Abstract
The base moiety of 1-N-methyladenosine can be protected with a chloroacetyl group for incorporation of this modified nucleoside into DNA and RNA. Carefully controlled anhydrous conditions are needed for deprotection of the oligonucleotides.
INTRODUCTION
Modified nucleosides offer an overwhelming variety of chemical and potential biological functions. More than 100 modified nucleosides have been discovered in nature (1). The structures of these nucleosides are well known but the contribution of modified nucleosides to the functional chemistry, structure and biological activity of RNAs largely remains to be discovered. Indeed, most of these modifications occur in various RNAs and they are introduced after the polymerisation reaction. Nucleic acid modifications have been linked to control of gene expression at both the levels of transcription and translation (2). Modified nucleosides have a function in RNA–RNA and RNA–protein interactions, in the dynamics of the translation process and in nucleic acids evolution. The interaction of one modified nucleoside with another may stabilise novel nucleic acid structures.
tRNA displays the largest variety of modified nucleosides. The study of the physico-chemical properties of these nucleosides and their influence on the translation process may provide an insight into the RNA world and give new impetus to the design of nucleic acids with novel functions (3). Post-transcriptional modifications in natural tRNA are important in stabilising the cloverleaf conformation of tRNAs and in some cases the modified nucleoside is involved in directing cloverleaf folding (4). A particular role in this folding has been attributed to 1-methyladenosine (m1A), obtained by post-transcriptional methylation of adenosine by methyl-1-adenosine transferase (5,6). Approximately 25% of all tRNAs have m1A at position 58 in the T loop, while m1A also often occurs at position 14 in the D loop (7). Methylation at position A9 is specific for mitochondrial tRNAs (8).
The function of modified nucleosides is studied by comparison of the properties of modified tRNAs (purified from wild-type organisms) with that of unmodified tRNAs (obtained by in vitro transcription) and of partially modified tRNAs (from modification mutants). These studies have allowed to demonstrate that m1A is a key element for correct folding of human mitochondrial tRNALys (4,9). It was observed that unmodified human mitochondrial tRNALys does not adopt the cloverleaf structure, but folds into a bulged hairpin, including an extended amino acid acceptor stem, an extra large loop instead of the T stem and loop and an anticodon-like domain (4,9). Introduction of a single methyl group (m1A9) is sufficient to induce cloverleaf folding of this tRNA (9), which is the first evidence of a role of m1A in RNA folding.
Methylation of adenosine at position 1 produces a drastic functional change in the nucleobase. 1-Methyladenosine (pKa ∼8.25) is a much stronger base than adenosine (pKa ∼3.5) (10). N-1 methylation excludes participation of the adenine base in canonical Watson–Crick base pairing, provides a positive charge to the nucleobase and also alters the hydrophobicity of the base, the stacking properties (11), the ordering of water molecules and the chelation properties. The base may become involved in non-canonical hydrogen bonding, in electrostatic interactions and, in general, it may contribute to the conformational dynamics of the tRNA. As an example, a reverse Hoogsteen base pair is formed between m1A58 and T54 in tRNAPhe (12).
Because 1-methyladenosine is a basic nucleoside, which easily undergoes Dimroth rearrangement to yield N6-methyladenosine, its incorporation in oligonucleotides by chemical means is not easy. This incorporation might be achieved either by working in a non-basic medium during the whole process of oligonucleotide synthesis and deprotection or by working in an anhydrous medium. Here we describe the synthesis of the phosphoramidite of protected 1-methyladenosine and its successful incorporation into DNA as well as RNA.
MATERIALS AND METHODS
NMR spectra were recorded using a Gemini 200 NMR spectrometer and a Varian 500 Unity spectrometer at 300 K. Chemical shifts were measured relative to the solvent signals. The coupling constants (J) are given in Hz. The signals were assigned using double resonance techniques.
Mass spectrometry and exact mass measurements of the building blocks were performed in a quadruple/orthogonal acceleration time-of-flight tandem mass spectrometer (Q-Tof-2; Micromass, Manchester, UK) equipped with a standard electrospray ionisation (ESI) interface. Samples were dissolved in a H2O:2-propanol mixture (1:1) and infused at 3 µl min–1. Oligonucleotides were dissolved in a mixture of acetonitrile:water (1:1), the final concentration being ∼15 pmol µl–1. Electrospray spectra were acquired in negative ion mode (–ESI) in a quadrupole/orthogonal acceleration time-of-flight tandem mass spectrometer (Q-Tof-2) equipped with a standard ESI interface. Samples were infused at 3 µl min–1. A collision energy of 120 eV was used for acquisition of the fragment ion spectra. Column chromatography was performed on silica gel (0.06–0.20 mm). TLC was carried out on Kieselgel 260 F (Fluka) with detection by UV light using the following systems: methylene chloride:methanol, 1:1 (A); methylene chloride:methanol, 9:1 (B); methylene chloride:methanol, 95:5 (C).
5′-O-monomethoxytrityl-1-methyladenosine (2)
A mixture of the hydroiodide salt of 1-methyladenosine (1) (4.1 g, 10 mmol) and monomethoxytrityl chloride (4.63 g, 15 mmol) in dry pyridine (30 ml) was stirred in the dark for 2 days at 20°C. To the suspension was added CH2Cl2 (200 ml) and the organic layer was subsequently washed with water (50 ml), 10% aqueous sodium bicarbonate (30 ml), 5% aqueous Na2S2O3 (30 ml) and water (2 × 10 ml). The organic layer was dried over Na2SO4, evaporated in vacuo and co-evaporated with toluene (3 × 30 ml). The residue was dissolved in acetone (20 ml) and precipitated into ether (500 ml). The mixture was kept at 0°C overnight, the precipitate filtered, washed with ether (30 ml) and dried to give 2 as a slightly yellow powder. Yield 3.76 g (68%). Rf 0.15 (A). Analysis: calculated for C31H31N5O5, C 67.26, H 5.64, N 12.65; found, C 67.05, H 5.41, N 12.56. +ESI MS: calculated for C31H31N5O5 + H+ 554.2403; found 554.2390.
1H NMR (CDCl3): 8.19 s (1H, H-8), 7.81 s (1H, H-2), 7.39–7.21 m (12H, Ph), 6.76 d (2H, J = 8.8, PhOMe), 5.97 d (1H, J1′,2′ = 6.0, H-1′), 4.79 dd (1H, J2′,3′ = 5.0, H-2′), 4.35 m (2H, H-3′,4′), 3.69 s (3H, OMe), 3.57 s (3H, NMe), 3.40 dd (1H, J5′a,4′ = 3.0, J5′a,5′b = –10.1, H-5′a), 3.35 dd (1H, J5′b,4′ = 4.0, H-5′b).
N6-methyl-5′-O-monomethoxytrityladenosine (4)
A solution of nucleoside 2 (277 mg, 0.5 mmol) in 2 M methylamine in methanol (5 ml) was kept for 8 days at 20°C and evaporated in vacuo to dryness. The residue was purified by column chromatography on silica gel (20 g). The column was washed with methylene chloride and methylene chloride:methanol 99:1 and then eluted with methylene chloride:methanol 98:2 to give 4 as a foam. Yield 250 mg (90%). Rf 0.50 (B). +ESI MS: calculated for C31H31N5O5 + H+ 554.2403; found 554.2463.
1H NMR (CDCl3): 8.32 s (1H, H-2), 8.01 s (1H, H-8), 7.31–7.18 m (12H, Ph), 6.75 d (2H, J = 8.8, PhOMe), 5.95 d (1H, J1′,2′ = 5.9, H-1′), 4.72 dd (1H, J2′,3′ = 5.2, H-2′), 4.39 m (2H, H-3′,4′), 3.76 s (3H, OMe), 3.44 dd (1H, J5′a,4′ = 3.1, J5′a,5′b = –10.3, H-5′a), 3.28 dd (1H, J5′b,4′ = 2.8, H-5′b), 3.20 s (3H, NMe).
N6-benzoyl-5′-O-monomethoxytrityl-1-methyladenosine (5a)
To a cold (0°C) solution of nucleoside 2 (277 mg, 0.5 mmol) in dry pyridine (3 ml) was added trimethylsilyl chloride (0.32 ml, 2.5 mmol) and the solution was stirred at 0°C for 45 min. Then benzoyl chloride (0.29 ml, 2.5 mmol) was added and the mixture was stirred at 0°C for 3 h. The reaction was quenched by addition of aqueous 10% sodium bicarbonate (5 ml) and the mixture was stirred at 0°C for 30 min and then at 20°C for 1.5 h. To the suspension were added CH2Cl2 (30 ml) and water (10 ml), and the organic layer was subsequently washed with 10% aqueous sodium bicarbonate (10 ml) and water (10 ml). The organic layer was dried over Na2SO4, evaporated in vacuo and co-evaporated with toluene (3 × 10 ml). The residue was purified by column chromatography on silica gel (20 g). The column was washed with methylene chloride and methylene chloride:methanol 97.5:2.5 and then eluted with methylene chloride:methanol 95:5 to give 5a as a foam. Yield 240 mg (73%). Rf 0.59 (B). +ESI MS: calculated for C38H35N5O6 + Na+ 680.2485; found 680.2506.
1H NMR (CDCl3): 8.13 m (2H, Bz), 7.86 s (1H, H-8), 7.70 s (1H, H-2), 7.41–7.17 m (15H, Bz, Ph), 6.75 d (2H, J = 8.8, PhOMe), 5.83 d (1H, J1′,2′ = 5.1, H-1′), 4.51 dd (1H, J2′,3′ = 4.8, H-2′), 4.26 m (2H, H-3′,4′), 3.72 s (3H, OMe), 3.64 s (3H, NMe), 3.35 dd (1H, J5′a,4′ = 3.7, J5′a,5′b = –10.3, H-5′a), 3.26 dd (1H, J5′a,4′ = 4.4, H-5′b).
13C NMR (CDCl3): 177.25 (C=O), 158.67 (Ph), 147.20 (C-2), 145.32 (C-6), 144.07 (C-4), 138.61 (Ph), 135.64 (C-8), 135.12, 132.18, 130.42, 129.78, 128.38, 128.14, 127.90 and 127.02 (Bz, Ph), 122.25 (C-5), 113.18 (Ph), 88.93 (C-1′), 86.71 (C-O), 84.62 (C-4′), 74.85 (C-2′), 71.54 (C-3′), 63.62 (C-5′), 55.15 (Ome), 36.97 (NMe).
N6-acetyl-5′-O-monomethoxytrityl-1-methyladenosine (5b)
Analogous acetylation of 2 with acetic anhydride yielded (70%) 5b as a foam. Rf 0.56 (B). +ESI MS: calculated for C33H33N5O6 + Na+ 618.2329; found 618.2347.
1H NMR (CDCl3): 7.83 s (1H, H-8), 7.78 s (1H, H-2), 7.40–7.20 m (12H, Ph), 6.79 d (2H, J = 8.8, PhOMe), 5.88 d (1H, J1′,2′ = 5.1, H-1′), 4.61 t (1H, J2′,3′ = 5.1, H-2′), 4.39dd (1H, J3′,4′ = 3.7, H-3′), 4.28 ddd (1H, J4′,5′a = 3.3, J4′,5′b = 3.4, H-4′), 3.77 s (3H, OMe), 3.48 s (3H, NMe), 3.40 dd (1H, J5′a,5′b = –10.3, H-5′a), 3.30 dd (1H, H-5′b), 2.33 (3H, Ac).
13C NMR (CDCl3): 184.90 (C=O), 158.70 (Ph), 147.32 (C-2), 144.68 (C-6), 144.07 (C-4), 138.52 (Ph), 135.15 (C-8), 130.42, 128.38, 127.90, 127.05 (Ph), 121.65 (C-5), 113.21 (Ph), 89.08 (C-1′), 86.74 (C-O), 84.71 (C-4′), 74.97 (C-2′), 71.54 (C-3′), 63.62 (C-5′), 55.18 (Ome), 36.48 (NMe), 26.56 (Ac).
N6-chloroacetyl-5′-O-monomethoxytrityl-1-methyladenosine (5c)
Method A. Analogous acetylation of 2 with chloroacetic anhydride yielded (58%) 5c as a foam. Rf 0.58 (B). Analysis: calculated for C33H32ClN5O6, C 62.90, H 5.12, N 11.11; found, C 62.81, H 4.91, N 10.93. +ESI MS: calculated for C33H32ClN5O6 + H+ 630.2119; found 630.2139.
1H NMR (CDCl3): 7.89 s (1H, H-8), 7.86 s (1H, H-2), 7.34–7.20 m (12H, Ph), 6.79 d (2H, J = 8.8, Ph), 5.89 d (1H, J1′,2′ = 5.1, H-1′), 4.66 dd (1H, J2′,3′ = 5.0, H-2′), 4.39 m (3H, H-3′, ClCH2), 4.31 ddd (1H, J4′,3′ = 3.6, J4′,5′a = J4′,5′b = 3.7, H-4′), 3.78 s (3H, OMe), 3.60 s (3H, NMe), 3.42 dd (1H, J5′a,5′b = –10.6, H-5′a), 3.31 dd (1H, H-5′b).
13C NMR (CDCl3): 177.98 (C=O), 158.64 (Ph), 147.90 (C-2), 147.23 (C-6), 145.62 (C-4), 139.61 (Ph), 135.06 (C-8), 130.39, 128.32, 127.84, 126.99 (Ph), 121.92 (C-5), 113.15 (Ph), 88.69 (C-1′), 86.68 (C-O), 84.47 (C-4′), 74.66 (C-2′), 71.23 (C-3′), 63.59 (C-5′), 55.15 (Ome), 45.95 (ClCH2), 36.79 (NMe).
Method B. To a cold (–20°C) solution of nucleoside 2 (1.66 g, 3 mmol) in a mixture of dry pyridine (3 ml) and dichloroethane (30 ml) was added chloroacetic anhydride (2.05 g, 12 mmol) and the solution was kept at 0°C for 30 min. To the solution were added CH2Cl2 (50 ml) and water (30 ml) and the organic layer was subsequently washed with 10% aqueous sodium bicarbonate (20 ml) and water (20 ml). The organic layer was dried over Na2SO4, evaporated in vacuo (bath temperature <30°C) to a volume of ∼5 ml, then 2 M ammonia in methanol (15 ml) was added and the solution was kept at 0°C for 30 min.
The mixture was evaporated in vacuo and co-evaporated with toluene (30 ml). The residue was purified by column chromatography on silica gel (100 g). The column was washed with methylene chloride and methylene chloride:methanol 99:1 and 98:2 and then eluted with methylene chloride:methanol 97:3 to give 5c as a foam. Yield 1.5 g (80%).
1,N6-dimethyl-5′-O-monomethoxytrityladenosine (6a)
A solution of nucleoside 5a (50 mg, 0.076 mmol) in 2 M methylamine in methanol (5 ml) was kept for 5 days at 20°C and evaporated in vacuo to dryness. To the residue dry ether (10 ml) was added and after 24 h at 20°C the precipitate was filtered, washed with ether and dried to afford 6a as a white powder. Yield 40 mg (93%). Rf 0.13 (A). +ESI MS: calculated for C32H33N5O5 + H+ 568.2560; found 568.2588.
1H NMR (CDCl3): 7.85 s (1H, H-8), 7.55 s (1H, H-2), 7.37–7.19 m (12H, Ph), 6.80 d (2H, J = 8.8, PhOMe), 5.90 d (1H, J1′,2′ = 6.5, H-1′), 4.71 dd (1H, J2′,3′ = 5.3, H-2′), 4.36 m (2H, H-3′,4′), 3.78 s (3H, OMe), 3.57 s (3H, 6NMe), 3.43 dd (1H, J5′a,4′ = 3.0, J5′a,5′b = –10.5, H-5′a), 3.39 s (3H, 1NMe), 3.27 dd (1H, J5′b,4′ = 3.0, H-5′b).
1-Ethyl-N6-methyl-5′-O-monomethoxytrityladenosine (6b)
A solution of nucleoside 5a (50 mg, 0.076 mmol) in 2 M ethylamine in methanol (5 ml) was kept for 8 days at 20°C and evaporated in vacuo to dryness. The residue was dissolved in acetone (5 ml) and hexane (10 ml) was added to a slight turbidity. After 24 h at 20°C the precipitate was filtered, washed with a mixture of acetone:hexane 1:3 and dried to give 6b as a white powder. Yield 33 mg (75%). Rf 0.14 (A). +ESI MS: calculated for C33H35N5O5 + H+ 582.2716; found 582.2692.
1H NMR (CDCl3): 7.83 s (1H, H-8), 7.58 s (1H, H-2), 7.36–7.23 m (12H, Ph), 6.79 d (2H, J = 8.8, PhOMe), 5.87 d (1H, J1′,2′ = 6.3, H-1′), 4.68 dd (1H, J2′,3′ = 5.4, H-2′), 4.35 m (2H, H-3′,4′), 4.11 dq (1H, JH,Me = 7.3, JH,H = –14.1, NCHHMe), 3.96 dq (1H, JH,Me = 7.3, NCHHMe), 3.77 s (3H, OMe), 3.43 dd (1H, J5′a,4′ = 3.0, J5′a,5′b = –10.5, H-5′a), 3.41 s (3H, NMe), 3.26 dd (1H, J5′b,4′ = 3.2, H-5′b), 1.27 t (3H, CH2Me).
Silylation of nucleoside 5c
To a solution of 5c (1.260 g, 2 mmol) and imidazole (340 mg, 5 mmol) in dry 1,2-dichloroethane (20 ml) was added t-butyldimethylsilyl chloride (362 mg, 2.4 mmol) and the mixture was kept at 20°C for 16 h. During this time fine crystals of imidazole hydrochloride were formed. A mixture of CH2Cl2 (30 ml) and 10% aqueous sodium bicarbonate (10 ml) was added and the organic layer was washed with water (10 ml), dried over Na2SO4 and evaporated in vacuo to dryness. The residue was purified by column chromatography on silica gel (30 g) using hexane:EtOAc (a gradient from 5:1 to 1:2) to yield 10% (170 mg) of the 2′,3′-O-bis-TBDMS derivative, 36% (540 mg) of 7 and 41% (620 mg) of the 3′-O-TBDMS derivative as white foams.
N6-chloroacetyl-5′-O-monomethoxytrithyl 2′,3′-O-bis-t-butyldimethylsilyl-1-methyladenosine
Rf 0.91 (C). +ESI MS: calculated for C45H60ClN5O6Si2 + H+ 858.3849; found 858.3837.
1H NMR (CDCl3): 7.96 s (1H, H-8), 7.80 s (1H, H-2), 7.45–7.27 m (12H, Ph), 6.84 d (2H, J = 8.8, PhOMe), 5.90 d (1H, J1′,2′ = 5.1, H-1′), 4.63 dd (1H, J2′,3′ = 4.4, H-2′), 4.44 s (2H, ClCH2), 4.23 ddd (1H, J4′,3′ = 3.7, J4′,5′a = 4,0, J4′,5′b = 4.4, H-4′), 4.15 dd (1H, H-3′), 3.81 s (3H, OMe), 3.63 s (3H, NMe), 3.47 dd (1H, J5′a,5′b = –10.6, H-5′a), 3.29 dd (1H, H-5′b), 0.84 s (9H, iBu), 0.81 s (9H, iBu), 0.02 s (3H, Me), –0.04 s (3H, Me), –0.07 (3H, Me), –0.20 (3H, Me).
13C NMR (CDCl3): 178.40 (C=O), 158.79 (Ph), 147.08 (C-2), 146.41 (C-6), 144.12 (C-4), 139.49 (Ph), 135.18 (C-8), 130.45, 128.44, 127.96, 127.14 (Ph), 122.62 (C-5), 113.27 (Ph), 88.59 (C-1′), 86.93 (C-O), 84.59 (C-4′), 75.48 (C-2′), 72.42 (C-3′), 63.34 (C-5′), 55.24 (Ome), 45.98 (ClCH2), 36.63 (NMe), 25.68 (Me), 25.59 (Me), 17.88 (C-Si), 17.73 (C-Si), –4.55 (Me), –4.76 (Me), –4.92 (Me), –5.13 (Me).
N6-chloroacetyl-5′-O-monomethoxytrityl-2′-O-t-butyldimethylsilyl-1-methyladenosine (7)
Rf 0.77 (C). Analysis: calculated for C39H46ClN5O6Si, C 62.93, H 6.23, N 9.41; found, C 62.77, H 6.09, N 9.25. +ESI MS: calculated for C39H46ClN5O6Si + H+ 744.2984; found 744.3000.
1H NMR (CDCl3): 7.95 s (1H, H-8), 7.78 s (1H, H-2), 7.47–7.25 m (12H, Ph), 6.83 d (2H, J = 8.8, PhOMe), 5.91 d (1H, J1′,2′ = 5.5, H-1′), 4.86 dd (1H, J2′,3′ = 4.8, H-2′), 4.44 s (2H, ClCH2), 4.30 ddd (3H, J3′,4′ = J3′,OH = 3.7, H-3′), 4.23 ddd (1H, J4′,5′a = 2.9, J4′,5′b = 3.7, H-4′), 3.80 s (3H, OMe), 3.62 s (3H, NMe), 3.49 dd (1H, J5′a,5′b = –10.6, H-5′a), 3.34 dd (1H, H-5′b), 2.65 d (1H, 3′-OH), 0.86 s (9H, iBu), 0.02 s (3H, Me), –0.10 (3H, Me).
13C NMR (CDCl3): 178.43 (C=O), 158.80 (Ph), 146.93 (C-2), 146.68 (C-6), 145.35 (C-4), 139.25 (Ph), 135.09 (C-8), 130.48, 128.41, 127.96, 127.14 (Ph), 122.47 (C-5), 113.24 (Ph), 88.20 (C-1′), 86.90 (C-O), 84.16 (C-4′), 75.76 (C-2′), 71.36 (C-3′), 63.43 (C-5′), 55.21 (Ome), 45.98 (ClCH2), 36.66 (NMe), 25.47 (Me), 17.79 (C-Si), –5.01 (Me), –5.25 (Me).
N6-chloroacetyl-5′-O-monomethoxytrithyl-3′-O-t-butyldimethylsilyl-1-methyladenosine
Rf 0.65 (C). +ESI MS: calculated for C39H46ClN5O6Si + H+ 744.2984; found 744.3036.
1H NMR (CDCl3): 7.99 s (1H, H-8), 7.87 s (1H, H-2), 7.41–7.26 m (12H, Ph), 6.83 d (2H, J = 8.8, PhOMe), 5.90 d (1H, J1′,2′ = 5.1, H-1′), 4.58 dd (1H, J2′,3′ = 5.1, H-2′), 4.47 dd (1H, J3′,4′ = 3.7, H-3′), 4.43 s (2H, ClCH2), 4.16 ddd (1H, J4′,5′a = 3.7, J4′,5′b = 4.0, H-4′), 3.80 s (3H, OMe), 3.63 s (3H, NMe), 3.49 dd (1H, J5′a,5′b = –10.6, H-5′a), 3.25 dd (1H, H-5′b), 0.88 s (9H, iBu), 0.07 s (3H, Me), –0.01 (3H, Me).
13C NMR (CDCl3): 178.22 (C=O), 158.76 (Ph), 147.26 (C-2), 146.75 (C-6), 145.38 (C-4), 139.16 (Ph), 135.06 (C-8), 130.36, 128.38, 127.93, 127.78 and 127.14 (Ph), 122.50 (C-5), 113.24 (Ph), 89.08 (C-1′), 86.83 (C-O), 84.35 (C-4′), 74.60 (C-2′), 71.72 (C-3′), 62.71 (C-5′), 55.21 (Ome), 45.92 (ClCH2), 36.63 (NMe), 25.56 (Me), 17.85 (C-Si), –4.82 (Me), –5.04 (Me).
Synthesis of phosphoramidite building block 8
About 1 mmol modified nucleoside 7, protected at the primary alcohol, was treated with dry N,N-diisopropylethylamine (3 equiv.) and 2-cyanoethyl-N,N-diisopropylchlorophophoramidite (1.5 equiv.) in dry dichloromethane (10 ml) and stirred at room temperature for 30 min. The reaction was quenched by addition of water (3 ml) and further stirred for 15 min. The mixture was washed with 5% sodium bicarbonate solution (30 ml) and saturated NaCl solution (3 × 30 ml), dried and evaporated. Column chromatography with n-hexane/acetone/triethylamine as eluent afforded the amidite. The product thus obtained was dissolved in dry dichloromethane (2 ml) and precipitated by dropwise addition to cold (–70°C) n-hexane (100 ml). The product was isolated, washed with n-hexane, dried and used as such for DNA synthesis. Yield: 63%. Rf (hexane:acetone:TEA 49:49:2): 0.51 (Rf for 7: 0.47). Analysis for C48H63N7O7Si1Cl1P1: [M+H]+, calculated 944.40625, found 944.4075; [M+Na]+, calculated 966.38821, found 966.3929. 31P NMR (p.p.m. external reference = H3PO4 capillar) 149.83, 151.66.
Solid phase oligonucleotide synthesis
Oligonucleotide synthesis was done on an ABI 392 DNA synthesiser (Applied Biosystems) at the 1.5 µmol scale using the phosphoramidite approach and the standard protocol, except for an increase in the amidite concentration for 8 from 0.1 to 0.12 M. Analogous to oligoribonucleotides, coupling of the modified monomer was allowed to proceed for 10 min. The oligomers were deprotected and cleaved from the solid support by treatment with an anhydrous solution of 2 M ammonia in MeOH for 60 h at room temperature. These reaction conditions are sufficient to cleave the N2-isobutyryl protecting group of the guanine base. The solution was decanted, the filtrate evaporated (SpeedVac), and the residue treated with 1.5 ml of a 1 M TBAF solution for 18 h at room temperature. The reaction was quenched by addition of 1 ml of a 1.5 M NH4OAc solution and was desalted twice on a NAP25 column and lyophilised. Purification was achieved on a Mono-Q HR10/10 anion exchange column (Pharmacia) at a flow rate of 2 ml min–1 with the following gradient system: A, 0.1 M KH2PO4, pH 6.4, 0.05 M NaCl; B, 0.1 M KH2PO4, pH 6.4, 0.9 M NaCl. The gradient used depended on the oligonucleotide. The low pressure liquid chromatography system consisted of a Merck-Hitachi L 6200.A intelligent pump, a Mono Q-HR 10/10 column (Pharmacia), a Uvicord SII UV detector (Pharmacia-LKB) and a recorder. The product-containing fraction was desalted on a NAP25 column. For oligoribonucleotides we used a 10 min coupling time throughout as prescribed. For purification a perchlorate gradient at a flow rate of 2 ml min–1 was used: A, 0.01 M NaClO4, 20 mM Tris–HCl, pH 6.8, containing 10% CH3CN; B, 0.6 M NaClO4, 20 mM Tris–HCl, pH 6.8, containing 10% CH3CN; 0 to 50% B.
Melting temperatures
Oligomers were dissolved in 0.1 M NaCl, 0.02 M potassium phosphate pH 7.5, 0.1 mM EDTA. The concentration was determined by measuring the absorbance at 260 nm at 80°C. The following extinction coefficients were used: dA, ɛ = 15 000; 1-methyladenosine, ɛ = 12 000; dT, ɛ = 8500; dG, ɛ = 12 500; dC, ɛ = 7500; U, ɛ = 10 000. The concentration in all experiments was ∼4 µM for each strand except for the hairpins, where Tm was verified at different concentrations (2–15 µM) to verify the hairpin mode of complexation. Melting curves were determined with a Cary 100Bio spectrophotometer. Cuvettes were maintained at constant temperature by means of water circulation through the cuvette holder. The temperature of the solution was measured with a thermistor directly immersed in the cuvette. Temperature control and data acquisition were done automatically with an IBM-compatible computer. The samples were heated and cooled at a rate of 0.2°C min–1, and little difference could be observed between the heating and cooling melting curves. Melting temperatures were evaluated by plotting the first derivative of the absorbance versus temperature curve.
Mass spectrometry
Prior to acquisition of spectra, the oligonucleotides were converted to their triethylammonium salt by reversed phase HPLC using TEAB buffer and elution with CH3CN. The mass spectrometer was tuned to a resolution of ∼7000 (measured at 50% intensity) in order to obtain monoisotopic mass values. The m/z values were deconvoluted using the MaxEnt algorithm of the processing software (MassLynx; Micromass, Manchester, UK). In order to check whether a Dimroth rearrangement of m1A occurred, the base anion was released from the oligonucleotide by in-source fragmentation. The fragment ion spectrum of the released base was then compared with those obtained from m1A- and m6A-containing nucleosides.
The position of the incorporated m1A was verified by digestion of the oligonucleotide using RNase T1 and analysis of the oligonucleotide mixture using capillary LC-MS (CapLC; Waters, Milford, MA). The chromatographic system was adapted from Apffel et al. (13). A 300 µm × 15 cm 3 µm RP C18 column (PepMap; LC Packings, San Francisco, CA) was used as the stationary phase. The fragments obtained from CID experiments agree with the theoretical values, confirming the correct sequence.
RESULTS
Synthesis of protected 1-methyladenosine
In the course of our ongoing programme on the synthesis of oligonucleotides containing minor tRNA components (14–16), we incorporated 1-methyladenosine (1) into oligonucleotides. It is known that, under alkaline conditions, m1A rearranges to N6-methyladenosine (3) (10,17). Also, m1A is difficult to handle due to its polar character (10,11). Oligonucleotides containing m1A (2) were prepared via ligation of its 3′,5′-mononucleoside biphosphate derivative to a synthetic 8mer corresponding to the 5′-end of a tRNA and an in vitro transcribed 64mer corresponding to the 3′-part of the tRNA (9). 1-Methyladenosine itself can be easily prepared on the preparative scale by methylation of adenosine (17). Before exploring the protecting group strategy, we examined the stability of 1 under different conditions of deblocking of acyl groups which are used in oligonucleotide synthesis (18,19). In 25% aqueous ammonia, the half-time of Dimroth rearrangement of 1 to give 4 was 36 h at room temperature. However, 1-methyladenosine was stable for 3 days at room temperature in 2 M ammonia in methanol and for 2 days at room temperature in 1 M Bu4NF in THF. These reaction conditions can be used to remove all common protecting groups employed in oligonucleotide synthesis (18,19).
Direct tritylation of the hydroiodide salt of 1-methyladenosine gave 2 in 68% yield. It should be mentioned that the monomethoxytrityl compound 2 is still rather polar (TLC Rf 0.15 in methylene chloride:methanol, 1:1). Several examples are described in the literature for amidine protection (20–22). For example, 1,3-dimethylcytosine can be protected using acetic anhydride and the acetyl group can be cleaved by sodium methoxide (23). Therefore, we investigated different acyl protecting groups for the amidine function of 2, starting with the benzoyl group, which is most commonly used in oligonucleotide synthesis. A transient protection strategy (24) was used for the preparation of 5a (Scheme 1). Unfortunately, the benzoyl group was found to be too stable towards ammonia in methanol (Table 1). Cleavage does not occur within the time limits used during oligonucleotide synthesis. Therefore, the more base-labile acetyl and chloroacetyl (25) protecting groups were evaluated. Compounds 5a–c were prepared in good yields. The preparation of 5c was optimised using chloroacetic anhydride in pyridine as acylating agent with subsequent cleavage of the O-chloroacetyl group with ammonia. Protection of the amidine function substantially decreased the polarity of N-protected 1-methyladenosines, the values of 5a–c (Rf 0.56–0.59, methylene chloride:methanol, 9:1) are nearly the same as for 5′-O-monomethoxytrityl-N6-benzoyladenosine (Rf 0.61).
Scheme 1. (a) MMTrCl/Py; (b) TMSCl/Py; (c) BzCl or Ac2O or (ClCH2CO)2O/Py; (d) MeNH2 or EtNH2/MeOH; (e) tBuMe2SiCl-imidazole.
Table 1. Results of the reaction of 5a–c with 2 M ammonia and 2 M ethylamine in methanol at 20°C.
| Substrate | 2 M NH3 in MeOHa | 2 M EtNH2 in MeOHa | ||||
|---|---|---|---|---|---|---|
| t | Complete solvolysis | Product formedb | t | Complete solvolysis | Product formedb | |
| 5a | 7 days | ND | 2 | 1 day | 5 days | 6b |
| 5b | 1 day | 5 days | 2 | 5 h | 25 h | 2 + 6bc |
| 5c | 4 h | 20 h | 2 | 50 min | 4 h | 2 + 6bd |
aThe reagent was used in at least 10-fold excess.
bStructure of the products formed are given in Scheme 1.
cRatio 1:4.
dRatio 1:1.
Deblocking of 5b and c was readily accomplished in 2 M NH3 in methanol. Solvolysis of 5a–c with 2 M EtNH2 in MeOH proceeded much faster. However, a new reaction product was formed. Reaction of 5a with EtNH2 in MeOH revealed the formation of 6b. In the case of 5b and c a mixture of 2 and 6b was formed. To obtain further insight into this transformation the reaction was repeated using 2 M MeNH2 in MeOH. In this case solvolysis of 5a gave 6a. These results may be explained by nucleophilic attack of alkylamines at the C-6-position of 1-methyladenosine derivatives, followed by elimination of the acylamide.
It should be mentioned that 2 rearranged to 5′-O-monomethoxytrityl-N6-methyladenosine (4) in 2 M MeNH2 in near quantitative yield at 20°C for 6 days. This reaction is much slower in the case of 2 M EtNH2 in MeOH (20 days). It may be concluded that ammonia in MeOH is the method of choice for the deprotection of N6-acyl derivatives of 1-methyladenosine. The structures of 6a and b were confirmed by mass and NMR spectroscopy. Full assignments of the 13C NMR spectra are presented in Table 2.
Table 2. 13C NMR chemical shifts (p.p.m.) of 2, 4, 6a and 6b in CDCl3 at 27°C.
| Chemical shifta | Compound | |||
|---|---|---|---|---|
| 2 | 4 | 6a | 6b | |
| C-1′ | 88.68 | 90.73 | 89.41 | 89.63 |
| C-2′ | 74.61 | 75.97 | 76.05 | 75.86 |
| C-3′ | 71.56 | 72.33 | 72.71 | 72.60 |
| C-4′ | 84.86 | 85.95 | 85.52 | 85.53 |
| C-5′ | 63.87 | 63.58 | 6 3.78 | 63.75 |
| C-6 | 153.06 | 155.27 | 147.30 | 145.91 |
| C-4 | 144.00 | 147.42 | 143.26 | 143.15 |
| C-5 | 121.37 | 119.97 | 123.06 | 122.71 |
| C-2 | 147.24 | 152.59 | 147.85 | 147.99 |
| C-8 | 139.93 | 137.72 | 135.06 | 135.09 |
| 1-Me | 37.67 | 36.84 | 36.90 | |
| 6-R | 27.31 | 37.20 | 43.59 | |
| 16.95 | ||||
| NOEDIF | ↓1-Me→ | ↓6-Me→ | ↓H-2→ | ↓1-Me→ |
| H-2↑(2%) | H-2(no NOE) | 1-Me↑(2%) | H-2↑(2%) | |
| GHMBC | 1-Me→H-2 | C-5→H-8 | 1-Me→H-2 | 1-Me→H-2 |
| C-5→H-8 | C-8→H-1′ | C-5→H-8 | C-5→H-8 | |
| C-8→H-1′ | C-8→H-1′ | C-8→H-1′ | ||
| C-6→1-Me C-2→1-Me | C-6→6-Me + 1-Me | C-6→6-CH2 + 1-Me | ||
| C-2→1-Me | C-2→1-Me | |||
aTypical shifts of the monomethoxytrityl group: 158.70, 144.08, 135.18, 130.42, 128.38, 127.90, 127.05, 113.23, 86.83 and 55.24.
The t-butyldimethylsilyl (TBDMS) group was used for protection of the 2′-hydroxyl function. Several methods for its introduction were tested: THF–AgNO3 (18), DMF–imidazole (19), Py–imidazole (26) and dichloroethane–imidazole (Table 3). In all cases the ratio of 2′- to 3′-isomers was near 1:1. Separation of isomers was achieved by column chromatography on silica gel using EtOAc/hexane (27) as the solvent system. For the preparation of 7 the last method may be recommended due to the easier work-up procedure.
Table 3. Conditions and yields for silylation of 5c.
| Conditions | 2′:3′-O-bis-TBDMS | 2′-TBDMS (7) | 3′-TBDMS |
|---|---|---|---|
| THF–AgNO3–Py | 13% | 32% | 32% |
| DMF–imidazole | 18% | 31% | 40% |
| Py–imidazole | 8% | 29% | 29% |
| Dichloroethane–imidazole | 10% | 36% | 41% |
Oligonucleotide synthesis
Using standard procedures, 7 was converted to the corresponding phosphoramidite 8, which was used for the synthesis of different oligonucleotides. First, the incorporation of 1-methyladenosine into oligodeoxyribonucleotides was investigated [using the sequence d(GCACGTAATXGTCC), where m1A is incorporated at position X], as well as its effect on hybridisation. The following protecting groups were used for the natural bases: N6-benzoyl for adenine; N2-isobutyryl for guanine; N4-acetyl for cytosine. For the hybridisation studies, we selected a tetradecamer forming a hairpin structure and incorporated 1-methyladenosine either at position A3, A7, A8 or A10 (Table 4). In addition, m1A was incorporated into a single-stranded sequence once within a dA stretch and once in a random sequence (Table 5). Oligonucleotides were synthesised in an Applied Biosystems apparatus using standard coupling conditions with a 10 min coupling time for the modification, with a yield of 90% according to trityl analysis. Deblocking of the base protecting groups and the cyanoethyl group was carried out with 2 M NH3 in MeOH (60 h at room temperature) followed by 18 h TBAF treatment at room temperature. The oligonucleotides were purified on a Mono Q anion exchange column at neutral pH and their molecular weight was confirmed by mass spectrometry. Anion exchange chromatography showed a shift in Rf value of the oligonucleotides containing 1-methyladenosine due to the presence of a positive charge on the modified base. Table 4 gives the Tm value of the hairpin oligonucleotide after incorporation of m1A. Incorporation of 1-methyladenosine into the loop region stabilised its structure by 1–3°C. On the other hand, the presence of 1-methyladenosine in the stem position resulted in complete loss of complexation, which is easily explained by the disruption of Watson–Crick base pairing and by the reduced stacking properties of m1A (11).
Table 4. Thermal stability of DNA hairpin oligonucleotide following incorporation of m1A at position A3, A7, A8 or A10 and mass spectrometric results, taking into account that m1A is a ribonucleoside.
| Sequence | Tm |
|---|---|
| 5′-d(GGACGTAATAGTCC) | 55.5°C |
| 5′-d(GGm1ACGTAATAGTCC) | n.d. |
| 5′-d(GGACGTm1AATAGTCC) | 58.5°C |
| 5′-d(GGACGTAm1ATAGTCC) | 56.5°C |
| 5′-d(GCACGTAATm1AGTCC) | 51.0°C |
n.d., no melting point detected.
Table 5. Thermal stability of DNA duplexes after incorporation of m1A.
| 5′-CCTTTTXTTTTCC-3′ | 5′-GGCGCCGXCGGTG-3′ | |||||||
|---|---|---|---|---|---|---|---|---|
| 3′-GGAAAAYAAAAGG-5′ | 3′-CCGCGGCYGCCAC-5′ | |||||||
| Y | X | |||||||
| C | T | A | G | C | T | A | G | |
| dA | 26 | 43 | 28 | 30 | 58 | 69 | 61 | 66 |
| W | 30 | 34 | 29 | 34 | 62 | 62 | 63 | 62 |
| dI | 40 | 32 | 37 | 32 | 68 | 64 | 70 | 65 |
| m1A | 24 | 30 | 28 | 28 | 58 | 62 | 59 | 60 |
Oligonucleotide concentration 4 µm, 0.1 M NaCl. W, 5-nitroindole 2′-deoxyribofuranose analogue. Tm values for natural base pairing are: 73°C for CG and GC; 70°C for AT and TA. All nucleosides are deoxyribonucleotides. No stable base pairing occurs with C, T, A and G.
The Tm values of these oligonucleotides did not change as a function of oligonucleotide concentration between 2 and 15 µM (data not shown), indicating the presence of a hairpin structure. The absence of an involvement of 1-methyladenosine in base pairing was confirmed by incorporation of 1-methyladenosine in the middle position of the sequence 5′-GGAAAAm1AAAAAGG-3′ and in the sixth position of the sequence 5′-CACCGm1ACGGCGCC-3′. The presence of C, T, A or G in the complementary sequence opposite 1-methyladenosine resulted in every case in a large decrease in Tm and in very similar duplex stabilities of all the analogue duplexes, as shown in Table 5, excluding m1A, which acts as a universal nucleoside (28).
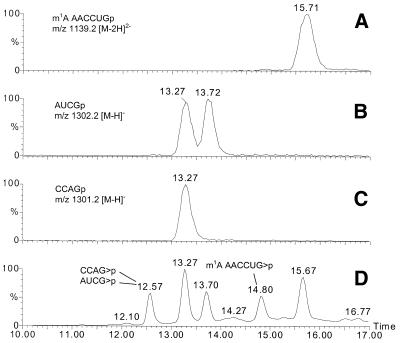
Finally, we incorporated 1-methyladenosine in an RNA sequence. A 17mer was selected corresponding with the T stem–loop sequence of tRNAMet from Schizosaccharomyces pombe (29). With the latter sequence, 1-methyladenosine was incorporated at position 10, where it occurs naturally. After oligonucleotide synthesis, the base-labile groups were removed with 2 M NH3 in MeOH and the 2′-O-t-butyldimethylsilyl groups were removed with 1 M TBAF in THF as before. The oligonucleotide was purified on a Mono Q column with a NaClO4 gradient in Tris buffer pH 6.8. In order to prove correct incorporation of m1A and to show that no Dimroth rearrangement occurred during deprotection, mass spectrometry was used together with enzymatic hydrolysis of the oligonucleotide followed by subsequent HPLC analysis of the fragments. The position of m1A in the 17mer was demonstrated by enzymatic hydrolysis of the oligonucleotide with RNase T1 followed by LC-MS analysis of the hydrolysis products. The theoretical fragments from this digestion are CCAGp, Gp, AUCGp, m1AAACCUGp and G-OH. Figure 1 shows the chromatograms obtained by capillary LC-MS analysis of the digestion mixture. The mononucleoside and mononucleotide elute early and are not shown in the figure. The unmodified tetramers CCAGp and AUCGp elute at 13.27 and 13.72 min, respectively. The peak at 13.27 min in Figure 1B (trace for AUCGp) corresponds to the 13C isotope-containing species CCAGp, which has a molecular mass 1 Da less than AUCGp. The m1A-modified oligonucleotide elutes at 15.71 min. Additional chromatographic peaks are visible in the UV trace which can be assigned to oligonucleotides containing a cyclic phosphate at their 3′-end. The enzymatic hydrolysis with RNase T1 proceeds through a 2′,3′-cyclic intermediate and an incomplete reaction explains the presence of these products.
Figure 1.
Capillary LC-MS analysis of the oligonucleotides obtained by enzymatic hydrolysis of the 17mer with RNase T1. (A–C) Reconstructed ion chromatograms for the theoretical m/z values and (D) UV trace at 260 nm with annotation of the other components. >p indicates a cyclic phosphate at the 3′-end.
The position of the methyl group on the AAACCUGp oligonucleotide was determined by an LC-MS/MS experiment in which fragments were generated by in-source fragmentation. The expected theoretical m/z values were determined and are listed in Table 6. In particular, the ions assigned to the fragments a2-B, a3-B and a4-B, together with the corresponding d1, d2 and d3 ions, confirm that the methylated adenine is present as the first residue at the 5′-end and that the subsequence m1AAACCUGp is present in the 17mer.
Table 6. Calculated and experimental m/z values for fragment ions of m1AAACCUGp.
| Fragment ion assignment | m/z | |
|---|---|---|
| Calculated | Found | |
| C– anion | 110.04 | 110.05 |
| U– anion | 111.02 | 111.04 |
| A– anion | 134.05 | 134.06 |
| m1A– anion | 148.06 | 148.07 |
| G– anion | 150.04 | 150.04 |
| a2-B– | 456.09 | 456.13 |
| a3-B– | 785.14 | 785.16 |
| a4-B– | 1114.19 | 1114.20 |
| d1– | 342.06 | 342.07 |
| d2– | 671.11 | 671.12 |
| d3– | 1000.16 | 1000.21 |
| d4– | 1305.20 | 1305.20 |
| w1– | 442.01 | 442.05 |
| w2– | 748.04 | 748.08 |
| y1– | 362.05 | 362.07 |
| y2– | 668.07 | 668.09 |
| y3– | 973.11 | 973.13 |
| y4– | 1278.16 | 1278.23 |
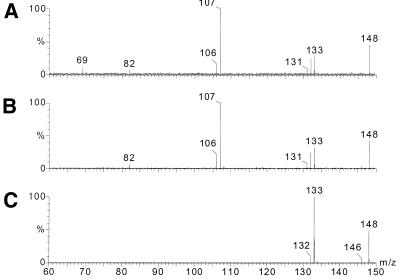
The identity of the methylated base in the oligonucleotide was checked by an LC-MS/MS/MS experiment. The base was released from the oligonucleotide by in-source fragmentation and the base anion (m/z 148) was subsequently fragmented. This fragment ion spectrum was then compared with spectra obtained in a similar way with the same collision energy (20 eV) from 1-methyladenosine and 6-methyladenosine. In Figure 2 the fragment ion spectrum for the methylated adenine from the oligonucleotide (Fig. 2A) is plotted together with the fragment ion spectra of the base anions from m1A (Fig. 2B) and m6A (Fig. 2C). The results clearly show that the spectrum obtained from the oligonucleotide corresponds to 1-methyladenine and, hence, no Dimroth rearrangement has taken place during synthesis.
Figure 2.
–ESI fragment ion spectra of the m/z 148 fragment recorded with collision energy set at 20 eV for (A) the base released from 5′-CCAGGAUCGm1AAACCUGG-3′ with 85 V cone voltage, (B) the m1A base anion released from 1-methyladenosine with 50 V cone voltage and (C) the m6A base anion released from 6-methyladenosine with 50 V cone voltage.
CONCLUSION
1-Methyladenosine can be protected at the base moiety with a chloroacetyl group and converted into a phosphoramidite building block for oligonucleotide synthesis. 1-Methyladenosine was successfully incorporated into DNA and RNA. Deblocking of base-labile protecting groups should be done under anhydrous conditions (2 M NH3 in MeOH). Mass spectrometry was used to prove correct incorporation of m1A into RNA. The modification destabilises a duplex but slightly improves the stability of a hairpin when incorporated into the loop.
Acknowledgments
ACKNOWLEDGEMENTS
Financial support from the RFBR, INTAS, NATO and KUL Research Council (GOA) is gratefully acknowledged. S.M. thanks the Katholieke Universiteit Leuven for a postdoctoral fellowship.
REFERENCES
- 1.Rozenski J., Crain,P.F. and McCloskey,J. (1999) The RNA modification database: 1999 update. Nucleic Acids Res., 27, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agris P.F. (1996) The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol., 53, 79–129. [DOI] [PubMed] [Google Scholar]
- 3.Gesteland R.F. and Atkins,J.F. (eds) (1993) The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 4.Helm M., Brulé,H., Degoul,F., Cepanec,C., Leroux,J.-P., Giegé,R. and Florentz,C. (1998) The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res., 26, 1636–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson J., Phan,L. and Hinnebusch,A.G. (2000) The Gcd10p/Gcd14p complex is the essential two-subunit tRNA (1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamazaki N., Hori,H., Ozawa,K., Nakanishi,S., Ueda,T., Kumagai,I., Watanabe,K. and Nishikawa,K. (1992) Purification and characterization of tRNA(adenosine-1-)-methyltransferase from Thermus thermophilus HB27. Nucleic Acids Symp. Ser., 27, 141–142. [PubMed] [Google Scholar]
- 7.Sierzputowska-Gracz H., Gopal,H.D. and Agris,P.F. (1986) Comparative structural analysis of 1-methyladenosine, 7-methylguanosine, ethenoadenosine and their protonated salts IV: 1H, 13C and 15N NMR studies at natural isotope abundance. Nucleic Acids Res., 14, 7783–7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helm M., Giegé,R. and Florentz,C. (1999) A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNA. Biochemistry, 38, 13338–13346. [DOI] [PubMed] [Google Scholar]
- 10.Macon J.B. and Wolfenden,R. (1968) 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry, 7, 3453–3458. [DOI] [PubMed] [Google Scholar]
- 11.Takeuchi Y., Tazawa,I. and Inoue,Y. (1982) Intramolecular stacking association of three dinucleoside monophosphates containing naturally-occurring 1-methyladenosine residue(s): m1ApA, Apm1A and m1Apm1A. Bull. Chem. Soc. Jpn, 55, 3598–3602. [Google Scholar]
- 12.Agris P.F., Sierzputowska-Gracz,H. and Smith,C. (1986) Transfer RNA contains sites of localized positive charge: carbon NMR studies of [13C]methyl-enriched Escherichia coli and yeast tRNAPhe. Biochemistry, 25, 5126–5131. [DOI] [PubMed] [Google Scholar]
- 13.Apffel A. and Hancock,W.S. (1997) Analysis of oligonucleotides by HPLC-electrospray ionization mass spectrometry. Anal. Chem., 69, 1320–1325. [DOI] [PubMed] [Google Scholar]
- 14.Boudou V., Langridge,J., Van Aerschot,A., Hendrix,C., Millar,A., Weiss,P. and Herdewijn,P. (2000) Synthesis of the anticodon hairpin tRNAfMet containing N-{[9-(b-D-ribofuranosyl)-9H-purin-6-yl]carbamoyl}-L-threonine (=N6-{{[(1S,2R)-1-carboxy-2-hydroxypropyl]amino}-carbonyl}adenosine, t6A). Helv. Chim. Acta, 83, 152–161. [Google Scholar]
- 15.Luyten I., Esnouf,R.M., Mikhailov,S.N., Efimtseva,E.V., Michiels,P., Heus,H.A., Hilbers,C.W. and Herdewijn,P. (2000) Solution structure of a RNA decamer duplex, containing 9-[2-O-(b-D-ribofuranosyl)-b-D-ribofuranosyl]adenine, a special residue in lower eukaryotic initiator tRNAs. Helv. Chim. Acta, 83, 1278–1289. [Google Scholar]
- 16.Rodionov A.A., Efimtseva,E.V., Mikhailov,S.N., Rozenski,J., Luyten,I. and Herdewijn,P. (2000). Synthesis and properties of O-β-D-ribofuranosyl-(1″-2′)-adenosine-5″-O-phosphate and its derivatives. Nucl. Nucl. Nucleic Acids, 19, 1847–1859. [DOI] [PubMed] [Google Scholar]
- 17.Jones J.W. and Robins,R.K. (1963) Purine nucleosides. III. Methylation studies of certain naturally occurring purine nucleosides. J. Am. Chem. Soc., 85, 193–201. [Google Scholar]
- 18.Sonveaux E. (1993) Protecting groups in oligonucleotide synthesis. In Agrawal,S. (ed.), Methods in Molecular Biology: Protocols for Oligonucleotide Conjugates and Analogs, Vol. 26, pp. 1–71. [DOI] [PubMed]
- 19.Damha M.J. and Ogilvie,K.K. (1993) Oligoribonucleotide synthesis. In Agrawal,S. (ed.), Methods in Molecular Biology: Protocols for Oligonucleotides and Analogs, Vol. 20, pp. 1–114. [DOI] [PubMed]
- 20.McOmie J.F.W. (1973) Protective Groups in Organic Chemistry. Plenum Press, New York, NY, pp. 403–413.
- 21.Greene T.W. and Wuts,P.G.M. (1991) Protective Groups in Organic Synthesis. John Wiley & Son, New York, NY.
- 22.Kocienski P.J. (1994) Protective Groups. George Thieme Verlag, Stuttgart, Germany.
- 23.Kenner G.W., Reese,C.B. and Todd,A. (1955) The acylation of 3-methylcytosine. J. Chem. Soc., 855–859. [Google Scholar]
- 24.Ti G.S., Gaffney,B.L. and Jones,R.A. (1982) Transient protection: efficient one-flask syntheses of protected deoxynucleosides. J. Am. Chem. Soc., 104, 1316–1319. [Google Scholar]
- 25.Reese C.B., van Boom,J.H., de Leeuw,H.P.M., Nagel,J. and de Rooy,J.F.M. (1975) Synthesis of oligoribonucleotides preparation of ribonucleoside 2′-acetal 3′-esters by selective deacylation. J. Chem. Soc. [Perkin 1], 934–942. [DOI] [PubMed] [Google Scholar]
- 26.Flockerzi D., Silber,G., Charubala,R., Schlosser,W., Varma,R.S., Creegan,F. and Pfleiderer,W. (1981) Synthese und Eigenschaften von 2′-O- und 3′-O-(Tert-Butyl Dimethylsilyl)-5′-O-(4-Methoxytrityl)- sowie 2′,3′-bis-O-(Tert-Butyldimethylsilyl)ribonucleosiden—ausgangssubstanzen für Oligoribonucleotid-Synthesen. Liebigs Ann. Chem., 1568–1585. [Google Scholar]
- 27.Beigelman L., Matulic-Adamiac,J., Karpeisky,A., Haerbel,P. and Sweedler,D. (2000) Base-modified phosphoramidite analogs of pyrimidine ribonucleosides for RNA structure-activity studies. Methods Enzymol., 317, 39–65. [DOI] [PubMed] [Google Scholar]
- 28.Van Aerschot A., Rozenski,J., Loakes,D., Pillet,N., Schepers,G. and Herdewijn,P. (1995) An acyclic 5-nitroindazole nucleoside analogue as ambiguous nucleoside. Nucleic Acids Res., 23, 4363–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keith G., Heitzler,J., el Adlouni,C., Glasser,A.-L., Fix,C., Desgrès,J. and Dirheimer,G. (1993) The primary structure of cytoplasmic initiator tRNAMet from Schizosaccarhomyces pombe. Nucleic Acids Res., 21, 2949. [DOI] [PMC free article] [PubMed] [Google Scholar]