Abstract
Background
Genetic variants in APOL1 are a major contributor to the increased risk of kidney disease in people of recent African ancestry.
Summary
Two alleles in the APOL1 gene, referred to as G1 and G2, confer increased risk of kidney disease under a recessive model of risk inheritance. Disease risk is inherited as a recessive trait: People with genotypes G1/G1, G2/G2, and G1/G2 (i.e., a risk allele from each parent) have increased risk for what we refer to here as APOL1-associated kidney disease. In the USA, about 13% of the self-identified African-American population has a high-risk genotype. As we discuss below, APOL1 is an unusual disease gene. Most studies to date have suggested that the G1 and G2 variants have toxic, gain-of-function effects on the encoded protein.
Key Message
In this article, we review key concepts critical to understanding APOL1-associated kidney disease, emphasizing ways in which it is highly atypical for a human disease-causing gene.
Keywords: APOL1, Kidney, Genetic, FSGS
Introduction
Two alleles in the APOL1 gene, each leading to an altered apolipoprotein-L1 protein sequence, confer greatly increased risk of kidney disease under a recessive model of risk inheritance [1, 2]. In what is now commonly used nomenclature, G1, the first of these alleles, refers to a pair of coding polymorphisms (or cSNPs, p.S342 and p.I384M) that are almost always inherited together; G2, the second disease-associated APOL1 allele, refers to a six nucleotide deletion that leads to the in-frame loss of two amino acids (p.N388 and p.Y389; see Fig. 1). We refer to the alleles lacking G1 or G2 as G0, which we view as preferable to the term “wild type.” For our purposes, we can limit ourselves largely to discussions of G0, G1, and G2, although there are additional coding and noncoding variants in APOL1 that further subdivide these haplotypes. (Note that one group has given the name G3 to another particular APOL1 haplotype that lacks the G1 or G2 defining variants − this allele is not associated with kidney disease [3].)
Fig. 1.
APOL1 protein and genotypes. Major APOL1 domains are named based on their role in trypanolysis. The APOL1 risk variants are located near the C-terminus in the serum resistance antigen (SRA)-binding domain. The membrane-addressing domain is a pH-sensitive switch that facilitates membrane insertion sequences of the G0, G1, and G2 APOL1 alleles. The SRA-binding domain forms a coiled-coil structure with a leucine zipper motif. Among African Americans, the allele frequency of G1 is approximately 23%, and G2 is approximately 15%. Two risk alleles (one inherited from each parent) are required for the APOL1 high-risk genotype that greatly increases risk of kidney disease.
The APOL1 locus and the specific variants associated with kidney disease were identified by studies designed to identify genetic contributions to the large racial disparity in rates of kidney disease (specifically, the high rate among African Americans). Now that the specific variants driving disease risk have been identified, we know that the G1 and G2 variants are more accurately described as ancestry-associated than race-associated. These variants arose in sub-Saharan Africa. Because people who identify as Black or African American are enriched for west African ancestry, they have much higher rates of these alleles than other commonly defined racial or ethnic groups.
Disease risk is inherited as a recessive trait: People with genotypes G1/G1, G2/G2, and G1/G2 (i.e., a risk allele from each parent) are the people at risk for what we refer to as APOL1-associated kidney disease (or, alternatively, APOL1-mediated kidney disease). In the USA, about 13% of the self-identified African-American population has a high-risk genotype. Among African-Americans with nondiabetic kidney disease, this number is on the order of 50% (see details below), illustrating the enormous impact of this gene.
APOL1 is but one contributor to the racial disparity in rates of kidney disease. Furthermore, an individual need not identify as African-American to inherit disease-associated APOL1 risk genotypes. We view it as important progress that we are able to better dissociate ancestry-associated and genetically driven kidney disease from other, largely social determinates of kidney health.
As we hope to illustrate below, APOL1 is an unusual disease gene. We review some key concepts critical to understanding APOL1-associated kidney disease, with an emphasis on ways in which it is atypical for a human disease-causing gene (see Table 1).
Table 1.
Unusual features of APOL1-associated kidney disease
| Common variants with large effect size |
| Both recessive and gain of function |
| Wide spectrum of associated phenotypes |
| Unique genetic testing issues |
| No clear role for APOL1 in normal kidney physiology |
Recessive Gain of Function
APOL1-associated kidney disease (also often referred to as APOL1-associated nephropathy, APOL1-mediated kidney disease) follows an essentially recessive pattern of inheritance. Individuals with two risk alleles (genotypes G1/G1, G1/G2, and G2/G2) are the individuals at high risk of kidney disease. Odds ratios vary widely, depending on the specific kidney phenotype being considered. For focal segmental glomerulosclerosis (FSGS), perhaps the prototypical form of APOL1-asociated kidney injury, odds ratios are approximately 15- to 20-fold higher for those people with high-risk APOL1 genotypes than those without. There appears to be a very small increase in disease risk for people heterozygous for risk alleles, but this has not been observed in all studies, and appears to be most relevant for the specific phenotypes that are most strongly associated with APOL1 genotype, such as HIV-associated nephropathy (HIVAN) and COVID19-associated nephropathy [4, 5]. Even when this heterozygous-associated disease risk has been observed, the effect is dwarfed in comparison to the risk associated with two risk alleles. For all practical purposes, it the genotypes G0/G0, G0/G1, and G0/G2 can all be considered “low risk.”
As a general rule, we tend to think of recessive diseases as childhood onset, loss-of-function diseases. By contrast, we tend to think of gain-of-function mutations as causing dominant diseases. While this is an oversimplification, it holds true for most inherited diseases. To contrast APOL1-associated kidney disease with other inherited kidney diseases, Bartter's syndrome is a collection of mostly recessive disease characterized by loss of an essential thick ascending limb ion transport function [6]. By contrast, the one subtype of Bartter syndrome known to be inherited as a dominant trait is caused by gain-of-function mutations in the calcium sensing receptor (CASR): mutations make the encoded receptor hyperactive rather than inactive [7].
Why do we think that APOL1-associated nephropathy is not the result of loss of normal APOL1 function, similar to what is seen in most recessive disorders? One piece of evidence comes from an observation in a single human [8]. A healthy man in rural India was described several years ago who developed a Trypansoma evansi infection and was found to have inactivating (loss-of-function) mutations in the APOL1 gene that prevented production of a normal APOL1 protein. While his susceptibility to this infection was presumably due to a loss of the anti-infective properties of APOL1, he had normal kidney disease (confirmed again many years later), demonstrating that APOL1 is not required for normal kidney function or kidney development [9].
Furthermore, APOL1 is a relatively new and rapidly changing gene [10]. Most organisms lack an APOL1 gene. This includes most mammals, including our favorite mammalian models the rat and the mouse. Chimpanzees, our closest relative, lack a functional APOL1 gene. The absence of APOL1 is clearly compatible with the presence of normal mammalian kidneys. Those primates with no APOL1 do not have worse kidney function than primates with an APOL1 gene.
Are the G1 and G2 alleles gain-of-function? Many groups of investigators have expressed APOL1 in various types of cultured cells. In most (but not all) cases, expression of the G1 and G2 forms of APOL1 in cultured cells cause increased cell death [11−18]. In studies using various cell types, including HEK293 cells, conditionally immortalized and primary podocytes, and various methods of APOL1 expression, apoptotic, autophagic, and pyroptotic cell death have been reported, generally much greater with the G1 and G2 forms of APOL1 than G0 [19].
What is the nature of this gain-of-function effect? Specifically, what are the mechanisms by which the G1 and G2 variants alter the function of kidney cells? APOL1 has an N-terminal pore-forming domain and a BH3- only domain, which in theory may be capable of driving increased cytotoxicity, but cell death has not been shown to be caused by these functional elements. It is widely thought that the podocyte is the main target of APOL1 toxicity, given the strong association between podocyte alterations and FSGS. However, the wide spectrum of APOL1 kidney disease presentation suggests that other cell types may also be primary targets of APOL1 toxicity. A variety of models of APOL1-mediated cell toxicity have been proposed.
We should not assume that one and only one pathway drives APOL1-associated toxicity. The presence of G1 and G2 variants may alter the biology of APOL1 in multiple ways. Human APOL1 has no essential role in kidney function that has been identified, and it is therefore not easy to point to one particular activity in the kidney that might be gained or lost in the presence of genetic variation.
The APOL1 protein can function as a cell membrane pH-gated pore or channel. This was first observed in the context of trypanosomes, in which human APOL1 can form membrane pores [20, 21]. APOL1 was reported to form anion channels in lysosomal membranes, leading to membrane depolarization and osmotic swelling, and ultimately trypanosome death [21, 22]. Studies of APOL1-induced membrane permeability to anions and cations have been somewhat inconsistent, though more recent evidence favors the primacy of cation flux, especially at neutral pH values. Thomson and Finkelstein showed that recombinant APOL1 protein produces solely cation conductances in lipid bilayers. Induction of this activity through membrane insertion was found to require acid pH, and was subsequently enhanced by raising the pH to 7.1 [23].
The ability of APOL1 to form channels in trypanosome membranes suggested that APOL1 may also form channels in human cells. Expression of either G1 or G2 forms of APOL1 in HEK293 cells was observed to cause cell swelling and cell death, much more than with G0-expressing cells, which was still more than seen in control cells. This cytotoxicity was preceded by G1 or G2 APOL1-induced net efflux of intracellular potassium [13]. Studies by Bruno and Edwards showed that APOL1 can confer pH-sensitive chloride permeability to phospholipoid membranes. At higher pH, potassium permeability is activated [24]. Giovanazzo et al. [25] demonstrated a population of active cation channels at the plasma membrane of human cells expressing the G1 or G2 APOL1 variants. Additional studies have shown that a transmembrane region close to the APOL1 C terminus functions as the pore-lining segment of the channel [26]. Localization of APOL1 at both the plasma membrane and the endoplasmic reticulum (ER) of podocytes has been convincingly demonstrated [27]. Studies of APOL1 membrane topology in the plasma membrane suggest that the N- and C-termini are extracellular or intraluminal (in intracellular organelles) [28, 29]. Other studies have suggested that the orientation of APOL1 in cell membranes may be variable, perhaps depending in part on the specific splice variant [30].
In trypanosomes, APOL1 has been shown to induce mitochondrial membrane permeabilization, suggesting that similar effects may be present in human cells [22]. Using HEK293 cells, Ma et al. [31] found that G1 and G2 expression led to impaired mitochondrial function and a large reduction in respiration, and mitochondrial membrane potential. Shah et al. [32] found that the APOL1 G0 and risk variant proteins shared the same import pathway into the mitochondria, but once inside, risk variant APOL1 was prone to forming higher-order oligomers and lead to opening of the mitochondrial permeability transition pore.
As noted, APOL1 associates with various cell membranes, including the podocyte ER [18, 30, 33]. Some studies have found an increase in ER stress in cells expressing APOL1 risk variants. Wen et al. found that podocytes expressing G1 or G2 APOL1 demonstrated biochemical indicators of ER stress as well as impaired processing of the slit diaphragm proteins podocin and nephrin. ER stress inhibitors reduced these effects [34]. Similarly, inhibition of ER stress was found to rescue APOL1-mediated toxicity in a drosophila model of APOL1-associated disease [35]. APOL1 may adopt both C-terminal in and C-terminal out conformations in the ER membrane [30].
APOL1 encodes a protein associated with circulating lipids. A functionally important interaction with intracellular lipids thus seems quite plausible. A recent study reported that APOL1 G1 expression in a bacterial artificial chromosome (BAC) transgenic mouse model results in increased triglyceride and cholesterol ester in kidney cortex [36]. The presence of an APOL1 risk variant appeared to increase the susceptibility of mice to lipid-dependent podocyte injury. Another study in these mice found that the presence of APOL1-G1 and APOL1-G2 risk variants in mice impaired reverse cholesterol transport through decreased expression of cholesterol efflux transporters in macrophages and postulated a role for these cells in driving kidney inflammation [37]. While a large fraction of risk variant APOL1 (G1 and G2) localizes to the ER, a significant proportion of wild-type APOL1 (G0) was found to localize to lipid droplets (LD) in one recent study [38]. Treatment of cells that promote LD formation with oleic acid shifted the localization of G1 and G2 from the ER to LDs, with accompanying reduction of autophagic flux and cytotoxicity.
Uzereau et al. reported that both truncation of the C-terminal helix of APOL1 and deletion of APOL3 induce reorganization of podocyte actomyosin activity. APOL3 was also reported to stimulate PI(4)P synthesis by PI4KB. The G1 and G2 forms of APOL1 were found to exhibit increased binding to APOL3, leading to a model whereby these variants inhibit APOL3 stimulation of PI4KB, with downstream consequences for cytoskeletal function [39]. This is consistent with the observation that kidney disease risk in humans associates with a null mutation in APOL3, independent of APOL1 genotype [40].
APOL1 Is a New Gene
Most human disease genes are shared across mammalian species. For example, the podocyte disease genes NPHS1, NPHS2, INF2, and ACTN4 all have very close homologs among mammals. APOL1 is different. APOL1 is one of the six APOL genes on human chromosome 22 that arose from a series of gene duplication events [41, 42]. APOL1 appeared late − as APOL1 is found only in primates, it likely arose less than 50 million years ago. Humans have six APOL genes. APOL1 is the best studied of these, and the only family member with a secreted as well as intracellular form [41, 43]. Some species (mice, for example) have more APOL genes than do humans. The human APOL1 and APOL3 proteins interact physically [39, 40]. Multiple members of the human ApoL1 protein family function as ion channels, albeit with somewhat different properties [44]. The genetic and functional interactions of these other APOLs with APOL1 have not been fully assessed.
An N-terminal signal peptide, present on some splice variants, allows for APOL1 export from the cell [10]. In contrast to APOL2 through 6, APOL1 circulates at high levels in blood, in association with HDL [45]. APOL1 is expressed widely. The circulating APOL1 is made chiefly by the liver [46]. Most of the APOL1 splice variants do not encode signal peptides. It is generally thought that it is the intracellularly expressed APOL1 that is responsible for mediating disease [19].
The G1 and G2 alleles are found only in people with recent (i.e., <10,000 years) west sub-Saharan African ancestry [1]. These risk variants show increased activity against certain African trypanosomes, suggesting that they provided a survival advantage compared with the G0 forms. In fact, examination of populations of modern G0, G1, and G2 haplotypes suggests that positive natural selection has been acting on the G1 and G2 alleles [47, 48].
The APOL1 high-risk variants are both common and powerful. This is unusual. People with heterozygosity for G1 or G2 exhibit enhanced activity against trypanosome infection, which is presumed to account for the fairly quick rise in the frequency of these variants over the past 5,000–10,000 years [1, 49]. Presumably, this led to a survival advantage in regions of Africa where the pathogenic trypanosomes are common.
This is reminiscent of sickle cell disease, another ancestry-associated human disorder, where homozygosity for a specific hemoglobin (HBB) mutation leads to sickle cell disease, but heterozygosity provides some protection against malaria [50]. In the case of sickle cell disease, there is evidence for balancing selection, such that equilibrium was reached a few thousand years after the mutation originated [51]. The APOL1 G1 and G2 variants are thought to have arisen ∼5,000–10,000 years ago, but in the case of APOL1, it is not known if equilibrium has been reached and selection is now balanced [52]. In most part of the world, APOL1 variants are more common than the sickle mutation in HBB. However, sickle cell disease is essentially Mendelian, with a much higher penetrance than seen for APOL1-associated kidney disease. In the case of individuals with two variant APOL1 risk alleles, most will never develop ESRD or overt kidney disease.
Common, Large Effect
The vast majority of common genetic variants that affect risk of common human disease phenotypes are in noncoding DNA sequence. Each of these has, individually, a very small effect on overall risk [53]. This holds for common phenotypes of interest to nephrologists, such as diabetic nephropathy and hypertension [54, 55]. (We note that there are also very rare Mendelian forms of hypertension and diabetes.) Large-effect gene variants are typically rare, because of natural selection. As a general rule, highly penetrant disease variants that affect the risk of a serious disease (as in Mendelian diseases) do not reach high frequency. By contrast, APOL1 has a very large effect on disease risk and is common, particularly in people of recent African ancestry. In the case of APOL1, there are both positive and negative selective pressures.
In Africa, where these risk variants originated, these alleles are present with very high frequency in West Africa, in contrast lower frequencies in east and southern Africa [52]. Some groups in parts of West Africa have combined risk variant allele frequencies exceeding 50% [56]. Despite the high frequency, effect sizes are large in studies in groups with significant recent African ancestry. The effect size is particularly strong in HIVAN. Before effective HIV therapy became widespread, approximately 50% of people with high-risk APOL1 genotypes and HIV infection developed HIVAN. It is noteworthy that in eastern African countries like Ethiopia, the G1 and G2 frequencies are much rarer than in West Africa and HIVAN rates have historically been very low [57].
Although the APOL1-associated effect sizes are large, most people with high-risk APOL1 genotypes do not develop overt kidney disease. Thus, other factors besides APOL1 genotype must be necessary for disease to develop (modifiers or “second hits”). These may include genetic and nongenetic factors (Table 2). The best-defined modifier of disease is a high interferon state, discussed below.
Table 2.
Modifiers (“second hits”) that may be involved in the development of overt kidney disease in people with high-risk APOL1 genotypes
| Category | Factor | Notes | Reference(s) |
|---|---|---|---|
| Genetic | APOL1 | Rare APOL1 duplications may modify the high-risk-associated phenotype. APOL1 risk-variant toxicity is affected by APOL1 haplotype | [58, 59] |
|
| |||
| Genetic | APOL3 | Deletion mutations in APOL3 increase risk of nephropathy. APOL1 and APOL3 interact physically | [40] |
|
| |||
| Genetic | UBD | APOL1 and UBD are coregulated in the glomerulus. UBD genotypes associated with lower UBD expression may correlate with the risk of APOL1-nephropathy | [60, 61] |
|
| |||
| Genetic | GSTM1 | Individuals APOL1-nephropathy and null GSTM1 alleles have increased rate kidney disease progression | [62] |
|
| |||
| Pharmacologic | Therapeutic IFN | FSGS and collapsing nephropathy have been seen almost exclusively in people with high-risk APOL1 genotypes | [63, 64] |
|
| |||
| Infectious | HIV | HIVAN is extremely rare in people without high-risk APOL1 genotypes | [65, 66] |
|
| |||
| Infectious | COVID-19 | COVAN has been reported almost exclusively in people with high-risk APOL1 genotypes | [67] |
|
| |||
| Inflammatory | Lupus | Greater risk of ESKD and collapsing lupus nephritis | [68, 69] |
Many Phenotypes, Immune and Nonmmune
High-risk APOL1 genotypes associate with a large spectrum of different kidney disease phenotypes. Two risk variants confer an odds ratio of approximately 7–10 for hypertension-associated ESKD, approximately 17 for FSGS, and approximately 29–89 for HIV nephropathy [65, 70]. It is striking that the G1 and G2 alleles are strong risk factors for what are thought to be a largely vascular disease (hypertension-attributed ESKD) which is neither infectious nor immune mediated, and diseases of the podocyte with clear infectious etiologies such as HIV-associated nephropathy. It thus seems that these different disease presentations are driven by similar mechanisms. Thus, for many purposes, it is better to think of these entities as different manifestations of APOL1-associated nephropathy spectrum than as separate diseases. APOL1 risk genotypes are associated with higher rates of ESKD in individuals with lupus nephritis [71]. Collapsing glomerulopathy can also be driven by therapeutic interferon administration. This effect is found almost exclusively in high-risk APOL1 individuals [63, 64].
COVID-19-associated collapsing glomerulopathy (recently named COVAN) is the newest addition to the list of APOL1-associated diseases [5]. The vast majority of cases of COVAN have been reported in individuals with two APOL1 risk alleles. At least one case has been observed in a heterozygous individual. Interestingly, one case of COVAN has been observed in the engrafted kidney of a transplant patient, where the kidney came from a donor who carried a low-risk APOL1 genotype [67].
Interferons markedly upregulate the expression of APOL1 RNA [11, 63]. APOL1 also responds to lipopolysaccharide, toll-like receptor agonists, tumor necrosis factor, and other cytokines [63, 72]. One likely model of disease pathogenesis is that both the high-risk APOL1 genotype and high APOL1 expression act together to cause overt disease. Associations between virus infection and APOL1-associated kidney disease also support the notion that viruses can activate APOL1 expression, leading to kidney injury [73, 74].
The effect of APOL1 risk genotype varies by age. For young people, when rates of nephropathy are typically low, the odds ratios conferred by APOL1 variants are very large, as is observed with FSGS. In later adulthood, APOL1 genotype confers a much lower odds ratio for incident kidneys, in part because the baseline disease rate is higher at older age, and because a substantial fraction of high-risk genotype people has already developed kidney disease. For example, in the Atherosclerosis Risk in Communities (ARIC) study that included participants between ages 45 and 64 years without baseline CKD, APOL1 risk genotypes confer an odds ratio for incident CKD of approximately 1.5 [75, 76]. Much higher odds ratios for APOL1 tend to be observed with younger populations and in studies with concrete end points (e.g., biopsy-proven FSGS or ESKD). While the risk of ESKD associated with high-risk genotypes was initially reported in African Americans, these genotypes are also highly associated with kidney disease in Africans, Caribbean Islanders, and South Americans with significant recent African ancestry [56, 57, 77, 78].
While APOL1 genotype is associated with a wide range of kidney phenotypes, it is interesting and perhaps surprising that it does not associate with incipient kidney disease in diabetics [79]. However, APOL1 risk genotypes do associate with faster rates of CKD progression, whether caused by diabetic nephropathy or some other etiology [80]. Similarly, APOL1 genotype is not associated with the risk of IgA nephropathy [81]. These observations suggest that not all kidney-modifying factors serve as “second hits” in the development of APOL1-assicated kidney disease.
The role of APOL1 in mediating the risk of nonrenal phenotypes is less clear. Risk of preeclampsia (which is arguably a kidney disease) has been studied as a function of APOL1. Mothers carrying fetuses with high-risk APOL1 genotype were found to have a higher risk of preeclampsia than those with low-risk genotype fetuses [82]. APOL1 high-risk genotype has been reported to associate with various cardiovascular phenotypes, including atherosclerotic disease, elevated blood pressure, coronary artery disease, and cerebral vascular disease [83−86]. The relatively weak and inconsistent associations that have been reported may be mediated by the effect of APOL1 on increasing the incidence of subclinical kidney disease, rather than the cardiovascular system per se.
Implications for Modeling Disease
The development of animal models of APOL1-associated kidney disease is not as straightforward as for many inherited human diseases. Typically, the high homology between most human and mouse genes allows human point mutations to be put in the relevant mouse gene. This is not possible for APOL1, as mice lack an APOL1 homology. This issue is present for the other commonly used model organisms as well. Investigators have therefore used a variety of creative strategies in different organisms to model aspects of disease.
Drosophila lack a mammalian kidney-like excretory organ, but cells known as nephrocytes that are found around the heart and esophagus have podocyte-like properties and lead to sequestration rather than excretion of filtrate [87]. Two independent studies reported experimental expression of APOL1 in drosophila nephrocytes, reporting that expression of risk variant forms of APOL1 lead to hypertrophy of these cells, together with an increased rate of cell death [15, 16].
Recent studies in a similar model found that expression of risk variant APOL1 elevates the endocytic function of garland cell nephrocytes together with signs of cell death. G0 APOL1 had a milder effect. Endoplasmic reticulum (ER) swelling and induction of chaperones suggested an ER stress response. Inhibition of ER stress decreased this APOL1-mediated cell death of the nephrocytes [35].
Bruggeman et al. [88] developed a transgenic mouse model where a nephrin promoter was used to drive podocyte-specific expression of the G0 or G2 form of APOL1. G2 transgenic mice lacked an overt phenotype early in life but had a lower podocyte density at old age. Studies crossing these mice with the well-established Tg26 model of HIV-associated nephropathy suggested that the G0 form of APOL1 may provide some protection to podocytes in this setting that is absent from the G2 mice, raising the possibility of loss of some kidney protective activity in G2 [89].
Ryu et al. [37] reported the development of transgenic mice that carry a BAC containing the entire human APOL1 genomic region. At 6 months of age, these mice (APOL1 G0 and G1) did not demonstrate an overt kidney phenotype. Administration of interferon gamma (IFN-γ) induced proteinuria only in the G1 mice, but not G0 mice.
Beckerman et al. [90] showed that mice with podocyte-specific expression of a G0 or G2 APOL1 cDNA, but not G0, developed albuminuria and FSGS-like histology. Disease severity is correlated with the level of expression of the risk allele. Expression of the risk-variant APOL1 was found to interfere with endosomal trafficking.
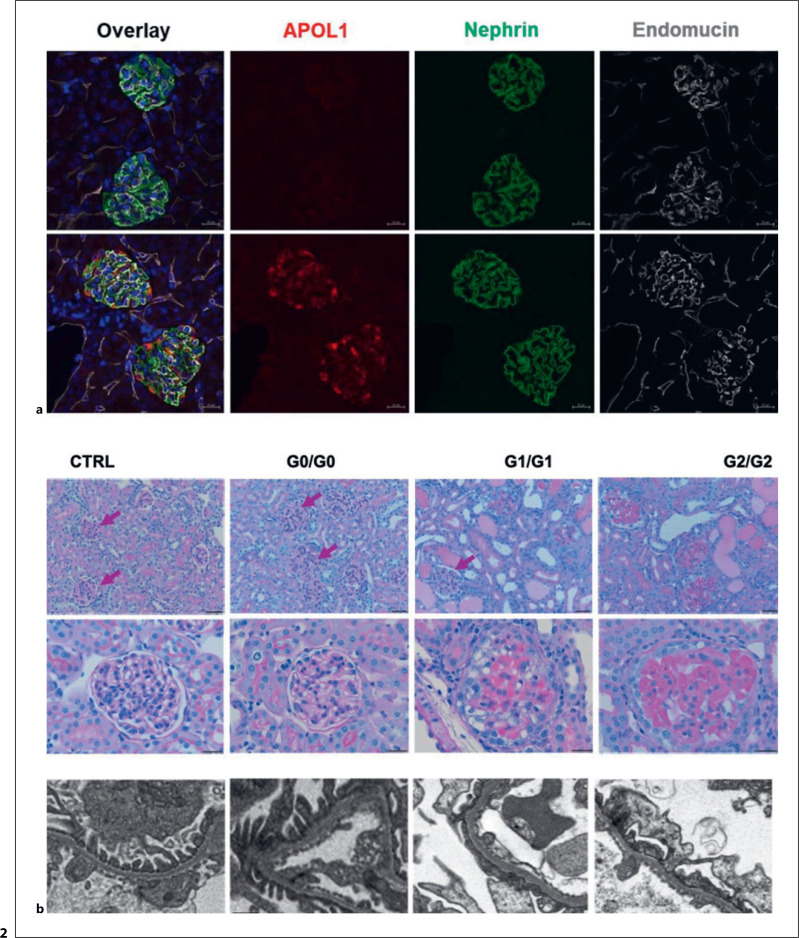
Our group recently developed co-isogenic BAC transgenic mice harboring either G0, G1, or G2 forms of human APOL1 [91]. Expression of IFN-γ resulted in upregulation of APOL1 protein in the kidneys and robust induction of proteinuria and glomerulosclerosis in G1/G1 and G2/G2 but not G0/G0 mice. Disease was greater in G2/G2 mice. In these studies, neither heterozygous (G1/G0 or G2/G0) risk variant mice nor hemizygous (G1/−, G2/−) mice had significant kidney injury in response to IFN-γ, suggesting that the lack of significant disease in humans heterozygous for G1 or G2 is not due to G0 rescue of G1 or G2 toxicity, in contradiction to a common hypothesis. Studies using additional mice with multiple copies of G2 supported the notion that disease is directly related to the level of risk variant APOL1 expression. The observation that these mice that are genetically identical with the exception of differences in the G0- versus G1- versus G2-defining variants should lay to rest any lingering doubts about the strong causal relationship between APOL1 genotype and kidney disease (Fig. 2).
Fig. 2.
Changing otherwise genetically identical APOL1 BAC transgenic mice from G0 to G1 or G2 transgenes confers susceptibility to IFN-γ-induced injury. a IFN-γ induces glomerular expression of human APOL1 in transgenic mice. b IFN-γ induces marked glomerular injury in APOL1 G1 and G2 transgenic mice but not in G0 transgenic mice nor FVB strain control mice. Adapted from [91], licensed under a Creative Commons Attribution 4.0 International License: https://creativecommons.org/licenses/by/4.0/.
Issues of Race and Ancestry
APOL1 risk genotypes, and APOL1-associated nephropathy, are highly associated with both self-reported Black race and sub-Saharan African ancestry. The association of APOL1 variants with racial identification simply reflects the fact that people who identify as Black are overwhelmingly of recent African ancestry. APOL1 should not be thought of as “the Black (or African American) kidney disease gene.” Most Black individuals in the USA do not have a high-risk APOL1 genotype. APOL1 is not the only cause of kidney disease in people of recent African ancestry. Discrimination against minority groups and the persistent disparities in social determinants of health are major contributors to the racial disparity in kidney disease and kidney disease treatment outcomes [92, 93]. The race-based disparities in kidney health and the ancestry-based effects of APOL1 genotype undoubtedly synergize to the detriment of many individuals.
The fact that most APOL1 studies to date have been conducted in Black patients has been criticized [94]. However, this simply reflects the fact that these APOL1 variants are so much more common in people who identify as Black than in other groups without known west African ancestry. According to the Gnomad database of genetic variants (at the time of this writing), the allele frequencies of G1 and G2 among African Americans are 0.1402 (G1, S342G variant) and 0.2276 (G2) [95]. By contrast, allele frequencies are 0.006772 (G1) and 0.0057 (G2) among people listed as Latino/Admixed, and less than 0.0001 in populations listed as European, Asian, and Ashkenazi. Among subsets of people in these populations with phenotypes known to associate with APOL1 (like FSGS), the frequency is higher, reflecting the relative risk associated with APOL1 [96]. Clinical studies aiming to efficiently recruit appropriate subjects naturally focus on Black, African, and African-American populations, as well as other specific populations known to have significant recent west African ancestry. It may prove to be the case that other variants related to ancestry may affect the severity of APOL1-associated kidney disease, but the individual effects of these variants, if they exist, are small [97]. G1 and G2 behave differently from G0 in a variety of model systems that allow APOL1 genotypes to be isolated as the only variable [98].
Unique Genetic Testing Issues
Is there any reason to genotype people for APOL1 variants? Most people with high-risk genotypes do not develop overt kidney disease. And people with high-risk genotypes can of course develop other forms of kidney disease (e.g., ANCA-mediated glomerulonephritis, polycystic kidney disease).
We believe that there are strong arguments for more routine testing for these risk alleles in people presenting with kidney disease. While the presence of a high-risk APOL1 genotype does not rule out the possibility of some other major disease driver (e.g., interstitial nephritis, vasculitis, drug toxicity), the absence of a high-risk genotype in someone with no other identifiable risk factors may make clinicians more likely to pursue an aggressive diagnostic workup. While there are no known therapies specific for APOL1-associated kidney disease, such therapies are under development [99, 100] (APOL1 trial paper, this issue of this journal). If proven effective specifically for APOL1-associated disease, APOL1 testing will be needed as a standard part of a kidney disease evaluation.
Testing in people without any evidence of kidney disease is trickier [101]. As noted elsewhere, the majority of people with high-risk APOL1 genotypes (∼80%) do not develop kidney disease. An argument for testing is that it may make people with high-risk genotypes more vigilant about their overall health [102]. Routine screening testing for proteinuria is currently not recommended in current primary care practice guidelines [103]. However, if treatments for APOL1-associated and non-APOL1-associated kidney diseases continue to improve, there will be an increased value in early diagnosis, and this recommendation may need reassessment [104].
The strongest case for APOL1 testing in asymptomatic people is in the kidney transplant setting [105]. Kidneys from APOL1 high-risk donors fare worse than those with low-risk genotypes [106, 107], but still seem preferable to dialysis in ESKD patients. A bigger concern is the risk to living kidney donors. It has been argued that people with high-risk APOL1 genotypes may be at increased risk of kidney disease and kidney failure after donation. Limited data support this notion, though the effect size is likely not large [108]. The donor risk is surely greater in younger individuals, who have not yet proven themselves to be resistant to kidney disease, in contrast to older individuals with APOL1 high-risk kidney disease without evidence of kidney disease. Unlike Mendelian kidney diseases such as polycystic kidney disease where related prospective donors are routinely tested for the presence of recipient's genetic lesion, the best approach is less clear with APOL1. The ongoing APOLLO trial may help clarify some of these issues [109].
Therapeutics
The atypical features of APOL1 and its variants have therapeutic implications. Because APOL1 is a dispensable gene, at least in parts of the globe where African trypanosomes are not present, it can presumably be inactivated without major deleterious consequences. The activity of many genes and gene products often must be finely regulated − both too much activity and not enough activity can be harmful, making therapeutic manipulation tricky. In the case of APOL1-associated kidney disease, we want to inactivate the toxic gain-of-function activity of the variant APOL1. It may not be necessary to coax G1 and G2 APOL1 to behave like G0. Simply inactivating all APOL1 may be effective. This is the strategy being pursued by some pharmaceutical companies [99] (Vertex trial paper − this issue). Treatment of APOL1 G1 mice an antisense oligonucleotide targeting APOL1 mRNA, prior to IFN- γ challenge, inhibited kidney APOL1 expression and protected against IFN-γ-induced proteinuria [99]. Similarly, Yang et al. [100] administered antisense oligonucleotide targeting APOL1 to podocyte-specific G2 transgenic mice. They reported reduction of APOL1 levels in the kidney and protection from albuminuria and kidney failure.
It is certainly possible that APOL1 has cellular functions that have not been identified, and that the G1 and/or G2 variants may exhibit loss of such functions, in addition to the gain-of-function effects discussed earlier. Thus, individuals treated with an APOL1 inactivation therapy will need careful monitoring for unexpected adverse effects. Nevertheless, we believe that such effects are likely to be minor, given the points about gene dispensability raised earlier.
Conclusion
APOL1-associated kidney disease is among the most common contributors to rates of chronic kidney disease worldwide. Because APOL1 genotype seems to require other factors to lead to overt kidney disease, it is hard to attribute any individual's kidney disease to APOL1 alone. This should not stop us from recognizing APOL1-associated kidney disease as a specific entity that requires unique approaches to treatment and management. These approaches may include preventive measures to modify nongenetic contributors to risk of APOL1-associated nephropathy, as well as therapeutic approaches specifically targeting the APOL1 pathway.
Conflict of Interest Statement
D. Friedman and M. Pollak are inventors on patents related to APOL1, own equity in Apolo1bio, and receive research funding from and have consulted for Vertex.
Funding Sources
Work on APOL1 and APOL1-associated nephropathy in the authors' laboratories was supported by grants from the NIH (NIMHD), the U.S. Department of Defense, Vertex Pharmaceuticals, and the Ellison Foundation.
Author Contributions
This paper was written by Martin Pollak and David Friedman.
Funding Statement
Work on APOL1 and APOL1-associated nephropathy in the authors' laboratories was supported by grants from the NIH (NIMHD), the U.S. Department of Defense, Vertex Pharmaceuticals, and the Ellison Foundation.
References
- 1.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128(3):345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko WY, Rajan P, Gomez F, Scheinfeldt L, An P, Winkler CA, et al. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am J Hum Genet. 2013;93(1):54–66. doi: 10.1016/j.ajhg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goyal R, Singhal PC. APOL1 risk variants and the development of HIV-associated nephropathy. FEBS J. 2021;288(19):5586–5597. doi: 10.1111/febs.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.May RM, Cassol C, Hannoudi A, Larsen CP, Lerma EV, Haun RS, et al. A multi-center retrospective cohort study defines the spectrum of kidney pathology in Coronavirus 2019 Disease (COVID-19) Kidney Int. 2021;100(6):1303–1315. doi: 10.1016/j.kint.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Florea L, Caba L, Gorduza EV. Genetic heterogeneity in bartter syndrome clinical and practical importance. Front Pediatr. 2022;10:908655. doi: 10.3389/fped.2022.908655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tőke J, Czirjak G, Enyedi P, Toth M. Rare diseases caused by abnormal calcium sensing and signalling. Endocrine. 2021;71(3):611–617. doi: 10.1007/s12020-021-02620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanhollebeke B, Truc P, Poelvoorde P, Pays A, Joshi PP, Katti R, et al. Human Trypanosoma evansi infection linked to a lack of apolipoprotein L-I. N Engl J Med. 2006;355(26):2752–2756. doi: 10.1056/NEJMoa063265. [DOI] [PubMed] [Google Scholar]
- 9.Johnstone DB, Shegokar V, Nihalani D, Rathore YS, Mallik L, Ashish, et al. APOL1 null alleles from a rural village in India do not correlate with glomerulosclerosis. PLoS One. 2012;7(12):e51546. doi: 10.1371/journal.pone.0051546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monajemi H, Fontijn RD, Pannekoek H, Horrevoets AJG. The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics. 2002;79(4):539–546. doi: 10.1006/geno.2002.6729. [DOI] [PubMed] [Google Scholar]
- 11.Zhaorigetu S, Wan G, Kaini R, Jiang Z, Hu CA. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4(8):1079–1082. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng D, Weckerle A, Yu Y, Ma L, Zhu X, Murea M, et al. Biogenesis and cytotoxicity of APOL1 renal risk variant proteins in hepatocytes and hepatoma cells. J Lipid Res. 2015;56(8):1583–1593. doi: 10.1194/jlr.M059733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olabisi OA, Zhang JY, VerPlank L, Zahler N, DiBartolo S, 3rd, Heneghan JF, et al. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc Natl Acad Sci U S A. 2016;113(4):830–837. doi: 10.1073/pnas.1522913113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uzureau S, Coquerelle C, Vermeiren C, Uzureau P, Van Acker A, Pilotte L, et al. Apolipoproteins L control cell death triggered by TLR3/TRIF signaling in dendritic cells. Eur J Immunol. 2016;46(8):1854–1866. doi: 10.1002/eji.201546252. [DOI] [PubMed] [Google Scholar]
- 15.Kruzel-Davila E, Shemer R, Ofir A, Bavli-Kertselli I, Darlyuk-Saadon I, Oren-Giladi P, et al. APOL1-Mediated cell injury involves disruption of conserved trafficking processes. J Am Soc Nephrol. 2017;28(4):1117–1130. doi: 10.1681/ASN.2016050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y, Zhu JY, Richman A, Zhang Y, Xie X, Das JR, et al. APOL1-G1 in nephrocytes induces hypertrophy and accelerates cell death. J Am Soc Nephrol. 2017;28(4):1106–1116. doi: 10.1681/ASN.2016050550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carney EF. APOL1 risk variants induce opening of the mitochondrial permeability transition pore. Nat Rev Nephrol. 2019;15(12):730. doi: 10.1038/s41581-019-0222-8. [DOI] [PubMed] [Google Scholar]
- 18.Granado D, Muller D, Krausel V, Kruzel-Davila E, Schuberth C, Eschborn M, et al. Intracellular APOL1 risk variants cause cytotoxicity accompanied by energy depletion. J Am Soc Nephrol. 2017;28(11):3227–3238. doi: 10.1681/ASN.2016111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman DJ, Pollak MR. APOL1 and kidney disease from genetics to biology. Annu Rev Physiol. 2020;82:323–342. doi: 10.1146/annurev-physiol-021119-034345. [DOI] [PubMed] [Google Scholar]
- 20.Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422(6927):83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, Nolan DP, Lins L, Homble F, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309(5733):469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 22.Vanwalleghem G, Fontaine F, Lecordier L, Tebabi P, Klewe K, Nolan DP, et al. Coupling of lysosomal and mitochondrial membrane permeabilization in trypanolysis by APOL1. Nat Commun. 2015;6:8078. doi: 10.1038/ncomms9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomson R, Finkelstein A. Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers relevance to trypanosome lysis. Proc Natl Acad Sci U S A. 2015;112(9):2894–2899. doi: 10.1073/pnas.1421953112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruno J, Pozzi N, Oliva J, Edwards JC. Apolipoprotein L1 confers pH-switchable ion permeability to phospholipid vesicles. J Biol Chem. 2017;292(44):18344–53. doi: 10.1074/jbc.M117.813444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giovinazzo JA, Thomson RP, Khalizova N, Zager PJ, Malani N, Rodriguez-Boulan E, et al. Apolipoprotein L-1 renal risk variants form active channels at the plasma membrane driving cytotoxicity. Elife. 2020;9:e51185. doi: 10.7554/eLife.51185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaub C, Verdi J, Lee P, Terra N, Limon G, Raper J, et al. Cation channel conductance and pH gating of the innate immunity factor APOL1 are governed by pore-lining residues within the C-terminal domain. J Biol Chem. 2020;295(38):13138–49. doi: 10.1074/jbc.RA120.014201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scales SJ, Gupta N, De Maziere AM, Posthuma G, Chiu CP, Pierce AA, et al. Apolipoprotein L1-specific antibodies detect endogenous APOL1 inside the endoplasmic reticulum and on the plasma membrane of podocytes. J Am Soc Nephrol. 2020;31(9):2044–2064. doi: 10.1681/ASN.2019080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta N, Wang X, Wen X, Moran P, Paluch M, Hass PE, et al. Domain-specific antibodies reveal differences in the membrane topologies of apolipoprotein L1 in serum and podocytes. J Am Soc Nephrol. 2020;31(9):2065–2082. doi: 10.1681/ASN.2019080830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaub C, Lee P, Racho-Jansen A, Giovinazzo J, Terra N, Raper J, et al. Coiled-coil binding of the leucine zipper domains of APOL1 is necessary for the open cation channel conformation. J Biol Chem. 2021;297(3):101009. doi: 10.1016/j.jbc.2021.101009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller D, Schmitz J, Fischer K, Granado D, Groh AC, Krausel V, et al. Evolution of renal-disease factor APOL1 results in cis and trans orientations at the endoplasmic reticulum that both show cytotoxic effects. Mol Biol Evol. 2021;38(11):4962–4976. doi: 10.1093/molbev/msab220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma L, Chou JW, Snipes JA, Bharadwaj MS, Craddock AL, Cheng D, et al. APOL1 renal-risk variants induce mitochondrial dysfunction. J Am Soc Nephrol. 2017;28(4):1093–1105. doi: 10.1681/ASN.2016050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah SS, Lannon H, Dias L, Zhang JY, Alper SL, Pollak MR, et al. APOL1 kidney risk variants induce cell death via mitochondrial translocation and opening of the mitochondrial permeability transition pore. J Am Soc Nephrol. 2019;30(12):2355–2368. doi: 10.1681/ASN.2019020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruzel-Davila E, Bavli-Kertselli I, Ofir A, Cheatham AM, Shemer R, Zaknoun E, et al. Endoplasmic reticulum-translocation is essential for APOL1 cellular toxicity. iScience. 2022;25(1):103717. doi: 10.1016/j.isci.2021.103717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen H, Kumar V, Lan X, Shoshtari SSM, Eng JM, Zhou X, et al. APOL1 risk variants cause podocytes injury through enhancing endoplasmic reticulum stress. Biosci Rep. 2018;38(4) doi: 10.1042/BSR20171713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerstner L, Chen M, Kampf LL, Milosavljevic J, Lang K, Schneider R, et al. Inhibition of endoplasmic reticulum stress signaling rescues cytotoxicity of human apolipoprotein-L1 risk variants in Drosophila. Kidney Int. 2022;101(6):1216–1231. doi: 10.1016/j.kint.2021.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge M, Molina J, Ducasa GM, Mallela SK, Varona Santos J, Mitrofanova A, et al. APOL1 risk variants affect podocyte lipid homeostasis and energy production in focal segmental glomerulosclerosis. Hum Mol Genet. 2021;30(3–4):182–197. doi: 10.1093/hmg/ddab022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryu JH, Ge M, Merscher S, Rosenberg AZ, Desante M, Roshanravan H, et al. APOL1 renal risk variants promote cholesterol accumulation in tissues and cultured macrophages from APOL1 transgenic mice. PLoS One. 2019;14(4):e0211559. doi: 10.1371/journal.pone.0211559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun J, Zhang JY, Wilkins MS, Subramanian B, Riella C, Magraner JM, et al. Recruitment of APOL1 kidney disease risk variants to lipid droplets attenuates cell toxicity. Proc Natl Acad Sci U S A. 2019;116(9):3712–3721. doi: 10.1073/pnas.1820414116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uzureau S, Lecordier L, Uzureau P, Hennig D, Graversen JH, Homble F, et al. APOL1 C-terminal variants may trigger kidney disease through interference with APOL3 control of actomyosin. Cell Rep. 2020;30(11):3821–36 e13. doi: 10.1016/j.celrep.2020.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skorecki KL, Lee JH, Langefeld CD, Rosset S, Tzur S, Wasser WG, et al. A null variant in the apolipoprotein L3 gene is associated with non-diabetic nephropathy. Nephrol Dial Transpl. 2018;33(2):323–330. doi: 10.1093/ndt/gfw451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith EE, Malik HS. The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res. 2009;19(5):850–858. doi: 10.1101/gr.085647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman DJ. A brief history of APOL1 a gene evolving. Semin Nephrol. 2017;37(6):508–513. doi: 10.1016/j.semnephrol.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Vanhollebeke B, Pays E. The function of apolipoproteins L. Cell Mol Life Sci. 2006;63(17):1937–1944. doi: 10.1007/s00018-006-6091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pant J, Giovinazzo JA, Tuka LS, Pena D, Raper J, Thomson R. Apolipoproteins L1-6 share key cation channel-regulating residues but have different membrane insertion and ion conductance properties. J Biol Chem. 2021;297(2):100951. doi: 10.1016/j.jbc.2021.100951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duchateau PN, Pullinger CR, Orellana RE, Kunitake ST, Naya-Vigne J, O'Connor PM, et al. Apolipoprotein L a new human high density lipoprotein apolipoprotein expressed by the pancreas. Identification, cloning, characterization, and plasma distribution of apolipoprotein L. J Biol Chem. 1997;272(41):25576–82. doi: 10.1074/jbc.272.41.25576. [DOI] [PubMed] [Google Scholar]
- 46.Shukha K, Mueller JL, Chung RT, Curry MP, Friedman DJ, Pollak MR, et al. Most ApoL1 is secreted by the liver. J Am Soc Nephrol. 2017;28(4):1079–1083. doi: 10.1681/ASN.2016040441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang M, Huang X, Li R, Xu H, Jin L, He Y. Detecting recent positive selection with high accuracy and reliability by conditional coalescent tree. Mol Biol Evol. 2014;31(11):3068–3080. doi: 10.1093/molbev/msu244. [DOI] [PubMed] [Google Scholar]
- 48.Cuadros-Espinoza S, Laval G, Quintana-Murci L, Patin E. The genomic signatures of natural selection in admixed human populations. Am J Hum Genet. 2022;109(4):710–726. doi: 10.1016/j.ajhg.2022.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper A, Ilboudo H, Pius Alibu V, Ravel S, Enyaru J, Weir W, et al. APOL1 renal risk variants have contrasting resistance and susceptibility associations with African trypanosomiasis. Elife. 2017;6:e25461. doi: 10.7554/eLife.25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones TR. Quantitative aspects of the relationship between the sickle-cell gene and malaria. Parasitol Today. 1997;13(3):107–111. doi: 10.1016/s0169-4758(96)10083-1. [DOI] [PubMed] [Google Scholar]
- 51.Shriner D, Rotimi CN. Whole-genome-sequence-based haplotypes reveal single origin of the sickle allele during the holocene wet phase. Am J Hum Genet. 2018;102(4):547–556. doi: 10.1016/j.ajhg.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A. 2014;111(20):E2130–E9. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17(9):502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 54.Pollak MR, Friedman DJ. The genetic architecture of kidney disease. Clin J Am Soc Nephrol. 2020;15(2):268–275. doi: 10.2215/CJN.09340819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Friedman DJ, Pollak MR. Genetics of kidney failure and the evolving story of APOL1. J Clin Invest. 2011;121(9):3367–3374. doi: 10.1172/JCI46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulasi II, Tzur S, Wasser WG, Shemer R, Kruzel E, Feigin E, et al. High population frequencies of APOL1 risk variants are associated with increased prevalence of non-diabetic chronic kidney disease in the Igbo people from south-eastern Nigeria. Nephron Clin Pract. 2013;123(1–2):123–128. doi: 10.1159/000353223. [DOI] [PubMed] [Google Scholar]
- 57.Behar DM, Kedem E, Rosset S, Haileselassie Y, Tzur S, Kra-Oz Z, et al. Absence of APOL1 risk variants protects against HIV-associated nephropathy in the Ethiopian population. Am J Nephrol. 2011;34(5):452–459. doi: 10.1159/000332378. [DOI] [PubMed] [Google Scholar]
- 58.Ruchi R, Genovese G, Lee J, Charoonratana VT, Bernhardy AJ, Alper SL, et al. Copy number variation at the APOL1 locus. PLoS One. 2015;10(5):e0125410. doi: 10.1371/journal.pone.0125410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lannon H, Shah SS, Dias L, Blackler D, Alper SL, Pollak MR, et al. Apolipoprotein L1 (APOL1) risk variant toxicity depends on the haplotype background. Kidney Int. 2019;96(6):1303–1307. doi: 10.1016/j.kint.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sampson MG, Robertson CC, Martini S, Mariani LH, Lemley KV, Gillies CE, et al. Integrative genomics identifies novel associations with APOL1 risk genotypes in Black NEPTUNE subjects. J Am Soc Nephrol. 2016;27(3):814–823. doi: 10.1681/ASN.2014111131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang JY, Wang M, Tian L, Genovese G, Yan P, Wilson JG, et al. UBD modifies APOL1-induced kidney disease risk. Proc Natl Acad Sci U S A. 2018;115(13):3446–3451. doi: 10.1073/pnas.1716113115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bodonyi-Kovacs G, Ma JZ, Chang J, Lipkowitz MS, Kopp JB, Winkler CA, et al. Combined effects of GSTM1 null allele and APOL1 renal risk alleles in CKD progression in the african American study of kidney disease and hypertension trial. J Am Soc Nephrol. 2016;27(10):3140–3152. doi: 10.1681/ASN.2015050487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nichols B, Jog P, Lee JH, Blackler D, Wilmot M, D'Agati V, et al. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int. 2015;87(2):332–342. doi: 10.1038/ki.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Markowitz GS, Nasr SH, Stokes MB, D'Agati VD. Treatment with IFN-{alpha}, -{beta}, or -{gamma} is associated with collapsing focal segmental glomerulosclerosis. Clin J Am Soc Nephrol. 2010;5(4):607–615. doi: 10.2215/CJN.07311009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22(11):2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kasembeli AN, Duarte R, Ramsay M, Naicker S. African origins and chronic kidney disease susceptibility in the human immunodeficiency virus era. World J Nephrol. 2015;4(2):295–306. doi: 10.5527/wjn.v4.i2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shetty AA, Tawhari I, Safar-Boueri L, Seif N, Alahmadi A, Gargiulo R, et al. COVID-19-Associated glomerular disease. J Am Soc Nephrol. 2021;32(1):33–40. doi: 10.1681/ASN.2020060804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larsen CP, Beggs ML, Saeed M, Walker PD. Apolipoprotein L1 risk variants associate with systemic lupus erythematosus-associated collapsing glomerulopathy. J Am Soc Nephrol. 2013;24(5):722–725. doi: 10.1681/ASN.2012121180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;66(2):390–396. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kasembeli AN, Duarte R, Ramsay M, Mosiane P, Dickens C, Dix-Peek T, et al. APOL1 risk variants are strongly associated with HIV-associated nephropathy in Black South Africans. J Am Soc Nephrol. 2015;26(11):2882–2890. doi: 10.1681/ASN.2014050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hiraki LT. APOL1 gene - implications for systemic lupus erythematosus. J Rheumatol. 2020;47(8):1155–1157. doi: 10.3899/jrheum.200038. [DOI] [PubMed] [Google Scholar]
- 72.Nystrom SE, Li G, Datta S, Soldano KL, Silas D, Weins A, et al. JAK inhibitor blocks COVID-19 cytokine-induced JAK/STAT/APOL1 signaling in glomerular cells and podocytopathy in human kidney organoids. JCI Insight. 2022;7(11):e157432. doi: 10.1172/jci.insight.157432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Velez JCQ, Caza T, Larsen CP. COVAN is the new HIVAN the re-emergence of collapsing glomerulopathy with COVID-19. Nat Rev Nephrol. 2020;16(10):565–567. doi: 10.1038/s41581-020-0332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muehlig AK, Gies S, Huber TB, Braun F. Collapsing focal segmental glomerulosclerosis in viral infections. Front Immunol. 2021;12:800074. doi: 10.3389/fimmu.2021.800074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tin A, Grams ME, Estrella M, Lipkowitz M, Greene TH, Kao WHL, et al. Patterns of kidney function decline associated with APOL1 genotypes results from AASK. Clin J Am Soc Nephrol. 2016;11(8):1353–1359. doi: 10.2215/CJN.12221115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grams ME, Rebholz CM, Chen Y, Rawlings AM, Estrella MM, Selvin E, et al. Race, APOL1 risk, and eGFR decline in the general population. J Am Soc Nephrol. 2016;27(9):2842–2850. doi: 10.1681/ASN.2015070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kramer HJ, Stilp AM, Laurie CC, Reiner AP, Lash J, Daviglus ML, et al. African ancestry-specific alleles and kidney disease risk in hispanics/latinos. J Am Soc Nephrol. 2017;28(3):915–922. doi: 10.1681/ASN.2016030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Riella C, Siemens TA, Wang M, Campos RP, Moraes TP, Riella LV, et al. APOL1-Associated kidney disease in Brazil. Kidney Int Rep. 2019;4(7):923–929. doi: 10.1016/j.ekir.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22(11):2098–2105. doi: 10.1681/ASN.2011050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parsa A, Kao WHL, Xie D, Astor BC, Li M, Hsu C, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369(23):2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papeta N, Kiryluk K, Patel A, Sterken R, Kacak N, Snyder HJ, et al. APOL1 variants increase risk for FSGS and HIVAN but not IgA nephropathy. J Am Soc Nephrol. 2011;22(11):1991–1996. doi: 10.1681/ASN.2011040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reidy KJ, Hjorten RC, Simpson CL, Rosenberg AZ, Rosenblum SD, Kovesdy CP, et al. Fetal-not maternal-APOL1 genotype associated with risk for preeclampsia in those with african ancestry. Am J Hum Genet. 2018;103(3):367–376. doi: 10.1016/j.ajhg.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mukamal KJ, Tremaglio J, Friedman DJ, Ix JH, Kuller LH, Tracy RP, et al. APOL1 genotype, kidney and cardiovascular disease, and death in older adults. Arterioscler Thromb Vasc Biol. 2016;36(2):398–403. doi: 10.1161/ATVBAHA.115.305970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nadkarni GN, Coca SG. APOL1 and blood pressure changes in young adults. Kidney Int. 2017;92(4):793–795. doi: 10.1016/j.kint.2017.05.030. [DOI] [PubMed] [Google Scholar]
- 85.Akinyemi R, Tiwari HK, Arnett DK, Ovbiagele B, Irvin MR, Wahab K, et al. APOL1, CDKN2A/CDKN2B, and HDAC9 polymorphisms and small vessel ischemic stroke. Acta Neurol Scand. 2018;137(1):133–141. doi: 10.1111/ane.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hughson MD, Hoy WE, Mott SA, Puelles VG, Bertram JF, Winkler CL, et al. APOL1 risk alleles are associated with more severe arteriosclerosis in renal resistance vessels with aging and hypertension. Kidney Int Rep. 2016;1(1):10–23. doi: 10.1016/j.ekir.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Helmstadter M, Huber TB, Hermle T. Using the Drosophila nephrocyte to model podocyte function and disease. Front Pediatr. 2017;5:262. doi: 10.3389/fped.2017.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bruggeman LA, Wu Z, Luo L, Madhavan SM, Konieczkowski M, Drawz PE, et al. APOL1-G0 or APOL1-G2 transgenic models develop preeclampsia but not kidney disease. J Am Soc Nephrol. 2016;27(12):3600–3610. doi: 10.1681/ASN.2015111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bruggeman LA, Wu Z, Luo L, Madhavan S, Drawz PE, Thomas DB, et al. APOL1-G0 protects podocytes in a mouse model of HIV-associated nephropathy. PLoS One. 2019;14(10):e0224408. doi: 10.1371/journal.pone.0224408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beckerman P, Bi-Karchin J, Park ASD, Qiu C, Dummer PD, Soomro I, et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat Med. 2017;23(4):429–438. doi: 10.1038/nm.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McCarthy GM, Blasio A, Donovan OG, Schaller LB, Bock-Hughes A, Magraner JM, et al. Recessive, gain-of-function toxicity in an APOL1 BAC transgenic mouse model mirrors human APOL1 kidney disease. Dis Model Mech. 2021;14(8):dmm048952. doi: 10.1242/dmm.048952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Powe NR. The pathogenesis of race and ethnic disparities targets for achieving health equity. Clin J Am Soc Nephrol. 2021;16(5):806–808. doi: 10.2215/CJN.12640820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cerdena JP, Tsai J, Grubbs V. APOL1 Black race and kidney disease turning attention to structural racism. Am J Kidney Dis. 2021;77(6):857–860. doi: 10.1053/j.ajkd.2020.11.029. [DOI] [PubMed] [Google Scholar]
- 94.Gopalakrishnan C, Patorno E. Time to end the misuse of race in medicine cases from nephrology. BMJ. 2021;375:n2435. doi: 10.1136/bmj.n2435. [DOI] [PubMed] [Google Scholar]
- 95.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kopp JB, Winkler CA, Zhao X, Radeva MK, Gassman JJ, D'Agati VD, et al. Clinical features and histology of apolipoprotein L1-associated nephropathy in the FSGS clinical trial. J Am Soc Nephrol. 2015;26(6):1443–1448. doi: 10.1681/ASN.2013111242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khan A, Turchin MC, Patki A, Srinivasasainagendra V, Shang N, Nadukuru R, et al. Genome-wide polygenic score to predict chronic kidney disease across ancestries. Nat Med. 2022;28(7):1412–1420. doi: 10.1038/s41591-022-01869-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kruzel-Davila E, Skorecki K. Dilemmas and challenges in apolipoprotein L1 nephropathy research. Curr Opin Nephrol Hypertens. 2019;28(1):77–86. doi: 10.1097/MNH.0000000000000462. [DOI] [PubMed] [Google Scholar]
- 99.Aghajan M, Booten SL, Althage M, Hart CE, Ericsson A, Maxvall I, et al. Antisense oligonucleotide treatment ameliorates IFN-gamma-induced proteinuria in APOL1-transgenic mice. JCI Insight. 2019;4(12):e126124. doi: 10.1172/jci.insight.126124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang YW, Poudel B, Frederick J, Dhillon P, Shrestha R, Ma Z, et al. Antisense oligonucleotides ameliorate kidney dysfunction in podocyte-specific APOL1 risk variant mice. Mol Ther. 2022;30(7):2491–2504. doi: 10.1016/j.ymthe.2022.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Young BA, Blacksher E, Cavanaugh KL, Freedman BI, Fullerton SM, Kopp JB, et al. Apolipoprotein L1 testing in african Americans involving the community in policy discussions. Am J Nephrol. 2019;50(4):303–311. doi: 10.1159/000502675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nadkarni GN, Fei K, Ramos MA, Hauser D, Bagiella E, Ellis SB, et al. Effects of testing and disclosing ancestry-specific genetic risk for kidney failure on patients and health care professionals a randomized clinical trial. JAMA Netw Open. 2022;5(3):e221048. doi: 10.1001/jamanetworkopen.2022.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Samal L, Linder JA. The primary care perspective on routine urine dipstick screening to identify patients with albuminuria. Clin J Am Soc Nephrol. 2013;8(1):131–135. doi: 10.2215/CJN.12681211. [DOI] [PubMed] [Google Scholar]
- 104.Kopp JB, Winkler CA. Genetic testing for APOL1 genetic variants in clinical practice finally starting to arrive. Clin J Am Soc Nephrol. 2020;15(1):126–128. doi: 10.2215/CJN.01810219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Doshi MD, Gordon EJ, Freedman BI, Glover C, Locke JE, Thomas CP. Integrating APOL1 kidney-risk variant testing in live kidney donor evaluation an expert panel opinion. Transplantation. 2021;105(10):2132–2134. doi: 10.1097/TP.0000000000003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reeves-Daniel AM, DePalma JA, Bleyer AJ, Rocco MV, Murea M, Adams PL, et al. The APOL1 gene and allograft survival after kidney transplantation. Am J Transpl. 2011;11(5):1025–1030. doi: 10.1111/j.1600-6143.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Freedman BI, Julian BA, Pastan SO, Israni AK, Schladt D, Gautreaux MD, et al. Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am J Transpl. 2015;15(6):1615–1622. doi: 10.1111/ajt.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Doshi MD, Ortigosa-Goggins M, Garg AX, Li L, Poggio ED, Winkler CA, et al. APOL1 genotype and renal function of Black living donors. J Am Soc Nephrol. 2018;29(4):1309–1316. doi: 10.1681/ASN.2017060658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Freedman BI, Moxey-Mims MM, Alexander AA, Astor BC, Birdwell KA, Bowden DW, et al. APOL1 long-term kidney transplantation outcomes network (APOLLO) design and rationale. Kidney Int Rep. 2020;5(3):278–288. doi: 10.1016/j.ekir.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]