image data can be used to assess biochemical or physiological attributes of plants; thus, such data have been increasingly applied in plant genetics and management studies because of their associations with target phenotypes, such as water content (550–1750 nm) (Ge et al., 2016), plant nutrient status (350–2500 nm) (Mahajan et al., 2014; Nigon et al., 2021), disease susceptibility (Thomas et al., 2017), yield (400–900 nm) (Yang et al., 2021), and biomass (380–850 nm) (Krause et al., 2019). For example, hyperspectral patterns genetically correlated with target phenotypes can potentially aid genomic prediction (Krause et al., 2019; Sandhu et al., 2021). In addition, hyperspectral phenotypes can be used for genetic inference studies, such as genome‐wide association (GWA), heritability, and genetic correlation analyses, to investigate associations between hyperspectral bands and genome (Barnaby et al., 2020; Feng et al., 2017; Sun et al., 2019; Wu et al., 2021).
Bayesian whole‐genome regression models are useful for GWA studies because they implicitly account for population structure and the multiple‐testing problem of classical single‐marker linear mixed models by simultaneously fitting all markers (Fernando et al., 2017; Wolc & Dekkers, 2022). Despite the increasing use of high‐throughput phenotyping through the compilation of hundreds or thousands of phenotypes, a limited number of whole‐genome regression studies have integrated hyperspectral data into genetic inference research (Barnaby et al., 2020; Sun et al., 2019; Yoosefzadeh‐Najafabadi et al., 2021). Moreover, how hyperspectral wavelength data are associated with PGPB responses in maize remains elusive because it is challenging to interpret changes in hyperspectral reflectance patterns with regard to plant biological processes. The objectives of this study were (1) to investigate whether hyperspectral reflectance values are under genomic control, (2) to determine whether variations in hyperspectral reflectance values are correlated with growth‐related traits at the genomic level, (3) to identify specific genomic regions associated with wavelengths that can be used to study maize growth‐related traits under PGPB inoculation, and (4) develop an open‐source a Shiny web application to explore multiphenotype GWA results interactively. We employed Bayesian whole‐genome GWA methods to identify possible candidate genes associated with growth‐related traits and hyperspectral reflectance bands.
2. MATERIALS AND METHODS
2.1. PGPB experiment
A public tropical maize association panel comprising 360 inbred lines was used to examine genetic basis of PGPB responses under greenhouse conditions. The inbred lines were evaluated under nitrogen stress with (B+) and without (B−) PGPB inoculation. Each experimental unit consisted of one pot containing one plant. Maize seeds were co‐inoculated with the PGPB strains Bacillus thuringiensis RZ2MS9, Delftia sp. RZ4MS18 (Batista et al., 2018, 2021), Pantoea agglomerans 33.1 (Quecine et al., 2012), and Azospirillum brasilense Ab‐v5 (Hungria et al., 2010). Each of the PGPB species was adjusted to the concentration of 108 colony‐forming units/mL. B− management constituted treatment with liquid Luria–Bertani medium only. A plot, including three seeds in an individual plastic bag, was inoculated with 1 mL of the respective management (B+ or B−), homogenized, and seeded. Irrigation, weed control, and application of fertilizer (excluding nitrogen) were carried out according to crop requirements. After germination, each plot was thinned to only one plant per plot. The plants were evaluated when most had six expanded leaves, that is, approximately 33 days after sowing. Manually measured phenotypes included plant height (PH), stalk diameter (SD), and shoot dry mass (SDM). A total of 1512 plots (plants) were evaluated under B+ and B− conditions (756 B+ and 756 B−). Detailed information on the experimental design can be found in Yassue (2021, 2022).
2.2. Genomic data
The genomic data used in this study have been previously published (Fritsche‐Neto et al., 2020; Yassue et al., 2021). In brief, the 360 inbred lines were genotyped using the genotyping‐by‐sequencing method, followed by a two‐enzyme (PstI and MseI) protocol (Poland et al., 2012; Sim et al., 2012). Deoxyribonucleic acid was extracted from leaves using cetyltrimethylammonium bromide (Doyle & Doyle, 1987). Single nucleotide polymorphism (SNP) calling was performed using TASSEL 5.0 (Bradbury et al., 2007) with B73 (B73‐RefGen_v4) as a reference genome. SNP markers were removed if the call rate was < 90%, nonbiallelic, or if the minor allele frequency was < 5%. Missing marker codes were imputed using Beagle 5.0 software (Browning et al., 2018). Markers with pairwise linkage disequilibrium > 0.99 were removed using the SNPRelate R package (Zheng et al., 2012). The average of linkage disequilibrium decay was 185 kbp considering a conservative cutoff of 0.10. In total, 13,826 SNPs were retained after quality control. Of the 360 inbred lines, 179 and 181 were collected from the plant breeding program from the Luiz de Queiroz College of Agriculture‐University of São Paulo (ESALQ‐USP) and from the Instituto de Desenvolvimento Rural do Paraná (IAPAR), respectively. Admixture analysis identified the presence of 18 groups (Yassue et al., 2022).
2.3. Hyperspectral imaging and processing
Leaves of B+ and B− plants were collected, and hyperspectral images were recorded using a benchtop Pika L. camera system (Resonon, Bozeman, MT, USA). The middle portion of the last completely expanded leaf was used as the region of interest for hyperspectral imaging. A dark room with an additional light supply was used to minimize light variation. Radiometric calibration was performed according to the manufacturer's instructions. For each plant, a hyperspectral cube image was produced, which contained 150 bands with wavelengths in the 386–1021 nm range. Image processing through the Spectral Python (Boggs, 2014) module was performed by applying a mask to remove the background from the image, and the mean reflectance of each pixel was used for further analysis. Hyperspectral imaging and processing details are described in Yassue et al. (2022). In addition, 131 hyperspectral indices (mathematical band combinations) were calculated based on the mean reflectance value for each wavelength using the R package hsdar (Lehnert et al., 2019). These hyperspectral indices have been reported to be associated with a variety of phenotypes, such as nutrient and chlorophyll content, pigments, photosynthesis, and water content (Gitelson et al., 2014; Ranjan et al., 2012; Zarco‐Tejada et al., 2004, 2005). A summary of the hyperspectral indices and their correlations are presented in the supporting information Table S1 and Figure S1, respectively. In total, 281 hyperspectral phenotypes were used in the analysis. Of these, 150 were single‐band reflectance and 131 were hyperspectral indices.
2.4. Univariate BayesC
BayesC (Kizilkaya et al., 2010) was applied to estimate the markers effect and variance components using the following model.
where is the vector of phenotypes (manually measured or hyperspectral phenotypes) for the kth replication, lth block nested within replication, and ith genotype; is the overall mean; and are the fixed effects for replication and block nested within replication, respectively; is the incidence matrices of marker covariates for each SNP coded as 0, 1, or 2; and is the jth marker effect. The prior of was as follows:
where is the common marker genetic variance, is a mixture proportion set to 0.99, and is the vector of residuals. A Gaussian prior was assigned to the vector of residuals, and a flat prior was assigned to , , and . The scaled inverse distribution was assigned to and with the degrees of freedom equal to 4 and choosing the scale parameter such that the prior mean of the variance equals half of the phenotypic variance. The variance components obtained from univariate BayesC were used to estimate genomic heritability , where is the genomic heritability, and are the additive genomic and residual variances, respectively, and is the number of replication (2).
2.5. Bivariate BayesC
A recent study showed that hyperspectral image data can be used to perform phenomic prediction for PH, SD, SDM with reasonable accuracy (Yassue et al., 2022). In the current study, we investigated if these hyperspectral reflectance values are under genetic control and whether they are associated with PH, SD, and SDM. Bivariate BayesC was used to investigate whether they are controlled by the same genomic regions by estimating the genetic correlation between manually measured and hyperspectral phenotypes. The model description follows that of univariate BayesC with some modification. Here, is the vector of manually measured and hyperspectral phenotypes, and the marker effect of trait for locus followed
The jth marker effect can be reparameterized as , where is a diagonal matrix with elements indicating whether the jth marker effect for trait is zero or nonzero and follows a multivariate normal distribution with null mean and covariance matrix , where , , and () are marker genetic variance for trait 1, marker genetic variance for trait 2, and marker genetic covariance between traits 1 and 2, respectively, and the residuals were assumed independently and identically distributed multivariate normal vectors with null mean and covariance matrix (Cheng et al., 2018). The covariance matrices, and , were assigned an inverse Wishart prior distribution with and , respectively. We assumed all possible combinations for , namely, (0,0), (0,1), (1,0), and (1,1) having nonzero probability.
2.6. Bayesian GWA analysis
The aforementioned BayesC was used to perform GWA analyses of hyperspectral reflectance values and manually measured phenotypes. Candidate markers were selected based on their posterior inclusion probabilities. The posterior inclusion probability indicates the probability of inclusion of a given marker in the model (Fernando & Garrick, 2013). According to a previous study (Fan et al., 2011), a posterior inclusion probability threshold of 0.10 was used for manually measured phenotypes, and 0.50 was used as a more conservative threshold for hyperspectral GWA. All Bayesian analyses were fit using 60,000 Markov chain Monte Carlo samples, 6000 burn‐ins, and a thinning rate of 60 implemented in JWAS (Cheng et al., 2018). Model convergence was assessed using trace plots of the posterior means of the parameters. For each selected SNP associated with hyperspectral patterns, genes within an interval of 50 kilobase pair (kbp) upstream and downstream of the SNP were explored using the MaizeMine V1.3 server (Shamimuzzaman et al., 2020). One challenge was the large number of generated Manhattan plots owing to the three manually measured phenotypes, 150 hyperspectral wavelengths, and 131 hyperspectral indices. Instead of including all Manhattan plots as supporting information, we developed a Shiny web application using the R package shiny (Chang et al., 2021), which provides functions for constructing interactive web applications. This application allows users to interactively explore all possible genome‐to‐phenome association combinations that are not elaborated here.
3. RESULTS
3.1. Estimates of genomic heritability and correlation
The averages of PH were 15.38 and 14.52 cm, SD were 10.70 and 10.11 mm, and SDM were 2.18 and 1.94 g for B+ and B−, respectively. The genomic heritability estimates of manually measured PH, SD, and SDM were 0.61, 0.60, and 0.30 in B+ plants and 0.57, 0.39, and 0.28 in B− plants, respectively. PH had the highest genomic heritability estimates, whereas SDM had the lowest. The means (and standard deviations) of genomic heritability estimates for hyperspectral wavelengths and indices were 0.45 (0.06) and 0.40 (0.08) for B+ and 0.44 (0.04) and 0.43 (0.08) for B−, respectively (Figure 1). The individual hyperspectral wavelengths that explained the largest genomic heritability estimates were 645 and 649 nm (B+) and 512 and 507 nm (B−), respectively. The estimates of genomic heritability showed a decreasing tendency as wavelength increased. Among the hyperspectral indices, NDNI and NPQI for B− and EVI for B+ showed the highest genomic heritability estimates. Although the mean of genomic heritability estimates of hyperspectral indices was lower than that of hyperspectral wavelengths, some indices explained more of the genetic variance than individual wavelengths. Overall, similar genomic heritability estimates were observed in B+ and B− plants for most wavelengths or indices.
FIGURE 1.
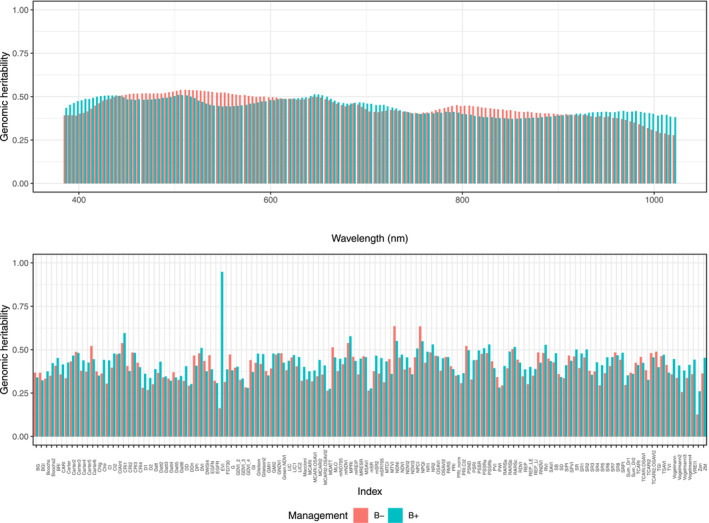
Genomic heritability for 150 hyperspectral reflectance values and 131 hyperspectral indices without (B−) and with (B+) plant growth‐promoting bacteria inoculation.
The genomic correlation estimates between hyperspectral wavelengths and manually measured traits were largely positive (Figure 2). Genomic correlation estimates of PH ranged from −0.34 to 0.34. Positive correlations were observed for both B− and B+ at wavelengths from 400–700 nm, whereas negative correlations were observed only for B+ > 700 nm. In absolute terms, the hyperspectral wavelengths that provided the largest genomic correlation estimates were 578 nm (0.286) and 398 nm (0.193) for B− and B+, respectively. The extent of genomic correlation estimates was low in SD. The estimates were mostly positive, except for the beginning (<400 nm) and end (>900 nm) of wavelengths. The hyperspectral wavelengths showed positive correlations throughout the entire spectral range, except at the start of the SDM wavelength. In particular, higher correlations were observed at 700–1000 nm for B−. The hyperspectral wavelengths that provided the largest genomic correlation estimates with SDM were 817 nm (0.392) and 734 nm (0.235) for B− and B+, respectively. In contrast, genomic correlation estimates varied markedly across hyperspectral indices. The extent of the correlation estimates was lower regarding SD, compared with those of PH and SDM. The hyperspectral indices that provided the largest genomic correlation estimates with PH were D2 (0.339) and RDVI (−0.292) for B− and B+, respectively, whereas those with SDM were RARSb (−0.357) and NPQI (−0.340) for B− and B+, respectively.
FIGURE 2.
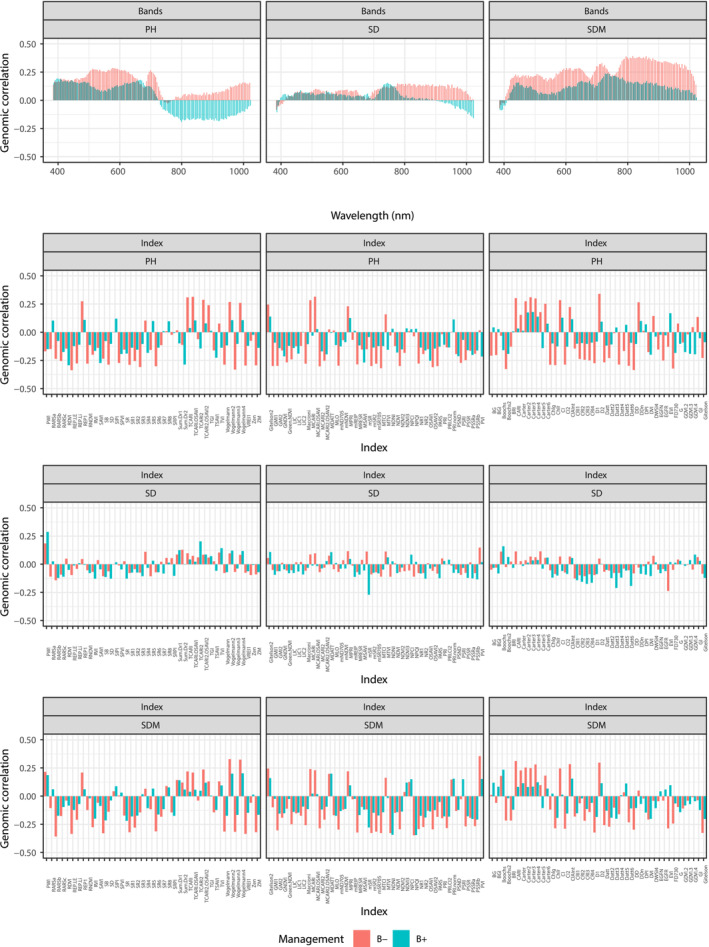
Genomic correlations between manually measured phenotypes and hyperspectral reflectance values and hyperspectral indices under without (B−) and with (B+) plant growth‐promoting bacteria inoculation.
3.2. GWA analyses for growth‐related and hyperspectral traits
A total of 86 SNPs were selected from the BayesC analysis using the posterior inclusion probability threshold of 0.10 for PH, SD, and SDM (Figure 3 and supporting information Tables S2–S6). This number was higher than a previous study (Yassue et al., 2022) that used a non‐Bayesian whole‐genome regression model likely because BayesC has a higher statistical power to detect genetic signals. There were eleven common SNPs between the current study and Yassue et al. (2022) (Table S7). PH showed the highest number of selected markers (21 and 24 for B+ and B−, respectively), whereas SDM had the lowest (5 for B+); no SDM markers were detected in B−. No overlapping SNPs were identified across manually measured phenotypes, whereas only four SNPs were selected for both PGPB inoculation conditions (B− and B+), indicating that PGPB may alter plant growth patterns and the genomic regions controlling them. A conservative posterior inclusion probability threshold of 0.50 was used to find SNPs associated with the hyperspectral‐derived phenotypes and candidate genes. Of the 25 detected SNPs, five SNPs were associated with at least five different hyperspectral phenotypes. Gene annotation of each selected SNP within an interval of 100 kbp showed the presence of genes that have been previously reported as related to growth‐related phenotypes or responses to abiotic stressors (Table 1). The hyperspectral indices Chlg, CRI2, CRI3, CRI4, Datt6, GMI1, PARS, SD, and SR3 were associated with genes nrt2, nrt2.2, and Zm00001d054060 on chromosome 4. The index CRI1 was associated with gene Zm00001d012924 on chromosome 5, and the EVI index was associated with genes Zm00001d029820 and Zm00001d007843 on chromosomes 1 and 2, respectively, and 100275163 on chromosome 6. In addition, hyperspectral wavelengths from 398 to 434 nm were associated with gene Zm00001d012719 on chromosome 8.
FIGURE 3.
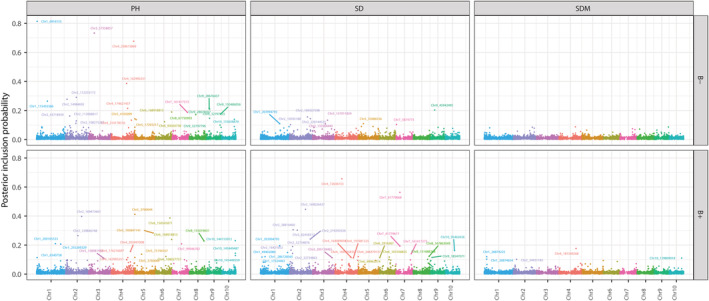
Genome‐wide association analysis of manually measured phenotypes without (B−) and with (B+) plant growth‐promoting bacteria inoculation. Plant height (PH), stalk diameter (SD), and shoot dry mass (SDM).
TABLE 1.
Selected single nucleotide polymorphisms markers based on BayesC using the posterior inclusion probability threshold of 0.50 for 281 hyperspectral phenotypes under with (B+) or without (B−) plant growth‐promoting bacteria inoculation.
| Management | Chr a | Marker ID b | Pheno c | MAF d | PIP e | NG f | Candidate gene |
|---|---|---|---|---|---|---|---|
| B− | 4 | Chr4_164335549 | 1 | 0.27 | 0.508 | 2 | |
| B− | 8 | Chr8_174418506 | 5 | 0.17 | 0.551 | 9 | |
| B+ | 1 | Chr1_69298773 | 1 | 0.06 | 0.981 | 6 | |
| B+ | 1 | Chr1_80258452 | 1 | 0.36 | 0.820 | 3 | |
| B+ | 1 | Chr1_88464412 | 1 | 0.18 | 0.661 | 7 | Zm00001d029820 |
| B+ | 1 | Chr1_113763491 | 1 | 0.13 | 0.894 | 5 | |
| B+ | 1 | Chr1_173938755 | 1 | 0.06 | 0.966 | 5 | |
| B+ | 2 | Chr2_181732219 | 1 | 0.27 | 0.549 | 8 | |
| B+ | 2 | Chr2_240687544 | 1 | 0.08 | 0.851 | 4 | Zm00001d007843 |
| B+ | 2 | Chr2_214786010 | 14 | 0.46 | 0.602 | 9 | |
| B+ | 4 | Chr4_245633076 | 9 | 0.38 | 0.585 | 22 | nrt2, nrt2.2 |
| Zm00001d054060 | |||||||
| B+ | 4 | Chr4_206259028 | 1 | 0.06 | 0.559 | 8 | |
| B+ | 5 | Chr5_2005858 | 1 | 0.32 | 0.581 | 15 | Zm00001d012924 |
| B+ | 5 | Chr5_213988333 | 1 | 0.33 | 0.604 | 7 | |
| B+ | 6 | Chr6_108027757 | 5 | 0.48 | 0.519 | 10 | |
| B+ | 6 | Chr6_9906627 | 1 | 0.07 | 0.633 | 12 | 100275163, 100192849 |
| B+ | 7 | Chr7_22239405 | 1 | 0.15 | 0.570 | 2 | |
| B+ | 8 | Chr8_177771765 | 1 | 0.05 | 0.826 | 13 | |
| B+ | 8 | Chr8_179214567 | 10 | 0.48 | 0.593 | 11 | Zm00001d012719 |
| B+ | 9 | Chr9_127613348 | 1 | 0.32 | 0.510 | 3 | |
| B+ | 10 | Chr10_88567864 | 1 | 0.10 | 0.954 | 6 | |
| B+ | 10 | Chr10_118526736 | 1 | 0.27 | 0.569 | 3 | |
| B+ | 10 | Chr10_122822696 | 1 | 0.13 | 0.969 | 12 | |
| B+ | 10 | Chr10_137486546 | 1 | 0.08 | 0.996 | 9 | |
| B+ | 10 | Chr10_145821750 | 1 | 0.16 | 0.544 | 6 |
Chromosome number.
Each marker ID is comprised of chromosome ID and marker location that are separated by the underscore sign.
The number of phenotypes influenced by the marker.
Minor allele frequency.
Average of posterior inclusion probability for the selected phenotypes.
Number of genes within the gene interval.
3.3. Integration of GWA analyses
Eight SNPs influenced both the manually measured and hyperspectral phenotypes, which were visualized in a phenome‐wide association plot (Figure 4). In general, most SNPs were identified in B+ plants. Two SNPs, Chr4_181569268 and Chr6_108027757, exhibited a strong association with a wide range of hyperspectral phenotypes, along with PH and SDM in B+ management. A list of candidate genes influencing both manually measured and hyperspectral phenotypes revealed that they may play an important role in nitrogen uptake and plant responses to biotic and abiotic stressors (Table 2). A Shiny web application was developed to explore the multiphenotype GWA results interactively (https://github.com/vt-ads/ShinyGWASPheWAS).
FIGURE 4.
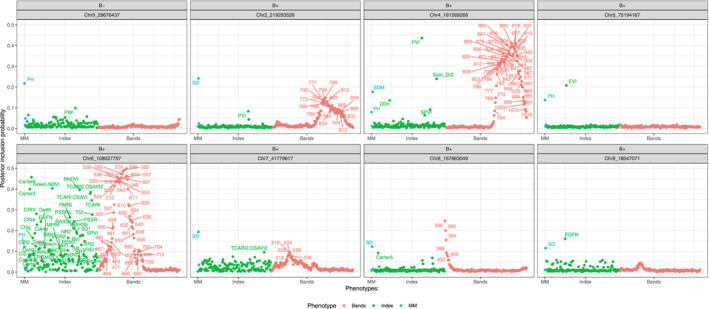
Phenome‐wide association analysis plot of selected eight markers having influence on both manually measured and hyperspectral phenotypes without (B−) and with (B+) plant growth‐promoting bacteria inoculation. Plant height (PH), stalk diameter (SD), shoot dry mass (SDM), hyperspectral bands (Bands); hyperspectral index (Index); and manually measured (MM). The abbreviations of hyperspectral indices are defined in Table S1.
TABLE 2.
List of candidate genes influencing both manually measured phenotypes and hyperspectral phenotypes.
| Management | Chr a | Marker ID b | Phenotypes | MAF c | NG d | Candidate gene |
|---|---|---|---|---|---|---|
| B− | 9 | Chr9_28676437 | PH | 0.23 | 8 | |
| B+ | 2 | Chr2_219293526 | SD | 0.36 | 11 | 103647869 |
| B+ | 4 | Chr4_181569268 | SDM | 0.33 | 19 | Zm00001d052164, |
| Zm00001d052165 | ||||||
| B+ | 5 | Chr5_75194167 | PH | 0.18 | 9 | |
| B+ | 6 | Chr6_108027757 | PH | 0.49 | 10 | |
| B+ | 7 | Chr7_41779617 | SD | 0.26 | 12 | Zm00001d006916 |
| B+ | 8 | Chr8_167863049 | SD | 0.44 | 10 | LOC100216557 |
| B+ | 9 | Chr9_18547071 | SD | 0.20 | 10 | 100274563 |
Note: Selected single nucleotide polymorphisms markers and their candidate genes influencing both manually measured phenotypes and hyperspectral phenotypes under with (B+) or without (B−) plant growth‐promoting bacteria inoculation.
Chromosome number.
Each marker ID is comprised of chromosome ID and marker location that are separated by the underscore sign.
Minor allele frequency.
Number of genes within the gene interval.
4. DISCUSSION
The utility of hyperspectral imaging technology for phenotyping has recently gained increasing attention in genetic studies because such data can capture the resonance of certain physicochemical compounds in plants and associated with genomic regions (Barnaby et al., 2020; Feng et al., 2017; Sun et al., 2019; Wu et al., 2021). Growth‐related traits, such as PH, SD, and SDM, are complex traits controlled by many genes with small individual effects. Therefore, we expected that hyperspectral imagery‐based data would shed light on the underlying genetic factors and help assess the variability of maize to aid in identifying candidate genes. However, translating hyperspectral reflectance values or indices into a biological context, such as metabolic, morphological, or functional changes, can be difficult and time‐consuming.
This study used GWA analysis of manually measured phenotypes, single‐band reflectance, and hyperspectral indices to investigate the genetic basis of responses to PGPB. BayesC, which performs variable selection, was applied for GWA analysis. The posterior inclusion probability of each marker was used to identify the relevant SNPs. The preference for using posterior inclusion probability instead of window posterior probability of association (Fernando et al., 2017) was due to the low marker density and unequal distribution of SNPs across the genome in the maize population. There is a major difference between the current study and Yassue et al. (2022). The objective of Yassue et al. (2022) was to use hyperspectral data to compare phenomic and genomic prediction and investigate the potential use of hyperspectral information to classify inoculation status. Overall, the main focus was placed on predictive modeling. In contrast, the objective of the current study was to understand the genetic basis of hyperspectral phenotypes. The novelty of the current work is the integration of hyperspectral phenotypes in GWA analysis in maize under different PGPB inoculation status. In other words, Yassue et al. (2022) used hyperspectral data as predictors to perform prediction and classification tasks, whereas the present work used hyperspectral data as phenotypes for inferential tasks.
4.1. Estimates of genomic heritability and correlation
The hyperspectral phenotypes showed a similar range of genomic heritability estimates relative to that of manually measured phenotypes. For most hyperspectral‐derived phenotypes, the heritability estimates varied from 0.30 to 0.50, indicating that hyperspectral data can capture genetic variation. In addition, the genetic correlation between the manually measured and hyperspectral phenotypes indicated that the same sets of genes probably influenced these responses.
The relatively higher genomic correlation estimates for PH in the spectral range of 400–700 nm (visible spectrum) of B− plants may indicate an association between PH and leaf pigments, such as carotenoids, chlorophyll a and b, and nitrogen concentrations (Ayala‐Silva & Beyl, 2005; Zhao et al., 2003). PH and other early growth traits are positively associated with the final dry matter yield and hyperspectral reflectance values (Capolupo et al., 2015; Freeman et al., 2007; Prey et al., 2018; Strigens et al., 2012; Williams et al., 2017; Xie et al., 2020) because more vigorous plants tend to be higher. Our previous work demonstrated that SDM, SD, and PH could be predicted with reasonable accuracy using hyperspectral image data (Yassue et al., 2022). The ability of the hyperspectral phenotypes to predict growth‐related phenotypes is likely due to some shared genetic association between them. It is known that hyperspectral phenotypes are related to other types of phenotypes, such as photosynthesis, nutrient uptake, and morphological traits (Krause et al., 2019; Ge et al., 2016; Mahajan et al., 2014; Nigon et al., 2021; Sandhu et al., 2021; Thomas et al., 2017) that were not evaluated in this study but are correlated with plant growth and vigor.
Similarly, higher genomic correlation estimates were observed for SDM at 700–1000 nm (near‐infrared). The association between near‐infrared spectra and plant biomass in maize was reported in maize in a previous study (Ma et al., 2020). Wavelengths in this range have also been reported to indicate nitrogen content in rapeseed (Müller et al., 2008) and wheat (Hansen & Schjoerring, 2003).
4.2. GWA analysis
Overall, more SNP associations were observed in B+ than in B− plants. However, the genomic heritability estimates were similar between the two managements. This suggests that the genetics underlying hyperspectral responses differ between the two managements. GWA analysis showed that SNP Chr4_245633076 was associated with nine hyperspectral phenotypes pointing to three candidate genes (nrt2, nrt2.2, and Zm00001d054060). These genes have been previously reported as part of transporter gene families and as candidates for nitrate uptake along the primary maize root (Liu et al., 2009; Sorgonà et al., 2011; Wang et al., 2020). The genes Zm00001d029820 and Zm00001d012924 are involved in plant development and environmental stress conditions (Zhang et al., 2020; Zhu et al., 2021). The candidate gene Zm00001d007843 may affect kernel size (Zhou et al., 2021), and gene Zm00001d012719 is a candidate transcription factor mediating plant responses to abiotic stressors (Vendramin et al., 2020). We did not directly evaluate the phenotypes related to previously reported candidate genes. Nevertheless, hyperspectral signatures may be of use for indirectly assessing these phenotypes. Furthermore, eight SNPs were detected in both the manually measured and hyperspectral phenotypes. Genes Zm00001d052164 and Zm00001d052165 on chromosome 4 regulate nitrogen assimilation (Wang et al., 2020), and Zm00001d006916 is a candidate gene responsible for autophagy, which may play a role regarding responses to abiotic stressors (Tang & Bassham, 2018). The LOC100216557 gene is associated with resilience of maize to aphids and may be responsible for plant defense responses and stress tolerance (Srivastava et al., 2018), and 103647869 is a candidate gene for resistance to Aspergillus flavus infection or aflatoxin contamination (Liu et al., 2021). The gene 100274563 is associated with ear weight per plant (Zhou et al., 2020). The results found in this study must be interpreted with caution because of the low density of SNPs and the relatively small number of lines used.
The Shiny web application with an interactive interface is a powerful tool for the visualization and interpretation of GWA analysis of multiple phenotypes. Two‐way Manhattan plots can be used to investigate all associations across traits and managements, including nonsignificant results that are not elaborated on here. In addition, phenome‐wide association plots can be used to identify and visualize markers with mutual influence across hundreds or thousands of phenotypes. The Shiny application can be easily extended to other high‐throughput phenotyping data, such as longitudinal, fluorescence, and thermal data.
5. CONCLUSIONS
The hyperspectral signatures captured some genetic variability in the maize diversity panel and were associated with growth‐related traits under PGPB inoculation. GWA analysis of hyperspectral data identified genomic regions that influenced both manually measured phenotypes and hyperspectral bands. In addition, a Shiny web application for multiple‐phenotype GWA was developed.
AUTHOR CONTRIBUTIONS
Rafael Massahiro Yassue: Conceptualization; data curation; formal analysis; investigation; methodology; visualization; writing—original draft; and writing—review and editing. Giovanni Galli: Investigation; methodology; and writing—review and editing. Chun‐Peng James Chen: Methodology and writing—review and editing. Roberto Fritsche‐Neto: Conceptualization; funding acquisition; supervision; and writing—review and editing. Gota Morota: Conceptualization; methodology; funding acquisition; supervision; writing—original draft; and writing—review and editing.
CONFLICT OF INTEREST STATEMENT
The authors declare that there is no conflict of interest.
Supporting information
PLD3_492_SupplementalMaizeHyperspectralGWAS.pdf
ACKNOWLEDGMENTS
The authors acknowledge Pedro Takao Yamamoto and Fernando Henrique Iost Filho for their support in collecting the hyperspectral images.
Massahiro Yassue, R. , Galli, G. , James Chen, C.‐P. , Fritsche‐Neto, R. , & Morota, G. (2023). Genome‐wide association analysis of hyperspectral reflectance data to dissect the genetic architecture of growth‐related traits in maize under plant growth‐promoting bacteria inoculation. Plant Direct, 7(4), e492. 10.1002/pld3.492
Funding information This study was supported in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior ‐ Brasil (CAPES) ‐ Finance Code 001, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), grant nos. #17/24327‐0, #19/04697‐2, and #2017/19407‐4 from São Paulo Research Foundation (FAPESP), International Business Machines Corporation (IBM, Brasil), and Virginia Polytechnic Institute and State University.
Contributor Information
Roberto Fritsche‐Neto, Email: r.fritscheneto@irri.org.
Gota Morota, Email: morota@vt.edu.
DATA AVAILABILITY STATEMENT
Genomic data used herein are available at https://data.mendeley.com/datasets/5gvznd2b3n/3.
REFERENCES
- Ayala‐Silva, T. , & Beyl, C. A. (2005). Changes in spectral reflectance of wheat leaves in response to specific macronutrient deficiency. Advances in Space Research, 35(2), 305–317. 10.1016/j.asr.2004.09.008 [DOI] [PubMed] [Google Scholar]
- Barnaby, J. Y. , Huggins, T. D. , Lee, H. , McClung, A. M. , Pinson, S. R. M. , Oh, M. , Bauchan, G. R. , Tarpley, L. , Lee, K. , Kim, M. S. , & Edwards, J. D. (2020). Vis/NIR hyperspectral imaging distinguishes subpopulation, production environment, and physicochemical grain properties in rice. Scientific Reports, 10(1). 10.1038/s41598-020-65999-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista, B. D. , Dourado, M. N. , Figueredo, E. F. , Hortencio, R. O. , Marques, J. P. R. , Piotto, F. A. , Bonatelli, M. L. , Settles, M. L. , Azevedo, J. L. , & Quecine, M. C. (2021). The auxin‐producing bacillus thuringiensis RZ2ms9 promotes the growth and modifies the root architecture of tomato (solanum lycopersicum cv. micro‐tom). Archives of Microbiology, 206, 3–42. 10.1007/s00203-021-02361-z [DOI] [PubMed] [Google Scholar]
- Batista, B. D. , Lacava, P. T. , Ferrari, A. , Teixeira‐Silva, N. S. , Bonatelli, M. L. , Tsui, S. , Mondin, M. , Kitajima, E. W. , Pereira, J. O. , Azevedo, J. L. , & Quecine, M. C. (2018). Screening of tropically derived, multi‐trait plant growth‐ promoting rhizobacteria and evaluation of corn and soybean colonization ability. Microbiological Research, 206, 33–42. 10.1016/j.micres.2017.09.007 [DOI] [PubMed] [Google Scholar]
- Boggs, T. (2014). Spectral python (spy). URL: https://www.spectralpython.net
- Bradbury, P. J. , Zhang, Z. , Kroon, D. E. , Casstevens, T. M. , Ramdoss, Y. , & Buckler, E. S. (2007). TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics, 23(19), 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Browning, B. L. , Zhou, Y. , & Browning, S. R. (2018). A one‐penny imputed genome from next‐generation reference panels. The American Journal of Human Genetics, 103(3), 338–348. 10.1016/j.ajhg.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capolupo, A. , Kooistra, L. , Berendonk, C. , Boccia, L. , & Suomalainen, J. (2015). Estimating plant traits of grasslands from UAV‐acquired hyperspectral images: A comparison of statistical approaches. ISPRS International Journal of Geo‐Information, 4(4), 2792–2820. 10.3390/ijgi4042792 [DOI] [Google Scholar]
- Chang, W. , Cheng, J. , Allaire, J. J. , Sievert, C. , Schloerke, B. , Xie, Y. , Allen, J. , McPherson, J. , Dipert, A. , & Borges, B. (2021). Shiny: Web application framework for r. https://CRAN.R-project.org/package=shiny. R package version 1.7.1.
- Cheng, H. , Fernando, R. L. , & Garrick, D. J. (2018). JWAS: Julia implementation of whole‐genome analysis software. Proceedings of the World Congress on Genetics Applied to Livestock Production, 11.859. Auckland, New Zealand. [Google Scholar]
- Cheng, H. , Kizilkaya, K. , Zeng, J. , Garrick, D. , & Fernando, R. (2018). Genomic prediction from multiple‐trait Bayesian regression methods using mixture priors. Genetics, 209(1), 89–103. 10.1534/genetics.118.300650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant, S. , Duffy, B. , Nowak, J. , Clément, C. , & Barka, E. A. (2005). Use of plant growth‐promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Applied and environmental microbiology, 71(9), 4951–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Benedetto, N. A. , Corbo, M. R. , Campaniello, D. , Cataldi, M. P. , Bevilacqua, A. , Sinigaglia, M. , & Flagella, Z. (2017). The role of plant growth promoting bacteria in improving nitrogen use efficiency for sustainable crop production: A focus on wheat. AIMS microbiology, 3(3), 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J. J. , & Doyle, J. L. (1987). A rapid dna isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19, 11–15. http://worldveg.tind.io/record/33886 [Google Scholar]
- Egamberdiyeva, D. (2007). The effect of plant growth promoting bacteria on growth and nutrient uptake of maize in two different soils. Applied Soil Ecology, 36(2–3), 184–189. 10.1016/j.apsoil.2007.02.005 [DOI] [Google Scholar]
- Fan, B. , Onteru, S. K. , Du, Z.‐Q. , Garrick, D. J. , Stalder, K. J. , & Rothschild, M. F. (2011). Genome‐wide association study identifies loci for body composition and structural soundness traits in pigs. PloS one, 6(2), e14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, H. , Guo, Z. , Yang, W. , Huang, C. , Chen, G. , Fang, W. , Xiong, X. , Zhang, H. , Wang, G. , Xiong, L. , & Liu, Q. (2017). An integrated hyperspectral imaging and genome‐wide association analysis platform provides spectral and genetic insights into the natural variation in rice. Scientific Reports, 7(1), 4401. 10.1038/s41598-017-04668-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando, R. L. , & Garrick, D. (2013). Bayesian methods applied to GWAS, Genome‐wide association studies and genomic prediction: Springer, pp. 237–274. [DOI] [PubMed] [Google Scholar]
- Fernando, R. , Toosi, A. , Wolc, A. , Garrick, D. , & Dekkers, J. (2017). Application of whole‐genome prediction methods for genome‐wide association studies: A Bayesian approach. Journal of Agricultural, Biological and Environmental Statistics, 22(2), 172–193. [Google Scholar]
- Freeman, K. W. , Girma, K. , Arnall, D. B. , Mullen, R. W. , Martin, K. L. , Teal, R. K. , & Raun, W. R. (2007). By‐plant prediction of corn forage biomass and nitrogen uptake at various growth stages using remote sensing and plant height. Agronomy Journal, 99(2), 530–536. 10.2134/agronj2006.0135 [DOI] [Google Scholar]
- Fritsche‐Neto, R. , Galli, G. , Gotardi, L. F. , Sabadin, J. F. G. , Borsato‐Junior, R. , Yassue, R. M. , Gevartosky, R. , Costa‐Neto, G. M. F. , Souza, P. H. , Morosini, J. S. , Espolador, F. G. , Barbosa, P. A. M. , & Garbuglio, D. (2020). TCGA: A tropical corn germplasm assembly for genomic prediction and high‐throughput phenotyping studies. Mendeley Data. https://data.mendeley.com/datasets/5gvznd2b3n/3 [Google Scholar]
- Ge, Y. , Bai, G. , Stoerger, V. , & Schnable, J. C. (2016). Temporal dynamics of maize plant growth, water use, and leaf water content using automated high throughput RGB and hyperspectral imaging. Computers and Electronics in Agriculture, 127, 625–632. 10.1016/j.compag.2016.07.028 [DOI] [Google Scholar]
- Gitelson, A. A. , Peng, Y. , & Huemmrich, K. F. (2014). Relationship between fraction of radiation absorbed by photosynthesizing maize and soybean canopies and NDVI from remotely sensed data taken at close range and from MODIS 250m resolution data. Remote Sensing of Environment, 147, 108–120. 10.1016/j.rse.2014.02.014 [DOI] [Google Scholar]
- Hansen, P. M. , & Schjoerring, J. K. (2003). Reflectance measurement of canopy biomass and nitrogen status in wheat crops using normalized difference vegetation indices and partial least squares regression. Remote Sensing of Environment, 86(4), 542–553. 10.1016/s0034-4257(03)00131-7 [DOI] [Google Scholar]
- Hungria, M. , Campo, R. J. , Souza, E. M. , & Pedrosa, F. O. (2010). Inoculation with selected strains of azospirillum brasilense and a. Lipoferum improves yields of maize and wheat in brazil. Plant and Soil, 331(1–2), 413–425. 10.1007/s11104-009-0262-0 [DOI] [Google Scholar]
- Kizilkaya, K. , Fernando, R. L. , & Garrick, D. J. (2010). Genomic prediction of simulated multibreed and purebred performance using observed fifty thousand single nucleotide polymorphism genotypes. Journal of Animal Science, 88(2), 544–551. [DOI] [PubMed] [Google Scholar]
- Krause, M. R. , González‐Pérez, L. , Crossa, J. , Pérez‐Rodríguez, P. , Montesinos‐López, O. , Singh, R. P. , Dreisigacker, S. , Poland, J. , Rutkoski, J. , Sorrells, M. , Gore, M. A. , & Mondal, S. (2019). Hyperspectral reflectance‐derived relationship matrices for genomic prediction of grain yield in wheat. G3 Genes|Genomes|Genetics, 9(4), 1231–1247. 10.1534/g3.118.200856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurance, W. F. , Sayer, J. , & Cassman, K. G. (2014). Agricultural expansion and its impacts on tropical nature. Trends in Ecology & Evolution, 29(2), 107–116. 10.1016/j.tree.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Lehnert, L. W. , Meyer, H. , Obermeier, W. A. , Silva, B. , Regeling, B. , Thies, B. , & Bendix, J. (2019). Hyperspectral data analysis in R: The hsdar package. Journal of Statistical Software, 89(12), 1–23. [Google Scholar]
- Liu, J. , Chen, F. , Olokhnuud, C. , Glass, A. D. M. , Tong, Y. , Zhang, F. , & Mi, G. (2009). Root size and nitrogen‐uptake activity in two maize (Zea mays) inbred lines differing in nitrogen‐use efficiency. Journal of Plant Nutrition and Soil Science, 172(2), 230–236. 10.1002/jpln.200800028 [DOI] [Google Scholar]
- Liu, H. , Wu, H. , Wang, Y. , Wang, H. , Chen, S. , & Yin, Z. (2021). Comparative transcriptome profiling and co‐expression network analysis uncover the key genes associated with early‐stage resistance to Aspergillus flavus in maize. BMC Plant Biology, 21(1), 216. 10.1186/s12870-021-02983-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, D. , Maki, H. , Neeno, S. , Zhang, L. , Wang, L. , & Jin, J. (2020). Application of non‐linear partial least squares analysis on prediction of biomass of maize plants using hyperspectral images. Biosystems Engineering, 200, 40–54. 10.1016/j.biosystemseng.2020.09.002 [DOI] [Google Scholar]
- Mahajan, G. R. , Sahoo, R. N. , Pandey, R. N. , Gupta, V. K. , & Kumar, D. (2014). Using hyperspectral remote sensing techniques to monitor nitrogen, phosphorus, sulphur and potassium in wheat (Triticum aestivum l.) Precision Agriculture, 15(5), 499–522. 10.1007/s11119-014-9348-7 [DOI] [Google Scholar]
- Mantelin, S. (2003). Plant growth‐promoting bacteria and nitrate availability: Impacts on root development and nitrate uptake. Journal of Experimental Botany, 55(394), 27–34. 10.1093/jxb/erh010 [DOI] [PubMed] [Google Scholar]
- Müller, K. , Böttcher, U. , Meyer‐Schatz, F. , & Kage, H. (2008). Analysis of vegetation indices derived from hyperspectral reflection measurements for estimating crop canopy parameters of oilseed rape (Brassica napus l.) Biosystems Engineering, 101(2), 172–182. 10.1016/j.biosystemseng.2008.07.004 [DOI] [Google Scholar]
- Nigon, T. , Paiao, G. D. , Mulla, D. J. , Fernández, F. G. , & Yang, C. (2021). The influence of aerial hyperspectral image processing workflow on nitrogen uptake prediction accuracy in maize. Remote Sensing, 14(1), 132. 10.3390/rs14010132 [DOI] [Google Scholar]
- Olanrewaju, O. S. , Glick, B. R. , & Babalola, O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World Journal of Microbiology and Biotechnology, 33(11), 197. 10.1007/s11274-017-2364-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pii, Y. , Mimmo, T. , Tomasi, N. , Terzano, R. , Cesco, S. , & Crecchio, C. (2015). Microbial interactions in the rhizosphere: Beneficial influences of plant growth‐promoting rhizobacteria on nutrient acquisition process. A review. Biology and Fertility of Soils, 51(4), 403–415. 10.1007/s00374-015-0996-1 [DOI] [Google Scholar]
- Poland, J. A. , Brown, P. J. , Sorrells, M. E. , & Jannink, J.‐L. (2012). Development of high‐density genetic maps for barley and wheat using a novel two‐enzyme genotyping‐by‐sequencing approach. PLoS ONE, 7(2), e32253. 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prey, L. , von Bloh, M. , & Schmidhalter, U. (2018). Evaluating RGB imaging and multispectral active and hyperspectral passive sensing for assessing early plant vigor in winter wheat. Sensors, 18(9), 2931. 10.3390/s18092931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quecine, M. C. , Araújo, W. L. , Rossetto, P. B. , Ferreira, A. , Tsui, S. , Lacava, P. T. , Mondin, M. , Azevedo, J. L. , & Pizzirani‐Kleiner, A. A. (2012). Sugarcane growth promotion by the endophytic bacterium Pantoea agglomerans 33.1. Applied and Environmental Microbiology, 78(21), 7511–7518. 10.1128/aem.00836-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan, R. , Chopra, U. K. , Sahoo, R. N. , Singh, A. K. , & Pradhan, S. (2012). Assessment of plant nitrogen stress in wheat (Triticum aestivum l.) through hyperspectral indices. International Journal of Remote Sensing, 33(20), 6342–6360. 10.1080/01431161.2012.687473 [DOI] [Google Scholar]
- Sandhu, K. , Patil, S. S. , Pumphrey, M. , & Carter, A. (2021). Multitrait machine‐ and deep‐learning models for genomic selection using spectral information in a wheat breeding program. The Plant Genome, 14(3), e20119. 10.1002/tpg2.20119 [DOI] [PubMed] [Google Scholar]
- Shamimuzzaman, M. , Gardiner, J. M. , Walsh, A. T. , Triant, D. A. , Tourneau, J. J. L. , Tayal, A. , Unni, D. R. , Nguyen, H. N. , Portwood, J. L. , Cannon, E. K. S. , Andorf, C. M. , & Elsik, C. G. (2020). MaizeMine: A data mining warehouse for the maize genetics and genomics database. Frontiers in Plant Science, 11, 592730. 10.3389/fpls.2020.592730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim, S.‐C. , Durstewitz, G. , Plieske, J. , Wieseke, R. , Ganal, M. W. , Deynze, A. V. , Hamilton, J. P. , Buell, C. R. , Causse, M. , Wijeratne, S. , & Francis, D. M. (2012). Development of a large SNP genotyping array and generation of high‐density genetic maps in tomato. PLoS ONE, 7(7), e40563. 10.1371/journal.pone.0040563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, V. K. , Singh, A. K. , Singh, P. P. , & Kumar, A. (2018). Interaction of plant growth promoting bacteria with tomato under abiotic stress: A review. Agriculture, Ecosystems & Environment, 267, 129–140. 10.1016/j.agee.2018.08.020 [DOI] [Google Scholar]
- Sorgonà, A. , Lupini, A. , Mercati, F. , di Dio, L. , Sunseri, F. , & Abenavoli, M. R. (2011). Nitrate uptake along the maize primary root: An integrated physiological and molecular approach. Plant, Cell & Environment, 34(7), 1127–1140. 10.1111/j.1365-3040.2011.02311.x [DOI] [PubMed] [Google Scholar]
- Srivastava, R. , Li, Z. , Russo, G. , Tang, J. , Bi, R. , Muppirala, U. , Chudalayandi, S. , Severin, A. , He, M. , Vaitkevicius, S. I. , Lawrence‐Dill, C. J. , Liu, P. , Stapleton, A. E. , Bassham, D. C. , Brandizzi, F. , & Howell, S. H. (2018). Response to persistent ER stress in plants: A multiphasic process that transitions cells from prosurvival activities to cell death. The Plant Cell, 30(6), 1220–1242. 10.1105/tpc.18.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strigens, A. , Grieder, C. , Haussmann, B. I. G. , & Melchinger, A. E. (2012). Genetic variation among inbred lines and testcrosses of maize for early growth parameters and their relationship to final dry matter yield. Crop Science, 52(3), 1084–1092. 10.2135/cropsci2011.08.0426 [DOI] [Google Scholar]
- Sun, D. , Cen, H. , Weng, H. , Wan, L. , Abdalla, A. , El‐Manawy, A. I. , Zhu, Y. , Zhao, N. , Fu, H. , Tang, J. , Li, X. , Zheng, H. , Shu, Q. , Liu, F. , & He, Y. (2019). Using hyperspectral analysis as a potential high throughput phenotyping tool in GWAS for protein content of rice quality. Plant Methods, 15(1), 54. 10.1186/s13007-019-0432-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, J. , & Bassham, D. C. (2018). Autophagy in crop plants: What's new beyond arabidopsis? Open Biology, 8(12), 180162. 10.1098/rsob.180162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S. , Kuska, M. T. , Bohnenkamp, D. , Brugger, A. , Alisaac, E. , Wahabzada, M. , Behmann, J. , & Mahlein, A.‐K. (2017). Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. Journal of Plant Diseases and Protection, 125(1), 5–20. 10.1007/s41348-017-0124-6 [DOI] [Google Scholar]
- Vendramin, S. , Huang, J. , Crisp, P. A. , Madzima, T. F. , & McGinnis, K. M. (2020). Epigenetic regulation of ABA‐induced transcriptional responses in maize. G3 Genes|Genomes|Genetics, 10(5), 1727–1743. 10.1534/g3.119.400993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Xu, J. , Ge, M. , Ning, L. , Hu, M. , & Zhao, H. (2020). High‐resolution profile of transcriptomes reveals a role of alternative splicing for modulating response to nitrogen in maize. BMC Genomics, 21(1), 353. 10.1186/s12864-020-6769-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, D. , Britten, A. , McCallum, S. , Jones, H. , Aitkenhead, M. , Karley, A. , Loades, K. , Prashar, A. , & Graham, J. (2017). A method for automatic segmentation and splitting of hyperspectral images of raspberry plants collected in field conditions. Plant Methods, 13(1), 74. 10.1186/s13007-017-0226-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolc, A. , & Dekkers, J. (2022). Application of Bayesian genomic prediction methods to genome‐wide association analyses. Genetics Selection Evolution, 54(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Feng, H. , Wu, D. , Yan, S. , Zhang, P. , Wang, W. , Zhang, J. , Ye, J. , Dai, G. , Fan, Y. , Li, W. , Song, B. , Geng, Z. , Yang, W. , Chen, G. , Qin, F. , Terzaghi, W. , Stitzer, M. , Li, L. , Xiong, L. , Yan, J. , Buckler, E. , Yang, W. , & Dai, M. (2021). Using high‐throughput multiple optical phenotyping to decipher the genetic architecture of maize drought tolerance. Genome Biology, 22(1), 185. 10.1186/s13059-021-02377-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Y. , Wang, C. , Yang, W. , Feng, M. , Qiao, X. , & Song, J. (2020). Canopy hyperspectral characteristics and yield estimation of winter wheat (Triticum aestivum) under low temperature injury. Scientific Reports, 10(1), 244. 10.1038/s41598-019-57100-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. , Nigon, T. , Hao, Z. , Paiao, G. D. , Fernández, F. G. , Mulla, D. , & Yang, C. (2021). Estimation of corn yield based on hyperspectral imagery and convolutional neural network. Computers and Electronics in Agriculture, 184, 106092. 10.1016/j.compag.2021.106092 [DOI] [Google Scholar]
- Yassue, R. M. , Carvalho, H. F. , Gevartosky, R. , Sabadin, F. , Souza, P. H. , Bonatelli, M. L. , Azevedo, J. L. , Quecine, M. C. , & Fritsche‐Neto, R. (2021). On the genetic architecture in a public tropical maize panel of the symbiosis between corn and plant growth‐promoting bacteria aiming to improve plant resilience. Molecular Breeding, 41(10), 63. 10.1007/s11032-021-01257-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassue, R. M. , Galli, G. , Fritsche‐Neto, R. , & Morota, G. (2022). Classification of plant growth‐promoting bacteria inoculation status and prediction of growth‐related traits in tropical maize using hyperspectral image and genomic data. Crop Science, 63, 88–100. 10.1002/csc2.20836 [DOI] [Google Scholar]
- Yassue, R. M. , Galli, G. , Junior, R. B. , Cheng, H. , Morota, G. , & Fritsche‐Neto, R. (2022). A low‐cost greenhouse‐based high‐throughput phenotyping platform for genetic studies: A case study in maize under inoculation with plant growth‐promoting bacteria. The Plant Phenome Journal, 5, e20043. 10.1101/2021.08.12.456112 [DOI] [Google Scholar]
- Yoosefzadeh‐Najafabadi, M. , Torabi, S. , Tulpan, D. , Rajcan, I. , & Eskandari, M. (2021). Genome‐wide association studies of soybean yield‐related hyperspectral reflectance bands using machine learning‐mediated data integration methods. Frontiers in Plant Science, 12, 777028. 10.3389/fpls.2021.777028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarco‐Tejada, P. J. , Miller, J. R. , Morales, A. , Berjón, A. , & Agüera, J. (2004). Hyperspectral indices and model simulation for chlorophyll estimation in open‐canopy tree crops. Remote Sensing of Environment, 90(4), 463–476. 10.1016/j.rse.2004.01.017 [DOI] [Google Scholar]
- Zarco‐Tejada, P. J. , Ustin, S. L. , & Whiting, M. L. (2005). Temporal and spatial relationships between within‐field yield variability in cotton and high‐spatial hyperspectral remote sensing imagery. Agronomy Journal, 97(3), 641–653. 10.2134/agronj2003.0257 [DOI] [Google Scholar]
- Zhang, Z. , Qu, J. , Li, F. , Li, S. , Xu, S. , Zhang, R. , Xue, J. , & Guo, D. (2020). Genome‐wide evolutionary characterization and expression analysis of SIAMESE‐RELATED family genes in maize. BMC Evolutionary Biology, 20(1), 91. 10.1186/s12862-020-01619-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D. , Reddy, K. R. , Kakani, V. G. , Read, J. J. , & Carter, G. A. (2003). Corn (Zea mays l.) growth, leaf pigment concentration, photosynthesis and leaf hyperspectral reflectance properties as affected by nitrogen supply. Plant and Soil, 257(1), 205–218. 10.1023/a:1026233732507 [DOI] [Google Scholar]
- Zheng, X. , Levine, D. , Shen, J. , Gogarten, S. M. , Laurie, C. , & Weir, B. S. (2012). A high‐performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics, 28(24), 3326–3328. 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q. , Fu, Z. , Liu, H. , Wang, J. , Guo, Z. , Zhang, X. , Tian, R. , Liu, Y. , Qu, J. , Li, W. , Yan, J. , & Tang, J. (2021). Mining novel kernel size‐related genes by pQTL mapping and multi‐omics integrative analysis in developing maize kernels. Plant Biotechnology Journal, 19(8), 1489–1491. 10.1111/pbi.13634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z. , Li, G. , Tan, S. , Li, D. , Weiß, T. M. , Wang, X. , Chen, S. , Würschum, T. , & Liu, W. (2020). A QTL atlas for grain yield and its component traits in maize (Zea mays). Plant Breeding, 139(3), 562–574. 10.1111/pbr.12809 [DOI] [Google Scholar]
- Zhu, J. , Zhou, Y. , Li, J. , & Li, H. (2021). Genome‐wide investigation of the phospholipase c gene family in Zea mays. Frontiers in Genetics, 11, 611414. 10.3389/fgene.2020.611414 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PLD3_492_SupplementalMaizeHyperspectralGWAS.pdf
Data Availability Statement
Genomic data used herein are available at https://data.mendeley.com/datasets/5gvznd2b3n/3.


