Background.
We studied the variation in molecular T cell–mediated rejection (TCMR) activity in kidney transplant indication biopsies and its relationship with histologic lesions (particularly tubulitis and atrophy-fibrosis) and time posttransplant.
Methods.
We examined 175 kidney transplant biopsies with molecular TCMR as defined by archetypal analysis in the INTERCOMEX study (ClinicalTrials.gov #NCT01299168). TCMR activity was defined by a molecular classifier.
Results.
Archetypal analysis identified 2 TCMR classes, TCMR1 and TCMR2: TCMR1 had higher TCMR activity and more antibody-mediated rejection (“mixed”) activity and arteritis but little hyalinosis, whereas TCMR2 had less TCMR activity but more atrophy-fibrosis. TCMR1 and TCMR2 had similar levels of molecular injury and tubulitis. Both TCMR1 and TCMR2 biopsies were uncommon after 2 y posttransplant and were rare after 10 y, particularly TCMR1. Within late TCMR biopsies, TCMR classifier activity and activity molecules such as IFNG fell progressively with time, but tubulitis and molecular injury were sustained. Atrophy-fibrosis was increased in TCMR biopsies, even in the first year posttransplant, and rose with time posttransplant. TCMR1 and TCMR2 both reduced graft survival, but in random forests, the strongest determinant of survival after biopsies with TCMR was molecular injury, not TCMR activity.
Conclusions.
TCMR varies in intensity but is always strongly related to molecular injury and atrophy-fibrosis, which ultimately explains its effect on survival. We hypothesize, based on the reciprocal relationship with hyalinosis, that the TCMR1-TCMR2 gradient reflects calcineurin inhibitor drug underexposure, whereas the time-dependent decline in TCMR activity and frequency after the first year reflects T-cell exhaustion.
INTRODUCTION
T cell–mediated rejection (TCMR) is the fundamental reaction of the mammalian host to allogeneic tissue and was the principal barrier to successful organ transplantation before effective immunosuppressive drugs (ISDs) were available. Biopsy-proven TCMR was used as the endpoint for the development of ISDs1 and substantially decreased in frequency with the introduction of calcineurin inhibitor (CNI)/mycophenolate-based protocols,2 leaving the main rejection phenotype as antibody-mediated rejection (AMR).3 Nevertheless, TCMR continues to be diagnosed in approximately 10% of indication biopsies,4 and histologic TCMR-related changes negatively impact survival.5,6 TCMR often reflects underimmunosuppression: nonadherence,7 ISD “minimization,”8 or dose reduction during infections such as BK nephropathy (BKN). BKN often develops a TCMR-like process as ISD minimization continues, directed at alloantigens, BK antigens, or both.9 Hyalinosis of the glomerular afferent arteriole, a marker for CNI exposure, is less than expected in TCMR biopsies, consistent with underimmunosuppression as a risk for TCMR.10,11 Although TCMR becomes uncommon after 5 to 10 y, unlike AMR,12 late TCMR (usually defined as after 1 y posttransplant) is associated with worse outcomes.13,14
Histologic diagnosis of TCMR largely depends on tubulitis (t-lesions) and interstitial inflammation (i-lesions). Pollak15 described tubulitis in the native kidney in 1975 as the invasion of tubules by mononuclear cells, usually accompanied by interstitial inflammation. Sibley et al16 in 1983 and Verani et al17 in 1984 described tubulitis in the transplanted kidney. In 1985, Solez et al18 recognized that the mononuclear cells in tubulitis lie between or beneath tubular epithelial cells inside the tubular basement membrane. Tubulitis was a major feature of the Banff classification in 199319 and remains the key histologic lesion in TCMR. Intimal arteritis (v-lesion) is less common and can be caused by TCMR, AMR, or injury.20
The present analysis aimed to study the variation in TCMR incidence, molecular activity, and histologic features with time posttransplant and the relative impact of these features on graft survival. We defined TCMR molecularly to permit us to assess its relationship with the diagnostic histologic lesions, particularly tubulitis. We characterized molecular TCMR using genome-wide microarray measurements interpreted by the Molecular Microscope Diagnostic System (MMDx) algorithms.21,22 Archetypal analysis (AA) recognized 2 TCMR classes21,22: TCMR1 and TCMR2. TCMR1 was formerly called “mixed” but was renamed TCMR1 because some TCMR1 biopsies lack AMR activity. We defined TCMR activity by the molecular TCMR classifier23 and its associated molecules such as IFNG and LAG3.24
MATERIALS AND METHODS
Study Population
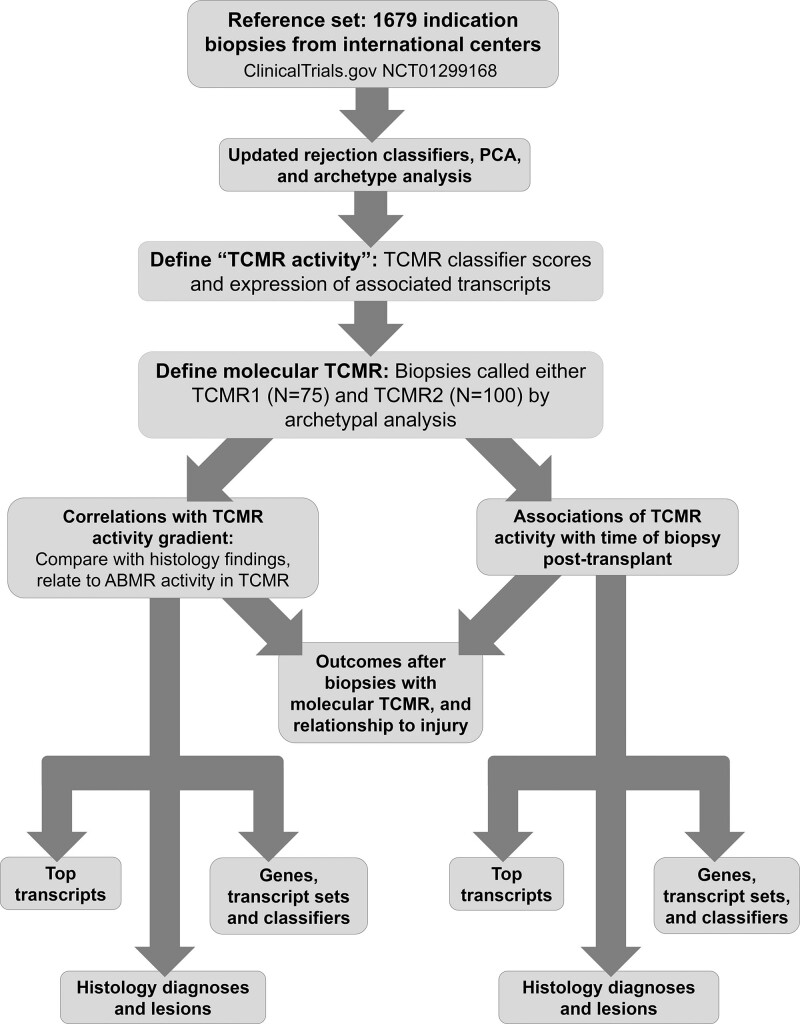
We studied 1679 prospectively collected indication biopsies obtained with consent from 19 established centers under local institutional review board–approved protocols (ClinicalTrials.gov #NCT01299168; Table S1, SDC, http://links.lww.com/TP/C603). The population was previously described22,25-27 and is summarized in Table S2 (SDC, http://links.lww.com/TP/C603). Histologic diagnoses were assigned by the local pathologist following standard-of-care (SOC) Banff guidelines per study protocol. Donor-specific antibody (DSA) assessment was assigned per local SOC. The research plan for these analyses is shown in Figure 1.
FIGURE 1.
Research plan. AMR, antibody-mediated rejection; PCA, principal component analysis; TCMR, T cell–mediated rejection.
Rederiving Molecular Features of Rejection
All algorithms previously derived in the 1208 biopsies21 were updated as recently published,22 using AA to assign 6 rejection archetype scores to each biopsy in the 1679 population, with 228 death-censored graft failures within 3 y postbiopsy. We used the scores assigned by an ensemble of 7 rejection classifiers to visualize the rejection states in principal component analysis (PCA) and assign AA groups.21,22,28 All classifier algorithms were previously developed, trained, and tested on class comparisons comparing an abnormal condition with a more normal condition. The rejection classifiers were those predicting histologic diagnoses of AMR (AMRProb) or TCMR (TCMRProb) or histologic lesions ptc (ptc>0Prob), g (g>0Prob), cg (cg>0Prob), i (i>1Prob), and t (t>1Prob).
This report focuses on the 175 biopsies in the 2 TCMR-related archetype groups, TCMR1 and TCMR2, compared with the no rejection (NR) group, and the relationships between molecular TCMR disease activity, time posttransplant, and histologic lesions. TCMR disease activity was defined as the TCMR molecular classifier score and associated transcript expression.
Principal Component Analysis
PCA was used to visualize the rejection states using classifier algorithms previously described.21,22 PCA was therefore based on a 1679 (samples) × 7 (variables) data matrix, using the “PCA” function in the R “FactoMineR” package.29
Transcript Sets
Details regarding transcript sets used in these analyses are shown in Table S3 (SDC, http://links.lww.com/TP/C603). Transcript sets were used as input in some analyses and to interpret results.
Statistical Analyses
All t tests for differential gene expression were performed using the "wilcox.test" function in base R.30
Survival Analyses
Survival analyses compared select biopsy subpopulations, that is, different archetypally assigned or histology classes. These analyses were done using 1 randomly selected biopsy per patient (N = 1153) and the “survfit” function from the “survival” package in R.31
Time Course of Archetypal Biopsy Assignment
Splines were used to show nonlinear relationships between variables. Three “knots” were selected for splines in this article, and smooth curves fit based on within-knot data with constraints so that curves between segments were joined. Overfitting was avoided by restricted cubic splines using only linear trend lines for the segments beyond the left-most and right-most knots, thereby reducing the influence of the extreme ends of these distributions where fewer data points are available. Splines were generated using the R package “rms.”32
Diagnoses
The dominant molecular diagnosis in each biopsy was assigned by its highest rejection archetype score: NR, TCMR1, TCMR2, early AMR (EAMR), fully developed AMR (FAMR), and late-stage AMR (LAMR). In addition, all MMDx report results are signed out by an expert to recognize additional details beyond the dominant diagnosis. All molecular diagnoses were assigned with no knowledge of local histologic findings, DSA, and C4d results. SOC histologic findings were also recorded by the local centers.
RESULTS
Patient Population
We defined TCMR molecularly to establish its relationship with various histologic features, particularly tubulitis, atrophy-fibrosis, and hyalinosis. The present analysis focuses on biopsies assigned to the TCMR1 and TCMR2 archetype groups, that is, whose dominant molecular phenotype was TCMR objectively assigned by AA. This included some biopsies that also had some AMR-related disease activity in TCMR2 (eg, mixed rejection). Note that TCMR1 and TCMR2 are archetype-assigned clusters that designate the dominant phenotype of the biopsy, but they both can include elements of a second rejection phenotype—AMR. We recognize this complexity in the MMDx diagnoses, where we distinguish “TCMR” from “mixed,” that is, biopsies that have molecular TCMR and molecular AMR classifier scores.
Table 1 presents the molecular report signouts and histology diagnoses in all 175 archetypal TCMR1/2 biopsies. MMDx report signouts called 57 mixed (TCMR1 = 44 and TCMR2 = 13). Histology called 26 mixed (TCMR1 = 18 and TCMR2 = 8). DSA was increased in the TCMR with mixed features but was often absent, like AMR generally.22 Details of the AMR groups were reported previously.22 Archetype assignments recognize the dominant rejection state of the biopsy automatically assigned by highest score. Molecular report signouts often recognize additional details in archetype assignments because they consider both archetype scores and binary classifiers (Table S4, SDC, http://links.lww.com/TP/C603).
TABLE 1.
Histologic and MMDx diagnoses and DSA in the TCMR1 vs TCMR2 biopsy groups in the 175 TCMR cohort
| Rejection archetype group | |||||
|---|---|---|---|---|---|
| All TCMR(N = 175) | TCMR1(N = 75) | TCMR2(N = 100) | P TCMR1 vs TCMR2 | ||
| MMDx signout diagnoses | |||||
| Rejection-related | AMR | 5 (3%) | 0 | 5 (5%) | 0.07 |
| Possible AMR | 0 | 0 | 0 | 1.00 | |
| Mixed | 57 (33%) | 44 (59%) | 13 (13%) | <0.0001 | |
| TCMR | 103 (59%) | 31 (41%) | 72 (72%) | <0.0001 | |
| Possible TCMR | 10 (6%) | 0 | 10 (10%) | 0.005 | |
| No rejection | 0 | 0 | 0 | 1.00 | |
| Histology diagnoses | |||||
| Rejection-related | AMR | 8 (5%) | 4 (5%) | 4 (4%) | 0.73 |
| Transplant glomerulopathy | 3 (2%) | 0 | 3 (3%) | 0.26 | |
| AMR suspected | 2 (1%) | 1 (1%) | 1 (1%) | 1.00 | |
| Mixed | 26 (15%) | 18 (24%) | 8 (8%) | 0.003 | |
| TCMR | 73 (42%) | 39 (52%) | 34 (34%) | 0.02 | |
| TCMR/BK | 3 (2%) | 2 (3%) | 1 (1%) | 0.61 | |
| Borderline rejection | 12 (7%) | 2 (3%) | 10 (10%) | 0.07 | |
| No rejection | BK nephropathy virus | 24 (14%) | 3 (4%) | 21 (21%) | 0.001 |
| No major abnormalities | 4 (2%) | 1 (1%) | 3 (3%) | 0.64 | |
| Othersa | 20 (11%) | 5 (7%) | 15 (15%) | 0.09 | |
| DSA status | |||||
| N in all TCMR (% of N = 142 tested) | N in TCMR1(% of N = 59 tested) | N in TCMR2 (% of N = 83 tested) | P TCMR1 vs TCMR2 | ||
| DSA positive | 51 (36%) | 24 (41%) | 27 (33%) | 0.32 | |
| DSA negative | 91 (64%) | 35 (59%) | 56 (67%) | ||
| DSA missing/unknown | 33 | 16 | 17 | ||
Others includes diabetic nephropathy, glomerulonephritis, fibrosis and atrophy (IFTA), calcineurin inhibitor toxicity, C4d deposition without morphologic evidence for active rejection, donor origin vascular disease, pyelonephritis, systemic infection/diarrhea, and bacterial infection.
Chi-square was performed on the comparison of TCMR1 vs TCMR2 for each histologic diagnosis.
Bold values indicate significant difference between TCMR1 and TCMR2.
AMR, antibody-mediated rejection; DSA, donor-specific antibody; MMDx, Molecular Microscope Diagnostic System; TCMR, T cell–mediated rejection.
TCMR Archetypes in the 1679 Biopsy Population
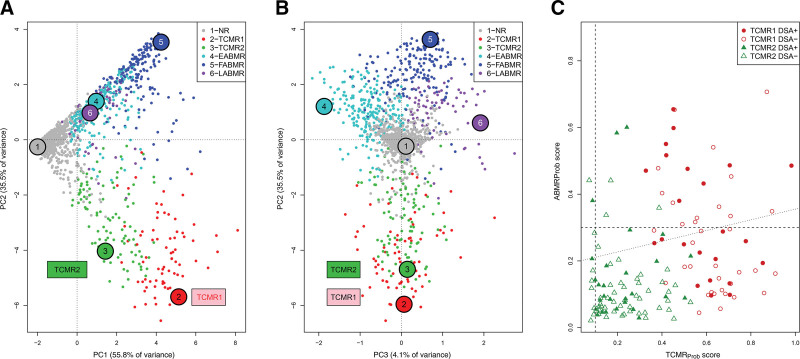
Figure 2A and B shows the biopsy distribution colored by the 1679 archetype group assignment.22 Principal component 1 (PC1) separated NR from all rejections, whereas principal component 2 (PC2) separated TCMR from AMR (Figure 2A). TCMR1 biopsies had slightly more negative PC2 scores than TCMR2 biopsies, reflecting rejection intensity. Principal component 3 (PC3) defined the stages of AMR (Figure 2B).
FIGURE 2.
Visualizing TCMR1 and TCMR2 archetypal groups. The 1679 biopsies are shown distributed by their rejection classifiers scores in PCA and colored by their archetype assignment, with y-axis PC2 and x-axis (A) PC1, and (B) PC3. A, TCMR1 and TCMR2 show a gradient across PC1, and TCMR1 is lower than TCMR2 in PC2. B, PC3 separates AMR stages but does not separate TCMR1 and TCMR2. C, TCMR1 and TCMR2 biopsies distributed by their AMR activity (y-axis, AMRProb classifier scores) vs their TCMR activity (x-axis, TCMRProb classifier scores). AMR activity was correlated with TCMR activity (Spearman correlation coefficient = 0.35, P = 1.8E–6). The biopsies from DSA-positive patients are indicated. AMR, antibody-mediated rejection; AMRProb, AMR-probability classifier; EAMR, early-stage molecular AMR; FAMR, fully developed molecular AMR; LAMR, late-stage molecular AMR; NR, no rejection; PC1, principal component 1; PC2, principal component 2; PC3, principal component 3; PCA, principal component analysis; TCMR, T cell–mediated rejection; TCMRProb, TCMR-probability classifier.
Within archetypal TCMR1/2 biopsies, the molecular AMRProb classifier scores (y-axis) rose as the TCMRProb classifier scores rose (x-axis, Spearman correlation = 0.35, P = 1.8E–6, Figure 2C). Thus, within biopsies with molecular TCMR, the intensity of TCMR activity (the classifier score) correlates with the probability of accompanying AMR activity.
The Gradient in TCMR-related Disease Activity Within the 175 TCMR1/2 Biopsies
The scores for histologic and molecular features are outlined in Table 2 for biopsies with TCMR1, TCMR2, and biopsies with NR as a comparator. Compared with TCMR2, TCMR1 had more molecular TCMR activity (ie, increased TCMR-related classifier scores), more frequent AMR histology lesions (g- and ptc-lesions), and increased molecular AMR classifier scores. TCMR1 had very little hyalinosis, compared with TCMR2 or NR biopsies. TCMR2 had more atrophy-fibrosis, more fibrous intimal thickening (cv), and more arteriolar hyalinosis (ah). However, even in TCMR2 biopsies, hyalinosis was less than in biopsies with NR. Despite their differences, TCMR1 and TCMR2 had similar levels of tubulitis, interstitial infiltrate, and molecular injury (eg, Injury repair response-associated transcripts scores), all of which were elevated compared with NR biopsies.
TABLE 2.
Differences between TCMR1 and TCMR2 in 25 scores for histology lesions and molecular scores
| No rejection (N = 1040) | TCMR1 (N = 75) | TCMR2(N = 100) | P value for TCMR1 vs TCMR2 | ||
|---|---|---|---|---|---|
| 9 histology lesion scores | |||||
| TCMR-related | t (tubulitis) | 0.30 | 2.19 | 2.00 | 0.09 |
| i (interstitial infiltrate) | 0.32 | 2.16 | 1.92 | 0.16 | |
| All rejection-related | v (vasculitis) | 0.01 | 0.37 | 0.07 | 0.0002 |
| AMR-related | g (glomerulitis) | 0.27 | 0.50 | 0.18 | 0.01 |
| ptc (capillaritis) | 0.25 | 0.32 | 0.60 | 0.01 | |
| Atrophy-fibrosis-related | ci (scarring) | 1.12 | 1.19 | 1.77 | 0.0003 |
| ct (atrophy) | 1.03 | 1.09 | 1.69 | 0.0001 | |
| cv (intimal thickening) | 0.90 | 0.55 | 1.04 | 0.0011 | |
| ah (arteriolar hyalinosis) | 1.00 | 0.21 | 0.73 | <0.0001 | |
| 16 transcript set and molecular classifier scores | |||||
| TCMR-related classifiers | TCMR (TCMRProb) | 0.03 | 0.60 | 0.24 | <0.0001 |
| i-score (i>1Prob) classifier | 0.06 | 0.84 | 0.65 | <0.0001 | |
| t-score (t>1Prob) classifier | 0.06 | 0.83 | 0.62 | <0.0001 | |
| All-rejection-related | RejProb classifier | 0.12 | 0.85 | 0.54 | <0.0001 |
| AMR-related | DSA-selective transcripts (DSAST) | 0.07 | 0.33 | 0.17 | <0.0001 |
| NK cell burden (NKB) | 0.36 | 1.01 | 0.80 | 0.001 | |
| AMR-related classifier (AMRProb) | 0.08 | 0.30 | 0.13 | <0.0001 | |
| Macrophage-related | Alternatively activated macrophage (AMAT1) | 0.40 | 1.53 | 1.23 | <0.0001 |
| Constitutive macrophage (QCMAT) | 0.31 | 1.46 | 1.11 | <0.0001 | |
| Recent injury-related | Fibrillar collagen (FICOL) | 1.11 | 1.61 | 1.58 | 0.72 |
| Injury-repair induced, day 3 (IRITD3) | 0.04 | 0.22 | 0.17 | 0.01 | |
| Injury-repair induced, day 5 (IRITD5) | 0.33 | 0.64 | 0.59 | 0.09 | |
| Injury/repair associated (IRRAT30) | 0.26 | 1.13 | 0.99 | 0.04 | |
| GFRprob | 0.32 | 0.60 | 0.52 | 0.05 | |
| Atrophy-fibrosis related | Fibrosis (ci>1Prob) classifier | 0.31 | 0.40 | 0.53 | 0.001 |
| Atrophy (ct>1 Prob) classifier | 0.26 | 0.30 | 0.46 | <0.0001 | |
Wilcoxon t test was performed between TCMR1 and TCMR2 biopsies for each molecular classifier.
Gray shading indicates significant difference P < 0.01 between TCMR1 and TCMR2.
Bolding indicates the higher value (between TCMR1 or TCMR2) when P < 0.05.
AMR, antibody-mediated rejection; AMRProb, AMR-probability classifier; ci > 1Prob, ci>1-probability classifier; ct > 1Prob, ct > 1-probability classifier; DSA, donor-specific antibody; GFR, glomerular filtration rate; i > 1Prob, i > 1-probability classifier; NK, natural killer; t > 1Prob, t > 1-probability classifier; TCMR, T cell–mediated rejection; TCMRProb, TCMR-probability classifier.
Effects of Time Posttransplant on the Frequency of Molecular TCMR Within 1679 Biopsies
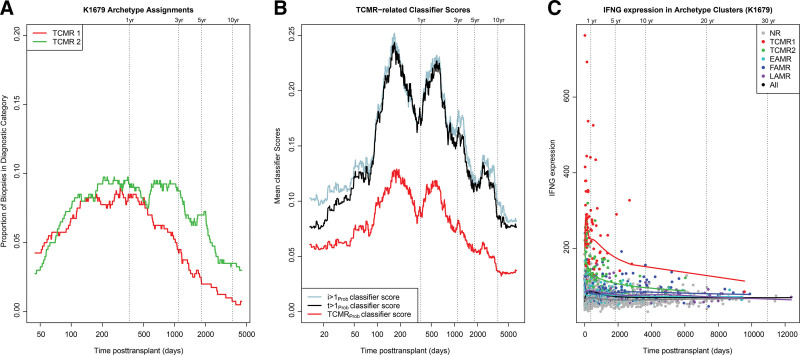
Consistent with previous findings,12 the frequency of biopsies assigned to TCMR1 and TCMR2 archetypes rose after 60 d, plateaued for about 3 y, then both declined to very low levels after 10 y (Figure 3A and Table 3). The fact that TCMR1 and TCMR2 both rose, peaked, and declined showed that the TCMR1-TCMR2 gradient in disease activity and the other differences between TCMR1 and TCMR2 were not primarily because of time posttransplant. TCMR1 in particular was very rare after 10 y, occurring in only 1 of 228 biopsies (0.4%).
FIGURE 3.
Rolling averages for the relationships between time posttransplant and the TCMR molecular classes and features in 1679 biopsies. Rolling averages over time posttransplant (A) showing the proportion of biopsies assigned TCMR1 and TCMR2 archetypes and (B) showing the TCMR-related classifier scores. (C) IFNG expression in archetype clusters over time posttransplant (shown as a linear scale). EAMR, early-stage molecular AMR; FAMR, fully developed molecular AMR; LAMR, late-stage molecular AMR; NR, no rejection; TCMR, T cell–mediated rejection.
TABLE 3.
Distribution of TCMR episodes (TCMR1 and TCMR2) over time intervals posttransplant
| No. of biopsies per time interval posttransplant (% of total in interval)(N out of 1679) | ||||||
|---|---|---|---|---|---|---|
| <2 m (N = 270) | 2 m–1 y(N = 437) | 1–5 y(N = 489) | 5–10 y(N = 247) | >10 y(N = 228) | Totals per row | |
| TCMR1 | 8 (3%) | 35 (8%) | 26 (5%) | 4 (2%) | 1 (0.4%) | 75 |
| TCMR2 | 6 (2%) | 38 (7%) | 40 (8%) | 11 (4%) | 4 (2%) | 100 |
| All TCMR (TCMR1 + TCMR2) | 8 (5%) | 73 (17%) | 66 (13%) | 15 (6%) | 5 (2%) | 175 |
TCMR, T cell–mediated rejection.
As a measure of TCMR activity over time within all 1679 biopsies, the TCMR-related classifiers are plotted in Figure 3B. The TCMRProb classifier showed 2 peaks before a steady decline. The i-lesion and t-lesion classifiers (i>1Prob and t>1Prob) trained on the histology i- and t-scores, respectively, showed similar patterns.
We studied the expression of IFNG, the classical cytokine induced by activation of effector T cells over time in the 1679 population. We plotted the biopsies on a linear x-axis to visualize the actual relationships between time and IFNG expression (Figure 3C). Biopsies were colored by their archetype group membership, and splines were calculated to represent the moving average of IFNG expression within each group. Among all biopsies, very high IFNG activity was an exclusive feature of the early TCMR1 biopsies. The second highest IFNG expression was in early TCMR2 biopsies. In both TCMR1 and TCMR2, high IFNG expression ceased after the first 2 y. Even FAMR (blue symbols) had much lower expression of IFNG, although higher than in NR biopsies. Therefore, high IFNG expression in early TCMR reflected TCMR intensity, not concomitant AMR.
Effect of Time Posttransplant on Disease Activity in Biopsies With TCMR
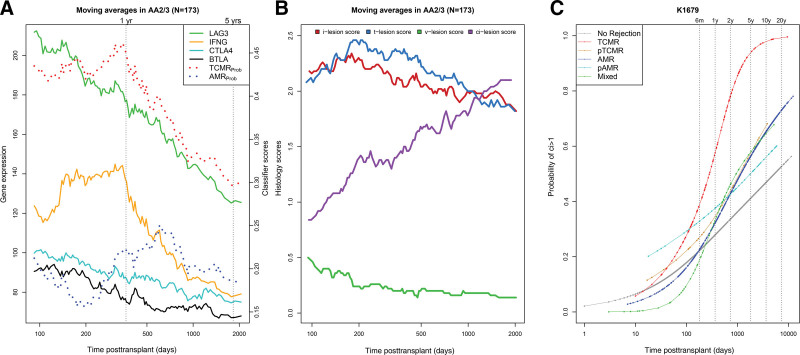
Within TCMR biopsies, we studied expression of IFNG and 3 other T cell–activation features—LAG3, CTLA4, and BTLA4—in TCMR1/2 biopsies over log time, compared with the TCMRProb classifier (Figure 4A). TCMRProb classifier scores (ie, TCMR activity) were high until 1 y posttransplant before progressively declining. The moving averages of IFNG and LAG3 expression also showed a steady decline after 1 y; CTLA4 and BTLA showed similar patterns despite lower overall expression. Despite the decline, the mean TCMRProb was still elevated in late TCMR (0.3), approximately 3 times its diagnostic threshold.
FIGURE 4.
Relationships between time posttransplant and the moving average scores for molecular and histologic features of the biopsies. Two biopsies were missing date of transplant and were excluded from the time courses. (A) Molecular features and TCMR-associated gene expression within 175 TCMR biopsies. (B) Histologic lesions within 175 TCMR biopsies. (C) The probability of ci lesion scores >1 per archetype group over time posttransplant, as calculated in logistic regression. AMR, antibody-mediated rejection; pAMR, possible antibody-mediated rejection; TCMR, T cell–mediated rejection; pTCMR, possible T cell–mediated rejection.
The AMRProb classifier score in TCMR biopsies peaked and plateaued between 1 and 3 y. We assessed the effect of time on moving average scores for histologic lesions (i, t, v, and ci) in TCMR biopsies (Figure 4B). The i- and t-lesion scores remained relatively high around their diagnostic thresholds of 2.0 even in late TCMR, whereas v-lesions became rare. Histologic fibrosis (ci lesion scores) within TCMR steadily increased with time.
Molecular TCMR Is Associated With Increasing Fibrosis
Fibrosis increases with time in kidney transplant biopsies, even those with NR.33 To determine whether TCMR further increased fibrosis, we used regression analysis to compare the probability of fibrosis among different molecular rejection groups (Figure 4C). The probability of ci lesion scores >1 increased with time in all biopsies but was highest in TCMR biopsies.
The mean and median ci- and ct-scores and the ci>1Prob and ct>1Prob classifiers in biopsies before and after 1 y are detailed in Table 4. The histology ci- and ct-scores, molecular ci>1Prob, and ct>1Prob classifier scores were all significantly higher in TCMR versus NR biopsies, even in the first year posttransplant. Transcript sets reflecting recent or ongoing parenchymal injury were also higher in TCMR. Therefore, biopsies with TCMR consistently display parenchymal damage, with increased scores for molecular injury and atrophy-fibrosis.
TABLE 4.
Comparing ci and ct lesion scores and classifiers in archetypal TCMR1 + 2 and NR before and after 1 y posttransplant
| Biopsies <1 y posttransplant | Biopsies ≥1 y posttransplant | ||||||
|---|---|---|---|---|---|---|---|
| TCMR175(N = 87) | No rejection(N = 513) | Wilcoxon test P value comparing TCMR and NR | TCMR175(N = 88) | No rejection(N = 526) | Wilcoxon test P value comparing TCMR and NR | ||
| Atrophy-fibrosis lesion scores | Mean (median) ci lesion score | 1.08 (1.0) | 0.81 (1.0) | 0.025 | 1.86 (2.0) | 1.39 (1.0) | 0.0001 |
| Mean (median) ct lesion score | 1.03 (1.0) | 0.72 (1.0) | 0.0078 | 1.83 (2.0) | 1.34 (1.0) | 4.2E–5 | |
| Atrophy-fibrosis classifiers | Mean (median) ci classifier | 0.40 (0.36) | 0.22 (0.15) | 6.6E–11 | 0.54 (0.53) | 0.41 (0.34) | 6.3E–6 |
| Mean (median) ct classifier | 0.32 (0.24) | 0.17 (0.10) | 9.5E–9 | 0.46 (0.43) | 0.35 (0.27) | 5.4E–5 | |
NR, no rejection; TCMR, T cell–mediated rejection.
Top Transcripts Correlating With Time in Molecular TCMR Biopsies
We studied the top 20 genes that increased and decreased with time posttransplant in TCMR biopsies. Among the top genes that decreased with time in TCMR1/2 were LAG3, IFNG-inducible chemokines CXCL9, CXCL10, and CXCL11, and IFNG-inducible genes such as GBP1, ANKRD22, and IDO1 (Table 5). IFNG also declined with time posttransplant (Spearman correlation coefficient –0.30, P = 0.00008), although it was not among the top 20 by P value. The top genes that increased in expression with time reflected atrophy-fibrosis,34,35 including immunoglobulin transcripts representing plasma cells and mast cell transcript CPA3 (Table 6). There was also increased expression of SPAG4, a gene expressed in injured renal tubule epithelial cells, inducible by hypoxia-inducible factor-1.36 Acknowledging that the peak-plateau-decline pattern can make correlations misleading, we confirmed these findings in 86 biopsies >1 y posttransplant. The results were similar: decline in TCMR activity features (eg, LAG3) and rise in atrophy-fibrosis features (eg, immunoglobulin transcripts; Tables S5 and S6, SDC, http://links.lww.com/TP/C603).
TABLE 5.
Top 20 genes by Spearman correlation decreased (negatively correlated) with time posttransplant within molecular TCMR biopsies (archetypes TCMR1/2; N = 175)
| Gene symbol | Gene name | Transcript set | Spearman correlation with time posttransplanta |
|---|---|---|---|
| CXCL9 | Chemokine (C-X-C motif) ligand 9 | IFNG-inducible | –0.50 |
| GBP1 | Guanylate binding protein 1, interferon-inducible | IFNG-inducible | –0.45 |
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | IFNG-inducible | –0.44 |
| LAG3 | Lymphocyte-activation gene 3 | T cell–activation gene; TCMR-RAT | –0.44 |
| ANKRD22 | Ankyrin repeat domain 22 | IFNG-inducible | –0.44 |
| APOL4 | Apolipoprotein L, 4 | IFNG-inducible | –0.43 |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | IFNG-inducible | –0.42 |
| NUSAP1 | Nucleolar and spindle associated protein 1 | Macrophage; injury-induced (IRITD5) | –0.42 |
| IDO1 | Indoleamine 2,3-dioxygenase 1 | IFNG-inducible | –0.41 |
| GBP4 | Guanylate binding protein 4 | IFNG-inducible | –0.41 |
| BATF2 | Basic leucine zipper transcription factor, ATF-like 2 | IFNG-inducible | –0.41 |
| RGL1 | Ral guanine nucleotide dissociation stimulator-like 1 | Macrophage gene | –0.40 |
| SMCO4 | Single-pass membrane protein with coiled-coil domains 4 | IFNG-inducible (macrophages) | –0.40 |
| TAP1 | Transporter 1, ATP-binding cassette, subfamily B (MDR) | IFNG-inducible | –0.40 |
| FBXO6 | F-box protein 6 | IFNG-inducible | –0.40 |
| FAM72A | Family with sequence similarity 72, member A | T cell–activation gene; TCMR-RAT | –0.39 |
| MOB1A | MOB kinase activator 1A | Injury-induced (cIRIT) | –0.39 |
| GABBR1 | Gamma-aminobutyric acid B receptor, 1 | All rejection | –0.39 |
| IL31RA | Interleukin 31 receptor A | IFNG-inducible | –0.39 |
| RRM2 | Ribonucleotide reductase M2 | Injury-induced, IRIT5 | –0.39 |
Pseudogenes have been deleted.
PBTs are listed on our home page https://www.ualberta.ca/medicine/institutes-centres-groups/atagc/research/gene-lists.
All P values <0.001.
PBT, pathogenesis-based transcript; TCMR, T cell–mediated rejection.
TABLE 6.
Top 20 genes by Spearman correlation increased (positively correlated) with time posttransplant within molecular TCMR biopsies (archetypes TCMR1/2; N = 175)
| Gene symbol | Gene name | PBT | Spearman correlation with time posttransplanta |
|---|---|---|---|
| IGHG1 | Immunoglobulin heavy constant gamma 1 (G1m marker) | IGT | 0.51 |
| IGHG1 | Immunoglobulin heavy constant gamma 1 (G1m marker) | IGT | 0.50 |
| IGHG1 | Immunoglobulin heavy constant gamma 1 (G1m marker) | IGT | 0.50 |
| IGHG1 | Immunoglobulin heavy constant gamma 1 (G1m marker) | IGT | 0.50 |
| IGHG1 | Immunoglobulin heavy constant gamma 1 (G1m marker) | IGT | 0.50 |
| IGHG1 | Immunoglobulin heavy constant gamma 1 (G1m marker) | IGT | 0.50 |
| IGHG3 | Immunoglobulin heavy constant gamma 3 (G3m marker) | IGT | 0.49 |
| IGKC | Immunoglobulin κ constant | IGT | 0.48 |
| IGKV1–39 | Immunoglobulin κ variable 1–39 (gene) | IGT | 0.48 |
| RGS13 | Regulator of G-protein signaling 13 | B cells, mast cells | 0.48 |
| CPA3 | Carboxypeptidase A3 (mast cell) | MCAT mast cells | 0.47 |
| TPSAB1 | Tryptase alpha | 0.47 | |
| IGH | Immunoglobulin heavy locus | IGT | 0.47 |
| SPAG4b | Sperm-associated antigen 4 | RPTECs; induced by HIF1 | 0.46 |
| IGKV1–5 | Immunoglobulin κ variable 1–5 | IGT | 0.46 |
| TPSAB1 | Tryptase alpha | 0.46 | |
| IGHG1 | Immunoglobulin heavy constant gamma 1 (G1m marker) | IGT | 0.46 |
| IGK | Immunoglobulin κ locus | IGT | 0.46 |
| MS4A2 | Membrane-spanning 4-domains, subfamily A, member 2 | Mast cells | 0.45 |
| FCRL5 | Fc receptor-like 5 | BAT (B cells) | 0.45 |
PBTs are listed on our home page https://www.ualberta.ca/medicine/institutes-centres-groups/atagc/research/gene-lists.
aAll P values <0.001.
SPAG4 is expressed in renal epithelial cell line and highly increased in injured kidneys.
HIF1, hypoxia-inducible factor-1; IGT, immunoglobulin transcript; PBT, pathogenesis-based transcript; RPTEC, renal proximal tubular epithelial cell; TCMR, T cell–mediated rejection.
The Correlation of Time Posttransplant With Histologic and Molecular Scores in TCMR Biopsies
Table 7 lists the correlations of histologic and molecular scores (the same scores as in Table 2) with time posttransplant in TCMR1/2 biopsies after 1 y posttransplant. Within TCMR1/2 biopsies, the defining histologic lesions of TCMR—the t- and i-scores—showed no significant change over time, that is, remained near diagnostic levels even in late TCMR biopsies, whereas v-lesions in late TCMR became uncommon. Atrophy-fibrosis–related changes were increased in later TCMR biopsies.
TABLE 7.
Correlation between histology lesion scores, molecular transcript set scores, and molecular classifier scores with time of biopsy posttransplant in molecular TCMR biopsies >1 y (N = 86)a
| Spearman correlation with time posttransplant | P | ||
|---|---|---|---|
| 9 histology lesion scores | |||
| TCMR related | t (tubulitis) | –0.17 | 0.17 |
| i (interstitial infiltrate) | –0.14 | 0.27 | |
| AMR related | g (glomerulitis) | –0.13 | 0.29 |
| ptc (capillaritis) | –0.15 | 0.25 | |
| All rejection related | v (vasculitis) | –0.21 | 0.10 |
| Atrophy-fibrosis related | ci (scarring) | 0.30 | 0.02 |
| ct (atrophy) | 0.34 | 0.01 | |
| cv (intimal thickening) | 0.32 | 0.01 | |
| ah (hyalinosis) | 0.65 | <0.001 | |
| 16 transcript set scores and classifier scores | |||
| TCMR related classifiers | TCMR (TCMRProb) | –0.27 | 0.01 |
| i-score (i > 1 Prob) classifier | –0.32 | 0.003 | |
| t-score (t > 1 Prob) classifier | –0.37 | 0.0006 | |
| All rejection related | RejectionProb classifier | –0.29 | 0.01 |
| AMR related | DSA-selective transcripts (DSAST) | –0.07 | 0.53 |
| NK cell burden (NKB) | 0.06 | 0.61 | |
| AMR (AMRProb) classifier | –0.17 | 0.11 | |
| Macrophage relatedb | Alternatively activated macrophage (AMAT1) | –0.08 | 0.45 |
| Constitutive macrophage (QCMAT) | –0.15 | 0.17 | |
| Recent injury relatedc | Fibrillar collagen (FICOL) | 0.07 | 0.55 |
| Injury-repair induced, day 3 (IRITD3) | 0.09 | 0.41 | |
| Injury-repair induced, day 5 (IRITD5) | 0.12 | 0.28 | |
| Injury/repair associated (human kidney) (IRRAT30) | 0.14 | 0.19 | |
| Low GFRprob | 0.13 | 0.23 | |
| Atrophy-fibrosis related | Fibrosis (ci > 1 Prob) | 0.34 | 0.002 |
| Atrophy (ct > 1 Prob) | 0.33 | 0.002 | |
PBTs are listed on our home page https://www.ualberta.ca/medicine/institutes-centres-groups/atagc/research/gene-lists.
Shading and bold indicate significant association of the score with time P < 0.05.
All macrophage-related classifier scores were significantly different between TCMR biopsies (N = 175) and NR biopsies (N = 1040) (P < 10–16).
All injury-associated classifier scores were significantly different between TCMR biopsies (N = 175) and NR biopsies (N = 1040) (P < 10–11).
AMR, antibody-mediated rejection; ci > 1Prob, ci > 1-probability classifier; ct > 1Prob, ct > 1-probability classifier; DSA, donor-specific antibody; GFR, glomerular filtration rate; NK, natural killer; PBT, pathogenesis-based transcript; TCMR, T cell–mediated rejection.
Among molecular scores, TCMR activity scores declined, and atrophy-fibrosis scores increased. However, recent molecular injury and macrophage transcript sets (which are abnormal in all TCMR) showed little change with time. Like tubulitis, parenchymal injury scores were consistently elevated even in late TCMR biopsies. AMR-related scores and AMRProb in TCMR biopsies did not decrease significantly. In summary, with increasing time posttransplant, molecular TCMR showed decreased molecular TCMR activity and increased atrophy/fibrosis but consistently displayed molecular injury and tubulitis.
Checkpoint Transcripts
We considered whether the gradient in disease activity between TCMR1-TCMR2 and the decline in TCMR activity over time might be associated with increased expression of immunologic checkpoint transcripts in the biopsies. Table 8 shows the correlations of selected checkpoint transcripts with time posttransplant in TCMR biopsies and compares their expression in TCMR1, TCMR2, and NR. No checkpoint transcripts increased with time: all were associated with TCMR activity and thus decreased in expression at least slightly with time, 3 significantly (LAG, CTLA4, and BTLA). All checkpoints were higher in TCMR1 than TCMR2. Declining TCMR activity over time and differences in TCMR activity between TCMR1 and TCMR2 were not associated with higher expression of checkpoint transcripts in the biopsy.
TABLE 8.
Expression of checkpoint transcripts in TCMR archetypes (N = 175) in relationship with time posttransplant and to class comparison between TCMR1 and TCMR2
| Checkpoints and ligands | Correlation with time posttransplant | Comparing expression in TCMR1 vs TCMR2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Rank by P | Spearman correlation coefficient | P | Rank by P | TCMR1 | TCMR2 | No rejection | P | Adjusted P | |
| TIGITa | 11 643 | –0.13 | NS | 66 | 165 | 94 | 43 | 9.41E–16 | 7.05E–13 |
| BTLA | 3193 | –0.20 | 0.008 | 5446 | 91 | 70 | 19 | 0.004 | 0.04 |
| CTLA4 | 2351 | –0.22 | 0.004 | 275 | 58 | 39 | 22 | 3.11E–11 | 5.59E–09 |
| PDCD1 | 12 630 | –0.12 | NS | 433 | 101 | 82 | 75 | 1.05E–09 | 1.20E–07 |
| CD160b | 11 579 | –0.13 | NS | 3607 | 72 | 53 | 25 | 0.001 | 0.01 |
| LAG3 | 32 | –0.44 | 1.0E–09 | 33 | 232 | 132 | 66 | 7.48E–18 | 1.12E–14 |
| CD244/2B4 | 32 205 | –0.05 | NS | 768 | 54 | 43 | 29 | 6.80E–08 | 4.38E–06 |
| HAVCR2/TIM3 | 35 399 | –0.04 | NS | 336 | 42 | 29 | 20 | 1.37E–10 | 2.02E–08 |
| TNFSF9/CD137 | 7967 | –0.15 | NS | 7 | 72 | 35 | 17 | 8.75E–21 | 6.03E–17 |
Bold and shaded are checkpoints that do not decrease in expression significantly with time but are higher in TCMR1 than TCMR2.
CD160 is included because it is a marker for exhausted T cells.
NS, not significant; TCMR, T cell–mediated rejection.
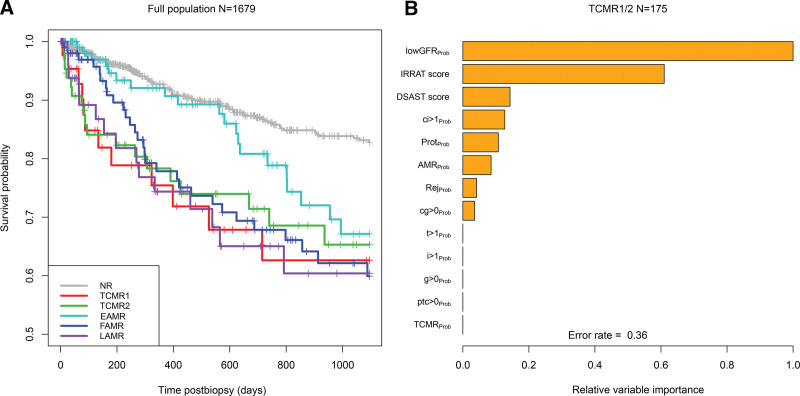
Kidney Graft Survival After TCMR Diagnoses in the Biopsy
We investigated 3 y postbiopsy graft loss in the 1679 cohort, selecting 1 random biopsy per group (Figure 5). In Figure 5A, TCMR1 and TCMR2 both showed increased probability of graft loss compared with NR. (EAMR showed relatively little graft loss in the first 2 y, as previously reported.21,22 We assessed the relative importance of various molecular scores in the biopsy for predicting short-term (3 y) graft survival after the diagnosis of TCMR (Figure 5B). The strongest molecular feature predicting graft survival was recent or ongoing parenchymal injury, that is, the lowGFRProb classifier score and the Injury repair response-associated transcripts transcript set, both measures of recent injury. Thus, in biopsies with molecular TCMR, the TCMR activity-related features were relatively unimportant once parenchymal injury features were included in the multivariable random forest analysis.
FIGURE 5.
Survival analysis during 3 y postbiopsy (days), with 1 random biopsy per patient. (A) Actuarial survival curves by archetype group. (B) Random forests showing the variable importance (including molecular and histologic features) in the prediction of 3 y postbiopsy graft survival. TCMR, T cell–mediated rejection.
Associations With Intimal Arteritis
Because molecular diagnoses are not influenced by the presence of histologic v-lesions, we could study the relationship between associations between v-lesions and molecular diagnoses (Table 9). In 1337 biopsies in which v-lesions could be scored, v-lesions >0 were recorded in 51 (4%), mostly v1. By archetypes, most v-lesions were in biopsies with TCMR1, FAMR, or NR biopsies. This is compatible with the previous conclusion that v-lesions can reflect AMR, TCMR, or injury.20,37
TABLE 9.
Number of biopsies with v-lesions in N = 1679 biopsies that could be scored for v-lesions (histology not shown because it uses v-lesions for classification)
| v-lesion score | ||||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | All v > 0 | ||
| Rejection archetype groups | No rejection | 816 | 8 | 1 | 0 | 9 |
| TCMR1 | 44 | 12 | 2 | 2 | 16 | |
| TCMR2 | 70 | 3 | 1 | 0 | 4 | |
| EAMR | 169 | 5 | 1 | 0 | 6 | |
| FAMR | 130 | 9 | 4 | 1 | 14 | |
| LAMR | 57 | 1 | 1 | 0 | 2 | |
| Total | 1286 | 38 | 10 | 3 | 51 | |
EAMR, early-stage molecular AMR; FAMR, fully developed molecular AMR; LAMR, late-stage molecular AMR; TCMR, T cell–mediated rejection.
Impact of BKN
BKN often has TCMR-like changes, directed at alloantigens, viral antigens, or both.9,38-43 In the 175 TCMR biopsies, 3 TCMR1 and 21 TCMR2 biopsies were diagnosed locally as BKN. The conclusions from the abovementioned analyses did not change when BKN biopsies were excluded. Details of the BKN biopsies have been published previously.9
DISCUSSION
We studied the variation in molecular and histologic features in molecular TCMR to understand the determinants of intensity and the effect of time posttransplant. Defining TCMR molecularly using automatically assigned archetypes permitted us to study the relationships between molecular TCMR activity (as defined by the TCMR classifier scores) and histologic lesions without using the lesions to make the TCMR diagnosis. In 175 molecular TCMR biopsies, there was a strong gradient of disease activity between TCMR1 and TCMR2: TCMR1 had higher TCMR activity, more AMR activity (mixed rejection), and more frequent v-lesions but little hyalinosis, whereas TCMR2 had more atrophy-fibrosis. However, TCMR1 and TCMR2 had similar molecular injury and tubulitis. Time had strong effects on the frequency of TCMR as previously described12 and also affected disease activity in the TCMR biopsies that did occur. TCMR biopsies after 1 y posttransplant showed progressively less molecular TCMR classifier activity and decreased expression of effector T-cell activity molecules such as IFNG and LAG3. TCMR was associated with increased probability of atrophy-fibrosis even within the first year. However, like the TCMR1-TCMR2 gradient, molecular injury and tubulitis were consistently present in TCMR, with tubulitis remaining at diagnostic levels (t2) even in late TCMR with atrophy-fibrosis. TCMR increased the probability of graft loss, and in random forests, the risk of loss after biopsies with TCMR correlated most strongly with recent or ongoing parenchymal injury, not TCMR activity. We conclude that TCMR biopsies display a reciprocal relationship between TCMR molecular activity and the extent of hyalinosis and fibrosis and that time posttransplant beyond 1 y is associated with attenuated molecular activity and increased fibrosis in TCMR. Nevertheless, molecular injury and tubulitis were always present in TCMR, and injury and atrophy-fibrosis, not TCMR activity, determined the risk of graft failure with 3 y postbiopsy.
The striking reciprocal relationship between TCMR activity, hyalinosis, and fibrosis suggests that underexposure to CNIs for a considerable time before the biopsy likely contributes to the TCMR1-TCMR2 gradient. This can only be proven by detailed studies of drug exposure in the months before biopsy and is not feasible in routine clinical practice. Single-drug levels at the time of indication biopsy are not informative because low levels, once detected at a clinic visit, will often be corrected before the biopsy.
It is a testament to the robustness of the original Banff definition of TCMR that, despite their heterogeneity, all molecular TCMRs, even late TCMR, manifest i- and t-lesions. Tubulitis, with its accompanying interstitial mononuclear infiltrate, emerges as a reflection of the parenchymal injury induced by TCMR and a universal feature of molecular TCMR regardless of time posttransplant. Increasing atrophy-fibrosis can interfere with assignment of t-scores. Nevertheless, mean tubulitis scores were at diagnostic levels even late posttransplant, accompanied by molecular features of recent or ongoing injury: injury-induced transcript sets, macrophage transcripts, and the lowGFRProb injury classifier.
These results cannot resolve the debate over whether tubulitis in atrophic tubules should be interpreted as a “chronic-active TCMR” phenotype, separate from the usual TCMR definition based on tubulitis. The concept of chronic-active TCMR based on inflammation in atrophy-fibrosis lesions in biopsies lacking tubulitis was introduced in Banff 201544 but has been controversial.44-48 Definitions of chronic-active TCMR were revised in Banff 201748 to require moderate or severe tubulitis and will be revisited in Banff 2022.46,47,49 Inflammation in atrophy-fibrosis lesions is a general feature of progressive nephron injury, even in primary renal diseases in native kidneys, and strongly related to future transplant failure.50,51 Our finding that mean tubulitis scores remain at or near diagnostic levels in late molecular TCMR is reassuring, but it would be useful to identify and validate histologic features indicating TCMR when the atrophy-fibrosis is too extensive for reliable tubulitis assessment. Calibrating candidate criteria against molecular TCMR activity scores could be useful in this process.
The cognate T cells that generate effector T cells in the secondary lymphoid organs are programmed to undergo exhaustion when they face persistent antigen,12,52-60 and we believe that this probably contributes to the time-related decline in TCMR activity, as well as TCMR frequency. Immunologic checkpoints are involved in this adaptation because checkpoint inhibition to treat cancer in transplant patients frequently triggers intense TCMR.61-63 The observation that checkpoint gene expression in the biopsy does not correlate with low TCMR activity or increase with time does not imply that checkpoints are not operating, given the complexity of the mechanisms that are implicated in the exhaustion phenotype.64,65 Moreover, analysis of an indication biopsy cohort has a limited ability to draw conclusions about the role of checkpoints because the patients with the strongest influence of checkpoints (ie, who have NR) will presumably not get rejection and thus will not be represented in an indication biopsy population. We also emphasize that we have not been able to find specific features of T cell exhaustion in the biopsies in this study.
TCMR is usually thought of as “episodes,” but its association with fibrosis suggests that TCMR is often a smoldering process for long periods before it is recognized, injuring nephrons (tubulitis) and driving atrophy-fibrosis. Multiple TCMR episodes increase the risk of graft loss,5 raising the possibility of unrecognized TCMR operating in patients between episodes. We need to determine whether our current treatments fully reverse cognate T cell–mediated inflammation and whether such treatments then arrest nephron loss and atrophy-fibrosis. It is also possible that, even if effector T cell activity is sterilized, the damaged nephron epithelium is programmed to progress to failure, reminiscent of the changes that occur in skin epithelia where injury programs epigenetic changes in stem cells to “remember” inflammation.66
The deleterious impact of TCMR on graft survival in the 1679 population differed somewhat from earlier findings in the first 703 biopsies,12 which showed a strong impact of AMR and mixed rejection but less impact of “pure” TCMR on survival. Greater numbers and longer follow-up in the present cohort make the damaging effect of pure TCMR clear.
There is strong agreement between MMDx and histology, despite histology using different fragments of tissue. Each uses a sample that is sufficient to give an estimate of the rejection state in the kidney, and there is no need for MMDx to read the same tissue pieces as histology. MMDx requires less tissue (0.3–0.5 mm of a core) because molecular changes (eg, IFNG effects) are more diffuse than histologic changes, and we have outlined reasons for confidence in MMDx when the 2 differ.4 Variation between pieces in MMDx measurements is much less than the variation between pathologists, because of the interobserver variation (“noise”) in histology assessments,1 and disagreement between MMDx and histology is about what is expected from this noise. It is reassuring that MMDx provides significantly better agreement with external measurements such as donor-derived cell-free DNA.67-69
The strong association of TCMR with parenchymal injury and atrophy-fibrosis and the crucial role of injury rather than TCMR activity in determining outcome after TCMR biopsies underscores the need to emphasize prevention of TCMR rather than simply trying to suppress TCMR activity by treatment. The increased atrophy-fibrosis within TCMR is presumably a consequence of the nephron injury that is manifested as increased molecular injury-induced transcripts and histologically as tubulitis. TCMR directly damages the nephron epithelium, accompanied by major structural changes such as loss of cadherins.70 This contrasts with AMR, a microcirculation disease that usually spares the parenchyma until glomerular damage accumulates and triggers shutdown, giving EAMR its relatively benign short-term prognosis.21,22 We believe that nephrons that have experienced TCMR injury and tubulitis may be programmed for irreversible shutdown (atrophy), again stressing the need for prevention. Based on the finding of under-hyalinosis in TCMR1 and even in TCMR2 to some extent, this suggests a renewed emphasis on maintaining adequate immunosuppression. However, we recognize the fact that hyalinosis has poor κ values in histology71 and that many confounders (eg, associations with aging, glomerulonephritis, glomerular sclerosis, and atrophy-fibrosis) limit the usefulness of under-hyalinosis in histologic diagnosis of individual biopsies.
ACKNOWLEDGMENTS
The authors thank our valued clinicians in the INTERCOMEX study group who partnered with us for this study by contributing biopsies and feedback (Harold Yang, Seth Narins, Carmen Lefaucheur, Alexandre Loupy, Bertram Kasiske, Arthur Matas, and Arjang Djamali).
Supplementary Material
Footnotes
A full list of INTERCOMEX Investigators is included in Table S1 (SDC, http://links.lww.com/TP/C603).
P.F.H. holds shares in Transcriptome Sciences Inc (TSI), a University of Alberta research company dedicated to developing molecular diagnostics, and is supported in part by a licensing agreement between TSI and Thermo Fisher and by a research grant from Natera. P.F.H. is a consultant to Natera. The other authors declare no conflicts of interest.
This research has been supported at times by grants from Genome Canada, Canada Foundation for Innovation, the University of Alberta Hospital Foundation, the Alberta Ministry of Advanced Education and Technology, the Mendez National Institute of Transplantation Foundation, and Industrial Research Assistance Program. Partial support was also provided by funding from a licensing agreement with the One Lambda division of Thermo Fisher. P.F.H. held a Canada Research Chair in Transplant Immunology until 2008 and currently holds the Muttart Chair in Clinical Immunology.
K.S.M-T. edited and reviewed the article and was responsible for data analysis and interpretation. G.A.B., J.B., G.E., F.E., G.G., M.M., O.V., and A.P-P. contributed biopsies and edited and reviewed the article. K.S. edited and reviewed the article. P.F.H. was the principal investigator, edited and reviewed the article, and was responsible for data interpretation and study design.
CEL files are available on the Gene Expression Omnibus website (GSE124203).
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Halloran PF. Immunosuppressive drugs for kidney transplantation. N Engl J Med. 2004;351:2715–2729. [DOI] [PubMed] [Google Scholar]
- 2.Ekberg H, Tedesco-Silva H, Demirbas A, et al. ; ELITE-Symphony Study. Reduced exposure to calcineurin inhibitors in renal transplantation. N Engl J Med. 2007;357:2562–2575. [DOI] [PubMed] [Google Scholar]
- 3.Einecke G, Sis B, Reeve J, et al. Antibody-mediated microcirculation injury is the major cause of late kidney transplant failure. Am J Transplant. 2009;9:2520–2531. [DOI] [PubMed] [Google Scholar]
- 4.Madill-Thomsen K, Perkowska-Ptasińska A, Böhmig GA, et al. ; MMDx-Kidney Study Group. Discrepancy analysis comparing molecular and histology diagnoses in kidney transplant biopsies. Am J Transplant. 2020;20:1341–1350. [DOI] [PubMed] [Google Scholar]
- 5.Rampersad C, Balshaw R, Gibson IW, et al. The negative impact of T cell-mediated rejection on renal allograft survival in the modern era. Am J Transplant. 2022;22:761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosio FG, Lager DJ, Lorenz EC, et al. Significance and implications of capillaritis during acute rejection of kidney allografts. Transplantation. 2010;89:1088–1094. [DOI] [PubMed] [Google Scholar]
- 7.Sellarés J, de Freitas DG, Mengel M, et al. Understanding the causes of kidney transplant failure: the dominant role of antibody-mediated rejection and nonadherence. Am J Transplant. 2012;12:388–399. [DOI] [PubMed] [Google Scholar]
- 8.Hricik DE, Formica RN, Nickerson P, et al. ; Clinical Trials in Organ Transplantation-09 Consortium. Adverse outcomes of tacrolimus withdrawal in immune-quiescent kidney transplant recipients. J Am Soc Nephrol. 2015;26:3114–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halloran PF, Madill-Thomsen KS, Böhmig GA, et al. ; INTERCOMEX Investigators. A 2-fold approach to polyoma virus (BK) nephropathy in kidney transplants: distinguishing direct virus effects from cognate T cell-mediated inflammation. Transplantation. 2021;105:2374–2384. [DOI] [PubMed] [Google Scholar]
- 10.Einecke G, Reeve J, Halloran PF. Hyalinosis lesions in renal transplant biopsies: time-dependent complexity of interpretation. Am J Transplant. 2017;17:1346–1357. [DOI] [PubMed] [Google Scholar]
- 11.Einecke G, Reeve J, Halloran PF. A molecular biopsy test based on arteriolar under-hyalinosis reflects increased probability of rejection related to under-immunosuppression. Am J Transplant. 2018;18:821–831. [DOI] [PubMed] [Google Scholar]
- 12.Halloran PF, Chang J, Famulski K, et al. Disappearance of T cell-mediated rejection despite continued antibody-mediated rejection in late kidney transplant recipients. J Am Soc Nephrol. 2015;26:1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burke JF, Jr, Pirsch JD, Ramos EL, et al. Long-term efficacy and safety of cyclosporine in renal-transplant recipients. N Engl J Med. 1994;331:358–363. [DOI] [PubMed] [Google Scholar]
- 14.Meier-Kriesche HU, Steffen BJ, Hochberg AM, et al. Long-term use of mycophenolate mofetil is associated with a reduction in the incidence and risk of late rejection. Am J Transplant. 2003;3:68–73. [DOI] [PubMed] [Google Scholar]
- 15.Ooi BS, Jao W, First MR, et al. Acute interstitial nephritis. A clinical and pathologic study based on renal biopsies. Am J Med. 1975;59:614–628. [DOI] [PubMed] [Google Scholar]
- 16.Sibley RK, Rynasiewicz J, Ferguson RM, et al. Morphology of cyclosporine nephrotoxicity and acute rejection in patients immunosuppressed with cyclosporine and prednisone. Surgery. 1983;94:225–234. [PubMed] [Google Scholar]
- 17.Verani RR, Flechner SM, Van Buren CT, et al. Acute cellular rejection or cyclosporine a nephrotoxicity? A review of transplant renal biopsies. Am J Kidney Dis. 1984;4:185–191. [DOI] [PubMed] [Google Scholar]
- 18.Beschorner WE, Burdick JF, Williams GM, et al. The presence of leu-7 reactive lymphocytes in renal-allografts undergoing acute rejection. Transplant Proc. 1985;17:618–622. [Google Scholar]
- 19.Solez K, Axelsen RA, Benediktsson H, et al. International standardization of criteria for the histologic diagnosis of renal allograft rejection: the Banff working classification of kidney transplant pathology. Kidney Int. 1993;44:411–422. [DOI] [PubMed] [Google Scholar]
- 20.Salazar ID, Merino López M, Chang J, et al. Reassessing the significance of intimal arteritis in kidney transplant biopsy specimens. J Am Soc Nephrol. 2015;26:3190–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reeve J, Böhmig GA, Eskandary F, et al. ; MMDx-Kidney study group. Assessing rejection-related disease in kidney transplant biopsies based on archetypal analysis of molecular phenotypes. JCI Insight. 2017;2:94197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halloran PF, Madill-Thomsen KS, Pon S, et al. ; INTERCOMEX Investigators. Molecular diagnosis of ABMR with or without donor-specific antibody in kidney transplant biopsies: differences in timing and intensity but similar mechanisms and outcomes. Am J Transplant. 2022;22:1976–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeve J, Sellarés J, Mengel M, et al. Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am J Transplant. 2013;13:645–655. [DOI] [PubMed] [Google Scholar]
- 24.Venner JM, Famulski KS, Badr D, et al. Molecular landscape of T cell-mediated rejection in human kidney transplants: prominence of CTLA4 and PD ligands. Am J Transplant. 2014;14:2565–2576. [DOI] [PubMed] [Google Scholar]
- 25.Madill-Thomsen KS, Böhmig GA, Bromberg J, et al. ; INTERCOMEX Investigators. Donor-specific antibody is associated with increased expression of rejection transcripts in renal transplant biopsies classified as no rejection. J Am Soc Nephrol. 2021;32:2743–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halloran PF, Reeve J, Akalin E, et al. Real time central assessment of kidney transplant indication biopsies by microarrays: the INTERCOMEX study. Am J Transplant. 2017;17:2851–2862. [DOI] [PubMed] [Google Scholar]
- 27.Reeve J, Böhmig GA, Eskandary F, et al. ; INTERCOMEX MMDx-Kidney Study Group. Generating automated kidney transplant biopsy reports combining molecular measurements with ensembles of machine learning classifiers. Am J Transplant. 2019;19:2719–2731. [DOI] [PubMed] [Google Scholar]
- 28.Reeve J, Madill-Thomsen KS, Halloran PF, et al. Using ensembles of machine learning classifiers to maximize the accuracy and stability of molecular biopsy interpretation. Am J Transplant. 2019;19(S3): 452–453. [DOI] [PubMed] [Google Scholar]
- 29.Lê S, Josse J, Husson F. FactoMineR: AnRPackage for multivariate analysis. J Stat Software. 2008;25:18. [Google Scholar]
- 30.Smythe GK. limma: linear models for microarray data. Gentleman RHW, Carey VJ, Irizarry RA, Dudoit S, eds. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; 2005:398–420. [Google Scholar]
- 31.Therneau T. A package for survival analysis in R. Published 2020. Available at https://CRAN.R-project.org/package=survival. Accessed March 1, 2022.
- 32.Harrell F. E., Jr. rms: regression modeling strategies. R package version 6.0-0. 2020. Available at https://CRAN.R-project.org/package=rms [computer program]. Accessed March 6, 2022.
- 33.Venner JM, Famulski KS, Reeve J, et al. Relationships among injury, fibrosis, and time in human kidney transplants. JCI Insight. 2016;1:e85323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Einecke G, Reeve J, Mengel M, et al. Expression of B cell and immunoglobulin transcripts is a feature of inflammation in late allografts. Am J Transplant. 2008;8:1434–1443. [DOI] [PubMed] [Google Scholar]
- 35.Mengel M, Reeve J, Bunnag S, et al. Molecular correlates of scarring in kidney transplants: the emergence of mast cell transcripts. Am J Transplant. 2009;9:169–178. [DOI] [PubMed] [Google Scholar]
- 36.Shoji K, Murayama T, Mimura I, et al. Sperm-associated antigen 4, a novel hypoxia-inducible factor 1 target, regulates cytokinesis, and its expression correlates with the prognosis of renal cell carcinoma. Am J Pathol. 2013;182:2191–2203. [DOI] [PubMed] [Google Scholar]
- 37.Reeve J, Einecke G, Mengel M, et al. Diagnosing rejection in renal transplants: a comparison of molecular- and histopathology-based approaches. Am J Transplant. 2009;9:1802–1810. [DOI] [PubMed] [Google Scholar]
- 38.Hirsch HH, Brennan DC, Drachenberg CB, et al. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation. 2005;79:1277–1286. [DOI] [PubMed] [Google Scholar]
- 39.Johnston O, Jaswal D, Gill JS, et al. Treatment of polyomavirus infection in kidney transplant recipients: a systematic review. Transplantation. 2010;89:1057–1070. [DOI] [PubMed] [Google Scholar]
- 40.Masutani K, Shapiro R, Basu A, et al. Putative episodes of T-cell-mediated rejection in patients with sustained BK viruria but no viremia. Transplantation. 2012;94:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmid H, Nitschko H, Gerth J, et al. Polyomavirus DNA and RNA detection in renal allograft biopsies: results from a European multicenter study. Transplantation. 2005;80:600–604. [DOI] [PubMed] [Google Scholar]
- 42.Stervbo U, Nienen M, Hecht J, et al. Differential diagnosis of interstitial allograft rejection and BKV nephropathy by T-cell receptor sequencing. Transplantation. 2020;104:e107–e108. [DOI] [PubMed] [Google Scholar]
- 43.Trydzenskaya H, Sattler A, Müller K, et al. Novel approach for improved assessment of phenotypic and functional characteristics of BKV-specific T-cell immunity. Transplantation. 2011;92:1269–1277. [DOI] [PubMed] [Google Scholar]
- 44.Loupy A, Haas M, Solez K, et al. The banff 2015 kidney meeting report: current challenges in rejection classification and prospects for adopting molecular pathology. Am J Transplant. 2017;17:28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halloran PF, Chang J, Famulski KS. Inflammation in scarred areas (i-IFTA) is a reflection of parenchymal injury (response to wounding) not T cell-mediated rejection. Am J Transplant. 2018;18(S4): 328–328.28766890 [Google Scholar]
- 46.Halloran PF, Matas A, Kasiske BL, et al. Molecular phenotype of kidney transplant indication biopsies with inflammation in scarred areas. Am J Transplant. 2019;19:1356–1370. [DOI] [PubMed] [Google Scholar]
- 47.Helgeson ES, Mannon R, Grande J, et al. i-IFTA and chronic active T cell-mediated rejection: a tale of 2 (DeKAF) cohorts. Am J Transplant. 2021;21:1866–1877. [DOI] [PubMed] [Google Scholar]
- 48.Haas M, Loupy A, Lefaucheur C, et al. The Banff 2017 kidney meeting report: revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018;18:293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naesens M, Haas M, Loupy A, et al. Does the definition of chronic active T cell-mediated rejection need revisiting? Am J Transplant. 2021;21:1689–1690. [DOI] [PubMed] [Google Scholar]
- 50.Nankivell BJ, Shingde M, Keung KL, et al. The causes, significance and consequences of inflammatory fibrosis in kidney transplantation: the Banff i-IFTA lesion. Am J Transplant. 2017;18:364–376. [DOI] [PubMed] [Google Scholar]
- 51.Lefaucheur C, Gosset C, Rabant M, et al. T cell-mediated rejection is a major determinant of inflammation in scarred areas in kidney allografts. Am J Transplant. 2018;18:377–390. [DOI] [PubMed] [Google Scholar]
- 52.Manohar S, Thongprayoon C, Cheungpasitporn W, et al. Systematic review of the safety of immune checkpoint inhibitors among kidney transplant patients. Kidney Int Rep. 2020;5:149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdel-Wahab N, Safa H, Abudayyeh A, et al. Checkpoint inhibitor therapy for cancer in solid organ transplantation recipients: an institutional experience and a systematic review of the literature. J Immunother Cancer. 2019;7:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mroue A, Moujaess E, Kourie HR, et al. Exploring the knowledge gap of immune checkpoint inhibitors in chronic renal failure: a systematic review of the literature. Crit Rev Oncol Hematol. 2021;157:103169. [DOI] [PubMed] [Google Scholar]
- 55.Adam BA, Murakami N, Reid G, et al. Gene expression profiling in kidney transplants with immune checkpoint inhibitor-associated adverse events. Clin J Am Soc Nephrol. 2021;16:1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mejia CD, Frank AM, Singh P, et al. Immune checkpoint inhibitor therapy-associated graft intolerance syndrome in a failed kidney transplant recipient. Am J Transplant. 2021;21:1322–1325. [DOI] [PubMed] [Google Scholar]
- 57.Nguyen LS, Ortuno S, Lebrun-Vignes B, et al. Transplant rejections associated with immune checkpoint inhibitors: a pharmacovigilance study and systematic literature review. Eur J Cancer. 2021;148:36–47. [DOI] [PubMed] [Google Scholar]
- 58.Venkatachalam K, Malone AF, Heady B, et al. Poor outcomes with the use of checkpoint inhibitors in kidney transplant recipients. Transplantation. 2020;104:1041–1047. [DOI] [PubMed] [Google Scholar]
- 59.Jose A, Yiannoullou P, Bhutani S, et al. Renal allograft failure after ipilimumab therapy for metastatic melanoma: a case report and review of the literature. Transplant Proc. 2016;48:3137–3141. [DOI] [PubMed] [Google Scholar]
- 60.Lesouhaitier M, Dudreuilh C, Tamain M, et al. Checkpoint blockade after kidney transplantation. Eur J Cancer. 2018;96:111–114. [DOI] [PubMed] [Google Scholar]
- 61.Zwald FO. Transplant-associated cancer in the era of immune checkpoint inhibitors: primum non nocere. Am J Transplant. 2020;20:2299–2300. [DOI] [PubMed] [Google Scholar]
- 62.d’Izarny-Gargas T, Durrbach A, Zaidan M. Efficacy and tolerance of immune checkpoint inhibitors in transplant patients with cancer: a systematic review. Am J Transplant. 2020;20:2457–2465. [DOI] [PubMed] [Google Scholar]
- 63.Lipson EJ, Bagnasco SM, Moore J, Jr, et al. Tumor regression and allograft rejection after administration of anti-PD-1. N Engl J Med. 2016;374:896–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Im SJ, Ha SJ. Re-defining T-cell exhaustion: subset, function, and regulation. Immune Netw. 2020;20:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blank CU, Haining WN, Held W, et al. Defining ‘T cell exhaustion’. Nat Rev Immunol. 2019;19:665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naik S, Larsen SB, Gomez NC, et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature. 2017;550:475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Halloran PF, Reeve J, Madill-Thomsen KS, et al. ; Trifecta Investigators. The trifecta study: comparing plasma levels of donor-derived cell-free DNA with the molecular phenotype of kidney transplant biopsies. J Am Soc Nephrol. 2022;33:387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gupta G, Moinuddin I, Kamal L, et al. Correlation of donor-derived cell-free DNA with histology and molecular diagnoses of kidney transplant biopsies. Transplantation. 2022;106:1061–1070. [DOI] [PubMed] [Google Scholar]
- 69.Xiao H, Gao F, Pang Q, et al. Diagnostic accuracy of donor-derived cell-free DNA in renal-allograft rejection: a meta-analysis. Transplantation. 2021;105:1303–1310. [DOI] [PubMed] [Google Scholar]
- 70.Halloran PF. T cell-mediated rejection of kidney transplants: a personal viewpoint. Am J Transplant. 2010;10:1126–1134. [DOI] [PubMed] [Google Scholar]
- 71.Furness PN, Taub N; Convergence of European Renal Transplant Pathology Assessment Procedures (CERTPAP) Project. International variation in the interpretation of renal transplant biopsies: report of the CERTPAP project. Kidney Int. 2001;60:1998–2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.