Abstract
We have used nucleotide analog interference mapping and site-specific substitution to determine the effect of 2′-deoxynucleotide substitution of each nucleotide in the VS ribozyme on the self-cleavage reaction. A large number of 2′-hydroxyls (2′-OHs) that contribute to cleavage activity of the VS ribozyme were found distributed throughout the core of the ribozyme. The locations of these 2′-OHs in the context of a recently developed helical orientation model of the VS ribozyme suggest roles in multi-stem junction structure, helix packing, internal loop structure and catalysis. The functional importance of three separate 2′-OHs supports the proposal that three uridine turns contribute to local and long-range tertiary structure formation. A cluster of important 2′-OHs near the loop that is the candidate region for the active site and one very important 2′-OH in the loop that contains the cleavage site confirm the functional importance of these two loops. A cluster of important 2′-OHs lining the minor groove of stem–loop I and helix II suggests that these regions of the backbone may play an important role in positioning helices in the active structure of the ribozyme.
INTRODUCTION
The importance of the 2′-hydroxyl (2′-OH) in the stabilization of RNA tertiary structure has been known since early studies on the crystal structures of tRNA molecules. Within the crystal structure of tRNAPhe, almost half of the 2′-OH moieties of non-helical nucleotides are involved in hydrogen bonds to the nitrogenous bases, phosphate oxygens or other ribose groups (1–3). These stabilize sharp turns, such as the uridine turn, help bulge out certain nucleotides from nucleotide stacks (3) and stabilize the close approach of backbone strands (1). In addition, the 2′-OH was observed to be a ligand for heavy metal ions (which are thought, in some cases, to bind at magnesium ion binding sites) and for hydrated magnesium (1,4).
More recent ribozyme crystal structures have supported an important role for the 2′-OH in tertiary structure. The crystal structure of the P4–P6 domain revealed that of the two major tertiary interactions that knit together the domain, the tetraloop–receptor interaction and the A-rich bulge interaction, they both involve ‘ribose zippers’, a network of hydrogen bonding mediated by at least two 2′-OHs as well as a purine N3 or pyrimidine O2 (5). Additionally, the 2′-OH of A114 in the J4–J5 junction region of P4–P6 was observed to be involved in an intermolecular crystal contact, thought to be analogous to the interaction of this region with P1, the substrate helix of the group I ribozyme reaction (5). Confirming the functional importance of this 2′-OH in a reaction analogous to the second step of splicing, substitution of A114 with 2′-deoxyadenosine results in inhibition of activity in a nucleotide analog interference mapping (NAIM) experiment (6).
A number of ribose zippers are observed in the crystal structures of the hepatitis delta virus (HDV) and hairpin ribozymes (7,8). Two ribose zippers are observed adjacent to the active site of the HDV ribozyme and are thought to help stabilize the core (8). 2′-Hydroxyls are also seen in hydrogen bonding patterns other than the ribose zipper: the 2′-OH of C75, the catalytic base of the HDV ribozyme, hydrogen bonds to the phosphate of G76 and the 2′-OH of U20 forms a hydrogen bond to the O4 of C75 (8).
Four 2′-OHs (A10, G11, A24 and C25) at the interface of loops A and B of the hairpin ribozyme were identified as functionally important by measuring the effect of site-specific 2′-deoxynucleotide substitution on the kcat of the cleavage reaction (9); three of these were also identified by a NAIM analysis of the ligation reaction performed under somewhat different conditions (10). Molecular modeling that took advantage of these results predicted two ribose zippers between the four nucleotides (11); one of the actual ribose zippers observed in the crystal structure of a hairpin ribozyme–inhibitor complex is identical to the prediction, while the other is similar (8), indicating that the functional importance of 2′-OHs can sometimes be used to accurately predict structure. It is interesting to note, however, that while the ribose zippers of the hairpin ribozyme are functionally important in the context of a minimal hairpin ribozyme (9) and lend thermodynamic stability to the docked form of this minimal ribozyme (12), they do not further stabilize the four-way junction form of the hairpin ribozyme nor are they functionally important for cleavage in this form (12), although it is in this form that they were observed in the crystal structure (8). This demonstrates that the stabilizing effects of hydrogen bonds may be context dependent (12).
The VS ribozyme performs an enzymatic cleavage reaction that produces the same termini (5′-OH and 2′,3′-cyclic phosphate) as the hammerhead, hairpin and HDV ribozymes (13,14), however it is structurally distinct from these other small nucleolytic ribozymes (15). The secondary structure of G11, a construct containing the minimal contiguous sequence and very little additional sequence (16), has been determined previously by chemical modification structure probing and site-directed mutagenesis (15) (Fig. 1A). The cis cleaving VS ribozyme can be split into a substrate molecule, consisting of stem–loop I, and a ribozyme molecule, consisting of helices II–VI; the ribozyme can cleave the substrate in trans in a multiple turnover reaction (17). The substrate and ribozyme domains interact, at least in part, through the formation of a kissing interaction between loop I and loop V (18). One role of this kissing interaction is to rearrange the secondary structure of stem–loop I from an inactive conformation to an active one (19) (Fig. 1A). The kissing interaction also has another role in promoting efficient self-cleavage, by an unknown mechanism (19).
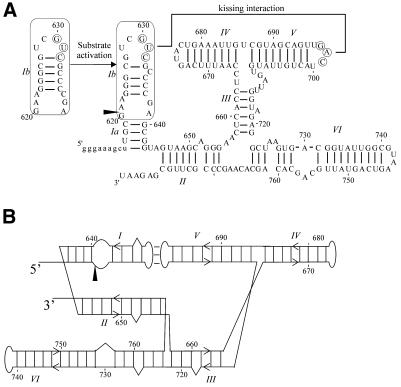
Figure 1.
(A) Secondary structure of G11. G11, a deletion construct of the VS ribozyme (16) consists of six helices (15) and undergoes a site-specific cleavage reaction at the phosphodiester indicated by the arrowhead. The minimal substrate domain is boxed. A 3 bp kissing interaction forms between the circled nucleotides in loops I and V, as indicated (18). In the absence of magnesium, the substrate exists in an inactive conformation; in the presence of magnesium and the kissing interaction, the secondary structure of the substrate is rearranged to form the active conformation (19,36). (B) Alternative representation of the secondary structure of G11 that accommodates structural and functional constraints (21,22). The cleavage site is indicated by the arrowhead.
A helical orientation model of the VS ribozyme has been developed that is consistent with the kissing interaction (18), a UV-inducible crosslink between helices II and VI (D.De Abreu and R.A.Collins, unpublished results), chemical modification interference analysis (20), phosphorothioate interference mapping (21), deletion analysis (22) and hydroxyl radical protection (23) (Fig. 1B). This model is also consistent with FRET analysis of an isolated three-way junction corresponding to the II–III–VI junction of the ribozyme (24). Recently, some structural features, including a uridine turn, of the III–IV–V junction have been elucidated (25). Little is yet known about the details of the three-dimensional structure of the VS ribozyme, but both base and backbone functional groups are likely to be involved in stabilization of the functional structure of the VS ribozyme. Within base paired regions of the ribozyme, most base identities are unimportant, whereas maintenance of the base pairs is required for activity (15,19,24,26); this suggests a potential role for backbone functional groups in tertiary interactions between base paired regions. In regions that are single stranded in the secondary structure, a large number of bases have been shown to be important for activity by site-directed mutagenesis and chemical modification interference studies (15,20,24,26; V.D.Sood and R.A.Collins, unpublished results). These regions are likely to form non-canonical structures which may also involve backbone functional groups. As only eight pro-Rp phosphate oxygens are functionally important for the cleavage reaction of G11 (21), and as the 2′-OH plays a large role in RNA tertiary structure stabilization and activity in many other RNAs (see above), we suspected that this backbone functional group would play a role in the self-cleavage activity of the VS ribozyme.
To identify all important 2′-OH groups in the VS ribozyme, we employed a combination of 2′-deoxynucleotide NAIM analysis on the cis cleavage reaction and site-specific 2′-deoxynucleotide substitution of the substrate for the trans cleavage reaction. A large number of important 2′-OHs were identified, highlighting the importance of this functional group in local and long-range tertiary structure stabilization in the VS ribozyme.
MATERIALS AND METHODS
Synthesis of 2′-deoxyphosphorothioate-modified RNA
Clone G11 (16) digested with SspI was transcribed using a mutant T7 RNA polymerase, Y639F (27), to incorporate dNTPαS into transcripts. A 100 µl in vitro transcription reaction contained 5 µg G11 DNA digested with SspI, 40 mM Tris–HCl pH 8, 5 mM DTT, 0.5 mM each rNTP, 125 µM of a particular dNTPαS (dATPαS, dCTPαS, dGTPαS and TTPαS, Amersham; dUMPαS, Glen Research) and 2.5 mM MgCl2; this low concentration of magnesium minimizes cleavage of the precursor ribozyme (28) and minimizes 3′-end heterogeneities caused by T7 RNA polymerase (23) during transcription. Transcription was carried out at 37°C for 30–60 min. The RNA was extracted with phenol/chloroform/isoamyl alcohol (25:24:1), precipitated with 300 mM sodium acetate and 2.5 vol ethanol, then redissolved in formamide loading buffer (80% v/v formamide, 35 mM EDTA, 0.5× TBE, 1% w/v xylene cyanol, 1% w/v bromphenol blue). The RNA was electrophoresed on 4% polyacrylamide–8.3 M urea gels, visualized by UV shadowing, cut from the gel, minced and eluted into 2 ml of diethylpyrocarbonate (DEPC)-treated water for 1 h at 65°C. The RNA was precipitated twice, 3′-end-labeled with 5′-[32P]pCp (29), purified by electrophoresis as described above and redissolved in DEPC-treated water. No more than 5% cleavage was typically seen after transcription and end-labeling, as estimated by the intensity of precursor and cleavage product bands during UV shadowing
NAIM analysis
Partially dAMPαS, dCMPαS, dGMPαS or dUMPαS substituted, 3′-end-labeled G11 transcripts were allowed to self-cleave at 37°C in solutions containing ∼50 nM RNA in cleavage buffer (40 mM Tris–HCl pH 8, 50 mM KCl, 2 mM spermidine, 25 mM MgCl2) for 10 min, at which time the bulk population was ∼50% cleaved. TMPαS-substituted RNA was also synthesized (before dUTPαS became commercially available) and subjected to the same self-cleavage conditions and NAIM analysis; the results were identical to the results obtained with dUMPαS (data not shown). Cleavage was stopped by placing reactions on ice and immediately precipitating. After precipitation, the pellets were dissolved in formamide loading dye. The precursor (Pre, uncleaved fraction) and downstream cleavage product (D, cleaved fraction) were separated by electrophoresis, visualized by autoradiography and purified as described above. Cleaved and uncleaved fractions of substituted transcripts were treated with iodine to cleave the phosphorothioate bonds (30), precipitated and dissolved in loading dye. Bands were detected by electrophoresis on 4 or 8% polyacrylamide–8.3 M urea gels followed by autoradiography. Gels were also exposed to PhosphorImager screens and band intensity was quantified using ImageQuant 3.3 (Molecular Dynamics).
To correct for possible loading differences, iodine cleavage bands were normalized to the average intensity of a non-interfering band, as described previously (21,31). Effects of nucleotide analog substitution at each position were quantified by dividing normalized band intensity in the cleaved fraction by normalized band intensity in the uncleaved fraction. Substitutions that inhibited self-cleavage activity resulted in a ratio (cleaved:uncleaved) between 0 and 1, while enhancement of activity resulted in a ratio >1. These ratios were plotted on a log scale to show inhibitory effects (ratios between 0 and 1) more clearly. Each experiment was performed 8–10 times and subjected to Student’s t-test to determine significance of the effect of nucleotide analog substitution at the α = 0.05 (95%) confidence level.
Trans cleavage of site-specifically modified stem–loop I by Rz646
Clone Rz646 (D.De Abreu and R.A.Collins, unpublished results), corresponding to VS nucleotides 646–881, was digested with SspI, transcribed in vitro with T7 RNA polymerase and purified from a 4% polyacrylamide–8.3 M urea gel as described above. Oligonucleotides corresponding to nucleotides 608–645 of G11 were synthesized containing either all ribonucleotides or site-specific 2′-deoxynucleotides at, separately, positions A621, A622, G623–C626 or C634–C637 (purchased from Dharmacon, Boulder, CO). An aliquot of 10 pmol each oligonucleotide was 5′-end-labeled with 12 pmol [γ-32P]ATP using 5 U T4 polynucleotide kinase (NEB) in the presence of 70 mM Tris–HCl pH 7.6, 10 mM MgCl2 and 5 mM DTT for 60 min at 37°C, followed by purification from a 16% polyacrylamide–8.3 M urea gel. The labeled oligonucleotides were incubated at a concentration of no more than 4 nM, in the presence of 200 nM Rz646, 50 mM Tris–HCl pH 8, 25 mM KCl, 2 mM spermidine and 50 mM MgCl2 at 30°C. Aliquots were removed into 2 vol of formamide loading buffer at various times and electrophoresed on 20% polyacrylamide–8.3 M urea gels, exposed to PhosphorImager screens and quantified using ImageQuant. The reactions were fitted to a first order exponential equation to obtain the pseudo first order rate constant. The mean rate constant from 4–7 time courses is given.
RESULTS
Identification of important 2′-OHs by NAIM analysis
To identify 2′-OHs that are important for self-cleavage by G11, we employed a NAIM protocol (32) using 2′-deoxyphosphorothioate nucleotides (dNMPαS). Since the sites of phosphorothioate inhibition of G11 self-cleavage are already known (21), the phosphorothioate can be used as a tag for detection of nucleotide analog incorporation, while any effects on cleavage activity that were not seen with the phosphorothioate-only NAIM analysis may be ascribed to the 2′-deoxy modification. Using a mutant T7 RNA polymerase that only poorly discriminates against 2′-deoxynucleotides (27), we synthesized and 3′-end-labeled pools of G11 randomly substituted at a low frequency with dAMPαS, dCMPαS, dGMPαS or dUMPαS and performed NAIM analysis on the self-cleavage reaction of these modified G11 pools. The results of the dNMPαS NAIM are shown in Figure 2 and Table 1.
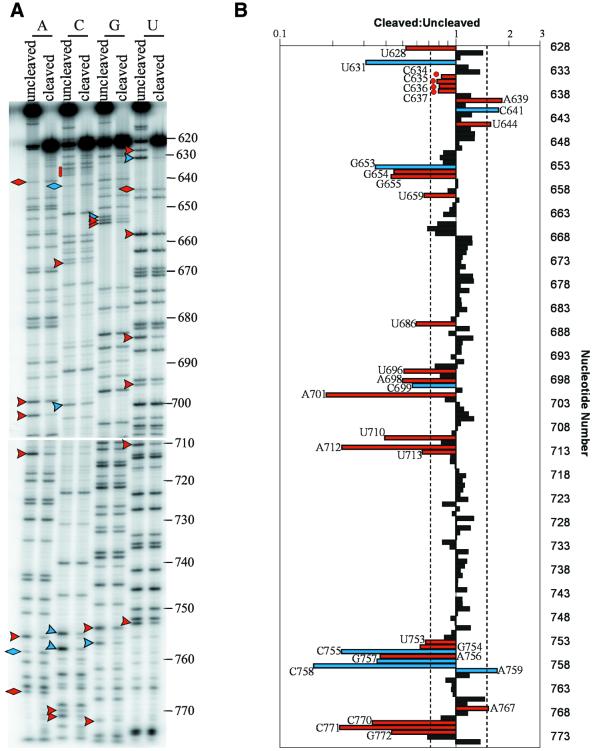
Figure 2.
NAIM analysis of the effects of 2′-deoxynucleotide substitution on G11 self-cleavage. (A) Populations of dNMPαS-substituted G11 were allowed to self-cleave for 10 min, purified, iodine treated and electrophoresed on 8% polyacrylamide–8 M urea gels. Representative autoradiograms are shown. Sites of inhibition are indicated with arrowheads and enhancements with diamonds. Blue symbols indicate sites of phosphorothioate only effects (21) and red symbols indicate new effects that are due to the absence of the 2′-OH. Red circles indicate important 2′-OHs in stem–loop I whose substitutions cause a small (<1.5-fold) but reproducible effect. (B) The results of the dNMPαS NAIM analysis were quantified using ImageQuant. The means of the ratios of the normalized band intensities in the cleaved to uncleaved fractions are represented graphically on a log scale. Dashed lines indicate 1.5-fold inhibitions or enhancements. Ratios that are not significantly different from 1 at the 95% confidence level as determined by Student’s t-test are shown as black bars; those that are significantly different from 1 and are >1.5-fold in magnitude are shown as colored bars, corresponding to the colors in (A). C634–C637 are significantly different from 1 but <1.5-fold in magnitude and therefore were also investigated in a different assay (see Fig. 3) and are marked with red circles. The P value for each significant effect was between 0.008 and 2.6 × 10–10, with most effects having a P value of <1 × 10–3.
Table 1. Summary of 2′-deoxynucleotide inhibitions and enhancements of G11 self-cleavage.
| Nucleotide | Sample size | Cleaved:uncleaved | SD |
|---|---|---|---|
| U628 | 10 | 0.52 | 0.26 |
| C634 | 10 | 0.83 | 0.14 |
| C635 | 10 | 0.78 | 0.12 |
| C636 | 10 | 0.81 | 0.21 |
| C637 | 10 | 0.80 | 0.16 |
| A639 | 8 | 1.83 | 0.72 |
| U644 | 10 | 1.57 | 0.24 |
| G654 | 8 | 0.45 | 0.09 |
| G655 | 8 | 0.43 | 0.08 |
| U659 | 10 | 0.66 | 0.34 |
| U686 | 10 | 0.60 | 0.33 |
| U696 | 10 | 0.51 | 0.21 |
| A698 | 10 | 0.50 | 0.08 |
| A701 | 10 | 0.18 | 0.09 |
| U710 | 10 | 0.40 | 0.14 |
| A712 | 10 | 0.23 | 0.12 |
| U713 | 10 | 0.65 | 0.18 |
| U753 | 9 | 0.67 | 0.25 |
| G754 | 8 | 0.63 | 0.12 |
| A756 | 10 | 0.37 | 0.08 |
| A767 | 10 | 1.60 | 0.63 |
| C770 | 10 | 0.34 | 0.21 |
| C771 | 10 | 0.22 | 0.14 |
| G772 | 8 | 0.43 | 0.23 |
All effects of 2′-deoxynucleotide substitution that were found to be statistically significant at the 95% confidence level (red symbols in Fig. 1) are listed. The ratio of cleaved:uncleaved was determined as described in the Materials and Methods. The average of 8–10 experiments (sample size) is given with one standard deviation (SD).
Most of the effects that were observed in the phosphorothioate NAIM (21) were also observed upon dNMPαS substitution (Fig. 2, blue symbols), with the exception of the phosphorothioate inhibition at G640. One explanation for this apparent discrepancy is that the 2′-OH of G640 is involved in an inhibitory interaction while the phosphate is involved in a functionally positive interaction, such that substitution of the phosphate results in a decrease in cleavage rate, while removal of the 2′-OH results in an increase in cleavage rate; simultaneous substitution of the phosphate and removal of the 2′-OH may cancel each other out, resulting in no overall effect on cleavage rate. The location of G640 near a previously identified inhibitory structural element (20,22) is consistent with the idea that its 2′-OH is involved in an inhibitory interaction. In addition to the previously observed phosphorothioate effects, a number of new effects due to dNMPαS substitution were observed (Fig. 2, red symbols) and these are most likely due to absence of the 2′-OH group. Most effects <1.5-fold in magnitude were not statistically significant effects and were not pursued further, as they are probably not important to the cleavage reaction. The inhibitions at C634–C637 in stem–loop I, however, while <1.5-fold in magnitude, were highly reproducible and were confirmed to be inhibitory by site-specific substitution (see below) and thus are highlighted in Figure 2 (red circles). Also observed were 17 statistically significant 2′-deoxynucleotide inhibitions and three enhancements >1.5-fold in magnitude (Fig. 2, red arrowheads and diamonds, respectively).
Trans cleavage of stem–loop I containing site-specific 2′-deoxy modifications
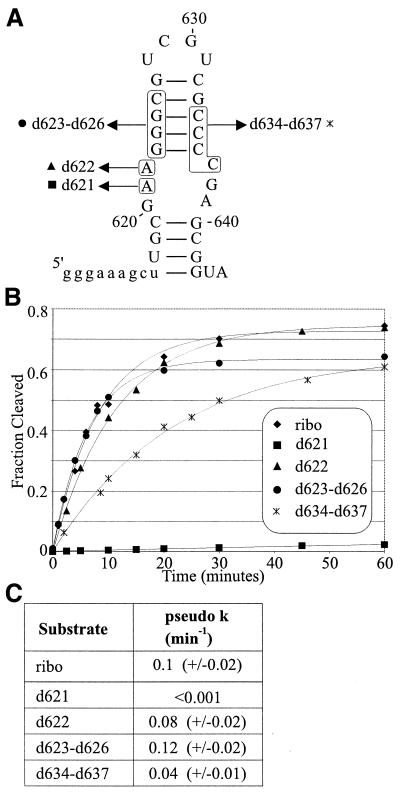
Some positions in stem–loop I could not be examined for an effect of 2′-deoxy substitution by the NAIM method. These are: A621, because the iodine cleavage product co-migrates with G11D, the product of ribozyme cleavage; A622, because phosphorothioate substitution alone abolishes cleavage activity (21); and G623–C626, because these bands migrate together in a compression artifact, due to the stability of the GC-rich stem (17). Finally, C634–C637 displayed highly reproducible inhibition in the NAIM analysis, however the magnitude of the effect was small. In order to determine or confirm the effect of 2′-deoxy substitution at these groups of nucleotides, oligonucleotides corresponding to stem–loop I were synthesized with site-specific 2′-deoxynucleotide modifications at each of these positions (Fig. 3A). These were cleaved in trans by Rz646, a trans-acting ribozyme consisting of nucleotides 646–783, and the results are shown in Figure 3B and C. The time course of cleavage and the relative pseudo first order rate constants indicate that, while substitution at A622 or G623–C626 has no effect under these conditions, substitutions of 2′-deoxynucleotides at C634–C637 is moderately deleterious for cleavage and substitution of a 2′-deoxynucleotide at A621 is extremely deleterious, resulting in a pseudo first order rate constant that is at least 100-fold decreased from the all-ribonucleotide version of the substrate.
Figure 3.
The effect of site-specific 2′-deoxynucleotide substitution in stem–loop I on cleavage by a trans-acting ribozyme. (A) Substrates for trans cleavage. An oligonucleotide corresponding to nucleotides 608–645 of G11 was synthesized in either an all-ribonucleotide version (ribo) or with 2′-deoxynucleotide substitutions (d) as indicated by the boxed sequences. The symbols next to each boxed substitution correspond to the graph in (B). (B) Time course of cleavage of stem–loop I substrates by Rz646. The substrates shown in (A) were incubated in the presence of 200 nM Rz646, a trans-acting ribozyme consisting of nucleotides 646–783 of G11. The fraction cleaved is plotted as a function of time. The curves represent the fit to a first order exponential equation. (C) Pseudo first order rate constants of cleavage were obtained by fitting time courses of cleavage to a first order exponential equation and the average is given with the standard deviation for each substrate used.
DISCUSSION
We employed a combination of dNMPαS NAIM analysis and site-specific substitution with 2′-deoxynucleotides to obtain a complete map of all 2′-OHs that are important for site-specific cleavage by the VS ribozyme, summarized in Figure 4. There are a large number of 2′-OHs that contribute to the cleavage activity of the VS ribozyme; these are all located within the region defined by deletion mutagenesis to be the core of the ribozyme (22). In contrast to phosphorothioate inhibitions (21), which are all located in single-stranded regions, 2′-deoxynucleotide inhibitions are not uniformly located within any one type of secondary structure element, but distributed throughout helices, hairpin loops, internal loops and multi-stem junctions.
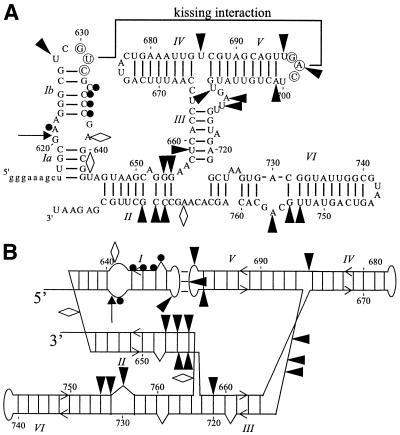
Figure 4.
(A) Superimposed on the secondary structure of G11 (15) are nucleotides that display an inhibitory effect on self-cleavage when substituted with 2′-deoxynucleotides in a NAIM experiment (filled arrowheads) and those that display a stimulatory effect (open diamonds). Nucleotides that were tested using site-specific 2′-deoxynucleotide substitutions in a trans cleavage assay and which display inhibitory effects are indicated by filled circles. The cleavage site is indicated by the arrow. (B) Important 2′-OHs are highlighted on the alternative representation of the secondary structure model. Symbols are as in (A).
At three positions in the ribozyme (A639, U644 and A767) 2′-deoxynucleotide substitution actually causes an enhancement of self-cleavage (Fig. 1). This may be due to the removal of an inhibitory hydrogen bond by dNMPαS substitution and, indeed, A639 and U644 are located in a known inhibitory region of G11 (15,22). Conversely, any of these riboses may adopt an unusual sugar pucker, for example a 2′-endo conformation, which is more easily adopted by 2′-deoxynucleotides than by ribonucleotides (33); unusual sugar puckers have been observed to be important in other RNAs in regions of non-canonical structure (6,34) and this may also be the case in the VS ribozyme.
Two functionally important 2′-OHs occur in loops I and V, at U628 and U696, just upstream of the nucleotides involved in the loop I–loop V kissing interaction. Both of these uridine nucleotides occur in triplets that conform to the consensus sequence of U-turns (18) and the presence of the functionally important 2′-OHs is consistent with a U-turn hypothesis. U-turns are often found in kissing interactions (3,35), where they stabilize a solvent-exposed A-form conformation of the following nucleotides, thereby pre-organizing those nucleotides for kissing interaction formation. The classic example of this is the U-turn in the anticodon loop of tRNAPhe, which organizes the anticodon nucleotides for base pairing to the codon (3). Another functionally important 2′-OH was found at U710 in the III–IV–V junction; U710 has also been shown to be involved in a U-turn that is required to organize the III–IV–V junction structure (25). There are three additional 2′-OHs in the III–IV–V junction that likely contribute to a network of interactions that stabilize the correct junction structure.
In an A-form helix the 2′-OHs line the minor groove along one face of the helix and are available for hydrogen bonding interactions (33). Within stem–loop I, four important 2′-OHs were identified at C634–C637. These 2′-OHs may be involved in the conformational rearrangement that occurs in stem–loop I (36), perhaps by stabilizing the cleavable conformation. Another possibility (not mutually exclusive with the first) is that the 2′-OHs of C634–C637 may be involved in the docking of stem–loop I into the ribozyme. In the model of G11 that orients the helices (Fig. 4B), helices II and VI are approximately parallel to each other and to stem–loop I. The presence of clusters of important 2′-OHs in helix II (nucleotides 654, 655 and 770–772) and helix VI (U753, G754 and A756) suggests a role for these 2′-OHs in helix packing interactions with each other and with stem–loop I. Consistent with a role in helix packing, these regions overlap with regions of strong protection from hydroxyl radical cleavage (23), as would be expected in a tightly packed core stabilized by many hydrogen bonds. Many of the hydroxyl radical protections are weaker in the absence of stem–loop I (23), suggesting that stem–loop I binds in the cleft created by helices II and VI; this interaction may be mediated by the 2′-OHs of all three helices.
It has previously been proposed that helix VI contributes to the active site of the ribozyme (21,26). There are a number of important 2′-OHs in both helix VI (at U753, G754 and A756) and the loop in stem–loop I that contain the cleavage site (at A621). Most of these have only a moderate effect on cleavage activity when substituted with 2′-deoxynucleotides, ranging from ∼1.5- to 2.7-fold inhibition, suggesting that these 2′-OHs are not individually essential for catalysis. These modest reductions in activity are consistent with these 2′-OHs forming hydrogen bonds that help stabilize local loop structures or form long-range hydrogen bonds important for interaction between stem–loop I and helix VI. At A621 in stem–loop I, adjacent to the ribozyme cleavage site, there is little detectable cleavage of a substrate containing a 2′-deoxynucleotide at this position after 60 min in a >50-fold ribozyme excess. This suggests an important role for this 2′-OH in site-specific cleavage by the VS ribozyme. As with the other important 2′-OHs in stem–loop I and helix VI, it is possible that the 2′-OH of A621 might help stabilize the correct structure of the cleavage site loop by interactions with other functional groups in the loop, or it may be required to mediate docking of the scissile phosphodiester into the active site of the ribozyme. A third possibility, by analogy to the 2′-OH of the guanosine nucleophile in the Tetrahymena group I intron ribozyme, which is thought to coordinate a metal ion that can stabilize the transition state of that transesterification reaction (37,38), is that the 2′-OH of A621 is involved in transition state stabilization by coordinating a catalytically important metal ion. Irrespective of the specific roles of each 2′-OH, the clusters of important 2′-OHs in the loop containing the cleavage site and near the candidate active site of the ribozyme support the functional importance of both these regions and, along with the important 2′-OHs in the base paired regions of stem I and helix II, suggest that a network of hydrogen bonds helps determine the active structure of the ribozyme and may contribute to catalysis.
The hydrogen bond donor and acceptor capabilities of the 2′-OH make this functional group extremely versatile in mediating tertiary interactions in a number of RNAs. In the hairpin (8), hammerhead (39–42) and HDV ribozymes (7), a small number of 2′-OHs in the catalytic cores of each ribozyme make functional contributions. In the VS ribozyme, however, we have identified a large number of 2′-OHs that are important for cleavage activity. The larger size of this ribozyme may require more tertiary interactions to stabilize the three-dimensional fold, consistent with the larger numbers of 2′-OHs that contribute to catalysis, structure, stability and substrate binding in the group I (5,6,43–47) and group II (48) introns. At least one 2′-OH in the VS ribozyme is located near enough to the cleavage site to be a likely candidate for participation in catalysis. The location of other important 2′-OHs in specific regions of the VS ribozyme suggest local interactions, as in the three U-turns, and long-range tertiary interactions, as in the proposed packing of the helical regions in the helical orientation model. The identification of important 2′-OHs provides a large number of functional constraints with which to build and analyze models of the VS ribozyme structure.
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. Zamel, D. Jaikaran and S. Hiley for comments on the manuscript. V.D.S. was supported by Fonds FCAR (Formation de Chercheurs et l’Aide à la Recherche), the University of Toronto and the Ontario Provincial Government. This work was supported by a grant from the Canadian Institutes of Health Research to R.A.C.
REFERENCES
- 1.Quigley G.J., Seeman,N.C., Wang,A.H., Suddath,F.L. and Rich,A. (1975) Yeast phenylalanine transfer RNA: atomic coordinates and torsion angles. Nucleic Acids Res., 2, 2329–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quigley G.J., Wang,A.H., Seeman,N.C., Suddath,F.L., Rich,A., Sussman,J.L. and Kim,S.H. (1975) Hydrogen bonding in yeast phenylalanine transfer RNA. Proc. Natl Acad. Sci. USA, 72, 4866–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quigley G.J. and Rich,A. (1976) Structural domains of transfer RNA molecules. Science, 194, 796–806. [DOI] [PubMed] [Google Scholar]
- 4.Jack A., Ladner,J.E., Rhodes,D., Brown,R.S. and Klug,A. (1977) A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J. Mol. Biol., 111, 315–328. [DOI] [PubMed] [Google Scholar]
- 5.Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- 6.Ortoleva-Donnelly L., Szewczak,A.A., Gutell,R.R. and Strobel,S.A. (1998) The chemical basis of adenosine conservation throughout the Tetrahymena ribozyme. RNA, 4, 498–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferré-d’Amare A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- 8.Rupert P.B. and Ferré-d’Amare,A.R. (2001) Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature, 410, 780–786. [DOI] [PubMed] [Google Scholar]
- 9.Chowrira B.M., Berzal-Herranz,A., Keller,C.F. and Burke,J.M. (1993) Four ribose 2′-hydroxyl groups essential for catalytic function of the hairpin ribozyme. J. Biol. Chem., 268, 19458–19462. [PubMed] [Google Scholar]
- 10.Ryder S.P. and Strobel,S.A. (1999) Nucleotide analog interference mapping of the hairpin ribozyme: implications for secondary and tertiary structure formation. J. Mol. Biol., 291, 295–311. [DOI] [PubMed] [Google Scholar]
- 11.Earnshaw D.J., Masquida,B., Muller,S., Sigurdsson,S.T., Eckstein,F., Westhof,E. and Gait,M.J. (1997) Inter-domain cross-linking and molecular modelling of the hairpin ribozyme. J. Mol. Biol., 274, 197–212. [DOI] [PubMed] [Google Scholar]
- 12.Klostermeier D. and Millar,D.P. (2001) Tertiary structure stability of the hairpin ribozyme in its natural and minimal forms: different energetic contributions from a ribose zipper motif. Biochemistry, 40, 11211–11218. [DOI] [PubMed] [Google Scholar]
- 13.Saville B.J. and Collins,R.A. (1990) A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell, 61, 685–696. [DOI] [PubMed] [Google Scholar]
- 14.Saville B.J. and Collins,R.A. (1991) RNA-mediated ligation of self-cleavage products of a Neurospora mitochondrial plasmid transcript. Proc. Natl Acad. Sci. USA, 88, 8826–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beattie T.L., Olive,J.E. and Collins,R.A. (1995) A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl Acad. Sci. USA, 92, 4686–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo H.C., De Abreu,D.M., Tillier,E.R., Saville,B.J., Olive,J.E. and Collins,R.A. (1993) Nucleotide sequence requirements for self-cleavage of Neurospora VS RNA. J. Mol. Biol., 232, 351–361. [DOI] [PubMed] [Google Scholar]
- 17.Guo H.C. and Collins,R.A. (1995) Efficient trans-cleavage of a stem–loop RNA substrate by a ribozyme derived from Neurospora VS RNA. EMBO J., 14, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rastogi T., Beattie,T.L., Olive,J.E. and Collins,R.A. (1996) A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J., 15, 2820–2825. [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen A.A. and Collins,R.A. (2001) Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc. Natl Acad. Sci. USA, 98, 7730–7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beattie T.L. and Collins,R.A. (1997) Identification of functional domains in the self-cleaving Neurospora VS ribozyme using damage selection. J. Mol. Biol., 267, 830–840. [DOI] [PubMed] [Google Scholar]
- 21.Sood V.D., Beattie,T.L. and Collins,R.A. (1998) Identification of phosphate groups involved in metal binding and tertiary interactions in the core of the Neurospora VS ribozyme. J. Mol. Biol., 282, 741–750. [DOI] [PubMed] [Google Scholar]
- 22.Rastogi T. and Collins,R.A. (1998) Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J. Mol. Biol., 277, 215–224. [DOI] [PubMed] [Google Scholar]
- 23.Hiley S.L. and Collins,R.A. (2001) Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J., 20, 5461–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafontaine D.A., Norman,D.G. and Lilley,D.M. (2001) Structure, folding and activity of the VS ribozyme: importance of the 2-3-6 helical junction. EMBO J., 20, 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sood V.D. and Collins,R.A. (2001) Functional equivalence of the uridine turn and the hairpin as building blocks of tertiary structure in the Neurospora VS ribozyme. J. Mol. Biol., 313, 1013–1019. [DOI] [PubMed] [Google Scholar]
- 26.Lafontaine D.A., Wilson,T.J., Norman,D.G. and Lilley,D.M. (2001) The A730 loop is an important component of the active site of the VS ribozyme. J. Mol. Biol., 312, 663–674. [DOI] [PubMed] [Google Scholar]
- 27.Sousa R. and Padilla,R. (1995) A mutant T7 RNA polymerase as a DNA polymerase. EMBO J., 14, 4609–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins R.A. and Olive,J.E. (1993) Reaction conditions and kinetics of self-cleavage of a ribozyme derived from Neurospora VS RNA. Biochemistry, 32, 2795–2799. [DOI] [PubMed] [Google Scholar]
- 29.Bruce A.G. and Uhlenbeck,O.C. (1978) Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res., 5, 3665–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gish G. and Eckstein,F. (1988) DNA and RNA sequence determination based on phosphorothioate chemistry. Science, 240, 1520–1522. [DOI] [PubMed] [Google Scholar]
- 31.Christian E.L. and Yarus,M. (1992) Analysis of the role of phosphate oxygens in the group I intron from Tetrahymena. J. Mol. Biol., 228, 743–758. [DOI] [PubMed] [Google Scholar]
- 32.Strobel S.A. and Shetty,K. (1997) Defining the chemical groups essential for Tetrahymena group I intron function by nucleotide analog interference mapping. Proc. Natl Acad. Sci. USA, 94, 2903–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saenger W. (1984) In Cantor,C.R. (ed.), Principles of Nucleic Acid Structure. Springer-Verlag, New York, NY, p. 61.
- 34.Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Szewczak,A.A., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) RNA tertiary structure mediation by adenosine platforms. Science, 273, 1696–1699. [DOI] [PubMed] [Google Scholar]
- 35.Franch T., Petersen,M., Wagner,E.G., Jacobsen,J.P. and Gerdes,K. (1999) Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol., 294, 1115–1125. [DOI] [PubMed] [Google Scholar]
- 36.Andersen A.A. and Collins,R.A. (2000) Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell, 5, 469–478. [DOI] [PubMed] [Google Scholar]
- 37.Shan S., Yoshida,A., Sun,S., Piccirilli,J.A. and Herschlag,D. (1999) Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc. Natl Acad. Sci. USA, 96, 12299–12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shan S.O. and Herschlag,D. (1999) Probing the role of metal ions in RNA catalysis: kinetic and thermodynamic characterization of a metal ion interaction with the 2′-moiety of the guanosine nucleophile in the Tetrahymena group I ribozyme. Biochemistry, 38, 10958–10975. [DOI] [PubMed] [Google Scholar]
- 39.Perreault J.P., Labuda,D., Usman,N., Yang,J.H. and Cedergren,R. (1991) Relationship between 2′-hydroxyls and magnesium binding in the hammerhead RNA domain: a model for ribozyme catalysis. Biochemistry, 30, 4020–4025. [DOI] [PubMed] [Google Scholar]
- 40.Yang J.H., Usman,N., Chartrand,P. and Cedergren,R. (1992) Minimum ribonucleotide requirement for catalysis by the RNA hammerhead domain. Biochemistry, 31, 5005–5009. [DOI] [PubMed] [Google Scholar]
- 41.Williams D.M., Pieken,W.A. and Eckstein,F. (1992) Function of specific 2′-hydroxyl groups of guanosines in a hammerhead ribozyme probed by 2′ modifications. Proc. Natl Acad. Sci. USA, 89, 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimayama T., Nishikawa,F., Nishikawa,S. and Taira,K. (1993) Nuclease-resistant chimeric ribozymes containing deoxyribonucleotides and phosphorothioate linkages. Nucleic Acids Res., 21, 2605–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bevilacqua P.C. and Turner,D.H. (1991) Comparison of binding of mixed ribose-deoxyribose analogues of CUCU to a ribozyme and to GGAGAA by equilibrium dialysis: evidence for ribozyme specific interactions with 2′ OH groups. Biochemistry, 30, 10632–10640. [DOI] [PubMed] [Google Scholar]
- 44.Pyle A.M., Murphy,F.L. and Cech,T.R. (1992) RNA substrate binding site in the catalytic core of the Tetrahymena ribozyme. Nature, 358, 123–128. [DOI] [PubMed] [Google Scholar]
- 45.Strobel S.A., Ortoleva-Donnelly,L., Ryder,S.P., Cate,J.H. and Moncoeur,E. (1998) Complementary sets of noncanonical base pairs mediate RNA helix packing in the group I intron active site. Nature Struct. Biol., 5, 60–66. [DOI] [PubMed] [Google Scholar]
- 46.Ortoleva-Donnelly L., Kronman,M. and Strobel,S.A. (1998) Identifying RNA minor groove tertiary contacts by nucleotide analogue interference mapping with N2-methylguanosine. Biochemistry, 37, 12933–12942. [DOI] [PubMed] [Google Scholar]
- 47.Szewczak A.A., Ortoleva-Donnelly,L., Ryder,S.P., Moncoeur,E. and Strobel,S.A. (1998) A minor groove RNA triple helix within the catalytic core of a group I intron. Nature Struct. Biol., 5, 1037–1042. [DOI] [PubMed] [Google Scholar]
- 48.Boudvillain M. and Pyle,A.M. (1998) Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J., 17, 7091–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]