Abstract
Metabolites produced by commensal gut microbes impact host health through their recognition by the immune system and their influence on numerous metabolic pathways. Notably, the gut microbiota can both transform and synthesize lipids as well as breakdown dietary lipids to generate products with these modulatory properties. While lipids have largely been consigned to structural roles, particularly in cell membranes, recent research has led to an increased appreciation of their signaling activities with potential impacts on host health and physiology. This review focuses on studies that highlight the functions of bioactive lipids in mammalian physiology, with a special emphasis on immunity and metabolism.
Description of Review- In Brief
Synthesis, ingestion and absorption of lipids is essential for human life, and is a process mediated by the gut microbiome. Alterations in lipid signaling pathways are behind numerous diseases. Here, Brown and colleagues describe the recent literature on the lipids microbiome species can synthesize, and their function on host physiology.
Introduction
Members of the gut microbiome create an enormous number of small molecules both through de novo biosynthesis and modification of host and dietary substrates. These small molecules have a major impact on all aspects of mammalian physiology in health and disease1. Much of our understanding about how these microbial metabolites influence host physiology has centered on molecules that can be confidently annotated, which is a small fraction of what can be detected in gut tissue and stool. In the absence of defined functions, the study of these small molecules has been organized by structural class. Lipids form an exclusionary class, defined largely by what they are not—not amino acids or peptides, not nucleic acid bases, DNA or RNA, not sugars, polysaccharides or complex carbohydrates. Lipids are what molecules are left over that are not soluble in water and encompass a bewildering variety of structures and functions. In this review, we will follow a conventional classification of lipids as alkyl chain-based fatty acids and their derivatives, with a focus on lipids that are found in the bacterial membrane bilayer. Other important microbial-derived lipids, including short-chain fatty acids and secondary metabolites of host bile acids, are found in stool but are not components of the bacterial outer membrane and not included in this review.
The role of lipid signaling in host-microbiome interactions has been understudied, but this inattention has been quickly dissipating over the past 5 years as chemical and lipidomic techniques have improved and our understanding of the functional importance of lipids in human health has increased2. Recent studies have shown that lipids biotransformed and biosynthesized de novo by the gut microbiome have important structural and signaling functions that impact host cells through both metabolic and immunological pathways3. Microbial-derived lipids can be directly sensed by the host to modulate innate and adaptive immune pathways and to regulate metabolic pathways, all of which can influence the progression of chronic inflammation, autoimmune diseases, cardiovascular disease and metabolic syndrome4. Additionally, changes in the lipid composition of host cell membranes induced by microbes can impact signaling pathways, as the optimal performance of membrane protein activity is a function of the surrounding lipid environment5. Microbiome species also play an important role in biotransformation, detoxification, and digestion of dietary lipids, and the resulting downstream products can affect local tissue and systemic immunity and metabolism in the host6.
Here, we review the recent literature in this area, which are collectively changing the way we think about how host and microbiome interact as an integrated community, a holobiont. Our focus is on bacterial-derived lipids in the gut microbiome, however it should be appreciated that commensal fungal species produce a wide variety of lipid metabolites that can interact with the host and principles reviewed here can be applied to fungi7. Moreover, although lipids have important functions in microbial physiology, membrane dynamics, and microbe-microbe interactions, we focus on their impact on mammalian cells as well as human health and disease. We also address how lipid-mediated interactions can help explain the dramatic rise in inflammatory and autoimmune diseases seen in industrialized countries over the past 50 years, where the microbiome plays a large role in pathogenesis8. Finally, we highlight outstanding questions in the field and further work needed to understand this fascinating and understudied area of biology.
1. Biosynthesis of lipids by the gut microbiome and their host interactions
Bacterial lipids have long been studied for their essential roles in maintaining the structural integrity of the membrane, facilitating energy generation through the electron transport chain, providing a suitable environment for outer membrane proteins, and protecting the cell from exogenous insults. The lipid signature of each bacterial species is unique and reflects both the genetically encoded biosynthetic machinery and the lifestyle of the bacteria; however, the majority of what is known about gut bacterial lipid biosynthesis derives from work on one species, Escherichia coli 9. Notably, the lipid content of E. coli is 10% of the total dry weight of the cell; from a biomass perspective, much of the microbiome metabolic content comprises lipids. Interest in identifying lipids from gut microbiome species of different phyla aside from Proteobacteria—such as Bacteroidetes, Firmicutes, Verrucomicrobia, and Actinobacteria—as well as from commensal fungal species has been recently renewed, buoyed by new metagenomic and metabolomic techniques paired with advances in machine-learning for data analysis.
To date, our understanding of the major lipid classes found in bacterial membranes relies upon studies of a small number of model bacterial organisms. These major classes include phospholipids, such as phosphoethanolamine (PE), phosphoserine (PS), phosphocholine (PC), phosphoinositol (PI) and phosphoglycerol (PG); glycerolipids, such as diacylglycerol (DAG) and triacylglycerol (TAG); and cardiolipins (CL). There are also saccharolipids, such as lipopolysaccharides (LPS), which are large acylated lipid moieties (e.g. lipid A) with a variety of head groups (e.g. sugars) attached. Other lipid classes are characteristic of specific bacteria phyla or taxa. These include sphingolipids, which are synthesized mainly by commensal Bacteroidetes strains, such as sphinganines, dihydroceramide (DHCer), ceramide phosphoethanolamine (CerPE), ceramide phosphoinositol (CerPI), alpha-galactosylceramide (alpha-GC) and deoxysphingolipids10,11,12. A subset anaerobic gut microbiome species synthesize plasmalogens; however, these are not restricted to a particular taxonomic group but produced by various members of Bacteroidetes and Firmicutes13. Sulfonolipids have been identified in some gut bacteria as well, including Bacteroides, Alistipes and Flavobacterium strains14. Each lipid class has a unique architecture, thus confering different structural features and functions to the bacterial membrane. Many are also signaling molecules that can be sensed by host pattern-recognition receptors, such as toll-like receptors (TLRs), NOD-like receptors (NLRs), C-type lectin receptors (CLRs), and G-protein coupled receptors (GPCRs)15. An overview of lipid classification and nomenclature can be found elsewhere16.
While bacterial and host lipid compositions are similar—for example, both use PE and PG in their membranes—there are subtle differences in their biosynthesis and in their recognition by innate immune receptors. The acyl chains of bacterial lipids can be odd- or even-numbered17, whereas those of mammalian lipids are even-numbered. Bacterial lipids have iso-branching at the tails of the acyl chains, which affects host receptor binding capacity and affinity18 and regulates sensing by the innate immune system19. Many bacterial lipids harbor a more diverse range of acyl chain carbon bonding saturations or desaturations than mammalian lipids, and numerous bacterial lipids have 3 or more acyl chains. The mammalian innate immune system likely evolved to be able to sense these unique and diverse biochemical features at a complexity that is only beginning to be unraveled20. The B and T cell arm of the immune system also has evolved to recognize bacterial lipids from the microbiome, as natural IgA can be specific for lipids and many subsets of T cells contain T-cell receptors (TCRs) which can recognize bacterial lipids bound by CD1 proteins21.
The diversity of lipids able to be synthesized by bacteria is enormous22. We have only begun to appreciate this diversity in the gut microbiome, as a large majority of lipids detected in the stool by metabolomic techniques remain unannotated and have unknown function23. Large efforts are underway to generate lipidomic profiles of cultured microbiome strains and piece together lipid classes from previous structural data combined with mass spectrometry. Many of these efforts center around host lipid annotations but recently have begun to address the unknowns in the bacterial community lipidome24,25,26,27. In the following sections, we will discuss the major lipid classes found in the gut microbiome and provide examples of their roles in host signaling (Figure 1).
Figure 1: Membrane lipids biosynthesized by the gut microbiome and their known host signaling functions.
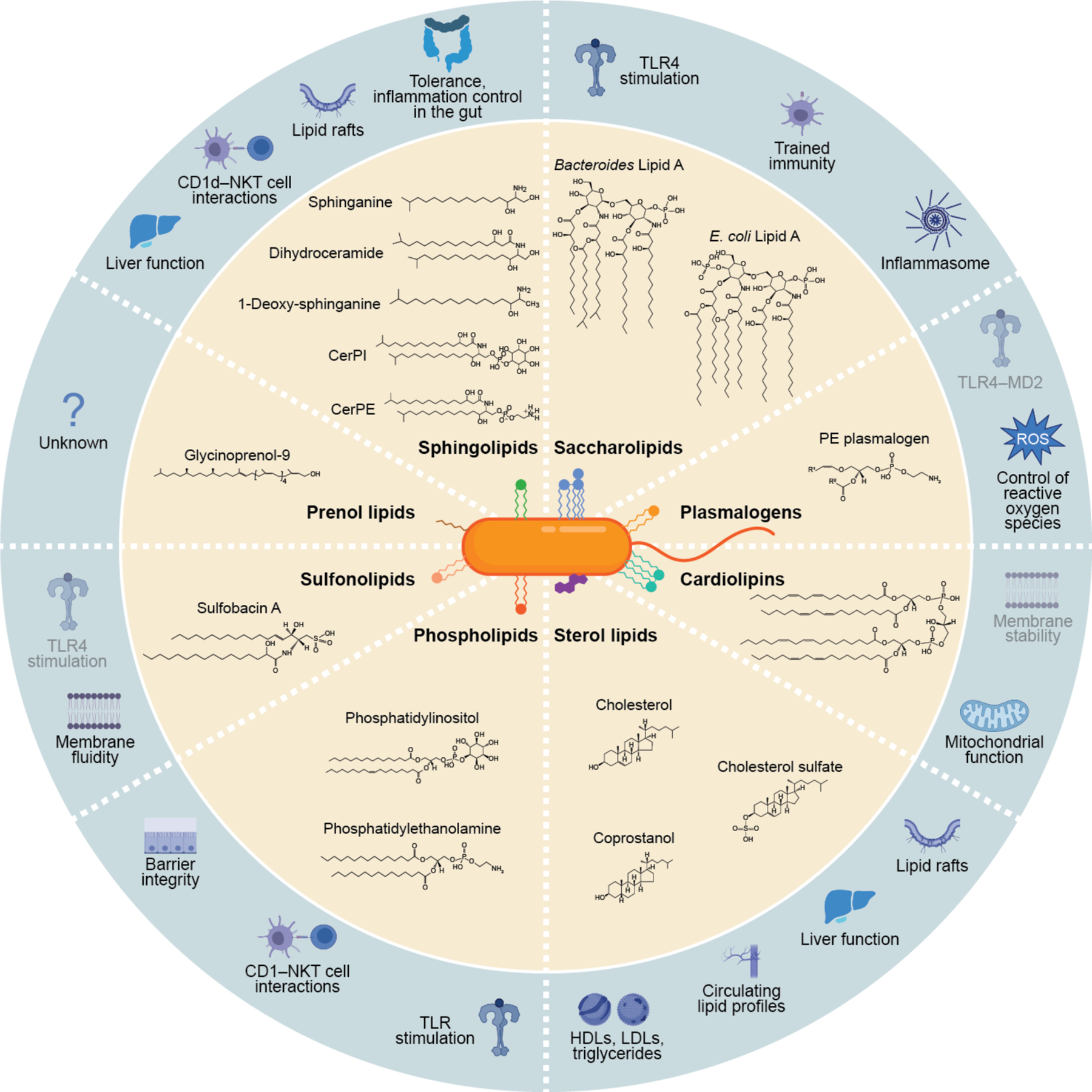
An overview of the diversity of membrane lipids known to be biosynthesized by the gut microbiome, which includes sphingolipids, saccharolipids, plasmalogens, cardiolipins, sterol lipids, phospholipids, sulfonolipids, and prenol lipids. Representative structures of each lipid are shown in the inner ring (beige) and putative host functions for each lipids are in the outer ring (blue). Functions with little experimental evidence are shown in a faded blue color in the outer ring.
Phospholipids
Bacterial phospholipids are highly diverse and the most abundant lipids recovered from the cell membranes of well-studied bacteria. Their biosynthetic pathways have largely been resolved in the model organism E. coli 28. PE is the predominant membrane phospholipid known to be widely synthesized by gut bacteria. Despite this, little is known about how PE or other phospholipids from commensals interact with the host. Screening of microbiome metabolites on host cells have not uncovered a wide variety of phospholipids capable of stimulating cellular responses. For example, large GPCR screens have failed to recognize microbiome phospholipids as agonists to orphan or known receptors29,30,31. A recent study, however, leveraged a bioactivity-guided fractionation approach to screen for immunoregulatory molecules produced by the common gut bacteria Akkermansia muciniphila and identified a diacyl phosphatidylethanolamine with two branched chains (a15:0-i15:0 PE) that activates a pattern recognition receptor (PRR) heterodimer consisting of TLR2-TLR132. This interaction had unique signaling properties compared to common TLR2 agonists, including dampening of the pro-inflammatory IL-12/IL-23 response in human monocytes, that hinted at molecular mechanisms underlying the known effects of Akkermansia on human physiology32.
Bacteria can tightly control the biochemical and biophysical properties of their membrane phospholipids, allowing them to thrive in a wide range of environments28. This includes altering acyl-chain length, iso-branching, modifying degree of unsaturation, and diversifying head groups (most commonly adding ethanolamine, glycerol, inositol, or choline to generate PE, PG, PI, or PC, respectively). How such modifications in response to environmental pressures in the gut may impact signaling capability to the host immune system is unknown. Data collected on eukaryotic lipids suggest that iso-branching can dictate binding strength to protein receptors33. Studies in pathogens suggest that oxidation, iso-branching, and saturation of microbial lipids can influence recognition by host receptors34.
Phospholipid levels in host cells are altered in germ-free mice compared to conventional mice35, suggesting that the gut microbiome influences properties of mammalian membranes as well. Changes in membrane phospholipid chemistry can result in increased intestinal permeability, allowing for bacterial dissemination in the host that has many pathological consequences36. Future studies are needed to address a potential role for microbiome-synthesized phospholipids in intestinal barrier function. In the meantime, we can draw some conclusions from work on pathogens that bacterial phospholipids would modulate immunity. Mycobacterium tuberculosis and Listeria phospholipids, generally PGs, are recognized by natural killer (NK) T cells37, through binding to the nonclassical MHC molecule CD1d that presents lipid antigens. Whether commensal phospholipids have similar roles in CD1 signaling or presentation to T cells is unknown, as is the biochemical determinants of CD1 loading of microbe versus host phospholipids.
Recent work determined that gut bacteria can synthesize PIs, a capability previously attributed only to fungi10,38. Given the role for PI in interactions between bacteria and plant innate immunity as well as inositol signaling in autophagy, it is reasonable to hypothesize that there is a host-specific role for these lipids. A small component of the eukaryotic membrane, inositol signaling has a large effect on many important biological signaling pathways related to bacterial sensing, including GPCR signaling39. Whether host enzymes can break down and incorporate PIs into tissue metabolic pathways is still unknown.
Saccharolipids
Saccharolipids are a type of bacterial glycolipid in which fatty acids are covalently linked to a sugar backbone, positioning the acyl chains in a geometrically defined fashion. These structures can be hosted in membrane bilayers and are ubiquitous in Gram-negative bacteria, with the best characterized being LPS40. Saccharolipids are not produced by eukaryotes and thus are recognized by the innate immune system as MAMPs (microbe-associated molecular patterns). In a classical view, bacterial LPS activate the PRR TLR4 on host cells to initiate pro-inflammatory responses. Activation, however, depends on the acyl chain structure of the lipid A component41, and recent work revealed altered lipid A structures in different bacteria compared to the intensely studied E. coli LPS42. This diversity originates in the variable biosynthetic machinery used to synthesize the lipid A portion of LPS and impacts how the host is able to sense the metabolite43. LPS isolated from microbiome strains of Bacteroides can signal both through TLR2 and TLR4, and does not stimulate as potent an inflammatory response due to its altered lipid A acyl chains44. Inhibition and immunosuppression by Prevotella and Bacteroides LPS has also been reported45. These structural differences can impact early-life systemic immune priming, immune development, and trained immunity46,47.
Saccharolipids, including LPS, can also signal through CLRs on mammalian cells. Lectin-receptor signaling has important roles in regulating gut barrier function and initiating anti-fungal or anti-bacterial immunity. Signaling through many CLRs also regulates the inflammasome, mediating the release of IL-1 family cytokines and alarmins19,48. There is at present no evidence for saccharolipids in the microbiome other than LPS analogs. Further structural analysis of different LPS from microbiome strains is warranted, as well as a detailed look at C-type lectin engagement by lipidated sugars.
Sphingolipids
Sphingolipids can be synthesized de novo by both prokaryotes and eukaryotes in related pathways, suggesting convergent evolution. In eukaryotes, the presence of membrane sphingolipids is ubiquitous, while only a small number of bacterial taxa have the enzymatic ability to make them. Bacteroidetes of the mammalian gut microbiome are unique in their capability to synthesize sphingolipids de novo using serine-palmitoyltransferase (spt) to catalyze the committed step of forming 3-ketosphinganine from serine and an acyl-ACP thioester. Sphingolipid-deficient Bacteroides species are viable but display a growth defect, altered membrane structure, and increased susceptible to oxidative stress49. Lipidomic studies on spt-null Bacteroides strains have uncovered hundreds of putative spt-dependent sphingolipid metabolites, many of which remain unannotated10,50. The most common that have been characterized are sphinganine, DHCer, CerPE, CerPI, alpha-GC, and serine dipeptide sphingolipids10,11,12. CerPI is one of the most abundant sphingolipids in Bacteroides, with an important role conferring bacterial fitness in the gut38. Mammalian receptor signaling functions, however, have not been elucidated for these lipids. Recent efforts have begun to elucidate the biosynthetic pathways required to synthesize sphingolipids, including annotation of the keto-sphinganine synthase51, dihydroceramide synthase52, and ceramide phosphoinositol synthase38. CerPI has also been detected in Bacteroides outer membrane vesicles (OMVs), as well as other phosphoinositol based sphingolipids53. The spt enzyme is also capable of incorporating glycine or alanine to synthesize glycine-based sphingolipids or deoxysphingolipids, respectively, with impact on host signaling yet to be discovered10,54. Bacteroides do make an analogue of 1-deoxysphinganine, which is known to alter inflammasome signaling by inhibiting ceramide synthase55.
Studies of how Bacteroidetes sphingolipids from the microbiome could interact with the host began with the observation that members of the phylum produce a lipid analogous to alpha-GC from Sphingomonas species, a previously known CD1 binding lipid. Two early reports first identified that a B. fragilis-derived alpha-GC binds to mouse CD1d and the TCR on natural killer (NK) T cells56,57. A recent report described the biochemical structure required for efficient alpha-GC-CD1d binding and functional consequence of alpha-GC recognition by the TCR58. When injected into mice, these sphingolipids induced an anti-inflammatory effect and decreased the number of colonic natural killer (NK) T cells58. Whether alpha-GC is the only sphingolipid capable of engaging CD1d receptors is a key question to address given the importance of CD1d-restricted, lipid-specific immune cells in various inflammatory diseases.
Further evidence that microbiome sphingolipids impact host inflammatory and metabolic pathways was provided when colonization of germ-free mice with sphingolipid-deficient bacteria resulted in gut inflammation and changes in host ceramide pools10. Bacteroides sphingolipids interact with the innate immune system, where presence of sphingolipids in the outer membrane of Bacteroides favors a tolerant immune response10. Additional studies in Bacteroidetes showed that sphingolipids in OMVs act as agonists for TLR2 signaling in macrophages and are important in limiting inflammatory signaling59.
In humans, stool sphingolipids from the host are the most significantly increased metabolite class in patients with inflammatory bowel disease (IBD), while microbiome sphingolipids are significantly decreased10. These data suggest a direct connection between host and microbial sphingolipids, which was confirmed in a recent study showing that Bacteroides sphinganine can be imported by host epithelial cells and enter sphingolipid metabolic pathways, altering the host sphingolipidome and liver function12. The gut-liver connection related to sphingolipid signaling was further assessed, and the presence of microbiome sphingolipids was sufficient to reverse fatty liver disease in mice. This correlated with trafficking of a serine dipeptide DHCer sphingolipid to the liver54. Thus, microbiome sphingolipids not only exert local effects but also systemic effects by trafficking to extra-intestinal organs and altering host sphingolipid signaling. The combination of co-evolutionary links, disease associations, and direct biochemical crosstalk between host and microbiome sphingolipids makes for a fascinating area of exploration for future studies.
These studies and others have also highlighted the huge diversity of sphingolipids able to be synthesized by Bacteroidetes, including CerPI which was shown one of the most common and abundant sphingolipids in Bacteroides and is important for bacterial fitness in the gut38, and sphinganine-1-phosphate which similar to host sphingosine-1 phosphate60. However, a mammalian receptor signaling function was not elucidated for either of these lipids. Mammalian sphingolipid signaling is necessary for many inflammatory and cell survival pathways, playing important roles in numerous metabolic and inflammatory diseases61, as well as providing essential signals for cell trafficking, neuronal function in the brain and lipid raft protein function62. Even small changes in host sphingolipid pools can have large effects on host physiology and the downstream impact of microbiome sphingolipid integration in these pathways and their bioactivity is still largely underexplored63. The combination of disease associations, co-evolutionary links and direct biochemical crosstalk between host and microbiome sphingolipids makes it a fascinating area of exploration for future studies.
Sulfonolipids
Sulfonolipids are structurally related to phosphorylated sphingolipids (essentially sulfur analogues of phosphoceramides) and generally have been found in members of the Bacteroidetes phylum such as Alistipes, Odoribacter, Flavobacterium, and Chryseobacterium. Although sulfonolipids have been known for decades to be biosynthesized in a manner similar to Bacteroidetes sphingolipids, a recent study discovered the first enzymatic step in their synthesis: the cysteate acyl-acyl carrier protein transferase sulA64. Relatively little is known about their function. In Flavobacteria species, sulfonolipids enable the bacteria to perform gliding motility65.
The main sulfonolipid that has been characterized is sulfobacin, and its only known interaction with the host is binding to von Willebrand factor receptors66. There is also evidence that sulfobacin is a TLR4 agonist that binds to MD-2 (unpublished), which physically associates with and potentiates TLR4 signaling, and sulfonolipids from Chryseobacterium were shown to have strong pro-inflammatory effects on murine macrophages14. Sulfonolipids with a glycan head group were also found to be pro-inflammatory in human dendritic cells67. Dietary intake in mice has a strong effect on sulfonolipid synthesis by Alistipes, with high-fat diet fed mice displaying a significantly increased abundance of gut sulfobacin68. In humans, Alistipes is the most common human gut microbiome species known to produce sulfonolipids and is significantly depleted in the stool of patients with IBD69, although it is generally not considered an opportunistic pathogen or pro-inflammatory species. A bacterially produced sulfonolipid had previously been identified as the molecular signal for a unicellular eukaryote to form a structured colony70.
Cardiolipins and Plasmalogens
Cardiolipins and plasmalogens are additional examples of lipid classes that can be synthesized de novo by both prokaryotes and eukaryotes in related pathways, again suggesting convergent evolution. Cardiolipins are lipid dimers consisting of two phosphatidyl groups bridged by a glycerol. Host cardiolipins are signature lipids found only in the inner membrane of mitochondria, which is interesting from an evolutionary perspective given their hypothesized role as an endosymbiont fusion of bacteria and eukaryote9. As critical lipids for many physiological roles of mitochondria such as energetic ATP production and mitochondrial protein functions71, dysregulation of cardiolipins is implicated in numerous diseases including aging, metabolic syndrome, heart failure, and cancer72. Host cardiolipins can also regulate immunity and cell death pathways, as exposure to the immune system activates the NLRP3 inflammasome73 and caspases74, and engages CD1 molecules to be presented to NKT cells75, likely as a way of sensing mitochondrial damage.
In bacteria, cardiolipins seem to play a role in stabilizing membranes, as deficiency in cardiolipin synthase renders bacteria more vulnerable to osmotic stress and it accumulates at the poles in bacterial membranes76. Which microbiome strains contain cardiolipins is generally unknown, although they have been reported to be enriched in Streptococcus species and E. coli 77. Little is known about bacterial cardiolipin signaling in host immunity. One study examining LPS signaling antagonists in the microbiome discovered cardiolipins as major metabolites able to reduce LPS binding to CD14 and MD-2 in the TLR4 signaling pathway78,79. Another study discovered a lipid molecule from Acinetobacter baumannii thought to be a cardiolipin that induced inflammation and cell death in mammalian cells through a TLR2-mediated pathway80. Future studies assessing how cardiolipins derived from the microbiome might regulate, alter, or mimic host cardiolipin pathways will be important.
Plasmalogens are glycerophospholipids that contain a vinyl ether bond instead of an ester bond. Plasmalogens have many important functions, including protecting cells from oxidative stress, and are found at the highest concentrations in neuronal and cardiovascular cell membranes81. While the full extent of their function in humans is unknown, defects in plasmalogen synthesis and abundance underlies many neurological diseases including Alzheimer’s82. Plasmalogens are also important mediators of reactive oxygen species and thus are thought to play a role in triggering or resolving chronic inflammation83.
A subset of bacteria from the gut microbiome are capable of synthesizing plasmalogens84. Recently, the enzymes responsible for plasmalogen synthesis were discovered in Clostridium, and homologues of these biosynthetic enzymes map to many different gut microbiome species13. Whether these microbial plasmalogens enter host metabolic pathways and impact human disease remains to be addressed by future studies.
2. Biotransformation of dietary lipids by the microbiome
Ingestion of dietary lipids is essential to human life. From birth to an early age, the human diet consists of breast milk, which is composed of ~55% lipids based on caloric content85. The saturation level and lipid species in breast milk triglycerides is an important variable in the healthy development of infants86. Older children and adults typically consume a diet with fewer calories from fat, dropping to an average of 32% of calories based on the most recent data in the United States87. Most lipids are capable of being synthesized by eukaryotic cells themselves, thus providing a secondary source aside from diet. The exceptions are omega-3 and omega-6 fatty acids, for which mammals do not encode the enzymes required for de novo synthesis and their precursors must derive from the diet88. In general, dietary lipids provide many health benefits to organs and cells in the body, in cell regeneration, protein signaling, energy balance, membrane stability, and homeostatic maintenance of metabolic pathways89.
The gut microbiome exists along the entirety of the mammalian GI tract and encounters all dietary lipids ingested. Enzymes within the gut microbiome thus act like a second liver to break down, transform, and detoxify dietary components, which can result in both beneficial and detrimental impacts on host health. As liver enzymes function to breakdown dietary and exogenous lipids, microbiome enzymes play a similar role in the gut, however most of the functional by-products of gut microbial enzymes, however, remain unknown. It is clear from metabolomic studies comparing stool from germ-free and conventional mice that lipid profiles, absorption, and abundances are dramatically altered by the presence of a microbiome. Below we discuss how major lipid classes ingested by humans are metabolized and bio-transformed by the gut microbiome and the downstream impact on host immune and metabolic pathways (Figure 2). As our diets contain a wide variety of lipids, this is by no means an exhaustive list of all physiologically important dietary lipids.
Figure 2: Biotransformation of dietary lipids by microbiome enzymes.
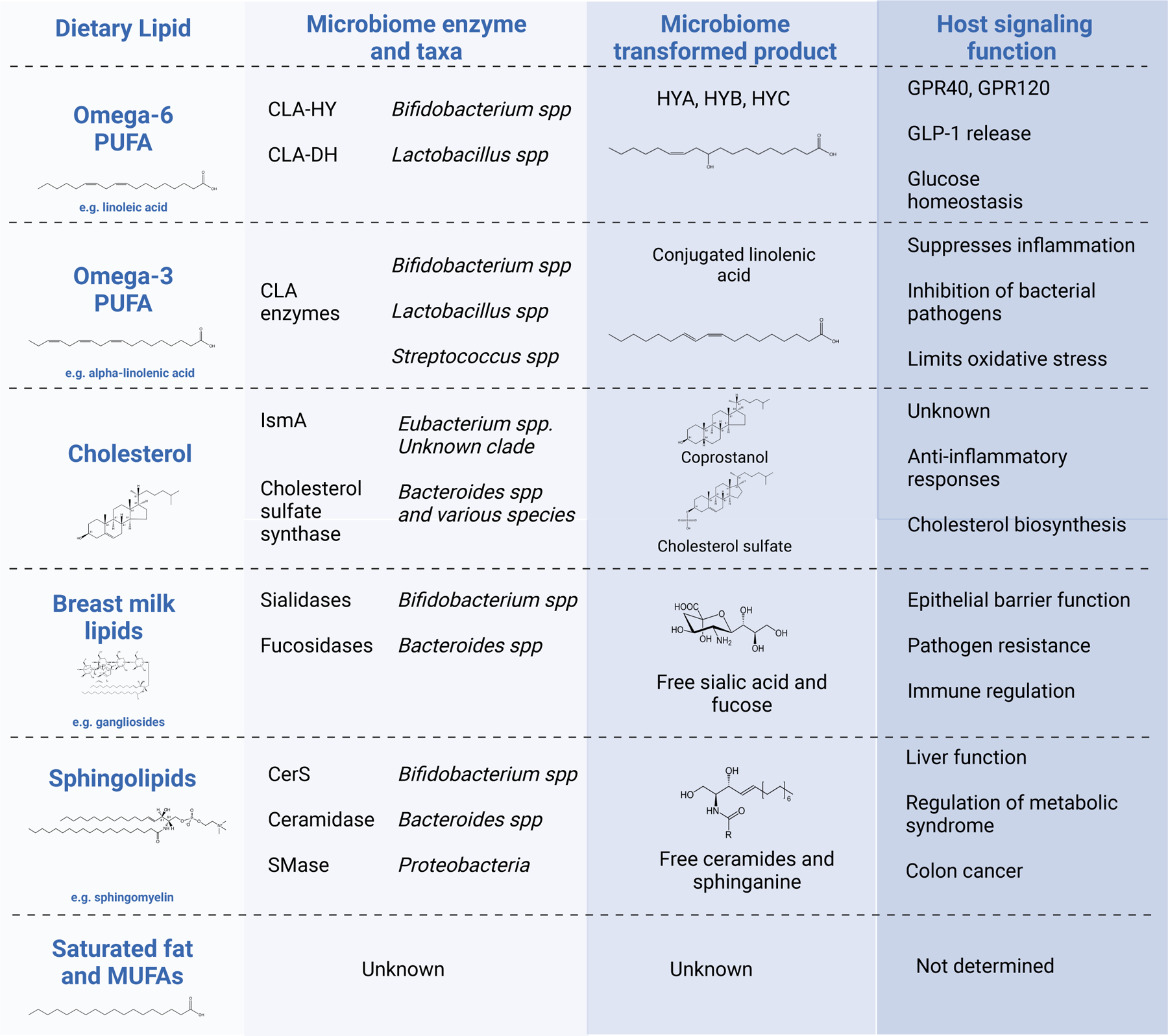
Examples of dietary lipids substrates utilized by gut bacteria and enzymatically converted. These are examples where the enzyme class, species and resulting biotransformed product are known and many more are likely to exist. The host signaling function of the microbiome-transformed lipid product is listed, with references given throughout the manuscript text for each example.
Sphingolipids
From an early age, humans are exposed to exogenous sphingolipids in the diet through breast milk, followed by consumption of dairy, meat, eggs, and many plant species90. Common classes of sphingolipids ingested include ceramides, sphingomyelins, cerebrosides, and gangliosides. There are no dietary guidelines for sphingolipids; nonetheless, complex sphingolipids are hydrolyzed and absorbed throughout the gastrointestinal tract, where they serve important roles regulating cellular growth, differentiation, immunity, and metabolism91. Sphingolipids are also important for brain function and nerve growth92, although the exact impact of or requirement for dietary sphingolipids is unknown89. Gangliosides, which play an essential role in brain cell signaling, are only found in animal fat as plants lack the enzymes to synthesize these lipids93.
In the microbiome, Bacteroidetes strains not only synthesize sphingolipids but also bio-transform dietary sphingolipids through the uptake of simple sphingoid-bases from the diet94. They also possess many glycan-degradation enzymes that can break down ingested gangliosides95. These include sialidases, which can break down sialic acid-containing metabolites (e.g. gangliosides) and release free sialic acid that aids in immunity and prevention of infection96,97. Surprisingly, Bifidobacterium strains, which do not encode the enzyme necessary for sphingolipid synthesis, can import and utilize sphingolipids to generate DHCer94. Early-life metagenomic cohorts revealed that Bifidobacterium also encode enzymes predicted to break down complex sphingolipids98. Studies so far are limited, however, one can surmise there are direct connections between the enzymatic activity of the microbiome and circulating sphingolipid species. At a local level in the gut, increased ingestion of complex sphingolipids has been shown to improve barrier function and reduce bacterial toxin exposure99, processes which can be influenced by known enzymatic functions of the microbiome.
The impact of dietary sphingolipid processing on mammalian biology is just starting to be uncovered. These interactions are of particular importance in the case of breast milk, where sphingolipids are enriched. Sphingolipid biosynthesis pathways are generally upregulated in the early-life microbiome, and this upregulation is a predictor of health100. Ingestion of breast milk versus infant formula—with the former having a more evolutionary consistent sphingolipid content—has a large downstream impact on growth and development101,102,103. Bacteroidetes and Bifidobacterium are highly abundant early-life microbiome strains, and their abilities to influence sphingolipid metabolism could impact human development. A recent study discovered a strong link between the presence of gut Bacteroides in the infant gut and enhanced neurodevelopment, as assessed by the Bayley Scale of Infant Development104. As the gut microbiota-brain axis is becoming a more accepted paradigm105, the roles of dietary and gut-derived sphingolipids in brain development should be explored further.
Cholesterol
Cholesterol is an essential building block of steroid hormones and cellular membranes in eukaryotic cells, and mammals are capable of synthesizing cholesterol without the requirement of dietary cholesterol. Signaling by and regulation of these sterol lipids can influence many immune and metabolic pathways important for human health and disease106.
Circulating cholesterol is utilized as an important biomarker of human health107, and the amount of free cholesterol available for absorption in the gut can influence circulating cholesterol levels108. The idea that gut microbiome metabolism of cholesterol impacts serum cholesterol levels was proposed almost 100 years ago109,110, yet relatively few studies investigated this connection until a recent report definitively confirmed this phenomenon in the human microbiome111. This study discovered the microbial enzyme IsmA in a previously uncultured Firmicute species of the gut microbiome that catalyzes the conversion of cholesterol to cholestenone and ultimately coprostanol111. Moreover, the activity and presence of this enzyme in humans impacts their total serum cholesterol levels111. The impact of this enzyme is clinically relevant, given that it affects serum cholesterol concentrations with an odds ratio equivalent to ezetimibe, an FDA-approved small molecule inhibitor of the intestinal cholesterol transporter and clinically-validated approach to lowering blood cholesterol112.
Biotransformation of dietary cholesterol by gut microbiome strains has recently been shown to be more prevalent than previously thought. Numerous commensal microbes, including Bacteroides, are capable of sulfonating cholesterol113,114. The Bacteroides gene cluster that harbors the sulfotransferase enzyme impacts serum cholesterol levels in mice, adding another variable to the ability of dietary cholesterol to be absorbed into the circulation113. This gene cluster can also sulfate steroid hormones such as vitamin D3 analogues, isoallo-lithocholic acid, coprostanol, and other dietary sterols such as ß-sitosterol114. In IBD and gut inflammation, the gene cluster is significantly deceased, which is of interest given the immune functions recently attributed to sterol signaling in T cells115.
Cholesterol-metabolizing microbial enzymes are not distributed equally among human populations; thus, some variability in cholesterol, cholesterol derivatives, and blood lipid panels may be explained by the subject’s gut metagenome. Although sulfated sterol derivatives such as secondary bile acids have been associated with healthy aging116, the biological functions of coprostanol and sulfated cholesterol metabolites from the microbiome are not yet fully elucidated and represent an exciting avenue for future investigation. As there are hundreds of possible modifications that could be added to sterol-backbones, one can imagine many other cholesterol-derivatives capable of being synthesized by the microbiome utilizing dietary cholesterol. Circulating triglycerides combined with HDL and LDL levels are commonly utilized as indicators of metabolic health. However, to what extent circulating cholesterol directly impacts the development of cardiovascular disease, compared to other pathological mechanisms, is controversial, as correlations are weak. Future studies are warranted to assess these pathways in therapeutic interventions, potentially as combination therapies.
Polyunsaturated fatty acids
Humans are capable of synthesizing all the fatty acids needed except linoleic and alpha-linolenic acids88, which must be derived from diet. These are the precursors to omega-6 and omega-3 polyunsaturated fatty acids (PUFAs), respectively. The downstream metabolic products of both omega-6 and omega-3 PUFAs are essential for membrane components and important for regulating inflammation117. Omega-3 fatty acids have anti-inflammatory effects118,119, while omega-6 fatty acids are pro-inflammatory. Each, however, can be oxidized easily and contribute to general oxidative stress through lipid peroxidation120. The blood omega-6:omega-3 ratio is an important marker for cardiovascular health, and a high omega-6 reading can be a sign of metabolic syndrome121. Omega-6 fatty acid intake can increase insulin resistance, a common mechanism underlying metabolic syndrome122. Evidence supports the importance of maintaining a balanced ratio of omega-6:omega-3 fatty acid intake to protect against metabolic syndrome, cardiovascular disease, and cancer. Consumption of omega-3 and omega-6 fatty acids is unbalanced in the Western diet121,123, with more than 10-times the amount of omega-6 fatty acids than more ancestral diets free of processed food124. The linoleic acid content of human adipose tissue and breast milk has been steadily increasing over the past 100 years125,126, which can negatively impact childhood development127.
Enzymes from the gut microbiome can bio-transform these lipids before they enter host metabolic pathways and thus modulate effects on host lipid metabolism. Bifidobacterium and Lactobacillus species contain CLA-HY, an enzyme that converts linoleic acid to conjugated linoleic acid and then to a molecule that can bind GPR40 and GPR120 to produce an anti-inflammatory signal and limit the amount of linoleic acid converted to downstream products128. CLA enzymes are common amoung human microbiomes and may contribute to variability in susceptibility to metabolic syndrome and obesity128. Dietary PUFAs can also be saturated by common gut microbial enzymes, limiting the number of double bonds and oxidative potential129.
Functionally different microbiome communities have been shown to alter the inflammatory potential of dietary PUFAs130. One study found that high concentrations of omega-6 in the gut can kill bacteria commonly associated with health and promote the growth of bacteria with inflammatory disease associations such as Proteobacteria131. Further, common PUFA metabolites differ significantly in the gut microbiomes from patients with IBD69, and their ingestion from ultra-high processed foods increases IBD risk132. Another study discovered that increased ingestion of linoleic acid in soybean oil alters the balance of liver sphingolipid metabolites, and the presence of a gut microbiome impacted which liver sphingolipids changed as well as their saturation levels133.
Numerous evidence exists that increased ingestion of omega-3 fatty acids in the diet can have an inflammation resolving and anti-inflammatory effect134. Ingestion of fish oil and food sources high in omega-3 has been shown to improve systemic inflammation for humans with inflammatory conditions such as arthritis by randomized, double blinded studies135. Dietary omega-3 fatty acids can alter microbiome composition in humans and mice, and this has been shown to have positive, anti-inflammatory effects on the host136. A possible mechanism by which omega-3 PUFA might beneficially impact host metabolism is through the production of conjugated fatty acids by the gut microbiome. Conjugated isomers of alpha-linolenic acid have reported anti-inflammatory, anti-carcinogenic and anti-obesogenic properties137. In vitro studies have shown that Bifidobacterium, Lactobacillus and Propionibacteria strains are capable of bio-transforming the omega-3 fatty acid alpha-linolenic acid into conjugated linolenic acid isomers138,139,140. Furthermore, conjugated linoleic acid isomers and non-conjugated metabolites are increased in the colonic contents of conventional mice compared to germ-free mice, indicating that the gut microbiome contributes to omega-3 fatty acid metabolism in vivo129. Short-term feeding of mice with alpha-linolenic acid metabolites derived from the gut microbiome affects intestinal immune homeostasis141. More work is needed to confirm the contribution of the gut microbiome to these metabolites and their beneficial effects in humans.
Ultimately, uptake and metabolism of PUFAs depends not only on dietary intake but also on which microbes exist in the gut. Modulation of dietary PUFAs by gut microbiome enzymes could be an important factor in susceptible to inflammatory diseases, metabolic syndrome and cardiovascular disease. This is an understudied area; there are likely many other examples of microbial modification of PUFAs into bioactive metabolites that could influence human health.
Saturated and monounsaturated fats
Triglycerides and phospholipids in plant and animal fats consumed by mammals are commonly categorized based on the saturation level of the acyl chain; this imparts important biochemical properties including redox potential and ability to enter inflammatory pathways. Saturated fatty acids such as stearic and palmitic acids (with 18-and 16-carbon backbones, respectively) are the most abundant in the mammalian diet. Once thought to be harmful and contribute to cardiovascular disease, the most recent data on saturated fatty acids do not support this87. The most ingested monounsaturated fatty acid is the omega-9 fatty acid oleic acid, which is generally accepted to be neutral or beneficial to human health. Research on gut microbiome strains or enzymes capable of bio-transforming or metabolizing saturated or monounsaturated fats is limited. One study showed stearic and oleic acids could selectively alter the growth of certain bacterial strains in vitro131. Mouse work with controlled diets has shown that the saturation level of the dietary fatty acid content can have large effects on mitochondria function, gut permeability, gut motility, and gut microbiome composition, with oxylipin PUFAs from soybean oil resulting in increased obesity142. The saturation level of fatty acids in the membranes of cells and mitochondria impacts cellular function and homeostasis6,124. Given these effects, it will be important to understand the full impact of gut microbial strains on dietary saturated and monounsaturated fatty acids as well as on interactions with host pathways, as it is clear they play a role in personalized responses to diet and inflammatory triggers6.
3. Host-microbiome lipid metabolism and inflammation
Chronic inflammation underlies numerous diseases that plague Westernized societies, including metabolic syndrome and autoimmunity. The quantity, balance, and types of lipids humans consume has been dramatically altered in the past 50–100 years in comparison to the hundreds of thousands of years of human history124. As our genes are slow at adapting to this rapid change, there becomes an even greater reliance on the metagenomic component within commensal microbes to evolve to dietary shifts. It is well accepted that alterations in the amount and types of dietary fat can result in increased systemic inflammation. As a further consequence, lipids directly synthesized by the mammalian microbiome can be potent stimulators of mucosal and systemic immunity, and this initiation of inflammation can feed back into altered lipid absorption and metabolism.
The vast majority of lipids are absorbed in the small intestine, a region of the gut where microbial-derived lipids come in direct contact with the immune system143. In intestinal epithelial cells, there exists a trade-off between expending energy towards immunity versus metabolism, with the adaptive IgA response preventing microbial-derived lipids from over stimulating the innate immune system, which would inhibit lipid absorption144. Lipid absorption can also promote microbiome-mediated gut inflammation through accumulation of pro-inflammatory dietary lipids in epithelial cells, a process facilitated by scavenger receptor CD36 and regulated by T cell signaling145,146. In T cells, lipid profile and metabolism maintains regulatory cells and the propensity to express a Th17 program115. Sterol derivatives from the microbiome have been shown to directly impact these pathways, influencing the T cell balance that is disrupted in gut inflammation147. Finally, early-life exposure to dietary and microbial-derived lipids is crucial to priming, training and development of the mammalian immune system, with implications for disease susceptibility later in life148.
Overall, interactions between i) dietary lipid profiles, ii) microbiome enzymatic functions, and iii) host genetics account for major variations in susceptibility to chronic inflammation induced by changes in lipid metabolism (Figure 3). Of these three variables, lipid products generated by microbiome enzymes and their impact on systemic inflammation and circulating lipid signaling are the least characterized. Many studies connected genes and genetic polymorphisms to specific diseases and circulating lipid profiles in humans; however, much less is known about the variation in microbiome enzymatic functions between individuals that drives clinical outcomes. We are beginning to understand that microbial-derived lipids and their impact on lipid metabolism have a systemic role in host biological functions across many organs. Deep metabolomic profiling on the serum of healthy humans has uncovered thousands of unknown metabolites of bacterial origin, many predicted to be lipids25. Another recent study discovered that microbiome signatures in the gut were as important as diet or host genetics in determining the circulating lipid profile in healthy humans149. Each of these lipidome profiles is unique to an individual over time, and changes could either initiate or resolve inflammation. For example, by-products of linoleic and arachidonic acid metabolism (eicosanoids and resolvins) can have a profound impact on either the initiation or resolution of inflammation in mammals, and microbiome enzymes in the gut can control their circulation134. Polymorphisms in FADS1 and FADS2, fatty acid desaturase genes linked to increased risk for many inflammatory diseases, also control circulating eicosanoid levels150; thus, the combination of microbiome and host genetics must be taken into consideration for risk of inflammation driven by dysregulated lipid metabolism. In many cases, the impact of environmental factors outweighs host genetic susceptibility in the initiation of chronic inflammatory conditions such as IBD or rheumatologic diseases151. Experimental gut inflammation can be triggered by excessive intake of simple carbohydrates, PUFAs, and food additives common in a Western diet124,151, which could account for the large variation in outcomes. Paired metagenomic and metabolomic studies of the human microbiome combined with human genetics are beginning to tease apart these complex connections. Such cohorts exist for IBD, where lipid metabolism pathways are of high importance in the risk for inflammation152.
Figure 3: The function of gut microbiome enzymes responding to dietary lipids are a key step to explain the variation in risk for metabolic syndrome and autoimmunity across human populations.
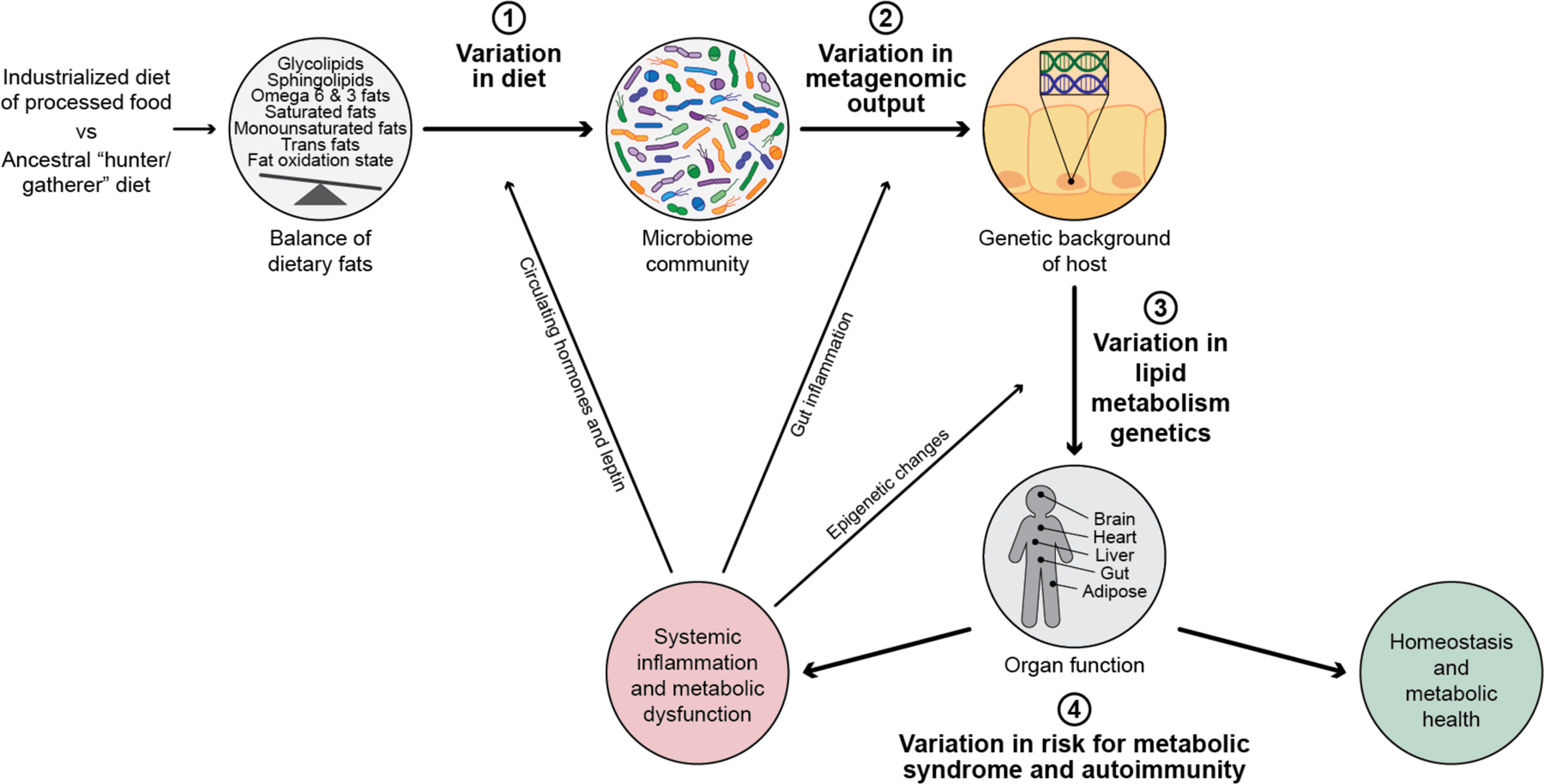
Metabolic syndrome and autoimmunity are diseases of more recent occurrence, and are seen at much higher levels in human populations eating an industrialized and processed food rich diet. This is compared to an ancestral diet which was common among hunter gatherer populations most of human history. Ingestion of dietary lipids are essential for human health and have a large role in all aspects of mammalian biology, and this lipid profiling we ingest has rapidly been changing through industrialization. As the incidence of these disease rise dramatically, it is clear there is variation from human to human on their risk profile for developing systemic inflammation and metabolic dysfunction. This variation in susceptibility for disease is multifactorial and can be explained by 3 main variables as summarized here in this figure; 1) Variation in diet consumed, 2) Variation in metagenomic output by the microbiome and 3) variation in activity and function of metabolism genes from each individual’s genome. Ultimately the outcome of these 3 variables results in; 4) the variation of risk for metabolic syndrome and autoimmunity. The percentage of each of these variables contributing risk is unknown. Human genetic risk explains some variance, however there is a large environmental component to risk of these conditions. The connection between dietary lipid input, and metagenomic output of the microbiome interacting with genetics are crucial, understudied variances in risk for onset of metabolic syndrome and autoimmunity. Systematic inflammation, once initiated, can influence every variable in the process, including what humans crave (e.g. leptin signaling), inflammation-induced ecological shift in microbiome function and epigenetic changes in host gene expression (e.g. stress related), as visually shown here by the arrows.
Aside from in the gut, microbiome lipid metabolism may impact chronic inflammation in the brain, potentially resulting in many neurodegenerative diseases. The brain is composed primarily of lipids, and whether microbiome lipid metabolism influences brain lipid chemistry is unknown. Approaching neurological disorders like Parkinson’s disease as conditions that begin in the gut is an idea taking hold within the field153. A holistic study of brain inflammation that includes examining gut microbial-derived lipids may yield interesting bioactive metabolites important in regulating brain function. The brain is rich in sphingolipids and plasmalogens, both known to be produced in the gut by microbes. NKT cells serve important roles in maintaining immune tolerance in the brain by responding to the lipid antigenic environment. Germ-free mice display many neurological traits, including lower anxiety and behavioral changes154, many of which can be induced upon gut colonization with different microbes. Future studies should systematically assess which microbial-derived lipids impact the gut-brain axis.
4. Future directions in microbiome lipid research
The role of the gut microbiome role in lipid metabolism, both consumption and production, is emerging as a major determinant of human health and disease. Future efforts should expand the modest inroads we have made linking the diverse array of lipids to biological functions and apply our understandings to human diseases. Microbiome enzymes that influence health can either be inhibited or introduced depending on the context, and subjects can be screened using metabolomic and genetic analyses to link metabolic signatures to microbiome population distributions on an individual basis. We can use these data for more informed analyses of prevention, diagnosis, and treatment. More generally, we should incorporate the knowledge that many of the lipids circulating in our bodies are of microbial origin, and our responses to them contribute greatly to clinical outcomes.
These endeavors require two systematic efforts. The first is large-scale annotation of microbiome metabolites. Expanding the tiny fraction of observed metabolomic features that can currently be annotated will require a large effort to purify microbiome metabolites to be run as standards for mass spectrometry. Studies using tandem mass spectrometry analysis to fingerprint microbiome metabolites and generate shared databases will be of great use to the field25,155. Computational and machine learning approaches may provide a more scalable method of lipid identification, as the retention times and masses of lipids can be better predicted from putative, theoretical structures than from other more complex metabolites.
The second major effort is linking lipids to microbes and host-microbe interactions. To this end, we need a more complete catalog of lipidomic profiles from cultured, tissue-associated microbes that correlate with health and disease. Further, genetic manipulation of microbial enzymes mediating lipid metabolism will be important. To date, genetic tools to manipulate strains and delete genes have only been developed for a small number of gut bacteria, limiting hypothesis testing about which enzymes are required for lipid synthesis and what their function on host biology could be. To expand our knowledge of microbiome lipid-host interactions we require gene deletion capabilities in more important taxa for human health such as Akkermansia, Bifidobacteria, Lachnospiraceae and Clostridiales strains. Flow cytometry-based methods that can screen for the capability of bacterial communities to interact with particular lipids based on click-chemistry may also help deconvolute which bacterial enzymes are capable of bio-transforming or synthesizing lipids. Pairing these tools with multi’omic human cohort studies and germ-free mouse experiments can better illuminate the roles of microbial-derived lipids in host physiology.
In the host, a number of orphan receptors (such as G protein-coupled receptors, or GPCRs) expressed by immune and epithelial cells in the gut may interact with yet-unknown microbial-derived lipids30. Moreover, many more microbial membrane lipids are likely recognized by CD1d than the one Bacteroides sphingolipid described. Uncovering new lipid-receptor interactions will require large, unbiased screening efforts of purified microbiome lipids.
OMVs are the major delivery system for microbiome small molecules to host cells156. In what form microbiome lipids are sensed and delivered to host eukaryotic cells requires further study. To that end, we need a better understanding of the lipidome of OMVs secreted by the microbiome. With a better understanding of OMV-mediated lipid secretion and delivery, we can begin to understand which microbiome lipids are delivered and sensed by the host. How microbial-derived lipids delivered by OMVs could be integrated into host lipid pathways or stably exist in extraintestinal organs such as the brain or liver should be assessed. It is tempting to speculate OMVs are capable of delivering lipids systemically throughout the body, with evidence showing this could result in many immunomodulatory properties including resistance to viruses157. Overall, as more key functions of the microbiome are being recognized, the unique lipid fingerprints of microbes colonizing the human gut should be considered in studies assessing health and disease.
Funding and Acknowledgements
We would like to acknowledge Theresa Reimels for her work editing the manuscript. This work was supported by grants from the Helmsley Foundation, the Center for Microbiome Informatics and Therapeutics (CMIT) at MIT and the National Institutes of Health (NIH) U19 AI110495, DK127171, HL157717 and P30 DK043351 to R.J.X.
R.J.X. is a member of the Scientific Advisory Board at Nestlé, founder of Jnana and Celsius Therapeutics and board member of MoonLake Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koppel N, Rekdal VM, and Balskus EP (2017). Chemical transformation of xenobiotics by the human gut microbiota. Science (80) 356, 1246–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichelmann F, Sellem L, Wittenbecher C, Jäger S, Kuxhaus O, Prada M, Cuadrat R, Jackson KG, Lovegrove JA, and Schulze MB (2022). Deep Lipidomics in Human Plasma: Cardiometabolic Disease Risk and Effect of Dietary Fat Modulation. Circulation 146, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamichhane S, Sen P, Alves MA, Ribeiro HC, Raunioniemi P, Hyötyläinen T, and Orešič M (2021). Linking Gut Microbiome and Lipid Metabolism: Moving beyond Associations. Metabolites 11, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon H, Shaw JL, Haigis MC, and Greka A (2021). Lipid metabolism in sickness and in health: Emerging regulators of lipotoxicity. Mol. Cell 81, 3708–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohr MW, Narasimhulu CA, Rudeski-Rohr TA, and Parthasarathy S (2020). Negative Effects of a High-Fat Diet on Intestinal Permeability: A Review. Adv. Nutr 11, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chadaideh KS, and Carmody RN (2021). Host-microbial interactions in the metabolism of different dietary fats. Cell Metab 33, 857–872. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Xia Y, He F, Zhu C, and Ren W (2021). Intestinal mycobiota in health and diseases: from a disrupted equilibrium to clinical opportunities. Microbiome 2021 91 9, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown EM, Kenny DJ, and Xavier RJ (2019a). Gut Microbiota Regulation of T Cells During Inflammation and Autoimmunity. Annu. Rev. Immunol 37, 599–624. [DOI] [PubMed] [Google Scholar]
- 9.Sohlenkamp C, and Geiger O (2016a). Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev 40, 133–159. [DOI] [PubMed] [Google Scholar]
- 10.Brown EM, Ke X, Hitchcock D, Jeanfavre S, Avila-Pacheco J, Nakata T, Arthur TD, Fornelos N, Heim C, Franzosa EA, et al. (2019b). Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis. Cell Host Microbe 25, 668–680.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heaver SL, Johnson EL, and Ley RE (2018). Sphingolipids in host-microbial interactions. Curr. Opin. Microbiol 43, 92–99. [DOI] [PubMed] [Google Scholar]
- 12.Johnson EL, Heaver SL, Waters JL, Kim BI, Bretin A, Goodman AL, Gewirtz AT, Worgall TS, and Ley RE (2020a). Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels. Nat. Commun 2020 111 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson DR, Cassilly CD, Plichta DR, Vlamakis H, Liu H, Melville SB, Xavier RJ, and Clardy J (2021). Plasmalogen Biosynthesis by Anaerobic Bacteria: Identification of a Two-Gene Operon Responsible for Plasmalogen Production in Clostridium perfringens. ACS Chem. Biol 16, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou L, Tian HY, Wang L, Ferris ZE, Wang J, Cai M, Older EA, Raja MRK, Xue D, Sun W, et al. (2022). Identification and biosynthesis of pro-inflammatory sulfonolipids from an opportunistic pathogen Chryseobacterium gleum. ACS Chem. Biol 17, 1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morozumi S, Ueda M, Okahashi N, and Arita M (2022). Structures and functions of the gut microbial lipidome. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 1867, 159110. [DOI] [PubMed] [Google Scholar]
- 16.Fahy E, Subramaniam S, Alex Brown H, Glass CK, Merrill AH, Murphy RC, H Raetz CR, Russell DW, Seyama Y, Shaw W, et al. (2005). A comprehensive classification system for lipids1. J. Lipid Res 46, 839–861. [DOI] [PubMed] [Google Scholar]
- 17.Řezanka T, and Sigler K (2009). Odd-numbered very-long-chain fatty acids from the microbial, animal and plant kingdoms. Prog. Lipid Res 48, 206–238. [DOI] [PubMed] [Google Scholar]
- 18.Parsons JB, and Rock CO (2013). Bacterial Lipids: Metabolism and Membrane Homeostasis. Prog. Lipid Res 52, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander-Floyd J, Bass AR, Harberts EM, Grubaugh D, Buxbaum JD, Brodsky IE, Ernst RK, and Shin S (2022). Lipid A Variants Activate Human TLR4 and the Noncanonical Inflammasome Differently and Require the Core Oligosaccharide for Inflammasome Activation. Infect. Immun 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst RK, and Chandler CE (2017). Bacterial lipids: powerful modifiers of the innate immune response. F1000Research 6, 1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porcelli SA, and Modlin RL (2003). THE CD1 SYSTEM: Antigen-Presenting Molecules for T Cell Recognition of Lipids and Glycolipids 10.1146/Annurev.Immunol.17.1.297 17, 297–329. [DOI] [PubMed] [Google Scholar]
- 22.Sohlenkamp C, and Geiger O (2016b). Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev 40, 133–159. [DOI] [PubMed] [Google Scholar]
- 23.Proctor LM, Creasy HH, Fettweis JM, Lloyd-Price J, Mahurkar A, Zhou W, Buck GA, Snyder MP, Strauss JF, Weinstock GM, et al. (2019). The Integrative Human Microbiome Project. Nat 2019. 5697758 569, 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, Koellensperger G, Huan T, Uritboonthai W, Aisporna AE, et al. (2018). METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem 90, 3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han S, Van Treuren W, Fischer CR, Merrill BD, DeFelice BC, Sanchez JM, Higginbottom SK, Guthrie L, Fall LA, Dodd D, et al. (2021). A metabolomics pipeline for mechanistic interrogation of the gut microbiome. Nature 595, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liebisch G, Fahy E, Aoki J, Dennis EA, Durand T, Ejsing CS, Fedorova M, Feussner I, Griffiths WJ, Köfeler H, et al. (2020). Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res 61, 1539–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang Z, Chong J, Zhou G, De Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques PÉ, Li S, and Xia J (2021). MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 49, W388–W396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang YM, and Rock CO (2008). Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol 2008. 63 6, 222–233. [DOI] [PubMed] [Google Scholar]
- 29.Cohen LJ, Esterhazy D, Kim SH, Lemetre C, Aguilar RR, Gordon EA, Pickard AJ, Cross JR, Emiliano AB, Han SM, et al. (2017). Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 549, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colosimo DA, Kohn JA, Luo PM, Piscotta FJ, Han SM, Pickard AJ, Rao A, Cross JR, Cohen LJ, and Brady SF (2019). Mapping Interactions of Microbial Metabolites with Human G-Protein-Coupled Receptors. Cell Host Microbe 26, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pándy-Szekeres G, Esguerra M, Hauser AS, Caroli J, Munk C, Pilger S, Keseru GM, Kooistra AJ, and Gloriam DE (2022). The G protein database, GproteinDb. Nucleic Acids Res 50, D518–D525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae M, Cassilly CD, Liu X, Park SM, Tusi BK, Chen X, Kwon J, Filipčík P, Bolze AS, Liu Z, et al. (2022). Akkermansia muciniphila phospholipid induces homeostatic immune responses. Nat 2022. 6087921 608, 168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace M, Green CR, Roberts LS, Lee YM, McCarville JL, Sanchez-Gurmaches J, Meurs N, Gengatharan JM, Hover JD, Phillips SA, et al. (2018). Enzyme promiscuity drives branched-chain fatty acid synthesis in adipose tissues. Nat. Chem. Biol 2018. 1411 14, 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatituri RVV, Watts GFM, Bhowruth V, Barton N, Rothchild A, Hsu FF, Almeida CF, Cox LR, Eggeling L, Cardell S, et al. (2013). Recognition of microbial and mammalian phospholipid antigens by NKT cells with diverse TCRs. Proc. Natl. Acad. Sci. U. S. A 110, 1827–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manca C, Boubertakh B, Leblanc N, Deschênes T, Lacroix S, Martin C, Houde A, Veilleux A, Flamand N, Muccioli GG, et al. (2020). Germ-free mice exhibit profound gut microbiota-dependent alterations of intestinal endocannabinoidome signaling. J. Lipid Res 61, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ammendolia DA, Bement WM, and Brumell JH (2021). Plasma membrane integrity: implications for health and disease. BMC Biol 2021. 191 19, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf BJ, Tatituri RVV, Almeida CF, Le Nours J, Bhowruth V, Johnson D, Uldrich AP, Hsu F-F, Brigl M, Besra GS, et al. (2015). Identification of a Potent Microbial Lipid Antigen for Diverse NKT Cells. J. Immunol 195, 2540–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heaver SL, Le HH, Tang P, Baslé A, Mirretta Barone C, Vu DL, Waters JL, Marles-Wright J, Johnson EL, Campopiano DJ, et al. (2022). Characterization of inositol lipid metabolism in gut-associated Bacteroidetes. Nat. Microbiol 2022. 77 7, 986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balla T (2013). Phosphoinositides: Tiny Lipids With Giant Impact on Cell Regulation. Physiol. Rev 93, 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raetz CRH, and Whitfield C (2002). Lipopolysaccharide Endotoxins. Annu. Rev. Biochem 71, 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park BS, Song DH, Kim HM, Choi BS, Lee H, and Lee JO (2009). The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195. [DOI] [PubMed] [Google Scholar]
- 42.Okahashi N, Ueda M, Matsuda F, and Arita M (2021). Analyses of Lipid A Diversity in Gram-Negative Intestinal Bacteria Using Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry. Metabolites 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anandan A, and Vrielink A (2020). Structure and function of lipid A-modifying enzymes. Ann. N. Y. Acad. Sci 1459, 19–37. [DOI] [PubMed] [Google Scholar]
- 44.Vatanen T, Kostic AD, D’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hämäläinen AM, et al. (2016). Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165, 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.d’Hennezel E, Abubucker S, Murphy LO, and Cullen TW (2017). Total Lipopolysaccharide from the Human Gut Microbiome Silences Toll-Like Receptor Signaling. MSystems 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Netea MG, Joosten LAB, Latz E, Mills KHG, Natoli G, Stunnenberg HG, O’Neill LAJ, and Xavier RJ (2016). Trained immunity: A program of innate immune memory in health and disease. Science (80-. ). 352, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitroulis I, Ruppova K, Wang B, Chen LS, Grzybek M, Grinenko T, Eugster A, Troullinaki M, Palladini A, Kourtzelis I, et al. (2018). Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell 172, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Wise L, and Fukuchi KI (2020). TLR4 Cross-Talk With NLRP3 Inflammasome and Complement Signaling Pathways in Alzheimer’s Disease. Front. Immunol 11, 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ana D, Na C, Bielawski J, Hannun YA, and Kasper DL (2011). Membrane sphingolipids as essential molecular signals for Bacteroides survival in the intestine. Proc. Natl. Acad. Sci. U. S. A 108, 4666–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson EL, Heaver SL, Waters JL, Kim BI, Bretin A, Goodman AL, Gewirtz AT, Worgall TS, and Ley RE (2020b). Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels. Nat. Commun 2020. 111 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee MT, Le HH, Besler KR, and Johnson EL (2022b). Identification and characterization of 3-ketosphinganine reductase activity encoded at the BT_0972 locus in Bacteroides thetaiotaomicron. J. Lipid Res 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stankeviciute G, Tang P, Ashley B, Chamberlain JD, Hansen MEB, Coleman A, D’Emilia R, Fu L, Mohan EC, Nguyen H, et al. (2022). Convergent evolution of bacterial ceramide synthesis. Nat. Chem. Biol 18, 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sartorio MG, Valguarnera E, Hsu F-F, and Feldman MF (2022). Lipidomics Analysis of Outer Membrane Vesicles and Elucidation of the Inositol Phosphoceramide Biosynthetic Pathway in Bacteroides thetaiotaomicron. Microbiol. Spectr 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le HH, Lee MT, Besler KR, and Johnson EL (2022a). Host hepatic metabolism is modulated by gut microbiota-derived sphingolipids. Cell Host Microbe 30, 798–808.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duan J, and Merrill AH (2015). 1-Deoxysphingolipids Encountered Exogenously and Made de Novo: Dangerous Mysteries inside an Enigma. J. Biol. Chem 290, 15380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, and Kasper DL (2014). Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M, Sonnenburg JL, Comstock LE, Bluestone JA, and Fischbach MA (2013). Production of α-Galactosylceramide by a Prominent Member of the Human Gut Microbiota. PLOS Biol 11, e1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oh SF, Praveena T, Song H, Yoo JS, Jung DJ, Erturk-Hasdemir D, Hwang YS, Lee CWC, Le Nours J, Kim H, et al. (2021). Host immunomodulatory lipids created by symbionts from dietary amino acids. Nat 2021. 6007888 600, 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rocha FG, Ottenberg G, Eure ZG, Davey ME, and Gibson FC (2021). Sphingolipid-Containing Outer Membrane Vesicles Serve as a Delivery Vehicle To Limit Macrophage Immune Response to Porphyromonas gingivalis. Infect. Immun 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ranjit DK, Moye ZD, Rocha FG, Ottenberg G, Nichols FC, Kim H-M, Walker AR, Frank C Gibson I, and Davey ME. (2022). Characterization of a Bacterial Kinase That Phosphorylates Dihydrosphingosine to Form dhS1P. Microbiol. Spectr 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacEyka M, and Spiegel S (2014). Sphingolipid metabolites in inflammatory disease. Nat 2014. 5107503 510, 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spiegel S, and Milstien S (2011). The outs and the ins of sphingosine-1-phosphate in immunity. Nat. Rev. Immunol 11, 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hannun YA, and Obeid LM (2008). Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol 2008. 92 9, 139–150. [DOI] [PubMed] [Google Scholar]
- 64.Radka CD, Miller DJ, Frank MW, and Rock CO (2022). Biochemical characterization of the first step in sulfonolipid biosynthesis in Alistipes finegoldii. J. Biol. Chem 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abbanat DR, Leadbetter ER, Godchaux W, and Escher A (1986). Sulphonolipids are molecular determinants of gliding motility. Nature 324, 367–369. [Google Scholar]
- 66.Kamiyama T, Umino T, Itezono Y, Nakamura Y, Satoh T, and Yokose K (1995). Sulfobacins A and B, novel von Willebrand factor receptor antagonists. II. Structural elucidation. J. Antibiot. (Tokyo) 48, 929–936. [DOI] [PubMed] [Google Scholar]
- 67.Manzo E, Cutignano A, Pagano D, Gallo C, Barra G, Nuzzo G, Sansone C, Ianora A, Urbanek K, Fenoglio D, et al. (2017). A new marine-derived sulfoglycolipid triggers dendritic cell activation and immune adjuvant response. Sci. Rep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walker A, Pfitzner B, Harir M, Schaubeck M, Calasan J, Heinzmann SS, Turaev D, Rattei T, Endesfelder D, Castell WZ, et al. (2017). Sulfonolipids as novel metabolite markers of Alistipes and Odoribacter affected by high-fat diets. Sci. Reports 2017. 71 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, et al. (2019). Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol 4, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alegado RA, Brown LW, Cao S, Dermenjian RK, Zuzow R, Fairclough SR, Clardy J, and King N (2012). A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. Elife 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Claypool SM, and Koehler CM (2012). The Complexity of Cardiolipin in Health and Disease. Trends Biochem. Sci 37, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chicco AJ, and Sparagna GC (2007). Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am. J. Physiol. - Cell Physiol 292, 33–44. [DOI] [PubMed] [Google Scholar]
- 73.Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, Sadler JJ, Knepper-Adrian V, Han R, Qiao L, et al. (2013). Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Elliott EI, Miller AN, Banoth B, Iyer SS, Stotland A, Weiss JP, Gottlieb RA, Sutterwala FS, and Cassel SL (2018). Cutting Edge: Mitochondrial Assembly of the NLRP3 Inflammasome Complex Is Initiated at Priming. J. Immunol 200, 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pizzuto M, and Pelegrin P (2020). Cardiolipin in Immune Signaling and Cell Death. Trends Cell Biol 30, 892–903. [DOI] [PubMed] [Google Scholar]
- 76.Schlame M (2008). Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J. Lipid Res 49, 1607–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rosch JW, Hsu FF, and Caparon MG (2007). Anionic Lipids Enriched at the ExPortal of Streptococcus pyogenes. J. Bacteriol 189, 801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Balasubramanian K, Maeda A, Lee JS, Mohammadyani D, Dar HH, Jiang JF, Croix CMS, Watkins S, Tyurin VA, Tyurina YY, et al. (2015). Dichotomous roles for externalized cardiolipin in extracellular signaling: Promotion of phagocytosis and attenuation of innate immunity. Sci. Signal 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coats SR, Hashim A, Paramonov NA, To TT, Curtis MA, and Darveau RP (2016). Cardiolipins Act as a Selective Barrier to Toll-Like Receptor 4 Activation in the Intestine. Appl. Environ. Microbiol 82, 4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tiku V, Kew C, Kofoed EM, Peng Y, Dikic I, and Tan MW (2022). Acinetobacter baumannii Secretes a Bioactive Lipid That Triggers Inflammatory Signaling and Cell Death. Front. Microbiol 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Braverman NE, and Moser AB (2012). Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta - Mol. Basis Dis 1822, 1442–1452. [DOI] [PubMed] [Google Scholar]
- 82.Su XQ, Wang J, and Sinclair AJ (2019). Plasmalogens and Alzheimer’s disease: A review. Lipids Health Dis 18, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bozelli JC, Azher S, and Epand RM (2021). Plasmalogens and Chronic Inflammatory Diseases. Front. Physiol 12, 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mawatari S, Sasuga Y, Morisaki T, Okubo M, Emura T, and Fujino T (2020). Identification of plasmalogens in Bifidobacterium longum, but not in Bifidobacterium animalis. Sci. Reports 2020. 101 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mazzocchi A, D’Oria V, De Cosmi V, Bettocchi S, Milani GP, Silano M, and Agostoni C (2018). The Role of Lipids in Human Milk and Infant Formulae. Nutrients 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Innis SM (2011). Dietary Triacylglycerol Structure and Its Role in Infant Nutrition. Adv. Nutr 2, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee JH, Duster M, Roberts T, and Devinsky O (2022a). United States Dietary Trends Since 1800: Lack of Association Between Saturated Fatty Acid Consumption and Non-communicable Diseases. Front. Nutr 8, 1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Das U (2006). Essential Fatty acids - a review. Curr. Pharm. Biotechnol 7, 467–482. [DOI] [PubMed] [Google Scholar]
- 89.Schoeler M, and Caesar R (2019). Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord 2019. 204 20, 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vesper H, Schmelz EM, Nikolova-Karakashian MN, Dillehay DL, Lynch DV, and Merrill AH (1999). Sphingolipids in Food and the Emerging Importance of Sphingolipids to Nutrition. J. Nutr 129, 1239–1250. [DOI] [PubMed] [Google Scholar]
- 91.Merrill AH (2011). Sphingolipid and Glycosphingolipid Metabolic Pathways in the Era of Sphingolipidomics. Chem. Rev 111, 6387–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hussain G, Wang J, Rasul A, Anwar H, Imran A, Qasim M, Zafar S, Kamran SKS, Razzaq A, Aziz N, et al. (2019). Role of cholesterol and sphingolipids in brain development and neurological diseases. Lipids Health Dis 18, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Palmano K, Rowan A, Guillermo R, Guan J, and McJarrow P (2015). The Role of Gangliosides in Neurodevelopment. Nutrients 7, 3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee MT, Le HH, and Johnson EL (2021). Dietary sphinganine is selectively assimilated by members of the mammalian gut microbiome. J. Lipid Res 62, 100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Koropatkin NM, Cameron EA, and Martens EC (2012). How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol 10, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lewis AL, and Lewis WG (2012). Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell. Microbiol 14, 1174–1182. [DOI] [PubMed] [Google Scholar]
- 97.Rueda R (2007). The role of dietary gangliosides on immunity and the prevention of infection. Br. J. Nutr 98 Suppl 1. [DOI] [PubMed] [Google Scholar]
- 98.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni HV, Metcalf GA, et al. (2018). Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nilsson Å (2016). Role of Sphingolipids in Infant Gut Health and Immunity. J. Pediatr 173, S53–S59. [DOI] [PubMed] [Google Scholar]
- 100.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, Lernmark Å, Hagopian WA, Rewers MJ, She JX, et al. (2018). The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Forbes JD, Azad MB, Vehling L, Tun HM, Konya TB, Guttman DS, Field CJ, Lefebvre D, Sears MR, Becker AB, et al. (2018). Association of Exposure to Formula in the Hospital and Subsequent Infant Feeding Practices With Gut Microbiota and Risk of Overweight in the First Year of Life. JAMA Pediatr 172, e181161–e181161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gregory KE, Samuel BS, Houghteling P, Shan G, Ausubel FM, Sadreyev RI, and Walker WA (2016). Influence of maternal breast milk ingestion on acquisition of the intestinal microbiome in preterm infants. Microbiome 4, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. (2012). Human gut microbiome viewed across age and geography. Nat 2012. 4867402 486, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tamana SK, Tun HM, Konya T, Chari RS, Field CJ, Guttman DS, Becker AB, Moraes TJ, Turvey SE, Subbarao P, et al. (2021). Bacteroides-dominant gut microbiome of late infancy is associated with enhanced neurodevelopment. Gut Microbes 13, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morais LH, Schreiber HL, and Mazmanian SK (2020). The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol 2020. 194 19, 241–255. [DOI] [PubMed] [Google Scholar]
- 106.Shimano H, and Sato R (2017). SREBP-regulated lipid metabolism: convergent physiology — divergent pathophysiology. Nat. Rev. Endocrinol 2017. 1312 13, 710–730. [DOI] [PubMed] [Google Scholar]
- 107.Luo J, Yang H, and Song BL (2019). Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol 2019. 214 21, 225–245. [DOI] [PubMed] [Google Scholar]
- 108.Gérard P (2020). The crosstalk between the gut microbiota and lipids. OCL 27, 70. [Google Scholar]
- 109.Rosenheim O, and Websteb TA (1935). Precursors of Coprosterol and the Bile Acids in the Animal Organism. Nat 1935. 1363438 136, 474–474. [Google Scholar]
- 110.Eyssen HJ, Parmentier GG, Compernolle FC, de Pauw G, and Piessens‐ Denef M (1973). Biohydrogenation of sterols by Eubacterium ATCC 21,408--Nova species. Eur. J. Biochem 36, 411–421. [DOI] [PubMed] [Google Scholar]
- 111.Kenny DJ, Plichta DR, Shungin D, Koppel N, Hall AB, Fu B, Vasan RS, Shaw SY, Vlamakis H, Balskus EP, et al. (2020). Cholesterol Metabolism by Uncultured Human Gut Bacteria Influences Host Cholesterol Level. Cell Host Microbe 28, 245–257.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bays HE, Neff D, Tomassini JE, and Tershakovec AM (2008). Ezetimibe: cholesterol lowering and beyond. Expert Rev. Cardiovasc. Ther 6, 447–470. [DOI] [PubMed] [Google Scholar]
- 113.Le HH, Lee MT, Besler KR, Comrie JMC, and Johnson EL (2022b). Characterization of interactions of dietary cholesterol with the murine and human gut microbiome. Nat. Microbiol 2022. 79 7, 1390–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yao L, D’Agostino GD, Park J, Hang S, Adhikari AA, Zhang Y, Li W, Avila-Pacheco J, Bae S, Clish CB, et al. (2022). A biosynthetic pathway for the selective sulfonation of steroidal metabolites by human gut bacteria. Nat. Microbiol 2022. 79 7, 1404–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shyer JA, Flavell RA, and Bailis W (2020). Metabolic signaling in T cells. Cell Res 2020. 308 30, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, Suda W, Takeshita K, Sasaki T, Okamoto S, et al. (2021). Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nat 2021. 5997885 599, 458–464. [DOI] [PubMed] [Google Scholar]
- 117.Saini RK, and Keum YS (2018). Omega-3 and omega-6 polyunsaturated fatty acids: Dietary sources, metabolism, and significance - A review. Life Sci 203, 255–267. [DOI] [PubMed] [Google Scholar]
- 118.Innes JK, and Calder PC (2018). Omega-6 fatty acids and inflammation. Prostaglandins. Leukot. Essent. Fatty Acids 132, 41–48. [DOI] [PubMed] [Google Scholar]
- 119.Shahidi F, and Ambigaipalan P (2018). Omega-3 Polyunsaturated Fatty Acids and Their Health Benefits 10.1146/Annurev-Food-111317-095850 9, 345–381. [DOI] [PubMed] [Google Scholar]
- 120.Foret MK, Lincoln R, Do Carmo S, Cuello AC, and Cosa G (2020). Connecting the “Dots”: From Free Radical Lipid Autoxidation to Cell Pathology and Disease. Chem. Rev 120, 12757–12787. [DOI] [PubMed] [Google Scholar]
- 121.Mariamenatu AH, and Abdu EM (2021). Overconsumption of Omega-6 Polyunsaturated Fatty Acids (PUFAs) versus Deficiency of Omega-3 PUFAs in Modern-Day Diets: The Disturbing Factor for Their “Balanced Antagonistic Metabolic Functions” in the Human Body. J. Lipids 2021, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Glass CK, and Olefsky JM (2012). Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab 15, 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sokoła-Wysoczańska E, Wysoczański T, Wagner J, Czyż K, Bodkowski R, Lochyński S, and Patkowska-Sokoła B (2018). Polyunsaturated Fatty Acids and Their Potential Therapeutic Role in Cardiovascular System Disorders—A Review. Nutrients 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Christ A, Lauterbach M, and Latz E (2019). Western Diet and the Immune System: An Inflammatory Connection. Immunity 51, 794–811. [DOI] [PubMed] [Google Scholar]
- 125.Ballard O, and Morrow AL (2013). Human Milk Composition: Nutrients and Bioactive Factors. Pediatr. Clin. North Am 60, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guyenet SJ, and Carlson SE (2015). Increase in Adipose Tissue Linoleic Acid of US Adults in the Last Half Century. Adv. Nutr 6, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bernard JY, Armand M, Garcia C, Forhan A, De Agostini M, Charles MA, and Heude B (2015). The association between linoleic acid levels in colostrum and child cognition at 2 and 3 y in the EDEN cohort. Pediatr. Res 2015. 776 77, 829–835. [DOI] [PubMed] [Google Scholar]
- 128.Miyamoto J, Igarashi M, Watanabe K, Karaki S ichiro, Mukouyama H., Kishino S., Li X, Ichimura A., Irie J, Sugimoto Y, et al. (2019). Gut microbiota confers host resistance to obesity by metabolizing dietary polyunsaturated fatty acids. Nat. Commun 2019. 101 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Druart C, Bindels LB, Schmaltz R, Neyrinck AM, Cani PD, Walter J, Ramer-Tait AE, and Delzenne NM (2015). Ability of the gut microbiota to produce PUFA-derived bacterial metabolites: Proof of concept in germ-free versus conventionalized mice. Mol. Nutr. Food Res 59, 1603–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kaliannan K, Wang B, Li XY, Kim KJ, and Kang JX (2015). A host-microbiome interaction mediates the opposing effects of omega-6 and omega-3 fatty acids on metabolic endotoxemia. Sci. Reports 2015. 51 5, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fornelos N, Franzosa EA, Bishai J, Annand JW, Oka A, Lloyd-Price J, Arthur TD, Garner A, Avila-Pacheco J, Haiser HJ, et al. (2020). Growth effects of N-acylethanolamines on gut bacteria reflect altered bacterial abundances in inflammatory bowel disease. Nat. Microbiol 5, 486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lo CH, Khandpur N, Rossato SL, Lochhead P, Lopes EW, Burke KE, Richter JM, Song M, Ardisson Korat AV, Sun Q, et al. (2022). Ultra-processed Foods and Risk of Crohn’s Disease and Ulcerative Colitis: A Prospective Cohort Study. Clin. Gastroenterol. Hepatol 20, e1323–e1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Di Rienzi SC, Johnson EL, Waters JL, Kennedy EA, Jacobson J, Lawrence P, Wang DH, Worgall TS, Brenna JT, and Ley RE (2021). The microbiome affects liver sphingolipids and plasma fatty acids in a murine model of the Western diet based on soybean oil. J. Nutr. Biochem 97, 108808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schulze MB, Minihane AM, Saleh RNM, and Risérus U (2020). Intake and metabolism of omega-3 and omega-6 polyunsaturated fatty acids: nutritional implications for cardiometabolic diseases. Lancet Diabetes Endocrinol 8, 915–930. [DOI] [PubMed] [Google Scholar]
- 135.Hahn J, Cook NR, Alexander EK, Friedman S, Walter J, Bubes V, Kotler G, Lee IM, Manson JAE, and Costenbader KH (2022). Vitamin D and marine omega 3 fatty acid supplementation and incident autoimmune disease: VITAL randomized controlled trial. BMJ 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Costantini L, Molinari R, Farinon B, and Merendino N (2017). Impact of Omega-3 Fatty Acids on the Gut Microbiota. Int. J. Mol. Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yuan GF, Chen XE, and Li D (2014). Conjugated linolenic acids and their bioactivities: a review. Food Funct 5, 1360–1368. [DOI] [PubMed] [Google Scholar]
- 138.Gorissen L, Raes K, Weckx S, Dannenberger D, Leroy F, De Vuyst L, and De Smet S (2010). Production of conjugated linoleic acid and conjugated linolenic acid isomers by Bifidobacterium species. Appl. Microbiol. Biotechnol 87, 2257–2266. [DOI] [PubMed] [Google Scholar]
- 139.Hennessy AA, Barrett E, Paul Ross R, Fitzgerald GF, Devery R, and Stanton C (2012). The production of conjugated α-linolenic, γ-linolenic and stearidonic acids by strains of bifidobacteria and propionibacteria. Lipids 47, 313–327. [DOI] [PubMed] [Google Scholar]
- 140.Ogawa J, Kishino S, Ando A, Sugimoto S, Mihara K, and Shimizu S (2005). Production of conjugated fatty acids by lactic acid bacteria. J. Biosci. Bioeng 100, 355–364. [DOI] [PubMed] [Google Scholar]
- 141.Ohue-Kitano R, Yasuoka Y, Goto T, Kitamura N, Park SB, Kishino S, Kimura I, Kasubuchi M, Takahashi H, Li Y, et al. (2018). α-Linolenic acid-derived metabolites from gut lactic acid bacteria induce differentiation of anti-inflammatory M2 macrophages through G protein-coupled receptor 40. FASEB J 32, 304–318. [DOI] [PubMed] [Google Scholar]
- 142.Deol P, Fahrmann J, Yang J, Evans JR, Rizo A, Grapov D, Salemi M, Wanichthanarak K, Fiehn O, Phinney B, et al. (2017). Omega-6 and omega-3 oxylipins are implicated in soybean oil-induced obesity in mice. Sci. Rep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Iqbal J, and Hussain MM (2009). Intestinal lipid absorption. Am. J. Physiol. - Endocrinol. Metab 296, E1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shulzhenko N, Morgun A, Hsiao W, Battle M, Yao M, Gavrilova O, Orandle M, Mayer L, Macpherson AJ, McCoy KD, et al. (2011). Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med 17, 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen Y, Zhang J, Cui W, and Silverstein RL (2022b). CD36, a signaling receptor and fatty acid transporter that regulates immune cell metabolism and fate. J. Exp. Med 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang H, Franco F, Tsui YC, Xie X, Trefny MP, Zappasodi R, Mohmood SR, Fernández-García J, Tsai CH, Schulze I, et al. (2020). CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol 2020. 213 21, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hang S, Paik D, Yao L, Kim E, Jamma T, Lu J, Ha S, Nelson BN, Kelly SP, Wu L, et al. (2019). Bile acid metabolites control TH17 and Treg cell differentiation. Nat 2019. 5767785 576, 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Okada H, Kuhn C, Feillet H, and Bach JF (2010). The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clin. Exp. Immunol 160, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen L, Zhernakova DV, Kurilshikov A, Andreu-Sánchez S, Wang D, Augustijn HE, Vich Vila A, Study LC, Weersma RK, Medema MH, et al. (2022a). Influence of the microbiome, diet and genetics on inter-individual variation in the human plasma metabolome. Nat. Med 2022. 1–11. [DOI] [PMC free article] [PubMed]
- 150.Koletzko B, Reischl E, Tanjung C, Gonzalez-Casanova I, Ramakrishnan U, Meldrum S, Simmer K, Heinrich J, and Demmelmair H (2019). FADS1 and FADS2 Polymorphisms Modulate Fatty Acid Metabolism and Dietary Impact on Health. Annu. Rev. Nutr 39, 21–44. [DOI] [PubMed] [Google Scholar]
- 151.Adolph TE, Meyer M, Schwärzler J, Mayr L, Grabherr F, and Tilg H (2022). The metabolic nature of inflammatory bowel diseases. Nat. Rev. Gastroenterol. Hepatol 2022. 1–15. [DOI] [PubMed]
- 152.Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, Morgan XC, and Huttenhower C (2015). Sequencing and beyond: integrating molecular “omics” for microbial community profiling. Nat. Rev. Microbiol 13, 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lionnet A, Leclair-Visonneau L, Neunlist M, Murayama S, Takao M, Adler CH, Derkinderen P, and Beach TG (2018). Does Parkinson’s disease start in the gut? Acta Neuropathol 135. [DOI] [PubMed] [Google Scholar]
- 154.Heijtz RD, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, Hibberd ML, Forssberg H, and Pettersson S (2011). Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U. S. A 108, 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dekkers KF, Sayols-Baixeras S, Baldanzi G, Nowak C, Hammar U, Nguyen D, Varotsis G, Brunkwall L, Nielsen N, Eklund AC, et al. (2022). An online atlas of human plasma metabolite signatures of gut microbiome composition. Nat. Commun 2022. 131 13, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Caruana JC, and Walper SA (2020). Bacterial Membrane Vesicles as Mediators of Microbe-Microbe and Microbe-Host Community Interactions. Front. Microbiol doi: 10.3389/fmicb.2020.00432 [DOI] [PMC free article] [PubMed]
- 157.Erttmann SF., Swacha P., Aung KM., Brindefalk B., Jiang H, Härtlova A., Uhlin BE., Wai SN., Gekara NO. (2022). The gut microbiota prime systemic antiviral immunity via the cGAS-STING-IFN-I axis. Immunity 55, 847–861. [DOI] [PubMed] [Google Scholar]


