Abstract
In Senegal, Coxiella burnetii, which causes Q fever, has often been identified in ticks and humans near livestock, which are considered to be reservoirs and main sources of infection. We describe the emergence of C. burnetii in rodents, not previously known to carry this pathogen, and describe 2 new genotypes.
Keywords: Coxiella burnetii, Q fever, bacteria, airborne diseases, rodents, Senegal, zoonoses
Coxiella burnetii is a causative agent of Q fever, a worldwide zoonosis. The disease may be acute (relatively benign) or chronic (with a wide range of clinical manifestations that can lead to high human mortality) (1). Humans are infected by inhaling contaminated environmental dust and aerosol particles from the birth products of infected animals, as well as through direct contact with milk, urine, or feces containing C. burnetii (2,3). Humans are not considered natural hosts of C. burnetii (4). A wide spectrum of animals can serve as hosts (4), but the reservoirs are livestock, mainly sheep, cattle, and goats (3), which are also the main sources of human infection (1).
In Senegal, C. burnetii has been reported in humans (4–6) and ticks (4). It has been isolated from rodent-associated soft ticks (Ornithodoros sonrai) and detected in several species of hard ticks collected from ruminants (4). Our previous study of zoonotic pathogens in rodents collected in 2017 revealed no presence of C. burnetii in rodent populations from the Ferlo region in northern Senegal (7). However, in this study, we tested rodent samples collected during 2019–2020 from the same region and found high prevalence of a new C. burnetii genotype, which might indicate an ongoing epizootic outbreak.
We screened 125 rodent samples for C. burnetii; the rodents were collected in the Ferlo region in northern Senegal near Widou Thiengoly (15.99°N, 15.32°W) under framework agreements between the French National Research Institute for Development and Senegal (7). None of the rodent species investigated were listed as protected with the International Union for Conservation of Nature or the Convention on International Trade in Endangered Species of Wild Fauna and Flora. Handling procedures were performed under Centre de Biologie Pour la Gestion des Populations agreement no. D-34-169-1 for experiments on wild animals and followed the guidelines of the American Society of Mammologists (8).
Rodents sampled belonged to the species Arvicanthis niloticus (n = 29), Desmodilliscus braueri (n = 3), Gerbillus nancillus (n = 9), G. nigeriae (n = 71), Jaculus jaculus (n = 4), Taterillus spp. (probably T. pygargus) (n = 8), and Xerus erythropus (n = 1). We extracted DNA from the spleen as described elsewhere (7) and stored it at −20°C. We detected bacterial DNA using C. burnetii–specific quantitative real-time PCR with primers and probes targeting IS1111 and IS30A spacers (4). For positive samples with a cycle threshold value <38, we first amplified 3 pairs of intergenic spacer primers, Cox2F/R, Cox5F/R, and Cox18F/R (5). Multispacer sequence typing (MST) genotyping of C. burnetii strains using sequences from the amplification of these 3 primer pairs revealed a potential new genotype. We amplified the other 7 primer pairs, Cox20F/R, Cox22F/R, Cox37F/R, Cox51F/R, Cox56F/R, Cox57F/R, and Cox61F/R, to describe this genotype.
Overall, 22.4% (28/125 for IS1111) and 19.2% (24/125 for IS30A) of rodents screened were positive for C. burnetii–specific quantitative PCR: Desmodilliscus braueri (33.3%; 1/3), G. nancillus (33.3%; 3/9), G. nigeriae (28.2%; 20/71), Jaculus jaculus (25%; 1/4), and Taterillus spp. (37.5%, 3/8). We found no Arvicanthis niloticus or Xerus erythropus rodents positive for C. burnetii. We performed complete MST genotyping of positive C. burnetii strain samples, and the sequences obtained from the primer pairs showed 100% identity for all positive samples. Nevertheless, 1 sample showed an insertion of 5 nucleotide bases in the amplified sequence of Cox56 spacer (Table), indicating the presence of a probable variant. All of these sequences demonstrated the presence of >1 new genotypes of C. burnetii according to the BLAST database (https://ifr48.timone.univ-mrs.fr/mst/coxiellaburnetii/blast.html). Phylogenetic analysis based on concatenated sequences of spacers revealed that the new genotypes, MST75 (major) and MST76 (with an insertion), are close together and cluster with other genotypes, including those already found in Senegal, such as MST19, and those infecting animals and humans, such as MST16, MST17, MST20, and MST61 (Figure).
Table. Characterization of new Coxiella burnetii MST75 and MST 76 genotypes described in study of new genotype of C. burnetii causing epizootic Q fever outbreak in rodents, northern Senegal*.
| Species | No. | Is1111 positive, no. (%) | Profile of spacers |
Genotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cox2 | Cox5 | Cox18 | Cox20 | Cox22 | Cox37 | Cox51 | Cox56 | Cox57 | Cox61 | ||||
|
Arvicanthis niloticus
|
29 |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
|
Desmodilliscus braueri
|
3 |
1 (33.3) |
3 |
14 |
6 |
6 |
5 |
4 |
10 |
10 |
13 |
5 |
MST75 |
|
Gerbillus nancillus
|
9 |
3 (33.3) |
3 |
14 |
6 |
6 |
5 |
4 |
10 |
10 |
13 |
5 |
MST75 |
|
G. nigeriae† |
71 |
20 (28.2) |
3 |
14 |
6 |
6 |
5 |
4 |
10 |
20/21‡ |
13 |
5 |
MST75/ MST76‡ |
|
Jaculus jaculus
|
4 |
1 (25) |
3 |
14 |
6 |
6 |
5 |
4 |
10 |
10 |
13 |
5 |
MST75 |
|
Taterillus spp. |
8 |
3 (37.5) |
3 |
14 |
6 |
6 |
5 |
.4 |
10 |
10 |
13 |
5 |
MST75 |
|
Xerus erythropus
|
1 |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
| Total | 125 | 28 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
*MST, multispacer sequence typing; NA, not applicable. †Invasive (expanding) species. ‡Genotype MST76 has the same profile as MST75 except for Cox 56.
Figure.
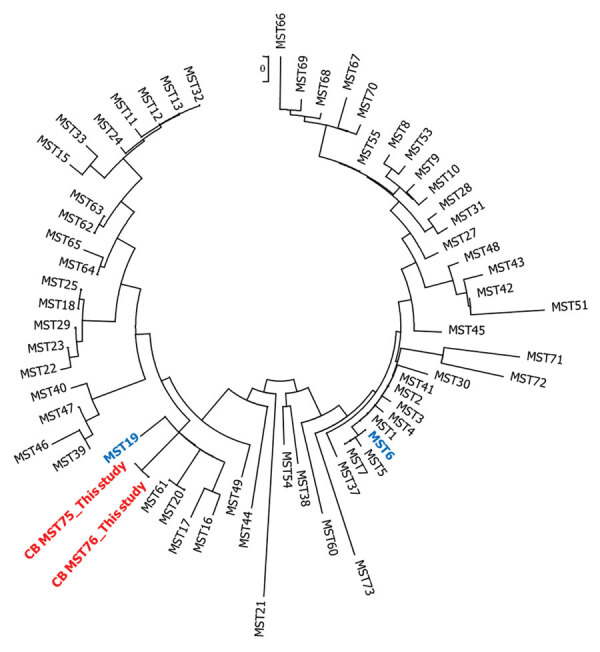
Neighbor-joining circular unrooted tree showing the relationship between Coxiella burnetii genotypes described in study of new genotype of C. burnetii causing epizootic Q fever outbreak in rodents, northern Senegal. MST75 and MST76 (red) were compared with genotypes already found in Senegal, MST19 and MST6 (blue), and other genotypes. The analysis involved 64 nt sequences. All positions containing gaps and missing data were eliminated. There were a total of 4,692 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (https://www.megasoftware.net). MST, multispacer sequence typing.
Our finding of C. burnetii MST75 and MST76 genotypes in the Ferlo rodent community suggests the emergence of a Q fever epizootic outbreak. Previously identified C. burnetii strains in Senegal were related to the proximity of livestock near the villages of Dielmo and Ndiop (9). In Ferlo, a previous study conducted on rodents sampled in 2017 did not find C. burnetii (7), indicating a relatively recent, possibly still ongoing epizootic outbreak. High C. burnetii prevalences (28%–38%) were observed in different species of gerbilline rodents, including G. nigeriae, which has recently colonized northern Senegal and is now the dominant species in outdoor rodent communities of Ferlo (10). The possibility of animal transmission from farms located near the rodent sampling area should also be explored. Our study shows the emergence in Senegal of new C. burnetii genotypes in susceptible animals, such as rodents (1), which might be a source of human infections. Although the pathogenicity of these new genotypes for humans is yet unknown, our findings signal the urgent need for epidemiologic surveillance for C. burnetii infection in humans in Senegal and neighboring countries.
Acknowledgments
We thank Philippe Gauthier for precise taxonomic identification of some rodent specimens using barcoding (based on cytb sequences), and Fabien Flirden for his technical support on molecular biology.
This study was supported by the Institut Hospitalo-Universitaire Méditerranée Infection, the French National Research Agency under the Investissements d’avenir programme, (reference ANR-10-IAHU-03), and the fourth Make Our Planet Great Again (MOPGA 4) program. This work was cofunded by the Labex Dispositif de Recherche Interdisciplinaire sur les Interactions Hommes-Milieux, and the Investissements d’Avenir program (ANR-11-LABX-0010), which is managed by the Agence nationale de la recherche.
Biography
Dr. Mangombi-Pambou is a postdoctoral researcher at the Institut Hospitalo Universitaire Méditérranée Infection in Marseille, France. His research interests include infectious diseases of zoonotic origin and the ecology and role of rodents in the dynamics of zoonotic diseases.
Footnotes
Suggested citation for this article: Mangombi-Pambou J, Granjon L, Labarrere C, Kane M, Niang Y, Fournier P-E, et al. New genotype of Coxiella burnetii causing epizootic Q fever outbreak in rodents, northern Senegal. Emerg Infect Dis. 2023 May [date cited]. https://doi.org/10.3201/eid2905.221034
References
- 1.Angelakis E, Raoult D. Q fever. Vet Microbiol. 2010;140:297–309. 10.1016/j.vetmic.2009.07.016 [DOI] [PubMed] [Google Scholar]
- 2.Tissot-Dupont H, Raoult D. Q fever. [ix.]. Infect Dis Clin North Am. 2008;22:505–14, ix. 10.1016/j.idc.2008.03.002 [DOI] [PubMed] [Google Scholar]
- 3.Vanderburg S, Rubach MP, Halliday JEB, Cleaveland S, Reddy EA, Crump JA. Epidemiology of Coxiella burnetii infection in Africa: a OneHealth systematic review. PLoS Negl Trop Dis. 2014;8:e2787. 10.1371/journal.pntd.0002787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mediannikov O, Fenollar F, Socolovschi C, Diatta G, Bassene H, Molez J-F, et al. Coxiella burnetii in humans and ticks in rural Senegal. PLoS Negl Trop Dis. 2010;4:e654. 10.1371/journal.pntd.0000654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angelakis E, Mediannikov O, Socolovschi C, Mouffok N, Bassene H, Tall A, et al. Coxiella burnetii-positive PCR in febrile patients in rural and urban Africa. Int J Infect Dis. 2014;28:107–10. 10.1016/j.ijid.2014.05.029 [DOI] [PubMed] [Google Scholar]
- 6.Sokhna C, Mediannikov O, Fenollar F, Bassene H, Diatta G, Tall A, et al. Point-of-care laboratory of pathogen diagnosis in rural Senegal. PLoS Negl Trop Dis. 2013;7:e1999. 10.1371/journal.pntd.0001999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahmana H, Granjon L, Diagne C, Davoust B, Fenollar F, Mediannikov O. Rodents as hosts of pathogens and related zoonotic disease risk. Pathogens. 2020;9:202. 10.3390/pathogens9030202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sikes RS, Gannon WL; Animal Care and Use Committee of the American Society of Mammologists. Guidelines of the American Society of Mammologists for the use of wild mammals in research. J Mammal. 2011;92:235–53. 10.1644/10-MAMM-F-355.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diouf FS, Ndiaye EHI, Hammoud A, Diamanka A, Bassene H, Ndiaye M, et al. Detection of Coxiella burnetii and Borrelia spp. DNA in cutaneous samples and in household dust in rural areas, Senegal. Vector Borne Zoonotic Dis. 2021;21:659–66. 10.1089/vbz.2020.2723 [DOI] [PubMed] [Google Scholar]
- 10.Granjon L, Ba K, Diagne C, Ndiaye A, Piry S, Thiam M. The Ferlo small rodent community: historical trends and characteristics of the current population [in French]. In: Boëtsch G, Duboz P, Guissé A, Sarr P, editors. The Great Green Wall, an African response to climate change [in French]. Paris: Centre national de la recherche scientifique; 2019. p. 1–19. [Google Scholar]


