Abstract
Infectious hepatitis type A and type E are caused by phylogenetically distinct single-stranded, positive-sense RNA viruses that were once considered to be non-enveloped. However, studies show that both are released nonlytically from hepatocytes as ‘quasi-enveloped’ virions cloaked in host membranes. These virion types predominate in the blood of infected individuals and mediate virus spread within the liver. They lack virally encoded proteins on their surface and are resistant to neutralizing anti-capsid antibodies induced by infection, yet they efficiently enter cells and initiate new rounds of virus replication. In this Review, we discuss the mechanisms by which specific peptide sequences in the capsids of these quasi-enveloped virions mediate their endosomal sorting complexes required for transport (ESCRT)-dependent release from hepatocytes through multivesicular endosomes, what is known about how they enter cells, and the impact of capsid quasi-envelopment on host immunity and pathogenesis.
Subject terms: Viral infection, Virus-host interactions, Viral hepatitis
In this Review, Lemon and colleagues describe quasi-enveloped virions, how they enter and how they are released from the human host cell, and how they affect host immunity and pathogenesis.
Introduction
Hepatitis A virus (HAV) and hepatitis E virus (HEV) are small, positive-strand RNA viruses that cause enterically transmitted hepatitis in humans. The likely cause of disease outbreaks described in antiquity, both viruses were discovered 40–50 years ago by immune electron microscopic examination of faecal material from experimentally infected humans1,2. However, only now are we beginning to understand unique aspects of their replication cycles and how they interact with host cells. Although phylogenetically distinct, both viruses are hepatotropic and replicate within hepatocytes, which are the major cell type within the liver3–5. Newly produced virions are released from the liver into the bloodstream, causing a viraemia, and into the biliary tract, resulting in faecal shedding of virus (Fig. 1a). Both viruses spread primarily by faecal–oral transmission, resulting in both sporadic infections and large outbreaks due to contaminated food or water6,7.
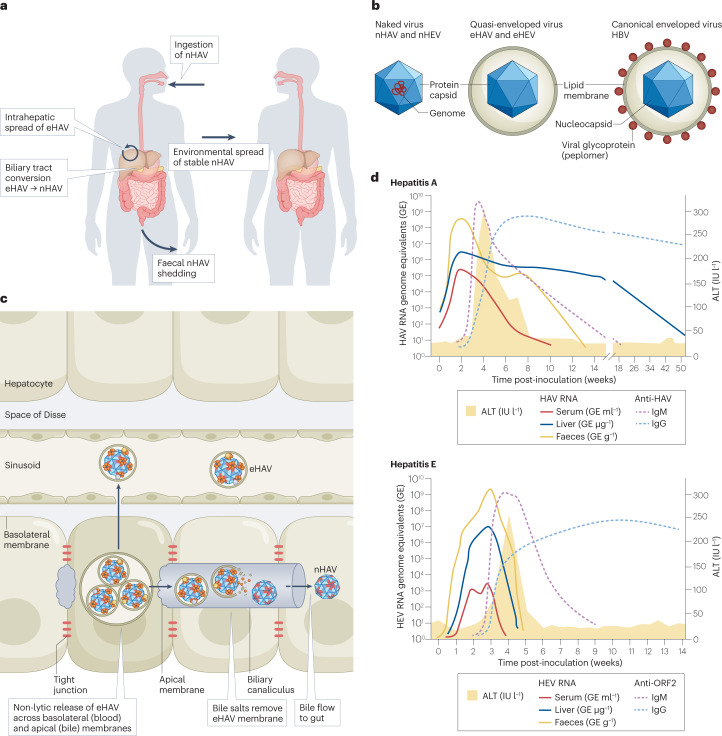
Fig. 1. Pathogenesis of enterically transmitted hepatitis A and hepatitis E virus.
a, Hepatitis A virus (HAV) life cycle, showing per-oral infection with naked HAV (nHAV), in-host spread of quasi-enveloped HAV (eHAV) and faecal shedding of nHAV resulting in environmental transmission to naive hosts. nHAV particles shed in faeces are produced by bile acid conversion of eHAV released from hepatocytes. The hepatitis E virus (HEV) life cycle is similar (not shown). b, Basic structures of naked versus quasi-enveloped hepatitis viruses and a canonical enveloped virus (hepatitis B virus (HBV)), showing the absence of virus-encoded proteins on the surface of quasi-enveloped virions11,31. c, Liver architecture, showing basolateral release of eHAV from polarized hepatocytes into blood flowing through hepatic sinusoids and apical release into a bile canaliculus. The quasi-envelope is stripped from eHAV by bile acids, resulting in faecal shedding of nHAV24. Events are similar in hepatitis E (not shown). d, Virological and serological markers in acute hepatitis A (top) and hepatitis E (bottom). ALT, alanine aminotransferase; IgG, immunoglobulin G; IgM, immunoglobulin M; IU l−1, international units per litre.
When first identified, both HAV and HEV were considered to be non-enveloped — that is, to lack an outer lipid layer and to be released from infected cells as ‘naked’ virions in which the RNA genome is encapsidated within a protein shell (the ‘capsid’) protecting it from the environment. However, the lifecycles of these viruses are much more complicated. Both viruses are now known to be released from infected cells with an outer lipid layer formed by membranes derived from the interior of the host cell8,9. Although infectious, these membrane-cloaked, ‘quasi-enveloped’ virions of HAV and HEV (‘eHAV’ and ‘eHEV’, respectively)10 lack any virally encoded proteins on their surface, a feature distinguishing them from canonical enveloped viruses such as coronaviruses or hepatitis B virus (Fig. 1b). They share some attributes of exosomes, a special class of small extracellular vesicles (EVs) that originate from multivesicular endosomes (MVEs; also known as multivesicular bodies) and may function as vehicles for intercellular transfer of proteins and regulatory RNAs11,12. Unlike other viruses that are shed from infected cells in larger types of EVs (Box 1), the structural proteins of HAV and HEV contain conserved sequence motifs that mediate interactions of the capsid with endosomal sorting complexes required for transport (ESCRT). Quasi-envelopment thus results from an active and highly selective sorting process that involves the budding of capsids into MVEs, followed by their release from the cell upon fusion of MVE and plasma membranes. Extracellular quasi-enveloped virions contain capsids that differ compositionally from the naked, non-enveloped virus shed in the faeces of infected individuals and represent a distinct type of infectious particle.
This Review focuses on recent gains in our understanding of how specific proteins expressed by these viruses — namely, the pX polypeptide of HAV and the small ORF3 protein of HEV — interact with the host cell to drive nonlytic release of quasi-enveloped virions, and how these extracellular infectious particles enter naive cells to establish infection in the absence of virally encoded proteins on their surface. These questions are important given increasing evidence that pathogenic viruses from many other virus families conventionally considered to be ‘non-enveloped’ are released from infected cells in EVs without lysing the cell (Box 1). Recent progress in developing small-animal models of HAV and HEV infection13,14, and in understanding mechanisms underlying the liver injury caused by these viruses15,16, have been reviewed elsewhere and will not be discussed in detail.
Box 1 Quasi-enveloped virus or virus shed in extracellular vesicles?
Many canonical ‘non-enveloped’ viruses other than hepatitis A virus (HAV) and hepatitis E virus (HEV) have been shown to be released from infected cells within extracellular vesicles (EVs)127–132. This can engender confusion over terminology — are these viruses ‘quasi-enveloped’ like the enteric hepatitis viruses (see the figure, part a), or are they simply viruses shed from cells in EVs (see the figure, parts b and c)? Most of these other viruses, including picornaviruses (enteroviruses and cardioviruses), noroviruses, rotaviruses and reoviruses, have RNA genomes and a cytoplasmic replication cycle, but John Cunningham polyomavirus, a DNA virus, is also released in EVs127–132. These virus-containing EVs vary in size depending on the mechanisms of biogenesis, none of which are clearly defined but probably include secretory autophagy (see the figure, part b) and microvesicle shedding from the plasma membrane (see the figure, part c). Enterovirus-containing EVs are substantially larger (300–920 nm diameter) than quasi-enveloped HAV (eHAV) or HEV (eHEV), contain numerous capsids and originate in autophagosomes127,128. Enterovirus capsids assembling on membranes are engulfed by phagophores activated by autophagy proteins (ATG5, ATG12, ATG16 and lipidated LC3) and become trapped within double-membrane autophagosomes120,133. This entrapment is probably a passive event, linked to autophagic signalling supporting genome replication, and is not known to be driven by any specific capsid signal. Autophagosomes loaded with assembled virus particles along with a mix of nonstructural viral proteins (nsPs) and both positive-strand and negative-strand viral RNAs traffic to the plasma membrane, where fusion of the outer autophagosome membrane results in release of single-membrane vesicles to the external environment before lysis of the cell120,131,134,135. This contrasts with the biogenesis of eHAV and eHEV, whereby conserved sequence motifs in structural polypeptides (pX and ORF3) function mechanistically like the ‘late domains’ of canonical enveloped viruses, mediating capsid interactions with endosomal complexes required for transport (ESCRT) and driving an active sorting process leading to budding of the capsid into the multivesicular endosome (MVE)8,93,121,122 (see the figure, part a, and Fig. 3). Sorting is highly specific and selective. eHAV virions contain only capsid proteins and no nonstructural viral proteins11. An additional distinguishing feature is that the capsids exported in eHAV and eHEV differ compositionally from naked extracellular virions (nHAV and nHEV, respectively), to which they are converted by bile acids following release from the hepatocyte (Fig. 2). It is uncertain whether other viruses are released through a similar process of quasi-envelopment, but EVs containing norovirus are similar in size to quasi-enveloped hepatitis viruses and thus could have a similar MVE-related origin.
Quasi-envelopment has a central role in the pathogenesis of HAV and HEV infections, accounting for most, if not all, virus released into the bloodstream. En bloc delivery of multiple enterovirus genomes along with other cellular and viral proteins present in large autophagosome-related EVs (see the figure, part b) may enhance infectivity and potentially modulate virus replication128,129,131, but a similarly central role in viral pathogenesis remains to be demonstrated for these virus-containing EVs. (ILV, intraluminal vesicle.)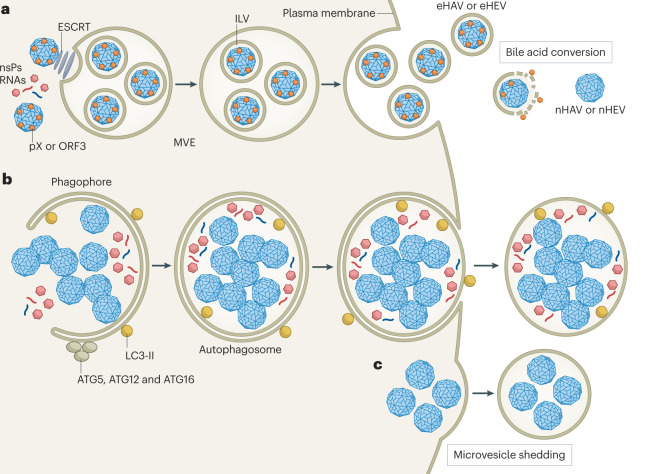
Life cycles and pathogenesis
Infection with either HAV or HEV typically results in only transient inflammatory liver injury, but both viruses can cause fulminant and even fatal hepatic failure17,18. Each poses a substantial threat to public health. HEV infects various animal species, most prominently swine, and zoonotic transmission to humans is not uncommon (Box 2). Fulminant hepatitis is strongly associated with one particular genotype (genotype 1) in pregnant women7. HEV infection can also persist in immunocompromised individuals, potentially causing chronic hepatitis and life-threatening cirrhosis19. Although viruses closely related to HAV infect many mammalian species (Box 2), the host range of HAV is limited, and zoonotic transmission does not contribute to its spread. Unlike HEV, HAV has not been associated with chronic hepatitis or cirrhosis. Inactivated HAV vaccines are highly effective, but outbreaks of acute hepatitis due to HAV continue to occur in both Europe and North America20,21, resulting in over 27,000 hospitalizations and more than 400 deaths in the USA since 2016 (Centers for Disease Control and Prevention).
Hepatocytes are polarized cells of epithelial origin with basolateral membranes facing onto the space of Disse, which communicates with the systemic circulation, and with apical membranes abutting bile canaliculi through which bile acids produced in hepatocytes flow to the gut (Fig. 1c). eHAV and eHEV are released across the basolateral membrane of hepatocytes, spilling into the bloodstream and causing a viraemia comprising primarily membrane-cloaked, yet still infectious virions10,22. Naked HEV virions (nHEV) lacking a quasi-envelope are also present in blood in some infected individuals, particularly late in infection when there is liver injury, but rarely represent more than 20% of circulating virus23. Only eHAV virions have been found in blood from naturally infected humans or experimentally infected chimpanzees8. Greater quantities of newly replicated viral progeny are released across the apical hepatocyte membrane into the biliary tract. High concentrations of bile acids present in the proximal bile canaliculus strip the membrane from eHAV, resulting in faecal shedding of naked HAV (nHAV)24 (Fig. 1d). Most evidence suggests that the majority of faecally shed nHAV is produced within hepatocytes and shed through the biliary tract25. No evidence for HAV replication was found within the small or large intestine in a mouse model of human hepatitis A in which there is extensive faecal shedding of virus26. nHEV particles shed in faeces may be similarly produced by bile acid conversion of eHEV released from hepatocytes across the apical membrane27,28. However, productive infection of primary human enterocytes has been described recently29, suggesting that HEV may be less strictly hepatotropic than HAV. Neurological complications, including Guillain–Barré syndrome, and renal injury also have been reported in patients with HEV infection19.
How these viruses first reach the liver is not well understood. Except in zoonotic transmission of HEV following ingestion of inadequately cooked liver or meat, or rare cases of transfusion-transmitted HAV or HEV infection, the initial infecting virion is likely to be a naked particle. Both HAV and HEV antigens have been detected within epithelial cells of intestinal crypts29,30 and, as noted above, primary enterocytes support productive HEV infection29. Nonetheless, it remains debatable whether a primary site of replication exists within the gastrointestinal tract for either virus. Regardless, once hepatocytes are infected, subsequent spread within the liver is likely due to quasi-enveloped virions only. This makes it important to understand how both naked and quasi-enveloped virions are able to enter cells and establish new infections, as well as the mechanisms by which viral progeny are sorted and exported from hepatocytes as quasi-enveloped virus.
HAV and HEV were the first ‘non-enveloped’ viruses found to be released from cells as quasi-enveloped virions8,9,31. This dual lifestyle, with both naked and membrane-cloaked extracellular particles, offers several distinct advantages. New viral progeny exit cells by usurping normal physiological pathways mediating the release of exosomes, leading to their release from the cell across the plasma membrane without cell lysis. This allows for productive HAV or HEV infection without direct cell damage and results in extended incubation periods (several weeks or more) during which virus is shed in faeces in the absence of liver injury (Fig. 1d). During this phase of the infection, the virus circulates within the body and spreads within the liver completely cloaked in host membranes (Fig. 1a). The membranes sequester viral antigens from the host immune system, adding to the stealthy nature of these infections (Box 3). By contrast, high residual infectivity of the naked particle after loss of the lipid coat within the biliary tract allows for faecal shedding of very stable virus. This facilitates transmission through the environment and provides for both person-to-person spread and large, common-source disease outbreaks (Fig. 1a).
Box 2 Zoonotic potential of enterically transmitted hepatitis viruses.
Bayesian phylogenies of the protein-coding regions of the hepatitis A virus (HAV) and hepatitis E virus (HEV) genomes are scaled similarly for genetic distance (see the figure). Hepatoviruses (see the figure, part a) constitute one of seven genera in the subfamily Heptrevirinae of the family Picornaviridae. Multiple serologically indistinguishable genotypes (gts) make up the Hepatovirus A species, which causes hepatitis in humans and nonhuman primates. Other distinct species (Hepatovirus B-I) infect various mammals, including seals, marsupials, rodents and bats35,136. Bat viruses are hepatotropic and serologically related to human HAV, and some show evidence of past host-species jumps35. Such host-species swaps could be favoured by the lack of a requirement for a specific protein receptor for viral entry, as gangliosides, which function as HAV entry receptors, are widely present in nature67. Despite significant divergence in their amino acid sequences, the pX proteins of both human and bat viruses recruit endosomal complexes required for transport (ESCRT) to drive vesicle-mediated cargo export when expressed in human cells93. Similarly, like the 3ABC protease of the human virus, the 3ABC proteases of bat viruses can cleave the human innate immune adapter mitochondrial antiviral signalling protein (MAVS) and disrupt interferon signalling137. Despite these findings, zoonotic transmission of hepatoviruses has yet to be demonstrated. Hepeviruses (see the figure, part b) are classified within the subfamily Orthohepevirinae, which comprises four of the five genera in the family Hepeviridae, two of which, Paslahepevirus and Rocahepevirus (previously Orthohepevirus A and Orthohepevirus C), infect humans. Members of the genus Paslahepevirus account for most cases of hepatitis E: gt1 and gt2 have been found only in humans, are mainly transmitted by contaminated water and are most prevalent in developing countries. Gt3 and gt4 are commonly found in swine and other mammalian species, have been associated with zoonotic transmission (usually due to ingestion of uncooked or inadequately cooked meat or liver) and can cause persistent infection in immunocompromised persons18. Human infections have also been documented with gt7 (camel) and a Rocahepevirus found in rats138,139. Figure adapted with permission from ref. 140, CSH.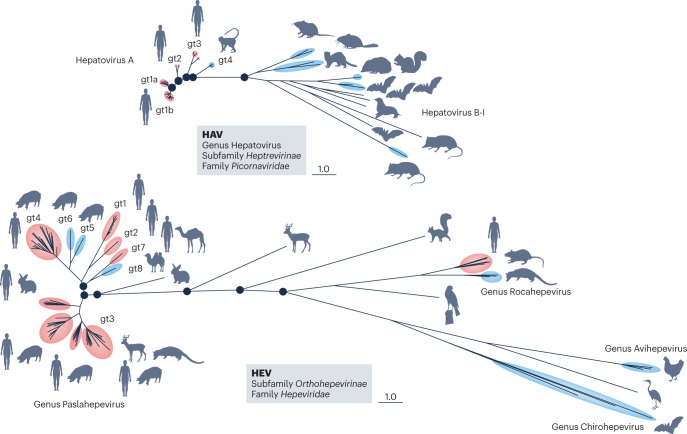
Box 3 The impact of quasi-envelopment on host immune responses.
Immune responses to hepatitis A virus (HAV) and hepatitis E virus (HEV) infection are incompletely understood, although evidence increasingly points to the importance of CD4+ T cells and neutralizing antibodies in virus control141. The release of viruses with quasi-envelopes is unlikely to affect cell-intrinsic or adaptive T cell responses to infection, but has profound implications for B cell immunity. Neutralizing antibodies recognize epitopes on the surface of the viral capsid52,142. These epitopes are not accessible to antibodies in either quasi-enveloped HAV (eHAV) or quasi-enveloped HEV (see the figure), neither of which is neutralized when incubated with antibodies before inoculation onto cells8,31. How then do antibodies in immune serum globulin protect against symptomatic hepatitis A when given 1–2 weeks after exposure, a phenomenon recognized over 75 years ago143? This is best explained by neutralization of the virus within endolysosomes following degradation of the quasi-envelope8. eHAV entry is relatively slow, requiring hours for trafficking to the endolysosome and degradation of the quasi-envelope. Neutralizing IgG and IgA antibodies that are taken up into endosomes can block replication when added to cells as late as 4–6 h after removal of an eHAV inoculum (see the figure). This interval can be extended by treating cells with an inhibitor of lysosomal acid lipase, which delays quasi-envelope degradation68. By contrast, antibodies are without effect when added to cells even immediately after endocytosis of naked virus. IgM antibodies fail to neutralize eHAV in this context, probably because they are unstable in the endolysosome8. Whether a specific immunoglobulin receptor is required to deliver antibody to the endolysosome for neutralization is unknown. Post-endocytosis neutralization of quasi-enveloped HEV has not been demonstrated, but seems likely. Sequestration of critical neutralization epitopes by the quasi-envelope may also delay the development of antibodies by making them inaccessible to programme B cell development.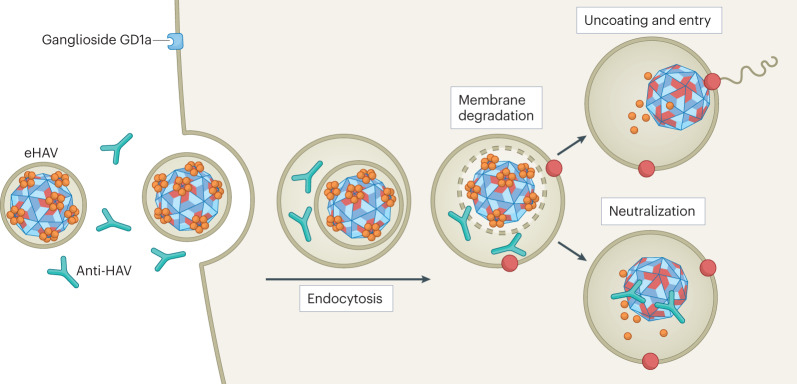
Genome replication and capsid assembly
Human strains of HAV represent one (Hepatovirus A) of nine viral species classified in the genus Hepatovirus of the family Picornaviridae (Box 2). The HAV capsid structure is distinct from that of other common picornaviral pathogens of humans, such as enteroviruses, and has features similar to those of distantly related viruses that infect insects32. HEV strains infecting humans demonstrate greater genetic diversity than the human-infecting strains of HAV (Box 2). Most are classified within the genus Paslahepevirus of the family Hepeviridae, but members of the genus Rocahepevirus also infect humans. These viruses may have had their origins in an ancient recombination event involving members of alphavirus-like and picornavirus-like superfamilies33,34. Multiple genotypes exist for both HAV and HEV, and in the case of HEV these define both routes of transmission and the potential to cause human disease (Box 2). Overall, there is little antigenic diversity among human strains of these viruses, all of which fall into single serotypes. In both cases, phylogenetically related nonhuman viruses infect various mammalian species35–37 (Box 2).
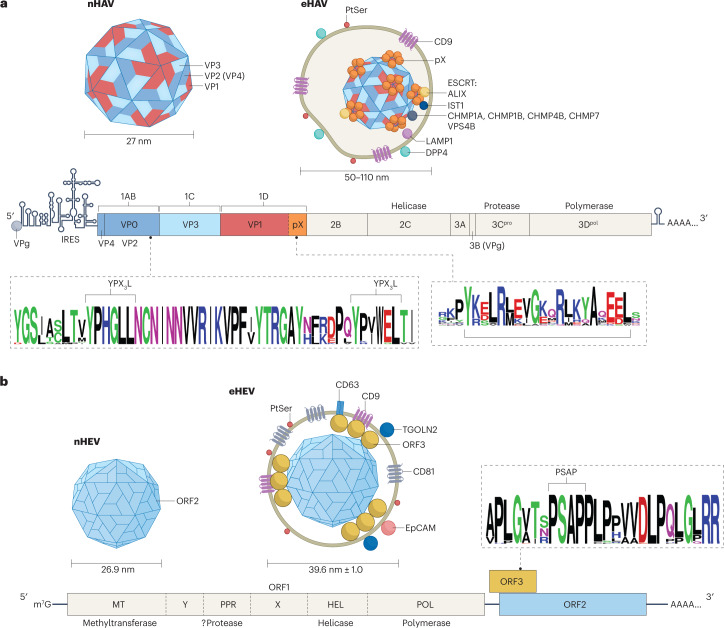
The genomes of both HAV and HEV are single-stranded, messenger-sense RNAs 7.2–7.5 kb in length with 3′ poly(A) tails (Fig. 2a,b). Beyond these similarities, however, genome organization and mechanisms of translation and RNA replication could hardly be more different. Like other picornaviruses, the HAV genome lacks a 5′ 7-methylguanosine cap and has a single large open reading frame (ORF) (Fig. 2a). Translation is initiated in a cap-independent fashion by an internal ribosome entry site located within a lengthy (~735 nucleotides) 5′ untranslated RNA segment. This results in synthesis of a large polyprotein that is proteolytically processed into multiple nonstructural proteins involved in genome replication and structural proteins that assemble into a stable capsid that packages newly produced genomic RNA. In contrast, the HEV genome is capped at its 5′ end and possesses three ORFs (Fig. 2b). The longest of these (ORF1) contains sequence motifs suggesting enzymatic activities typically associated with plus-strand RNA viruses, including a methyltransferase, a helicase, an RNA-dependent RNA polymerase and a papain-like protease. Surprisingly, however, it remains uncertain whether the ORF1 product is processed into smaller, functionally distinct nonstructural proteins, like the polyproteins of other positive-strand RNA viruses38–40. Only a single 190-kDa polyprotein was detected in cells transfected with a replicating subgenomic RNA with epitope tags in ORF139. The second longest ORF (ORF2; Fig. 2b) encodes the capsid protein, a secreted variant of which is found in high abundance in the bloodstream where it may decoy neutralizing antibodies41–43. Different ORF2 isoforms have been suggested to result from either leaky ribosome scanning or variable topology of an N-terminal signal sequence41,44. ORF2 is overlapped by a third ORF (ORF3; Fig. 2b) encoding a small, multifunctional phosphoprotein that appears to play a key role in virus release. In addition, a short fourth ORF has been described in genotype 1 HEV that overlaps ORF1 and appears to be translated under control of an upstream internal ribosome entry site during conditions of endoplasmic reticulum stress45.
Fig. 2. Genome organization and structures of naked and quasi-enveloped hepatitis A and hepatitis E virions.
a, Hepatitis A (HAV) RNA (7.5 kb) contains a lengthy 5′ untranslated region with secondary structure essential for genome replication and an internal ribosomal entry site (IRES) that initiates cap-independent translation of a single long open reading frame (ORF) encoding a large polyprotein. The polyprotein is processed by the HAV protease 3Cpro into three proteins that form the capsid, VP0 (also known as 1AB; subsequently processed into VP4 (1A) and VP2 (1B)), VP3 (1C) and VP1pX (1D), and six nonstructural proteins that mediate genome replication, 2B, 2C, 3A, 3B, 3Cpro and 3Dpol. The 8-kDa pX segment is present in quasi-enveloped HAV (eHAV), but cleaved from VP1 upon loss of the membrane, and it is not present in naked HAV (nHAV)8. Proteomics studies show that programmed cell death 6-interacting protein (ALIX), vacuolar protein sorting-associated protein IST1 homologue (IST1, a component of the endosomal complexes required for transport (ESCRT)-III complex), and multiple charged multivesicular body proteins (CHMPs) such as CHMP1A, CHMP1B, CHMP4B and CHMP7 (also components of ESCRT-III) are physically associated with eHAV11. b, Hepatitis E virus (HEV) RNA (7.2 kb) has a 5′ 7-methylguanosine (m7G) cap and three ORFs that are translated in a cap-dependent manner. ORF1 encodes a multifunctional polyprotein; whether this is processed into smaller proteins in infected cells is uncertain. ORF2 and ORF3 are translated from a subgenomic RNA. ORF2 encodes the capsid protein, but also produces a secreted protein from an in-frame start codon that is thought to decoy neutralizing antibody. ORF3 protein associates with membranes and recruits TSG101, a component of ESCRT-I; it is not present in nHEV104,121,122. An additional ORF4 exists within the ORF1 coding region of genotype 1 virus (not shown). WebLogos show conserved domains in VP2 and pX91,93 proteins of HAV and in ORF3 protein of HEV (sequences from 40 paslahepeviruses) that recruit ESCRT during capsid quasi-envelopment. Naked and quasi-enveloped virions are shown at the top of each panel at the left and right, respectively, with associated host proteins and lipids11,22,59. EpCAM, epithelial cell adhesion molecule (or CD326 antigen); LAMP1, lysosome-associated membrane protein 1; PtSer, phosphatidylserine; TGOLN2, trans-Golgi network integral membrane protein 2.
Molecular details of the RNA replication cycle are not well characterized for either HAV or HEV. Like most positive-strand RNA viruses, their genomes are synthesized within membranous organelles derived from the endoplasmic reticulum or Golgi. HAV replication is thought to be mechanistically similar to that of other, better-studied picornaviruses, with protein-primed RNA synthesis leading to transcription of a genome-length, negative-strand intermediate that subsequently templates multiple rounds of positive-strand RNA synthesis46. Unlike other picornaviruses, however, HAV RNA synthesis is surprisingly dependent upon host-encoded terminal nucleotidyltransferases47. The HEV genome is similarly transcribed into a negative-strand RNA intermediate that templates positive-strand RNA synthesis, but a second, dicistronic RNA that directs the synthesis of ORF2 and ORF3 proteins is also produced48. Little is known about how transcription of the genomic and subgenomic RNAs is regulated. To a large extent, this gap in knowledge can be attributed to the lack of cell culture systems supporting robust replication of the virus49. Recent advances using stem cell-derived hepatocytes or better cell culture support medium may allow more efficient replication and facilitate future studies of molecular events in the viral life cycle28,50.
Although similar in size, the structures of the capsids of these viruses are also very different. The HAV capsid assembles from 60 copies each of three major structural proteins — VP0 (also known as 1AB), VP3 (1C) and VP1pX (1D) — following processing of the polyprotein by the 3C protease (Fig. 2a). As in other picornaviruses, VP0 undergoes additional processing into VP4 (1A) and VP2 (1B) after the RNA genome is packaged. It is not known whether these assembly events precede or occur coincident with interactions of the capsid with endosomal membranes leading to its quasi-envelopment and export (see below). Following the release of eHAV and degradation of its surrounding membrane, VP1pX is cleaved again by an unknown host protease, resulting in removal of the C-terminal 8 kDa (pX segment) from the capsid8. This results in the presence of four mature capsid proteins (VP1, VP2, VP3 and VP4) in the naked extracellular capsid, which is extraordinarily stable at low pH and high temperature32,51. Model structures produced by X-ray crystallography and cryo-electron microscopy (cryo-EM) show the naked particle to be a pseudo T = 3 icosahedron of approximately 27 nm diameter with a relatively featureless surface32,52. The capsid lacks the deep canyon surrounding the fivefold axis of symmetry existing in enteroviruses and has a prominent domain swap in VP2 found also in parechoviruses and insect dicistroviruses32,53. There are no similar studies of the quasi-enveloped capsid.
The HEV capsid is 30–33 nm in diameter, slightly larger than HAV, and comprises 180 copies of the ORF2 protein assembled into a T = 3 icosahedron54 (Fig. 2b). The ORF2 protein contains an N-terminal endoplasmic reticulum-targeting signal sequence55. However, ORF2 protein present in capsids lacks either 13 or 15 N-terminal amino acids, due either to translation initiating at an internal start codon or possibly to cleavage by an unknown intramembrane protease that disrupts the signal peptide sequence41,42,44. Thus, like the capsid proteins of HAV, there are no membrane interaction domains in the mature HEV capsid proteins. The naked nHEV virion is not as stable as nHAV, but is still well suited for faecal–oral transmission and spread to naive hosts56. Atomic-level resolution models exist for recombinant virus-like particles, but not for bona fide infectious nHEV or eHEV capsids.
Quasi-enveloped virions of HAV and HEV
Most virus released into supernatant fluids of cell cultures infected with a low-passage, noncytopathic strain of HAV is quasi-enveloped24. A minor population of naked extracellular virions is likely to represent cryptic cell lysis or loss of the membrane from eHAV after egress. Electron microscopy reveals one to three capsids enclosed in vesicles with diameters ranging from 50 to 110 nm8 (Fig. 2a). As described above, these capsids contain intact VP1pX rather than the processed VP1 present in extracellular naked particles8,11. eHAV bands at a density of 1.08–1.12 g/cm3 in iodixanol gradients, compared with 1.28 g/cm3 for nHAV, and is not bound by anti-capsid antibodies in the absence of detergent8. The host protein composition of eHAV closely matches that of exosomes, including CD63 antigen and epithelial cell adhesion molecule (EpCAM), although eHAV virions are relatively enriched for CD9, dipeptidyl peptidase 4 (DPP4) and proteins associated with ESCRT11. Only genome-length RNA is detected in northern blots of purified eHAV57. Most of this RNA seems to be fully encapsidated. Like exosomes, eHAV vesicles are enriched in sphingomyelins and ceramides (S.M.L., unpublished work), and they display phosphatidylserine on their surface57.
eHEV virions released from infected PLC/PRF/5 human hepatoma cells have a buoyant density similar to those of eHAV (1.11 g/cm3) but are morphologically different (Fig. 2b). eHEV virions are smaller (~40 nm diameter) and more uniform in appearance than eHAV, with the membrane adhering more tightly to the capsid22. Only a single capsid is present in each eHEV virion, highlighting a key difference from eHAV and suggesting an integrated process for capsid assembly and quasi-envelopment. eHEV contains both ORF2 and ORF3 proteins, whereas non-enveloped nHEV capsids contain only ORF2 protein58. The ORF3 protein thus resembles pX in being associated only with quasi-enveloped and not naked virions. This is consistent with critical roles for both ORF3 protein and pX in interactions with ESCRT during capsid quasi-envelopment. Like eHAV, no viral antigens are present on the surface of the eHEV quasi-envelope11,31. The host proteins associated with eHEV membranes have yet to be studied at the same level of detail as eHAV, but immunoprecipitation studies suggest a similar complement of exosome-associated proteins, including CD9 and EpCAM22. Consistent with originating in MVEs, the membrane surrounding eHEV virions contains the trans-Golgi network protein TGOLN2 (ref. 59). As with eHAV, phosphatidylserine is displayed on the surface of eHEV virions22.
The relative specific infectivities (infectious units per RNA genome equivalent) of quasi-enveloped versus naked virus particles can vary substantially depending upon the host cell substrate, probably reflecting different requirements for attachment and entry of the two virion types (see below). The specific infectivity of eHAV approximates that of the naked virion in Huh-7 human hepatoma cells8. Similarly, the specific infectivity of eHEV has been estimated to be about 50% that of nHEV in HepG2 human hepatoma cells60, although such measurements are difficult given low overall infectivity in cell culture. Both types of quasi-enveloped virions have been shown to be infectious in vivo61,62.
Cell entry
Cell entry of naked viruses
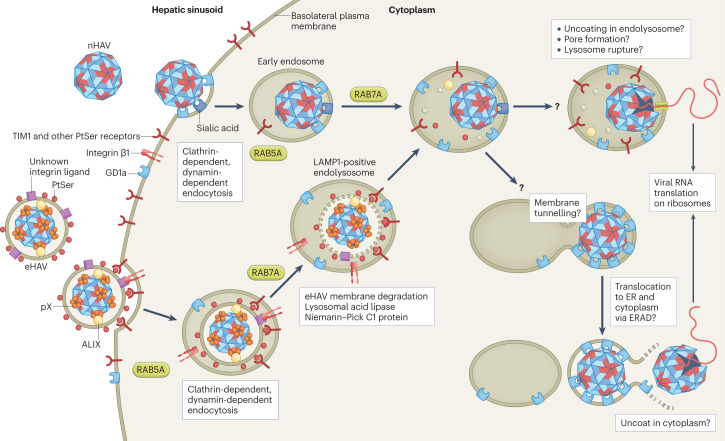
Non-enveloped viruses typically enter host cells through interactions of the capsid with receptor molecules on the cell surface that initiate endocytosis and, in some cases, uncoating of the viral genome63. These interactions are generally virus-specific, with different receptor molecules often used by otherwise closely related viruses. The transmembrane T cell immunoglobulin mucin receptor 1 (TIM1, also known as CD365) was suggested decades ago to function as a receptor for HAV leading to its currently approved name: hepatitis A virus cellular receptor 1 (HAVCR1)64. However, more recent studies with targeted CRISPR deletion show that TIM1 facilitates entry only of quasi-enveloped virions, most probably by binding phosphatidylserine on the vesicle surface61,65,66. Double-knockout Tim1−/−Ifnar1−/− mice are readily infected with HAV61. No specific protein receptor has yet been shown to be required for HAV entry.
By contrast, genome-wide CRISPR screens have revealed the synthesis of gangliosides (sphingolipids with carbohydrate headgroups containing one or more sialic acid moieties) to be essential for HAV infection65,67. Gene depletion and confocal microscopy studies show that nHAV enters hepatoma cells via clathrin-dependent, dynamin-dependent endocytosis, co-localizing sequentially with the small GTPases RAB5A and RAB7A as it traffics from early to late endosomes68 (Fig. 3). Endocytosis is blocked by depletion of integrin β1, but no specific α-integrin binding partner has been identified. In cells genetically deficient in ganglioside synthesis, nHAV continues to undergo endocytosis and traffics to lysosomal-associated membrane protein 1 (LAMP1)-positive late endolysosomes, where its trafficking is arrested without evidence of capsid disassembly67. Adding gangliosides to the medium, even hours after infection, rescues entry, and the virus can then be observed to uncoat its genome and begin to express viral proteins67. Gangliosides are thus essential for a late step in viral entry, which includes uncoating of the genome and its transport across the endolysomal membrane to the cytoplasm, where it can begin to be translated on ribosomes to produce viral proteins (Fig. 3).
Fig. 3. Cellular entry of naked and quasi-enveloped hepatitis A virus.
Both virion types undergo clathrin-dependent endocytosis driven by interactions between distinct ligands, including phosphatidylserine (PtSer) on the quasi-enveloped hepatitis A virus (eHAV) surface and cellular PtSer receptors such as T cell immunoglobulin mucin receptor 1 (TIM1)68. Endocytosis of naked HAV (nHAV) is inhibited by either sialidase or trypsin treatment of the cell, suggesting that gangliosides and proteins (possibly sialylated) are involved67. Integrin β1, presumably in association with different α-integrins, is required for endocytosis of both virion types. Both virion types traffic through early (RAB5A-positive) and late (RAB7A-positive) endosomal compartments. Subsequent steps that involve uncoating and genome release into the cytoplasm require endolysosomal gangliosides, preferably GD1a (ref. 67). These late steps are delayed for eHAV, which must traffic first to lysosomal-associated membrane protein 1 (LAMP1)-positive endolysosomes, where the quasi-envelope is degraded by lysosomal enzymes and cholesterol transporters, such as lysosomal acid lipase and Niemann–Pick C1 protein. Like nHAV entry, eHAV entry is dependent upon endosomal gangliosides. Subsequent steps in entry are not well understood, but progressive binding of the now-naked capsid to membrane-bound gangliosides may result in tunnelling of the capsid into the membrane. The trigger for capsid disassembly is not known, nor is it known whether uncoating occurs in the endolysosomal lumen or following transport of the capsid across the endolysosomal membrane to the cytoplasm. Entry of naked hepatitis E virus is not dependent upon RAB5A or RAB7A, but entry of quasi-enveloped hepatitis E virus is similar to eHAV entry and also involves trafficking to endolysosomes for degradation of the quasi-envelope60. Receptors have not been identified for the hepatitis E virus capsid. Steps in entry that are not understood are indicated by a question mark. ER, endoplasmic reticulum; ERAD, endoplasmic reticulum-associated degradation pathway.
nHAV capsids bind gangliosides immobilized on a solid-phase support, and preincubating nHAV with gangliosides blocks infection67. The disialoganglioside GD1a is most active in such experiments, with a 50% inhibitory concentration (IC50) of 1.25 µM. Infection is also blocked partially by pretreating cells with sialidase67. However, trypsin treatment similarly impairs infection, suggesting that proteins expressed on the cell surface — possibly sialylated glycoproteins — may contribute to endocytosis of nHAV.
It is not known whether HAV uncoating initiates within the lumen of the endosome, as it does with other picornaviruses69. Although PLA2G16, a cellular A2 phospholipase, is essential for this late step in entry of many picornaviruses, it is not required and may even inhibit entry of nHAV68. The nHAV capsid is maximally stable at the low pH of endolysosomes, and it is not destabilized by the binding of GD1a32,67. The exceptional stability of the capsid at high temperatures or low pH raises questions as to how its disassembly initiates. Whereas a high-resolution X-ray structure provides no clues as to where a cellular receptor might interact to trigger uncoating32, a cryo-EM study shows that the capsid can be destabilized by binding of a potent neutralizing antibody near the twofold axis of symmetry52. However, it is not clear how this relates to uncoating during viral entry, and the final steps in nHAV entry remain to be defined. One intriguing possibility is that entry involves transport of the intact capsid into the cytoplasm, with uncoating triggered by interactions with a cytoplasmic protein32 (Fig. 3). Although unprecedented for a picornavirus, progressive binding of multiple membrane-bound gangliosides to polyomavirus capsids has been shown to promote a considerable inward curvature of the membrane, resulting eventually in the particle tunnelling into the membrane70.
Less is known about the entry of nHEV particles. A putative receptor-binding site has been mapped to a region in the ORF2 protein with polysaccharide-binding activity in recombinant HEV-like particles71,72, but no specific receptor molecule has been identified. Entry of nHEV is at least partially dependent upon clathrin-dependent, dynamin-dependent endocytosis, but unlike nHAV it does not require either RAB5A or RAB7A60. This suggests that nHEV uncoating is initiated early in an endocytic pathway.
Cell entry of quasi-enveloped viruses
The membrane surrounding the viral capsid poses an additional barrier that must be overcome for both eHAV and eHEV to enter cells and deliver their RNA genome to the cytoplasm, where it can be translated on ribosomes. The entry of canonical enveloped viruses typically involves membrane fusion mediated by a virally encoded surface glycoprotein (a ‘peplomer’)73. No such proteins exist in either eHEV or eHAV. Most evidence supports a model in which these quasi-enveloped virions enter cells via endocytosis followed by degradation of their membranes within the endolysosome.
The initial attachment and endocytosis of quasi-enveloped virions is probably mediated by multiple nonspecific interactions. In the case of eHAV, these include interactions of the virion with phosphatidylserine receptors such as TIM161,66 (Fig. 3). TIM1 is widely expressed, particularly in the kidney, and facilitates the attachment and entry of various conventional enveloped viruses74. The density of such receptors on the cell surface may determine the relative efficiency with which quasi-enveloped viruses bind to and enter cells. For example, eHAV binding to GL37 African green monkey kidney cells, which express very high levels of TIM1, is more efficient than nHAV binding at 4 °C, whereas the binding efficiencies are reversed in hepatoma cells expressing much less TIM1 (ref. 66). In addition to TIM1, other phosphatidylserine receptors such as TIM4 or AXL (tyrosine-protein kinase receptor UFO) may facilitate endocytosis. eHAV subsequently undergoes clathrin-dependent endocytosis and, to a lesser degree, caveolin-dependent endocytosis, and can be seen to traffic through RAB5A-positive (early) and RAB7A-positive (late) endosomes to LAMP1-positive endolysosomes68 (Fig. 3). This may be driven in part by membrane interactions with TIM1, as TIM1 family proteins are known to carry their cargo from the cell surface to the lysosome75. Interestingly, like nHAV, the endocytosis of eHAV is also dependent upon integrin β1 (ref. 68). The ligand in eHAV that binds integrin β1 is unknown but is almost certainly distinct from the nHAV ligand. eHEV entry also involves clathrin-mediated endocytosis and, in sharp contrast to nHEV, similar RAB5A-dependent and RAB7A-dependent trafficking to endolysosomes60. However, roles have yet to be defined for phosphatidylserine receptors or integrins in eHEV entry.
Subsequent steps in entry of both quasi-enveloped viruses are facilitated by constitutive cellular mechanisms that mediate the degradation of membrane lipids in lysosomes76 (Fig. 3). Confocal microscopy of cells infected with eHAV virions labelled with an irreversible membrane dye shows that the eHAV membrane is degraded in LAMP1-positive endolysosomes68. The eHEV membrane is similarly degraded in endolysosomes60. The time required for trafficking of the virus to the endolysosome and degradation of the quasi-envelope renders the entry kinetics of eHAV and eHEV significantly slower than those of their naked counterparts8,60,61,68. Lysosomal acid lipase (LAL) and Niemann–Pick C1 protein (NPC1) contribute to the degradation of the eHAV and eHEV quasi-envelopes60,68. NPC1 is an essential receptor required for Ebola virus entry, during which it interacts directly with the viral glycoprotein in lysosomes77,78. Its role is very different in eHAV entry, during which it is likely to scavenge cholesterol from the quasi-envelope membrane60,68 (Fig. 3). Degradation of the quasi-envelope is rate-limiting in the entry of eHAV (and probably eHEV) and can be slowed by lalistat 1, a small-molecule LAL inhibitor68. Lysosomal proteases may also contribute to quasi-enveloped virus entry, either by processing VP1pX to VP1 following removal of the quasi-envelope79 or by degrading the HEV ORF3 protein. However, it is not known whether either of these actions is required for subsequent steps in entry. ORF3 has been reported to possess ion channel activity80, but this seems unlikely to be important for entry.
eHAV continues to be taken up by endocytosis in cells lacking gangliosides but, like the naked particle, fails to uncoat its genome67. This suggests that the capsid interacts with the same endosomal ganglioside receptors as nHAV following its liberation from the quasi-envelope (Fig. 3). As with nHAV, the trigger for capsid disassembly and uncoating is not known, nor is it certain that uncoating occurs within the endolysosomal lumen. However, disassembly of the eHAV capsid occurs in temporal association with a loss of endolysosomal membrane integrity68. This suggests that the virus induces pores in the endolysosomal membrane, similar to pores forming in endosomal membranes during the entry of some picornaviruses81. Structural rearrangements occurring during disassembly of the enterovirus capsid externalize its VP4 protein, which forms pores within the endosomal membrane through which the viral RNA may pass82. The much-smaller VP4 protein of HAV has pore-forming activity in vitro83, but whether it is similarly externalized during uncoating of the capsid is not known. Interestingly, breaches in endosomal membranes consistent with pore formation were not observed during entry of nHAV particles68. This does not necessarily indicate that distinct mechanisms exist for nHAV versus eHAV in traversing the membrane and could simply reflect more rapid membrane repair at sites breached by nHAV earlier in the endolysosomal pathway.
Like eHAV, how and where the eHEV capsid uncoats is not known. The endolysosome may serve only as a transitional compartment for both eHEV and eHAV within which the virus loses its quasi-envelope. For example, following endocytosis, non-enveloped human papillomavirus virions hijack retromers to traffic from the late endosome to the trans-Golgi network, after which they are released from the endoplasmic reticulum via the endoplasmic reticulum-associated degradation (ERAD) pathway84. Polyomavirus follows a similar complicated entry pathway85. Speculatively, it could be the same for both eHEV and eHAV (Fig. 3). Overexpressed HEV ORF2 proteins translocate to the cytoplasm via the endoplasmic reticulum-associated degradation pathway86. Moreover, ganglioside GD1a, which binds to the HAV capsid and serves as an endosomal receptor67, has been shown to act as a signal for sorting endolysosomal polyomavirus to the endoplasmic reticulum following its endocytosis85.
An alternative model for eHAV entry involving fusion rather than degradation of the eHAV membrane was proposed recently87. Biochemical and cryo-EM studies provide evidence that exosomes originating from MVEs can fuse with low efficiency to endosomal membranes following endocytosis88,89. However, there is no direct experimental evidence supporting fusion of the quasi-envelope and endosomal membranes. Moreover, a fusion model fails to explain why the membrane-degrading activities of LAL and NPC1 are required for quasi-enveloped viral entry60,68. Fusion would also deliver viral capsids or RNA directly to the cytoplasm88,89, making it difficult to understand why endosomal gangliosides are essential for eHAV infection67.
A second controversy concerns the extent to which non-encapsidated RNA contributes to the infectivity of eHAV87. Although some non-encapsidated RNA could be transmitted between cells within exosomes, as described for hepatitis C virus90, the requirement for endosomal ganglioside receptors also argues against non-encapsidated RNAs being primarily responsible for the infectivity of these virions. As important, anti-capsid antibodies effectively neutralize infectivity when added to cells following endocytosis of the quasi-enveloped virus, most probably interacting with the capsid after degradation of the eHAV membrane (Box 3)8,68. Membrane-bound capsids are also readily visualized in purified eHAV preparations8, and interactions of the capsid with ESCRT are essential for the biogenesis of infectious eHAV91–93, as described in the following section.
ESCRT-dependent release
Quasi-enveloped viruses and the ESCRT system
Despite major differences in the mechanisms responsible for viral protein synthesis and RNA transcription, mechanisms underlying the biogenesis and release of eHAV and eHEV seem to be quite similar. Both HAV and HEV capsids have been identified within cytoplasmic vesicles with morphology consistent with MVEs in infected liver tissue or cell cultures9,94,95. These observations are consistent with other data indicating that the biogenesis of quasi-enveloped virions involves the sorting of viral capsids into MVEs in an ESCRT-dependent process mirroring the biogenesis of exosomes12 (Fig. 4). Importantly, quantitative proteomics studies of purified eHAV virions show that the sorting of intracellular HAV capsids into vesicles for export from the cell is highly specific and selective11. Extracellular eHAV vesicles contain only the capsid proteins of HAV and no detectable nonstructural viral proteins.
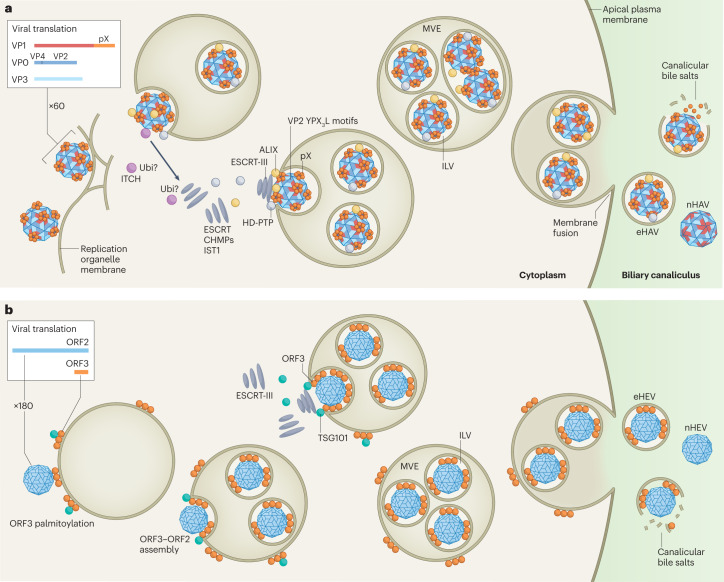
Fig. 4. Multivesicular endosome-dependent biogenesis of quasi-enveloped virions.
a, Hepatitis A virus (HAV) capsids assemble from 60 copies of three major capsid proteins in close association with the limiting membrane of endosomes into which they bud. An 8-kDa C-terminal extension of the VP1 protein, pX, recruits two endosomal complexes required for transport (ESCRT)-associated proteins — programmed cell death 6-interacting protein (ALIX) and tyrosine-protein phosphatase non-receptor type 23 (HD-PTP) — as well as the ubiquitin ligase ITCH. This induces assembly of ESCRT-III complexes containing multiple charged multivesicular body proteins (CHMPs) and vacuolar protein sorting-associated protein IST1 homologue (IST1) that pinch off the membrane, creating an intraluminal vesicle (ILV) that contains single or multiple HAV capsids93. Ubiquitin (Ubi) tags conjugated to viral or ESCRT-associated proteins by ITCH may facilitate this process. ALIX-interacting late domains also exist within the VP2 capsid protein (YPX3L motifs)91. Multivesicular endosomes (MVEs) containing multiple HAV-laden ILVs traffic to the apical (shown) and basolateral (not shown) membranes, where fusion leads to the release of quasi-enveloped HAV (eHAV) into the biliary tract and sinusoidal blood, respectively. High concentrations of bile acids strip the membrane from eHAV in the bile canaliculus, resulting in faecal shedding of naked virus (nHAV). b, The biogenesis of quasi-enveloped hepatitis E virus (eHEV) is similar to eHAV, with capsids assembling from 180 copies of ORF2 protein interacting with ORF3 protein associated with the cytosolic leaflet of endosomal membranes via its palmitoylated N terminus121. ORF3 protein recruits the ESCRT-I protein TSG101 to initiate ESCRT-dependent membrane scission and ILV formation104,122. Additional interactions of ORF2 or ORF3 proteins with other ESCRT-associated proteins seem likely to occur but have not been identified. eHEV ILVs generally contain only a single capsid, possibly reflecting a more integrated process of capsid assembly and quasi-envelopment. Fusion of the MVE and plasma membranes and release of HEV virions is similar to HAV.
The ESCRT system is responsible for constricting and severing membranes in cellular events involving deformation of membranes in an outward direction from the cytosol, in reverse of the topology of endocytosis96,97. Distinct ESCRT complexes act sequentially to sort and load cargo into vesicles within the lumen of MVEs that ultimately become exosomes12,96,98. Monoubiquitylation or K63 ubiquitin chain formation typically mark cargo destined for MVEs, and ESCRT-0, ESCRT-I and ESCRT-II complexes contain multiple ubiquitin-binding domains that bind such cargo99. ESCRT-I and ESCRT-II represent core ESCRT machinery that sequester cargo at the endosomal membrane, deforming the membrane and creating an inward bud97,100. ESCRT-III oligomers are recruited by ESCRT-II and assemble into a complex that constricts the neck of the budding membrane and mediates membrane scission97,101, releasing the buds into the endosomal lumen as intraluminal vesicles (ILVs) to form MVEs. In later steps, deubiquitinases remove ubiquitin moieties and the ATPase VPS4A/B dissociates and recycles the ESCRT-III oligomers. The Bro1 domain-containing paralogs — programmed cell death 6-interacting protein (ALIX) and tyrosine-protein phosphatase non-receptor type 23 (HD-PTP) — are accessory ESCRT proteins that feed into ESCRT-III complexes in parallel with ESCRT-I and ESCRT-II, and interactions with them provide alternative pathways for cargo selection100,102,103. MVEs may either fuse with lysosomes for degradation of their cargo, or traffic to the plasma membrane where fusion releases ILVs as extracellular exosomes12. Efficient release of eHAV from infected cells requires both ALIX and HD-PTP, as well as several ESCRT-III components and VPS4B, all indicating a crucial role for ESCRT in quasi-envelopment of the capsid8,11,93. Similarly, eHEV release requires TSG101, a component of ESCRT-I, and VPS4104.
Biogenesis of quasi-enveloped HAV virions
ESCRT complexes are essential for the egress of many canonical enveloped viruses from infected cells105,106. The budding of human HIV-1 from the plasma membrane of infected cells requires specific ‘late domains’ in its p6Gag protein conforming to peptide motifs that bind ALIX (YPX1–3L) and TSG101 (PTAP)107,108. Fusing p6Gag to the C terminus of a de novo-designed protein that self-assembles into a 60-copy, 25-nm dodecahedron results in release of these ‘nanocages’ from transfected cells in exosome-like vesicles109. Both ALIX and TSG101 are required for efficient HIV-1 budding, and both late domains are required for release of the nanocage protein in EVs109. Nanocage release also requires a myristoylation signal specifying addition of myristic acid to the N-terminal glycine of the protein, identifying membrane association as a second critical element required for export of the particle. Nanocages containing the hepatovirus pX sequence (Fig. 2a) in lieu of p6Gag are similarly released from cells in an ESCRT-dependent manner, revealing the existence of a potent export signal within pX that is capable of functionally substituting for both p6Gag late domains93. pX similarly directs the cellular release of GFP in exosomes when fused to its C terminus92. A 24-amino acid segment within the centre of pX is both necessary and sufficient for nanocage release (Fig. 2a), and viruses with mutations in this segment are impaired in egress from infected cells93. This pX sequence is conserved across a broad range of hepatovirus species identified in small mammals, including bats (Box 2), suggesting that nonlytic release of quasi-enveloped virus is a shared attribute of hepatoviruses infecting host species separated by over 40 million years of evolution93.
ALIX has a crucial role in eHAV release. The protein contains an N-terminal, boomerang-shaped Bro1 domain, a central V domain with a grooved conformation and a C-terminal proline-rich PRD domain107,110,111. Whereas the YPX1–3L late domain in p6Gag binds to the V domain of ALIX112, pX binds to the Bro1 domains of both ALIX and its paralog, HD-PTP93. Many canonical enveloped viruses are dependent upon ALIX for release from infected cells, but eHAV is thus far unique in also requiring HD-PTP93. Despite closely related N-terminal domain architecture, ALIX and HD-PTP differ functionally. ALIX contributes to numerous ESCRT-related processes, including cytokinesis, nuclear envelope reformation, endolysosomal repair and MVE formation113,114. By contrast, HD-PTP functions selectively at endosomes, where it facilitates the recycling of activated epidermal growth factor receptor (EGFR) by sorting it into ILVs during MVE formation102,103. Unlike HIV-1, eHAV release does not require the ESCRT-I component TSG1018. However, HD-PTP has binding sites for STAM2, which is also an ESCRT-0 and ESCRT-I component115, providing an alternative mechanism for early ESCRT recruitment. Super-resolution microscopy shows that pX co-localizes with ALIX and HD-PTP, primarily at sites on intracellular membranes but also, to a limited extent, on the plasma membrane of infected cells93.
Together with recruitment of the two Bro1 domain proteins, pX also interacts directly with the NEDD4-family E3 ubiquitin ligase ITCH116. Although there is no evidence that ITCH ubiquitylates pX, the release of eHAV from infected cells is inhibited by chemical compounds that disrupt interactions between NEDD4 ligases and their target proteins116. RNAi depletion of ITCH also impairs eHAV release. It is possible that ITCH facilitates eHAV release by ubiquitylating the capsid, thus providing a signal for ESCRT-I recruitment. Alternatively, it could promote eHAV release by ubiquitylating and thereby activating an ESCRT component99,117,118.
In addition to the ESCRT-adaptor functions of pX, the VP2 capsid protein contains conserved tandem YPX3L motifs, which conform to late domains in canonical enveloped viruses that bind ALIX8,91 (Fig. 2a). Mutations within these motifs impair eHAV release but also interfere with capsid assembly, making them difficult to study91. An X-ray crystallographic model of detergent-treated nHAV particles show the YPX3L motifs are largely buried beneath the surface of the capsid, where they cannot bind ALIX32,53. A certain degree of ‘breathing’ occurs in the structures of other picornaviral capsids, resulting in externalization of otherwise internal VP4 polypeptide sequence119. Although this has not been documented with HAV, such dynamic changes in structure might explain how the VP2 late domains interact with ALIX.
The variable number of HAV capsids present within quasi-enveloped virions and the loose appearance of the membranes surrounding the capsids (Fig. 2a) suggest that capsid assembly might occur before and possibly independent of the membrane interactions required for quasi-envelopment and export. The amino terminus of VP4, normally buried within the interior of the HAV capsid, has been shown to associate with membranes independent of myristoylation83,93. Breathing of the capsid might enable the VP4 protein to provide for the membrane association suggested by the nanocage studies93,109. Alternatively, HAV capsids could assemble directly on intracellular membranes, as shown recently for enteroviral capsids by cryo-electron tomography120. Whether this is also true for HAV is unknown, but it raises the intriguing possibility that quasi-envelopment might be coupled temporally and spatially with HAV capsid assembly, possibly involving an assembly intermediate in which both the amino terminus of VP4 and the VP2 late domains are accessible for membrane interactions. It remains to be shown whether the structure of the intracellular HAV capsid that buds into the MVE is identical to that present in extracellular nHAV, as is generally assumed.
Biogenesis of quasi-enveloped HEV virions
Although not studied in as much detail, the biogenesis of eHEV is similar to that of eHAV in many respects. As indicated above, both TSG101 and VPS4 are required for efficient release of eHEV, indicating that eHEV release is ESCRT-dependent104. Like pX, the HEV ORF3 protein is found associated only with quasi-enveloped virions, not with naked virions58. Also, and again like pX, HEV ORF3 is not present on the exterior of the eHEV membrane. Palmitoylation of ORF3 mediates its association with the cytosolic leaflet of membranes and is essential for eHEV release121. It seems likely that capsids assembled from nonglycosylated ORF2 protein interact with palmitoylated ORF3 on the cytosolic surface of endosomal membranes, then bud into the endosome to form an ILV with ORF3 functioning to recruit ESCRT to mediate membrane scission (Fig. 4). The presence of only a single HEV capsid in each quasi-enveloped virion and the close proximity of the eHEV membrane to the capsid contrasts with eHAV and suggests the possibility that capsid assembly and quasi-envelopment in HEV are more tightly linked spatially or temporally than in HAV. The HEV ORF3 protein contains a PSAP motif (Fig. 2b) near its C terminus similar to the PTAP late domain in p6Gag that binds TSG101104,122. ORF3 is conserved among human strains but highly variable among hepeviruses infecting other animal species. The PSAP motif is not present in rat HEV and thus may have evolved relatively recently. It exists near the middle of the ferret HEV ORF3 sequence, but virions in ferret blood do not appear to be quasi-enveloped123.
Most HEV is released from the apical (canalicular) surface of polarized HepG2 hepatocytes124,125, and ORF3 co-localizes with the ORF2 capsid protein in vesicular structures near the apical surface of these cells124. Likewise, in HEV-infected human liver chimeric mice, ORF3 is found close to the apical membrane and within biliary canaliculi126. A mutant HEV lacking ORF3 expression replicated transiently in such mice, but failed to be shed into faeces, further confirming a key role for ORF3 in virus release125.
Conclusions
Despite being phylogenetically and structurally distinct, HAV and HEV have evolved common life cycles and similar mechanisms for extracellular release. The discovery of quasi-enveloped viruses blurred old distinctions between non-enveloped and enveloped viruses and has profoundly changed the way we think about these infections. Despite the absence of virus-encoded proteins on their surface, these virions are infectious and efficiently spread infection in vivo. Quasi-envelopment of the viral capsid isolates capsid-associated antigens from the immune system, probably delaying immune system recognition and explaining why neutralizing antiviral antibodies do not appear until several weeks after infection (Fig. 1d). It also prevents the neutralization of cell-free viruses once such antibodies are present. Much has been learned about the mechanisms underlying the biogenesis of quasi-enveloped virions, but questions remain concerning the recruitment of ESCRT, particularly by the HEV capsid. We have yet to determine what drives MVEs containing viral capsids to the plasma membrane for release, rather than to lysosomes with which MVEs often fuse for degradation of their cargo. Many questions also remain concerning how these viruses enter cells, including the trigger for uncoating of the capsids and how their RNA genomes are delivered across the endolysosomal membrane to the cytoplasm. Antibody-mediated post-endocytotic neutralization of these enterically transmitted hepatitis viruses (Box 3) is likely to be important in the pathogenesis and control of both infections, but it also remains incompletely understood. The study of these viruses has been a particularly fruitful field of research in virology in recent years, and further investigation is likely to yield additional surprises along with a better understanding of their life cycle and pathogenesis.
Acknowledgements
Research in the authors’ laboratories has been supported in part by grants from the U.S. National Institutes of Health: R01-AI103083, R01-AI131685 and R01-AI150095 (S.M.L.) and R01-AI139511 and R21-AI159735 (Z.F.).
Glossary
- Autophagy
A cellular process by which the cytoplasmic content becomes engulfed in a double-membrane vesicle (autophagosome) that fuses either with a lysosome for degradation or with the plasma membrane for secretion of its cargo.
- Bile canaliculus
A small channel connecting the apical membrane of hepatocytes with larger biliary tract ductules and ducts that drain bile from the liver to the small intestine.
- Endoplasmic reticulum-associated protein degradation
A cellular degradative pathway that extracts misfolded proteins from within the lumen of the endoplasmic reticulum for proteasome-mediated degradation in the cytoplasm.
- Endosomal sorting complexes required for transport (ESCRT)
Cytoplasmic multi-protein complexes that function sequentially to mediate membrane budding in an outward direction from the cytoplasm followed by abscission.
- Exosomes
Small extracellular vesicles (~50–150 nm across) resulting from fusion of multivesicular endosomes with the plasma membrane that transport various cargoes (proteins, RNA) between cells.
- Gangliosides
Glycosphingolipids with a sialic acid-containing carbohydrate head group fused to a sphingoid or a ceramide lipid moiety though a glycosidic linkage.
- Intraluminal vesicles (ILVs)
Small cargo-laden vesicles within multivesicular endosomes that are released as exosomes following fusion of the multivesicular endosome membrane with the plasma membrane of the cell.
- Lysosome
A cytoplasmic organelle containing degradative machinery capable of recycling proteins and lipids following fusion with autophagosomes or late endosomes.
- Microvesicle
A large (~200–1000 µm diameter) extracellular vesicle that sheds directly from the plasma membrane of cells.
- Multivesicular endosomes (MVEs)
Late endosomes containing multiple intraluminal vesicles loaded with cytoplasmic cargo that can fuse either with lysosomes for degradation of cargo or with the plasma membrane for release of cargo-laden intraluminal vesicles as exosomes.
- Nanocages
Virus-sized dodecahedrons formed by 60 copies of a self-assembling de novo-designed protein expressed within cells.
- Phagophores
Double-membrane structures that engulf cytoplasmic contents during autophagy.
- Phosphatidylserine
Glycerophospholipid component of the cell membrane normally present only within the cytoplasmic leaflet of the plasma membrane, but typically present on the exterior surface of enveloped viruses.
- Uncoating
Process by which a viral capsid disassembles and releases its RNA or DNA genome.
Author contributions
A.D., E.E.R.-S., X.Y., Z.F. and S.M.L. researched data for the article. A.D., E.E.R.-S., X.Y., C.M.W., Z.F. and S.M.L. substantially contributed to discussion of content, wrote the article and reviewed and edited the manuscript before submission.
Peer review
Peer review information
Nature Reviews Microbiology thanks Jacques Izopet, Eike Steinmann and the other, anonymous, reviewers for their contribution to the peer review of this work.
Competing interests
SML is co-inventor on a pending patent application related to antiviral compounds with activity against hepatitis A virus. The other authors have no competing interests to declare.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related Links
Centers for Disease Control and Prevention: https://www.cdc.gov/hepatitis/outbreaks/2017March-HepatitisA.htm
Contributor Information
Zongdi Feng, Email: zongdi.Feng@nationwidechildrens.org.
Stanley M. Lemon, Email: smlemon@med.unc.edu
References
- 1.Balayan MS, et al. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route. Intervirology. 1983;20:23–31. doi: 10.1159/000149370. [DOI] [PubMed] [Google Scholar]
- 2.Feinstone SM, Kapikian AZ, Purcell RH. Hepatitis A: detection by immune electron microscopy of a viruslike antigen associated with acute illness. Science. 1973;182:1026–1028. doi: 10.1126/science.182.4116.1026. [DOI] [PubMed] [Google Scholar]
- 3.Lanford RE, et al. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc. Natl Acad. Sci. USA. 2011;108:11223–11228. doi: 10.1073/pnas.1101939108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu C, et al. Pathogenesis of hepatitis E virus and hepatitis C virus in chimpanzees: similarities and differences. J. Virol. 2010;84:11264–11278. doi: 10.1128/JVI.01205-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purcell RH, et al. Pathobiology of hepatitis E: lessons learned from primate models. Emerg. Microbes Infect. 2013;2:e9. doi: 10.1038/emi.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobsen KH. Globalization and the changing epidemiology of hepatitis A virus. Cold Spring Harb. Perspect. Med. 2018 doi: 10.1101/cshperspect.a031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rein DB, Stevens GA, Theaker J, Wittenborn JS, Wiersma ST. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology. 2012;55:988–997. doi: 10.1002/hep.25505. [DOI] [PubMed] [Google Scholar]
- 8.Feng Z, et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagashima S, et al. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J. Gen. Virol. 2014;95:2166–2175. doi: 10.1099/vir.0.066910-0. [DOI] [PubMed] [Google Scholar]
- 10.Feng Z, Hirai-Yuki A, McKnight KL, Lemon SM. Naked viruses that aren’t always naked: quasi-enveloped agents of acute hepatitis. Annu. Rev. Virol. 2014;1:539–560. doi: 10.1146/annurev-virology-031413-085359. [DOI] [PubMed] [Google Scholar]
- 11.McKnight KL, et al. Protein composition of the hepatitis A virus quasi-envelope. Proc. Natl Acad. Sci. USA. 2017;114:6587–6592. doi: 10.1073/pnas.1619519114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirai-Yuki A, Whitmire JK, Joyce M, Tyrrell DL, Lemon SM. Murine models of hepatitis A virus infection. Cold Spring Harb. Perspect. Med. 2019 doi: 10.1101/cshperspect.a031674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayed IM, Meuleman P. Updates in hepatitis E virus (HEV) field; lessons learned from human liver chimeric mice. Rev. Med. Virol. 2020;30:e2086. doi: 10.1002/rmv.2086. [DOI] [PubMed] [Google Scholar]
- 15.Wang M, Feng Z. Mechanisms of hepatocellular injury in hepatitis A. Viruses. 2021 doi: 10.3390/v13050861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yadav KK, Kenney SP. Hepatitis E virus immunopathogenesis. Pathogens. 2021 doi: 10.3390/pathogens10091180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemon SM, Ott JJ, Van Damme P, Shouval D. Type A viral hepatitis: a summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J. Hepatol. 2018;68:167–184. doi: 10.1016/j.jhep.2017.08.034. [DOI] [PubMed] [Google Scholar]
- 18.Kamar N, Dalton HR, Abravanel F, Izopet J. Hepatitis E virus infection. Clin. Microbiol. Rev. 2014;27:116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamar N, Pischke S. Acute and persistent hepatitis E virus genotype 3 and 4 infection: clinical features, pathogenesis, and treatment. Cold Spring Harb. Perspect. Med. 2018 doi: 10.1101/cshperspect.a031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmeister MG, et al. Hepatitis A person-to-person outbreaks: epidemiology, morbidity burden, and factors associated with hospitalization-multiple states, 2016–2019. J. Infect. Dis. 2021;223:426–434. doi: 10.1093/infdis/jiaa636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ndumbi P, et al. Hepatitis A outbreak disproportionately affecting men who have sex with men (MSM) in the European Union and European Economic Area, June 2016 to May 2017. Eur. Surveill. 2018 doi: 10.2807/1560-7917.Es.2018.23.33.1700641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagashima S, et al. Characterization of the quasi-enveloped hepatitis E virus particles released by the cellular exosomal pathway. J. Virol. 2017 doi: 10.1128/jvi.00822-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costafreda MI, Sauleda S, Rico A, Piron M, Bes M. Detection of nonenveloped hepatitis E virus in plasma of infected blood donors. J. Infect. Dis. 2022;226:1753–1760. doi: 10.1093/infdis/jiab589. [DOI] [PubMed] [Google Scholar]
- 24.Hirai-Yuki A, Hensley L, Whitmire JK, Lemon SM. Biliary secretion of quasi-enveloped human hepatitis A virus. MBio. 2016;7:e01998–01916. doi: 10.1128/mBio.01998-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen JM, Lemon SM. Comparative pathology of hepatitis A virus and hepatitis E virus infection. Cold Spring Harb. Perspect. Med. 2018 doi: 10.1101/cshperspect.a033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirai-Yuki A, et al. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science. 2016;353:1541–1545. doi: 10.1126/science.aaf8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto H. Culture systems for hepatitis E virus. J. Gastroenterol. 2013;48:147–158. doi: 10.1007/s00535-012-0682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dao Thi VL, et al. Stem cell-derived polarized hepatocytes. Nat. Commun. 2020;11:1677. doi: 10.1038/s41467-020-15337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marion O, et al. Hepatitis E virus replication in human intestinal cells. Gut. 2020;69:901–910. doi: 10.1136/gutjnl-2019-319004. [DOI] [PubMed] [Google Scholar]
- 30.Asher LVS, et al. Pathogenesis of hepatitis A in orally inoculated owl monkeys (Aotus trivergatus) J. Med. Virol. 1995;47:260–268. doi: 10.1002/jmv.1890470312. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi M, et al. Hepatitis E virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. J. Clin. Microbiol. 2010;48:1112–1125. doi: 10.1128/JCM.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, et al. Hepatitis A virus and the origins of picornaviruses. Nature. 2015;517:85–88. doi: 10.1038/nature13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology. 2015;479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly AG, Netzler NE, White PA. Ancient recombination events and the origins of hepatitis E virus. BMC Evol. Biol. 2016;16:210. doi: 10.1186/s12862-016-0785-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drexler JF, et al. Evolutionary origins of hepatitis A virus in small mammals. Proc. Natl Acad. Sci. USA. 2015;112:15190–15195. doi: 10.1073/pnas.1516992112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pavio N, Meng XJ, Doceul V. Zoonotic origin of hepatitis E. Curr. Opin. Virol. 2015;10:34–41. doi: 10.1016/j.coviro.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 37.Meng X-J. Expanding host range and cross-species infection of hepatitis E virus. PLoS Pathog. 2016;12:e1005695. doi: 10.1371/journal.ppat.1005695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suppiah S, Zhou Y, Frey TK. Lack of processing of the expressed ORF1 gene product of hepatitis E virus. Virol. J. 2011;8:245. doi: 10.1186/1743-422X-8-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szkolnicka D, et al. Recombinant hepatitis E viruses harboring tags in the ORF1 protein. J. Virol. 2019 doi: 10.1128/jvi.00459-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar M, Hooda P, Khanna M, Patel U, Sehgal D. Development of BacMam induced hepatitis E virus replication model in hepatoma cells to study the polyprotein processing. Front. Microbiol. 2020;11:1347. doi: 10.3389/fmicb.2020.01347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin X, et al. Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection. Proc. Natl Acad. Sci. USA. 2018;115:4773–4778. doi: 10.1073/pnas.1721345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montpellier C, et al. Hepatitis E Virus lifecycle and identification of 3 forms of the ORF2 capsid protein. Gastroenterology. 2018;154:211–223.e8. doi: 10.1053/j.gastro.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 43.Meister TL, et al. A ribavirin-induced ORF2 single-nucleotide variant produces defective hepatitis E virus particles with immune decoy function. Proc. Natl Acad. Sci. USA. 2022;119:e2202653119. doi: 10.1073/pnas.2202653119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hervouet K, et al. An arginine-rich motif in the ORF2 capsid protein regulates the hepatitis E virus lifecycle and interactions with the host cell. PLoS Pathog. 2022;18:e1010798. doi: 10.1371/journal.ppat.1010798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair VP, et al. Endoplasmic reticulum stress induced synthesis of a novel viral factor mediates efficient replication of genotype-1 hepatitis E virus. PLoS Pathog. 2016;12:e1005521. doi: 10.1371/journal.ppat.1005521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKnight KL, Lemon SM. Hepatitis A virus genome organization and replication strategy. Cold Spring Harb. Perspect. Med. 2018 doi: 10.1101/cshperspect.a033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, et al. The ZCCHC14/TENT4 complex is required for hepatitis A virus RNA synthesis. Proc. Natl Acad. Sci. USA. 2022;119:e2204511119. doi: 10.1073/pnas.2204511119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kenney SP, Meng XJ. Hepatitis E virus genome structure and replication strategy. Cold Spring Harb. Perspect. Med. 2019 doi: 10.1101/cshperspect.a031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meister TL, Bruening J, Todt D, Steinmann E. Cell culture systems for the study of hepatitis E virus. Antivir. Res. 2019;163:34–49. doi: 10.1016/j.antiviral.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 50.Todt D, et al. Robust hepatitis E virus infection and transcriptional response in human hepatocytes. Proc. Natl Acad. Sci. USA. 2020;117:1731–1741. doi: 10.1073/pnas.1912307117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siegl G, Weitz M, Kronauer G. Stability of hepatitis A virus. Intervirology. 1984;22:218–226. doi: 10.1159/000149554. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, et al. Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site. Proc. Natl Acad. Sci. USA. 2017;114:770–775. doi: 10.1073/pnas.1616502114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart DI, Ren J, Wang X, Rao Z, Fry EE. Hepatitis A virus capsid structure. Cold Spring Harb. Perspect. Med. 2018 doi: 10.1101/cshperspect.a031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xing L, et al. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 2010;285:33175–33183. doi: 10.1074/jbc.M110.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zafrullah M, Ozdener MH, Kumar R, Panda SK, Jameel S. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J. Virol. 1999;73:4074–4082. doi: 10.1128/JVI.73.5.4074-4082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emerson SU, Arankalle VA, Purcell RH. Thermal stability of hepatitis E virus. J. Infect. Dis. 2005;192:930–933. doi: 10.1086/432488. [DOI] [PubMed] [Google Scholar]
- 57.Feng Z, et al. Human pDCs preferentially sense enveloped hepatitis A virions. J. Clin. Invest. 2015;125:169–176. doi: 10.1172/JCI77527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi M, et al. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch. Virol. 2008;153:1703–1713. doi: 10.1007/s00705-008-0179-6. [DOI] [PubMed] [Google Scholar]
- 59.Nagashima S, et al. The membrane on the surface of hepatitis E virus particles is derived from the intracellular membrane and contains trans-Golgi network protein 2. Arch. Virol. 2014;159:979–991. doi: 10.1007/s00705-013-1912-3. [DOI] [PubMed] [Google Scholar]
- 60.Yin X, Ambardekar C, Lu Y, Feng Z. Distinct entry mechanisms for nonenveloped and quasi-enveloped hepatitis E viruses. J. Virol. 2016;90:4232–4242. doi: 10.1128/JVI.02804-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Das A, et al. TIM1 (HAVCR1) Is not essential for cellular entry of either quasi-enveloped or naked hepatitis A virions. mBio. 2017 doi: 10.1128/mBio.00969-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sayed IM, et al. Transmission of hepatitis E virus infection to human-liver chimeric FRG mice using patient plasma. Antivir. Res. 2017;141:150–154. doi: 10.1016/j.antiviral.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 63.Kumar CS, Dey D, Ghosh S, Banerjee M. Breach: host membrane penetration and entry by nonenveloped viruses. Trends Microbiol. 2017 doi: 10.1016/j.tim.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 64.Kaplan G, et al. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J. 1996;15:4282–4296. doi: 10.1002/j.1460-2075.1996.tb00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kulsuptrakul J, Wang R, Meyers NL, Ott M, Puschnik AS. A genome-wide CRISPR screen identifies UFMylation and TRAMP-like complexes as host factors required for hepatitis A virus infection. Cell Rep. 2021;34:108859. doi: 10.1016/j.celrep.2021.108859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Das A, Maury W, Lemon SM. TIM1 (HAVCR1): an essential “receptor” or an “accessory attachment factor” for hepatitis A virus? J. Virol. 2019 doi: 10.1128/jvi.01793-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das A, et al. Gangliosides are essential endosomal receptors for quasi-enveloped and naked hepatitis A virus. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0727-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rivera-Serrano EE, Gonzalez-Lopez O, Das A, Lemon SM. Cellular entry and uncoating of naked and quasi-enveloped human hepatoviruses. eLife. 2019;8:e43983. doi: 10.7554/eLife.43983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J. Virol. 2013;87:3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ewers H, et al. GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 2010;12:11–18. doi: 10.1038/ncb1999. [DOI] [PubMed] [Google Scholar]
- 71.He S, et al. Putative receptor-binding sites of hepatitis E virus. J. Gen. Virol. 2008;89:245–249. doi: 10.1099/vir.0.83308-0. [DOI] [PubMed] [Google Scholar]
- 72.Guu TS, et al. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl Acad. Sci. USA. 2009;106:12992–12997. doi: 10.1073/pnas.0904848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harrison SC. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moller-Tank S, Maury W. Phosphatidylserine receptors: enhancers of enveloped virus entry and infection. Virology. 2014;468-470:565–580. doi: 10.1016/j.virol.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Balasubramanian S, Kota SK, Kuchroo VK, Humphreys BD, Strom TB. TIM family proteins promote the lysosomal degradation of the nuclear receptor NUR77. Sci. Signal. 2012;5:ra90. doi: 10.1126/scisignal.2003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolter T, Sandhoff K. Lysosomal degradation of membrane lipids. FEBS Lett. 2010;584:1700–1712. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 77.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang H, et al. Ebola viral glycoprotein bound to its endosomal receptor Niemann–Pick C1. Cell. 2016;164:258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morace G, Kusov Y, Dzagurov G, Beneduce F, Gauss-Muller V. The unique role of domain 2A of the hepatitis A virus precursor polypeptide P1-2A in viral morphogenesis. BMB Rep. 2008;41:678–683. doi: 10.5483/BMBRep.2008.41.9.678. [DOI] [PubMed] [Google Scholar]
- 80.Ding Q, et al. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc. Natl Acad. Sci. USA. 2017;114:1147–1152. doi: 10.1073/pnas.1614955114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fuchs R, Blaas D. Uncoating of human rhinoviruses. Rev. Med. Virol. 2010;20:281–297. doi: 10.1002/rmv.654. [DOI] [PubMed] [Google Scholar]
- 82.Panjwani A, et al. Capsid protein VP4 of human rhinovirus induces membrane permeability by the formation of a size-selective multimeric pore. PLoS Pathog. 2014;10:e1004294. doi: 10.1371/journal.ppat.1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shukla A, Padhi AK, Gomes J, Banerjee M. The VP4 peptide of hepatitis A virus ruptures membranes through formation of discrete pores. J. Virol. 2014;88:12409–12421. doi: 10.1128/JVI.01896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang P, Monteiro da Silva G, Deatherage C, Burd C, DiMaio D. Cell-penetrating peptide mediates intracellular membrane passage of human papillomavirus L2 protein to trigger retrograde trafficking. Cell. 2018;174:1465–1476.e13. doi: 10.1016/j.cell.2018.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qian M, Cai D, Verhey KJ, Tsai B. A lipid receptor sorts polyomavirus from the endolysosome to the endoplasmic reticulum to cause infection. PLoS Pathog. 2009;5:e1000465. doi: 10.1371/journal.ppat.1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Surjit M, Jameel S, Lal SK. Cytoplasmic localization of the ORF2 protein of hepatitis E virus is dependent on its ability to undergo retrotranslocation from the endoplasmic reticulum. J. Virol. 2007;81:3339–3345. doi: 10.1128/JVI.02039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Costafreda MI, Abbasi A, Lu H, Kaplan G. Exosome mimicry by a HAVCR1-NPC1 pathway of endosomal fusion mediates hepatitis A virus infection. Nat. Microbiol. 2020;5:1096–1106. doi: 10.1038/s41564-020-0740-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bonsergent E, et al. Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nat. Commun. 2021;12:1864. doi: 10.1038/s41467-021-22126-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morandi MI, et al. Extracellular vesicle fusion visualized by cryo-electron microscopy. PNAS Nexus. 2022;1:pgac156. doi: 10.1093/pnasnexus/pgac156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Longatti A, Boyd B, Chisari FV. Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J. Virol. 2015;89:2956–2961. doi: 10.1128/JVI.02721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gonzalez-Lopez O, et al. Redundant late domain functions of tandem VP2 YPX3L motifs in nonlytic cellular egress of quasi-enveloped hepatitis A virus. J. Virol. 2018;92:1308–1318. doi: 10.1128/JVI.01308-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang W, et al. Hepatitis A virus structural protein pX interacts with ALIX and promotes the secretion of virions and foreign proteins through exosome-like vesicles. J. Extracell. Vesicles. 2020;9:1716513. doi: 10.1080/20013078.2020.1716513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shirasaki T, et al. Nonlytic cellular release of hepatitis A virus requires dual capsid recruitment of the ESCRT-associated Bro1 domain proteins HD-PTP and ALIX. PLoS Pathog. 2022;18:e1010543. doi: 10.1371/journal.ppat.1010543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shimizu YK, et al. Detection of hepatitis A antigen in human liver. Infect. Immun. 1982;36:320–324. doi: 10.1128/iai.36.1.320-324.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cromeans T, Humphrey C, Sobsey M, Fields H. Use of immunogold preembedding technique to detect hepatitis A viral antigen in infected cells. Am. J. Anat. 1989;185:314–320. doi: 10.1002/aja.1001850225. [DOI] [PubMed] [Google Scholar]
- 96.Migliano SM, Wenzel EM, Stenmark H. Biophysical and molecular mechanisms of ESCRT functions, and their implications for disease. Curr. Opin. Cell Biol. 2022;75:102062. doi: 10.1016/j.ceb.2022.01.007. [DOI] [PubMed] [Google Scholar]
- 97.Remec Pavlin M, Hurley JH. The ESCRTs — converging on mechanism. J. Cell Sci. 2020 doi: 10.1242/jcs.240333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Frankel EB, Audhya A. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin. Cell Dev. Biol. 2018;74:4–10. doi: 10.1016/j.semcdb.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Korbei B. Ubiquitination of the ubiquitin-binding machinery: how early ESCRT components are controlled. Essays Biochem. 2022;66:169–177. doi: 10.1042/EBC20210042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schoneberg J, Lee IH, Iwasa JH, Hurley JH. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell. Biol. 2017;18:5–17. doi: 10.1038/nrm.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tabernero L, Woodman P. Dissecting the role of His domain protein tyrosine phosphatase/PTPN23 and ESCRTs in sorting activated epidermal growth factor receptor to the multivesicular body. Biochem. Soc. Trans. 2018;46:1037–1046. doi: 10.1042/BST20170443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Desrochers G, Kazan JM, Pause A. Structure and functions of His domain protein tyrosine phosphatase in receptor trafficking and cancer (1) Biochem. Cell. Biol. 2019;97:68–72. doi: 10.1139/bcb-2017-0322. [DOI] [PubMed] [Google Scholar]
- 104.Nagashima S, et al. Tumour susceptibility gene 101 and the vacuolar protein sorting pathway are required for the release of hepatitis E virions. J. Gen. Virol. 2011;92:2838–2848. doi: 10.1099/vir.0.035378-0. [DOI] [PubMed] [Google Scholar]
- 105.Vietri M, Radulovic M, Stenmark H. The many functions of ESCRTs. Nat. Rev. Mol. Cell. Biol. 2020;21:25–42. doi: 10.1038/s41580-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 106.Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Freed EO. Viral late domains. J. Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Votteler J, et al. Designed proteins induce the formation of nanocage-containing extracellular vesicles. Nature. 2016;540:292–295. doi: 10.1038/nature20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fisher RD, et al. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 111.Kim J, et al. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhai Q, et al. Structural and functional studies of ALIX interactions with YPXnL late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 113.Bissig C, Gruenberg J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol. 2014;24:19–25. doi: 10.1016/j.tcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 114.Skowyra ML, Schlesinger PH, Naismith TV, Hanson PI. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science. 2018 doi: 10.1126/science.aar5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gahloth D, et al. The open architecture of HD-PTP phosphatase provides new insights into the mechanism of regulation of ESCRT function. Sci. Rep. 2017;7:9151. doi: 10.1038/s41598-017-09467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shirasaki T, et al. Nonlytic quasi-enveloped hepatovirus release is facilitated by pX protein interaction with the E3 ubiquitin ligase ITCH. J. Virol. 2022;96:e0119522. doi: 10.1128/jvi.01195-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ziegler CM, et al. NEDD4 family ubiquitin ligases associate with LCMV Z’s PPXY domain and are required for virus budding, but not via direct ubiquitination of Z. PLoS Pathog. 2019;15:e1008100. doi: 10.1371/journal.ppat.1008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chung HY, et al. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin J, et al. Structure of the Fab-labeled “breathing” state of native poliovirus. J. Virol. 2012;86:5959–5962. doi: 10.1128/JVI.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dahmane S, et al. Membrane-assisted assembly and selective secretory autophagy of enteroviruses. Nat. Commun. 2022;13:5986. doi: 10.1038/s41467-022-33483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gouttenoire J, et al. Palmitoylation mediates membrane association of hepatitis E virus ORF3 protein and is required for infectious particle secretion. PLoS Pathog. 2018;14:e1007471. doi: 10.1371/journal.ppat.1007471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nagashima S, et al. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J. Gen. Virol. 2011;92:269–278. doi: 10.1099/vir.0.025791-0. [DOI] [PubMed] [Google Scholar]
- 123.Li TC, et al. Genetic and physicochemical analyses of a novel ferret hepatitis E virus, and clinical signs of infection after birth. Infect. Genet. Evol. 2017;51:153–159. doi: 10.1016/j.meegid.2017.03.026. [DOI] [PubMed] [Google Scholar]
- 124.Capelli N, et al. Vectorial release of hepatitis E virus in polarized human hepatocytes. J. Virol. 2019 doi: 10.1128/jvi.01207-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sari G, et al. The viral ORF3 protein Is required for hepatitis E virus apical release and efficient growth in polarized hepatocytes and humanized mice. J. Virol. 2021;95:e0058521. doi: 10.1128/JVI.00585-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Allweiss L, et al. Human liver chimeric mice as a new model of chronic hepatitis E virus infection and preclinical drug evaluation. J. Hepatol. 2016;64:1033–1040. doi: 10.1016/j.jhep.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 127.Robinson SM, et al. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 2014;10:e1004045. doi: 10.1371/journal.ppat.1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen YH, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Santiana M, et al. Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell Host Microbe. 2018;24:208–220.e8. doi: 10.1016/j.chom.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.van der Grein SG, et al. Picornavirus infection induces temporal release of multiple extracellular vesicle subsets that differ in molecular composition and infectious potential. PLoS Pathog. 2019;15:e1007594. doi: 10.1371/journal.ppat.1007594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang JE, et al. Complexity and ultrastructure of infectious extracellular vesicles from cells infected by non-enveloped virus. Sci. Rep. 2020;10:7939. doi: 10.1038/s41598-020-64531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Morris-Love J, et al. JC polyomavirus uses extracellular vesicles to infect target cells. MBio. 2019 doi: 10.1128/mBio.00379-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Richards AL, Jackson WT. How positive-strand RNA viruses benefit from autophagosome maturation. J. Virol. 2013;87:9966–9972. doi: 10.1128/JVI.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Taylor MP, Kirkegaard K. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy. 2007;4:286–289. doi: 10.4161/auto.5377. [DOI] [PubMed] [Google Scholar]
- 135.Bird SW, Maynard ND, Covert MW, Kirkegaard K. Nonlytic viral spread enhanced by autophagy components. Proc. Natl Acad. Sci. USA. 2014;111:13081–13086. doi: 10.1073/pnas.1401437111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Anthony SJ, et al. Discovery of a novel hepatovirus (phopivirus of seals) related to human hepatitis A virus. mBio. 2015 doi: 10.1128/mBio.01180-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Feng H, et al. Hepatovirus 3ABC proteases and evolution of mitochondrial antiviral signaling protein (MAVS) J. Hepatol. 2019;71:25–34. doi: 10.1016/j.jhep.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee GH, et al. Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology. 2016;150:355–357.e3. doi: 10.1053/j.gastro.2015.10.048. [DOI] [PubMed] [Google Scholar]
- 139.Andonov A, et al. Rat hepatitis E virus linked to severe acute hepatitis in an immunocompetent patient. J. Infect. Dis. 2019;220:951–955. doi: 10.1093/infdis/jiz025. [DOI] [PubMed] [Google Scholar]
- 140.Sander AL, Corman VM, Lukashev AN, Drexler JF. Evolutionary origins of enteric hepatitis viruses. Cold Spring Harb. Perspect. Med. 2018 doi: 10.1101/cshperspect.a031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Walker CM. Adaptive immune responses in hepatitis A virus and hepatitis E virus infections. Cold Spring Harb. Perspect. Med. 2018 doi: 10.1101/cshperspect.a033472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ping LH, Lemon SM. Antigenic structure of human hepatitis A virus defined by analysis of escape mutants selected against murine monoclonal antibodies. J. Virol. 1992;66:2208–2216. doi: 10.1128/jvi.66.4.2208-2216.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gellis SS, et al. The use of human immune serum globulin (gamma globulin) in infectious (epidemic) hepatitis in the Mediterranean theater of operations. I. Studies on prophylaxis in two epidemics of infectious hepatitis. JAMA. 1945;128:1062–1063. doi: 10.1001/jama.1945.02860320004002. [DOI] [Google Scholar]