Abstract
Nonsense mutations are responsible for around 10% of cases of genetic diseases, including cystic fibrosis. 2,6-diaminopurine (DAP) has recently been shown to promote efficient readthrough of UGA premature stop codons. In this study, we show that DAP can correct a nonsense mutation in the Cftr gene in vivo in a new CF mouse model, in utero, and through breastfeeding, thanks, notably, to adequate pharmacokinetic properties. DAP turns out to be very stable in plasma and is distributed throughout the body. The ability of DAP to correct various endogenous UGA nonsense mutations in the CFTR gene and to restore its function in mice, in organoids derived from murine or patient cells, and in cells from patients with cystic fibrosis reveals the potential of such readthrough-stimulating molecules in developing a therapeutic approach. The fact that correction by DAP of certain nonsense mutations reaches a clinically relevant level, as judged from previous studies, makes the use of this compound all the more attractive.
Keywords: cystic fibrosis; nonsense mutation; readthrough molecule; 2,6-diaminopurine; mouse model
Graphical abstract
No treatment is currently available for patients with genetic diseases caused by a nonsense mutation. Leroy et al. show the potential of 2,6-diaminopurine for the treatment of these diseases, taking cystic fibrosis as an example.
Introduction
Cystic fibrosis (CF) is the most frequent rare disease, with about 1 case per 3,000 births in Europe and the United States. This autosomal recessive pathology is related to malfunction or absence of the CF transmembrane conductance regulator (CFTR), a channel transporting chloride ions from the intracellular to the extracellular environment. CFTR is a membrane protein that is primarily expressed by epithelial cells for regulating mucus production. About 2,000 mutations have been identified in CF, including delF508 in nearly 70%–80% of CF cases (www.orpha.net). Fortunately, for this mutation in particular, treatments with two molecules (lumacaftor-ivacaftor, or Orkambi, or tezacaftor-ivacaftor, or Symdeko) or three molecules (elexacaftor-tezacaftor-ivacaftor, or Trikafta) are available to patients from the manufacturer Vertex Pharmaceuticals. As these CFTR modulators restore CFTR folding and CFTR potentiation, there remains a large group of CF patients that do not respond to these treatments due to mutations that result in complete absence of CFTR protein. About 10% of the CF patients carry mutations that result in nonsense mutations. Nonsense mutations are responsible for about 10% of genetic disease cases, including CF.1 The presence of a nonsense mutation results in low expression levels of the mutant mRNA, due to activation of nonsense-mediated mRNA decay (NMD), an mRNA surveillance mechanism that degrades mRNAs harboring a premature termination codon (PTC).2,3,4,5,6,7 Strategies for rescuing the expression of genes carrying a nonsense mutation include NMD inhibition,8,9,10,11,12,13 gene therapy,14 gene editing,15,16,17 antisense oligonucleotides,18,19,20 or PTC readthrough, a mechanism leading to the introduction of an amino acid at the PTC position during translation.21,22,23 Molecules stimulating PTC readthrough include aminoglycosides (e.g., G418 or ELX-02) and non-aminoglycosides (e.g., ataluren or amlexanox).24,25,26 Although ataluren and ELX-02 have reached the clinical trial stage, their efficacy is still debatable,27,28 and currently there are no treatments available for patients with a nonsense mutation, hence the need to identify novel molecules capable of efficiently and safely correcting nonsense mutations in pathological contexts.
Recently, the purine derivative 2,6-diaminopurine (DAP) has been shown to correct UGA nonsense mutations efficiently by activating PTC readthrough.29 In human cells, DAP rescues the expression and functions of genes carrying UGA nonsense mutations more efficiently than G418. DAP has also been shown to correct UGA nonsense mutations in vivo after oral exposure of mice to DAP.29 DAP causes readthrough by inhibiting the activity of FTSJ1, a transmethylase that modifies certain tRNAs post-transcriptionally, including that carrying the amino acid tryptophan. Hence, DAP-induced readthrough leads to incorporation of a tryptophan at the position of the PTC UGA.29 Therefore, DAP is particularly suitable for correcting mutations that convert a tryptophan-encoding codon to a PTC. An example is W1282X, accounting for 18% of CF-causing nonsense mutations.27 This makes it worthwhile to evaluate the capacity of DAP to correct UGA nonsense mutations in several pathological contexts. Here, in animal models of CF pathology, CF-patient-derived organoids, and patient cells carrying a UGA nonsense mutation in the CFTR gene, we provide evidence in favor of using DAP to restore CFTR function.
Results
Pharmacokinetics of DAP in mouse
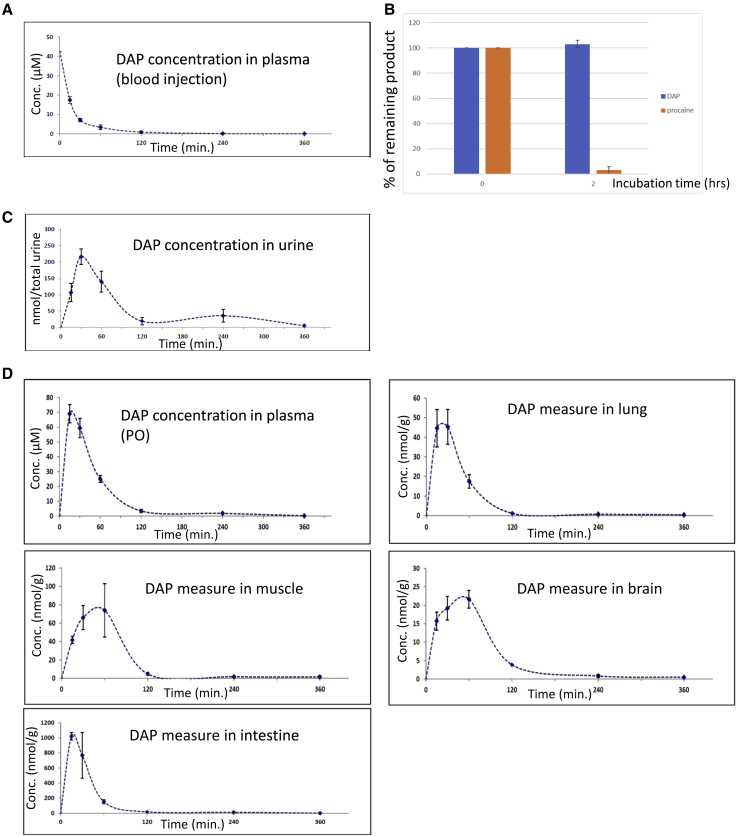
DAP has been shown to correct UGA nonsense mutations in human cells.29 It is also reported to be stable in hepatic extract, where no degradation was observed even after 4 h of incubation. To evaluate further the possibility of using DAP in vivo, several pharmacokinetic parameters were measured. First, the DAP concentration in plasma was measured at different time points after intravenous DAP injection at 8.1 mg/kg (Figure 1A). This concentration was found to decrease rapidly, DAP being undetectable 2 h post-injection. These results indicate that DAP is either rapidly degraded in plasma or naturally eliminated. To test these hypotheses, the stability of DAP was measured in plasma (Figure 1B). The DAP concentration was found to remain unchanged over a 2-h incubation period, unlike procaine, used as a positive control. As this suggests that DAP could be rapidly eliminated via the urinary tract, its presence in urine was measured at different time points post-injection (Figure 1C). The amount of DAP detected in urine samples was found to peak 30 min after intravenous injection, indicating that DAP is cleared very quickly by this route. It was not possible to establish a concentration, because the volume of urine taken from the bladder at the time of sacrifice depended on the last time the animal had urinated. During injection, about 1.35 μmol of DAP was introduced into the body, of which about 40% was found in the urine. This 40% is an underestimate, as we cannot exclude that the mouse might have urinated between two time points. It appears, however, that not all of the DAP is removed by this route and that a certain amount is temporarily retained within different tissues.
Figure 1.
Pharmacokinetic parameters of DAP in CD-1 mouse
(A) DAP plasma levels after blood injection of DAP at 8.1 mg/kg. Plasma concentrations of DAP were measured 30, 60, 120, 240, and 360 min post-injection. (B) Measure of DAP stability in plasma. DAP or procaine (positive control) was incubated for 2 h in plasma to assess its stability. (C) Measure by mass spectrometry of the concentration of DAP in urine after DAP injection. (D) DAP plasma levels after PO exposure of 29 mg/kg DAP (upper left), and biodistribution of DAP after PO exposure. DAP was measured by mass spectrometry in lung, muscle, brain tissues, and small intestine. Three mice were used for each time point. Error bars show the SD.
Since DAP has previously been shown to correct a nonsense mutation in vivo following per os (PO) exposure,29 a very convenient way to administer medication, the DAP plasma level was measured under these exposure conditions (Figure 1D). Consistent with the previous results,29 DAP was detected in plasma after PO exposure. Its blood concentration peaked at about 70 μM 15–30 min after exposure, corresponding to the necessary absorption time. It dropped to 25 μM 1 h after PO exposure and to 1.8 μM 4 h after PO exposure (Table 1). This suggests that readthrough should still be promoted 4 h after PO exposure, since our previously reported results show that DAP readthrough can be measured by western blotting at DAP concentrations as low as 1.6 μM.29
Table 1.
Measurement of the DAP concentration (in μM) as a function of time (in hours) after per os exposure
| Hour after per os exposure | Concentration (μM) |
|---|---|
| 0.25 | 69 ± 8 |
| 0.5 | 59 ± 8 |
| 1 | 25 ± 3 |
| 2 | 3.4 ± 1 |
| 4 | 1.8 ± 0.5 |
| 6 | 0.15 ± 0.02 |
The next step was therefore to measure the amount of DAP present in different tissues to assess its biodistribution (Figure 1D). The quantity of DAP was thus measured in muscle, lung, brain, and intestine tissues following DAP PO exposure at 29 mg/kg. The quantity of DAP measured in each tissue was divided by the tissue mass. DAP was detected in all four tested tissues, with a very strong exposure of the small intestines. This suggests that DAP undergoes broad tissue distribution. The peak level was reached 15–30 min post-exposure, in keeping with the concentration curve of DAP in plasma (Figure 1D). After peaking, DAP decreased rapidly in all tissues but the brain, where the decrease was slower, suggesting transient accumulation in this tissue (Figure 1D). The fact that DAP can reach the brain also indicates that it crosses the blood-brain barrier. It should be noted that the biodistribution of DAP measured after PO exposure is closely similar to the biodistribution of DAP measured in the same organs after blood injection, indicating that the route of exposure does not appear to influence the biodistribution of DAP (Figure S1). Overall, these results show that once introduced into the blood, DAP can reach different tissues without being massively retained there. Overall, the results of Figure 1, in combination with previously reported data, indicate that DAP can be used in vivo to correct nonsense mutations.29
Characterization of a new mouse model of CF carrying a nonsense mutation in Cftr
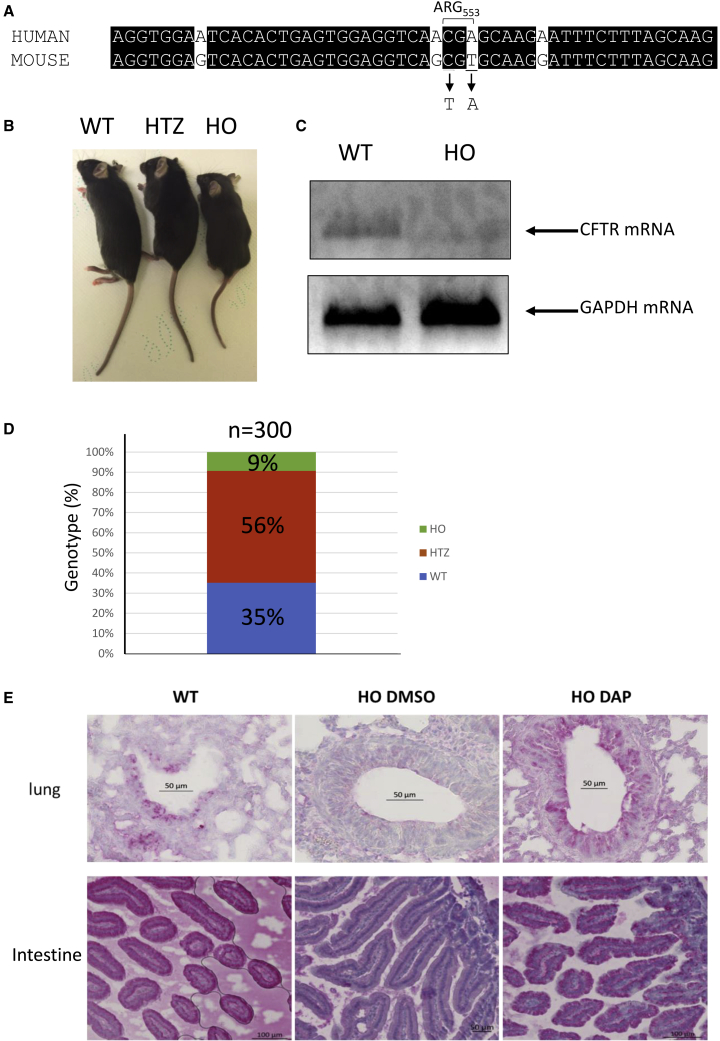
As at the start of this project there existed no mouse model carrying an endogenous nonsense mutation in the mouse Cftr gene, the Cre-LOX system was used to substitute a UGA nonsense mutation for arginine codon 553 in the mouse Cftr gene30 (Figure 2A). This new mouse model was named CFTR-NS. In keeping with previous reports that Cftr-knockout mice are smaller than their heterozygous and wild-type counterparts, we observed this also with the CFTR-NS model (Figure 2B).31,32 CFTR mRNA is not detectable by RT-PCR in the lung of CFTR-NS mice, unlike wild-type mice, likely due to NMD (Figure 2C). In addition, most Cftr-knockout mice die less than 2 months after birth and are reported to develop intestinal obstructions during the first weeks of life. In our model, genotyping performed 2 weeks after birth showed that mice homozygous for the mutation (referred hereafter as HO) were underrepresented: only about 9% showed this genotype, as opposed to the expected 25%. This genotype would thus appear to be lethal (Figure 2D). The percentage of pups with a heterozygous genotype was 56%, as expected, and about 35% of the pups displayed a homozygous wild-type genotype, this disequilibrium reflecting the low proportion of HO pups. Figure S2 shows examples of growth curves of wild-type (WT) and HO mice surviving more than 1 month after birth. The curves show that the absence of CFTR leads to delayed growth and to a growth reduction of about 30%–40%, as previously reported.31 Last, the absence of CFTR was assessed by immunohistochemistry (IHC) applied to lung and intestinal tissues, reported to express CFTR.33 In WT mice, but not HO mice treated with DMSO, CFTR staining was clearly detected in cells forming the bronchi and bronchioles of lung tissue and in cells forming the intestinal villi (Figure 2E). Overall, the results of Figure 2 demonstrate that CFTR-NS is a valid mouse model for studying the in vivo efficacy of nonsense mutation correctors.
Figure 2.
The CFTR-NS mouse model
(A) Alignment of the human and mouse sequences in the region containing arginine 553. The two nucleotides changed in the murine sequence to generate the UGA stop codon are underlined. (B) Picture showing a homozygous wild-type mouse (WT), a mouse heterozygous for the nonsense mutation (HTZ), and a mouse homozygous for the nonsense mutation (HO). (C) Measurement of CFTR mRNA level in the lungs of wild-type (WT) and CFTR-NS (HO) mice by RT-PCR. Measurement of the GAPDH mRNA level serves as a loading control. (D) Proportion of the three genotypes at weaning. (E) Detection of CFTR (in red) by IHC in lung (around the bronchi) and intestinal tissues (duodenum) from WT and HO mice exposed PO to DMSO or DAP. All the IHCs are representative of at least five mice.
In CFTR-NS mice, DAP corrects the nonsense mutation in the Cftr gene
A previous report has shown that DAP can rescue expression of a gene carrying a UGA nonsense mutation in xenografted cells in mouse.29 To demonstrate that DAP can correct endogenous nonsense mutations in the Cftr gene, 4-week-old CFTR-NS mice were exposed PO daily to 10% DMSO solution or 1 mg DAP for 3 days before collection of their lungs and intestines to check for Cftr expression (Figure 2E). In WT mice, red staining of the CFTR protein appeared highly concentrated around the bronchi and bronchioles, on the central lumen side. CFTR was also detected in cells forming the intestinal villi, again on the side of the central lumen. In contrast, DMSO-treated HO mice displayed no specific labeling. DAP-treated lung and intestinal tissues from HO mice, on the other hand, did show specific staining of CFTR. This result, consistent with the biodistribution data of Figure 1D, indicates that in mice exposed PO to DAP for 3 days, DAP can diffuse through their bodies and correct a UGA nonsense mutation in the endogenous Cftr gene.
DAP restores Cftr expression in utero
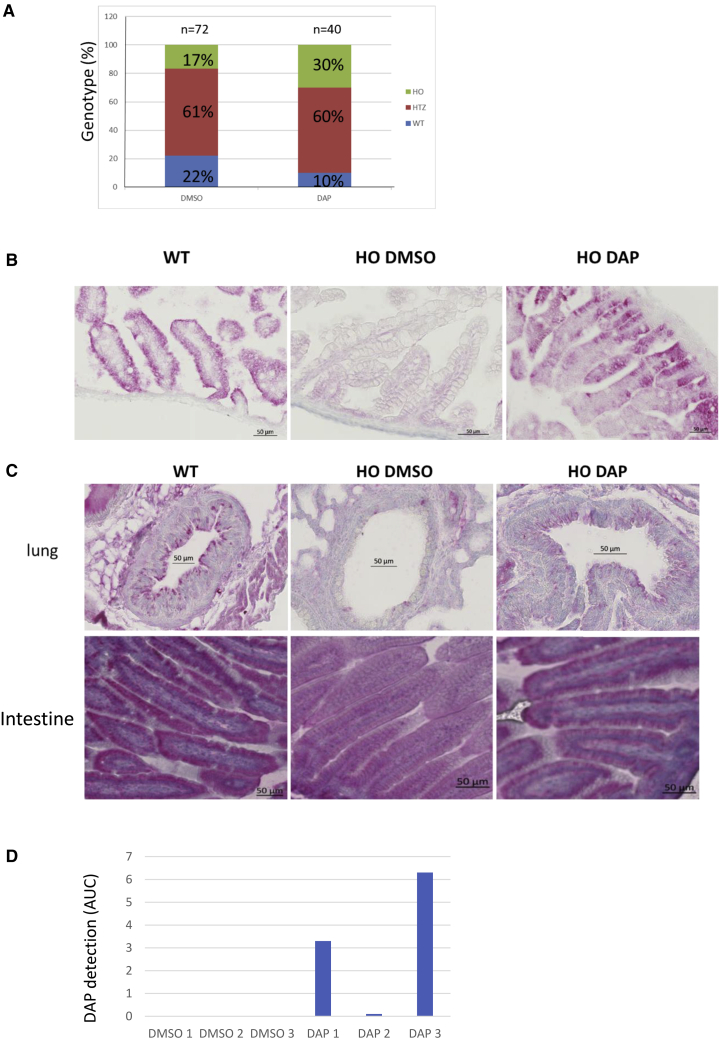
Since nearly all HO CFTR-NS mice die before adulthood, it was necessary to devise a method for testing the ability of DAP to restore CFTR function. Figure 2D shows that the HO genotype is underrepresented among pups, indicating that CFTR plays a role during development. Figure 1D shows that DAP crosses the blood-brain barrier, and this suggests that it might also cross the placental barrier. The idea was thus to attempt to restore the expected proportion of HO genotypes, i.e., about 25%, by crossing heterozygous (HTZ) mice and treating only the female in each mating pair. Mating female HTZ mice were exposed PO daily to either 10% DMSO solution or 1 mg DAP. Genotyping of the newborn pups showed DAP treatment to increase the proportion of pups with the HO genotype from 17% to 30%. This suggests that intrauterine mortality due to the absence of CFTR was prevented by DAP treatment of the mother during gestation (Figure 3A). It is interesting to note that these data were obtained from eight pairs, of which the female was treated PO with either DMSO or DAP over a period of approximately 10 months without showing any sign of toxicity. To check that the restored presence of CFTR was responsible for restoring the proportion of HO genotypes to the expected level, an IHC assay was performed on intestinal tissues from pups on their day of birth (their lungs were not yet clearly identifiable at that time) (Figure 3B). While no CFTR was detected in HO pups whose mother had been exposed to DMSO, those whose mother had been exposed to DAP showed a staining pattern similar to that of WT newborns. This indicates that DAP can cross the placental barrier and correct a UGA nonsense mutation in utero by allowing Cftr expression. The results in Figures 3A and 3B show that DAP can reach the fetus to restore CFTR protein synthesis and function when the heterozygous mother is exposed PO daily to this compound.
Figure 3.
CFTR rescue by DAP in the CFTR-NS mouse model
(A) Proportions of the three genotypes among live newborns. Mothers were exposed to DMSO or DAP by daily PO exposure during gestation. The counts are from four mothers treated with DMSO and four mothers treated with DAP for one to five litters and the total number of newborn mice analyzed is indicated on the top of each bar. A statistical analysis resulted in a p value lower than 0.01, indicating a strong trust in these results (Monte-Carlo method). (B) CFTR (in red) in the intestine (duodenum), detected by IHC applied to tissue from WT and HO newborns, from breastfed pups whose mothers were exposed to DMSO or DAP by daily PO exposure during and after gestation, and from adults (WT and HO) exposed to 3 days of DMSO or DAP by daily PO exposure. (C) CFTR (in red) detected by IHC in lung (around the bronchi) and intestinal (duodenum) tissues from 2-week-breastfed WT and HO pups whose mothers were exposed PO daily to DMSO or DAP. All the IHCs are representative of at least five mice. (D) Measurement of the presence of DAP in the contents of the stomach of newborns. Each sample is a mix of the contents of the entire litter.
Rescue of Cftr expression in newborns
It remains very challenging to expose mice PO during their first 2 weeks of life. Since DAP can cross both the blood-brain barrier and the placental barrier (Figures 1 and 3B), we hypothesized that it might be possible to treat newborns via breastfeeding. For this, PO exposure of heterozygous mothers was prolonged for 2 weeks after birth of their offspring, and then CFTR IHC was performed on lung and intestinal tissues from the pups. Unlike the tissues from pups of DMSO-treated mothers, those obtained from pups of DAP-treated mothers displayed CFTR staining (Figure 3C). These results led to the conclusion that newborn mice can be exposed to DAP by treating the nursing mother.
To validate the hypothesis that DAP can be transmitted to newborns through maternal milk, the presence of DAP was sought in maternal milk. For this, six mothers who had never been previously exposed to DAP were exposed PO to DMSO or DAP on the day of the birth of their pups. Newborns were sacrificed 1 h later. The stomach contents of newborns were then collected, pooled by litter, and analyzed by mass spectrometry. The result indicates that DAP is not found in the stomachs of newborns whose mothers have been exposed PO to DMSO but, on the other hand, is detected in the stomachs of newborns whose mothers have been exposed PO to the DAP 1 h before the sacrifice (Figure 3D). The variability of the quantity of DAP detected is linked to the fact that the interval between the moment of the last feed and the sacrifice is also very variable and cannot be controlled, since newborn mice drink milk according to their need. This result demonstrates that DAP can be transmitted to newborns via breastfeeding.
DAP restores CFTR synthesis and function in organoids derived from CFTR-NS mouse cells carrying UGA nonsense mutations
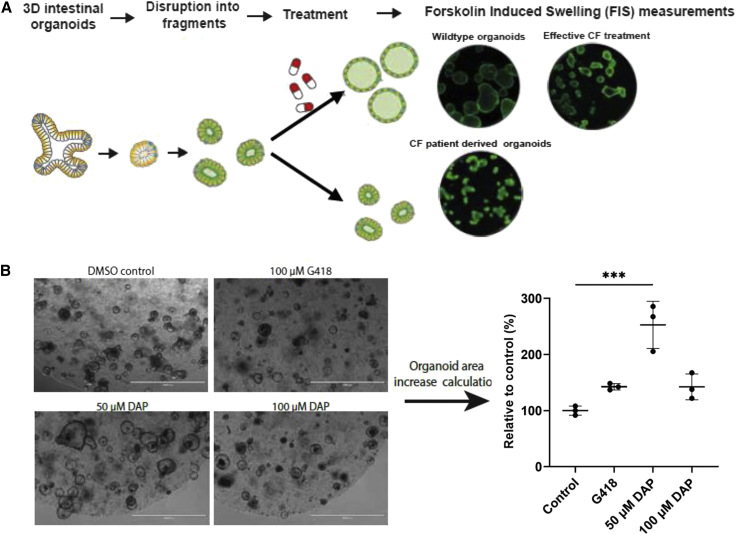
The next step was to evaluate the functional rescue of CFTR more directly than by calculating the genotype ratio. For this purpose, organoids derived from CFTR-NS mouse intestinal stem cells were established. Organoid models have been established from many organ types, both healthy and diseased, and have been shown to mimic the tissue from which they were derived.34 By measuring organoid swelling under specific conditions, the function of CFTR can be quantified35,36 (Figure 4A). Murine organoids were incubated in the presence of DMSO, DAP (50 or 100 μM), or 100 μM G418 (positive control) for 48 h before quantifying forskolin-induced swelling (FIS) as a readout for CFTR function (Figure 4B). At 100 μM, DAP led to CFTR channel function as efficiently as G418 at the same concentration, which has previously shown to be effective in an organoid model.27 At 50 μM, however, DAP proved to be about twice as effective as at 100 μM. These results show that CFTR rescued by DAP leads to the synthesis of functional CFTR in a mouse organoid model and that 50 μM is optimal in this test. They importantly confirm, consistent with the results in Figure 3, the ability of DAP to read through the R553X UGA PTC in mouse cells.
Figure 4.
DAP rescues CFTR function in mouse organoids
(A) Schematic representation of the forskolin-induced swelling (FIS) assay, in which CFTR function can be assessed in mouse- or patient-derived intestinal organoids. (B) Organoids derived from CFTR-NS mouse intestinal cells were exposed to DMSO, DAP, or G418, and CFTR function was assessed by FIS. All the results are representative of three experiments. The p values were calculated by one-way ANOVA: ∗∗∗p < 0.005.
DAP restores the function of CFTR in patient cells and in human bronchial epithelial cell lines carrying a UGA nonsense mutation
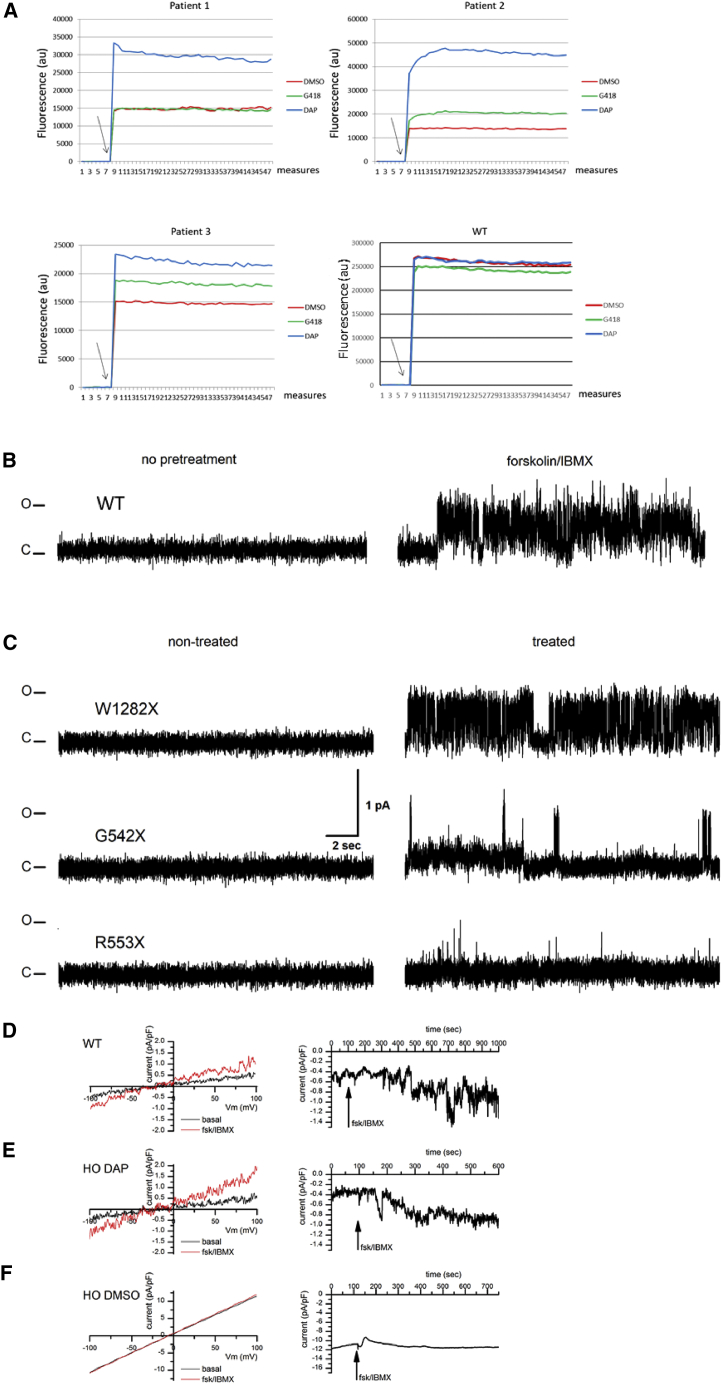
To test the ability of DAP to rescue CFTR function in yet another preclinical model, patient cells were collected from the nose epithelium and incubated for 24 h with DMSO, G418 at 600 μM, or DAP at 100 μM (optimal concentration determined previously in cell line29) in an SPQ (6-Methoxy-N-(3-Sulfopropyl)Quinolinium, Inner Salt) assay.37 Cells homozygous for either G542X or W1282X were tested. In this test, and for both mutations, DAP at 100 μM demonstrated higher functional rescue than G418 at 600 μM (Figure 5A). To exclude the possibility that the fluorescence increase observed with DAP in particular might be a non-specific effect, WT nasal cells were incubated with DMSO, DAP, or G418 under the same conditions as the CF cells. When CFTR channel activators (forskolin and IBMX (3-Isobutyl-1-methylxanthine)) were added, the fluorescence increased regardless of the treatment (DMSO, DAP, or G418). This indicates that the variation in fluorescence observed with the CF cells was indeed due to correction of the nonsense mutation and not to a non-specific effect of DAP in particular. It is worth noting that the effect of nonsense mutation correctors differed according to the nonsense mutation (compare the effects on G542X and W1282X) and, for the same mutation, according to the patient from whom the cells were derived (compare patients 2 and 3). An influence of the genetic background on CF treatment efficacy has been reported previously.38,39 The results of Figure 5A show that DAP restores CFTR function more efficiently than G418 in CF patient cells carrying a UGA nonsense mutation.
Figure 5.
Rescue of CFTR function in CF patient cells and in 16HBE14o− cells carrying a nonsense mutation in both Cftr alleles
(A) CFTR function was assessed in CF patient cells by means of an SPQ assay after DMSO (red), G418 (green), or DAP (blue) treatment. Arrows indicate addition of the forskolin-cAMP cocktail to the cell medium. The tested genotypes are W1282X/W1282X (patient 1), G542X/G542X (patients 2 and 3), and WT (lower right). (B) Sample single-channel activity of the CFTR Cl− currents in the 16HBE14o− expressing wild-type (WT) CFTR without (left) or following pretreatment with 40 μM forskolin and 400 μM IBMX for 30 min, prior to patching the cells (right). Note the absence of activity in patches made to non-stimulated cells and the presence of a characteristic CFTR activity following forskolin/IBMX treatment. (C) Single-channel activity of CFTR Cl− currents in 16HBE14o− cells expressing mutant CFTR. To stimulate CFTR currents, cells were pretreated with 40 μM forskolin and 400 μM IBMX for 30 min, prior to patching the cells. Sample traces were acquired in excised outside patches of forskolin/IBMX-pretreated cells expressing, from the top to the bottom, W1282X, G542X, and R553X CFTR. Note the absence of activity in the non-treated (left side) cells and presence of the characteristic CFTR activity in the cells treated with DAP. C and O denote closed and open states of the ion channel. (D–F) Whole-cell currents in isolated cells from WT (D), HO DAP (E), and HO DMSO (F) mice. Whole-cell currents were acquired in NMDG-Cl-based solutions as described. After whole-cell mode was established, the cells were left undisturbed for 5 min to allow replacement of intracellular K+ and Na+ with NMDG, to register basal current levels with Cl− as the major ion current carrier. This was followed by application of 40 μM forskolin and 400 μM IBMX to stimulate CFTR activity. Panels compare representative IV (current-voltage) relationships (left) as well as the time course of the CFTR activity stimulation with forskolin/IBMX (right). Note an increase in current following forskolin/IBMX application in WT and HO DAP mice and the absence of the effect in the HO DMSO case.
To reinforce the previous results on the effect of DAP on the restoration of CFTR channel function, single-channel patch-clamp experiments were undertaken. These types of experiments make it possible to isolate an individual CFTR channel and to evaluate its capacity to allow ion transport after stimulation. For this, 16HBE14o− cells, in which a nonsense mutation was introduced by CRISPR-Cas9, were used. The CFTR ion channel is an anionic channel that is known to have a distinctive single-channel activity pattern, characterized by a relatively small single-channel conductance of approximately 9–10 pS, bursting behavior, and sensitivity to ATP, as well as phosphorylation by the PKA catalytic subunit on the cytoplasmic side in order to be gated.40,41,42 First, to confirm the presence of CFTR ion channels, as well as to establish the optimal observation conditions, patch-clamp data were acquired in the WT 16HBE14o− cells. For patch-clamp experiments, cells were immersed in a solution containing 150 mM N-methyl-D-glucamine chloride (NMDG-Cl) as the principal component, with a symmetric composition of the pipette solution. Patches were made to the cells in the middle of the established clusters, surrounded on all sides by other cells. To ensure robust CFTR activity, cells were pretreated with 40 μM forskolin and 400 μM IBMX for 30 min, prior to patching.40,41 Recordings were made in cell-attached patches, as well as excised inside-out patches, which allowed us to record activity under symmetric ionic conditions, where the only conductive ion present was Cl−, thus allowing us to isolate anionic currents carried by CFTR ion channels. Under such conditions, a characteristic CFTR can be observed in over half of the traces following the forskolin pretreatment in cell-attached as well as excised inside-out patches with a characteristic single-channel conductance of 9.6 ± 0.5 pS (Figure 5B, n = 8 of 14 patches). To confirm the sensitivity of the observed activity to ATP, we measured CFTR activity in excised inside-out patches made to cells that were not pretreated with forskolin/IBMX. In such patches CFTR activity could be observed at very low levels following the patch excision, while application to the cytoplasmic side of the patch membrane of 25 nM PKA catalytic subunit + 1 mM Mg-ATP typically restored the characteristic CFTR currents (Figure S3, n = 7).
Having established the conditions under which robust CFTR activity could be observed in WT 16HBE14o− cells, we performed a series of experiments aimed at determining the presence of such activity in the non-treated 16HBE14o− cells modified via the CRISPR-Cas9 procedure to contain various mutants introducing premature stop codons. Figure 5C compares such activity in non-treated modified cells vs. cells treated for 24 h with DAP. While the non-treated cells universally lacked the characteristic CFTR activity, such activity could be restored in the treated W1282X mutant cells, where the treatment was expected to reintroduce the matching amino acid, restoring the proper protein sequence (Figure 5C). For the other two mutants, namely G542X and R553X, treatment with DAP restored some functionality of the CFTR channel, since the channel was found alternately in the open or closed state (Figure 5C).
Further, to establish the physiological relevance of the restoration of the CFTR expression, we studied whole-cell CFTR currents in cells isolated from the duodenum of 11 CFTR-NS mice under conditions where Cl− was the only ion current carrier (NMDG-Cl-based extracellular and intracellular solutions as described). The recordings done on the cells isolated from control (WT) as well as HO DAP mice showed a small but significant whole-cell current evoked by application of 40 μM forskolin and 400 μM IBMX. Cells isolated from WT mice yielded 0.81 ± 0.19 pA/pF increase in currents (Figure 5D, n = 2), while cells isolated from HO DAP mice yielded an increase of 0.86 ± 0.16 pA/pF (Figure 5E, p = 0.0046, n = 4). The HO DMSO-treated mice had a significant amount of tissue degradation, with the isolation procedure yielding a very small number of cells, the majority of which were not viable in culture, consistent with the well-established importance of the CFTR ion channel for cell survival. Repeated attempts to patch these cells yielded only a single successful recording. Application of forskolin/IBMX in this patch did not produce any significant change in the whole-cell current (Figure 5F).
DAP restores CFTR synthesis and function in organoids derived from patient cells carrying a UGA nonsense mutation
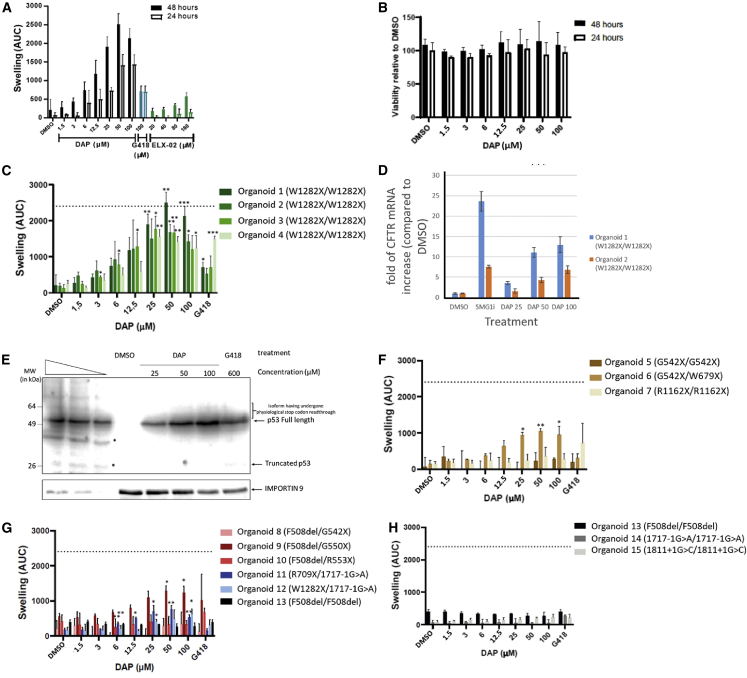
To complete the demonstration of the ability of DAP to read through UGA PTCs, 15 patient-derived intestinal organoids (PDIOs) were exposed to increasing doses of DAP (Figures 6, S4, and S5). DMSO was used as a negative control and did not promote any organoid swelling. G418 and ELX-02 were used as positive controls for the correction of nonsense mutations. First, to determine the best experimental conditions for observing a functional effect of DAP on the PDIOs, dose-response curves were constructed after two incubation times (24 and 48 h). To ensure that a positive effect of DAP could be detected, PDIOs homozygous for the nonsense mutation W1282X were used, since DAP causes a tryptophan to be introduced at the site of the nonsense mutation and can thus restore synthesis of a WT CFTR protein. The results show that CFTR function was restored in a dose-dependent manner, becoming significant from 25 μM. The measured effect was also greater when the PDIOs were treated for 48 h than for 24 h (Figure 6A). Interestingly, as for murine organoids (Figure 4), the concentration causing optimal restoration of CFTR function was 50 μM. The comparison of the effect obtained with DAP compared with G418 or with the molecule in clinical trial, ELX-02, shows that DAP is more effective in correcting the nonsense mutation in this line of organoid. alamarBlue and propidium iodide staining, used to measure viability for each condition in Figure 4B, showed the absence of DAP toxicity toward the organoids (Figures 6B and S5). In subsequent assays, a 48-h incubation time was used to optimize the window of opportunity to detect effects.
Figure 6.
DAP rescues CFTR function in patient-cell-derived organoids
(A) FIS assay on organoids derived from rectal cells of CF patients homozygous for the W1282X mutation. Organoids were exposed to DMSO, DAP, G418, or ELX-02 for 24 or 48 h. (B) Cell viability measured by alamarBlue incorporation into organoids derived from rectal cells of CF patients homozygous for the W1282X mutation. The organoids were exposed to DMSO or DAP. (C) FIS assay on four such organoids exposed to DMSO or DAP. (D) Measurement by qRT-PCR of the level of CFTR mRNA in organoids 1 and 2 to evaluate the efficacy of NMD in the presence of DMSO, an inhibitor of NMD (SMG1i), or DAP at 25, 50, or 100 μM. (E) Western blot demonstrating the absence of physiological stop codon readthrough by DAP at 25, 50, or 100 μM. Calu-6 cells were exposed to DMSO, DAP, or G418 previously to extract protein and perform a western blot analysis. The three leftmost lanes are serial dilutions of U2OS cell extract. Asterisks show degradation products or non-specific bands. The place to which the isoforms having undergone a readthrough of the physiological stop codon should migrate is indicated on the right side of the gel by a bracket. (F) FIS assay on CF patient-cell-derived organoids carrying UGA nonsense mutation on both CFTR alleles. (G) FIS assay on CF patient-cell-derived organoids carrying a UGA nonsense mutation on one CFTR allele and a different mutation from a nonsense mutation (F508del or 1717-1G>A) on the other CFTR allele. (H) FIS assay on CF patient-cell-derived organoids carrying a mutation different from UGA nonsense mutation on both CFTR alleles (F508del or 1717-1G>A). The dotted line corresponds to the amplitude of swelling obtained with organoids homozygous for the F508del mutation upon treatment with lumacaftor-ivacaftor. Error bar shows the SD, p values were calculated with Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, n.s., not significant. Averages of three experiments with three technical replicates each are shown.
To further characterize the capacity of DAP to restore CFTR function in W1282X/W1282X PDIOs, FIS was assessed in three additional homozygous W1282X PDIOs (Figures 6C and S4). In all four PDIOs, DAP proved able to induce functional rescue of CFTR in a dose-dependent manner. DAP-induced CFTR rescue even reached the level obtained in F508del/F508del PDIOs with lumacaftor-ivacaftor (dotted line). This is an important benchmark, as lumacaftor-ivacaftor is clinically effective. Within-genotype variation was observed, organoid line 1 being most responsive to DAP and organoid lines 2/3/4 reaching similar areas under the curve. In all organoid lines except organoid line 4, DAP treatment at 50 μM restored CFTR function more effectively than G418. This function-restoring action of DAP in the presence of the W1282X mutation is consistent with its mode of action, which leads to incorporation of a tryptophan at the position of the nonsense mutation. Hence, the protein synthesized after treatment with DAP is the WT protein. Since the concentrations used are greater than those that were previously used in the cell lines and for which no inhibition of NMD had been measured,29 the effectiveness of NMD was measured in two PDIOs by measuring the level of mRNA CFTR by qRT-PCR (Figure 6D). The results show in particular that above 25 μM, DAP induces an inhibition of NMD. This is probably not the main effect of suppression of UGA stop codons, since it is observed that the functional restoration of CFTR at 100 μM is less effective than the effect at 50 μM, even if the inhibition of NMD is greater at 100 μM DAP than at 50 μM DAP (Figures 6C and 6D).
To confirm the physiological stop codon integrity, Calu-6 cells carrying a premature UGA stop codon in the TP53 gene were treated with DMSO, DAP, or G418. The use of Calu-6 cells presents several advantages, including that the analysis of the TP53 gene by western blot is very efficient and that the TP53 gene ends its open reading phase with a physiological stop codon, UGA. A long exposure of the western blot analysis does not result in the detection of isoforms of size greater than the isoform terminating at the physiological stop codon, indicating that DAP does not result in readthrough of physiological stop codons (Figure 6E).
Other mutations were then tested to assess the ability of DAP to rescue the function of CFTR in PDIOs carrying other UGA nonsense mutations on both CFTR alleles (Figure 6F) or on one CFTR allele (Figure 6G) or other types of mutations as negative controls (Figure 6H). Although the rescue was of lower magnitude than with the W1282X mutation, DAP proved able to restore CFTR function in PDIOs heterozygous for the UGA nonsense mutation G550X, R709X, or W679X. Under these conditions, however, it proved unable to suppress a biallelic R1162X mutation, a biallelic G542X mutation, or a monoallelic G542X or R553X mutation associated with F508del. As expected, DAP failed to suppress a biallelic F508del or 1717-1G>A splice mutation (Figure 6H). Overall, the results of Figures 6 and S4 show that DAP can restore CFTR function in PDIOs with various UGA nonsense mutations in the CFTR gene.
Discussion
The study presented here aims to prioritize DAP for further clinical development by studying its efficacy and pharmacokinetics in preclinical models. The models used include a new mouse model of CF, patient cells, PDIOs, and human bronchial epithelial cells. The pharmacokinetic data obtained show that DAP appears in the blood after oral administration and is then rapidly eliminated in the urine, but that a fraction is distributed to various tissues such as the lungs, muscles, and brain (Figure 1). This distribution suggests that DAP can correct nonsense mutations in different tissues, which is crucial for a pathology such as CF, in which the absence of the CFTR channel influences the functioning of several epithelia and tissues. Interestingly, DAP crosses the blood-brain barrier. One might thus consider DAP for correcting nonsense mutations involved in neurological diseases. DAP also crosses the placental barrier, as shown in the new mouse model CFTR-NS carrying the R553X nonsense mutation (Figures 2 and 3). In this in vivo model, DAP is shown here to correct a UGA nonsense mutation in the Cftr gene and to reverse the phenotype associated with it (lethality, absence of CFTR). However, even though DAP clearly crosses the placental barrier and corrects the nonsense mutation in utero, treatment in utero is probably not possible with this molecule, because the presence of DAP was associated with a reduction in the number of pups per litter, suggesting a possible deleterious effect of DAP during gestation. Yet we have been unable, so far, to verify statistically an influence of DAP on the number of newborns (Figure 3).
Another demonstration that DAP might be used therapeutically in CF patient cells or in patients with CF linked to the presence of a UGA nonsense mutation is provided by the present experiments with PDIOs (Figures 5 and 6). In these study models, it is possible to measure the function of the CFTR channel and thus to evaluate the ability of DAP to correct functionally a nonsense mutation in the CFTR gene. Importantly, as several UGA nonsense mutations lead to loss of a tryptophan, the use of DAP seems particularly suitable. For example, the W1282X mutation represents 1.2% of the mutations found in CF.43 On the W1282X mutation, DAP was more effective than G418 or ELX-02. However, it cannot be excluded that the ELX-02 molecule purchased from MedChemExpress may be less active than the molecule from the Eloxx Pharmaceuticals laboratories. A comparison of these two sources of ELX-02 by an independent laboratory would be a good way to verify this point.
In several PDIOs from patients homozygous for the W1282X mutation, DAP was found to rescue the function of the CFTR channel as potently as does lumacaftor-ivacaftor, from Vertex Pharmaceuticals, in the case of the F508del mutation. This result is suggestive of clinical efficacy (Figure 6C). On other mutations the effect of DAP is weaker, likely because DAP-promoted introduction of a tryptophan at the PTC position might have an impact, at least partially, on the function of CFTR, consistent with the patch-clamp results obtained on human bronchial epithelial cells (Figure 5B).
Each of the four preclinical models used in this study has its specificities and experimental conditions liable to cause variations in responses to molecules. For example, the R553X mutation responds to DAP in mouse organoids, but the same mutation in PDIOs does not. Likewise, the G542X mutation responds to DAP in patient cells or in 16HBE14o− cells but not in PDIOs. These differences can be linked to experimental specificities of each model, the sensitivity of each assay, and the genotype of each individual, previously reported to influence the response to a drug.38,39 Overall, the results of this study demonstrate that DAP can be viewed as a good drug candidate, so far, to correct UGA nonsense mutations.
Materials and methods
CFTR-NS mouse model
The CFTR-NS mouse was developed at the Institut Clinique de la Souris (ICS). Both point mutations were introduced at the same time, using the Cre-Lox system and homologous recombination in embryonic stem cells before being introduced into embryos with the C57BL/6N genetic background. The leftover LoxP site is located in intron 12 and allowed genotyping. The mouse strain is maintained in the heterozygous state. All in vivo experiments complied with all relevant ethical regulations for animal testing and research and were authorized by the Ethic Committee 075 under the number 12971-2018010815336092.
Collecting primary intestinal epithelial mouse cells
Five centimeters of duodenum was cut into 1-mm pieces after removing all the stool. The pieces were washed five times with PBS before being incubated for 20 min at 37°C in the presence of 200 U/mL collagenase and 100 U/mL hyaluronidase. After 5 min centrifugation at 100 × g and 4°C, the pellet was resuspended in DMEM containing 2% fetal bovine serum (FBS) and 2% sorbitol. The solution was centrifuged for 5 min at 250 × g at room temperature, the pellet was resuspended in DMEM containing 10% FBS, and the cells were placed in culture dishes.
Molecule preparation
DAP was purchased from Sigma-Aldrich (ref. no. 247847) and dissolved at 100 mM in DMSO (cell and organoid treatments) or at 5 mg/mL in 10% DMSO/90% PBS (mouse treatments). ELX-02 was purchased from MedChemExpress (ref. no. HY-114231B) and dissolved in DMSO.
Collecting primary epithelial cells from CF patients
All experimentation using human tissues described herein was approved by the medical ethics committee of the University Medical Center Utrecht (UMCU; TcBio 14-008 and TcBio 16-586). Informed consent for tissue collection and for generation, storage, and use of organoids was obtained from all participating patients. Biobanked intestinal organoids are stored and cataloged (https://huborganoids.nl/) at the Hubrecht Organoid Technology foundation (http://hub4organoids.eu) and can be requested from info@hub4organoids.eu. Collection of patient tissues and data was performed according to the guidelines of the European Network of Research Ethics Committees (EUREC) and to European, national, and local law.
Mouse intestinal organoid culture and characterization of CFTR function
Crypts were isolated from small intestinal biopsies of mice as previously described.44 Crypts were plated in 50% Matrigel and developed into organoid structures within 3 weeks. The same medium was used as for the human intestinal organoids, with the exception that the end volume of WNT-conditioned medium was 75% instead of 50%. Organoids were incubated in a humidified chamber with 5% CO2 at 37°C, the medium was refreshed every 2–3 days, and organoids were passaged 1:4 every 5–7 days. For assessing restoration of CFTR function, organoids were preincubated with DAP or G418 for 48 h prior to forskolin addition (5 μM). After 1 h of forskolin stimulation, 10× bright-field pictures were taken using an EVOS inverted microscope. Pictures were assessed in a blinded manner, by quantifying the size of 10 organoids in each picture using ImageJ. For each condition, the average of three replicates was calculated, and results were normalized to the negative control (DMSO treated).
Human intestinal organoid cultures
Crypts were isolated from biopsies of people with CF as previously described.45 Briefly, organoids were cultured in 50% Matrigel (Corning; 356255) in advanced DMEM/F12 medium supplemented with penicillin and streptomycin, 10 mM HEPES, Glutamax, B27 (all from Invitrogen), 1 μM N-acetylcysteine (Sigma), and the following growth factors: 50 ng/mL mouse epidermal growth factor (mEGF), 50% Wnt3a-conditioned medium (WCM), 10% Noggin-conditioned medium (NCM), 20% Rspo1-conditioned medium (RCM), 10 μM nicotinamide (Sigma), 500 nM A83-01 (Tocris), and 10 μM SB202190 (Sigma). Organoids were incubated in a humidified chamber under 5% CO2 at 37°C. The medium was refreshed every 2–3 days and the organoids were passaged 1:4 every 7–10 days.
Functional assessment of CFTR and measurement of DAP toxicity in PDIOs derived from CF patient cells
Prior to measuring CFTR function in FIS assays, PDIOs were grown for at least 3 weeks after crypt isolation or thawing. They were split as previously described45 and plated in 3-μL basement extract membrane drops on 96-well plates. After a 5-min incubation at 37°C, the medium with DMSO, DAP, or G418 was added to the wells. After a 48-h incubation, forskolin (5 μM) and calcein (3 μM) were added to all wells. PDIO swelling was monitored for 60 min with a Zeiss LSM 710 confocal microscope. Total organoid surface area per well was quantified with Zen software on the basis of calcein staining, and the area under the curve over time was calculated as described in Vonk et al.45 After the FIS experiments on PDIOs of donor 1, the medium was replaced with 100 μL alamarBlue stock solution diluted 1:10 in DMEM without phenol red. Plates were incubated at 37°C for 4 h, after which fluorescence (excitation wavelength 470 nm, emission wavelength 530 nm) was measured with a CLARIOstar multimode microplate reader. Viability was normalized to negative controls (PDIOs treated with 0.1% DMSO) and positive controls (PDIOs treated with 10% DMSO for 24 h).
RNA isolation from PDIOs and SYBR-quantitative real-time PCR
Forty-eight hours prior to RNA isolation, DAP and SMG1i were added to the culture medium at 25/50/100 and 0.3 μM, respectively. RNA was isolated for two homozygous W1282X PDIOs, using the NucleoSpin RNA kit (BIOKE) according to the manufacturer’s protocol. RNA yield was measured by a NanoDrop spectrophotometer, and extracted mRNA was used for cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s protocol. YWHAZ, GAPDH, and CFTR regions were amplified in a two-step quantitative real-time PCR SYBR green reaction (CFX-384 real-time PCR; Bio-Rad) in a total assay volume of 10 μL. Primers sequences were the following: YWHAZ (forward, 5′-CTGGAACGGTGAAGGTGACA-3′; reverse, 5′-AAGGGACTTCCTGTAACAATGCA-3′), GAPDH (forward, 5′-TGCACCACCAACTGCTTAGC-3′; reverse, 5′-GGCATGGACTGTGGTCATGAG-3′), and CFTR (forward, 5′-CAACATCTAGTGAGCAGTCAGG-3′; reverse, 5′-CCCAGGTAAGGGATGTATTGTG-3′).
Samples were incubated during the PCR as follows: 2 min at 95°C and 39 cycles of 30 s at 95°C, 30 s at 59°C. Relative gene expression of CFTR was first normalized against the housekeeping genes GAPDH and YWHAZ, after which CFTR expression was normalized against control samples using the comparative 2−ΔΔCT method. Melt peaks were analyzed to confirm amplification of a single product. Two biological replicate experiments were performed with three technical replicates per experiment.
RNA isolation from CFTR-NS mouse lung
Total RNA was extracted from approximately one-fifth of a lung using RNAzol and the supplier-provided protocol. Reverse transcription was done using SuperScript II (Invitrogen) and random hexamers. CFTR cDNA and GAPDH cDNA were then amplified by 35 cycles of PCR using the following oligonucleotides: CFTR sense, 5′-GACGAGTTCTAAAACAAGCC-3′; CFTR antisense, 5′-TACCCATACCCATATGAACG-3′; GAPDH sense, 5′-CATTGACCTCAACTACATGG-3′; GAPDH antisense, 5′-GCCATGCCAGTGAGCTTCC-3′. PCR amplifications were loaded on a 1% agarose gel containing ethidium bromide.
Western blotting
After 24 h of treatment, 2 × 106 Calu-6 cells were harvested and proteins extracted in a lysis buffer containing 5% SDS, 50 mM Tris, and 20 mM EDTA. The equivalent of 2.5 × 105 cells was subjected to 10% SDS-PAGE before transfer of the proteins to a nitrocellulose membrane. The membranes were incubated overnight at 4°C in the presence of a 1/200 dilution of anti-p53 antibody (DO1; Santa Cruz Biotechnology, Dallas, TX, USA) or 1/1,000 dilution of anti-IMPORTIN9 antibody (Abcam, Cambridge, UK). After three washes of the membrane in TBS Tween, the membranes were exposed to a solution of peroxidase-coupled secondary antibody for detection of mouse- or rabbit-raised antibody (Jackson ImmunoResearch, Suffolk, UK). Antibodies were then detected with SuperSignal West Femto maximum sensitivity substrate (Pierce Biotechnology, Rockford, IL, USA).
Immunohistochemistry
Lung and intestinal tissues were fixed in 4% paraformaldehyde (PFA) or in Carnoy solution (60% methanol, 30% chloroform, 10% acetic acid) before being embedded in paraffin and sectioned into 5-μm lung slices or 10-μm intestine slices. For detection of CFTR in adults and 2-week-old mice, the sections were incubated with blocking buffer consisting of goat serum and anti-mouse IgG before incubating with the anti-CFTR antibody (Abcam ab234037 for lung or LS-Bio LS-C14758 for intestine) at 1:100 dilution. For detection of CFTR in the intestines of newborns, a blocking step with Bloxall (Vector Laboratories) was performed before incubation with the goat serum, and anti-CFTR antibody from Santa Cruz (sc-376683) was used at 1:100 dilution. Mayer’s hemalum counterstain was then applied to all slices.
Pharmacokinetic (PK) data
Bioavailability
A PK study was performed using 6-week-old CD-1 mice. A dose of 29 mg/kg DAP solution (10% DMSO/90% PBS) was administered by oral gavage. Blood was taken by intracardiac puncture 0.5, 1, 2, 4, 6, and 24 h after gavage. Blood samples were then placed into an EDTA-coated tube and centrifuged at 4°C, 12,000 × g for 10 min, to collect plasma, which was stored frozen at −80°C until analysis. For this, a volume of 400 μL of plasma was mixed with 1 mL of acetonitrile to precipitate the proteins and extract the DAP. Samples were vortexed for 5 min before being sonicated for 1 min. The proteins were pelletized by centrifugation at 15,000 × g for 5 min at 16°C. One milliliter of supernatant was then evaporated, and the dry matter was resuspended in 100 μL of water containing 1% trifluoroacetic acid before being analyzed by LC-MS/MS mass spectrometry.
Biodistribution
DAP was measured in lung, muscle, and brain from male CD-1 mice. DAP (8.1 mg/kg; maximum solubility in 5% DMSO/95% PBS) was injected intravenously into 18 mice. Three mice were sacrificed at 15, 30, 60, 120, 240, and 360 min. For each time point, plasma, lung, muscles, intestine, and brain were collected and stored at −80°C before proceeding with the analysis. Tissues were collected after perfusion of the mice with 50 mL 0.9% NaCl solution to remove the blood. The lungs, tibialis anterior, small intestine, and brain were crushed in 400 μL water before adding 800 μL acetonitrile. The ground material was vortexed for 5 min and then sonicated for 1 min prior to centrifugation for 5 min at 15,000 × g and 16°C. The supernatant (120 μL) was evaporated and the dry matter resuspended in 120 μL water containing 1% trifluoroacetic acid. The sample was then analyzed by LC-MS/MS. During this experiment, urine was collected from the animal’s bladder and stored at −80°C while awaiting analysis. The urine (50–300 μL) was diluted with 200 μL water. Each sample was placed on an SPE cartridge conditioned with 1 mL methanol and then with 1 mL water before washing the cartridge with 1 mL water and eluting with 800 μL methanol. The eluate was vacuum evaporated, the dry matter obtained was resuspended in 500 μL water containing 1% trifluoroacetic acid, and the samples were then treated as described above.
Measurement of DAP in stomach contents
The stomach contents of newborns were collected using a syringe after opening the stomach. The proteins in the samples were precipitated with acetonitrile. The supernatant was analyzed by mass spectrometry after adding 9 vol of water containing 1% trifluoroacetic acid. The samples were analyzed by UHPLC coupled to a Shimadzu LC-MS 8030 triple quadrupole.
Patch-clamp recordings
CFTR activity was acquired using the patch-clamp technique in cell-attached and inside-out configurations. To ensure proper CFTR expression, only cells within confluent islands and completely surrounded by other cells were patched. In some cultures where seals were difficult to make, the cells were treated with trypsin for 30 s. Currents were recorded via an Axopatch 200B amplifier and digitized using the Digidata 1322 digitizer using the pClamp software (Molecular Devices, San Jose, CA, USA). All patch-clamp recordings were made at 20°C–22°C. Patch pipettes were fabricated from borosilicate glass capillaries (World Precision Instruments, Sarasota, FL, USA) on a horizontal puller (Sutter Instruments, Novato, CA, USA) and had a resistance in the range of 3–5 MΩ. The standard bath and pipette solutions contained 150 mM NMDG-Cl, 2.5 mM CaC12, 2.5 mM MgCl2, and 10 mM HEPES, with pH adjusted to 7.3 with HCl. To ensure robust activation of CFTR ion channels, cells were treated with 40 μM forskolin and 400 μM IBMX for 30 min prior to patching. Bath solution was supplemented with 1 mM Mg-ATP to maintain CFTR activity in the excised inside-out patches. In the experiments where online PKA/ATP effects were verified, cells were immersed in the base solution not supplemented by ATP and patched without forskolin/IBMX pretreatment. Instead, 25 nM catalytic domain of PKA + 1 mM Mg-ATP was added to the bath online during data acquisition.
Whole-cell recordings were performed on the cells isolated from the WT and HO DAP- and HO DMSO-treated mice as described above. Isolated cells were immersed in the NMDG-Cl-based solution identical to that used for single-channel recordings. To eliminate K+ and Na+ channel involvement, the pipette solution was also based on the same NMDG-Cl solution, with free Ca2+ buffered to ∼100 nM by EGTA, resulting in the following composition: 150 mM NMDG-Cl, 2.5 mM MgCl2, 0.2 mM Ca2+, 0.5 mM EGTA, and 10 mM HEPES, with pH adjusted to 7.3 with HCl. Whole-cell recordings were performed using the ramp protocol with cells briefly held (for 50 ms) at −100 mV, followed by ramp from −100 to +100 mV, followed by another brief holding at +100 mV for 50 ms. Such ramps were applied once every second, with cells held at −40 mV between ramps to stabilize their condition. After the whole-cell mode was established, the cells were left undisturbed for 5 min to register basal current levels, followed by extracellular application of 40 μM forskolin and 400 μM IBMX to stimulate CFTR activity. Under such conditions, with Cl− being the only ion current carrier, current deviation following the forskolin/IBMX application produced symmetric deviation of current at both +100 and −100 mV, with linear IV response during the ramp phase.
Statistics
Data are represented as means ± SD. One-way ANOVAs were performed to compare mean FIS efficacies upon treatment with the various compounds (cocktails). Differences were considered statistically significant at p < 0.05. Data analysis was performed with GraphPad Prism 7.0 software (San Diego, CA, USA).
Data availability
All data presented in this work are available from the authors upon request.
Acknowledgments
The authors would like to thank Dr. Anne Tsicopoulos and Dr. Quentin Thommen for helpful discussions and advice on IHC and statistical analysis. The authors would also like to deeply thank the Cystic Fibrosis Foundation, for the donation of 16HBE14o− cells carrying nonsense mutations, and Professor Dieter Gruenert for the donation of 16HBE14o− WT cells. F.L. is supported by funding from Vaincre la Mucoviscidose, the Association Française contre les Myopathies, the Agence Nationale de la Recherche, the Fondation Les Ailes, and the Fondation Maladies Rares. Canther Laboratory is part of ONCOLille Institute. This work is supported by a grant from Contrat de Plan Etat-Région CPER Cancer 2015–2020.
Author contributions
Conceptualization, C.L., S.S., J.B., G.S., and F.L.; methodology, C.L., S.S., N.C.E., V.P., R.K., P.G., S.A., and C. Bailly; investigation, C.L., S.S., V.P., N.C.E., R.K., P.G., S.A., C. Bourban and C. Bailly; funding acquisition, N.P., J.B., and F.L.; project administration, F.L.; supervision, P.G., S.A., C. Bailly, D. Hannebique, A.P., P.R., D. Hubert, M.G., M.C., S.R., J.B., and F.L.; writing – original draft, C.L., S.S., G.S., and F.L.; writing – review & editing, all authors.
Declaration of interests
J.B. is inventor on a patent related to the FIS assay and received financial royalties from 2017 onward. S.A., C. Bailly, S.R., and F.L. are inventors on a patent demonstrating that DAP is a readthrough molecule useful for the treatment of genetic diseases related to nonsense mutations.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2023.01.014.
Supplemental information
References
- 1.Mort M., Ivanov D., Cooper D.N., Chuzhanova N.A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 2.Gupta P., Li Y.R. Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay. Mol. Biol. Rep. 2018;45:39–55. doi: 10.1007/s11033-017-4139-7. [DOI] [PubMed] [Google Scholar]
- 3.Lejeune F. Nonsense-mediated mRNA decay at the crossroads of many cellular pathways. BMB Rep. 2017;50:175–185. doi: 10.5483/BMBRep.2017.50.4.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurosaki T., Maquat L.E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. Feb. 2016;129:461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karousis E.D., Nasif S., Muhlemann O. Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip. Rev. RNA. 2016;7:661–682. doi: 10.1002/wrna.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hug N., Longman D., Caceres J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nogueira G., Fernandes R., Garcia-Moreno J.F., Romao L. Nonsense-mediated RNA decay and its bipolar function in cancer. Mol. Cancer. 2021;20:72. doi: 10.1186/s12943-021-01364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand S., Cougot N., Mahuteau-Betzer F., Nguyen C.H., Grierson D.S., Bertrand E., Tazi J., Lejeune F. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J. Cell Biol. 2007;178:1145–1160. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang Y., Low W.K., Xu J., Gehring N.H., Dietz H.C., Romo D., Liu J.O. Inhibition of nonsense-mediated mRNA decay by the natural product pateamine A through eukaryotic initiation factor 4AIII. J. Biol. Chem. 2009;284:23613–23621. doi: 10.1074/jbc.M109.009985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhuvanagiri M., Lewis J., Putzker K., Becker J.P., Leicht S., Krijgsveld J., Batra R., Turnwald B., Jovanovic B., Hauer C., et al. 5-azacytidine inhibits nonsense-mediated decay in a MYC-dependent fashion. EMBO Mol. Med. 2014;6:1593–1609. doi: 10.15252/emmm.201404461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin L., Grigoryan A., Wang D., Wang J., Breda L., Rivella S., Cardozo T., Gardner L.B. Identification and characterization of small molecules that inhibit nonsense mediated RNA decay and suppress nonsense p53 mutations. Cancer Res. 2014;74:3104–3113. doi: 10.1158/0008-5472.CAN-13-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotham V.J., Hobbs M.C., Burgin R., Turton D., Smythe C., Coldham I. Synthesis and activity of a novel inhibitor of nonsense-mediated mRNA decay. Org. Biomol. Chem. 2016;14:1559–1563. doi: 10.1039/c5ob02482j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Usuki F., Yamashita A., Higuchi I., Ohnishi T., Shiraishi T., Osame M., Ohno S. Inhibition of nonsense-mediated mRNA decay rescues the phenotype in Ullrich's disease. Ann. Neurol. 2004;55:740–744. doi: 10.1002/ana.20107. [DOI] [PubMed] [Google Scholar]
- 14.Bandara R.A., Chen Z.R., Hu J. Potential of helper-dependent Adenoviral vectors in CRISPR-cas9-mediated lung gene therapy. Cell Biosci. 2021;11:145. doi: 10.1186/s13578-021-00662-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnamurthy S., Traore S., Cooney A.L., Brommel C.M., Kulhankova K., Sinn P.L., Newby G.A., Liu D.R., McCray P.B. Functional correction of CFTR mutations in human airway epithelial cells using adenine base editors. Nucleic Acids Res. 2021;49:10558–10572. doi: 10.1093/nar/gkab788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos L., Mention K., Cavusoglu-Doran K., Sanz D.J., Bacalhau M., Lopes-Pacheco M., Harrison P.T., Farinha C.M. Comparison of Cas9 and Cas12a CRISPR editing methods to correct the W1282X-CFTR mutation. J. Cyst Fibros. 2021;21:181–187. doi: 10.1016/j.jcf.2021.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Erwood S., Laselva O., Bily T.M.I., Brewer R.A., Rutherford A.H., Bear C.E., Ivakine E.A. Allele-specific prevention of nonsense-mediated decay in cystic fibrosis using homology-independent genome editing. Mol. Ther. Methods Clin. Dev. 2020;17:1118–1128. doi: 10.1016/j.omtm.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michaels W.E., Pena-Rasgado C., Kotaria R., Bridges R.J., Hastings M.L. Open reading frame correction using splice-switching antisense oligonucleotides for the treatment of cystic fibrosis. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2114886119. e2114886119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y.J., Sivetz N., Layne J., Voss D.M., Yang L., Zhang Q., Krainer A.R. Exon-skipping antisense oligonucleotides for cystic fibrosis therapy. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2114858118. e2114858118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oren Y.S., Avizur-Barchad O., Ozeri-Galai E., Elgrabli R., Schirelman M.R., Blinder T., Stampfer C.D., Ordan M., Laselva O., Cohen-Cymberknoh M., et al. Antisense oligonucleotide splicing modulation as a novel Cystic Fibrosis therapeutic approach for the W1282X nonsense mutation. J. Cyst Fibros. 2021;21:630–636. doi: 10.1016/j.jcf.2021.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Palma M., Lejeune F. Deciphering the molecular mechanism of stop codon readthrough. Biol. Rev. Camb Philos. Soc. 2021;96:310–329. doi: 10.1111/brv.12657. [DOI] [PubMed] [Google Scholar]
- 22.Martins-Dias P., Romao L. Nonsense suppression therapies in human genetic diseases. Cell Mol. Life Sci. 2021;78:4677–4701. doi: 10.1007/s00018-021-03809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venturini A., Borrelli A., Musante I., Scudieri P., Capurro V., Renda M., Pedemonte N., Galietta L.J. Comprehensive analysis of combinatorial pharmacological treatments to correct nonsense mutations in the CFTR gene. Int. J. Mol. Sci. 2021;22:11972–11993. doi: 10.3390/ijms222111972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Hilarion S., Beghyn T., Jia J., Debreuck N., Berte G., Mamchaoui K., Mouly V., Gruenert D.C., Déprez B., Lejeune F. Rescue of nonsense mutations by amlexanox in human cells. Orphanet J. Rare Dis. 2012;7:58. doi: 10.1186/1750-1172-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brasell E.J., Chu L.L., Akpa M.M., Eshkar-Oren I., Alroy I., Corsini R., Gilfix B.M., Yamanaka Y., Huertas P., Goodyer P. The novel aminoglycoside, ELX-02, permits CTNSW138X translational read-through and restores lysosomal cystine efflux in cystinosis. PLoS One. 2019;14:e0223954. doi: 10.1371/journal.pone.0223954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welch E.M., Barton E.R., Zhuo J., Tomizawa Y., Friesen W.J., Trifillis P., Paushkin S., Patel M., Trotta C.R., Hwang S., et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 27.de Poel E., Spelier S., Suen S.W.F., Kruisselbrink E., Graeber S.Y., Mall M.A., Weersink E.J.M., van der Eerden M.M., Koppelman G.H., van der Ent C.K., Beekman J.M. Functional restoration of CFTR nonsense mutations in intestinal organoids. J. Cyst Fibros. 2021;21:246–253. doi: 10.1016/j.jcf.2021.09.020. [DOI] [PubMed] [Google Scholar]
- 28.Konstan M.W., VanDevanter D.R., Rowe S.M., Wilschanski M., Kerem E., Sermet-Gaudelus I., DiMango E., Melotti P., McIntosh J., De Boeck K., ACT CF Study Group Efficacy and safety of ataluren in patients with nonsense-mutation cystic fibrosis not receiving chronic inhaled aminoglycosides: the international, randomized, double-blind, placebo-controlled Ataluren Confirmatory Trial in Cystic Fibrosis (ACT CF) J. Cyst Fibros. 2020;19:595–601. doi: 10.1016/j.jcf.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trzaska C., Amand S., Bailly C., Leroy C., Marchand V., Duvernois-Berthet E., Saliou J.M., Benhabiles H., Werkmeister E., Chassat T., et al. 2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations. Nat. Commun. 2020;11:1509. doi: 10.1038/s41467-020-15140-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sauer B., Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snouwaert J.N., Brigman K.K., Latour A.M., Malouf N.N., Boucher R.C., Smithies O., Koller B.H. An animal model for cystic fibrosis made by gene targeting. Science. 1992;257:1083–1088. doi: 10.1126/science.257.5073.1083. [DOI] [PubMed] [Google Scholar]
- 32.McHugh D.R., Steele M.S., Valerio D.M., Miron A., Mann R.J., LePage D.F., Conlon R.A., Cotton C.U., Drumm M.L., Hodges C.A. A G542X cystic fibrosis mouse model for examining nonsense mutation directed therapies. PLoS One. 2018;13:e0199573. doi: 10.1371/journal.pone.0199573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doucet L., Mendes F., Montier T., Delépine P., Penque D., Férec C., Amaral M.D. Applicability of different antibodies for the immunohistochemical localization of CFTR in respiratory and intestinal tissues of human and murine origin. J. Histochem. Cytochem. 2003;51:1191–1199. doi: 10.1177/002215540305100909. [DOI] [PubMed] [Google Scholar]
- 34.Drost J., Clevers H. Organoids in cancer research. Nat. Rev. Cancer. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 35.Dekkers J.F., Wiegerinck C.L., de Jonge H.R., Bronsveld I., Janssens H.M., de Winter-de Groot K.M., Brandsma A.M., de Jong N.W., Bijvelds M.J., Scholte B.J., et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 36.Liu J., Walker N.M., Ootani A., Strubberg A.M., Clarke L.L. Defective goblet cell exocytosis contributes to murine cystic fibrosis-associated intestinal disease. J. Clin. Invest. 2015;125:1056–1068. doi: 10.1172/JCI73193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansoura M.K., Biwersi J., Ashlock M.A., Verkman A.S. Fluorescent chloride indicators to assess the efficacy of CFTR cDNA delivery. Hum. Gene Ther. 1999;10:861–875. doi: 10.1089/10430349950018274. [DOI] [PubMed] [Google Scholar]
- 38.Cutting G.R., Kasch L.M., Rosenstein B.J., Tsui L.C., Kazazian H.H., Jr., Antonarakis S.E. Two patients with cystic fibrosis, nonsense mutations in each cystic fibrosis gene, and mild pulmonary disease. N. Engl. J. Med. 1990;323:1685–1689. doi: 10.1056/NEJM199012133232407. [DOI] [PubMed] [Google Scholar]
- 39.Kerem B.S., Zielenski J., Markiewicz D., Bozon D., Gazit E., Yahav J., Kennedy D., Riordan J.R., Collins F.S., Rommens J.M. Identification of mutations in regions corresponding to the two putative nucleotide (ATP)-binding folds of the cystic fibrosis gene. Proc. Natl. Acad. Sci. USA. 1990;87:8447–8451. doi: 10.1073/pnas.87.21.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haws C., Krouse M.E., Xia Y., Gruenert D.C., Wine J.J. CFTR channels in immortalized human airway cells. Am. J. Physiol. 1992;263:L692–L707. doi: 10.1152/ajplung.1992.263.6.L692. [DOI] [PubMed] [Google Scholar]
- 41.Sheppard D.N., Welsh M.J. Structure and function of the CFTR chloride channel. Physiol. Rev. 1999;79:S23–S45. doi: 10.1152/physrev.1999.79.1.S23. [DOI] [PubMed] [Google Scholar]
- 42.Winter M.C., Sheppard D.N., Carson M.R., Welsh M.J. Effect of ATP concentration on CFTR Cl- channels: a kinetic analysis of channel regulation. Biophys. J. 1994;66:1398–1403. doi: 10.1016/S0006-3495(94)80930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Population variation of common cystic fibrosis mutations. The cystic fibrosis genetic analysis consortium. Hum. Mutat. 1994;4:167–177. doi: 10.1002/humu.1380040302. [DOI] [PubMed] [Google Scholar]
- 44.Sato T., Clevers H. Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 2013;945:319–328. doi: 10.1007/978-1-62703-125-7_19. [DOI] [PubMed] [Google Scholar]
- 45.Vonk A.M., van Mourik P., Ramalho A.S., Silva I.A.L., Statia M., Kruisselbrink E., Suen S.W.F., Dekkers J.F., Vleggaar F.P., Houwen R.H.J., et al. Protocol for application, standardization and validation of the forskolin-induced swelling assay in cystic fibrosis human colon organoids. STAR Protoc. 2020;1:100019. doi: 10.1016/j.xpro.2020.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this work are available from the authors upon request.