Abstract
Emfourin (M4in) is a protein metalloprotease inhibitor recently discovered in the bacterium Serratia proteamaculans and the prototype of a new family of protein protease inhibitors with an unknown mechanism of action. Protealysin-like proteases (PLPs) of the thermolysin family are natural targets of emfourin-like inhibitors widespread in bacteria and known in archaea. The available data indicate the involvement of PLPs in interbacterial interaction as well as bacterial interaction with other organisms and likely in pathogenesis. Arguably, emfourin-like inhibitors participate in the regulation of bacterial pathogenesis by controlling PLP activity. Here, we determined the 3D structure of M4in using solution NMR spectroscopy. The obtained structure demonstrated no significant similarity to known protein structures. This structure was used to model the M4in–enzyme complex and the complex model was verified by small-angle X-ray scattering. Based on the model analysis, we propose a molecular mechanism for the inhibitor, which was confirmed by site-directed mutagenesis. We show that two spatially close flexible loop regions are critical for the inhibitor–protease interaction. One region includes aspartic acid forming a coordination bond with catalytic Zn2+ of the enzyme and the second region carries hydrophobic amino acids interacting with protease substrate binding sites. Such an active site structure corresponds to the noncanonical inhibition mechanism. This is the first demonstration of such a mechanism for protein inhibitors of thermolysin family metalloproteases, which puts forward M4in as a new basis for the development of antibacterial agents relying on selective inhibition of prominent factors of bacterial pathogenesis belonging to this family.
Keywords: metalloprotease, protease inhibitor, protein complex, protein structure, gram-negative bacteria, inhibition mechanism, nuclear magnetic resonance (NMR), emfourin, protealysin, thermolysin-like protease
Protein inhibitors of proteases (PIPs) appear to be one of the key regulatory factors of proteolytic activity in all living systems. In certain significant cases, the biological functions and mechanisms of PIP action are relatively well studied (e.g., (1, 2, 3, 4, 5, 6, 7)). However, the bulk of PIPs remain largely underexplored. For instance, the MEROPS database of peptidases and their protein inhibitors (www.ebi.ac.uk/merops/) (8) includes 1,142,788 sequences and 4431 identifiers of peptidases but as low as 131,636 sequences and 686 identifiers of the inhibitors (as of February 21, 2023). The physiological targets remain unknown for most inhibitors and protein inhibitors are not known for most proteases. For instance, 354 (∼80%) out of 444 human proteases in MEROPS have no annotated inhibitors and 13 (∼14%) out of 94 inhibitors have no annotated targets (9). Among prokaryotes, the biological function of inhibitors is known in rare cases (10). Considering the critical role of proteases in the regulation of numerous biochemical pathways, the identification of new endogenous PIPs and their natural targets as well as the elucidation of the physiological function of the protease-inhibitor pairs seem critical for the understanding of protease biology and cellular regulation in general.
Emfourin (M4in) is a novel protein inhibitor of metalloproteases recently described by us in the bacterium Serratia proteamaculans. M4in is the prototype of a new family I104 in the MEROPS database (www.ebi.ac.uk/merops/cgi-bin/famsum?family=I104). The genes encoding representatives of the family, emfourin-like inhibitors (ELIs), are widespread in bacteria and occur in archaea. In all these species, the ELI genes are immediately downstream of genes encoding protealysin-like proteases (PLPs), metalloproteases of the MEROPS family M4 (https://www.ebi.ac.uk/merops/cgi-bin/famsum?family=M4) and putative natural targets of ELIs. In many cases, the protease and inhibitor genes form a bicistronic operon. The biological role of the ELI-PLP association currently remains unclear. These proteins are assumed to participate in yet unidentified system of interbacterial competition where PLPs are toxic effectors and ELIs function as immunity proteins protecting sister cells (11). At the same time, published data indicate the involvement of PLPs in bacterial interaction with higher organisms and possibly in pathogenesis (12, 13, 14, 15, 16, 17, 18). Yet, the available data are fragmentary and the role of PLPs in the pathogenesis as well as the molecular mechanisms of their action remain obscure. It is not improbable that ELIs function in cooperation with PLPs in this context as well. Anyway, the PLP–ELI pairs can exemplify the coevolution of proteases and their inhibitors and their investigation can uncover new mechanisms of proteolytic regulation.
To date, M4in is the only characterized protein in the ELI family. M4in is a potent slow-binding competitive inhibitor of protealysin (PLN), a metalloprotease of S. proteamaculans and a natural target of M4in. It was established that M4in interacts with the active site region of PLN; however, the inhibition mechanism remains to be elucidated. Experiments with thermolysin demonstrated that M4in can inhibit other M4 peptidases that are not PLPs (11). This further substantiates the investigation of ELIs. M4 proteases are synthesized by significant human pathogens Pseudomonas aeruginosa, Staphylococcus aureus, Staphylococcus epidermidis, Burkholderia cenocepacia, Enterococcus faecalis, Legionella pneumophila, Clostridium perfringens, and several pathogenic Vibrio species (19), which proposes their protein inhibitors as a promising base for the development of new-generation antibacterial agents (20, 21).
Here, solution NMR spectroscopy was used to determine the 3D structure of M4in. This structure has no significant similarity to known PIP structures, which confirms that ELIs are a separate family. The obtained M4in structure allowed us to generate a model of M4in–PLN interaction and to propose a mechanism of the inhibitor effect. This mechanism was corroborated by site-directed mutagenesis. The data obtained indicate that the ELI inhibition is critically governed by the Asp residue coordinating the zinc ion in the enzyme active site as well as the hydrophobic amino acid residues interacting with the enzyme region that determines its substrate specificity.
Results
NMR structure of emfourin
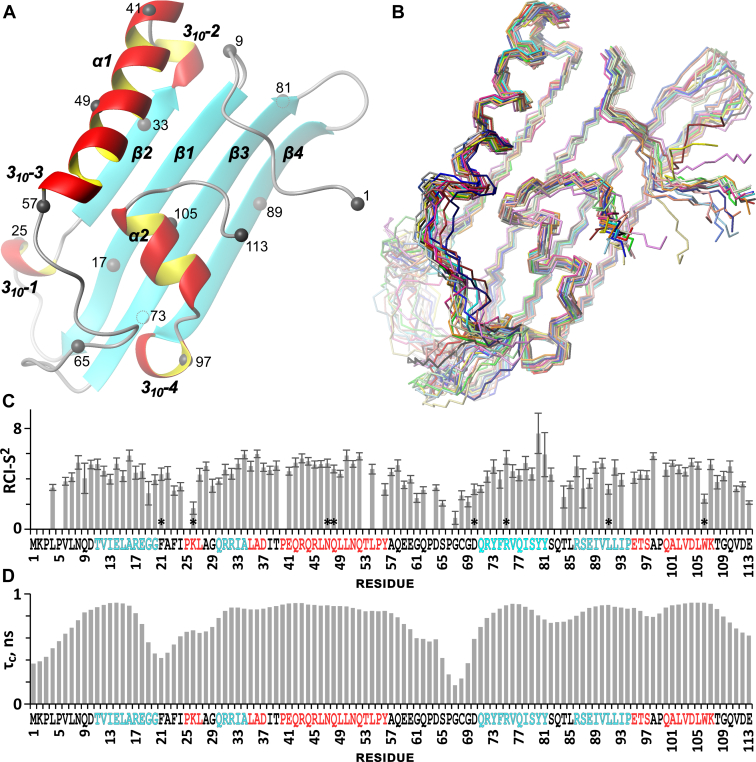
Figure 1 shows the overlay of the 20 structures deposited in the Protein Data Bank (PDB) under the numbers ranged by target function (PDB ID: 6ZYG) and the cartoon of the representative M4in structure selected from the ensemble. Table 1 summarizes experimental restraints used for structure calculations and structural statistics. The experimentally determined secondary structure of M4in sufficiently corresponds to that predicted from chemical shifts (Fig. S1, C and D). The major element of the M4in molecule is the β-sheet composed of four antiparallel β-strands (Fig. 1): β1, Thr11-Gly20; β2, Gln30-Ala34; β3, Gln71-Tyr81; and β4, Arg86-Pro94. (The limits of the secondary structure elements correspond to the ensemble model 1 with the lowest target function.) One of the β-sheet surfaces interacts via hydrophobic contacts with two α-helices: the long α1 (Pro40-Thr53) going into a loop of the 310-helix (310-3 in Figs. 1 and S1; for most of the 20 structures, 310-3 includes Leu54-Tyr56 region; however, certain 310-helices start with Gln52 or Thr53) and the short α2 (which includes the Gln100-Lys107 area in most of these structures but sometimes ends with Leu105). In both cases, the main chain transition from β-sheet to α-helix occurs within a short region via an insertion of an extra turn of the 310-helix (310-2, Leu35-Asp37, and 310-4, Glu95-Ser97). The α2 helix is almost parallel to the β-sheet plane and the helix position is further stabilized by the hydrogen bond Trp106-NHε1…O=Gly29. M4in features the presence of the dynamically reconfigurable (according to the NOE and TALOS-N data, see Experimental procedures for details) 310-2 helix (Pro25-Leu27), whose rearrangement likely affects the neighboring regions. The presence of antiparallel β-strands together with α- and 310-helices in M4in molecule assigns the 3D fold of this compact globule to the α+β type (22).
Figure 1.
Spatial structure and mobility of the M4in molecule.A, the cartoon representation of the ensemble model with the lowest CYANA target function (first in the deposited set, PDB ID 6ZYG). The gray spheres indicate Cα atoms of every fourth residue. B, the overlay of 20 calculated molecular models of M4in. The structures were aligned by atoms of the main chain rigid regions 6 to 18, 31 to 60, and 71 to 112. This projection corresponds to that in figure A. C, the distribution of rotational correlation time (τc, ns). RMSDs are shown as error bars. The residues with overlapping spectral signals are indicated by asterisks (their deviation can exceed the specified value). D, random coil index order parameter (RCI-S2) calculated by TALOS-N (its values reflect the mobility of the main chain atoms in the millisecond range). The amino acid sequence in C and D is colored according to the secondary structure as in A. PDB, Protein Data Bank.
Table 1.
Structural statistics: input and output data
| Initial data for structure calculation | |
|---|---|
| Total number of unique NOE constraints (upper/lower) | 1719/30,996 |
| Intraresidue | 974/103 |
| Interresidue | 745/30,893 |
| sequential (|i-j| = 1) | 467/632 |
| medium-range (1<|i-j|≤4) | 144/2596 |
| long-range (|i-j|>4) | 134/27,665 |
| Upper/lower constraints on hydrogen bonds | 120/120 (na = 60) |
| Upper/lower constraints on disulfide bonds | 0/0 (n = 0) |
| Constraints on dihedral angles, total | 1049 (n = 458) |
| Without usage of CYANA macro ramaacob | 946 (n = 436) |
| angle φ of the main chain | 84 (n = 84) |
| angle ψ of the main chain | 88 (n = 88) |
| angle χ1 of a side chain in a certain conformation | 24 (n = 24) |
| angle χ1 of a side chain with two possible conformations | 10 (n = 5) |
| angle χ2 of a side chain in a certain conformation | 1 (n = 1) |
| angles χ1 (n = 68), χ2 (52), χ21 (19), χ22 (19), χ3 (12), χ31 (21), χ32 (14), χ4 (4), and χ5 (3) for the formation of a hindered conformation at sp3-hybridized atoms, OH, CH3, and NH3+ groups (three intervals for each angle) | 636 (n = 212) |
| Additional constraints on the angles φ and ψ of some residues from flexible regions, which, according to a preliminary calculation, fell into energetically unfavorable areas on the Ramachandran map (obtained with ramaacob) | 103 (n = 22) |
| angle φ of the main chain | 49 (n = 10) |
| angle ψ of the main chain | 54 (n = 12) |
| Structure calculation quality parameters (20 models), PDB ID | 6ZYG |
|---|---|
| Target function (Å2, mean ± RMSDc)d | 7.11 ± 0.99 |
| Number of violations (mean ± RMSDc)d | |
| of upper/lower distance restraints (>0.2 Å) | 13 ± 4/3 ± 2 |
| of upper/lower distance restraints (>0.3 Å) | 5 ± 2/1 ± 1 |
| per sum of van der Waals radii (>0.2 Å, >0.3 Å) | 5 ± 1, 0 ± 1 |
| of dihedral angle restraints (>5o, >8o) | 5 ± 2, 2 ± 2 |
| Averaged pairwise RMSDc among models (Å, mean ± RMSD)e | |
| calculated for all backbone atoms | 1.65 ± 0.34 |
| calculated for all heavy atoms | 2.58 ± 0.36 |
| calculated for backbone atoms of “rigid” regions (6..18, 31..60, 71..112) | 1.02 ± 0.21 |
| calculated for heavy atoms of “rigid” regions (6..18, 31..60, 71..112) | 1.95 ± 0.21 |
| Ramachandran map analysisf, % residues | |
| in most favored regions | 80.4 |
| in additionally allowed regions | 19.1 |
| in generously allowed regions | 0.2 |
| in disallowed regions | 0.4 |
Number of bonds or angles.
CYANA macro for φ, ψ angle restrictions within favorable areas on the Ramachandran map.
Root mean square deviation.
Calculated in CYANA.
Calculated by MOLMOL.
Calculated in CYANA accordingly with categories defined by the program Procheck (90).
The structure of M4in is stabilized by hydrogen bonds; their full list used in the calculation as well as the method for their identification are given in Fig. S1, A and B, and in Supporting information Methods. In the case of the donor amino groups of Gln9 and Glu41, the only possible hydrogen bond acceptors are the carbonyl groups of side chains of the same residues. Considering the steric hindrance of such hydrogen bond formation and hence their lability, the standard (strict) constraints were set for interatomic distances to form the more stable Gln9-NH…Oε1-Gln9 bond (for which the chemical shift temperature coefficient of amide proton Δδ1HN/ΔT ∼−1.5 ppb/K) and a more labile hydrogen bond involving the Glu41 amino group (Δδ1HN/ΔT ∼ 3.0 ppb/K) was left open. Based on the NOE contacts, the conformation of some Pro residues was set to “up” for Pro3, Pro25, Pro40, Pro63, and Pro94; “down” for Pro66; and left unclear for Pro5, Pro55, and Pro99.
Based on the rotational correlation time (τc, ns) estimated from 15N CSA/dipolar cross-correlated transverse relaxation NMR spectrum (23) as well as on the random coil index order parameter (reflecting the mobility of the main chain atoms in the millisecond range), high mobility regions relative to other ones can be recognized in the M4in molecule. Apart from the N- and C-terminal regions (Met1-Asn8 and Thr108-Glu113, respectively), such regions include Glu18-Gly29 and Gly61-Asp70 (Fig. 1, C and D).
All NMR spectra of M4in demonstrate multiple peaks of several (at least two) minor conformations with the relative intensity of about 1/8 of the corresponding major conformation signal. About half of minor signals are in the vicinity of the major conformation peaks. M4in has nine proline residues and some of them (Pro3, Pro5, Pro25, Pro63, and Pro66) reside within the dynamic regions of the molecule, which suggests conformational rearrangements caused by cis-trans isomerization of the peptide bonds proceeding two prolines; this could give rise to two minor conformations with the relative proportion of ∼10% each and a third one (undetectable) with the intensity of ∼1%.
The comparison of the M4in spatial structure against the PDB using the Dali web server (http://ekhidna2.biocenter.helsinki.fi/dali/) (24) demonstrated no significant structural similarity (Dali Z-score > 8 (25)) between M4in and other known structures. At the same time, this comparison reveals a fair amount of “gray area” structures with the Dali Z-score from 2 to 4.6, i.e., having topological similarity to M4in. In most cases, the similarity is limited to the β-structure with a fold similar (usually partially) to the β-sheet of M4in. At the same time, proteins of the mago nashi family demonstrate similarity not only in all four β-sheet strands but also in both turns of the α2 helix and have a similar molecule topology in general. The Dali results include 19 structures of mago nashi proteins within different complexes (with the Z-score from 3.2 to 3.8). The highest Dali Z-scores are observed for the human mago nashi protein homolog (PDB ID 2hyi, chain G, Dali Z-score = 3.8) and Drosophila melanogaster mago nashi protein (PDB ID 2x1g, chain B, Dali Z-score = 3.7) (Fig. S2). Eukaryotic proteins of the mago nashi family (http://pfam.xfam.org/family/PF02792) are one of the core elements of the exon junction complex, which in turn is a universal messenger ribonucleoprotein component that is assembled during splicing and binds to mRNAs upstream of exon-exon junctions (26). Thus, the proteins most structurally similar to M4in have no significant amino acid similarity with it, have different functions, and belong to remote biological taxa.
Overall, M4in demonstrates no structural similarity with known PIPs, which confirms the recognition of the family of ELIs and indicates that ELIs do not belong to any known PIP clans (according to the MEROPS classification).
Model of emfourin–PLN interaction
The PLN–M4in interaction was studied by small-angle X-ray scattering (SAXS) of their complex in solution. The SAXS experiments demonstrated (in accordance with the previous data (11)) that PLN and M4in are monomeric in solution throughout the concentration range used (see Supporting information Results, Fig. 2A, and Table S1 for details).
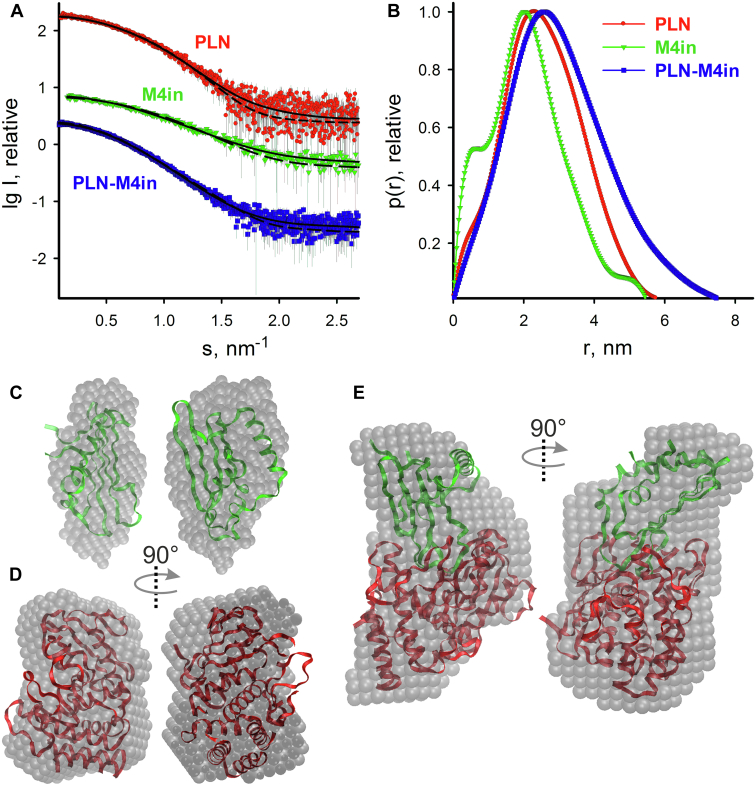
Figure 2.
Experimental X-ray scattering pattern, distance distribution functions, and SAXS-based structural models of protealysin (PLN), emfourin (M4in), and PLN–M4in complex.A, small-angle X-ray scattering patterns of PLN–M4in complex (blue curve), PLN (red curve), and M4in (green curve). Experimental data are displayed as dots with error bars; the scattering from the ab initio model by DAMMIN, as solid lines; and the calculated curves from the atomic models computed by CRYSOL from the crystallographic model of PLN (PDB ID: 2VQX), NMR model of M4in (PDB ID: 6ZYG), and the rigid body model of PLN–M4in complex (obtained by ROSIE), as dashed lines. The plots display the logarithm of the scattering intensity as a function of the momentum transfer. The curves are shifted down by one logarithmic unit for clarity. B, the distance distribution functions estimated by GNOM. The color scheme is the same as in A. C, ab initio bead model of M4in obtained by DAMMIN (gray semitransparent spheres) superimposed on the NMR M4in model in ribbons format (green). The right view was obtained by a 90° clockwise rotation around the vertical axis. D, ab initio bead model of PLN obtained by DAMMIN (gray semitransparent spheres) superimposed on the crystallographic model of PLN in ribbons format (red). The right was obtained by a 90° clockwise rotation around the vertical axis. E, ab initio bead model of the PLN–M4in complex obtained by DAMMIN (gray semitransparent spheres) superimposed on the rigid body model of PLN–M4in complex obtained by ROSIE (red and green structures of PLN and M4in, respectively, in ribbons format). The right was obtained by a 90° clockwise rotation around the vertical axis. The figures were generated using the VMD program (89). PDB, Protein Data Bank.
The SAXS analysis of the PLN–M4in solution (Table S1) yields the experimental Vp = (71 ± 5) nm3, which is consistent with the protein complex (that has a 1:1 stoichiometry) in solution. One can see that the values of Rg (2.35 ± 0.05 nm) and Dmax (7.6 ± 0.2 nm) for the PLN–M4in complex are significantly higher than those for standalone PLN and M4in calculated from their atomic structures (PDB ID: 2VQX, Rg = 1.9 nm and Dmax = 6.1 nm for PLN; PDB ID: 6ZYG, Rg = 1.7 nm and Dmax = 5.4 nm for M4in). The overall parameters of PLN–M4in solution indicate an elongated structure of the protein complex and its p(r) function has an asymmetric tail (Fig. 2B). The 1:1 stoichiometry of the PLN–M4in complex coincides with the previous SAXS data collected at the X-ray lab source (11).
The macromolecular shapes of individual proteins and the PLN–M4in complex have been reconstructed by ab initio modeling using only the experimental X-ray scattering data. The ab initio low-resolution models displayed in Figure 2, C–E have been generated by DAMMIN (see Experimental procedures for details). The ab initio low-resolution shape of M4in (Fig. 2C) fits the experimental data with χ2 = 1.16 (Fig. 2A, solid line). The model has an elongated ellipsoidal shape. The scattering curve from the NMR-derived model (PDB ID: 6ZYG) calculated by CRYSOL also agrees with the experimental data (χ2 = 1.20, (Fig. 2A, dashed line). Both models overlap (Fig. 2C). A typical low-resolution shape of the PLN protein reconstructed ab initio (Fig. 2D) fits the experimental data with χ2 = 0.99 (Fig. 2A, solid line). The scattering curve from the crystallographic model (PDB ID: 2VQX) calculated by CRYSOL also agrees with the experimental data (χ2 = 1.15, Fig. 2A, dashed line). Both models overlap (Fig. 2D). The ab initio low-resolution model of the PLN–M4in complex (Fig. 2E) obtained by DAMMIN fits the experimental data with χ2 = 0.97 (Fig. 2A, solid line). The model has an elongated shape with a moderate twisting along the long axis.
We used high-resolution atomic models of PLN (PDB ID: 2VQX) and M4in (PDB ID: 6ZYG) to build the rigid-body model of the PLN–M4in complex for comparison with the SAXS data. The positions and orientations of M4in with respect to PLN were refined using the SASREF program. Originally, several runs without contact restrains were performed, and in ∼10% of cases, M4in contacted the active region of PLN (involving the following PLN residues: Tyr126, Glu129, Tyr130, GlnQ131, Asn132, Phe134, Trp135, Asn136, Asp146, Ile149, Phe150, Phe153, Val159, His162, Ala163 corresponding to Glu163 in the active protein, His166, Tyr176, Asp259, His264, Leu265, Leu216, Leu226, Arg227, Tyr236, Pro245, and Asn260; numbering after the PLN precursor). Importantly, the latter models allowed M4in contact with the active site of PLN either by the region containing Asp70 and Phe21 of M4in (∼20% of cases) or by the opposite part embracing the N- and C-termini of M4in (∼80% of cases).
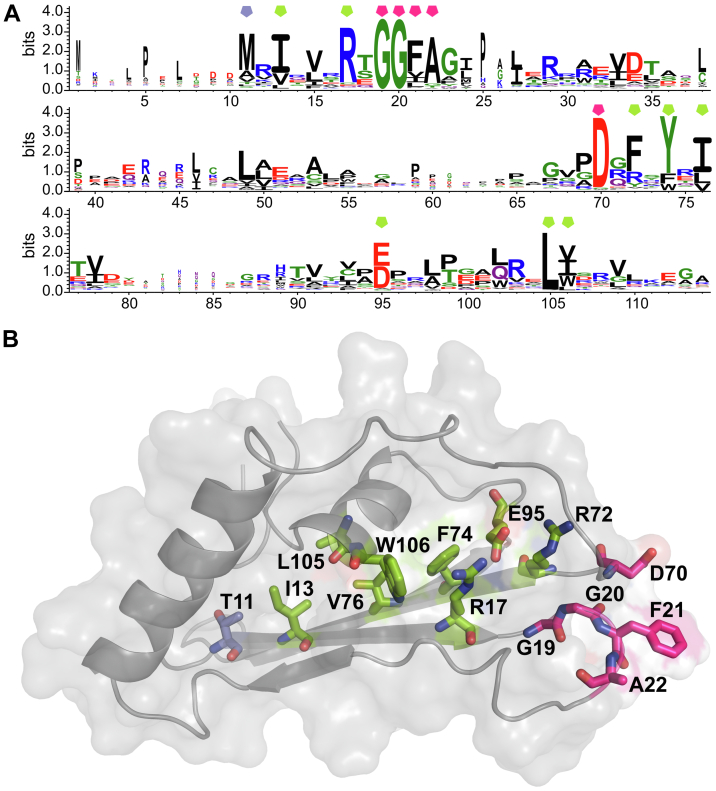
Since the SAXS analysis of the PLN–M4in complex structure allowed no conclusion about the inhibitor–enzyme interface, sequences of ELIs were compared. Nine out of 14 most conserved amino acid residues (Fig. 3A) proved buried in the protein globule in M4in (Fig. 3B). Five of them (Ile13, Phe74, Val76, Leu105, and Trp106 in M4in; numbering after M4in) are hydrophobic and are likely incorporated into the hydrophobic core of the protein. Arg17 and Glu95 (roughly half of ELIs have Asp at position 95) form an intramolecular ionic bond (the distance between oxygen atoms in the γ-carboxyl group of Glu95 and nitrogen atoms of the guanidine group of Arg17 is at least 2.7 Å in the structures calculated from NMR data); in addition, the carboxyl group of Glu95 interacts with the guanidine group of Arg72 (with the distance of at least 2.7 Å). The hydrocarbon part of the side radical of Arg72 as well as the Phe residue present at this position in most ELIs is apparently incorporated into the hydrophobic core of the molecule. Thus, the residues 13, 17, 72, 74, 76, 95, 105, and 106 have a structural function and are likely not involved in the interaction of ELIs with proteases. The conserved positions in ELIs include Met11 (Thr in M4in). The N-terminus of 2/3 of ELIs is ten residues shorter than M4in; consequently, position 11 corresponds to the first residue of the polypeptide chain. Thus, this residue buried in M4in and involved in an intramolecular hydrogen bond cannot be considered as a possible element of the protein–protein interface.
Figure 3.
Conserved amino acids in emfourin-like inhibitors.A, LOGO presentation of the consensus sequence of emfourin-like inhibitors. Colored pentagons indicate conserved residues discussed in the text; the colors are as in panel B. Numbering after emfourin. В, localization of the most conserved amino acids specified in panel A in emfourin. Buried residues are marked green, surface amino acids presumably forming the inhibitor active site are marked pink; and Thr11 corresponding to the start methionine in 2/3 of ELIs sequences is marked blue-gray. Oxygen and nitrogen atoms are marked red and blue, respectively.
The other five conserved amino acid residues (Gly19, Gly20, Phe21, Ala22, and Asp70) form a compact cluster at the surface of M4in corresponding to one of two regions capable of interacting with the active site of PLN according to the SAXS data. The side chains of Phe21, Ala22, and Asp70 are exposed to the solvent in the majority of structural models (PDB ID: 6ZYG) calculated from the NMR data; i.e., these residues can be involved in protein–protein interactions. It should be noted that, according to NMR studies, the 18 to 29 and 61 to 70 regions are the most flexible parts of the molecule (Fig. 1, C and D). Essentially, the hydrophobic Phe and Ala residues correspond to the PLN substrate specificity (27, 28) and Asp can form coordination bonds with metal ions. Thus, Phe21 and Ala22 presumably occupy the S′1 and S′2 substrate-binding sites of the enzyme (as designated by Schechter and Berger (29)) and Asp70 interacts with the zinc ion in the active site to displace the catalytic water, thus, inactivating the enzyme.
On the basis of these observations, we decided to check our hypothesis and impose the contact restrains during the refinement SASREF process between the active region of PLN and the Asp70 residue of M4in, in particular, between Zn2+ in the active site of PLN and the oxygen atom of Asp70 of M4in (the distance between these atoms should not exceed 2.2 Å, otherwise the penalty terms are added). The obtained models neatly fitted the experimental SAXS data, which confirms our hypothesis; however, these models contained a significant degree of overlap between side chains in the contact region (that is not taken into account during SASREF modeling).
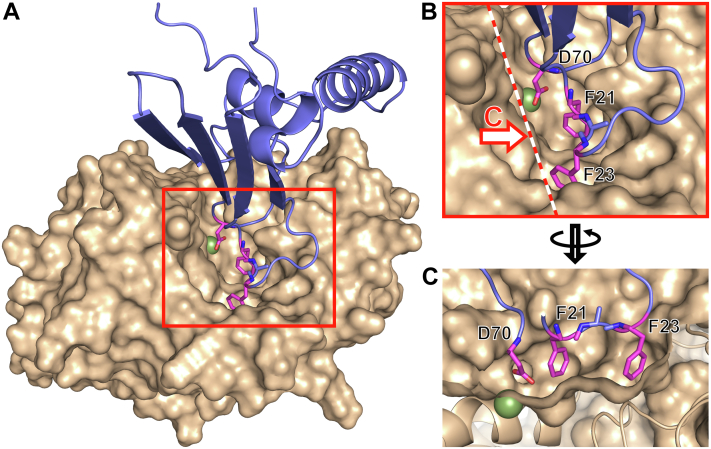
Based on the assumption of the interaction between the β-carboxyl group of Asp70 and the zinc atom of the PLN active site as well as of side chains of Phe21 and Ala22 and the S′1 and S′2 sites of the enzyme, we modeled the PLN–M4in complex using the local protein–protein docking approach. The docking was performed for all models of M4in structures calculated from the NMR data. Since the positions of the loops harboring residues 70, 21, and 22 as well as of the corresponding side chains varied from model to model, the docking results were also different. However, the geometry of the region containing Asp70, Phe21, and Ala22 in the 3D structure of M4in corresponded to that of the PLN active site cleft in all cases. The best results were observed for the model 15 of 6ZYG (Fig. 4, Supporting information 1, and Supporting information 2). In this case, the interacting molecule parts have complementary geometry and the side chain positions of the M4in residues 70 and 21 correspond to the above assumption. In addition, the side chain of Phe23 in M4in (which is not conserved in other ELIs) occupies the hydrophobic pocket of PLN in this model (Fig. 4), which is occupied by Met14 of the propeptide, an intramolecular inhibitor, in the PLN precursor molecule (28). This allows us to propose that Phe23 can substantially contribute to the inhibitor–protease interaction, at least for M4in and PLN.
Figure 4.
Molecular docking model of emfourin (blue cartoons) and protealysin (wheat surface) interaction.A, general view of the complex. B, magnified region of the active site; dashed line marks the section plane for panel C. C, view on the plane indicated by the dashed line from the arrow side in panel B. Asp70, Phe21, and Phe23 are indicated in magenta; the green sphere is the Zn2+ of the protealysin active site. The figure was generated using the PyMOL Molecular Graphics System.
Thus, the molecular docking results give credence to the putative Asp70-Zn2+/Phe21-S′1 interaction. It is further confirmed by the good agreement of the generated docking model with the SAXS analysis of the PLN–M4in complex. The best fit of such a refined model with χ2 = 1.14 is displayed in Figure 2A (dashed line). The docking model closely corresponds to the ab initio envelope of the PLN–M4in complex (Fig. 2E) and qualitatively agrees with the previous two-phase ab initio model of the PLN–M4in complex (11).
Site-directed mutagenesis and production of emfourin variants
Asp70, Phe21, Ala22, and Phe23 were selected as targets of site-directed mutagenesis of M4in. The generated M4in mutants are listed in Table 2. All these variants had a C-terminal His6-tag to simplify protein isolation and the Cys68-Ser substitution prohibiting the formation of inactive covalent dimers (11). The M4in variants were expressed in Escherichia coli and purified to electrophoretic homogeneity using metal-chelate affinity chromatography and gel permeation chromatography (GPC) (The purification process is shown in Fig. S3 and Table S2).
Table 2.
 |
Abbreviations: ND, not determined; SE, standard error.
The gray shading indicates amino acid substitutions.
In the case of D70A, F21S, F21S-A22S, A22S-F23S, and F21S-A22S-F23S, two independent protein samples were isolated. Three independent experiments were conducted for each sample; the constant values were calculated for all six experiments. In other cases, the constant values were calculated for three independent experiments for each protein sample.
Rounded to integers.
It should be noted that a fair amount of self-aggregates were detected apart from the monomeric M4in in GPC fractionation of samples after metal-chelate affinity chromatography. The proportion of aggregates varied for different inhibitor variants. It was about 20% for the reference protein with the His6-tag and Cys68-Ser substitution (M4inH6) as well as the F21S, A22S, F23S, F21S-A22S, A22S-F23S, F21S-F23S, and F21S-A22S-F23S variants (for designations see Table 2). In all variants with the Asp70 modification (D70A, D70S, D70N, D70E, and D70A-F21S-A22S-F23S), the aggregate proportion was much higher, up to 50%. Upon storage, the monomeric protein samples after GPC gradually formed aggregates. Thus, M4in with an extra His6-tag at the C-terminus tends to aggregate. To exclude the effect of aggregation on the studied properties of the proteins, the monomer fractions after high-performance GPC were switched to an appropriate buffer by gel filtration on desalting columns and immediately used to determine the CD spectra and activity.
The CD spectra for all M4in mutant proteins (Fig. S4) were quite similar (the relative contributions of α-helices, β-sheets, turns, and random coils were 32 ± 3, 23 ± 9, 19 ± 5, and 27 ± 3%, respectively) and mostly consistent with those predicted for the ensemble of M4in NMR structures reported here (the relative contributions of α-helices, β-sheets, turns, and random coils were 28 ± 2; 30 ± 2, 4 ± 2, and 38 ± 1%, respectively). The coincidence of the spectra shows that the mutations induced no significant changes in the 3D structure of M4in.
Inhibition of PLN activity by emfourin variants
The effect of M4in mutants on the proteolytic activity of PLN was analyzed using the internally quenched fluorescent peptide substrate Abz-RSVIK(Dnp) reported previously (30). The hydrolysis kinetics was determined for two substrate concentrations (30 and 90 μM) and six concentrations of each M4in variant. The concentration ranges for M4in variants were selected based on the inhibitory activity of each protein (Fig. S5). The characteristics of M4in variants are presented in Table 2.
The data obtained demonstrate that the introduced His6-tag has no effect on the inhibitory properties of M4in.
Noteworthily, some of the modifications (D70S, D70A, D70N, D70E, F21S-A22S, F21S-F23S, and F21S-A22S-F23S) switch the M4in inhibition pattern from slow binding to the classical fast inhibition (Table 2 and Fig. S5). (In slow-binding inhibition, the equilibrium between enzyme, inhibitor, and enzyme–inhibitor complex is established slowly on a time scale of seconds to minutes (31).) This inhibition switch largely correlates with the increased inhibition constant (Ki): it is observed after this parameter increases at least 10-fold. However, this trend is not corroborated by the D70A-F21S-A22S-F23S variant with the lowest inhibitory activity and the slow binding (Table 2).
All Asp70 substitutions significantly affect the M4in inhibitor activity: the calculated inhibition constants (Ki) increase by ∼2 to 3 orders of magnitude (Table 2). These effect increases in the series Glu→Asn→Ser→Ala, which agrees with the assumed coordination bond between the Asp70 side chain and Zn2+ of the enzyme active site. The Asp70-Glu substitution (D70E) has the least effect, apparently, due to the side chain elongation along with the retention of the charge and the capacity for zinc ion coordination. Asp70 replacement with uncharged Asn (D70N) has a greater effect on the inhibitory properties, which can be attributed to the suppressed electrostatic interaction. Asp70 replacement with the polar Ser (D70S), whose side chain cannot form coordination bonds, further increases the Ki. Finally, the substitution with a hydrophobic side group with the minimal affinity for Zn2+ shows the greatest drop in the M4in inhibitor activity.
Single Phe21, Ala22, or Phe23 substitutions for Ser (F21S, A22S, and F23S, respectively, in Table 2) had a relatively weak effect on M4in inhibitory activity, Ki increased 1.5 to 6 times. The double substitutions (F21S-F23S, A22S-F23S, and F21S-A22S) induced more significant changes, Ki increased up to 130 times. It should be noted that the analysis of double mutants revealed the synergetic effect of F21S and А22S as well as of F21S and F23S (the Ki increase was much higher than their independent action). An opposite effect was observed for the A22S and F23S combination (the effect was lower than expected). The maximum effect (a 400-fold increase in Ki) was observed for the combination of all three substitutions (F21S-A22S-F23S). Thus, the cluster of hydrophobic amino acids 21 to 23 significantly contributes to M4in activity, which corroborates the proposed interaction of the 21 to 23 region with the S′ substrate-binding region of the protease.
Yet, the contribution of individual amino acids to the interaction can hardly be unambiguously evaluated conceivably due to the mutual influence of neighboring residues and the decreased overall hydrophobicity of the 21 to 23 region after polar Ser introduction. Nevertheless, the comparative analysis of single and double substitutions (Table 2) suggests that Ala22 has the least impact on the inhibitor–protease interaction, while Phe21 and Phe23 equally contribute to the binding. Notice that ELIs always have a hydrophobic amino acid at position 21, commonly Phe; position 22 is typically occupied by Ala or more rarely Gly and Thr. Position 23 is less conserved with Gly being the most common residue (Fig. 3). Thus, the protease interaction with amino acids corresponding to positions 21 and 22 in M4in is similarly realized in nearly all ELI-PLP pairs, while the role of the residue at position 23 can vary. Considering that this position is often occupied by Gly, the main chain flexibility or conformation rather than the interaction of the side group of residues 23 with the substrate-binding site of the protease can be critical in many cases.
Thus, our data indicate that Asp70 and the 21 to 23 cluster of hydrophobic residues in M4in directly interact with the protease active site and can be two key elements of the inhibitor active site. Hence, simultaneous inactivation of both these elements should dramatically decrease M4in activity. Indeed, a combination of four mutations Asp70-Ala, Phe21-Ser, Ala22-Ser, and Phe23-Ser (D70A-F21S-A22S-F23S) tremendously increased the Ki by more than four orders of magnitude. Yet, this increase is roughly ten times lower than could be expected for an independent effect of the triple F21S-A22S-F23S mutation and the D70A substitution.
Overall, one can conclude that M4in and likely other ELIs do not imply the “canonical mechanism” (also known as the “standard” or “Laskowski” mechanism) with the substrate-like interaction between PIP and enzyme (32, 33).
Discussion
This study presents the first 3D structure of the recently discovered protein inhibitor of M4 peptidases, M4in, which provided the basis for the investigation of its molecular mechanism. It was shown that two spatially close flexible loop regions are critical for the inhibitor–protease interaction. The former region includes aspartic acid residue forming a coordination bond with Zn2+ of the enzyme active site and the latter carries hydrophobic amino acids corresponding to the protease substrate specificity and interacting with the subsites of its S′ region (as designated by Schechter and Berger (29)). Such active site structure corresponds to the noncanonical mechanism of action typical for proteinaceous metalloprotease inhibitors.
The canonical mechanism also known as the Laskowski mechanism is primarily typical for protein inhibitors of serine proteases. The protein–inhibitor binding in this case is similar to the substrate binding. The enzyme active site interacts with reactive loop, which is hydrolyzed by the protease. But unlike substrate, the cleaved inhibitor remains in the active site since the rigid structure of the inhibitor protein preserves the spatial structure of the reactive loop unaltered (34). Interestingly, metallopeptidase inhibitors utilizing the canonical Laskowski mechanism were found for the M4 family, which includes PLPs, natural ELI targets. These are the insect metallopeptidase inhibitor (IMPI) from Galleria mellonella (21, 35, 36), which belongs to the I8 MEROPS inhibitor family, and Streptomyces metalloproteinase inhibitor from Streptomyces nigrescens (37) of the I36 family. Apparently, the canonical mechanism towards M4 peptidases also applies to the inhibitors of the I16 family produced by streptomycetes, such as Streptomyces subtilisin inhibitor, alkaline protease inhibitor-2c’, and plasminostreptin (38, 39). Other known inhibitors of M4 peptidases belong to the I39 family of α2-macroglobulins. These proteins affect a wide range of endopeptidases irrespective of their catalytic type as molecular traps that do not block the active site but restrict substrate access to the peptidases in a molecular-size-dependent way (40). Thus, M4in is the first M4 peptidases inhibitor utilizing the true noncanonical mechanism.
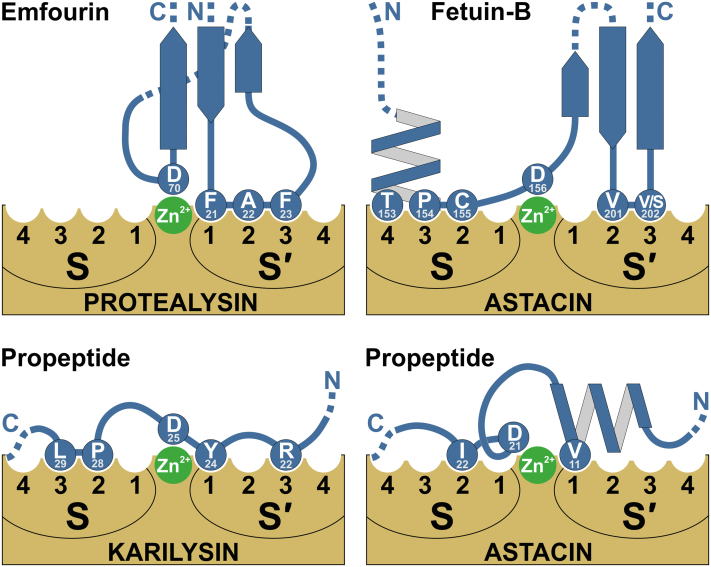
The noncanonical mechanism of action of metallopeptidase inhibitors is implemented, as with M4in, by means of the group coordinating the zinc ion in the enzyme active site and correct positioning of this group is provided by accessory interactions often including the S and S′ substrate-binding regions of the protease. At the same time, zinc coordination most commonly relies on the terminal amino acids of the inhibitor. In endopeptidases inhibitors, these are N-terminal residues commonly interacting with the metal ion via the carbonyl oxygen and terminal amino group (similar to tissue inhibitors of metalloproteinases, TIMPs, of the MEROPS family I35 (41, 42, 43) and apron from P. aeruginosa of the I38 family (44)) or occasionally via the carbonyl oxygen alone (as in the metallopeptidase inhibitor of Erwinia chrysanthemi of family I38 (45)). In metallopeptidase inhibitors of families I37, I44, I46, I68, and I92, the side carboxyl group of the inhibitor C-terminal residues interacts with the zinc ion (46). Zn2+ coordination by a side chain of a nonterminal amino acid has been reported only for inhibitors of the astacin family of metalloendopeptidases: fetuin-Bs (FBs) of subfamily I25С (47, 48) and sizzled from Xenopus laevis of family I93 (49). In both cases as well as in PLN inhibition by M4in, the β-carboxyl group of aspartic acid interacts with the zinc ion.
The 3D structures of the FB complexes with archetypal crayfish astacin have been determined for the mouse (PDB ID 6HT9 (47)) and human (PDB ID 6SAZ (48)) FBs, which makes it possible to compare the mechanisms of action of FBs and M4in. As mentioned above, in both cases the side carboxyl group of Asp forming the coordination bond with the zinc ion of the enzyme active site is the key element (Fig. 5). In both cases, Asp positioning is realized through the interaction between the loop amino acids (that are spatially close to Asp) and the substrate-binding sites in a way similar to the substrate residues. In both cases, the whole construct is mounted on similarly disposed three antiparallel β strands. Arguably, the active site idea is generally similar in M4in and FBs, although particular realizations differ significantly. Namely, M4in interacts with the S′1, S′2, and S′3 sites of the enzyme (apparently, other ELIs interact only with S′1 and S′2). FBs also interact with the S′ region but within the S′2 and S′3 sites as well as with the nonprimed side of the substrate-binding site, thus occupying the S2, S3, and S4 sites. Consequently, the active site of M4in is more compact; however, the inhibition constants in M4in and FBs fall within the subnanomolar range (47, 48) despite the smaller number of contacts between the M4in active site and the enzyme. Regarding the differences, FBs are composed of two cystatin-like domains (unlike the single-domain M4in) and the region with amino acids binding the S2-S4 subsites and the downstream crucial Asp are in the interdomain linker; they also have disulfide bonds (unlike M4in). Overall, this indicates that the similarity of the active sites of FBs and M4in results from convergent evolution coming from the similarity of the active sites of metalloproteinases rather than from the common origin of the inhibitors.
Figure 5.
Schematic interaction of metalloproteases with autonomous protein inhibitors and propeptides with Asp coordinating a zinc ion in the active site.
Apart from autonomous protein inhibitors, the activity of cognate enzymes can be suppressed by propeptides. Their structure and the mechanisms of latency in metallopeptidase zymogens are diverse (50). Predominantly, the inhibition relies on the coordination of zinc ion in the active site. Similar to autonomous PIPs, N- or C-terminal amino acids of the propeptide can interact with Zn2+; however, zinc coordination by a side chain of a nonterminal amino acid is more common. This propeptide mechanism is called the zinc-ligand switch since the side chain interacting with Zn2+ displaces the catalytic water, one of the zinc ligands in the active site of metalloproteases. Depending on specific amino acids involved in zinc coordination, they are referred to as Cys-, Asn-, Glu-, His-, and Asp-switches.
The Asp-switch inhibition was demonstrated for astacin (PDB ID 3LQ0 (51)) and meprin β (PDB ID 4GWM (52)) of the M12 family as well as for karilysin (PDB ID 4R3V (53)) of the M10 family. The propeptides utilize a different way to approach zinc than autonomous PIPs. The propeptide region resides in the active site cleft in reverse orientation to a substrate. In this case, the side chains of certain amino acids on both sides of Asp interact with the substrate-binding sites in the S and S′ regions (Fig. 5). Moreover, extra nonsubstrate-like interactions take place between the propeptide and the enzyme in the active site cleft.
Thus, the inhibition mechanism based on the catalytic water displacement from the active site of metalloproteases involving zinc coordination by side groups of different amino acids is universal for both autonomous PIPs and propeptides. However, it is the major mechanism in propeptides (50), while it is quite rare in autonomous PIPs and includes only the Asp-mediated variant. This disproportion should have a reason incidental to the evolution of propeptides as nonautonomous protein modules, which is indicated by the critical differences in the interaction of autonomous PIPs and propeptides with the enzyme active site specified above. However, the incidence of particular mechanisms can be rectified as more metalloprotease inhibitors are described.
As concerns the M4in inhibition mechanism, it should be noted that the targets of ELIs are peptidases of the M4 family produced in a variety of bacterial pathogens of humans and these peptidases are considered potential drug targets (19). In conditions of increasing bacterial resistance to antibiotics, such alternative targets become increasingly attractive; however, attempts to develop conventional small-molecule drugs against new targets often fail due to the low-target selectivity of such drugs (54). Therapeutic proteins can possibly solve this problem.
To date, no selective low-molecular–weight inhibitors of M4 peptidases, factors of bacterial pathogenesis, are known. This substantiated the development of novel antimicrobial drugs against infections caused by P. aeruginosa and S. aureus based on IMPI from G. mellonella, the only PIP of M4 peptidases with a thoroughly described mechanism of action (20, 21, 55). Our data propose M4in as a new framework for the development of highly selective therapeutic inhibitors of M4 peptidases with critical differences in the mechanism relative to the IMPI-based approach, which can extend the repertoire of new-age antibacterial agents.
Experimental procedures
Production of PLN and emfourin
Mature active PLN carrying a C-terminal His6-tag and M4in without the tag carrying the Cys68-Ser mutation were overproduced in E. coli and purified as described (11, 56). The recombinant 13C- and 15N-labeled M4in without the tag carrying the Cys68-Ser substitution was produced as described (57).
NMR experiments and data processing
NMR sample of recombinant 0.37 mM 13C,15N-labeled M4in with 18 mM phosphate (Na2HPO4/NaH2PO4) and 0.05% NaN3, pH 6.5, was prepared in 10% D2O/90% H2O. A set of 2D and 3D NMR spectra (including various variants of 3D NOESY spectra) were previously recorded and 1H, 13C, and 15N resonance NMR signals were assigned (57). In addition, some NMR spectra were collected for molecular structure and dynamics analysis. These were obtained in spin-echo difference 2D 13C-{15N} 1H-13C-constant time-heteronuclear single quantum coherence (CT-HSQC) (58), 2D 13C-{13CO} 1H/13C-CT-HSQC (59), and 2D 13C-{13CA} 1H-13C-CT-HSQC (60) experiments for J-couplings measurement; moreover, 2D 1H-15N-CLEANEX spectrum (61) and several 1H-15N-HSQC spectra at different temperatures (298 K, 300 K, 303 K, 305 K, 308 K) were acquired for hydrogen bonds recognition. The 15N CSA/dipolar cross-correlated transverse relaxation experiments were acquired in an interleaved manner for the reference and attenuated spectra using a 2D 1H-15N-ct-TROSY-HSQC-based pulse sequence (23) with the constant period of 26.9 ms and the relaxation period of 10.8 ms to characterize backbone mobility. All these experiments were recorded at 303 K (unless otherwise indicated) on a 600 MHz AVANCE III spectrometer (Bruker BioSpin) and a 700 MHz Varian NMR-system spectrometer (Varian-Agilent) both equipped with 5 mm pulsed-field gradient triple-resonance cryoprobes.
NMR spectra analysis and NOESY cross-peak integration were performed in the CARA (62) program. All NMR spectra of M4in revealed multiple peaks corresponding to the major and several minor protein conformations, the latter with an occupancy of ∼10% or less (see Results). The set of M4in structures was calculated in CYANA (63) with the constraints on interproton distances estimated from the NOE connectivities observed for the major protein conformation as well as on the angles φ and ψ derived from the 1H,13C,15N-chemical shifts by the TALOS-N software (64) (see Supporting information Methods for more details).
The NOE data analysis did not allow us to unambiguously define the angle between the β-layer and the large helix Pro40-Tyr56 due to the proximity of chemical shifts of some methyl groups in the studied protein. This angle could have two slightly different values depending on the interpretation of the NOE contacts; hence, two sets of structures with slightly wider and slightly narrower angles between the long helix and the β-layer were calculated. Then each of these structures from two sets was fitted into the experimental SAXS curve with the calculation of the χ2 value. The set-averaged value of χ2 turned out to be somewhat lower in the variant with the smaller angle describing a more compact molecule that corresponds the main conformation of the protein in solution. This allowed us to clarify the assignment of some NOESY interresidue cross-peaks.
For the Pro25-Leu27 region, a contradiction was found between the chemical shift values corresponding to the 310-helix turn according to TALOS-N and the NOE data, which indicates the absence of this helix. To eliminate the collision, the resulting set of structures was calculated based on the assumption of a dynamically reconfigurable turn of the helix.
For the characterization of the M4in intramolecular dynamics, the effective rotational correlation times τc were evaluated for the backbone amide groups based on the 15N CSA/dipolar cross-correlated transverse relaxation experiment. The τc values were calculated from the peak intensities of the reference and attenuated spectra (23) and the uncertainties were estimated from the noise RMSD.
As a result of the M4in spatial structure calculation, protons of many methylene and amino groups as well as the carbons of methyl groups were stereo assigned. Refined assignments have been deposited in the Biological Magnetic Resonance Bank (65) as entry ID 34546. The 1H-15N-HSQC and 1H-13С-HSQC NMR spectra with a complete assignment including the stereo assignment are given in Figs. S6 and S7.
Sequence analysis
The described set of 908 ELI sequences (11) was used. Sequences were aligned using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/) (66). Fifteen sequences with incomplete N- or C-terminal regions were excluded from the alignment. In the remaining alignment, the regions missing in M4in (GenBank accession no. ARS03586) were removed. The resulting set of aligned sequences was used to generate the logo by WebLogo version 3.7.4 (http://weblogo.threeplusone.com) (67).
Molecular docking
Molecular docking was conducted by the Rosetta online server (ROSIE) using the local protein–protein docking protocol (rosie.rosettacommons.org/docking2) (68). Docking partners included each of the 20 M4in models deposited in the PDB (ID: 6ZYG) as well as the 3D structure model of mature PLN based on the crystal structure of PLN precursor (PDB ID: 2VQX) generated by the deletion of the propeptide region and including the 51 to 341 residues (numbering corresponds to the precursor sequence). The starting positions for docking were obtained by positioning M4in structure models relative to that of mature PLN so that the Asp70 side chain of M4in was close to Zn2+ of PLN catalytic site and the side chain of Phe21 of M4in was directed toward the S1′-S2′ substrate-binding site (28) of the enzyme. The mature PLN structure and the starting positions of partners were created using the PyMOL Molecular Graphics System, Version 2.4 (Schrödinger).
SAXS measurements and data processing
The M4in without the histidine tag and with the Cys68-Ser mutation was used for SAXS analysis. Synchrotron radiation SAXS experiments were performed at the P12 beamline of EMBL at the PETRA III storage ring, DESY Hamburg (69). The samples were loaded using a robotic sample changer (70) into a flow-through capillary 1.7 mm in diameter. The solutions of PLN, M4in, and the PLN–M4in complex (with 1:1 stoichiometry) were measured at solute concentration ranges 0.63 to 10.0 mg/ml, 1.0 to 16.0 mg/ml, and 3.0 to 12.0 mg/ml, respectively, in 50 mM Tris–HCl buffer, pH 7.3. The data were recorded using a Pilatus 2M detector (DECTRIS) with 20 × 0.05 s exposure time at the sample-detector distance of 3.0 m and wavelength of 0.124 nm covering the momentum transfer range from 0.02 nm−1 to 7.0 nm−1. The temperature was kept at 20 °C. The data collection and reduction were performed using the BECQUEREL software (71) and the SASFLOW pipeline (72), including the comparison of frames for radiation damage, averaging, and buffer subtraction. The averaged frames were normalized to the transmitted beam using a beam-stop with an integrated photodiode (73). No measurable radiation damage was detected by comparing successive time frames. The difference data were extrapolated to zero solute concentration following standard procedures. All data manipulations were performed using the PRIMUS program (74) of the ATSAS package (75).
The radius of gyration Rg of solute and the forward scattering I(0) were evaluated from the Guinier approximation (76) at small angles (s < 1.3/Rg) (assuming that the intensity follows the equation I(s) = I(0) exp(−1/3(sRg)2) as well as from the entire scattering pattern by the GNOM program (77) using indirect Fourier transformation. In the latter case, the distance distribution functions p(r) and the maximum particle dimensions Dmax were also computed.
The excluded volume of the hydrated protein molecule (Vp) was calculated using the Porod approximation (78) in which the intensity I(s) was modified by subtraction of an appropriate constant from each data point to force the s−4 decay of the intensity at higher angles following the Porod’s law (78) for homogeneous particles.
The DAMMIN program (79) was employed to construct low-resolution ab initio bead models of PLN–M4in complex and individual proteins that best fitted the experimental scattering data. DAMMIN employs a simulated annealing procedure to build a compact bead configuration inside a sphere with the diameter Dmax that fits the experimental data I(s) to minimize the discrepancy between experimental and calculated intensities (χ2 fit quality factor).
The mature PLN model generated from the high-resolution crystallographic atomic model of unprocessed PLN precursor (PDB ID: 2VQX) and the NMR model of M4in (PDB ID: 6ZYG) as described above were used to reconstruct the rigid body model of the PLN–M4in complex using the SASREF program (80). The latter program employs a simulated annealing protocol to generate an interconnected assembly of subunits without steric clashes fitting the scattering data. The scattering amplitudes from the atomic coordinates of the proteins were calculated by CRYSOL (81). The superposition of ab initio and atomic models was performed using the SUPALM program (82).
Site-directed mutagenesis and production of emfourin variants
The plasmids pD70A (encodes variant of M4in with the Asp70-Ala substitution), pD70S (Asp70-Ser), pD70N (Asp70-Asn), pD70E (Asp70-Glu), pF21S (Phe21-Ser), pA22S (Ala22-Ser), pF23S (Phe23-Ser), pF21S-F23S (Phe21-Ser and Phe23-Ser), pF21S-A22S (Phe21-Ser and Ala22-Ser), pA22S-F23S (Ala22-Ser and Phe23-Ser), pF21S-A22S-F23S (Phe21-Ser, Ala22-Ser, and Phe23-Ser), and pD70A-F21S-A22S-F23S (Asp70-Ser, Phe21-Ser, Ala22-Ser, and Phe23-Ser) were derived from the pM4in-His/S plasmid encoding the M4in variant with the Cys68-Ser mutation and a C-terminal His6-tag (11) using PCR and standard molecular cloning techniques. A detailed description of the construction procedure is provided in Supporting information Methods and Table S3.
E. coli BL-21 (DE3) cells were transformed with pM4in-His/S, pD70A, pD70S, pD70N, pD70E, pF21S, pA22S, pF23S, pF21S-A22S, pA22S-F23S, pF21S-F23S, pF21S-A22S-F23S, or pD70A-F21S-A22S-F23S and grown in 250 ml of auto-induction medium (10 g/l peptone, 5 g/l yeast extract, 5 ml/l glycerol, 0.5 g/l glucose, 2 g/l lactose, 7.1 g/l Na2HPO4, 6.8 g/l KH2PO4, 3.3 g/l (NH4)2SO4, and 0.2 g/l MgCl2) containing 100 μg/ml ampicillin under vigorous agitation (220 rpm) at 37 °C for 2 h and then at 16 °C for 96 h. The cell suspension at A600 = 12 was centrifuged at 4000g and 4 °C for 15 min. The pellet was frozen and stored at −20 °C. A pellet aliquot corresponding to 100 ml of culture was resuspended in 15 ml of 50 mM Tris–HCl buffer, pH 8.0. The resulting suspension was ultrasonicated (Omni Sonic Ruptor 4000) at 4 °C, 30% power, and 30% pulser for 12 min. After centrifugation (9000g, 4 °C, 15 min), the supernatant was loaded onto a 1.5 × 1 cm column with Ni-NTA XPure Agarose Resin (UBPBio) equilibrated with the same buffer. The column was washed with 6 ml of the same buffer and with a step gradient of 10, 100, and 250 mM imidazole in 30 ml of the same buffer at a flow rate of 0.5 ml/min. Proteins were eluted with 100 mM imidazole in the same buffer. The fractions with the highest content of the protein (according to electrophoresis) were combined and concentrated to a volume of 2 ml by ultrafiltration on an Amicon Ultra-15 centrifugal filter device (Millipore). A 0.5 ml aliquot of the resulting sample was loaded onto a Superdex 75 Increase 10/300 GL column (GE Healthcare) equilibrated with 50 mM Tris–HCl containing 0.15 M NaCl, pH 7.4 and eluted with the same buffer at a flow rate of 0.5 ml/min. The fractions containing the target protein (according to electrophoresis) were combined and concentrated to a volume of 2 ml by ultrafiltration on an Amicon Ultra-15 centrifugal filter device (Millipore). The concentrate was loaded onto a PD-10 Desalting Column (GE Healthcare) equilibrated with 10 mM Tris–HCl, pH 7.4 and eluted with the same buffer. The obtained sample of the target protein was centrifuged (9000g, 4 °C, 5 min) and immediately used for activity analysis and registration of CD spectra. The purification process of pM4in variants is shown in Fig. S3 and Table S2.
Hydrolysis of Abz-RSVIK(Dnp) by PLN in the presence of emfourin variants and the determination of inhibition constant
The inhibition assay was performed using an internally quenched fluorescent peptide substrate, 2-aminobenzoyl-L-arginyl-L-seryl-L-valyl-L-isoleucyl-L-(ε-2,4-dinitrophenyl)lysine (Abz-RSVIK(Dnp)) (30) as described (11, 83) with minor modifications. The PLN concentration in the reaction mixture was 50 pM and the Abz-RSVIK(Dnp) concentration was 30 or 90 μM. The concentrations of M4in variants varied depending on their activities and ranged from 50 pM to 10 μM (Fig. S5). The concentrations of PLN, M4in variants, and the substrate were determined spectrophotometrically as described (11). The reactions were performed in 50 mM Tris–HCl, pH 7.4. Three independent measurements were made for each pair of substrate and inhibitor concentrations. Time-related fluorescence changes were monitored on a CLARIOstar Plus microplate reader (BMG Labtech). For slow-binding inhibition, the experimental kinetic curves were fitted by the integrated rate equation and the inhibition constants were calculated as described elsewhere (11). For classical inhibition, the inhibition constants were obtained using the Dixon method (84, 85). Data analysis was performed using GraphPad Prism version 8.0 (GraphPad Software, www.graphpad.com).
CD spectroscopy
For CD spectroscopy-based folding verification, the proteins were diluted to 0.16 mg/ml in 10 mM Tris–HCl, pH 7.4, and centrifuged (9000g, 4 °C, 5 min) prior to all measurements. The CD spectra were registered on a Chirascan VX Spectrophotometer (Applied Photophysics) in cuvettes of 0.05 cm optical path. All spectra were analyzed by the DichroWeb software (dichroweb.cryst.bbk.ac.uk) using the CDSSTR method and the reference dataset 4 (86). PDBMD2CD server was used to predict CD spectra from protein structure (87).
Data availability
All data are contained within the article. Assigned resonance NMR signals have been deposited in the Biological Magnetic Resonance Bank with the accession code 34546. Atomic coordinates have been deposited in the PDB with the accession code 6ZYG. The SAXS data were deposited in the SASBDB database (88) with the accession code SASDHN7.
Supporting information
This article contains supporting information (57, 91).
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
The work was carried out using the equipment of the Center of Cellular and Gene Technology at the Institute of Molecular Genetics of the National Research Centre “Kurchatov Institute.” Modeling of the emfourin–protealysin complex, site-directed mutagenesis of emfourin, and characterization of mutant variants were performed with the financial support of the Russian Science Foundation (project no. 22-24-00135).
Author contributions
T. N. B., P. V. K., E. V. B., and I. V. D. data curation; T. N. B., I. M. B., P. V. K., A. M. V., E. V. B., and I. V. D. formal analysis; T. N. B., I. M. B., K. N. C., M. A. K., P. V. K., A. M. V., E. V. B., and I. V. D. investigation; T. N. B., I. M. B., K. N. C., M. A. K., P. V. K., A. M. V., E. V. B., and I. V. D. validation; T. N. B., I. M. B., K. N. C., P. V. K., A. M. V., and I. V. D. writing–original draft; T. N. B., I. M. B., K. N. C., M. A. K., P. V. K., A. M. V., A. S. A., S. V. K., E. V. B., and I. V. D. writing–review and editing; T. N. B., I. M. B., P. V. K., A. M. V., and I. V. D. visualization; I. M. B., K. N. C., M. A. K., P. V. K., D. M. L., and I. V. D. methodology; P. V. K., A. M. V., D. M. L., A. S. A., S. V. K., and E. V. B. resources; P. V. K. and D. M. L. software; S. V. K. and I. V. D. supervision; P. V. K., S. V. K., and I. V. D. funding acquisition.
Reviewed by members of the JBC Editorial Board. Edited by Wolfgang Peti
Supporting information
References
- 1.Cabral-Pacheco G.A., Garza-Veloz I., Castruita-De la Rosa C., Ramirez-Acuna J.M., Perez-Romero B.A., Guerrero-Rodriguez J.F., et al. The roles of matrix metalloproteinases and their inhibitors in human diseases. Int. J. Mol. Sci. 2020;21:9739. doi: 10.3390/ijms21249739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilkinson D.J. Serpins in cartilage and osteoarthritis: what do we know? Biochem. Soc. Trans. 2021;49:1013–1026. doi: 10.1042/BST20201231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sillen M., Declerck P.J. A narrative review on plasminogen activator inhibitor-1 and its (patho)physiological role: to target or not to target? Int. J. Mol. Sci. 2021;22:2721. doi: 10.3390/ijms22052721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Acunto E., Fra A., Visentin C., Manno M., Ricagno S., Galliciotti G., et al. Neuroserpin: structure, function, physiology and pathology. Cell. Mol. Life Sci. 2021;78:6409–6430. doi: 10.1007/s00018-021-03907-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly-Robinson G.A., Reihill J.A., Lundy F.T., McGarvey L.P., Lockhart J.C., Litherland G.J., et al. The serpin superfamily and their role in the regulation and dysfunction of serine protease activity in COPD and other chronic lung diseases. Int. J. Mol. Sci. 2021;22:6351. doi: 10.3390/ijms22126351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nugteren S., Samsom J.N. Secretory leukocyte protease inhibitor (SLPI) in mucosal tissues: protects against inflammation, but promotes cancer. Cytokine Growth Factor Rev. 2021;59:22–35. doi: 10.1016/j.cytogfr.2021.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Luo S., Tong L. Structure and function of the separase-securin complex. Subcell Biochem. 2021;96:217–232. doi: 10.1007/978-3-030-58971-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortelny N., Butler G.S., Overall C.M., Pavlidis P. Protease-inhibitor interaction predictions: lessons on the complexity of protein-protein interactions. Mol. Cell. Proteomics. 2017;16:1038–1051. doi: 10.1074/mcp.M116.065706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kantyka T., Rawlings N.D., Potempa J. Prokaryote-derived protein inhibitors of peptidases: a sketchy occurrence and mostly unknown function. Biochimie. 2010;92:1644–1656. doi: 10.1016/j.biochi.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chukhontseva K.N., Berdyshev I.M., Safina D.R., Karaseva M.A., Bozin T.N., Salnikov V.V., et al. The protealysin operon encodes emfourin, a prototype of a novel family of protein metalloprotease inhibitors. Int. J. Biol. Macromol. 2021;169:583–596. doi: 10.1016/j.ijbiomac.2020.12.170. [DOI] [PubMed] [Google Scholar]
- 12.Kyostio S.R., Cramer C.L., Lacy G.H. Erwinia carotovora subsp. carotovora extracellular protease: characterization and nucleotide sequence of the gene. J. Bacteriol. 1991;173:6537–6546. doi: 10.1128/jb.173.20.6537-6546.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Held K.G., LaRock C.N., D'Argenio D.A., Berg C.A., Collins C.M. A metalloprotease secreted by the insect pathogen Photorhabdus luminescens induces melanization. Appl. Environ. Microbiol. 2007;73:7622–7628. doi: 10.1128/AEM.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsaplina O.A., Efremova T.N., Kever L.V., Komissarchik Y.Y., Demidyuk I.V., Kostrov S.V., et al. Probing for actinase activity of protealysin. Biochemistry (Mosc.) 2009;74:648–654. doi: 10.1134/s0006297909060091. [DOI] [PubMed] [Google Scholar]
- 15.Bozhokina E.S., Tsaplina O.A., Efremova T.N., Kever L.V., Demidyuk I.V., Kostrov S.V., et al. Bacterial invasion of eukaryotic cells can be mediated by actin-hydrolysing metalloproteases grimelysin and protealysin. Cell Biol. Int. 2011;35:111–118. doi: 10.1042/CBI20100314. [DOI] [PubMed] [Google Scholar]
- 16.Tsaplina O., Efremova T., Demidyuk I., Khaitlina S. Filamentous actin is a substrate for protealysin, a metalloprotease of invasive Serratia proteamaculans. FEBS J. 2012;279:264–274. doi: 10.1111/j.1742-4658.2011.08420.x. [DOI] [PubMed] [Google Scholar]
- 17.Eshwar A.K., Wolfrum N., Stephan R., Fanning S., Lehner A. Interaction of matrix metalloproteinase-9 and Zpx in Cronobacter turicensis LMG 23827(T) mediated infections in the zebrafish model. Cell. Microbiol. 2018;20 doi: 10.1111/cmi.12888. [DOI] [PubMed] [Google Scholar]
- 18.Khaitlina S., Bozhokina E., Tsaplina O., Efremova T. Bacterial actin-specific endoproteases grimelysin and protealysin as virulence factors contributing to the invasive activities of Serratia. Int. J. Mol. Sci. 2020;21:4025. doi: 10.3390/ijms21114025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adekoya O.A., Sylte I. The thermolysin family (M4) of enzymes: therapeutic and biotechnological potential. Chem. Biol. Drug Des. 2009;73:7–16. doi: 10.1111/j.1747-0285.2008.00757.x. [DOI] [PubMed] [Google Scholar]
- 20.Eisenhardt M., Schlupp P., Hofer F., Schmidts T., Hoffmann D., Czermak P., et al. The therapeutic potential of the insect metalloproteinase inhibitor against infections caused by Pseudomonas aeruginosa. J. Pharm. Pharmacol. 2019;71:316–328. doi: 10.1111/jphp.13034. [DOI] [PubMed] [Google Scholar]
- 21.Mendes S.R., Eckhard U., Rodriguez-Banqueri A., Guevara T., Czermak P., Marcos E., et al. An engineered protein-based submicromolar competitive inhibitor of the Staphylococcus aureus virulence factor aureolysin. Comput. Struct. Biotechnol. J. 2022;20:534–544. doi: 10.1016/j.csbj.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levitt M., Chothia C. Structural patterns in globular proteins. Nature. 1976;261:552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- 23.Chill J.H., Louis J.M., Baber J.L., Bax A. Measurement of 15N relaxation in the detergent-solubilized tetrameric KcsA potassium channel. J. Biomol. NMR. 2006;36:123–136. doi: 10.1007/s10858-006-9071-4. [DOI] [PubMed] [Google Scholar]
- 24.Holm L. Using Dali for protein structure comparison. Methods Mol. Biol. 2020;2112:29–42. doi: 10.1007/978-1-0716-0270-6_3. [DOI] [PubMed] [Google Scholar]
- 25.Holm L., Kääriäinen S., Wilton C., Plewczynski D. Using Dali for structural comparison of proteins. Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0505s14. Chapter 5:Unit 5.5. [DOI] [PubMed] [Google Scholar]
- 26.Schlautmann L.P., Gehring N.H. A day in the life of the exon junction complex. Biomolecules. 2020;10:866. doi: 10.3390/biom10060866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demidyuk I.V., Gromova T.Y., Kostrov S.V. In: Handbook of Proteolytic Enzymes. 3rd Ed. Rawlings N.D., Salvesen G., editors. Academic Press; Oxford: 2013. Protealysin; pp. 507–602. [Google Scholar]
- 28.Demidyuk I.V., Gromova T.Y., Polyakov K.M., Melik-Adamyan W.R., Kuranova I.P., Kostrov S.V. Crystal structure of the protealysin precursor: insights into propeptide function. J. Biol. Chem. 2010;285:2003–2013. doi: 10.1074/jbc.M109.015396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 30.Karaseva M.A., Chukhontseva K.N., Lemeskina I.S., Pridatchenko M.L., Kostrov S.V., Demidyuk I.V. An internally quenched fluorescent peptide substrate for protealysin. Sci. Rep. 2019;9:14352. doi: 10.1038/s41598-019-50764-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golicnik M., Stojan J. Slow-binding inhibition: a theoretical and practical course for students. Biochem. Mol. Biol. Educ. 2004;32:228–235. doi: 10.1002/bmb.2004.494032040358. [DOI] [PubMed] [Google Scholar]
- 32.Bode W., Huber R. Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 1992;204:433–451. doi: 10.1111/j.1432-1033.1992.tb16654.x. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski M., Jr., Kato I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 34.Farady C.J., Craik C.S. Mechanisms of macromolecular protease inhibitors. Chembiochem. 2010;11:2341–2346. doi: 10.1002/cbic.201000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arolas J.L., Botelho T.O., Vilcinskas A., Gomis-Ruth F.X. Structural evidence for standard-mechanism inhibition in metallopeptidases from a complex poised to resynthesize a peptide bond. Angew. Chem. Int. Ed. Engl. 2011;50:10357–10360. doi: 10.1002/anie.201103262. [DOI] [PubMed] [Google Scholar]
- 36.Wedde M., Weise C., Kopacek P., Franke P., Vilcinskas A. Purification and characterization of an inducible metalloprotease inhibitor from the hemolymph of greater wax moth larvae, Galleria mellonella. Eur. J. Biochem. 1998;255:535–543. doi: 10.1046/j.1432-1327.1998.2550535.x. [DOI] [PubMed] [Google Scholar]
- 37.Hiraga K., Seeram S.S., Tate S., Tanaka N., Kainosho M., Oda K. Mutational analysis of the reactive site loop of Streptomyces metalloproteinase inhibitor, SMPI. J. Biochem. 1999;125:202–209. doi: 10.1093/oxfordjournals.jbchem.a022260. [DOI] [PubMed] [Google Scholar]
- 38.Kumazaki T., Kajiwara K., Kojima S., Miura K., Ishii S. Interaction of Streptomyces subtilisin inhibitor (SSI) with Streptomyces griseus metallo-endopeptidase II (SGMP II) J. Biochem. 1993;114:570–575. doi: 10.1093/oxfordjournals.jbchem.a124218. [DOI] [PubMed] [Google Scholar]
- 39.Tsuyuki H., Kajiwara K., Fujita A., Kumazaki T., Ishii S. Purification and characterization of Streptomyces griseus metalloendopeptidases I and II. J. Biochem. 1991;110:339–344. doi: 10.1093/oxfordjournals.jbchem.a123582. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Ferrer I., Marrero A., Gomis-Rüth F.X., Goulas T. In: Macromolecular Protein Complexes: Structure and Function. Harris J.R., Marles-Wright J., editors. Springer International Publishing; Cham: 2017. α2-Macroglobulins: structure and function; pp. 149–183. [Google Scholar]
- 41.Gomis-Rüth F.X., Maskos K., Betz M., Bergner A., Huber R., Suzuki K., et al. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature. 1997;389:77–81. doi: 10.1038/37995. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez-Catalan C., Bode W., Huber R., Turk D., Calvete J.J., Lichte A., et al. Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor. EMBO J. 1998;17:5238–5248. doi: 10.1093/emboj/17.17.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wisniewska M., Goettig P., Maskos K., Belouski E., Winters D., Hecht R., et al. Structural determinants of the ADAM inhibition by TIMP-3: crystal structure of the TACE-N-TIMP-3 complex. J. Mol. Biol. 2008;381:1307–1319. doi: 10.1016/j.jmb.2008.06.088. [DOI] [PubMed] [Google Scholar]
- 44.Feltzer R.E., Gray R.D., Dean W.L., Pierce W.M., Jr. Alkaline proteinase inhibitor of Pseudomonas aeruginosa. Interaction of native and N-terminally truncated inhibitor proteins with pseudomonas metalloproteinases. J. Biol. Chem. 2000;275:21002–21009. doi: 10.1074/jbc.M002088200. [DOI] [PubMed] [Google Scholar]
- 45.Létoffé S., Delepelaire P., Wandersman C. Characterization of a protein inhibitor of extracellular proteases produced by Erwinia chrysanthemi. Mol. Microbiol. 1989;3:79–86. doi: 10.1111/j.1365-2958.1989.tb00106.x. [DOI] [PubMed] [Google Scholar]
- 46.Covaleda G., del Rivero M.A., Chávez M.A., Avilés F.X., Reverter D. Crystal structure of novel metallocarboxypeptidase inhibitor from marine mollusk Nerita versicolor in complex with human carboxypeptidase A4. J. Biol. Chem. 2012;287:9250–9258. doi: 10.1074/jbc.M111.330100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuppari A., Korschgen H., Fahrenkamp D., Schmitz C., Guevara T., Karmilin K., et al. Structure of mammalian plasma fetuin-B and its mechanism of selective metallopeptidase inhibition. IUCrJ. 2019;6:317–330. doi: 10.1107/S2052252519001568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guevara T., Korschgen H., Cuppari A., Schmitz C., Kuske M., Yiallouros I., et al. The C-terminal region of human plasma fetuin-B is dispensable for the raised-elephant-trunk mechanism of inhibition of astacin metallopeptidases. Sci. Rep. 2019;9:14683. doi: 10.1038/s41598-019-51095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bu Q., Li Z., Zhang J., Xu F., Liu J., Liu H. The crystal structure of full-length sizzled from Xenopus laevis yields insights into Wnt-antagonistic function of secreted frizzled-related proteins. J. Biol. Chem. 2017;292:16055–16069. doi: 10.1074/jbc.M117.791756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arolas J.L., Goulas T., Cuppari A., Gomis-Ruth F.X. Multiple architectures and mechanisms of latency in metallopeptidase zymogens. Chem. Rev. 2018;118:5581–5597. doi: 10.1021/acs.chemrev.8b00030. [DOI] [PubMed] [Google Scholar]
- 51.Guevara T., Yiallouros I., Kappelhoff R., Bissdorf S., Stocker W., Gomis-Ruth F.X. Proenzyme structure and activation of astacin metallopeptidase. J. Biol. Chem. 2010;285:13958–13965. doi: 10.1074/jbc.M109.097436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arolas J.L., Broder C., Jefferson T., Guevara T., Sterchi E.E., Bode W., et al. Structural basis for the sheddase function of human meprin beta metalloproteinase at the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16131–16136. doi: 10.1073/pnas.1211076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopez-Pelegrin M., Ksiazek M., Karim A.Y., Guevara T., Arolas J.L., Potempa J., et al. A novel mechanism of latency in matrix metalloproteinases. J. Biol. Chem. 2015;290:4728–4740. doi: 10.1074/jbc.M114.605956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Craik D.J., Fairlie D.P., Liras S., Price D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 55.Eisenhardt M., Dobler D., Schlupp P., Schmidts T., Salzig M., Vilcinskas A., et al. Development of an insect metalloproteinase inhibitor drug carrier system for application in chronic wound infections. J. Pharm. Pharmacol. 2015;67:1481–1491. doi: 10.1111/jphp.12452. [DOI] [PubMed] [Google Scholar]
- 56.Gromova T.Y., Demidyuk I.V., Kozlovskiy V.I., Kuranova I.P., Kostrov S.V. Processing of protealysin precursor. Biochimie. 2009;91:639–645. doi: 10.1016/j.biochi.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 57.Bozin T.N., Chukhontseva K.N., Lesovoy D.M., Filatov V.V., Kozlovskiy V.I., Demidyuk I.V., et al. NMR assignments and secondary structure distribution of emfourin, a novel proteinaceous protease inhibitor. Biomol. NMR Assign. 2021;15:361–366. doi: 10.1007/s12104-021-10030-x. [DOI] [PubMed] [Google Scholar]
- 58.Vuister G.W., Wang A.C., Bax A. Measurement of three-bond nitrogen-carbon J couplings in proteins uniformly enriched in nitrogen-15 and carbon-13. J. Am. Chem. Soc. 1993;115:5334–5335. [Google Scholar]
- 59.Grzesiek S., Vuister G.W., Bax A. A simple and sensitive experiment for measurement of JCC couplings between backbone carbonyl and methyl carbons in isotopically enriched proteins. J. Biomol. NMR. 1993;3:487–493. doi: 10.1007/BF00176014. [DOI] [PubMed] [Google Scholar]
- 60.MacKenzie K.R., Prestegard J.H., Engelman D.M. Leucine side-chain rotamers in a glycophorin A transmembrane peptide as revealed by three-bond carbon-carbon couplings and 13C chemical shifts. J. Biomol. NMR. 1996;7:256–260. doi: 10.1007/BF00202043. [DOI] [PubMed] [Google Scholar]
- 61.Hwang T.L., van Zijl P.C., Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J. Biomol. NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 62.Keller R.L.J. CANTINA Verlag; Goldau, Switzerland: 2004. The Computer Aided Resonance Assignment Tutorial, Cantina, Goldau. [Google Scholar]
- 63.Güntert P. Automated NMR protein structure calculation. Prog. Nucl. Magn. Reson. Spectrosc. 2003;43:105–125. [Google Scholar]
- 64.Shen Y., Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ulrich E.L., Akutsu H., Doreleijers J.F., Harano Y., Ioannidis Y.E., Lin J., et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lyskov S., Chou F.C., Conchuir S.O., Der B.S., Drew K., Kuroda D., et al. Serverification of molecular modeling applications: the Rosetta online server that includes everyone (ROSIE) PLoS One. 2013;8 doi: 10.1371/journal.pone.0063906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blanchet C.E., Spilotros A., Schwemmer F., Graewert M.A., Kikhney A., Jeffries C.M., et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY) J. Appl. Crystallogr. 2015;48:431–443. doi: 10.1107/S160057671500254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Round A., Felisaz F., Fodinger L., Gobbo A., Huet J., Villard C., et al. BioSAXS sample changer: a robotic sample changer for rapid and reliable high-throughput X-ray solution scattering experiments. Acta Crystallogr. D Biol. Crystallogr. 2015;71:67–75. doi: 10.1107/S1399004714026959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hajizadeh N.R., Franke D., Svergun D.I. Integrated beamline control and data acquisition for small-angle X-ray scattering at the P12 BioSAXS beamline at PETRAIII storage ring DESY. J. Synchrotron Radiat. 2018;25:906–914. doi: 10.1107/S1600577518005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franke D., Kikhney A.G., Svergun D.I. Automated acquisition and analysis of small angle X-ray scattering data. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrom. Detect. Assoc. Equip. 2012;689:52–59. [Google Scholar]
- 73.Blanchet C.E., Hermes C., Svergun D.I., Fiedler S. A small and robust active beamstop for scattering experiments on high-brilliance undulator beamlines. J. Synchrotron Radiat. 2015;22:461–464. doi: 10.1107/S160057751402829X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS - a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 75.Manalastas-Cantos K., Konarev P.V., Hajizadeh N.R., Kikhney A.G., Petoukhov M.V., Molodenskiy D.S., et al. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 2021;54:343–355. doi: 10.1107/S1600576720013412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guinier A. La diffraction des rayons X aux tres petits angles; application a l'etude de phenomenes ultramicroscopiques. Ann. Phys. 1939;12:161–237. [Google Scholar]
- 77.Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 78.Porod G. In: Small-Angle X-Ray Scattering. Glatter O., Kratky O., editors. Academic Press; London: 1982. General theory; pp. 17–51. [Google Scholar]
- 79.Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petoukhov M.V., Svergun D.I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Svergun D.I., Barberato C., Koch M.H.J. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 82.Konarev P.V., Petoukhov M.V., Svergun D.I. Rapid automated superposition of shapes and macromolecular models using spherical harmonics. J. Appl. Crystallogr. 2016;49:953–960. doi: 10.1107/S1600576716005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berdyshev I.M., Karaseva M.A., Demidyuk I.V. Assay for protealysin-like protease inhibitor activity. Bio Protoc. 2022;12 doi: 10.21769/BioProtoc.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Butterworth P.J. The use of Dixon plots to study enzyme inhibition. Biochim. Biophys. Acta. 1972;289:251–253. doi: 10.1016/0005-2744(72)90074-5. [DOI] [PubMed] [Google Scholar]
- 85.Dixon M. The determination of enzyme inhibitor constants. Biochem. J. 1953;55:170–171. doi: 10.1042/bj0550170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sreerama N., Woody R.W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 87.Drew E.D., Janes R.W. PDBMD2CD: providing predicted protein circular dichroism spectra from multiple molecular dynamics-generated protein structures. Nucleic Acids Res. 2020;48:W17–W24. doi: 10.1093/nar/gkaa296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kikhney A.G., Borges C.R., Molodenskiy D.S., Jeffries C.M., Svergun D.I. SASBDB: towards an automatically curated and validated repository for biological scattering data. Protein Sci. 2020;29:66–75. doi: 10.1002/pro.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 90.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 91.Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., et al. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article. Assigned resonance NMR signals have been deposited in the Biological Magnetic Resonance Bank with the accession code 34546. Atomic coordinates have been deposited in the PDB with the accession code 6ZYG. The SAXS data were deposited in the SASBDB database (88) with the accession code SASDHN7.