Abstract
Uterine fibroids are exceedingly common benign tumours of the female reproductive system and cause severe symptoms, including acute pain, bleeding, and infertility. Fibroids are frequently associated with genetic alterations affecting mediator complex subunit 12 (MED12), fumarate hydratase (FH), high mobility group AT-hook 2 (HMGA2) and collagen, type IV alpha 5 and alpha 6 (COL4A5-COL4A6). Recently, we reported MED12 exon 2 mutations in 39 out of 65 uterine fibroids (60%) from 14 Australian patients. The aim of this study was to evaluate the status of FH mutations in MED12 mutation-positive and mutation-negative uterine fibroids. FH mutation screening of altogether 65 uterine fibroids and corresponding adjacent normal myometrium (n = 14) was carried out by Sanger sequencing. Three out of 14 patients displayed somatic mutations in FH exon 1 in addition to harbouring MED12 mutation in uterine fibroids. This study is the first to report that the mutations in MED12 and FH co-exist in uterine fibroids of Australian women.
Keywords: ANM, adjacent normal myometrium FH, fumarate hydratase MED12, mediator complex subunit 12, leiomyoma
Impact statement
What is already known on this subject: The majority of fibroids harbor mutations as high as 60% in relation to the most common genetic driver in uterine fibroid that is MED12.
What the results of this study add: Sanger sequencing has identified that three out of 14 patients displayed somatic mutations in FH exon 1 and these patients were also harboring MED12 mutations in their fibroid.
What the implications are of these findings for clinical practice and/or further research: The events of both MED12 and FH mutations co-occur in uterine fibroid sets the foundation for further functional studies to investigate the relationship between MED12 and FH in leiomyogenesis. The impact of this study may guide the development of new approaches for managing millions of patients affected by this disease.
Introduction
Uterine leiomyomas (also termed fibroids) are extremely common tumours of the female reproductive tract affecting as high as 70% of women by the age of 50 (1). The clinical symptoms attributable to fibroids include pelvic discomfort, profuse menstrual bleeding, reduced fertility, and pregnancy complications (2). Fibroids are the leading indication for hysterectomy among US women accounting for over 200,000 procedures performed annually (3). An improved understanding of the aetiology and pathogenesis that drive fibroid growth may guide the development of new treatment options for millions of patients who suffer from the symptoms of fibroids.
In recent years, significant progress has been made in understanding the genetic basis of uterine leiomyogenesis through whole genome sequencing studies (4, 5). There are currently four well-studied genetic alterations that are present in human uterine leiomyomas, namely, mediator complex subunit 12 (MED12) mutation, high mobility group AT-hook 2 (HMGA2) overexpression, biallelic fumarate hydratase (FH) inactivation and collagen, type IV alpha 5 (COL4A5) and collagen, type IV alpha 6 (COL4A6) deletion (5, 6). Mutations in the MED12 gene are the most common and present in approximately 70% of patients (5).
A previous study has shown that MED12 and FH aberrations are mutually exclusive, suggesting that they are independent genetic events that occur in the fibroid formation (7). MED12 belongs to a highly conserved multiprotein complex Mediator that involves in the processing of RNA polymerase II transcription (8). Oncogenic anomalies in the N-terminal part of MED12 are the most common and occur in nearly 70% of uterine fibroid (4). Fibroids may also harbour germline mutations of the FH, which encodes the tricarboxylic cycle enzyme fumarase, predisposing women to the hereditary leiomyomatosis and renal cell cancer (HLRCC) syndrome (9).
Recently, we identified 3% and 60% of uterine fibroids harbour specific mutations in MED12 exon 1 and exon 2 respectively of 65 uterine fibroids studied that were collected from Australian women (10). The majority of the mutations were predominantly missense mutations affecting a codon encoding glycine 44 in exon 2, which is a known mutational hotspot in MED12. In this study, we examined the status of FH in Australian patients with MED12 mutations and found three patients (3/14 patients) with co-existing FH and MED12 mutations in the same fibroid. Histopathological examination was performed to determine the significance of these co-existing mutations on fibroid growth.
Materials and methods
Subjects
Sixty-five uterine fibroids of different sizes from 14 patients, and 14 ANM from the same patients (1 ANM per patient), were obtained following hysterectomy procedures at the John Hunter Hospital, Newcastle, NSW, Australia. The human tissue collection and experimentation were approved in accordance with the guidelines of the Institutional Human Research Ethics Committee at the University of Newcastle. The collected tissues were processed as described in (11). We categorized fibroids into three groups based on their tumour size: small (diameter <2 cm), medium (diameter 2 cm–4 cm), and large (diameter >4 cm) (10).
Mutation screening
Small portion of fresh frozen fibroid or ANM tissues (approximately 25 mg) were used for genomic DNA extraction. Isolation of genomic DNA was performed using the QIAamp DNA Mini kit (Qiagen) according to the manufacturer's instructions. DNA from fibroid and ANM tissue samples were amplified with Immolase™ DNA polymerase (Bioline) using ten designated intronic pairs of PCR primers that target the 10 FH exons, as described previously (12). 5 µl of PCR product was run on 1.5% agarose gel (Bioline) to verify specificity to the reaction, and the rest of the products were purified using QIAquick PCR Purification Kit (Qiagen) according to the manufacturer's instructions. The purified PCR products were sequenced using the Sanger method, as previously described (10). Sequence chromatographs were analysed for somatic mutations in 10 FH exons using the Mutation Surveyor software (Softgenetics, State College, PA, USA).
Histological and immunohistochemical (IHC) analysis
Collected fibroid or ANM tissues were fixed for 20 h in neutral buffered formalin, followed by embedding them into paraffin blocks. Six micron thick tissue sections were obtained. Tissue was cleared in xylene and rehydrated in increasing concentrations of ethanol and, finally, water. Haematoxylin and eosin staining was performed using the standard protocol. The IHC protocol followed is described by us in (11). Primary antibody, Ki67 (1:800, ab16667, Abcam, USA), and biotinylated secondary goat anti-rabbit (ready to use; Biogenex, CA, USA), were used. Stained slides were imaged at high resolution on the Aperio Scanscope slide scanner. The gain and exposure time were set constant across tissue samples. Analysis for intensity and the number of cells was done using the Halo Image analysis software (Indica labs, NM, USA).
Statistics
All indicated values are in mean ± SEM and were subjected to unpaired Student's t-test to assess differences between MED12 and MED12 + FH groups with a p-value threshold set to less than 0.05 for statistical significance. All analyses were carried out on Graph Pad Prism 6.0.
Results
MED12 and FH genetic alterations co-exist in uterine fibroids
To determine the status of FH in uterine fibroids, we performed Sanger sequencing and screened for FH mutations in 14 Australian patients. 13 out of 65 fibroids from 14 patients were positive for FH mutations (13/65; 20%; Table 1). In particular, we identified three out of these 14 patients harboured the FH mutation-positive (namely ULM4, ULM7, and ULM8). We have screened all these patients for MED12 aberrations and identified that 60% fibroids (39/65) were positive for MED12 mutations/deletions. The remaining 26 fibroids were found to be MED12 mutation-negative (10).
Table 1.
Summary of somatic MED12 and FH mutations observed in fibroids of various sizes from three patients. Fibroids were classified as small (diameter <2 cm), medium (diameter 2 cm–4 cm), and large (diameter >4 cm) based on their size.
| MED12 mutation status | FH status | ||||||
|---|---|---|---|---|---|---|---|
| Index | Patient | Age (years) | Sample ID | Description | Exon 1 | Exon 2 | Exon 1 |
| 1 | 1 | 56 | ULM 4.1 | Large fibroid | wt | c.115_135del, p.L39_F45del | c.147G > A, p.L28L |
| 2 | 1 | 56 | ULM 4.2 | Fibroid large (2nd site) | wt | c.115_135del, p.L39_F45del | c.147G > A, p.L28L |
| 3 | 1 | 56 | ULM 4.3 | Small fibroid within Large | wt | c.114_134del, p.L39_F45del | c.147G > A, p.L28L |
| 4 | 1 | 56 | ULM 4.4 | Large fibroid intact | wt | c.115_135del, p.L39_F45del | c.147G > A, p.L28L |
| 5 | 1 | 56 | ULM 4.5 | Adjacent normal myometrium | wt | wt | c.147G > A, p.L28L |
| 6 | 1 | 56 | ULM 4.6 | Small fibroid | wt | c.114_134del, p.L39_F45del | c.147G > A, p.L28L |
| 7 | 2 | 56 | ULM 7.1 | Small fibroid | wt | c.131G > T, p.G44V | wt |
| 8 | 2 | 56 | ULM 7.2 | Very small fibroid | wt | c.131G > A, p.G44D | wt |
| 9 | 2 | 56 | ULM 7.3 | Medium fibroid | wt | c.131G > A, p.G44D | wt |
| 10 | 2 | 56 | ULM 7.4 | Medium II fibroid | wt | c.130G > C, p.G44R | wt |
| 11 | 2 | 56 | ULM 7.5 | Very small fibroid | wt | wt | wt |
| 12 | 2 | 56 | ULM 7.5.1 | Very small fibroid | wt | wt | wt |
| 13 | 2 | 56 | ULM 7.6 | Large fibroid | wt | c.126_132del, p.K42_G44del | c.64A > C, p.M1L |
| 14 | 2 | 56 | ULM 7.7 | Adjacent normal myometrium | wt | wt | wt |
| 15 | 2 | 56 | ULM 7.8 | Large fibroid | wt | c.130G > A, p.G44S | wt |
| 16 | 3 | 58 | ULM 8.1 | Adjacent normal myometrium | wt | wt | c.80G > C, p.R6P; c.86T > C, p.L8P; c.88G > C, p.A9P; c.106G > C, p.V15L |
| 17 | 3 | 58 | ULM 8.2 | Large fibroid | wt | wt | wt |
| 18 | 3 | 58 | ULM 8.3 | Small fibroid | wt | c.131G > A, p.G44D | c.80G > C, p.R6P |
| 19 | 3 | 58 | ULM 8.4 | Smallest subserosal fibroid | wt | c.131G > A, p.G44D | wt |
| 20 | 3 | 58 | ULM 8.5 | Large fibroid sub mucosal | wt | wt | c.88G > C, p.A9P |
| 21 | 3 | 58 | ULM 8.6 | Normal adjacent to large fibroid | wt | wt | c.88G > C, p.A9P |
| 22 | 3 | 58 | ULM 8.7 | Midsize fibroid | wt | wt | wt |
| 23 | 3 | 58 | ULM 8.8 | Intral mural midsize fibroid | wt | wt | c.88G > C, p.A9P |
| 24 | 3 | 58 | ULM 8.9 | Mid size intra mural fibroid | wt | wt | c.86T > C, p.L8P; c.88G > C, p.A9P; c.106G > C, p.V15L |
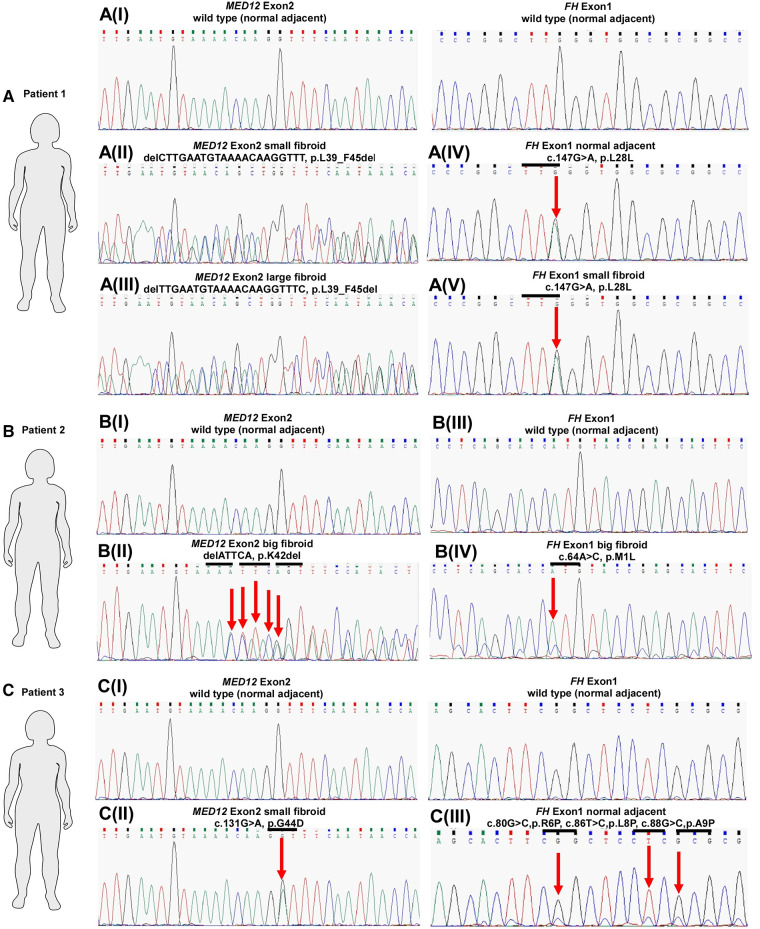
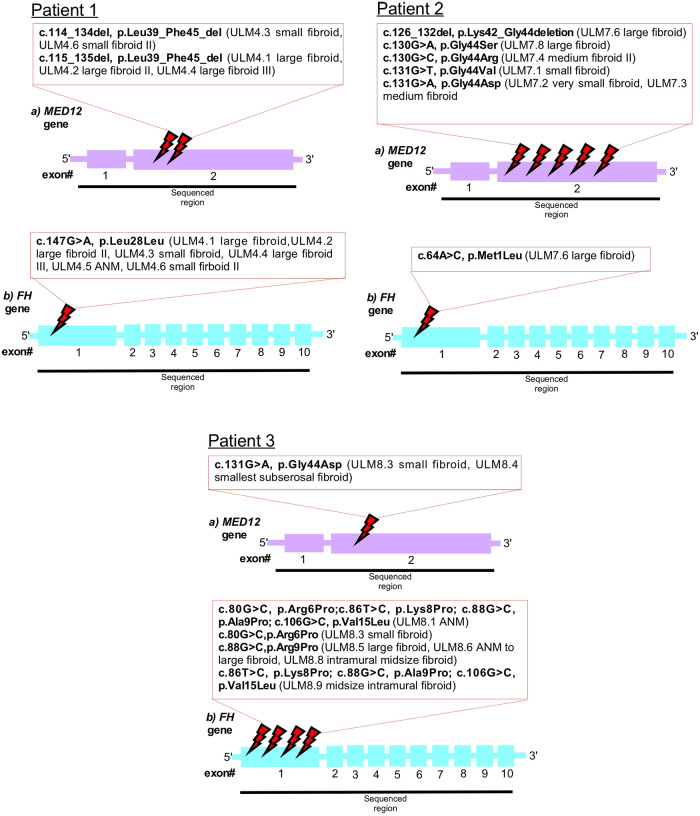
Our genetic analysis highlighted that multiple fibroids and ANM within patient one (ULM4) displayed silent mutations, all affecting codon 28 in exon 1 of FH (Figures 1, 2 and Table 1). Patient one had five fibroids predominantly affected with a deleterious mutation (p.L39_F45del) in MED12 exon 2. No MED12 mutations were present in the patient's ANM tissue DNA (Table 1).
Figure 1.
MED12 and FH mutation status in three patients with uterine leiomyomas. Sequence chromatograms of selected fibroids and corresponding ANM samples in terms of MED12 vs. FH mutation are shown. Codon in MED12 and FH is highlighted by the horizontal bars above the traces. Arrows indicate mutated bases.
Figure 2.
Schematic representation of the MED12 (A) and FH (B) genes. Mutated bases are indicated in the red box. Intronic regions are not drawn.
In Patient two, only one missense mutation was detected in codon M1L of FH exon 1 in the large fibroid (sample ULM7.6) (Figure 1 and Table 1). More specifically, this fibroid tissue displayed deletion mutation (p.K42_G44del) in MED12 exon 2. Other anomalies in MED12 exon 2 (including G44D, G44R, G44S and G44V) are also present in other fibroids but they are not affected by FH (Table 1 and Figure 2).
In Patient three, we observed multiple mutations affecting codon 6, 8, 9 and 15 in FH exon 1 and all of these mutations were also present in the patient's ANM (sample ULM8.1). Missense mutations were also found across various size tumours and these include R6P and A9P (Table 1 and Figure 2). Another hot spot was detected in codons L8P, A9P and V15L in medium size intra mural fibroid (sample ULM 8.9) (Table 1 and Figure 2). Interestingly, one (sample ULM8.3) out of these five tumours with FH mutations also had a missense mutation, affecting codon 44 in exon 2 of MED12 (Figures 1, 2 and Table 1). We sought to analyse the mutation in exon 1 as well across the three patients, and no changes were observed. All of the mutations residing in FH exon 1 had not been previously reported. This analysis confirmed that mutations in FH and MED12 co-exist for the first time in uterine fibroids in Australian women.
Histopathological analysis of uterine fibroids with MED12 and FH mutations
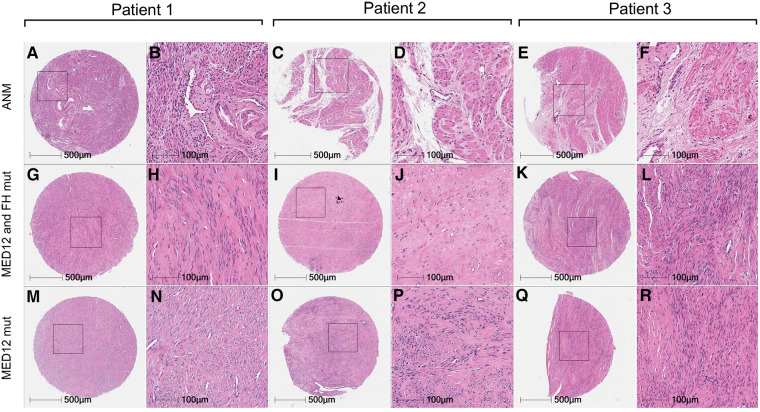
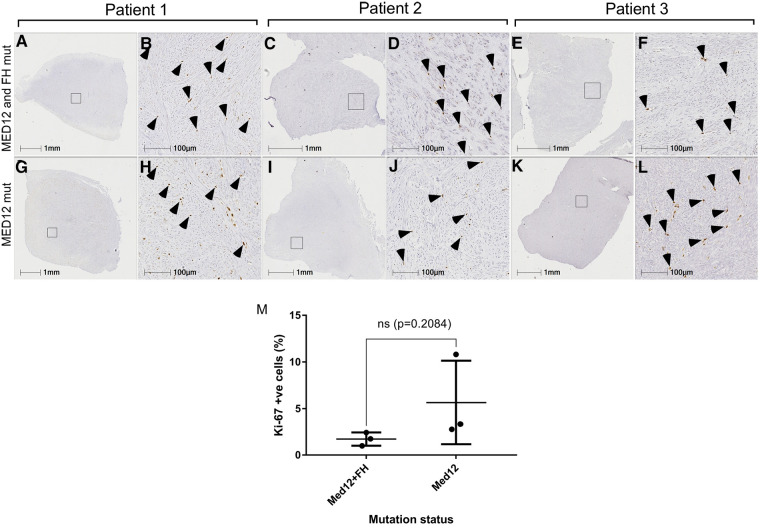
Histological analysis of all fibroids and ANM was performed to reaffirm the clinical diagnosis in patients. All fibroids collected showed an atypical lack of organised muscle bundles, higher amounts of extracellular matrix deposition, compressed muscle fibers, and the presence of single cell type (smooth muscle cells), which are hallmarks of leiomyomas, compared to the defined vasculature and smooth muscle bundles in ANM (Figure 3). No significant differences in the number of smooth muscle cells and amount of extracellular matrix deposition were seen between MED12 mutation-positive (Figures 3M–R) and MED12 + FH mutation-positive (Figures 3M,G–L) fibroids. To understand if driver mutations in more than one gene would give a distinguishable growth advantage, the number of mitotic entities per 20X microscopic field was analysed for both groups by immunohistochemical staining for Ki-67 (Figure 4). The Ki-67+ve cells were capped at below 10% (characteristic of benign tumours). There was a slight increase in the number of mitotic entities per visible field, which was statistically insignificant (p-value = 0.2084).
Figure 3.
Comparative histology of uterine leiomyomas with paired adjacent normal myometrium. Haematoxylin and eosin staining of uterine leiomyomas from three patients with mutations in MED12 only (M–R) vs. MED12 + FH (G–L) in comparison with adjacent normal myometrium (A–E) from the same patients.
Figure 4.
Mitotic index of MED12 mutation vs. MED12 and FH mutation leiomyomas. Ki-67 staining for MED12 mutation (G–L) and MED12 and FH mutation (A–F). Arrowheads indicate the Ki-67 positive cells. No significant difference was observed in the percentage of Ki-67 positive cells between these two groups (M).
Discussion
Uterine fibroids are benign tumours that affect the reproductive health of millions of women (13–15) It is a main cause of infertility and hysterectomy (3, 15). The majority of the tumours (more than 70%) are identified by specific mutations in MED12 exon 2 irrespective of ethnicity (16). On the other hand, there are other driver mutations, including HMGA2, FH and COL4A5-COL4A6 (6). While these four gene alterations are shown to be mutually exclusive in some patients (17, 18), there have been other reports involving HMGA1 or 7q aberrations of CUX1 gene that co-occur with MED12 mutations (17). Holzmann and colleagues described a patient with MED12 mutations and HMGA12 rearrangements in her leiomyomas (19). Similarly, a recent study in the United States confirmed a patient carrying both driver mutations affecting FH and MED12 in a single fibroid (20).
The aim of this study was to search for somatic mutations in FH gene in MED12 mutation-positive and mutation-negative uterine fibroids of Australian women. Altogether 65 uterine fibroids and corresponding ANM tissues from 14 patients were successfully analysed using Sanger sequencing method. Three out of 14 patients displayed somatic mutations in FH exon 1 with co-existing mutations in MED12.
In Patient One, a deletion mutation L39_F45 in MED12 exon 2 were detected in all of the fibroid tissues except the ANM (Table 1 and Figure 2). This agrees with previous studies that reported two mutation types in MED12 exon 2 (including missense and inframe insertion-deletion mutations) identified in the human tumours (4, 16). Our genetic analysis investigation on MED12 exon 2 mutations also found both. The effects of this mutation (L39_F45 deletion), which is located at the beginning of MED12 region, are likely to impair protein-protein interactions between MED12 and Cdk8-Cdk19 and Cyclin C, which ultimately leads to the inhibition of the transcription. This statement was supported by previous expression profiling and functional studies that mutant MED12 protein showed a weak binding affinity with Cdk8 and Cyclin C compared to wild-type MED12 (19, 20). Apart from the deletion mutation that exists in this patient, we found another silent mutation affecting codon 28 in exon 1 of FH in this patient's normal tissue DNA. A similar change (L28L) in this region of FH exon 1 was present in all fibroid tissue. This is a novel and rare type of mutation in FH that has not been reported before in uterine fibroids and other FH-related diseases. Taken together (L39_F45 deletion in MED12 and L28L missense in FH found in this patient), we suspect a potential involvement of these mutations that promote tumorigenesis through their distinct mechanisms.
In Patient Two, we found three missense mutations affecting codon G44D, G44R, G44S and a deletion mutation K42_G44 in MED12 exon 2 in the medium and large size fibroid, respectively (Table 1 and Supplementary Figure S1). All of these changes were clustered around codon 44, which is the frequent hotspot for MED12 mutation, and again, this N-terminal site (start of 100 amino acids, encoded by exon 1 and 2 in MED12) of MED12 is essential for protein-protein interaction. Notably, one (sample ULM7.6) out of these four MED12 mutation-positive fibroid also have missense mutation (M1L) in exon 1 of FH. In the case of this patient, it sets the foundation for further investigation to analyse the protein expression patterns in FH mutation-positive and mutation-negative uterine fibroids and relate to MED12. The outcome of this investigation will lead to the identification of protein expression patterns associated with MED12 and FH as biomarkers for diagnosing uterine fibroids. For FH-negative mutation fibroids (in ULM7.1, ULM7.2, ULM7.3, ULM7.4 and ULM7.8) observed in this patient, the presence of other genetic mutations such as MED12 are still active in the subset of these fibroids, and the MED12 mutation alone can drive tumour formation (Table 1 and Figure 2). This evidence is in line with a mouse model study that the expression of missense mutant Med12 can be the sole cause of uterine fibroids (23). Consequently, the impact of other driver genetic mutations, including HMGA2 and COL4A5-COL4A6, on fibroid protein expression requires further investigation.
In Patient Three, we saw majority of the various size fibroids and ANM harbouring either missense and multiple mutations affecting codon R6P, L8P, A9P and V15L in FH exon 1 (Table 1). Again, this is the third patient in our study which reported mutation in the N-terminal of FH exon 1. This first exon of FH encodes the mitochondrial signal peptide of fumarase that targets the proteins to the mitochondria (24). It is tempting to speculate that the functional consequences of mutation in this FH region may potentially abolish mitochondrial targeting and import and subsequently lead to the loss of enzymatic activity. Ongoing and future research should explore this particular region exon 1 of FH and relate to the uterine fibroid. As opposed to the FH exon 1 mutation, we detected two small fibroids with a missense mutation, affecting codon 44 in exon 2 of MED12, and one of them carries the FH mutation (Figure 1 and Table 1).
Combining these three-patient case studies proves that MED12 and FH mutations co-exist, possibly driving and competing for the same roles in tumorigenesis of uterine fibroid. From the immunopathological analysis carried out on our cohort, no distinct mitotic advantage was seen between fibroids with mutations in MED12 only or MED12 + FH genes. Alternatively, these two genes, MED12 and FH, are also likely to act independently irrespective of MED12 or FH pathway, as discussed by another group. Various germline FH mutations have been reported previously in women patients associated with HLRCC syndrome (25). Missense mutations appear to be more frequently detected mutations in FH than frameshift, nonsense and splice site in the HLRCC patients (26). This is, to our knowledge the first evidence and genetic annotation of fibroids in Australian women that harbor both somatic MED12 and FH mutation.
Funding Statement
This work is in part supported by funding from the National Health and Medical Research Council, the Australian Research Council, and the Cancer Institute NSW.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving human participants were reviewed and approved by University of Newcastle Human Research Ethics Committee. The patients/participants provided their written informed consent to participate in this study.
Author contributions
PST: Designed research. MFBJ, PBN, Y-AK, PB: Performed research. RJS, PN: Contributed new reagents. MFBJ: Analyzed data. MFBJ, PST: Wrote the paper. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frph.2023.1081092/full#supplementary-material.
References
- 1.Commandeur AE, Styer AK, Teixeira JM. Epidemiological and genetic clues for molecular mechanisms involved in uterine leiomyoma development and growth. Hum Reprod Update. (2015) 21:593–615. 10.1093/humupd/dmv030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navarro A, Bariani MV, Yang Q, Al-Hendy A. Understanding the impact of uterine fibroids on human endometrium function. Front Cell Dev Biol. (2021) 9:633180. 10.3389/fcell.2021.633180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardozo ER, Clark AD, Banks NK, Henne MB, Stegmann BJ, Segars JH. The estimated annual cost of uterine leiomyomata in the United States. Am J Obstet Gynecol. (2012) 206:211.e1–9. 10.1016/j.ajog.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, et al. MED12, The mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. (2011b) 334:252–5. 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- 5.Mehine M, Kaasinen E, Makinen N, Katainen R, Kampjarvi K, Pitkanen E, et al. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med. (2013) 369:43–53. 10.1056/NEJMoa1302736. [DOI] [PubMed] [Google Scholar]
- 6.Mehine M, Kaasinen E, Heinonen H-R, Mäkinen N, Kämpjärvi K, Sarvilinna N, et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc Natl Acad Sci USA 113, 1315–20. 10.1073/pnas.1518752113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kampjarvi K, Makinen N, Mehine M, Valipakka S, Uimari O, Pitkanen E, et al. MED12 mutations and FH inactivation are mutually exclusive in uterine leiomyomas. Br J Cancer. (2016) 114:1405–11. 10.1038/bjc.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark AD, Oldenbroek M, Boyer TG. Mediator kinase module and human tumorigenesis. Crit Rev Biochem Mol Biol. (2015) 50:393–426. 10.3109/10409238.2015.1064854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. (2002) 30:406–10. 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 10.Jamaluddin MFB, Ko YA, Kumar M, Brown Y, Bajwa P, Nagendra PB, et al. Proteomic profiling of human uterine fibroids reveals upregulation of the extracellular matrix protein periostin. Endocrinology. (2018) 159:1106–18. 10.1210/en.2017-03018. [DOI] [PubMed] [Google Scholar]
- 11.Ko YA, Jamaluddin M, Adebayo M, Bajwa P, Scott R, Dharmarajan A, et al. Extracellular Matrix (ECM) activates beta-catenin signaling in uterine fibroids. Reproduction. (2017) 155:61–71. 10.3109/10409238.2015.1064854 [DOI] [PubMed] [Google Scholar]
- 12.Kiuru M, Lehtonen R, Arola J, Salovaara R, Jarvinen H, Aittomaki K, et al. Few FH mutations in sporadic counterparts of tumor types observed in hereditary leiomyomatosis and renal cell cancer families. Cancer Res. (2002) 62:4554–7. [PubMed] [Google Scholar]
- 13.Baird DD, Dunson DB, Hill MC, Cousins D, Schectman JM. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am J Obstet Gynecol. (2003) 188:100–7. 10.1067/mob.2003.99. [DOI] [PubMed] [Google Scholar]
- 14.Stewart EA, Laughlin-Tommaso SK, Catherino WH, Lalitkumar S, Gupta D, Vollenhoven B. Uterine fibroids. Nat Rev Dis Primers. (2016) 2:16043. 10.1038/nrdp.2016.43. [DOI] [PubMed] [Google Scholar]
- 15.Cheng LC, Li HY, Gong QQ, Huang CY, Zhang C, Yan JZ. Global, regional, and national burden of uterine fibroids in the last 30 years: estimates from the 1990 to 2019 global burden of disease study. Front Med. (2022) 9:1003605. 10.3389/fmed.2022.1003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makinen N, Heinonen HR, Moore S, Tomlinson IP, Van Der Spuy ZM, Aaltonen LA. MED12 exon 2 mutations are common in uterine leiomyomas from South African patients. Oncotarget. (2011a) 2:966–9. 10.18632/oncotarget.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markowski DN, Bartnitzke S, Loning T, Drieschner N, Helmke BM, Bullerdiek J. MED12 mutations in uterine fibroids–their relationship to cytogenetic subgroups. Int J Cancer. (2012) 131:1528–36. 10.1002/ijc.27424. [DOI] [PubMed] [Google Scholar]
- 18.Hodge JC, Kim TM, Dreyfuss JM, Somasundaram P, Christacos NC, Rousselle M, et al. Expression profiling of uterine leiomyomata cytogenetic subgroups reveals distinct signatures in matched myometrium: transcriptional profilingof the t(12;14) and evidence in support of predisposing genetic heterogeneity. Hum Mol Genet. (2012) 21:2312–29. 10.1093/hmg/dds051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holzmann C, Markowski DN, Bartnitzke S, Koczan D, Helmke BM, Bullerdiek J. A rare coincidence of different types of driver mutations among uterine leiomyomas (UL). Mol Cytogenet. (2015) 8:76. 10.1186/s13039-015-0177-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bateman NW, Tarney CM, Abulez T, Soltis AR, Zhou M, Conrads K, et al. Proteogenomic landscape of uterine leiomyomas from hereditary leiomyomatosis and renal cell cancer patients. Sci Rep. (2021) 11(1):9371. 10.1038/s41598-021-88585-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turunen M, Spaeth JM, Keskitalo S, Park MJ, Kivioja T, Clark AD, et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep. (2014) 7:654–60. 10.1016/j.celrep.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kampjarvi K, Park MJ, Mehine M, Kim NH, Clark AD, Butzow R, et al. Mutations in exon 1 highlight the role of MED12 in uterine leiomyomas. Hum Mutat. (2014) 35:1136–41. 10.1002/humu.22612. [DOI] [PubMed] [Google Scholar]
- 23.Mittal P, Shin Y-H, Yatsenko SA, Castro CA, Surti U, Rajkovic A. MED12 gain-of-function mutation causes leiomyomas and genomic instability. J Clin Invest. (2015) 125:3280–4. 10.1172/JCI81534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen H, Engelbrecht J, Brunak S, Von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. (1997) 10:1–6. 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Bayley J-P, Launonen V, Tomlinson IPM. The FH mutation database: an online database of fumarate hydratase mutations involved in the MCUL (HLRCC) tumor syndrome and congenital fumarase deficiency. BMC Med Genet. (2008) 9:20. 10.1186/1471-2350-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardie B, Remenieras A, Kattygnarath D, Bombled J, Lefevre S, Perrier-Trudova V, et al. Novel FH mutations in families with hereditary leiomyomatosis and renal cell cancer (HLRCC) and patients with isolated type 2 papillary renal cell carcinoma. J Med Genet. (2011) 48:226–34. 10.1136/jmg.2010.085068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.