Abstract

Primary succession in mine tailings is a prerequisite for tailing vegetation. Microorganisms, including bacteria, fungi, and protists, play an important role in this process in the driving force for improving the nutritional status. Compared to bacteria and fungi, protist populations have rarely been investigated regarding their role in mine tailings, especially for those inhabiting tailings associated with primary succession. Protists are the primary consumers of fungi and bacteria, and their predatory actions promote the release of nutrients immobilized in the microbial biomass, as well as the uptake and turnover of nutrients, affecting the functions of the wider ecosystems. In this study, three different types of mine tailings associated with three successional stages (original tailings, biological crusts, and Miscanthus sinensis grasslands) were selected to characterize the protistan community diversity, structure, and function during primary succession. Some members classified as consumers dominated the network of microbial communities in the tailings, especially in the original bare land tailings. The keystone phototrophs of Chlorophyceae and Trebouxiophyceae showed the highest relative abundance in the biological crusts and grassland rhizosphere, respectively. In addition, the co-occurrences between protist and bacterial taxa demonstrated that the proportion of protistan phototrophs gradually increased during primary succession. Further, the metagenomic analysis of protistan metabolic potential showed that abundances of many functional genes associated with photosynthesis increased during the primary succession of tailings. Overall, these results suggest that the primary succession of mine tailings drives the changes observed in the protistan community, and in turn, the protistan phototrophs facilitate the primary succession of tailings. This research offers an initial insight into the changes in biodiversity, structure, and function of the protistan community during ecological succession on tailings.
Keywords: mine tailings, primary succession, protistan community, biological crusts, Miscanthus sinensis, metagenomic analysis, interaction networks, keystone taxa
Introduction
Tailings are slurry wastes from ore processing and contain abundant amounts of metal(loid)s, sulfates, as well as other minerals and salts. Tailings are also considered to be the major anthropogenic cause of soil erosion.1,2 Around 5–7 billion tons of waste tailings are produced annually, causing damage to extensive areas of land resources and mined lands.3,4 Therefore, large land areas have been set aside for storing tailings in Australia, China, Europe, Peru, South Africa, and the USA.5,6 However, the tailings not only cover a large amount of mining land resource area and waste precious resources but also cause catastrophic environmental events due to the leaching of toxic metal(loid)s.7 Therefore, it is crucial to improve the management of tailings using appropriate management and treatment strategies.8
Bioremediation provides a promising remediation strategy at contaminated tailing sites because some microorganisms may contribute substantially to a wide range of ecological services, including nitrogen fixation,9 carbon fixation,10 and metal(loid) cycling,11 thereby affecting the ecosystem of tailing sites and promoting tailing vegetation. For instance, Meiothermus and Thiobacillus from alkaline tailings have shown potential as nitrogen- and carbon-fixing organisms as well as in metal transformation and pollution tolerance; these bacteria were even beneficial for phytoremediation by providing nutrients for plants.10 Some bacteria that evolved to fix nitrogen autotrophically in response to nutrient deficiencies (e.g., nitrogen and carbon) were often detected in tailings, which could play important roles in facilitating nutrient acquisition by plants and ecological succession in tailings.6 In addition, some ectomycorrhizal fungi could exude ergosterol and low-molecular-mass organic acids that help to solubilize tailings; these fungi exhibited a potential to improve the survival rate, growth, and health of white spruce seedlings in tailings.12 However, a growing number of researches have mainly focused on the communities of fungi and bacteria in tailing habitats, but little research has been done on the protistan community, which limits our overall understanding of the ecological roles of the microbiome in tailings.
As a major group of microbes, protists are diverse in taxonomy and rich in function.13−15 Protists are the main consumers of fungi and bacteria,13,16 and their predation activities facilitate the release of nutrients immobilized in the microbial biomass while promoting nutrient uptake as well as turnover, influencing the functions of a wide range of ecosystems.17−21 In addition, protists can also facilitate nutrient recycling by regulating the fixation of carbon and degradation of organic matter.22,23 Therefore, it is fair to propose that protists play an important role in the ecological succession of tailings. However, only limited studies have focused on the protists inhabiting mine tailings.24,25 Therefore, it is crucial and necessary to elucidate the role of the protistan community in tailings, which can provide new insights into the bioremediation of tailings.
Mine tailings can be colonized by vegetation as a consequence of primary succession, which is desirable for stabilizing the tailing surfaces and has been the subject of intensive research recently.26,27 However, the microbial community dynamics in the primary succession of tailings have received less attention and are more poorly understood than those involved in the establishment and succession of natural vegetation.28,29 Notably, the composition, function, and shift of the protistan community associated with ecosystem development in mine tailings have been rarely documented. Therefore, in this study, we selected three different types of tailings (e.g., mine tailings contaminated with antimony (Sb), lead/zinc (Pb/Zn), and gold (Au)) and collected samples in three different stages of primary succession (e.g., original tailings from bare land, tailings of biological crusts, and tailings from the plant rhizosphere). The 18S rRNA and 16S rRNA analysis, shotgun metagenomics, and multivariable statistics were performed to (i) investigate the changes in the composition of taxonomic and functional communities of protists, (ii) elucidate interactions between protistan and bacterial communities, and (iii) detect the most important functional protists found during the primary succession of tailings.
Materials and Methods
Tailing Sample Collection and Geochemical Analyses
Tailing samples were collected from mining sites across Southern China. The sampling information is provided (Figure S1 and Table S1). In brief, the samples were collected from three metal(loid)-contaminated mining areas containing Sb (Xikuangshan), Pb/Zn (Fankou), and Au (Huangjia). Three sample types (e.g., original tailings from bare land (BL), tailings of biological crusts (BC), and tailings from the Miscanthus sinensis rhizosphere (MS)) were collected. A total of 98 samples were collected; these were transported to the laboratory within 24 h and stored at 4 °C. For analyzing pH, 2 g of freeze-dried tailings was fully mixed with 10 mL of distilled water, and then pH was determined using an HQ30d pH meter (Hach, CO, USA). To analyze the total contents of S and P, the mixture was centrifugated and measured by ion chromatography on an ICS-40 system (DIONEX, Sunnyvale, USA). The total concentrations of metal(loid)s in tailing samples were measured by digesting the tailings with a mixture of HNO3 and HCl (1:3) using ICP-OES (Vista MPX, Varian, USA).30
DNA Extraction and Illumina MiSeq Sequencing of 18S rRNA
Genomic DNA was extracted from tailings with a DNeasy Powersoil kit (QIAGEN, Dresden, Germany) following the manufacturer’s instructions.31,32 The DNA quality and quantity were measured using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).33 The 18S rRNA V9 was amplified on the ABI 2720 thermocycler (Applied Biosystems, Carlsbad, CA, USA) using the primer pair 1380F (5′-CCCTGCCHTTTGTACACAC-3′) and 1510R (5′-CCTTCYGCAGGTTCACCTAC-3′).34 The 16S rRNA gene (V4–V5 region) was amplified with a 515F/806R primer pair.35 Amplicons were quantitated using an FLX800T spectrophotometer (BioTek Instruments, Inc., Winooski, Vermont, USA) and pooled equimolarly for paired-end sequencing on an Illumina MiSeq platform (Illumina, Inc., San Diego, CA, USA) at Personal Biotechnology (Shanghai, China). QIIME2 was applied to analyze the sequencing reads. Protistan and bacterial representative amplicon sequence variants (ASVs) were assigned against the Protist Ribosomal Reference (PR2) database36 and SILVA database,37 respectively. The trophic functional groups, including parasites, consumers, and phototrophs, were assigned at the genus level according to previous reports (Table S2).38−40
Shotgun Metagenomic Sequencing
Three samples (BL = 1, BC = 1, and MS = 1) were selected for shotgun metagenomic sequencing at Personal Biotechnology (Shanghai, China). The characterization of functional genes of the protistan community was conducted according to a previous description.41 The sequences were quality-controlled by the Trimmomatic software.42 The clean reads were evaluated for quality using a FastQC toolkit, and the community composition was classified using Kraken2.43 The reference databases (e.g., bacteria, fungi, protozoa, and archaea) were searched using the parameters ″–paired −use-names −use-mpa-style −report-zero-counts″. Furthermore, the output files of Kraken2 were adjusted using the highly accurate statistical method Bracken to gain the final read count table by the ″kreort2mpa.py″ function.44 The protist-classified reads were filtered using the Kraken2 database with the parameter ″–classified-out″41 and were assembled to contigs using Megahit with parameters ″–k-min 21 −k-max 121 −k-step 10″.45 Finally, the gained contigs were subjected to the annotation of genes using the KofamKOALA web server (https://www.genome.jp/tools/kofamkoala/) against the KEGG database with default parameters.46 The count number of KEGG annotation was filtered for downstream comparison. The raw data were submitted to the NCBI database (Accession No. PRJNA793085).
Statistical Analysis
The alpha diversity was calculated using the QIIME2 microbiome bioinformatics platform. Principal coordinate analysis (PCoA) was done using ade4 v1.7.13, and permutational multivariate analysis of variance was taken out with the vegan package. Statistical analysis was performed by a Student’s t test in the SPSS software v.20. The niche breadth and generalist/specialist species were calculated based on the previous descriptions.47−49 The generalist species can thrive in a wide range of environmental conditions and utilize a variety of different resources. However, specialist species are able to only thrive in a limited range of environmental conditions or have a narrow diet.50,51 The interactions of ″microbe–microbe″ and ″environment–microbe″ were analyzed using a co-occurrence network.52,53 A connected link denotes a high and significant Spearman’s correlation between variables. The size of each node is positively related to the count of connections–degree. A larger node represents a node with more connections. The thickness of each link is positively proportional to the absolute Spearman value. The network plots were modeled by the interactive Gephi platform.54 Redundancy analysis (RDA) was conducted by OmicStudio (https://www.omicstudio.cn/tool).
Results
Geochemical Analyses
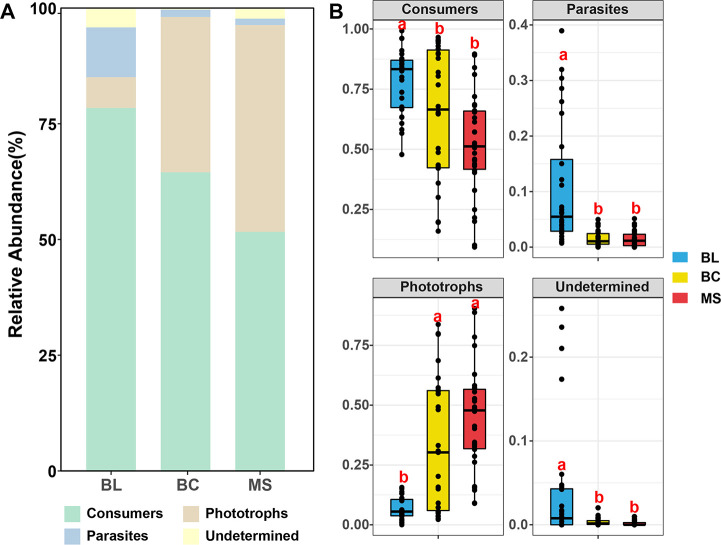
This study analyzed the total concentration of Al, As, Cr, Mn, Pb, Sb, Si, Zn, pH, K, P, and S (Figure 1). As, Sb, pH, and S concentrations decreased as the primary succession progressed from BL to BC and finally to MS. However, contrary patterns of K and P were observed. Lower Al and Mn contents were observed in BC compared to those in BL or MS. The highest Si concentration was observed in BC followed by MS and BL. In addition, the total concentration of Cr, Pb, and Zn showed no significant difference between the series.
Figure 1.
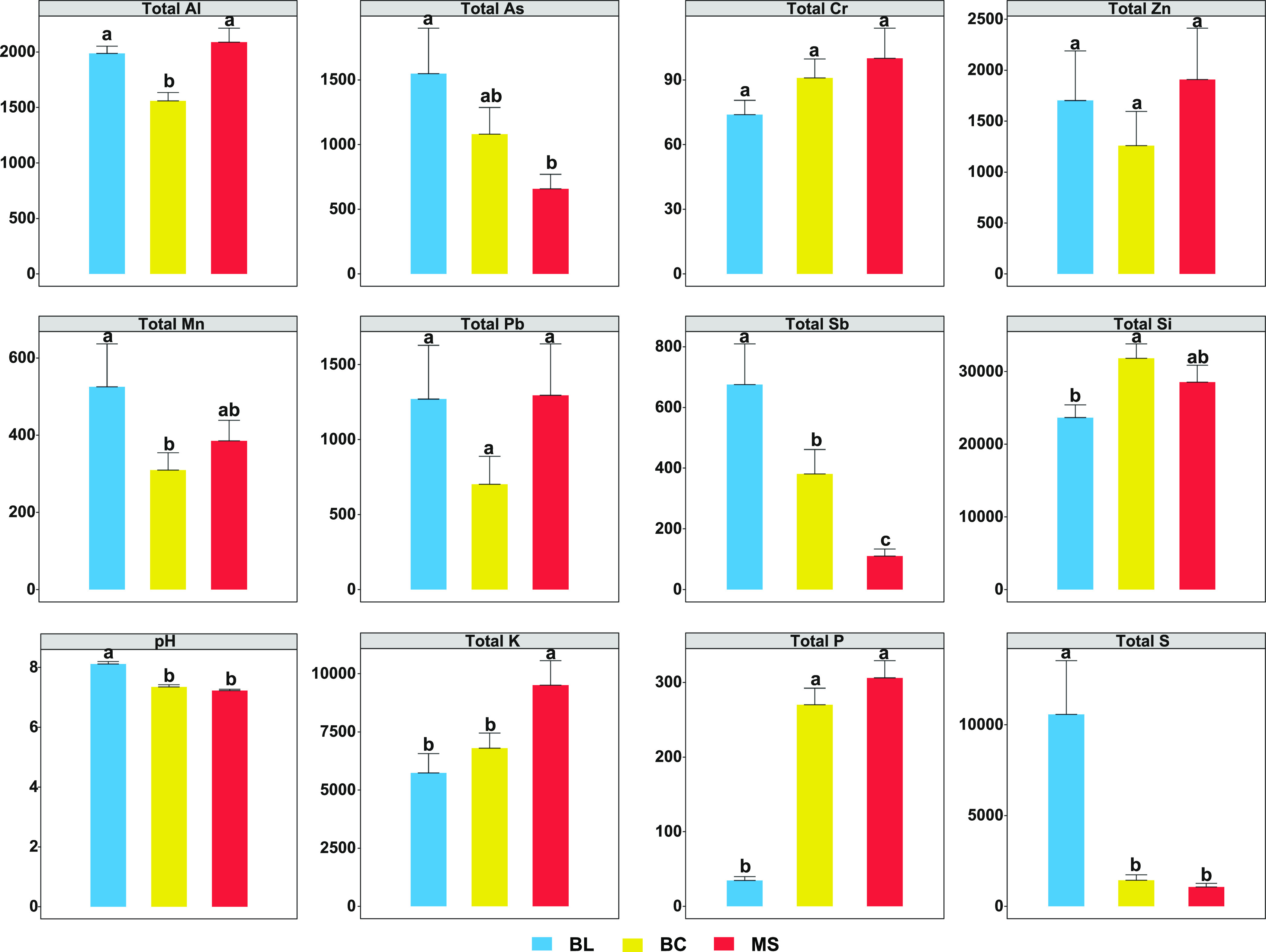
Geochemical parameters of samples. Different letters (a, b, c) indicate significant difference based on a least significant difference (LSD) test (p < 0.05). BL: original tailings from bare land, BC: tailings of biological crusts, and MS: tailings from the Miscanthus sinensis rhizosphere.
Characterization of the Overall Protistan Community
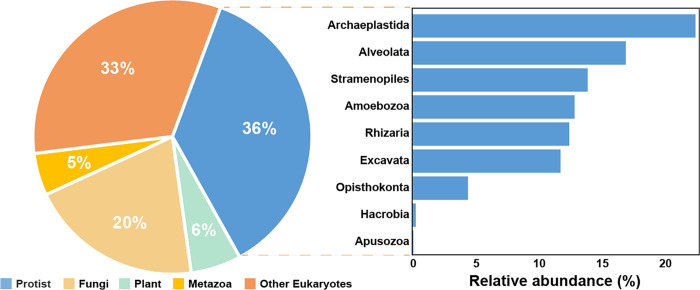
Based on the analysis of eukaryotic sequences, protists were found to be the most dominant eukaryotes (36%) (Figure 2). Sequences of Archaeplastida were the most abundant supergroup of protists followed by Alveolata, Stramenopiles, Amoebozoa, Rhizaria, Excavata, Opisthokonta, Hacrobia, and Apusozoa (Figure 1).
Figure 2.
Compositions of eukaryotic communities in mine tailing samples.
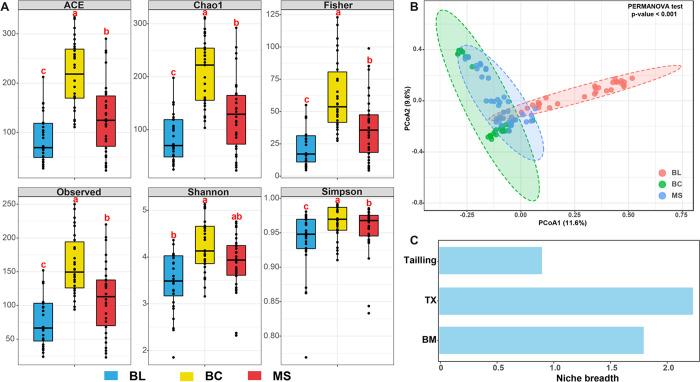
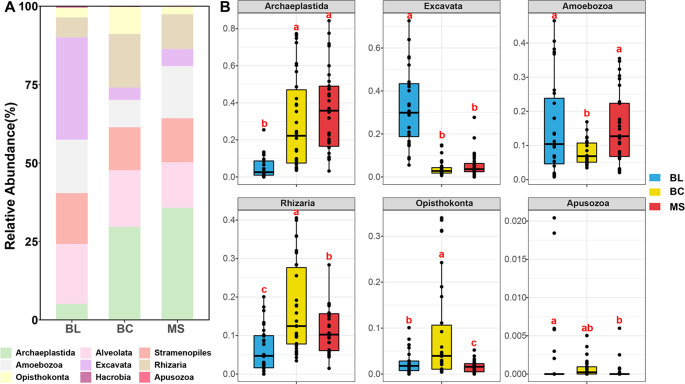
For the whole protistan community, the alpha diversities (ACE, Chao1, Fisher, Observed, Shannon, and Simpson) were the highest in BC followed by BL and MS (Figure 3A). Protistan community compositions varied significantly among different stages of tailings during primary succession, as evidenced by the obvious separation between the cluster of BL samples and the clusters of BC and MS samples (p < 0.001) (Figure 3B). In addition, the different niche breadths of BL, BC, and MS furthermore demonstrated the differences of the protistan communities among the three stages of tailings during primary succession (Figure 3C). Community composition analysis indicated that Excavata sequences were the most abundant in the BL samples, while Archaeplastida sequences became the most abundant in BC and MS (Figure 4A). In the primary succession samples, significant differences were observed in the supergroup (Figure 4B). The relative abundance of Archaeplastida was 4.8- and 6.0-fold higher in BC and MS samples than in BL, while the relative abundance of Excavata was 0.9- and 0.8-fold lower, respectively. In addition, the highest and lowest relative abundance of Rhizaria and Amoebozoa was observed in the BC samples, respectively.
Figure 3.
Comparisons of (A) the diversity indices, (B) the principal coordinate analysis based on the Bray–Curtis distance, and (C) the niche breadth of the whole protistan community. Different letters (a, b, c) indicate significant difference based on a least significant difference (LSD) test (p < 0.05). BL: original tailings from bare land, BC: tailings of biological crusts, and MS: tailings from the Miscanthus sinensis rhizosphere.
Figure 4.
Relative abundances at the supergroup level: (A) bar graph and (B) box plot. Different letters (a, b, c) indicate significant difference based on a least significant difference (LSD) test (p < 0.05). BL: original tailings from bare land, BC: tailings of biological crusts, and MS: tailings from the Miscanthus sinensis rhizosphere.
Functional Protistan Community Compositions
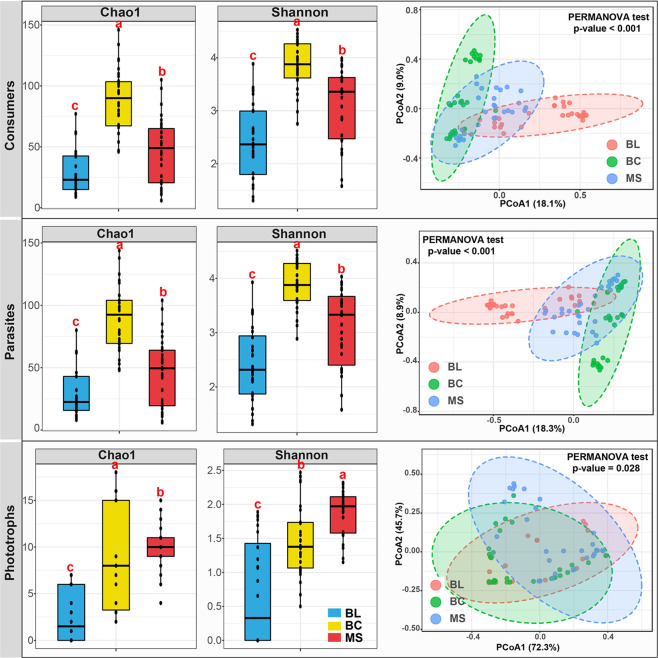
The assignment of the taxonomic profiles to their individual functions illuminated the impacts of primary succession on the potential functions of protists. The protistan community was functionally divided into consumers, parasites, and phototrophs (Figure 5). The alpha diversities and PCoA revealed significant variations in the functional community composition of protists among the primary succession of the three types of tailings. Thereby, the protistan functional community composition was furthermore summarized (Figure 6A). The consumers were observed to be the most dominant functional group in all tailing samples. The relative abundances of consumers and parasites were significantly higher in BL samples than those in BC and MS samples, while the relative abundances of phototrophs were significantly lower in BL than those in BC and MS samples (Figure 6B).
Figure 5.
Comparisons of the diversity indices and the PCoA based on the Bray–Curtis distance of the functional protistan communities. Different letters (a, b, c) indicate significant difference based on a least significant difference (LSD) test (p < 0.05). BL: original tailings from bare land, BC: tailings of biological crusts, and MS: tailings from the Miscanthus sinensis rhizosphere.
Figure 6.
The relative abundances of functional protists. (A) Bar graph and (B) box plots. Different letters (a, b, c) indicate significant difference based on a least significant difference (LSD) test (p < 0.05). BL: original tailings from bare land, BC: tailings of biological crusts, and MS: tailings from the Miscanthus sinensis rhizosphere.
Interaction Networks and Key Functional Protists
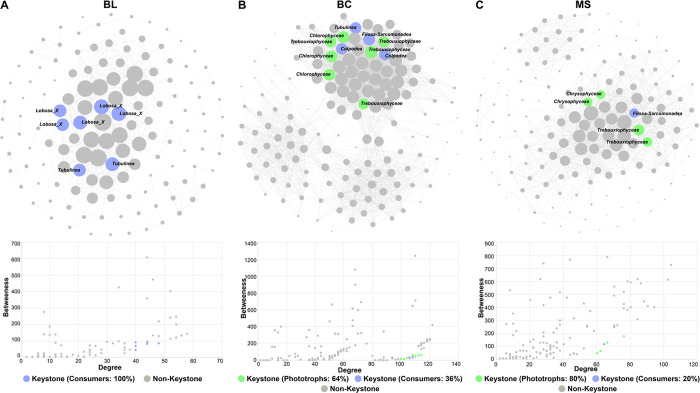
The interaction networks of the functional community of protists were generated. Consistent with the functional community composition, the consumers accounted for more than 60% in each network of BL, BC, and MS samples (Figure S2). Importantly, the ratio of phototrophs was increasing over time with the primary succession of tailings (from BL to MS). In addition, the proportion of positive links occupied more than 90% in all three networks. The keystone taxa were identified according to the criteria of the nodes with high degrees and low betweenness centralities.55 The keystone taxa are highly connected microorganisms, which individually or within a community have a significant impact on the structure and function of the microbial communities regardless of their abundance across space and time.52 These taxa play a unique and critical role in the microbiome, and their removal can lead to great changes in the structure and function of microbial communities.55 In BL, all identified keystone taxa were consumers, including Lobosa_X and Tubulinea (Figure 7A and Table S3). In BC (Figure 7B and Table S4) and MS (Figure 7C and Table S5), the identified keystone taxa were classified as consumers and phototrophs, in which the keystone phototrophs become dominant (BC: 64%, MS: 80%). The keystone phototrophs were classified as Chlorophyceae and Trebouxiophyceae at the class level. The highest relative abundance of Chlorophyceae was observed in BC; the relative abundance of Trebouxiophyceae was increasing with the primary succession of tailings (Figure S3).
Figure 7.
Co-occurrence network analysis of protist–protist showing the biological interactions in (A) BL, (B) BC, and (C) MS. Edges show significant connections. The size of each node is proportional to the number of connections to it. The scatter plot below the network plot shows criteria used to select the keystone taxa. BL: original tailings from bare land, BC: tailings of biological crusts, and MS: tailings from the Miscanthus sinensis rhizosphere.
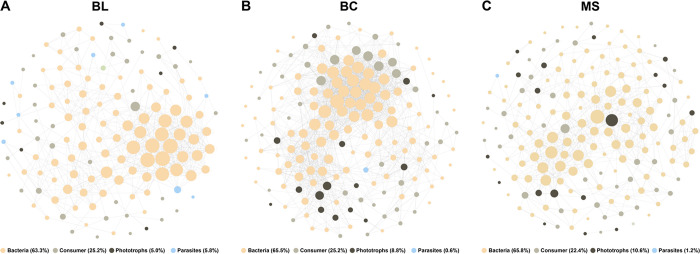
Furthermore, the interactions between protistan and bacterial communities were analyzed to assess the ecological roles of protists in soil microbial communities (Figure 8). The percentage of consumers was reasonably similar in all three networks, while the ratio of phototrophs increased (from 5.0 to 10.6%) during the primary succession of tailings. Generalist species have wider fundamental niches than specialist species, leading to the strong competitiveness of generalists.56 The largest generalists of phototrophic protists were observed in MS followed by BC and BL (Figure 9). Taken together, these results suggest that phototrophic protists are becoming more and more competitive over time and may play important ecological functions during the primary succession of the tailings.
Figure 8.
The co-occurrence network of protist–bacteria showing the biological interactions in (A) BL, (B) BC, and (C) MS. Edges only show significant connections. The size of each node is proportional to the number of connections to it. BL: original tailings from bare land, BC: tailings of biological crusts, and MS: tailings from the Miscanthus sinensis rhizosphere.
Figure 9.
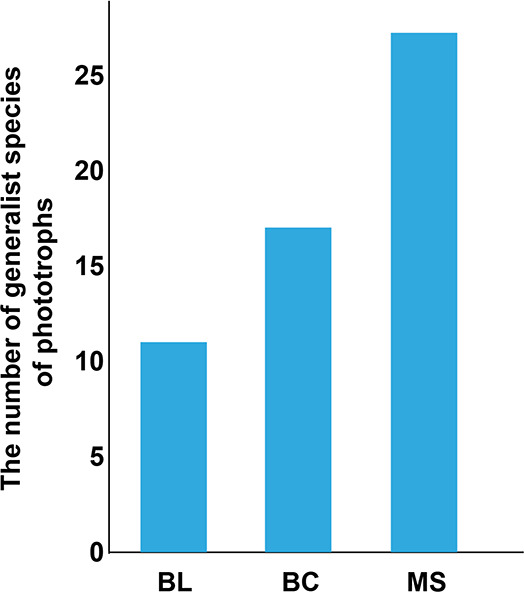
The number of generalist species of phototrophs in BL, BC, and MS. BL: original tailings from bare land, BC: tailings of biological crusts, and MS: tailings from the Miscanthus sinensis rhizosphere.
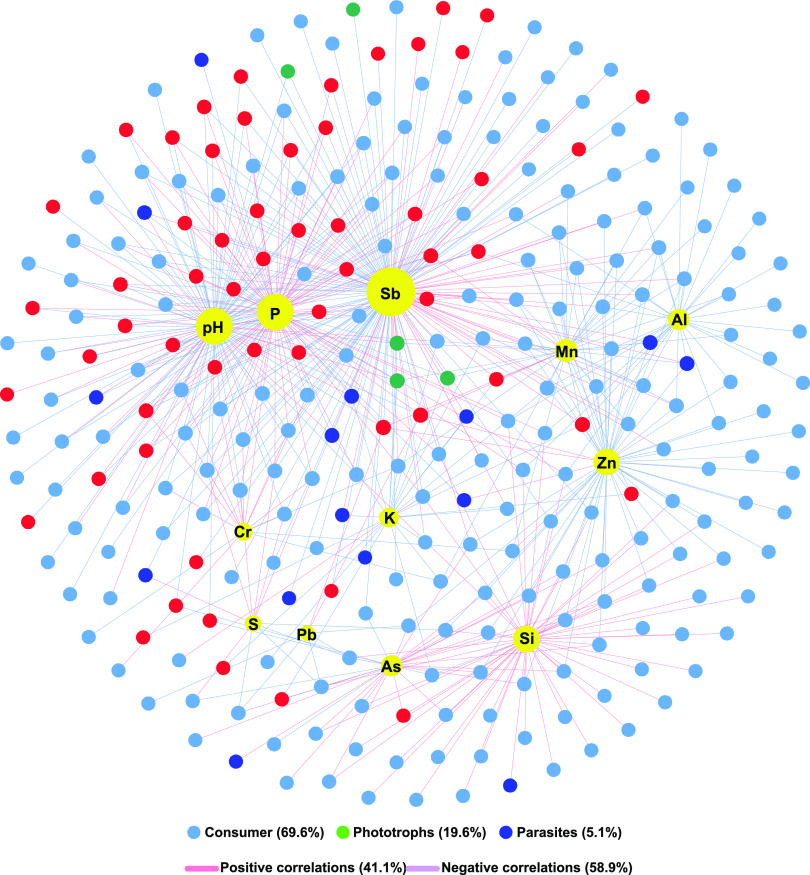
The interactions between the geochemical parameters and the functional community of protists are shown in Figure 10. The Sb, pH, and P contents were the main factors, which affected 55.7% of ASVs. Other geochemical parameters including Si, Zn, and Mn also had great influences on the functional community. The above results were also demonstrated by the RDA analysis (Figure S4).
Figure 10.
Co-occurrence network of environment–microbe showing the correlations between the geochemical parameters and the relative abundances (ASV level).
Functional Genes of the Protistan Community
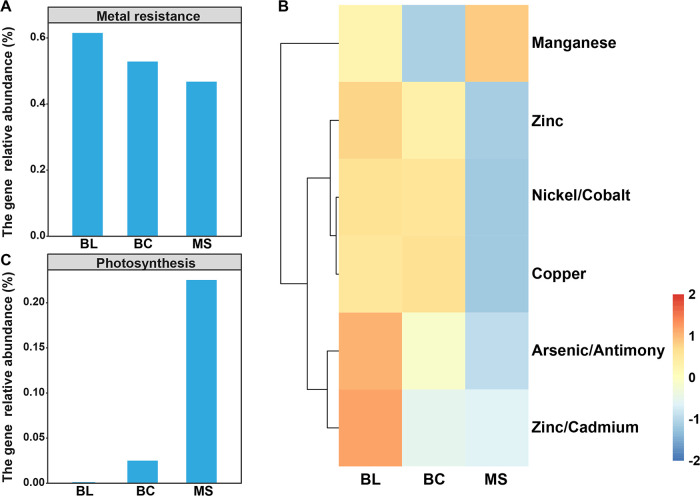
The reads classified as protists from the metagenomes were retained for the annotation of functional genes (Figure 11). In general, BL samples had the highest relative abundance of genes associated with metal tolerance (Figure 11A), including Zn, nickel, cobalt, copper, As, Sb, and cadmium, except for manganese (Figure 11B), which indicated that the presence of vegetation may provide a better niche for protists. Importantly, we focused on genes associated with photosynthesis (Figure 11C). The highest relative abundance was observed in MS followed by BC and BL, which further demonstrated that phototrophic protists may play important roles during the primary succession of mine tailings.
Figure 11.
Metagenomic analysis indicates the metabolic potentials of the protistan community inhabiting mine tailings. (A) Total genes providing metal tolerance, (B) individual genes providing metal tolerance, and (C) photosynthesis genes. BL: original tailings from bare land, BC: tailings of biological crusts, and MS: tailings from the Miscanthus sinensis rhizosphere.
Discussion
Compared to bacteria, archaea, and fungi, our understanding of protists in environmental samples is limited, especially for those inhabiting tailings. Protists may play a key role in the primary succession of mine tailings by adjusting the food web and carbon flow. In this study, three different types of tailings including those with Sb, Pb/Zn, and Au contamination from mines with three remarkably different stages (original tailings from bare land, biological crusts, and tailings from the rhizosphere of Miscanthus sinensis grasslands) were selected to investigate the protistan community structure and function associated with ecosystem development in mine tailings. The microbial interactions, keystone of functional protists, and functional genes were also evaluated.
Dominant Protists in the Tailings
Clear differences were observed among the protistan community in the three stages of tailings as described above (Figure 3), suggesting that the primary succession of tailings serves as a strong selection force in shaping the protistan community composition. This result is in line with prior reports that different stages of primary succession of tailings typically harbor unique microbiomes, though the protistan community was excluded in these studies,5,29,57,58 except that a dynamic and diverse eukaryotic community containing protists was described in oil sand tailings at Northern Alberta.59
Our research indicated that the protistan community in tailing samples was predominated by supergroups Archaeplastida, Excavata, Amoebozoa, and Rhizaria (Figure 4). A significantly enriched Excavata group was observed in the BL samples (Figure 4). Excavata showed highly unusual mitochondrial, nuclear, and chloroplast genomes.60 The phylogenetic relationship of Excavata members indicated that these members are more diverse than previously thought.60 Some Excavata have been isolated from various environmental samples such as soils, sediments, and AMD,41,60 but only a few have been found in the tailing samples.59 Therefore, our results suggest that the protist Excavata might play important roles in the tailings, especially in bare land.
The relative abundances of Archaeplastida and Rhizaria were significantly higher in the BC and MS samples compared to BL (Figure 4). The Archaeplastida, containing a large proportion of the large seaweeds, algae, and plants that support life on earth, are part of the main evolutionary lineages of the photosynthetic species and are critical to the world’s ecosystems and human welfare and health.61 Some members of Archaeplastida may even invade the land and set the stage for the evolutionary movement onto the land of many life groups.61 Therefore, it is reasonable that the Archaeplastida would be enriched during ecosystem development on mine tailings. Fine-tuning between microbe genomic traits and plant immune response is a well-known method used to mediate microbial colonization.62 Interestingly, in the Rhizaria supergroup, the cell-surface G-protein-coupled receptor mediated signaling pathway was found to be significantly enriched.63 The cell-surface G-protein-coupled receptor could sense various extracellular signals from the plant host and has been shown to facilitate colonization by microorganisms.64 Thus, the enriched Rhizaria may have a better colonization capacity in plants, even in biological crusts. Further studies are necessary to elucidate molecular plant–protists (even biological crusts–protists) and the interactions leading to the selective recruitment of some functional protists in the plant rhizosphere and biological crusts.
Functional Protists in the Tailings
Taxonomically related protists can serve different functions from microbial predators (consumers) to plant parasites and pathogens, and similarly, organisms that belong to different taxonomic groups may have developed to share similar patterns of nutritional feeding.13 Therefore, studying not only the taxonomy but also the potential function of protists would provide invaluable information.15,65,66 In addition, the co-occurrence network, in ecosystem studies, has become an ever more important tool for exploring the symbiosis modes of the microbial community and identifying keystone taxa.9,11,67 It has been reported that the biotic interactions of microorganisms are the most essential factors in the microbial community composition and function.68 The identification of keystone taxa is also critical because these taxa are essential for regulating the structure, composition, and functions of microbial communities52 and for facilitating the revegetation because they may be beneficial for plant growth and development.69
Methods for grouping protists by function and analyzing interactions within and between functional groups offered a better visualization of the correlation between protistan community members. In all three stages of primary succession of tailings, most interactions and nodes were linked to consumers (predators) (Figure S2), and some consumers were identified as the keystone taxa (Figure 7). It has been reported that predator–prey interactions increase the biotic network complexity and stability,70 and some protists belonging to consumer groups have been indicated to be the keystone taxa in the microbial networks.66 This is not surprising because consumers influence almost all microorganisms within their network.15 Populations of microorganisms preyed upon by consumers may be drastically reduced in size below the detection limit.15,71 Other microbes that have not preyed (i.e., nonpreferred or predation-resistant microbes) may benefit from the release of nutrients from the protist-preyed microbial biomass and/or obtain a competitive advantage due to protistan predation on their strong competitors.15,72,73 Therefore, our results suggest that consumers may have an important role in driving the microbial community network in mine tailings, especially in the stage of bare land.
The phototrophic protists may play the most important roles in the primary succession of mine tailings based on the identification of keystone phototroph taxa (Figure 7) and genes associated with photosynthesis (Figure 11C). Most metal(loid)s can impair photosynthesis in microorganisms.74 However, siderophores can protect microorganisms against various types and levels of metal(loid) toxicity.75 Siderophores can not only chelate Fe(III) with an extremely high affinity but also chelate numerous other metal(loid)s with variable affinities.76,77 With a high affinity for iron and other metal(loid)s and their substantial production and secretion into the extracellular medium, siderophores apparently provide a kind of extracellular protection for bacteria by blocking external metal(loid)s from entering bacteria and helping bacteria to avoid the diffusion of metal(loid)s through porins into the bacteria.78 Some microorganisms isolated from biological soil have been reported to produce siderophores.74 The rhizospheric microorganisms and the plant itself could produce siderophores. These secreted siderophores could also be used by other microorganisms.74,79 Therefore, our results may imply that the niches of biological crusts and the rhizosphere provided better environments for protistan phototrophs than bare land in mine tailings. Further research will be needed to illuminate the molecular mechanisms leading to the selective recruitment of protistan phototrophs in biological crusts and plant rhizospheres as well as to understand the ecological roles of protistan phototrophs in tailings.
To date, the ecology of protists in mining areas has been less studied, and the shift of the protist community across various geochemical gradients in mining areas is scarce. Protistan communities were investigated on a terrace contaminated by acid mine drainage (AMD) (AMD study hereafter).41 The intrusion of AMD created a sharp geochemical gradient along the terrace. In this study (Tailing study hereafter), the primary succession changes also created a sharp geochemical gradient. Therefore, these two studies provided an opportunity to investigate the response of protists inhabiting mining areas to a sharp geochemical shift, and their responses were compared here. In the AMD study, pH was identified as the major driving force, but Sb and P were identified as the major environmental factors in the Tailing study. Different geochemical conditions enriched different dominant protistan taxa. It was observed that Leishmania, Plasmodium, and Besnoitia were the most abundant protistan genera in AMD contaminated terrace, while Archaeplastida, Alveolata, Stramenopiles, Amoebozoa, and Excavata were the dominant taxa in the tailings. In addition, both the metabolic potentials of the innate protistan communities were investigated in the two studies, providing an in-depth analysis to understand the response of innate protists to various geochemical changes. In AMD studies, the enrichment of many functional genes suggested the unique metabolic potential of protistan communities to adapt to the extreme AMD environment. For example, genes for acid stress (e.g., kdpA) and metal resistance (e.g., ASNA1, ArsR, and mmtH) were detected in the AMD study. Like the AMD study, genes for metal resistance were also prevailed in the Tailing study. But genes for photosynthesis increased from BC to MS, suggesting that phototrophic protists may play an important role in tailing vegetation. The comparison of two mining contaminated sites suggested that the geochemical conditions substantially shaped the innate protistan communities. Innate protistan communities may develop various survival strategies such as metal resistance, acid stress resistance, and photosynthesis to respond to extreme geochemical conditions.
Conclusions
In summary, this study provided novel insights and proof that primary succession within mine tailings induces changes in the alpha diversity, structure, composition, and biotic interaction of the protistan community based on 18S rRNA sequencing. Furthermore, taxonomic profiles were analyzed as they relate to functionality. Some members classified as consumers showed that they served vertical roles and dominated the network of microbial communities in mine tailings, especially in the bare land stage. Some phototrophic protists (e.g., Chlorophyceae and Trebouxiophyceae) could be selectively enriched in the biological crusts and plant rhizosphere, which may be beneficial for the successful colonization of biological crusts and pioneer plants on mine tailings. Overall, these results suggest that the primary succession of mine tailings is a strong driver of the protistan community, facilitating the functioning of the tailing ecosystem.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 32161143018, 42107133, U21A2035, 41907212, and 42107285), GDAS’ Project of Science and Technology Development (Grants 2020GDASYL-20200102014, 2021GDASYL-20210103048, 2019GDASYL-0102002-1, 2019GDASYL-0102002-4, and 2022GDASZH-2022010203), China Postdoctoral Science Foundation (Grants 2021M690745 and 2021M700890), and Guangdong Introducing Innovative and Entrepreneurial Talents (Grant 2017GC010570). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsenvironau.1c00066.
Sampling locations; co-occurrence network analysis of protist–protist; relative abundance of keystone taxa of phototrophs; and redundancy analysis (Figures S1–S4) (PDF)
Sample information of sites; trophic functional groups at the genus level; and node table for protist–protist interaction network in bare land/biological/M. sinensis rhizosphere (Tables S1–S5 (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Wilkinson B.; McElroy B. The impact of humans on continental erosion and sedimentation. Geol. Soc. Am. Bull. 2007, 119, 140. 10.1130/B25899.1. [DOI] [Google Scholar]
- Li X.; You F.; Bond P. L.; Huang L. Establishing microbial diversity and functions in weathered and neutral Cu–Pb–Zn tailings with native soil addition. Geoderma 2015, 247-248, 108–116. 10.1016/j.geoderma.2015.02.010. [DOI] [Google Scholar]
- Edraki M.; Baumgartl T.; Manlapig E.; Bradshaw D.; Franks D. M.; Moran C. J. Designing mine tailings for better environmental, social and economic outcomes: a review of alternative approaches. J. Cleaner Prod. 2014, 84, 411–420. 10.1016/j.jclepro.2014.04.079. [DOI] [Google Scholar]
- Wang L.; Ji B.; Hu Y.; Liu R.; Sun W. A review on in situ phytoremediation of mine tailings. Chemosphere 2017, 184, 594–600. 10.1016/j.chemosphere.2017.06.025. [DOI] [PubMed] [Google Scholar]
- Maltsev Y.; Maltseva S.; Maltseva I. Diversity of cyanobacteria and algae during primary succession in iron ore tailing dumps. Microb. Ecol. 2021, 83, 408–423. 10.1007/s00248-021-01759-y. [DOI] [PubMed] [Google Scholar]
- Sun X.; Kong T.; Häggblom M. M.; Kolton M.; Li F.; Dong Y.; Huang Y.; Li B.; Sun W. Chemolithoautotropic diazotrophy dominates the nitrogen fixation process in mine tailings. Environ. Sci. Technol. 2020, 54, 6082–6093. 10.1021/acs.est.9b07835. [DOI] [PubMed] [Google Scholar]
- Xu D. M.; Zhan C. L.; Liu H. X.; Lin H. Z. A critical review on environmental implications, recycling strategies, and ecological remediation for mine tailings. Environ. Sci. Pollut. Res. Int. 2019, 26, 35657–35669. 10.1007/s11356-019-06555-3. [DOI] [PubMed] [Google Scholar]
- Adiansyah J.; Rosano M.; Vink S.; Keir G. A framework for a sustainable approach to mine tailings management: Disposal strategies. J. Cleaner Prod. 2015, 108, 1050. 10.1016/j.jclepro.2015.07.139. [DOI] [Google Scholar]
- Li Y.; Lin H.; Gao P.; Yang N.; Xu R.; Sun X.; Li B.; Xu F.; Wang X.; Song B.; Sun W. Synergistic impacts of arsenic and antimony co-contamination on diazotrophic communities. Microb. Ecol. 2021, 1–15. 10.1007/s00248-021-01824-6. [DOI] [PubMed] [Google Scholar]
- Sun W.; Xiao E.; Häggblom M.; Krumins V.; Dong Y.; Sun X.; Li F.; Wang Q.; Li B.; Yan B. Bacterial survival strategies in an alkaline tailing site and the physiological mechanisms of dominant phylotypes as revealed by metagenomic analyses. Environ. Sci. Technol. 2018, 52, 13370–13380. 10.1021/acs.est.8b03853. [DOI] [PubMed] [Google Scholar]
- Li Y.; Zhang M.; Xu R.; Lin H.; Sun X.; Xu F.; Gao P.; Kong T.; Xiao E.; Yang N.; Sun W. Arsenic and antimony co-contamination influences on soil microbial community composition and functions: Relevance to arsenic resistance and carbon, nitrogen, and sulfur cycling. Environ. Int. 2021, 153, 106522 10.1016/j.envint.2021.106522. [DOI] [PubMed] [Google Scholar]
- Azaiez A.; Beaudoin Nadeau M.; Bertrand A.; Khasa D. P. In vitro selection of ecologically adapted ectomycorrhizal fungi through production of fungal biomass and metabolites for use in reclamation of biotite mine tailings. Mycologia 2018, 110, 1017–1032. 10.1080/00275514.2018.1520036. [DOI] [PubMed] [Google Scholar]
- Geisen S.; Mitchell E.; Adl S.; Bonkowski M.; Dunthorn M.; Ekelund F.; Fernández L.; Jousset A.; Krashevska V.; Singer D.; Spiegel F.; Walochnik J.; Lara E. Soil protists: A fertile frontier in soil biology research. FEMS Microbiol. Rev. 2018, 42, 293. 10.1093/femsre/fuy006. [DOI] [PubMed] [Google Scholar]
- Adl S.; Bass D.; Lane C.; Luke J.; Schoch C.; Alexey S.; Dr. Agatha S.; Berney C.; Brown M.; Burki F.; Ardenas P.; O I.; Chistyakova L.; del Campo J.; Dunthorn M.; Edvardsen B.; Eglit Y.; Guillou L.; Hampl V.; Zhang Q. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. 10.1111/jeu.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asiloglu R.; Samuel S. O.; Sevilir B.; Akca M. O.; Acar Bozkurt P.; Suzuki K.; Murase J.; Turgay O. C.; Harada N. Biochar affects taxonomic and functional community composition of protists. Biol. Fert. Soils 2021, 57, 15–29. 10.1007/s00374-020-01502-8. [DOI] [Google Scholar]
- Geisen S. The bacterial-fungal energy channel concept challenged by enormous functional versatility of soil protists. Soil Biol. Biochem. 2016, 102, 22–25. 10.1016/j.soilbio.2016.06.013. [DOI] [Google Scholar]
- Kuikman P.; Veen J. The impact of Protozoa on the availability of bacterial nitrogen to plants. Biol. Fert. Soils 1989, 8, 13–18. 10.1007/BF00260510. [DOI] [Google Scholar]
- Bonkowski M. Protozoa and plant growth: The microbial loop in soil revisited. New Phytol. 2004, 162, 617–631. 10.1111/j.1469-8137.2004.01066.x. [DOI] [PubMed] [Google Scholar]
- Amaral Zettler L. A.; Gómez F.; Zettler E.; Keenan B. G.; Amils R.; Sogin M. L. Eukaryotic diversity in Spain’s river of fire. Nature 2002, 417, 137–137. 10.1038/417137a. [DOI] [PubMed] [Google Scholar]
- Ishii Y.; Shimada M. Learning predator promotes coexistence of prey species in host-parasitoid systems. Proc. Natl. Acad. Sci. USA 2012, 109, 5116–5120. 10.1073/pnas.1115133109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakoç C.; Clark A.; Chatzinotas A. Diversity and coexistence are influenced by time-dependent species interactions in a predator-prey system: Coexistence in a predator-prey system. Ecol. Lett. 2020, 23, 983. 10.1111/ele.13500. [DOI] [PubMed] [Google Scholar]
- Jassey V. E. J.; Signarbieux C.; Hättenschwiler S.; Bragazza L.; Buttler A.; Delarue F.; Fournier B.; Gilbert D.; Laggoun-Dëfarge F.; Lara E.; T. E. Mills R.; Mitchell E. A. D.; Payne R. J.; Robroek B. J. M. An unexpected role for mixotrophs in the response of peatland carbon cycling to climate warming. Sci. Rep. 2015, 5, 16931. 10.1038/srep16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S.; Dibbern D.; Moll J.; Huenninghaus M.; Koller R.; Krüger D.; Marhan S.; Urich T.; Wubet T.; Bonkowski M.; Buscot F.; Lueders T.; Kandeler E. Resource partitioning between bacteria, fungi, and protists in the detritusphere of an agricultural soil. Front. Microbiol. 2016, 7, 1524. 10.3389/fmicb.2016.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi-Mehrabad A.; He Z.; Tamas I.; Sharp C. E.; Brady A. L.; Rochman F. F.; Bodrossy L.; Abell G. C.; Penner T.; Dong X.; Sensen C. W.; Dunfield P. F. Methanotrophic bacteria in oilsands tailings ponds of northern Alberta. ISME J. 2013, 7, 908–921. 10.1038/ismej.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar M.; Richardson E.; Tan B.; Walker G.; Dunfield P. F.; Bass D.; Nesbø C.; Foght J.; Dacks J. B. Next-generation sequencing assessment of eukaryotic diversity in oil sands tailings ponds sediments and surface water. J. Eukaryot Microbiol. 2016, 63, 732–743. 10.1111/jeu.12320. [DOI] [PubMed] [Google Scholar]
- Shu W.; Ye Z.; Zhang Z.; Lan C. Y.; Wong M. Natural colonization of plants on five lead/zinc mine tailings in southern china. Restor. Ecol. 2005, 13, 49–60. 10.1111/j.1526-100X.2005.00007.x. [DOI] [Google Scholar]
- Marrs R. H.; Bradshaw A. D. Primary succession on man-made wastes: the importance of resource acquisition. Primary Success. Land 1993, 221–248. [Google Scholar]
- Hery M.; Philippot L.; Mériaux E.; Poly F.; Roux X.; Navarro E. Nickel mine spoils revegetation attempts: Effect of pioneer plants on two functional bacterial communities involved in the N-cycle. Environ. Microbiol. 2005, 7, 486–498. 10.1111/j.1462-2920.2005.00705.x. [DOI] [PubMed] [Google Scholar]
- Huang L.-N.; Tang F.-Z.; Song Y.-S.; Wan C.-Y.; Wang S.-L.; Liu W.; Shu W. Biodiversity, abundance, and activity of nitrogen-fixing bacteria during primary succession on a copper mine tailings. FEMS Microbiol. Ecol. 2011, 78, 439–450. 10.1111/j.1574-6941.2011.01178.x. [DOI] [PubMed] [Google Scholar]
- Sun X.; Song B.; Xu R.; Zhang M.; Gao P.; Lin H.; Sun W. Root-associated (rhizosphere and endosphere) microbiomes of the Miscanthus sinensis and their response to the heavy metal contamination. J. Environ. Sci. 2021, 104, 387–398. 10.1016/j.jes.2020.12.019. [DOI] [PubMed] [Google Scholar]
- Li Y.; Guo L.; Häggblom M. M.; Yang R.; Li M.; Sun X.; Chen Z.; Li F.; Su X.; Yan G.; Xiao E.; Zhang H.; Sun W. Serratia spp. are responsible for nitrogen fixation fueled by As(III) Oxidation, a novel biogeochemical process identified in mine tailings. Environ. Sci. Technol. 2022, 56, 2033–2043. 10.1021/acs.est.1c06857. [DOI] [PubMed] [Google Scholar]
- Sun X.; Kong T.; Li F.; Häggblom M. M.; Kolton M.; Lan L.; Lau Vetter M. C.; Dong Y.; Gao P.; Kostka J. E.; Li B.; Sun W. Desulfurivibrio spp. mediate sulfur-oxidation coupled to Sb (V) reduction, a novel biogeochemical process. ISME J. 2022, 1547–1556. 10.1038/s41396-022-01201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Wang M.; Chen S. Application of N2-fixing Paenibacillus triticisoli BJ-18 changes the compositions and functions of the bacterial, diazotrophic, and fungal microbiomes in the rhizosphere and root/shoot endosphere of wheat under field conditions. Biol. Fert. Soils 2021, 57, 347–362. 10.1007/s00374-020-01528-y. [DOI] [Google Scholar]
- Amaral-Zettler L. A.; McCliment E. A.; Ducklow H. W.; Huse S. M. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 2009, 4, e6372 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G.; Kuczynski J.; Stombaugh J.; Bittinger K.; Bushman F. D.; Costello E. K.; Fierer N.; Peña A. G.; Goodrich J. K.; Gordon J. I.; Huttley G. A.; Kelley S. T.; Knights D.; Koenig J. E.; Ley R. E.; Lozupone C. A.; McDonald D.; Muegge B. D.; Pirrung M.; Reeder J.; Sevinsky J. R.; Turnbaugh P. J.; Walters W. A.; Widmann J.; Yatsunenko T.; Zaneveld J.; Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L.; Bachar D.; Audic S.; Bass D.; Berney C.; Bittner L.; Boutte C.; Burgaud G.; de Vargas C.; Decelle J.; Del Campo J.; Dolan J. R.; Dunthorn M.; Edvardsen B.; Holzmann M.; Kooistra W. H.; Lara E.; Le Bescot N.; Logares R.; Mahé F.; Massana R.; Montresor M.; Morard R.; Not F.; Pawlowski J.; Probert I.; Sauvadet A. L.; Siano R.; Stoeck T.; Vaulot D.; Zimmermann P.; Christen R. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013, 41, D597–D604. 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C.; Pruesse E.; Yilmaz P.; Gerken J.; Schweer T.; Yarza P.; Peplies J.; Glöckner F. O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen B.-A.; Chen Q.; Hu H. Oxytetracycline and ciprofloxacin exposure altered the composition of protistan consumers in an agricultural soil. Environ. Sci. Technol. 2020, 54, 9556–9563. 10.1021/acs.est.0c02531. [DOI] [PubMed] [Google Scholar]
- Oliverio A. M.; Geisen S.; Delgado-Baquerizo M.; Maestre F. T.; Turner B. L.; Fierer N. The global-scale distributions of soil protists and their contributions to belowground systems. Sci. Adv. 2020, 6, eaax8787 10.1126/sciadv.aax8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A.; Jiao X. Y.; Chen Q.; Trivedi P.; Li Z.; Li F.; Zheng Y.; Lin Y.; Hu H. W.; He J. Z. Fertilization alters protistan consumers and parasites in crop-associated microbiomes. Environ. Microbiol. 2021, 23, 2169–2183. 10.1111/1462-2920.15385. [DOI] [PubMed] [Google Scholar]
- Xu R.; Zhang M.; Lin H.; Gao P.; Yang Z.; Wang D.; Sun X.; Li B.; Wang Q.; Sun W. Response of soil protozoa to acid mine drainage in a contaminated terrace. J. Hazard. Mater. 2022, 421, 126790 10.1016/j.jhazmat.2021.126790. [DOI] [PubMed] [Google Scholar]
- Bolger A. M.; Lohse M.; Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D. E.; Lu J.; Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.; Breitwieser F.; Thielen P.; Salzberg S. Bracken: Estimating species abundance in metagenomics data. Peer J. Comput. Sci. 2017, 3, e104 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- Li D.; Liu C.-M.; Luo R.; Sadakane K.; Lam T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Aramaki T.; Blanc-Mathieu R.; Endo H.; Ohkubo K.; Kanehisa M.; Goto S.; Ogata H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S. N.; Kolasa J.; Cottenie K. Contrasts between habitat generalists and specialists: an empirical extension to the basic metacommunity framework. Ecology 2009, 90, 2253–2262. 10.1890/08-0851.1. [DOI] [PubMed] [Google Scholar]
- Salazar G., EcolUtils: Utilities for community ecology analysis. R package version 0.1. Github, In 2018.
- Zhang J.; Zhang B.; Liu Y.; Guo Y.; Shi P.; Wei G. Distinct large-scale biogeographic patterns of fungal communities in bulk soil and soybean rhizosphere in China. Sci. Total Environ. 2018, 644, 791–800. 10.1016/j.scitotenv.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Marvier M.; Kareiva P.; Neubert M. G. Habitat destruction, fragmentation, and disturbance promote invasion by habitat generalists in a multispecies metapopulation. Risk Anal. Int. J. 2004, 24, 869–878. 10.1111/j.0272-4332.2004.00485.x. [DOI] [PubMed] [Google Scholar]
- Sriswasdi S.; Yang C.-C.; Iwasaki W. Generalist species drive microbial dispersion and evolution. Nat. Commun. 2017, 8, 1162. 10.1038/s41467-017-01265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S.; Schlaeppi K.; Van der Heijden M. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 1. 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- Li Y.; Yang R.; Guo L.; Gao W.; Su P.; Xu Z.; Xiao H.; Ma Z.; Liu X.; Gao P.; Li B.; Sun X.; Yan G.; Sun W. The composition, biotic network, and assembly of plastisphere protistan taxonomic and functional communities in plastic-mulching croplands. J. Hazard. Mater. 2022, 430, 128390 10.1016/j.jhazmat.2022.128390. [DOI] [PubMed] [Google Scholar]
- Newman M. E. J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577. 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D.; Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014, 5, 219. 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B.; Hayek L.-A. C. Distinguishing relative specialist and generalist species in the fossil record. Mar. Micropaleontol. 2015, 119, 7–16. 10.1016/j.marmicro.2015.07.001. [DOI] [Google Scholar]
- Santini T. C.; Raudsepp M.; Hamilton J.; Nunn J. Extreme geochemical conditions and dispersal limitation retard primary succession of microbial communities in gold tailings. Front. Microbiol. 2018, 9, 2785. 10.3389/fmicb.2018.02785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin Y.; Goberna M.; Verdú M.; Navarro-Cano J. A. Successional trajectories of soil bacterial communities in mine tailings: The role of plant functional traits. J. Environ. Manage. 2019, 241, 284–292. 10.1016/j.jenvman.2019.04.023. [DOI] [PubMed] [Google Scholar]
- Richardson E.; Bass D.; Smirnova A.; Paoli L.; Dunfield P.; Dacks J. B. Phylogenetic estimation of community composition and novel eukaryotic lineages in base mine lake: an oil sands tailings reclamation site in Northern Alberta. J. Eukaryot. Microbiol. 2020, 67, 86–99. 10.1111/jeu.12757. [DOI] [PubMed] [Google Scholar]
- von der Heyden S.; Chao E. E.; Vickerman K.; CAVALIER-SMITH T. Ribosomal RNA phylogeny of bodonid and diplonemid flagellates and the evolution of Euglenozoa. J. Eukaryot. Microbiol. 2004, 51, 402–416. 10.1111/j.1550-7408.2004.tb00387.x. [DOI] [PubMed] [Google Scholar]
- McCourt R., Archaeplastida: Diversification of red algae and the green plant lineage. In Encyclopedia of Evolutionary Biology, Kliman R. M., Ed. Academic Press: Oxford, 2016. pp. 101–106. [Google Scholar]
- Trivedi P.; Leach J. E.; Tringe S. G.; Sa T.; Singh B. K. Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- Bi K.; Chen T.; He Z.; Gao Z.; Zhao Y.; Liu H.; Fu Y.; Xie J.; Cheng J.; Jiang D. Comparative genomics reveals the unique evolutionary status of Plasmodiophora brassicae and the essential role of GPCR signaling pathways. Phytopathology Res. 2019, 1, 12. 10.1186/s42483-019-0018-6. [DOI] [Google Scholar]
- Xu X.-H.; Su Z.-Z.; Wang C.; Kubicek C. P.; Feng X.-X.; Mao L.-J.; Wang J.-Y.; Chen C.; Lin F.-C.; Zhang C.-L. The rice endophyte Harpophora oryzae genome reveals evolution from a pathogen to a mutualistic endophyte. Sci. Rep. 2014, 4, 5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G.; Zaneveld J.; Caporaso J. G.; McDonald D.; Knights D.; Reyes J. A.; Clemente J. C.; Burkepile D. E.; Thurber R. L. V.; Knight R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W.; Jousset A.; Guo S.; Karlsson I.; Zhao Q.; Wu H.; Kowalchuk G. A.; Shen Q.; Li R.; Geisen S. Soil protist communities form a dynamic hub in the soil microbiome. ISME J. 2018, 12, 634–638. 10.1038/ismej.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Lin H.; Gao P.; Yang N.; Xu R.; Sun X.; Li B.; Xu F.; Wang X.; Song B.; Sun W. Variation in the diazotrophic community in a vertical soil profile contaminated with antimony and arsenic. Environ. Pollut. 2021, 291, 118248 10.1016/j.envpol.2021.118248. [DOI] [PubMed] [Google Scholar]
- Bascompte J.; Stouffer D. B. The assembly and disassembly of ecological networks. Philos. Trans. R. Soc., B 2009, 364, 1781–1787. 10.1098/rstb.2008.0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.; Xu R.; Dong Y.; Li F.; Tao W.; Kong T.; Zhang M.; Qiu L.; Wang X.; Sun W. Investigation of the ecological roles of putative keystone taxa during tailing revegetation. Environ. Sci. Technol. 2020, 54, 11258–11270. 10.1021/acs.est.0c03031. [DOI] [PubMed] [Google Scholar]
- Allesina S.; Tang S. Stability criteria for complex ecosystems. Nature 2012, 483, 205–208. 10.1038/nature10832. [DOI] [PubMed] [Google Scholar]
- Rosenberg K.; Bertaux J.; Krome K.; Hartmann A.; Scheu S.; Bonkowski M. Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana. ISME J. 2009, 3, 675–684. 10.1038/ismej.2009.11. [DOI] [PubMed] [Google Scholar]
- Griffiths B. Microbial-feeding nematodes and protozoa in soil: Their effectson microbial activity and nitrogen mineralization in decomposition hotspots and the rhizosphere. Plant Soil 1994, 164, 25–33. 10.1007/BF00010107. [DOI] [Google Scholar]
- Saleem M.; Fetzer I.; Dormann C. F.; Harms H.; Chatzinotas A. Predator richness increases the effect of prey diversity on prey yield. Nat. Commun. 2012, 3, 1–7. 10.1038/ncomms2287. [DOI] [PubMed] [Google Scholar]
- Szivák I.; Behra R.; Sigg L. MEtal-induced reactive oxygen species production in chlamydomonas reinhardtii (Chlorophyceae). J. Phycol. 2009, 45, 427–435. 10.1111/j.1529-8817.2009.00663.x. [DOI] [PubMed] [Google Scholar]
- Schalk I. J.; Hannauer M.; Braud A. New roles for bacterial siderophores in metal transport and tolerance. Environ. Microbiol. 2011, 13, 2844–2854. 10.1111/j.1462-2920.2011.02556.x. [DOI] [PubMed] [Google Scholar]
- Hernlem B. J.; Vane L. M.; Sayles G. D. Stability constants for complexes of the siderophore desferrioxamine B with selected heavy metal cations. Inorg. Chim. Acta 1996, 244, 179–184. 10.1016/0020-1693(95)04780-8. [DOI] [Google Scholar]
- Neubauer U.; Nowack B.; Furrer G.; Schulin R. Heavy metal sorption on clay minerals affected by the siderophore desferrioxamine B. Environ. Sci. Technol. 2000, 34, 2749–2755. 10.1021/es990495w. [DOI] [Google Scholar]
- Zelaya-Molina L. X.; Hernández-Soto L. M.; Guerra-Camacho J. E.; Monterrubio-López R.; Patiño-Siciliano A.; Villa-Tanaca L.; Hernández-Rodríguez C. Ammonia-oligotrophic and diazotrophic heavy metal-resistant Serratia liquefaciens strains from pioneer plants and mine tailings. Microb. Ecol. 2016, 72, 324–346. 10.1007/s00248-016-0771-3. [DOI] [PubMed] [Google Scholar]
- Stintzi A.; Barnes C.; Xu J.; Raymond K. Microbial iron transport via a siderophore shuttle: A membrane ion transport paradigm. Proc. Natl. Acad. Sci. USA 2000, 97, 10691–10696. 10.1073/pnas.200318797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.