Abstract
Targeted protein degradation has emerged as a powerful therapeutic modality that uses molecular glues or heterobifunctional small-molecules to induce proximity between E3 ubiquitin ligases and target proteins to ubiquitinate and degrade specific disease-relevant proteins of interest. However, in many diseases proteins are aberrantly ubiquitinated and degraded to drive disease pathology, including ion channels in channelopathies such as cystic fibrosis or tumor suppressors in cancer. In these cases, targeted protein stabilization (TPS), rather than degradation, of the actively degraded target using a small-molecule would be therapeutically beneficial. Here, we present the Deubiquitinase-Targeting Chimera (DUBTAC) platform for targeted stabilization of specific proteins degraded in a ubiquitin-dependent fashion. Using chemoproteomic approaches, we discovered the covalent ligand EN523 that targets a non-catalytic allosteric cysteine C23 in the K48 ubiquitin-specific deubiquitinase OTUB1. We then developed a heterobifunctional DUBTAC consisting of our EN523 OTUB1 recruiter linked to lumacaftor, a drug used to treat cystic fibrosis that binds ΔF508-CFTR. We demonstrated proof-of-concept of TPS by showing that this DUBTAC robustly stabilized ΔF508-CFTR in human cystic fibrosis bronchial epithelial cells in an OTUB1-dependent manner. We also demonstrated stabilization of the tumor suppressor kinase WEE1 in hepatoma cells. Our study underscores the utility of chemoproteomics-enabled covalent ligand discovery approaches to develop new induced proximity-based therapeutic modalities and introduces the DUBTAC platform for TPS.
Keywords: activity-based protein profiling, targeted protein stabilization, deubiquitinase targeting chimeras, DUBTAC, cysteine, covalent ligand, chemoproteomics, induced proximity, deubiquitinase, cystic fibrosis, CFTR, WEE1, OTUB1
Editorial summary
We have developed the Deubiquitinase Targeting Chimera (DUBTAC) platform for targeted protein stabilization. We have discovered a covalent recruiter against the deubiquitinase OTUB1 that we have linked to the mutant ΔF508-CFTR targeting cystic fibrosis drug Lumacaftor to stabilize mutant CFTR protein in cells.
Introduction
One of the biggest challenges in developing new disease therapies is that more than 85 % of the proteome is considered “undruggable,” meaning these proteins are devoid of well-characterized, functional binding pockets or “ligandable hotspots” that small-molecules can bind to modulate the protein’s function-1,2. Engaging the mostly undruggable proteome to uncover new disease therapies not only requires technological innovations that facilitate rapid discovery of ligandable hotspots across the proteome, but also demands new therapeutic modalities that alter protein function through novel mechanisms. Targeted protein degradation (TPD) tackles the undruggable proteome by targeting specific proteins for ubiquitination and proteasomal degradation. One major class of small-molecule effectors of TPD, proteolysis-targeting chimeras (PROTACs), employ heterobifunctional molecules that consist of an E3 ligase recruiter linked to a protein-targeting ligand to induce the formation of ternary complexes that bring together an E3 ubiquitin ligase and the target protein as a neo-substrate 3–5. PROTACs have enabled the targeted and specific degradation of numerous disease-causing proteins in cells 3,6. New approaches for TPD have also arisen that exploit endosomal and lysosomal degradation pathways with Lysosome Targeting Chimeras (LYTACs) or autophagy with Autophagy Targeting Chimeras (AUTACs) 7,8. While TPD with PROTACs, LYTACs, or AUTACs enables the targeted degradation of potentially any protein target within, at the surface, or outside of the cell, these induced-proximity approaches are limited to degradation of proteins. New approaches for chemically induced proximity beyond degradation have also been developed in recent years, including targeted phosphorylation with Phosphorylation-Inducing Chimeric Small-Molecules (PHICS) and targeted dephosphorylation, but no small-molecule based induced proximity approaches exist for targeted deubiquitination and subsequent stabilization of proteins 9,10.
The active ubiquitination and degradation of proteins is the root cause of several classes of diseases, including many tumor suppressors in cancer (e.g. TP53, CDKN1A, CDN1C, BAX), mutated and misfolded proteins such as ΔF508-CFTR in cystic fibrosis, or glucokinase in pancreatic cells in Maturity-onset diabetes of the young type 2 (MODY2). In these cases a targeted protein stabilization (TPS) therapeutic strategy, rather than degradation, would be beneficial 11–14. Analogous to TPD, we hypothesized that TPS could be enabled by the discovery of a small-molecule recruiter of a deubiquitinase (DUB) that could be linked to a protein-targeting ligand to form a chimeric molecule which would induce the deubiquitination and stabilization of proteins of interest. We call this heterobifunctional stabilizer a Deubiquitinase Targeting Chimera (DUBTAC) (Figure 1a). In this study, we report the discovery of a covalent recruiter for the K48 ubiquitin-chain specific DUB OTUB1 which when linked to a protein-targeting ligand stabilizes an actively degraded target protein to demonstrate proof-of-concept for the DUBTAC platform.
Figure 1. DUBTAC platform.
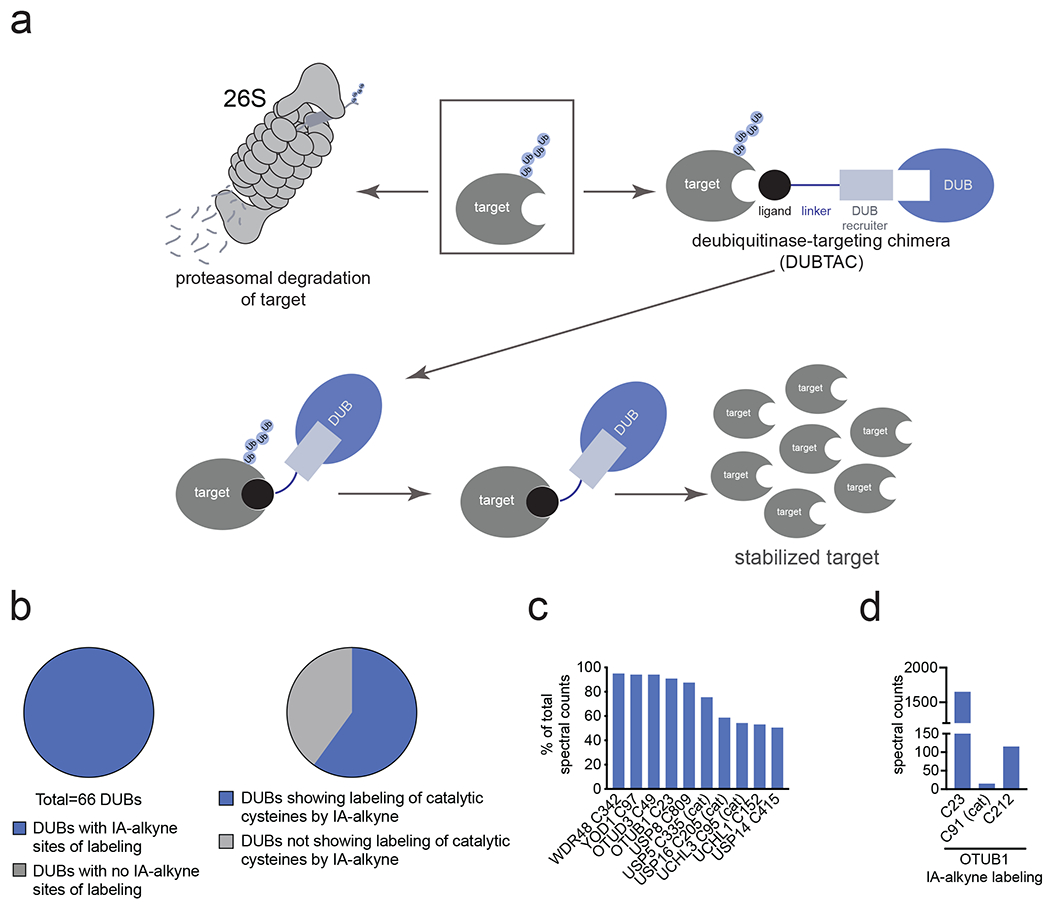
(a) DUBTACs are heterobifunctional molecules consisting of a protein-targeting ligand linked to a DUB recruiter via a linker. DUBTACs are ideally used for stabilizing the levels of actively ubiquitinated proteins that are degraded by the proteasome. When treated in cells, a DUBTAC will induce the proximity of a DUB with a target protein to remove polyubiquitin chains to prevent the protein from undergoing proteasome-mediated degradation, thereby stabilizing and elevating the level of the actively degraded protein. (b) Out of 65 DUBs mined in our research group’s aggregate chemoproteomic datasets of cysteine-reactive probe labeling with IA-alkyne in various complex proteomes, we identified probe-modified cysteines across all 100 % of the 65 DUBs. This is shown in the first pie chart. Among the 65 DUBs that showed probe-modified cysteines, 40 of these DUBs showed >10 aggregate spectral counts across our chemoproteomic datasets. 24 DUBs, or 60 %, of these 40 DUBs showed labeling of the DUB catalytic or active site cysteines. (c) Mining the DUB data, we identified 10 DUBs wherein there was one probe-modified cysteine that represented >50 % of the total aggregate spectral counts for probe-modified cysteine peptides for the particular DUB. 7 of those 10 DUBs do not target a known catalytic cysteine whereas 3 do target the catalytic cysteine (abbreviated by cat). (d) Analysis of aggregate chemoproteomic data for OTUB1 IA-alkyne labeling showing that C23 is the dominant site labeled by IA-alkyne compared to the catalytic (cat) C91. Chemoproteomic data analysis of DUBs across aggregated datasets can be found in Table S1.
Results
Identifying Allosteric Ligandable Sites within DUBs
To enable the DUBTAC platform, our first goal was to identify a small-molecule recruiter that targeted an allosteric site on a DUB without inhibiting DUB function, since the recruitment of a functional DUB would be required to deubiquitinate and stabilize the target protein. While many DUBs possess well-defined active sites bearing a catalytic and highly nucleophilic cysteine, there have not yet been systematic evaluations of allosteric, non-catalytic, and ligandable sites on DUBs that could be pharmacologically targeted to develop a DUB recruiter. Chemoproteomic platforms such as activity-based protein profiling (ABPP) have proven to be powerful approaches to map proteome-wide covalently ligandable sites directly in complex proteomes. ABPP utilizes reactivity-based amino acid-specific chemical probes to profile reactive, functional, and potentially ligandable sites directly in complex proteomes 2,15. When used in a competitive manner, pre-treatment with libraries of unmodified covalent small-molecule ligands can be used to screen for competion of probe binding to recombinant protein or complex proteomes to enable covalent ligand discovery against potential ligandable sites revealed by the reactivity-based probe 2,16. Previous studies have shown that isotopic tandem orthogonal proteolysis-ABPP (isoTOP-ABPP) platforms for mapping sites of labeling with reactivity-based probes using quantitative proteomic approaches can identify hyper-reactive, functional, and ligandable cysteines 15,16.
To identify DUB candidates that possess potential ligandable allosteric cysteines, we analyzed our research group’s aggregate chemoproteomic data of proteome-wide sites modified by reactivity-based probes collected since the start of our laboratory. Specifically, we mined our collective chemoproteomic data of cysteine-reactive alkyne-functionalized iodoacetamide (IA-alkyne) probe labeling sites from isoTOP-ABPP experiments from 455 distinct experiments in human cell line proteomes for total aggregate spectral counts identified for each probe-modified site across the DUB family. We postulated that probe-modified cysteines within DUBs that showed the highest spectral counts aggregated over all chemoproteomic datasets, compared to those sites within the same DUB that showed lower spectral counts, may represent more reactive and potentially more ligandable cysteines. Caveats to this premise include cysteines that might be located in regions within a protein sequence that does not yield suitable tryptic peptides with respect to ionization and compatibility with mass spectrometry-based sequencing, and labeling of surface-exposed cysteines that may not be part of binding pockets. However, we conjectured that the aggregate chemoproteomics data would still yield candidate allosteric ligandable sites within DUBs that could be prioritized for covalent ligand screening. We initially mined our aggregate chemoproteomic data for 66 members of the cysteine protease family of DUBs—including ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin domain proteases (MJDs) and ovarian tumor proteases (OTU)—since they encompass the majority of DUB superfamilies. Interestingly, we found probe-modified cysteines all of these DUB enzymes (Figure 1b; Table S1). Consistent with our aggregate chemoproteomic data of probe-modified sites being enriched in functional sites within DUBs, among the 40 DUBs that showed a total of >10 aggregate spectral counts of probe-modified peptides, 24 of those DUBs, 60 %, showed labeling of the DUB catalytic cysteine (Figure 1b). We next prioritized this list of 40 DUBs to identify suitable candidates for TPS. We prioritized DUBs where the dominant probe-modified cysteine was: 1) located at an allosteric site and not the catalytic cysteine such that we could target the identified cysteine with a covalent ligand while retaining the catalytic activity of the DUB; 2) in a dominantly identified probe-labeled peptide compared to other probe-modified sites within the same DUB, which - even without correcting for compatibility with MS/MS-based peptide identification - would indicate a high degree of reactivity and potential covalent ligandability of the identified allosteric cysteine compared to the catalytic site; and 3) frequently identified in chemoproteomics datasets which would indicate the general accessibility of the cysteine in complex proteomes. We found 10 DUBs where one probe-modified cysteine represented >50 % of the spectral counts of all modified cysteines for the particular protein, of which 7 of these DUBs showed primary sites of probe-modification that did not correspond to the catalytic cysteine (Figure 1c). Of these 10 DUBs, OTUB1 C23 was captured with >1000 total aggregate spectral counts compared to <500 aggregate spectral counts for the other DUBs (Extended Data Figure 1a). In our aggregated chemoproteomic data, the tryptic peptide encompassing OTUB1 C23 was the dominant peptide labeled by IA-alkyne with >1500 total spectral counts, compared to 15 spectral counts for the peptide encompassing the catalytic C91, and 115 spectral counts for C212 (Figure 1d).
OTUB1 is a highly expressed DUB that specifically cleaves K48-linked polyubiquitin chains—the type of ubiquitin linkage that destines proteins for proteasome-mediated degradation—and C23 represents a known ubiquitin substrate recognition site that is distant from the catalytic C91 (Extended Data Figure 1b) 17–20. Interestingly, C23 is also predicted to be in an intrinsically disordered region of the protein based on Predictor of Natural Disordered Regions (PONDR) analysis of OTUB1 (Extended Data Figure 1b) 21,22. Given our analysis of chemoproteomic data, the properties of OTUB1, and the location of C23, we chose OTUB1 as our candidate DUB for covalent ligand screening using gel-based ABPP approaches to discover an OTUB1 recruiter.
Discovering and Characterizing a Covalent Recruiter Against OTUB1
We performed a gel-based ABPP screen in which we screened 702 cysteine-reactive covalent ligands against labeling of pure OTUB1 protein with a rhodamine-functionalized cysteine-reactivity iodoacetamide (IA-rhodamine) probe (Figure 2a; Extended Data Figure 1b; Table S2). Through this screen, we identified the acrylamide EN523 as a top hit (Figure 2b). We confirmed that EN523 dose-responsively displaced IA-rhodamine labeling of OTUB1 without causing any protein aggregation or precipitation (Figure 2c). We next performed liquid chromatography-tandem mass spectrometry analysis (LC-MS/MS) of tryptic peptides from EN523 bound to OTUB1 and showed that EN523 selectively targets C23, with no detectable modification of the catalytic C91 (Figure 2d). Following these data, we performed an in vitro reconstituted OTUB1 deubiquitination activity assay monitoring monoubiquitin release from di-ubiquitin and demonstrated that EN523 does not inhibit OTUB1 deubiquitination activity (Figure 2e) 23. These studies were performed in the presence of OTUB1-stimulating Ubiquitin-conjugating enzyme E2 D1 (UBE2D1), an E2 ubiquitin ligase that engages in a complex with OTUB1 to stimulate OTUB1 activity.
Figure 2. Discovery of covalent ligands that target OTUB1.
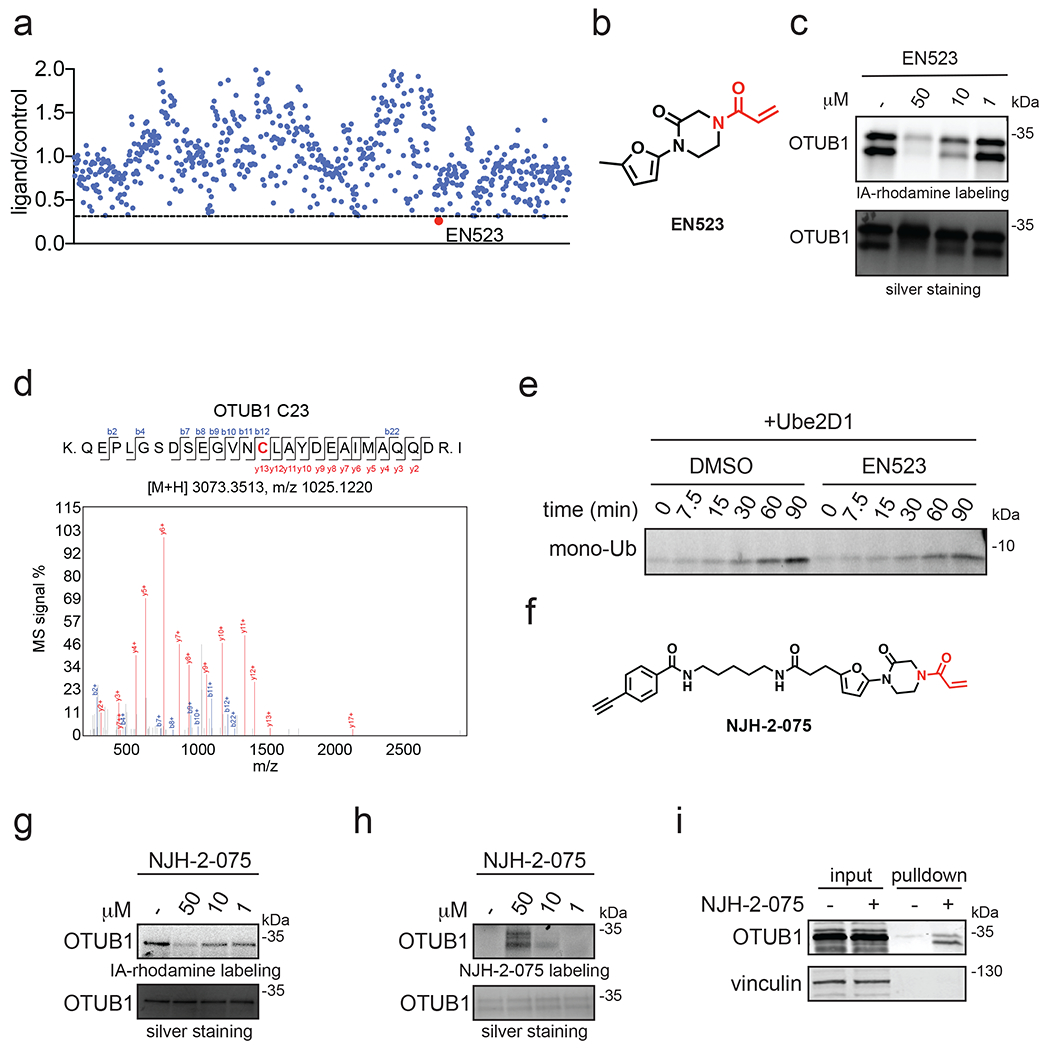
(a) Covalent ligand screen of a cysteine-reactive library competed against IA-rhodamine labeling of recombinant OTUB1 to identify binders to OTUB1 by gel-based ABPP. Vehicle DMSO or cysteine-reactive covalent ligands (50 μM) were pre-incubated with OTUB1 for 30 min at room temperature prior to IA-rhodamine labeling (500 nM, 30 min room temperature). OTUB1 was then separated by SDS/PAGE and in-gel fluorescence was assessed and quantified. Gel-based ABPP data and quantification of in-gel fluorescence shown in Extended Data Figure 1b and Table S2. EN523 annotated in red was the top hit that showed the greatest inhibition of OTUB1 IA-rhodamine labeling. (b) Structure of EN523 shown with cysteine-reactive acrylamide highlighted in red. (c) Gel-based ABPP confirmation showing dose-responsive inhibition of IA-rhodamine binding of OTUB1. Vehicle DMSO or EN523 were pre-incubated with recombinant OTUB1 for 30 min at 37 °C prior to IA-rhodamine labeling (500 nM, 30 min room temperature). OTUB1 was then separated by SDS/PAGE and in-gel fluorescence was assessed. Also shown is silver staining showing protein loading. Shown is a representative gel of n=3 biologically independent samples/group. (d) LC-MS/MS data showing EN523-modified adduct on C23 of OTUB1. OTUB1 (10 μg) recombinant protein was incubated with EN523 (50 μM) for 30 min, after which the protein was precipitated and digested with trypsin and tryptic digests were analyzed by LC-MS/MS to identify modified sites. (e) OTUB1 DUB activity monitored by cleavage of K48 diubiquitin. Recombinant OTUB1 were pre-incubated with DMSO or EN523 (50 μM) for 1 h. After pre-incubation, OTUB1 was added to a mixture of diubiquitin and UBE2D1. The appearance of mono-Ub was monitored by Western blotting. (f) Structure of alkyne-functionalized EN523 probe—NJH-2-075. (g) Gel-based ABPP of NJH-2-075. Vehicle DMSO or NJH-2-075 were pre-incubated with OTUB1 for 30 min at 37 °C prior to IA-rhodamine labeling (500 nM, 30 min room temperature). OTUB1 was then separated by SDS/PAGE and in-gel fluorescence was assessed. Also shown is silver staining showing protein loading. (h) NJH-2-075 labeling of recombinant OTUB1. OTUB1 (0.5 μg) was labeled with DMSO or NJH-2-075 for 1.5 h at 37° C, after which rhodamine-azide was appended by CuAAC, OTUB1 was separated by SDS/PAGE and in-gel fluorescence was assessed. Also shown is silver staining showing protein loading. (i) NJH-2-075 engagement of OTUB1 in HEK293T cells. HEK293T cells were treated with DMSO vehicle or NJH-2-075 (50 μM) for 2 h, after which cell lysates were subjected to CuAAC with biotin picolyl azide and NJH-2-075 labeled proteins were subjected to avidin pulldown, eluted, separated by SDS/PAGE, and blotted for OTUB1 and vinculin. Both input lysate and pulldown levels are shown. Gels or blots shown in (c, e, g, h, i) are representative of n=3 biologically independent samples/group.
We next used NMR analysis to further characterize EN523 binding to OTUB1. A 13C-HMQC spectrum of OTUB1 revealed the presence of a homogenous and mostly folded protein with well dispersed ILVA methyl group resonances (Extended Data Figure 2a). EN523 treatment of OTUB1 led to subtle but significant chemical shift perturbations (CSPs) (Extended Data Figure 2b). As we did not assign any peaks in the OTUB1 spectrum, we could not determine the exact binding site of the small molecule. However, almost all affected resonances had stronger intensities than the average signal strengths and had chemical shifts that are close to the random coil values of the respective amino acids. These observations suggest that the amino acids giving rise to these resonances are located in unfolded sections of the protein (in agreement with our PONDR data) predicting that C23 (the site of EN523 binding) belongs to an intrinsically disordered region of the protein. Next, we investigated if the covalent binding of EN523 to C23 prevents the interaction of OTUB1 with the ubiquitin-loaded and free form of ubiquitin-conjugating enzyme E2 D2 (UBE2D2). As the latter protein activates OTUB1 DUB activity, we wanted to make sure that EN523 did not interfere with this protein-protein-interaction. We mixed OTUB1 with ubiquitinylated or free UBE2D2 and compared the chemical shifts of OTUB1 residues in the presence and absence of EN523. Binding of either form of the conjugating enzyme to OTUB induces strong peak perturbations (Extended Data Figure 2c, 2d). These shift differences were almost identical for samples with and without EN523. The only differences we detected were shift changes for peaks which are affected by compound binding. These results indicated that EN523 does not interfere with binding of UBE2D2 to OTUB1.
We also explored structure-activity relationships (SAR) of our OTUB1 recruiter. Consistent with the necessity of the reactive acrylamide warhead for interacting with C23 of OTUB1, a non-reactive acetamide version of EN523, NJH-2-080, showed loss in binding against OTUB1 (Extended Data Figure 3). Replacing the methylfuran substituent with a benzoimidazolone, benzothiophene, benzofuran, phenyloxazole, or imidazopyridine, but not methylimidazole, still retained potency against OTUB1 (Extended Data Figure 3). We also explored preliminary SAR of the piperazinone core as well. Dimethyl and methyl piperazinone substitutions with a tert-butyl propionate extension from the furan still maintained potency against OTUB1. We also found that the (R)-methylpiperazinone derivative was more potent than the (S)-methylpiperazinone derivative of EN523, indicating that we may be able to achieve stereochemically specific interactions with OTUB1 (Extended Data Figure 3). These data also demonstrated that extension off the furan may present an optimal exit vector for synthesis of DUBTACs. Overall, while the SAR showed room for flexibility within the EN523 core scaffold, we did not identify significantly more potent OTUB1 ligands and thus we chose to further pursue EN523 as our OTUB1 recruiter for follow-up studies.
An alkyne-functionalized probe of EN523—NJH-2-075—was then synthesized with the goal of assessing whether this ligand engaged OTUB1 in cells (Fig. 2f). NJH-2-075 retained binding to OTUB1 in vitro, as shown by: 1) gel-based ABPP demonstrating competition of NJH-2-075 against IA-rhodamine labeling of recombinant OTUB1 and; 2) direct labeling of recombinant OTUB1 by NJH-2-075 visualized by copper-catalyzed azide-alkyne cycloaddition (CuAAC) of azide-functionalized rhodamine to NJH-2-075-labeled OTUB1 (monitored by in-gel fluorescence) (Fig 2g–2h). We demonstrated NJH-2-075 engagement of OTUB1 in cells by enrichment of endogenous OTUB1 through NJH-2-075 as compared with vehicle treatment in HEK293T cells, ex situ CuAAC of azide-functionalized biotin to NJH-2-075-labeled proteins and subsequent avidin-enrichment and blotting for OTUB1 (Fig. 2i). We further showed that an unrelated protein vinculin was not enriched by NJH-2-075 (Fig. 2i). Collectively, these data highlighted EN523 as a promising covalent OTUB1 ligand that targeted a non-catalytic and allosteric C23 on OTUB1 without inhibiting OTUB1 deubiquitination activity and engaged OTUB1 in cells.
Showing Proof-of-Concept of the DUBTAC Platform with Mutant CFTR
To demonstrate the feasibility of using EN523 as an OTUB1 recruiting module of a heterobifunctional DUBTAC, we identified the mutant ΔF508-CFTR chloride channel as a proof-of-concept case where protein stabilization would be therapeutically desirable. ΔF508, a frameshift mutation caused by deletion at codon 508 in exon 10 of CFTR, resulting in the absence of a phenylalanine residue, is the most common mutation that induces the Cystic Fibrosis phenotype 24. This mutation causes the protein to become conformationally unstable leading to K48 polyubitiquination and degradation prior to trafficking from the endoplasmic reticulum to the cell surface 14,24. Previous studies have demonstrated the feasibility of stabilizing mutant CFTR not only by genetic and pharmacological inhibition of the cognate E3 ligase RNF5, but in a recent study also through targeted recruitment of DUBs using a genetically encoded and engineered DUB targeted to CFTR using a CFTR-targeting nanobody 25–27. Importantly for our work, suitable CFTR-targeting small-molecule ligands exist. Lumacaftor, a drug for cystic fibrosis developed by Vertex Pharmaceuticals, acts as a chemical chaperone for ΔF508-CFTR and corrects its misfolding, leading to increased trafficking of ΔF508-CFTR to the cell membrane and partial restoration of protein function 28. Despite lumacaftor’s chaperoning activity, the vast majority of ΔF508-CFTR is still actively ubiquitinated and degraded, making the potential of a synergistic effect via DUBTAC-induced deubiquitination a therapeutically attractive option. This potential synergy was also the reason to focus on lumacaftor instead of the channel potentiator ivacaftor.
With this in mind, we synthesized DUBTACs linking the OTUB1 recruiter EN523 to the CFTR chaperone lumacaftor with two different C3 or C5 alkyl linkers, NJH-2-056 and NJH-2-057 (Figure 3a–3b). We confirmed that these two DUBTACs still engaged recombinant OTUB1 in vitro by gel-based ABPP (Figure 3c–3d). We used CFBE41o-4.7 human bronchial epithelial cells expressing ΔF508-CFTR, a human cystic fibrosis bronchial epithelial cell line, as a model system to test our DUBTACs. We first showed that EN523 did not alter OTUB1 protein levels in these cells (Extended Data Figure 4a). We also demonstrated that the alkyne-functionalized EN523 probe, NJH-2-075 still engaged OTUB1 in this cell line (Extended Data Figure 4b). The DUBTACs were then tested in these cells alongside lumacaftor or EN523 treatment alone. While neither NJH-2-056, lumacaftor, nor EN523 treatment altered mutant CFTR levels, we observed a robust and significant increase in CFTR protein levels with NJH-2-057 treatment (Figure 3e–3f). This stabilization was dose-responsive, with clear stabilization occurring with 8 μM, and time-dependent with stabilization at 10 μM evident starting at 16 h (Extended Data Figure 5). We further confirmed that the stabilized protein was CFTR using three additional commercially available CFTR antibodies (Extended Data Figure 6) and showed that the DUBTAC-stabilized CFTR band was attenuated upon CFTR knockdown (Extended Data Figure 7).
Figure 3. DUBTAC against mutant CFTR.
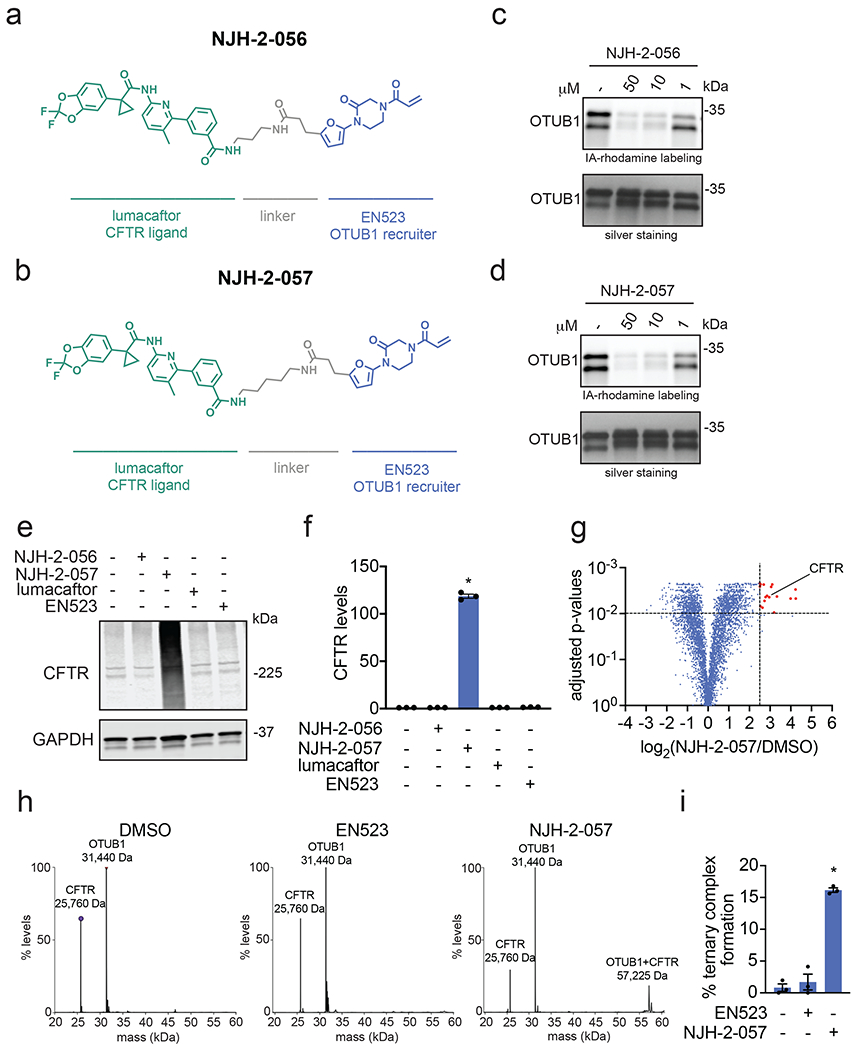
(a, b) Structures of NJH-2-056 and NJH-2-057; these DUBTACs against mutant CFTR protein are formed by linking CFTR ligand lumacaftor to OTUB1 recruiter EN523 through C3 and C5 alkyl linkers, respectively. (c, d) Gel-based ABPP analysis of NJH-2-056 and NJH-2-057 against OTUB1. Vehicle DMSO or DUBTACs were preincubated with recombinant OTUB1 for 30 min at 37 °C prior to addition of IA-rhodamine (100 nM) for 30 min at room temperature. OTUB1 was run on SDS/PAGE and in-gel fluorescence was assessed. Protein loading was assessed by silver staining. (e) Effect of DUBTACs on mutant CFTR levels. CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO, NJH-2-056 (10 μM), NJH-2-057 (10 μM), lumacaftor (10 μM), or EN523 (10 μM) for 24 h, and mutant CFTR and loading control GAPDH levels were assessed by Western blotting. (f) Quantification of the experiment described in (e). (g) TMT-based quantitative proteomic profiling of NJH-2-057 treatment. CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or NJH-2-057 (10 μM) for 24 h. Data shown are from n=3 biologically independent samples/group. Full data for this experiment can be found in Table S3. (h) Native MS analysis of DUBTAC-mediated ternary complex formation. OTUB1 (2 μM) and the CFTR-nucleotide binding domain (2 μM) were incubated with DMSO vehicle, EN523 (50 μM), or NJH-2-057 (50 μM) in 150 mM ammonium acetate with MgCl2 (100 μM) and ATP (100 μM). Representative mass spectra from n=3 biologically independent samples/group are shown. (i) percentage of ternary complex formation assessed by measuring the CFTR-OTUB1 complex formed in the experiment described in (h). Gels shown in (c, d, e) are representative of n=3 biologically independent samples/group. Data in (f, i) show individual biological replicate values and average ± sem from n=3 biologically independent samples/group. Statistical significance was calculated with unpaired two-tailed Student’s t-tests in (f, i) compared to vehicle-treated controls and is expressed as *p<0.05.
We also explored the dependence of CFTR stabilization on linker length and composition (Extended Data Figure 8). DUBTACs bearing C5 and C6 alkyl linkers, but not C3 and C4 alkyl linkers, stabilized CFTR. Interestingly, none of the DUBTACs bearing PEG linkers were able to stabilize CFTR (Extended Data Figure 8). We also made eight additional CFTR DUBTACs bearing more rigid heterocycle-containing linkers to determine whether these compounds that may be less flexible and potentially more drug-like may perform better in stabilizing mutant CFTR (Extended Data Figure 9). Interestingly, nearly all these DUBTACs increased CFTR levels with substantially improved response from GL-03, bearing a [7,5] fused saturated diamine linker, compared to NJH-2-075 (Extended Data Figure 9). Consistent with the necessity of the cysteine-reactive warhead in binding to OTUB1, we also demonstrated that a non-reactive propiolamide version of NJH-2-057, NJH-2-106, was incapable of stabilizing CFTR (Extended Data Figure 10). The alkyne probe NJH-2-075, which contains an identical linker to NJH-2-057 but exchanges an ethynylphenyl group for lumacaftor, also did not induce CFTR stabilization (Extended Data Figure 10). The CFTR smear that we observed in the blot with NJH-2-057 treatment is consistent with previous studies investigating CFTR and in line with our observations with the proteasome inhibitor bortezomib, likely representing a combination of differential glycosylation states, other forms of ubiquitination on CFTR that may not be removed by OTUB1 (e.g. K63 ubiquitin chains), and previously observed anomalous migration of CFTR on SDS/PAGE due to the presence of SDS-resistant ternary structures within the protein (Extended Data Figure 11) 24,29. Based on the molecular weight of the darkest part of the CFTR blot >225 kDa, we conjectured that we are stabilizing the fully mature glycosylated form of mutant CFTR (Figure 3e).
To further validate our Western blot data for CFTR stabilization and to assess the proteome-wide activity of NJH-2-057, we performed a TMT-based quantitative proteomic analysis of NJH-2-057 treated CFBE41o-4.7 cells expressing ΔF508-CFTR. Satisfyingly, the proteomic analysis showed CFTR amongst the most robustly stabilized proteins (ratio of 7.8 comparing NJH-2-057 to vehicle treatment, Figure 3g; Table S3). While there were additional proteins with significant changes in abundance levels, we only observed 21 proteins that were significantly stabilized by >5-fold with an adjusted p-value <0.01 compared to vehicle-treated controls out of 4552 total quantified proteins (Figure 3g; Table S3). These observed changes in protein abundance levels by the DUBTAC appear to be DUBTAC-specific changes since these changes were not detected with EN523 or Lumacaftor treatment alone (Extended Data Figure 12; Table S3) and may represent compensatory changes occurring from elevations in CFTR levels in cells or could represent changes resulting from off-targets of the DUBTAC. Interestingly, among proteins elevated along with CFTR were several protein chaperones including Heat shock 70 kDa protein 6 (HSPA6), DnaJ Heat shock protein family Hsp40 Member B1 (DNAJB1), Heat shock 50 kDa protein 1A (HSPA1A), and DNAJ homolog subfamily B member 4 (DNAJB4). These changes could reflect potential compensatory on-target upregulation of protein chaperones in response to highly elevated levels of a relatively unstable mutant CFTR . Nonetheless, we did not observe widespread alterations in protein levels with DUBTAC treatment, suggesting that we were not substantially disrupting global protein turnover.
Having identified NJH-2-057 as a DUBTAC that was capable of stabilizing mutant CFTR in cells, we next sought to confirm the formation of a ternary complex between CFTR, NJH-2-057, and OTUB1 in vitro using recombinant protein and native mass spectrometry (MS)-based approaches (Figure 3h–3i). While the highest intensity signals corresponded to unmodified OTUB1 and the ΔF508-harboring CFTR nucleotide-binding domain used in this experiment, potentially indicating low levels of target engagement under these experimental conditions, we observed significant CFTR-OTUB1 complex formation with NJH-2-057 treatment, but not with DMSO vehicle or EN523 treatment (Figure 3h–3i). The predominantly observed mass for this complex corresponded to OTUB1 and CFTR but not the combined masses of OTUB1, CFTR, and NJH-2-057. This may be because the NJH-2-057 adduct on OTUB1 may be unstable to electrospray ionization and desolvation energy conditions required to be able to observe the protein complex, since we were also not able to observe the NJH-2-057 mass adduct on OTUB1. However, minor peaks indicated the presence of adducts consistent with either full NJH-2-057 or either MS-induced fragments or break-down products (Extended Data Figure 13). Nonetheless, given that this OTUB1-CFTR complex was only observed with DUBTAC treatment but not with DMSO or EN523 treatment, our data strongly suggests that the DUBTAC enables ternary complex formation.
To further confirm that the robust stabilization in mutant CFTR levels conferred by NJH-2-057 was due to the proposed on-target activity, we demonstrated that the stabilization of CFTR was attenuated by pre-treatment with either lumacaftor or EN523, indicating that the stabilization by the DUBTAC was due to targets engaged by both lumacaftor and EN523 (Figure 4a–4b). These data also indicate the necessity for ternary complex formation to stabilize CFTR levels. To further verify that the CFTR stabilization was dependent on OTUB1, OTUB1 knockdown significantly attenuated mutant CFTR stabilization by NJH-2-057 (Figure 4c–4d).
Figure 4. Characterizing the mechanism of the CFTR DUBTAC NJH-2-057.
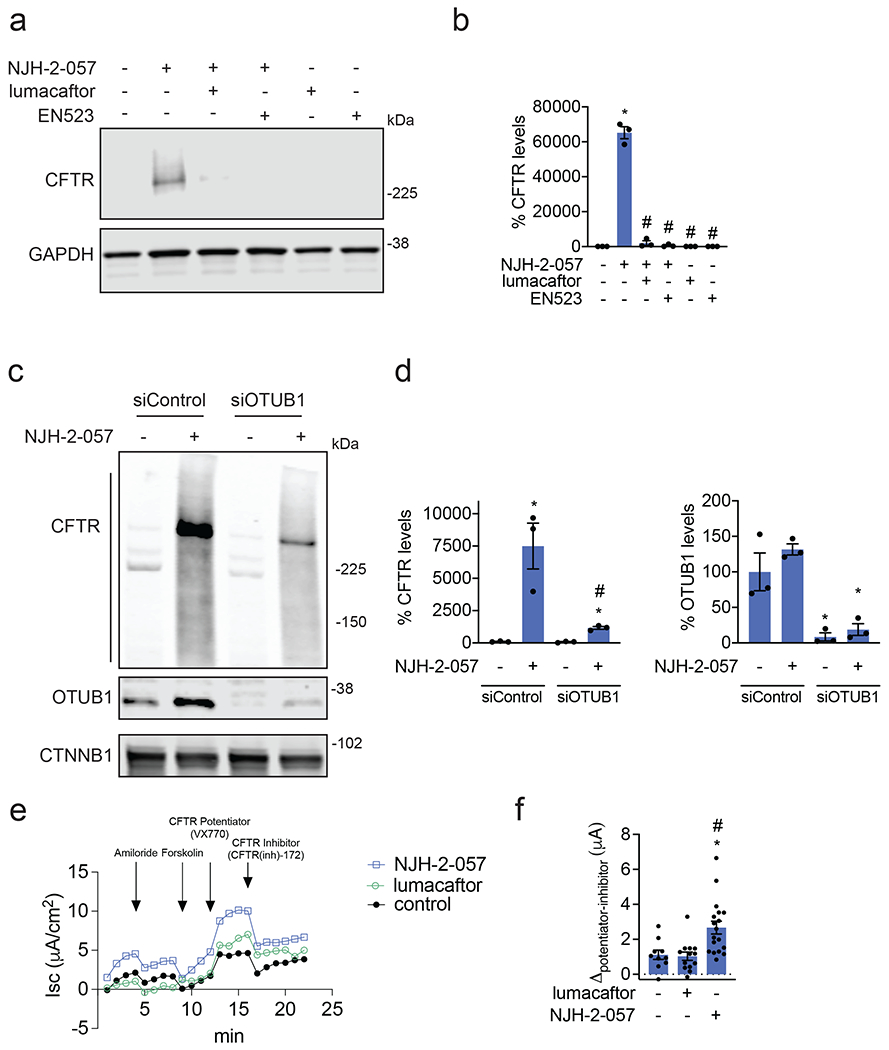
(a) Effect of lumacaftor or EN523 pre-incubation on NJH-2-057 DUBTAC-mediated stabilization of mutant CFTR levels. CFBE41o-4.7 cells expressing ΔF508-CFTR were pre-treated with vehicle DMSO, lumacaftor (100 μM), or EN523 (100 μM) for 1 h prior to treatment with NJH-2-057 (10 μM) for 24 h. Mutant CFTR and loading control GAPDH levels were assessed by Western blotting. (b) Quantification of the experiment described in (a). (c) Effect of OTUB1 knockdown on NJH-2-057 DUBTAC-mediated mutant CFTR stabilization. CFBE41o-4.7 cells expressing ΔF508-CFTR were transiently transfected with siControl or siOTUB1 oligonucleotides for 48 h prior to treatment of cells with vehicle DMSO or NJH-2-057 (10 μM) for 16 h. Mutant CFTR, OTUB1, and loading control GAPDH levels were assessed by Western blotting. (d) Levels of mutant CFTR and OTUB1 from the experiment described in (c). (e) Transepithelial conductance in primary human cystic fibrosis donor bronchial epithelial cells bearing the ΔF508-CFTR mutation. Cells were treated with DMSO vehicle, NJH-2-057 (10 μM), or lumacaftor (10 μM) 24 h prior to the TECC24 assay in which cells received four additional sequential treatments with a sodium channel inhibitor amiloride (10 μM), cAMP activator Forskolin (20 μM), a CFTR potentiator VX770 (0.5 μM), and finally with a CFTR inhibitor CFTR-Inh172 (30 μM). Shown are the average values from conductance from a single donor. Experiments were conducted in primary cells from two donors. (f) Changes in current between potentiator VX770 (Ivacaftor) treatment and the CFTR inhibitor treatment in the experiment described in (e) in two primary human cystic fibrosis donor bronchial epithelial cells bearing the ΔF508-CFTR mutation. Individual replicate data are shown in the bar graph from n=10 biologically independent samples in the DMSO vehicle treated group, n=13 biologically independent samples in the lumacaftor treated group, and n=18 biologically independent samples in the NJH-2-057 treated group. Gels shown in (a, c) are representative of n=3 biologically independent samples/group. Data in (b, d) show individual biological replicate values and average ± sem from n=3 biologically independent samples/group. Statistical significance in was calculated with unpaired two-tailed Student’s t-tests in (b, d, f) and is expressed as *p<0.05 compared to vehicle-treated control in (b, f) and vehicle-treated siControl in (d) and #p<0.05 compared to the NJH-2-057 treated group in (b) and NJH-2-057 treated siControl group for CFTR levels in (d).
We next performed a competitive isoTOP-ABPP study to assess the overall proteome-wide selectivity and cysteine-reactivity of NJH-2-057 treatment in CFBE41o-4.7 cells expressing ΔF508-CFTR (Extended Data Figure 12; Table S4). Out of 1270 IA-alkyne probe-modified peptides quantified in two out of three biological replicates, there were only 5 targets that showed isotopically light to heavy or control to NJH-2-057 treatment probe-modified peptide ratios of >4 with an adjusted p-value <0.05: VDAC2 C76, TUBB1 C201, RLF C744, and VDAC2 C47, and VDAC3 C66. None of these targets are part of the ubiquitin-proteasome system and as far as we can tell at this point unlikely to influence the activity of our DUBTAC (Extended Data Figure 14; Table S4). OTUB1 C23 was captured in our isoTOP-ABPP experiment, but only showed a ratio of 1.6 which would correspond to ~60% target occupancy (Table S3). This likely indicates that the observed CFTR stabilization by NJH-2-057 is occurring through relatively low levels of OTUB1 occupancy in cells which would also be in line with our in vitro labeling and native MS data. Activity of heterobifunctional molecules has also been reported previously in studies using covalent E3 ligase recruiters for targeted protein degradation applications showing that relatively minimal target occupancy of E3 ligases can still lead to robust degradation of target proteins due to the catalytic mechanism of action of the E3 ligases 30–33. We conjectured that a similar catalytic effect in a DUBTAC also leads to robust stabilization of the target protein with partial OTUB1 occupancy. It is important to note that the covalent installation of the DUBTAC on OTUB1 still allows for such a catalytic effect on CFTR deubiquitination.
Having shown stabilization of CFTR protein levels with our DUBTAC, we next sought to determine whether our DUBTAC-mediated increase in CFTR protein levels led to improved cell surface CFTR function (Figure 4e–4f). We measured transepithelial conductance in primary human cystic fibrosis donor bronchial epithelial cells bearing the ΔF508-CFTR mutation. These cells were pre-treated with vehicle, lumacaftor, or NJH-2-057 for 24 h prior to sequential treatments with a sodium channel inhibitor amiloride, cAMP activator Forskolin, and a CFTR potentiator VX770 (Ivacaftor) to fully activate CFTR function in cells. After chloride channel conductance was potentiated with VX770, the cells were treated with a CFTR inhibitor, CFTR(inh)-172, to show CFTR-dependence of any increases observed in transepithelial conductance. The difference in conductance between potentiator VX770 and CFTR inhibitor-treatment were quantified under the three different treatment conditions (vehicle, lumacaftor, or DUBTAC treatment) to ascertain the effects that our DUBTAC had on CFTR-mediated conductance compared to lumacaftor or vehicle treatment (Figure 4e–4f). Our studies showed that treatment of these primary cells with NJH-2-057 led to significant improvement in CFTR-dependent transepithelial conductance compared to lumacaftor or vehicle treatment, indicating that our CFTR DUBTAC not only elevated CFTR protein levels, but also functional CFTR at the cell surface leading to improved CFTR function (Figure 4e–4f).
Using DUBTACs to Stabilize WEE1
With our first proof-of-concept for a fully synthetic DUBTAC-mediated stabilization of mutant CFTR in hand, we next sought to show a second example of TPS with DUBTACs against another actively degraded target for which a well-validated ligand had been reported in the literature. We selected WEE1, a tumor suppressor kinase in non-malignant eukaryotic somatic cells that phosphorylates the cyclin-dependent kinase (CDK1)-cyclin B1 complex to inhibit cell cycle progression during S and G2 phases of mitosis and whose activity must be downregulated for mitotic progression to occur 34,35. One mechanism through which WEE1 activity is suppressed via ubiquitin-mediated proteasomal degradation 35,36. Clinical WEE1 inhibitors such as AZD1775 have been developed to be given in combination with DNA-damaging chemotherapy agents for inducing premature mitosis to exert anti-cancer effects, and WEE1 PROTACs using AZD1775 have also been developed to selectively degrade WEE1 in cancer cells 37. We first confirmed previously reported results that treatment of HEP3B hepatoma cancer cell lines with a proteasome inhibitor bortezomib stabilized WEE1 levels, confirming that WEE1 was regulated by ubiquitin-mediated proteasomal degradation in this cell line (Extended Data Figure 15a) 36. We next synthesized four DUBTACs linking AZD1775 to our OTUB1 recruiter EN523 through no linker (LEB-03-153), a C3 alkyl linker (LEB-03-144), a C5 alkyl linker (LEB-03-145), or a PEG linker (LEB-03-146) (Extended Data Figure 15b). LEB-03-144 and LEB-03-146 with the C3 alkyl linker and the PEG linker, respectively, showed significant WEE1 stabilization in HEP3B cells comparable to WEE1 levels observed with bortezomib treatment, whereas EN523 or AZD1775 treatment alone had no impact on WEE1 levels (Extended Data Figure 15c, 15d). While the therapeutic relevance of a WEE1 DUBTAC employing a WEE1 inhibitor remains to be seen, these data show additional mechanistic proof-of-concept for TPS using DUBTACs.
Conclusions
In this study, we discovered a covalent small-molecule recruiter EN523 for the K48-ubiquitin chain-specific DUB OTUB1. We demonstrated that this recruiter can be used incorporated into fully synthetic heterobifunctional DUBTACs by linking a DUB recruiter to protein targeting ligands to enable TPS of actively degraded target proteins in cells. We showed two successful examples of TPS with ΔF508-CFTR and WEE1. For ΔF508-CFTR, we also demonstrated that we not only heightened the levels of the mutant protein, but also improved cell surface chloride channel conductance of CFTR with our DUBTAC in combination with the potentiator ivacaftor, compared to lumacaftor and ivacaftor treatments. While we showed early validation of the DUBTAC platform here, there are many avenues for future exploration. These include further optimization of DUB recruiters against OTUB1 to improve their potency and proteome-wide selectivity, as well as the discovery of new recruiters against other candidate DUBs. For exploring optimization of CFTR DUBTACs, further improvement of the linker between lumacaftor and the DUB recruiter could improve potency and degree of CFTR stabilization. In addition, elucidating the mechanism, structural underpinnings, and kinetics in the formation of the ternary complex formed between CFTR and OTUB1 and understanding how CFTR is deubiquitinated by the DUBTAC will be important. Furthermore, better understanding of whether we are disrupting endogenous OTUB1 function would be important to understanding the mechanism and safety of DUBTACs.
Given our initial proof-of-concept for CFTR stabilization with a DUBTAC, there are many promising areas that could benefit from targeted deubiquitination of actively ubiquitinated and degraded proteins to provide therapeutic benefit. Targets that could benefit from a DUBTAC that already possess protein-targeting ligands include stabilizing BAX levels in the mitochondria to induce apoptosis, stabilizing STING for immunooncology applications, or stabilizing glucokinase in pancreatic cells for MODY2 38–41. Other targets that would benefit from a DUBTAC would be various tumor suppressors such as TP53, CDKN1A, CDKN1C, AXIN1, PTEN, and others that are actively ubiquitinated and degraded to maintain cancer cell proliferation 42. There are also many other genetic disorders beyond cystic fibrosis where mutations can lead to protein destabilization and ubiquitin-mediated degradation that can also be stabilized by DUBTACs for therapeutic benefit. These include glucocerebrosidase (GBA) mutations in Gaucher’s disease or Parkinson’s disease43 and phenylalanine hydroxylase (PAH) and fumarylacetoacetate hydroxylase (FAH) mutations in phenylketonuria 44. These disorders could directly benefit from TPS via DUBTACs. Additionally, targets for which pharmacological chaperones have been made as a potential therapeutic approach for destabilizing missense mutations could be coupled with the DUBTAC approach to improve therapeutic outcomes 45. In diseases caused by haploinsufficiency where loss of one copy of a gene leads to disease pathology, DUBTACs could potentially slow down the turnover rate of the protein to increase the levels of the protein to alleviate the disease 46. Finally, one can also envision DUBTAC platforms for other types of ubiquitin chains with roles beyond degradation such as signaling, protein localization, and modulation of protein-protein interactions. Overall, our study puts forth the discovery of DUB recruiters and shows proof-of-concept for the DUBTAC platform for TPS via induced proximity of a DUB with a target protein. In addition, our study underscores the utility of using chemoproteomics-enabled covalent ligand discovery platforms to facilitate development of unique induced proximity-based therapeutic modalities beyond targeted protein degradation.
Online Methods
Materials
Cysteine-reactive covalent ligand libraries were either previously synthesized and described or for the compounds starting with “EN” were purchased from Enamine, including EN523 30,33,47–50. Lumacaftor was purchased from Medchemexpress LLC.
Cell Culture
CFBE41o-4.7 ΔF508-CFTR Human CF Bronchial Epithelial cells were purchased from Millipore Sigma (SCC159). CFBE41o-4.7 ΔF508-CFTR Human CF Bronchial Epithelial cells were cultured in MEM (Gibco) containing 10% (v/v) fetal bovine serum (FBS) and maintained at 37 °C with 5% CO2.
Gel-Based ABPP
Recombinant OTUB1 (0.1μg/sample) was pre-treated with either DMSO vehicle or covalent ligand or DUBTACs at 37 °C for 30 min in 25 μL of PBS, and subsequently treated with of IA-Rhodamine (concentrations designated in figure legends) (Setareh Biotech) at room temperature for 1 h. The reaction was stopped by addition of 4×reducing Laemmli SDS sample loading buffer (Alfa Aesar). After boiling at 95 °C for 5 min, the samples were separated on precast 4–20% Criterion TGX gels (Bio-Rad). Probe-labeled proteins were analyzed by in-gel fluorescence using a ChemiDoc MP (Bio-Rad).
NJH-2-057 Probe Labeling of Recombinant OTUB1
Recombinant and pure OTUB1 protein (0.5 μg) per sample per replicate was suspended in 50 μL total PBS. 1 μL of either DMSO or NJH-2-075 (to give final concentrations of 50, 10, 1, and 0.1 μM) was added, followed by a 1.5 h incubation at 37 °C. Next, 7.8 μL of a solution composed of 9.4 μL of 5mM Azide-Fluor 545 (in DMSO), 112 μL of TBTA ligand (Stock 1.7 mM in 4 parts t-butanol + 1 part DMSO), 37.5 μL of 50 mM TCEP (in water), and 37.5 μL of 50 mM Copper (II) sulfate was added to each sample and the samples were incubated for 1 hour at room temperature. Following CuAAC, 30 μL of Laemmli Sample Buffer (4 x) was added to each sample, vortexed and boiled for 6 min at 95 °C. Samples were loaded on an SDS/PAGE gel and analyzed for in-gel fluorescence.
Deubiquitinase Activity Assay
Previously described methods were used to assess EN523 effects on OTUB1 activity 23. Recombinant OTUB1 (500 nM) was pre-incubated with DMSO or EN523 (50 μM) for 1 hr. To initiate assay pre-treated OTUB1 enzyme was mixed 1:1 with di-Ub reaction mix for final concentrations of 250 nM OTUB1, 1.5 μM di-Ub, 12.5 μM UBE2D1 and 5 mM DTT. The appearance of mono-Ub was monitored by Western blotting over time by removing a portion of the reaction mix and adding Laemmli’s buffer to terminaye reaction. Blot shown is a representative gel from n=3 biologically independent experiments/group.
Bio-NMR Analysis of EN523-OTUB1 Interactions
We recorded all NMR spectra on a Bruker 600 MHz spectrometer, equipped with a 5 mm QCI-F cryo probe with z-gradient, and kept the temperature constant at 298K during all experiments. To probe compound and E2 ligase binding to OTUB1, we recorded 1H-1D and 13C-SOFAST-HMQC experiments. We used 3 mm NMR tubes filled with 160 μL of 50 μM {U}-2H,1H/13C-methyl-Ile/Leu/Val/Ala(ILVA),{U}-15N labeled OTUB1, 25 mM d-Tris, pH 7.5, 150 mM NaCl, 5% D2O (to lock), 100 μM DSS (internal standard), 75 μM EN-523 (dissolved in 100% d6-DMSO; for compound binding study) and/or 100 μM E2 D2 / Ub-E2 D2 (for ligase binding studies). To allow for complete binding of the compound to OTUB1, we chose an incubation period of ~40 hours. We also recorded reference spectra with the adequate volumes of pure d6-DMSO and/or E2 buffer to compensate for solvent induced effects, and repeated experiments after 40 hours to make sure that any spectral changes were not related to protein oxidation.
Labeling of Endogenous OTUB1 in HEK293T Cells with NJH-2-075 Probe
One plate of 70% confluent HEK293T cells per condition per replicate were treated with either DMSO vehicle or NJH-02-075 (50 μM) for 2 hours. Cells were harvested by scraping, suspended in 600 μL of PBS, lysed by probe sonication, and centrifuged for 10 min at 5000 rpm to remove debris. Lysate was normalized to 3.1 mg/mL and 85 μL removed for Western blotting analysis of input. 500 μL of lysate was then incubated for 1 hour at room temperature with 10 μL of 5 mM biotin picolyl azide (in water), 10 μL of 50mM TCEP (in water), 30 μL TBTA ligand (Stock 1.7 mM in 4 parts t-butanol + 1 part DMSO), and 10 μL of 50 mM Copper (II) sulfate. Following CuAAC, precipitated proteins were washed 3 x with cold methanol and resolubilized in 200 μL 1.2% SDS/PBS. To ensure solubility, proteins were heated to 90 °C for 5 min following resuspension. 1 mL of PBS was then added to each sample, followed by 50 μL of high-capacity streptavidin beads. Samples were then incubated overnight on a rocker at 4 °C. The following morning the samples were warmed to room temperature, and non-specific binding proteins were washed away with 3 x PBS washes followed by 3 x water washes. Beads were then resuspended in 100 μL PBS and 30 μL Laemmli Sample Buffer (4 x) and boiled for 13 min at 95 °C. Samples were vortexed and loaded onto an SDS/PAGE gel along with saved input samples for Western blotting analysis.
Western Blotting
Proteins were resolved by SDS/PAGE and transferred to nitrocellulose membranes using the Trans-Blot Turbo transfer system (Bio-Rad). Membranes were blocked with 5% BSA in Tris-buffered saline containing Tween 20 (TBS-T) solution for 30 min at RT, washed in TBS-T, and probed with primary antibody diluted in recommended diluent per manufacturer overnight at 4°C. After 3 washes with TBS-T, the membranes were incubated in the dark with IR680- or IR800-conjugated secondary antibodies at 1:10,000 dilution in 5 % BSA in TBS-T at RT for 1 h. After 3 additional washes with TBST, blots were visualized using an Odyssey Li-Cor fluorescent scanner. The membranes were stripped using ReBlot Plus Strong Antibody Stripping Solution (EMD Millipore) when additional primary antibody incubations were performed. Antibodies used in this study were CFTR (Cell Signaling Technologies, Rb mAb #78335, Figures 3 and 4), CFTR (R&D Systems, Ms mAb, #MAB25031, Extended Data Figure 3), CFTR (Millipore, Ms mAb, #MAB3484, Extended Data Figure 3), CFTR (Prestige, Rb pAb, #HPA021939, Extended Data Figure 3), GAPDH (Proteintech, Ms mAb, #60004-1-Ig), OTUB1 (Abcam, Rb mAb, #ab175200, [EPR13028(B)]), and CTNNB1 (Cell Signaling Technologies, Rb mAb, #8480), and WEE1 (Cell Signaling Technologies, #4936).
Native Mass Spectrometry analysis of Ternary Complex Formation
Native mass spectrometry experiments were performed on a Thermo QE UHMR equipped with a nano-electrospray ionization source (Advion TriVersa NanoMate). Recombinant OTUB1 was first buffer exchanged into 150 mM ammonium acetate, 100 μM MgCl2, and 100 μM ATP at pH 6.7. 4 μM OTUB1 was then pre-incubated at room temperature for 24 hours with either DMSO, EN523 (100 μM), or NJH-2-057 (100 μM). After 24 hours, 4 μM CFTR, in the same buffer, was added to the OTUB1 solution, for final concentrations of 2 μM of each protein with either DMSO or 50 μM compound. The solution was then allowed to incubate for 30 minutes prior to analysis on the mass spectrometer. Mass spectra were recorded in positive ion mode with a mass range of 1000-8000 m/z. Each spectrum was then deconvoluted and relevant peaks were integrated to determine % ternary complex formed. All experiments were performed in triplicate.
IsoTOP-ABPP Chemoproteomic Experiments
IsoTOP-ABPP studies were done as previously reported 15,30,51. Our aggregate chemoproteomic data analysis of DUBs were obtained from 455 distinct isoTOP-ABPP experiments performed in the Nomura Research Group. These data are aggregated from various human cell lines, including 231MFP, A549, HeLa, HEK293T, HEK293A, UM-Chor1, PaCa2, PC3, HUH7, NCI-H460, THP1, SKOV3, U2OS, and K562 cells. Some of the chemoproteomic data have been previously reported as part of other studies 33,47–49,51–57. All of the isoTOP-ABPP datasets were prepared as described below using the IA-alkyne probe. Cells were lysed by probe sonication in PBS and protein concentrations were measured by BCA assay. Cells were treated for 4 h with either DMSO vehicle or EN4 (from 1,000x DMSO stock) before cell collection and lysis. Proteomes were subsequently labeled with IA-alkyne labeling (100 μM for DUB ligandability analysis and 200 μM for profiling cysteine-reactivity of NJH-2-057) for 1 h at room temperature. CuAAC was used by sequential addition of tris(2-carboxyethyl)phosphine (1 mM, Strem, 15-7400), tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (34 μM, Sigma, 678937), copper(II) sulfate (1 mM, Sigma, 451657) and biotin-linker-azide—the linker functionalized with a tobacco etch virus (TEV) protease recognition sequence as well as an isotopically light or heavy valine for treatment of control or treated proteome, respectively. After CuAAC, proteomes were precipitated by centrifugation at 6,500g, washed in ice-cold methanol, combined in a 1:1 control:treated ratio, washed again, then denatured and resolubilized by heating in 1.2% SDS–PBS to 80 °C for 5 min. Insoluble components were precipitated by centrifugation at 6,500g and soluble proteome was diluted in 5 ml 0.2% SDS–PBS. Labeled proteins were bound to streptavidin-agarose beads (170 μl resuspended beads per sample, Thermo Fisher, 20349) while rotating overnight at 4 °C. Bead-linked proteins were enriched by washing three times each in PBS and water, then resuspended in 6 M urea/PBS, and reduced in TCEP (1 mM, Strem, 15-7400), alkylated with iodoacetamide (18 mM, Sigma), before being washed and resuspended in 2 M urea/PBS and trypsinized overnight with 0.5 μg /μL sequencing grade trypsin (Promega, V5111). Tryptic peptides were eluted off. Beads were washed three times each in PBS and water, washed in TEV buffer solution (water, TEV buffer, 100 μM dithiothreitol) and resuspended in buffer with Ac-TEV protease (Invitrogen, 12575-015) and incubated overnight. Peptides were diluted in water and acidified with formic acid (1.2 M, Fisher, A117-50) and prepared for analysis.
IsoTOP-ABPP Mass Spectrometry Analysis
Peptides from all chemoproteomic experiments were pressure-loaded onto a 250 μm inner diameter fused silica capillary tubing packed with 4 cm of Aqua C18 reverse-phase resin (Phenomenex, 04A-4299), which was previously equilibrated on an Agilent 600 series high-performance liquid chromatograph using the gradient from 100% buffer A to 100% buffer B over 10 min, followed by a 5 min wash with 100% buffer B and a 5 min wash with 100% buffer A. The samples were then attached using a MicroTee PEEK 360 μm fitting (Thermo Fisher Scientific p-888) to a 13 cm laser pulled column packed with 10 cm Aqua C18 reverse-phase resin and 3 cm of strong-cation exchange resin for isoTOP-ABPP studies. Samples were analyzed using an Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) using a five-step Multidimensional Protein Identification Technology (MudPIT) program, using 0, 25, 50, 80 and 100% salt bumps of 500 mM aqueous ammonium acetate and using a gradient of 5–55% buffer B in buffer A (buffer A: 95:5 water:acetonitrile, 0.1% formic acid; buffer B 80:20 acetonitrile:water, 0.1% formic acid). Data were collected in data-dependent acquisition mode with dynamic exclusion enabled (60 s). One full mass spectrometry (MS1) scan (400–1,800 mass-to-charge ratio (m/z)) was followed by 15 MS2 scans of the nth most abundant ions. Heated capillary temperature was set to 200 °C and the nanospray voltage was set to 2.75 kV.
Data were extracted in the form of MS1 and MS2 files using Raw Extractor v.1.9.9.2 (Scripps Research Institute) and searched against the Uniprot human database using ProLuCID search methodology in IP2 v.3-v.5 (Integrated Proteomics Applications, Inc.) 58. Cysteine residues were searched with a static modification for carboxyaminomethylation (+57.02146) and up to two differential modifications for methionine oxidation and either the light or heavy TEV tags (+464.28596 or +470.29977, respectively). Peptides were required to be fully tryptic peptides and to contain the TEV modification. ProLUCID data were filtered through DTASelect to achieve a peptide false-positive rate below 5%. Only those probe-modified peptides that were evident across two out of three biological replicates were interpreted for their isotopic light to heavy ratios. For those probe-modified peptides that showed ratios greater than two, we only interpreted those targets that were present across all three biological replicates, were statistically significant and showed good quality MS1 peak shapes across all biological replicates. Light versus heavy isotopic probe-modified peptide ratios are calculated by taking the mean of the ratios of each replicate paired light versus heavy precursor abundance for all peptide-spectral matches associated with a peptide. The paired abundances were also used to calculate a paired sample t-test P value in an effort to estimate constancy in paired abundances and significance in change between treatment and control. P values were corrected using the Benjamini–Hochberg method.
Knockdown studies
RNA interference was performed using siRNA purchased from Dharmacon. CFBE41o-4.7 cells were seeded at 400,000 cells per 6 cm plate and allowed to adhere overnight. Cells were transfected with 33 nM of either nontargeting (ON-TARGETplus Non-targeting Control Pool, Dharmacon #D-001810-10-20) or anti-CFTR siRNA (Dharmacon, custom) using 8 μL of transfection reagent: either DharmaFECT 1 (Dharmacon #T-2001-02), DharmaFECT 4 (Dharmacon, T-2004-02) or Lipofectamine 2000 (ThermoFisher #11668027). Transfection reagent was added to OPTIMEM (ThermoFisher #31985070) media, allowed to incubate for 5 min at room temperature. Meanwhile siRNA was added to an equal amount of OPTIMEM. Solutions of transfection reagent and siRNA in OPTIMEM were then combined and allowed to incubate for 30 minutes at room temperature. These combined solutions were diluted with complete MEM to provide 33nM siRNA and 8 μL of transfection reagent per 4 mL MEM, and the media exchanged. Cells were incubated with transfection reagents for 24h, at which point the media replaced with media containing DMSO or 10 μM NJH-2-057 and incubated for another 24h. Cells were then harvested, and protein abundance analyzed by Western blotting.
Transepithelial Conductance Assays in Human Bronchial Epithelial Cells
Human bronchial epithelial cells (HBECs) from cystic fibrosis (CF) patients bearing the ΔF508-CFTR mutation were cultured at 37°C and 5% CO2 in Bronchial Epithelial Cell Growth Basal Medium (BEGM) with SingleQuots Supplements and Growth Factors (Lonza, #CC-3170). Cells were maintained in cell culture flasks (Corning, #430641U) for one week and media was replaced every two to three days. Cells were washed with Dulbecco’s phosphate buffered saline (Thermo Fisher Scientific, #14040141), trypsinized for five to ten minutes with 0.05% Trypsin-EDTA (Thermo Fisher Scientific, #25300120), after which Trypsin Neutralizing Solution (TNS, Thermo Fisher Scientific, #R002100) was added. Cells were pelleted at 300 x g for five minutes and resuspended in BEGM with Dulbecco’s modified Eagle medium (DMEM, Thermo Fisher Scientific, #11965092) and plated at one million cells per plate in 24-well transwell plates (Corning, #3526). Cells were grown submerged in BEGM with DMEM for one week with media changed every two to three days, at which time they were taken to air liquid interface (ALI) and grown another two weeks before being ready to use.
Cells were treated with either DMSO vehicle, 10 μM VX-809, or 10 μM DUBTAC 24 hours before the experiment. Cells were then submerged in Ham’s F12 buffer (Thermo Fisher Scientific, #21700075) with 20 mM HEPES (Thermo Fisher Scientific, #15630080) at pH 7.4 and mounted into the assay system. Transepithelial resistance was recorded using a 24-channel transepithelial current clamp amplifier (TECC-24, EP Design, Bertem, Belgium). Resistance measurements were taken at intervals of approximately six minutes. Four values were taken to determine baseline resistance, and another four measurements were taken after each of the following additions: 10 μM Amiloride (Millipore Sigma, #A7410) added apically, 20 μM Forskolin (Millipore Sigma, #F6886) added apically, and 0.5 μM VX-770 added both apically and basolaterally. CFTR Inhibitor 172 (Millipore Sigma, #219672) was then added and a final six measurements taken. Transepithelial conductance (G) was calculated from resistance measurements (G = 1/R). Chloride ion transport across the epithelial monolayer is mediated by CFTR, and activation or inhibition of functional CFTR therefore causes changes in transepithelial conductance. In this way, ΔG can be used to measure functional CFTR expression and the functional rescue of CFTR through compound addition.
Quantitative TMT Proteomics Analysis
Quantitative TMT-based proteomic analysis was performed as previously described 30. Acquired MS data was processed using Proteome Discoverer v. 2.4.0.305 software (Thermo) utilizing Mascot v 2.5.1 search engine (Matrix Science, London, UK) together with Percolator validation node for peptide-spectral match filtering 59. Data was searched against Uniprot protein database (canonical human sequences, EBI, Cambridge, UK) supplemented with sequences of common contaminants. Peptide search tolerances were set to 10 ppm for precursors, and 0.8 Da for fragments. Trypsin cleavage specificity (cleavage at K, R except if followed by P) allowed for up to 2 missed cleavages. Carbamidomethylation of cysteine was set as a fixed modification, methionine oxidation, and TMT-modification of N-termini and lysine residues were set as variable modifications. Data validation of peptide and protein identifications was done at the level of the complete dataset consisting of combined Mascot search results for all individual samples per experiment via the Percolator validation node in Proteome Discoverer. Reporter ion ratio calculations were performed using summed abundances with most confident centroid selected from 20 ppm window. Only peptide-to-spectrum matches that are unique assignments to a given identified protein within the total dataset are considered for protein quantitation. High confidence protein identifications were reported using a Percolator estimated <1% false discovery rate (FDR) cut-off. Differential abundance significance was estimated using ANOVA with Benjamini-Hochberg correction to determine adjusted p-values.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code Availability Statement
Data processing and statistical analysis algorithms from our lab can be found on our lab’s Github site: https://github.com/NomuraRG, and we can make any further code from this study available at reasonable request.
Chemical Synthesis and Characterization
Starting materials, reagents and solvents were purchased from commercial suppliers and were used without further purification unless otherwise noted. All reactions were monitored by TLC (TLC Silica gel 60 F254, Sepulco Millipore Sigma). Reaction products were purified by flash column chromatography using a Biotage Isolera with Biotage Sfar® or Silicycle normal-phase silica flash columns (5 g, 10 g, 25 g, or 40 g). 1H NMR and 13C NMR spectra were recorded on a 400 MHz Bruker Avance I spectrometer or a 600 MHz Bruker Avance III spectrometer equipped with a 5 mm 1H/BB Prodigy cryo-probe. Chemical shifts are reported in parts per million (ppm, δ) downfield from tetramethylsilane (TMS). Coupling constants (J) are reported in Hz. Spin multiplicities are described as br (broad), s (singlet), d (doublet), t (triplet), q (quartet) and m (multiplet).
General Procedure A:
Carboxylic acid (1.0 Eq.) was dissolved in DCM (0.1 M). An amine (1.25 Eq.) was added, followed by DIEA (4.0 Eq.), HOBt (0.2 Eq.) and EDCI (2.0 Eq.). The reaction mixture was stirred overnight at rt, water was added, and the mixture extracted three times with DCM. Combined organic extracts were washed with 1M HCl, washed with brine, dried over sodium sulfate, and concentrated. The crude product was purified by silica gel chromatography to provide the amide.
General Procedure B:
Boc-protected amine was dissolved in DCM (0.1 M), and TFA was added to give a 1:2 TFA:DCM ratio. The solution was allowed to stir for 1h. The volatiles were then evaporated, and the resulting oil redissolved in DCM and treated with aqueous saturated NaHCO3. The resulting mixture was then extracted with DCM three times, the combined organic extracts dried over Na2SO4, and concentrated to provide the amine without further purification.
General Procedure C:
Tert-butyl ester such as Intermediate 3 (30 mg, 0.086 mmol, 3.0 Eq) was dissolved in DCM (600 μL). TFA (300 μL) was added and the solution stirred for 1h. Volatiles were evaporated under vacuum, and DCM (1 mL) was added and evaporated to give the carboxylic acid intermediate, though some excess TFA remained. This intermediate was dissolved in DMF (500 μL) and DIEA (150 μL, 30 Eq.) and the appropriate amine (0.029 mmol, 1.0 Eq) were added, followed by HATU (30 mg, 0.079 mmol, 2.7 Eq.). The reaction mixture was allowed to stir for 1h at rt. Water was added, and the mixture extracted three times with EtOAc for CFTR DUBTACS, or 4:1 CHCl3:IPA for WEE1 DUBTACs. Organic extracts were combined, washed with brine, dried over sodium sulfate, and concentrated. Crude residues were purified by silica gel chromatography to provide the final compounds.
General Procedure D:
To a solution of the appropriate bromide dissolved in dioxane, N,N’-dimethylethylenediamine (0.25 eq), K2CO3 (3.0 eq), CuI (0.1 eq), and the appropriate amide coupling partner (1.0 eq) were added. The reaction mixture was degassed, the atmosphere exchanged for nitrogen, and stirred at 100 °C overnight. Saturated NH4Cl was added to the completed reaction mixture once cooled, which was stirred for 20 minutes, then filtered through celite and the celite pad was washed with EtOAc. The mixture was extracted with EtOAc three times, washed with brine twice, and dried by NaSO4, before concentration in vacuo. Resulting crude mixtures were purified via silica gel column chromatography.
General Procedure E:
The appropriate amine was dissolved in THF and water (2:1 THF:H2O) with potassium carbonate (3.0 eq). Benzyl chloroformate (1-2 eq) was added dropwise to the reaction mixture, which was then stirred vigorously overnight at room temperature. Water was added and the mixture extracted with EtOAc three times. Organic extracts were combined, washed with brine twice, concentrated and the resulting crude purified using flash column chromatography.
General Procedure F:
The coupled product was dissolved in DCM, followed by a dropwise addition of trifluoroacetic acid (1:2 TFA:DCM) until consumption of starting material was observed via TLC (15-30 min). The mixture was then washed with DCM twice and immediately used without further purification.
General Procedure G:
Pd/C (10% wt.) was added to a mixture of the Cbz-protected compound in EtOH (0.2 M), and the atmosphere was exchanged for H2 (balloon). The reaction mixture was stirred vigorously overnight, before being diluted with DCM, filtered through a syringe filter (0.45 um), concentrated, and purified using silica gel column chromatography.
General Procedure H:
The amine starting material was dissolved in DCM on ice. TEA (3.0 eq) and acryloyl chloride (1.5 eq) were then added to the reaction mixture until consumption of the starting material was observed by TLC (0.5 – 2 hrs). Water was added, and the reaction mixture was extracted with DCM three times. Organic extracts were combined, washed with H2O then brine, concentrated, and purified via silica gel column chromatography.
SYNTHESIS OF CFTR DUBTACs
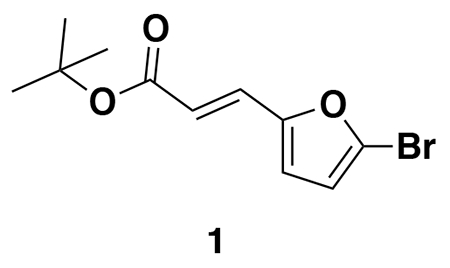
tert-butyl (E)-3-(5-bromofuran-2-yl)acrylate (1): tert-butyl diethylphosphonoacetate (971 mg, 0.908 mL, 3.85 mmol) was dissolved in THF (22 mL) and the solution cooled to 0 °C. Then, 5-bromofuran-2-carbaldehyde (613 mg, 3.50 mmol) was added portion-wise over 5 minutes. The reaction was stirred for 20 minutes at 0 °C as a gummy solid precipitated. Water was added and the resulting mixture was extracted with EtOAc three times. Combined organic extracts were washed with brine, dried over Na2SO4, and concentrated. The crude residue was purified by silica gel chromatography (0-15% EtOAc/Hex) to provide the title compound as a light yellow oil (782 mg, 2.86 mmol, 82%).
1H NMR (400 MHz, CDCl3) δ 7.26 (d, J = 15.7 Hz, 1H), 6.55 (d, J = 3.5 Hz, 1H), 6.42 (d, J = 3.4 Hz, 1H), 6.29 (d, J = 15.7 Hz, 1H), 1.55 (s, 9H).

benzyl (E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-3-oxopiperazine-1-carboxylate (2): tert-butyl (E)-3-(5-bromofuran-2-yl)acrylate (1.62 g, 5.94 mmol) was dissolved in dioxane (30 mL) and benzyl 3-oxopiperazine-1-carboxylate (1.4 g, 5.94 mmol), K2CO3 (2.46 g, 17.8 mmol), N,N′-dimethyldiaminoethane (0.167 mL, 1.49 mmol), and CuI (114 mg, 0.59 mmol) were added. The mixture was stirred under nitrogen at reflux for 40 h, then cooled to rt. 5 mL saturated aq. NH4Cl was added and the mixture stirred for 30 min. Then the mixture was diluted in EtOAc, filtered through celite, water was added, the mixture partitioned, and the aqueous layer extracted with EtOAc. The extracts were combined, washed with brine, dried over Na2SO4, concentrated, and purified by silica gel chromatography (0-35% EtOAc/Hex) to provide the title compound as an orange oil (1.95 g, 4.59 mmol, 77%).
LC/MS: [M+2H-tBu]+ m/z calc. 371.18, found 373.1.
1H NMR (400 MHz, DMSO) δ 7.45 – 7.24 (m, 6H), 6.98 (s, 1H), 6.57 (s, 1H), 6.08 (dd, J = 15.7, 3.4 Hz, 1H), 5.14 (dd, J = 4.4, 2.3 Hz, 2H), 4.22 (s, 2H), 4.01 (s, 2H), 3.77 (s, 2H), 1.47 (s, 9H).
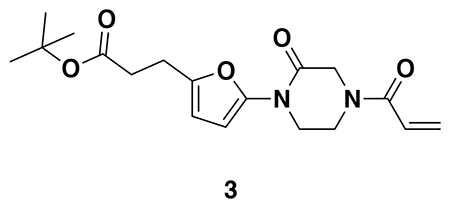
tert-butyl 3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoate (Intermediate (3): benzyl (E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-3-oxopiperazine-1-carboxylate (1.95 g, 4.59 mmol) was dissolved in EtOH (25 mL) and Pd/C (200 mg, 10% wt. Pd) was added. The reaction was placed under an atmosphere of H2 and stirred vigorously overnight, before being filtered through celite twice and then concentrated. The crude product was then redissolved in DCM (25 mL), cooled to 0 °C and treated with TEA (1.28 mL, 9.18 mmol) before a solution of acryloyl chloride (445 μL, 5.51 mmol) in DCM (5 mL) was added over 2 minutes. After stirring for 20 min, water was added, and the mixture extracted with DCM three times. Combined organic extracts were washed with brine, dried over Na2SO4, concentrated, and the resulting crude oil was purified by silica gel chromatography (0-75% EtOAc/Hex) to obtain the title compound as a light yellow oil (846 mg, 2.43 mmol, 53% over two steps).
1H NMR (400 MHz, CDCl3) δ 6.64 – 6.46 (m, 1H), 6.41 (dd, J = 16.7, 2.0 Hz, 1H), 6.29 (d, J = 3.2 Hz, 1H), 6.04 (d, J = 3.3 Hz, 1H), 5.82 (dd, J = 10.2, 2.0 Hz, 1H), 4.42 (d, J = 24.9 Hz, 2H), 4.06 – 3.82 (m, 4H), 2.88 (t, J = 7.8 Hz, 2H), 2.54 (d, J = 7.6 Hz, 2H), 1.44 (s, 9H).
LC/MS: [M+2H-tBu]+ m/z calc. 293.1, found 293.1.
Note: Intermediate 3 is prone to decomposition, likely through polymerization. Care should be taken to store at −20 °C. Attempts to dry thoroughly (e.g. leaving on vacuum at rt overnight) occasionally led to decomposition of ~50% of the material.
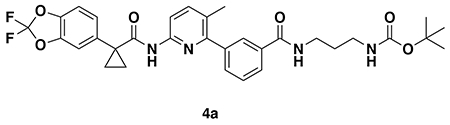
tert-butyl (3-(3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamido)propyl)carbamate (4a): Lumacaftor (3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzoic acid) (18 mg, 0.04 mmol), tert-butyl (3-aminopropyl)carbamate (14 mg, 0.08 mmol), DIEA (35 μL, 0.20 mmol), and HOBt (5.4 mg, 0.04 mmol) were dissolved in DCM (1 mL), followed by the addition of EDCI HCl (15 mg, 0.05 mmol). The reaction was stirred at rt for 2 days before water was added, the mixture partitioned, and the aqueous layer extracted with DCM twice. The combined organic extracts were washed with brine, dried over Na2SO4, concentrated, and the resulting crude oil was purified by silica gel chromatography (0-60% EtOAc/Hex) to obtain the 4a as a clear oil (23 mg, 0.038 mmol, 94%).
1H NMR (400 MHz, CDCl3) δ 8.13 (d, J = 8.4 Hz, 1H), 7.95 (s, 1H), 7.88 (d, J = 7.6 Hz, 1H), 7.74 (s, 1H), 7.62 (d, J = 8.5 Hz, 1H), 7.60 – 7.49 (m, 2H), 7.34 (s, 1H), 7.30 – 7.18 (m, 2H), 7.11 (d, J = 8.2 Hz, 1H), 4.96 (s, 1H), 3.54 (q, J = 6.2 Hz, 2H), 3.27 (q, J = 6.3 Hz, 2H), 2.31 (s, 3H), 1.78 (q, J = 3.9 Hz, 2H), 1.76 – 1.70 (m, 2H), 1.47 (s, 9H), 1.19 (q, J = 3.9 Hz, 2H).
LC/MS: [M+H]+ m/z calc. 609.24, found 609.3.
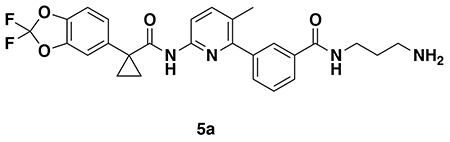
N-(3-aminopropyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (5a): Intermediate 4a (23 mg, 0.038 mmol) was dissolved in DCM (1 mL) and TFA (1 mL) was added and the solution stirred for 2 hours. The volatiles were then evaporated and the resulting oil redissolved in DCM and treated with aqueous saturated NaHCO3. The resulting mixture was then extracted with DCM three times, combined organic extracts dried over Na2SO4, concentrated to provide 5a (15 mg, 0.029 mmol, 78%) as a colorless oil which was used in the next step without further purification.
1H NMR (400 MHz, CDCl3) δ 10.73 (s, 1H), 8.96 (s, 1H), 8.66 (t, J = 5.7 Hz, 1H), 7.95 – 7.85 (m, 3H), 7.79 – 7.66 (m, 2H), 7.60 (d, J = 7.6 Hz, 1H), 7.56 – 7.49 (m, 2H), 7.41 – 7.30 (m, 2H), 3.33 (q, J = 6.4 Hz, 2H), 2.88 – 2.77 (m, 2H), 2.21 (s, 3H), 1.79 (p, J = 6.9 Hz, 2H), 1.52 (dd, J = 4.9, 2.5 Hz, 2H), 1.19 – 1.15 (m, 2H).
LC/MS: [M+H]+ m/z calc. 509.19, found 509.2.
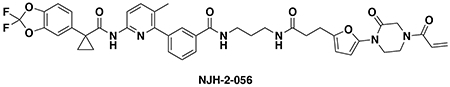
N-(3-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanamido)propyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (NJH-2-056): Intermediate 3 (tert-butyl 3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoate) (14 mg, 0.04 mmol) was dissolved in DCM (0.6 mL) and TFA (0.3 mL) was added and the solution was stirred for 1 h at rt until starting material was consumed as monitored by TLC. Volatiles were evaporated, DCM was added and evaporated again. The residue was dissolved in DCM (1.5 mL) and DIEA (140 μL, 0.80 mmol) was added followed by N-(3-aminopropyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (5.4 mg, 0.1 mmol). EDCI HCl (15 mg, 0.08 mmol) was then added, and the mixture stirred for 16h. Water was added and the resulting suspension was extracted with DCM three times. Combined organic extracts were washed with brine and dried over Na2SO4 before being concentrated. The crude residue was purified by silica gel chromatography (0-5% MeOH/DCM) to obtain NJH-2-056 (9.5 mg, 0.012 mmol, 30%) as a white powder following lyophilization from 1:1 water:acetonitrile (2 mL).
1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 8.4 Hz, 1H), 7.93 – 7.87 (m, 1H), 7.83 (dt, J = 7.5, 1.6 Hz, 1H), 7.72 (s, 1H), 7.59 (d, J = 8.5 Hz, 1H), 7.57 – 7.45 (m, 2H), 7.29 (s, 1H), 7.23 (dd, J = 8.2, 1.8 Hz, 1H), 7.19 (d, J = 1.7 Hz, 1H), 7.07 (d, J = 8.1 Hz, 1H), 6.50 (s, 1H), 6.43 – 6.33 (m, 2H), 6.19 (d, J = 3.2 Hz, 1H), 6.07 (d, J = 3.3 Hz, 1H), 5.81 (d, J = 10.1 Hz, 1H), 4.47 – 4.31 (m, 2H), 4.04 – 3.78 (m, 4H), 3.36 (q, J = 6.2 Hz, 2H), 3.32 – 3.23 (m, 2H), 2.96 (t, J = 7.2 Hz, 2H), 2.55 (t, J = 7.2 Hz, 2H), 2.26 (s, 3H), 1.74 (q, J = 3.9 Hz, 2H), 1.69 – 1.58 (m, 2H), 1.16 (q, J = 3.9 Hz, 2H).
13C NMR (151 MHz, CDCl3) δ 172.5, 171.8, 167.4, 165.0, 155.5, 149.8, 148.9, 145.0, 144.1, 143.6, 141.0, 140.2, 134.9, 134.6, 131.8, 131.7, 130.0, 128.5, 127.8, 127.0, 126.6, 126.5, 126.3, 112.9, 112.4, 110.2, 107.6, 101.3, 36.0, 35.9, 35.2, 31.2, 29.5, 24.4, 19.2, 17.2.
HRMS: [M+H]+ m/z calc. 783.2949, found 783.2954.
Synthesis of NJH-2-132
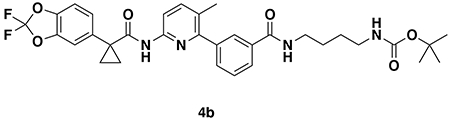
tert-butyl (4-(3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamido)butyl)carbamate (4b): Lumacaftor (100 mg, 0.22 mmol), tert-butyl (4-aminobutyl)carbamate were reacted according to General Procedure A and purified by silica gel chromatography (0-60% EtOAc/Hex) to obtain 4b as a clear colorless oil (128 mg, 0.20 mmol, 93%).
1H NMR (400 MHz, CDCl3) δ 8.14 (d, J = 8.4 Hz, 1H), 7.87 (s, 1H), 7.83 (d, J = 7.7 Hz, 1H), 7.74 (s, 1H), 7.63 (d, J = 8.5 Hz, 1H), 7.57 (dt, J = 7.7, 1.5 Hz, 1H), 7.51 (t, J = 7.6 Hz, 1H), 7.27 (dd, J = 8.1, 1.8 Hz, 1H), 7.23 (d, J = 1.7 Hz, 1H), 7.12 (d, J = 8.2 Hz, 1H), 6.58 (s, 1H), 4.68 (s, 1H), 3.52 (q, J = 6.4 Hz, 2H), 3.20 (q, J = 6.6 Hz, 2H), 2.29 (s, 3H), 1.79 (q, J = 3.9 Hz, 2H), 1.71 – 1.67 (m, 2H), 1.65 (s, 9H), 1.65 – 1.57 (m, 2H), 1.20 (q, J = 3.9 Hz, 2H).
LC/MS: [M+H]+ m/z calc. 623.3, found 623.3.
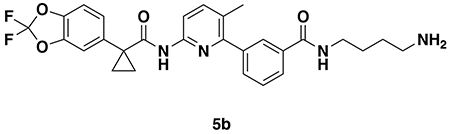
N-(4-aminobutyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (5b): 4b (128 mg, 0.20 mmol)was deprotected according to General Procedure B to provide the amine 5b (104 mg, 0.20 mmol, quant.) as a colorless oil.
1H NMR (400 MHz, CDCl3) δ 8.13 (dd, J = 8.4, 1.7 Hz, 1H), 7.85 (tt, J = 8.5, 1.8 Hz, 1H), 7.81 (dt, J = 7.6, 1.6 Hz, 1H), 7.73 (s, 1H), 7.62 (dd, J = 8.5, 2.1 Hz, 1H), 7.56 (ddt, J = 7.7, 2.9, 1.5 Hz, 1H), 7.50 (td, J = 7.6, 3.0 Hz, 1H), 7.27 (dd, J = 8.2, 1.8 Hz, 1H), 7.22 (t, J = 1.8 Hz, 1H), 7.11 (d, J = 8.1 Hz, 1H), 7.03 (d, J = 5.3 Hz, 1H), 3.57 – 3.46 (m, 2H), 3.27 (d, J = 6.7 Hz, 1H), 2.80 (t, J = 6.7 Hz, 1H), 2.28 (d, J = 2.5 Hz, 3H), 1.98 (d, J = 1.4 Hz, 1H), 1.86 (s, 1H), 1.79 (q, J = 3.9 Hz, 2H), 1.72 (dd, J = 8.1, 6.3 Hz, 1H), 1.63 – 1.53 (m, 1H), 1.20 (qd, J = 4.0, 1.1 Hz, 2H).
LC/MS: [M+H]+ m/z calc. 523.2, found 523.2.
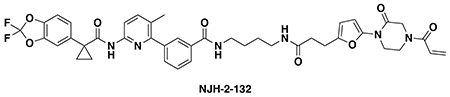
N-(4-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanamido)butyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (NJH-2-132): Intermediate 3 (tert-butyl 3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoate) (30 mg, 0.086 mmol) was dissolved in DCM (0.6 mL) and TFA (0.3 mL) was added and the solution stirred for 1 h until starting material was consumed. Volatiles were evaporated, DCM was added and evaporated again. The residue was dissolved in DCM (1.5 mL) and DIEA (150 μL, 0.86 mmol) was added followed by N-(4-aminobutyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (5b) (15 mg, 0.029 mmol). HATU (30mg, 0.079 mmol) was then added and the mixture stirred for 16h. Water was added and the resulting suspension was extracted with DCM three times. Combined organic extracts were washed brine and dried over sodium sulfate before being concentrated. The crude residue was purified by silica gel chromatography (0-5% MeOH/DCM) to obtain the title compound (9.5 mg, 0.012 mmol, 30%) as a white solid.
1H NMR (400 MHz, CDCl3) δ 8.12 (d, J = 8.4 Hz, 1H), 7.88 (t, J = 1.8 Hz, 1H), 7.84 (dt, J = 7.6, 1.6 Hz, 1H), 7.74 (s, 1H), 7.62 (d, J = 8.5 Hz, 1H), 7.56 (dt, J = 7.7, 1.5 Hz, 1H), 7.51 (t, J = 7.6 Hz, 1H), 7.26 (dd, J = 8.2, 1.7 Hz, 1H), 7.22 (d, J = 1.7 Hz, 1H), 7.11 (d, J = 8.2 Hz, 1H), 6.80 (s, 1H), 6.53 (d, J = 24.7 Hz, 1H), 6.41 (dd, J = 16.7, 2.0 Hz, 1H), 6.20 (d, J = 3.2 Hz, 1H), 6.07 (d, J = 3.3 Hz, 2H), 5.83 (dd, J = 10.2, 2.0 Hz, 1H), 4.38 (d, J = 28.2 Hz, 2H), 4.07 – 3.79 (m, 4H), 3.73 (tt, J = 9.8, 4.9 Hz, 1H), 3.45 (q, J = 6.4 Hz, 2H), 3.27 (q, J = 6.2 Hz, 2H), 3.20 (qd, J = 7.4, 3.4 Hz, 1H), 2.94 (q, J = 6.1, 5.0 Hz, 2H), 2.52 (t, J = 7.2 Hz, 2H), 2.28 (s, 3H), 1.77 (q, J = 3.9 Hz, 2H), 1.63 – 1.51 (m, 2H), 1.19 (q, J = 3.9 Hz, 2H).
13C NMR (151 MHz, CDCl3) δ 171.8, 167.4, 165.0, 155.5, 149.9, 148.9, 144.7, 144.1, 143.6, 141.0, 140.2, 134.9, 134.7, 133.4, 131.8, 131.7, 128.5, 127.6, 127.0, 126.6, 126.4, 112.9, 112.4, 110.2, 107.4, 101.2, 55.5, 43.5, 39.6, 39.0, 34.9, 31.2, 26.8, 26.7, 24.3, 19.2, 18.6, 17.2, 17.2, 12.5.
HRMS (ESI): m/z calc. 797.3032, found 797.3109.
Synthesis of NJH-2-057
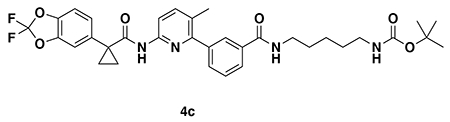
tert-butyl (5-(3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamido)pentyl)carbamate (4c): Lumacaftor (3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzoic acid) (181 mg, 0.40 mmol), tert-butyl (5-aminopentyl)carbamate (121 mg, 0.60 mmol), DIEA (350 μL, 2.00 mmol), and HOBt (54 mg, 0.4mmol) were dissolved in DCM (6 mL), followed by addition of EDCI HCl (153 mg, 0.50 mmol). The reaction was stirred at rt for 16 hours before water was added, the mixture partitioned, and the aqueous layer extracted with DCM twice. The combined organic extracts were washed with brine, dried over Na2SO4, concentrated, and the resulting crude oil was purified by silica gel chromatography (0-50% EtOAc/Hex) to obtain Intermediate 4c as a clear oil (240 mg, 0.38 mmol, 95%).
1H NMR (400 MHz, CDCl3) δ 8.14 (d, J = 8.4 Hz, 1H), 7.84 (s, 1H), 7.80 (dt, J = 7.6, 1.6 Hz, 1H), 7.73 (s, 1H), 7.63 (d, J = 8.5 Hz, 1H), 7.57 (dt, J = 7.7, 1.5 Hz, 1H), 7.51 (t, J = 7.6 Hz, 1H), 7.27 (dd, J = 8.1, 1.8 Hz, 1H), 7.23 (d, J = 1.7 Hz, 1H), 7.12 (d, J = 8.2 Hz, 1H), 6.25 (s, 1H), 3.17 (d, J = 6.8 Hz, 2H), 4.61 (s, 1H), 3.49 (q, J = 7.0, 6.8, 6.3 Hz, 2H), 2.29 (s, 3H), 1.79 (q, J = 3.9 Hz, 2H), 1.56 (q, J = 7.2 Hz, 2H), 1.46 (s, 11H), 1.36 – 1.27 (m, 2H), 1.20 (q, J = 3.9 Hz, 2H), 0.97 – 0.89 (m, 2H).
LC/MS [M+H]+ m/z calc. 637.28, found 637.3.
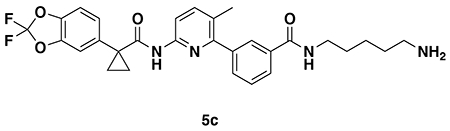
N-(5-aminopentyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (5c): Intermediate 4c (240 mg, 0.038 mmol) was dissolved in DCM (2 mL) and TFA (2 mL) was added and the solution stirred for 2 hours. The volatiles were then evaporated and the resulting oil redissolved in DCM and treated with aqueous saturated NaHCO3. The layers were separated and the aqueous layer was then extracted with DCM three times. The combined organic extracts were dried over Na2SO4, and concentrated to provide the title compound (184 mg, 0.34 mmol, 85% over two steps) as a colorless oil which was used in the next step without further purification.
1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 8.4 Hz, 1H), 7.80 (t, J = 1.8 Hz, 1H), 7.76 (dd, J = 7.7, 1.5 Hz, 1H), 7.69 (s, 1H), 7.59 (d, J = 8.5 Hz, 1H), 7.57 – 7.50 (m, 1H), 7.47 (t, J = 7.6 Hz, 1H), 7.23 (dd, J = 8.2, 1.7 Hz, 1H), 7.19 (d, J = 1.8 Hz, 1H), 7.08 (d, J = 8.2 Hz, 1H), 6.30 (s, 1H), 3.45 (q, J = 6.7 Hz, 2H), 2.74 (t, J = 6.8 Hz, 2H), 2.25 (s, 3H), 1.65 – 1.59 (m, 2H), 1.57 – 1.47 (m, 2H), 1.48 – 1.40 (m, 2H), 1.33 – 1.23 (m, 2H), 1.20 – 1.12 (m, 2H), 0.91 – 0.85 (m, 2H).
LC/MS [M+H]+ m/z calc. 537.22, found 537.2.
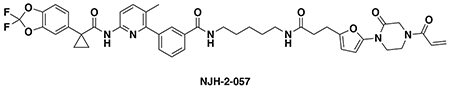
N-(5-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanamido)pentyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (NJH-2-057): Intermediate 3 (tert-butyl 3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoate) (70 mg, 0.20 mmol) was dissolved in DCM (1.0 mL) and TFA (0.8 mL) was added and the solution stirred for 1 h until starting material was consumed as monitored by TLC. The volatiles were evaporated, DCM was added and evaporated again. The residue was dissolved in DMF (1.5 mL) and DIEA (150 μL, 0.86 mmol) was added followed by intermediate 5c (N-(5-aminopentyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide) (54 mg, 0.1 mmol). HATU (152 mg, 0.4mmol) was then added and the mixture stirred for 1 h. Water was added, and the resulting suspension was extracted with DCM three times. Combined organic extracts were washed twice with 1M HCl twice, saturated NaHCO3, twice with 5% LiCl, brine, and dried over Na2SO4 before being concentrated. The crude residue was purified by silica gel chromatography (0-4% MeOH/DCM) to obtain the title compound (35 mg, 0.043 mmol, 43%) as a white powder following lyophilization from 1:1 water:acetonitrile (2 mL).
1H NMR (600 MHz, CDCl3) δ 8.11 (d, J = 8.4 Hz, 1H), 7.85 (t, J = 1.8 Hz, 1H), 7.81 (dt, J = 7.8, 1.5 Hz, 1H), 7.71 (s, 1H), 7.61 (d, J = 8.5 Hz, 1H), 7.55 (dt, J = 7.7, 1.4 Hz, 1H), 7.49 (t, J = 7.6 Hz, 1H), 7.25 (dd, J = 8.2, 1.8 Hz, 1H), 7.21 (d, J = 1.8 Hz, 1H), 7.10 (d, J = 8.2 Hz, 1H), 6.53 (s, 1H), 6.41 (dd, J = 16.7, 1.8 Hz, 2H), 6.22 (d, J = 3.3 Hz, 1H), 6.03 (d, J = 3.3 Hz, 1H), 5.82 (dd, J = 10.4, 1.8 Hz, 2H), 4.54 – 4.32 (m, 2H), 4.07 – 3.79 (m, 4H), 3.45 (q, J = 6.6 Hz, 2H), 3.24 (q, J = 6.6 Hz, 2H), 2.91 (t, J = 7.3 Hz, 2H), 2.46 (t, J = 7.3 Hz, 2H), 2.27 (s, 3H), 1.77 (q, J = 3.9 Hz, 2H), 1.65 – 1.59 (m, 2H), 1.52 (p, J = 7.0 Hz, 2H), 1.40 – 1.32 (m, 2H), 1.18 (q, J = 3.9 Hz, 2H).
13C NMR (151 MHz, CDCl3) δ 171.7, 167.4, 165.0, 155.5, 148.9, 144.8, 144.1, 143.6, 141.0, 140.2, 134.9, 134.8, 131.8, 128.4, 127.5, 127.0, 126.6, 126.6, 126.3, 112.9, 112.4, 110.2, 107.4, 100.9, 39.7, 39.1, 31.2, 29.0, 24.2, 23.7, 19.2, 17.2.
HRMS [M+H]+ m/z calc. 811.3262, found 811.3267.
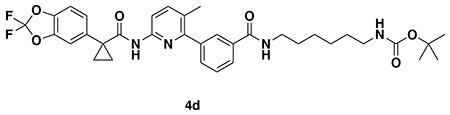
tert-butyl (6-(3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamido)hexyl)carbamate (4d): Lumacaftor (100 mg, 0.22 mmol) and tert-butyl (6-aminohexyl)carbamate were reacted according to General Procedure A and purified by silica gel chromatography (0-60% EtOAc/Hex) to obtain intermediate 4d (114 mg, 0.18 mmol, 80%) as a clear colorless oil.
1H NMR (400 MHz, CDCl3) δ 8.14 (d, J = 8.4 Hz, 1H), 7.86 (s, 1H), 7.82 (d, J = 7.7 Hz, 1H), 7.72 (s, 1H), 7.63 (d, J = 8.5 Hz, 1H), 7.57 (dt, J = 7.6, 1.5 Hz, 1H), 7.51 (t, J = 7.6 Hz, 1H), 7.27 (dd, J = 8.2, 1.8 Hz, 1H), 7.23 (d, J = 1.7 Hz, 1H), 7.12 (d, J = 8.1 Hz, 1H), 6.37 (s, 1H), 4.58 (s, 1H), 3.48 (q, J = 6.7 Hz, 2H), 3.17 (q, J = 6.7 Hz, 2H), 2.29 (s, 3H), 1.79 (q, J = 3.9 Hz, 2H), 1.69 – 1.64 (m, 1H), 1.58 – 1.49 (m, 1H), 1.46 (s, 9H), 1.45 – 1.38 (m, 6H), 1.20 (q, J = 3.9 Hz, 2H).
LC/MS [M+H]+ m/z calc. 651.3, found 651.2.
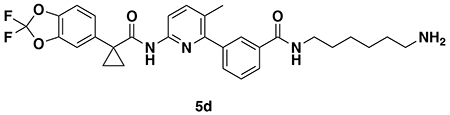
N-(6-aminohexyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide) (5d): 4d (114 mg, 0.18 mmol) was deprotected according to General Procedure B to provide the amine 5d (99 mg, 0.18 mmol, quant.) as a colorless oil.
1H NMR (400 MHz, CDCl3) δ 8.10 (d, J = 8.4 Hz, 1H), 7.78 (s, 1H), 7.74 (dt, J = 7.5, 1.6 Hz, 1H), 7.69 (s, 1H), 7.59 (d, J = 8.4 Hz, 1H), 7.53 (dt, J = 7.7, 1.5 Hz, 1H), 7.47 (t, J = 7.6 Hz, 1H), 7.22 (dd, J = 8.2, 1.8 Hz, 1H), 7.18 (d, J = 1.6 Hz, 1H), 7.07 (d, J = 8.2 Hz, 1H), 6.17 (s, 1H), 3.44 (td, J = 7.2, 5.8 Hz, 2H), 2.68 (t, J = 6.8 Hz, 2H), 2.24 (s, 3H), 1.99 (s, 1H), 1.81 (s, 1H), 1.74 (q, J = 3.9 Hz, 2H), 1.67 – 1.55 (m, 3H), 1.51 – 1.33 (m, 5H), 1.16 (q, J = 3.9 Hz, 2H).
LC/MS [M+H]+ m/z calc. 551.2, found 551.2.
N-(6-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanamido)hexyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (NJH-2-133): Intermediate 3 (30 mg, 0.086 mmol) was deprotected and coupled to intermediate 5d (16 mg, 0.029 mmol) following General Procedure C to provide NJH-2-133 (17.4 mg, 0.021 mmol, 73%) as a clear colorless oil.
1H NMR (400 MHz, CDCl3) δ 8.12 (d, J = 8.4 Hz, 1H), 7.89 – 7.79 (m, 2H), 7.73 (s, 1H), 7.62 (d, J = 8.5 Hz, 1H), 7.56 (dt, J = 7.7, 1.5 Hz, 1H), 7.51 (t, J = 7.6 Hz, 1H), 7.27 (dd, J = 8.2, 1.8 Hz, 1H), 7.22 (d, J = 1.7 Hz, 1H), 7.11 (d, J = 8.2 Hz, 1H), 6.54 (d, J = 31.0 Hz, 2H), 6.41 (dd, J = 16.8, 1.9 Hz, 1H), 6.25 (d, J = 3.3 Hz, 1H), 6.07 (d, J = 3.3 Hz, 1H), 5.98 (d, J = 39.7 Hz, 1H), 5.83 (dd, J = 10.3, 2.0 Hz, 1H), 4.42 (d, J = 21.6 Hz, 2H), 4.05 – 3.81 (m, 4H), 3.74 (p, J = 6.7 Hz, 2H), 3.45 (q, J = 6.7 Hz, 2H), 3.22 (dq, J = 13.2, 6.9 Hz, 3H), 2.94 (q, J = 6.4, 5.5 Hz, 2H), 2.52 (t, J = 7.4 Hz, 2H), 2.28 (s, 3H), 1.78 (q, J = 3.9 Hz, 2H), 1.61 (p, J = 6.9 Hz, 2H), 1.42 – 1.30 (m, 3H), 1.20 (q, J = 3.9 Hz, 2H).
13C NMR (151 MHz, CDCl3) δ 171.8, 167.3, 155.5, 149.7, 148.9, 144.7, 144.1, 143.6, 141.0, 140.2, 134.9, 134.9, 133.4, 131.7, 131.7, 130.0, 128.5, 127.5, 127.0, 126.6, 126.6, 126.4, 112.9, 112.4, 110.2, 107.3, 100.8, 55.6, 43.6, 39.6, 39.1, 34.8, 31.2, 29.4, 29.3, 26.0, 25.9, 24.2, 19.1, 18.6, 17.2, 12.5.
HRMS (ESI): [M+H]+ m/z calc. 825.3345, found 825.3425.
tert-butyl (2-(2-(3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamido)ethoxy)ethyl)carbamate (4e): Lumacaftor (100 mg, 0.22 mmol) and tert-butyl (2-(2-aminoethoxy)ethyl)carbamate (57 mg, 0.28 mmol) were reacted according to General Procedure A and purified by silica gel chromatography (0-60% EtOAc/Hex) to obtain intermediate 4e (122 mg, 0.19 mmol, 87%) as a clear colorless oil.
1H NMR (400 MHz, Chloroform-d) δ 8.14 (d, J = 8.4 Hz, 1H), 7.88 (t, J = 1.8 Hz, 1H), 7.81 (dt, J = 7.5, 1.6 Hz, 1H), 7.72 (s, 1H), 7.63 (d, J = 8.5 Hz, 1H), 7.59 (dt, J = 7.7, 1.5 Hz, 1H), 7.52 (t, J = 7.6 Hz, 1H), 7.27 (dd, J = 8.2, 1.8 Hz, 1H), 7.23 (d, J = 1.7 Hz, 1H), 7.12 (d, J = 8.2 Hz, 1H), 6.60 (s, 1H), 4.87 (s, 1H), 3.74 – 3.62 (m, 4H), 3.58 (t, J = 5.2 Hz, 2H), 3.41 – 3.31 (m, 2H), 2.29 (s, 3H), 1.79 (q, J = 3.9 Hz, 2H), 1.46 (s, 9H), 1.20 (q, J = 3.9 Hz, 2H).
LC/MS [M+H]+ m/z calc. 639.3, found 639.2.
N-(2-(2-aminoethoxy)ethyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (5e): Intermediate 4e (122 mg, 0.19 mmol) was deprotected according to General Procedure B to provide the amine 5e (102 mg, 0.19 mmol, quant.) as a colorless oil.
1H NMR (400 MHz, Chloroform-d) δ 8.14 (d, J = 8.4 Hz, 1H), 7.88 (t, J = 1.7 Hz, 1H), 7.85 (s, 1H), 7.77 (s, 1H), 7.63 (d, J = 8.5 Hz, 1H), 7.57 (dt, J = 7.7, 1.5 Hz, 1H), 7.51 (t, J = 7.6 Hz, 1H), 7.27 (dd, J = 8.1, 1.8 Hz, 1H), 7.23 (d, J = 1.7 Hz, 1H), 7.12 (d, J = 8.2 Hz, 1H), 6.91 (s, 0H), 3.70 (tdd, J = 7.9, 4.0, 1.2 Hz, 4H), 3.55 (t, J = 5.2 Hz, 2H), 2.91 (t, J = 5.2 Hz, 2H), 2.29 (s, 3H), 1.79 (q, J = 3.9 Hz, 2H), 1.20 (q, J = 3.9 Hz, 2H).
LC/MS: [M+H]+ m/z calc. 539.2 found 639.2.
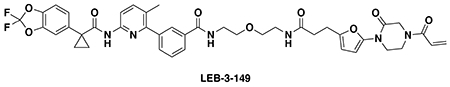
N-(2-(2-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanamido)ethoxy)ethyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (LEB-03-149). Intermediate 3 (30 mg, 0.086 mmol) was deprotected and coupled to intermediate 5e (23 mg, 0.043 mmol) following General Procedure C to provide LEB-03-149 (10.9 mg, 0.0134 mmol, 31% yield) as a white foam.
1H NMR (600 MHz, Chloroform-d) δ 8.10 (d, J = 8.4 Hz, 1H), 7.88 (t, J = 1.8 Hz, 1H), 7.82 (dt, J = 7.7, 1.5 Hz, 1H), 7.72 (s, 1H), 7.60 (d, J = 8.5 Hz, 1H), 7.55 (dt, J = 7.6, 1.4 Hz, 1H), 7.48 (t, J = 7.7 Hz, 1H), 7.25 (dd, J = 8.2, 1.8 Hz, 1H), 7.21 (d, J = 1.7 Hz, 1H), 7.09 (d, J = 8.2 Hz, 1H), 6.86 (s, 1H), 6.39 (dd, J = 16.7, 1.8 Hz, 1H), 6.20 (d, J = 3.3 Hz, 1H), 6.02 (d, J = 3.2 Hz, 1H), 5.81 (dd, J = 10.4, 1.8 Hz, 1H), 3.82 (s, 2H), 3.73 (hept, J = 6.6 Hz, 2H), 3.63 (d, J = 4.1 Hz, 4H), 3.53 (t, J = 5.1 Hz, 2H), 3.41 (q, J = 5.3 Hz, 2H), 3.19 (q, J = 7.4 Hz, 2H), 2.89 (t, J = 7.5 Hz, 2H), 2.47 (t, J = 7.3 Hz, 2H), 2.26 (s, 3H), 1.48 (t, J = 7.4 Hz, 3H), 1.18 (q, J = 3.9 Hz, 2H), 0.12 – 0.06 (m, 1H).
13C NMR (151 MHz, CDCl3) δ 171.77, 167.53, 165.03, 155.44, 149.75, 148.91, 144.71, 144.11, 143.59, 140.95, 140.22, 134.94, 134.55, 131.91, 131.68, 128.46, 127.72, 126.98, 126.64, 126.36, 112.96, 112.39, 110.21, 107.27, 100.91, 69.63, 69.50, 55.72, 53.43, 43.65, 39.83, 39.18, 34.69, 31.20, 24.08, 19.14, 17.18, 12.52.
HRMS (ESI): [M+H]+ m/z calc. 813.31, found 813.3055.
tert-butyl (2-(2-(2-(3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamido)ethoxy)ethoxy)ethyl)carbamate (4f): Lumacaftor (100 mg, 0.22 mmol) and tert-butyl (2-(2-(2-aminoethoxy)ethoxy)ethyl)carbamate (70 mg, 0.28 mmol) were reacted according to General Procedure A and purified by silica gel chromatography (0-80% EtOAc/Hex) to obtain intermediate 4f (127 mg, 0.19 mmol, 85%) as a clear colorless oil.
1H NMR (400 MHz, Chloroform-d) δ 8.14 (d, J = 8.4 Hz, 1H), 7.88 (s, 1H), 7.80 (d, J = 7.5 Hz, 1H), 7.77 – 7.72 (m, 1H), 7.63 (d, J = 8.4 Hz, 1H), 7.57 (d, J = 7.5 Hz, 1H), 7.52 (t, J = 7.6 Hz, 1H), 7.27 (dd, J = 8.2, 1.8 Hz, 1H), 7.23 (d, J = 1.7 Hz, 1H), 7.11 (d, J = 8.2 Hz, 1H), 6.74 (s, 1H), 5.02 (s, 1H), 3.75 – 3.61 (m, 8H), 3.56 (t, J = 5.4 Hz, 2H), 3.31 (d, J = 5.8 Hz, 2H), 2.28 (s, 3H), 1.79 (q, J = 3.9 Hz, 2H), 1.45 (s, 9H), 1.20 (q, J = 3.9 Hz, 2H).
LC/MS: [M+H]+ m/z calc. 683.3, found 683.3.
N-(2-(2-(2-aminoethoxy)ethoxy)ethyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (5f): Intermediate 4f (127 mg, 0.19 mmol) was deprotected according to General Procedure B to provide the amine 5f (111 mg, 0.19 mmol, quant.) as a colorless oil.
1H NMR (400 MHz, Chloroform-d) δ 8.13 (d, J = 8.4 Hz, 1H), 7.89 (t, J = 1.7 Hz, 1H), 7.83 (dt, J = 7.7, 1.5 Hz, 1H), 7.78 (s, 1H), 7.63 (d, J = 8.5 Hz, 1H), 7.56 (dt, J = 7.7, 1.5 Hz, 1H), 7.51 (t, J = 7.6 Hz, 1H), 7.27 (dd, J = 8.2, 1.7 Hz, 1H), 7.23 (d, J = 1.7 Hz, 1H), 7.12 (d, J = 8.2 Hz, 1H), 7.08 (s, 1H), 3.73 – 3.62 (m, 9H), 3.51 (t, J = 5.2 Hz, 2H), 2.82 (t, J = 5.1 Hz, 2H), 2.28 (s, 3H), 1.79 (q, J = 3.9 Hz, 2H), 1.20 (q, J = 3.9 Hz, 2H).
LC/MS: [M+H]+ m/z calc. 583.2, found 583.3.
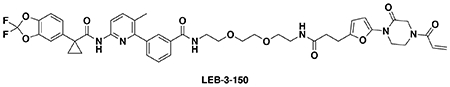
N-(2-(2-(2-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanamido)ethoxy)ethoxy)ethyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (LEB-03-150). Intermediate 3 (30 mg, 0.086 mmol) was deprotected and coupled to intermediate 5f (25 mg, 0.043 mmol) following General Procedure C to provide LEB-03-150 (11.6 mg, 0.0134 mmol, 31% yield) as a clear colorless oil.
1H NMR (600 MHz, Chloroform-d) δ 8.11 (d, J = 8.4 Hz, 1H), 7.86 (tt, J = 1.8, 1.2 Hz, 1H), 7.79 (ddd, J = 7.7, 1.8, 1.2 Hz, 1H), 7.72 (s, 1H), 7.62 – 7.58 (m, 1H), 7.55 (ddd, J = 7.6, 1.7, 1.2 Hz, 1H), 7.48 (td, J = 7.7, 0.6 Hz, 1H), 7.25 (dd, J = 8.2, 1.8 Hz, 1H), 7.21 (d, J = 1.7 Hz, 1H), 7.09 (d, J = 8.2 Hz, 1H), 6.83 (d, J = 5.8 Hz, 1H), 6.41 (dd, J = 16.7, 1.8 Hz, 1H), 6.24 (d, J = 3.2 Hz, 1H), 6.18 (s, 1H), 6.05 (dd, J = 3.3, 1.0 Hz, 1H), 5.82 (dd, J = 10.5, 1.8 Hz, 1H), 5.32 (s, 1H), 4.40 (d, J = 39.8 Hz, 2H), 3.94 (d, J = 47.9 Hz, 1H), 3.85 (s, 2H), 3.70 – 3.58 (m, 7H), 3.50 (dd, J = 5.6, 4.8 Hz, 2H), 3.39 (q, J = 5.4 Hz, 2H), 2.93 (t, J = 7.5 Hz, 2H), 2.47 (t, J = 7.5 Hz, 2H), 2.25 (s, 3H), 2.19 (s, 1H), 1.76 (q, J = 3.8 Hz, 2H), 1.47 (d, J = 12.2 Hz, 1H), 1.18 (p, J = 3.8 Hz, 2H).
13C NMR 151 MHz, CDCl3) δ 171.78, 171.54, 167.31, 164.98, 155.46, 148.91, 144.68, 144.12, 143.60, 140.94, 140.25, 134.93, 134.64, 131.88, 131.68, 128.46, 127.72, 126.98, 126.63, 126.51, 126.34, 113.00, 112.39, 110.19, 107.18, 100.77, 70.23, 70.18, 69.80, 55.62, 53.43, 43.58, 39.81, 39.16, 34.67, 31.20, 30.92, 23.97, 19.13, 17.19, 12.47, 1.02.
HRMS (ESI): [M+H]+ m/z calc. 857.33, found 857.3319.
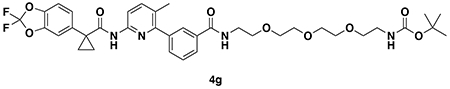
tert-butyl (1-(3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)phenyl)-1-oxo-5,8,11-trioxa-2-azatridecan-13-yl)carbamate (4g): Lumacaftor (100 mg, 0.22 mmol) and tert-butyl (2-(2-(2-(2-aminoethoxy)ethoxy)ethoxy)ethyl)carbamate (82 mg, 0.28 mmol) were reacted according to General Procedure A and purified by silica gel chromatography (0-100% EtOAc/Hex) to obtain intermediate 4g (139 mg, 0.19 mmol, 87%) as a colorless oil.
1H NMR (400 MHz, Chloroform-d) δ 8.14 (d, J = 8.4 Hz, 1H), 7.89 (s, 1H), 7.82 (d, J = 7.5 Hz, 1H), 7.73 (s, 1H), 7.63 (d, J = 8.5 Hz, 1H), 7.57 (d, J = 7.5 Hz, 1H), 7.51 (t, J = 7.6 Hz, 1H), 7.27 (dd, J = 8.2, 1.8 Hz, 1H), 7.23 (d, J = 1.7 Hz, 1H), 7.12 (d, J = 8.2 Hz, 1H), 6.80 (s, 1H), 3.73 – 3.66 (m, 9H), 3.64 (dd, J = 6.1, 3.2 Hz, 2H), 3.59 (dd, J = 6.1, 3.2 Hz, 2H), 3.50 (t, J = 5.1 Hz, 2H), 3.30 (d, J = 5.7 Hz, 2H), 2.28 (s, 3H), 1.79 (q, J = 3.9 Hz, 2H), 1.46 (s, 9H), 1.20 (q, J = 3.9 Hz, 2H).
LC/MS: [M+H]+ m/z calc. 727.3, found 727.2.

N-(2-(2-(2-(2-aminoethoxy)ethoxy)ethoxy)ethyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (5g): Intermediate 4g (139 mg, 0.19 mmol) was deprotected according to General Procedure B to provide the amine 5g (119 mg, 0.19 mmol, quant.) as a colorless oil.
1H NMR (400 MHz, Chloroform-d) δ 8.13 (d, J = 8.4 Hz, 1H), 7.93 (t, J = 1.8 Hz, 1H), 7.87 (dt, J = 7.6, 1.6 Hz, 1H), 7.76 (s, 1H), 7.62 (d, J = 8.5 Hz, 1H), 7.60 (s, 1H), 7.55 (dt, J = 7.7, 1.5 Hz, 1H), 7.50 (t, J = 7.6 Hz, 1H), 7.27 (dd, J = 8.2, 1.7 Hz, 1H), 7.23 (d, J = 1.7 Hz, 1H), 7.12 (d, J = 8.2 Hz, 1H), 3.73 – 3.63 (m, 9H), 3.61 (dt, J = 6.0, 1.8 Hz, 4H), 3.48 – 3.43 (m, 2H), 2.82 – 2.75 (m, 2H), 2.29 (s, 3H), 1.79 (q, J = 3.9 Hz, 2H), 1.20 (q, J = 3.9 Hz, 2H).
LC/MS: [M+H]+ m/z calc. 627.3, found 627.3.
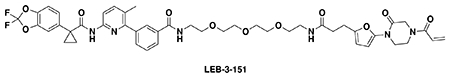
N-(15-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)-13-oxo-3,6,9-trioxa-12-azapentadecyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide (LEB-03-151). Intermediate 3 (30 mg, 0.086 mmol) was deprotected and coupled to intermediate 5g (27 mg, 0.043 mmol) following General Procedure C to provide LEB-03-151 (13.7 mg, 0.0152 mmol, 35% yield) as a colorless oil.
1H NMR 1H NMR (600 MHz, Chloroform-d) δ 8.10 (d, J = 8.5 Hz, 1H), 7.87 (t, J = 1.8 Hz, 1H), 7.80 (dt, J = 7.8, 1.5 Hz, 1H), 7.73 (s, 1H), 7.60 (d, J = 8.5 Hz, 1H), 7.54 (dt, J = 7.7, 1.4 Hz, 1H), 7.48 (t, J = 7.7 Hz, 1H), 7.25 (dd, J = 8.2, 1.8 Hz, 1H), 7.21 (d, J = 1.7 Hz, 1H), 7.09 (d, J = 8.2 Hz, 1H), 6.93 (d, J = 6.0 Hz, 1H), 6.41 (dd, J = 16.7, 1.8 Hz, 1H), 6.25 (d, J = 3.2 Hz, 1H), 6.05 (d, J = 3.3 Hz, 1H), 5.82 (dd, J = 10.4, 1.8 Hz, 1H), 4.41 (d, J = 35.7 Hz, 2H), 3.95 (d, J = 50.4 Hz, 3H), 3.85 (s, 2H), 3.70 – 3.62 (m, 8H), 3.62 – 3.57 (m, 2H), 3.57 – 3.52 (m, 2H), 3.47 (dd, J = 5.6, 4.6 Hz, 2H), 3.39 (q, J = 5.3 Hz, 2H), 2.93 (t, J = 7.6 Hz, 2H), 2.47 (t, J = 7.6 Hz, 2H), 2.25 (s, 3H), 2.19 (s, 1H), 1.76 (q, J = 3.9 Hz, 2H), 1.18 (q, J = 3.9 Hz, 2H).
13C NMR (151 MHz, CDCl3) δ 171.78, 171.50, 167.25, 164.97, 155.52, 148.90, 144.64, 144.13, 143.60, 140.92, 140.21, 134.93, 134.65, 133.37, 131.81, 131.68, 129.98, 128.40, 127.78, 126.98, 126.62, 126.59, 126.35, 112.96, 112.38, 110.20, 107.12, 100.70, 70.43, 70.38, 70.18, 70.07, 69.85, 69.82, 53.43, 39.81, 39.19, 34.59, 31.20, 30.92, 23.93, 19.13, 17.18.
HRMS (ESI): [M+H]+ m/z calc. 901.36, found 901.3584.
Synthesis of WEE1 DUBTACs:
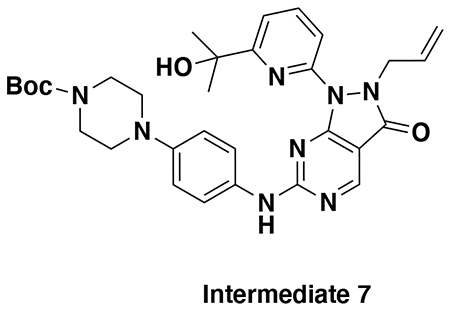
tert-butyl 4-(4-((2-allyl-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-3-oxo-2,3-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino)phenyl)piperazine-1-carboxylate (Intermediate 7). Commercially available Intermediate 6 2-allyl-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-6-(methylthio)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one (250 mg, 0.7 mmol) was dissolved in 7mL of toluene and cooled to 0°C. meta-Chloroperoxybenzoic acid (190 mg, 0.77 mmol) was added to the reaction mixture on ice, and the reaction mixture was warmed to room temperature and stirred for 1 hour. N,N-Diisopropylethylamine (365 μL, 2.1 mmol) and 1-Piperazinecarboxylic acid, 4-(4-aminophenyl)-, 1,1-dimethylethyl ester (232 mg, 0.84 mmol) were then added slowly and the reaction mixture was stirred overnight. The reaction mixture was extracted in EtOAc, washed 3X with brine, and dried on silica. Purification by flash column chromatography (DCM/Hexane 5:95) yielded Intermediate 7 (0.445 mmol, 64% yield).
1H NMR (400 MHz, Chloroform-d) δ 8.99 (s, 1H), 7.95 (t, J = 7.9 Hz, 1H), 7.80 (dd, J = 8.1, 0.8 Hz, 1H), 7.44 (dd, J = 7.7, 0.8 Hz, 1H), 5.83 – 5.65 (m, 1H), 5.13 – 5.04 (m, 1H), 4.97 (dq, J = 17.1, 1.4 Hz, 1H), 4.85 (dt, J = 6.2, 1.4 Hz, 2H), 3.80 (s, 1H), 2.63 (s, 3H), 1.63 (s, 6H).
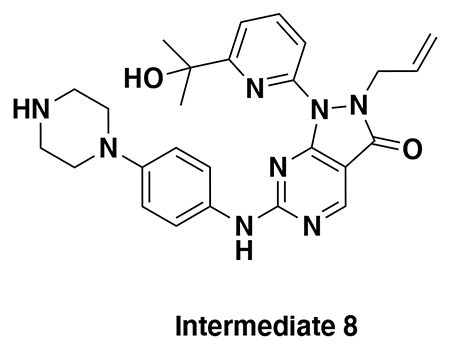
2-allyl-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-6-((4-(piperazin-1-yl)phenyl)amino)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one (Intermediate 8). Intermediate 7 (261 mg, 0.445 mmol) was dissolved in 4mL of DCM and cooled to 0°C. 1mL of trifluoroacetic acid was added dropwise on ice. The reaction mixture was stirred at room temperature for 1 hour, then extracted in DCM, washed 3X with brine, and dried on silica. Purification by flash column chromatography (DCM/Hexane 5:95) yielded Intermediate 8 (0.398 mmol, 89% yield).
1H NMR (400 MHz, Chloroform-d) δ 8.84 (s, 1H), 7.86 (t, J = 7.9 Hz, 1H), 7.75 (d, J = 8.1 Hz, 1H), 7.48 (d, J = 8.5 Hz, 2H), 7.34 (d, J = 7.6 Hz, 1H), 6.93 (d, J = 9.0 Hz, 2H), 5.78 – 5.59 (m, 1H), 5.04 (d, J = 10.2 Hz, 1H), 4.94 (d, J = 17.0 Hz, 1H), 4.74 (d, J = 6.2 Hz, 2H), 3.94 (s, 1H), 3.60 (t, J = 5.1 Hz, 4H), 3.11 (t, J = 5.1 Hz, 4H), 2.05 (s, 1H), 1.59 (s, 6H), 1.49 (s, 9H).
Synthesis of LEB-03-153
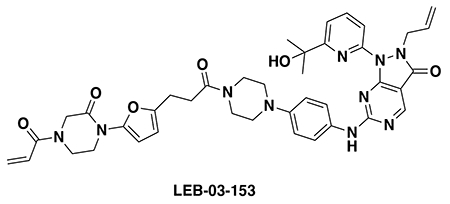
6-((4-(4-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoyl)piperazin-1-yl)phenyl)amino)-2-allyl-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one (LEB-03-153). LEB-03-139 (0.0449 mmol) was dissolved in 3mL of DCM and the reaction mixture was cooled on ice. 1mL of trifluoroacetic acid was added dropwise and the solution was warmed to room temperature and stirred for 1 hour. The deprotected amine salt was washed twice with DCM and dried under vacuum. Immediately following deprotection, the crude product was dissolved in 0.5 mL DMF and deprotected intermediate 3 (.0898 mmol) was added to the mixture, followed by DIPEA (0.449 mmol) and HATU (0.0898 mmol). The reaction was stirred for 30 minutes before water was added. The mixture was extracted three times with EtOAc, and combined organic extracts were washed with brine, dried over sodium sulfate, and concentrated. Purification by flash column chromatography (MeOH:DCM 8:92) yielded LEB-03-153 as a light-yellow solid (12.9 mg, 0.0169 mmol, 38% yield).
1H NMR (600 MHz, Chloroform-d) δ 8.76 (s, 1H), 7.80 (t, J = 7.9 Hz, 1H), 7.66 (d, J = 8.0 Hz, 1H), 7.43 (d, J = 8.4 Hz, 2H), 7.29 (d, J = 7.6 Hz, 1H), 7.19 (s, 1H), 6.87 – 6.82 (m, 2H), 6.45 (s, 1H), 6.34 (dd, J = 16.7, 1.8 Hz, 1H), 6.20 (d, J = 3.2 Hz, 1H), 6.01 (d, J = 3.2 Hz, 1H), 5.74 (t, J = 11.1, 10.6 Hz, 1H), 5.67 – 5.59 (m, 1H), 5.23 (s, 1H), 4.97 (dd, J = 9.8, 0.8 Hz, 1H), 4.87 (dd, J = 17.4, 0.8 Hz, 1H), 4.67 (d, J = 6.2 Hz, 2H), 4.38 (s, 1H), 4.31 (s, 1H), 3.73 (t, J = 5.2 Hz, 2H), 3.55 (t, J = 5.1 Hz, 2H), 3.06 (t, J = 5.2 Hz, 4H), 2.92 (t, J = 7.8 Hz, 2H), 2.62 (d, J = 8.4 Hz, 2H), 1.59 (s, 4H), 1.52 (s, 6H).
13C NMR (151 MHz, Chloroform-d) δ 169.96, 165.90, 165.00, 162.18, 161.36, 161.26, 156.36, 150.16, 147.68, 147.51, 144.67, 138.85, 131.56, 131.29, 126.30, 119.07, 117.20, 116.21, 116.12, 107.23, 101.12, 72.46, 50.15, 49.85, 49.49, 47.67, 45.40, 41.64, 31.55, 30.56, 23.71.
HRMS (ESI): [M+H]+ m/z calc. 761.35, found 761.3522.
Synthesis of LEB-03-144
2-allyl-6-((4-(4-(3-aminopropyl)piperazin-1-yl)phenyl)amino)-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one (LEB-03-140). Intermediate 8 (40 mg, 0.0823 mmol) was dissolved in 0.5 mL of DMF. tert-butyl (3-bromopropyl)carbamate (24 mg, 1.2 eq, 0.0987 mmol) and potassium carbonate (34 mg, 3.0 eq, 0.247 mmol) were added to the mixture, and the reaction was warmed to 50°C and stirred overnight. Water was added, the mixture extracted three times with EtOAc, combined organic extracts were washed with brine, and dried over sodium sulfate, and concentrated. Purification by flash column chromatography (EtOAc:Hexanes 50:50) yielded the boc-protected intermediate. This was immediately dissolved in 3mL of DCM and the reaction mixture was cooled on ice. 1 mL of trifluoroacetic acid was added dropwise and the solution was warmed to room temperature and stirred for 1 hour. The deprotected amine TFA salt was washed twice with DCM and dried under vacuum to yield LEB-03-140 (33 mg, 0.0497 mmol, 60% yield over two steps) as a yellow oil.
1H NMR (300 MHz, Chloroform-d) δ 8.80 (s, 1H), 7.92 (t, J = 7.9 Hz, 1H), 7.63 (d, J = 8.0 Hz, 1H), 7.54 (d, J = 8.4 Hz, 3H), 6.90 (d, J = 8.9 Hz, 2H), 5.75 – 5.54 (m, 1H), 5.05 (d, J = 10.2 Hz, 1H), 4.89 (d, J = 17.1 Hz, 1H), 4.75 (d, J = 6.2 Hz, 2H), 3.66 (s, 1H), 3.43 (s, 9H), 3.28 (q, J = 9.4, 8.5 Hz, 2H), 3.19 (s, 1H), 3.06 (t, J = 7.1 Hz, 2H), 2.23 (d, J = 8.2 Hz, 3H), 1.59 (s, 6H).
LC/MS: [M+H]+ m/z calc. 544.3, found 544.3.
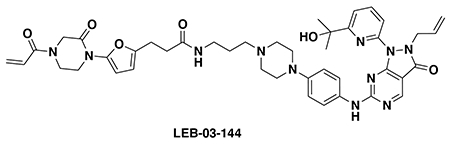
3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)-N-(3-(4-(4-((2-allyl-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-3-oxo-2,3-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino)phenyl)piperazin-1-yl)propyl)propenamide (LEB-03-144). Intermediate 3 (19 mg, 0.0558 mmol) and LEB-3-140 (0.0497 mmol) were reacted according to general procedure C. After hydrolysis, deprotected 3 and LEB-3-140 were dissolved in DMF (0.5 mL), followed by DIPEA (43 μL, 0.249 mmol) and HATU (23 mg, 0.0596 mmol). The reaction was stirred for 30 minutes. Water was added and the mixture extracted three times with 4:1 CHCl3:IPA. Combined organic extracts were washed with brine, and dried over sodium sulfate, and concentrated. Purification by prep TLC (10% MeOH in DCM) yielded LEB-03-144 as a light-yellow solid (8.1 mg, 0.0099 mmol, 20% yield).
1H NMR (600 MHz, DMSO) δ 10.07 (s, 1H), 8.75 (s, 1H), 7.97 (s, 1H), 7.83 (t, J = 5.6 Hz, 1H), 7.68 (d, J = 8.1 Hz, 1H), 7.54 (d, J = 7.7 Hz, 1H), 7.51 (s, 2H), 6.85 (d, J = 8.6 Hz, 2H), 6.80 – 6.72 (m, 1H), 6.16 – 6.08 (m, 2H), 6.04 (d, J = 3.2 Hz, 1H), 5.71 – 5.66 (m, 1H), 5.64 – 5.55 (m, 1H), 5.24 (s, 1H), 4.92 (d, J = 10.2 Hz, 1H), 4.76 (d, J = 17.0 Hz, 1H), 4.61 (d, J = 6.0 Hz, 2H), 4.27 (d, J = 93.6 Hz, 2H), 3.95 – 3.63 (m, 4H), 3.02 (q, J = 6.4 Hz, 6H), 2.73 (t, J = 7.5 Hz, 2H), 2.31 (t, J = 7.5 Hz, 2H), 2.24 (t, J = 7.2 Hz, 2H), 1.55 – 1.47 (m, 2H), 1.39 (s, 2H), 1.17 (s, 6H), 0.80 – 0.74 (m, 2H).
13C NMR (151 MHz, DMSO) δ 171.0, 168.0, 164.6, 161.6, 156.5, 150.1, 139.3, 132.7, 131.3, 128.8, 118.7, 116.8, 115.9, 106.9, 100.5, 72.8, 55.9, 53.2, 49.2, 47.6, 47.1, 46.9, 42.5, 37.4, 34.7, 33.8, 31.4, 30.9, 29.5, 26.9, 25.3, 24.0, 22.6, 22.5, 14.4.
HRMS (ESI): [M+H]+ m/z calc. 818.41, found 818.4101.
Synthesis of LEB-3-145
2-allyl-6-((4-(4-(5-aminopentyl)piperazin-1-yl)phenyl)amino)-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one (LEB-03-141). Intermediate 8 (40 mg, 0.0823 mmol) was dissolved in 0.5 mL of DMF. tert-butyl (5-bromopentyl)carbamate (26 mg, 1.2 eq, 0.0987 mmol) and potassium carbonate (34 mg, 3.0 eq, 0.247 mmol) were added to the mixture, and the reaction was warmed to 50°C and stirred overnight. Water was added, the mixture extracted three times with EtOAc, combined organic extracts were washed with brine, and dried over sodium sulfate, and concentrated. Purification by flash column chromatography (EtOAc:Hexanes 50:50) yielded boc-protected intermediate. This was immediately dissolved in 3mL of DCM and the reaction mixture was cooled on ice. 1mL of trifluoroacetic acid was added dropwise and the solution was warmed to room temperature and stirred for 1 hour. The deprotected amine TFA salt was washed twice with DCM and dried under vacuum to yield LEB-03-141 (21 mg, 0.0307 mmol, 37% yield over two steps) as a yellow oil.
1H NMR (300 MHz, Chloroform-d) δ 8.80 (s, 1H), 8.12 (s, 1H), 7.94 (t, J = 7.9 Hz, 1H), 7.64 (d, J = 8.0 Hz, 1H), 7.56 (d, J = 8.2 Hz, 3H), 6.92 (d, J = 8.8 Hz, 2H), 5.67 (dd, J = 16.8, 10.4 Hz, 1H), 5.07 (d, J = 10.2 Hz, 1H), 4.90 (d, J = 17.1 Hz, 1H), 4.76 (d, J = 6.2 Hz, 2H), 3.69 (s, 1H), 3.55 – 3.47 (m, 8H), 3.22 (s, 1H), 3.19 – 2.89 (m, 4H), 1.91 – 1.66 (m, 4H), 1.61 (s, 6H), 1.51 (s, 2H), 1.27 (s, 1H).
LC/MS: [M+H]+ m/z calc. 572.3, found 572.3.
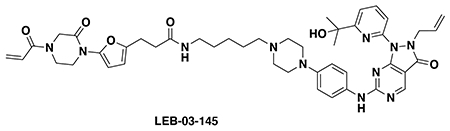
3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)-N-(5-(4-(4-((2-allyl-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-3-oxo-2,3-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino)phenyl)piperazin-1-yl)pentyl)propanamide (LEB-03-145). Intermediate 3 (19 mg, 0.0558 mmol) and LEB-2-141 (21 mg, 0.0307 mmol) were coupled according to general procedure C. After hydrolysis, deprotected 3 and LEB-2-141 were dissolved in DMF (0.5 mL), followed by DIPEA (27 μL, 0.153 mmol) and HATU (14 mg, 0.0368 mmol). Water was added and the mixture extracted three times with 4:1 CHCl3:IPA. Combined organic extracts were washed with brine, and dried over sodium sulfate, and concentrated. Purification by prep TLC (8% MeOH in DCM) yielded LEB-03-145 as a light-yellow solid (10.1 mg, 0.0119 mmol, 39% yield).
1H NMR (600 MHz, DMSO-d6) δ 10.15 (s, 1H), 8.83 (s, 1H), 8.05 (s, 1H), 7.86 (t, J = 5.6 Hz, 1H), 7.78 – 7.72 (m, 1H), 7.61 (d, J = 7.7 Hz, 2H), 7.58 (s, 2H), 6.92 (d, J = 8.7 Hz, 2H), 6.88 – 6.76 (m, 1H), 6.24 – 6.19 (m, 1H), 6.10 (d, J = 3.2 Hz, 1H), 5.76 (q, J = 9.8, 8.3 Hz, 1H), 5.72 – 5.61 (m, 1H), 5.36 – 5.26 (m, 1H), 5.00 (dq, J = 10.3, 1.3 Hz, 1H), 4.84 (dq, J = 17.2, 1.5 Hz, 1H), 4.69 (d, J = 6.0 Hz, 2H), 4.43 (s, 1H), 4.27 (s, 1H), 3.95 (d, J = 5.8 Hz, 1H), 3.86 (s, 1H), 3.82 – 3.73 (m, 2H), 3.13 – 3.08 (m, 4H), 3.08 – 3.01 (m, 2H), 2.80 (t, J = 7.5 Hz, 2H), 2.38 (t, J = 7.5 Hz, 2H), 2.30 (t, J = 7.4 Hz, 2H), 1.47 (s, 6H), 1.45 – 1.37 (m, 2H), 1.26 – 1.21 (m, 6H), 0.89 – 0.81 (m, 2H).
13C NMR (151 MHz, DMSO) δ 170.97, 168.04, 161.64, 156.46, 150.07, 139.28, 132.67, 128.77, 118.72, 115.93, 106.92, 100.44, 72.78, 58.33, 53.28, 49.17, 47.57, 47.06, 46.88, 42.46, 38.88, 33.80, 30.92, 29.54, 29.48, 29.16, 26.48, 24.81, 24.00, 22.56, 14.42.
HRMS (ESI): [M+H]+ m/z calc. 846.44, found 846.4395.
Synthesis of LEB-3-146
2-allyl-6-((4-(4-(2-(2-(2-aminoethoxy)ethoxy)ethyl)piperazin-1-yl)phenyl)amino)-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one (LEB-03-142). Intermediate 8 (40 mg, 0.0823 mmol) was dissolved in 0.5 mL of DMF. tert-butyl (2-(2-(bromomethoxy)ethoxy)ethyl)carbamate (31 mg, 1.2 eq, 0.0987 mmol) and potassium carbonate (34 mg, 3.0 eq, 0.247 mmol) were added to the mixture, and the reaction was warmed to 50°C and stirred overnight. Water was added, the mixture extracted three times with EtOAc, combined organic extracts were washed with brine, and dried over sodium sulfate, and concentrated. Purification by flash column chromatography (EtOAc:Hexanes 50:50) yielded boc-protected intermediate. This was immediately dissolved in 3mL of DCM and the reaction mixture was cooled on ice. 1mL of trifluoroacetic acid was added dropwise and the solution was warmed to room temperature and stirred for 1 hour. The deprotected amine TFA salt was washed twice with DCM and dried under vacuum to yield LEB-03-142 (28 mg, 0.0389 mmol, 47% yield).
1H NMR (300 MHz, Chloroform-d) δ 10.99 (s, 1H), 8.74 (s, 1H), 8.25 (s, 1H), 7.97 (t, J = 7.9 Hz, 1H), 7.60 (t, J = 8.9 Hz, 2H), 7.50 (d, J = 8.5 Hz, 2H), 6.87 (d, J = 8.7 Hz, 2H), 5.66 (ddd, J = 16.5, 10.3, 5.6 Hz, 1H), 5.07 (d, J = 10.2 Hz, 1H), 4.90 (d, J = 17.1 Hz, 1H), 4.75 (d, J = 6.3 Hz, 4H), 3.87 (d, J = 4.6 Hz, 4H), 3.76 – 3.69 (m, 4H), 3.65 (s, 4H), 3.39 – 3.10 (m, 8H), 1.61 (s, 6H).
LC/MS: [M+H]+ m/z calc. 618.3, found 618.3.
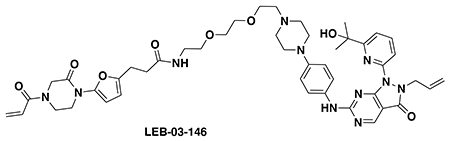
3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)-N-(2-(2-(2-(4-(4-((2-allyl-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-3-oxo-2,3-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino)phenyl)piperazin-1-yl)ethoxy)ethoxy)ethyl)propanamide (LEB-03-146). Intermediate 3 (19 mg, 0.0558 mmol) and LEB-3-142 (28 mg, 0.0389 mmol) were coupled according to general procedure C. After hydrolysis, deprotected 3 and LEB-3-142 were dissolved in DMF (0.5 mL), followed by DIPEA (34 μL, 0.195 mmol) and HATU (18 mg, 0.0466 mmol). The reaction was stirred for 30 minutes. Water was added and the mixture extracted three times with 4:1 CHCl3:IPA. Combined organic extracts were washed with brine, and dried over sodium sulfate, and concentrated. Purification by prep TLC (8% MeOH in DCM) yielded LEB-03-146 as a light-yellow solid (8.3 mg, 0.0093 mmol, 17% yield).
1H NMR (600 MHz, DMSO-d6) δ 8.83 (s, 1H), 8.05 (s, 1H), 7.97 (t, J = 5.8 Hz, 1H), 7.76 (s, 1H), 7.61 (d, J = 7.8 Hz, 1H), 7.58 (s, 3H), 6.92 (d, J = 8.5 Hz, 2H), 6.81 (d, J = 12.8 Hz, 1H), 6.23 – 6.15 (m, 2H), 6.10 (d, J = 3.2 Hz, 1H), 5.76 (d, J = 7.0 Hz, 2H), 5.67 (ddt, J = 16.5, 10.8, 6.0 Hz, 1H), 5.31 (s, 1H), 5.04 – 4.97 (m, 1H), 4.87 – 4.80 (m, 1H), 4.69 (s, 2H), 4.42 (s, 1H), 4.26 (s, 1H), 3.94 (s, 1H), 3.85 (s, 1H), 3.77 (d, J = 24.8 Hz, 2H), 3.56 (t, J = 5.8 Hz, 2H), 3.54 – 3.49 (m, 6H), 3.42 (t, J = 5.9 Hz, 2H), 3.22 (q, J = 5.8 Hz, 2H), 3.09 (d, J = 5.8 Hz, 4H), 2.79 (t, J = 7.6 Hz, 2H), 2.58 (t, J = 4.8 Hz, 4H), 2.44 – 2.36 (m, 4H), 1.47 (s, 6H), 0.86 (d, J = 7.4 Hz, 1H).
13C NMR (151 MHz, DMSO) δ 171.31, 168.04, 161.64, 156.46, 150.02, 139.28, 132.68, 128.76, 118.72, 115.93, 106.93, 100.44, 72.78, 70.12, 70.04, 69.58, 68.89, 57.72, 53.63, 49.14, 47.07, 46.88, 42.46, 39.07, 33.65, 30.92, 29.49, 23.89, 14.42.
HRMS (ESI): [M+H]+ m/z calc. 892.45, found 892.4454.
Synthesis of LEB-3-147
2-allyl-6-((4-(4-(2-(2-(2-(2-aminoethoxy)ethoxy)ethoxy)ethyl)piperazin-1-yl)phenyl)amino)-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one (LEB-03-143). Intermediate 8 (40 mg, 0.0823 mmol) was dissolved in 0.5 mL of DMF. tert-butyl (2-(2-(2-(bromomethoxy)ethoxy)ethoxy)ethyl)carbamate (35 mg, 1.2 eq, 0.0987 mmol) and potassium carbonate (34 mg, 3.0 eq, 0.247 mmol) were added to the mixture, and the reaction was warmed to 50°C and stirred overnight. Water was added, the mixture extracted three times with EtOAc, combined organic extracts were washed with brine, and dried over sodium sulfate, and concentrated. Purification by flash column chromatography (EtOAc:Hexanes 50:50) yielded boc-protected intermediate. This was immediately dissolved in 3mL of DCM and the reaction mixture was cooled on ice. 1mL of trifluoroacetic acid was added dropwise and the solution was warmed to room temperature and stirred for 1 hour. The deprotected amine TFA salt was washed twice with DCM and dried under vacuum to yield LEB-03-143 (22 mg, 0.0279 mmol, 34% yield) as a yellow oil.
1H NMR (300 MHz, Chloroform-d) δ 11.44 (s, 1H), 8.71 (s, 1H), 8.10 (s, 4H), 8.00 (t, J = 7.9 Hz, 1H), 7.65 (d, J = 8.0 Hz, 1H), 7.57 (t, J = 7.4 Hz, 3H), 6.91 (d, J = 8.6 Hz, 2H), 5.69 (ddt, J = 16.5, 10.1, 6.2 Hz, 1H), 5.11 (d, J = 10.1 Hz, 1H), 4.92 (d, J = 17.1 Hz, 1H), 4.78 (d, J = 6.3 Hz, 2H), 3.98 – 3.77 (m, 5H), 3.77 – 3.63 (m, 9H), 3.33 (d, J = 59.7 Hz, 8H), 1.64 (s, 6H), 1.29 (s, 1H).
LC/MS: [M+H]+ m/z calc. 662.3, found 662.4
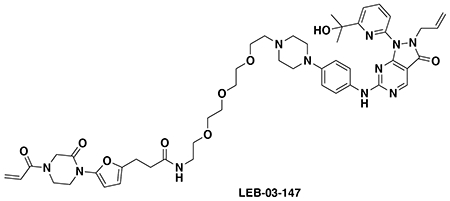
3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)-N-(2-(2-(2-(2-(4-(4-((2-allyl-1-(6-(2-hydroxypropan-2-yl)pyridin-2-yl)-3-oxo-2,3-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino)phenyl)piperazin-1-yl)ethoxy)ethoxy)ethoxy)ethyl)propanamide (LEB-03-147). Intermediate 3 (19 mg, 0.0558 mmol) and LEB-3-143 (22 mg, 0.0279 mmol) were coupled according to general procedure C. After hydrolysis, deprotected 3 and LEB-3-142 were dissolved in DMF (0.5 mL), followed by DIPEA (49 μL, 0.279 mmol) and HATU (21 mg, 0.0558 mmol). The reaction was stirred for 30 minutes. Water was added and the mixture extracted three times with 4:1 CHCl3:IPA. Combined organic extracts were washed with brine, and dried over sodium sulfate, and concentrated. Purification by prep TLC (8% MeOH in DCM) yielded LEB-03-147 as a light-yellow solid (10.0 mg, 0.0107 mmol, 19% yield).
1H NMR (600 MHz, DMSO-d6) δ 10.14 (s, 1H), 8.83 (s, 1H), 8.05 (s, 1H), 7.97 (t, J = 5.6 Hz, 1H), 7.76 (d, J = 8.1 Hz, 1H), 7.61 (d, J = 7.4 Hz, 1H), 6.92 (d, J = 8.8 Hz, 2H), 6.87 – 6.75 (m, 1H), 6.21 (d, J = 3.2 Hz, 1H), 6.11 – 6.06 (m, 1H), 5.76 (s, 2H), 5.67 (ddt, J = 16.3, 10.2, 6.0 Hz, 1H), 5.32 (s, 1H), 5.00 (dq, J = 10.2, 1.4 Hz, 1H), 4.84 (dq, J = 17.1, 1.5 Hz, 1H), 4.69 (d, J = 6.0 Hz, 2H), 4.26 (s, 1H), 3.95 (s, 1H), 3.77 (d, J = 24.9 Hz, 2H), 3.59 – 3.48 (m, 9H), 3.41 (t, J = 5.9 Hz, 2H), 3.21 (q, J = 5.8 Hz, 2H), 3.09 (d, J = 5.3 Hz, 4H), 2.83 – 2.76 (m, 2H), 2.57 (t, J = 5.0 Hz, 4H), 2.51 (p, J = 1.9 Hz, 9H), 1.47 (s, 6H).
13C NMR (151 MHz, DMSO-d6) δ 171.30, 164.62, 156.47, 150.02, 147.70, 139.28, 132.68, 128.76, 118.72, 116.75, 115.93, 106.92, 100.44, 72.78, 70.26, 70.23, 70.17, 70.08, 69.59, 68.87, 57.72, 55.38, 53.62, 49.15, 47.54, 47.07, 46.87, 42.45, 39.05, 33.64, 30.92, 23.88.
HRMS (ESI): [M+H]+ m/z calc. 936.47, found 936.4723.
Synthesis of NJH-2-106
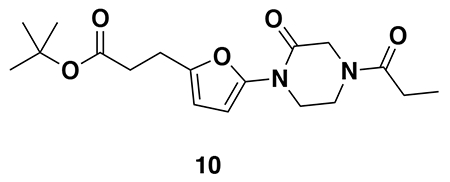
tert-butyl 3-(5-(2-oxo-4-propionylpiperazin-1-yl)furan-2-yl)propanoate (10): Intermediate 2 (benzyl (E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-3-oxopiperazine-1-carboxylate) (85 mg, 0.20 mmol) was dissolved in EtOH (5 mL) and Pd/C (10 mg, 10% wt.) was added. The atmosphere was exchanged for hydrogen (balloon) and the mixture was stirred vigorously overnight. After 16h, the suspension was diluted with DCM and filtered through Celite to remove Pd/C, then concentrated. The crude residue was redissolved in DCM (2 mL), and TEA (83 μL, 0.60 mmol) was added. The solution was then cooled to 0 °C and propionyl chloride (25 μL, 0.31 mmol) was added and the mixture stirred for 30 min at 0 °C. Water was added and the mixture was extracted with DCM three times. Organic extracts were combined, washed with brine, dried over sodium sulfate, and concentrated. The crude residue was purified by silica gel chromatography to provide Intermediate 10 (48 mg, 0.14 mmol, 69% yield over two steps) as a white solid.
1H NMR (600 MHz, CDCl3) δ 6.28 (d, J = 3.2 Hz, 1H), 6.04 (d, J = 3.2 Hz, 1H), 4.40 (s, 1H), 4.29 (s, 1H), 3.91 (dt, J = 30.8, 5.3 Hz, 2H), 3.85 – 3.78 (m, 2H), 2.88 (t, J = 7.6 Hz, 2H), 2.54 (t, J = 7.5 Hz, 2H), 2.43 – 2.34 (m, 2H), 1.44 (s, 9H), 1.19 (q, J = 6.9 Hz, 3H).
LC/MS: [M+H]+ m/z calc. 351.2, found 351.2.
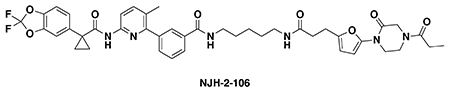
3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)-N-(5-(3-(5-(2-oxo-4-propionylpiperazin-1-yl)furan-2-yl)propanamido)pentyl)benzamide (NJH-2-106): Intermediate 10 (15 mg, 0.043 mmol) and Intermediate 5c (15 mg, 0.029 mmol) were reacted according to General Procedure C to provide NJH-2-106 (18 mg, 0.022 mmol, 76%) as a clear colorless oil.
1H NMR (400 MHz, CDCl3) δ 8.10 (d, J = 8.4 Hz, 1H), 7.82 (s, 1H), 7.79 (d, J = 7.6 Hz, 1H), 7.72 (s, 1H), 7.59 (d, J = 8.5 Hz, 1H), 7.52 (d, J = 7.7 Hz, 1H), 7.46 (t, J = 7.6 Hz, 1H), 7.22 (dd, J = 8.1, 1.8 Hz, 1H), 7.19 (d, J = 1.7 Hz, 1H), 7.07 (d, J = 8.1 Hz, 1H), 6.51 – 6.39 (m, 1H), 6.22 – 6.15 (m, 1H), 6.02 – 5.97 (m, 1H), 5.88 – 5.76 (m, 1H), 4.31 (d, J = 50.6 Hz, 2H), 3.96 – 3.71 (m, 4H), 3.42 (q, J = 6.6 Hz, 2H), 3.21 (q, J = 6.5 Hz, 2H), 2.92 – 2.82 (m, 2H), 2.44 (t, J = 7.4 Hz, 2H), 2.41 – 2.29 (m, 2H), 2.24 (s, 3H), 1.74 (q, J = 3.9 Hz, 2H), 1.64 – 1.56 (m, 2H), 1.53 – 1.43 (m, 2H), 1.38 – 1.26 (m, 2H), 1.21 – 1.09 (m, 5H).
13C NMR (151 MHz, CDCl3) δ 206.9, 172.2, 171.8, 167.3, 155.4, 149.8, 148.9, 144.9, 144.1, 143.6, 141.0, 140.1, 134.9, 134.8, 131.7, 131.7, 128.4, 127.5, 127.0, 126.6, 113.0, 112.4, 110.2, 107.3, 100.9, 53.4, 49.3, 47.2, 42.4, 39.7, 39.1, 38.7, 34.9, 31.2, 29.0, 26.5, 24.2, 23.8, 19.1, 17.2, 9.0.
HRMS (ESI): [M+H]+ m/z calc. 813.3345, found 813.3422.
Synthesis of NJH-2-080:
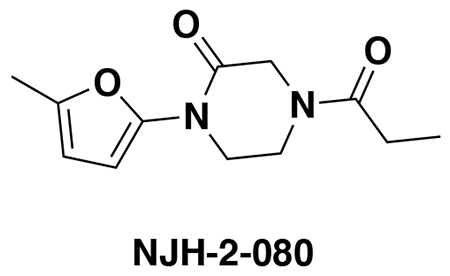
1-(5-methylfuran-2-yl)-4-propionylpiperazin-2-one (NJH-2-080): 1-(5-methylfuran-2-yl)piperazin-2-one (30 mg, 0.17 mmol) was dissolved in DCM (2 mL). The solution was cooled to 0 °C and TEA (69 μL, 0.50 mmol) and propionyl chloride (21 μL, 0.25 mmol) were added. After stirring at 0 °C for 30 min, water was added, and the reaction extracted three times with DCM. Organic extracts were combined, washed with brine, dried over sodium sulfate, and concentrated. The crude residue was purified by silica gel chromatography (0-100% EtOAc/Hex) to provide NJH-2-080 (17.3 mg, 0.073 mmol, 43%) as a white solid.
1H NMR (600 MHz, CDCl3) δ 6.25 (d, J = 3.2 Hz, 1H), 6.00 (d, J = 2.2 Hz, 1H), 4.41 (s, 1H), 4.29 (s, 1H), 3.97 – 3.86 (m, 2H), 3.82 (t, J = 5.5 Hz, 2H), 2.45 – 2.34 (m, 2H), 2.27 (s, 3H), 1.23 – 1.16 (m, 3H). 13C NMR (151 MHz, CDCl3) δ 172.2, 163.4, 147.5, 144.4, 107.2, 101.0, 49.3, 46.9, 38.8, 26.5, 13.4, 9.0.
13C NMR (151 MHz, CDCl3) δ 172.2, 163.4, 147.5, 144.4, 107.2, 101.0, 49.3, 46.9, 38.8, 26.5, 13.4, 9.0.
HRMS (ESI): [M+H]+ m/z calc. 259.1160, found 259.1053.
SYNTHESIS OF NJH-2-075
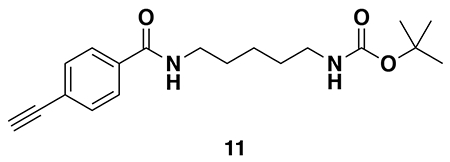
tert-butyl (5-(4-ethynylbenzamido)pentyl)carbamate (11): 4-ethynylbenzoic acid (27 mg, 0.19 mmol), N-Boc-1,5-diaminopentane (47 mg, 0.23 mmol), HOBt (26 mg, 0.19 mmol), and DIEA (165 μL, 0.95 mmol) were dissolved in DCM (1.5 mL) and EDCI•HCl (73 mg, 0.38 mmol) was added. After stirring the mixture for 16h at rt, water was added, the mixture partitioned, and the aqueous phase extracted with DCM. Combined organic extracts were washed with brine and dried over Na2SO4, concentrated, and the crude residue was purified by silica gel chromatography (0-50% EtOAc/Hex) to obtain the Boc-protected amine 11 (27 mg, 0.082 mmol, 43%) as a white solid.
1H NMR (300 MHz, CDCl3) δ 7.78 (d, J = 8.3 Hz, 2H), 7.59 (d, J = 8.7 Hz, 2H), 6.32 (s, 1H), 4.63 (s, 1H), 3.50 (td, J = 7.0, 5.7 Hz, 2H), 3.23 (s, 1H), 3.18 (q, J = 6.5 Hz, 2H), 1.70 (d, J = 7.5 Hz, 2H), 1.62 – 1.52 (m, 2H), 1.46 (s, 11H).
LC/MS [M+H]+ m/z calc. 331.19, found 331.1.
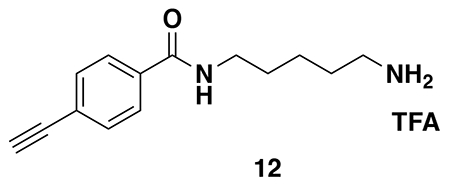
N-(5-aminopentyl)-4-ethynylbenzamide (12): tert-butyl (5-(4-ethynylbenzamido)pentyl)carbamate 11 (27 mg, 0.082 mmol) was dissolved in DCM (1 mL) and TFA (0.5 mL) was added. After stirring at rt for 2h, the mixture was diluted in DCM and evaporated repeatedly to remove volatiles and provide the amine as a TFA salt and an orange oil (32 mg, 0.096 mmol, 117%), which was used without further purification.
1H NMR (400 MHz, DMSO) δ 8.55 (t, J = 5.7 Hz, 1H), 7.84 (d, J = 8.2 Hz, 2H), 7.63 (s, 2H), 7.57 (d, J = 8.1 Hz, 2H), 4.39 (s, 1H), 3.26 (q, J = 6.6 Hz, 2H), 2.83 – 2.74 (m, 2H), 1.62 – 1.48 (m, 4H), 1.40 – 1.32 (m, 2H).
LC/MS [M+H]+ m/z calc. 231.14, found 231.1.
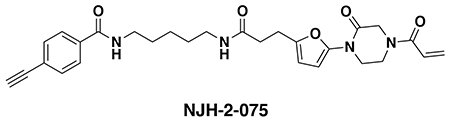
N-(5-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanamido)pentyl)-4-ethynylbenzamide (NJH-2-075): Intermediate 3, tert-butyl 3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoate, (20 mg, 0.057 mmol) was dissolved in DCM (0.5 mL) and treated with TFA (0.25 mL). The mixture was stirred at rt for 45 minutes until the starting material was consumed, followed by dilution with DCM and evaporation to remove volatiles. The carboxylic acid was then dissolved in DMF, and intermediate 12 N-(5-aminopentyl)-4-ethynylbenzamide TFA (22 mg, 0.062 mmol), DIEA (50 μL, 0.29 mmol), and HATU (43 mg, 0.11 mmol) were added. After stirring the mixture at rt for 1 h, water was added. The resulting suspension was extracted three times with DCM. Combined organic extracts were washed brine and dried over Na2SO4, concentrated, and the crude residue was purified by silica gel chromatography (0-4% MeOH/DCM) to obtain NJH-2-075 (7.6 mg, 0.016 mmol, 27%) as a pale yellow oil.
1H NMR (300 MHz, CDCl3) δ 7.82 (d, J = 8.3 Hz, 2H), 7.58 (d, J = 8.3 Hz, 2H), 6.77 – 6.50 (m, 2H), 6.43 (dd, J = 16.7, 2.1 Hz, 1H), 6.24 (d, J = 3.2 Hz, 1H), 6.06 (d, J = 3.3 Hz, 1H), 5.93 (s, 1H), 5.86 (dd, J = 10.1, 2.1 Hz, 1H), 4.44 (d, J = 17.4 Hz, 2H), 4.01 (s, 2H), 3.91 – 3.84 (m, 2H), 3.46 (q, J = 6.6 Hz, 2H), 3.32 – 3.19 (m, 3H), 2.93 (t, J = 7.2 Hz, 2H), 2.50 (t, J = 7.3 Hz, 2H), 1.72 – 1.61 (m, 2H), 1.60 – 1.46 (m, 2H), 1.44 – 1.35 (m, 2H).
13C NMR (151 MHz, DMSO) δ 171.0, 165.7, 164.6, 150.1, 135.2, 132.1, 128.8, 127.9, 124.7, 106.9, 100.5, 83.4, 83.1, 38.9, 33.8, 29.3, 29.2, 24.3, 24.0.
HRMS [M+H]+ m/z calc. 380.1586, found 380.1581.
Synthesis of EN523 Analogs:
Synthesis of EZ-1-032
1-(1-acryloylpiperidin-4-yl)-1,3-dihydro-2H-benzo[d]imidazol-2-one (EZ-1-032): 1-(piperidin-4-yl)-1,3-dihydro-2H-benzo[d]imidazol-2-one (50 mg, 0.23 mmol) was acylated via general procedure H and the crude residue was purified by silica gel chromatography (0 to 20% MeOH/DCM) to afford the title compound as a clear yellow oil (11.8 mg, 0.043 mmol, 19%).
1H NMR (400 MHz, DMSO) δ 10.87 (s, 1H), 7.29 – 7.17 (m, 1H), 7.05 – 6.95 (m, 3H), 6.88 (ddd, J = 16.1, 10.5, 3.3 Hz, 1H), 6.16 (d, J = 2.4 Hz, 1H), 5.70 (dd, J = 10.4, 2.4 Hz, 1H), 4.61 (d, J = 13.1 Hz, 1H), 4.44 (tt, J = 12.0, 3.9 Hz, 1H), 4.21 (d, J = 13.8 Hz, 1H), 3.21 (t, J = 13.3 Hz, 1H), 2.76 (t, J = 12.9 Hz, 1H), 2.34 – 2.07 (m, 2H), 1.75 (d, J = 12.4 Hz, 2H).
13C NMR (151 MHz, DMSO) δ 164.8, 154.2, 129.7, 129.0, 129.0, 127.7, 121.1, 120.9, 109.3, 109.0, 50.3, 45.1, 41.6, 29.9, 29.0.
HRMS (ESI): [M+H]+ m/z calc. 272.14, found 272.1394.
Synthesis of EZ-1-038
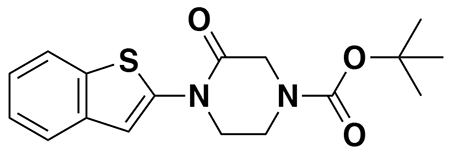
tert-butyl 4-(benzo[b]thiophen-2-yl)-3-oxopiperazine-1-carboxylate (EZ-1-035): 2-bromobenzo[b]thiophene (100 mg, 0.47 mmol) was coupled to tert-butyl 3-oxopiperazine-1-carboxylate (93.5 mg, 0.47 mmol) via general procedure D and the crude residue was purified by silica gel chromatography (0 to 100% EtOAc/hexane) to yield a pale yellow solid (22.3 mg, 0.116 mmol, 14%).
1H NMR (400 MHz, CDCl3) δ 7.81 (d, J = 7.8 Hz, 1H), 7.72 (d, J = 7.7 Hz, 1H), 7.30 (s, 2H), 6.92 (s, 1H), 4.40 (s, 2H), 4.01 (t, J = 5.4 Hz, 2H), 3.92 (t, J = 5.4 Hz, 2H), 1.54 (s, 9H).
LC/MS: [M+H]+ m/z calc. 333.1, found 333.1
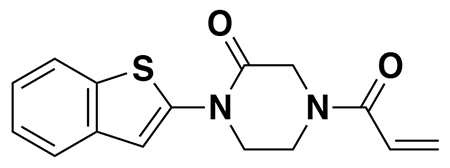
4-acryloyl-1-(benzo[b]thiophen-2-yl)piperazin-2-one (EZ-1-038): tert-butyl 4-(benzo[b]thiophen-2-yl)-3-oxopiperazine-1-carboxylate (EZ-1-035) (18 mg, 0.05 mmol) was deprotected and acylated via general procedures F and H respectively. The crude residue was purified by silica gel chromatography (0 to 100% EtOAc/Hex) to afford the title compound as a pale yellow solid (6.6 mg, 0.023 mmol, 46%).
1H NMR (400 MHz, DMSO) δ 7.86 (d, J = 7.9 Hz, 1H), 7.74 (t, J = 7.2 Hz, 1H), 7.45 – 7.32 (m, 1H), 7.28 (q, J = 6.8 Hz, 1H), 7.11 (s, 1H), 6.98 – 6.77 (m, 1H), 6.21 (d, J = 16.7 Hz, 1H), 5.83 – 5.74 (m, 1H), 4.50 (d, J = 68.5 Hz, 2H), 4.18 – 3.91 (m, 4H).
13C NMR (151 MHz, DMSO) δ 164.7, 142.0, 136.7, 136.2, 128.9, 128.0, 124.9, 123.9, 122.8, 122.1, 108.0, 49.2, 48.4, 47.6, 46.8.
HRMS (ESI): [M+Na]+ m/z calc. 309.0674, found 309.0667.
Synthesis of EZ-1-045
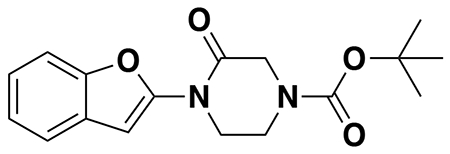
tert-butyl 4-(benzofuran-2-yl)-3-oxopiperazine-1-carboxylate (EZ-1-044): 2-bromobenzofuran (200 mg, 1.02 mmol) was coupled with tert-butyl 3-oxopiperazine-1-carboxylate (204.24 mg, 1.02 mmol) via general procedure D and purified by silica gel chromatography (0 to 50% EtOAc/hexane) to yield a yellow solid (44.3 mg, 0.14 mmol, 14%).
1H NMR (400 MHz, CDCl3) δ 7.65 – 7.52 (m, 1H), 7.48 – 7.39 (m, 1H), 7.26 (dd, J = 6.0, 3.3 Hz, 2H), 6.96 (d, J = 1.2 Hz, 1H), 4.35 (s, 2H), 4.19 – 4.05 (m, 2H), 3.86 (d, J = 5.6 Hz, 2H), 1.53 (d, J = 1.6 Hz, 9H).
LC/MS: [M+H]+ m/z calc. 316.1, found 316.2
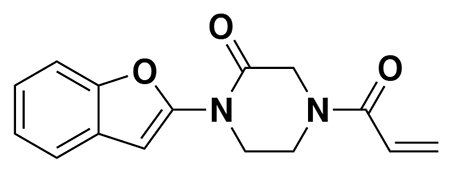
4-acryloyl-1-(benzofuran-2-yl)piperazin-2-one (EZ-1-045): tert-butyl 4-(benzofuran-2-yl)-3-oxopiperazine-1-carboxylate (EZ-1-044) (44.3 mg, 0.14 mmol) was deprotected and acylated via general procedures F and H and purified by silica gel chromatography (0 to 50% EtOAc/hexane) to afford the title compound as a pale yellow solid (9.3 mg, 0.034 mmol, 25%).
1H NMR (300 MHz, CDCl3) δ 7.63 – 7.51 (m, 1H), 7.43 (dt, J = 7.1, 3.8 Hz, 1H), 7.29 (td, J = 6.3, 2.8 Hz, 2H), 6.98 (d, J = 1.0 Hz, 1H), 6.57 (d, J = 9.8 Hz, 1H), 6.47 (dd, J = 16.7, 2.2 Hz, 1H), 5.88 (dd, J = 10.1, 2.2 Hz, 1H), 4.52 (s, 2H), 4.24 – 3.92 (m, 4H).
13C NMR (151 MHz, DMSO) δ 165.0, 150.1, 149.5, 129.0, 128.8, 128.1, 123.9, 123.9, 121.2, 111.1, 94.6, 49.5, 47.1, 46.6, 42.4.
HRMS (ESI): [M+H]+ m/z calc. 271.1004, found 271.1078.
Synthesis of EZ-1-055
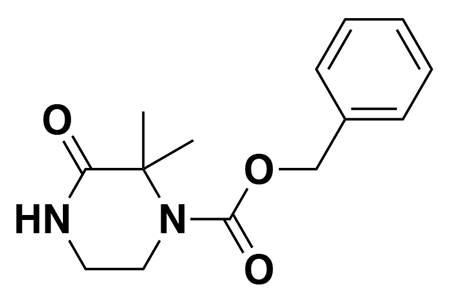
benzyl 2,2-dimethyl-3-oxopiperazine-1-carboxylate (EZ-1-050): 3,3-dimethylpiperazin-2-one (400 mg, 3.12 mmol) was protected with benzyl chloroformate via general procedure E and purified by silica gel chromatography (0 to 10% MeOH/DCM) to yield a fluffy white powder (492.1 mg, 1.88 mmol, 60%).
1H NMR (300 MHz, CDCl3) δ 7.41 (s, 5H), 6.02 (s, 1H), 5.19 (s, 2H), 3.87 – 3.74 (m, 2H), 3.49 – 3.35 (m, 2H), 1.75 (s, 6H).
LC/MS: [M+H]+ m/z calc. 263.1, found 263.1.
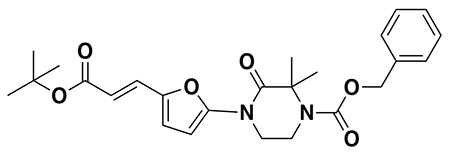
benzyl (E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-2,2-dimethyl-3-oxopiperazine-1-carboxylate (EZ-1-052): tert-butyl (E)-3-(5-bromofuran-2-yl)acrylate (Intermediate 2) (104 mg, 0.38 mmol) and benzyl 2,2-dimethyl-3-oxopiperazine-1-carboxylate (EZ-1-050) (100 mg, 0.38 mmol) were coupled via general procedure D and purified by silica gel chromatography (0 to 50% EtOAc/hexane) to yield a clear yellow oil that solidified upon standing (133.7 mg, 0.29 mmol, 77%).
1H NMR (400 MHz, CDCl3) δ 7.43 (d, J = 5.1 Hz, 6H), 6.66 (q, J = 3.6 Hz, 2H), 6.12 (d, J = 15.6 Hz, 1H), 5.22 (s, 2H), 4.04 – 3.98 (m, 2H), 3.91 (d, J = 5.2 Hz, 2H), 1.80 (s, 6H), 1.56 (d, J = 4.0 Hz, 9H).
LC/MS: [M+H]+ m/z calc. 455.2, found 455.2
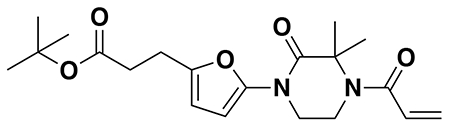
tert-butyl 3-(5-(4-acryloyl-3,3-dimethyl-2-oxopiperazin-1-yl)furan-2-yl)propanoate (EZ-1-055): benzyl (E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-2,2-dimethyl-3-oxopiperazine-1-carboxylate (EZ-1-052) (30 mg, 0.066 mmol) was deprotected and acylated via general procedures G and H and purified by silica gel chromatography (0-70% EtOAc/hexane) to afford the title compound as a clear colorless oil (7.2 mg, 0.019 mmol, 29% over two steps).
1H NMR (400 MHz, CDCl3) δ 6.51 (ddd, J = 16.8, 10.6, 2.3 Hz, 1H), 6.29 (t, J = 2.9 Hz, 1H), 6.23 (dt, J = 16.8, 2.1 Hz, 1H), 6.03 (d, J = 3.2 Hz, 1H), 5.70 (dt, J = 10.5, 2.1 Hz, 1H), 3.88 (dd, J = 6.4, 3.4 Hz, 2H), 3.78 (dd, J = 6.1, 3.6 Hz, 2H), 2.87 (t, J = 7.6 Hz, 2H), 2.54 (td, J = 7.9, 2.3 Hz, 2H), 1.83 (d, J = 2.3 Hz, 6H), 1.44 (d, J = 2.3 Hz, 9H).
13C NMR (151 MHz, DMSO) δ 171.6, 171.1, 166.3, 149.1, 146.2, 131.5, 127.2, 107.2, 99.7, 80.4, 63.6, 47.5, 42.7, 28.2, 23.8, 23.5.
HRMS (ESI): [M+Na]+ m/z calc. 399.1896, found 399.1883.
Synthesis of EZ-1-058
Benzyl 2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-049): 3-methylpiperazin-2-one (400 mg, 3.5 mmol) was protected with benzyl chloroformate via general procedure E and purified by silica gel chromatography (0 to 10% MeOH/DCM) to yield a pale yellow solid (123.9 mg, 0.5 mmol, 14%).
1H NMR (300 MHz, CDCl3) δ 7.36 (s, 5H), 5.96 (s, 1H), 5.16 (s, 2H), 4.69 (s, 1H), 4.18 (s, 1H), 3.47 (d, J = 12.1 Hz, 1H), 3.27 (d, J = 12.2 Hz, 2H), 1.46 (d, J = 7.1 Hz, 3H).
LC/MS: [M+H]+ m/z calc. 249.1, found 249.1.
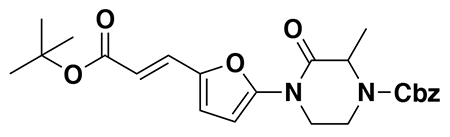
benzyl (E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-051): Benzyl 2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-049) (60 mg, 0.24 mmol) and tert-butyl (E)-3-(5-bromofuran-2-yl)acrylate (EZ-1-048) (66 mg, 0.24 mmol) were coupled via general procedure D and purified by silica gel chromatography (0 to 50% EtOAc/hexane) to yield a yellow solid (69.3 mg, 0.16 mmol, 66%).
1H NMR (400 MHz, CDCl3) δ 7.42 (s, 5H), 7.32 – 7.24 (m, 1H), 6.70 – 6.62 (m, 2H), 6.12 (d, J = 15.4 Hz, 1H), 5.23 (d, J = 2.5 Hz, 2H), 4.89 (s, 1H), 4.35 (s, 1H), 4.00 (d, J = 13.9 Hz, 2H), 3.50 (s, 1H), 1.72 – 1.49 (m, 12H).
LC/MS: [M+H]+ m/z calc. 441.2, found 441.2.
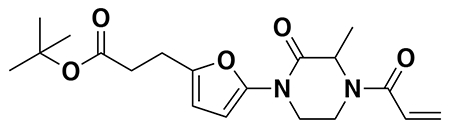
tert-butyl 3-(5-(4-acryloyl-3-methyl-2-oxopiperazin-1-yl)furan-2-yl)propanoate (EZ-1-058): benzyl (E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-051) (52.3 mg, 0.12 mmol) was deprotected and acylated via general procedures G and H and purified by silica gel chromatography (0-100% EtOAc/hexane) to yield the title compound as a clear colorless oil (17.9 mg, 0.05 mmol, 42% over two steps).
1H NMR (400 MHz, CDCl3) δ 6.66 – 6.51 (m, 1H), 6.46 (d, J = 16.7 Hz, 1H), 6.32 (d, J = 3.2 Hz, 1H), 6.07 (dd, J = 3.2, 1.0 Hz, 1H), 5.84 (d, J = 9.8 Hz, 1H), 4.74 (s, 1H), 4.23 – 3.23 (m, 4H), 2.91 (t, J = 7.6 Hz, 2H), 2.57 (dd, J = 8.2, 6.9 Hz, 3H), 1.63 (s, 3H), 1.47 (s, 9H).
13C NMR (151 MHz, DMSO) δ 171.6, 167.6, 164.2, 149.5, 145.9, 128.7, 128.2, 107.2, 100.7, 80.4, 60.2, 54.5, 52.0, 48.2, 33.4, 28.2, 23.5, 17.0.
HRMS (ESI): [M+Na]+ m/z calc. 385.1739, found 385.1728.
Synthesis of EZ-1-067
tert-butyl 3-oxo-4-(2-phenyloxazol-5-yl)piperazine-1-carboxylate (EZ-1-066): 5-bromo-2-phenyloxazole (50 mg, 0.22 mmol) was coupled with tert-butyl 3-oxopiperazine-1-carboxylate (44.7 mg, 0.22 mmol) via general procedure D and purified by silica gel chromatography (0 to 60% EtOAc/hexane) to yield a white solid (40.4 mg, 0.117 mmol, 54%).
1H NMR (400 MHz, CDCl3) δ 8.05 – 7.98 (m, 2H), 7.49 (dd, J = 5.7, 1.8 Hz, 3H), 7.38 (s, 1H), 4.36 (s, 2H), 4.04 (t, J = 5.4 Hz, 2H), 3.89 (t, J = 5.3 Hz, 2H), 1.55 (s, 9H).
LC/MS: [M+H]+ m/z calc. 344.2, found 344.1.
4-acryloyl-1-(2-phenyloxazol-5-yl)piperazin-2-one (EZ-1-067): tert-butyl 3-oxo-4-(2-phenyloxazol-5-yl)piperazine-1-carboxylate (EZ-1-066) (40.4 mg, 0.117 mmol) was deprotected and acylated via general procedures F and H and purified by silica gel chromatography (0 to 80% EtOAc/hexane) to afford the title compound as a yellow solid (34.6 mg, 0.116 mmol, 45% over two steps)
1H NMR (300 MHz, CDCl3) δ 8.01 (dd, J = 6.8, 3.0 Hz, 2H), 7.54 – 7.46 (m, 3H), 7.39 (s, 1H), 6.59 (s, 1H), 6.54 – 6.42 (m, 1H), 5.90 (d, J = 11.6 Hz, 1H), 4.53 (s, 2H), 4.10 (s, 4H).
13C NMR (151 MHz, DMSO) δ 164.6, 155.1, 146.6, 130.8, 129.6, 128.9, 128.3, 128.1, 127.1, 125.9, 116.2, 49.4, 47.2, 46.9.
HRMS (ESI): [M+H]+ m/z calc. 298.1113, found 298.1187.
Synthesis of EZ-1-068
phenyl (R)-2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-062): (R)-3-methylpiperazin-2-one (100 mg, 0.88 mmol) was protected with benzyl chloroformate (186 μL, 0.876 mmol) via general procedure E and purified by silica gel chromatography (0 to 100% EtOAc/hexane) to yield a white solid (47.2 mg, 0.25 mmol, 22%).
1H NMR (400 MHz, CDCl3) δ 6.15 (s, 1H), 5.21 (s, 2H), 4.73 (s, 1H), 4.24 (s, 1H), 3.51 (d, J = 12.5 Hz, 1H), 3.31 (d, J = 12.6 Hz, 2H), 1.50 (d, J = 7.0 Hz, 3H).
LC/MS: [M+H]+ m/z calc. 248.1, found 248.1.
benzyl (R,E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-064): phenyl (R)-2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-062) (44.6 mg, 0.18 mmol) was coupled to tert-butyl (E)-3-(5-bromofuran-2-yl)acrylate (EZ-1-048) (49.1 mg, 0.18 mmol) via general procedure D and purified by silica gel chromatography (0 to 35% EtOAc/hexane) to yield a clear yellow oil (56.7 mg, 0.13 mmol, 72%).
1H NMR (400 MHz, CDCl3) δ 7.42 (d, J = 5.3 Hz, 5H), 7.30 (s, 1H), 6.74 – 6.62 (m, 2H), 6.12 (d, J = 15.6 Hz, 1H), 5.23 (d, J = 2.3 Hz, 2H), 4.89 (s, 1H), 4.34 (s, 1H), 4.02 (s, 2H), 3.49 (s, 1H), 1.61 (s, 3H), 1.56 (d, J = 5.5 Hz, 9H).
LC/MS: [M+H]+ m/z calc. 441.2, found 441.2.
tert-butyl (R)-3-(5-(4-acryloyl-3-methyl-2-oxopiperazin-1-yl)furan-2-yl)propanoate (EZ-1-068): benzyl (R,E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-064) (31.2 mg, 0.07 mmol) was deprotected and acylated via general procedures F and H and purified by silica gel chromatography (0 to 100% EtOAc/hexane) to afford the title compound as a yellow solid (18.9 mg, 0.052 mmol, 68% over two steps).
1H NMR (300 MHz, CDCl3) δ 6.65 – 6.40 (m, 2H), 6.33 (d, J = 3.4 Hz, 1H), 6.08 (d, J = 3.3 Hz, 1H), 5.90 – 5.81 (m, 1H), 4.76 (s, 1H), 3.93 – 3.34 (m, 4H), 2.92 (t, J = 7.5 Hz, 2H), 2.58 (dd, J = 8.3, 6.8 Hz, 2H), 2.22 (s, 3H), 1.48 (s, 9H).
13C NMR (151 MHz, DMSO) δ 171.6, 167.6, 164.2, 149.4, 145.9, 128.7, 128.2, 107.2, 100.6, 80.4, 52.0, 48.2, 47.2, 33.4, 28.2, 23.5.
HRMS (ESI): [M+Na]+ m/z calc. 385.1739, found 385.1730.
Synthesis of EZ-1-071
benzyl (S)-2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-063): (S)-3-methylpiperazin-2-one (100mg , 0.88mmol) was protected with benzyl chloroformate (149.4 mg, 0.88 mmol) via general procedure E and purified by silica gel chromatography (0 to 100% EtOAc/hexane) to yield a white solid (89.4 mg, 0.36 mmol, 41%).
1H NMR (400 MHz, CDCl3) δ 7.40 (d, J = 4.6 Hz, 5H), 6.13 (s, 1H), 5.21 (s, 2H), 4.72 (s, 1H), 4.24 (s, 1H), 3.53 (s, 1H), 3.31 (d, J = 12.5 Hz, 2H).
LC/MS: [M+H]+ m/z calc. 248.1, found 248.1.
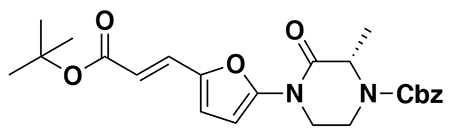
benzyl (S,E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-069): benzyl (S)-2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-063) (41.6 mg, 0.17 mmol) was coupled to tert-butyl (E)-3-(5-bromofuran-2-yl)acrylate (EZ-1-048) (46.8 mg, 0.17 mmol) via general procedure D and purified by silica gel chromatography (0 to 50% EtOAc/hexane) to yield a clear yellow oil (41.3 mg, 0.09 mmol, 56%).
1H NMR (300 MHz, CDCl3) δ 7.45 – 7.37 (m, 5H), 7.31 (d, J = 1.3 Hz, 1H), 6.72 – 6.61 (m, 2H), 6.12 (d, J = 15.6 Hz, 1H), 5.23 (d, J = 1.3 Hz, 2H), 4.90 (s, 1H), 4.34 (s, 1H), 4.05 – 3.92 (m, 2H), 3.49 (s, 1H), 1.62 (s, 3H), 1.56 (d, J = 3.1 Hz, 9H).
LC/MS: [M+H]+ m/z calc. 441.2, found 441.2.
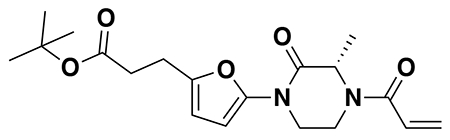
tert-butyl (S)-3-(5-(4-acryloyl-3-methyl-2-oxopiperazin-1-yl)furan-2-yl)propanoate (EZ-1-071): benzyl (S,E)-4-(5-(3-(tert-butoxy)-3-oxoprop-1-en-1-yl)furan-2-yl)-2-methyl-3-oxopiperazine-1-carboxylate (EZ-1-069) (35.4 mg, 0.08 mmol) was deprotected and acylated via general procedures F and H and purified by silica gel chromatography (0 to 100% EtOAc/hexane) to afford the title compound as a clear colorless oil (16.9 mg, 0.047 mmol, 58% over two steps).
1H NMR (400 MHz, CDCl3) δ 6.63 – 6.41 (m, 2H), 6.32 (d, J = 3.4 Hz, 1H), 6.07 (d, J = 3.5 Hz, 1H), 5.85 (d, J = 10.3 Hz, 1H), 4.77 (s, 2H), 3.88 (s, 2H), 3.34 (s, 1H), 2.91 (t, J = 7.5 Hz, 2H), 2.58 (dt, J = 8.8, 5.2 Hz, 2H), 1.74 (s, 3H), 1.48 (d, J = 4.0 Hz, 9H).
13C NMR (151 MHz, DMSO) δ 171.6, 164.2, 149.5, 145.9, 128.7, 128.1, 107.2, 100.7, 80.4, 54.4, 52.0, 48.2, 33.4, 28.2, 23.5.
HRMS (ESI): [M+Na]+ m/z calc. 385.1739, found 385.1726.
Synthesis of EZ-1-073
tert-butyl 4-(imidazo[1,2-a]pyridin-2-yl)-3-oxopiperazine-1-carboxylate (EZ-1-072): 2-bromoimidazo[1,2-a]pyridine (50 mg, 0.25 mmol) was coupled to tert-butyl 3-oxopiperazine-1-carboxylate (50.8 mg, 0.25 mmol) via general procedure D and purified by silica gel chromatography (0 to 80% EtOAc/hexane) to yield a clear colorless oil (35.7 mg, 0.11 mmol, 45%).
1H NMR (400 MHz, CDCl3) δ 8.33 (s, 1H), 8.15 (d, J = 6.7 Hz, 1H), 7.54 (d, J = 9.1 Hz, 1H), 7.22 (ddd, J = 8.7, 6.9, 1.4 Hz, 1H), 6.85 (td, J = 6.8, 1.3 Hz, 1H), 4.34 (s, 2H), 4.31 (t, J = 5.5 Hz, 2H), 3.83 (t, J = 5.4 Hz, 2H), 1.53 (s, 9H).
LC/MS: [M+H]+ m/z calc. 317.2, found 317.2.
4-acryloyl-1-(imidazo[1,2-a]pyridin-2-yl)piperazin-2-one (EZ-1-073): tert-butyl 4-(imidazo[1,2-a]pyridin-2-yl)-3-oxopiperazine-1-carboxylate (EZ-1-072) (23.4 mg, 0.074 mmol) was deprotected and acylated via general procedures F and H and purified by silica gel chromatography (0 to 100% EtOAc/hexane) to afford the title compound as an off white solid (3.8 mg, 0.014 mmol, 19% over two steps).
1H NMR (400 MHz, CDCl3) δ 8.32 (s, 1H), 8.15 (d, J = 6.9 Hz, 1H), 7.55 (d, J = 9.0 Hz, 1H), 7.24 (t, J = 7.9 Hz, 1H), 6.87 (t, J = 6.8 Hz, 1H), 6.62 (s, 1H), 6.46 (d, J = 16.7 Hz, 1H), 5.86 (d, J = 10.5 Hz, 1H), 4.53 (d, J = 23.7 Hz, 2H), 4.38 (s, 2H), 4.04 (d, J = 33.0 Hz, 2H).
HRMS (ESI): [M+H]+ m/z calc. 271.1117, found 271.1190.
Synthesis of EZ-1-085
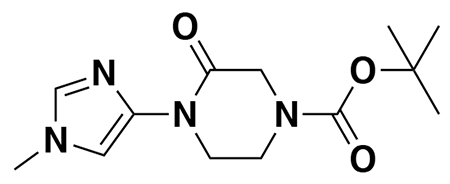
tert-butyl 4-(1-methyl-1H-imidazol-4-yl)-3-oxopiperazine-1-carboxylate (EZ-1-084): 4-bromo-1-methyl-1H-imidazole (155 μL, 1.55 mmol) was coupled to tert-butyl 3-oxopiperazine-1-carboxylate (311 mg, 1.55 mmol) via general procedure D and the crude residue was purified by silica gel chromatography (0-100% EtOAc/Hex) to yield a pale yellow solid (412 mg, 1.47 mmol, 95%).
1H NMR (400 MHz, CDCl3) δ 7.58 – 7.50 (m, 1H), 7.39 – 7.26 (m, 1H), 4.27 (d, J = 9.5 Hz, 2H), 4.18 – 4.06 (m, 3H), 3.80 – 3.61 (m, 4H), 1.51 (d, J = 4.1 Hz, 9H).
LC/MS: [M+H]+ m/z calc. 281.2, found 281.2.
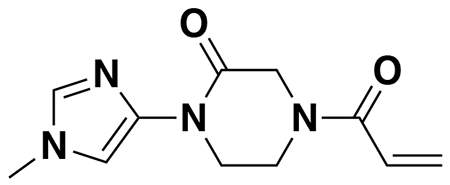
4-acryloyl-1-(1-methyl-1H-imidazol-4-yl)piperazin-2-one (EZ-1-085): tert-butyl 4-(1-methyl-1H-imidazol-4-yl)-3-oxopiperazine-1-carboxylate (EZ-1-084) (100 mg, 0.36 mmol) was deprotected and acylated via general procedures F and H and the crude residue was purified by silica gel chromatography (0 to 10% MeOH/DCM) to afford the title compound as a white solid (27.7 mg, 0.12 mmol, 33%).
1H NMR (300 MHz, CDCl3) δ 7.54 (s, 1H), 7.27 (s, 1H), 6.58 (s, 1H), 6.44 (dd, J = 16.7, 2.0 Hz, 1H), 5.88 – 5.81 (m, 1H), 4.46 (d, J = 16.0 Hz, 2H), 4.18 (s, 2H), 3.99 (d, J = 23.0 Hz, 2H), 3.73 (s, 3H).
13C NMR (151 MHz, DMSO) δ 163.4, 162.9, 138.9, 133.9, 128.6, 128.5, 128.2, 46.9, 44.9, 42.6, 33.7.
HRMS (ESI): [M+H]+ m/z calc. 235.1117, found 235.1190.
Synthesis of EZ-1-099
ethyl 5-(tributylstannyl)isoxazole-3-carboxylate (EZ-1-090): To a solution of ethyl-2-chloro-2(hydroxyiminoacetate) (481 mg , 3.17 mmol) dissolved in anhydrous DCM (15 mL), potassium carbonate (482.5mg, 3.5mmol) and tributyl(ethynyl)stannane (872 μL, 3.17 mmol) were added and stirred at room temperature overnight. The reaction was then quenched with water, extracted with DCM and dried over anhydrous sodium sulfate. The organic layer was purified via silica gel column chromatography (0 to 10% EtOAc/hexane) to give the product as a clear colorless watery oil (753 mg, 1.75 mmol, 55%).
1H NMR (400 MHz, CDCl3) δ 6.84 (s, 1H), 4.48 (q, J = 7.1 Hz, 2H), 1.70 – 1.10 (m, 27H), 0.94 (s, 3H).
Ethyl 5-bromoisoxazole-3-carboxylate (EZ-1-091): Br2 (134 μL , 2.62 mmol) was added to a solution of ethyl 5-(tributylstannyl)isoxazole-3-carboxylate (EZ-1-090) (753 mg , 1.74 mmol) and sodium carbonate (203 mg, 1.91 mmol) dissolved in DCM (10 mL), and stirred at room temperature overnight. The reaction mixture was then quenched with saturated sodium thiosulfate (8 mL) before extracting with DCM and washing with brine. The organic layer was dried over anhydrous sodium sulfate and purified via silica gel column chromatography (0 to 15% EtOAc/hexane) to produce a clear colorless oil (241.8 mg, 1.1 mmol, 63%) that crystallized upon standing.
1H NMR (400 MHz, CDCl3) δ 6.76 (s, 1H), 4.49 (q, J = 7.1 Hz, 2H), 1.47 (dt, J = 9.6, 6.9 Hz, 3H).
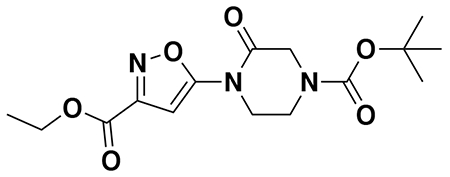
Ethyl 5-(4-(tert-butoxycarbonyl)-2-oxopiperazin-1-yl)isoxazole-3-carboxylate (EZ-1-097): Anhydrous dioxane (3 mL) was added to a vial flushed with N2 containing ethyl 5-bromoisoxazole-3-carboxylate (EZ-1-091) (94.6 mg, 0.43 mmol), tert-butyl 3-oxopiperazine-1-carboxylate (0.43mmol, 86.1mg), cesium carbonate (280.2 mg, 0.86 mmol), Xantphos (19 mg, 0.032 mmol), Pd(dba)3 (10 mg, 0.011 mmol) and the suspension was degassed. The reaction mixture was stirred at 90 °C overnight. The product was extracted with EtOAc, washed with brine, and purified via silica gel column chromatography (0 to 75% EtOAc/hexane) to afford a clear yellow oil (14 mg, 0.04 mmol, 9.6%).
1H NMR (400 MHz, CDCl3) δ 4.48 (q, J = 7.1 Hz, 2H), 4.38 (s, 2H), 4.13 (q, J = 5.5 Hz, 2H), 3.91 – 3.84 (m, 2H), 1.54 (d, J = 2.8 Hz, 9H), 1.46 (t, J = 7.1 Hz, 3H).
LC/MS: [M+H]+ m/z calc. 340.1, found 340.2
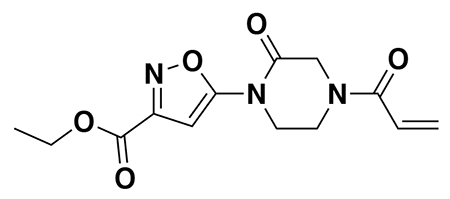
Ethyl 5-(4-acryloyl-2-oxopiperazin-1-yl)isoxazole-3-carboxylate (EZ-1-099):
Ethyl 5-(4-(tert-butoxycarbonyl)-2-oxopiperazin-1-yl)isoxazole-3-carboxylate (EZ-1-097) (14 mg , 0.04 mmol) was deprotected and acylated via general procedures F and H respectively and the crude residue was purified by silica gel chromatography (0 to 100% EtOAc/hexane) to afford the title compound as a clear colorless oil (5.0 mg, 0.017 mmol, 42%).
1H NMR (400 MHz, CDCl3) δ 7.01 (s, 1H), 6.57 (s, 1H), 6.47 (dd, J = 16.8, 2.0 Hz, 1H), 5.90 (dd, J = 10.1, 2.0 Hz, 1H), 4.56 (s, 2H), 4.48 (q, J = 7.1 Hz, 2H), 4.18 (d, J = 5.3 Hz, 2H), 4.07 (s, 2H), 1.46 (t, J = 7.1 Hz, 3H).
13C NMR (151 MHz, DMSO) δ 173.4, 144.1, 143.7, 135.1, 121.6, 119.5, 118.2, 117.4, 64.5, 44.7, 27.3, 16.5, 9.9.
HRMS (ESI): [M+Na]+ m/z calc. 316.0909, found 316.0907.
GL-01: N-(6-(3-(4-(((1-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoyl)pyrrolidin-3-yl)oxy)methyl)piperidine-1-carbonyl)phenyl)-5-methylpyridin-2-yl)-1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamide
1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 8.3 Hz, 1H), 7.68 (s, 1H), 7.59 (d, J = 8.4 Hz, 1H), 7.47 – 7.42 (m, 3H), 7.39 (d, J = 1.8 Hz, 1H), 7.23 (dd, J = 8.2, 1.8 Hz, 1H), 7.19 (d, J = 1.7 Hz, 1H), 7.08 (d, J = 8.1 Hz, 1H), 6.52 (s, 1H), 6.40 (dd, J = 16.7, 2.0 Hz, 1H), 6.26 (dd, J = 3.2, 1.2 Hz, 1H), 6.05 (d, J = 3.2 Hz, 1H), 5.81 (dd, J = 10.2, 2.0 Hz, 1H), 4.72 (s, 1H), 4.48 – 4.34 (m, 2H), 4.10 – 3.75 (m, 6H), 3.66 – 3.58 (m, 1H), 3.54 – 3.39 (m, 3H), 3.34 – 3.22 (m, 2H), 3.05 – 2.87 (m, 3H), 2.81 – 2.70 (m, 1H), 2.60 – 2.50 (m, 2H), 2.26 (s, 3H), 2.12 – 1.95 (m, 2H), 1.94 – 1.71 (m, 5H), 1.21 – 1.04 (m, 4H)
13C NMR (101 MHz, CDCl3) δ 171.78, 170.23, 170.02, 169.92, 164.97, 150.29, 148.82, 144.50, 144.13, 143.61, 141.08, 136.23, 134.92, 134.24, 131.69, 130.09, 128.33, 127.73, 127.69, 127.02, 126.67, 126.31, 112.85, 112.44, 110.20, 107.03, 100.93, 78.63, 73.53, 73.39, 52.05, 50.93, 44.59, 43.69, 42.18, 39.06, 36.74, 36.70, 33.03, 32.80, 31.66, 31.21, 29.66, 23.29, 19.25, 17.23.
19F: (376 MHz, CDCl3) δ −49.51, −49.52
HRMS (TOF, ES+): m/z calcd for C48H51F2N6O9 (M+H)+ 893.3686; found 893.3688
GL-02: N-(6-(3-(4-((1-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoyl)azetidin-3-yl)oxy)piperidine-1-carbonyl)phenyl)-5-methylpyridin-2-yl)-1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamide
1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 8.4 Hz, 1H), 7.68 (s, 1H), 7.59 (d, J = 8.4 Hz, 1H), 7.50 – 7.42 (m, 3H), 7.39 (dt, J = 7.0, 1.8 Hz, 1H), 7.23 (dd, J = 8.2, 1.8 Hz, 1H), 7.19 (d, J = 1.7 Hz, 1H), 7.09 (d, J = 8.2 Hz, 1H), 6.52 (s, 1H), 6.40 (dd, J = 16.7, 2.0 Hz, 1H), 6.26 (d, J = 3.3 Hz, 1H), 6.05 (d, J = 3.3 Hz, 1H), 5.82 (dd, J = 10.3, 2.0 Hz, 1H), 4.50 – 4.34 (m, 3H), 4.27 – 4.16 (m, 2H), 4.12 – 3.81 (m, 7H), 3.71 – 3.54 (m, 2H), 3.45 (d, J = 21.9 Hz, 1H), 3.22 (s, 1H), 2.96 – 2.86 (m, 2H), 2.39 (t, J = 7.6 Hz, 2H), 2.26 (s, 3H), 1.88 (s, 1H), 1.80 – 1.72 (m, 3H), 1.51 (s, 2H), 1.17 (q, J = 3.9 Hz, 2H)
13C NMR (101 MHz, CDCl3) δ 171.78, 171.58, 169.98, 164.97, 163.22, 155.21, 149.95, 148.84, 144.63, 144.12, 143.61, 141.12, 135.90, 134.94, 134.24, 131.69, 130.25, 129.98, 129.15, 128.43, 127.63, 127.01, 126.69, 126.29, 112.89, 112.46, 110.20, 107.21, 101.04, 73.90, 65.51, 58.03, 55.96, 49.46, 46.72, 44.80, 39.10, 31.96, 31.21, 30.09, 23.25, 19.22, 17.25.
19F: (376 MHz, CDCl3) δ −49.52
HRMS (TOF, ES+): m/z calcd for C46H47F2N6O9 (M+H)+ 865.3373; found 865.3416
GL-03: N-(6-(3-((3aR,8aS)-2-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoyl)decahydropyrrolo[3,4-d]azepine-6-carbonyl)phenyl)-5-methylpyridin-2-yl)-1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamide
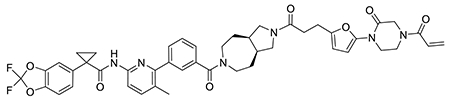
1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 8.4 Hz, 1H), 7.66 (s, 1H), 7.59 (d, J = 8.4 Hz, 1H), 7.48 – 7.42 (m, 3H), 7.38 (dt, J = 6.3, 2.0 Hz, 1H), 7.23 (dd, J = 8.2, 1.8 Hz, 1H), 7.19 (d, J = 1.7 Hz, 1H), 7.08 (d, J = 8.2 Hz, 1H), 6.52 (s, 1H), 6.40 (dd, J = 16.7, 2.0 Hz, 1H), 6.25 (d, J = 1.8 Hz, 1H), 6.05 (d, J = 3.3 Hz, 1H), 5.81 (dd, J = 10.2, 2.0 Hz, 1H), 4.52 – 4.28 (m, 2H), 4.06 – 3.79 (m, 5H), 3.74 – 3.45 (m, 4H), 3.37 – 3.25 (m, 2H), 3.22 – 3.11 (m, 1H), 2.95 (t, J = 7.6 Hz, 2H), 2.56 (t, J = 8.3 Hz, 3H), 2.53 – 2.38 (m, 2H), 2.25 (s, 3H), 2.11 – 1.97 (m, 1H), 1.85 – 1.72 (m, 5H), 1.16 (q, J = 3.9 Hz, 2H)
13C NMR (101 MHz, CDCl3) δ 171.77, 170.94, 170.04, 164.97, 163.22, 155.29, 150.26, 148.85, 144.58, 144.11, 143.60, 141.07, 140.07, 136.55, 134.94, 134.23, 131.68, 130.08, 129.14, 128.43, 127.48, 126.99, 126.70, 126.53, 126.31, 112.89, 112.46, 110.20, 107.11, 100.98, 52.48, 51.83, 51.57, 51.14, 49.46, 47.76, 43.17, 42.94, 40.49, 39.10, 32.86, 31.20, 30.18, 23.39, 19.24, 17.26.
19F: (376 MHz, CDCl3) δ −49.55
HRMS (TOF, ES+): m/z calcd for C46H47F2N6O8 (M+H)+ 849.3423; found 849.3475
GL-04: N-(6-(3-(4-(2-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanamido)ethyl)piperidine-1-carbonyl)phenyl)-5-methylpyridin-2-yl)-1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamide
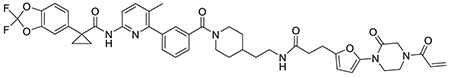
1H NMR (400 MHz, CDCl3) δ 8.10 (s, 1H), 7.82 – 7.54 (m, 2H), 7.49 – 7.42 (m, 3H), 7.39 (dd, J = 5.4, 3.2 Hz, 1H), 7.23 (dd, J = 8.2, 1.7 Hz, 1H), 7.19 (d, J = 1.7 Hz, 1H), 7.09 (d, J = 8.1 Hz, 1H), 6.52 (s, 1H), 6.41 (dd, J = 16.7, 2.0 Hz, 1H), 6.20 (d, J = 3.2 Hz, 1H), 6.05 (d, J = 3.2 Hz, 1H), 5.82 (dd, J = 10.2, 2.0 Hz, 1H), 5.61 (s, 1H), 4.67 (s, 1H), 4.51 – 4.32 (m, 2H), 4.05 – 3.72 (m, 5H), 3.25 (q, J = 6.9 Hz, 2H), 2.99 – 2.87 (m, 3H), 2.73 (s, 1H), 2.48 (t, J = 7.3 Hz, 2H), 2.26 (s, 3H), 1.83 – 1.72 (m, 3H), 1.57 – 1.47 (m, 2H), 1.42 (q, J = 7.1 Hz, 2H), 1.23 – 1.00 (m, 4H)
13C NMR (101 MHz, CDCl3) δ 171.45, 169.80, 164.99, 149.89, 148.76, 144.92, 144.14, 143.64, 136.32, 134.87, 134.24, 131.69, 130.07, 129.17, 128.37, 127.68, 127.09, 126.69, 126.26, 112.93, 112.46, 110.21, 107.37, 101.10, 47.99, 42.42, 39.06, 36.98, 36.12, 35.02, 33.69, 32.67, 31.74, 31.26, 24.26, 19.18, 17.25.
19F: (376 MHz, CDCl3) δ −49.50
HRMS (TOF, ES+): m/z calcd for C45H47F2N6O8 (M+H)+ 837.3423, found 837.3448
GL-05: N-(6-(3-(4-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoyl)piperazine-1-carbonyl)phenyl)-5-methylpyridin-2-yl)-1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamide
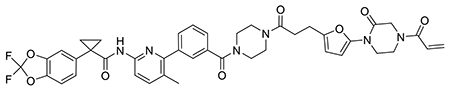
1H NMR (400 MHz, CDCl3) δ 8.08 (d, J = 8.4 Hz, 1H), 7.65 (s, 1H), 7.59 (d, J = 8.4 Hz, 1H), 7.54 – 7.45 (m, 3H), 7.40 (dt, J = 7.4, 1.6 Hz, 1H), 7.23 (dd, J = 8.2, 1.7 Hz, 1H), 7.19 (d, J = 1.7 Hz, 1H), 7.10 (d, J = 8.2 Hz, 1H), 6.52 (s, 1H), 6.41 (dd, J = 16.7, 2.0 Hz, 1H), 6.26 (d, J = 3.3 Hz, 1H), 6.07 (d, J = 3.3 Hz, 1H), 5.82 (dd, J = 10.2, 2.0 Hz, 1H), 4.48 – 4.36 (m, 2H), 4.05 – 3.83 (m, 4H), 3.80 – 3.37 (m, 8H), 2.97 (dd, J = 8.8, 6.4 Hz, 2H), 2.65 (d, J = 9.4 Hz, 2H), 2.26 (s, 3H), 1.74 (q, J = 3.9 Hz, 2H), 1.16 (q, J = 3.9 Hz, 2H)
13C NMR (101 MHz, CDCl3) δ 171.75, 170.24, 170.14, 164.97, 155.06, 149.93, 148.92, 144.72, 144.13, 143.62, 141.11, 140.28, 135.04, 134.94, 134.23, 131.68, 130.70, 129.99, 129.14, 128.49, 128.00, 126.94, 126.84, 126.62, 126.29, 112.94, 112.44, 110.26, 107.31, 101.08, 49.46, 46.83, 39.06, 31.56, 31.19, 23.60, 19.28, 17.23.
19F: (376 MHz, CDCl3) δ −49.54
HRMS (TOF, ES+): m/z calcd for C42H41F2N6O8 (M+H)+ 795.2954, found 795.2943
GL-06: N-(1-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoyl)piperidin-4-yl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)-N-methylbenzamide

1H NMR (400 MHz, CDCl3) δ 8.10 (s, 1H), 7.76 – 7.55 (m, 1H), 7.51 – 7.43 (m, 3H), 7.39 (d, J = 6.9 Hz, 1H), 7.23 (dd, J = 8.2, 1.7 Hz, 1H), 7.19 (d, J = 1.7 Hz, 1H), 7.08 (d, J = 8.2 Hz, 1H), 6.50 (s, 1H), 6.41 (dd, J = 16.7, 2.0 Hz, 1H), 6.27 (d, J = 3.2 Hz, 1H), 6.07 (d, J = 3.2 Hz, 1H), 5.82 (dd, J = 10.2, 2.0 Hz, 1H), 4.77 (s, 2H), 4.50 – 4.32 (m, 2H), 4.09 – 3.71 (m, 6H), 3.17 (s, 1H), 2.96 (t, J = 7.7 Hz, 2H), 2.93 – 2.75 (m, 3H), 2.66 (s, 2H), 2.27 (s, 3H), 1.81 – 1.72 (m,3H), 1.59 (s, 2H), 1.37 – 1.28 (m, 1H), 1.18 (s, 2H)
13C NMR (101 MHz, CDCl3) δ 169.70, 164.97, 150.17, 144.62, 144.16, 143.65, 131.69, 130.14, 128.39, 126.68, 126.30, 112.46, 110.21, 107.16, 100.94, 69.02, 49.37, 44.78, 39.08, 31.50, 29.72, 23.72, 19.21, 17.26.
19F: (376 MHz, CDCl3) δ −49.56
HRMS (TOF, ES+): m/z calcd for C44H45F2N6O8 (M+H)+ 823.3267; found 823.3247
GL-07: N-((1-(1-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoyl)piperidin-4-yl)-1H-1,2,3-triazol-4-yl)methyl)-3-(6-(1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamido)-3-methylpyridin-2-yl)benzamide
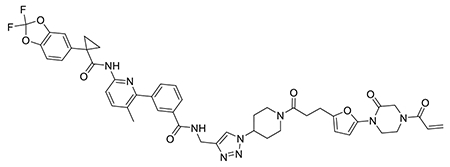
1H NMR (400 MHz, CDCl3) δ 8.18 (br s, 1H), 8.01 – 7.59 (m, 5H), 7.58 – 7.44 (m, 3H), 7.23 (dd, J = 8.2, 1.8 Hz, 1H), 7.18 (d, J = 1.7 Hz, 1H), 7.08 (d, J = 8.2 Hz, 1H), 6.50 (d, J = 11.6 Hz, 1H), 6.39 (dd, J = 16.7, 1.9 Hz, 1H), 6.25 (d, J = 3.2 Hz, 1H), 6.07 (d, J = 3.2 Hz, 1H), 5.80 (dd, J = 10.3, 1.9 Hz, 1H), 4.78 – 4.67 (m, 3H), 4.63 (tt, J = 11.3, 4.1 Hz, 1H), 4.47 – 4.33 (m, 2H), 4.07 – 3.79 (m, 5H), 3.22 (ddd, J = 14.2, 11.9, 2.8 Hz, 1H), 2.97 (td, J = 7.6, 2.8 Hz, 2H), 2.90 – 2.78 (m, 1H), 2.68 (q, J = 7.4 Hz, 2H), 2.41 – 2.13 (m, 5H), 2.01 – 1.84 (m, 2H), 1.77 (q, J = 4.0 Hz, 2H), 1.21 (s, 2H)
13C NMR (101 MHz, CDCl3) δ 169.91, 167.08, 164.98, 150.02, 144.71, 144.20, 143.75, 134.23, 131.97, 131.69, 129.94, 129.15, 128.68, 126.71, 126.33, 120.54, 112.46, 110.24, 107.36, 101.06, 57.82, 49.45, 46.82, 44.13, 40.51, 35.57, 32.74, 32.09, 31.98, 31.45, 23.78, 19.02, 17.43.
19F: (376 MHz, CDCl3) δ −49.46
HRMS (TOF, ES+): m/z calcd for C46H46F2N9O8 (M+H)+ 890.3437; found 890.3433.
GL-08: N-(6-(3-(2-(3-(5-(4-acryloyl-2-oxopiperazin-1-yl)furan-2-yl)propanoyl)-2,8-diazaspiro[4.5]decane-8-carbonyl)phenyl)-5-methylpyridin-2-yl)-1-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)cyclopropane-1-carboxamide
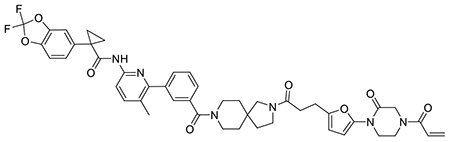
1H NMR (400 MHz, CDCl3) δ 8.11 (d, J = 8.4 Hz, 1H), 7.83 (s, 1H), 7.61 (d, J = 8.5 Hz, 1H), 7.49 – 7.43 (m, 3H), 7.42 – 7.38 (m, 1H), 7.25 – 7.20 (m, 1H), 7.19 (t, J = 1.7 Hz, 1H), 7.08 (dd, J = 8.2, 3.9 Hz,1H), 6.52 (s, 1H), 6.44 – 6.36 (m, 1H), 6.25 (dd, J = 3.2, 2.2 Hz, 1H), 6.05 (t, J = 2.5 Hz, 1H), 5.82 (d, J = 1.5 Hz, 1H), 4.49 – 4.36 (m, 2H), 4.06 – 3.76 (m, 5H), 3.62 – 3.17 (m, 7H), 2.96 (t, J = 7.6 Hz, 2H), 2.57 (dd, J = 8.7, 6.4 Hz, 2H), 2.26 (s, 3H), 2.11 (d, J = 15.1 Hz, 2H), 1.91 – 1.78 (m, 2H), 1.75 (q, J = 3.8 Hz, 2H), 1.54 – 1.39 (m, 2H), 1.17 (q, J = 4.1 Hz, 2H)
13C NMR (101 MHz, CDCl3) δ 171.83, 170.42, 170.19, 169.97, 169.92, 164.98, 154.97, 150.26, 148.76, 144.60, 144.12, 143.62, 141.43, 139.60, 135.93, 135.85, 134.85, 131.69, 130.31, 130.23, 128.45, 127.68, 127.12, 126.82, 126.68, 126.29, 113.10, 112.42, 110.20, 107.11, 101.12, 56.64, 54.66, 44.71, 44.04, 41.63, 39.62, 36.60, 33.97, 33.08, 32.74, 31.23, 29.72, 23.40, 19.18, 17.27.
19F: (376 MHz, CDCl3) δ −49.52
HRMS (TOF, ES+): m/z calcd for C46H47F2N6O8 (M+H)+ 849.3423; found 849.3419
Extended Data
Extended Data Figure 1. Primary covalent ligand screen against OTUB1.
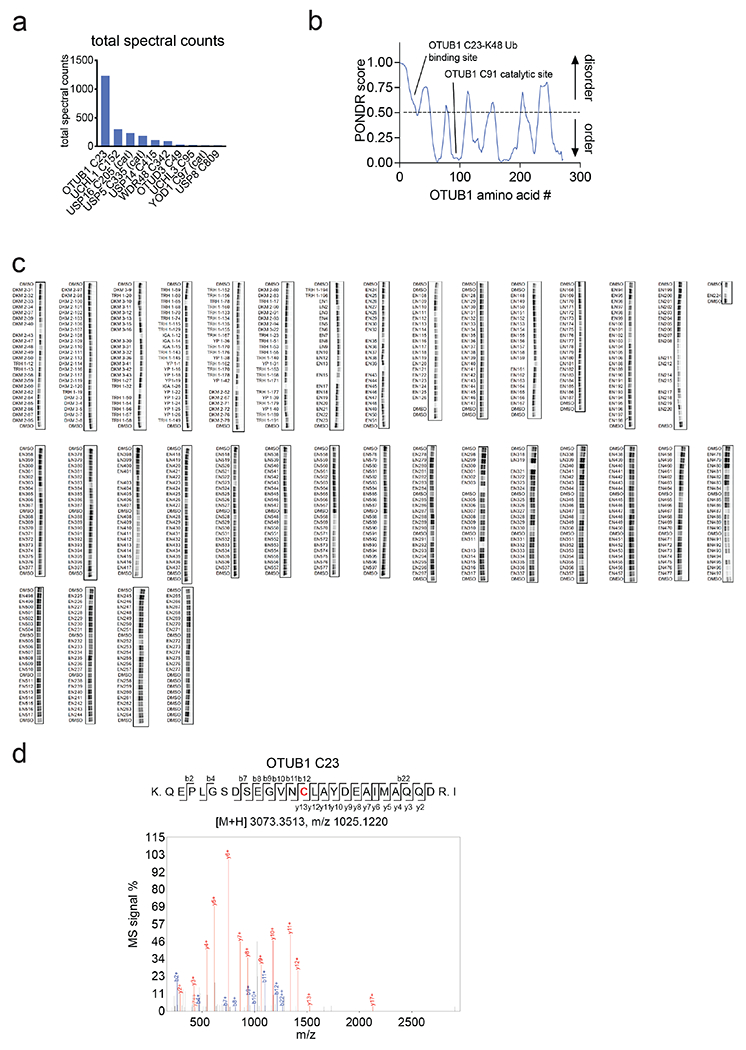
(a) Analysis of aggregate chemoproteomic data for DUBs. Top 10 candidate DUBs described in Figure 1c for total aggregate spectral counts of the particular probe-modified cysteine found in our aggregate chemoproteomic data showing OTUB1 C23 appears far more frequently in chemoproteomic datasets compared to the other DUBs. (b) C23 belongs to an intrinsically disordered region within OTUB1 as assessed by PONDR. (c) Covalent ligand screen of cysteine-reactive libraries competed against IA-rhodamine labeling of recombinant OTUB1 to identify binders to OTUB1 by gel-based ABPP. Vehicle DMSO or cysteine-reactive covalent ligands (50 μM) were pre-incubated with OTUB1 for 30 min at room temperature prior to IA-rhodamine labeling (500 nM, 30 min room temperature). OTUB1 was then separated by SDS/PAGE and in-gel fluorescence was assessed and quantified.
Extended Data Figure 2. NMR analysis of OTUB1, EN523, and UBE2D2.
(a) 13C-HMQC spectrum of OTUB1 labeled on methyl groups of isoleucine, alanine, valine and leucine residues. The presence of peaks with negative proton chemical shifts indicates that the protein is properly folded. (b) Overlay of HMQC spectra of apo-OTUB1 (black) and EN523-bound OTUB1 (red). While both spectra are mostly identical, we identified small but clear chemical shift perturbations of alanine, isoleucine, valine and leucine peaks. Some of these signal changes are shown in the respective blow-up boxes. (c) Overlay of HMQC spectra of apo-OTUB1 (black), UBE2D2 bound OTUB1 (red) and EN523/UBE2D2-bound OTUB1 (blue). The strong chemical shift perturbations (CSPs) are evidence of specific interactions between OTUB1 and the ubiquitinylated ubiquitin-conjugating enzyme. The lack of significant differences between spectra recorded in the presence and absence of EN523 prove that the covalent ligand does not interfere with the protein-protein interaction. Differing peak shift pattern are only seen for peaks directly affected by compound binding (see inlay for blow-up of Ala region). (d) Overlay of HMQC spectra of apo-OTUB1 (black), Ub-UBE2D2 bound OTUB1 (red) and EN523/Ub-UBE2D2-bound OTUB1 (blue). The strong CSPs are evidence of specific interactions between OTUB1 and the ubiquitin-conjugating enzyme. The lack of significant differences between spectra recorded in the presence and absence of EN523 prove that the covalent ligand does not interfere with the protein-protein interaction. Differing peak shift pattern are only seen for peaks directly affected by compound binding (see inlay for blow-up of Ala region).
Extended Data Figure 3. Structure-activity relationships of EN523 analogs with OTUB1.
Gel-based ABPP analysis EN523 analogs against OTUB1. Vehicle DMSO or EN523 analogs were pre-incubated with recombinant OTUB1 for 30 min at 37 °C prior to IA-rhodamine labeling (100 nM, 30 min room temperature). OTUB1 was then separated by SDS/PAGE and in-gel fluorescence was assessed. Also shown is silver staining showing protein loading. Shown are representative gels of n=3 biologically independent samples/group.
Extended Data Figure 4. EN523 does not alter ORUB1 levels and NJH-2-075 engages OTUB1 in CFBE41o-4.7 cells expressing ΔF508-CFTR.
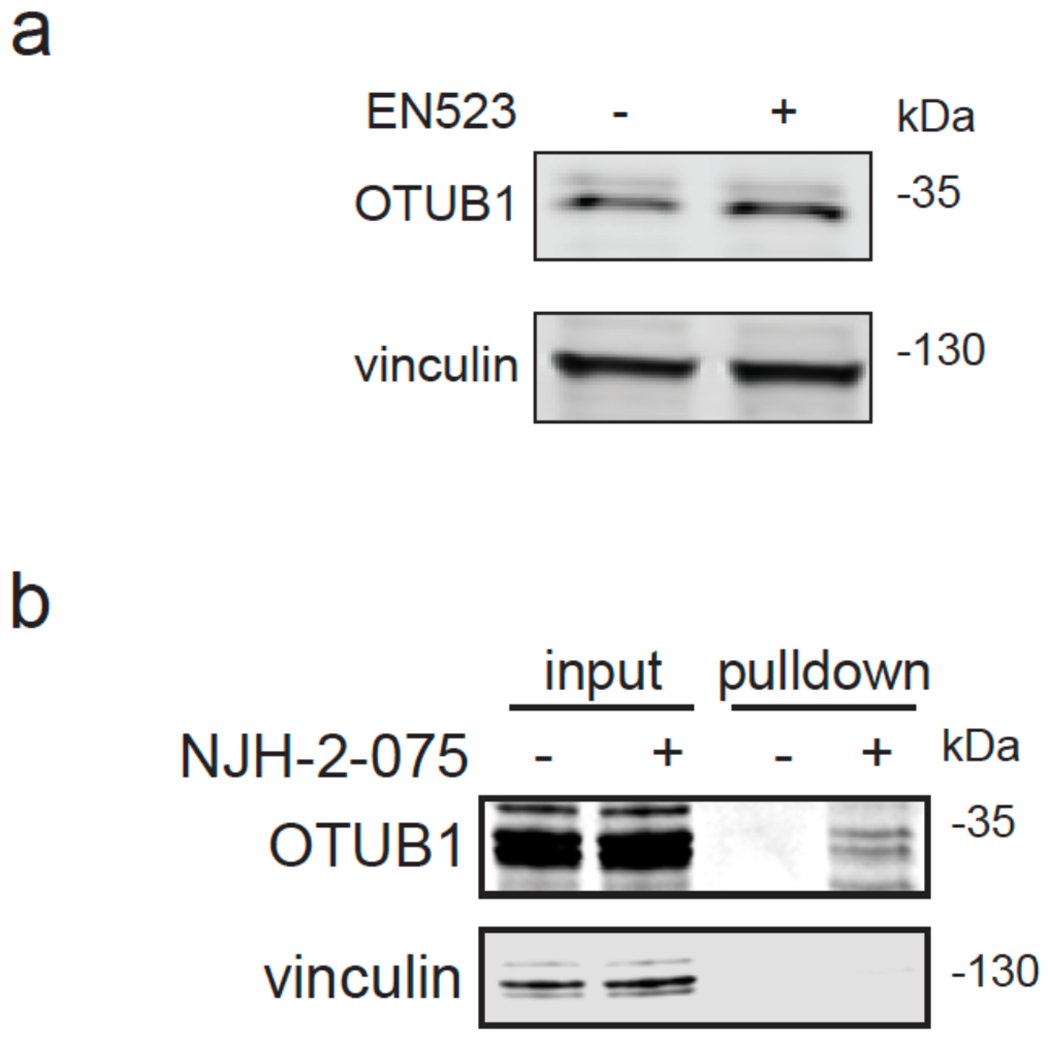
(a) CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or EN523 (10 μM) for 24 h and OTUB1 and loading control vinculin levels were assessed by Western blotting. (b) NJH-2-075 engagement of OTUB1 in CFBE41o-4.7 cells expressing ΔF508-CFTR. Cells were treated with DMSO vehicle or NJH-2-075 (50 μM) for 2 h, after which cell lysates were subjected to CuAAC with biotin picolyl azide and NJH-2-075 labeled proteins were subjected to avidin pulldown, eluted, separated by SDS/PAGE, and blotted for OTUB1 and vinculin. Both input lysate and pulldown levels are shown. Blots shown are representative blots from n=3 biologically independent samples/group.
Extended Data Figure 5. Effect of DUBTACs on mutant CFTR levels.
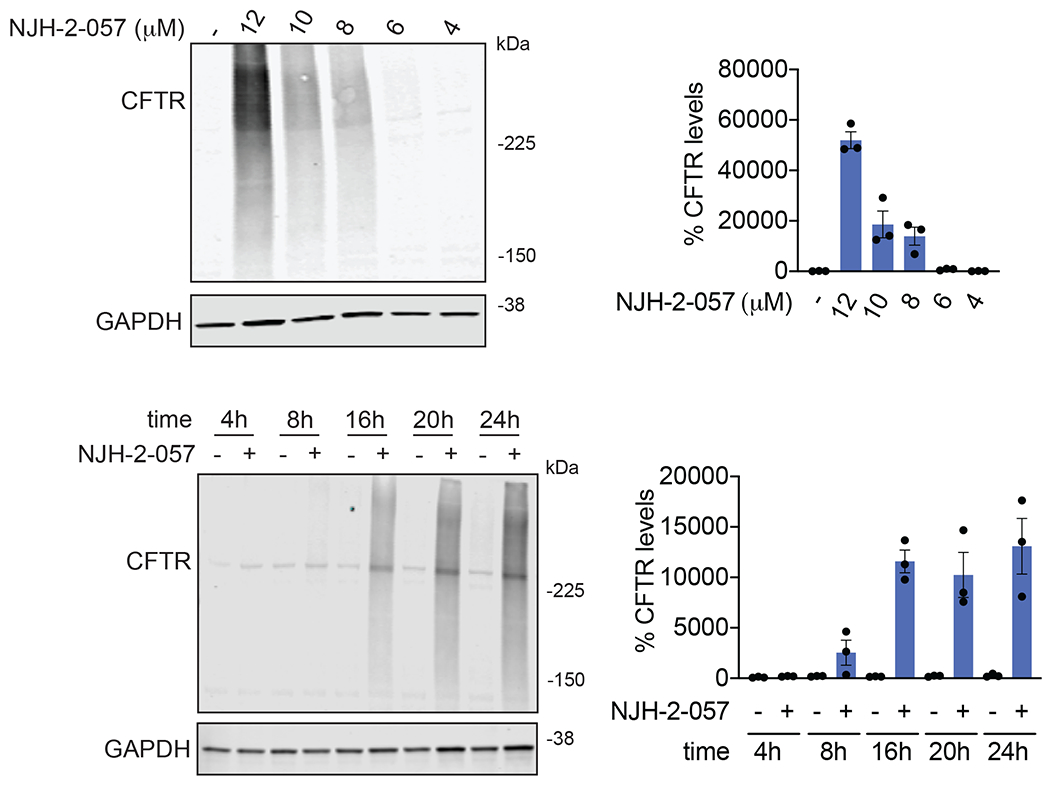
CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or NJH-2-057 and CFTR and loading control GAPDH levels were assessed by Western blotting. For dose-response studies, NJH-2-057 was treated for 24 h. For time-course studies, NJH-2-057 was treated at 10 μM. Dose-response and time-course data gels are representative of n=3 biologically independent samples/group and are quantified in the bar graphs to the right. Data in bar graphs show individual biological replicate values and average ± sem from n=3 biologically independent samples/group.
Extended Data Figure 6. Effect of DUBTACs on mutant CFTR levels.
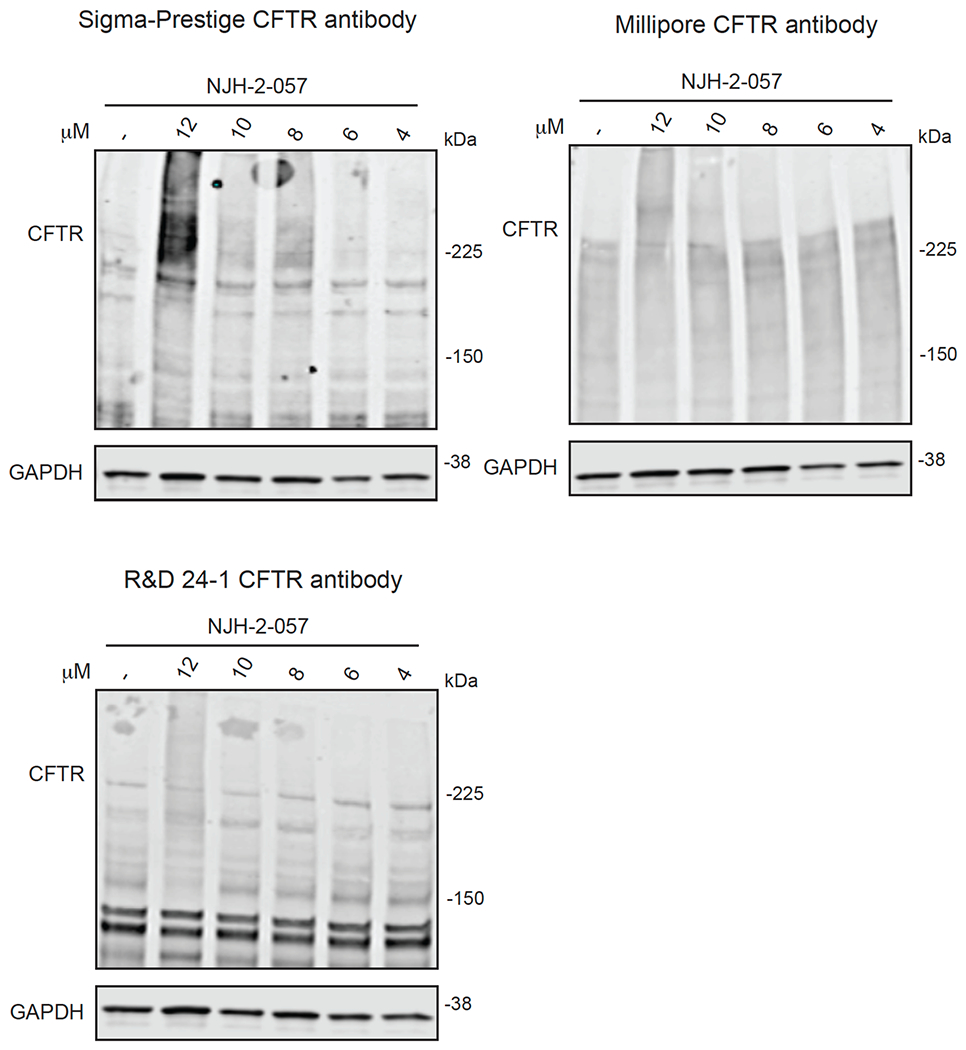
CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or NJH-2-057 and CFTR and loading control GAPDH levels were assessed by Western blotting using three different antibodies against CFTR from the ones used for the main figures. NJH-2-057 was treated for 24 h. Gels are representative of n=3 biologically independent samples/group.
Extended Data Figure 7. Effect of DUBTACs on mutant CFTR levels in siControl and siCFTR cells.
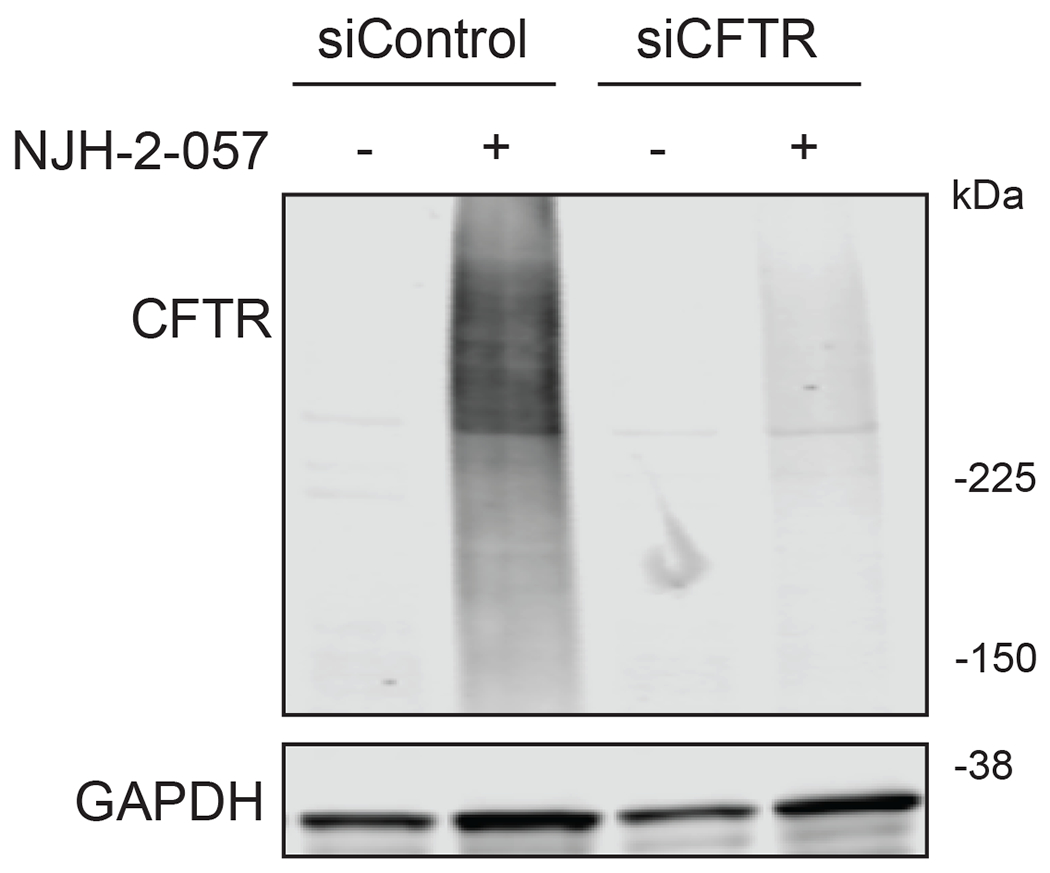
CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or NJH-2-057 (10 μM) for 24 h and CFTR and loading control GAPDH levels were assessed by Western blotting. Blot is representative of n=3 biologically independent samples/group.
Extended Data Figure 8.
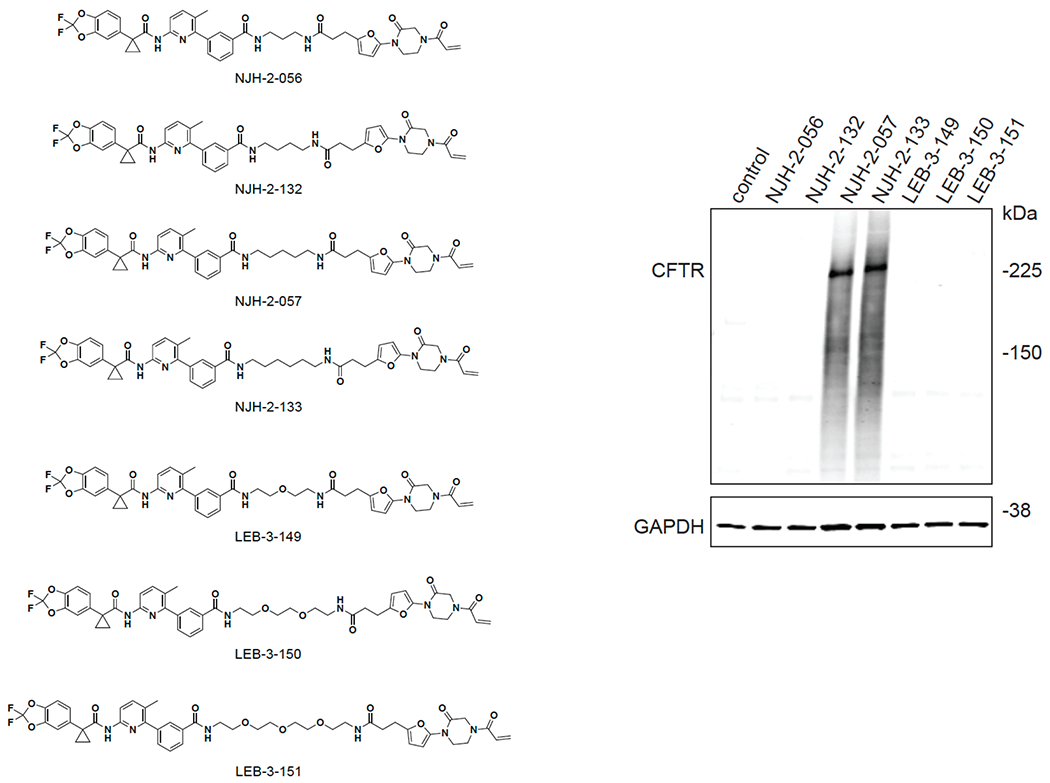
CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or DUBTACs (10 μM) for 24 h and CFTR and loading control GAPDH levels were assessed by Western blotting. Blot is representative of n=3 biologically independent samples/group.
Extended Data Figure 9.
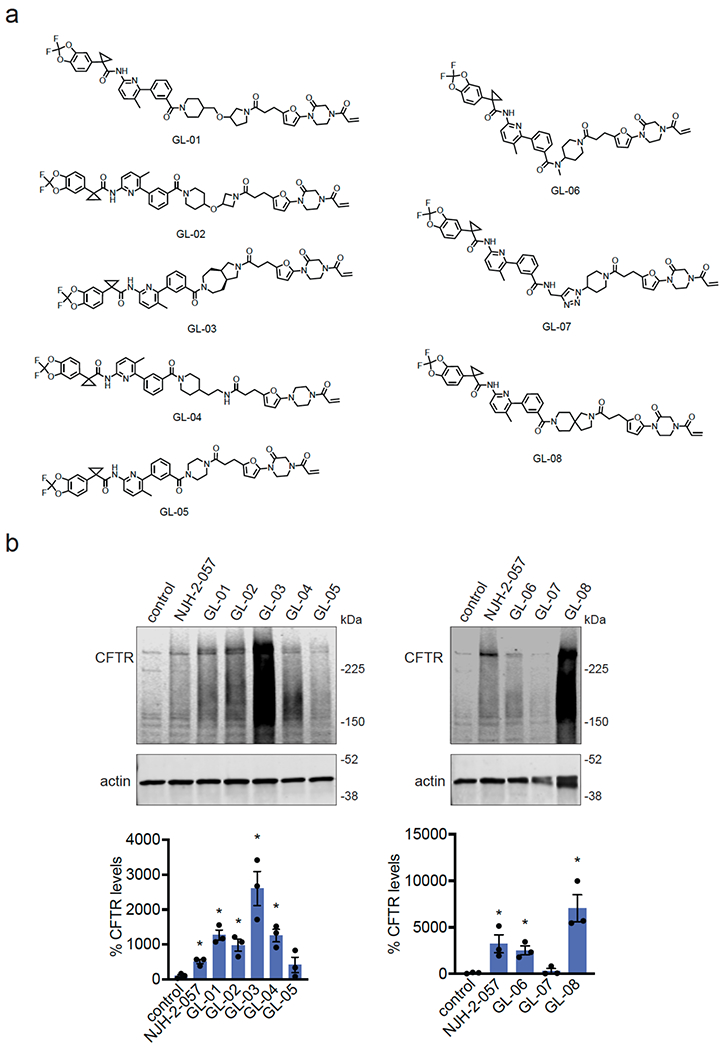
CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or DUBTACs (10 μM) for 24 h and CFTR and loading control actin levels were assessed by Western blotting. Blot is representative of n=3 biologically independent samples/group. Bar graphs show quantification of CFTR levels shown as individual biological replicate data and average ± sem. Statistical significance was calculated with unpaired two-tailed Student’s t-tests compared to vehicle-treated controls and is expressed as *p<0.05.
Extended Data Figure 10.
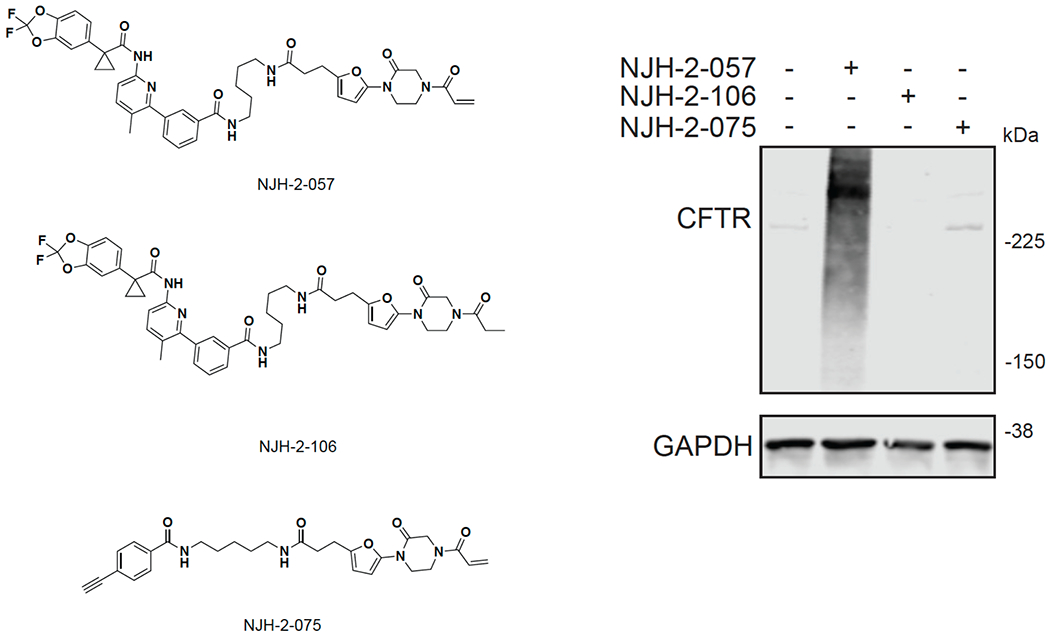
CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or compounds (10 μM) for 24 h and CFTR and loading control GAPDH levels were assessed by Western blotting. Blot is representative of n=3 biologically independent samples/group.
Extended Data Figure 11. Effect of bortezomib and lumacaftor on mutant CFTR levels.
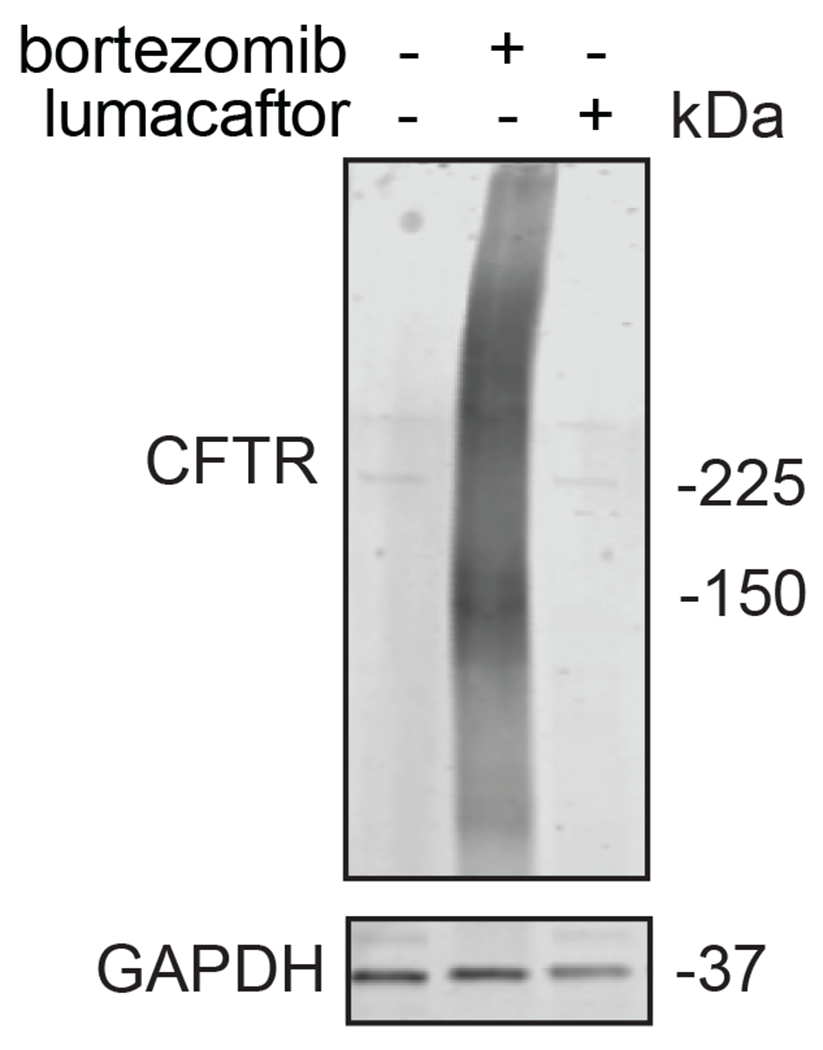
CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO, bortezomib (1 μM), or lumacaftor (1 μM) for 24 h and CFTR and loading control GAPDH levels were assessed by Western blotting. The gel shown is representative of n=3 biologically independent samples/group.
Extended Data Figure 12.
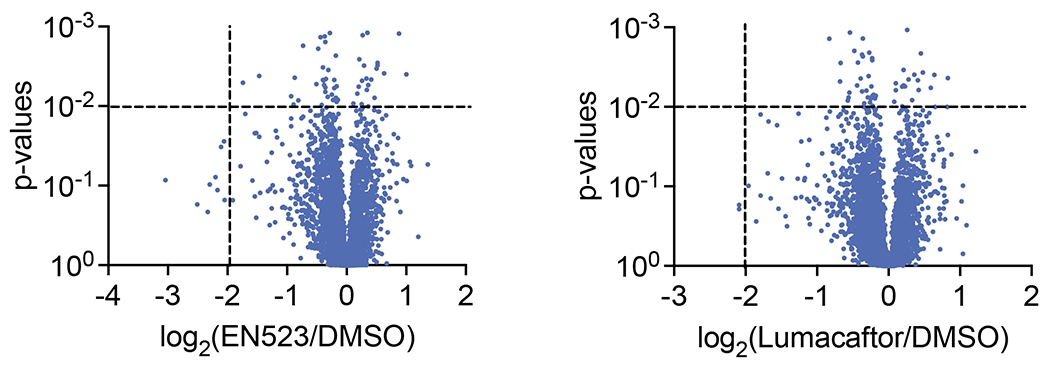
TMT-based quantitative proteomic profiling of EN523 or Lumacaftor treatment. CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or EN523 or Lumacaftor (10 μM) for 24 h. Data shown are from n=3 biologically independent samples/group. Full data for this experiment can be found in Table S3.
Extended Data Figure 13.
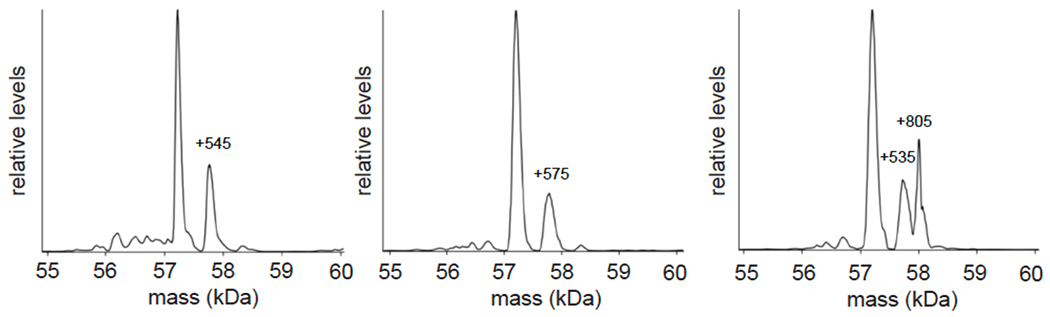
Native MS analysis of DUBTAC-mediated ternary complex formation. OTUB1 (2 μM) and the CFTR-nucleotide binding domain (2 μM) were incubated with DMSO vehicle, EN523 (50 μM), or NJH-2-057 (50 μM) in 150 mM ammonium acetate with MgCl2 (100 μM) and ATP (100 μM). Zoomed in spectra of just the ternary complex is shown for each biologically independent replicate showing the minor peaks next to the OTUB1-CFTR complex mass of 57,225 Da.
Extended Data Figure 14. IsoTOP-ABPP analysis of NJH-2-057.
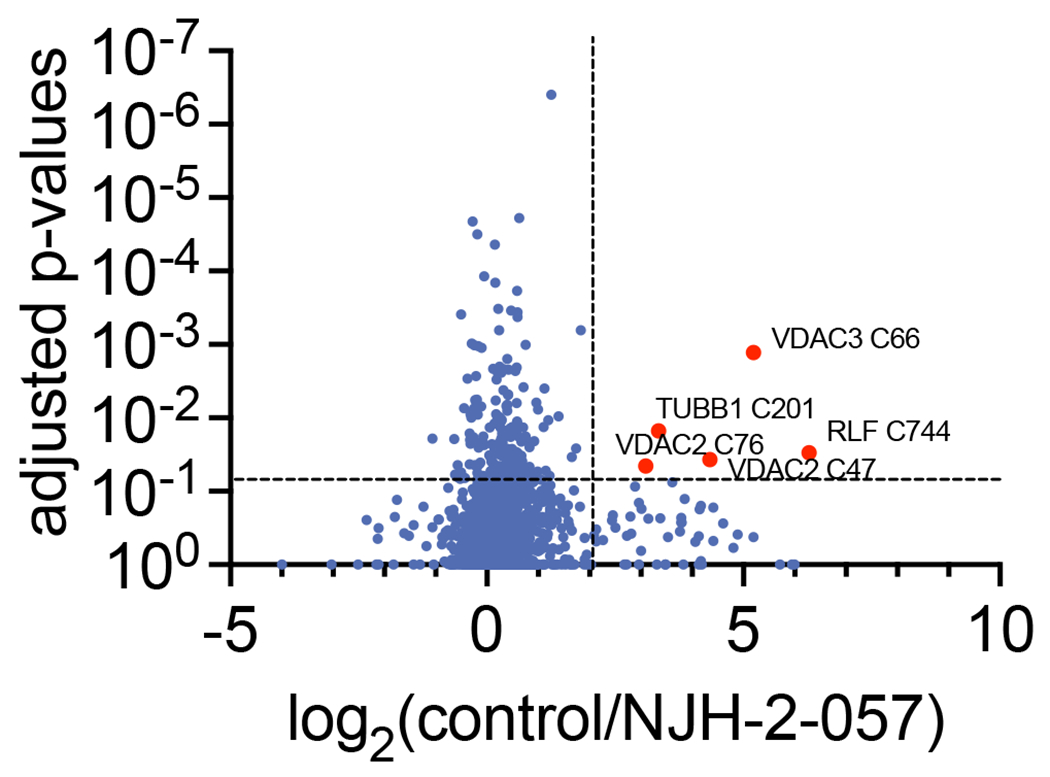
CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or NJH-2-057 for 8 h. Resulting cell lysates were labeled with IA-alkyne (200 μM) for 1 h and taken through the isoTOP-ABPP procedure. Shown in red are the probe-modified peptides that showed isotopically light/heavy or control/NJH-2-57 ratios >4 with adjusted p-values <0.05. The data are from n=3 biologically independent samples/group. The full isoTOP-ABPP dataset can be found in Table S4.
Extended Data Figure 15. WEE1 DUBTAC.
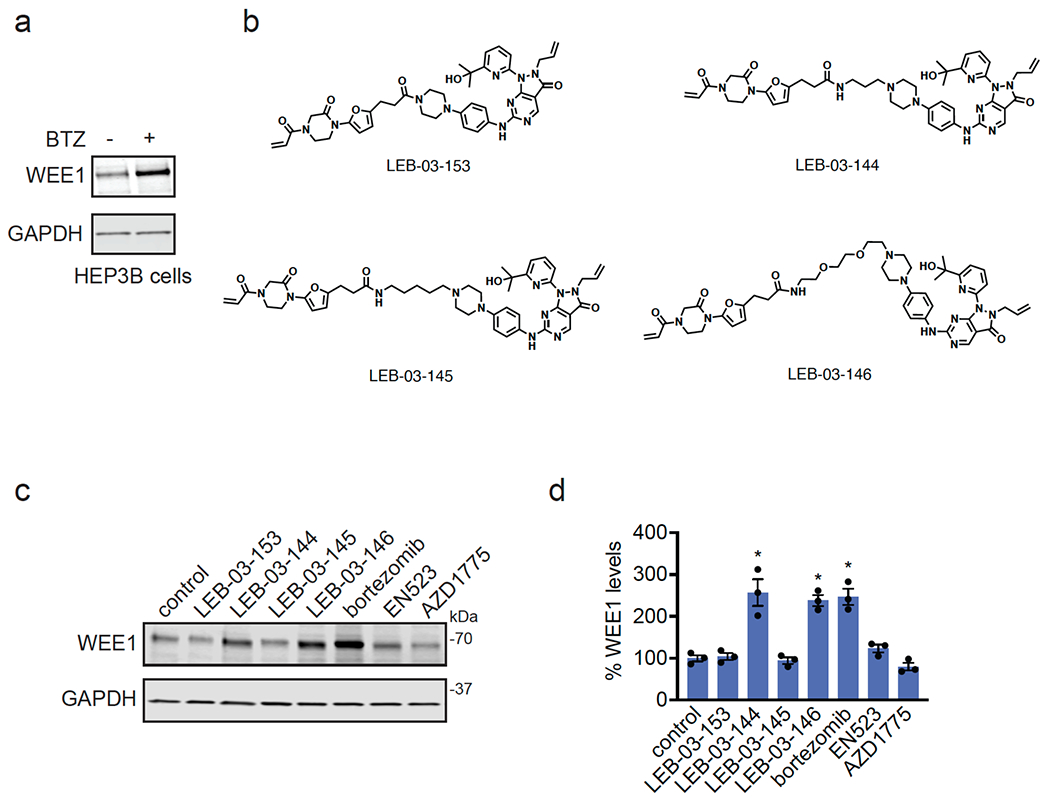
(a) HEP3B cells were treated with DMSO vehicle or bortezomib (1 μM) for 24 h. WEE1 and loading control GAPDH levels were assessed by Western blotting. (b) Structures of four WEE1 DUBTACs linking AZD1775 to the OTUB1 recruiter EN523 through four different linkers. (c) HEP3B cells were treated with DMSO vehicle, the four DUBTACs, bortezomib, EN523, or AZD1775 at 1 μM for 24 h. WEE1 and loading control GAPDH levels were assessed by Western blotting. Blots shown in (a) and (b) are representative blots from n=3 biologically independent samples/group. Data in bar graphs show individual biological replicate values and average ± sem from n=3 biologically independent samples/group.
Supplementary Material
Table S1. Chemoproteomic analysis of DUBs. Table of all probe-modified cysteines identified for 65 DUBs from 455 distinct isoTOP-ABPP experiments. This data is from our research group’s aggregate chemoproteomic experiments from various human cell lines wherein cell lysates were labeled with an alkyne-functionalized iodoacetamide (IA-alkyne) probe and taken through the isoTOP-ABPP procedure. Shown are the DUBs, probe-modified peptides, the site of the modified cysteine, the aggregate spectral counts for each particular probe-modified site identified across 455 experiments, and the total numbers of experiments for which the probe-modified peptide was observed.
Table S2. Structures of covalent ligands screened against OTUB1. Covalent ligand screen of cysteine-reactive libraries competed against IA-rhodamine labeling of recombinant OTUB1 to identify binders to OTUB1 by gel-based ABPP. Vehicle DMSO or cysteine-reactive covalent ligands (50 μM) were pre-incubated with OTUB1 for 30 min at room temperature prior to IA-rhodamine labeling (500 nM, 30 min room temperature). OTUB1 was then separated by SDS/PAGE and in-gel fluorescence was assessed and quantified. Gel-based ABPP data is shown in Extended Data Figure 1a. Quantification of gel-based ABPP data normalized to average of DMSO vehicle-treated controls in each gel is shown in Tab 1.
Table S4. IsoTOP-ABPP analysis of NJH-2-057. CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or NJH-2-057 (10 μM) for 8 h. Resulting cell lysates were labeled with IA-alkyne (200 μM) for 1 h and taken through the isoTOP-ABPP procedure Shown in red are the probe-modified peptides that showed isotopically light/heavy or control/NJH-2-57 ratios >4 with adjusted p-values <0.05. The data are from n=3 biologically independent samples/group.
Table S3. TMT-based quantitative proteomic profiling of NJH-2-057 treatment. CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or NJH-2-057 (Tabs 1-2) or EN523 or Lumacaftor (Tabs 3-4) (10 μM) for 24 h. Data shown are from n=3 biologically independent samples/group.
Scheme 1.
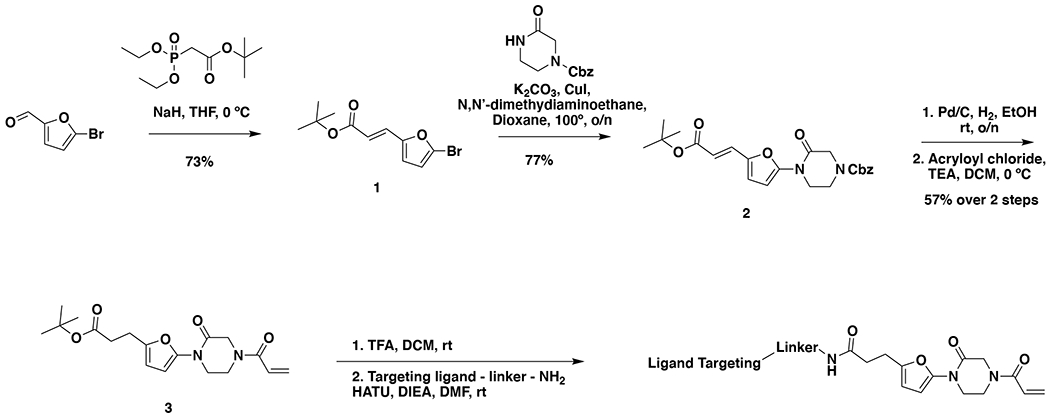
General scheme describing synthetic route to bifunctional DUBTACs containing EN523 as an OTUB1 recruiter.
Acknowledgement
We thank the members of the Nomura Research Group and Novartis Institutes for BioMedical Research for critical reading of the manuscript. This work was supported by Novartis Institutes for BioMedical Research and the Novartis-Berkeley Center for Proteomics and Chemistry Technologies (NB-CPACT) for all listed authors. This work was also supported by the Nomura Research Group and the Mark Foundation for Cancer Research ASPIRE Award for DKN, NJH, LB, JNS, CCW, and BB. This work was also supported by grants from the National Institutes of Health (R01CA240981 for DKN) and the National Science Foundation Graduate Fellowship (for LB). We also thank Drs. Hasan Celik, Alicia Lund, and UC Berkeley’s NMR facility in the College of Chemistry (CoC-NMR) for spectroscopic assistance. Instruments in the CoC-NMR are supported in part by NIH S10OD024998.
Competing Financial Interests Statement
JAT, JMK, LMM, DD, MH, MS, SMB, GL, RVM, LWP, DJR, FW, AOF, DF are employees of Novartis Institutes for BioMedical Research. This study was funded by the Novartis Institutes for BioMedical Research and the Novartis-Berkeley Center for Proteomics and Chemistry Technologies. DKN is a co-founder, shareholder, and adviser for Frontier Medicines and Vicinitas Therapeutics. DKN is also on the scientific advisory board of The Mark Foundation for Cancer Research and Photys Therapeutics.
References
- 1.Dixon SJ & Stockwell BR Identifying druggable disease-modifying gene products. Curr. Opin. Chem. Biol 13, 549–555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spradlin JN, Zhang E & Nomura DK Reimagining Druggability Using Chemoproteomic Platforms. Acc. Chem. Res 54, 1801–1813 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Nalawansha DA & Crews CM PROTACs: An Emerging Therapeutic Modality in Precision Medicine. Cell Chem. Biol 27, 998–1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burslem GM & Crews CM Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell 181, 102–114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schapira M, Calabrese MF, Bullock AN & Crews CM Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov 18, 949–963 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Farnaby W, Koegl M, McConnell DB & Ciulli A Transforming targeted cancer therapy with PROTACs: A forward-looking perspective. Curr. Opin. Pharmacol 57, 175–183 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Banik SM et al. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 584, 291–297 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi D et al. AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol. Cell 76, 797–810.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Yamazoe S et al. Heterobifunctional Molecules Induce Dephosphorylation of Kinases–A Proof of Concept Study. J. Med. Chem 63, 2807–2813 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Siriwardena SU et al. Phosphorylation-Inducing Chimeric Small Molecules. J. Am. Chem. Soc 142, 14052–14057 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Sabapathy K & Lane DP Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol 15, 13–30 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Abbas T & Dutta A p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer 9, 400–414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B & Dou QP Bax degradation by the ubiquitin/proteasome-dependent pathway: Involvement in tumor survival and progression. Proc. Natl. Acad. Sci. U. S. A 97, 3850–3855 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward CL, Omura S & Kopito RR Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Weerapana E et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Backus KM et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiener R, Zhang X, Wang T & Wolberger C The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483, 618–622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakada S et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Que LT, Morrow ME & Wolberger C Comparison of Cross-Regulation by Different OTUB1:E2 Complexes. Biochemistry 59, 921–932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.French ME, Koehler CF & Hunter T Emerging functions of branched ubiquitin chains. Cell Discov. 7, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunker AK et al. Intrinsically disordered protein. J. Mol. Graph. Model 19, 26–59 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Garner, null, Romero, null, Dunker, null, Brown, null & Obradovic, null. Predicting Binding Regions within Disordered Proteins. Genome Inform. Workshop Genome Inform. 10, 41–50 (1999). [PubMed] [Google Scholar]
- 23.Wiener R et al. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol 20, 1033–1039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riordan JR CFTR function and prospects for therapy. Annu. Rev. Biochem 77, 701–726 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Kanner SA, Shuja Z, Choudhury P, Jain A & Colecraft HM Targeted deubiquitination rescues distinct trafficking-deficient ion channelopathies. Nat. Methods 17, 1245–1253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomati V et al. Genetic Inhibition Of The Ubiquitin Ligase Rnf5 Attenuates Phenotypes Associated To F508del Cystic Fibrosis Mutation. Sci. Rep 5, 12138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sondo E et al. Pharmacological Inhibition of the Ubiquitin Ligase RNF5 Rescues F508del-CFTR in Cystic Fibrosis Airway Epithelia. Cell Chem. Biol 25, 891–905.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Lopes-Pacheco M CFTR Modulators: The Changing Face of Cystic Fibrosis in the Era of Precision Medicine. Front. Pharmacol 10, 1662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rath A, Glibowicka M, Nadeau VG, Chen G & Deber CM Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. U. S. A 106, 1760–1765 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spradlin JN et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat. Chem. Biol 15, 747–755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Crowley VM, Wucherpfennig TG, Dix MM & Cravatt BF Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. Nat. Chem. Biol 15, 737–746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X et al. DCAF11 Supports Targeted Protein Degradation by Electrophilic Proteolysis-Targeting Chimeras. J. Am. Chem. Soc 143, 5141–5149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward CC et al. Covalent Ligand Screening Uncovers a RNF4 E3 Ligase Recruiter for Targeted Protein Degradation Applications. ACS Chem. Biol 14, 2430–2440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghelli Luserna di Rorà A, Cerchione C, Martinelli G & Simonetti G A WEE1 family business: regulation of mitosis, cancer progression, and therapeutic target. J. Hematol. Oncol.J Hematol Oncol 13, 126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith A, Simanski S, Fallahi M & Ayad NG Redundant ubiquitin ligase activities regulate wee1 degradation and mitotic entry. Cell Cycle Georget. Tex 6, 2795–2799 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto O et al. Inhibition of proteasome-dependent degradation of Wee1 in G2-arrested Hep3B cells by TGF beta 1. Mol. Carcinog 36, 171–182 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Li Z et al. Development and Characterization of a Wee1 Kinase Degrader. Cell Chem. Biol 27, 57–65.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavathiotis E, Reyna DE, Bellairs JA, Leshchiner ES & Walensky LD Direct and selective small-molecule activation of proapoptotic BAX. Nat. Chem. Biol 8, 639–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pryde DC et al. The discovery of potent small molecule activators of human STING. Eur. J. Med. Chem 209, 112869 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Ramanjulu JM et al. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 564, 439–443 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Zorn JA & Wells JA Turning enzymes ON with small molecules. Nat. Chem. Biol 6, 179–188 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Chen L, Liu S & Tao Y Regulating tumor suppressor genes: post-translational modifications. Signal Transduct. Target. Ther 5, 1–25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riboldi GM & Di Fonzo AB GBA, Gaucher Disease, and Parkinson’s Disease: From Genetic to Clinic to New Therapeutic Approaches. Cells 8, E364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarodaya N, Suresh B, Kim K-S & Ramakrishna S Protein Degradation and the Pathologic Basis of Phenylketonuria and Hereditary Tyrosinemia. Int. J. Mol. Sci 21, E4996 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liguori L et al. Pharmacological Chaperones: A Therapeutic Approach for Diseases Caused by Destabilizing Missense Mutations. Int. J. Mol. Sci 21, E489 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartha I, di Iulio J, Venter JC & Telenti A Human gene essentiality. Nat. Rev. Genet 19, 51–62 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Bateman LA et al. Chemoproteomics-enabled covalent ligand screen reveals a cysteine hotspot in reticulon 4 that impairs ER morphology and cancer pathogenicity. Chem. Commun. Camb. Engl 53, 7234–7237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung CY-S et al. Covalent targeting of the vacuolar H+-ATPase activates autophagy via mTORC1 inhibition. Nat. Chem. Biol 15, 776–785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boike L et al. Discovery of a Functional Covalent Ligand Targeting an Intrinsically Disordered Cysteine within MYC. Cell Chem. Biol 28, 4–13.e17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo M et al. Chemoproteomics-enabled discovery of covalent RNF114-based degraders that mimic natural product function. Cell Chem. Biol 28, 559–566.e15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grossman EA et al. Covalent Ligand Discovery against Druggable Hotspots Targeted by Anti-cancer Natural Products. Cell Chem. Biol 24, 1368–1376.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts AM et al. Chemoproteomic Screening of Covalent Ligands Reveals UBA5 As a Novel Pancreatic Cancer Target. ACS Chem. Biol 12, 899–904 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Counihan JL, Wiggenhorn AL, Anderson KE & Nomura DK Chemoproteomics-Enabled Covalent Ligand Screening Reveals ALDH3A1 as a Lung Cancer Therapy Target. ACS Chem. Biol 13, 1970–1977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berdan CA et al. Parthenolide Covalently Targets and Inhibits Focal Adhesion Kinase in Breast Cancer Cells. Cell Chem. Biol 26, 1027–1035.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spradlin JN et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat. Chem. Biol 15, 747–755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isobe Y et al. Manumycin polyketides act as molecular glues between UBR7 and P53. Nat. Chem. Biol 16, 1189–1198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo M et al. Chemoproteomics-enabled discovery of covalent RNF114-based degraders that mimic natural product function. Cell Chem. Biol (2021) doi: 10.1016/j.chembiol.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu T et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics 129, 16–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Käil L, Canterbury JD, Weston J, Noble WS & MacCoss MJ Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Chemoproteomic analysis of DUBs. Table of all probe-modified cysteines identified for 65 DUBs from 455 distinct isoTOP-ABPP experiments. This data is from our research group’s aggregate chemoproteomic experiments from various human cell lines wherein cell lysates were labeled with an alkyne-functionalized iodoacetamide (IA-alkyne) probe and taken through the isoTOP-ABPP procedure. Shown are the DUBs, probe-modified peptides, the site of the modified cysteine, the aggregate spectral counts for each particular probe-modified site identified across 455 experiments, and the total numbers of experiments for which the probe-modified peptide was observed.
Table S2. Structures of covalent ligands screened against OTUB1. Covalent ligand screen of cysteine-reactive libraries competed against IA-rhodamine labeling of recombinant OTUB1 to identify binders to OTUB1 by gel-based ABPP. Vehicle DMSO or cysteine-reactive covalent ligands (50 μM) were pre-incubated with OTUB1 for 30 min at room temperature prior to IA-rhodamine labeling (500 nM, 30 min room temperature). OTUB1 was then separated by SDS/PAGE and in-gel fluorescence was assessed and quantified. Gel-based ABPP data is shown in Extended Data Figure 1a. Quantification of gel-based ABPP data normalized to average of DMSO vehicle-treated controls in each gel is shown in Tab 1.
Table S4. IsoTOP-ABPP analysis of NJH-2-057. CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or NJH-2-057 (10 μM) for 8 h. Resulting cell lysates were labeled with IA-alkyne (200 μM) for 1 h and taken through the isoTOP-ABPP procedure Shown in red are the probe-modified peptides that showed isotopically light/heavy or control/NJH-2-57 ratios >4 with adjusted p-values <0.05. The data are from n=3 biologically independent samples/group.
Table S3. TMT-based quantitative proteomic profiling of NJH-2-057 treatment. CFBE41o-4.7 cells expressing ΔF508-CFTR were treated with vehicle DMSO or NJH-2-057 (Tabs 1-2) or EN523 or Lumacaftor (Tabs 3-4) (10 μM) for 24 h. Data shown are from n=3 biologically independent samples/group.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.


