Abstract
Purpose
The goal of this study is to include transcytolemmal water exchange into MR cell size imaging using the IMPULSED model for more accurate characterization of tissue cellular properties (e.g., apparent volume fraction of intracellular space ) and quantification of indicators of transcytolemmal water exchange.
Methods
We propose a heuristic model that incorporates transcytolemmal water exchange into a multi-compartment diffusion-based method (IMPULSED) that was developed previously to extract microstructural parameters (e.g., mean cell size and apparent volume fraction of intracellular space ) assuming no water exchange. For ≤ 5 ms, the water exchange can be ignored, and the signal model is the same as the IMPULSED model. For ≥ 30 ms, we incorporated the modified Kärger model that includes both restricted diffusion and exchange between compartments. Using simulations and previously published in vitro cell data, we evaluated the accuracy and precision of model-derived parameters and how they are dependent on SNR and imaging parameters.
Results
The joint model provides more accurate values for cell sizes ranging from 10 to 12 microns when water exchange is fast (e.g., intracellular water pre-exchange lifetime ≤ 100 ms) than IMPULSED, and reduces the bias of IMPULSED-derived estimates of , especially when water exchange is relatively slow (e.g., >200 ms). Indicators of transcytolemmal water exchange derived from the proposed joint model are linearly correlated with ground truth values and can detect changes in cell membrane permeability induced by saponin treatment in Murine erythroleukemia (MEL) cancer cells.
Conclusion
Our results suggest this joint model not only improves the accuracy of IMPULSED-derived microstructural parameters, but also provides indicators of water exchange which are usually ignored in diffusion models of tissues.
Keywords: transcytolemmal water exchange, diffusion MRI, diffusion time, cell size, membrane permeability, water lifetime, temporal diffusion spectroscopy
Introduction
There is continuing interest in characterizing tissues using multi-compartment diffusion MRI (dMRI) models that use images acquired with multiple b values and diffusion times. Techniques such as AxCaliber [1, 2], ActiveAx [3, 4], VERDICT (vascular, extracellular and restricted diffusion for cytometry in tumors) [5-7], IMPULSED (imaging microstructural parameters using limited spectrally edited diffusion) [8-10], and POMACE (pulsed and oscillating gradient MRI for assessment of cell size and extracellular space) [11] have been developed to provide estimates of tissue microstructural properties, including cell size, apparent volume fraction of intracellular space, and cell density. Among these methods, VERDICT and IMPULSED have been successfully implemented in patients with prostate [6] and breast [10] cancer, respectively, within clinically feasible scan times. Moreover, IMPULSED derived spherical cell sizes have been comprehensively validated using computer simulations in silico, cells in vitro, and animals in vivo [8, 9]. These models use simple geometries such as cylinders and spheres to represent cells and assume that transcytolemmal water exchange between intra- and extracellular compartments can be ignored, so that closed-form mathematical expressions can be derived to predict measured dMRI signals. However, the assumption of negligible water exchange may not hold true in cases where transcytolemmal water exchange increases significantly, such as in disorders including Parkinson's [12] and Alzheimer diseases [13], or tumors undergoing treatments [14, 15], when the average intracellular water pre-exchange lifetime may be comparable to typical diffusion times and cannot be ignored. Our recent studies [16, 17] have shown that IMPULSED can still provide accurate estimates of mean cell size independent of practical transcytolemmal water exchange rates, but IMPULSED-derived apparent volume fractions of intracellular spaces and cell densities are underestimated, and such biases become greater with faster water exchange [8, 9, 16, 17].
The neglect of transcytolemmal water exchange effects not only limits the accuracy of fitted IMPULSED parameters, but it also fails to reveal important biophysical information including cell membrane permeability and intracellular water pre-exchange lifetime . Note that is dependent on via the relation of for a sphere, where is the cell diameter and is the averaged intracellular diffusion coefficient [18]. provides a convenient metric to evaluate whether transcytolemmal exchange is fast or slow compared with diffusion time, and has been measured in numerous studies [19-21]. Both and have been suggested as indicators of disease progression and treatment response [22, 23]. The Kärger model [24] describes dMRI signals of two Gaussian compartments undergoing exchange and has been commonly used to estimate or [25, 26]. It has also been modified to described non-Gaussian diffusion by considering the effect of restricted diffusion and the effect of restriction size, which we refer to as the modified Kärger model. The modified Kärger model provides a reasonable description of the diffusion-weighted signal when the exchange is relatively slow, but in the case of fast exchange the model fails [27]. This slow water exchange limit depends on multiple tissue cellular properties, including , intrinsic intracellular water diffusion coefficient , and , and thus should be determined on a case-by-case basis. Moreover, the modified Kärger model usually needs high signal-to-noise ratio (SNR) to extract parameters reliably, which limits its usage in practice [28]. Apparent exchange rate (AXR) imaging [14, 29, 30] employs double diffusion encoding imaging sequences (DDE) [31] and provides an apparent exchange rate AXR = , where is the apparent volume fraction of extracellular space. This makes it less specific about the origin of measured variations. The recently reported diffusion time-dependent kurtosis imaging [32] provides an interesting means to estimate water exchange times in a clinically-feasible manner [32], but it does not provide important information on other parameters such as , , and . For more comprehensive characterization of microstructure, it is possible to combine different experiments that probe different parameters, but this in turn significantly increases the total scan time.
In this study, we propose a model that incorporates transcytolemmal water exchange into the IMPULSED method [8-10]. IMPULSED uses a broad range of diffusion times and b values to provide comprehensive information on microstructure at different length scales using different gradient waveforms. In practice, IMPULSED combines pulsed gradient spin echo (PGSE) and oscillating gradient spin echo (OGSE) acquisitions to cover a broader range of effective diffusion times (e.g., 5 – 100 ms) for quantitative characterization of microstructure at length scales that are clinically relevant (e.g, 10 – 20 μm corresponding to many cell sizes). Strictly speaking, diffusion time is usually not well-defined unless a PGSE with short gradient pulses is used. However, effective diffusion time has been widely used in practice to interpret experiments. For PGSE, , where Δ is the separation of the diffusion gradients and is the duration of the diffusion gradients. For OGSE, , where is the frequency of the oscillating diffusion gradients. For such a broad range of , the influences of transcytolemmal water exchange on MR diffusion measurements are dependent on [16], specifically:
For ≤ 5 ms using a 50 Hz oscillating gradient, the influence of transcytolemmal water exchange (typically > 30 ms) is negligible since . Therefore, the previous IMPULSED method is still valid and the influence of water exchange on dMRI signals may be ignored.
For ≥ 30 ms using PGSE, transcytolemmal water exchange significantly affects dMRI signals so it cannot be ignored. Although the original Kärger model describes diffusion-weighted signals of two Gaussian compartments with exchange, a modified Kärger model has been developed to describe non-Gaussian diffusion by taking into account the effect of restricted diffusion and the effect of cell size [18, 33]. It is noted that for the PGSE measurements performed in this study, the narrow (gradient) pulse approximation (i.e., and ) is valid so the modified Kärger model can be used.
By such a means, transcytolemmal water exchange can be included in the IMPULSED method which, in turn, may improve the estimates of IMPULSED-derived cellular properties and provide information on transcytolemmal water exchange.
This report first describes our joint model in detail. Then, we use computer simulations to (i) validate the assumption that water exchange can be ignored for OGSE acquisitions with ≤ 5 ms for cell sizes ranging from 10 – 20 μm; and (ii) investigate the accuracy of model-derived parameters. Last but not least, by re-analyzing in vitro cell data that have been reported in a published study [17], we show that not only can the proposed method reduce the bias of IMPULSED-derived values of due to transcytolemmal water exchange, but also it detects changes in cell membrane permeability in response to treatments.
Theory
Solid tissues are modeled as two compartments in which water diffuses within each and exchanges between them. Cells are modeled as spheres and T2 relaxation is assumed homogeneous [34, 35]. The analytic expressions describing hindered/restricted diffusion in extra/intra-cellular compartments are the same as in the IMPULSED method [8, 9], but the effect of water diffusion is incorporated differently in different ranges.
For (≤ 5 ms) acquired with OGSE sequences with oscillating frequency ≥ 50 Hz, the influence of the transcytolemmal water exchange between the two compartments is minimal so that the previous IMPULSED formulation which ignores water exchange is valid [8]. The details of IMPULSED have been reported and comprehensively validated with histology previously [9, 36, 37]. Briefly, the total measured MRI signals are expressed as the sum of the fractions experiencing restricted diffusion within impermeable spherical cells and hindered diffusion in the extracellular spaces, namely,
| [1] |
where is the apparent volume fraction of intracellular space, and and are the diffusion-weighted signal magnitudes per volume from the intra- and extracellular spaces, respectively. Analytical expressions of and acquired by OGSE sequences have been reported previously [9, 10, 38, 39] and are provided in the Appendix. For PGSE sequences with ≥ 30 ms, diffusion signals are described by inserting the analytical expressions for hindered/restricted diffusion (used in IMPULSED) into the modified Kärger model, which is an approach first proposed in [25]. Sequence parameters are chosen to fulfill the narrow pulse approximation ( and ) so the modified Kärger model is valid. The evolution of the transverse magnetization in intra and extracellular compartments can be described by the modified Kärger model [25, 40]:
| [2] |
With the equilibrium condition:
The solution to the above differential equations is presented as follows [25]:
| [3] |
where and are the initial magnetizations in the intra and extracellular compartments. Apparent intracellular diffusion coefficient is a function of cell size , averaged intracellular diffusion coefficient without the influences of cell membranes, and other ‘timing’ parameters of the gradient waveform (such as , , and , but not to confused with the time ‘t’ in equation [2] and [3] which represents the time axis of a pulse sequence). is calculated in two steps. First, we calculate the diffusion-weighted signal magnitudes per volume from an impermeable sphere () acquired by PGSE sequences using expressions that have been reported previously [9, 36, 37] and are provided in the Appendix. Second, we define assuming a Gaussian phase approximation. Extracellular diffusion coefficient is approximated as a constant independent of because the range of used in this study is narrow.
To calculate the intra/extracellular signals at the echo time, the PGSE sequence is split into three intervals using the narrow pulse approximation (as shown in Supplemental Figure S1), including (i) from the excitation (time = 0) to the first gradient (); (ii) between two gradient pulses ( to ); and (iii) from the second gradient to the data collection ( to TE). During the first and third intervals, there are only water exchange effects. During the second part, there are both diffusion and exchange effects. Using Eq. [2], signals from intra and extracellular spaces at and TE can be calculated successively. Note that signals at the end of each interval are used as the initial signals to calculate signals at the end of the next interval.
In summary, the current joint model modifies the IMPULSED model by including transcytolemmal water exchange in the signals with ≥ 30 ms. Five parameters (cell size , intracellular diffusion coefficient , apparent extracellular diffusion coefficient , apparent volume fraction of intracellular space , and intracellular water pre-exchange lifetime ) can be obtained simultaneously by fitting the proposed joint model to dMRI signals involving multiple values and . Cell membrane permeability can be calculated by for a sphere [18].
Methods
Computer simulations:
An improved finite difference method [41] was used in the simulations which incorporates the cell transmembrane permeability as described in [42]. Finite difference simulations [17] were performed on modeled tissues consisting of closely packed spherical cells positioned on a three-dimensional face-centered cubic structure with physiologically relevant parameters found from previous experiments in vivo [9]: = 61.8%, = 2 μm2/ms, and = 1.56 μm2/ms, and homogeneous T2 relaxation times due to the minimal difference in T2 between intra and extracellular spaces, as reported previously. Eleven different values of cell diameter evenly distributed from 10 to 20 μm, with each evaluated for 16 different ’s: 50, 60, 70, 80, 90, 100, 125, 150, 200, 250, 300, 350, 400, 450, 500, and ms, covered the range of cell sizes and intracellular water pre-exchange lifetimes that have been reported in several biological tissues in health and disease. Bipolar gradients with = 30, 50, 70, 100 ms with a gradient rise time 0.9 ms, and oscillating gradients with 50 Hz ( = 5 ms), were used to generate diffusion MRI signals [10]. PGSE signals with eleven b values evenly distributed between 0 and 3 ms/μm2 were simulated and OGSE signals with nine b values evenly distributed between 0 and 2 ms/μm2 were also analyzed. To evaluate limits of what may be feasible, the maximum gradient strength and slew rate considered are within the range achieved in recent OGSE imaging studies with specialized human brain gradient coils [43, 44]. After noise-free dMRI signals were calculated for each cell diameter, Rician noise to achieve three different SNR levels (20, 50, and 100) was added, and then the noisy signals were used for data fitting. This process was repeated 50 times to evaluate both accuracy and precision of model-derived parameters.
Retrospective in vitro cell study:
Li et al. [17] investigated how transcytolemmal water exchange affects the IMPULSED-derived parameters using murine erythroleukemia (MEL) cancer cells with different permeabilities induced by saponin treatment. In their study, values for cells treated with different concentrations of saponin were measured using the conventional constant gradient (CG) method governed by the Kärger model [18, 28]. For the same cell samples, microstructural properties, such as cell size and proton volume fraction of intracellular space, were obtained by fitting the IMPULSED model to multi- and multi- and dMRI data. Correlations between and IMPULSED-derived parameters were then established. In the current study, we fit the proposed joint model to the multi- and multi- dMRI cell data in vitro that were reported in Li et al., and then compare joint model-derived values to those derived from the CG method. Also, we compare the microstructural properties derived from the joint model and IMPULSED methods. A brief description about the experimental design is given below and summarized in Table 1. More details about cell preparation, MR imaging protocol can be found in [8, 17].
Table 1.
A summary of the retrospective in vitro cell study, including the imaging acquisitions, data and models used for analysis, and results of three different analyses. Note the details of data acquisitions were reported previously [17] for four groups (n=6 for each group) of cell pellets prepared from MEL cells with different membrane permeabilities induced by saponin treatment. Analysis II and III applied the joint model and IMPULSED model, respectively, to the OGSE and PGSE diffusion data set with ranging from ~ 3.1 to 50.7 ms.
| Acquisitions | Analysis | Outcome |
|---|---|---|
| Constant gradient (CG) experiments using STEAM sequences gradient strength = 50 mT/m =10 ms The minimum was 20 ms and the maximum was 426, 426, 223, and 121.5 ms for cell samples with saponin concentrations of 0, 0.01%, 0.025% and 0.05%, respectively | I. fit the modified Kärger model to STEAM data (published in [17]) | |
|
II. fit the joint model to the OGSE and PGSE data | , , , |
| III. fit the IMPULSED to the OGSE and PGSE data | , , , , |
MEL cells were fixed with 4% paraformaldehyde and divided into four groups (n=6) at a cell density 4.2×107 cell/ml, each treated with 0%, 0.01%, 0.025% and 0.05% (w/v) -saponin at room temperature for 30 minutes to induce a range of cell membrane permeability. Each group of cells was centrifuged at 2000g for 2 minutes in a 0.65-mL Eppendorf tube to obtain a tight cell pellet for MR imaging. Small aliquoted samples were spotted on glass slides and imaged directly by light microscopy to estimate cell diameters. MR diffusion measurements using stimulated echo (STEAM), PGSE, and OGSE sequences, were performed on a 4.7T MRI spectrometer. The sample temperature was maintained at ~ 17°C using a cooling water circulation system.
In Li et al. [17] , and other cellular properties were estimated in two separate experiments. was estimated using constant gradient (CG) experiments. With a stimulated echo (STEAM) sequence, diffusion weighting was achieved by keeping gradient duration = 10 ms, gradient strength = 50 mT/m, and varying the gradient separation in 30 increments. The minimum was 20 ms and the maximum was 426, 426, 223, and 121.5 ms for cell samples with saponin concentrations of 0, 0.01%, 0.025% and 0.05%, respectively. was estimated as , where is the ADC of the slowly decaying component determined by linear regression using high b-value data points only. Other cellular properties, including cell size , , , and , were estimated by fitting the IMPULSED signal model to a combination of one PGSE ( = 4/52 ms) and two OGSE ( = 40 and 80 Hz, corresponding to = 6.25 and 3.125 ms, respectively) signals from cell pellets. For PGSE acquisitions, diffusion gradients were applied simultaneously on three axes with 11 gradient strengths varying linearly from 0 to 200 mT/m. For OGSE acquisitions, nine b values evenly distributed between 0 and 2 ms/μm2 were used with a maximum gradient strength of 300 mT/m, which is still achievable on special human brain gradient coils [45].
Data analysis:
IMPULSED was used to fit dMRI data to estimate four parameters () while the joint model was used to derive five parameters () simultaneously from the same data set. The constraints for fitting parameters were based on physiologically relevant values [17, 46]: 0 ≤ ≤ 30 μm, 0 ≤ ≤ 1, 0 ≤ ≤ 3.0 μm2/ms, 0 ≤ ≤ 3.0 μm2/ms, and 0 ≤ ≤ 1000 ms. Initial values for each fitting parameter were randomly selected from their ranges. Each fitting was repeated 50 times to evaluate both accuracy and precision of model-derived parameters. All the data processing was performed using a MATLAB-based in-house software package that is available online (https://github.com/jzxu0622/mati).
Results
Computer simulations
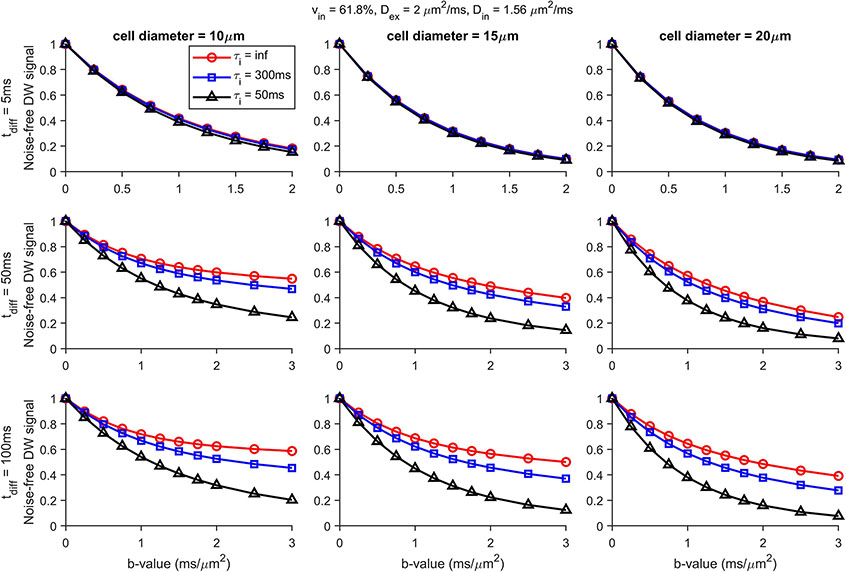
Figure 1 shows the simulated dMRI signals for different cell sizes (10, 15, and 20 μm), values (5, 50, and 100 ms) and values (50, 300, and ms). The effect of transcytolemmal water exchange increases with decreasing , increasing , and decreasing cell sizes, as expected. For all the cell sizes, there are significant differences in the simulated dMRI signals with = 50 and 100 ms among three ’s. By contrast, the simulated 50Hz OGSE signals are almost the same for all the cell sizes, and ’s, confirming the negligible influences of transcytolemmal water exchange on DWI signals for typical tumor cells when ≤ 5 ms.
Figure 1.
Simulated diffusion signals with diffusion times = 5, 50, and 100 ms for three cell diameters (10, 15, and 20 μm) and intracellular water pre-exchange lifetime ’s (50, 300, and ms).
The performance of the proposed joint model was evaluated firstly for the noise-free simulated signals with three ’s (5, 30, and 70 ms), which are readily achievable on current human scanners using a combination of OGSE (50 Hz, corresponding to = 5 ms) and PGSE acquisitions. Supplemental Figure S2 displays the fitting errors using either IMPULSED or the joint model for each b-value data point from three sets of simulated signals ( = 10, 15, and 20 μm, and = 50 ms). Fitting errors of IMPULSED are > 5% for data points with b value > 1 ms/μm2, while those of the joint model are less than 0.5%.
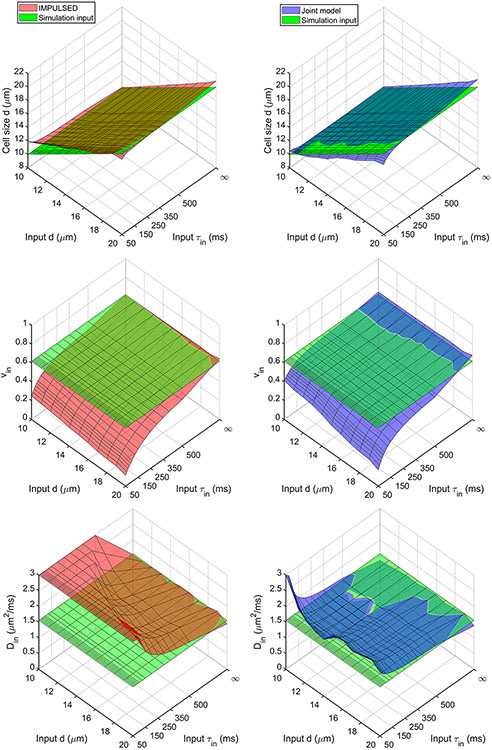
In Figure 2, the fitted four parameters (, , and ) compared with input values dependent on and are displayed for our proposed joint model and IMPULSED. The relative biases are displayed in Supplemental Figure S3. Note that there is only input in the simulations so the fitting errors of were not calculated, although it is expected that . The joint model provides accurate estimates of for >100 ms and all the cell sizes with error < 5%, while IMPULSED provides accurate estimates of for >100 ms and > 15 μm with error < 5%. When is between 50 and 100 ms, IMPULSED overestimates the sizes of small cells (e.g., for = 10 μm and = 50 ms, the error is ~ 20%) and underestimates the sizes of large cells (e.g., for = 20 μm and = 50 ms, the error is ~ −10%). The joint model-derived is less affected by the fast water exchange (e.g., when = 50 ms, the error is ~ 5% and −5% for = 10 and 20 μm, respectively). Consistent with previous computer simulation and experimental results [17], IMPULSED significantly underestimated by ignoring water exchange and this bias increases with faster exchange rates. The joint model improves the estimate of , particularly for relatively small cells (e.g., 10 ≤ ≤ 15 μm). IMPULSED provides only approximate fits for with errors ~ 10-30% for 15 ≤ ≤ 20 μm and > 300 ms, while errors from the joint model are less than 30% for all and values except for the combinations of very small and very short .
Figure 2.
Simulated dependence of the fitting errors of fitted parameters (d, , and ) on input d and values for IMPULSED and the joint model.
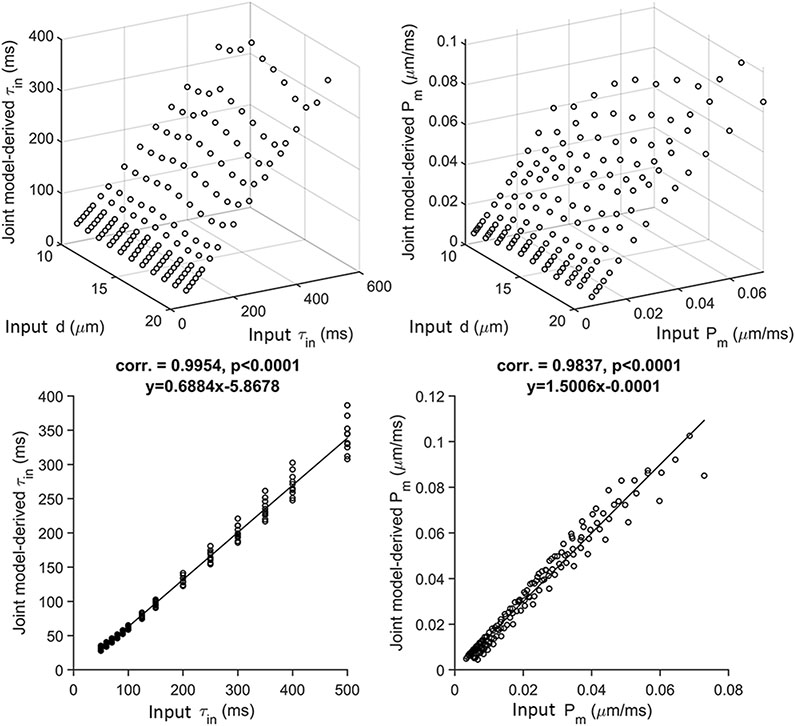
The joint model-derived and are highly linearly correlated with input values (Pearson correlation coefficient = 0.99 and 0.98 with p<0.01 and slope of the regression line = 0.69 and 1.50, for and , respectively) (Figure 3). Also, these linear correlations are independent of the cell size , as shown in the Supplemental Figure S10.
Figure 3.
Correlations between joint model-derived and from the simulated noise-free diffusion signals with = 5, 30, and 70 ms and input values. Subfigures at the second row are the projections of 3D scatter plots at the first row along axis ‘’. The solid lines represent the regression lines.
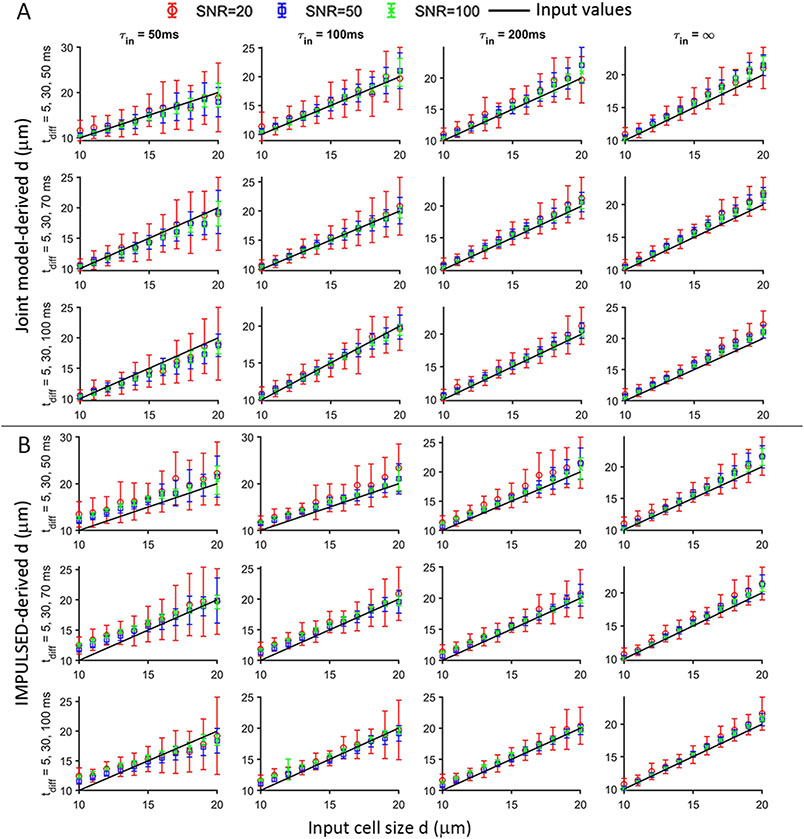
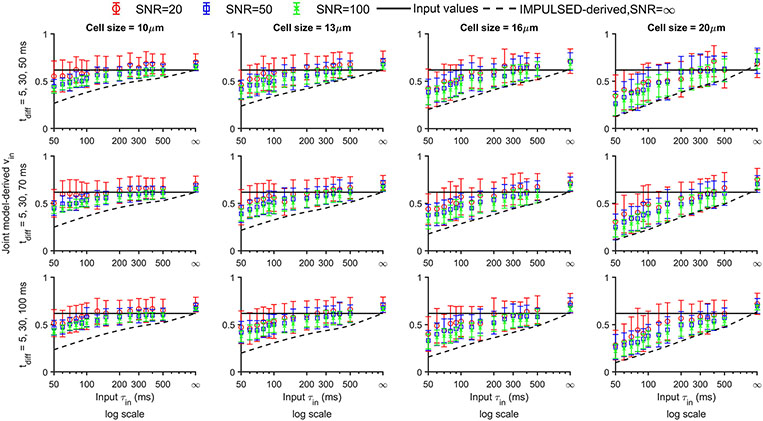
The performance of the proposed joint model was also evaluated and compared with IMPULSED for noisy simulated signals. Figures 4 shows the simulated dependence of the accuracy and precision of joint model and IMPULSED-derived on and ranges (the longest = 50, 70, and 100, respectively), and SNR levels (20, 50, and 100). The average fitted values using the joint model are very close to input values for most and . The joint model provides more accurate values for small cells (e.g., 10 ≤ ≤ 12) with fast water exchange (e.g., ≤ 100 ms) than IMPULSED alone. Variations of fitted increase with decreasing SNR and increasing cell size but are almost not affected by changing longest .
Figure 4.
Simulated dependence of the accuracy and precision of joint model and IMPULSED-derived on input cell size , (50, 100, 200ms, and for four columns, respectively), ranges (the longest = 50, 70, and 100 for three rows, respectively), and SNRs (SNR of b0 signal = 20, 50 and 100 for red, blue, and green symbols, respectively). Standard deviations of the fitted d were calculated from 50 sets of simulated diffusion data. Black solid lines represent input values.
As shown in Figure 5, ignoring water exchange leads to an underestimation of . The joint model reduces the bias especially when water exchange is relatively slow (e.g., >200 ms), but with large variations. Average values of fitted and using the joint model show good agreement with ground truth values (supplemental Figure S4&5). Variations of fitted and increase with decreasing SNR, increasing cell size, and decreasing longest . In Supplemental Figure S6, it is shown that IMPULSED provides reasonable fits of only for large cells and very slow water exchange. For other cases, IMPULSED-derived reaches the boundary of the constraint for (= 3 μm2/ms), indicating an inability to properly infer . The joint model provides better fits of for most and values, suggesting better sensitivity to by including water exchange into the model.
Figure 5.
Simulated dependence of the accuracy and precision of joint model-derived on , , ranges (the longest = 50, 70, and 100 for three rows, respectively), and SNRs (SNR of b0 signal = 20, 50 and 100 for red, blue, and green symbols, respectively). Standard deviations of the fitted were calculated from 50 sets of simulated diffusion data. Black solid lines represent input values. Black dash lines represent IMPULSED-derived from simulated noise-free diffusion data.
Retrospective in vitro cell experiment
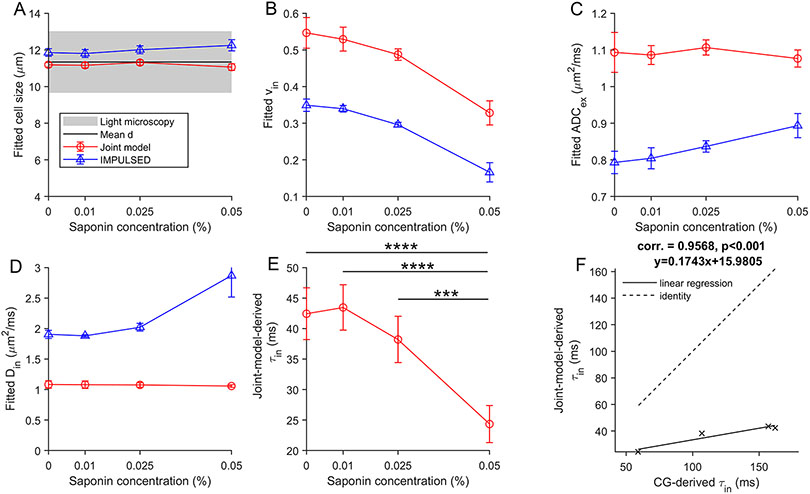
The average MEL cell diameter was measured as 11.3±1.68 μm using light microcopy. Since MR-derived cell sizes are volume weighted [47, 48], the volume weighted cell diameter was calculated as 12.11 μm. The CG-experiments provided values of 161.8, 157.8, 106.6, and 59.4 ms for 0, 0.01, 0.025, and 0.05% concentrations of saponin, respectively.
Representative diffusion signals and their fits using IMPULSED/joint model from four MEL cell pellets treated with 0%, 0.01%, 0.025%, and 0.05% saponin, respectively, are shown in Supplemental Figure S7. The fitting errors for each b value and each are displayed in Supplemental Figure S8, indicating that the joint model provides a better fit to the data than IMPULSED alone. Figure 6(A-E) summarizes the IMPULSED/joint model derived parameters (mean ± standard deviation) for four groups of MEL cell pellets (n=6) treated with 0%, 0.01%, 0.025%, and 0.05% saponin, respectively. Cell sizes measured by light microscopy are included in Figure 6A as well. Consistent with our simulation results, the joint model-derived ’s are slightly smaller than IMPULSED-derived ’s but both are very close to the values measured using light microscopy. The joint model-derived ’s are larger than IMPULSED-derived ’s, and both decrease with increasing saponin concentrations, indicating more severe negative impact of water exchange. The joint model-derived ’s are higher than IMPULSED-derived ’s which is consistent with our simulation results (Figure 2), and both are within physiologically reasonable ranges. The joint model-derived ’s are about 1 μm2/ms independent of saponin concentrations while IMPULSED-derived ’s increase with increasing saponin concentrations (almost reaching the upper boundary of the fitting constraint for when the saponin concentration is 0.05%). The average joint model-derived values decrease signaificantly (p<0.001) when the concentration of saponin increases from 0.01% to 0.05%. In Figure 6F, the joint model-derived values show good correlation with those measured using CG experiments.
Figure 6.
Summary of fitting parameters vs in different studies. Error bars in each sub-figure denote across-sample STD (n=6). The gray band represents microscopy-derived mean cell diameter ± STD of all cells. ***P < .001 and ****P < .0001, as measured by one-way analysis of variance (ANOVA) with a Bonferroni posttest.
Discussion
Both our joint model and IMPULSED provide accurate estimates of cell size independent of water exchange when > 100 ms. The joint model improves quantification of especially for small cells (e.g., 10 ≤ ≤ 12) when is between 50 and 100 ms. Meanwhile, the joint model significantly reduces the underestimation of compared with IMPULSED. Because both models were fit to the same dMRI data, this may provide new insights into how microstructural information is probed using IMPULSED with multi- and multi- dMRI data. can be reliably fit with or without the consideration of water exchange, and water exchange has negligible influence on OGSE measurements with short , so the estimation of using IMPULSED may be mainly determined by OGSE measurements with short . Likely, may be mainly determined by PGSE measurements with long and larger values when water exchange plays a dominant role. Although in-depth investigation is still needed, these findings could assist us better design dMRI experiments with emphasis on specific parameters.
This improvement could lead to significant enhanced performance for MRI-derived cell size as an indicator of effective treatment in solid tumors. Decreased cell sizes have been associated with treatment response due to several factors, such as cell shrinkage during apoptosis [49] and T cell infiltration [50]. Increased cell membrane permeability, or decreased water pre-exchange lifetime, have also been suggested as indicators of treatment response in many types of cancer [51] [52]. In Figure 4, IMPULSED overestimates cell size when is between 50 and 100 ms, which may potentially lower the sensitivity for detecting treatment-induced cell shrinkage.
Measurements of transcytolemmal water exchange are of great interest in e.g., early assessment of treatment response of solid tumors. First, alone has been suggested as an early indicator of pathological complete response of breast cancer patients undergoing neoadjuvant chemotherapy [51] and a prognostic marker for patients with head and neck cancer [52]. Second, or provides complementary information on tissue microstructure to the measurements of , , , and using IMPULSED. Previous studies have suggested IMPULSED-derived , , , and as indicators of treatment-induced apoptosis or mitotic-arrest in tumors [36, 49]. It is well-known that not only cell shrinkage ( decreases) but also membrane blebbing (resulting in an increase of or a decrease of ) are hallmarks of changes in cellular morphology during apoptosis. Simultaneous quantification of and may provide more comprehensive information on cellular changes and hence provide more specific assessment of apoptosis non-invasively.
It is possible to perform two separate experiments, e.g., IMPULSED [8, 9] and -dependent kurtosis [32], to get both and . However, this increases the total scan time, which is not desirable for e.g., clinical applications in which scan time limits are essential. A main advantage of the current joint model is that it keeps the same total scan time as in the IMPULSED method, which has been translated to human imaging on clinical scanners with relatively fast acquisitions (~ 7 mins of scan time) [10]. It is of interest to translate the current joint model to human imaging but that is out of the main scope of this study.
Our simulations show that although the joint model-derived and are still biased compared with input values, they are highly linearly correlated (Pearson correlation coefficient = 0.99, and 0.98, p<0.01, slope of the regression line = 0.69 and 1.50, for and , respectively) with the input values. In our retrospective in vitro cell study, the joint model-derived values are highly correlated (Pearson correlation coefficient = 0.96, p<0.01, slope of the regression line = 0.17) with the CG method-derived values. But the joint model-derived values are significantly smaller than those obtained by CG. The CG (constant gradient) method is a diffusion MRI-based method that has been developed for decades to measure an averaged transcytolemmal water exchange rate constant [18, 53]. The CG method is valid under two conditions, including a sufficiently long diffusion time () and a relatively slow transcytolemmal water exchange rate constant (). Note that very high b values are required in CG measurements with a fixed gradient strength (as well as the q value) so that the diffusion times are usually very long to meet the first condition. But the satisfaction of the second condition should be discussed on a case-by-case basis. A recent computer simulation study [54] suggests that the CG method provides a reasonable estimation of especially when > 100 ms. For < 100 ms, were overestimated by the CG method and the error depends on cell sizes. In our cell experiments, values measured by the CG method for cells treated with 0.05% and 0.025% saponin are < 100 ms and slightly larger than 100 ms, respectively, which may be larger than the real values. On the other hand, the joint model can only be applied to measure the slow water exchange which is expressed as , because of the involvement of the modified Kärger model at long . For MEL cells with an average cell size of ~ 11 μm, the joint model is valid when ≫ 30 ms. Therefore, it may be challenging to measure the real values for MEL cells treated with saponin using the joint model. In addition, the effect of transcytolemmal water exchange for OGSE acquisitions is ignored for the joint model. Efferts to elucidate how these limitations affect the accuracy of are ongoing. Although the Joint model-derived may underestimate the true values, the strong correlation between values derived from the CG method and joint model suggests that the Joint model-derived could be useful for monitoring temporal changes in water exchange. For one of the main applications of our cell size imaging technique, i.e., early detection of treatment response, quantifying changes in is more important than quantifying the absolute values of .
The simulation studies in the current work used the regular packing (face-center cubic) to construct modeled tissues. As shown previously [55], different types of cell packing can significantly affect the diffusion time dependence of extracellular diffusion. For example, a 3D disordered packing results in a power law diffusion-time-dependence of the extracellular water ( ) under the long diffusion time limit i.e., . However, it is unclear wheter water diffusion in tumors fullfills the long diffusion time limit. Also, considering the diffusion time dependence of extracellular diffusion leads to at least one more fitting parameters, which in turn will decreases the fitting precision of all other parameters. For a limited diffusion time range (5 - 100 ms) used in this study as well as other cancer applications of IMPULSED method, we have shown [10] that assuming a constant enhances the fitting precision of other parameters, such as cell size. Therefore, we hereby hypothesize that the change of cell packing has minor effects on IMPULSE/joint model-derived parameters and should not affect our conclusions obtained from the simulations.
As shown in Figure 6,, the joint model-derived detects changes in between 0.05% saponin treated and control MEL cells, however, it fails to distinguish controls from 0.025% saponin treated cells, suggesting the detection sensitivity is limited (Figure 6F). Note that the accuracy and precision of the joint model-derived and values depend on the SNR as well as the range of used in dMRI acquisitions. As shown in supplemental Figure S4&5, with SNR = 100 and the longest = 100 ms, the joint model-derived values for cell size between 10 and 16 μm and between 50 and 200 ms show a good agreement with input values with small variations. Including diffusion signals obtained with long could increase the sensitivity for quantifying smaller and longer .
Although this study tested the joint model using a single cell line, it should be valid for most cancer imaging. Most cancer cell sizes range from 10 to 15 μm, so the modified Kärger model is valid when ≫ 16 ms or ≫ 36 ms for a cell size of 10 or 15 μm, respectively, assuming is ~ 1.56 μm2/ms. These conditions are usually met in practice. For example, a previous in vitro study has reported = 119±14 ms for HeLa cancer cells with diameter of 10.1 ± 0.4 μm using inversion-recovery experiments [56]. A recent animal and human in vivo study reported cellular-interstitial water exchange time as 68 ms and 93 ms for GL261 and 4T1 animal brain tumors in vivo, and 70 ms and 106 ms for human breast tumors in vivo [32]. This corresponds to in the range of 226 and 353 ms if =30%. Also, numerical simulations have suggested that neurite/soma and neurite/neurite exchange occur at longer time scales (on the order of 100 ms or more) [57]. Studies using relaxation-based methods have suggested water exchange times of 100 – 150 ms in astrocyte and neuron cultures [20], in rat subcortical structures – presumably the striatum [58] and in rat perfused cortical cultures [59]. Therefore, our joint model could be applied in neurological applications.
Both the joint model and our computer simulations ignore the possible differences in relaxation properties between intra- and extracellular spaces, in line with most multi-compartment diffusion models [5, 8, 11]. This assumption is particularly pronounced in tumors. For example, a quantitative magnetization transfer (qMT) study suggests that the relatively higher transcytolemmal water exchange in tumors enables enough proton mixing and hence yields a homogenous T1 and MT in the intra- and extracellular spaces [35]. Moreover, a multi-echo T2 spectrum experiment showed that the majority (93.2 ± 6.2%) of total MRI signals arise from a single peak of T2 (76.4 ± 9.3 ms) in rodent brain tumors on 7 Tesla [34], suggesting small differences between intra- and extracellular spaces in tumors.
Conclusion
A heuristic joint model that combines the modified Kärger model and IMPULSED method has been proposed and evaluated using computer simulations and in vitro cell experiments. This joint model not only improves the accuracy of IMPULSED-derived microstructural parameters, such as cell size and apparent volume fraction of intracellular space , but also provides information on water exchange which cannot be obtained by IMPULSED alone. Although further validations using in vivo animal and human studies are needed, this joint model could potentially provide multiple indicators with potential for clinical applications.
Supplementary Material
Acknowledgements
This work was funded by NIH Grants/Award Numbers: R01CA109106, S10OD021771, and P30CA068485.
Abbreviations:
effective diffusion time
cell membrane permeability
intracellular water pre-exchange lifetime
averaged intra-/extra-cellular diffusion coefficient without the influences of cell membrane
apparent intra-/extra-cellular diffusion coefficient with the influences of cell membrane
- OGSE
oscillating gradient spin echo
- IMPULSED
imaging microstructural parameters using limited spectrally edited diffusion
Data Availability Statement
The data analysis code and sample data are available online at https://github.com/jzxu0622/mati.
References
- 1.Assaf Y, et al. , AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magn Reson Med, 2008. 59(6): p. 1347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barazany D, Basser PJ, and Assaf Y, In vivo measurement of axon diameter distribution in the corpus callosum of rat brain. Brain, 2009. 132(Pt 5): p. 1210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander DC, et al. , Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage, 2010. 52(4): p. 1374–89. [DOI] [PubMed] [Google Scholar]
- 4.Dyrby TB, et al. , Contrast and stability of the axon diameter index from microstructure imaging with diffusion MRI. Magn Reson Med, 2013. 70(3): p. 711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panagiotaki E, et al. , Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res, 2014. 74(7): p. 1902–12. [DOI] [PubMed] [Google Scholar]
- 6.Panagiotaki E, et al. , Microstructural characterization of normal and malignant human prostate tissue with vascular, extracellular, and restricted diffusion for cytometry in tumours magnetic resonance imaging. Investigative radiology, 2015. 50(4): p. 218–227. [DOI] [PubMed] [Google Scholar]
- 7.Bonet-Carne E, et al. , VERDICT-AMICO: Ultrafast fitting algorithm for non-invasive prostate microstructure characterization. NMR Biomed, 2019. 32(1): p. e4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang X, et al. , Quantification of cell size using temporal diffusion spectroscopy. Magn Reson Med, 2016. 75(3): p. 1076–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X, et al. , In vivo imaging of cancer cell size and cellularity using temporal diffusion spectroscopy. Magn Reson Med, 2017. 78(1): p. 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, et al. , Magnetic resonance imaging of mean cell size in human breast tumors. Magn Reson Med, 2020. 83(6): p. 2002–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynaud O, et al. , Pulsed and oscillating gradient MRI for assessment of cell size and extracellular space (POMACE) in mouse gliomas. NMR Biomed, 2016. 29(10): p. 1350–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volles MJ and Lansbury PT Jr., Vesicle permeabilization by protofibrillar alpha-synuclein is sensitive to Parkinson's disease-linked mutations and occurs by a pore-like mechanism. Biochemistry, 2002. 41(14): p. 4595–602. [DOI] [PubMed] [Google Scholar]
- 13.Moftakhar P, et al. , Aquaporin expression in the brains of patients with or without cerebral amyloid angiopathy. J Neuropathol Exp Neurol, 2010. 69(12): p. 1201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson M, et al. , Noninvasive mapping of water diffusional exchange in the human brain using filter-exchange imaging. Magn Reson Med, 2013. 69(6): p. 1573–81. [DOI] [PubMed] [Google Scholar]
- 15.Bailey C, Moosvi F, and Stanisz GJ, Mapping water exchange rates in rat tumor xenografts using the late-stage uptake following bolus injections of contrast agent. Magn Reson Med, 2014. 71(5): p. 1874–87. [DOI] [PubMed] [Google Scholar]
- 16.Li H, et al. , Time-Dependent Influence of Cell Membrane Permeability on MR Diffusion Measurements. Magn Reson Med, 2016. 75(5): p. 1927–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, et al. , Impact of transcytolemmal water exchange on estimates of tissue microstructural properties derived from diffusion MRI. Magn Reson Med, 2017. 77(6): p. 2239–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meier C, Dreher W, and Leibfritz D, Diffusion in compartmental systems. I. A comparison of an analytical model with simulations. Magn Reson Med, 2003. 50(3): p. 500–9. [DOI] [PubMed] [Google Scholar]
- 19.Zhao L, et al. , Intracellular water specific MR of microbead-adherent cells: HeLa cell intracellular water diffusion. Magn Reson Med, 2008. 59(1): p. 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang DM, et al. , Intracellular water preexchange lifetime in neurons and astrocytes. Magn Reson Med, 2018. 79(3): p. 1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J and Kim SG, Estimation of cellular-interstitial water exchange in dynamic contrast enhanced MRI using two flip angles. NMR Biomed, 2019. 32(11): p. e4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruggiero MR, et al. , Evidence for the Role of Intracellular Water Lifetime as a Tumour Biomarker Obtained by In Vivo Field-Cycling Relaxometry. Angew Chem Int Ed Engl, 2018. 57(25): p. 7468–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volles MJ, et al. , Vesicle permeabilization by protofibrillar alpha-synuclein: implications for the pathogenesis and treatment of Parkinson's disease. Biochemistry, 2001. 40(26): p. 7812–9. [DOI] [PubMed] [Google Scholar]
- 24.Kärger J, Pfeifer H, Wilfried H, Principles and application of self-diffusion measurements by nuclear magnetic resonance. Adv. Mag. Res, 1988. 12(1): p. 1–89. [Google Scholar]
- 25.Stanisz GJ, et al. , Water dynamics in human blood via combined measurements of T2 relaxation and diffusion in the presence of gadolinium. Magn Reson Med, 1998. 39(2): p. 223–33. [DOI] [PubMed] [Google Scholar]
- 26.Latt J, et al. , Diffusion-weighted MRI measurements on stroke patients reveal water-exchange mechanisms in sub-acute ischaemic lesions. NMR Biomed, 2009. 22(6): p. 619–28. [DOI] [PubMed] [Google Scholar]
- 27.Fieremans E, et al. , Monte Carlo study of a two-compartment exchange model of diffusion. NMR Biomed, 2010. 23(7): p. 711–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier C, Dreher W, and Leibfritz D, Diffusion in compartmental systems. II. Diffusion-weighted measurements of rat brain tissue in vivo and postmortem at very large b-values. Magn Reson Med, 2003. 50(3): p. 510–4. [DOI] [PubMed] [Google Scholar]
- 29.Lasic S, et al. , Apparent exchange rate mapping with diffusion MRI. Magn Reson Med, 2011. 66(2): p. 356–65. [DOI] [PubMed] [Google Scholar]
- 30.Lampinen B, et al. , Optimal experimental design for filter exchange imaging: Apparent exchange rate measurements in the healthy brain and in intracranial tumors. Magn Reson Med, 2017. 77(3): p. 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shemesh N, et al. , Conventions and nomenclature for double diffusion encoding NMR and MRI. Magn Reson Med, 2016. 75(1): p. 82–7. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, et al. , Measurement of cellular-interstitial water exchange time in tumors based on diffusion-time-dependent diffusional kurtosis imaging. NMR Biomed, 2021: p. e4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanisz GJ, et al. , An analytical model of restricted diffusion in bovine optic nerve. Magn Reson Med, 1997. 37(1): p. 103–11. [DOI] [PubMed] [Google Scholar]
- 34.Dortch RD, et al. , Evidence of multiexponential T2 in rat glioblastoma. NMR Biomed, 2009. 22(6): p. 609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li K, et al. , Influence of water compartmentation and heterogeneous relaxation on quantitative magnetization transfer imaging in rodent brain tumors. Magn Reson Med, 2016. 76(2): p. 635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang X, et al. , Early Detection of Treatment-Induced Mitotic Arrest Using Temporal Diffusion Magnetic Resonance Spectroscopy. Neoplasia, 2016. 18(6): p. 387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, et al. , Magnetic resonance imaging of mean cell size in human breast tumors. Magn Reson Med, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Does MD, and Gore JC, Quantitative characterization of tissue microstructure with temporal diffusion spectroscopy. J Magn Reson, 2009. 200(2): p. 189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gore JC, et al. , Characterization of tissue structure at varying length scales using temporal diffusion spectroscopy. NMR Biomed, 2010. 23(7): p. 745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karger J, Pfeifer H, and Heink W, Principles and Application of Self-Diffusion Measurements by Nuclear Magnetic Resonance. Advances in Magnetic and Optical Resonance, 1988. 12: p. 1–89. [Google Scholar]
- 41.Xu J, Does MD, and Gore JC, Numerical study of water diffusion in biological tissues using an improved finite difference method. Phys Med Biol, 2007. 52(7): p. N111–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang SN, et al. , An image-based finite difference model for simulating restricted diffusion. Magn Reson Med, 2003. 50(2): p. 373–82. [DOI] [PubMed] [Google Scholar]
- 43.Tan ET, et al. , Oscillating diffusion-encoding with a high gradient-amplitude and high slew-rate head-only gradient for human brain imaging. Magn Reson Med, 2020. 84(2): p. 950–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hennel F, Michael ES, and Pruessmann KP, Improved gradient waveforms for oscillating gradient spin-echo (OGSE) diffusion tensor imaging. NMR Biomed, 2020: p. e4434. [DOI] [PubMed] [Google Scholar]
- 45.Setsompop K, et al. , Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. Neuroimage, 2013. 80: p. 220–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang X, et al. , MR cell size imaging with temporal diffusion spectroscopy. Magn Reson Imaging, 2021. 77: p. 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afzali M, et al. , SPHERIOUSLY? The challenges of estimating sphere radius non-invasively in the human brain from diffusion MRI. Neuroimage, 2021. 237: p. 118183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olesen JL, et al. , Diffusion time dependence, power-law scaling, and exchange in gray matter. arXiv:2108.09983v1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang X, et al. , In vivo magnetic resonance imaging of treatment-induced apoptosis. Sci Rep, 2019. 9(1): p. 9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang X, et al. , MRI of tumor T cell infiltration in response to checkpoint inhibitor therapy. J Immunother Cancer, 2020. 8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Springer CS Jr., et al. , Intratumor mapping of intracellular water lifetime: metabolic images of breast cancer? NMR Biomed, 2014. 27(7): p. 760–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chawla S, et al. , Dynamic Contrast-Enhanced MRI-Derived Intracellular Water Lifetime (tau i ): A Prognostic Marker for Patients with Head and Neck Squamous Cell Carcinomas. AJNR Am J Neuroradiol, 2018. 39(1): p. 138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfeuffer J, et al. , Restricted diffusion and exchange of intracellular water: theoretical modelling and diffusion time dependence of 1H NMR measurements on perfused glial cells. NMR Biomed, 1998. 11(1): p. 19–31. [DOI] [PubMed] [Google Scholar]
- 54.Tian X, et al. , Evaluation and comparison of diffusion MR methods for measuring apparent transcytolemmal water exchange rate constant. J Magn Reson, 2017. 275: p. 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novikov DS, et al. , Revealing mesoscopic structural universality with diffusion. Proc Natl Acad Sci U S A, 2014. 111(14): p. 5088–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao L, et al. , Intracellular water-specific MR of microbead-adherent cells: the HeLa cell intracellular water exchange lifetime. NMR Biomed, 2008. 21: p. 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jelescu LO, et al. , Neurite Exchange Imaging (NEXI): A minimal model of diffusion in gray matter with inter-compartment water exchange. arXiv:2108.06121v1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quirk JD, et al. , Equilibrium water exchange between the intra- and extracellular spaces of mammalian brain. Magn Reson Med, 2003. 50(3): p. 493–9. [DOI] [PubMed] [Google Scholar]
- 59.Bai R, et al. , Fast, Na(+) /K(+) pump driven, steady-state transcytolemmal water exchange in neuronal tissue: A study of rat brain cortical cultures. Magn Reson Med, 2018. 79(6): p. 3207–3217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analysis code and sample data are available online at https://github.com/jzxu0622/mati.