Abstract
Increasing reports of invasive Streptococcus pyogenes infections mandate surveillance for toxigenic lineage M1UK. An allele-specific PCR was developed to distinguish M1UK from other emm1 strains. The M1UK lineage represented 91% of invasive emm1 isolates in England in 2020. Allele-specific PCR will permit surveillance for M1UK without need for genome sequencing.
Keywords: Streptococcus pyogenes, M1UK, emm1, scarlet fever, molecular diagnostics, PCR, bacteria, respiratory infections, streptococci, superantigen, allele-specific, emerging lineage, England, iGAS, group A Streptococcus
Upsurges in invasive group A Streptococcus (GAS) infections have been widely reported in England and elsewhere (1), emphasizing the need to examine the relationship between circulating S. pyogenes that cause pharyngitis and scarlet fever and cases of invasive disease. Although many factors, such as exposure history, underlying conditions, viral co-infection, and genetic susceptibility, might increase susceptibility to S. pyogenes infection, strain-specific virulence might also be crucial.
In England, where both scarlet fever and invasive S. pyogenes infections are notifiable, pronounced upsurges in scarlet fever were recorded over an 8-year period (2,3), but subsided during the COVID-19 pandemic. During the 2015–16 season, a notable increase in invasive infections was observed that had not been evident previously (4). Both scarlet fever and invasive infections were associated with the emergence of M1UK, a new sublineage of emm1 S. pyogenes (4) that appeared to outcompete the highly successful, contemporary epidemic emm1 M1global strain, which emerged and spread globally during the 1980s (5,6). Despite an unchanged phage repertoire, M1UK strains produce more superantigenic scarlet fever toxin SpeA (streptococcal pyrogenic exotoxin A) than contemporary M1global S. pyogenes strains (4).
emm1 S. pyogenes strains are highly virulent (5) and disproportionately associated with invasive infections; any increase in the prevalence of emm1 strains in persons with pharyngitis or scarlet fever is, therefore, a public health concern. Known distribution of M1UK is largely limited to those countries undertaking and reporting genome sequencing (Figure 1). M1UK has been identified in other countries in Europe, from a single isolate in Denmark (4) to dominant status in the Netherlands (7). The lineage has also been reported in North America; the Public Health Agency of Canada reported that 17/178 (10%) of emm1 isolates from 2016 were M1UK (8). This finding contrasts with a reported M1UK frequency of just 0%–2.8% of emm1 isolates in the United States, according to the Active Bacterial Core surveillance system of the US Centers for Disease Control and Prevention; however, the low US frequency was associated with severe infections (9). Of note, most reports used genomic data that were >5 years old, so a reappraisal of prevalence is needed. A recent study in Australia using data through 2020 indicated expansion of M1UK in Queensland and Victoria (10). The authors identified acquisition of an additional phage encoding superantigen genes ssa and spec and a single-nucleotide polymorphism (SNP) implicated in SpeA upregulation in the M1UK lineage. Multicountry increases in GAS infections (1) since pandemic restrictions were lifted underscore the importance of increasing global surveillance of lineages that have potentially enhanced fitness, such as M1UK.
Figure 1.
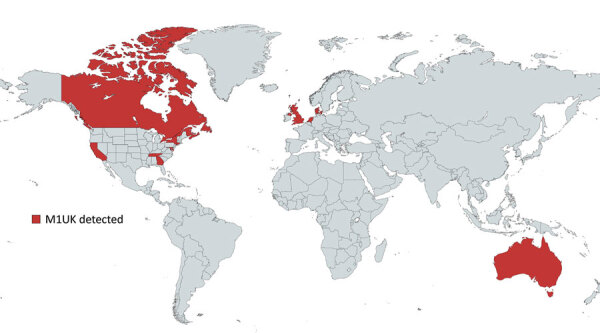
Countries and US states with reported M1UK Streptococcus pyogenes cases. Map created by using MapChart (https://www.mapchart.net) as part of a study of emerging invasive group A Streptococcus M1UK lineage detected by allele-specific PCR, England, 2020.
The Study
Genetic distinction between M1UK and M1global strains is possible by using whole-genome sequencing to detect the 27 SNPs that characterize the M1UK lineage (4), but sequencing technology is not available in all countries. We designed an allele-specific PCR (AS-PCR) method to detect M1UK-specific SNPs in the rofA, gldA, and pstB genes. We chose amplification targets to separate M1UK and M1global strains but also to identify strains from less common intermediate sublineages that had only 13 or 23 of the 27 M1UK-specific SNPs (4). We optimized PCR conditions for each pair of amplicons by using DNA from control strains for each lineage (Table; Appendix Figure). Collecting bacterial samples from patients was part of routine clinical care; collecting surplus samples after anonymizing patient information was approved by the West London National Research Ethics Committee (approval no. 06/Q0406/20).
Table. PCR primers and conditions used to differentiate M1global and M1UK Streptococcus pyogenes lineages in study of emerging invasive group A Streptococcus M1UK lineage detected by allele-specific PCR, England, 2020*.
| Target gene | Primer type† | Sequences‡ | PCR cycle conditions | Product, bp |
|---|---|---|---|---|
|
rofA
|
WT sequence | TGTTAATTGCTTGGTTAAATCA | 30 cycles of 95°C for 3 min, 45 s; 59.2°C for 30 s; 72°C for 1 min (final cycle: 5 min) |
278 |
| Forward-SNP | 5′-TGTTAATTGCTTGGTTAAAGtA-'3 | |||
| Forward-WT | 5′-TGTTAATTGCTTGGTTAAAGCA-'3 | |||
| Reverse |
5′-GCTCATCTCCTAACGGATTCTT-'3 |
|||
|
gldA
|
WT sequence | AGATGGGTTAGCAACATGG | 30 cycles of 95°C for 3 min, 45 s; 61.8°C for 30 s;72°C for 1 min (final cycle: 5 min) |
292 |
| Forward-SNP | 5′-AGATGGGTTAGCAACAAaG-'3 | |||
| Forward-WT | 5′-AGATGGGTTAGCAACAAGG-'3 | |||
| Reverse |
5′-GAATAGCACCTGTCAGCG-'3 |
|||
| pstB | WT sequence | GATAAATCAATCTTAGACCA | 30 cycles of 95°C for 3 min, 45 s; 50°C for 30 s; 72°C for 1 min (final cycle: 5 min) | 287 |
| Forward-SNP | 5′-GATAAATCAATCTTAGATaA-'3 | |||
| Forward-WT | 5′-GATAAATCAATCTTAGATCA-'3 | |||
| Reverse | 5′-CGTGAGGCTTGCTGCATTGAG-'3 |
*SNP, single-nucleotide polymorphism; WT, wild-type. †Forward primers were designed to detect either the targeted SNP (M1UK) or WT (M1global) sequences. ‡Lowercase bold letters in primer sequences denote the base complementary to the targeted SNP in the M1UK sequence. Underlined uppercase letters indicate an additional mismatched base introduced into primer sequences to increase primer specificity.
To evaluate allele-specific PCR, we tested whether the rofA and pstB primers correctly identified lineages of 27 newly genome-sequenced noninvasive emm1 S. pyogenes strains isolated during 2017–18 and collected by the infection bioresource at Imperial College. We artificially enriched the isolates for M1global strains to ensure adequate numbers of each lineage: 8/27 isolates were M1global, and 19/27 were M1UK. PCR amplification of rofA and pstB alleles from those isolates assigned 100% of strains to the correct lineage previously identified by sequencing (Appendix Table 1).
To evaluate the ability of AS-PCR to identify emm1 isolates from M1global, M1UK, and intermediate sublineages (4), we tested 16 strains from 2013–2016 that comprised 4 isolates each of M1global, M113snps, M123snps, and M1UK lineages (Appendix Table 2). SNPs were correctly detected in the rofA gene from all M113snps, M123snps, and M1UK isolates (Appendix Table 3). SNPs were also correctly detected in gldA from all M123snps and M1UK isolates but not M1global or M113snps isolates. Finally, SNPs in pstB were only identified in M1UK isolates. Thus, in all cases, SNP profiles determined by AS-PCR were consistent with strain-specific genome sequences.
In England, submission of all isolates from invasive infection is requested by the UK Health Security Agency reference laboratory for emm genotyping. emm1 isolates are routinely the dominant genotype among invasive sterile-site isolates, typically representing 20%–30% of invasive infections. During 2020, when incidence of common respiratory infections was reduced by COVID-19–related public health interventions, emm1 S. pyogenes frequency varied each month from 0%–24% of all invasive infections and decreased toward the end of the year. We subjected all 305 invasive emm1 S. pyogenes isolates from 2020 that were available for this study to AS-PCR (Appendix Table 4). AS-PCR identified M1UK-specific SNPs in rofA, gldA, and pstB in 278/305 (91.1%) of isolates, which were, therefore, assigned to the M1UK lineage. No SNPs were detected in the remaining 27 isolates, which were assigned to M1global; no intermediate lineage emm1 strains were identified in isolates collected during 2020 by using AS-PCR.
We performed Western blot analysis of 10 M1UK isolates identified by AS-PCR. We confirmed that SpeA production was similar to M1UK strains tested previously; however, we did not quantify SpeA production.
Conclusions
The longevity of emergent S. pyogenes lineages in a population is difficult to predict. Although an emm89emergent acapsular lineage has disseminated globally (11), an emergent emm3 SpeC-producing lineage, associated with upsurges in scarlet fever and invasive infections, ceased to be detectable within a few years (12). Taken together with previously reported genome-sequenced emm1 isolates (Figure 2), AS-PCR results indicated that the M1UK lineage continued to expand among invasive S. pyogenes isolates from 2016 to the end of 2020 in England.
Figure 2.
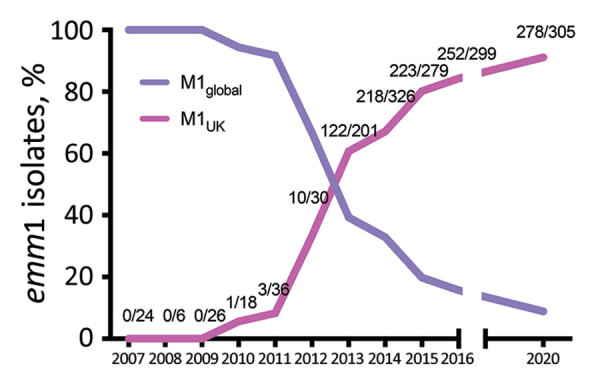
Prevalence of M1UK and M1global Streptococcus pyogenes lineages over time in study of emerging invasive group A Streptococcus M1UK lineage detected by allele-specific PCR, England, 2020. We determined percentages of emm1 isolates in England that belonged to M1UK or M1global lineages by using all available emm1 S. pyogenes genome sequences for 2007–2016 (4) and all available invasive isolates from 2020 that we tested by allele-specific PCR. Numbers on graph indicate number of isolates assigned as M1UK/total number sequenced for each year. Graph was adapted and updated from data previously described (4).
Increased invasive GAS activity in several countries (1) indicates a need for ongoing surveillance of novel lineages, given the potential public health effects. AS-PCR provides a readily available method to detect M1UK that is straightforward and, for screening purposes only, can be simplified by using only rofA primers to identify M1UK or associated sublineages. A limitation of our study is that the assay requires validation in reference laboratory settings. AS-PCR does not replace genome sequencing as the preferred method for surveillance of highly pathogenic bacteria, but sequencing is not widely available and is expensive.
emm1 strains have accounted for >50% of invasive infections in children in England during the 2022–23 season (13). Our results indicate that the M1UK lineage remained dominant in England and expanded to the end of 2020, and contact tracing in 2018 demonstrated a high frequency of secondary acquisition of M1UK in school outbreak settings (14). Given the recognized association between emm1 S. pyogenes and fatal outcome of invasive infections (15), enhanced surveillance for the M1UK sublineage is warranted. We conclude that AS-PCR is a readily available method to determine whether emm1 S. pyogenes isolates belong to the M1UK clade without need for genome sequencing and will improve surveillance of invasive GAS strains.
Additional information for emerging invasive group A Streptococcus M1UK lineage detected by allele-specific PCR, England, 2020.
Acknowledgments
We thank the National Institute for Health Research (NIHR) Imperial Biomedical Research Centre (BRC); NIHR Health Protection Research Unit (HPRU) in Healthcare-associated Infection and Antimicrobial Resistance at Imperial College London; Medical Research Council Centre for Molecular Bacteriology and Infection; scientific and biomedical staff from the NIHR BRC Colebrook laboratory, NIHR HPRU, NIHR BRC Imperial Genomics Facility, and Northwest London Pathology/Imperial College Healthcare Trust Diagnostic laboratory for contributing to the collection, biobanking, typing, and sequencing of bacterial isolates; and staff at the UK Health Security Agency reference laboratory who genotyped emm-invasive isolates used in this study.
This study was funded by UK Research and Innovation Medical Research Council (MR/P022669/1) and NIHR Imperial Biomedical Research Centre.
Biography
Dr. Zhi is a postdoctoral research associate at Imperial College London. Her research focuses on pathogenesis and vaccine prevention of Streptococcus pyogenes disease.
Footnotes
Suggested citation for this article: Zhi X, Li HK, Li H, Loboda Z, Charles S, Vieira A, et al. Emerging invasive group A Streptococcus M1UK lineage detected by allele-specific PCR, England, 2020. Emerg Infect Dis. 2023 May [date cited]. https://doi.org/10.3201/eid2905.221887
Data from this study were presented at the 21st Lancefield International Symposium on Streptococci and Streptococcal Diseases; Stockholm, Sweden; June 7–10, 2022.
References
- 1.World Health Organization. Increase in invasive group A streptococcal infections among children in Europe, including fatalities [cited 2022 Dec 14]. https://www.who.int/europe/news/item/12-12-2022-increase-in-invasive-group-a-streptococcal-infections-among-children-in-europe--including-fatalities
- 2.Turner CE, Pyzio M, Song B, Lamagni T, Meltzer M, Chow JY, et al. Scarlet fever upsurge in England and molecular-genetic analysis in north-west London, 2014. Emerg Infect Dis. 2016;22:1075–8. 10.3201/eid2206.151726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamagni T, Guy R, Chand M, Henderson KL, Chalker V, Lewis J, et al. Resurgence of scarlet fever in England, 2014-16: a population-based surveillance study. Lancet Infect Dis. 2018;18:180–7. 10.1016/S1473-3099(17)30693-X [DOI] [PubMed] [Google Scholar]
- 4.Lynskey NN, Jauneikaite E, Li HK, Zhi X, Turner CE, Mosavie M, et al. Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study. Lancet Infect Dis. 2019;19:1209–18. 10.1016/S1473-3099(19)30446-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, et al. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A. 2014;111:E1768–76. 10.1073/pnas.1403138111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–82. 10.1086/432514 [DOI] [PubMed] [Google Scholar]
- 7.Rümke LW, de Gier B, Vestjens SMT, van der Ende A, van Sorge NM, Vlaminckx BJM, et al. Dominance of M1UK clade among Dutch M1 Streptococcus pyogenes. Lancet Infect Dis. 2020;20:539–40. 10.1016/S1473-3099(20)30278-4 [DOI] [PubMed] [Google Scholar]
- 8.Demczuk W, Martin I, Domingo FR, MacDonald D, Mulvey MR. Identification of Streptococcus pyogenes M1UK clone in Canada. Lancet Infect Dis. 2019;19:1284–5. 10.1016/S1473-3099(19)30622-X [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Nanduri SA, Van Beneden CA, Beall BW. M1UK lineage in invasive group A streptococcus isolates from the USA. Lancet Infect Dis. 2020;20:538–9. 10.1016/S1473-3099(20)30279-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies MR, Keller N, Brouwer S, Jespersen MG, Cork AJ, Hayes AJ, et al. Detection of Streptococcus pyogenes M1UK in Australia and characterization of the mutation driving enhanced expression of superantigen SpeA. Nat Commun. 2023;14:1051. 10.1038/s41467-023-36717-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner CE, Abbott J, Lamagni T, Holden MTG, David S, Jones MD, et al. Emergence of a new highly successful acapsular group A Streptococcus clade of genotype emm89 in the United Kingdom. MBio. 2015;6:e00622. 10.1128/mBio.00622-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Shahib A, Underwood A, Afshar B, Turner CE, Lamagni T, Sriskandan S, et al. Emergence of a novel lineage containing a prophage in emm/M3 group A Streptococcus associated with upsurge in invasive disease in the UK. Microb Genom. 2016;2:e000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UK Health Security Agency. Group A streptococcal infections: second update on seasonal activity in England 2022 to 2023. [cited 2022 Dec 16]. https://www.gov.uk/government/publications/group-a-streptococcal-infections-activity-during-the-2022-to-2023-season/group-a-streptococcal-infections-second-update-on-seasonal-activity-in-england-2022-to-2023
- 14.Cordery R, Purba AK, Begum L, Mills E, Mosavie M, Vieira A, et al. Frequency of transmission, asymptomatic shedding, and airborne spread of Streptococcus pyogenes in schoolchildren exposed to scarlet fever: a prospective, longitudinal, multicohort, molecular epidemiological, contact-tracing study in England, UK. Lancet Microbe. 2022;3:e366–75. 10.1016/S2666-5247(21)00332-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson GE, Pondo T, Toews KA, Farley MM, Lindegren ML, Lynfield R, et al. Epidemiology of invasive group A streptococcal infections in the United States, 2005–2012. Clin Infect Dis. 2016;63:478–86. 10.1093/cid/ciw248 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information for emerging invasive group A Streptococcus M1UK lineage detected by allele-specific PCR, England, 2020.


