Abstract
Combinatorial fluorescence energy transfer (CFET) tags, constructed by exploiting energy transfer and combinatorial synthesis, allow multiple biological targets to be analyzed simultaneously. We here describe a multiplex single nucleotide polymorphism (SNP) assay based on single base extension (SBE) using CFET tags and biotinylated dideoxynucleotides (biotin-ddNTPs). A library of CFET-labeled oligonucleotide primers was mixed with biotin-ddNTPs, DNA polymerase and the DNA templates containing the SNPs in a single tube. The nucleotide at the 3′-end of each CFET-labeled oligonucleotide primer was complementary to a particular SNP in the template. Only the CFET-labeled primer that is fully complementary to the DNA template was extended by DNA polymerase with a biotin-ddNTP. We isolated the DNA extension fragments that carry a biotin at the 3′-end by capture with streptavidin-coated magnetic beads, while the unextended primers were eliminated. The biotinylated fluorescent DNA fragments were subsequently analyzed in a multicolor fluorescence electrophoresis system. The distinct fluorescence signature and electrophoretic mobility of each DNA extension product in the electropherogram coded the SNPs without the use of a sizing standard. We simultaneously distinguished six nucleotide variations in synthetic DNA templates and a PCR product from the retinoblastoma tumor suppressor gene. The use of CFET-labeled primers and biotin-ddNTPs coupled with the specificity of DNA polymerase in SBE offered a multiplex method for detecting SNPs.
INTRODUCTION
Single nucleotide polymorphisms (SNPs), the most common genetic variations found in the human genome, are important markers for identifying disease-associated loci and for pharmacogenetic studies (1,2). SNPs appear in the human genome with an average density of once every 1000 bp (3). A variety of approaches have been used to detect SNPs, such as DNA sequencing (4), mass spectrometry (5), gene chips (6), single-stranded conformational polymorphism (7), denaturing capillary electrophoresis (8) and electrochemical detection (9).
Single base extension (SBE) is the main technique used for detecting SNPs (10). In this method, a primer is annealed to a DNA template and extended by DNA polymerase with a single dideoxynucleotide that is complementary to the nucleotide at the variable site. Primer extension with DNA polymerase discriminates single nucleotide differences at polymorphic sites with high accuracy. Multiplex fluorescence-based primer extension has been used in mutation analysis of mitochondrial DNA (11,12). We recently developed a high fidelity DNA sequencing method using solid phase capturable biotinylated dideoxynucleotide terminators by detection with fluorescence (13) and mass spectrometry (14), eliminating false terminations. We also reported an approach for developing a large number of combinatorial fluorescence energy transfer (CFET) tags by exploiting energy transfer (ET) and combinatorial synthesis to tune the fluorescence emission signatures for multiplex biological assays (15).
We here describe an approach that couples CFET tags and biotinylated dideoxynucleotides (biotin-ddNTPs) in SBE for multiplex SNP detection. The goal is to simultaneously detect CFET-labeled SBE fragments generated in a single tube, with the tag at the 5′-end of the fragments giving a unique fluorescence signature for each of the multiplex SNPs and a biotin at the 3′-end for solid phase capture, allowing only the extended DNA fragments to be isolated and detected. The nucleotide at the 3′-end of each CFET-labeled oligonucleotide primer was complementary to a particular SNP in the template. After SBE of the CFET-labeled primer, we isolated the primer extension fragments that carry a biotin at the 3′-end by capture with streptavidin-coated magnetic beads. The biotinylated DNA fragments are subsequently released and loaded onto a multicolor laser-induced fluorescence electrophoresis system to obtain accurate SNPs, while the unextended CFET-labeled primers are eliminated. The use of biotin-ddNTPs thus allows true SNPs to be isolated and identified. Using three different fluorescent dye molecules, we constructed eight CFET tags with unique fluorescence signatures. A 1′,2′-dideoxynucleotide phosphate spacer is used to separate the donor–acceptor pair to tune the ET efficiency, producing unique fluorescence signatures in the CFET tags (15). The spacer also serves as a mobility tag to tune the mobility of CFET-labeled DNA for electrophoresis analysis. The previously reported multiplex SNP analysis methods using fluorescence primers require the use of a sizing standard in gel electrophoresis (16–19). Our method using CFET-labeled oligonucleotide primers and biotin-ddNTPs does not need a sizing standard because each of the SNPs is coded by a unique fluorescence signature and mobility in the electropherogram. We simultaneously detected six nucleotide variations in synthetic DNA templates and three nucleotide variations in a PCR product from the retinoblastoma tumor suppressor (RB1) gene. The use of CFET tag-labeled primer and biotin-ddNTPs coupled with the specificity of DNA polymerase for SBE provides a multiplex approach to conduct high throughput analyses of SNPs.
MATERIALS AND METHODS
Synthesis of CFET tags and oligonucleotides
The CFET tags were prepared by phosphoramidite chemistry in a DNA synthesizer and solution coupling synthesis. 6-Carboxyfluorescein (FAM) and N,N,N′,N′-tetramethyl-6-carboxyrhodamine (TAM) were incorporated into the oligonucleotides with fluorescein-dT and TAMRA-dT phosphoramidites (Glen Research, Sterling, VA), respectively. A modified thymidine having an amino linker (amino modifier C6-dT; Glen Research) was incorporated into the oligonucleotide for coupling with Alexa 594 N-hydroxysuccinimidyl (NHS) ester (Molecular Probes, Eugene, OR). The 1′,2′-dideoxyribose phosphate spacers were incorporated into the CFET tags with dSpacer phosphoramidite (Glen Research), which provided high coupling yield. To synthesize the CFET tag containing Alexa 594, 10–12 nmol FAM-labeled oligonucleotides containing an amino linker in 40 µl of 0.25 M Na2CO3/NaHCO3 buffer pH 9.0, were incubated for 10 h at room temperature with an ∼27-fold excess of the Alexa 594 NHS ester in 12 µl of anhydrous dimethylsulfoxide. Unreacted dye was removed by size exclusion chromatography on a PD-10 column (Amersham Pharmacia Biotech, Piscataway, NJ). The CFET tags were then purified by gel electrophoresis and desalted with an oligonucleotide purification cartridge.
PCR protocol for DNA template preparation
DNA template was obtained from human genomic DNA from a patient blood sample. A 278 nt sequence between nucleotide 156 615 and 156 892 of the RB1 gene was amplified by PCR in 20 µl reactions that contained the following components: 4 pmol forward and reverse primers (5′-ATG CTA CTT AAC AGC ATT ATA ATT AG-3′ and 5′-ATC AGT TAA CAA GTA AGT AGG GAG-3′), 1× PCR buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris–HCl), 0.2 mM dNTPs, 600 pg genomic DNA and 1 U Taq DNA polymerase (Amersham Pharmacia Biotech). The PCR conditions were 30 cycles of 95°C for 20 s, 57°C for 20 s, 72°C for 2 min. At the end of the PCR reaction, 20 µl of an enzymatic mixture containing 5 U shrimp alkaline phosphatase (SAP), 4 µl of 10× SAP buffer, 6 U Escherichia coli exonuclease I and 10 µl of water were added to the PCR reaction to degrade the excess primers and dNTPs. The reaction mixture was incubated at 37°C for 90 min before the enzymes were heat inactivated at 72°C for 30 min. The region of the PCR product sequence containing the SNPs (underlined) was: 5′-CAC AGT GTA TCG GCT AGC CTA TCT CCG GCT AAA TAC ACT TTG TGA ACG CCT TCT GTC TGA GCA CCC AGA ATT-3′. These SNPs were confirmed by DNA sequencing using energy transfer terminator chemistry and a MegaBACE 1000 capillary electrophoresis DNA sequencer (Amersham Pharmacia Biotech).
SBE assay using CFET tags and biotin-ddNTPs
For individual single base extension, 10 µl of SBE solution contained 0.5–2 pmol CFET-labeled oligonucleotide primer, 1–4 pmol DNA template, 2 pmol each biotinylated dideoxynucleoside triphosphate [biotin-11-dd(A,C,G,U)TP] (NEN Life Science, Boston, MA), 2 µl of Thermo Sequenase reaction buffer (260 mM Tris–HCl pH 9.5, 65 mM MgCl2) and 1 U Thermo Sequenase in its dilution buffer (Amersham Pharmacia Biotech). Extension was carried out at 54°C for 30 s. For multiplex SBEs, the contents of the reaction mixture were the same as those for individual SBEs described above except that 0.4–5 pmol CFET-labeled primers and 2–4 pmol DNA templates were used. For SBE on the PCR product, 0.5–2.8 pmol CFET-labeled primer and 0.68 pmol DNA template were used. The extension was performed at 94°C for 30 s and 54°C for 30 s.
Solid phase purification and laser-induced fluorescence detection
Fifty microliters of streptavidin-coated magnetic beads (Dynabeads M-280 Streptavidin; Dynal, Oslo, Norway) were washed with 2 × 50 µl of binding and washing (B&W) buffer (10 mM Tris–HCl, 1 mM EDTA, 2.0 M NaCl, pH 7.5) and then resuspensed in 10 µl of B&W buffer. The primer extension product mixture was then combined with the streptavidin-coated magnetic beads at 25°C for 0.5 h with occasional mixing. The supernatant was removed while the magnetic beads were immobilized with a magnet. The beads were then washed with 3 × 50 µl of B&W buffer. The DNA template was denatured from the extended primer by treatment with 50 µl of fresh 0.1 M NaOH at 25°C for 5 min. The reaction mixture was then washed with 2 × 50 µl of water. The extended primers were released from the magnetic beads in 98% formamide containing 10 mM EDTA at 94°C for 5 min. An aliquot of 0.3–0.8 µl of the supernatant was loaded onto a sequencing gel (5% Long Ranger, 6 M urea) for electrophoresis and fluorescence signature detection on an ABI 377 DNA sequencer (Applied Biosystems, Foster City, CA) with an argon laser for excitation (488/514 nm). Electrophoresis was conducted at 1000 V with 1× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.0).
RESULTS AND DISCUSSION
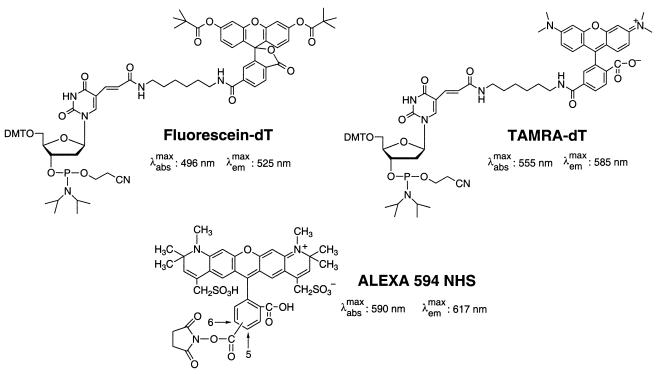
We used the following three fluorescent dyes to construct the CFET tags for labeling the oligonucleotide primers: 6-carboxyfluorescein (FAM or F), N,N,N′,N′-tetramethyl-6-carboxyrhodamine (TAM or T) and Alexa 594 (ALX or A). Figure 1 shows the chemical structures of the dye derivatives used for labeling the oligonucleotides. FAM was used as the donor, while TAM or ALX was selected as the acceptor in constructing the CFET tags. Since FAM (λemmax = 525 nm), TAM (λemmax = 585 nm) and ALX (λemmax = 617 nm) have distinct maximum fluorescence emission wavelengths, the CFET tags can be distinguished by a multicolor fluorescence detection system, such as the ABI 377 DNA sequencer, without any modification of its detector. ALX was used because it has higher fluorescence quantum yield and photostability than other fluorophores with similar emission maxima (20). A spacer (S) constructed from 1′,2′-dideoxyribose phosphates (21) was used to separate the donor and acceptor chromophores to tune the ET efficiency and also as a mobility tag to tune the electrophoretic mobility of the SBE fragments.
Figure 1.
Chemical structures of the dye derivatives, fluorescein-dT, TAMRA-dT and ALEXA 594 NHS ester, used to label the oligonucleotides.
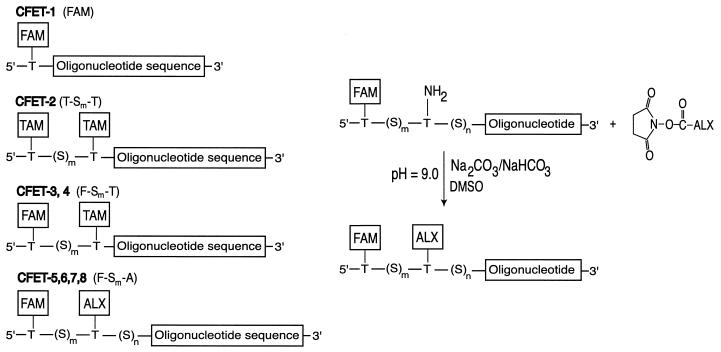
Schematics and the nomenclature of the CFET tags are shown in Figure 2 (left): CFET-1 consists of a single dye (FAM) covalently linked to an oligonucleotide primer, while CFET-2 (T-S4-T) is doubly labeled with TAM; CFET-3 and CFET-4 (F-Sm-T) and CFET-5, CFET-6, CFET-7 and CFET-8 (F-Sm-A) consist of two dyes (FAM and TAM, or FAM and ALX), where Sm indicates the number of dideoxyribose spacers between each donor–acceptor pair. The spacers (Sm,n) were also used as mobility tags in the electrophoretic analysis. A synthetic scheme for coupling ALX to the oligonucleotide is shown in Figure 2 (right) (see Materials and Methods for details).
Figure 2.
(Left) CFET tag-labeled oligonucleotides. The CFET tags are constructed from three chromophores FAM (F), TAM (T) and ALEXA 594 (A). The number of 1′,2′-dideoxyribose phosphate spacers (S) between the chromophores is indicated in the CFET tag designation: FAM (CFET-1); T-4-T (CFET-2, m = 4); F-9-T (CFET-3, m = 9); F-13-T (CFET-4, m = 13); F-5-A (CFET-5, m = 5, n = 13); F-7-A (CFET-6, m = 7, n = 10); F-10-A (CFET-7, m = 10, n = 11); F-13-A (CFET-8, m = 13, n = 8). Spacers (S)m,n are used to separate the chromophores to tune the ET efficiency and as mobility tags for electrophoresis analysis. The CFET-labeled oligonucleotides (20 bp) are designed to be complementary to the DNA template containing multiple SNPs. (Right) A synthetic scheme for the preparation of a CFET tag containing FAM and ALEXA.
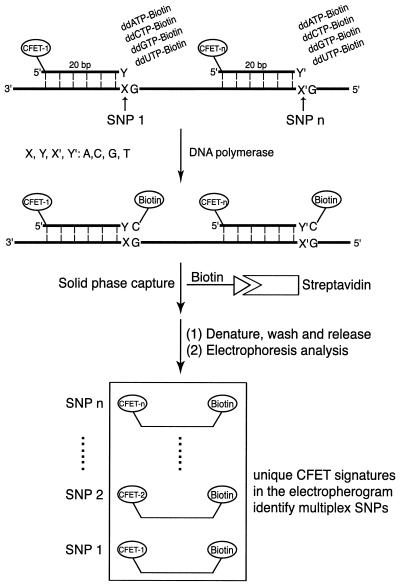
The multiplex SNP detection approach using CFET-labeled oligonucleotide primers and biotin-ddNTPs is shown in Figure 3. Upon hybridization to the DNA templates, each of the CFET-labeled primers that has a perfect match to the SNP site will incorporate a biotin-ddNTP at its 3′-end in the presence of DNA polymerase. Under our experimental conditions, no extension reaction occurred when there was a mismatch between the 3′-end of the CFET-labeled primers (Fig. 3, Y and Y′) and the nucleotides at the polymorphic sites (Fig. 3, X and X′) in the target DNA template. After SBE, the CFET-labeled primer extension products were immobilized on streptavidin-coated magnetic beads and the other components were washed away. The extension products were cleaved from the magnetic beads by denaturing the biotin–streptavidin interaction with formamide and analyzed with a fluorescence DNA sequencer. The CFET-labeled extension products were unambiguously detected due to their distinct mobilities and fluorescence signatures in the electropherogram. In the case of heterozygous genotypes, we used two CFET tags with different fluorescence signatures and electrophoretic mobilities to label the oligonucleotides corresponding to each allele. The CFET-labeled oligonucleotides, biotin-ddNTPs and DNA polymerase thus formed a high fidelity SNP identification system in which the nucleotide at the 3′-end of the oligonucleotide dictated its extension by incorporating a specific biotin-ddNTP, and the unique fluorescence signature in the electropherogram identified each of the corresponding SNPs.
Figure 3.
SBEs for multiplex SNP detection using CFET-labeled primers and biotin-ddNTPs. DNA template containing polymorphic sites is incubated with CFET-labeled primers, biotin-ddNTPs and DNA polymerase. The nucleotide at the 3′-end of each CFET-labeled primer is complementary to a particular SNP in the template. Only the fully complementary CFET-labeled primer was extended by DNA polymerase with a biotin-ddNTP. After solid phase capture and isolation of the biotinylated DNA extension fragments the SBE products are analyzed for their fluorescence signatures, each of which codes for a unique SNP.
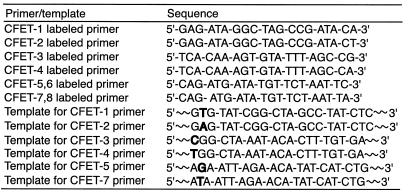
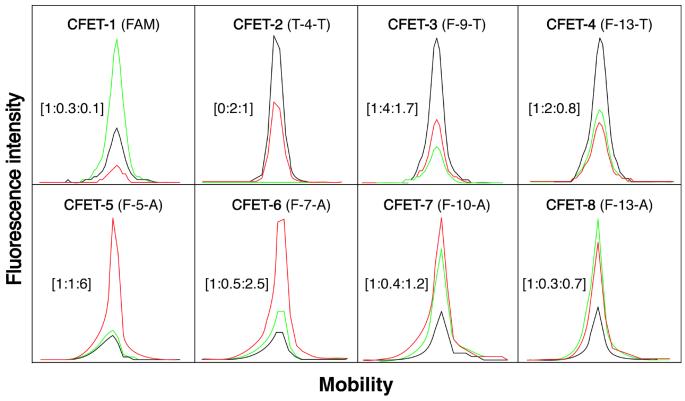
Eight unique fluorescence signatures of the CFET tags described in Figure 2 (see Table 1 for the oligonucleotide sequences) are shown in Figure 4. Excited by an argon laser at 488/514 nm and detected by an ABI 377 sequencer, the fluorescence emission signatures were distinct as a result of different fluorescence signal intensities measured in the three detection channels from each CFET tag (FAM channel, 530–541 nm, green; TAM channel, 581–591 nm, black; ALX channel, 610–620 nm, red). For clarity, the unique fluorescence emission signature of each CFET tag is denoted by a digital ratio (DR) of the signal intensity from each of the three detection channels (DR = green:black:red; the FAM signal is set to 1). In the FAM-labeled oligonucleotide (CFET-1), the chromophore was efficiently excited by the argon laser at 488/514 nm in the ABI 377 sequencer, displaying a dominant FAM emission (green) and a DR of 1:0.3:0.1. To achieve higher fluorescence emission intensity, we coupled two TAM molecules in T-4-T (CFET-2) in order to compensate for the fact that TAM cannot be excited efficiently by the argon laser. Accordingly, CFET-2 produced a unique fluorescence signature with a DR of 0:2:1 with a dominant TAM emission (black). Since FAM is not present in CFET-2, there was no signal detected from the FAM channel and, thus, the green signal is 0 in the signature of T-4-T. Coupling TAM to the oligonucleotide primer as an acceptor for FAM with separation distances of 9 and 13 dideoxyribose spacers produced F-9-T (CFET-3) and F-13-T (CFET-4). The ET efficiency decreased as the number of dideoxyribose spacers increased from 9 to 13, as indicated by the different fluorescence emission ratios in F-9-T (CFET-3) (DR = 1:4:1.7) and F-13-T (CFET-4) (DR = 1:2:0.8). A set of CFET tags with four distinct fluorescence signatures was thus created by fluorescently labeling the oligonucleotides with two individual dyes (FAM and TAM). Four additional CFET tags with unique fluorescence signatures were generated using FAM as the donor and ALX as the acceptor: the fluorescence signatures of F-5-A (CFET-5), F-7-A (CFET-6) and F-10-A (CFET-7) displayed considerable ALX emission (red) that was produced through ET from FAM to ALX after excitation at 488/514 nm. On the other hand, the fluorescence signature of F-13-A (CFET-8) had a higher FAM than ALX signal. Again, consistent with the ET principle, a greater separation distance between the donor and acceptor lowered the ET efficiency, as indicated by the different DR values in CFET-5 (DR = 1:1:6), CFET-6 (DR = 1:0.5:2.5), CFET-7 (DR = 1:0.4:1.2) and CFET-8 (DR = 1:0.3:0.7), respectively.
Table 1. Oligonucleotide sequences labeled by the eight CFET tags and the portion of DNA template sequences containing SNPs identified by the CFET tags (SNPs in bold).
Figure 4.
The eight unique fluorescence signatures of CFET tags generated by a fluorescence DNA sequencer (ABI 377). FAM channel (530–541 nm), green; TAM channel (581–591 nm), black; ALEXA channel (610–620 nm), red. The digital ratio denoting the fluorescence signature for each CFET tag from the three channels [green:black:red] is shown in brackets. The fluorescence signatures in the electropherogram were obtained using the eight CFET-labeled oligonucleotides described in Figure 2.
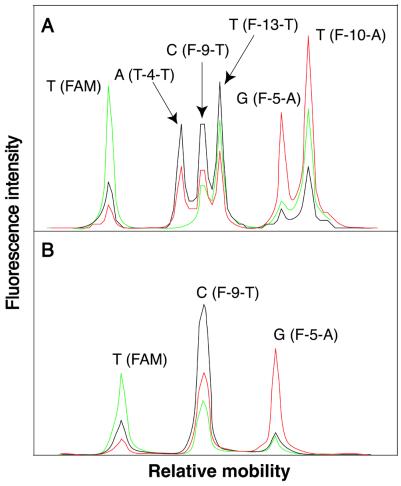
We used CFET-labeled oligonucleotide primers and biotin-ddNTPs in SBE for multiplex SNP detection. The reactions were carried out simultaneously in a single tube with six different CFET-labeled oligonucleotide primers directed to six nucleotide variations in synthetic templates mimicking exon 20 of the RB1 gene. Mutations affecting both alleles of the RB1 gene are a prerequisite for the development of retinoblastoma, which is a malignant tumor of the eye (22). Table 1 shows the sequences of the primers and synthetic templates used in this study. After extension and solid phase purification, the extension products were analyzed with an ABI 377 DNA sequencer, resulting in simultaneous detection of six nucleotide variations by the unique fluorescence signatures of the CFET-labeled extension products (Fig. 5A). The unique fluorescence signatures were spatially resolved in the electropherogram as a result of the different mobilities of the CFET-labeled extension products. In this model experiment, both CFET-1 (FAM) and CFET-2 (T-4-T) distinguished a mimic of the RB1 mutation V654E, detecting both the wild-type (T) and mutation (A) at the same location in the template. CFET-3 (F-9-T) and CFET-4 (F-13-T) identified a mutation, R661W, with a heterozygous genotype (C/T). CFET-5 (F-5-A) and CFET-7 (F-10-A) detected another mutation, E675X, with a heterozygous genotype (G/T).
Figure 5.
Electropheretogram of CFET-labeled SBE products for SNP identification in exon 20 of the RB1 gene. (A) Detection of six nucleotide variations from synthetic DNA templates. The FAM and T-4-T peaks detected a heterozygous genotype (T/A), while the F-9-T and F-13-T peaks identified a C/T heterozygous genotype. The F-5-A and F-10-A peaks identified another heterozygous genotype (G/T). (B) Detection of three homozygous genotypes (T, C and G) for a PCR product from the RB1 gene.
To further validate the SNP detection method using CFET-labeled oligonucleotides and biotin-ddNTPs, we used three CFET-labeled oligonucleotide primers (CFET-1, CFET-3 and CFET-5) to identify three SNPs in a PCR product amplified from exon 20 of the RB1 gene from patient genomic DNA. The extension reactions were performed in a single tube and the reaction products were analyzed with an ABI 377 DNA sequencer. Three individual homozygous genotypes (T, C and G), verified by DNA sequencing, were unambiguously identified by the three distinct fluorescence signatures from the CFET-labeled SBE products (Fig. 5B): T (FAM, CFET-1), C (F-9-T, CFET-3) and G (F-5-A, CFET-5). Thus, both heterozygous and homozygous genotypes can be identified unambiguously by the unique fluorescence signatures and electrophoretic mobilities of the CFET-labeled SBE products.
In summary, the SBE assay using CFET-labeled oligonucleotides and biotin-ddNTPs in conjunction with solid phase capture described here offers a fast and accurate method for high throughput SNP detection and analysis. This single tube homogeneous assay generated CFET-labeled extension fragments carrying a tag at the 5′-end to give unique fluorescence signatures for each of the multiplex SNPs and a biotin at the 3′-end for solid phase capture, allowing only the extended fragments to be detected. The unique fluorescence signatures, detected by a multicolor fluorescence DNA sequencer, were used to simultaneously identify multiplex SNPs in synthetic DNA templates and a PCR product from the RB1 gene. Compared to SNP detection methods that require a sizing standard in gel electrophoresis, our method is simple and accurate since each SNP is distinguished by a distinct fluorescence signature with a specific electrophoretic mobility. All the primers used in our experiments were 20 bp and this facilitates the hybridization and denaturing processes during analysis. It also simplifies the design of the primers when compared to other melting point-based hybridization assays for SNP analysis (23,24). The oligonucleotide ligation assay (OLA) has been widely used for genetic mutation analysis (25–27). SBE is faster than OLA because the DNA polymerase incorporates ddNTPs specifically in a fraction of a second (28,29). Also, primer design is needed only next to the polymorphic sites in the target DNA template, and the additional oligonucleotides that are required in OLA are unnecessary for SBE (30). The DNA polymerase, such as Thermo Sequenase, employed in SBE is as specific as the Taq DNA ligase used in OLA in distinguishing single nucleotide differences at a polymorphic site, and hence accurate discrimination of allelic variants is assured. Other laser-induced multicolor fluorescence electrophoresis systems, such as capillary array DNA sequencers (31) and miniaturized chip-based devices (32,33), can be used to separate the DNA extension fragments and detect the unique fluorescence signatures for high throughput SNP analyses.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr James Russo for the genomic DNA template and Dr Zengmin Li for helpful discussions. This research was supported by the National Science Foundation (Biophotonics Partnership Initiative Grant 86933) as well as funding from the Columbia Genome Center and Department of Chemical Engineering at Columbia University.
REFERENCES
- 1.Landegren U., Nilsson,M. and Kwok,P.-Y. (1998) Reading bits of genetic information: methods for single-nucleotide polymorphism analysis. Genome Res., 8, 769–776. [DOI] [PubMed] [Google Scholar]
- 2.Roses A.D. (2000) Pharmacogenetics and the practice of medicine. Nature, 405, 857–865. [DOI] [PubMed] [Google Scholar]
- 3.The International SNP Map Working Group (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- 4.Kwok P.-Y., Carlson,C., Yager,T.D., Ankener,W. and Nickerson,D.A. (1994) Comparative analysis of human DNA variations by fluorescence-based sequencing of PCR products. Genomics, 23, 138–144. [DOI] [PubMed] [Google Scholar]
- 5.Haff L.A. and Smirnov,I.P. (1997) Single-nucleotide polymorphism identification assays using a thermostable DNA polymerase and delayed extraction MALDI-TOF mass spectrometry. Genome Res., 7, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D.G., Fan,J.B., Siao,C.J., Berno,A., Young,P., Sapolsky,R., Ghandour,G., Perkins,N., Winchester,E., Spencer,J. et al. (1998) Large-scale identification, mapping and genotyping of single-nucleotide polymorphisms in the human genome. Science, 280, 1077–1082. [DOI] [PubMed] [Google Scholar]
- 7.Kozlowski P. and Krzyzosiak,W.J. (2001) Combined SSCP/duplex analysis by capillary electrophoresis for more efficient mutation detection. Nucleic Acids Res., 29, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wenz H.M., Robertson,J.M., Menchen,S., Oaks,F., Demorest,D.M., Scheibler,D., Rosenblum,B.B., Wike,C., Gilbert,D.A. and Efcavitch,J.W. (1998) High-precision genotyping by denaturing capillary electrophoresis. Genome Res., 8, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boon E.M., Ceres,D.M., Drummond,T.G., Hill,M.G. and Barton,J.K. (2000) Mutation detection by electrocatalysis at DNA-modified electrodes. Nat. Biotechnol., 18, 1096–1100. [DOI] [PubMed] [Google Scholar]
- 10.Syvänen A.C., Aalto-Steälä,K., Harju,L., Kontula,K. and Söderlund,H. (1990) A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics, 8, 684–692. [DOI] [PubMed] [Google Scholar]
- 11.Tully G., Sullvian,K.M., Nixon,P., Stones,R.E. and Gill,P. (1996) Rapid detection of mitochondrial sequence polymorphisms using multiplex solid-phase fluorescent minisequencing. Genomics, 34, 107–113. [DOI] [PubMed] [Google Scholar]
- 12.Fahy E., Nazarbaghi,R., Zomorrodi,M., Herrnstadt,C., Parker,W.D., Davis,R.E. and Ghosh,S.S. (1997) Multiplex fluorescence-based primer extension method for quantitative mutation analysis of mitochondrial DNA and its diagnostic application for Alzheimer’s disease. Nucleic Acids Res., 25, 3102–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju J. (1999) US patent no. 5 876 936.
- 14.Edwards J.R., Itagaki,Y. and Ju,J. (2001) DNA sequencing using biotinylated dideoxynucleotides and mass spectrometry. Nucleic Acids Res., 29, e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong A.K., Li,Z., Jones,G.S., Russo,J.J. and Ju,J. (2001) Combinatorial fluorescence energy transfer tags for multiplex biological assays. Nat. Biotechnol., 19, 756–759. [DOI] [PubMed] [Google Scholar]
- 16.Edwards A., Civitello,A., Hammond,H.A. and Caskey,C.T. (1991) DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am. J. Hum. Genet., 49, 746–756. [PMC free article] [PubMed] [Google Scholar]
- 17.Day D.J., Speiser,P.W., White,P.C. and Barany,F. (1995) Detection of steroid 21-hydroxylase alleles using gene-specific PCR and a multiplexed ligation detection reaction. Genomics, 29, 152–162. [DOI] [PubMed] [Google Scholar]
- 18.Igloi G.L. (2001) Simultaneous identification of mutations by dual-parameter multiplex hybridization in peptide nucleic acid-containing virtual arrays. Genomics, 74, 402–407. [DOI] [PubMed] [Google Scholar]
- 19.Fitness J. (2001) Ligation detection reaction (LDR): enabling multiplexed detection of known polymorphisms. NEB Transcript, 11, 10–11. [Google Scholar]
- 20.Panchuk-Voloshina N., Haugland,R.P., Bishop-Stewart,J., Bhalgat,M.K., Millard,P.J., Mao,F., Leung,W.-Y. and Haugland,R.P. (1999) Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. J. Histochem. Cytochem., 47, 1179–1188. [DOI] [PubMed] [Google Scholar]
- 21.Ju J., Glazer,A.N. and Mathies,R.A. (1996) Cassette labeling for facile construction of energy transfer fluorescent primers. Nucleic Acids Res., 24, 1144–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohmann D.R. (1999) RB1 gene mutations in retinoblastoma. Hum. Mutat., 14, 283–288. [DOI] [PubMed] [Google Scholar]
- 23.Bernard P.S., Pritham,G.H. and Wittwer,C.T. (1999) Color multiplexing hybridization probes using the apolipoprotein E locus as a model system for genotyping. Anal. Biochem., 273, 221–228. [DOI] [PubMed] [Google Scholar]
- 24.Nauck M., Wieland,H. and März,W. (1999) Rapid, homogeneous genotyping of the 4G/5G polymorphism in the promoter region of the PAII gene by fluorescence resonance energy transfer and probe melting curves. Clin. Chem., 45, 1141–1147. [PubMed] [Google Scholar]
- 25.Grossman P.D., Bloch,W., Brinson,E., Chang,C.C., Eggerding,F.A., Fung,S., Iovannisci,D.M., Woo,S. and Winn-Deen,E.S. (1994) High-density multiplex detection of nucleic acid sequences: oligonucleotide ligation assay and sequence-coded separation. Nucleic Acids Res., 22, 4527–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reyes A.A., Carrera,P., Cardillo,E., Ugozzoli,L., Lowery,J.D., Lin,C.-I.P., Go,M., Ferrari,M. and Wallace,R.B. (1997) Ligase chain reaction assay for human mutations: the sickle cell by LCR assay. Clin. Chem., 43, 40–44. [PubMed] [Google Scholar]
- 27.Eggerding F.A. (2000) Fluorescent oligonucleotide ligation technology for identification of ras oncogene mutations. Mol. Biotechnol., 14, 223–233. [DOI] [PubMed] [Google Scholar]
- 28.Tabor S. and Richardson,C.C. (1987) DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc. Natl Acad. Sci. USA, 84, 4767–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabor S. and Richardson,C.C. (1995) A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc. Natl Acad. Sci. USA, 92, 6339–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landegren U., Kaiser,R., Sanders,J. and Hood,L. (1988) A ligase-mediated gene detection technique. Science, 241, 1077–1080. [DOI] [PubMed] [Google Scholar]
- 31.Medintz I., Wong,W.W., Sensabaugh,G. and Mathies,R.A. (2000) High speed single nucleotide polymorphism typing of a hereditary haemochromatosis mutation with capillary array electrophoresis microplates. Electrophoresis, 21, 2352–2358. [DOI] [PubMed] [Google Scholar]
- 32.Woolley A.T., Sensabaugh,G.F. and Mathies,R.A. (1997) High-speed DNA genotyping using microfabricated capillary array electrophoresis chips. Anal. Chem., 69, 2181–2186. [DOI] [PubMed] [Google Scholar]
- 33.Buchholz B.A., Doherty,E.A.S., Albarghouthi,M.N., Bogdan,F.M., Zahn,J.M. and Barron,A.E. (2001) Microchannel DNA sequencing matrices with a thermally controlled “viscosity switch”. Anal. Chem., 73, 157–164. [DOI] [PubMed] [Google Scholar]