Abstract
There is a great demand for technologies to simultaneously measure mRNA levels from multiple genes. Here we report a new quantitative competitive PCR technology and demonstrate simultaneous quantification of mRNA from multiple genes. First, a sequential 2-fold dilution series containing equal amounts of gene-specific standard DNAs for 10–12 genes is prepared. Second, the serially diluted standard DNAs are individually added to equal amounts of tissue-derived cDNA and amplified with gene-specific primers for 19–26 PCR cycles. Each gene/standard DNA pair is amplified individually. All amplified DNA products (n = 80) are resolved by one microplate array diagonal gel electrophoresis using 5% polyacrylamide. Changes in mRNA levels of ∼15% can be detected by this technology. The mRNA levels from 10–12 genes were simultaneously quantified. mRNA levels were compared in RNA samples from rat liver, kidney and skeletal muscle. This quick, specific, sensitive, reproducible and yet inexpensive technique is ideal for simultaneously studying co-ordinate changes in mRNA levels from multiple genes.
INTRODUCTION
Many complex diseases, such as diabetes and hypertension, are likely to be caused by abnormal expression of multiple genes (1,2). It is estimated that the number of genes involved in multifactorial diseases is about one thousand (3). A change in expression of one gene is often caused by interaction of a number of factors on the promoter region of the gene, resulting in co-ordinate changes in mRNA levels of a number of responsive genes. Thus, there is a demand for a technology to simultaneously measure co-ordinate changes in mRNA levels from multiple genes.
There is no medium throughput technology so far offering simultaneous quantification of mRNA levels from multiple (i.e. 10–100) genes. The cDNA microarray (4) is a high throughput technology with respect to the number of genes analysed (>1000 genes), but low throughput (often one at a time) for the number of samples to be analysed. Real-time PCR (5–7) is a technology with low throughput in the number of genes (one or two) analysed, but high throughput (96 or more) in the number of samples analysed. An important concern relevant to both cDNA microarrays and real-time PCR is their requirement for expensive equipment and their high running costs, limiting accessibility to most laboratories. Another concern relevant to cDNA microarrays is the possibility of generating false signals due to non-specific hybridisation (i.e. low specificity), which may occur as arrays containing gene-specific oligos (∼30 bases long) often differing by only a few nucleotides are hybridised simultaneously with sample cDNA. The lack of sensitivity with cDNA microarrays (8) makes it difficult to analyse mRNA levels of low abundance, and limits its application. Quantitative competitive PCR (qcPCR) (9–12) offers excellent specificity, sensitivity and good reproducibility and low running costs. However, its low throughput in both the number of genes (usually one gene) and the number of samples (a few samples) to be simultaneously analysed makes qcPCR laborious for the analysis of multiple genes and multiple samples. In order to solve the low throughput problem of qcPCR, we have modified the method for undertaking conventional qcPCR. With our novel design, the throughput of current qcPCR in the number of genes and number of samples has been markedly increased. We refer to the novel qcPCR as medium throughput qcPCR (MT qcPCR). This technology retains the features of qcPCR of high specificity and sensitivity, with good reproducibility and low cost. Here we demonstrate both the novel design and a potential application, and show for the first time simultaneous mRNA quantification from 10–12 different genes using MT qcPCR.
MATERIALS AND METHODS
Design of PCR primers and synthesis of standard DNA competitors
The PCR primers were designed according to the following points. (i) Three primers were designed for each gene. Primer 1 contained the sense sequence and primer 5 the antisense sequence. An internal antisense primer sequence of ∼60 bp 3′ to the primer 5 sequence was defined. Thus, primer 3 contained the primer 5 sequence with the internal sequence attached to its 3′-end (11) (Table 1). (ii) The sizes were ∼385 bp for the standard (or competitor) and ∼445 bp for the target DNA bands. Thus, there was an ∼60 bp (∼16%) difference between the two products to be differentiated by gel electrophoresis. (iii) The percentage of A/T was similar to that of C/G in the primer sequences and the melting temperatures were similar (∼60°C). (iv) The sense and antisense primers spanned at least one intron if possible. (v) All primer sequences were searched against existing sequences submitted to GenBank using Blastn software at the National Center for Biological Information to ensure that each of the primer sequences did not form a match with sequences of other genes. The primer sequences for the 10 genes investigated are listed in Table 1. Synthesised primers were dissolved in 1× TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) at a concentration of 200 µM stock solution.
Table 1. The primer sequences for the 10 genes.
Primer 1 is the sense sequence, primers 3 and 5 are the antisense sequences.
To synthesise standard DNA, cDNA was synthesised from ∼500 ng total RNA (total RNA was prepared as described before; 13) prepared from rat liver tissues under the conditions specified below (see cDNA synthesis). Aliquots of the synthesised cDNA were amplified by PCR using a pair of gene-specific PCR primers (primers 1 + 3, Table 1). The amplified DNA products for all genes were gel purified using a Wizard PCR Preps DNA Purification System (Promega, Southampton, UK). The concentrations of standard DNA samples were quantified by spectrophotometry.
The gene-specific standard DNAs (gsDNAs) for the 10 genes were initially diluted to a concentration of 1 nM and mixed together in equal portions to give a concentration of 100 pM in the gsDNA mixture. The gsDNA mixture was further serially diluted 2-fold. A series of eight gsDNA solutions containing sequentially 2-fold diluted gsDNAs for 10 genes was used in the quantitative analysis.
cDNA synthesis
An aliquot of 1 µl of mRNA (467, 485 and 560 ng/µl for rat liver, kidney and skeletal muscle; Sigma) was added to 5 µl of H2O, denatured at 70°C for 5 min and added to a reverse transcription solution containing 0.5 mM dNTPs, 5 µM oligo(dT), 10 U/µl MMLV reverse transcriptase (Promega), 1 U/µl RNase inhibitor (Promega) and 1× reaction buffer containing 75 mM Tris–Cl, pH 8.3, 50 mM KCl, 10 mM DTT, 3 mM MgCl2 in a 20 µl reaction volume. The cDNA synthesis reaction was undertaken at 42°C for 60 min, followed by denaturing at 95°C for 5 min. Synthesised cDNA was diluted 100-fold with H2O to concentrations containing 4.67, 4.85 and 5.6 ng/µl rat liver, kidney and skeletal muscle mRNA.
PCR amplifications
An aliquot of 10 µl of diluted cDNA solution was mixed with 10 µl of gsDNA mixture containing equal concentrations (50 pM) of gsDNA for the 10 genes, 100 µl of 2× PCR ReadyMix Taq PCR Reaction Mix (1× PCR ReadyMix contains 1.5 U Taq polymerase, 10 mM Tris–Cl, 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 0.2 mM dNTP and stabilizers; Sigma) and 60 µl H2O (total volume 180 µl). Then 18 µl of the cDNA/gsDNA/PCR reagents mixture was added to each well in an array (A1–A10, Table 2) and to each well was added 2 µl (5 µM) of gene-specific PCR primers (in a 20 µl PCR reaction volume), followed by amplification for 19 cycles. In the same way, 10 µl of diluted cDNA solution was mixed with 10 µl of gsDNA mixture of reduced concentration (by half, 25 pM) and aliquoted to 10 tubes. To each aliquot was added gene-specific PCR primers, followed by amplification by PCR for 20 cycles (Table 2). The process was repeated until gsDNA of the lowest concentration was added to cDNA and amplified for 26 cycles (Table 2). The PCR conditions were: a preliminary incubation at 94°C for 4 min, followed by denaturing at 94°C for 50 s, annealing at 57°C for 50 s and extension at 72°C for 1 min for the specified number of PCR cycles (Table 2), with a final extension at 72°C for 7 min.
Table 2. Design of medium throughput qcPCR.
| A | B | C | D | E | F | G | H | ||
|---|---|---|---|---|---|---|---|---|---|
| Master mix preparation | |||||||||
| sDNAa (pM) | 2.50 | 1.25 | 0.63 | 0.31 | 0.16 | 0.08 | 0.04 | 0.02 | |
| PCR amplification | |||||||||
| 1 | ANG | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 |
| 2 | α-Fg | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 |
| … | … | … | … | … | … | … | … | … | |
| 10 | PPAR-α | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 |
asDNA (pM) refers to standard DNA concentration in each of the PCR reactions.
To measure mRNA levels from 10 genes, cDNA (10 µl) was added to gsDNA (10 µl) containing equal concentrations (50 pM) of gsDNA for the 10 genes and PCR reagents. The mixture of cDNA/gsDNA/PCR reagents was divided into equal aliquots and to each aliquot was added only one pair of gene-specific PCR primers (for ANG, α-Fg, …, PPAR-α) in a total of 20 µl reaction volume, followed by amplification for 19 PCR cycles (reactions in column A). In the same way, reduced amounts (by half) of gsDNA were added to equal amounts of cDNA and amplified by 20 PCR cycles (reactions in column B), and so on.
Data analysis
After PCR amplification was complete, 5 µl (6×) of DNA loading buffer was mixed with each PCR reaction and 5 µl of each mixture of PCR products was loaded on a pre-stained (1 µg/ml ethidium bromide in 1× TBE buffer for 20 min) microplate array diagonal gel electrophoresis (MADGE gel; 14) using 5% polyacrylamide (the protocol for preparing the MADGE gel can be found at http://research.bmn.com/tto/, P02068). After 70 min running at 150 V, the gel was destained in H2O for 30 min. Pictures were taken with a digital camera (UVP, Cambridge, UK), with the gel facing towards a UV transilluminator (glass support facing up). Images of the gels were saved onto a floppy disk and the fluorescence of the DNA bands was analysed using Phoretix 1 D advanced v.4.01 software (Phoretix, Newcastle upon Tyne, UK).
Statistics analysis
All statistical calculations were performed using SPSS software. Differences in mean value of mRNA levels between different target genes were examined by paired Student’s t-test. Results are presented as means ± SD.
RESULTS
Preparation of gsDNA
Each of the gsDNAs (as competitors during PCR amplification) for 10 genes was designed to share identical PCR binding sites with their target genes, but an ∼60 bp deletion was introduced (11). Thus, the PCR products amplified from standard and target DNAs could be differentiated by gel electrophoresis. Synthesised gsDNAs for all genes were gel purified, diluted to 1 nM and checked by PCR amplification to verify that the qualities of gsDNAs were acceptable and their concentrations were similar. The gsDNAs for all genes were further diluted and a 2-fold dilution series (concentrations of 50, 25, 12.5, 6.25, 3.13, 1.56, 0.78 and 0.39 pM) was used in the quantification assay.
Quantification of multiplex mRNA levels from target genes
To simultaneously quantify mRNA levels from 10 genes, 10 µl of diluted cDNA was mixed with 10 µl of gsDNA mixture of the highest concentration (50 pM), plus PCR reagents and H2O, and divided into 10 aliquots. To each of these aliquots was added gene-specific PCR primers for one gene and all 10 aliquots were amplified by PCR for 19 cycles in a 20 µl reaction volume (Table 2). In the same way, the same amount of cDNA was mixed with gsDNA mixture of a lower concentration (25 pM), divided into 10 aliquots and each aliquot amplified by 20 PCR cycles (PCR cycle number increased by one to compensate for the halved amount of gsDNA), and so on (Table 2). Amplified DNA products were resolved by MADGE (Fig. 1) in 5% polyacrylamide (14). With 19 PCR cycles (Fig. 1, column A), two bands amplified from both target and gsDNA from the α and β-fibrinogen, hepatic lipase, 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) and phosphoenolpyruvate carboxykinase (PEPCK) genes were visible (Fig. 1, A2, A3, A6, A7 and A9), suggesting that mRNA levels from these genes were greater than those from the other genes. DNA bands amplified by 20 PCR cycles showed a greater fluorescence of the target DNA bands (upper bands) relative to those of the gsDNA bands (lower bands) and the target DNA bands of the insulin receptor (IR) and peroxisome proliferator activated receptor-α (PPARα) were now visible (Fig. 1, B8 and B10). In general, as the number of PCR cycles increased, the ratio of target to standard DNA band fluorescence increased (from <1 to >1) and those amplified from mRNAs of lesser expressed genes became visible.
Figure 1.
Gel analysis of PCR products. Eighty samples were analysed in one MADGE containing 5% acrylamide. The sample loading wells (maximum 96) were indicated by crosses between eight columns (A–H) and 12 rows (1–12). The dotted arrow at A1 indicates the direction of electrophoresis of the samples. Each row represents one gene analysed (as indicated), while each column represents DNA amplified by the same number of PCR cycles. For example, samples in column A were amplified by 19 cycles, samples in column B by 20 cycles, and so on. The two samples loaded in wells E11 and F11 were used to measure the inter-assay CV for the gel. ANG, angiotensinogen; α-Fg, α-fibrinogen; β-Fg, β-fibrinogen; GK, glucokinase; GR, glucocorticoid receptor; HL, hepatic lipase; HSD1, 11β-hydroxysteroid dehydrogenase type 1; IR, insulin receptor; PEPCK, phosphoenolpyruvate carboxykinase; PPAR-α, peroxisome proliferator activated receptor-α.
Analysis of data obtained by PCR amplification
The gel was destained in H2O for 30 min before being photographed using a digital camera under a standard UV transilluminator and the DNA bands were quantified using Phoretix 1D advanced v.4.01 software. The fluorescence of DNA bands (units given by the Phoretix software are in volumes) was linear between 101 and 8600 volumes (r = 0.998). The intra-assay coefficient of variation (CV) for quantification of DNA bands on the gels was 5% (n = 15). The inter-assay CVs for MADGE gels were calculated using two different ratios of target and standard bands from two different samples loaded on different gels (Fig. 1, E11 and F11). A 7.84% inter-assay CV was obtained for samples whose value of the ratio between the upper and lower bands was 0.487 ± 0.04 (n = 8; Fig. 1, E11), while a 6.09% CV was found for the other sample, in which the value of the ratio was 1.80 ± 0.11 (n = 8; Fig. 1, F11).
The fluorescence of specific DNA bands was quantified using the Phoretix software and exported to Microsoft Excel to calculate the mRNA levels. mRNA abundance was calculated using the formula (11):
Ft/Fs = (St × Mt)/(Ss × Ms) 1
As values of Ft and Fs (fluorescence of the target and standard DNA bands), St and Ss (the size of the target and standard DNA bands) and the number of standard DNA molecules (Ms) were known, the number of target DNA molecules reverse transcribed from mRNA (Mt) could be calculated.
When the numbers of molecules of the two templates prior to PCR amplification were identical (i.e. Mt = Ms), the PCR amplification efficiency of the two templates was almost the same and reproducibility of the results was optimum. In our design, target DNA sizes (St) for the 10 genes were ∼445 bp and standard DNA sizes (Ss) were ∼385 bp. Therefore, when the fluorescence ratio of target to standard DNA bands (Ft/Fs) was ∼1.16 (445/385 = 1.16), the number of target DNA molecules was similar to the number of standard DNA molecules (i.e. Mt ≈ Ms, according to equation 1). Thus, Ft/Fs values close to 1.16 were used as an ideal ratio in the calculation of mRNA levels.
As the amount of standard DNAs added to the PCR was sequentially reduced by half for each gene from 19 to 26 PCR cycles, the expected values of Ft/Fs should be consecutively doubled according to equation 1, as the PCR amplification efficiency of the two templates was similar under our experimental conditions (J.Zhang and C.D.Byrne, manuscript in preparation). To reduce the variation that may occur due to data processing and to simplify the process, the mRNA level for each gene was calculated by taking the mean from three or four mRNA (Mt) values for the gene. Each of these three or four Mt values was calculated from Ft/Fs values that were close to both the ideal ratio (1.16) and to the expected values obtained from consecutive PCR cycles (e.g. hypothetical Ft/Fs values such as 0.58, 1.16, 2.32 and 4.64 obtained from 19, 20, 21 and 22 cycles). For a theoretical consideration of the calculation see the Appendix.
Using the above guidelines, the mRNA levels for 10 genes from liver mRNA were calculated and are listed in Table 3. The inter-assay CV for this technology was ∼15% (Table 3). The values of mRNA levels in Table 3 were calculated using the assumption that the efficiency of cDNA synthesis was 100%, as a DNA competitor was used. However, cDNA synthesis efficiency may be <100% (15) and, therefore, we present mRNA level values for relative comparison only.
Table 3. Quantification of mRNA levels measured using MT qcPCR technology.
| Liver | Kidney | Skeletal muscle | |||
|---|---|---|---|---|---|
| Mean ± SD | CV (%) | n | |||
| ANG | 1.17 ± 0.15 | 12.76 | 6 | 0.14 | 0.03 |
| α-Fg | 13.06 ± 1.40 | 10.73 | 8 | 0.40 | < 0.03 |
| β-Fg | 12.45 ± 2.36 | 18.95 | 8 | 0.15 | < 0.03 |
| GK | 0.20 ± 0.03 | N/A | 5 | < 0.05 | < 0.03 |
| GR | 0.24 ± 0.05 | N/A | 4 | 0.44 | 0.24 |
| HL | 16.45 ± 1.25 | 7.61 | 6 | 0.05 | < 0.03 |
| 11β-HSD1 | 3.61 ± 0.82 | 22.68 | 6 | 9.39 | 0.67 |
| IR | 1.76 ± 0.17 | N/A | 3 | 0.91 | 0.56 |
| PEPCK | 28.87 ± 5.85 | 20.27 | 7 | 66.63 | 0.51 |
| PPAR-α | 2.74 ± 0.62 | N/A | 5 | 1.84 | 0.67 |
The units of mRNA level are given in amol (1 amol = 6.023 × 105 molecules, from Avogadro’s constant, 1 mol = 6.023 × 1023 molecules). CV, coefficient of variation; N/A, not available.
Detection sensitivity of MT qcPCR
We have determined whether a 10% difference in mRNA levels was detectable using MT qcPCR. cDNA reverse transcribed from foetal liver total RNA was sequentially diluted by 10% (A–E, Table 4). The diluted cDNA concentrations (B–E) were 0.90, 0.81, 0.73 and 0.66 of the original concentration A containing 50 ng total RNA/µl. Equal volumes of cDNA from the 10% dilution series were co-amplified with the standard DNA mixture as described above (Table 2), using primers for the β-fibrinogen gene and 20–23 PCR cycles (Fig. 2). For each of these different cDNA concentrations, six repeat PCR reactions were undertaken to assess the intra-assay CV for MT qcPCR. The ratio Ft/Fs decreased in parallel with decreasing cDNA concentration (Table 4). mRNA levels of β-fibrinogen were calculated and are presented as amol/ng total RNA. Samples differing by 10% in the dilution series were analysed to determine whether it was possible to detect a 10% difference in mRNA copy number. For example, the mean of calculated mRNA copy number of six repeats undertaken at 20 cycles for sample A (Table 4) was compared with the mean calculated mRNA copy number of six repeats at 20 cycles for sample B by paired Student’s t-test. P < 0.001 indicated that the mean values of 0.95 and 0.83 amol/ng total RNA, respectively, were different from each other and that the 10% difference in mRNA copy number could be detected (see Table 4 for more t-test results).
Table 4. Determination of detection sensitivity of MT qcPCR.
*Concentrations refer to diluted concentrations (B–E) relative to the original concentration A.
Columns A–E represent different cDNA concentrations (the first row) of the dilution series. Mean (± SD) values of Ft/Fs ratios from six repeat reactions (Fig. 2) by PCR amplification with the same cycle number (e.g. 20 cycles) were calculated. mRNA copy numbers (amol/ng total RNA) were calculated from relevant Ft/Fs ratios using equation 1 and are presented for relative comparison purposes, assuming that the efficiency of cDNA synthesis was 100%.
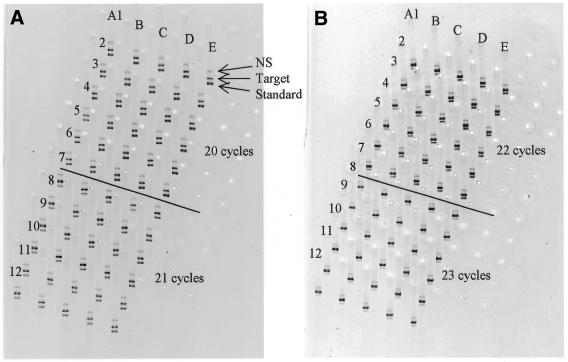
Figure 2.
Detection sensitivity of MT qcPCR. cDNA reverse transcribed from rat feotal liver total RNA was co-amplified with gsDNA by 20–23 cycles with primers for β-fibrinogen. For reactions amplified by 20 PCR cycles (A1–E6, A), equal amounts of gsDNA (1.25 pM final concentration) were added to each of these reactions. The amounts of cDNA added decreased by 10%, i.e. reactions in column B contained 90% of column A, reactions in column C contained 90% of column B, and so on. Six repeat PCRs were undertaken (A1–A6, B1–B6, etc.) to assess the intra-assay CV for MT qcPCR. In the same way, PCRs were set up and amplified by 21 cycles (in A7–E12, A), 22 cycles (A1–E6, B) and 23 cycles (A7–E12, B), with decreasing concentrations of gsDNA according to the number of PCR cycles amplified (0.63, 0.315 and 0.158 pM final concentrations for 21, 22 and 23 cycles, respectively). A non-specific band was observed for β-fibrinogen, which was specific to this sample, since this non-specific band was not observed in other samples (see for example Fig. 1). The non-specific band was most likely derived from amplification of contaminating genomic DNA.
We noted that the ratio Ft/Fs obtained from PCR amplification after 23 cycles was 26.12, which was obtained from cDNA of concentration A (Table 4). This value of Ft/Fs was far from our ideal ratio of ∼1.16 for this target and standard (see above, analysis of data obtained from PCR amplification) and much greater than had been anticipated (this was clearly illustrated by the dose–response curve, see Fig. 4). The calculated copy number (3.27 amol/ng total RNA) deviated markedly from the mean values (∼1 amol/ng total RNA) obtained from the other three PCR cycles. These data supported the notion that it was important to use a ratio Ft/Fs close to the ideal ratio (∼1.16 for this specific target and standard) for accurate measurement of mRNA levels.
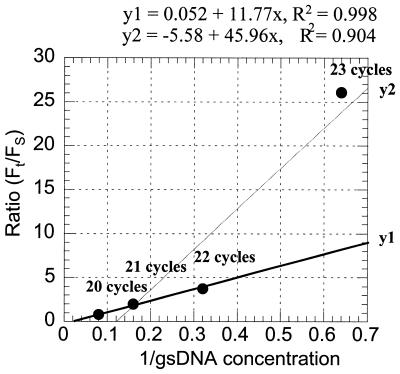
Figure 4.
Dose–response curve for MT qcPCR. Values of Ft/Fs ratios (listed in Table 4) were plotted against the inverted concentrations of gsDNA according to equation 1. As Ft/Fs ratios were obtained by PCR amplification with different cycle numbers, the cycle numbers are included in the plot.
We further determined whether a small difference in mRNA levels could be detected using MT qcPCR if the PCR cycle number was high (>30), as a high cycle number would be required to measure levels of poorly expressed mRNAs. We first diluted gsDNA from 25 pM (used for the 20 cycle PCR amplification reaction) to 39 fM concentration (a 641-fold dilution). The gsDNA (39 fM) was used for amplification by 30 PCR cycles. A clear DNA band was amplified and we estimated that the PCR amplification efficiency was ∼90% up to 30 cycles. As we anticipated that PCR amplification efficiency would decrease at high cycle numbers, we further diluted the gsDNA by a factor of 1.8 rather than 2. A series of six 1.8-fold dilutions was prepared with concentrations of 21.67 (31), 12.04 (32), 6.69 (33), 3.72 (34) and 2.06 fM (35), the number in parentheses indicating the proposed amplification cycle number for that gsDNA concentration. We amplified the first four of the gsDNAs by the proposed PCR cycle number and for each concentration of gsDNA eight repeats of PCR were undertaken (not shown). The values of the amplified DNA bands were similar between these gsDNAs (6612 ± 954, 5454 ± 737, 6924 ± 654 and 4861 ± 1028, n = 8 for each cycle, for those amplified by 30, 31, 32 and 33 cycles) and their CVs were acceptable (∼15%) even at high PCR cycle numbers. The PCR amplification efficiency was estimated to be ∼80%, as anticipated.
We then diluted the cDNA by ∼1000-fold, which was used to amplify β-fibrinogen by 20–23 PCR cycles, to determine whether it was possible to detect a small difference in mRNA copy number for genes of low expression. A serial dilution of 85, 70 and 55% of the diluted cDNA concentration A (a 15% decrease in concentration, Table 5) was prepared. We analysed β-fibrinogen mRNA by 30–33 PCR cycles as described above using the four cDNAs of 15% decreasing concentration (Fig. 3). Data were analysed as described above. Compared to the data obtained with 20–23 cycles of PCR amplification, the values of intra-assay CVs from those obtained by 30–33 PCR amplification cycles were increased [mean intra-assay CVs were 2.58 ± 1.01, 4.92 ± 3.2, 11.58 ± 7.79 and 11.54 ± 2.93 for 20–23 PCR cycles, respectively, n = 5 (Table 4); compared to 11.28 ± 5.43, 13.28 ± 6.16, 19.68 ± 7.85 and 23.68 ± 5.87 for 30–33 cycles, respectively, n = 4 (Table 5)]. The P values were all <0.05 when means of mRNA copy number calculated from results obtained from 30 cycles of amplification (Table 5) were compared, suggesting that it was possible to detect a 15% difference in mRNA levels. However, the P values were >0.05 when mRNA levels calculated from results obtained from 31–33 cycles of PCR amplification were compared, suggesting that a 15% difference in mRNA levels was not detectable. Thus, the 15% difference in mRNA levels was at the limit of detection. When statistical analysis was undertaken between means of groups with greater differences (e.g. column A versus column C and column B versus column D), P values were <0.05 (data not shown). Thus, the data suggest that it was possible to detect mRNA levels differing by >15%, even though expression levels were low and required a high PCR amplification cycle number.
Table 5. Determination of detection sensitivity of MT qcPCR for mRNA of low abundance.
*Concentrations refer to diluted concentrations (B–E) relative to the original concentration A.
Columns A–D represent different cDNA concentrations (the first row) of the dilution series. Mean (± SD) values of Ft/Fs ratios from six repeat reactions (Fig. 3) by PCR amplification with the same cycle number (e.g. 30 cycles) were calculated. mRNA copy numbers (amol/ng total RNA) were calculated from relevant Ft/Fs ratios using equation 1 and are presented for relative comparison purposes, assuming that the efficiency of cDNA synthesis was 100%.
Figure 3.
Detection sensitivity of MT qcPCR for mRNA of low abundance. Parallel dilutions of both cDNA reverse transcribed from foetal liver total RNA and gsDNA were co-amplified using primers for β-fibrinogen by 30–33 PCR cycles. The PCR reaction conditions were identical for all reactions. The final concentrations for gsDNA were 1.95, 1.084, 0.601 and 0.334 fM for reactions amplified by 30, 31, 32 and 33 PCR cycles, respectively. For reactions amplified by each different PCR cycle number (e.g. 30 cycles), the amounts of cDNA added decreased from column A to D (or E to H) by 15%, i.e. the concentrations for cDNA in column B, C and D were 85, 70 and 55% of that in column A. For each specific cDNA concentration (e.g. concentration A), six repeats of an identical concentration of cDNA were added to PCR reactions (A1–A6) to assess the intra-assay CV for MT qcPCR. PCR cycle numbers are 30 (A1–D6), 31 (E1–H6), 32 (A7–D12) and 33 (E7–H12).
Dose–response curve
The novelty of MT qcPCR over conventional quantitative competitive PCR is illustrated clearly by the dose–response curve, unique to MT qcPCR. The three key parameters (changes in the ratio Ft/Fs, changes in standard or target DNA concentration and changes in the number of PCR cycles) for quantitative competitive PCR were all included in the dose–response curve for MT qcPCR (Fig. 4). Mean ratios Ft/Fs calculated from six repeat PCR reactions for sample A obtained from each of the four different PCR cycles (Table 4) were plotted against the inverted concentrations of standard DNA mixtures (Fig. 4). As expected, a linear relationship was observed for the results of PCR amplification by 20, 21 and 22 cycles (r2 = 0.998; Fig. 4, regression line y1), suggesting that PCR amplification was appropriate. Indeed, the mRNA copy numbers calculated from the PCR results with amplification by the three different cycles were very similar (0.95, 1.00 and 0.94 amol/ng total RNA for 20, 21 and 22 cycles, Table 4). However, results from PCR amplification by 23 cycles were outside the linear regression curve y1, suggesting that target DNA templates were over-amplified relative to standard DNA templates, and the mRNA copy number was inappropriately calculated as 3.27 amol/ng total RNA (Table 5), which was an overestimate compared to the value calculated from the other three cycle numbers. Thus, from the dose–response curve the appropriateness of PCR amplification could be assessed.
A potential application of MT qcPCR in the measurement of mRNA levels from multiple genes involving multiple samples
mRNA patterns of gene expression from the 10 genes in different tissues. We illustrate the application of this technology for measuring mRNA levels from multiple genes in different tissues (rat kidney and skeletal muscle tissues).
We first semi-quantitatively assessed the mRNA levels from 10 genes in rat kidney and skeletal muscle tissues by amplifying the synthesised cDNA for 30 PCR cycles. DNA bands from nine expressed genes (except glucokinase) were visible in kidney tissue, and six (angiotensinogen, glucocorticoid receptor, 11 β-HSD1, insulin receptor, PEPCK and PPARα genes) were visible in skeletal muscle tissue.
We further more precisely quantitatively determined the mRNA levels from the above genes using MT qcPCR. The calculated mRNA levels for these genes are presented in Table 3.
mRNA levels from multiple genes involving multiple samples. We further illustrate utilisation of the MT qcPCR technique by measuring mRNA levels from multiple genes and multiple samples (12 genes × 8 samples, Fig. 5A–D) using MT qcPCR. cDNA synthesised from foetal liver total RNA from eight different rats (columns A–H, Fig. 5) was amplified with gene-specific PCR primers for 12 genes (rows 1–12, Fig. 5) by 20–23 PCR cycles (Fig. 5, gels A–D). Calculated mRNA abundances are presented in Table 6 (arbitrary units, ≈ ×1000 copies/ng total RNA).
Figure 5.
Simultaneous quantification of mRNA levels from multiple genes and multiple samples. Equal amounts of cDNA (containing ∼50 ng total RNA/reaction) reverse transcribed from total RNA from eight different rats (columns A–H) were amplified with gsDNA and PCR primers for 12 genes, by either 20 cycles (A), 21 cycles (B), 22 cycles (C) or 23 cycles (D). The PCR conditions were identical for all reactions and the amounts of gsDNA decreased by half from (A) to (D), as described in Materials and Methods. Reactions in each column (e.g. A1–A12) represent cDNA from one sample and each row represents one specific gene. The 12 gene-specific PCR primers were (from row 1 to row 12): acyl-CoA oxidase (ACO), CD36, diacylglycerol acyltransferase (DGAT), fatty acid synthase (FAS), 3-hydroxy-3-methylglutaryl-CoA reductase (HMG-CoA), lecithin-cholesterol acyltranserase (LCAT), long-chain acyl-CoA synthetase (LCAS), carnitine parmitoyl transferase I (CPT), hepatic lipase (HL), peroxisome proliferator-activated receptor-α (PPAR-α), peroxisome proliferator-activated receptor-γ (PPAR-γ) and insulin receptor (IR). Calculated mRNA copy numbers for the 12 genes are listed in Table 6.
Table 6. Simultaneous quantification of mRNA levels from multiple genes and multiple samples.
Columns A–H represent samples from eight different rats. Each row represents a specific gene analysed. mRNA levels were calculated as described above and are presened as arbitrary units (×1000 copies mRNA/ng total RNA).
Thus, we have demonstrated a potential application of MT qcPCR in simultaneous measurement of mRNA levels from multiple genes and from multiple samples (either from the same tissue or different tissues).
DISCUSSION
Here we demonstrate a novel MT qcPCR technology to simultaneously quantify mRNA levels from 10–12 genes. We have presented data showing that a small difference (∼15%) in mRNA levels can be detected using our MT qcPCR and we have applied this technology to simultaneous measurement of multiple genes either from multiple tissues or from multiple samples.
The novelty of MT qcPCR includes the unique design for undertaking qcPCR and our approach of combining qcPCR with MADGE. In our novel design we use a sequential dilution series of a standard DNA mixture containing iso-molar amounts of gsDNA molecules for each of the target genes added to a fixed amount of cDNA, in combination with a sequential increase in number of PCR amplification cycles. This is the best approach of the different experimental designs that we have tried, allowing simultaneous measurement of multiple mRNAs varying markedly in abundance. The novelty of MT qcPCR over conventional qcPCR is clearly illustrated by the unique dose–response curve, where three key parameters, i.e. changes in the ratio Ft/Fs, changes in standard or target DNA concentration and changes in PCR cycle number, are all included in the plot (Fig. 4). In conventional qcPCR only two of the three parameters are plotted, as the third parameter remains unchanged (either the number of PCR cycles or the relative ratio of target cDNA to standard DNA; 11). In the dose–response curve for MT qcPCR changes in all three parameters are analysed in one plot. Thus, this plot provides more comprehensive information on the PCR amplification process. Using this novel design, simultaneous quantification of mRNA levels from multiple genes is simple and quick by combining the technique with MADGE gel analysis of PCR products. With MT qcPCR, the low throughput problem associated with conventional qcPCR in both the number of genes and the number of samples that can be analysed has been solved. We estimate that the throughput efficiency is increased at least 10-fold using MT qcPCR over conventional qcPCR.
The reproducibility of MT qcPCR is acceptable. An intra-assay CV of 5% and inter-assay CV of 6–8% was noted for the MADGE gel. Approximately 15% differences in mRNA levels can be detected using MT qcPCR, if all PCR products are analysed on the same MADGE gel (Table 4). A slightly greater variation (∼15% inter-assay CV, Table 3) was observed when the same panel of genes was analysed on different gels. The detection sensitivity of MT qcPCR is still acceptable with poorly expressed messages, as a difference of >15% in mRNA levels can be detected (Table 5) with PCR amplification by 30–33 cycles. As PCR amplification efficiency is ∼80% between 30 and 33 cycles, we consider that there is the potential to increase the number of amplification cycles to 35 without marked loss of reproducibility. Since most poorly expressed mRNAs can be amplified with ∼35 cycles, MT qcPCR is, therefore, suitable for analysis of poorly expressed mRNA giving acceptable reproducibility.
The ratio Ft/Fs (the fluorescence values for the target versus standard DNA bands) are increased from low values (<1) to high values (>1) as the PCR cycle number increases (Fig. 1). Thus, for a specific gene there is always a specific cycle number that will give an ∼1:1 Ft/Fs ratio. This specific number of PCR cycles may vary from gene to gene, depending upon the level of mRNA expression. The greater the mRNA level, the lower the cycle number required. As values of the Ft/Fs ratio are important in the calculation of mRNA copy number, it should be noted from our dose–response curve (Fig. 4) results that it is better to use the three values of Ft/Fs closest to unity, as a value markedly greater than unity generates considerable variation in mRNA copy number (Fig. 4, 23 cycles, and Table 4). The mRNA level is calculated by taking the mean of mRNA levels calculated from the three Ft/Fs ratios, and this mean value is a good estimation of the true mRNA level for a specific gene.
The procedure for MT qcPCR has been greatly simplified compared to conventional qcPCR, because optimisation is needed for the gsDNA. The optimised concentrations for gsDNA to be amplified by different PCR cycles have been listed in Table 2. As only a few experiments are required to optimise the PCR conditions when different samples are used, analysis of mRNA levels from different samples can be undertaken as illustrated in Scheme 1. It takes only ∼1.5 h to synthesise cDNA and hands-on time for preparation of PCR reactions is ∼1 h. The analysis of data obtained from 96 PCR samples and calculation of mRNA levels can be finished within 20 min. Thus, results can be obtained quickly. Reproducible results have been obtained using optimised concentrations for gsDNA and relevant numbers of PCR amplifications (Table 2) with gsDNA for different genes [e.g. a fragment of DNA containing PCR primer sequences for 16 genes (see www.soton.ac.uk/~jzhang/ for details) was used in the analysis of mRNA levels (data not shown)].
Scheme 1.
The strengths of MT qcPCR over real-time PCR are as follows. (i) A greater number of genes can be analysed. The number of genes that can be analysed simultaneously using real-time PCR is limited to one or two genes (16,17). In contrast, 10 or more genes from multiple samples can be simultaneously analysed using MT qcPCR. The novel design of MT qcPCR can be further applied to simultaneous analysis of up to 384 or more genes, as 384-well PCR and 384- and 768-well MADGE plates (http://www.sgel.humgen.soton.ac.uk) are now available. (ii) A greater accuracy of calculated mRNA levels. Since MT qcPCR is a competitive PCR technology, mRNA levels can be expressed as copies/ng total RNA (this assumes that cDNA synthesis efficiency is 100% and is equivalent in all cDNA reactions; Tables 4 and 5). Even though the information provided in this manuscript is for relative comparison, useful information can be obtained regarding the relative levels of gene expression between different genes and different samples, particularly when mRNA levels differ markedly. Such information is not obtainable using real-time PCR. (iii) Misinterpretation of results obtained from real-time PCR may occur, whereas there is no such concern with MT qcPCR. In competitive PCR, target mRNA copy number is calculated using the ratio Ft/Fs of a known amount of added standard DNA that is co-amplified with the target DNA template within the same tube by the same PCR primer pair (equation 1). In contrast, in real-time PCR the mRNA levels are calculated relative to an unknown amount of RNA from a reference gene, assuming that the abundance of the reference gene does not change under the experimental conditions. Unfortunately, the mRNA abundance of typical housekeeping genes, such as dihydrofolate reductase, β-actin and glyceraldehyde 3-phosphate dehydrogenase, and 18S rRNA levels change under some experimental conditions (18–20). Thus, a change in mRNA levels relevant to a reference gene may be misinterpreted as a change in mRNA levels due to the experimental conditions. This potential error cannot be eliminated unless mRNA levels of a chosen reference gene have been shown not to change under the experimental conditions in question. Therefore, data obtained from multiple mRNA quantification using MT qcPCR (competitive PCR) is likely to be more reliable than data obtained from real-time PCR (non-competitive PCR). Furthermore, MT qcPCR does not require expensive equipment (compared with real-time PCR) and routine PCR reagents are required. Thus, MT qcPCR is accessible to most laboratories.
MT qcPCR is unique in that it combines the advantages of medium throughput in both the number of genes and the number of samples that can be simultaneously analysed with high specificity and sensitivity and good reproducibility. mRNA abundance in different tissues can be compared, as this comparison is based on the known amount of gsDNA used. The inter-assay CV is low (∼15%). Thus, we consider that MT qcPCR is ideal for simultaneously measuring changes in mRNA levels from several important genes along biochemical pathways.
In conclusion, we have demonstrated a novel medium throughput qcPCR technology that allows quantification of mRNA from multiple genes simultaneously, quickly, simply and inexpensively. This technique is highly reproducible and has many potential applications.
APPENDIX
A theoretical consideration of the calculation of Mt values is listed below.
As the amount of DNA products during PCR amplification accumulates, as per Wang et al. (9) and Zhang et al. (11), Mt(n) = Mt(1 + Et)n and Ms(n) = Ms(1 + Es)n, where Mt(n) and Ms(n) are the number of target and standard DNA molecules after n cycles of PCR amplification, Mt and Ms are the number of target and standard DNA molecules prior to PCR amplification and Et and Es are the PCR amplification efficiencies for the target and standard DNA templates.
The fluorescence (F) generated by DNA bands is directly proportional to the amounts and lengths of the DNA products, thus Ft(n) ∝ St × Mt(n) or Ft(n) ∝ St × Mt(1 + Et)n and Fs(n) ∝ Ss × Ms(n) or Fs(n) ∝ Ss × Ms(1 + Es)n and
Ft(n)/Fs(n) = [St × Mt(1 + Et)n]/[Ss × Ms(1 + Es)n] A1
or
Mt = [Ft(n) ×Ss(1 + Es)n × Ms]/[Fs(n) × St(1 + Et)n] A2
For the (n + 1) cycle reaction, the amount of standard DNA added to the reaction prior to PCR amplification was halved compared to those added to reactions amplified with n cycles.
According to equation A1, the value of Ft(n + 1)/Fs(n + 1) should increase by 2 if the PCR amplification efficiencies between the two templates are the same (i.e. Et = Es), as the amount of cDNA added to reactions amplified by (n + 1) cycles equals that added to reactions amplified by n cycles, thus Ft(n + 1)/Fs(n + 1) = 2 × Ft(n)/Fs(n) and
Mt = [Ft(n + 1) × Ss(1 + Es)n + 1 × Ms]/[Fs(n + 1) × St(1 + Et)n + 1 × 2]
= [Ft(n) × Ss(1 + Es)n × Ms]/[Fs(n) × St(1 + Et)n] A3
Comparing equation A3 to A2, the value of Mt calculated from equation A3 equals the Mt value calculated from equation A2 only when Et = Es, i.e. the PCR amplification efficiency between the two templates is identical, even though the efficiency may not be 100%. In the same way the Mt values can be calculated from results amplified by (n + 2) cycles. In order to reduce the variation due to data processing, we took the mean values of Mt calculated from between three and four Ft/Fs values obtained from three or four PCR cycles based upon the above theoretical considerations.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge advice from Mr Tom R. Gaunt on MADGE. J.Z. is currently supported by the School of Medicine, University of Southampton. C.D.B. is the Director of the Wellcome Trust Clinical Research Facility at Southampton University Trust and the work was supported by the Wellcome Trust.
REFERENCES
- 1.Guillausseau P.J., Tielmans,D., Virally-Monod,M. and Assayag,M. (1997) Diabetes: from phenotypes to genotypes. Diabetes Metab., 23 (suppl. 2), 14–21. [PubMed] [Google Scholar]
- 2.Shimkets R.A. and Lifton,R.P. (1996) Recent advances in the molecular genetics of hypertension. Curr. Opin. Nephrol. Hypertens., 5, 162–165. [DOI] [PubMed] [Google Scholar]
- 3.Drews J. (2000) Drug discovery: a historical perspective. Science, 287, 1960–1964. [DOI] [PubMed] [Google Scholar]
- 4.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 5.Gibson U.E., Heid,C.A. and Williams,P.M. (1996) A novel method for real time quantitative RT-PCR. Genome Res., 6, 995–1001. [DOI] [PubMed] [Google Scholar]
- 6.Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology, 11, 1026–1030. [DOI] [PubMed] [Google Scholar]
- 8.Carulli J.P., Artinger,M., Swain,P.M., Root,C.D., Chee,L., Tulig,C., Guerin,J., Osborne,M., Stein,G., Lian,J. and Lomedico,P.T. (1998) High throughput analysis of differential gene expression. J. Cell. Biochem., 30–31 (suppl.), 286–296. [PubMed] [Google Scholar]
- 9.Wang A.M., Doyle,M.V. and Mark,D.F. (1989) Quantitation of mRNA by the polymerase chain reaction. Proc. Natl Acad. Sci. USA, 86, 9717–9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker-Andre M. and Hahlbrock,K. (1989) Absolute mRNA quantification using the polymerase chain reaction (PCR). A novel approach by a PCR aided transcript titration assay (PATTY). Nucleic Acids Res., 17, 9437–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J., Desai,M., Ozanne,S.E., Doherty,C., Hales,C.N. and Byrne,C.D. (1997) Two variants of quantitative reverse transcriptase PCR used to show differential expression of alpha-, beta- and gamma-fibrinogen genes in rat liver lobes. Biochem. J., 321, 769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J. and Byrne,C.D. (1997) A novel highly reproducible quantitative competitve RT PCR system. J. Mol. Biol., 274, 338–352. [DOI] [PubMed] [Google Scholar]
- 13.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 14.Day I.N. and Humphries,S.E. (1994) Electrophoresis for genotyping: microtiter array diagonal gel electrophoresis on horizontal polyacrylamide gels, hydrolink, or agarose. Anal. Biochem., 222, 389–395. [DOI] [PubMed] [Google Scholar]
- 15.Newton C.R. and Graham,A. (1994) In Graham,J.M. and Billington,D. (eds), PCR. BIOS Scientific Publishers, Oxford, UK.
- 16.van Elden L.J., Nijhuis,M., Schipper,P., Schuurman,R. and van Loon,A.M. (2001) Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol., 39, 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eun A.J., Seoh,M. and Wong,S. (2000) Simultaneous quantitation of two orchid viruses by the TaqMan real-time RT-PCR. J. Virol. Methods, 87, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt E.E. and Merrill,G.F. (1991) Changes in dihydrofolate reductase (DHFR) mRNA levels can account fully for changes in DHFR synthesis rates during terminal differentiation in a highly amplified myogenic cell line. Mol. Cell. Biol., 11, 3726–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elder P.K., French,C.L., Subramaniam,M., Schmidt,L.J. and Getz,M.J. (1988) Evidence that the functional beta-actin gene is single copy in most mice and is associated with 5′ sequences capable of conferring serum- and cycloheximide-dependent regulation. Mol. Cell. Biol., 8, 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmittgen T.D. and Zakrajsek,B.A. (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods, 46, 69–81. [DOI] [PubMed] [Google Scholar]