Abstract
Background
Previous studies confirmed that most neoantigens predicted by algorithms do not work in clinical practice, and experimental validations remain indispensable for confirming immunogenic neoantigens. In this study, we identified the potential neoantigens with tetramer staining, and established the Co-HA system, a single-plasmid system coexpressing patient human leukocyte antigen (HLA) and antigen, to detect the immunogenicity of neoantigens and verify new dominant hepatocellular carcinoma (HCC) neoantigens.
Methods
First, we enrolled 14 patients with HCC for next-generation sequencing for variation calling and predicting potential neoantigens. Then, the Co-HA system was established. To test the feasibility of the system, we constructed target cells coexpressing HLA-A*11:01 and the reported KRAS G12D neoantigen as well as specific T-cell receptor (TCR)-T cells. The specific cytotoxicity generated by this neoantigen was shown using the Co-HA system. Moreover, potential HCC-dominant neoantigens were screened out by tetramer staining and validated by the Co-HA system using methods including flow cytometry, enzyme-linked immunospot assay and ELISA. Finally, antitumor test in mouse mode and TCR sequencing were performed to further evaluate the dominant neoantigen.
Results
First, 2875 somatic mutations in 14 patients with HCC were identified. The main base substitutions were C>T/G>A transitions, and the main mutational signatures were 4, 1 and 16. The high-frequency mutated genes included HMCN1, TTN and TP53. Then, 541 potential neoantigens were predicted. Importantly, 19 of the 23 potential neoantigens in tumor tissues also existed in portal vein tumor thrombi. Moreover, 37 predicted neoantigens restricted by HLA-A*11:01, HLA-A*24:02 or HLA-A*02:01 were performed by tetramer staining to screen out potential HCC-dominant neoantigens. HLA-A*24:02-restricted epitope 5'-FYAFSCYYDL-3' and HLA-A*02:01-restricted epitope 5'-WVWCMSPTI-3' demonstrated strong immunogenicity in HCC, as verified by the Co-HA system. Finally, the antitumor efficacy of 5'-FYAFSCYYDL-3'-specific T cells was verified in the B-NDG-B2mtm1Fcrntm1(mB2m) mouse and their specific TCRs were successfully identified.
Conclusion
We found the dominant neoantigens with high immunogenicity in HCC, which were verified with the Co-HA system.
Keywords: Liver Neoplasms; Antigens, Neoplasm; Immunotherapy; Immunoassay; Immunogenicity, Vaccine
WHAT IS ALREADY KNOWN ON THIS TOPIC
Though clinical trials testing neoantigen immunotherapy have yielded encouraging results in patients, it is hard to know which neoantigens are dominant in determining the responsiveness to hepatocellular carcinoma (HCC) immunotherapy treatment.
WHAT THIS STUDY ADDS
In this study, we constructed the Co-HA system, a single-plasmid system coexpressing patient human leukocyte antigen and antigen, to verify HCC neoantigens with substantial immunogenicity.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
The Co-HA system is a convenient molecular tool for verifying T-cell epitopes, and it could provide efficient immunotherapeutic targets for HCC.
Introduction
Liver cancer is the sixth most common malignancy and has the third highest mortality worldwide.1 Hepatocellular carcinoma (HCC) is the most common type of liver cancer and accounts for ~90% of cases.2 Surgery has long been the main curative therapy for HCC; however, recurrence remains a major obstacle, with recurrence rates as high as ~70% at 5 years.2 In recent years, patients with HCC have exhibited improved clinical responses to immunotherapy.3 In particular, peptide vaccines or adoptive cell therapies targeting tumor-associated antigens (TAAs), such as alpha-fetoprotein4 and glypican-3,5 have shown appropriate antitumor activity in HCC. Meanwhile, due to their limited specificity, such treatments may result in side effects in the non-cancerous portion of the liver. Therefore, new therapeutic strategies are needed to overcome these obstacles.
Compared with TAAs, tumor neoantigens,6 7 new peptides generated by somatic mutations from tumor tissues but not normal tissues, have better tumor specificity and stronger immunogenicity. Hence, tumors with foreign neoantigens can be well recognized and attacked by the immune system. In addition, clinical trials testing neoantigen immunotherapy have yielded encouraging results in patients with melanoma8 or lung cancer9 worldwide.
The tumor mutation burden (TMB) of HCC ranks 12th among 30 different types of tumors, and the median TMB is ~2.0 mutations/megabase,10 suggesting that there are neoantigens in HCC. Li et al found that a higher somatic mutation load and somatic mutation-derived neoantigen load in HCC are associated with a better clinical outcome.11 Yang et al found that patients with HCC with TP53 neoantigens have longer survival times and a stronger immune response than others.12 According to ClinicalTrials.gov (https://clinicaltrials.gov/ct2/home), several clinical trials testing HCC neoantigen vaccines have been ongoing since 2017. It has been reported that patients with HCC with neoantigen-induced T-cell responses have better clinical benefits than those without responsive neoantigens.13 14 Despite a good curative improvement in HCC treatment, research progress regarding HCC neoantigen vaccines has been significantly slower than that in melanoma and lung cancer vaccines. Ultimately, more work is needed to screen and verify HCC neoantigens efficiently.
Currently, prediction with computer algorithms is the main strategy for clinical neoantigen screening and is used to assess the affinity between neoantigens and human leukocyte antigens (HLAs).15 However, previous studies have confirmed that most neoantigens predicted by algorithms did not work in clinical practice.13 14 16 In a recent report, although researchers chose 6~20 personalized predicted neoantigen peptides with a mutated peptide affinity (MutAff) ≤500 nM for each patient with HCC, half of the patients did not achieve any immune response from neoantigen treatment, and their prognosis was poor.13 Ineffective neoantigen vaccinations are costly and squander the limited survival time of these patients with advanced HCC. Another clinical trial that used neoantigen-based dendritic cell vaccination and adoptive T-cell transfer immunotherapy demonstrated similar negative results.14 To overcome these issues, experimental validations in vitro are needed to identify the true neoantigens among the predicted options.
At present, three methods can be used to validate HCC neoantigens7: (1) Liquid chromatography–mass spectrometry (LC/MS); (2) tetramer staining; and (3) cytotoxicity testing. LC/MS can detect true neoantigens presented by specific HLAs. However, only 5 true neoantigens were detected among 5498 predicted neoantigens in HCC due to the inferior sensitivity of LC/MS.17 Tetramer staining can detect the frequency of the predicted neoantigen-specific T cells in patient tumors or peripheral blood.17 18 Cytotoxicity testing can indicate cytotoxic T lymphocyte (CTL)-mediated killing of target cells. T2 cells, as classical target cells, have been extensively applied to detect the immunogenicity of HLA-A*02-restricted peptides. However, HLAs are highly heterogeneous, and T2 cells cannot validate diverse HLA-restricted peptides.19
Considering the drawbacks of these methods, a more feasible approach must be established. Therefore, we innovatively established the Co-HA system, a single-plasmid system coexpressing patient HLA and antigen, to verify new dominant HCC neoantigens. This approach could provide efficient immunotherapeutic targets for HCC and is a convenient molecular tool for verifying T-cell epitopes.
Materials and methods
Patients and samples
We enrolled 14 patients with HCC who underwent curative resection (from January 2018 to August 2020), including 6 patients from the Peking University People’s Hospital and 8 patients from the Affiliated Hospital of Guilin Medical University. All cases were confirmed by pathology reports. This study involves animal subjects and was approved by the Research Ethics Committee of the Peking University People’s Hospital (ID: 2019PHE018). COS-7 cells and HEK293T cells were purchased from American Type Culture Collection. Huh-7 cells were purchased from Cell Resource Center, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences.
Construction of the KRAS G12D TCR-T cells
The starting vector was lenti-BSD-T2A-EGFP. The vector map is shown in online supplemental figure 1A (supplemental plasmid sequence NO. 1). The gene segment of EGFP in the vector was released from the vector by cleavage with restriction enzymes BfuAI and EcoRI, and the vector without EGFP was recovered from the gel. Meanwhile, we synthesized the gene segment of the KRAS G12D TCR, including TK412 Vβ, Cys mTRBC1, P2A, TK412 Vα and LVL_Cys mTRAC, as described by Wang et al.20 Gibson Assembly was then performed to assemble the T-cell receptor (TCR) gene segment into the viral vector without EGFP to construct the TCR plasmid lenti-BSD-T2A-TK412 (online supplemental figure 1A, online supplemental plasmid sequence NO. 2). The 3589 bp gene segment excised from the TCR plasmid by restriction enzymes NotI and BamHI was successfully verified (online supplemental figure 1B). The TCR-T lentivirus was produced according our lentiviral production and infection methods.21 Peripheral blood mononuclear cells (PBMCs) from healthy donors were infected with lentivirus. After 14 days of cultivation, TCR-T cells recognizing the KRAS G12D neoantigen were acquired. DNA was extracted from the TCR-T cells. Sanger sequencing was used to determine the key sequence of the TCR (online supplemental figure 1C).
jitc-2022-006334supp001.pdf (62.3MB, pdf)
Establishment of the Co-HA system
The Co-HA system is a single-plasmid system with coexpressed HLA and antigen (online supplemental figure 2). Here, we chose KRAS G12D neoantigen 5'-VVGADGVGK-3', a reported dominant neoantigen, as a positive control to show its establishment.
Construction of the HLA vectors: The KRAS G12D neoantigen is an HLA-A*11:01-restricted neoantigen.20 The HLA-A*11:01 vector was constructed with the lenti-PURO-T2A-EGFP vector (online supplemental figure 3A, online supplemental plasmid sequence NO. 3). The steps of constructing the HLA-A*11:01 vector were described as follows. (1) To prevent disturbing subsequent restriction enzyme digestion, the recognition site of restriction enzyme BsmBI on the lenti-PURO-T2A-EGFP vector was synonymously mutated. (2) We synthesized the gene segment including HLA-A*11:01, E2A, Tag-A*11:01, P2A and EGFP. The sequence of HLA-A*11:01 was acquired from the IPD-IMGT/HLA database (https://www.ebi.ac.uk/ipd/imgt/hla/), and the recognition site of restriction enzyme BsmBI in HLA-A*11:01 was synonymously mutated. (3) The gene segment of EGFP was excised by restriction enzymes BamHI and EcoRI, and the main part of the vector was recovered from the gel. Then, the synthesized gene segment was assembled into the main part of the vector by Gibson Assembly, and the HLA-A*11:01 vector was constructed and named lenti-PURO-T2A-(HLA-A*11:01)-E2A-(Tag-A*11:01)-P2A-EGFP (online supplemental figure 3A, online supplemental plasmid sequence NO. 4). (4) The 2611 bp gene segment excised from the HLA-A*11:01 vector was successfully verified (online supplemental figure 3B). (5) The lentivirus of the HLA-A*11:01 vector was produced. Then, COS-7 cells were infected with the lentivirus and cultured with 4 µg/mL puromycin for 1 week to establish stable cell lines. DNA was extracted from the COS-7 cells. The key sequence of the HLA-A*11:01 vector was analyzed with Sanger sequencing (online supplemental figure 3C). In addition, the expression of HLA-A*11:01 was detected by flow cytometry (FCM) (online supplemental figure 3D). We designed the recognition sites of the restriction enzymes BamHI and AfeI in the HLA-A*11:01 vector to replace the gene segment of the HLA with its tag to construct various HLA vectors. Thirty-nine HLA vectors (online supplemental table 1), including HLA-A*24:02 and HLA-A*02:01 (online supplemental figure 3D), were constructed for future use.
Construction of the target cells (online supplemental figure 4A): We first synthesized the gene segments of the KRAS G12D neoantigen (online supplemental table 2 SEQ ID NO. 1) and its paired wild type (WT) antigen (online supplemental table 2 SEQ ID NO. 2). Then, we excised the gene segment of EGFP by restriction enzyme BsmBI and recovered the HLA-A*11:01 vector without EGFP from the gel (online supplemental figure 4B). Gibson Assembly was performed to assemble the gene segments of the KRAS G12D neoantigen and its paired WT antigen into the HLA-A*11:01 vector without EGFP. The target plasmids lenti-(HLA-A*11:01)-KRAS-M and lenti-(HLA-A*11:01)-KRAS-W were constructed (online supplemental figure 4A, online supplemental plasmid sequence NO. 5–6). The 3290 bp gene segment excised from the target plasmids was successfully verified (online supplemental figure 4C). Subsequently, we established stable cell lines by COS-7 cells. DNA was extracted from the target cells. Sanger sequencing was further performed to analyze the key sequences (online supplemental figure 4D).
Other detailed methods, such as next-generation sequencing, variation calling, neoantigen prediction, and tetramer staining, were described in the online supplemental materials and methods.
Results
Characteristics of patients and samples
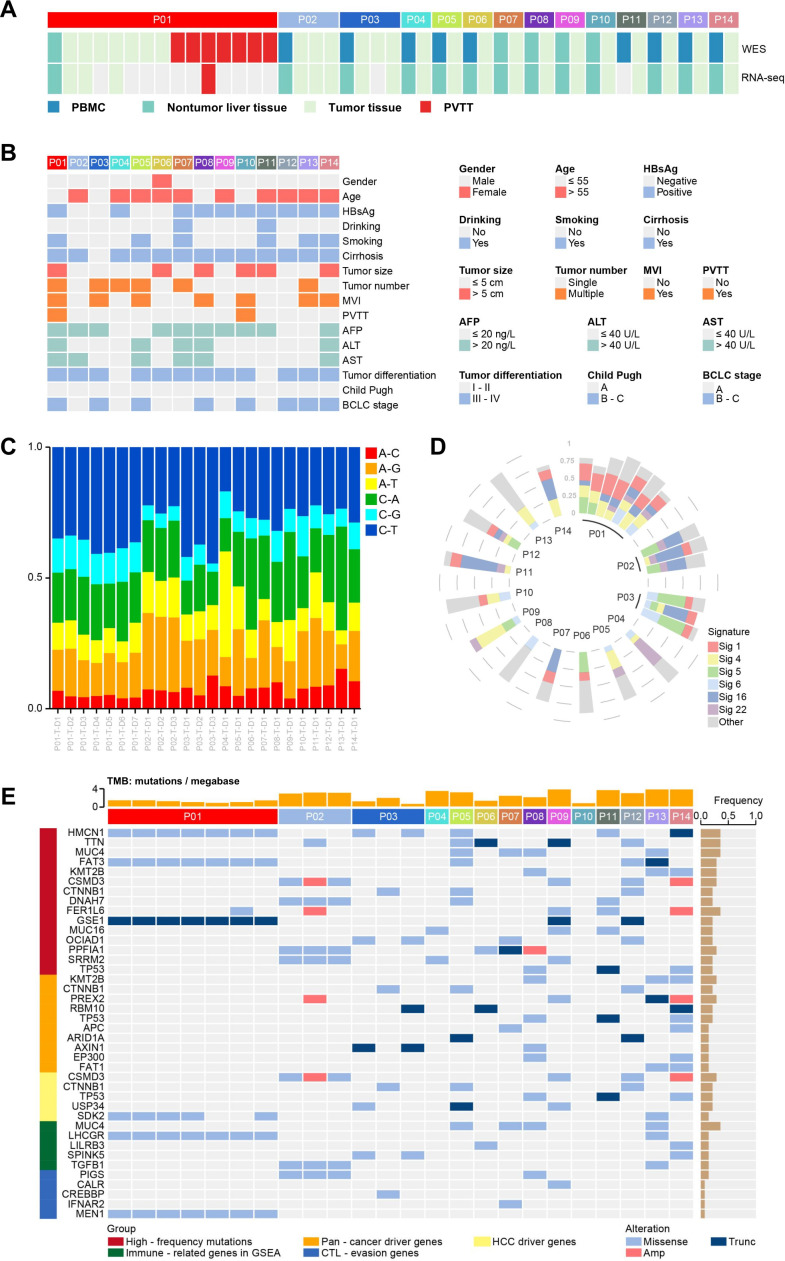
We obtained 45 samples from 14 patients with HCC, including tumor tissues, non-tumor liver tissues, portal vein tumor thrombi (PVTTs) and PBMCs (figure 1A). The basic clinicopathological characteristics of the 14 patients were listed in figure 1B. There was an obvious sex bias; 93% (13/14) of the patients were men, and 7% (1/14) were women. Most of the patients were older than 55 years (10/14) and had hepatitis B virus (HBV) infection (8/14). Fourteen per cent (2/14) of patients had PVTTs.
Figure 1.
Sequencing, clinical information and the mutation spectrum of the tumor tissues of 14 patients with HCC. (A) Illustration of WES and RNA sequencing performed on each sample from 14 patients with HCC. (B) Clinical characteristics of 14 patients with HCC. (C) The proportions of base substitution subtypes in samples. (D) The proportions of mutational signatures in samples. (E) The key mutated genes and the TMB in samples. HCC, hepatocellular carcinoma; PBMC, peripheral blood mononuclear cell; PVTT, portal vein tumor thrombus; TMB, tumor mutation burden; WES, whole-exome sequencing.
The mutation spectrum of HCC tumor tissues
First, whole-exome sequencing (WES) was performed on the 45 samples from 14 patients with HCC (figure 1A). One hundred per cent of the clean reads were successfully mapped to the reference genome (UCSChg19 release). Target regions of samples were covered with an average of 95.58% (ranging from 92.20% to 97.26%). Then, we used the Genome Analysis Toolkit Mutect222 to identify mutations, and the mutations with TLOD ≥10 were retained. TLOD means that tumor does not meet likelihood threshold. Furthermore, we used Variant Effect Predictor23 to predict mutation classifications and screened out nine mutation classifications as the set of somatic mutations, including Missense Mutation, In Frame Ins, In Frame Del, Non-sense Mutation, Non-stop Mutation, Splice Site, Frame Shift Del, Frame Shift Ins and Splice Region, totaling 2875 mutations.
We analyzed base substitution mutations and found that the main substitutions were C>T/G>A transitions in these 14 patients (figure 1C), consistent with The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) data set analyzed by the R project (online supplemental figure 5A). The results indicated that the occurrence and development of HCC may be related to C-base or G-base damage caused by some etiologies. The main mutational signatures were 4, 1 and 16 (figure 1D), consistent with those in the Catalogue Of Somatic Mutations In Cancer (COSMIC) database (https://cancer.sanger.ac.uk/cosmic) (online supplemental figure 5B). Signature 4 is associated with smoking. Signature 1 is the result of an endogenous mutational process initiated by spontaneous deamination of 5-methylcytosine. Signature 16 is a specific signature of liver cancer and exhibits an extremely strong transcriptional strand bias for T>C mutations.
The genes mutated at high frequencies among these 14 patients with HCC included HMCN1 (5/14), TTN (5/14), MUC4 (4/14), and TP53 (3/14). Pan-cancer driver genes,24 such as KMT2B and CTNNB1, HCC driver genes,25–29 such as SCMD3 and TP53, immune-related genes (according to Gene Set Enrichment Analysis, GSEA, https://www.gsea-msigdb.org/gsea/index.jsp, such as LHCGR and LILRB3) and CTL-evasion genes30 such as PIGS and CALR all had mutations (figure 1E). The results were similar to those in the TCGA-LIHC data set (online supplemental figure 5C). The mutation frequencies of HMCN1, TTN, MUC4 and TP53 were 8%, 28%, 10% and 35%, respectively, in the TCGA-LIHC data set (online supplemental figure 5C). In summary, these results indicated that our variation calling approach was reliable and could be used for HCC neoantigen prediction.
Potential neoantigen profiles
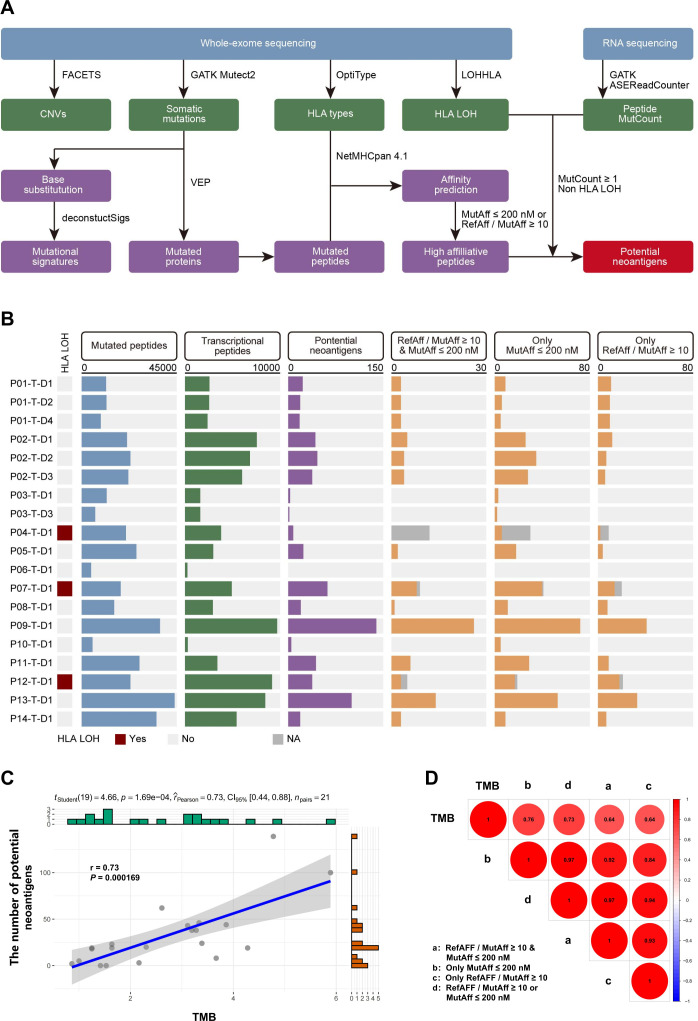
First, mutated DNA sequences from the 14 patients with HCC were translated and disassembled into potential mutated peptides. In total, 375,137 mutated peptides were identified in the tumor tissues, among which 82,501 could be detected by RNA sequencing. Then, the NetMHCpan-4.1 algorithm31 was used to calculate the affinity of the mutated peptides to the patients’ HLAs. The data were filtered based on MutAff ≤200 nM or referenced peptide affinity/MutAff ≥10 (online supplemental figures 6A-C, 7A-C). Finally, we excluded duplicate neoantigens and HLA loss of heterozygosity,32 and found 541 potential neoantigens (figure 2A, B).
Figure 2.
Potential neoantigen profiles of 14 patients with HCC. (A) Schematic of the approach for screening potential neoantigens using computer algorithms. (B) Potential HCC neoantigen profiles determined by different indexes and HLA LOH. (C) Correlation analysis between the number of potential neoantigens and the TMB. (D) Correlation analysis between the number of potential neoantigens with different indexes and the TMB. HCC, hepatocellular carcinoma; HLA, human leukocyte antigen; LOH, loss of heterozygosity; TMB, tumor mutation burden.
We found that a higher number of potential neoantigens was related to a higher TMB (figure 2C, D). The levels of potential neoantigens determined by different indexes showed strong correlations (figure 2D).
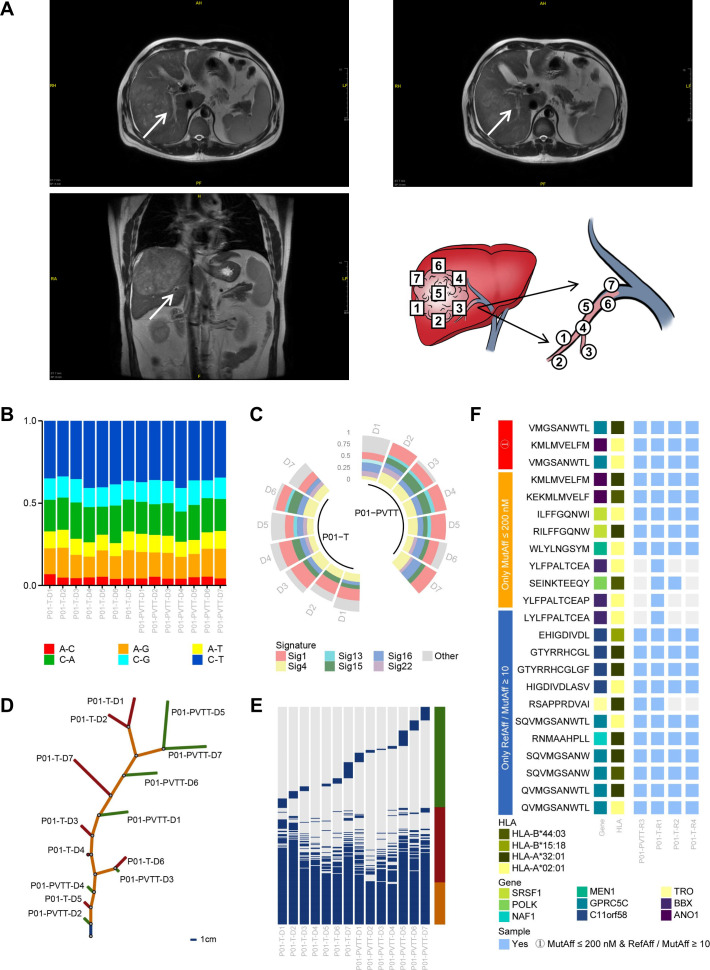
The goodness of fit of mutations and neoantigens between PVTTs and primary tumor tissues
Seven primary tumor tissues and seven PVTTs were collected from patient 01. The locations of these samples were shown in figure 3A. We further analyzed the WES data of these samples and found that mutations and neoantigens between tumor tissues and PVTTs had a substantial degree of consistency. The main substitutions were C>T/G>A transitions, and the main mutational signatures were 4, 1 and 16 (figure 3B, C) in both tumor tissues and PVTTs. The evolutionary tree showed that the tumor tissues and PVTTs shared the most mutations and had unique mutations (figure 3D, E). Furthermore, among the 23 potential neoantigens predicted from tumor tissue data, 19 were predicted from PVTT data (figure 3F). Therefore, we hypothesized that neoantigens in tumor tissues could be used for the prevention and treatment of advanced HCC with PVTTs and that neoantigens in PVTTs could play an important role in the prevention of tumor recurrence and metastasis.
Figure 3.
The goodness of fit of mutations and neoantigens between PVTTs and primary tumor tissues. (A) Illustration of the locations of multiple samples collected from the tumor tissue and PVTT. (B) Comparison of base substitution subtypes between PVTTs and primary tumor tissues. (C) Comparison of mutational signatures between PVTTs and primary tumor tissues. (D) Tumor evolutionary trees of patient 01. The blue trunk indicates mutations shared by all samples, the orange trunk indicates shared mutations that are not in all the samples, and the red and green branches indicate unique mutations in the samples. The 1 cm trunk indicates 40 mutations, and the 1 cm branch indicates 4 mutations. (E) Distribution of mutations in PVTTs and primary tumor. The blue pillars indicate mutations; the orange pillar indicates mutations shared by all samples; the red pillar indicates shared mutations that are not in all the samples, and the green pillar indicates unique mutations. (F) The potential neoantigens determined by different indexes in PVTTs and primary tumor tissues. HLA, human leukocyte antigen; PVTTs, portal vein tumor thrombi.
Validation of the Co-HA system
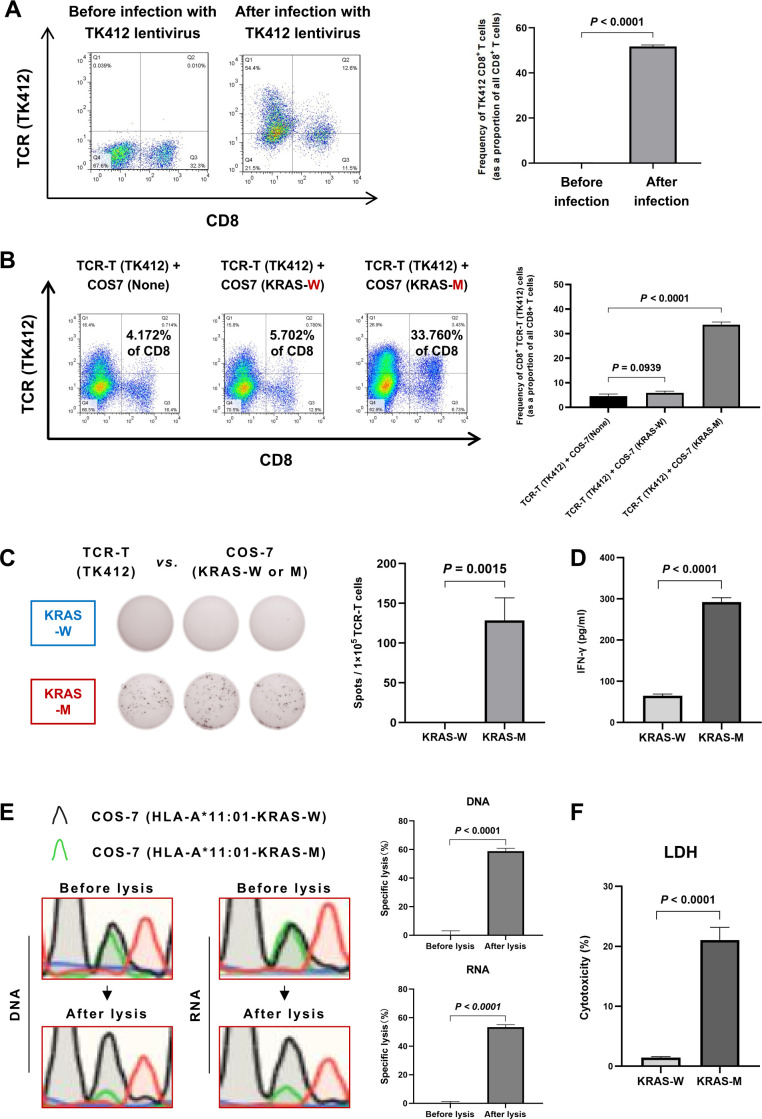
The establishment of the Co-HA system was shown in the materials and methods and the online supplemental materials and methods. In the study, the reported KRAS G12D neoantigen, a positive control, was used to test the feasibility of the Co-HA system. First, the expression of the KRAS TCR in T cells from healthy donors was successfully detected by FCM (figure 4A). Subsequently, the CD8+ TCR-T cells exhibited enhanced proliferation (33.61%±0.93%) when co-cultured with the COS-7 cells expressing the KRAS G12D neoantigen in comparison with those cultured with the COS-7 cells expressing the paired WT antigen (5.91%±0.53%) or WT COS-7 cells (4.58%±0.68%) in the Co-HA system (figure 4B). In addition, we performed a series of cytotoxicity tests to detect the immunogenicity of the KRAS G12D neoantigen in the Co-HA system. The average number of interferon (IFN)-γ spots (128.33±23.33 spots/well) produced by the coincubation of TCR-T cells and target cells expressing the KRAS G12D neoantigen was significantly greater than that of the WT antigen control detected by an enzyme-linked immunospot assay (ELISpot) in the Co-HA system (figure 4C). Meanwhile, ELISA results showed that the IFN-γ levels (292.37±8.31 pg/mL) produced by coincubation of the specific T cells and the target cells expressing the KRAS G12D neoantigen were significantly higher than those observed in the WT antigen control (64.82±3.37 pg/mL) in the Co-HA system (figure 4D).
Figure 4.
Validation of the Co-HA system. (A) Construction of the KRAS G12D TCR-T cells from healthy donors detected by FCM. (B) The specific proliferation levels of the KRAS G12D TCR-T cells detected by FCM after co-culture with the COS-7 cells expressing the relevant neoantigen in the Co-HA system. (C) The specific secretion of IFN-γ by TCR-T cells against the KRAS G12D neoantigen in the Co-HA system detected by ELISpot. (D) The specific secretion of IFN-γ by TCR-T cells against the KRAS G12D neoantigen in the Co-HA system detected by ELISA. (E) The specific lysis of the COS-7 cells generated by the KRAS G12D neoantigen in the Co-HA system detected by Sanger sequencing. (F) The LDH levels induced by TCR-T cells against the KRAS G12D neoantigen in the Co-HA system detected by ELISA. FCM, flow cytometry; ELISpot, enzyme-linked immunospot assay; IFN-γ, interferon-γ; LDH, lactate dehydrogenase; TCR, T-cell receptor.
In particular, we tried a novel functional assay to assess specific cell lysis induced by the cytotoxicity of T cells in the Co-HA system. Because of the different base compositions of the paired mutated and WT peptides, the relative number of the COS-7 cells expressing the KRAS G12D neoantigen and its paired WT peptide were recognizable by Sanger sequencing in the Co-HA system. The number of the COS-7 cells expressing the KRAS G12D neoantigen appeared relatively low when they were killed by T cells. The relative number of the COS-7 cells expressing its paired WT peptide remained nearly unchanged because they could not induce a strong immune response. By comparing the relative number of target cells expressing paired mutated or WT peptides before and after lysis, we could determine the percentage of specific cell lysis induced by the cytotoxicity of T cells. The specific lysis rates of the COS-7 cells generated by the KRAS G12D neoantigen in the Co-HA system were 58.84%±1.70% according to the DNA analysis and 53.35%±1.41% according to the RNA analysis (figure 4E). In addition, the level of lactate dehydrogenase (LDH) induced by cytotoxicity was significantly higher in the KRAS G12D neoantigen samples (21.05%±1.72%) than that in the WT antigen control (1.44%±0.13%) after cytotoxicity in the Co-HA system (figure 4F).
The above results showed that the strong immunogenicity of the KRAS G12D neoantigen could be verified in the Co-HA system, confirming the feasibility of the system.
Screening of potential HCC-dominant neoantigens by specific tetramers
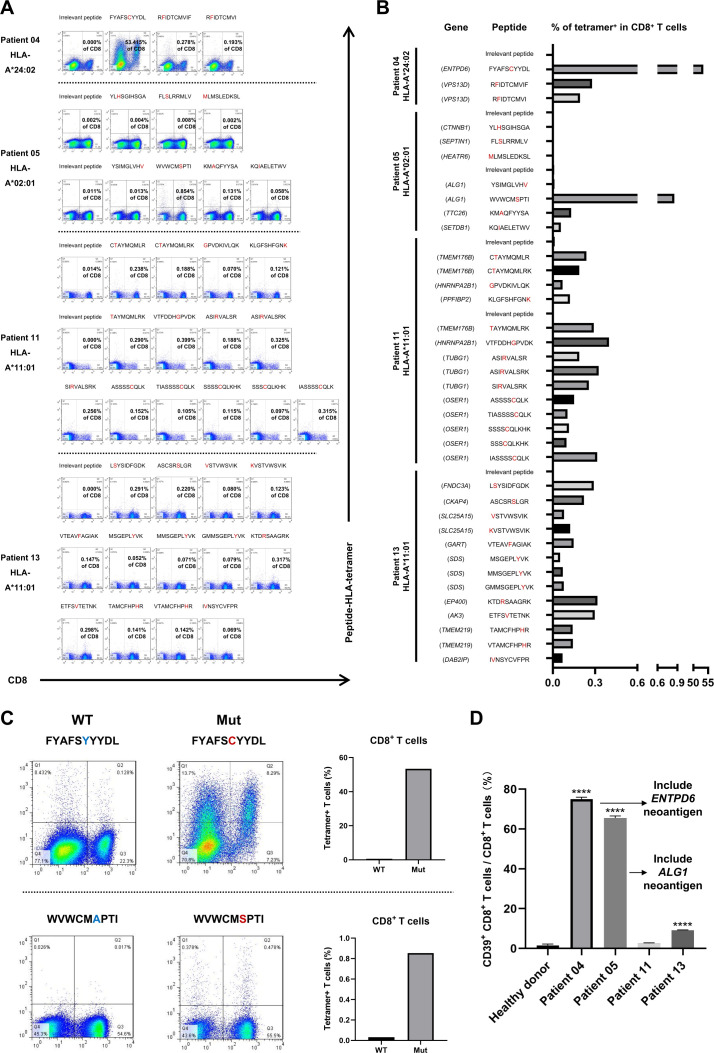
We used tetramer staining to preliminarily screen potential HCC-dominant neoantigens. Since HLA-A*11:01, HLA-A*24:02 and HLA-A*02:01 are prevalent in the population,33 we produced 37 predicted neoantigen peptides restricted by HLA-A*11:01, HLA-A*24:02 or HLA-A*02:01 and coincubated them with PBMCs from patients to promote specific T-cell proliferation. The paired linear sequences of mutated and WT peptides and their affinities are shown in online supplemental table 3. Then, we detected the frequency of the specific T cells by analyzing specific tetramers (figure 5A, B). The frequencies of the mutated peptide 5'-FYAFSCYYDL-3'-specific T cells and the mutated peptide 5'-WVWCMSPTI-3'-specific T cells were relatively high. In particular, the frequency of 5'-FYAFSCYYDL-3'-specific T cells (53.415%) was much greater than that of other reported dominant neoantigens. In contrast, specific T-cell reactivity against the paired WT peptides was weaker than that against the mutated peptides, particularly in CD8+ T cells (figure 5C). Thus, 5'-FYAFSCYYDL-3' may have excellent immunogenicity.
Figure 5.
Screening of new HCC-dominant neoantigens by specific tetramers. (A) Tetramer staining of PBMCs from patients after stimulation with predicted neoantigen peptides. The irrelevant peptides are isotype controls. The amino acids in the red font are mutation sites. (B) The frequency of the specific T cells after stimulation with predicted neoantigen peptides. (C) The frequency of the specific T cells in PBMCs of patient 04 after stimulation with potential dominant neoantigens and paired WT peptides. (D) The frequency of CD39+ CD8+ T cells after stimulation with predicted neoantigen peptides. HCC, hepatocellular carcinoma; PBMCs, peripheral blood mononuclear cells; WT, wild type.
Several lines of evidence have suggested that CD39+ CD8+ T cells may have better abilities to recognize neoantigens, and high-affinity neoantigens may trigger potent anti-HCC activity by activating CD39+ CD8+ T cells.34 35 In this study, there were potential dominant neoantigens with substantial immunogenicity in patient 04 and patient 05. Interestingly, the expression of CD39 on CD8+ T cells of patient 04 and patient 05 were dramatically higher than other patients (figure 5D, online supplemental figure 8A). The CD39+ CD8+ T cells induced by 5'-FYAFSCYYDL-3' neoantigen peptide were more than those induced by its paired WT peptide (online supplemental figure 8B). Importantly, almost all the tetramer+ 5'-FYAFSCYYDL-3' neoantigen-specific T cells sorted by FCM were CD39+ CD8+ T cells (online supplemental figure 8B).
In short, we found that 5'-FYAFSCYYDL-3' restricted by HLA-A*24:02 and 5'-WVWCMSPTI-3' restricted by HLA-A*02:01 were potential HCC-dominant neoantigens.
New HCC-dominant neoantigens verified by the Co-HA system demonstrate strong immunogenicity
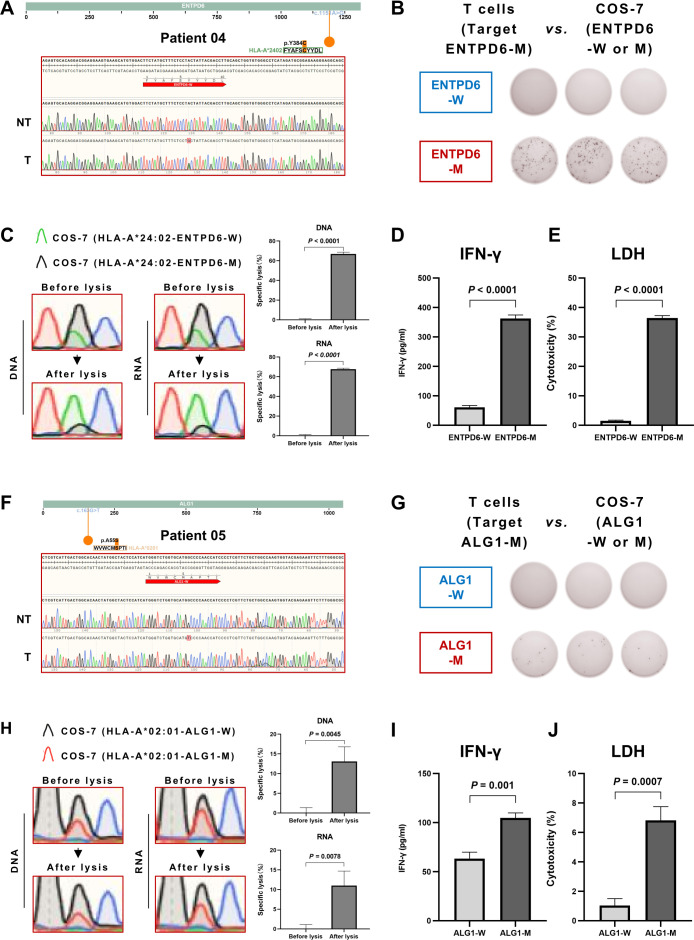
In figure 6A, we verified the transcriptional neoantigen 5'-FYAFSCYYDL-3' existed in the tumor tissue of patient 04 but not in the non-tumor liver tissue. Then, we conducted a series of tests to detect the immunogenicity of potential HCC-dominant neoantigens in the Co-HA system, as previously described for the positive control KRAS G12D neoantigen. We found the average number of IFN-γ spots (142±10.23 spots/well) produced by the coincubation of specific T cells and target cells expressing the neoantigen 5'-FYAFSCYYDL-3' was significantly greater than that of the WT antigen control in the Co-HA system, as demonstrated by ELISpot analysis (figure 6B). In particular, the specific lysis of the COS-7 cells generated by the neoantigen 5'-FYAFSCYYDL-3' in the Co-HA system was 66.93%±1.40% according to the DNA analysis and 67.71%±0.83% according to the RNA analysis (figure 6C). Meanwhile, ELISA results showed that the IFN-γ levels (362.47±9.80 pg/mL) produced by coincubation of the specific T cells and the target cells expressing the neoantigen 5'-FYAFSCYYDL-3' was significantly greater than that of the WT antigen control (60.99±4.92 pg/mL) in the Co-HA system (figure 6D). Finally, the level of LDH induced by cytotoxicity in the Co-HA system was significantly greater in the neoantigen 5'-FYAFSCYYDL-3' (36.45%±0.65%) than that produced with the WT antigen control (1.51%±0.18%) (figure 6E).
Figure 6.
Verification of new hepatocellular carcinoma-dominant neoantigens by the Co-HA system. (A) The transcriptional neoantigen 5'-FYAFSCYYDL-3' verified in the tumor tissues of patient 04 with Sanger sequencing. (B) The specific secretion of IFN-γ by T cells from PBMCs of patient 04 against the neoantigen 5'-FYAFSCYYDL-3' in the Co-HA system detected by ELISpot. (C) The specific lysis of the COS-7 cells generated by the neoantigen 5'-FYAFSCYYDL-3' in the Co-HA system detected by Sanger sequencing. (D) The specific secretion of IFN-γ by T cells from PBMCs of patient 04 against the neoantigen 5'-FYAFSCYYDL-3' in the Co-HA system detected by ELISA. (E) The LDH levels induced by the specific T cells against the neoantigen 5'-FYAFSCYYDL-3' in the Co-HA system detected by ELISA. (F) The transcriptional neoantigen 5'-WVWCMSPTI-3' verified in the tumor tissues of patient 05 with Sanger sequencing. (G) The specific secretion of IFN-γ by T cells from PBMCs of patient 05 against the neoantigen 5'-WVWCMSPTI-3' in the Co-HA system detected by ELISpot. (H) The specific lysis of the COS-7 cells generated by the neoantigen 5'-WVWCMSPTI-3' in the Co-HA system detected by Sanger sequencing. (I) The specific secretion of IFN-γ by T cells from PBMCs of patient 05 against the neoantigen 5'-WVWCMSPTI-3' in the Co-HA system detected by ELISA. (J) The LDH levels induced by the specific T cells against the neoantigen 5’-WVWCMSPTI-3’ in the Co-HA system detected by ELISA. HLA, human leukocyte antigen; ELISpot, enzyme-linked immunospot assay; IFN-γ, interferon-γ; LDH, lactate dehydrogenase; PBMCs, peripheral blood mononuclear cells.
Similarly, the transcriptional neoantigen 5'-WVWCMSPTI-3' was verified in the tumor tissue of patient 05 but not in the non-tumor liver tissue (figure 6F). The average number of IFN-γ spots (8.33±3.09 spots/well) produced by the coincubation of specific T cells and target cells expressing the neoantigen 5'-WVWCMSPTI-3' was significantly greater than that observed with the WT antigen control in the Co-HA system, as demonstrated by the ELISpot analysis (figure 6G). In particular, the specific lysis of the COS-7 cells generated by the neoantigen 5'-WVWCMSPTI-3' in the Co-HA system was 13.09%±3.03% according to the DNA analysis and 11.00%±3.01% according to the RNA analysis (figure 6H). Meanwhile, the ELISA results showed that the IFN-γ levels (104.74±4.21 pg/mL) produced by the coincubation of the specific T cells and the target cells expressing the neoantigen 5'-WVWCMSPTI-3' in the Co-HA system were significantly greater than those produced by the WT antigen control (63.32±5.34 pg/mL) (figure 6I). Finally, the level of LDH induced by cytotoxicity was significantly higher in neoantigen 5'-WVWCMSPTI-3' (6.81%±0.77%) than that produced by the WT antigen control (1.03%±0.39%) (figure 6J). In summary, the results indicated that 5'-FYAFSCYYDL-3' and 5'-WVWCMSPTI-3' were new HCC-dominant neoantigens with substantial immunogenicity.
Antitumor efficacy of ectonucleoside triphosphate diphosphohydrolase 6 neoantigen-specific T cells in vivo
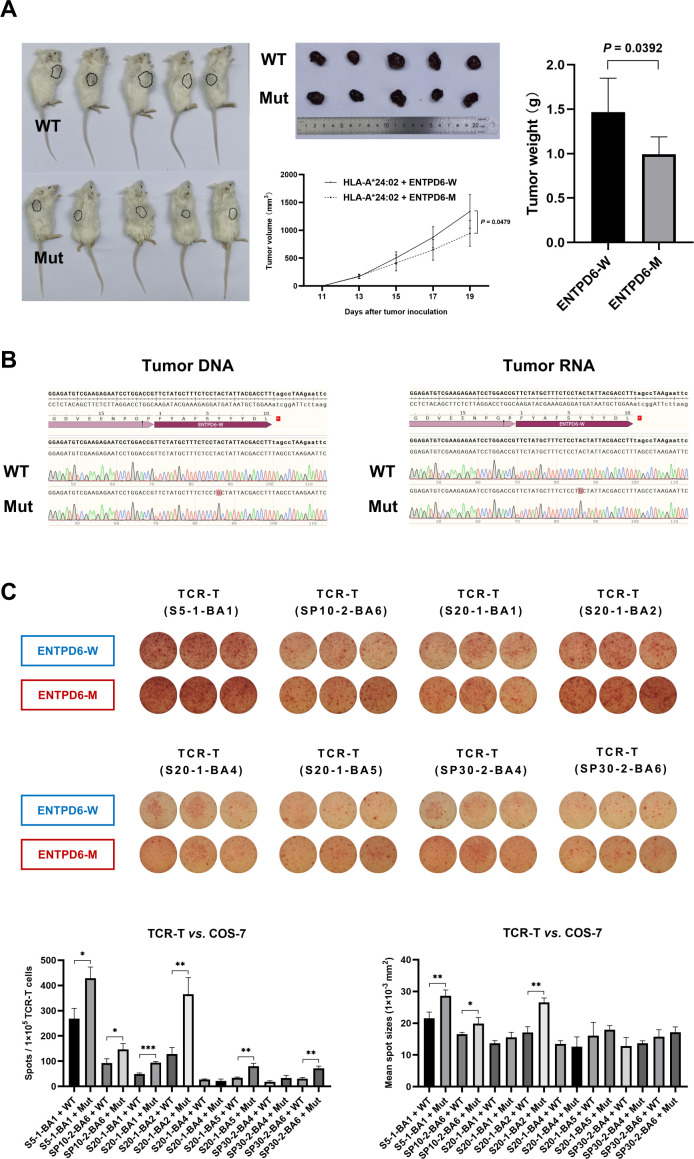
The B-NDG-B2mtm1Fcrntm1(mB2m)/Bcgen mouse model was used for evaluation of antitumor efficacy of ectonucleoside triphosphate diphosphohydrolase 6 (ENTPD6) neoantigen-specific T cells. The Huh-7 cells coexpressing HLA-A*24:02 and ENTPD6 neoantigen were selected as the target cells (online supplemental figure 9A), and then implanted into mice. When tumor size was approximately 150±50 mm3, they were treated with the neoantigen-specific T cells every 2 days. After three times of treatment, the volume and weight of tumors with neoantigens were both smaller than those of WT type (figure 7A). In addition, the key sequences of the ENTPD6 in tumors were successfully detected by Sanger sequencing (figure 7B). These results suggested that the neoantigen-specific T cells could kill tumor cells with the specific neoantigen and the dominant neoantigens could be ideal therapeutic targets for HCC immunotherapy.
Figure 7.
Antitumor efficacy of ENTPD6 neoantigen-specific T cells in vivo and their specific TCRs. (A) Evaluation of antitumor efficacy for the ENTPD6 neoantigen in the B-NDG-B2mtm1Fcrntm1(mB2m)/Bcgen mouse model. (B) The key sequences of the tumor analyzed with Sanger sequencing. (C) The specific secretion of IFN-γ by the potential TCR-T cells against the ENTPD6 neoantigen in the Co-HA system detected by ELISpot (*p<0.05; **p<0.01; ***p<0.001). ELISpot, enzyme-linked immunospot assay; ENTPD6, ectonucleoside triphosphate diphosphohydrolase 6; IFN-γ, interferon-γ; TCR, T-cell receptor.
Specific TCR of the ENTPD6 neoantigen
For neoantigens, the corresponding TCR is a necessary condition for activating T cells. So, the TCR sequencing was considered to further identify recognition of neoantigens. The ENTPD6 neoantigen-specific T cells from patient 04 were sorted by FCM after tetramer staining (online supplemental figure 10A, B). We captured a total of eight TCRs by Sanger sequencing (online supplemental table 4). The sequences of these TCRs were successfully verified by the IgBlast tool (https://www.ncbi.nlm.nih.gov/). Then, we constructed TCR-T cells that may be able to recognize the ENTPD6 neoantigen. The number and mean size of IFN-γ spots produced by the coincubation of cytotoxic TCR-T cells and target cells expressing the neoantigen were significantly greater than that of the WT antigen control detected by an ELISpot analysis in the Co-HA system (figure 7C). Finally, we found that the dominant clone of TCRs for ENTPD6 neoantigen was S20-1-BA2 (figure 7C).
Discussion
Immune checkpoint inhibitors (ICIs) have revolutionized cancer therapy and have demonstrated clinical efficacy in HCC immunotherapy in the CheckMate-040 trial36 and the IMbrave 150 trial.37 However, ~70% of patients with advanced HCC did not benefit from these treatments. Although ICIs can reverse tumor-induced immunosuppression and release T-cell-mediated antitumor responses,38 the dominant immunotherapeutic targets should also be considered when T cells are activated. Tumor neoantigens were excellent targets when combined with ICIs, which would strengthen T-cell cytotoxicity in HCC.7 However, due to the lack of an effective HCC neoantigen profile, the progress of HCC neoantigen clinical trials is slow. In this study, we established a systematic and reliable workflow for the screening and verification of HCC neoantigens.
The precondition for predicting neoantigens is effective variation calling. The base substitutions, mutation characteristics, and mutated genes in our 14 enrolled patients with HCC were mostly consistent with those in the TCGA-LIHC data set, COSMIC database (online supplemental figure 5A-C) and some previous reports.17 39 These characteristics were not identical as the number of samples was limited, and approximately 50% of the patients in the study had HBV; thus, our patient population was representative of the Asian population but not the global patient with HCC population. Therefore, the mutation frequencies of some high-frequency mutated genes were slightly different in our population from those in the TCGA-LIHC data set. For example, the mutation frequency of TP53 was 21.4% (3/14) in our population, but that in the TCGA-LIHC data set was 35.4% (109/308).
We predicted 541 potential neoantigens in tumors. Due to the high heterogeneity of neoantigens,6 7 most of these potential neoantigens have not been previously reported, and there was no overlap among the potential neoantigens in the 14 patients. We tried to detect 5'-FYAFSCYYDL-3' and 5'-WVWCMSPTI-3' in 80 HCC tumor tissues (online supplemental figure 11); only patient 04 and patient 05 had these new dominant HCC neoantigens. These results suggest that neoantigen vaccines should be individualized.
PVTT is the main form of intrahepatic HCC metastasis, with an incidence of 35%–50%.40 41 The evolutionary tree constructed in our study showed that most potential neoantigens in PVTTs were derived from tumor tissues, suggesting that HCC neoantigens identified in tumor tissues could be used as vaccines to prevent and treat PVTTs. Since PVTTs are related to HCC recurrence and metastasis, the neoantigens identified in PVTTs are crucial for preventing such events.
Experimental validations in vitro and in vivo are indispensable for neoantigen identification. Cheng et al only identified 1 positive neoantigen among 322 predicted HCC neoantigens by tetramer staining, and the frequency of specific T cells was 0.220% in CD8+ tumor-infiltrating lymphocytes (TILs).18 Dong et al coincubated predicted neoantigen peptides with PBMCs to promote specific T-cell proliferation, and the maximum percentage of tetramer-positive T cells was approximately 18%.17 With the same method, we found that the frequency of 5'-FYAFSCYYDL-3'-specific T cells (53.415%) was much greater than that of other neoantigen-specific T cells, suggesting that 5'-FYAFSCYYDL-3' may be a dominant neoantigen in HCC. Furthermore, neoantigen vaccines are mainly used for patients with advanced HCC. Most TILs in these patients have been exhausted due to the crosstalk between HCC and the immune system. However, specific T cells targeting neoantigens in PBMCs still exist. The ability to promote specific T-cell proliferation in PBMCs makes these neoantigens suitable for tumor vaccines, particularly peptide vaccines. Thus, we chose PBMCs from patients for screening potential HCC-dominant neoantigens.
Furthermore, the Co-HA system verified that the HLA-A*24:02-restricted epitope 5'-FYAFSCYYDL-3', produced by the mutated ENTPD6, and the HLA-A*02:01-restricted epitope 5'-WVWCMSPTI-3', produced by the mutated asparagine-linked glycosylation protein 1 homolog (ALG1), had strong immunogenicity in HCC. However, it was confirmed that two predicted neoantigens (5'-KMAQFYYSA-3' and 5'-KQIAELETWV-3') that generated low-frequency tetramer+ T cells could not trigger potent immune response in the Co-HA system (online supplemental figure 12A-F). It is well known that HCC-dominant neoantigens have excellent affinity and that their paired WT peptides are weak. The affinity of 5'-FYAFSCYYDL-3' was 82.16 nM, and the affinity of its paired WT peptide was 51.58 nM. The affinity of 5'-WVWCMSPTI-3' was 41.20 nM, and the affinity of its paired WT peptide was 72.26 nM. These results suggest that HCC-dominant neoantigens generally have excellent affinity; however, the affinity of their paired WT peptides may not be weak. As different HLAs show different distributions of binding affinity and promiscuity, it is common to screen neoantigens in terms of the rank of mutated peptide affinity (MutRank) ≤2 from a sample of random peptides.42 However, the MutRank of 5'-FYAFSCYYDL-3' is 1.957, and the MutRank of 5'-WVWCMSPTI-3' is 3.375. This result suggests that screening HCC-dominant neoantigens by MutRank ≤2 may be insufficient. ENTPD6 is similar to E-type nucleotidases, which mediate the catabolism of purine and pyrimidine.43 44 ALG1 encodes a transmembrane chitobiosyldiphosphodolichol β-mannosyltransferase, which mediates the biosynthesis of lipid-linked oligosaccharides.45 46 ENTPD6 and ALG1 were both remarkably overexpressed in several human cancers, including HCC (online supplemental figure 13A, B). Patients with HCC with high ENTPD6 and ALG1 expression levels were likely to have poor overall survival outcomes (online supplemental figure 13C). One study revealed that ENTPD6 might be a tumor suppressor gene in testicular cancer.44 In addition, the knockdown of ENTPD6 significantly desensitized pancreatic cancer cells to gemcitabine and cytosine arabinoside.47 These results illustrated that ENTPD6 and ALG1 might play an important role in HCC. The mutations and neoantigens generated from ENTPD6 and ALG1 may be the products of crosstalk between HCC and the immune system.
Adoptive transfer of T cells transduced with TCRs or chimeric antigen receptors (CARs) is a promising approach to cancer immunotherapy. Compared with CAR-T cells, TCR-T cells have more target options.48 In this study, the Co-HA system was used to verify that TCR-T cells targeting the reported KRAS G12D neoantigen restricted by HLA-A*11:01 could kill target cells. In addition, we identified that the dominant clone of TCRs for ENTPD6 neoantigen was S20-1-BA2 (figure 7C) by the Co-HA system. These results suggest that this system can be used to validate the feasibility of TCR-T cells.
The main novelty of the study is the development and validation of the Co-HA system. The design of the HLA vector of the Co-HA system is innovative: all functional regions, such as the HLA regions and neoantigen regions, are replaceable. Therefore, we could construct the prevalent HLA vectors with only one cutting and joining procedure. Then, the neoantigen DNA could be seamlessly assembled between the P2A DNA and termination codon, which is beneficial for its independent expression without the production of non-natural antigens. In general, peptides including HLA and neoantigen could be cleaved by the P2A peptide, which has the highest cleavage efficiency among 2A peptides.49 The Co-HA system is easy to replicate in other laboratories for verifying the function of different peptides or the TCR-T cells. In addition, two Chinese invention patents (ZL202111576586.2 and ZL202010634108.1) have been granted for the Co-HA system, which is being evaluated in clinical trials (ClinicalTrials.gov: NCT05105815).
The main limitation of our study is that only HLA-A*11:01-, HLA-A*24:02- or HLA-A*02:01-restricted neoantigens were identified, with limited patient numbers. An evaluation of a larger number of patients will be conducted in the future using the Co-HA system to develop an effective neoantigen profile for HCC immunotherapeutic targeting.
In conclusion, this study presents a convenient system for identifying HCC neoantigens and may promote personalized immunotherapies for HCC.
jitc-2022-006334supp002.pdf (408.5KB, pdf)
Acknowledgments
We thank Jiancheng Luo, Zhihao Wang, Xiaofei Ma, and Shan Zhang from Aiyimed, and Dong Jiang, Xueyan Wang, and Jianghua Wang from CorreGene for their assistance and suggestions on this project.
Footnotes
PC and DC contributed equally.
Contributors: Concept and design: HC, WL. Development of methodology: XX, PC, DC. Writing of the manuscript: PC, DC. Revision of the manuscript: XX, YY. Analysis and interpretation of NGS data: DB, WQ, LR. Acquisition of samples: JG, WQ, KD. Tetramer staining: PC, KD, SS. Immunogenicity test: DC, WX. Study supervision: HC, WL, XX. DC. Responsible for the overall content as guarantor: HC.
Funding: This work was supported by the National Key Sci-Tech Special Project of China (No. 2018ZX10302207), the Beijing Natural Science Foundation (No. 7222191), the Peking University Medicine Seed Fund for Interdisciplinary Research supported by ‘the Fundamental Research Funds for the Central Universities’ (BMU2022MX001 and BMU2021MX007), the Science and Technology Planning Project of Guilin (No. 20190218-1), the Peking University People’s Hospital Scientific Research Development Funds (RDX2020-06), Beijing Natural Science Foundation Haidian Origination and Innovation Joint Fund (L222007) and Qi-Min Project (grant number: NA.).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by the Research Ethics Committees of the Peking University People’s Hospital (ID: 2021PHB262-001) and the Research Ethics Committees of the Affiliated Hospital of Guilin Medical University (ID: 2021WJWZC14). Participants gave informed consent to participate in the study before taking part.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:6. 10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol 2022;19:151–72. 10.1038/s41571-021-00573-2 [DOI] [PubMed] [Google Scholar]
- 4.Galle PR, Foerster F, Kudo M, et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int 2019;39:2214–29. 10.1111/liv.14223 [DOI] [PubMed] [Google Scholar]
- 5.Sawada Y, Yoshikawa T, Ofuji K, et al. Phase II study of the GPC3-derived peptide vaccine as an adjuvant therapy for hepatocellular carcinoma patients. OncoImmunology 2016;5:e1129483. 10.1080/2162402X.2015.1129483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Repáraz D, Ruiz M, Llopiz D, et al. Neoantigens as potential vaccines in hepatocellular carcinoma. J Immunother Cancer 2022;10:e003978. 10.1136/jitc-2021-003978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P, Fang Q-X, Chen D-B, et al. Neoantigen vaccine: an emerging immunotherapy for hepatocellular carcinoma. World J Gastrointest Oncol 2021;13:673–83. 10.4251/wjgo.v13.i7.673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017;547:217–21. 10.1038/nature22991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Z, Li Q, Zhang R, et al. Personalized neoantigen pulsed dendritic cell vaccine for advanced lung cancer. Signal Transduct Target Ther 2021;6:26. 10.1038/s41392-020-00448-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu L, Jiang J, Zhan M, et al. Targeting neoantigens in hepatocellular carcinoma for immunotherapy: a futile strategy? Hepatology 2021;73:414–21. 10.1002/hep.31279 [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Chen G, Cai Z, et al. Profiling of hepatocellular carcinoma neoantigens reveals immune microenvironment and clonal evolution related patterns. Chin J Cancer Res 2021;33:364–78. 10.21147/j.issn.1000-9604.2021.03.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H, Sun L, Guan A, et al. Unique TP53 neoantigen and the immune microenvironment in long-term survivors of hepatocellular carcinoma. Cancer Immunol Immunother 2021;70:667–77. 10.1007/s00262-020-02711-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Z, Su X, Qiu L, et al. Personalized neoantigen vaccine prevents postoperative recurrence in hepatocellular carcinoma patients with vascular invasion. Mol Cancer 2021;20:164. 10.1186/s12943-021-01467-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng S, Chen S, Hu W, et al. Combination neoantigen-based dendritic cell vaccination and adoptive T-cell transfer induces antitumor responses against recurrence of hepatocellular carcinoma. Cancer Immunol Res 2022;10:728–44. 10.1158/2326-6066.CIR-21-0931 [DOI] [PubMed] [Google Scholar]
- 15.Lang F, Schrörs B, Löwer M, et al. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat Rev Drug Discov 2022;21:261–82. 10.1038/s41573-021-00387-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khodadoust MS, Olsson N, Wagar LE, et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 2017;543:723–7. 10.1038/nature21433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong L-Q, Peng L-H, Ma L-J, et al. Heterogeneous immunogenomic features and distinct escape mechanisms in multifocal hepatocellular carcinoma. J Hepatol 2020;72:896–908. 10.1016/j.jhep.2019.12.014 [DOI] [PubMed] [Google Scholar]
- 18.Cheng Y, Gunasegaran B, Singh HD, et al. Non-terminally exhausted tumor-resident memory HBV-specific T cell responses correlate with relapse-free survival in hepatocellular carcinoma. Immunity 2021;54:1825–40. 10.1016/j.immuni.2021.06.013 [DOI] [PubMed] [Google Scholar]
- 19.Jin X, Liu X, Zhou Z, et al. Identification of HLA-A2 restricted epitopes of glypican-3 and induction of CTL responses in HLA-A2 transgenic mice. Cancer Immunol Immunother 2022;71:1569–82. 10.1007/s00262-021-03096-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang QJ, Yu Z, Griffith K, et al. Identification of T-cell receptors targeting KRAS-mutated human tumors. Cancer Immunol Res 2016;4:204–14. 10.1158/2326-6066.CIR-15-0188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie X, Wang X, Liao W, et al. MYL6B, a myosin light chain, promotes mdm2-mediated p53 degradation and drives HCC development. J Exp Clin Cancer Res 2018;37:28. 10.1186/s13046-018-0693-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei S, Liu T, Ren X, et al. Benchmarking variant callers in next-generation and third-generation sequencing analysis. Brief Bioinform 2021;22:bbaa148. 10.1093/bib/bbaa148 [DOI] [PubMed] [Google Scholar]
- 23.Cunningham F, Allen JE, Allen J, et al. Ensembl 2022. Nucleic Acids Res 2022;50:D988–95. 10.1093/nar/gkab1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allegretti M, Fabi A, Buglioni S, et al. Tearing down the walls: FDA approves next generation sequencing (NGS) assays for actionable cancer genomic aberrations. J Exp Clin Cancer Res 2018;37:47. 10.1186/s13046-018-0702-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou S-L, Zhou Z-J, Hu Z-Q, et al. Genomic sequencing identifies WNK2 as a driver in hepatocellular carcinoma and a risk factor for early recurrence. J Hepatol 2019;71:1152–63. 10.1016/j.jhep.2019.07.014 [DOI] [PubMed] [Google Scholar]
- 26.Li X, Xu W, Kang W, et al. Genomic analysis of liver cancer unveils novel driver genes and distinct prognostic features. Theranostics 2018;8:1740–51. 10.7150/thno.22010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505–11. 10.1038/ng.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Totoki Y, Tatsuno K, Covington KR, et al. Trans-Ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267–73. 10.1038/ng.3126 [DOI] [PubMed] [Google Scholar]
- 29.Sawey ET, Chanrion M, Cai C, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell 2011;19:347–58. 10.1016/j.ccr.2011.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson KA, Sousa CM, Zhang X, et al. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature 2020;586:120–6. 10.1038/s41586-020-2746-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynisson B, Alvarez B, Paul S, et al. NetMHCpan-4.1 and netmhciipan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 2020;48:W449–54. 10.1093/nar/gkaa379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGranahan N, Rosenthal R, Hiley CT, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 2017;171:1259–71. 10.1016/j.cell.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou F, Cao H, Zuo X, et al. Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nat Genet 2016;48:740–6. 10.1038/ng.3576 [DOI] [PubMed] [Google Scholar]
- 34.Liu T, Tan J, Wu M, et al. High-Affinity neoantigens correlate with better prognosis and trigger potent antihepatocellular carcinoma (HCC) activity by activating CD39+CD8+ T cells. Gut 2021;70:1965–77. 10.1136/gutjnl-2020-322196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simoni Y, Becht E, Fehlings M, et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 2018;557:575–9. 10.1038/s41586-018-0130-2 [DOI] [PubMed] [Google Scholar]
- 36.Yau T, Kang Y-K, Kim T-Y, et al. Efficacy and safety of nivolumab plus ipilimumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib: the checkmate 040 randomized clinical trial. JAMA Oncol 2020;6:e204564. 10.1001/jamaoncol.2020.4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med 2020;382:1894–905. 10.1056/NEJMoa1915745 [DOI] [PubMed] [Google Scholar]
- 38.Fehlings M, Simoni Y, Penny HL, et al. Checkpoint blockade immunotherapy reshapes the high-dimensional phenotypic heterogeneity of murine intratumoural neoantigen-specific CD8+ T cells. Nat Commun 2017;8:562. 10.1038/s41467-017-00627-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Q, Zhu H, Dong L, et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell 2019;179:561–77. 10.1016/j.cell.2019.08.052 [DOI] [PubMed] [Google Scholar]
- 40.Zane KE, Makary MS. Locoregional therapies for hepatocellular carcinoma with portal vein tumor thrombosis. Cancers (Basel) 2021;13:5430. 10.3390/cancers13215430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerrito L, Annicchiarico BE, Iezzi R, et al. Treatment of hepatocellular carcinoma in patients with portal vein tumor thrombosis: beyond the known frontiers. World J Gastroenterol 2019;25:4360–82. 10.3748/wjg.v25.i31.4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boegel S, Castle JC, Kodysh J, et al. Bioinformatic methods for cancer neoantigen prediction. Prog Mol Biol Transl Sci 2019;164:25–60. 10.1016/bs.pmbts.2019.06.016 [DOI] [PubMed] [Google Scholar]
- 43.Turcot V, Lu Y, Highland HM, et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet 2018;50:26–41. 10.1038/s41588-017-0011-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tada Y, Yokomizo A, Shiota M, et al. Ectonucleoside triphosphate diphosphohydrolase 6 expression in testis and testicular cancer and its implication in cisplatin resistance. Oncol Rep 2011;26:161–7. 10.3892/or.2011.1274 [DOI] [PubMed] [Google Scholar]
- 45.González-Domínguez CA, Fiesco-Roa MO, Gómez-Carmona S, et al. ALG1-CDG caused by non-functional alternative splicing involving a novel pathogenic complex allele. Front Genet 2021;12:744884. 10.3389/fgene.2021.744884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marques-da-Silva D, Dos Reis Ferreira V, Monticelli M, et al. Liver involvement in congenital disorders of glycosylation (CDG). A systematic review of the literature. J Inherit Metab Dis 2017;40:195–207. 10.1007/s10545-016-0012-4 [DOI] [PubMed] [Google Scholar]
- 47.Fridley BL, Batzler A, Li L, et al. Gene set analysis of purine and pyrimidine antimetabolites cancer therapies. Pharmacogenet Genomics 2011;21:701–12. 10.1097/FPC.0b013e32834a48a9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Song X, Shen L, et al. Exploiting T cell signaling to optimize engineered T cell therapies. Trends Cancer 2022;8:123–34. 10.1016/j.trecan.2021.10.007 [DOI] [PubMed] [Google Scholar]
- 49.Kim JH, Lee S-R, Li L-H, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 2011;6:e18556. 10.1371/journal.pone.0018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-006334supp001.pdf (62.3MB, pdf)
jitc-2022-006334supp002.pdf (408.5KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.