Abstract
Treatment of HeLa cells with the DNA damaging agent, bleomycin, results in the formation of a nonenzymatic 5-methylene-2-pyrrolone histone covalent modification on lysine residues (KMP). KMP is much more electrophilic than other N-acyllysine covalent modification and post-translational modifications, including N-acetyllysine (KAc). Using histone peptides containing KMP we show that this modification inhibits the class I histone deacetylase, HDAC1, by reacting with a conserved cysteine (C261) located near the active site. HDAC 1 is inhibited by histone peptides whose corresponding N-acetylated sequences are known deacetylation substrates, but not one containing a scrambled sequence. The HDAC1 inhibitor, trichostatin A, competes with covalent modification by the KMP-containing peptides. HDAC1 is also covalently modified by a KMP-containing peptide in a complex milieu. These data indicate that peptides containing KMP are recognized by HDAC1 and are bound in the active site. The effects on HDAC1 indicate that KMP formation in cells may contribute to the biological effects of DNA damaging agents, such as bleomycin that form this nonenzymatic covalent modification.
Graphical Abstract
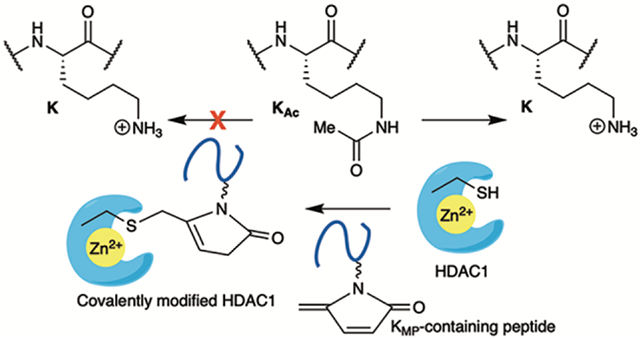
Introduction.
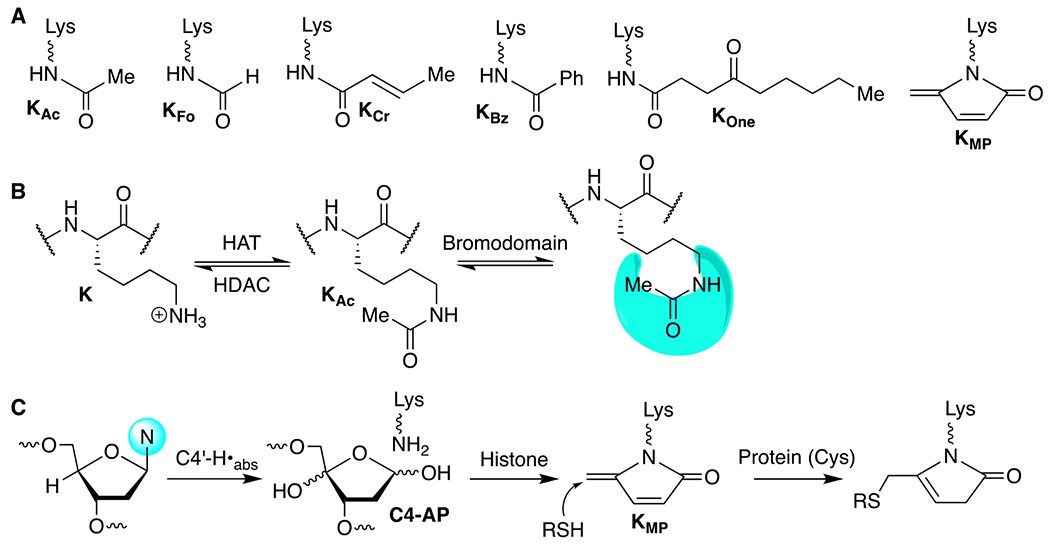
A variety of N-acyllysine post-translational modifications (PTMs) have been identified. Many are produced enzymatically (e.g. KAc, KCr), but some are formed nonenzymatically (e.g. KFo, KOne) (Scheme 1A) on histones and other proteins.1 N-acetyllysine (KAc) is an enzymatic PTM that affects chromatin structure and plays important roles in regulating cellular processes, including transcription.2, 3 Three families of proteins are involved in histone acetylation (Scheme 1B). Histone acetyl transferases (HATs) install KAc, which results in increased chromatin accessibility that is partly attributed to loss of positive charge, and a weakening of the interaction between DNA and histone proteins.2, 4 KAc sites are read in chromatin by bromodomain containing proteins, which interact with protein complexes that control processes such as transcription. Acetyl removal from lysine by histone deacetylases (HDACs) turns off gene expression and favors chromatin compaction. Regulation of histone acetylation is important in human health. For instance, HDAC overexpression and mistargeting occur in a variety of cancers.5
Scheme 1.
N-Acyllysine histone modifications.
HDAC1 is a member of the Class I family of histone deacetylases, which utilize Zn2+ to activate water for nucleophilic attack on N-acetyllysine (KAc).6 The list of N-acyllysine PTMs that are substrates for HDAC1 and other deacetylases is growing.7–9 The sirtuins, such as Sirt2, preferentially deacetylate N-acetyllysine in the histone H4 “basic patch” at H4K16. To a lesser extent, it also deacetylates H3K9Ac. SIRT2 displays a relatively broad N-acyllysine substrate-scope. For example, The 4-ketoamide histone adducts formed under oxidative stress during inflammation, such as KOne are substrates of Sirt2.10, 11 Sirt2 is also shown to remove benzoyl groups in histone benzol-lysine (KBz).12 Others such as the nonenzymatic covalent modification formylation (KFo), which is formed predominantly via initial reaction with formaldehyde is not reported to be an excellent substrate for HDACs.8,13 However, KFo could affect chromatin structure and gene expression. We recently reported HeLa cell treatment with bleomycin (BLM) results in formation of a DNA damage-induced, nonenzymatic covalent modification (NECM) of histones that contains an electrophilic 5-methylene-2-pyrrolone (KMP, Scheme 1A,C).14 Herein, we characterize the interaction between KMP-containing histone peptides and HDAC1. Our data suggest that KMP-containing histones could potentially covalently modify HDAC1 in cells and affect cellular processes.
KMP forms concomitantly with a single-strand DNA break via histone lysine attack on the oxidized abasic site (C4-AP, Scheme 1C).15, 16 The half-life for the formation of KMP in nucleosome core particles is as short as ~15 min. C4-AP is produced by ionizing radiation and several antitumor agents that oxidatively damage DNA, including bleomycin.17–20 KMP was detected via chemoproteomic analysis at 17 of 57 core histone lysine residues in HeLa cells treated with bleomycin.14 The sites at which the NECM are determined by C4-AP location, and it is likely that KMP distribution is even broader following treatment with more promiscuous DNA damaging agents, such as ionizing radiation. Of the 17 positions at which KMP was detected, 15 are also lysine acetylation sites.21 Various deacetylases act on the group of acetylated lysines that overlap with sites at which KMP is formed.6 We sought to address whether KMP affects HDAC1 activity.
Results and Discussion.
Peptide design and preparation.
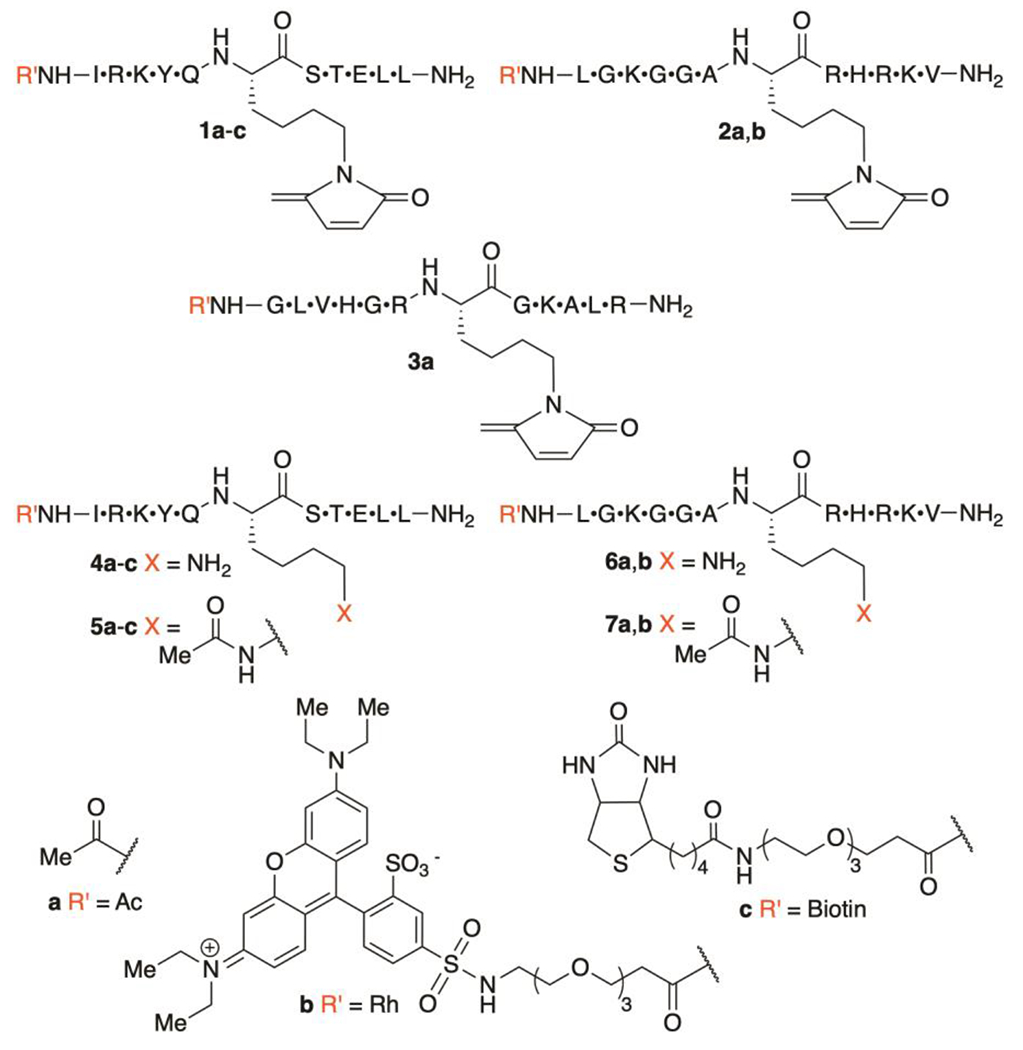
Histone H3K56 is a position at which KMP is formed within the globular region of the octameric core upon treatment of HeLa cell with bleomycin.14 In vivo acetylation at H3K56 (H3K56Ac) affects chromatin structure and regulates transcription.22, 23 Cellular levels of H3K56Ac are regulated by HDAC1.24 Similarly, histone H4K16Ac is an important acetylation site within the corresponding histone tail. H4K16Ac inhibits chromatin compaction and is also a substrate for HDAC1.25, 26 Consequently, we examined the interaction of histone peptides containing H3K56MP (1a) and H4K16MP (2a) with HDAC1 (Chart 1). A peptide comprised of the same amino acid composition as 2a, but with scrambled sequence (3a) was also prepared. Peptides containing KMP were synthesized via a method that distinguishes between multiple lysines using orthogonal ε-amine protecting groups in which the electrophilic 5-methylene-2-pyrrolone is introduced after peptide cleavage from the solid-phase support.14
Chart 1.
Histone peptides used in these experiments.
HDAC1 inhibition by KMP-containing histone peptides.
The possibility that KMP-containing histone peptides are recognized by HDAC1 was explored using a commercially available fluorescence assay kit (Scheme S1) that takes advantage of trypsin cleavage of deacylated lysine (Figure 1). HDAC1 activity was measured following preincubation with peptides 1a (Figure 1A) and 2a (Figure 1B). The peptides exhibited comparable levels of inhibition between 0.5 and 50 μM. HDAC1 activity was reduced by ~50% following preincubation with either peptide at the maximum concentration employed (50 μM). However, the corresponding peptides (50 μM) containing either lysine (4a, 6a) or N-acetyllysine (5a, 7a) had no effect on HDAC1 activity when incubated with the enzyme. Importantly, inhibition by the scrambled version of 2a (3, Figure 1C, S4) was clearly evident only at ≥ 25 μM of 3. There also was no statistical difference in the extent of inhibition in the presence of 25 or 50 μM of 3, and the level of activity retained was considerably higher than when HDAC1 was preincubated with 1a or 1b at these same concentrations. The smaller effect observed utilizing the scrambled sequence (3) supports the hypothesis that KMP-containing peptides based on the H3K56 and H4K16 histone modifications are not due to the promiscuity of the electrophile. It also eliminates inactivation of trypsin, which is employed in the assay, as the source of the observed HDAC1 inhibition.
Figure 1.
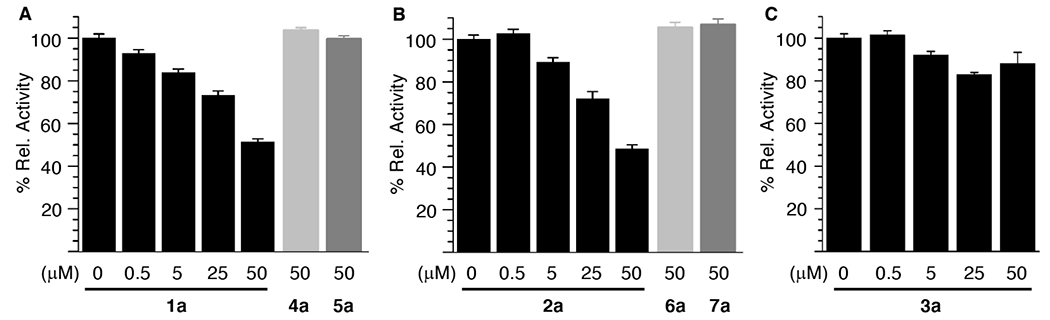
Effect of KMP-containing histone peptides on HDAC1 activity. A) Histone peptides containing H3K56 (1a, 4a, 5a) B) Peptides containing H4K16 (2a, 6a, 7a) C) Histone peptides containing H3K16 with sequence scrambled (3) [HDAC1] = 10 nM. 100% Relative activity was determined by comparing enzymatic activity with and without peptide. Data are presented as the ave. ± std. dev. (n=3). For indicators of statistical significance see Figure S4.
Covalent modification of HDAC1 by KMP-containing histone peptides.
Electrophoretic analysis under denaturing conditions using fluorescently labeled versions of 1a and 2a (1b, 2b) revealed that the peptides covalently modify HDAC1 (Figure 2A, S1). In contrast, the corresponding peptides containing free lysine (4b, 6b) or N-acetyllysine (5b, 7b) do not. Although HDAC1 contains 9 cysteine residues (Figure 2B), none play a direct role in catalysis. However, C261 and C273, which are conserved throughout the class I HDACs, are less than 10 Å from the zinc ion that activates water for nucleophilic attack on N-acetyllysine (Figure 2C).27 These residues, which are modified by electrophilic small molecules seemed like good candidates for reaction with KMP in histone peptides.27 Following incubation of HDAC1 separately with 1a (Figure 2D, Table S1) or 2a (Figure S2), the protein was digested with trypsin and analyzed by LC-MS/MS. Following incubation with the histone peptide H3K56MP (1a), we identified a modified peptide encompassing HDAC1 residues V248-R270 that contains C261. The expected fragment containing a modification on C261 that consists of the portion of the KMP-containing peptide that remains after trypsin treatment (y11, Figure 2D) was detected. Fragments that lack C261 (e.g. y9 and b13) are unmodified, consistent with incorporation of the modification at this amino acid position. Similarly, the same modified peptide that includes C261 within the HDAC1 residues V248-R270 was detected following incubation with H4K16MP (2a) (Figure S2). MS2 spectra identified fragment ions establishing the modification on C261 (y10-y19 and b15, Figure S2). Fragment ions lacking C261 (e.g. y3-y6 and b2-b11) further support the assignment of the modification at this position. In contrast, no peptides containing a modification at C273 were detected following incubation with 1a or 2a; nor was modification of any of the other 7 cysteine residues, some of which are surface exposed, observed.
Figure 2.
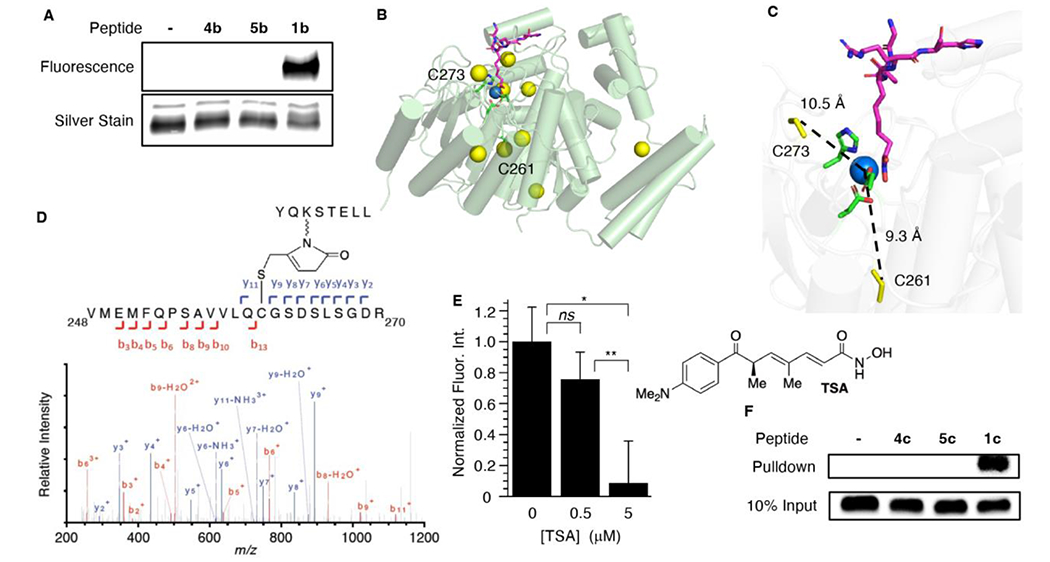
Covalent modification of HDAC1 by a KMP-containing peptide (H3K56MP). A) Covalent modification of HDAC1 (0.1 μM) by fluorescently labeled KMP-containing histone peptide (1b, 50 μM). B) X-ray crystal structure of HDAC1 bound with a peptide inhibitor (PDB: 5ICN), showing active site zinc (blue) and 9 cysteine residues (yellow). C) Active site showing the distance between Zn2+ and C261, C273. D) LC-MS/MS analysis of HDAC1 trypsin fragment V248-R270 that is covalently modified by a histone peptide containing H3K56MP (1a). E) Trichostatin A (TSA) competition for covalent modification of HDAC1 (100 nM) by KMP-containing histone peptide H3K56MP (1b, 5 μM). Average fluorescence intensity of reaction in the absence of TSA is normalized to 1.0. Data are presented as the ave. ± std. dev. (n=3). (ns) indicates not significantly different, (*) indicates p < 0.0005 and (**) indicates p < 0.005. F) Covalent modification of HDAC1 by histone peptide containing H3K56MP (1c, 10 μM) in HeLa cell nuclear lysate. Detection is carried out using anti-HDAC1 antibody. HDAC1 migrates at ~60 kDa on the gel (Figure S1).
Covalent modification of C261, which is proximal to the HDAC1 active site supports the hypothesis that peptides 1a and 2a are bound at or near this region of the enzyme.
Furthermore, the lack of detection of modification of any of the other 8 cysteine residues suggest that covalent modification is not due to nonselective, promiscuous reactions of the electrophilic KMP-containing histone peptides with the deacetylase.
An HDAC1 competitive inhibitor prevents covalent modification by a KMP-containing histone peptide.
LC-MS/MS analyses of trypsin digests are consistent with recognition of the KMP-containing histone peptides in a comparable manner as the corresponding N-acetyllysine molecules, and that KMP is positioned within or near the active site. This hypothesis was explored further by examining the ability of trichostatin A (TSA) to inhibit HDAC1 modification by the KMP-containing histone peptides. TSA is a small molecule competitive inhibitor of HDAC1 that binds in the N-acetyllysine binding site. Although 1b and 2b covalently bind HDAC1, we anticipated that the small molecule could decrease covalent modification by the KMP-containing histone peptides that interact with the active site due to its tight binding (KD = 12 nM).28 HDAC1 (0.1 μM) was preincubated (10 min) with 0.5 or 5 μM TSA prior to adding a KMP-containing histone peptide. The yield of covalently modified HDAC1 with the fluorescently labeled H3K56MP (1b, Figure 2E) or H4K16MP (2b, Figure S3) peptide decreased by similar amounts when preincubated with TSA and the decrease was dependent on TSA concentration. Preincubation with 5 μM TSA reduced covalent modification by ~90%. These data are consistent with peptide binding in the N-acetyllysine binding site. Importantly, the observation that only ~10% of the covalent modification remains after preincubation with TSA is consistent with non-promiscuous HDAC1 modification by the KMP-containing histone peptides. Although, we are unable to measure the binding constant for HDAC1 recognition of KMP-containing peptides, the fact that HDAC1 cross-linking persists even in the presence of TSA present at almost 1,000-times its KD suggests that peptides containing the modification are bound reasonably strongly by the enzyme. It also suggests that proteins containing KMP will also be effective at covalently modifying HDAC1 in cells.
Covalent modification of HDAC1 by a KMP-containing histone peptide in nuclear lysate.
The ability of biotinylated 1 (1c) to covalently modify HDAC1 in a complex milieu was examined using HeLa nuclear lysate. Lysate was incubated with 1c or the corresponding biotinylated free lysine (4c) or N-acetyllysine (5c) peptides (10 μM). After pulling down the biotinylated substrates using Neutravidin resin, vigorous washing and release, the eluted materials were interrogated via Western blot analysis using anti-HDAC1 antibody (Figure 2F). Consistent with the above experiments, HDAC1 was detected when lysate was incubated with KMP-containing histone peptide (1c) but not the unmodified (4c) or N-acetyllysine peptides (5c). Hence, covalent modification of HDAC1 in this complex milieu is consistent with a selective and strong interaction with the KMP modified peptide (1c).
Summary
KMP is one of a growing family of NECMs.29–33 Notably, KMP is part of a subset of NECMs that are sufficiently electrophilic to react further with nucleophilic amino acid residues. Covalent modification of HDAC1 by KMP-containing peptides reflects its greater electrophilicity than other N-acylated lysine histone modifications. Although such a reaction (Scheme 1C) is reversible in the presence high thiol concentration (20 mM), the adducts were stable under physiological conditions in our hands.34 The recalcitrance of KCr to react with thiols is reflected by the covalent capture of proteins containing it by phosphine-containing nucleophilic probes.35 However, an engineered Sirtuin containing an unnatural thiol amino acid does covalently capture KCr.36 N-formyllysine (KFo), which is refractory to repair by HDAC1, also does not react with cysteine residues in proteins.9, 13 Covalent modification of HDAC1 by KMP-modified histone proteins in cells could result in aberrant gene expression. KMP formation is a secondary effect in cells treated with bleomycin, a cytotoxic DNA damaging agent.14 To our knowledge, it is not known whether bleomycin treatment affects cellular HDAC1 activity or expression level. However, pulmonary fibrosis is a side reaction of bleomycin. Increased levels of HDAC2 (Class I histone deacetylase) and HDAC4 have been detected in mouse lung fibroblasts treated with bleomycin.37 Although it is not known whether KMP contributes to this effect, the observations described above indicate that DNA damaging agents that produce C4-AP could have downstream effects in cells that warrant investigation.
Supplementary Material
ACKNOWLEDGMENT
We are grateful for generous financial support from the National Institute of General Medical Science (GM-131736).
Footnotes
Supporting Information. Experimental details for all experiments. Scheme for HDAC1 activity assay, NMR spectra of compounds, LC-MS/MS data, peptide chromatograms. Supporting Information is available free of charge on the ACS Publications website.
Accession Codes. HDAC1 (UniProt entry: Q6IT96).
REFERENCES
- 1.Harmel R, and Fiedler D (2018) Features and regulation of non-enzymatic post-translational modifications, Nat. Chem. Biol 14, 244–252. [DOI] [PubMed] [Google Scholar]
- 2.Shahbazian MD, and Grunstein M (2007) Functions of Site-Specific Histone Acetylation and Deacetylation, Annu. Rev. Biochem 76, 75–100. [DOI] [PubMed] [Google Scholar]
- 3.Steunou A-L, Rossetto D, and Côté J (2014) Regulating Chromatin by Histone Acetylation, In Fundamentals of Chromatin (Workman JL, and Abmayr SM, Eds.), pp 147–212, Springer, New York, NY. [Google Scholar]
- 4.Li G, and Reinberg D (2011) Chromatin higher-order structures and gene regulation, Curr. Opin. Genet. Dev 21, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West AC, and Johnstone RW (2014) New and emerging HDAC inhibitors for cancer treatment, J. Clin. Invest 124, 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seto E, and Yoshida M (2014) Erasers of Histone Acetylation: The Histone Deacetylase Enzymes, Cold Spring Harbor Perspectives in Biol, 6:a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauve AA, Wolberger C, Schramm VL, and Boeke JD (2006) The Biochemistry of Sirtuins, Annu. Rev. Biochem 75, 435–465. [DOI] [PubMed] [Google Scholar]
- 8.Moreno-Yruela C, Zhang D, Wei W, Bæk M, Liu W, Gao J, Danková D, Nielsen AL, Bolding JE, Yang L, Jameson ST, Wong J, Olsen CA, and Zhao Y (2022) Class I histone deacetylases (HDAC1-3) are histone lysine delactylases, Sci. Adv 8, eabi6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madsen AS, Andersen C, Daoud M, Anderson KA, Laursen JS, Chakladar S, Huynh FK, Colaço AR, Backos DS, Fristrup P, Hirschey MD, and Olsen CA (2016) Investigating the Sensitivity of NAD+-dependent Sirtuin Deacylation Activities to NADH, J. Biol. Chem 291, 7128–7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin J, He B, Zhang X, Lin H, and Wang Y (2016) SIRT2 Reverses 4-Oxononanoyl Lysine Modification on Histones, J. Am. Chem. Soc 138, 12304–12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Y, Li X, Lin J, Hao Q, and Li XD (2017) Histone Ketoamide Adduction by 4-Oxo-2-nonenal Is a Reversible Posttranslational Modification Regulated by Sirt2, ACS Chem. Biol 12, 47–51. [DOI] [PubMed] [Google Scholar]
- 12.Huang H, Zhang D, Wang Y, Perez-Neut M, Han Z, Zheng YG, Hao Q, and Zhao Y (2018) Lysine benzoylation is a histone mark regulated by SIRT2, Nat. Commun 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edrissi B, Taghizadeh K, and Dedon PC (2013) Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-6-Formyllysine That Is Refractory to Histone Deacetylases, PLoS Genet. 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacinto MP, Fried SD, and Greenberg MM (2022) Intracellular Formation of a DNA Damage-Induced, Histone Post-Translational Modification Following Bleomycin Treatment, J. Am. Chem. Soc 144, 7600–7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett RAO, Swerdlow PS, and Povirk LF (1993) Spontaneous Cleavage of Bleomycin-Induced Abasic Sites in Chromatin and Their Mutagenicity in Mammalian Shuttle Vectors, Biochemistry 32, 3188–3195. [DOI] [PubMed] [Google Scholar]
- 16.Zhou C, Sczepanski JT, and Greenberg MM (2013) Histone Modification via Rapid Cleavage of C4’-Oxidized Abasic Sites in Nucleosome Core Particles, J. Am. Chem. Soc 135, 5274–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabow LE, Stubbe J, and Kozarich JW (1990) Identification and quantitation of the lesion accompanying base release in bleomycin-mediated DNA degradation, J. Am. Chem. Soc 112, 3196–3203. [Google Scholar]
- 18.Sugiyama H, Xu C, Murugesan N, and Hecht SM (1985) Structure of the alkali-labile product formed during iron(II)-bleomycin-mediated DNA strand scission, J. Am. Chem. Soc 107, 4104–4105. [Google Scholar]
- 19.Sugiyama H, Kawabata H, Fujiwara T, Dannoue Y, and Saito I (1990) Specific detection of C-4’ hydroxylated abasic sites generated by bleomycin and neocarzinostatin in DNA, J. Am. Chem. Soc 112, 5252–5257. [Google Scholar]
- 20.Dizdaroglu M (2014) Clemens von Sonntag and the early history of radiation-induced sugar damage in DNA, Int. J. Radiat. Biol 90, 446–458. [DOI] [PubMed] [Google Scholar]
- 21.Huang H, Sabari BR, Garcia BA, Allis CD, and Zhao Y (2014) SnapShot: Histone Modifications, Cell 159, 458–458.e451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan J, Pu M, Zhang Z, and Lou Z (2009) Histone H3-K56 acetylation is important for genomic stability in mammals, Cell Cycle 8, 1747–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mersfelder EL, and Parthun MR (2006) The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure, Nucleic Acids Res. 34, 2653–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dovey OM, Foster CT, and Cowley SM (2010) Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation, Proc. Natl. Acad. Sci. U S A 107, 8242–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, and Peterson CL (2006) Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions, Science 311, 844–847. [DOI] [PubMed] [Google Scholar]
- 26.Watson PJ, Millard CJ, Riley AM, Robertson NS, Wright LC, Godage HY, Cowley SM, Jamieson AG, Potter BVL, and Schwabe JWR (2016) Insights into the activation mechanism of class I HDAC complexes by inositol phosphates, Nature Comm. 7, 11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doyle K, and Fitzpatrick FA (2010) Redox Signaling, Alkylation (Carbonylation) of Conserved Cysteines Inactivates Class I Histone Deacetylases 1, 2, and 3 and Antagonizes Their Transcriptional Repressor Function*, J. Biol. Chem 285, 17417–17424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lauffer BEL, Mintzer R, Fong R, Mukund S, Tam C, Zilberleyb I, Flicke B, Ritscher A, Fedorowicz G, Vallero R, Ortwine DF, Gunzner J, Modrusan Z, Neumann L, Koth CM, Lupardus PJ, Kaminker JS, Heise CE, and Steiner P (2013) Histone Deacetylase (HDAC) Inhibitor Kinetic Rate Constants Correlate with Cellular Histone Acetylation but Not Transcription and Cell Viability, J. Biol. Chem 288, 26926–26943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galligan JJ, Wepy JA, Streeter MD, Kingsley PJ, Mitchener MM, Wauchope OR, Beavers WN, Rose KL, Wang T, Spiegel DA, and Marnett LJ (2018) Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks, Proc. Nat. Acad. Sci. USA 115, 9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Q, Omans ND, Leicher R, Osunsade A, Agustinus AS, Finkin-Groner E, D’Ambrosio H, Liu B, Chandarlapaty S, Liu S, and David Y (2019) Reversible histone glycation is associated with disease-related changes in chromatin architecture, Nature Comm. 10, 1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maksimovic I, and David Y (2021) Non-enzymatic Covalent Modifications as a New Chapter in the Histone Code, Trends Biochem. Sci 46, 718–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galligan JJ, and Marnett LJ (2017) Histone Adduction and Its Functional Impact on Epigenetics, Chem. Res. Toxicol 30, 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray DM, Jennings EQ, Maksimovic I, Chai X, Galligan JJ, David Y, and Zheng Q (2022) Chemical Labeling and Enrichment of Histone Glyoxal Adducts, ACS Chem. Biol 17, 756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhou X, Xie Y, Greenberg MM, Xi Z, and Zhou C (2017) Thiol Specific and Tracelessly Removable Bioconjugation via Michael Addition to 5-Methylene Pyrrolones, J. Am. Chem. Soc 139, 6146–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bos J, and Muir TW (2018) A Chemical Probe for Protein Crotonylation, J. Am. Chem. Soc 140, 4757–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji Y, Sun L, Chen Y, Qin H, and Xuan W (2022) Sirtuin-Derived Covalent Binder for the Selective Recognition of Protein Crotonylation, Angew. Chem. Int. Ed 61, e202205522. [DOI] [PubMed] [Google Scholar]
- 37.Huang SK, Scruggs AM, Donaghy J, Horowitz JC, Zaslona Z, Przybranowski S, White ES, and Peters-Golden M (2013) Histone modifications are responsible for decreased Fas expression and apoptosis resistance in fibrotic lung fibroblasts, Cell Death & Disease 4, e621–e621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


