Abstract
Caveolin-1 (CAV1) is a membrane-sculpting protein that oligomerizes to generate flask-shaped invaginations of the plasma membrane known as caveolae. Mutations in CAV1 have been linked to multiple diseases in humans. Such mutations often interfere with oligomerization and the intracellular trafficking processes required for successful caveolae assembly, but the molecular mechanisms underlying these defects have not been structurally explained. Here, we investigate how a disease-associated mutation in one of the most highly conserved residues in CAV1, P132L, affects CAV1 structure and oligomerization. We show that P132 is positioned at a major site of protomer–protomer interactions within the CAV1 complex, providing a structural explanation for why the mutant protein fails to homo-oligomerize correctly. Using a combination of computational, structural, biochemical, and cell biological approaches, we find that despite its homo-oligomerization defects P132L is capable of forming mixed hetero-oligomeric complexes with WT CAV1 and that these complexes can be incorporated into caveolae. These findings provide insights into the fundamental mechanisms that control the formation of homo- and hetero-oligomers of caveolins that are essential for caveolae biogenesis, as well as how these processes are disrupted in human disease.
Keywords: caveolin, caveolae, membrane protein, computational biology, structural biology, oligomerization, cancer, intracellular trafficking
Flask-shaped invaginations of the plasma membrane known as caveolae function as critical regulators of human health and disease (1, 2, 3). The integral membrane protein caveolin-1 (CAV1) is a major structural component of caveolae and is required for caveolae formation in nonmuscle cells (4). Its intrinsic ability to induce membrane curvature enables it to trigger the formation of caveolae-like structures even in bacteria (5, 6). In mammals, CAV1 and caveolae are widely distributed in many tissues where they function in mechanosensation, lipid homeostasis, signaling, endocytosis, and mechanoprotection (1, 7, 8, 9). Conversely, dysregulation of CAV1 and caveolae contributes to the development and progression of diseases such as cancer, asthma, pulmonary fibrosis, pulmonary arterial hypertension, chronic inflammatory respiratory diseases, and lipodystrophy (10, 11, 12, 13, 14, 15, 16).
Assembly of CAV1 into oligomeric complexes is an essential step in caveolae biogenesis, and defects in oligomerization can give rise to disease (17, 18, 19, 20, 21). Under normal conditions, CAV1 undergoes an initial oligomerization step that generates complexes ∼8S in size when analyzed by velocity gradient centrifugation, termed 8S complexes (20). 8S complexes then can form higher order 70S complexes that are ultimately incorporated into caveolae together with cavins and accessory proteins (20, 21, 22, 23, 24). Oligomerization of CAV1 depends on the presence of a region of the protein known as the oligomerization domain (18). Disruption of other regions of CAV1 can also interfere with its oligomerization, suggesting that the conformation of caveolins is highly optimized (19).
One important mutation known to interfere with oligomer formation is a proline (P) to leucine (L) mutation in one of the most highly conserved residues across caveolins, P132 (25, 26). Identified in 16% of primary human breast cancers, P132L was subsequently shown to impact cell migration, invasion, and metastasis (27, 28, 29). Although the prevalence of the P132L mutation in breast cancer was later questioned (30, 31, 32, 33), an identical mutation has been identified in lung adenocarcinomas (34), and a related P132A mutation has been identified as a genetic variant linked to amyotrophic lateral sclerosis (35). Furthermore, an equivalent mutation in caveolin-3 (CAV3), P105L (denoted as P104L in many studies), gives rise to muscular dystrophies both in humans and model organisms (36, 37, 38, 39). These findings highlight the importance of P132 in caveolin function in health and disease.
CAV1 P132L exhibits a number of defects compared to its WT counterpart. Instead of assembling into 8S oligomers, P132L forms a combination of monomers/dimers and high-molecular-weight complexes (19, 20, 40, 41). It also is retained intracellularly (19, 20, 40). Oligomerization and trafficking defects have likewise been observed for P105L mutants of CAV3 (37, 42). P132L has also been reported to disrupt the ability of WT CAV1 to traffic to the plasma membrane correctly, suggesting it can function as a dominant negative (40). P132L is thus a powerful tool to investigate how defects in trafficking and oligomerization of caveolins interfere with caveolae formation and other functions of the protein (5, 19, 20, 28, 43, 44, 45, 46, 47).
Despite the importance of P132 to caveolin biology, due to the lack of a high-resolution structure of CAV1, it has not been possible to study how this residue contributes to the structure and function of the protein. We have now determined a 3.5 Å cryo-EM structure of the human CAV1 8S complex (48). The complex consists of 11 CAV1 protomers organized into a disc with a central protruding β-barrel at the center (48). The structure uncovers extensive interprotomer interactions that extend along the entire length of each protomer, locked in place by a previously unidentified pin motif close to the N-terminus and a C-terminal β-barrel extending from the cytoplasmic face of the disc (48).
In the current study, we use the structure as a framework for investigating the molecular basis for how the P132L mutation alters CAV1 structure and oligomerization states. We propose a new structure-based model predicting how the P132L mutation disrupts homo-oligomerization. We also provide evidence that P132L can form hetero-oligomeric complexes with WT CAV1 and that these complexes are capable of supporting caveolae biogenesis. These findings provide new insights into the mechanisms that control caveolae biogenesis at a molecular level and the structural impact of disease-associated mutants on this process. They may also help lead to the development of therapeutic tools for the treatment of disease.
Results
P132 contributes to multiple protomer–protomer interfaces in the CAV1 complex
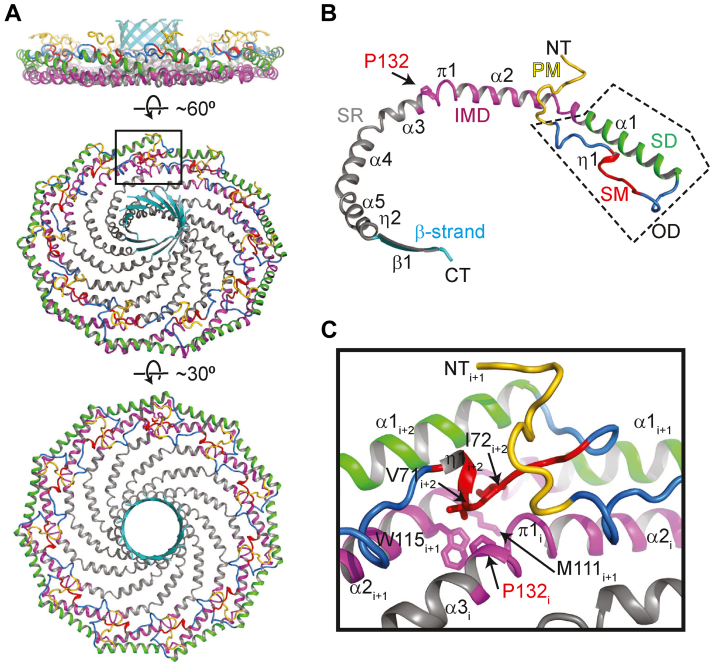
We first examined how P132 contributes to the structure of 8S complexes. Based on the cryo-EM structure, P132 falls in α-helix 3 of the highly hydrophobic region of CAV1 termed the “intramembrane domain” (IMD). In the cryo-EM structure, the IMD corresponds to a predominantly helical structure lying along the plane parallel to the membrane surface (48) (Fig. 1). The positioning of the IMD in the complex suggests it fulfills two distinct functions: forming a portion of the flat membrane-facing surface of the complex (Fig. 1A) and participating in numerous protomer–protomer interactions stabilizing the integrity of the complex. Within the IMD, P132 is located in a hydrophobic pocket between adjacent protomers, suggesting it primarily contributes to the packing of the complex rather than mediating membrane binding events (Fig. 1, A–C).
Figure 1.
P132 is located at a major protomer–protomer interface in the CAV1 complex.A, side, tilted, and en face views of the structure of the 8S CAV1 complex are shown in ribbon representation and colored as in panel B. B, structure of CAV1 protomer. The previously defined regions are labeled and colored: PM, pin motif (yellow); SM, signature motif (red); SD, scaffolding domain (green); IMD, intermembrane domain (purple); SR, spoke region (gray); and β-strand (cyan). The oligomerization domain (OD), which contains the SM and SD, is indicated by the dashed box. P132 is shown in stick representation and indicated by an arrow. C, zoomed up view of the boxed region in panel A. Select residues are shown in stick representation and labeled.
To probe the potential roles of P132 in supporting oligomerization, we examined its proximity to other protomers and conserved residues within this region. To facilitate this analysis, we refer to three adjacent protomers, i, i+1, and i+2 (Fig. 1C). The sidechain of P132 of chain i points into a hydrophobic pocket formed by M111 and the highly conserved W115 of the protomer i+1 on the membrane-facing side and V71 and I72 of the protomer i+2 on the cytoplasm-facing side. Notably, V71 and I72 are located in the signature motif of CAV1, the most strongly conserved region of the protein across members of the caveolin gene family (49, 50) (Fig. S1). Thus, P132 makes important contacts with other conserved hydrophobic residues in several adjacent protomers within the complex. Furthermore, there is little space in the hydrophobic pocket to accommodate amino acids with larger side chains without structural rearrangements on the backbone atoms on the neighboring protomer. Mutation of P132 could also potentially affect the angle of the kink located in the middle of the nearby π helix. Mutations at this location of the structure could thus potentially alter the ability of CAV1 protomers to tightly pack into a stable oligomer via multiple mechanisms.
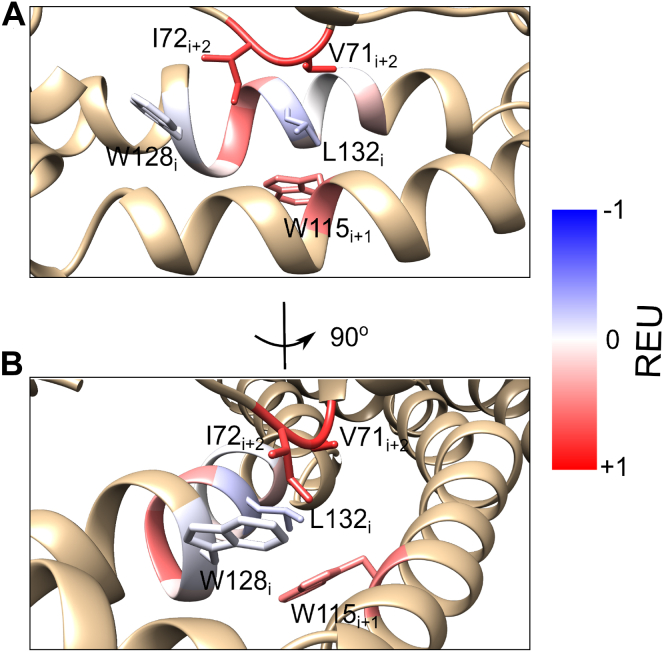
To explore the structural impact of the P132L mutation further, we used a computational approach to examine its effect on 12 neighboring residues that fall within 8 Å of this position (Table 1). This group includes four residues on either side of P132 on the same protomer (128–131 and 133–136 of chain i) and highly conserved residues on adjacent protomers i+2 (V71, I72) and i+1 (W115) in the structure (Fig. 2). For these studies, we used Rosetta to calculate the ddG for each of these residues induced by the P132L mutation (Table 1 and Fig. 2). Positive ddG values indicate that the respective mutation is not energetically favorable, while negative ddG values indicate that the mutation is energetically favorable. For this analysis, we focused primarily on local changes, without allowing for large-scale rearrangement of the CAV1 complex structure; the impact of the P132L mutation on the global stability of the CAV1 complex is analyzed further subsequently. Most residues within the vicinity of P132 had positive ddG values in response to the P132L mutation, suggesting this mutation is not favorable. Part of this effect was caused by the destabilization of the helix backbone due to the replacement of a restrained proline with a leucine residue as observed for the protomer i residues facing away from L132, and part of it was caused by the clashes induced by the bulkier leucine side chain that faced other hydrophobic residues in the vicinity. Bulky residues such as V71i+2, I72i+2, and W115i+1 showed the largest increase in ddG, which was also reflected by the movement of the V71i+2 and W115i+1 sidechains away from the leucine residue in the mutant structure. These calculations suggest P132L disrupts both the stability of the helix backbone and the hydrophobic packing of the residues in its vicinity. In combination, these disruptions would be predicted to alter the ability of mutant protomers to pack into an organized oligomeric complex.
Table 1.
Effect of P132L mutation on the predicted stability of adjacent residues
| Residue | Rosetta ddG | ConSurf score |
|---|---|---|
| W115i+1 | 0.51 | 5 |
| W128i | −0.09 | 9 |
| A129i | 0.10 | 4 |
| V130i | 0.55 | 6 |
| V131i | 0.41 | 6 |
| P132i | −0.21 | 9 |
| C133i | 0.10 | 5 |
| I134i | 0.01 | 5 |
| K135i | −0.05 | 6 |
| S136i | 0.22 | 5 |
| V71i+2 | 0.81 | 8 |
| I72i+2 | 0.74 | 8 |
Per-residue Rosetta energies of 12 amino acids within 8 Å of P132L were subtracted from that of the P132P mutation. Residues predicted to be most strongly impacted by the P132L mutation are highlighted in bold. ConSurf scores (84) are provided for each residue (range of 1–9, where 1 = variable, 5 = average, and 9 = highly conserved). See Figures 1 and 2 for the positioning of protomers i, i+1, and i+2 in the complex. All ddG units are in Rosetta Energy Units (REU).
Figure 2.
Impact of the P132L mutation on the stability of nearby residues.A, Rosetta ddG scores calculated for the residues within 8 Å of L132 are color coded on the mutant structure. Blue indicates the P132L mutation has a stabilizing effect on the residue, and red indicates P132L’s effect is destabilizing. B, as in A, except rotated 90 degrees.
Computational modeling predicts that substituting any other residue for P132 destabilizes CAV1 homo-oligomers
We next asked how the structural alterations introduced by the P132L mutation change the ability of CAV1 to homo-oligomerize. For comparison, we studied two other hydrophobic residues positioned at protomer-protomer interfaces, L122 and V130 (Fig. S2). Several naturally occurring genetic CAV1 variants in humans have been reported for both of these residues (Table 2). As an additional control, we studied F99. F99 is located at the outermost rim of the complex and is predicted to make contact with the membrane (Fig. S2).
Table 2.
Examples of naturally occurring genetic variants of CAV1
| Variant | Raw CADD | PHRED | Allele frequency | SIFT | SIFT category | PolyPhen | PolyPhen category | Rosetta ddG |
|---|---|---|---|---|---|---|---|---|
| P132L | 3.91 | 27.00 | 0.000003984 | 0.00 | Deleterious | 1.00 | Probably damaging | 3.78 |
| P132A | 3.50 | 25.00 | 0.0000239 | 0.00 | Deleterious | 1.00 | Probably damaging | 1.97 |
| P132S | 3.70 | 25.70 | 0.000003984 | 0.00 | Deleterious | 1.00 | Probably damaging | 2.86 |
| V130F | 3.99 | 27.60 | 0.000003984 | 0.00 | Deleterious | 0.67 | Possibly damaging | 4.20 |
| V130L | 3.32 | 24.40 | 0.000003984 | 0.04 | Deleterious | 0.33 | Benign | 1.70 |
| V130I | 3.16 | 24.00 | 0.000003984 | 0.31 | Tolerated | 0.33 | Benign | 1.37 |
| L122F | 2.91 | 23.40 | 0.000003982 | 0.01 | Deleterious | 0.13 | Benign | 2.50 |
| L122V | 2.13 | 20.90 | 0.000003982 | 0.16 | Tolerated | 0.03 | Benign | −1.70 |
CADD, PHRED, allele frequency, SIFT, and PolyPhen scores for each variant were obtained from Uniprot or gnomAD. Rosetta ddG values are from the current study and are reported in Rosetta Energy Units (REU).
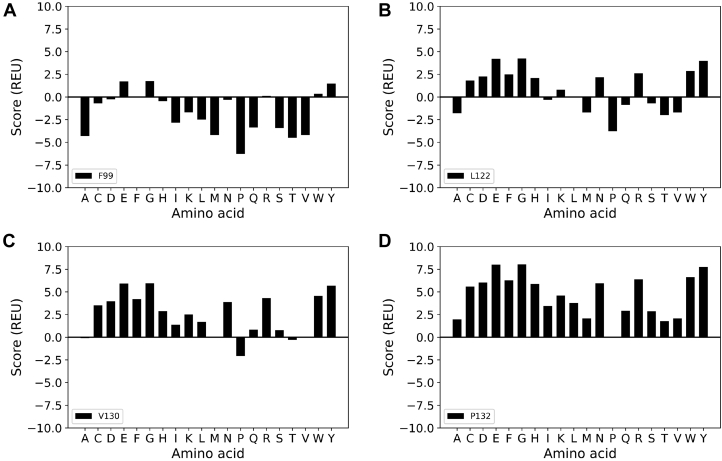
To investigate the contributions of these residues on the stability of CAV1 complexes, we used the cryo-EM 8S CAV1 structure as a template in Rosetta to analyze to how single amino acid substitutions at P132, F99, L122, and V130 are predicted to affect CAV1 oligomers. We computationally performed single amino acid substitutions at each position and calculated the corresponding ddG values relative to substitution with the identical residues (i.e., P132P) as a control (Fig. 3). Positive ddG values indicate that the respective mutation destabilizes the structure, while negative ddG values indicate that the mutation is stabilizing.
Figure 3.
Impact of single amino acid substitutions at P132, F99, L122, or V130 on ddG values for a single CAV1 protomer. Per-protomer Rosetta ddG scores were calculated for CAV1 complexes relaxed with 11-fold symmetry containing the indicated single amino acid substitutions at each of the indicated residues, relative to substitution with the WT residue at this site. Scores are reported in Rosetta Energy Units (REU). A, F99. B, L122. C, V130. D, P132.
Amino acid substitutions of F99 were predicted to yield ddG values that were either close to zero or negative, suggesting that this site can tolerate a variety of different residues (Fig. 3A). Substitutions of L122 yielded a combination of stabilizing and destabilizing effects, whereas substitutions of V130 were on average destabilizing. Compared to these other residues, the substitution of any amino acid at residue 132 resulted in the most highly destabilizing ddG values (Fig. 3). The extent of destabilization varied depending on the specific amino acid substitution. For example, P132T, P132M, and P132A were predicted to have relatively small effects, whereas P132G, P132E, and P132Y led to the highest ddG values indicating they are the most destabilizing (Fig. 3D). P132L had a moderate effect on the stability of the system. Thus, while any amino acid substitution at position 132 is predicted to destabilize the oligomer, the replacement of P132 with charged residues that would be buried in a hydrophobic environment, with bulky aromatic amino acids that can clash with other residues in the neighboring protomers, or with glycine that affects the helix backbone have particularly large destabilizing effects on the system.
We also compared the ddG scores for several of the naturally occurring CAV1 variants with commonly used bioinformatical metrics that report on the frequency and deleteriousness of amino acid substitutions, including CADD, PHRED, allele frequency, SIFT, and PolyPhen scores (Table 2). CADD and PHRED are metrics to measure the relative pathogenicity of human genetic variants (51). Allele frequency is the frequency at which a particular variant is observed among the tested populations. More deleterious mutations are expected to occur at a lower frequency than less deleterious mutations since these potentially have more disruptive effects. SIFT score is a normalized probability of observing a different amino acid at a given position within a range of 0 to 1 (52). Finally, PolyPhen scores predict the probability of a variant being damaging in terms of phenotype (53, 54). The allele frequencies were low for most variants. That said, the allele frequency of P132A was six times higher than the frequency of P132L, which may suggest a milder phenotype for P132A compared to P132L. There was no significant correlation between the SIFT or PolyPhen scores and the calculated ddG values. However, we observed a strong correlation (R2 = 0.85) between the calculated ddG values and the CADD and PHRED scores. Overall, these comparisons suggest that the ddG calculations can capture the destabilizing effects of various CAV1 mutations reported by other metrics.
Experimental analysis confirms that single amino acid substitutions to P132 cause CAV1 to accumulate in the Golgi complex
It is well established that P132L is trapped intracellularly as a consequence of its inability to oligomerize correctly (19, 20, 40). To test whether this is also the case for the other P132 mutants, we utilized a previously described caveolae reconstitution assay (26, 55, 56). In this assay, the CAV1 variants of interest are exogenously expressed in CAV1−/− mouse embryonic fibroblast (MEF) cells. Cell surface levels of cavin-1, another essential caveolae component, is normally low in these CAV1−/− MEFs due to the requirement for CAV1 to stabilize it at the plasma membrane (57). Re-expression of WT CAV1 leads to the recruitment of cavin-1 to cell surface puncta, providing a readout of successful caveolae assembly (26, 55, 56).
We selected a subset of CAV1 P132 single amino acid substitutions for this analysis, including P132I, P132K, P132Y, P132S, P132W, P132A, and P132T. For comparison, we studied several naturally occurring genetic variants of CAV1 to determine if they exhibit similar trafficking defects, including V130F, V130L, V130I, L122F, and L122V (Table 2), as well as a F99C mutant. To carry out these experiments, myc-tagged versions of WT or mutant CAV1 were each expressed individually in CAV1−/− MEFs. Cells were then fixed and immunostained for myc and endogenous cavin-1 (Figs. 4, S3 and S4).
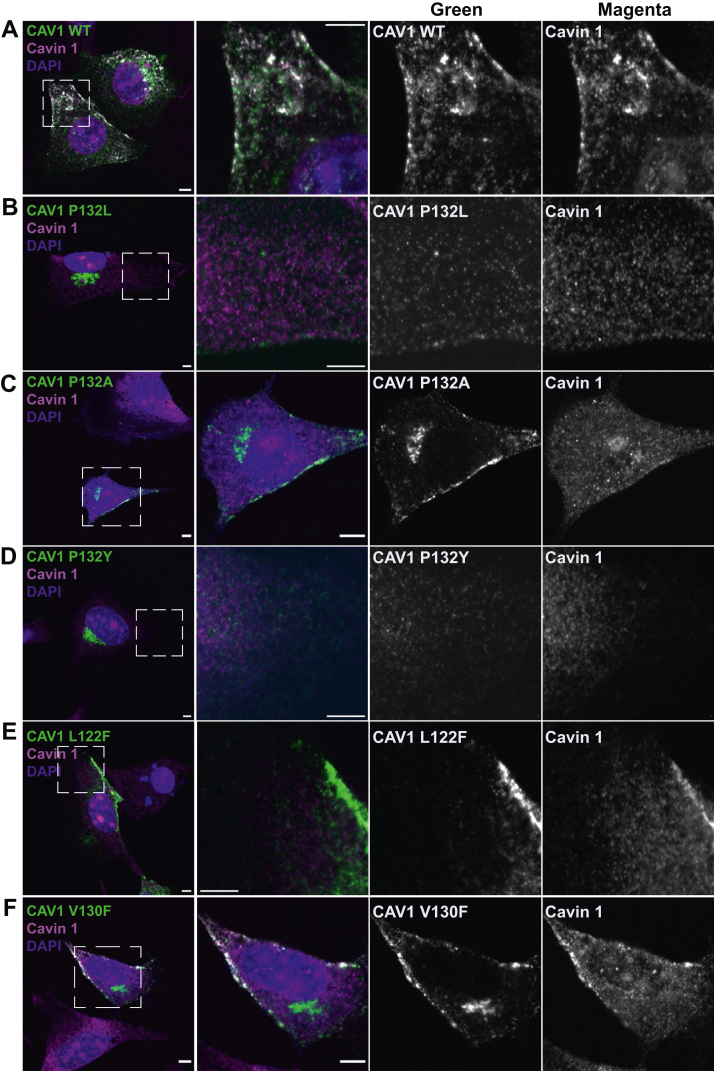
Figure 4.
Single amino acid substitutions to P132, but not to L122 or V130 cause CAV1 to accumulate in the Golgi complex in CAV1−/−MEFs. Representative confocal images are shown for CAV1−/− MEF cells expressing myc-tagged A, WT CAV1; B, P132L; C, P132A; D, P132Y; E, L122F, or F, V130F. Cells were allowed to express the indicated constructs for 24 h, fixed, and immunostained for endogenous cavin-1 (magenta) and myc (green) prior to imaging. Dashed boxes indicate the position of zoomed images. Bars represent 5 μm.
As expected, expression of WT CAV1 led to the formation of cell surface puncta positive for both CAV1 and cavin-1 (Fig. 4). A similar result was obtained for F99C (Fig. S4). L122 and V130 point mutants likewise were delivered to the plasma membrane and recruited cavin-1 although not as efficiently as WT CAV1 or F99C (Figs. 4 and S4). In contrast, all of the P132 mutants showed extensive perinuclear localization in Golgi-like compartments (Figs. 4 and S3). Interestingly, the P132A mutant also showed some plasma membrane staining and accompanying recruitment of cavin-1 (Fig. 4). These observations suggest that P132A has less severe trafficking and oligomerization defects than the other P132 mutants examined, in line with the existence of P132A as an exome variant in humans and its less destabilizing Rosetta ddG score compared to P132L (Table 2 and Fig. 3). P132I likewise showed some plasma membrane localization, although its Rosetta ddG score was relatively close to that of P132L (Figs. 3 and S3). Given the established disease connections of P132L, combined with its severe trafficking defects, for all further studies we focused on this mutant as a model.
Computational modeling predicts P132L can form mixed complexes with WT CAV1
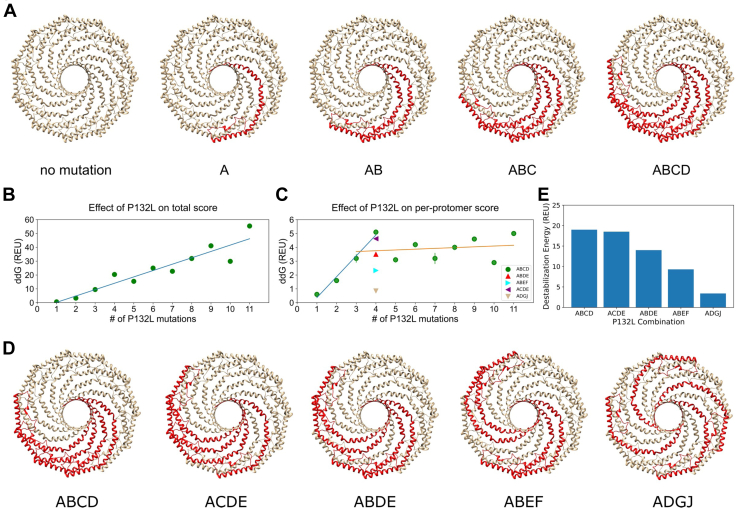
P132L was reported to function as a dominant negative, resulting in the trapping of WT CAV1 with P132L in internal compartments (40). One possible mechanism underlying this phenomenon is via oligomerization of P132L with WT CAV1. However, the dominant negative phenotype is dependent on overexpression, cell type, and tagging strategies (41, 58, 59). It thus remains unclear to what extent the stability and fate of CAV1 complexes are perturbed by the incorporation of P132L protomers. As a first step toward addressing this question, we introduced the P132L mutation in silico, systematically increasing the number of mutant protomers from 1 to 11 while correspondingly decreasing the number of WT copies of CAV1. For this initial analysis, mutant protomers were introduced at neighboring positions within the complex (Fig. 5A). We then calculated the destabilization caused by these mutations as a function of the increasing number of P132L mutations in terms of Rosetta Energy Units (REU), both in terms of the overall stability of the complex (Fig. 5B) and on a per-mutant basis (Fig. 5C).
Figure 5.
Computational predictions of the energetic cost of incorporating P132L protomers into mixed complexes.A, schematic of CAV1 complexes containing increasing numbers of P132L mutant protomers. In this scenario, mutant protomers are located directly adjacent to one another. B, Rosetta ddG for CAV1 complexes containing increasing numbers of P132L mutants located directly adjacent to one another as in panel A. C, as in B, except here, Rosetta ddG was normalized per number of P132L mutations. Lines are drawn to guide the eye to highlight differences in predicted stability when one to three copies of P132L (blue line) versus four or more (orange line) positioned directly adjacent to one another are present in a mixed complex with WT CAV1 (green circles). For comparison, per-protomer Rosetta ddG values are shown for mixed complexes containing four copies of P132L arranged at different spacings (colored triangles) as shown in panel D. D, schematic of CAV1 complexes containing four copies of P132L arranged at different spacings. E, Rosetta ddG values for CAV1 complexes containing four P132L protomers arranged at different spacings shown in panel D. REU, Rosetta energy units.
A single P132L mutation destabilized the complex by ∼0.6 REU. This low value implies that the amount of destabilization is minor and may be compensated by the rest of the system (Fig. 5C). However, as the number of copies of P132L was increased from one to four, the ddG calculated on a per-mutation basis increased drastically (Fig. 5C, blue line). The per-protomer ddGs then leveled off for complexes containing five or more P132L mutations at a value indicative of substantial destabilization (Fig. 5C, orange line).
We next examined the effects of spacing of the P132L mutants on the stability of the mixed complexes. We hypothesized that as the spacing between the P132L protomers increased, their overall impact on the stability of the complex might be less perturbing. To test this, we generated five different models. Each contained four copies of P132L and seven copies of WT CAV1, but differed in the arrangement of the mutant protomers (Fig. 5D). In the first model, the four P132L protomers were arranged side by side as described earlier. In the second model, two P132L protomers were placed side by side and were separated by a single WT protomer. The third model also contained two pairs of P132L protomers arranged side by side, in this case separated by two WT protomers. The fourth model consisted of three adjacent P132L protomers separated from a single mutant protomer by a single WT protomer. In the final model, individual P132L protomers were separated as far as possible from one another. In this configuration, each mutant protomer was flanked on either side by WT CAV1. Rosetta ddG calculations revealed that the most significant destabilization occurred when three or four mutant protomers were directly adjacent to one another (Models 1 and 2), followed by the models with pairs of P132L protomers separated by one or two WT CAV1 protomers (Models 3 and 4) (Fig. 5, C and E). The complex containing the largest spacing between the P132L protomers showed the lowest perturbation overall, corresponding to less than 1 REU per mutant protomer (Model 5) (Fig. 5, C and E). These findings suggest that, depending on their spacing, multiple copies of P132L can be incorporated into complexes with WT CAV1 with minimal disruption to the overall stability of the complex.
P132L and WT CAV1 co-assemble into 8S complexes in a heterologous Escherichia coli model system
We next set out to test the prediction of our computational analysis that P132L can be efficiently incorporated into CAV1 8S complexes using a heterologous E. coli model system. As we have shown, WT CAV1 expressed in E. coli assembles into 8S complexes that can be purified and analyzed biochemically and by electron microscopy (48, 60). Furthermore, E. coli-expressed P132L has been reported to exhibit oligomerization defects similar to those seen in mammalian cells (5), suggesting E. coli is a good platform to investigate the effect of the P132L mutation on CAV1.
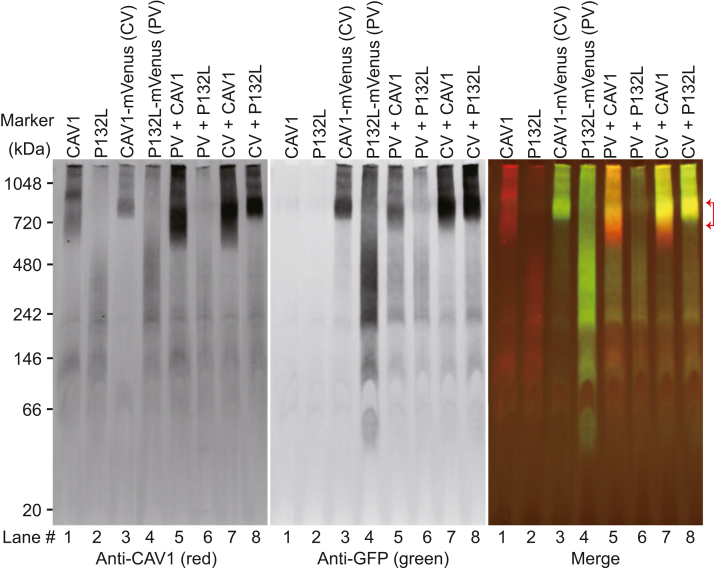
We first confirmed that P132L and WT CAV1 behave as expected using blue native PAGE (BN-PAGE) to monitor their oligomerization status (19, 41, 55, 56, 60). For these experiments, we expressed WT CAV1 or P132L, either individually or in combination, with and without fused mVenus tag (CAV1, P132L, CAV1-mVenus, and P132L-mVenus) in E. coli. Following the cell lysis and purification of total membranes, we checked the oligomerization status of the proteins using blue native-PAGE followed by Western blotting. Two antibodies were used to specifically detect CAV1: an anti-N-term CAV1 antibody that recognizes both WT and P132L, and an anti-GFP antibody, which only detects the protein fused to mVenus.
In blue native gels, WT CAV1-mVenus migrated as a high-molecular-weight band in a position expected for 8S complexes (Fig. 6) (60). In contrast, P132L migrated as a smear (Fig. 6), consistent with its reported oligomerization defects (5). Strikingly, however, when both proteins were coexpressed, P132L-mVenus comigrated in a high-molecular-weight band with WT CAV1 (Fig. 6, lane 5, red brackets). These results show that P132L CAV1 can form mixed 8S complexes with WT CAV1 when expressed in E. coli as predicted by our Rosetta calculations. The finding that the high molecular bands were not disrupted by coexpressing with P132L further implies that the presence of P132L does not grossly destabilize these complexes (Fig. 6, lane 5 and 8).
Figure 6.
P132L forms mixed 8S oligomers with WT CAV1 in Escherichia coli cells as assessed by BN-PAGE. Total membrane from E. coli cells expressing the indicated constructs was lysed in C12M and subjected to BN-PAGE followed by Western blotting for mVenus (GFP, green) and CAV1 (red). Lane numbers were labeled at the bottom. The range of high molecular bands is indicated with red brackets. CV, CAV1-mVenus; PV, P132L-mVenus.
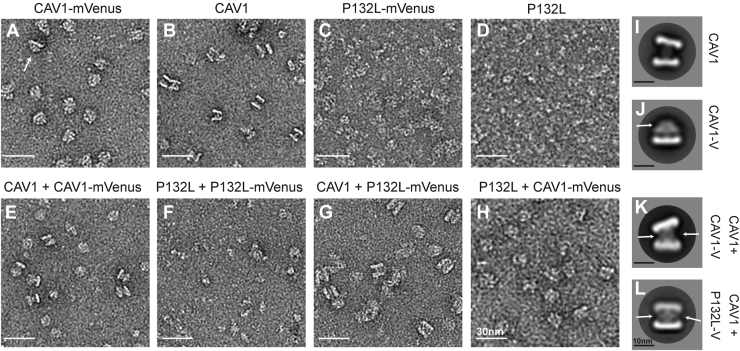
We wondered whether the structure of the oligomers was perturbed by the incorporation of the P132L mutant. To test this, we examined oligomers formed by P132L and the P132L/WT CAV1 mixed complexes using negative stain EM. For these experiments, WT CAV1, CAV1-mVenus, P132L, and P132LmVenus were expressed singly or in combination in E. coli, purified using nickel beads, and fractionated using FPLC (Fig. S5). Pooled samples were then negatively stained and imaged by EM.
Consistent with our previous results (60), CAV1-mVenus or CAV1 fractionated into two peaks, P1 and P2 (Fig. S5, A and C). We focused our analysis on the P1 fractions, which consist of 8S complexes (60). As reported previously, these complexes are disc-shaped and ∼15 nm in diameter (Figs. 7, A and B and S6, A and B). Examples of both en face views and side views of the disc-shaped 8S complexes can be seen in 2D average classes of CAV1 and CAV1-mVenus (Figs. 7, A and B and S6, A and B). CAV1 oligomers also form 8S complex “dimers” composed of two 8S complexes interacting with each other via their central protrusions (Fig. 7, B and I). CAV1-mVenus particles contain an additional fan-shaped density emanating from a central protrusion (β-barrel) visible in the side views consistent with the C-terminal mVenus tag (Fig. 7, A and J). Unlike CAV1, CAV1-mVenus complexes do not form dimers (Fig. 7A). However, in control experiments in which CAV1 and CAV1-Venus were co-expressed, dimeric complexes could be observed (Fig. 7E). 2D averages of the side views of CAV1/CAV1-Venus dimers had increased central density in the stalk region (Fig. 7K) when compared to CAV1 complexes alone (Fig. 7I), likely reflecting the presence of the mVenus tag.
Figure 7.
P132L forms mixed 8S complexes with WT CAV1 as assessed by negative stain EM. Purified CAV1 complexes were negatively stained and visualized by EM. Representative images of negatively stained P1 fractions of (A) CAV1-mVenus, (B) CAV1, (C) P132L-Venus, (D) P132L, (E) CAV1 + CAV1-mVenus, (F) P132L + P132L-mVenus, (G) CAV1 + P132L-mVenus, and (H) P132L + CAV1-mVenus. Scale bar represents 30 nm. Representative 2D averages of (I) CAV1 complexes, (J) CAV1-mVenus 8S complexes, (K) CAV1 + CAV1-mVenus mixed complexes, and (L) CAV1 + P132L-mVenus mixed complexes. The arrow marks regions of additional density from the Venus tag. Scale bars represent 10 nm.
P132L displayed a more complicated FPLC profile than WT CAV1 complexes, consisting of at least seven shoulders containing heterogeneous particles when examined by negative stain EM (Figs. 7D, S5B, and S6C). Consistent with their predicted inability to homo-oligomerize correctly, the P132L and P132L–mVenus complexes in the P1 fraction were not structurally well organized. They were thus not amenable for 2D averaging (Fig. 7, C and D).
Finally, we visualized mixed complexes formed by co-expression of WT-CAV1 and P132L-mVenus (Fig. 7G). 2D class averages (Fig. 7L) revealed the presence of discs with diameters comparable to those formed by WT CAV1 + WT CAV1-mVenus. Here again, increased density could be observed between dimers, again suggestive of the presence of mVenus. Taken together, these findings indicate that P132L and WT CAV1 are capable of co-assembling into 8S complexes similar in size and shape to those generated by WT CAV1. They further suggest that the presence of P132L does not disrupt the overall structure of at least a subset of 8S complexes.
P132L and WT CAV1 form mixed detergent-insoluble 8S complexes in mammalian cells.
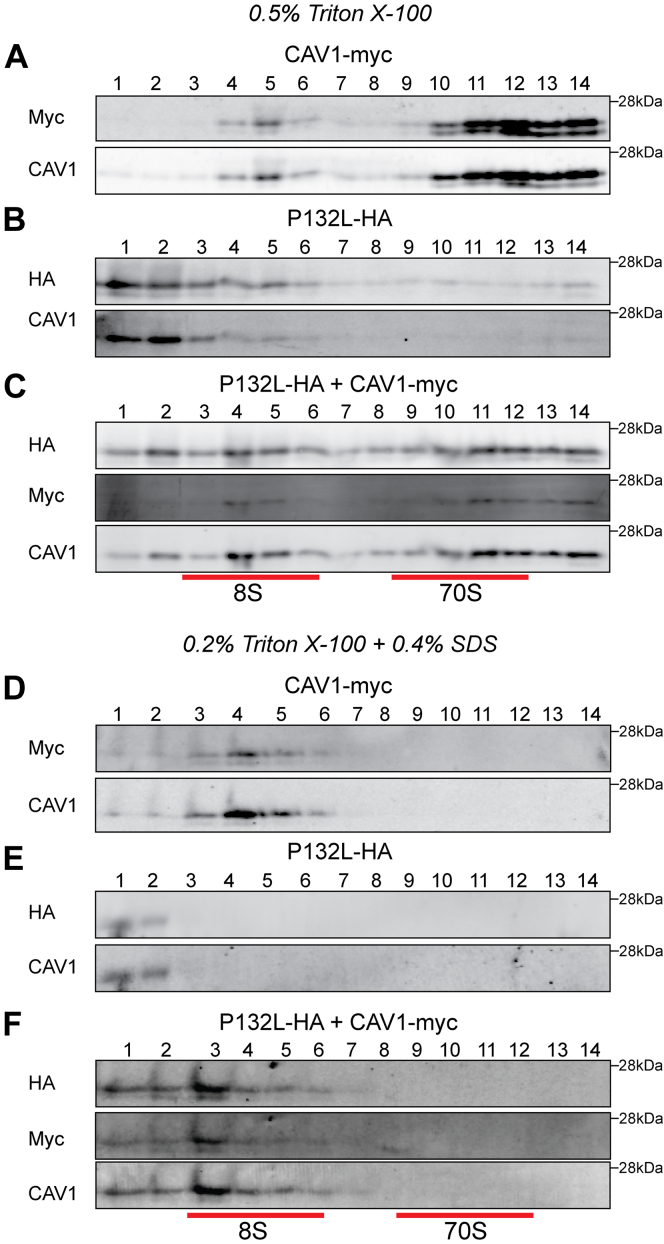
We next asked whether P132L has the potential to productively co-assemble with WT CAV1 into 8S complexes in mammalian cells. To test this, we used a biochemical approach to assay for 8S complex formation (20, 41, 55, 56). WT CAV1 and P132L constructs were expressed either individually or together in cells lacking endogenous caveolin expression, CAV1−/− MEF cells. After transfection with CAV1 constructs, the cells were allowed to express the protein for 16 h and then lysed with 0.5% TX-100. The lysates were then subjected to sucrose density centrifugation (20, 41, 55, 56). WT CAV1 was enriched in two peaks corresponding to the position of 8S and 70S complexes (Fig. 8A), whereas P132L was mostly fractionated as low-molecular-weight species (Fig. 8B). However, when co-expressed with WT CAV1, P132L was recruited into both 8S and 70S complexes (Fig. 8C). This strongly suggests that P132L and WT CAV1 form mixed 8S and 70S complexes.
Figure 8.
P132L forms mixed 8S and 70S complexes with WT CAV1 in CAV1−/−MEF cells. CAV1−/− MEF cells transiently transfected with (A and D) CAV1-myc, (B and E) P132L-HA, or (C and F) CAV1-myc plus P132L-HA were lysed in either 0.5% Triton-X-100 (A–C) or 0.2% Triton X-100 plus 0.4% SDS (D–F) at room temperature. Extracts were run through 10 to 40% sucrose velocity gradients and fractions were analyzed by SDS-PAGE/Western blot. Fraction numbers are indicated at the top of the blots and the positions of 8S and 70S complexes are indicated by red lines.
We next probed the stability of the mixed complexes. To do so, we subjected cells to lysis in 0.4% SDS plus TX-100 and then carried out sucrose density centrifugation (20, 56). This treatment causes the dissociation of 70S complexes while maintaining the integrity of 8S complexes of WT CAV1 (56). However, 8S complexes formed by F160X, another disease-associated mutation of CAV1, dissociate into smaller structures under these conditions (56). This approach can thus be used to evaluate the impact of disease-associated mutation on complex stability. We found the mixed 8S complexes formed by WT CAV1 and P132L remained intact under these more extreme lysis conditions, similar to the behavior of WT CAV1 (Fig. 8, D–F). This implies that the mixed WT/P132L 8S complexes are biochemically stable.
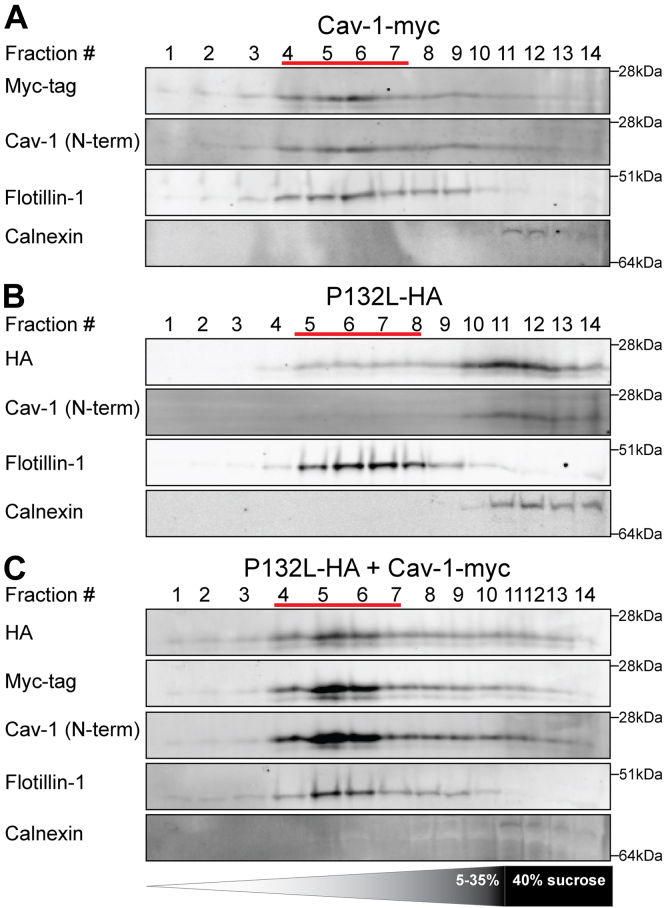
Another characteristic biochemical feature of WT CAV1 is that it associates with buoyant detergent-resistant membranes (DRMs) when properly incorporated into caveolae (19, 55, 56, 61). P132L, however, is typically excluded from DRMs (40, 41). We thus wondered whether the mixed complexes formed by WT CAV1 and P132L are detergent resistant. To address this question, CAV1−/− MEF cells overexpressing WT-CAV1 and P132L were extracted with 0.5% cold Triton X-100 followed by sucrose density fractionation (55, 56). WT CAV1 was associated primarily with DRM fractions, as expected (Fig. 9A). Although P132L localized to detergent soluble fractions when expressed on its own (Fig. 9B), it shifted to the DRM fractions upon co-expression with WT CAV1 (Fig. 9C). Thus, P132L can become incorporated together with WT CAV1 into detergent-resistant 8S complexes in mammalian cells.
Figure 9.
P132L is recruited into DRMs by WT CAV1 in CAV1−/−MEFs. DRMs were isolated from CAV1−/− MEF cells transiently transfected with (A) CAV1-myc, (B) P132L-HA, or (C) CAV1-myc plus P132L-HA. Fractions were analyzed by SDS-PAGE/Western blotting. Flotillin-1 and calnexin were used as markers for DRMs and detergent soluble fractions, respectively. Fraction numbers are indicated at the top of the blots and the positions of the DRM fractions are indicated by red lines.
P132L and WT CAV1 co-assemble into caveolae in mammalian cells.
Both 8S and 70S complexes function as the fundamental building blocks of caveolae (20). We, therefore, wondered whether mixed 8S complexes formed by P132L and WT CAV1 could support caveolae biogenesis. To address this question, we utilized CAV1−/− MEFs, this time to assay for caveolae formation.
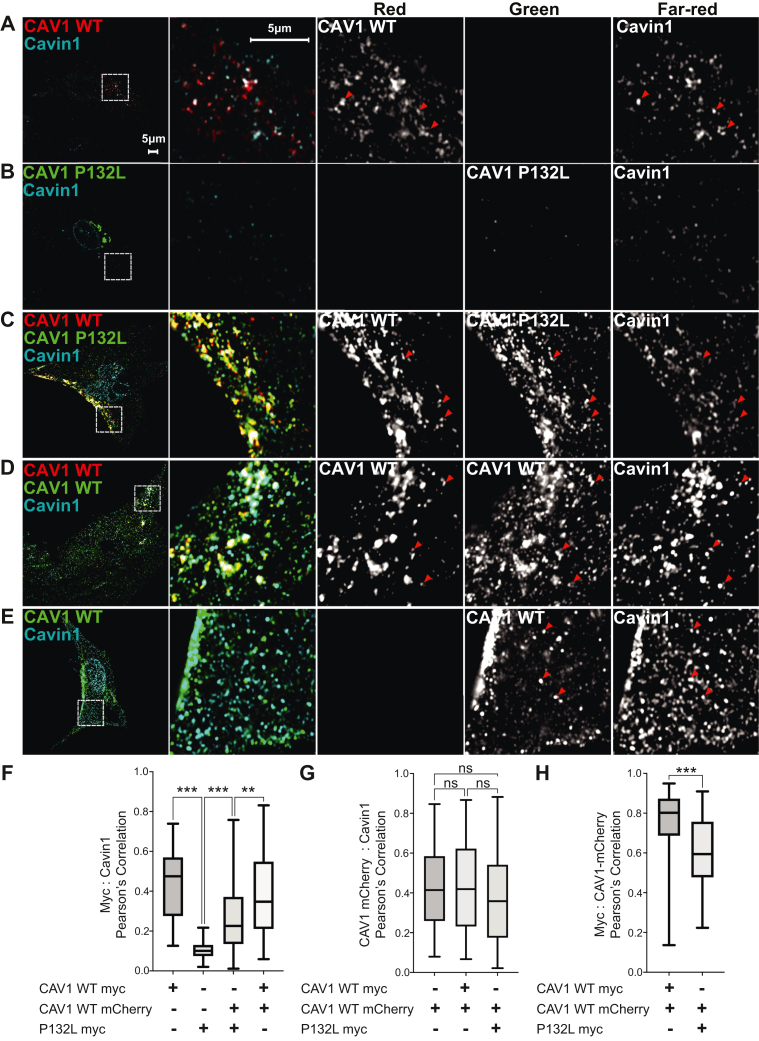
As expected, expression of WT CAV1 in CAV1−/− MEFs recruits endogenous cavin-1 to the plasma membrane, where it colocalizes with caveolin-positive puncta as visualized by AiryScan confocal microscopy (Fig. 10A) or TIRF microscopy (Fig. S7A). Similar results were obtained in cells expressing either CAV1-myc or CAV1-mCherry, or when both CAV1-myc and CAV1-mCherry were co-expressed (Fig. 10, A, D, E and H). In contrast, P132L localized primarily to the Golgi complex, and little P132L or Cavin-1 staining could be detected at the plasma membrane (Figs. 10B and S7B). We then asked whether WT CAV1 could recruit P132L into caveolae upon co-expression of the two proteins. In this analysis, P132L colocalized extensively with WT CAV1 and cavin-1 in puncta at the cell surface (Figs. 10C and S7C). We did note however that the level of colocalization of WT CAV1 with P132L was slightly lower than in cells co-expressing two different tagged forms of WT CAV1 (Fig. 10H).
Figure 10.
P132L localization defect is rescued by co-expression of WT CAV1 in CAV1−/−MEFs. Representative AiryScan images are shown for CAV1−/− MEF cells expressing (A) CAV1-mCherry, (B) P132L-myc, (C) CAV1-mCherry plus P132L-myc, (D) CAV1-mCherry plus CAV1-myc, or (E) CAV1-myc. Cells were allowed to express the indicated constructs for 24 h, fixed, and immunostained for endogenous cavin and myc-tagged constructs prior to imaging. Bar represents 5 μm. In (A–E), red arrowheads point to examples of colocalized puncta. F–H, Pearson’s correlation analysis (n = 90 ROIs from three independent experiments). A one-way ANOVA with Tukey's test (≥3 groups) or an unpaired Student's t test (2 groups) was used to calculate p-values. n.s., not significant.
In addition to the cavins, caveolae incorporate accessory proteins that help regulate their morphology and dynamics (24, 62, 63, 64, 65, 66). We tested whether two accessory proteins, PACSIN2 and EHD2, are found in mixed caveolae containing P132L. EHD2 and PACSIN2 partially colocalized with caveolae containing either WT CAV1 complexes or P132L/WT complexes (Figs. S8 and S9). We conclude that mixed 8S complexes formed by P132L and WT CAV1 can successfully assemble into caveolae that contain a normal complement of accessory proteins.
Discussion
Our results suggest the following model for how P132L impacts the structure, oligomerization, and function of CAV1. We propose that P132 normally contributes to hydrophobic packing between protomers along the outer ring of the 8S complex. The introduction of a leucine residue at P132 introduces additional bulk facing the residues in neighboring protomers. This causes clashes with other conserved hydrophobic residues of CAV1 and prevents homo-oligomers containing more than a few copies of P132L from forming. Despite its homo-oligomerization defects, P132L can form hetero-oligomers with WT CAV1. Our computational analysis suggests that multiple copies of P132L can be incorporated into 8S complexes with WT CAV1 without compromising the integrity of the complex. These complexes can be detected experimentally using multiple techniques, appear structurally similar to those formed exclusively by WT CAV1, are biochemically stable, and can even become incorporated into caveolae that recruit caveolae accessory proteins appropriately. How many P132Ls can productively oligomerize together with WT CAV1 likely depends on their arrangement in a given complex. Complexes containing widely spaced mutants are predicted to be energetically favored over those where the mutant protomers are closely apposed. Such destabilized complexes could in turn induce proteostatic stress (67). Alternatively, the presence of intracellular pools of incompletely oligomerized P132L could potentially interfere with the normal functions of caveolin by disrupting interactions with critical binding partners of CAV1. For example, CAV1 P132L is not recognized correctly by valosin-containing protein, a protein that cooperates with the ubiquitin-like–domain-containing protein UBXD1 to bind to ubiquitylated CAV1 oligomers and sort them to endolysosomes for degradation (43).
Based on structural analysis of CAV1 within the context of a complete 8S complex, we show that P132L primarily disrupts hydrophobic packing. This conflicts with previous work based on studies of a truncated form of CAV1 (residues 62–178) that suggested the P132L mutation alters the secondary structure of the IMD by extending an α-helix (44). We speculate the truncated form of the protein, which behaved as a monomer, may have undergone structural transitions that do not occur in the context of the fully assembled oligomer. Our results do agree, however, with the conclusion that no other hydrophobic residue can fully substitute for the function of P132 in this position (44). One partial exception is a P132A mutant. Although also predicted to be destabilizing, P132A could partially traffic to the plasma membrane and recruit cavin-1. However, it did so much less efficiently than WT CAV1. Another group studied a related construct (consisting of 5 alanine substitutions from residues 130–134) and also found it to behave more similarly to WT CAV1 than P132L (19). This could potentially be of physiological importance, as a P132A mutation has been identified as a genetic variant linked to ALS (35). We also observed that low levels of P132I could be delivered to the plasma membrane. For comparison to P132, we examined the impact of several point mutations to other hydrophobic resides including naturally occurring variants of L122 and V130 as controls. These variants were generally predicted to be energetically unfavorable. However, unlike the P132 mutants, neither the L122 nor V130 variants accumulated in the Golgi complex. While this does not exclude the possibility that these variants are perturbing at some level, it emphasizes these residues play a fundamentally different role in regulating the oligomerization and trafficking of CAV1 than P132 does despite their location at protomer interfaces.
We also found that P132L can become incorporated into caveolae together with WT CAV1. This finding was surprising in light of previous reports that expression of P132L cause WT CAV1 to become trapped intracellularly (40). However, these original studies were carried out in cells that express WT CAV1 endogenously and have a tendency to accumulate overexpressed CAV1 in the perinuclear region as a consequence of aggresome formation (58, 59). On the other hand, stable exogenous expression of P132L has also been reported to have no effect on caveolae formation in H1299 cells, a cell line expressing endogenous CAV1 (28). Taken together, these findings highlight the complexities of studying caveolins and the importance of studying the proteins under conditions that preserve their ability to oligomerize and assemble into caveolae.
The P132L mutation has been identified in several different patient samples including breast cancer and lung adenocarcinomas (25, 26, 34). In addition to P132L, a variety of other pathogenic mutations in caveolins have been identified in humans (12, 35, 39, 45, 55, 56, 68, 69, 70, 71, 72, 73, 74, 75, 76). The most direct equivalent to P132L is a P105L mutation in CAV3 associated with muscular dystrophy (69, 70). Muscle biopsies from patients harboring the P105L mutation with an autosomal form of dominant limb-girdle muscular dystrophy contain considerably decreased CAV3 levels compared to controls (69). Like CAV1 P132L, in mammalian heterologous expression systems, the P105L mutant protein accumulates in the perinuclear region and can trap WT CAV3 intracellularly (37, 77). However, the nature of the oligomerization defects appears to be somewhat different, as P105L CAV3 tended to form much larger oligomers than WT CAV3 (37). Although a high-resolution structure of CAV3 has yet to be determined, it seems likely that the P105L mutation plays a similar role in destabilizing the structure of CAV3 complexes, as is the case for CAV1. To this point, it was recently suggested that P105 contributes to hydrophobic packing based on the modeling of the structure of CAV3 using AlphaFold2 (67).
In contrast to P105L CAV3, disease-associated frameshift mutations in the C-terminal region of CAV1 appear to operate by a different mechanism than P132L. A mutation identified in patients with both familial and idiopathic forms of PAH, P158P, gives rise to a new C-terminus that is one residue longer than WT CAV1 and introduces a de novo ER retention signal (55, 78). A different frameshift mutation, F160X, was found in a patient with both PAH and CGL and leads to premature termination of the protein (56, 71, 72). Unlike P132L, F160X can form 8S-like complexes and even support caveolae assembly when expressed in CAV1−/− MEFs (56). However, mixed complexes formed by WT CAV1 and F160X in patient cells are destabilized compared to those formed by WT CAV1 (56). This destabilization likely results from the inability of F160X to form the central β-barrel of the 8S complex, evidenced by the loss of the central protrusion in these complexes when observed by negative stain EM (60). Both P158P and F160X can also form 8S-like complexes in E. coli (60). While disc shaped, these complexes are less regular than WT 8S oligomers, suggesting that disruption of the C-terminus interferes with packing and stability of the complex (60). This can be understood based on their position in the cryo-EM structure: P158 and F160 are both located close to the center of the complex between helix 5 and the beginning of the β-barrel (48). In contrast, the defects introduced by the P132L mutation are much more dramatic, highlighting the importance of this residue in controlling the overall oligomerization state of the protein.
In conclusion, we have now identified the importance of P132 in controlling the oligomerization of CAV1 and the impact of a disease mutation at this site. These findings provide a molecular framework for understanding how defects in caveolins ultimately influence the assembly of caveolar domains and new insights into the fundamental processes that control caveolae biogenesis.
Experimental procedures
Cell culture
CAV1−/− MEFs (KO MEFs) were obtained from ATCC and cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 1% Pen/Strep at 37 °C, and 5% CO2.
Constructs and transfections
CAV1-mCherry, CAV1-mycHis, and P132L-mycHis for mammalian cell expression and CAV1-His, P132L-His, CAV1-mVenusHis, and P132L-mVenusHis for E. coli expression were as described previously (41, 58, 60). The construction of C-terminus HA or myc-tagged CAV1 (WT and P132L mutant) expression constructs was based above mentioned CAV1-mycHis or P132L-mycHis plasmids using PCR. The primers used were as follows: CAV1/P132L-HA: CGGGATCCATGTCTGGGGGCAAATACGTAG and CGGAATTCTTAGCTAGCGTAGTCTGGGACGTCGTATGGGTATATTTCTTTCTGCAAGTTGATGCG; CAV1/P132L-myc: CGGGATCCATGTCTGGGGGCAAATACGTAG and CGGAATTCTTACAGATCCTCTTCTGAGATGAGTTTTTGTTCGGGCCCAAGCTTTATTTCTTTCTGCAAGTTGATGCG. Constructs were verified by sequencing.
Additional single-site mutations were introduced by the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent, #210518) using CAV1-myc plasmid as a template. The primers used (Integrated DNA Technologies) were as follows.
F99C: GTGACGAAATACTGGTGCTACCGCTTGCTGTCTGCCCTCTTTGG; P132I: GCACATCTGGGCAGTTGTAATCTGCATTAAGAGCTTCCTGATTGAG; P132K: GCACATCTGGGCAGTTGTAAAGTGCATTAAGAGCTTCCTGATTGAG; P132Y: GCACATCTGGGCAGTTGTATACTGCATTAAGAGCTTCCTGATTGAG; P132S: GCACATCTGGGCAGTTGTATCCTGCATTAAGAGCTTCCTGATTGAG; P132W: GCACATCTGGGCAGTTGTATGGTGCATTAAGAGCTTCCTGATTGAG; P132A: GCACATCTGGGCAGTTGTAGCGTGCATTAAGAGCTTCCTGATTGAG; P132T: GCACATCTGGGCAGTTGTAACCTGCATTAAGAGCTTCCTGATTGAG; L122F: GGCATTTACTTCGCCATTTTCTCTTTCCTGCACATCTGG; L122V: GGCATTTACTTCGCCATTGTCTCTTTCCTGCACATCTGG; V130F:CCTGCACATCTGGGCATTCGTACCATGCATTAAGAGC; V130I:CCTGCACATCTGGGCAATCGTACCATGCATTAAGAGC; V130L:CCTGCACATCTGGGCACTCGTACCATGCATTAAGAGC. Constructs were verified by sequencing (Genewiz).
Transient transfections were performed using Lipofectamine 2000 (Life Technologies) as per the manufacturer’s instructions. Cells were transfected 1 day before observation or biochemical analysis.
Antibodies
Mouse anti-GFP mAb (catalog number 632381) was obtained from Clontech. Mouse anti-mCherry mAb (catalog number NBP1-96752) was obtained from NOVUS. For Western blots, rabbit anti c-Myc pAb (catalog number sc-789) was obtained from Santa Cruz Biotechnology. Mouse anti c-Myc mAb (clone 9B11, #2276) and mouse anti HA mAb (6E2) (catalog number 2367) were obtained from Cell Signaling Technology. Rabbit anti-CAV1 pAb (catalog number 610060), mouse anti-flotillin-1 mAb (catalog number 610820), and mouse anti-calnexin mouse mAb (catalog number 610523) were purchased from BD Transduction Laboratories. Rabbit anti-6X His pAb (catalog number 137839), rabbit anti-cavin1 pAb (catalog number 76919), and goat anti-EHD2 pAb (catalog number 23935) were purchased from Abcam. Rabbit anti-PACSIN2 pAb (catalog number AP8088b) was purchased from Abgent. For Western blotting, fluorescently conjugated secondary antibodies and blocking buffer were obtained from LI-COR Biosciences. For immunofluorescence assays, Alexa-labeled secondary antibodies were obtained from Life Technologies.
Immunofluorescence microscopy of cells expressing single amino acid substitution mutants of CAV1
CAV1−/− MEFs were seeded at 150,000 cells/well of six-well plates containing glass coverslips and transfected with Lipofectamine per manufacturer’s protocol with each of the indicated constructs. After 15 h, cells were washed 2× in PBS and fixed for 15 min in 4% paraformaldehyde. Crosslinking was quenched via washing 3× in 100 mM glycine in PBS. Cells were blocked and permeabilized with blocking buffer (5% glycine in PBS containing 0.1% TX-100 and 5% goat serum) at room temperature (RT) for 1 h. Cells were costained with mouse anti-Myc (Cell Signaling Technology, 911B, 1:200 dilution in blocking buffer) and rabbit anti-cavin1 (Abcam, ab76919, 1:200 dilution in blocking buffer) antibody for 2 h at RT in a humid chamber, then washed 3× in PBS. Coverslips were incubated in Alexa 488 and Alexa 546-conjugated secondary antibodies (488 goat anti-mouse IgG, Life Technologies; 546 goat anti-rabbit IgG, Life Technologies) at 1:500 concentration for 1 h at RT. After washing 3× in PBS, coverslips were mounted using ProLong Gold Antifade reagent with DAPI (Cell Signaling Technology). Cell samples were imaged using a Zeiss LSM 880 confocal microscope with a Plan-Apochromat 63×/1.4 oil DIC M27 objective at RT. Signals from the green (Alexa 488) and red (Alexa 546) channels were collected with a 34-channel GAsP spectral detector. All images were collected at 16-bit depth. Contrasts were adjusted linearly with ImageJ and exported as TIFFs.
Immunofluorescence microscopy of cells expressing mixed caveolin complexes
CAV1−/− MEFs were seeded at 150,000 cells/plate in MatTek dishes and transfected with Lipofectamine as per the manufacturer’s instructions. Twenty four hours posttransfection, cells were rinsed 2× with PBS and fixed for 12 min in 4% paraformaldehyde in PBS at RT. After another 3× rinsing in PBS containing 100 mM glycine, the cells were blocked and permeabilized with blocking buffer (0.1% saponin in PBS containing 0.5% bovine serum albumin) at RT for 1 h. The cells were stained with anti-Myc antibody (9B11, 1:1000 dilution in blocking buffer) plus antibodies against respective caveolin accessory proteins as indicated in figures (rabbit anti-cavin1, 1:200, Abcam 76919; goat anti-EHD2, 1:200, Abcam 23935; rabbit anti-PACSIN2, 1:200, Abgent AP8088b) at 4 °C overnight. Glass-bottom dishes were then rinsed 3× with PBS containing 0.01% Triton X-100 and then incubated in a 1:500 dilution of Alexa 488 and Alexa 647-conjugated secondary antibodies (EHD2 sample: 488 donkey anti mouse IgG, 647 donkey anti goat IgG; cavin1 and PACSIN2 samples: 488 goat anti mouse IgG, 647 goat anti rabbit IgG). After another 3× rinse in PBS containing 0.01% Triton X-100, 1 ml PBS was added to each dish.
For AiryScan imaging, cell samples were imaged using a Zeiss LSM 880 confocal microscope with a Plan-Apochromat 63×/1.4 oil DIC M27 objective, in PBS, at RT. Signals from all three channels were collected with the AiryScan detector in super resolution, frame-switching mode. For quantitation, images were taken with the same objective and a 6× zoom factor, in confocal mode. Signals from the far-red channel (Alexa 647) were collected with a PMT detector, while signals from the green (Alexa 488) and red (Alexa 546) channels were collected with a 34-channel GAsP spectral detector. All images were taken in 16 bits format. Contrasts were adjusted linearly with ImageJ. Images were then exported as JPGs.
For total internal reflection fluorescence microscopy, samples were imaged using a Leica Thunder Imager equipped with a TIRF system and the LAX operating software, in 3-channel mode (647/546/488), at RT. An HC PL APO 100×/1.47 oil objective and a Leica-DFC9000GTC-VSC11976 camera were used and the penetration depth was 110 nm.
Colocalization analysis was performed as described before (56) using Macbiophotonics ImageJ with the "Intensity Correlation Analysis" plugin. In brief, images used for colocalization analysis were taken with a 6× zoom factor at 22.5 μm × 22.5 μm dimensions. For each set of comparisons, 90 images from at least three independent experiments were used for analysis. Pearson's correlation coefficients are reported as the mean ± standard error for all the images. A one-way ANOVA with Tukey's test (≥3 groups) or an unpaired Student's t test (2 groups) was used to calculate p-values.
Electrophoresis
Blue Native-PAGE was performed as described before (41) with the NativePAGE Bis-Tris Gel System (Life Technologies). Total membranes of E. coli were lysed at 4 °C for 30 min with lysis buffer [NativePAGE 1× Sample Buffer, complete protease inhibitor cocktail from Roche, and 2% C12 M (n-dodecyl-β-maltopyranoside) (Anatrace)], then spun at 13,100 rpm, 4 °C. The supernatant was used for the following analysis. 4 to 16% NativePAGE gels (Life Technologies) were used for the protein separation. Equal amounts of protein were loaded. The molecular weight was evaluated using NativeMark unstained protein standards.
SDS-PAGE electrophoresis was conducted by using Novex NuPAGE SDS-PAGE Gel System (Life Technologies). NuPAGE 4 to 12% Bis-Tris gels (Life Technologies) were used for the protein separation. See Blue Plus2 Pre-stained Protein was used to evaluate the molecular weight.
Western blotting
The electrophoretic transfer was conducted by using the Mini Trans-Blot Electrophoretic Transfer system (Bio-Rad). For Blue Native-PAGE, the polyvinylidene fluoride membranes (Millipore Sigma) were destained with methanol and washed with Tris-buffered saline buffer. An LI-COR Odyssey infrared imaging system (LI-COR Biosciences) was used for the signal detecting of blots. Quantification of Western blot images was performed using IMAGEJ.
Caveolin complexes fractionation
Caveolin complexes were fractionated by velocity gradient centrifugation as described before (20, 41). For each fractionation, 4 × 106 CAV1−/− MEFs were lysed for 20 min in 330 μl 0.5% TX-100 (or 0.4% SDS plus 0.2% TX-100) in TNE [100 mM NaCl, 20 m M Tris–HCl pH 7.5, 5 mM EDTA] buffer, supplemented with ‘Complete’ protease inhibitors cocktail (Roche) at RT. Postnuclear supernatants were prepared by conducting a 5-min centrifugation at 1100g. Three hundred microliters of the postnuclear supernatant were recovered and loaded onto linear 10 to 40% linear sucrose gradients. The sucrose solution was prepared with TNE buffer with protease inhibitors cocktail (Roche). Sucrose gradients were centrifuged in an SW55 rotor using an Optima LE-80K Ultracentrifuge (Beckman Coulter) for 5 h at 48,000 rpm and 4 °C. Fourteen equal volume (about 360 μl) fractions were harvested from the top and analyzed by SDS-PAGE/Western Blot with 10 μl loading from each fraction.
DRM fractionation
Caveolae-enriched DRMs were fractionated as described (41). Approximemtly, 1.6 × 107 CAV1−/− MEFs were suspended in 300 μl precooled 0.5% TX100 in TNE buffer, supplemented with “Complete” protease inhibitors cocktail (Roche). Cell suspensions were homogenized by passing 10 times through a precooled 1-ml syringe with a 27-gauge stainless steel needle (BD Biosciences). The homogenate was adjusted to about 40% sucrose by the addition of 700 μl of 60% sucrose prepared in TNE and placed at the bottom of an ultracentrifuge tube. A 5 to 30% linear sucrose gradient was formed above the homogenate and centrifuged at 40,100 rpm and 4 °C for 16 h in an SW55 rotor using an OptimaTM LE-80K Ultracentrifuge (Beckman Coulter). Fourteen 360 μl fractions were collected from the top and analyzed by SDS–PAGE/Western blot with an equal loading volume. Western blots were images and quantified as indicated above.
Expression of caveolin in E. coli and purification of CAV1 complexes
Protein expression and purification were conducted as described with minor modifications (60). In brief, caveolin proteins were expressed in E. coli BL21 using the auto-induction expression system (79). First, MDG starter of monoclonal bacteria was cultured at 37 °C and 250 rpm for 20 h, then auto-inducing ZYM-5052 media was used to enlarge the culture at 25 °C and 300 rpm for 24 h. E. coli cells were washed with 0.9% NaCl and then resuspended with buffer (200 mM NaCl and 20 mM Tris–HCl, pH 8.0). Bacterial cells were homogenized with a French press pressure homogenizer, and 1 mM PMSF and DTT were added just prior to homogenization. A 15 min centrifugation at 9000 rpm and 4 °C was conducted to remove large cell debris, then total membranes were pelleted at 40,000 rpm (Ti-45 rotor, Beckman Coulter) and 4 °C for 1 h. Membrane pellets were homogenized with Dounce tissue grinder in a buffer composed of 200 mM NaCl, 20 mM Tris–HCl (pH 8.0), and 1 mM DTT. To solubilize caveolin proteins from membranes, 10% C12 M (Anatrace) stock solution was mixed into membrane homogenate to a final concentration of 2%, and the mixture was slowly stirred for 2 h at 4 °C. Insoluble material was pelleted down by centrifugation at 42,000 rpm (Ti-50.2 rotor) for 35 min, and the supernatant was used for nickel Sepharose–based affinity purification. The caveolin-containing eluate was concentrated and further separated by size-exclusion chromatography using a Superpose 6 Increase 10/300 Gl column (GE Healthcare) in buffer containing 200 mM NaCl, 20 mM Tris–HCl (pH 8.0), 1 mM DTT, and 0.05% C12 M.
Negative stain EM and data processing
Negative stain EM was performed using established methods (80). In brief, 200-mesh copper grids covered with carbon-coated collodion film (EMS) were glow discharged for 30 s at 10 mA in a PELCO easiGlow glow discharge unit (Fresno). Aliquots (3.5 μl) of the purified sample were adsorbed to the grids and incubated for 1 min at RT. Samples were then washed with 2 drops of water and stained with two successive drops of 0.7% (w/v) uranyl formate (EMS) followed by blotting until dry. Samples were visualized on a Morgagni transmission electron microscope equipped with a field emission gun operating at an accelerating voltage of 100 keV (Thermo Fisher Scientific) at a nominal magnification of 22,000× (2.1 Å per pixel).
The negative stain datasets were collected using a Tecnai Spirit T12 transmission electron microscope operated at 120 keV (Thermo Fisher Scientific). Datasets were collected at a nominal magnification of × 26,000 (2.34 Å per pixel) except for the P132L-mVenus and P132L-mVenus + CAV1-His datasets, which were collected at a nominal magnification of ×42,000 (1.45 Å per pixel). Sample data were collected using Leginon software on a 4 k × 4 k Rio complementary metal-oxide semiconductor camera (Gatan) at −1.5-μm defocus value (81). Images were manually curated. All data processing was carried out in Relion 3.1.0 (82). About 1000 particles were picked manually and 2D classified. Clear resulting classes were selected and used as references for particle selection on all images. Particles were extracted with a 128-pixel (2.34 Å per pixel datasets) or 208-pixel (1.45 Å per pixel datasets) box size (30 nm by 30 nm boxes). The extracted particles were then 2D classified. The P132L-His and P132L-mVenus datasets consisted of 35,405 and 12,634 particles, respectively. The P132L-mVenus + P132L-His, P132L-His + CAV1-mVenus, and P132L-mVenus + CAV1-His datasets had 47,711, 51,339, and 12,015 particles. The CAV1-His, CAV1-mVenus, and CAV1-mVenus + CAV1-His datasets had 31,956, 9,370, and 42,197 particles.
Symmetric ddG calculations
The experimental CAV1 structure was used as the starting point for all the calculations (PDB 7SC0). Membrane coordinates of this structure were calculated with the PPM (Positioning of Proteins in Membranes) server (83). A single chain from this structure was used as the input for the symmetric FastRelax calculations in Rosetta 3.10. 11-fold symmetry files were created with Rosetta 3.10 based on the experimental structure, and this symmetry was imposed on the monomeric unit for all the following calculations. The position 132 was mutated into all 20 amino acids (including P132P) to screen for the effect of different mutations at this site. Specifically, the amino acid at this site was first mutated using the MutateResidue mover of Rosetta 3.10, followed by a FastRelax calculation to minimize the system with restraints on the system (0.5 Å deviation was allowed). All the calculations were run with the membrane score function mpframework_smooth_fa_2012. Scores are reported for a single protomer (i.e., total score/11). A similar approach was used to analyze point mutants of F99, L122, and V130. Bar graphs were generated with the matplotlib package of Python.
Asymmetric ddG calculations
Asymmetric ddG calculations were used to analyze varying numbers of P132L incorporated into the CAV1 complex. First, different numbers P132L mutations (1–11) were introduced at neighboring protomers, and the Rosetta energy differences with respect to the P132P self-mutation were calculated both for the overall system and per protomer (total score/number of P132L mutations). Only backbone motions and side chain repacking were allowed for the residues within 8 Å of position 132. Next, five different combinations bearing four P132L protomers were tested using the same protocol.
Data availability
Data not already contained in the manuscript are available upon request. Contact Anne Kenworthy at akk7hp@virginia.edu for more information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The University of Michigan Cryo-EM Facility (U-M Cryo-EM) has received generous support from the U-M Life Sciences Institute and the U-M Biosciences Initiative.
Author contributions
B. H., M. D. O., J. M., and A. K. K. conceptualization; B. H. and A. G. methodology; B. H., A. G., S. C., T. W., B. M., J. P., A. T., A. D., L. C., and Y. P. investigation; B. H., A. G., S. C., T. W., M. D. O., J. M., and A. K. K. writing–original draft; B. H., A. G., S. C., T. W., B. M., J. P., A. T., A. D., L. C., Y. P., H. S. M., E. K., M. D. O., J. M., and A. K. K. writing–review and editing; B. H., T. W., A. T., and A. D. validation; B. H., A. G., S. C., T. W., B. M., J. P., A. T., and E. K. visualization; A. G., S. C., T. W., and J. P. formal analysis; S. C. and J. P. data curation; E. K., M. D. O., J. M., and A. K. K. supervision; M. D. O., J. M., and A. K. K. project administration; M. D. O., J. M., and A. K. K. funding acquisition.
Funding and additional information
This work was supported by National Institutes of Health grant R01 HL144131 (A. K. K. and M. D. O.), NIH R01 HL111259 (A. K. K.), NIH R01 GM106720 (A. K. K.), National Institutes of Health grant S10OD020011 (M. D. O.), National Institutes of Health grant S10OD030275 (M. D. O.), National Institutes of Health grant T-32-GM007315 (S. C.), American Heart Association grant 90705 (S. C.), National Institutes of Health grant R01GM080403 (J. M.), National Institutes of Health grant R01HL122010 (J. M.), National Institutes of Health grant R01GM129261 (J. M.), and Humboldt Professorship of the Alexander von Humboldt Foundation (J. M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Karen Fleming
Footnotes
Present address for Angie Deng: Stanford Children’s Hospital, Palo Alto, CA, USA.
Supporting information
References
- 1.Parton R.G., del Pozo M.A. Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 2013;14:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- 2.Ariotti N., Parton R.G. SnapShot: caveolae, caveolins, and cavins. Cell. 2013;154:704–704.e1. doi: 10.1016/j.cell.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Hansen C.G., Nichols B.J. Exploring the caves: cavins, caveolins and caveolae. Trends Cell Biol. 2010;20:177–186. doi: 10.1016/j.tcb.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Razani B., Lisanti M.P. Caveolin-deficient mice: insights into caveolar function human disease. J. Clin. Invest. 2001;108:1553–1561. doi: 10.1172/JCI14611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walser P.J., Ariotti N., Howes M., Ferguson C., Webb R., Schwudke D., et al. Constitutive formation of caveolae in a bacterium. Cell. 2012;150:752–763. doi: 10.1016/j.cell.2012.06.042. [DOI] [PubMed] [Google Scholar]
- 6.Ariotti N., Rae J., Leneva N., Ferguson C., Loo D., Okano S., et al. Molecular characterization of caveolin-induced membrane curvature. J. Biol. Chem. 2015;290:24875–24890. doi: 10.1074/jbc.M115.644336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamaze C., Tardif N., Dewulf M., Vassilopoulos S., Blouin C.M. The caveolae dress code: structure and signaling. Curr. Opin. Cell Biol. 2017;47:117–125. doi: 10.1016/j.ceb.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Parton R.G., McMahon K.A., Wu Y. Caveolae: formation, dynamics, and function. Curr. Opin. Cell Biol. 2020;65:8–16. doi: 10.1016/j.ceb.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Andrade V., Bai J., Gupta-Rossi N., Jimenez A.J., Delevoye C., Lamaze C., et al. Caveolae promote successful abscission by controlling intercellular bridge tension during cytokinesis. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abm5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parton R.G. Caveolae: structure, function, and relationship to disease. Annu. Rev. Cell Dev Biol. 2018;34:111–136. doi: 10.1146/annurev-cellbio-100617-062737. [DOI] [PubMed] [Google Scholar]
- 11.Plucinsky S., Glover K. The C-terminal domain of caveolin-1 and pulmonary arterial hypertension: an emerging relationship. J. Rare Dis. Res. Treat. 2017;2:44–48. [Google Scholar]
- 12.Patni N., Garg A. Congenital generalized lipodystrophies-new insights into metabolic dysfunction. Nat. Rev. Endocrinol. 2015;11:522–534. doi: 10.1038/nrendo.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams T.M., Lisanti M.P. The caveolin genes: from cell biology to medicine. Ann. Med. 2004;36:584–595. doi: 10.1080/07853890410018899. [DOI] [PubMed] [Google Scholar]
- 14.Lamaze C., Torrino S. Caveolae and cancer: a new mechanical perspective. Biomed. J. 2015;38:367–379. doi: 10.4103/2319-4170.164229. [DOI] [PubMed] [Google Scholar]
- 15.Yin H., Liu T., Zhang Y., Yang B. Caveolin proteins: a molecular insight into disease. Front Med. 2016;10:397–404. doi: 10.1007/s11684-016-0483-6. [DOI] [PubMed] [Google Scholar]
- 16.Mathew R. Critical role of caveolin-1 loss/dysfunction in pulmonary hypertension. Med. Sci. (Basel) 2021;9:58. doi: 10.3390/medsci9040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monier S., Parton R.G., Vogel F., Behlke J., Henske A., Kurzchalia T.V. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol. Biol. Cell. 1995;6:911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sargiacomo M., Scherer P.E., Tang Z., Kubler E., Song K.S., Sanders M.C., et al. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren X., Ostermeyer A.G., Ramcharan L.T., Zeng Y., Lublin D.M., Brown D.A. Conformational defects slow Golgi exit, block oligomerization, and reduce raft affinity of caveolin-1 mutant proteins. Mol. Biol. Cell. 2004;15:4556–4567. doi: 10.1091/mbc.E04-06-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayer A., Stoeber M., Bissig C., Helenius A. Biogenesis of caveolae: stepwise assembly of large caveolin and cavin complexes. Traffic. 2010;11:361–382. doi: 10.1111/j.1600-0854.2009.01023.x. [DOI] [PubMed] [Google Scholar]
- 21.Han B., Copeland C.A., Tiwari A., Kenworthy A.K. Assembly and turnover of caveolae: what do we really know? Front Cell Dev Biol. 2016;4:68. doi: 10.3389/fcell.2016.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parton R.G., Tillu V., McMahon K.A., Collins B.M. Key phases in the formation of caveolae. Curr. Opin. Cell Biol. 2021;71:7–14. doi: 10.1016/j.ceb.2021.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Busija A.R., Patel H.H., Insel P.A. Caveolins and cavins in the trafficking, maturation, and degradation of caveolae: implications for cell physiology. Am. J. Physiol. Cell Physiol. 2017;312:C459–C477. doi: 10.1152/ajpcell.00355.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthaeus C., Taraska J.W. Energy and dynamics of caveolae trafficking. Front Cell Dev Biol. 2020;8:614472. doi: 10.3389/fcell.2020.614472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Z., Okamoto T., Boontrakulpoontawee P., Katada T., Otsuka A.J., Lisanti M.P. Identification, sequence, and expression of an invertebrate caveolin gene family from the nematode Caenorhabditis elegans. Implications for the molecular evolution of mammalian caveolin genes. J. Biol. Chem. 1997;272:2437–2445. doi: 10.1074/jbc.272.4.2437. [DOI] [PubMed] [Google Scholar]
- 26.Kirkham M., Nixon S.J., Howes M.T., Abi-Rached L., Wakeham D.E., Hanzal-Bayer M., et al. Evolutionary analysis and molecular dissection of caveola biogenesis. J. Cell Sci. 2008;121:2075–2086. doi: 10.1242/jcs.024588. [DOI] [PubMed] [Google Scholar]
- 27.Hayashi K., Matsuda S., Machida K., Yamamoto T., Fukuda Y., Nimura Y., et al. Invasion activating caveolin-1 mutation in human scirrhous breast cancers. Cancer Res. 2001;61:2361–2364. [PubMed] [Google Scholar]
- 28.Shatz M., Lustig G., Reich R., Liscovitch M. Caveolin-1 mutants P132L and Y14F are dominant negative regulators of invasion, migration and aggregation in H1299 lung cancer cells. Exp. Cell Res. 2010;316:1748–1762. doi: 10.1016/j.yexcr.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Bonuccelli G., Casimiro M.C., Sotgia F., Wang C., Liu M., Katiyar S., et al. Caveolin-1 (P132L), a common breast cancer mutation, confers mammary cell invasiveness and defines a novel stem cell/metastasis-associated gene signature. Am. J. Pathol. 2009;174:1650–1662. doi: 10.2353/ajpath.2009.080648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koike S., Kodera Y., Nakao A., Iwata H., Yatabe Y. Absence of the caveolin-1 P132L mutation in cancers of the breast and other organs. J. Mol. Diagn. 2010;12:712–717. doi: 10.2353/jmoldx.2010.090180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacroix-Triki M., Geyer F.C., Reis-Filho J.S. Caveolin-1 P132L mutation in human cancers: 1 CAVeat to be voiced. J. Mol. Diagn. 2010;12:562–565. doi: 10.2353/jmoldx.2010.100093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferraldeschi R., Latif A., Clarke R.B., Spence K., Ashton G., O'Sullivan J., et al. Lack of caveolin-1 (P132L) somatic mutations in breast cancer. Breast Cancer Res. Treat. 2012;132:1185–1186. doi: 10.1007/s10549-012-1981-0. [DOI] [PubMed] [Google Scholar]
- 33.Patani N., Lambros M.B., Natrajan R., Dedes K.J., Geyer F.C., Ward E., et al. Non-existence of caveolin-1 gene mutations in human breast cancer. Breast Cancer Res. Treat. 2012;131:307–310. doi: 10.1007/s10549-011-1761-2. [DOI] [PubMed] [Google Scholar]
- 34.Duregon E., Senetta R., Pittaro A., Verdun di Cantogno L., Stella G., De Blasi P., et al. CAVEOLIN-1 expression in brain metastasis from lung cancer predicts worse outcome and radioresistance, irrespective of tumor histotype. Oncotarget. 2015;6:29626–29636. doi: 10.18632/oncotarget.4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper-Knock J., Zhang S., Kenna K.P., Moll T., Franklin J.P., Allen S., et al. Rare variant Burden analysis within enhancers identifies CAV1 as an ALS risk gene. Cell Rep. 2020;33:108456. doi: 10.1016/j.celrep.2020.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minetti C., Sotgia F., Bruno C., Scartezzini P., Broda P., Bado M., et al. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat. Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- 37.Galbiati F., Volonte D., Minetti C., Chu J.B., Lisanti M.P. Phenotypic behavior of caveolin-3 mutations that cause autosomal dominant limb girdle muscular dystrophy (LGMD-1C). Retention of LGMD-1C caveolin-3 mutants within the golgi complex. J. Biol. Chem. 1999;274:25632–25641. doi: 10.1074/jbc.274.36.25632. [DOI] [PubMed] [Google Scholar]
- 38.Carbone I., Bruno C., Sotgia F., Bado M., Broda P., Masetti E., et al. Mutation in the CAV3 gene causes partial caveolin-3 deficiency and hyperCKemia. Neurology. 2000;54:1373–1376. doi: 10.1212/wnl.54.6.1373. [DOI] [PubMed] [Google Scholar]
- 39.Pradhan B.S., Proszynski T.J. A role for caveolin-3 in the pathogenesis of muscular dystrophies. Int. J. Mol. Sci. 2020;21:8736. doi: 10.3390/ijms21228736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee H., Park D.S., Razani B., Russell R.G., Pestell R.G., Lisanti M.P. Caveolin-1 mutations (P132L and null) and the pathogenesis of breast cancer: caveolin-1 (P132L) behaves in a dominant-negative manner and caveolin-1 (-/-) null mice show mammary epithelial cell hyperplasia. Am. J. Pathol. 2002;161:1357–1369. doi: 10.1016/S0002-9440(10)64412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han B., Tiwari A., Kenworthy A.K. Tagging strategies strongly affect the fate of overexpressed caveolin-1. Traffic. 2015;16:417–438. doi: 10.1111/tra.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galbiati F., Volonte D., Minetti C., Bregman D.B., Lisanti M.P. Limb-girdle muscular dystrophy (LGMD-1C) mutants of caveolin-3 undergo ubiquitination and proteasomal degradation. Treatment with proteasomal inhibitors blocks the dominant negative effect of LGMD-1C mutants and rescues wild-type caveolin-3. J. Biol. Chem. 2000;275:37702–37711. doi: 10.1074/jbc.M006657200. [DOI] [PubMed] [Google Scholar]
- 43.Ritz D., Vuk M., Kirchner P., Bug M., Schutz S., Hayer A., et al. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieth M.D., Lee J., Glover K.J. Probing the caveolin-1 P132L mutant: critical insights into its oligomeric behavior and structure. Biochemistry. 2012;51:3911–3918. doi: 10.1021/bi3001853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryu B.K., Lee M.G., Kim N.H., Lee K.Y., Oh S.J., Moon J.R., et al. Bidirectional alteration of Cav-1 expression is associated with mitogenic conversion of its function in gastric tumor progression. BMC Cancer. 2017;17:766. doi: 10.1186/s12885-017-3770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Syme C.A., Zhang L., Bisello A. Caveolin-1 regulates cellular trafficking and function of the glucagon-like Peptide 1 receptor. Mol. Endocrinol. 2006;20:3400–3411. doi: 10.1210/me.2006-0178. [DOI] [PubMed] [Google Scholar]
- 47.Ravid D., Leser G.P., Lamb R.A. A role for caveolin 1 in assembly and budding of the paramyxovirus parainfluenza virus 5. J. Virol. 2010;84:9749–9759. doi: 10.1128/JVI.01079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porta J.C., Han B., Gulsevin A., Chung J.M., Peskova Y., Connolly S., et al. Molecular architecture of the human caveolin-1 complex. Sci. Adv. 2022;8:eabn7232. doi: 10.1126/sciadv.abn7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scherer P.E., Okamoto T., Chun M., Nishimoto I., Lodish H.F., Lisanti M.P. Identification, sequence, and expression of caveolin-2 defines a caveolin gene family. Proc. Natl. Acad. Sci. U. S. A. 1996;93:131–135. doi: 10.1073/pnas.93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang Z., Scherer P.E., Okamoto T., Song K., Chu C., Kohtz D.S., et al. Molecular cloning of caveolin-3, a novel member of the caveolin gene family expressed predominantly in muscle. J. Biol. Chem. 1996;271:2255–2261. doi: 10.1074/jbc.271.4.2255. [DOI] [PubMed] [Google Scholar]
- 51.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7, Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Copeland C.A., Han B., Tiwari A., Austin E.D., Loyd J.E., West J.D., et al. A disease-associated frameshift mutation in caveolin-1 disrupts caveolae formation and function through introduction of a de novo ER retention signal. Mol. Biol. Cell. 2017;28:3095–3111. doi: 10.1091/mbc.E17-06-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han B., Copeland C.A., Kawano Y., Rosenzweig E.B., Austin E.D., Shahmirzadi L., et al. Characterization of a caveolin-1 mutation associated with both pulmonary arterial hypertension and congenital generalized lipodystrophy. Traffic. 2016;17:1297–1312. doi: 10.1111/tra.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hill M.M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S.J., et al. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 2008;132:113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanson C.A., Drake K.R., Baird M.A., Han B., Kraft L.J., Davidson M.W., et al. Overexpression of caveolin-1 is sufficient to phenocopy the behavior of a disease-associated mutant. Traffic. 2013;14:663–677. doi: 10.1111/tra.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tiwari A., Copeland C.A., Han B., Hanson C.A., Raghunathan K., Kenworthy A.K. Caveolin-1 is an aggresome-inducing protein. Sci. Rep. 2016;6:38681. doi: 10.1038/srep38681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han B., Porta J.C., Hanks J.L., Peskova Y., Binshtein E., Dryden K., et al. Structure and assembly of CAV1 8S complexes revealed by single particle electron microscopy. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song K.S., Tang Z.L., Li S.W., Lisanti M.P. Mutational analysis of the properties of caveolin-1. A novel role for the C-terminal domain in mediating homo-typic caveolin-caveolin interactions. J. Biol. Chem. 1997;272:4398–4403. doi: 10.1074/jbc.272.7.4398. [DOI] [PubMed] [Google Scholar]
- 62.Senju Y., Itoh Y., Takano K., Hamada S., Suetsugu S. Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J. Cell Sci. 2011;124:2032–2040. doi: 10.1242/jcs.086264. [DOI] [PubMed] [Google Scholar]
- 63.Moren B., Shah C., Howes M.T., Schieber N.L., McMahon H.T., Parton R.G., et al. EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Mol. Biol. Cell. 2012;23:1316–1329. doi: 10.1091/mbc.E11-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hubert M., Larsson E., Lundmark R. Keeping in touch with the membrane; protein- and lipid-mediated confinement of caveolae to the cell surface. Biochem. Soc. Trans. 2020;48:155–163. doi: 10.1042/BST20190386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ludwig A., Howard G., Mendoza-Topaz C., Deerinck T., Mackey M., Sandin S., et al. Molecular composition and ultrastructure of the caveolar coat complex. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matthaeus C., Sochacki K.A., Dickey A.M., Puchkov D., Haucke V., Lehmann M., et al. The molecular organization of differentially curved caveolae indicates bendable structural units at the plasma membrane. Nat. Commun. 2022;13:7234. doi: 10.1038/s41467-022-34958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morales-Paytuvi F., Ruiz-Mirapeix C., Fajardo A., Rae J., Bosch M., Enrich C., et al. Proteostatic regulation of caveolins avoids premature oligomerisation and preserves ER homeostasis. bioRxiv. 2022 doi: 10.1101/2022.04.24.489297. [preprint] [DOI] [Google Scholar]
- 68.Mercier I., Jasmin J.F., Pavlides S., Minetti C., Flomenberg N., Pestell R.G., et al. Clinical and translational implications of the caveolin gene family: Lessons from mouse models and human genetic disorders. Lab Invest. 2009;89:614–623. doi: 10.1038/labinvest.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shah D.S., Nisr R.B., Stretton C., Krasteva-Christ G., Hundal H.S. Caveolin-3 deficiency associated with the dystrophy P104L mutation impairs skeletal muscle mitochondrial form and function. J. Cachexia Sarcopenia Muscle. 2020;11:838–858. doi: 10.1002/jcsm.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woodman S.E., Sotgia F., Galbiati F., Minetti C., Lisanti M.P. Caveolinopathies: mutations in caveolin-3 cause four distinct autosomal dominant muscle diseases. Neurology. 2004;62:538–543. doi: 10.1212/wnl.62.4.538. [DOI] [PubMed] [Google Scholar]
- 71.Garg A., Kircher M., Del Campo M., Amato R.S., Agarwal A.K. Whole exome sequencing identifies de novo heterozygous CAV1 mutations associated with a novel neonatal onset lipodystrophy syndrome. Am. J. Med. Genet. A. 2015;167:1796–1806. doi: 10.1002/ajmg.a.37115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schrauwen I., Szelinger S., Siniard A.L., Kurdoglu A., Corneveaux J.J., Malenica I., et al. A frame-shift mutation in CAV1 is associated with a severe neonatal progeroid and lipodystrophy syndrome. PLoS One. 2015;10 doi: 10.1371/journal.pone.0131797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marsboom G., Chen Z., Yuan Y., Zhang Y., Tiruppathi C., Loyd J.E., et al. Aberrant caveolin-1-mediated Smad signaling and proliferation identified by analysis of adenine 474 deletion mutation (c.474delA) in patient fibroblasts: A new perspective on the mechanism of pulmonary hypertension. Mol. Biol. Cell. 2017;28:1177–1185. doi: 10.1091/mbc.E16-11-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim C.A., Delepine M., Boutet E., El Mourabit H., Le Lay S., Meier M., et al. Association of a homozygous nonsense caveolin-1 mutation with Berardinelli-Seip congenital lipodystrophy. J. Clin. Endocrinol. Metab. 2008;93:1129–1134. doi: 10.1210/jc.2007-1328. [DOI] [PubMed] [Google Scholar]
- 75.Cao H., Alston L., Ruschman J., Hegele R.A. Heterozygous CAV1 frameshift mutations (MIM 601047) in patients with atypical partial lipodystrophy and hypertriglyceridemia. Lipids Health Dis. 2008;7:3. doi: 10.1186/1476-511X-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karhan A.N., Zammouri J., Auclair M., Capel E., Apaydin F.D., Ates F., et al. Biallelic CAV1 null variants induce congenital generalized lipodystrophy with achalasia. Eur. J. Endocrinol. 2021;185:841–854. doi: 10.1530/EJE-21-0915. [DOI] [PubMed] [Google Scholar]
- 77.Sotgia F., Woodman S.E., Bonuccelli G., Capozza F., Minetti C., Scherer P.E., et al. Phenotypic behavior of caveolin-3 R26Q, a mutant associated with hyperCKemia, distal myopathy, and rippling muscle disease. Am. J. Physiol. Cell Physiol. 2003;285:C1150–1160. doi: 10.1152/ajpcell.00166.2003. [DOI] [PubMed] [Google Scholar]
- 78.Austin E.D., Ma L., LeDuc C., Berman Rosenzweig E., Borczuk A., Phillips J.A., 3rd, et al. Whole exome sequencing to identify a novel gene (caveolin-1) associated with human pulmonary arterial hypertension. Circ. Cardiovasc. Genet. 2012;5:336–343. doi: 10.1161/CIRCGENETICS.111.961888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 80.Ohi M., Li Y., Cheng Y., Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 82.Zivanov J., Nakane T., Scheres S.H.W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020;7:253–267. doi: 10.1107/S2052252520000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lomize M.A., Pogozheva I.D., Joo H., Mosberg H.I., Lomize A.L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data not already contained in the manuscript are available upon request. Contact Anne Kenworthy at akk7hp@virginia.edu for more information.