Abstract
Phage display technology has been applied in many fields of biological and medical sciences to study molecular interactions and especially in the generation of monoclonal antibodies of human origin. However, extremely low display level of antibody molecules on the surface of phage is an intrinsic problem of a phagemid-based display system resulting in low success rate of isolating specific binding molecules. We show here that display of single-chain antibody fragment (scFv) generated with pIGT3 phagemid can be increased dramatically by using a genetically modified Ex-phage. Ex-phage has a mutant pIII gene that produces a functional wild-type pIII in suppressing Escherichia coli strains but does not make any pIII in non-suppressing E.coli strains. Packaging phagemids encoding antibody-pIII fusion in F+ non-suppressing E.coli strains with Ex-phage enhanced the display level of antibody fragments on the surfaces of recombinant phage particles resulting in an increase of antigen-binding reactivity >100-fold compared to packaging with M13KO7 helper phage. Thus, the Ex-phage and pIGT3 phagemid vector provides a system for the efficient enrichment of specific binding antibodies from a phage display library and, thereby, increases the chance of obtaining more diverse antibodies specific for target antigens.
INTRODUCTION
The best characterized filamentous phage, M13, f1 and fd, which infect Escherichia coli have a single-stranded DNA genome which is encased in a long cylinder ∼7 nm wide by 900–2000 nm in length, and share 98% sequence homology (1). These phage have been modified to allow the display of foreign peptides or proteins on the surfaces by inserting specific oligonucleotides or entire coding genes into genes of phage capsid proteins such as pIII for oligovalent or pVIII for polyvalent expression of the fusions (2,3). The origin of phage display began by fusing EcoRI endonuclease to the minor capsid protein pIII, thereby displaying a EcoRI–gIII fusion protein on the surface of M13 virus particles. From this phage display, Smith (4) demonstrated that phage containing EcoRI–gIII fusion protein could be enriched >1000-fold over wild-type phage with an immobilized polyclonal antibody. Based on George Smith’s experiments (4) and advances in recombinant DNA technology, it became possible to build large libraries (i.e. <108) that display random peptides (5) or heterogeneous proteins such as antibody fragments (6). So far, a wide range of proteins and protein domains has been displayed on the phage particles for carrying out directed evolution of molecules. For example, stronger binding ligands for a receptor (7), novel enzyme inhibitors (8), novel DNA binding proteins (9), novel antagonists (10), potentially novel enzymes (11) or antibody fragments specific for various antigens (6,12,13) have been identified using a phage display technology. Judging from the numerous experimental data published up to now, it is certain that phage display is an important technique that rapidly made paths in the fields of immunology, biochemistry, protein engineering and cell biology, and has a great potential in massive functional analysis of novel genes and proteomes.
There are two types of vectors that have been used for the display of exogenous genes on the surfaces of filamentous phage. One is a phage vector (fUSE5, fAFF1, fd-CAT1 or fdtetDOG) (5,6,14,15) and the other is a phagemid vector (pHEN1, pComb3, pComb8 or pSEX) (15–17). In a phage vector system, peptides can be displayed as gIII fusion for oligovalent (14) or gVIII fusion for multivalent expressions (18) by cloning synthesized genes directly within the phage genome. Thus, a phage vector system provides a high display level of foreign peptides or protein fragments since all pIII molecules are originally presented as fusions. However, there is limitation in size of exogenous protein fragments fused with pIII (>100 amino acids) since the presence of a large foreign protein fragment at the N-terminal of pIII hinders the interaction of pIII with sex pili on bacterium that is absolutely required at the initial step of phage infection. In the case of pVIII, fusion with >10 amino acid residues compromises coat protein function in general, although recent publications have demonstrated that much larger protein fragments can be displayed as pVIII fusions (19,20).
For the display of larger molecules such as antibodies, therefore, a phagemid vector system is more suitable. In addition, this system has more advantages over a phage vector system including the more efficient ligation-transformation that allows creating libraries of larger size and relatively easy genetic manipulation for introducing special features into a phagemid. In a phagemid vector system, DNA of exogenous proteins are cloned into gIII (or gVIII) present within a phagemid vector, and the packaging of recombinant phagemid DNA and display of the fusions are provided by a helper phage such as M13KO7 or VCSM13. Thereby, the phagemid presents modified capsid proteins as fusions, and a helper phage supplies wild-type version of the coat proteins that is required for the successful reinfection of recombinant phage for amplification. The resulting phage particles display pIII from both wild-type gIII of the helper phage and the fusion pIII from the resident phagemid. This theoretically allows ‘oligovalent’ display of the foreign peptide and, thereby, should minimize avidity effects during affinity selection of the recombinant phage. In reality, however, the majority of pIII molecules present on the phage particles are wild-type pIII because of proteolytic degradation of the pIII fusion protein at the periplasmic space of E.coli. This implies that a large proportion of phage particles is actually ‘bald’ with respect to display of fusion in a phagemid display system, and the low display level results in low efficiency of isolating specific binding molecules from a library. In a phagemid display system, therefore, a new strategy to achieve high level display of pIII fusion protein on the surfaces of recombinant phage is needed for the successful isolation of diverse specific binders from a phage display library. To get around this problem, M13 helper phage with gIII deletion (M13δg3) (21,22) had been designed to observe the enhancement of display level. Recently, this strategy was further slightly modified. A packaging cell line (DH5α/pIII) was generated by inserting M13 gIII into the chromosome of DH5α cells, and high titer of hyperphage was produced by transformation of M13KO7ΔpIII helper phage DNA into DH5α/pIII cells (23). Mutation of the signal sequence and use of helper phage with trypsin-cleavable pIII coat protein also have been reported for improvement of the display of proteins on filamentous phage (24).
In this study, amber codons were introduced in the M13KO7 helper phage genome by site-directed mutagenesis so that the mutant helper phage, named Ex-phage, produce functional pIII in E.coli suppressor strains but not in non-suppressor strains. The phagemid vector, pIGT3, was also designed to produce the antibody fusion proteins fused with pIII in non-suppressing E.coli strains, and contained trypsin and enterokinase (EK) cleavage sites for proteolytic elution of phage. By packaging recombinant phage with Ex-phage, the display level of antibody molecules was increased drastically, thereby, increasing the efficiency of isolating specific binding phage antibodies from a phage display library by panning.
MATERIALS AND METHODS
Mutagenesis of M13KO7 helper phage genome
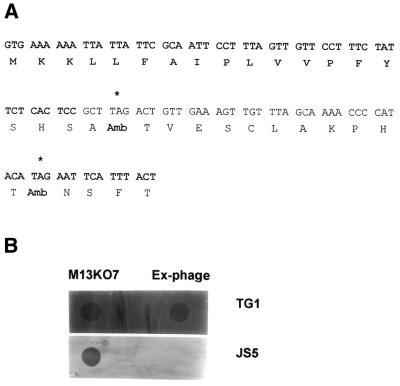
Amber codon (TAG) was introduced at the 5′ region of gIII of M13KO7 helper phage genome (Stratagene, USA) by site-directed mutagenesis using Mutan™-K enzyme and vector set (Takara, Japan) according to the manufacturer’s protocol using a synthetic oligonucleotide (1: 5′-TTCAACAGTCTAAGCGGAGTG-3′, the amber codon is underlined). Location of complementary sequences to oligonucleotides used in the experiment at the gIII are shown in Figure 1A. The mutant helper phage were identified by comparison of plaque formation on TG1 (suppressing strain) or JS5 cells (non-suppressing strain). Then the mutant phage that form clear plaques on TG1 cells but not on JS5 cells were isolated, single-stranded DNA was purified, and the second round of site-directed mutagenesis was performed using oligonucleotide 2 (5′-AAATGAATTCTATGTATGGGG-3′, the amber codon is underlined). The resulting mutant phage, named Ex-phage, has two amber codons at the 5′ region of gIII.
Figure 1.
Mutagenesis of M13KO7 helper phage genome. (A) gIII DNA sequence of Ex-phage. Two codons for Glu (GAA) were substituted to amber codons (TAG) at the 5′ region of gIII of M13KO7 helper phage genome by site-directed mutagenesis using Mutan™-K kit as described in the Materials and Methods. Leader sequence of gIII is denoted by bold. Asterisks show location of the amber codons. (B) Plaque formation by Ex-phage. After two consecutive site-directed mutageneses, several plaques were randomly isolated, and the phage were released in LB. Two microliters of phage suspension were spotted onto top agar LB plates containing TG1 or JS5 bacterial lawn, and incubated at 37°C overnight to determine phage propagation on these E.coli strains. M13KO7 helper phage was used as a control.
Construction of phagemid vectors, pIGT2 and pIGT3
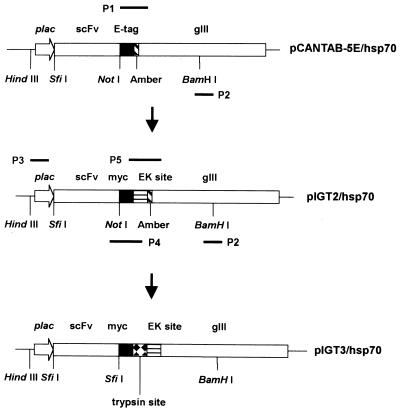
Construction of pIGT2. Standard cloning procedures and immunoblot after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) were carried out according to standard protocols (25), and flow chart of constructing pIGT2 and pIGT3 is shown in Figure 2. In pIGT2, E-tag of pCANTAB-5E (Amersham Pharmacia) was replaced with myc tag and an EK cleavage site was introduced into the vector. Briefly, 600 bp of gIII fragments between NotI and BamHI sites in pCANTAB-5E were obtained by PCR amplification. The sense primer (P1: 5′-GGGGCGGCCGCAGAACAAAAACTCATCTCAGAAGAGGATCTGTCTAGATAGGACGATGACGATAAGACTGTTGAAAGTTGTTTAGCAAAA-3′) was designed to contain NotI restriction enzyme site, myc tag, XbaI restriction enzyme site, an amber codon, EK cleavage site and the sequence complementary to 5′ region of gIII. Antisense primer was complementary to the middle of gIII region with BamHI restriction enzyme site (P2: 5′-ACGAATGGATCCTCATTAAAGCC-3′, the BamHI site is underlined). The pCANTAB-5E containing scFv specific for human HSP-70 (pCANTAB/hsp70) was isolated from a semi-synthetic scFv library by using a recombinant human HSP-70 protein for panning (H.Baek and S.Cha, unpublished data). Forty nanograms of pCANTAB/hsp70 were used as a template for PCR amplification using GeneAmp PCR system 9600 (Perkin Elmer, USA). The resulting 600 bp PCR product was treated with NotI/BamHI, and purified with Wizard DNA clean up kit (Promega, USA). The pCANTAB/hsp70 was restricted with the same set of restriction enzymes and purified with 1% low melting temperature agarose gel for eliminating the original 600 bp of NotI/BamHI DNA fragment including E-tag sequence. The resulting vector fragment and the PCR product were ligated together using T4 DNA ligase (Promega) at 16°C overnight, and transformed into HB2151 electro-competent cells using Gene Pulser (Bio-Rad). Bacterial colonies were randomly picked after incubating cells on 2 × YT/Amp plate at 37°C overnight, grown in LB/Amp in the presence of 1 mM isopropyl-β-d-thiogalactoside (IPTG) (Sigma Co., USA). Total cellular proteins were separated with 12% SDS–PAGE, and transferred to nitrocellulose membrane (Bio-Rad). The clones expressing a human anti-hsp-70 scFv fused with myc tag at its C-terminal were identified by immunoblot using the 9E10 anti-myc mAb (ATCC, USA) (26).
Figure 2.
Construction of pIGT2 and pIGT3: PCR amplifications for the construction of pIGT2 and pIGT3. In pIGT2, E-tag of pCABTAB-5E was replaced with myc tag using P1 and P2 PCR primers. P1 primer contains myc tag, EK cleavage site and sequence complementary to the 5′ region of gIII. P2 primer is complementary to the middle of gIII region containing BamHI restriction enzyme site. pIGT3 was generated by overlapping PCR using pIGT2 as a template. P3 primer is complementary to the upstream region of plac containing HindIII. P4 primer is complementary to VL FR4, myc tag and part of EK cleavage site, and contains SfiI restriction site next to VL FR4 and trypsin cleavage site next to myc tag as overhangings. P5 primer is complementary to EK cleavage site and 5′ region of gIII, but there is no TAG amber codon in front of gIII. Linking PCR to connect two PCR products was performed using P3 and P2 primers. Primer sequences are shown in the Materials and Methods.
Construction of pIGT3. pIGT3 was generated by replacing NotI restriction enzyme site of pIGT2 with SfiI, and introducing trpysin cleavage sequence between myc tag and EK cleavage site of pIGT2. Moreover, an amber codon in front of gIII was removed. Overlapping PCR was carried out for those modifications and pIGT2 was used as a PCR template as shown in Figure 2. The first PCR fragment (800 bp) for replacing NotI cloning site of pIGT2 with SfiI was obtained by using a sense primer (P3: 5′-GATTACGCCAAGCTTTGGAGC-3′, the HindIII restriction site is underlined) and an antisense primer (P4: 5′-CTCTTCTGAGATGTGTTTTTGTTCTTGGCCACGTCGGCCACGTTTGATTTCCACCTTGGTCCC-3′, the SfiI restriction site is underlined). The second PCR fragment (600 bp) for introducing trypsin cleavage sequence and removing an amber codon of pIGT2 was obtained by using a sense primer (P5: 5′-GAACAAAAACTCATCTCAGAAGAGGATCTGAAACGTGAAGACGATGACGATAAGACTGTTGAA-3′) and an antisense primer (P2: 5′-ACGAATGGATCCTCATTAAAGCC-3′, the BamHI site is underlined). The resulting two PCR fragments were linked together by overlapping PCR using mixture of 80 ng of each no. 1 PCR product and no. 2 PCR product as templates and primers (P3: 5′-GATTACGCCAAGCTTTGGAGC-3′; P2: 5′-ACGAATGGATCCTCATTAAAGCC-3′). The resulting 1400 bp PCR fragment was treated with HindIII/BamHI and cloned into pIGT2 in order to generate pIGT3 by substituting the original HindIII/BamHI fragment of pIGT2.
Phage quantification by ELISA
Serial dilutions of phage were coated in coating buffer (0.1 M NaHCO3 pH 9.6) were coated in microtiter plates (Falcon, USA) at 4°C overnight. After blocking the plate with 1% bovine serum albumin (BSA) (Sigma Co.) in PBS (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1 mM KH2PO4, pH 7.3), the bound phage were detected with anti-M13 antibody conjugated with horse radish peroxidase (HRPO) (Amersham Pharmacia). The signal was visualized with 2,2′-azino-di-(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) substrate and quantitated with ELISA reader (Bio-Rad). M13KO7 helper phage of known plaque forming units (p.f.u.) were used for standardization (23).
Immunoblot
High titers of M13KO7 helper phage and Ex-phage were prepared by infecting TG1 cells in 100 ml LB and incubated at 37°C for 6 h with vigorous agitation in a shaking incubator. To obtain recombinant phage, a human anti-hsp70 scFv gene that our laboratory obtained previously from a scFv phage display library constructed by using pCANTAB-5E vector was cloned into pIGT3, and JS5 cells carrying pIGT3-hsp70 phagemid were infected with either M13KO7 helper phage or Ex-phage preparation at an OD600 of 0.5 at a multiplicity of infection of 20 for 1 h in 50 ml LB containing ampicillin. Final concentration of 1 mM IPTG and 50 µg/ml kanamycin were added, and cultured at 30°C overnight. The recombinant phage were harvested by centrifugation and purified by PEG precipitation. Phage proteins were separated by loading approximately 1010 recombinant phage into each lane of 10% SDS–PAGE, and transferred to nitrocellulose membrane (Amersham Pharmacia). The membrane was blocked with 3% skimmed milk solution in PBS for 1 h at room temperature. Immunoblot was carried out with anti-gIII monoclonal antibody (mAb) (Mobitec, Germany), and goat anti-mouse IgG antibody conjugated with HRPO (Sigma Co.) was used for the secondary antibody. The signal was visualized on X-ray film (Roche, Germany) using ECL substrate (Amersham Pharmacia).
Determination of antigen-binding reactivity by phage ELISA
To determine antigen-binding specificity of recombinant phage particles, 100 ng of BSA, lysozyme (Sigma Co.), recombinant glutathione S-transferase (GST) or recombinant human HSP-70 in coating buffer (0.1 M NaHCO3 pH 9.6) were coated in microtiter plates (Falcon) at 4°C overnight. Recombinant GST protein was produced by growing DH-5α cells with pGEX vector (Amersham Pharmacia) in the presence of 1 mM IPTG, and affinity-purified by using glutathione agarose beads (Sigma Co.) Recombinant human HSP-70 protein was produced by growing BL21 (DE3) cells harboring pET28 vector (Invitrogen, USA) with human hsp-70 cDNA insert, and affinity-purified with Probond resin (Invitrogen). After blocking the plate with 1% BSA in PBS, 1010 scFv phage packaged with either M13KO7 or Ex-phage in 1% BSA solution were applied to each well for 1 h at room temperature. M13KO7 (1010) helper phage were used as negative control. After washing four times with PBS containing 0.1% Tween-20 (PBS–tween), the bound phage were detected with anti-M13 antibody conjugated with HRPO. The signal was visualized with ABTS substrate and quantitated with ELISA reader (Bio-Rad) at OD405. To observe sensitivity of antigen-binding reactivity of phage particles packaged with either M13KO7 or Ex-phage, ELISA was performed as described above except that serial dilutions of purified human HSP-70 fusion protein were used to coat microtiter plates.
Panning procedure
One hundred nanograms of recombinant human HSP-70 protein in coating buffer (0.1 M NaHCO3 pH 9.6) was coated in microtiter plates at 4°C overnight. Recombinant phage obtained by packaging pIGT3-hsp70 with either M13KO7 or Ex-phage were mixed with M13KO7 helper phage for non-specific backgrounds at 1:104, 1:106 or 1:108 ratio. After blocking the plate with 3% BSA in PBS, total 1010 phage from each diluting mixture were added into each well and incubated for 2 h at room temperature. Unbound phage were removed by washing with PBS–tween six times with vigorous pipetting (27), and bound phage were eluted by 1 µg/ml trypsin treatment (Sigma Co.) (23). Panning was repeated twice. The number of eluted phage was calculated by colony forming units (c.f.u.) on JS5 cells in LB plates containing 50 µg/ml ampicillin (LB/amp plate) (25), and amplified phage were quantitated by phage ELISA using M13KO7 helper phage as standards as described. Eluted phage from the first and the second round of panning were amplified in 50 ml of JS5 cells with either M13KO7 or Ex-phage superinfection and purified by PEG/NaCl precipitation. To determine the enrichment of antigen-specific phage after each round of panning, phage ELISA was performed with microplates coated with recombinant human HSP-70 protein. For polyclonal phage ELISA, 1010 of amplified phage after panning were added onto microtiter plates. For monoclonal phage ELISA, JS5 cells were infected with eluted phage for 20 min at room temperature, and spread onto LB/amp plates. After overnight incubation at 37°C, E.coli colonies were randomly picked and inoculated into 200 µl LB/amp in sterile 96-well plates (Corning, USA). Individual phage clones were obtained by superinfecting E.coli clones with either M13KO7 or Ex-phage at 30°C overnight. One hundred microliters of culture supernatant containing phage particles from the 96-well plates were added onto microtiter plates, and phage ELISA was performed as described. BSA was used as an antigen negative control and M13KO7 helper phage was used as a phage negative control.
RESULTS
Generation of Ex-phage and pIGT3 phagemid vector
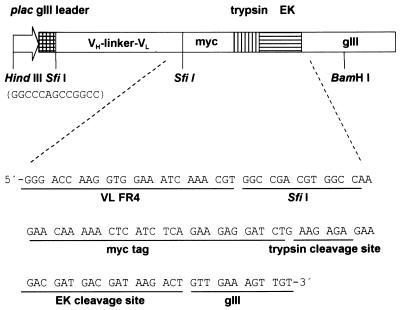
Initially, three independent site-directed mutageneses had been performed to replace Glu at the end of signal peptide, the first and the second Glu residues at the N-terminal of mature pIII to amber codons, respectively. Mutant M13KO7 with an amber codon located at the first Glu residue in the N-terminal of mature pIII still formed plaques in non-suppressing JS5 cells, and those with an amber codon at the end of signal peptide or the second Glu position showed a few revertants (data not shown). These results indicated that amber nonsense codon was somewhat leaky in JS5 cells. Therefore, one more round of site-directed mutagenesis was performed to introduce an amber codon to the second Glu residue at the N-terminal of mature pIII using the M13KO7 genome that already had the amber codon at the Glu site at the end of signal peptide of gIII. The resulting mutant phage that has two amber codons was named Ex-phage, and the location of amber codons in gIII of Ex-phage is shown in Figure 1A. The Ex-phage produced clear plaque in TG1 cells (supE genotype) but not in JS5 cells at all (Fig. 1B). This result demonstrated that Ex-phage can propagate in suppressing E.coli strains since amber codons are translated for Glu but not in non-suppressing E.coli strains because of premature stop of translation by two nonsense codons. There were no differences of yield of phage particles (p.f.u.) between M13KO7 and Ex-phage when they were amplified in TG1 cells, and high titer (1012 p.f.u./ml) of Ex-phage was easily obtained. The Ex-phage also formed plaques on XL-1 Blue (supE genotype) bacterial lawn but not on MV1184 cells (data not shown). In order to apply Ex-phage in phage display, pIGT2 and finally pIGT3 phagemid vectors were constructed by genetic modification of pCANTAB-5E as described in the Materials and Methods, yet pUC119 backbone of pCANTAB-5E was not altered (Fig. 2). In pIGT2, E-tag of pCABTAB-5E was replaced with myc tag by PCR using a PCR sense primer containing myc tag and EK cleavage sequences. pIGT3 was generated by overlapping PCR using pIGT2 as a template so that the amber codon located between scFv cloning sites and gIII was removed. For the trypsin protease elution of recombinant phage, a trypsin cleavage site was inserted in front of gIII in pIGT3. In addition, SfiI–NotI restriction endonuclease sites for cloning scFv genes in pIGT2 were changed to SfiI–SfiI, so that single digestion of the vector and scFv genes with SfiI could be used in the cloning step. Different nucleotide sequences for these two SfiI sites were utilized to reduce self-ligation of the vector, as shown in Figure 3. The general concept of using Ex-phage and pIGT3 for the enhanced display of scFv molecules on the surfaces of recombinant phage is depicted in Figure 4.
Figure 3.
Detailed diagram of pIGT3. Two different SfiI sites (GGCCCAGCCGGCC and GGCCGACGTGGCC) were utilized to reduce self-ligation of the pIGT3 during cloning process. Presence of myc tag facilitated the detection of scFv molecules, and trypsin and EK cleavage sites were introduced for elution of antigen-binding recombinant phage by protease treatment during panning.
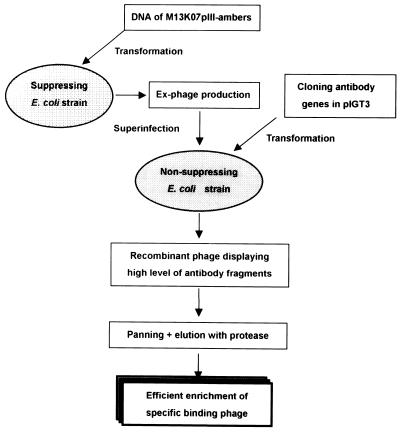
Figure 4.
Diagram of Ex-phage system. Ex-phage are obtained by infecting the mutant phage into TG1 suppressing E.coli strain, and used for packaging of single-chain antibody display phagemid pIGT3 in JS5 non-suppressing E.coli strain. The resulting recombinant phage carries several copies of the antibody molecules. The trypsin and EK cleavage sites linking the scFv and pIII can be used for protease elution after panning, and restoration of wild-type pIII enables the recombinant phage to reinfect E.coli.
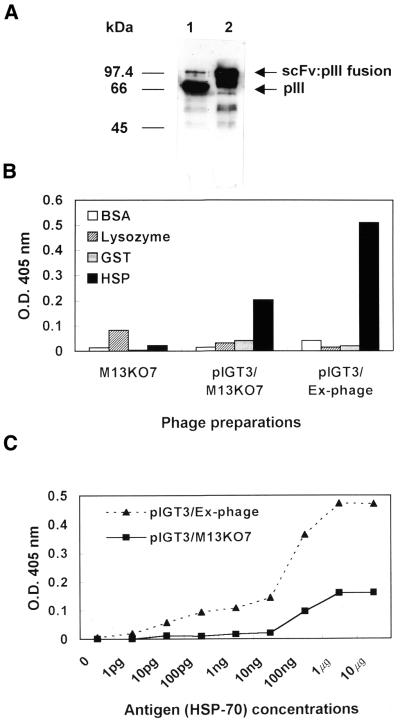
Immunoblot and phage ELISA
pIGT3 containing anti-hsp70 scFv-pIII fusion protein (pIGT3/hsp70) in JS5 cells were packaged with either Ex-phage (pIGT3/Ex-phage) or M13KO7 helper phage (pIGT3/M13KO7), and the assembly of functional phage antibody particles was monitored. Yields of pIGT3/Ex-phage and pIGT3/M13KO7 were determined by phage ELISA using anti-M13 polyclonal antibody. M13KO7 helper phage with known p.f.u. were used as standards. The number of phage particles produced by packaging with Ex-phage was ∼50-fold lower than phage preparations using M13KO7 helper phage mainly due to the absence of wild-type pIII during amplification in liquid culture (data not shown). To access potential effect of Ex-phage packaging on an increase of display level of the pIGT3 phage particles, the same number of pIGT3/Ex-phage and pIGT3/M13KO7 were analyzed by immunoblot using mouse mAb specific for pIII of M13. As expected, scFv:pIII fusion protein was the major form of pIII in pIGT3/Ex-phage, whereas most of pIII was wild-type in pIGT3/M13KO7 demonstrating dramatic increase of display level by Ex-phage (Fig. 5A). Densitometry analysis of the immunoblot (Fig. 5A) showed that only 5% of pIII were scFv:pIII fusion forms in pIGT3/M13KO7 indicating that only one out of four phage displayed only one scFv molecule on its surface, but three to four out of five pIII minor coat proteins were displayed as the scFv:pIII fusions on the surface of every pIGT3/Ex-phage. The presence of weak wild-type pIII in the pIGT/Ex-phage preparation seems to be due to the proteolytic breakdown. The effects of Ex-phage packaging on antigen-binding specificity and sensitivity were analyzed by phage ELISA. As shown in Figure 5B, pIGT3/Ex-phage and pIGT3/M13KO7 phage particles specifically reacted with human recombinant HSP-70 protein only, but not to BSA, lysozyme or recombinant GST protein. In addition, pIGT3/Ex-phage gave more than two times more signal to the HSP-70 protein compared to pIGT3/M13KO7 indicating that an increase of display level enhanced antigen-binding signal through avidity effect. In order to determine antigen-binding sensitivity of recombinant phage, different concentrations of human recombinant HSP-70 protein (0–10 µg) were coated onto microtiter plates, and 1010 of pIGT3/Ex-phage or pIGT3/M13KO7 were tested for antigen-binding reactivity by phage ELISA. pIGT3/Ex-phage bound at 100-fold lower concentration of the antigen compared with pIGT3/M13KO7 at the same ELISA signal (OD405 = 0.15), and gave positive signals at much smaller amounts of the antigen indicating that increase of displaying scFv:pIII fusion by Ex-phage directly enhanced antigen-binding sensitivity of recombinant phage particles (Fig. 5C). In accordance with Figure 5, packaging pIGT3/hsp70 with Ex-phage led to an increase of ELISA signal up to three times.
Figure 5.
Effects of Ex-phage packaging on antigen-binding reactivity of scFv displayed on recombinant phage. (A) Determination of scFv:pIII fusion protein expression by immunoblot. Recombinant phage particles were obtained by infecting JS5 cells carrying pIGT3-hsp70 phagemid with either M13KO7 helper phage (pIGT3/ M13KO7) (lane 1) or Ex-phage (pIGT3/ Ex-phage) (lane 2). Phage proteins were separated by loading approximately 1010 recombinant phage into each lane of 10% SDS–PAGE, and immunoblot was carried out with anti-gIII mAb to determine the amount of scFv:pIII fusion proteins displayed on the surfaces of phage particles. Goat anti-mouse IgG antibody conjugated with HRPO was used for the secondary antibody. (B) Antigen-binding specificity of recombinant phage packaged with either M13KO7 or Ex-phage. BSA, lysozyme (Sigma Co.), recombinant GST or recombinant human HSP-70 were coated in microtiters, and 1010 scFv phage packaged with either M13KO7 or Ex-phage were applied to each well for phage ELISA. The same amount of M13KO7 helper phage was used as a negative control. The bound phage were detected with anti-M13 antibody conjugated with HRPO. The binding signal was analyzed at OD405. (C) Antigen-binding sensitivity of phage particles packaged with either M13KO7 or Ex-phage. Phage ELISA was performed as described above except that serial dilutions of purified human HSP-70 fusion protein were used to coat microtiter plates.
Enhanced selective enrichment of antigen-specific phage by Ex-phage packaging
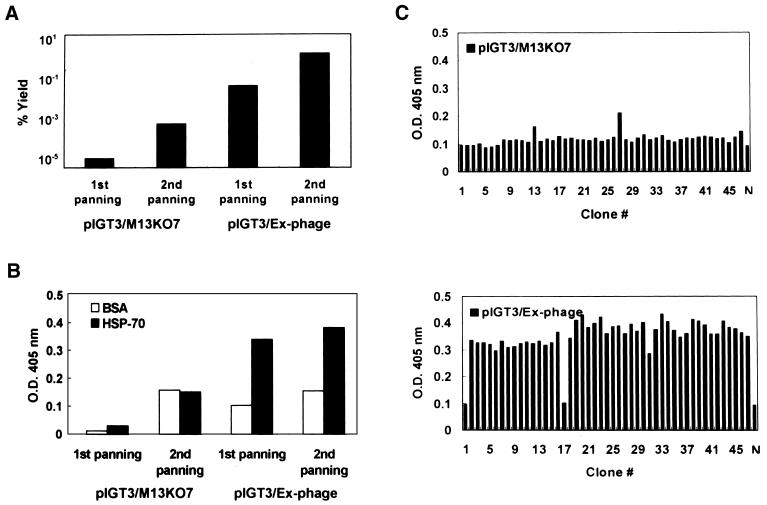
To demonstrate the potential of Ex-phage for the efficient selection of specific binders over the large proportion of non-specific binders, pIGT3/M13KO7 and pIGT3/Ex-phage were mixed with abundant number of M13KO7 helper phage at 1:104, 1:106 or 1:108 dilution ratio, and two rounds of panning were carried out. Both pIGT3/M13KO7 and pIGT3/Ex-phage did not show any enrichment at 1:108 dilution. Enhanced enrichment of antigen-specific phage was clearly observed in pIGT3/Ex-phage at 1:106 dilution (Fig. 6). Figure 6A shows the percentage yield after each round of panning. Percentage yields after the second panning of pIGT3/M13KO7 and pIGT3/Ex-phage were increased ∼100-fold from the first panning, suggesting that the selective enrichment might occur during two consecutive panning by both pIGT3/M13KO7 and pIGT3/Ex-phage. pIGT3/Ex-phage gave about 10 000 times higher percentage yield compared to pIGT3/M13KO7, probably due to the higher binding reactivity to the antigen. However, phage ELISA using amplified phage particles after panning indicated that an increase in precentage yield shown by pIGT3/M13KO7 was caused by non-specific binders, and only pIGT3/Ex-phage were selectively enriched among high background of M13KO7 helper phage by panning (Fig. 6B). This was clearly proved by the monoclonal phage ELISA in Figure 6C. Forty-five phage clones were randomly obtained by packaging with either M13KO7 helper phage or Ex-phage as described in the Materials and Methods. Among 45 pIGT3/M13KO7 phage clones, only two clones were positive to a human HSP-70 protein. On the other hand, 43/45 pIGT3/Ex-phage clones gave a specific binding signal to a human HSP-70 protein demonstrating that >95% of pIGT3/Ex-phage after the second panning were positives. This result demonstrated that presence of one antigen-specific phage out of 106 non-specific background phage could be successfully selected by Ex-phage packaging. None of pIGT3/M13KO7 and pIGT3/Ex-phage clones bound to BSA, and pIGT3/M13KO7 did not show any significant antigen-specific selection even at 1:104 dilution (data not shown).
Figure 6.
Enhancement of panning efficiency by Ex-phage package pIGT3/M13KO7 or pIGT3/Ex-phage were mixed with non-specific M13KO7 helper phage at 1:104, 1:106 or 1:108 ratio, and these phage mixtures (total 1010 phage from each diluting mixture) were applied for panning procedure with human HSP-70 protein as an antigen. Unbound phage were removed by washing with PBS–tween, and bound phage were eluted by trypsin treatment. Panning was repeated twice. Phage ELISA was performed to determine the enrichment of antigen-specific phage after each round of panning. (A) Percentage yield after panning. Input phage number was determined by phage ELISA, and the number of eluted phage (output) was determined by c.f.u. as described in the Materials and Methods. Percentage yield was calculated as (number of output phage/number of input phage) × 100. (B) Polyclonal phage ELISA on human HSP-70 protein after panning. Eluted phage from the first and the second round of panning were amplified and purified by PEG/NaCl precipitation. For polyclonal phage ELISA, 1010 of amplified phage after each round of panning were used for ELISA. BSA was used as a negative antigen control. (C) Monoclonal phage ELISA on human HSP-70 protein. Forty-five E.coli colonies were randomly picked after the second round of panning, and grown up in 96-well plates in the presence of M13KO7 or Ex-phage. One hundred microliters of culture supernatant containing recombinant phage particles were used in phage ELISA. M13KO7 helper phage was used as a negative phage control (N).
DISCUSSION
Phage display technology enables us to isolate human antibodies against almost any antigen by clonal affinity selection of antibodies that are readily applicable in in vivo diagnosis or therapy (28). However, isolation of antibodies specific for an antigen from a naive antibody library is not always successful because the number of antibody clones applied in a panning reaction is limited and only a minor fraction of total phage population displays antibody fragments in a phagemid vector system. Even in a phage vector system where all pIII are displayed as fusions, only 20–30% of the fusions are intact because of proteolytic degradation (29). In a naive antibody display library, specific binder antibodies are composed of very small fraction of the repertoire. It has been documented that a library of at least 107 size is enough to isolate specific phage antibodies, and more diverse antibody molecules (4000–10 000) can be obtained when a library of larger repertoire (1010) is used (30). This means that affinity selection has to go from >10 binders in 107 clones at the beginning to 1–10 binders to every 10 clones at the end, and there should be 105 magnitudes of enrichment for successful panning. McCafferty (29) reported that two factors, affinity and display level, directly affect panning efficiency. Existence of the displayed binder with high affinity in a phage display library improves enrichment even if display level is low. However, a major drawback of a naive or synthetic antibody library is that there is little chance of containing high affinity clones (31). Therefore, there is a low chance of finding specific binding clones from a naive or synthetic antibody library unless the repertoire of a library is so large that it contains good affinity binders or a phagemid vector system is modified to increase display level.
Herein, we developed a novel phagemid vector system combined with Ex-phage in order to enhance display level of fusions on phage particles. Basic strategy is similar to M13δg3 mutant helper phage (21,22) or M13KO7ΔpIII helper phage system (23). However, ours has three distinctive features. First, two other systems used M13 helper phage with gIII deletion, but ours used M13 helper phage with genetically modified gIII containing two amber codons. In M13δg3 rescue system, M13δg3 DNA was introduced in E.coli having plasmids that encode gIII for pIII supplementation, and the resulting infectious helper phage were used to rescue recombinant phagemid vectors. The use of this mutant helper phage restored the display level of the fusions to that achieved from phage vectors. Unfortunately, however, the titer of helper phage produced was so low (at most 109/l) that the M13δg3 system was impractical for general use. Recently, this strategy was further slightly modified. Instead of supplying pIII by a separate plasmid, a packaging cell line (DH5α/pIII) was generated by inserting M13 gIII into the chromosome of DH5α cells. Although yield was somewhat lower than M13KO7 helper phage, higher titer of hyperphage was produced by transformation of M13KO7ΔpIII helper phage DNA into DH5α/pIII cells followed by PEG precipitation. In our system, the Ex-phage can propagate in suppressing E.coli strains (e.g. supE genotype) such as XL-1 Blue cells (F′[proAB+ lacIq lacZΔM15 Tn10(tetr)]/supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 lac–) or TG1 cells (F′[traD36 proAB+ lacIq lacZΔM15)]/supE hsdΔ5 thi Δ(lac-proAB) just like conventional M13KO7 helper phage. Therefore, high titer of Ex-phage that is comparable to M13KO7 is easily obtained. However, no phage is produced in non-suppressing E.coli strains such as JS5 (F′[proAB+ lacIq lacZΔM15 Tn10(tetr)]/araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)χ74 (Strr) hsdR2 (rk–mk+) mcrA mcrB1) or MV1184 {F′[traD36 proAB+ lacIq lacZΔM15] ara Δ(lac-proAB) rpsL thi (φ80 lacZΔM15) Δ(srl-recA)306::Tn10(tetr)} because functional pIII is absent due to the presence of nonsence codons (TAG) at gIII.
Secondly, we generated a phage display library in non-suppressing E.coli strains, while others used cells with this genotype for producing soluble antibody molecules. In other words, package of recombinant phage was carried out in non-suppressing E.coli strains so that Ex-phage do not express any functional wild-type pIII any longer, and all antibody:pIII fusion protein are displayed on the surfaces of phage. In order for this, pIGT3 phagemid vector was designed to fuse a scFv gene and gIII without an amber codon between them. McCafferty (29) had demonstrated that antibodies with a wide range of affinity (10–18 000 nM) could be selected by increasing display level through either using a phage vector or mutant helper phage (M13δg3 rescue). Therefore, an increase of display level is advantageous in that a greater number of particles will have at least one copy of the fusion and so are capable of participating in binding with target molecules during selection procedure. Phage particles packaged with Ex-phage packaged the recombinant phage that displayed 70–80% of pIII as fusions, and achieved much higher display level of scFv:pIII than M13K07 helper phage. This may be due to the less proteolytic degradation of the fusions in our system. Different E.coli strains used in both systems may explain this difference. Ex-phage packaging also increased antigen-binding sensitivity >100-fold because of an increase in both number of total phage displaying antibodies and the number of scFv molecules per phage. The increase of antibody fragments presented on the surface of the packaged phagemids by Ex-phage also directly enhanced specific enrichment of binders from high non-specific backgrounds in accordance with M13δg3 and hyperphage systems. Indeed, high frequency (>95%) of antigen-specific phage clones could be selected over the presence of 106 times of non-specific backgrounds by two rounds of panning, which demonstrated at least 106 magnitudes of selective enrichment by Ex-phage packaging system.
Finally, EK site was added after trypsin site (KR) in the pIGT3 phagemid vector so that EK can be used instead of trypsin for phage elution. Protease elution a using trypsin cleavage site (KDIR) in front of gIII had been used previously because protease treatment restored wild-type pIII from antibody:pIII for the reinfection of recombinant phage after elution (32). In addition, use of protease elution is advantageous over an acid elution method in that mild elution condition may avoid adverse effect on reinfection efficiency caused by low pH shift, and high affinity binders that do not elute with acid buffer can be obtained. However, it had been suggested that a typsin treatment may cause a complication of panning especially when antigens of high protease sensitivity, whole cells or tissues are used for selection (33). Therefore, EK treatment for elution can avoid the possible complication of panning since EK is a more specific protease than trypsin.
Access to diverse antibodies to a target antigen can be crucial to finding the most useful candidates for the development of therapeutic antibody drugs. Since a phagemid vector system has very low display level of antibody:pIII fusions, there is a possibility that some useful antibodies can be missing during selection procedure. Furthermore, an increase in display level is especially critical for successful isolation of specific binders from a phage display library when the concentration of panning antigen is limited, as in the case of using cell surfaces or crude antigen preparations for panning. Our results show that Ex-phage offers high efficiency for isolating specific antibodies in a phage display with greater antigen-binding sensitivity that allows panning with smaller amount of antigens. Construction of phage display antibody libraries of either naive or synthetic origin with Ex-phage packaging may facilitate isolation of therapeutic antibodies specific for novel tumor antigens and diverse antibodies against proteomes by using crude antigens from cells, tissue samples or blots after two-dimensional gel electrophoresis for panning. Ex-phage-packaged libraries also may provide useful tools for high throughput analysis of large numbers of ORFs using automated robotics with high efficiency of enrichment, which, in particular, will be extremely valuable in post-functional genomics era.
REFERENCES
- 1.Hill D.F. and Petersen,G.B. (1982) Nucleotide sequence of bacteriophage f1 DNA. J. Virol., 44, 32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith G.P. (1988) Filamentous phages as cloning vectors. In Rodriquez,R.L. and Denhardt,D.T. (eds), Vectors: A Survey of Molecular Cloning Vectors and Their Uses. Butterworth, Boston, MA, pp. 61–84.
- 3.Hoess R.H. (1993) Phage display of peptides and protein domains. Curr. Opin. Struct. Biol., 3, 572–579. [Google Scholar]
- 4.Smith G.P. (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the surface of the virion. Science, 228, 1315–1317. [DOI] [PubMed] [Google Scholar]
- 5.Cwirla S.E., Peters,E.A., Barret,R.W. and Dover,W.J. (1990) Peptides on phage: a vast library of peptides for identifying ligands. Proc. Natl Acad. Sci. USA, 87, 6378–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCafferty J., Griffiths,A.D., Winter,G. and Chiswell,D.J. (1990) Phage antibodies: filamentous phage displaying antibody variable domains. Nature, 348, 552–554. [DOI] [PubMed] [Google Scholar]
- 7.Lowman H.B., Bass,S.H., Simpson,N. and Wells,J.A. (1991) Selecting high-affinity binding proteins by monovalent phage display. Biochemistry, 30, 10832–10838. [DOI] [PubMed] [Google Scholar]
- 8.Roberts B., Markland,W., Ley,A., Kent,R., White,D., Guterman,S. and Ladner,R. (1992) Directed evolution of a protein: selection of potent neutrophil elastase inhibitors displayed on M13 fusion phage. Proc. Natl Acad. Sci. USA, 89, 2429–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo Y. and Klug,A. (1994) Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interaction. Proc. Natl Acad. Sci. USA, 91, 11168–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin F., Toniatti,C., Salvati,A.L., Venturini,S., Ciliberto,G., Cortese,R. and Sollazo,M. (1994) The affinity-selection of a minibody polypeptide inhibitor of human interleukin-6. EMBO J., 13, 5303–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soumillion P., Jespers,L., Bouchet,M., Marchand-Brynaert,J., Winter,G. and Fastrez,J. (1994) Selection of beta-lactamase on filamentous bacteriophage by catalytic activity. J. Mol. Biol., 237, 415–422. [DOI] [PubMed] [Google Scholar]
- 12.Barbas C., Kang,A., Lerner,R. and Benkovic,S. (1991) Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl Acad. Sci. USA, 88, 7978–7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clackson T., Hoogenboom,H.R., Griffiths,A.D. and Winter,G. (1991) Making antibody fragments using phage display libraries. Nature, 352, 624–628. [DOI] [PubMed] [Google Scholar]
- 14.Scott J.K. and Smith,G.P. (1990) Searching for peptide ligands with an epitope library. Science, 249, 386–390. [DOI] [PubMed] [Google Scholar]
- 15.Hoogenboom H., Griffiths,A., Johnson,K., Chisswell,D., Hudson,P. and Winter,G. (1991) Multi-subunit proteins on the surfaces of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res., 19, 4133–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gram H., Marconi,L.A., Barbas,C.F., Collet,T.A., Lerner,R.A. and Kang,A.S. (1992) In vitro selection and affinity maturation of antibodies from a naïve combinatorial immunoglobulin library. Proc. Natl Acad. Sci. USA, 89, 3576–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs P., Dübel,S., Breitling,F., Braunagel,M., Klewinghaus,I. and Little,M. (1992) Recombinant human monoclonal antibodies: basic principles of the immune system transferred to E. coli. Cell Biophys., 21, 81–91. [DOI] [PubMed] [Google Scholar]
- 18.Greenwood J., Willis,A.E. and Perham,R.N. (1991) Multiple display of foreign peptides on a filamentous bacteriophage. J. Mol. Biol., 220, 821–827. [DOI] [PubMed] [Google Scholar]
- 19.Sidhu S.S. (2000) Phage display in pharmaceutical biotechnology. Curr. Opin. Biotechnol., 11, 610–616. [DOI] [PubMed] [Google Scholar]
- 20.Sidhu S.S., Weiss,G.A. and Wells,J. A (2000) High copy display on phage for functional selections. J. Mol. Biol., 296, 487–495. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths A.D., Malmqvist,M., Marks,J.D., Bye,J.M., Embleton,M.J., McCafferty,J., Gorick,B.D., Hughes-Jones,N.C., Hoogenboom,H.R. and Winter,G.P. (1993) Human anti-self antibodies with high specificity from phage display libraries. EMBO J., 12, 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakonjac J., Jovanovic,G. and Model,P. (1993) Filamentous phage infection-mediated gene expression: construction and propagation of the gIII deletion mutant helper phage R408d3. Gene, 198, 99–103. [DOI] [PubMed] [Google Scholar]
- 23.Rondot S., Koch,J., Breitling,F. and Dübel,S. (2001) A helper phage to improve single-chain antibody presentation in pahge display. Nat. Biotechnol., 19, 75–78. [DOI] [PubMed] [Google Scholar]
- 24.Jestin J., Volioti,G. and Winter,G. (2001) Improving the display of proteins on filamentous phage. Res. Microbiol., 152, 187–191. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning. A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Evan G.I., Lewis,G.K., Ramsay,G. and Bishop,J.M. (1985) Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol, 5, 3610–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chae J., Choi,J., Lim,H. and Cha,S. (2001) generation of a murine single chain Fv (scFv) antibody specific for cucumber mosaic virus (CMV) using a phage display library. Mol. Cell, 11, 7–12. [PubMed] [Google Scholar]
- 28.Griffiths A.D. and Duncan,A.R. (1998) Strategies for selection of antibodies by phage display. Curr. Opin. Biotechnol., 9, 102–108. [DOI] [PubMed] [Google Scholar]
- 29.McCafferty J. (1996) Phage display: factors affecting panning efficiency. In Kay,B.K., Winter,J. and McCafferty,J. (eds), Phage Display of Peptides and Proteins: A Laboratory Manual. Academic Press Inc., San Diego, CA, pp. 261–276.
- 30.Griffiths A.D., Williams,S.C., Hartley,O., Tomlinson,I.M., Waterhouse,P., Crosby,W.L., Kontermann,R.E., Jones,P.T., Low,N.M. and Allison,T.J. (1994) Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J., 13, 3245–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoogenboom H. and Charmes,P. (2000) Natural and designer binding sites made by phage display technology. Immunol. Today, 21, 371–378. [DOI] [PubMed] [Google Scholar]
- 32.Breitling F., Dübel,S., Seehaus,T., Klewinghaus,I. and Little,M. (1991) A surface expression vector for antibody screening. Gene, 104, 147–153. [DOI] [PubMed] [Google Scholar]
- 33.Diaz-Perales A., Linacero,R. and Vazquez,A.M. (2001) New series of vectors for phage display and prokaryotic expression of proteins. Biotechniques, 30, 720–726. [PubMed] [Google Scholar]