Abstract
Mother-to-child transmission is a major route for infections in newborns. Vaccination in mothers to leverage the maternal immune system is a promising approach to vertically transfer protective immunity. During infectious disease outbreaks, such as the 2016 Zika virus (ZIKV) outbreak, rapid availability of vaccines can prove critical in reducing widespread disease burden. The recent successes of mRNA vaccines support their evaluation in pregnant animal models to justify their use in neonatal settings. Here we evaluated immunogenicity of self-amplifying replicon (repRNA) vaccines, delivered with our clinical-stage LION nanoparticle formulation, in pregnant rabbits using ZIKV and HIV-1 as model disease targets. We showed that LION/repRNA vaccines induced robust antigen-specific antibody responses in adult pregnant rabbits that passively transferred to newborn kits in utero. Using a matrixed study design, we further elucidate the effect of vaccination in kits on the presence of pre-existing maternal antibodies. Our findings showed that timing of maternal vaccination is critical in maximizing in utero antibody transfer, and subsequent vaccination in newborns maintained elevated antibody levels compared with no vaccination. Overall, our results support further development of the LION/repRNA vaccine platform for maternal and neonatal settings.
Keywords: mother-to-child transmission, self-amplifying RNA, nanoparticle emulsion, HIV-1, ZIKV, maternal transfer, humoral immunity, LION formulation, pregnant rabbit model
Graphical abstract
Challenges remain to adopt mRNA vaccines in preventing mother-to-child-transmission, a major cause of new infections in infants. Khandhar et al. report immunogenicity of self-replicating RNA vaccines using a pregnant rabbit model. The authors show that vaccination in pregnant rabbits was well tolerated and promoted passive transfer of antibodies to newborns.
Introduction
The recent success of mRNA vaccines in response to the COVID-19 pandemic is a catalyst to develop mRNA vaccines targeting other infectious diseases. The safety profile of mRNA vaccines expressing SARS-CoV-2 spike now extends beyond healthy adults, with the US Food and Drug Administration authorizing the Pfizer/BioNTech and Moderna mRNA vaccines for children aged 6 months and older.1 Moreover, preliminary findings in pregnant persons showed no obvious detrimental safety signals.2 The combined safety and efficacy data justify evaluation of mRNA-based maternal and pediatric vaccines against viruses presenting a significant risk of transmission in perinatal settings.
Here we evaluated self-amplifying replicon RNA (repRNA) as a maternal vaccine in pregnant New Zealand White (NZW) rabbits. RepRNA vaccines encode viral replicases that amplify expression of an encoded gene of interest by 10- to 100-fold over non-replicating mRNA,3 thus providing dose-sparing and manufacturing advantages.4,5 Two self-amplifying RNA COVID-19 vaccines were recently evaluated in phase I trials in the United States, both delivered with lipid nanoparticle (LNP) delivery vehicles.6,7 Arcturus Therapeutics announced 55% efficacy in preventing symptomatic disease following two 5-μg doses of their repRNA vaccine encoding the original SARS-CoV-2 spike in the context of Delta and Omicron variants in circulation.8 In comparison, Moderna’s mRNA-1273 vaccine, which also encodes the original SARS-CoV-2 spike, provided 44% efficacy against the Omicron variant with two 100-μg doses.9 In contrast to LNP formulations, which encapsulate the RNA, we developed a proprietary delivery technology called LION—a 2°C–8°C stable oil-in-water nanoparticle emulsion, which electrostatically binds and protects anionic nucleic acids such as repRNA. Upon complexing, the LION/repRNA complex is stable as a liquid for weeks at 2°C–8°C, can be stored long-term frozen below −60°C, or can be lyophilized and stored long term at 2°C–8°C. Moreover, because LION is stored independent of repRNA it has plug-and-play functionality, allowing for rapid evaluation of new repRNA vaccine constructs such as those we recently developed to address emerging SARS-CoV-2 variants.10,11 Our LION/repRNA vaccine targeting SARS-CoV-212 received emergency use authorization in India,13 and is currently in phase I trials in South Korea, Brazil, and the United States with multiple variants under investigation.14,15,16
The ongoing clinical trials with LION/repRNA, and pre-clinical evidence of safety and efficacy, motivated us to evaluate this platform for maternal vaccination. We evaluated HIV-1 and Zika virus (ZIKV) as model pathogens because of their significant role in causing infections in newborns following mother-to-child transmission (MTCT). The ZIKV outbreak in 2016 had high rates of microcephaly and other birth defects in newborns delivered by mothers infected during pregnancy.17 The rapid spread of ZIKV and uncertainty of future outbreaks highlights the need to evaluate rapid-response RNA platforms for use in maternal and pediatric settings.
We previously tested a ZIKV repRNA vaccine encoding the precursor membrane (prM) and envelope (E) antigens (RNA-prM/E) in mice and guinea pigs, and demonstrated protection against ZIKV infection in mice immunized with low-nanogram-level doses.18 Here, we evaluated RNA-prM/E in pregnant rabbits, assessing both safe and effective immunization of the mother as well as maternal transfer of immunity to kits. MTCT is a major contributor to new HIV-1 infections in infants,19,20,21 resulting in an estimated 150,000 new infections in 2020 in children aged 0–9 years.22 To evaluate anti-HIV-1 responses, we screened repRNA constructs expressing various conformations of the BG505-derived Envelope glycoprotein (Env), including membrane and soluble trimers. Owing to the immense genetic diversity in HIV-1 viruses and their Env glycoproteins,23 novel iterations of engineered Env trimers will likely be needed to provide broad protection within each HIV-1 subtype. Leading strategies in HIV-1 vaccine development focus on inducing broadly neutralizing antibodies (bnAbs) targeting the viral pre-fusion Env trimer that mediates cell entry. Native Env is a membrane-bound trimer of heterodimers, each heterodimer consisting of a transmembrane (gp41) and a surface unit (gp120) associated by weak non-covalent forces. Subunit vaccines based on engineered Env trimers contain stabilizing mutations to preserve the native quaternary structure and optimally present neutralizing epitopes. Soluble trimers belonging to the “SOSIP” family, such as BG505 SOSIP.664—currently in phase I clinical trials (NCT03699241)24—contain trimer-stabilizing cysteine mutations to covalently bridge cleaved gp120 and truncated gp41 units. Cleavage-independent engineered trimers, which include the native flexibly linked trimers25 and uncleaved pre-fusion optimized trimers,26 are likely more amenable to in vivo expression from nucleic-acid-based vaccines.27 Following immunogenicity-based screening in C57BL/6 mice and young adult female NZW rabbits, we advanced an repRNA construct encoding the soluble BG505 SOSIP.664 trimer with a glycine-serine single-chain flexible linker replacing the furin cleavage site (RNA-SOSIP-scfl) to evaluate immunogenicity in pregnant rabbits. We report vaccine-specific induction of antibody responses in pregnant does along with significant maternal antibody transfer to newborn kits. Furthermore, we show that the LION/repRNA platform can induce vaccine-specific antibody responses in kits even in the context of pre-existing maternal antibodies against the same antigen.
Results
Characterization of in vitro transcribed repRNA constructs expressing HIV-1 Env trimer
The adaptability of mRNA is well suited to address the challenge of producing an effective HIV-1 vaccine, as diverse antigens with complex conformations can be screened and downselected for evaluation in clinical trials. We synthesized repRNA constructs encoding distinct conformations of Env derived from the HIV-1 BG505 isolate, a subtype A T/F virus isolated from an infant in a mother-to-infant-transmission study.28 Figure 1A shows a summary of synthesized constructs and in vitro confirmation of protein expression by western blotting. The relevant genes of interest were cloned into the subgenome (SG) of the TC-83 strain of Venezuelan equine encephalitis virus, substituting for the alphavirus structural genes. Construct A encoded a full-length HIV-1 Gag gene (NCBI reference sequence: NP_057850.1) and was used as a negative control to measure non-specific responses against Env. Construct B encoded the full-length unmodified Env gp160 gene derived from the BG505 isolate (GenBank: DQ208458.1). Construct C encoded the sequence of a cleavage-independent BG505 SOSIP.664 gp140 trimer reported by Georgiev et al.,29 which replaces the R6 furin cleavage site with a 15-amino-acid single-chain flexible linker (scfl). The trimer-stabilizing mutations (A501C, T605C, and I559P), a glycan epitope optimizing substitution (T332N), and deletion of the membrane-proximal external region (MPER) were preserved. The polycistronic constructs D, E, F, and G were designed to co-express Gag and Env gp160 to further evaluate in vivo expression and secretion of Gag-based virus-like particles (VLPs) displaying Env trimers. Construct D encoded Gag and Env gp160 genes separated by an internal ribosomal entry site (IRES) sequence derived from encephalomyocarditis virus. In construct E, the gene order was switched compared with construct D, such that the Gag gene was downstream of the IRES. Construct F employed an alternative co-expression strategy, substituting a T2A ribosome skipping peptide sequence for the IRES in construct D. Lastly, we synthesized construct G with the aim of enhancing Env incorporation in Gag VLPs by using the host protein TIP47 (tail-interacting protein of 47 kDa; NCBI reference sequence: NP_005808.3), which has previously been shown to simultaneously bind with the cytoplasmic and matrix domains in Env and Gag, respectively. Consequently, overexpression of TIP47 was shown to increase Env packaging in HIV-1 virions.30
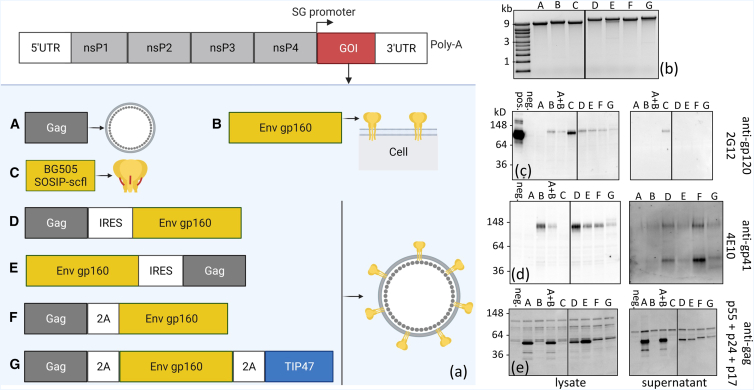
Figure 1.
Summary of in vitro transcribed repRNA constructs encoding HIV-1 Env genes and confirmation of protein expression by western blotting
(a) Schematic of the TC-83 replicon structure showing the subgenome encoding the gene of interest (GOI). HIV-1 genes were cloned in the TC-83 subgenome, and repRNA constructs were synthesized by in vitro transcribed reactions. (b) In vitro transcribed RNA constructs were analyzed by agarose gel electrophoresis to verify molecular weight and molecule integrity. (c–e) Western blot analyses of cell lysates and supernatants collected from BHK21 cells transfected with synthesized constructs. Gels were labeled with anti-gp120 human monoclonal antibody (mAb) 2G12, anti-gp41 human mAb 4E10 or anti-HIV-1 rabbit polyclonal against Gag polyprotein. pos., recombinant BG505 SOSIP.664 used as a positive control; neg., repRNA-encoding RFP as a negative control; A + B, co-transfection with constructs A and B. All samples were run on the same RNA (b) or protein (c–e) gel. Where applicable, black dividing lines are shown to exclude an irrelevant sample run in between constructs C and D.
In vitro transcribed repRNA constructs were characterized by denaturing gel electrophoresis. Visualization of a dominant single band near the target size, ranging from ∼9 kb (construct A) to ∼13 kb (construct G), provided a qualitative measure of repRNA purity and integrity (Figure 1B). Protein expression was confirmed by western blot analysis of lysate and supernatant obtained from transfection of BHK-21 cells. Human monoclonal antibodies 2G12 (Figure 1C) and 4E10 (Figure 1D), which bind to neutralizing epitopes on gp120 and gp41 subunits, respectively, were used to confirm Env expression, and polyclonal anti-Gag was used to confirm Gag expression. To control for non-specific binding, an repRNA construct expressing red fluorescent protein (RFP) was used. All constructs encoding full-length Env, solely or in combination with Gag, were labeled by both 2G12 and 4E10, confirming Env expression in cell lysates. As expected, the soluble BG505 SOSIP.664 scfl gp140 trimer was not detected by 4E10, which binds to MPER in the gp41 subunit, a region deleted in the stabilized trimer. Secretion of the soluble SOSIP trimer was confirmed by positive 2G12 labeling in the supernatant. Finally, detection of a primary band at ∼55 kDa, corresponding to full-length Gag polyprotein, confirmed Gag co-expression with Env in cells transfected with the polycistronic constructs D, E, F, and G.
Immunogenicity screening in mice and rabbits to downselect repRNA constructs expressing HIV-1 Env immunogens
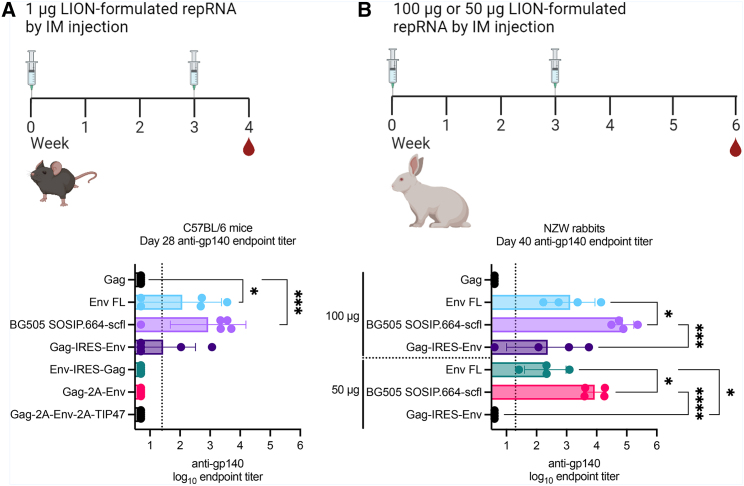
To inform downselection of repRNA constructs encoding Env immunogens, we compared vaccine-induced antibody production in C57Bl/6 mice (6- to 8-week-old females; n = 5/group). Mice were immunized by the intramuscular route twice 3 weeks apart on days 0 and 21 with 1 μg of repRNA complexed to LION formulation. The Gag-expressing repRNA was used to measure non-specific anti-Env responses in mice. Groups immunized with repRNA constructs encoding Env-IRES-Gag, Gag-2A-Env, or Gag-2A-Env-2A-TIP47 were all seronegative for anti-gp140 immunoglobulin G (IgG). Groups immunized with repRNA encoding full-length Env (Env FL), BG505 SOSIP.664-scfl, or Gag-IRES-Env were all partially seropositive with mean (SD) log10 endpoint reciprocal dilution titers of 2.1 (1.3), 2.9 (1.3), and 1.4 (1.1), respectively.
Guided by the immunogenicity data in mice, we advanced Env FL, BG505 SOSIP.664-scfl, and Gag-IRES-Env to a dose-escalation immunogenicity study in young adult female NZW rabbits. Rabbits were immunized twice, 3 weeks apart (on days 0 and 21) with 50 μg or 100 μg of LION-formulated repRNA by intramuscular injection. One group received 100 μg of repRNA encoding Gag to account for non-specific binding antibody responses arising from the repRNA non-structural proteins. Serum anti-gp140 IgG on day 40, approximately 3 weeks after the second immunization, are shown in Figure 2D. All three candidate repRNA constructs resulted in dose-dependent increase in mean endpoint titer. In both 100-μg and 50-μg cohorts, mean titer induced by BG505 SOSIP.664-scfl expressing repRNA was significantly higher than Env FL or Gag-IRES-Env expressing repRNA constructs. Furthermore, the mean titer for the group receiving 100 μg of repRNA encoding BG505 SOSIP.664-scfl was significantly higher than that for the corresponding 50-μg group (Mann-Whitney two-tailed p = 0.029). Based on the immunogenicity screening studies in mice and rabbits, we advanced repRNA encoding the BG505 SOSIP.664-scfl trimer at the 100-μg dose for evaluation as a maternal vaccine candidate in pregnant rabbits. The Zika repRNA vaccine encoded ZIKV prM and E antigens, whose selection was based on previously published reports demonstrating single-dose immunogenicity and efficacy in several pre-clinical models.18,31
Figure 2.
Comparison of anti-gp140 IgG responses induced by various HIV-1 repRNA constructs in mouse and rabbit models
Serum anti-gp140 IgG responses in (A) female C57Bl/6 mice (6–8 weeks old; n = 5/group) and (B) young adult female New Zealand White (NZW) rabbits (approximately 28 weeks old; n = 4/group) after receiving two doses 3 weeks apart. (A) Mice in each group were immunized by intramuscular injection with 1 μg of LION-formulated repRNA construct indicated on the vertical axis. Mice immunized with repRNA expressing Env FL, BG505 SOSIP.664-scfl, or Gag-IRES-Env were partially seropositive and advanced to rabbit immunogenicity study shown in (B). NZW rabbits were immunized by intramuscular injection with 100 μg or 50 μg of each candidate repRNA formulated with LION. Serum IgG responses in mice and rabbits against the BG505 SOSIP.664 gp140 recombinant protein were measured by ELISA. Individual and mean (SD) endpoint titers are shown. Mean endpoint titer from each group in (A) was log10 transformed and compared against the Gag group by ordinary one-way ANOVA and Dunnett’s multiple comparisons test. Mean endpoint titer from each group in (B) was log10 transformed and compared against every other group by ordinary one-way ANOVA and Tukey’s multiple comparisons test. Statistically significant differences in (A) are only shown for groups with at least one seropositive animal and in (B) only shown for each dose cohort. Summary of p values: ∗ < 0.05; ∗∗∗ < 0.001; ∗∗∗∗ < 0.0001.
Both RNA-SOSIP-scfl and RNA-prM/E induced robust antigen-specific antibodies but distinct neutralizing antibody responses in pregnant rabbits
Next, we characterized antibody responses in pregnant does immunized with repRNA vaccines encoding HIV-1 or ZIKV antigens. Young adult female NZW rabbits (28 weeks old; n = 5/group) were immunized with 100 μg of repRNA encoding BG505 SOSIP.664 scfl (RNA-SOSIP-scfl) or ZIKV prM/E (RNA-prM/E). To compare anti-gp140 responses induced by RNA-SOSIP-scfl with a subunit protein vaccine, we immunized one group with recombinant BG505 SOSIP.664 gp140 protein. The fully cleaved and disulfide stabilized BG505 SOSIP.664 protein is affinity purified using the 2G12 bnAb and is predominantly composed of gp140 trimers in the native-like conformation. Rabbits in the BG505 SOSIP.664 protein group, hereafter referred to as Protein-SOSIP, received 30 μg of protein combined with the MF59-like squalene emulsion adjuvant AddaVax (Invivogen). All groups were compared with an unvaccinated group receiving a sterile saline injection.
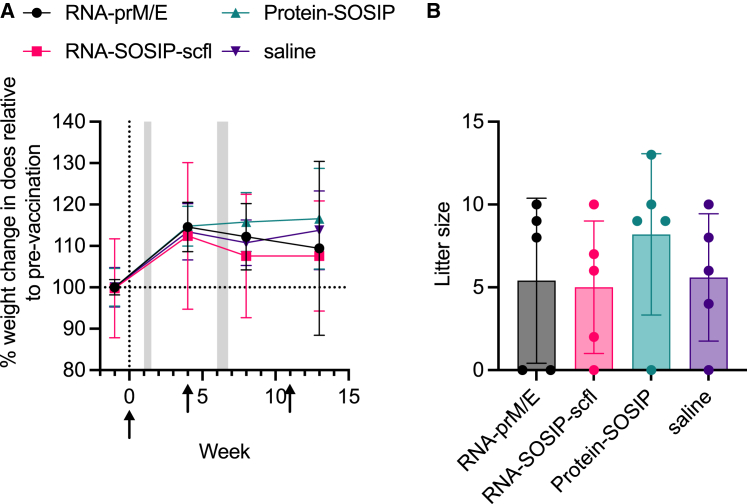
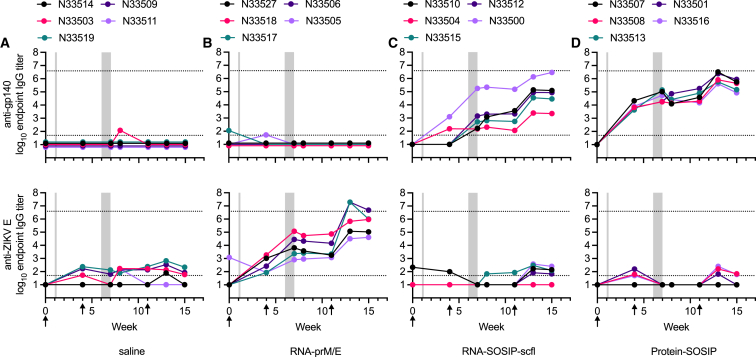
As illustrated in the study timeline (Figure 3A), does were mated 1 week after the first immunization and received a booster 4 weeks later, approximately 2 weeks before delivery. A third immunization was administered on week 11 following primary immunization, approximately 1 week before kits were weaned. As a preliminary measure of tolerogenicity, we monitored mean weight change in pregnant does and recorded their litter sizes (Figure 4). There were no significant differences in percent weight change or litter size between the unvaccinated (saline) group and any of the vaccinated groups. Binding antibody response kinetics in individual does from each group is shown in Figure 5. Groups receiving RNA-SOSIP-scfl or Protein-SOSIP were seropositive for anti-gp140 IgG, and the group receiving RNA-prM/E was seropositive for anti-ZIKV E IgG, confirming induction of vaccine-dependent antigen-specific antibodies.
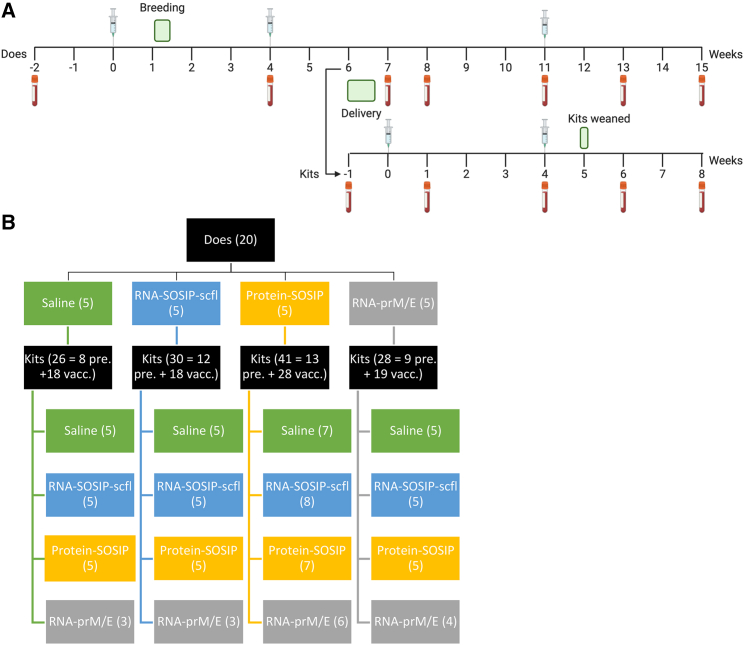
Figure 3.
Overview of timeline and study design conducted in pregnant rabbits and newborn kits
(A) Study timeline to evaluate maternal vaccination in pregnant NZW female rabbits and newborn kits. (B) Matrixed study design showing allocation of kits from each litter to vaccination group. Numbers in parentheses indicate sample size.
Figure 4.
Mean (±SD) (A) weight change in pregnant rabbits and (B) corresponding litter sizes as a measure of tolerogenicity. There were no significant differences in litter sizes between groups compared by ordinary one-way ANOVA and Dunnett's multiple comparisons test.
Figure 5.
Vaccine-induced serum IgG kinetics in NZW female adult rabbits
Rabbits (n = 5 per group) received (A) saline, (B) RNA-prM/E, (C) RNA-SOSIP-scfl-scfl, or (D) Protein-SOSIP. Animals in both RNA groups (B and C) received 100 μg of repRNA formulated with LION and in the protein group (D) received 50 μg of recombinant BG505-SOSIP.664 trimer adjuvanted with the squalene emulsion adjuvant AddaVax. Sera were assayed by antigen-specific ELISA to measure binding antibodies against (top panels) BG505 SOSIP.664 gp140 or (bottom panels) ZIKV E protein. Plotted lines track the binding antibody response of a single animal corresponding to the unique tag number in the legend. Gray shaded regions mark periods when rabbits were mated (week 1) and when kits were born (week 6–7). Arrows indicate immunization time points.
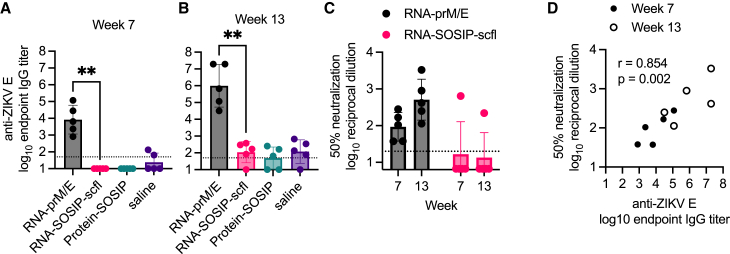
Next, we assessed neutralizing antibody levels for the repRNA-based vaccines. ZIKV neutralizing antibody levels were measured using the xCELLigence cellular impedance system and reported as reciprocal dilution of serum resulting in 50% reduction of ZIKV-induced cytopathic effect (CPE) in Vero E6 cells compared with cells treated with virus alone. The RNA-prM/E vaccine induced robust binding and neutralizing antibody responses after the second and third immunizations (Figures 6A–6C). There was a significant positive correlation between mean anti-ZIKV E IgG and ZIKV neutralizing antibody titer on weeks 7 and 13 (Pearson’s r = 0.85; p = 0.002) (Figure 6D).
Figure 6.
RNA-prM/E induced antigen-specific binding and neutralizing antibody responses in pregnant rabbits
Sera collected on weeks 7 and 13 after primary immunization were assayed for (A and B) anti-ZIKV E IgG responses by ELISA and (C) ZIKV (PRVABC56 isolate) neutralization by measuring cytopathic effect (CPE) in Vero E6 cells using the xCELLigence system. (D) Correlation plot between binding and neutralizing responses showing Pearson’s correlation coefficient (r value) and significance of correlation (p value). Mean endpoint titer between RNA-prM/E and RNA-SOSIP-scfl groups was compared using non-parametric Mann-Whitney test (∗∗p < 0.005).
Contrary to the RNA-prM/E-immunized rabbits, immunizing does with RNA-SOSIP-scfl resulted in poor neutralizing antibodies. Mean binding anti-gp140 titer for the RNA-SOSIP-scfl group trended lower than the Protein-SOSIP group (Figure 7A), but the difference was not statistically different (Mann-Whitney two-tailed p values of 0.151 and 0.056 for weeks 7 and 13, respectively). However, neutralizing antibody response induced by RNA-SOSIP-scfl was significantly lower than in the Protein-SOSIP group (Figure 7B). Autologous neutralization was measured as percent inhibition of BG505.T332N pseudovirus replication in TZM-bl cells at a 1:50 serum dilution. Sera from rabbits immunized with Protein-SOSIP had detectable neutralizing antibodies on week 7, and significantly increased levels 2 weeks after the second booster on week 13. In comparison, only two of five animals in the RNA-SOSIP-scfl group had neutralizing antibodies on week 13.
Figure 7.
Summary of serum HIV-1 antibody responses in pregnant NZW rabbits on week 7 (3 weeks post second immunization) and week 13 (2 weeks post third immunization)
(A) Individual and mean endpoint binding IgG titers against BG505 SOSIP.664 gp140 were measured by ELISA. (B) Individual and mean neutralizing antibodies shown as percent neutralization of autologous BG505.T332N pseudovirus in TZM-bl cells. RNA-SOSIP-scfl and Protein-SOSIP groups were compared using Mann-Whitney test (∗p < 0.05; ns, not significant).
In summary, both LION-formulated repRNA vaccines—RNA-SOSIP-scfl and RNA-prM/E—induced antigen-specific binding antibody titers in pregnant rabbits, but only RNA-prM/E-immunized animals showed a significant positive correlation between binding and neutralizing antibodies.
Maternal antibody transfer is directly proportional to vaccine-induced immunogenicity during pregnancy in rabbits
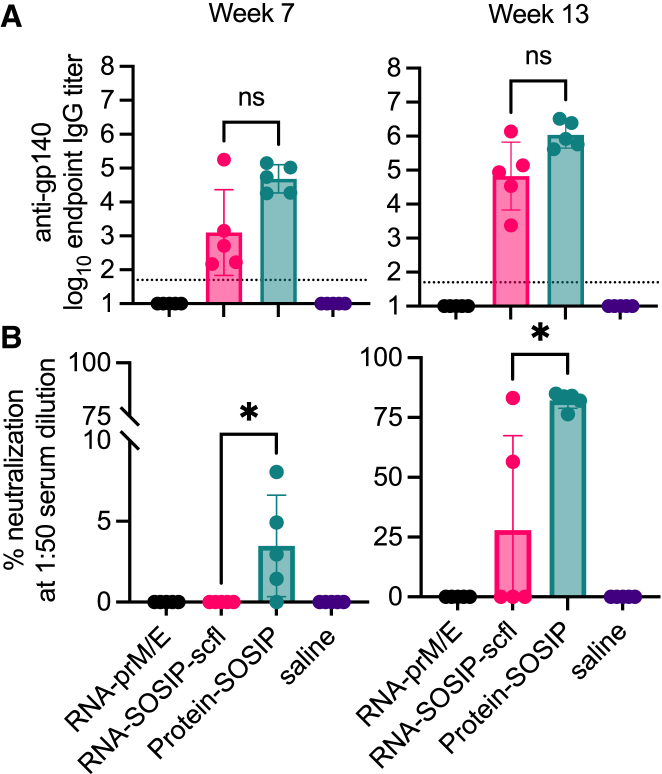
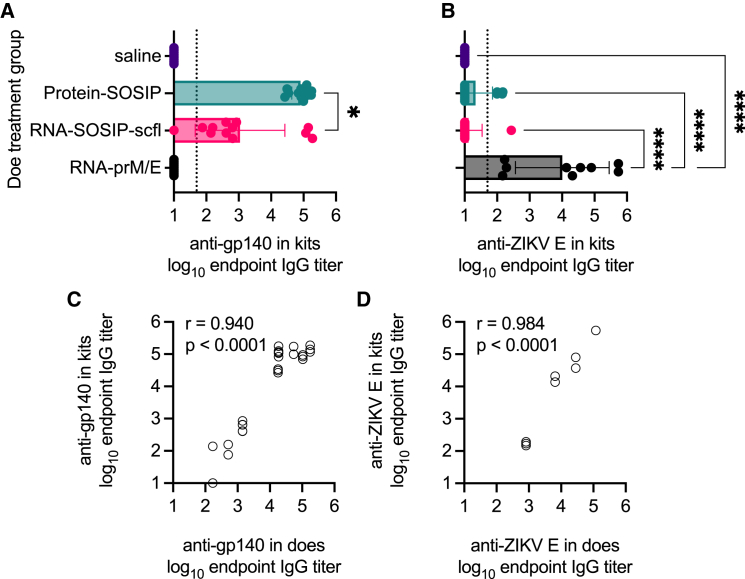
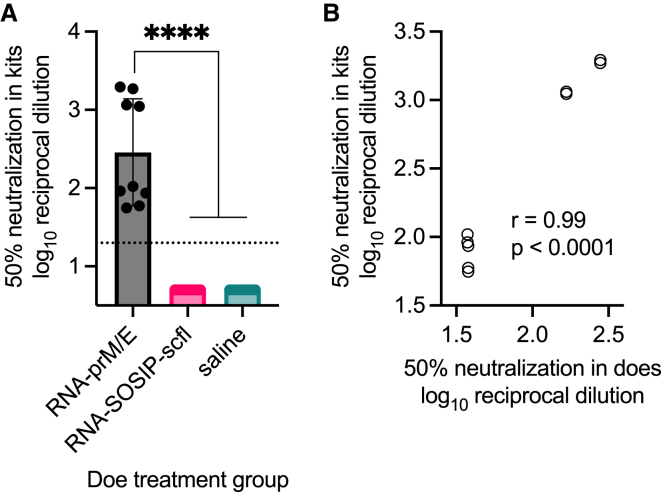
Given that mothers could be immunized safely and effectively, we asked whether vaccine-induced antibodies in pregnant does passively transferred to newborn kits. Kits were born approximately 2–3 weeks after does received their second immunization. At least two kits from each litter were euthanized and their sera collected to assay for antigen-specific antibodies. Kits delivered by does receiving RNA-SOSIP-scfl or Protein-SOSIP were nearly universally (24 of 25 evaluated) seropositive for anti-gp140 binding IgG antibodies, while kits from does receiving saline or RNA-prM/E were all seronegative (Figure 8A). Similarly, the mean anti-ZIKV E antibody level in kits delivered by does vaccinated with RNA-prM/E was significantly higher than all groups (Figure 8B). There was a significant positive correlation in antigen-specific IgG antibody levels between vaccinated does and their corresponding litters at time of birth (Figures 8C and 8D, p < 0.0001). The Pearson correlation coefficient was 0.94 for RNA-SOSIP-scfl or Protein-SOSIP groups and 0.98 for the RNA-prM/E group (Figures 8C and 8D). Furthermore, we observed that kits delivered by pregnant does vaccinated with RNA-prM/E contained neutralizing antibodies against ZIKV (Figure 9) and that the level of neutralizing antibodies in newborns was significantly and directly proportional to the level in corresponding does (neutralizing antibodies against HIV-1 were not measured). In summary, strong positive correlations in antibody responses between kits and does at the time of delivery across both HIV-1 (binding) and ZIKV (binding and neutralizing) vaccines indicate that maternal antibodies are likely transferred to kits in utero.
Figure 8.
Newborn kits acquired antibodies in utero from vaccinated does
Individual and mean (±SD) (A) anti-gp140 and (B) anti-ZIKV E IgG titers in kits near birth showed that passively acquired antibodies are antigen specific and depend on the vaccine status of does. In (A), log10 transformed mean endpoint titers of RNA-SOSIP-scfl and Protein-SOSIP groups were compared by Mann-Whitney test (∗two-tailed p value <0.05). Mean endpoint titer of each group in (B) not immunized with a ZIKV E antigen was compared with the mean IgG titer in the RNA-prM/E group by ordinary one-way ANOVA and Dunnett’s multiple comparisons test (∗∗∗∗p < 0.0001). Vaccine-induced (C) anti-gp140 and (D) anti-ZIKV E antibodies showed strong positive correlation between week-7 IgG levels in does and corresponding kits at time of birth.
Figure 9.
Newborns from RNA-prM/E vaccinated does acquired ZIKV neutralizing antibodies in utero that were directly proportional to neutralizing antibody levels in does
(A) Sera from newborns collected at time of delivery between weeks 6 and 7 were assayed for ZIKV (PRVABC56 isolate) neutralization by measuring CPE in Vero E6 cells using the xCELLigence system. Mean serum dilution resulting in 50% neutralization between indicated groups was log10 transformed and compared by ordinary one-way ANOVA and Tukey’s multiple comparisons test (∗∗∗∗p < 0.0001). (B) Correlation plot showing Pearson’s correlation coefficient (r value) and significance of correlation (p value) between neutralizing responses in does and kits.
Vaccination increased antigen-specific antibody responses in kits regardless of vaccination history in does
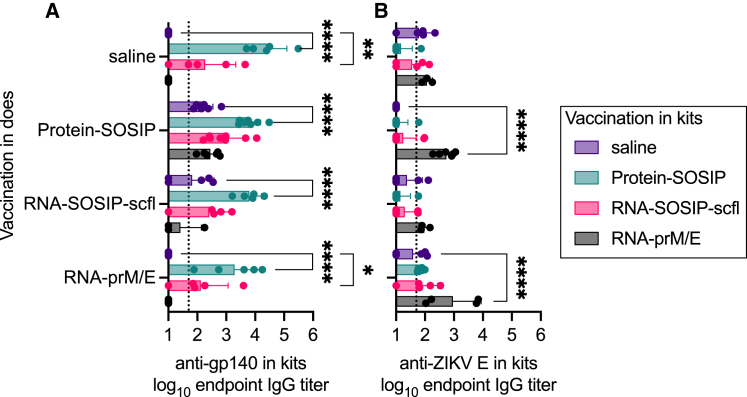
We next sought to evaluate vaccine-induced antibody responses in kits with or without passively acquired antigen-specific maternal antibodies. We implemented a matrixed study design (Figure 3B) by allocating kits from each litter to receive either saline, RNA-prM/E, RNA-SOSIP-scfl, or Protein-SOSIP, allowing us to evaluate the effect of each vaccine in the context of maternal vaccination history. Kits were immunized approximately 1 week after birth and received two immunizations, 4 weeks apart, of 50 μg of LION-formulated repRNA (RNA-SOSIP-scfl or RNA-prM/E), or 15 μg of Protein-SOSIP or an equivalent volume of sterile saline. A summary of antigen-specific serum IgG responses in kits 4 weeks after booster immunization is presented in Figure 10. Vaccination with Protein-SOSIP or RNA-SOSIP-scfl effectively increased anti-gp140 antibodies in kits lacking maternal antibodies—offspring of does receiving saline—from undetectable levels to a mean log10 endpoint titer of 4.4 (0.7) and 2.3 (1.1), respectively (Figure 10A). In comparison, vaccination with RNA-prM/E did not produce a significant increase in anti-ZIKV E in kits lacking maternal antibodies (Figure 10B). Kits with passively acquired maternal antibodies sustained their mean antigen-specific antibody titer 4 weeks after receiving the second dose of an antigen-matched vaccine compared with unmatched vaccine or saline. For instance, the mean anti-gp140 titer in kits delivered by Protein-SOSIP vaccinated does and subsequently vaccinated with Protein-SOSIP or RNA-SOSIP-scfl was 3.8 (0.4) or 2.9 (0.6), respectively, compared with 2.2 (0.3) or 2.4 (0.3) in kits from the same cohort receiving saline or RNA-prM/E, respectively (Figure 10A). The trend continued in kits delivered by RNA-SOSIP-scfl vaccinated does, with mean anti-gp140 titer of 3.8 (0.4) or 2.4 (0.8) after receiving Protein-SOSIP or RNA-SOSIP-scfl compared with 1.8 (0.8) or 1.4 (0.7) after receiving saline or RNA-prM/E. The mean anti-ZIKV E titer in kits delivered by RNA-prM/E vaccinated does was also higher after receiving RNA-prM/E, 3.0 (1.0), compared with receiving saline, 1.6 (0.5), Protein-SOSIP, 1.9 (0.1), or RNA-SOSIP-scfl, 1.9 (0.6). Finally, there was no statistically significant difference in overall vaccine-induced antigen-specific antibody responses between kits from unvaccinated and vaccinated does regardless of the vaccine delivered to the kits (Figure 11).
Figure 10.
Vaccination in kits induced higher mean antibody titers compared with unvaccinated kits regardless of doe vaccination status
Antibody responses shown here are from 4 weeks after booster immunization. Each cohort of newborns delivered by does in the same group (vertical axis labels) was randomized and received RNA-prM/E, RNA-SOSIP-scfl, Protein-SOSIP, or saline (boxed legend). Mean (±SD) and individual endpoint binding IgG titers in kits measured by ELISA against (A) gp140 and (B) ZIKV E antigens. Statistical analyses performed on log10 transformed data. Means of vaccinated and unvaccinated (saline) groups were compared by two-way ANOVA and Dunnett’s multiple comparisons test. Only statistical significance is shown (∗p < 0.05, ∗∗p < 0.005, ∗∗∗∗p < 0.0001).
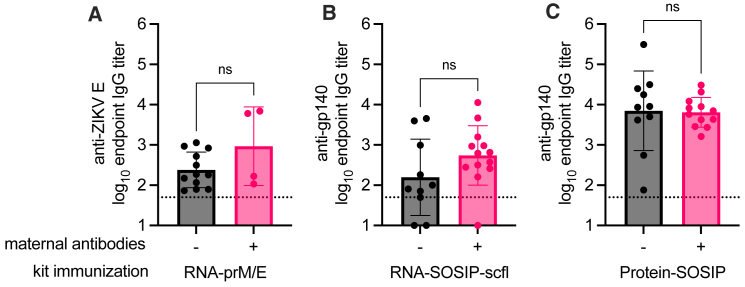
Figure 11.
Comparison of vaccine-induced responses in the context of pre-existing maternal antibodies
Serum anti-gp140 IgG endpoint titer in kits 4 weeks after receiving a booster immunization of (A) RNA-prM/E, (B) RNA-SOSIP-scfl or (C) Protein-SOSIP vaccine. Kits birthed from does receiving saline or RNA-prM/E are grouped together as negative (−) for maternal antibodies, and those birthed from does receiving RNA-SOSIP-scfl or Protein-SOSIP are grouped together as positive (+) for pre-existing maternal antibodies. Data were log10 transformed and compared by Mann-Whitney test to measure statistical differences, which were not statistically significant (ns, p > 0.05).
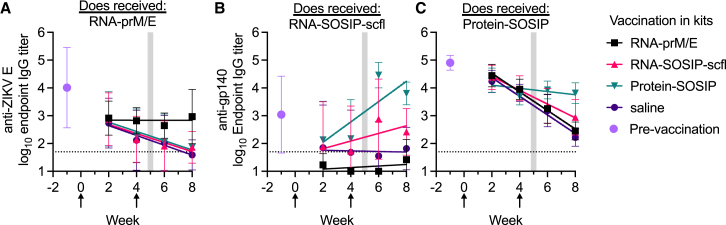
Vaccination maintained elevated antibody levels in kits with maternal antibodies compared with mock or no vaccination
To assess the waning of passively acquired maternal antibodies in kits, we calculated the linear rate of decay in mean log10 antibody titer over time (log10/week) by least-squares regression analysis (Figure 12). The mean (SD) serum antibody titer in newborn kits euthanized at time of birth for each group is shown for reference. Kits were weaned between 4.5 and 5.5 weeks after birth, highlighted by the gray shaded region. Maternally acquired anti-ZIKV E antibodies decreased at a rate of −0.18 log10/week in unimmunized kits, which was similar in RNA-SOSIP-scfl (−0.16 log10/week) and Protein-SOSIP (−0.17 log10/week) vaccinated kits. In comparison, the mean anti-ZIKV E titer in kits vaccinated with RNA-prM/E showed no measurable change over time. We observed a similar trend in kits delivered by RNA-SOSIP-scfl or Protein-SOSIP vaccinated does. Mean anti-gp140 maternal antibodies in kits delivered by RNA-SOSIP-scfl vaccinated does and then receiving saline or the unmatched RNA-prM/E vaccine rapidly waned to the lower limit of detection in approximately 3 weeks after birth, but two doses of RNA-SOSIP-scfl or Protein-SOSIP reversed antibody waning, with mean titer increasing at a rate of 0.14 log10/week or 0.37 log10/week, respectively. Maternal antibodies in both unvaccinated and RNA-prM/E vaccinated kits from Protein-SOSIP vaccinated does waned at a rate of −0.33 log10/week; in vaccinated kits, however, the rate of decay was slower: down to −0.25 log10/week in RNA-SOSIP-scfl and −0.06 log10/week in Protein-SOSIP vaccinated kits. Taken together, these data show that vaccination in kits with pre-existing maternal antibodies resulted in prolonged antibody detection in sera compared with kits receiving mock or no vaccination.
Figure 12.
Vaccination slowed passive waning of maternal antibodies
Mean (±SD) IgG kinetics in kits with maternal antibodies against (A) ZIKV E or (B and C) gp140. Lines show fits by least-squares regression analysis. Mean (±SD) pre-existing antibody levels measured in euthanized newborn kits are shown to indicate the pre-vaccination antibody levels in each cohort of kits. Data arrows indicate weeks when kits in each cohort received RNA-prM/E, RNA-SOSIP-scfl, Protein-SOSIP, or saline (labels in legend). Vertical shaded regions indicate time frame when kits were weaned.
Discussion
MTCT is a major pathway for infections in newborns. We evaluated the ability of our repRNA vaccine platform to induce antibody responses in pregnant NZW rabbits at levels sufficient to allow subsequent maternal transfer to newborn kits. Using two distinct model vaccine targets (HIV-1 and ZIKV), we showed that repRNA immunization at a relatively high dose (100 μg per immunization) was well tolerated in pregnant rabbits with no detrimental impact on litter size compared with the unimmunized group. RepRNA vaccines were also immunogenic, mounting robust antigen-specific antibody responses in pregnant does. The magnitude of circulating antibodies in pregnant does positively and significantly correlated with the magnitude of serum antibodies in newborn kits delivered approximately 2 weeks after the second immunization. Antibody kinetics in pregnant does did, however, indicate that a second booster (third immunization) resulted in a significant boost in responses. These data suggest that timing and frequency of immunization could be adjusted as a means to optimize maternal transfer of antibodies. Our results demonstrate that maternal vaccination with repRNA vaccines can passively transfer neutralizing antibodies to newborns, although protection against infection was not measured in this study. In a maternal HIV-1 vaccination study in non-human primates, Eudailey et al. showed that passively transferred neutralizing antibodies from mother to infants were not enough to protect from repeat oral simian HIV challenges, likely due to rapid waning in passively acquired antibodies.32 Thus, in addition to optimizing maternal vaccination timing to maximize passive antibody transfer, active immunization in newborns might be needed to maintain overall antibody response in infants after birth. We evaluated vaccine-induced antibody responses in kits in the context of pre-existing maternal antibodies. Regardless of vaccination status in does, we found that repRNA vaccines in newborn kits were immunogenic and successfully induced antigen-specific antibody responses. Second, our results demonstrate that vaccinating kits with pre-existing maternal antibodies prevented rapid waning of serum antibody levels relative to unvaccinated kits, supporting the implementation of both pre- and post-natal vaccination to effectively transition newborns from passively acquired to vaccine-induced immunity.
In addition to elucidating strategies for maternal vaccination, this study highlighted the importance of target-specific optimization of antigens expressed from mRNA vaccines. The ZIKV repRNA vaccine encoding prM and E proteins elicited robust binding and neutralizing antibodies in does. On the other hand, only the recombinant protein (Protein-SOSIP vaccine) version of BG505 SOSIP.664 trimer induced both binding and autologous neutralizing antibody responses. The repRNA version of the soluble trimer (RNA-SOSIP-scfl), which replaced the furin cleavage site with a single-chain flexible linker, induced robust binding responses but only two out of five (40%) rabbits had detectable levels of autologous neutralizing antibodies in serum. Unlike ZIKV prM and E proteins, which self-assemble in the cytosol before budding to mimic ZIKV virions, the complex quaternary structure of secreted BG505 SOSIP.664 is achieved in the supernatant. In vitro expressed BG505 SOSIP.664 timer is affinity purified using the conformation-specific 2G12 monoclonal bnAb to exclude a subpopulation of gp140 monomers or uncleaved trimers,24,33 which is not possible to do for proteins expressed in vivo from mRNA vaccines. Expression of native, membrane-bound Env or engineered outer domains (eODs) anchored to protein nanoparticles may be more suitable for in vivo expression by nucleic acid vaccines. Curiously, few previous studies of mRNA-based HIV-1 vaccines reported neutralizing antibody responses. Aldon et al. evaluated polyethyleneimine-formulated self-amplifying RNA, comparing immunogenicity of soluble and membrane-bound Env trimer in mice, guinea pigs, rabbits, and non-human primates, but did not report neutralizing antibody responses.34 Melo et al. used an LNP-formulated self-amplifying RNA vaccine expressing eOD-GT8—a nanoparticle protein displaying the engineered outer domain of Env—and, although they did not report neutralizing antibody responses, they showed robust germinal center formation due to sustained antigen expression in draining lymph nodes.35 More recently, Zhang et al. demonstrated induction of neutralization antibody responses with LNP-formulated mRNA encoding membrane-bound Env.36 Based on results presented here and previously published reports, we are evaluating both humoral and cellular responses induced by membrane and nanoparticle anchored Env trimers delivered with our LION/repRNA platform.
Conclusions
We demonstrate the potential of our LION-formulated repRNA platform for maternal and neonatal vaccination in a rabbit model. LION is a mature cationic emulsion delivery technology in different phases of clinical evaluation across multiple countries as a component within repRNA SARS-CoV-2 vaccines. There is a scarcity in immunogenicity and safety evaluation of RNA-based vaccines in pregnant animal models to support clinical development of next-generation RNA vaccines against pathogens of high MTCT risk. Using ZIKV and HIV-1 as model pathogens needing maternal vaccination to protect against MTCT, we demonstrated robust induction of antibody responses in pregnant rabbits, in utero transfer of maternal antibodies to newborn kits, and successful induction of vaccine-induce immunogenicity in kits regardless of vaccination history in mothers. Our study also highlights the importance of timing in vaccination of pregnant mothers to maximize vertical transfer of immunity. Additional studies are required to determine if RNA-based maternal vaccines can afford protection against infection by MTCT to newborns.
Materials and methods
RNA production and qualification
DNA plasmid templates were linearized by restriction digest using Not-I enzyme (NEB) and purified by phenol-chloroform extraction followed by sodium acetate precipitation. RNA was transcribed using recombinantly produced T7 polymerase, pyrophosphatase, RNAse inhibitor, DNAse enzymes (Aldevron), and reaction buffer. RNA was subsequently capped with Vaccinia virus capping enzyme (Aldevron) using guanosine triphosphate and S-adenosyl-methionine (New England Biolabs) to add a cap-0 structure. RNA was purified using LiCl precipitation and quantified by nanodrop measurement. RNA integrity and size were confirmed using denaturing gel electrophoresis.
Transfections and western blots
Three micrograms of each RNA construct were complexed using Lipofectamine 2000 (Invitrogen) per manufacturer’s instructions and added to 6-well tissue culture plates. BHK-21 cells were then added to each well, and cells and supernatants were harvested 24 h post transfection. Supernatants containing Gag VLPs were concentrated using ultracentrifugation through 20% sucrose cushions at 100,000 RCF followed by resuspension in PBS. Cell lysates, supernatants, and concentrated Gag VLPs were separated on denaturing SDS-PAGE gels (Invitrogen) under reducing and non-reducing conditions followed by immunoblotting using anti-gp120 (2G12 from Polymun Scientific), anti-gp41 (4E10). The following reagents were obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: anti-human immunodeficiency virus (HIV)-1 gp41 monoclonal antibody (4E10), ARP-10091 (contributed by DAIDS/NIAID), or an anti-Gag (cat. #ab63917 from Abcam) antibody.
Animals and husbandry
A total of 35 female 6- to 8-week-old C57Bl/6 mice (Charles River Laboratories) were used for the immunogenicity screening study and were maintained in specific pathogen-free conditions. For the dose-response rabbit immunogenicity study, a total of 28 (∼8–9 months old) and for the maternal vaccine study, a total of 20 young adult female (approximately 28 weeks of age and 3.0–4.0 kg) and six proven-breeder male NZW rabbits (HsdHra:(NZW) SPF) were purchased from Envigo Global Services (Denver, PA). Rabbits were singly housed in standard, commercially available, non-ventilated rabbit racks (EuroCage; Allentown, Allentown, NJ, USA). The doe with litter was maintained in the double cage unit until the time of weaning (approximately 7 weeks of age). Males were only used for breeding purposes. All procedures for the care and use of mice were approved by Infectious Disease Research Institute’s Institutional Animal Care and Use Committee (IACUC). All procedures for the care and use of rabbits were approved by Duke University’s IACUC.
LION formulation
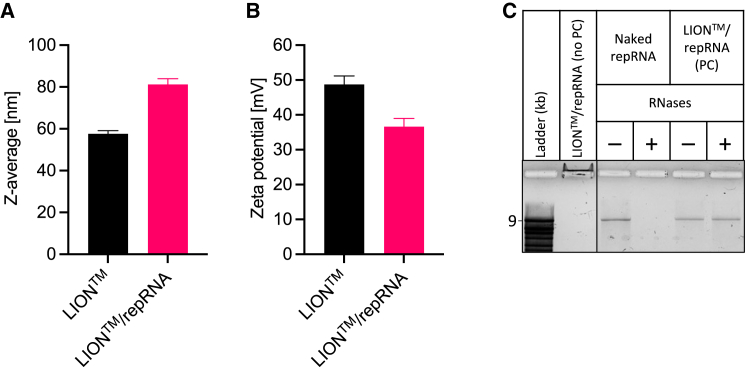
LION is HDT Bio’s cationic oil-in-water nanoparticle emulsion optimized for intramuscular delivery of nucleic acids. The average hydrodynamic size and zeta potential (Figure 13) is 60 nm (intensity-weighted Z-average diameter) and +50 mV (1:100 dilution in unbuffered Milli-Q water), respectively. After mixing with repRNA, particle size increases and zeta potential decreases, indicating the formation of LION/repRNA complexes driven by electrostatic interaction between the quaternary ammonium cations of DOTAP on the surface of LION nanoparticles and phosphate anions in the repRNA backbone. Electrophoresis of the LION/repRNA complex in a 1% agarose gel shows that repRNA is completely bound to LION, with no free repRNA detected. Bound repRNA was observed migrating to the cathode (negative electrode) due to the net positive charge of the complex. Moreover, LION protected repRNA from RNase-catalyzed hydrolysis, as shown in Figure 13C, as the complex structure likely creates a steric barrier that inhibits RNases from binding with repRNA. The manufacturing process of LION has been detailed previously;12 in brief, it involves iterative passaging an oil-in-water suspension containing squalene, the cationic lipid DOTAP chloride, and non-ionic surfactants sorbitan monostearate and polysorbate 80 through a high-pressure microfluidizer (Microfluidics M110P) equipped with a 75-μm Y-type diamond interaction chamber until the target hydrodynamic diameter is achieved. The final bulk LION nanoemulsion was aseptically filtered using a sterile 0.2-μm media filter unit (Nalgene) and stored at 2°C–8°C.
Figure 13.
Physicochemical characteristics of LION formulation and LION/repRNA complexed vaccine
Mean (±SD) (A) z-average particle size and (B) zeta potential characteristics before and after mixing LION and repRNA to form LION/repRNA complex at N:P ratio of 15. (C) Denaturing agarose gel electrophoresis was performed to assess binding of repRNA with LION, shown in lane labeled “LION™/repRNA complex (no PC),” where PC denotes phenol-chloroform extraction. As observed, no free repRNA was detected, and bound repRNA migrated toward the negatively charged electrode due to the net positive charge of the LION/repRNA complex. Also shown is the role of LION in repRNA protection against RNase-catalyzed hydrolysis. Naked (unformulated) repRNA or LION/repRNA complex was incubated in nuclease-free water (−) or RNases (+), followed by proteinase K inhibition and extraction with phenol-chloroform (PC). Compared with naked repRNA, LION-formulated repRNA was protected from RNases. All samples were run on the same gel.
Immunizations
RNA vaccines were formulated with LION at a nitrogen/phosphate (N:P) molar ratio of 15. In brief, repRNA and LION were diluted separately to 2× target dose in nuclease-free water or 20% sucrose/10 mM sodium citrate (pH 6.0) buffer, respectively, and combined 1:1 by volume. The mixed vaccine was allowed to equilibrate at least 30 min on ice before injection. The BG505 SOSIP.664 protein was donated by the Trimer production core at Cornell and at a stock 1 mg/mL concentration. Prior to injection, the protein was diluted to 120 ng/μL in 2× PBS and then mixed with an equal volume of AddaVax adjuvant (cat. #vac-adx-10 from Invivogen) resulting in 60 ng/μL final protein concentration. Mice were immunized by intramuscular injection of 50 μL of vaccine in the thigh. Adult female rabbits were immunized via the intramuscular route. Each rabbit received four intramuscular injections of 125 μL (500 μL in total), two in each quadriceps muscle. Kits were immunized starting at approximately 1 week of age onward and because of their smaller size, a total of two intramuscular injections of 125 μL, one in each quadriceps muscle, were administered.
Breeding
Adult female rabbits were bred by live cover. In brief, does were placed in the male’s cage and after visualization of a successful mounting noted by the male falling over, the doe was returned to her cage. Males were limited to breeding 10–12 does per week and received at least two rest days per week.
Blood collection
Blood was collected from the retro-orbital sinus of immunized mice. For rabbits, adult females were sedated with acepromazine (1–2 mg/kg) delivered intramuscularly, and an analgesic cream (lidocaine 2.5% and prilocaine 2.5% cream) was applied topically over the central ear arteries prior to blood collection from this vessel. Owing to the small size, the lateral saphenous vein was also used as a collection site for the rabbit kits while under isoflurane-induced anesthesia. Terminal, intracardiac blood collection while under deep anesthesia with ketamine and xylazine (ketamine: 30–40 mg/kg + xylazine: 3–5 mg/kg, intramuscularly) was also performed on kits at the time of euthanasia. Blood was collected in serum separator tubes and spun for serum isolation per manufacturer’s recommendation. Serum was then stored at −20°C until further analyses.
Euthanasia
In accordance with the 2020 AVMA Guidelines for the Euthanasia of Animals, rabbits were fully anesthetized with a dose of ketamine and xylazine (ketamine: 30–40 mg/kg + xylazine: 3–5 mg/kg, intramuscularly) prior to euthanasia with an overdose of Euthasol (pentobarbital sodium and phenytoin sodium administered via intracardiac or intraperitoneal routes in small kits; at least 1 mL per 2 kg).
Enzyme-linked immunosorbent assay
Antigen-specific antibody responses were measured using ELISA. In brief, Corning high-binding 384-well plates were coated with 2 μg/mL of either BG505 SOSIP.664 protein (donated by the Trimer Core at Weill Cornell Medicine) or ZIKV E protein (Meridian Life Science) diluted in PBS to measure HIV- and ZIKV-specific antibodies, respectively. Plates were blocked with PBS + 0.05% Tween 20 + 2% non-fat dry milk +1% goat serum prior to the addition of an eight-point 5-fold series of serum dilutions made in assay buffer (PBS + 0.05% Tween 20 + 1% non-fat dry milk +1% goat serum) starting with a 1:50 dilution. Goat anti-rabbit IgG-horseradish peroxidase secondary antibody (Southern Biotech #4030-05) was added at a 1:16,000 dilution in assay buffer, and plates were developed with SureBlue TMB peroxidase substrate (Seracare) followed by quenching with TMB Stop Solution (Seracare). Optical density at each dilution was measured at 405 nm and fit to a four-parameter logistic regression model using GraphPad Prism software. Endpoint reciprocal dilution titers were calculated from the best-fit parameters by interpolating the x value (dilution) at y value (absorption cutoff) 3 standard deviations above media background.
HIV pseudovirus neutralization assay
HIV-1 pseudovirus assays were performed with BG505 as described previously (PMID: 32470041). TZM-bl cells (expressing CD4 and CCR5, obtained from the NIH AIDS Reagent Program) were maintained in DMEM complete (Gibco) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were plated at a density of 4,000 cells per well 24 h prior to addition of virus and plasma. Heat-inactivated plasma was tested at a dilution of 1:50 in a total volume of 60 μL per well in triplicate wells. Pre-bleed plasma was used as an entry control to account for serum-associated background. Equal volume of diluted BG505 pseudovirus (in the NL4-3.Luc.RE backbone) was added to diluted plasma and incubated at 37°C for 1.5 h. Prior to the addition of the plasma/virus mixture, cells were treated at 37°C with 2 μg/mL polybrene for 30 min. The polybrene was then aspirated from the cells and replaced with 100 μL of plasma-pseudovirus mixture. Plates were incubated at 37°C for 72 h, medium was aspirated, and cells were lysed to measure luciferase activity using Steady-Glo Luciferase Reagent (Promega, Madison, WI). The percent neutralization at 1:50 was determined as [(RLUpre − RLUpost)/RLUpre] × 100, where RLUpre is the average RLU for pre-bleed plasma and RLUpost is the average RLU for immune samples.
ZIKV neutralization assay
To quantify ZIKV neutralizing antibody responses in animal sera, we used a real-time cell analysis platform (xCELLigence; Agilent) that monitors changes to monolayers of cells, such as virus-induced cytopathic effect. After recording a background reading of cell-culture medium in a 96-well E-plate (Agilent), Vero cells were seeded in each well and the plate returned to the plate reader with impedance measurements recorded every 15 min. The next day, serial dilutions of heat-inactivated sera were incubated with ZIKV (PRVABC56 isolate from BEI Resources) and added to the E-plate of Vero cells along with naive-serum and medium-only controls. Measurements were recorded every 15 min for 72–84 h, and percent neutralization was calculated based on areas under the impedance curves, normalized to the media-only control with background subtraction based on the naive-serum controls.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
This research was supported in part by NIAID contract #75N93019C00008, NIAID UW CFAR grant #2P30AI027757-31, and NIAID grant 1R61AI161811-01. The authors thank Drs. Malcolm Duthie and Darrick Carter for providing a critical review of the manuscript and their suggestions. The authors also thank Drs. John Moore and Albert Cupo from the Trimer Core at Weill Cornell Medicine for donating the BG505 SOSIP.664 trimer protein.
Author contributions
A.P.K. designed the LION nanoparticle formulation, wrote the original draft of the manuscript, and conceptualized the project. A.P.K., C.D.L., and H.F.S. conceived the maternal immunization experimentation. C.D.L. and H.F.S. performed the rabbit experiments. J.A. synthesized and characterized all RNA constructs. K.K., S.R., and B.J.B. developed and performed ELISAs. J.H.E. and N.L.W. performed ZIKV neutralization assays. N.D.S. directed the HIV pseudovirus neutralization assay. All authors reviewed and edited the original draft of the manuscript.
Declaration of interests
A.P.K., J.A., S.R., K.K., B.J.B., N.L.W., and J.H.E. have equity interest in HDT Bio and are listed as inventors on patents covering the LION formulation.
References
- 1.CDC COVID-19 vaccine recommendations for children and teens. https://www.cdc.gov/coronavirus/2019-ncov/vaccines/vaccine-safety-children-teens.html
- 2.Shimabukuro T.T., Kim S.Y., Myers T.R., Moro P.L., Oduyebo T., Panagiotakopoulos L., Marquez P.L., Olson C.K., Liu R., Chang K.T., et al. Preliminary findings of mRNA Covid-19 vaccine safety in pregnant persons. N. Engl. J. Med. 2021;384:2273–2282. doi: 10.1056/nejmoa2104983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim D.Y., Atasheva S., McAuley A.J., Plante J.A., Frolova E.I., Beasley D.W.C., Frolov I. Enhancement of protein expression by alphavirus replicons by designing self-replicating subgenomic RNAs. Proc. Natl. Acad. Sci. USA. 2014;111:10708–10713. doi: 10.1073/pnas.1408677111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H., et al. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erasmus J.H., Archer J., Fuerte-Stone J., Khandhar A.P., Voigt E., Granger B., Bombardi R.G., Govero J., Tan Q., Durnell L.A., et al. Intramuscular delivery of replicon RNA encoding ZIKV-117 human monoclonal antibody protects against zika virus infection. Mol. Ther. Methods Clin. Dev. 2020;18:402–414. doi: 10.1016/j.omtm.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollock K.M., Cheeseman H.M., Szubert A.J., Libri V., Boffito M., Owen D., Bern H., O’Hara J., McFarlane L.R., Lemm N.-M., et al. Safety and immunogenicity of a self-amplifying RNA vaccine against COVID-19: COVAC1, a phase I, dose-ranging trial. EClinicalMedicine. 2021;44:101262. doi: 10.2139/ssrn.3859294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Low J.G., Alwis R. de, Chen S., Kalimuddin S., Leong Y.S., Mah T.K.L., Yuen N., Tan H.C., Zhang S.L., Sim J.X.Y., et al. A phase 1/2 randomized, double-blinded, placebo controlled ascending dose trial to assess the safety, tolerability and immunogenicity of ARCT-021 in healthy adults. medRxiv. 2021 doi: 10.1101/2021.07.01.21259831. Preprint at. [DOI] [Google Scholar]
- 8.Arcturus Theraputics Holdings Inc. Arcturus announces self-amplifying COVID-19 mRNA vaccine candidate ARCT-154 meets primary efficacy endpoint in phase 3 study. 2022. https://ir.arcturusrx.com/news-releases/news-release-details/arcturus-announces-self-amplifying-covid-19-mrna-vaccine
- 9.Tseng H.F., Ackerson B.K., Luo Y., Sy L.S., Talarico C.A., Tian Y., Bruxvoort K.J., Tubert J.E., Florea A., Ku J.H., et al. Effectiveness of mRNA-1273 against SARS-CoV-2 Omicron and Delta variants. Nat. Med. 2022;28:1063–1071. doi: 10.1038/s41591-022-01753-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawman D.W., Meade-White K., Archer J., Leventhal S.S., Wilson D., Shaia C., Randall S., Khandhar A.P., Krieger K., Hsiang T.-Y., et al. SARS-CoV2 variant-specific replicating RNA vaccines protect from disease and pathology and reduce viral shedding following challenge with heterologous SARS-CoV2 variants of concern. Elife. 2022;11:e75537. doi: 10.7554/elife.75537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawman D.W., Meade-White K., Clancy C., Archer J., Hinkley T., Leventhal S.S., Rao D., Stamper A., Lewis M., Rosenke R., et al. Replicating RNA platform enables rapid response to the SARS-CoV-2 Omicron variant and elicits enhanced protection in naïve hamsters compared to ancestral vaccine. Ebiomedicine. 2022;83:104196. doi: 10.1016/j.ebiom.2022.104196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erasmus J.H., Khandhar A.P., O’Connor M.A., Walls A.C., Hemann E.A., Murapa P., Archer J., Leventhal S., Fuller J.T., Lewis T.B., et al. An Alphavirus -derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci. Transl. Med. 2020;12:eabc9396. doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.New York Times Coronavirus vaccine tracker. https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html
- 14.HDT Bio Corp. press release Brazil’s SENAI CIMATEC doses first healthy volunteers in phase 1 trial of HDT Bio’s RNA COVID-19 vaccine. 2022. https://www.hdt.bio/news-blog/brazils-senai-cimatec-doses-first-healthy-volunteers-in-phase-1-trial-of-hdt-bios-rna-covid-19-vaccine
- 15.HDT Bio Corp. press release HDT Bio doses first healthy volunteer in US phase 1 trial of next generation COVID-19 variant RNA vaccine. 2022. https://www.hdt.bio/news-blog/hdt-bio-initiates-us-phase-1-trial-of-covid-19-variant-rna-vaccine
- 16.HDT Bio Corp. press release HDT Bio partner quratis doses first healthy volunteers in phase 1 trial of HDT Bio’s RNA COVID-19 vaccine in South Korea. 2021. https://www.hdt.bio/news-blog/hdt-bio-partner-quratis-doses-first-healthy-volunteers-in-phase-1-trial-of-hdt-bios-rna-covid-19-vaccine-in-south-korea
- 17.Antoniou E., Orovou E., Sarella A., Iliadou M., Rigas N., Palaska E., Iatrakis G., Dagla M. Zika virus and the risk of developing microcephaly in infants: a systematic review. Int. J. Environ. Res. Public Health. 2020;17:3806. doi: 10.3390/ijerph17113806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erasmus J.H., Khandhar A.P., Guderian J., Granger B., Archer J., Archer M., Gage E., Fuerte-Stone J., Larson E., Lin S., et al. A nanostructured lipid carrier for delivery of a replicating viral RNA provides single, low-dose protection against zika. Mol. Ther. 2018;26:2507–2522. doi: 10.1016/j.ymthe.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman G., Mazanderani A.H. In: HIV Infection in Children and Adolescents. Bobat R., editor. Springer; 2020. Diagnosis of HIV in children and adolescents; pp. 15–22. [DOI] [Google Scholar]
- 20.Fouda G.G., Cunningham C.K., Permar S.R. Infant HIV-1 vaccines: supplementing strategies to reduce maternal-child transmission. Jama. 2015;313:1513–1514. doi: 10.1001/jama.2015.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen J. Far from over. Science. 2018;360:1162–1163. doi: 10.1126/science.360.6394.1162. [DOI] [PubMed] [Google Scholar]
- 22.UNAIDS Unaids data 2021. 2021. https://www.unaids.org/sites/default/files/media_asset/JC3032_AIDS_Data_book_2021_En.pdf
- 23.Lynch R.M., Shen T., Gnanakaran S., Derdeyn C.A. Appreciating HIV type 1 diversity: subtype differences in env. AIDS Res. Hum. Retroviruses. 2009;25:237–248. doi: 10.1089/aid.2008.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders R.W., Derking R., Cupo A., Julien J.-P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Peña A.T., Korzun J., et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. Plos Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkar A., Bale S., Behrens A.-J., Kumar S., Sharma S.K., de Val N., Pallesen J., Irimia A., Diwanji D.C., Stanfield R.L., et al. Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer. Nat. Commun. 2018;9:1956. doi: 10.1038/s41467-018-04272-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong L., He L., de Val N., Vora N., Morris C.D., Azadnia P., Sok D., Zhou B., Burton D.R., Ward A.B., et al. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat. Commun. 2016;7:12040. doi: 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulp D.W., Steichen J.M., Pauthner M., Hu X., Schiffner T., Liguori A., Cottrell C.A., Havenar-Daughton C., Ozorowski G., Georgeson E., et al. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat. Commun. 2017;8:1655. doi: 10.1038/s41467-017-01549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X., Parast A.B., Richardson B.A., Nduati R., John-Stewart G., Mbori-Ngacha D., Rainwater S.M.J., Overbaugh J. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 2006;80:835–844. doi: 10.1128/jvi.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgiev I.S., Joyce M.G., Yang Y., Sastry M., Zhang B., Baxa U., Chen R.E., Druz A., Lees C.R., Narpala S., et al. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 env. J. Virol. 2015;89:5318–5329. doi: 10.1128/jvi.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Vergès S., Camus G., Blot G., Beauvoir R., Benarous R., Berlioz-Torrent C. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc. Natl. Acad. Sci. USA. 2006;103:14947–14952. doi: 10.1073/pnas.0602941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eudailey J.A., Dennis M.L., Parker M.E., Phillips B.L., Huffman T.N., Bay C.P., Hudgens M.G., Wiseman R.W., Pollara J.J., Fouda G.G., et al. Maternal HIV-1 env vaccination for systemic and breast milk immunity to prevent oral SHIV acquisition in infant macaques. Msphere. 2018;3 doi: 10.1128/msphere.00505-17. e00505-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dey A.K., Cupo A., Ozorowski G., Sharma V.K., Behrens A.-J., Go E.P., Ketas T.J., Yasmeen A., Klasse P.J., Sayeed E., et al. cGMP production and analysis of BG505 SOSIP.664, an extensively glycosylated, trimeric HIV-1 envelope glycoprotein vaccine candidate. Biotechnol. Bioeng. 2018;115:885–899. doi: 10.1002/bit.26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aldon Y., McKay P.F., Moreno Herrero J., Vogel A.B., Lévai R., Maisonnasse P., Dereuddre-Bosquet N., Haas H., Fábián K., Le Grand R., et al. Immunogenicity of stabilized HIV-1 Env trimers delivered by self-amplifying mRNA. Mol. Ther. Nucleic Acids. 2021;25:483–493. doi: 10.1016/j.omtn.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melo M., Porter E., Zhang Y., Silva M., Li N., Dobosh B., Liguori A., Skog P., Landais E., Menis S., et al. Immunogenicity of RNA replicons encoding HIV env immunogens designed for self-assembly into nanoparticles. Mol. Ther. 2019;27:2080–2090. doi: 10.1016/j.ymthe.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang P., Narayanan E., Liu Q., Tsybovsky Y., Boswell K., Ding S., Hu Z., Follmann D., Lin Y., Miao H., et al. A multiclade env–gag VLP mRNA vaccine elicits tier-2 HIV-1-neutralizing antibodies and reduces the risk of heterologous SHIV infection in macaques. Nat. Med. 2021;27:2234–2245. doi: 10.1038/s41591-021-01574-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.