Abstract
Multi-locus sequence typing (MLST) is widely used to investigate genetic relationships among eukaryotic taxa, including parasitic pathogens. MLST analysis workflows typically involve construction of alignment-based phylogenetic trees – i.e., where tree structures are computed from nucleotide differences observed in a multiple sequence alignment (MSA). Notably, alignment-based phylogenetic methods require that all isolates/taxa are represented by a single sequence. When multiple loci are sequenced these sequences may be concatenated to produce one tree that includes information from all loci. Alignment-based phylogenetic techniques are robust and widely used yet possess some shortcomings, including how heterozygous sites are handled, intolerance for missing data (i.e., partial genotypes), and differences in the way insertions-deletions (indels) are scored/treated during tree construction. In certain contexts, ‘haplotype-based’ methods may represent a viable alternative to alignment-based techniques, as they do not possess the aforementioned limitations. This is namely because haplotype-based methods assess genetic similarity based on numbers of shared (i.e., intersecting) haplotypes as opposed to similarities in nucleotide composition observed in an MSA. For haplotype-based comparisons, choosing an appropriate distance statistic is fundamental, and several statistics are available to choose from. However, a comprehensive assessment of various available statistics for their ability to produce a robust haplotype-based phylogenetic reconstruction has not yet been performed. We evaluated seven distance statistics by applying them to extant MLST datasets from the gastrointestinal parasite Cyclospora cayetanensis and two species of pathogenic nematode of the genus Strongyloides. We compare the genetic relationships identified using each statistic to epidemiologic, geographic, and host metadata. We show that Barratt’s heuristic definition of genetic distance was the most robust among the statistics evaluated. Consequently, it is proposed that Barratt’s heuristic represents a useful approach for use in the context of challenging MLST datasets possessing features (i.e., high heterozygosity, partial genotypes, and indel or repeat-based polymorphisms) that confound or preclude the use of alignment-based methods.
Keywords: Phylogeny, Genotyping, Eukaryotes, Parasites, Strongyloides, Cyclospora
1. Introduction
Multi-locus sequence typing (MLST) is used widely to explore genetic relationships among eukaryotic taxa. MLST analysis workflows typically involve construction of alignment-based phylogenetic trees or haplotype networks – i.e., where tree/network structures are computed based on nucleotide differences observed in a multiple sequence alignment (MSA). Notably, alignment-based techniques require that all isolates/taxa are represented by a single sequence. When multiple loci are sequenced the sequences from each locus are often concatenated to produce a single tree (Barratt et al., 2017; Ellis et al., 2021; Kamvar et al., 2015; Leigh et al., 2015; Martins et al., 2020). In the context of eukaryotic pathogens, these workflows are used routinely to investigate relationships between genotype and geography (Martins et al., 2020), genotype and host species (Jaleta et al., 2017), to identify sources of exposure during outbreaks (Hlavsa et al., 2017), and to improve our understanding of evolutionary relationships among related taxa (Barratt et al., 2017; Ellis et al., 2021; Kaufer et al., 2017).
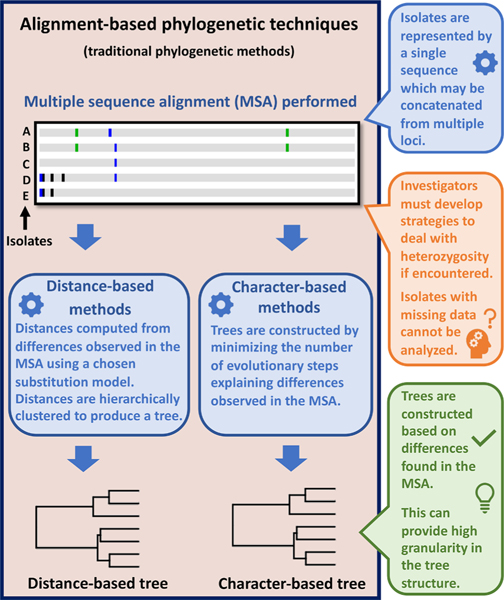
Alignment-based phylogenetic techniques are generally divided into two types; distance-based and character-based methods. Distance-based methods require selection from a range of substitution models to compute genetic distances (Rzhetsky and Nei, 1995; Som, 2006). A hierarchical clustering method such as UPGMA, WPGMA, or Neighbor-Joining (Lin, 1982; Saitou and Nei, 1987) is then used to generate a tree from these distances, where isolate pairs separated by small distances are placed on branch tips that share a node, and each node represents a common ancestor (Mount, 2008a). Character-based tree-building, using maximum parsimony and maximum-likelihood for example, aims to minimize the number of evolutionary steps explaining nucleotide differences observed in the MSA (Kannan and Wheeler, 2012; Mount, 2008b; Munjal et al., 2019). Irrespective of the method, the underlying objective of alignment-based approaches is to score and subsequently visualize relationships between isolates based on genetic differences observed in an MSA, where each isolate is represented by a single sequence (Fig. 1).
Fig. 1.
Overview of alignment-based phylogenetic workflows. Alignment-based (i.e., distance-based or character-based) phylogenetic methods generate tree structures based on nucleotide differences observed between isolates in an MSA, where each isolate must be represented by a single sequence. Consequently, heterozygosity is confounding for alignment-based methods. MLST analysis workflows often involve concatenating multiple sequenced loci into one sequence, as alignment-based methods require a single continuous, homologous sequence for each isolate. Therefore, if a sequence cannot be obtained for one or more genotyping loci for some isolates, these isolates must be excluded, or the concatenated sequence of all isolates may be truncated to maintain consistency across all isolates. An advantage of alignment-based methods is that tree structures reflect differences observed at each nucleotide position in the alignment, providing good granularity.
Despite being robust and widely used, alignment-based phylogeny has limitations, including intolerance for heterozygosity and missing data (i.e., partial genotypes), and inconsistent treatment of insertions and deletions (indels). In sexually reproducing pathogens (e.g., many parasites), two alleles may be detected at one locus. This is confounding for alignment-based methods, which require one sequence. To force compatibility, investigators may delete heterozygous bases (Lischer et al., 2014), or replace them with International Union of Pure and Applied Chemistry (IUPAC) codes (Johnson, 2010), noting that some substitution models ignore IUPAC codes (Rzhetsky and Nei, 1995; Som, 2006). Investigators may arbitrarily select one allele, exclude heterozygous loci, or exclude heterozygous isolates, all of which excludes valuable data. Alignment-based phylogeny also requires that isolates are sequenced at precisely the same loci/locus. In practice, material for DNA extraction may be limited by specimen volume, miniscule organism size, or inappropriate specimen storage causing DNA degradation (Barratt et al., 2019a; Bozidis et al., 2021; Nguyen et al., 2019). These factors may reduce DNA yields allowing sequencing of a subset of target loci for some isolates. Differences in indel handling by different sequence aligners and substitution models impacts alignment-based phylogeny with varying effects (Ashkenazy et al., 2014; Jordan and Goldman, 2012; Larkin et al., 2007; Mount, 2008a; Redelings and Suchard, 2007; Rubio-Largo et al., 2018). Indels may lead to poor alignments (Castresana, 2000; Talavera et al., 2007; Tan et al., 2015), and some substitution models ignore indels while others treat them as substitutions (Mount, 2008a). Treating short gaps (one or two bases) as substitutions may be helpful, but treating large gaps as substitutions (e.g., large retrotransposon insertions) may lead to nonsensical results. Low-complexity microsatellite repeats cause alignment gaps, yet these polymorphisms are used extensively for assessing kinship between parasite isolates (Martins et al., 2020; Plucinski and Barratt, 2021), so indel exclusion in this context makes little sense.
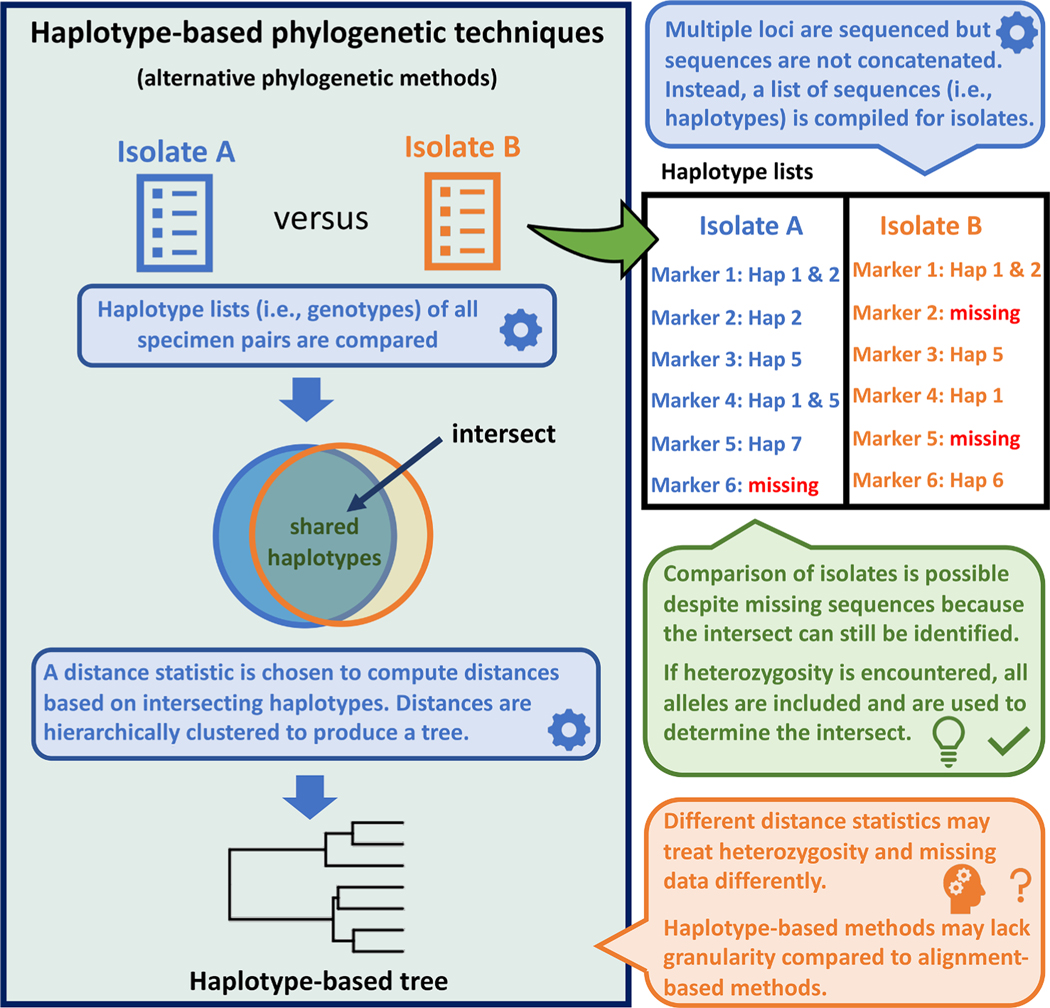
For datasets with features that are not amenable to alignment-based phylogeny, alternative approaches exist that may represent a viable option in certain contexts. The key difference between these alternatives and alignment-based methods is that they are not alignment-based – they are ‘haplotype-based’ (i.e., hap-based), meaning that distances are computed from numbers of shared (i.e., intersecting) haplotypes observed between isolate pairs and are not based on nucleotide similarities observed in an MSA. For hap-based methods, investigators assign haplotypes a unique identifier (i.e., a name/number), and a list of haplotype identifiers (i.e., a genotype) is generated for each isolate. Hap-based methods use the intersect of these lists to define the level of similarity between pairs by computing a distance; isolates sharing many haplotypes are separated by smaller distances, while those sharing few (or no) haplotypes are separated by larger distances. As hap-based methods compare lists of sequence identifiers and not the sequences themselves, the nature of the polymorphism (i.e., whether haplotypes are distinguished by repeat length, the presence of indels, or SNP’s) is inconsequential. These lists may include multiple alleles of the same locus – i.e., heterozygosity is tolerated (Fig. 2). Incomplete lists (i.e., due to some isolates missing a sequence for some markers) may also be compared, understanding that comparisons become increasingly tenuous as the number of missing loci increases (Nascimento et al., 2020).
Fig. 2.
Overview of haplotype-based phylogenetic workflows and their advantages. Haplotype-based phylogenetic workflows produce a tree structure based on numbers of intersecting haplotypes. Isolates are represented by a list of haplotypes (i.e., their genotype), including loci possessing multiple alleles. For this reason, heterozygosity is not a confounding factor. Because distances are computed from the number of intersecting haplotypes, isolates with data missing for a small number of loci may still be retained for analysis, understanding that comparisons become increasingly tenuous as the number of missing values increases. Haplotype-based tree structures may lack granularity compared to alignment-based trees because haplotype-based methods consider haplotype matches in a binary manner: isolates either share a haplotype or they do not, and the fact that some haplotypes may be more similar in sequence than others is not considered during distance computation. However, the granularity of haplotype-based phylogenetic reconstructions can be increased by sequencing genotyping markers possessing certain features (discussed later in this paper). Importantly, the statistic selected for distance computation is the foundation of a haplotype-based method.
Notably, the statistic used for distance computation is the foundation of hap-based approaches. Applicable distance statistics include Jaccard distances (JD), Bray-Curtis dissimilarity (BC), Jensen-Shannon divergence (JSD), Euclidean distances (ED), Manhattan distances (MD), Plucinski’s Bayesian (PB) definition of genetic distance, and Barratt’s heuristic (BH) definition of genetic distance (Barratt et al., 2019a; Barratt and Sapp, 2020; Houghton et al., 2020; Kartal et al., 2020; Nascimento et al., 2020; Pettengill et al., 2016). These statistics each produce distances that can be clustered for visualization of genetic relationships as a tree. However, while some of these statistics have been applied to pathogen-derived MLST datasets previously (Barratt and Sapp, 2020; Houghton et al., 2020; Nascimento et al., 2020) a comprehensive assessment of their ability to produce a robust hap-based phylogeny has not yet been performed.
We evaluated seven distance statistics (JD, BC, JSD, ED, MD, PB, and BH) for their ability to facilitate a robust hap-based phylogenetic reconstruction in the context of parasite-derived MLST datasets possessing features that preclude the use of alignment-based methods. We applied these statistics to three extant MLST datasets of varying size and complexity. The largest dataset comprised 1137 genotypes of the protozoan parasite Cyclospora cayetanensis; the etiological agent of human cyclosporiasis (Barratt et al., 2021). A second moderately sized dataset included 704 genotypes of the parasitic worm Strongyloides stercoralis (Barratt and Sapp, 2020). The third and smallest dataset comprised 133 isolates of Strongyloides fuelleborni, including 18 isolates of an unclassified Strongyloides species (Barratt and Sapp, 2020). These datasets were clustered using each of the seven statistics, and clustering performance was assessed using epidemiologic, geographic, and host metadata accompanying the datasets as a reference for expected clustering outcomes.
2. Materials and methods
2.1. Selection of MLST datasets
We utilized a publicly available MLST dataset for C. cayetanensis generated by the United States (U.S.) Centers for Disease Control and Prevention (CDC), the Public Health Agency of Canada, and certain U.S. State public health departments, as part of ongoing C. cayetanensis genotyping performed during 2018, 2019, and 2020 (Barratt et al., 2021; Casillas et al., 2018; Nascimento et al., 2020; Anonymous, 2018; Anonymous, 2019a; Anonymous, 2020; Anonymous, 2019b). Briefly, this C. cayetanensis dataset comprised 1137 genotypes with high heterozygosity, some repeat-based polymorphisms, and many isolates with a partial genotype (Barratt et al., 2021). These isolates had been sequenced at eight markers as previously described (Barratt et al., 2021; Nascimento et al., 2020; Barratt et al., 2021), including six nuclear markers and two mitochondrial markers (Table 1). Illumina data from these isolates were accessed under NCBI BioProject Number PRJNA578931, and each isolates’ genotype was ascertained using bioinformatic workflows previously described (Nascimento et al., 2020). The two Strongyloides datasets analyzed here were compiled from data already published in GenBank. Notably, these Strongyloides datasets are identical to those described by Barratt and Sapp (Barratt and Sapp, 2020). This included a dataset of moderate size and complexity comprised 704 isolates of the parasitic nematode Strongyloides stercoralis (Barratt and Sapp, 2020) and a smaller, low-complexity dataset comprising 133 isolates of Strongyloides fuelleborni, including 18 isolates of an unclassified Strongyloides species (Barratt and Sapp, 2020).
Table 1.
Summarized characteristics for each parasite-derived MLST dataset included in the present evaluation.
| General dataset characteristics | |||
|---|---|---|---|
|
|
|||
| Cyclospora cayetanensis | Strongyloides stercoralis | Strongyloides fuelleborn | |
| Number of specimens/isolates | 1137 | 704 | 133 † |
| Number of loci | 8 (6 nuclear, 2 mitochondrial)α | 3 (2 nuclear, 1 mitochondrial)β | 3 (2 nuclear, 1 mitochondrial)β |
| Sum of Shannon entropies | 4.45 bans ‡ | 2.59 bans ‡ | 2.42 bans ‡ |
| Missing data (partial genotypes) | |||
| Isolates with 4 markers not sequenced | 54 (4.7%) | NA (only three markers in dataset) | NA (only three markers in dataset) |
| Isolates with 3 markers not sequenced | 59 (5.2%) | NA (only three markers in dataset) | NA (only three markers in dataset) |
| Isolates with 2 markers not sequenced | 104 (9.1%) | 533 (75.7%) | 86 (64.7%) |
| Isolates with 1 marker not sequenced | 206 (18.1%) | 75 (10.7%) | 42 (31.6%) |
| Isolates with all markers sequenced | 714 (62.8%) | 96 (13.6%) | 5 (3.8%) |
| Heterozygosity | |||
| Number of heterozygous specimens | More than 1 allele present in ~ 90% of specimens for at least one of the six nuclear loci sequenced. | More than 1 allele present in ~ 18% of specimens for nuclear markers. | No heterozygosity |
| Metadata available for assessment of clustering performance | |||
| Metadata available for assessment of clustering performance | 552 of the 1137 isolates possess links to epidemiologically-defined outbreak clusters of cyclosporiasis that occurred in the USA and/or Canada in 2018, 2019, or 2020. | The geographic origin of each isolate is known, as is the host species from which the isolate was collected. There is pre-existing knowledge on the population structure of S. stercoralis based on alignment-based phylogenetic methods. The results obtained in this study can be compared to what is expected based on previous phylogenetic analyses. | The geographic origin of each isolate is known, as is the host species from which the isolate was collected. |
Includes 18 isolates of a Strongyloides fuelleborni-like nematode collected from Bornean slow lorises.
Calculations of the sum of Shannon entropies (i.e., as a measure of dataset complexity) are provided in Supplementary File S1.
Markers sequenced as part of the C. cayetanensis MLST panel include the MSR and Mitochondrial Junction loci (mitochondrial markers), and the CDS1, CDS2, CDS3, CDS4, 360i2 and 378 loci (nuclear loci) (Nascimento et al., 2020).
Markers sequenced as part of the Strongyloides MLST panel include the HVR-I and HVR-IV regions of the 18S rDNA (nuclear loci), and the cox1 locus (a mitochondrial locus) (Barratt and Sapp, 2020).
Selection of these MLST datasets was driven partly by the availability of high-quality metadata accompanying these MLST genotypes, in addition to the large size of the MLST datasets themselves which would support the validity of any conclusions drawn. As discussed in the methods below, various types of metadata (i.e., epidemiologic linkages, host information, and geographic information – see Table 1) were used for assessment of genetic clustering performance as they served as a reference for expected clustering outcomes. Therefore, the existence of such metadata as an accompaniment to the MLST data was considered an important prerequisite for inclusion of the selected datasets in this analysis. These MLST datasets were also favored because they are derived from organisms that are widely disparate from one another taxonomically (worms and protozoa). Consequently, the use of these datasets would support that any conclusions drawn should be relatively generalizable across a diverse range of taxa. Finally, Strongyloides sp. and C. cayetanensis represent an interesting use-cases for haplotype-based phylogenetic methods as isolates of these organisms are subject to heterozygosity; their life cycles each possess sexual stages. Heterozygosity has been observed at the 18S rDNA locus of some Strongyloides sp. isolates (Barratt and Sapp, 2020; Zhou et al., 2019), and these heterozygous isolates are included in the present analysis as a demonstration that haplotype-based methods can accommodate heterozygosity. The sexual cycle of the unicellular protozoan C. cayetanensis occurs exclusively within the intestine of an infected human host. As a consequence of ongoing sexual reproduction throughout the course of an infection, C. cayetanensis infections characteristically comprise a genetically heterogenous population of parasites, as reflected in the MLST genotypes generated from these infections, which tend to be complex (Barratt et al., 2021; Barratt et al., 2019a; Barratt et al., 2019b; Nascimento et al., 2020). This genetic heterogeneity makes MLST data from C. cayetanensis another interesting use-case for haplotype-based approaches.
2.2. Origin of datasets
The Strongyloides sp. datasets described in Table 1 were originally compiled by Barratt and Sapp (Barratt and Sapp, 2020), from MLST data that was publicly available via the NCBI nucleotide database. For the vast majority of the Strongyloides sp. MLST genotypes included in this study, the data were generated via isolation of individual worms from various host species using methods described elsewhere (Jaleta et al., 2017; Janwan et al., 2020; Sanpool et al., 2019; Schär et al., 2014; Thanchomnang et al., 2019; Zhou et al., 2019). Subsequently, DNA was extracted from each individual worm for downstream PCR, targeting various combinations of the mitochondrial cox1 locus, hypervariable region 1 of the 18S rDNA locus (HVR-I) and/or hypervariable region 4 (HVR-IV) of the 18S rDNA locus. These PCR products were then sequenced using Sanger technology (Jaleta et al., 2017; Janwan et al., 2020; Sanpool et al., 2019; Schär et al., 2014; Thanchomnang et al., 2019; Zhou et al., 2019). A smaller portion of the Strongyloides sp. genotypes included in this analysis were generated from publicly available Illumina reads (whole genome shotgun reads published in the NCBI SRA database) generated from individually isolated worms, where complete haplotypes for HVR-I, HVR-IV, and cox1were extracted from this data (Kikuchi et al., 2016). In other cases, Strongyloides sp. genotypes were generated by PCR amplification of HVR-I, HVR-IV, and cox1 from DNA extracted directly from fecal specimens collected from infected hosts (Barratt et al., 2019b; Beknazarova et al., 2019). These amplicons were subsequently sequenced on the Illumina MiSeq platform (Barratt et al., 2019b; Beknazarova et al., 2019). Metadata accompanying each genotype was extracted from the original publications describing the MLST data, and/or from the GenBank flatfiles available for the sequences published in NCBI. For full details of how the Strongyloides sp. datasets were compiled please refer to the study by Barratt and Sapp (Barratt and Sapp, 2020).
The C. cayetanensis dataset was compiled from publicly available data generated by the US Centers for Disease Control and Prevention and partnering public health laboratories from 2018 to 2020, as part of routine genotyping performed in support of C. cayetanensis outbreak investigations (Barratt et al., 2021; Nascimento et al., 2020; Barratt et al., 2021). As part of the C. cayetanensis genotyping procedure employed at CDC, DNA is extracted from human fecal specimens and eight markers are amplified from these extracts. A brief description of these markers is provided in the footnotes of Table 1. These eight amplicons are subjected to deep sequencing on the Illumina MiSeq platform, and all underlying alleles/haplotypes are identified at each marker using a custom bioinformatic pipeline to generate the final MLST genotype of each isolate (Barratt et al., 2021). Importantly, the use of deep amplicon facilitates detection of multiple alleles for each marker, and multiple alleles are frequently observed within a single infection, particularly at nuclear markers (Barratt et al., 2021; Nascimento et al., 2020; Barratt et al., 2021). Illumina data for isolates included in this analysis can be accessed under NCBI BioProject PRJNA578931. Epidemiologic information accompanying these C. cayetanensis genotypes were available from the published manuscripts originally describing these MLST data (Barratt et al., 2021; Nascimento et al., 2020; Barratt et al., 2021).
2.3. Data formatting and distance computations
Haplotypes were assigned unique identifiers following previously established conventions for each dataset (Barratt and Sapp, 2020; Nascimento et al., 2020). Next, the haplotype lists (i.e., genotypes) generated for each isolate were formatted to a haplotype data sheet (Supplementary File S1); a condensed format for presenting haplotype data described here: https://github.com/Joel-Barratt/Eukaryotyping. Computation of Jaccard (JD), Euclidean (ED), and Manhattan distances (MD), Bray-Curtis dissimilarity (BC), and Jensen-Shannon divergence (JSD) was performed via the phyloseq R package using the haplotype data sheets as input. To do this, each unique haplotype was defined as an operational taxonomic unit (OTU) to generate phyloseq objects using the otu_table and tax_table functions. The distance function in the phyloseq package was used to compute a distance matrix using each of these five statistics listed above. Calculation of Plucinski’s Bayesian (PB) and Barratt’s heuristic distances (BH) was performed as previously described (Barratt and Sapp, 2020; Nascimento et al., 2020), using the algorithms and instructions available here: https://github.com/Joel-Barratt/Eukaryotyping. When computing PB distances, investigators must first select a value for epsilon (Nascimento et al., 2020). For the Strongyloides datasets, epsilon was set to 0.05 as was done in the original study describing these data (Barratt and Sapp, 2020). For C. cayetanensis, epsilon was set to 0.3072 as previously described (Nascimento et al., 2020). For an explanation of epsilon please refer to the original description of the PB algorithm (Barratt et al., 2021; Nascimento et al., 2020). The BH and PB statistics were originally designed as an ensemble (Barratt et al., 2019a; Nascimento et al., 2020), so two additional matrices were computed for each dataset; one by taking the average of the BH and PB matrices, and the other by mapping distances generated using the PB statistic to the empiric distribution of distances computed using the BH statistic, and the average of the resulting pairs was taken. The latter procedure constitutes the Barratt-Plucinski ensemble (Nascimento et al., 2020). Consequently, nine distance matrices were generated for each dataset. Each matrix (n = 27) was clustered using Ward’s method to produce a hierarchical tree for each matrix (Nascimento et al., 2020). Matrices are provided in Supplementary Files S3 to S5. For generation of figures, hierarchical trees were rendered using the ggtree R package. Custom images and annotations were added using the GNU Image Manipulation Program (GIMP).
2.4. Tree dissection
Using the cutree R function, each of the 27 hierarchical trees was dissected into partitions. The C. cayetanensis hierarchical trees were each dissected empirically into 46 partitions. Hierarchical trees generated for the Strongyloides datasets were empirically dissected into 6 partitions. For all datasets, the partition membership of each isolate was noted for assessment of clustering performance against expected clustering outcomes. Notably, despite selecting an empirical partition number when dissecting each hierarchical tree, isolates would be assigned to partitions containing sets of closely related isolates regardless, facilitating a relative comparison of clustering performance across all distance statistics.
2.5. Assessment of clustering performance
Molecular phylogeny aims to predict evolutionary/kinship relationships between isolates/taxa using genetic information as input. Given the nature of this objective, it is difficult to test the validity of a phylogenetic reconstruction because evolutionary processes are difficult to observe. Therefore, to validate clustering performance here, we utilized external metadata accompanying each MLST dataset to predict clustering outcomes that likely constitute ‘ground truth’. It is generally accepted that members of a species (or related taxa) collected from one geographic location are more likely to be closely related compared to isolates collected from disparate regions based on the well-described phenomenon of allopatric speciation (Barratt and Sapp, 2020). Epidemiologic data may also be used to assess the validity of clustering outcomes; the field of molecular epidemiology relies on the fact that epidemiologically-linked isolates are often genetically similar as they are derived from a common source (van Belkum et al., 2007). The use of such metadata to establish some degree of ‘ground truth’ is also commonly used in the field of machine learning to assess performance (Goodswen et al., 2021).
The C. cayetanensis dataset was accompanied by previously defined epidemiologic links for 552 of the 1137 genotyped isolates included in this analysis (Casillas et al., 2018; Nascimento et al., 2020; Anonymous, 2018; Anonymous, 2019a; Anonymous, 2020; Anonymous, 2019b). Therefore, to assess clustering performance for the C. cayetanensis dataset, we compared observed isolate partition memberships following dissection of hierarchical trees to their epidemiologic linkage, to determine the level of concordance between them. For a quantitative assessment, we classified clustering results obtained for each isolate as either a true positive, false positive, true negative, or false negative, using the definitions in Table 2. From these classifications we calculated various performance metrics including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy, as previously described (Nascimento et al., 2020). Each metric was weighted by the ratio of genotyped isolates in each epidemiologic cluster to the total number of genotyped isolates with epidemiologic links (n = 552) so that larger epidemiologic clusters (i.e., with more genotyped isolates) would contribute more to the final values. Performance metrics were calculated in this way for each of the nine C. cayetanensis matrices.
Table 2.
Comparison of Cyclospora isolate cluster memberships identified using Barratt’s heuristic to epidemiologic data available for 552 isolates.
| Epidemiologic clusters (year of outbreak) | Total isolates from epi-cluster | Mode Partition Number † | True Positives (TP) | True Negatives (TN) | False Positives (FP) | False Negatives (FN) | Sensitivity: | Specificity: | PPV: | NPV: | Accuracy: |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Distributor A - Type 1 (2019) | 13 | 30 | 13 | 539 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Distributor A - Type 17 (2019) | 43 | 1 | 39 | 509 | 0 | 4 | 90.7% | 100.0% | 100.0% | 99.2% | 99.3% |
| Distributor A - Type 18 (2019) | 14 | 29 | 9 | 537 | 1 | 5 | 64.3% | 99.8% | 90.0% | 99.1% | 98.9% |
| Distributor A - Type 3 (2019) | 18 | 7 | 18 | 530 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Pre-packaged salad mix 2020_001 (2020) | 140 | 6 | 127 | 410 | 2 | 13 | 90.7% | 99.5% | 98.4% | 96.9% | 97.3% |
| Pre-packaged salad mix 2020_003 (2020) | 79 | 14 | 75 | 473 | 0 | 4 | 94.9% | 100.0% | 100.0% | 99.2% | 99.3% |
| Prepackaged salad 002 (2020) | 8 | 4 | 8 | 525 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Restaurant A (2019) | 13 | 4 | 13 | 525 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Restaurant B (2019) | 13 | 13 | 11 | 539 | 0 | 2 | 84.6% | 100.0% | 100.0% | 99.6% | 99.6% |
| Restaurant C (2019) | 6 | 5 | 6 | 546 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Restaurant C (Herb 2) Associated Cluster (2018) | 2 | 19 | 2 | 550 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Restaurant D (2019) | 13 | 11 | 13 | 539 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Salad Chain A - 2020_025 (2020) | 4 | 7 | 4 | 530 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Supplier X - Restaurants A & B (Herb 1) Associated Cluster (2018) | 6 | 4 | 6 | 525 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Temporospatial Cluster A (2018) | 8 | 46 | 8 | 544 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| TN/GA/VA Mexican-style restaurant / cilantro sub-cluster (2020) | 10 | 3 | 10 | 542 | 0 | 0 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Vendor A (2018) | 99 | 17 | 88 | 453 | 0 | 11 | 88.9% | 100.0% | 100.0% | 97.6% | 98.0% |
| Vendor B (2018) | 63 | 32 | 51 | 489 | 0 | 12 | 81.0% | 100.0% | 100.0% | 97.6% | 97.8% |
| Adjusted overall result: | 90.8% | 99.9% | 99.4% | 98.3% | 98.5% |
Note: Partition memberships used to calculate these metrics are available in Supplementary File S2.
The most common partition number to which specimens with this epidemiologic linkage were assigned. Note that partition numbers are arbitrary.
TP: Number of specimens assigned to the correct partition number.
TN: Number of specimens with different epidemiologic linkage not assigned to the same partition.
FP: Number of specimens with different epidemiologic linkage assigned to the same partition. Note that there are some exceptions to this classification. The two epidemiologic clusters shaded yellow were both assigned to partition number 7. These were not considered as FP classifications for one another as the outbreaks occurred in different years and were seemingly caused by the same genotype. Three epidemiologic clusters are shaded blue and each are assigned to partition number 4. These were not considered as FP classifications for one another as the outbreaks were caused by parasites of the same genotype and occurred in different years.
FN: Number of specimens from this epidemiologic cluster not assigned to the correct partition.
PPV: Positive Predictive Value.
NPV: Negative predictive Value.
For S. stercoralis we evaluated clustering performance in a qualitative manner, based on previous studies confirming that certain S. stercoralis types preferentially infect specific host species. Two major lineages of S. stercoralis are known (lineages A and B) (Ko et al., 2020; Nagayasu et al., 2017), where lineage B infects dogs exclusively and is typically found in parts of South East Asia (Jaleta et al., 2017). Possession of haplotype B at the HVR-IV locus (see footnotes of Table 1) is a defining feature of S. stercoralis lineage B: this lineage invariably possesses haplotype B (Barratt et al., 2019b; Barratt and Sapp, 2020; Beknazarova et al., 2019; Jaleta et al., 2017). Lineage A of S. stercoralis invariably possesses haplotype A at HVR-IV and reportedly infects humans and dogs (Barratt and Sapp, 2020; Jaleta et al., 2017), though sub-populations within lineage A may preferentially infect dogs while others may prefer humans (Barratt and Sapp, 2020). These observations relating genotype to host specificity are supported by numerous studies (Barratt et al., 2019b; Barratt and Sapp, 2020; Beknazarova et al., 2019; Jaleta et al., 2017; Ko et al., 2020; Nagayasu et al., 2017). The S. fuelleborni dataset was evaluated in a similar fashion to the S. stercoralis dataset, where results were expected to reflect previous observations associating host preference and geographic range with specific genotypes (Barratt and Sapp, 2020). Specifically, the isolates of S. fuelleborni were derived from defined geographic regions and hosts, and includes six major groups; 1) isolates from Japanese macaques, 2) isolates from long-tailed macaques from Thailand & Laos, and one from an Indian human, 3) isolates from Tanzanian primates (including humans), 4) isolates from primates in Central/West Africa (including humans), 5) isolates from monkeys and orangutans in Malaysian Borneo, and finally 6) isolates of an undefined Strongyloides species collected from Bornean slow lorises (Barratt and Sapp, 2020). Given the relatively isolated geographic regions from which these six groups are derived, they were expected to cluster based on their geographic/host origin as previously observed (Barratt and Sapp, 2020). Host, and/or geographic metadata associated with each Strongyloides isolate are provided in Supplementary File S2.
Comparison of cluster memberships
The qualitative and quantitative performance assessment described above was used to determine the distance statistic resulting in the most robust haplotype-based phylogenetic reconstruction. Once this was established, we used the Rand index and adjusted Rand index to compare clustering similarity between the most robust method and all other methods for each of the three MLST datasets. Note that the rand index is a measure of clustering similarity computed in a similar way to accuracy (i.e., [(TP + TN)/(TP + TN + FP + FN)] – see Table 2 (Rand, 1971). The adjusted Rand Index is a variation of the Rand Index that accounts for the possibility that some clustering similarity observed could be due to random chance. Calculation of the Rand Index and adjusted Rand Index was performed using the rand.index and adj.rand.index functions respectively, provided with the fossil R package.
3. Results
3.1. The Cyclospora cayetanensis MLST dataset
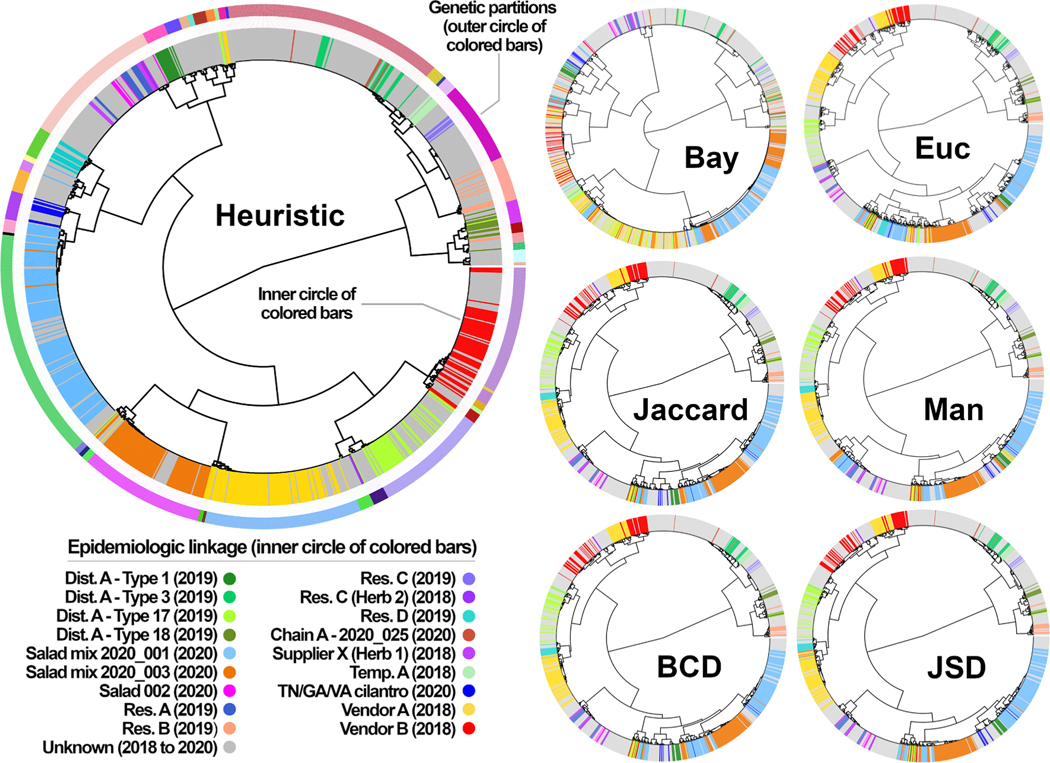
Following hierarchical clustering of the nine C. cayetanensis distance matrices generated, each hierarchical tree was divided into 46 partitions. The 552 isolates possessing epidemiologic links were then classified as either a true positive, true negative, false positive, or false negative based on the partition number to which they were assigned (1 to 46; see Table 2). Weighted values of sensitivity, specificity, PPV, NPV, and accuracy, were 90.8%, 99.9%, 99.4%, 98.3% and 98.5%, respectively using Barratt’s heuristic, which performed more robustly than the other statistics (Table 3). Euclidean distances performed with the lowest sensitivity (53.99%), the lowest accuracy (97.34%), and the lowest NPV (97.34%) (Table 3). Plucinski’s Bayesian distances performed with the lowest specificity (99.19%) and lowest PPV (83.60%), noting that these values are still high and reflect good performance. A qualitative assessment of performance via manual examination of cluster dendrograms generated for the C. cayetanensis dataset using each statistic reflected the quantitative metrics (Table 3), where isolates with the same epidemiologic linkage were more frequently assigned to the same partition when distances were computed using Barratt’s heuristic (Fig. 3). Comparison of partition memberships using the Rand Index and adjusted Rand Index showed that cluster memberships obtained using Plucinski’s method more closely resembled those obtained using Barratt’s method compared to the other statistics (Table 4).
Table 3.
Comparison of results obtained for each metric applied to the Cyclospora dataset.
| Barratt’s heuristic distances | Plucinski’s Bayesian distances | Jaccard distances | Bray-Curtis dissimilarity | Euclidean distances | Manhattan distances | Jensen-Shannon divergence | Ensemble approach † | Average of Barratt’s & Plucinski’s distances | |
|---|---|---|---|---|---|---|---|---|---|
| Classification of clustering results based on definitions in Table 2 | |||||||||
| True Positives | 501 | 372 | 383 | 382 | 298 | 382 | 394 | 492 | 481 |
| True Negatives | 9305 | 8960 | 9298 | 9300 | 9304 | 9296 | 9298 | 9308 | 9304 |
| False Positives | 3 | 73 | 15 | 15 | 8 | 19 | 15 | 4 | 10 |
| False Negatives | 51 | 180 | 169 | 170 | 254 | 170 | 158 | 60 | 71 |
| Performance metrics calculated based on above classifications | |||||||||
| Sensitivity | 90.76% | 67.39% | 69.38% | 69.20% | 53.99% | 69.20% | 71.38% | 89.13% | 87.14% |
| Specificity | 99.97% | 99.19% | 99.84% | 99.84% | 99.91% | 99.80% | 99.84% | 99.96% | 99.89% |
| Positive Predictive Value | 99.40% | 83.60% | 96.23% | 96.22% | 97.39% | 95.26% | 96.33% | 99.19% | 97.96% |
| Negative Predictive value | 99.45% | 98. 03% | 98.21% | 98.20% | 97.34% | 98.20% | 98.33% | 99.36% | 99.24% |
| Accuracy | 99.45% | 97.36% | 98.13% | 98.13% | 97.34% | 98. 08% | 98.25% | 99.35% | 99.18% |
A normalization procedure described previously where the Plucinski’s Bayesian distances were mapped to the empiric distribution of distances calculated using Barratt’s heuristic and the average of the resulting pairs was taken (Nascimento et al., 2020).
Note: True positive, true negative, false positive, and false negative classifications were made according to the definitions provided in Table 2.
Fig. 3.
Cluster dendrograms showing the population structure predicted for the C. cayetanensis dataset using each of seven distance statistics. Seven distance matrices computed from 1137 C. cayetanensis genotypes were clustered using Ward’s method to generate the dendrograms shown. A partition number of 46 was used to dissect each dendrogram for calculation of the metrics in Table 2, Table 3, and Table 4. The largest dendrogram was generated using Barratt’s heuristic, where the outer circle of colored bars shows the boundary between each of the 46 partitions. The inner circle of bars on the larger dendrogram is color coded to indicate genotypes epidemiologically linked to clusters of cyclosporiasis. The color coding on the smaller dendrograms also reflects the epidemiologic linkage of the various genotypes. Examination of each dendrogram shows that genotypes labelled with the same color more frequently cluster within the same partition when Barratt’s heuristic definition of genetic distance is used to compute a distance matrix. Heuristic: Barratt’s heuristic, Bay: Plucinski’s Bayesian distances, Euc: Euclidean distances, Jaccard: Jaccard distances, Man: Manhattan distances, BCD: Bray-Curtis Dissimilarity, JSD: Jensen-Shannon Divergence, Dist: Distributor, Res: Restaurant, Temp: Temporo-spatial cluster.
Table 4.
Rand Index calculated for comparing partition memberships obtained using Barratt’s to all other metrics.
| Datasets: | Cyclospora cayetanensis | Strongyloides stercoralis | Strongyloides fuelleborni † | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Rand Index | Adjusted Rand Index | Rand Index | Adjusted Rand Index | Rand Index | Adjusted Rand Index | |
| Barratt’s heuristic | 1 | 1 | 1 | 1 | 1 | 1 |
| Plucinski’s Bayesian | 0.93419333 | 0.570332115 | 0.92141029 | 0.97397921 | 0.73221673 | 0.90453406 |
| Jaccard distances | 0.87506813 | 0. 035431205 | 0.84566455 | 0.94575601 | 1 | 1 |
| Bray-Curtis dissimilarity | 0.87525394 | 0. 035538072 | 0.84566455 | 0.94575601 | 1 | 1 |
| Jensen-Shannon divergence | 0.87415146 | 0. 036197724 | 0.76180456 | 0.91162873 | 1 | 1 |
| Euclidean distances | 0.8893911 | 0. 030285313 | 0.91104675 | 0.97078268 | 0.71708547 | 0.89735703 |
| Manhattan distances | 0.8753422 | 0. 033919177 | 0.84566455 | 0.94575601 | 1 | 1 |
Includes genotypes from 18 isolates of a S. fuelleborni-like species collected from Bornean slow lorises.
Note: Based on the Rand Index and Adjusted Rand index the cluster memberships obtained using Plucinski’s Bayesian definition of genetic distances most closely resemble those obtained using Barratt’s heuristic definition of genetic distance for two of the three datasets.
3.2. The Strongyloides stercoralis MLST dataset
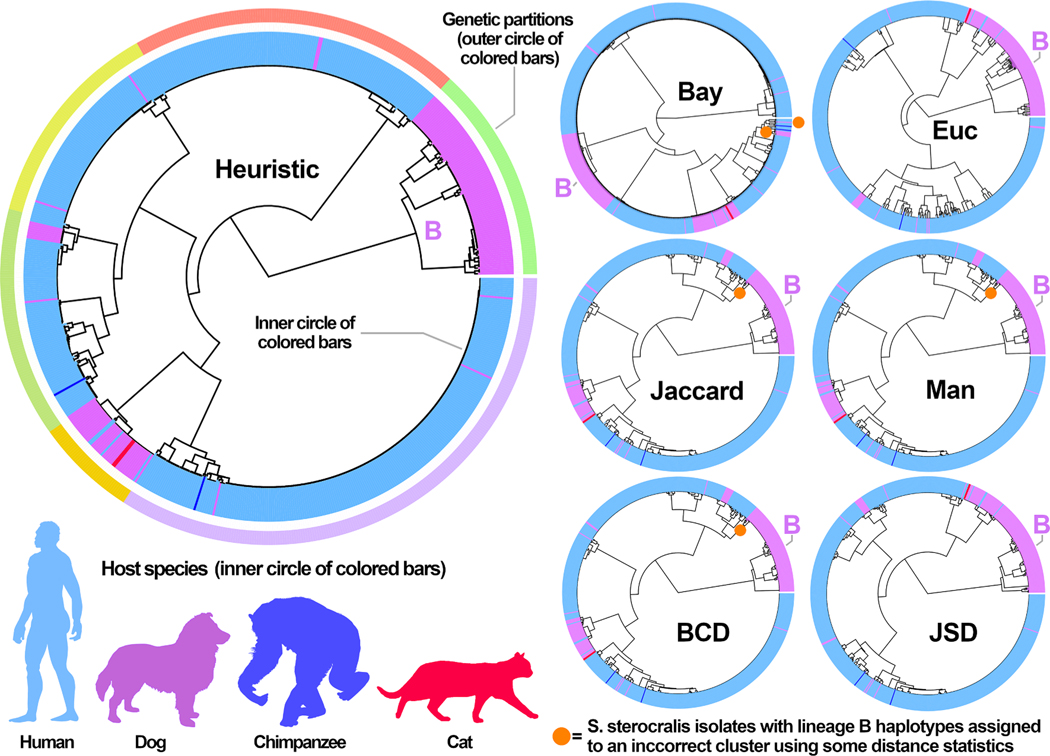
Prior studies on the population structure of S. stercoralis provide a strong indication that two major lineages of S. stercoralis exist (lineage A and lineage B) and that these two lineages are monophyletic (Ko et al., 2020; Nagayasu et al., 2017). Isolates belonging to lineage B invariably possess haplotype B at their HVR-IV locus, while members of lineage A possess haplotype A at their HVR-IV locus (Jaleta et al., 2017). Of the seven distance statistics evaluated, Barratt’s heuristic, Jansen-Shannon divergence, and Euclidean distances correctly placed all S. stercoralis isolates possessing haplotype B within a cluster exclusively comprising S. stercoralis isolates belonging to lineage B (Fig. 4). The four other distance statistics incorrectly excluded six isolates of S. stercoralis possessing haplotype B from the lineage B cluster, assigning these isolates to a cluster containing all lineage A isolates. Comparison of cluster memberships obtained when each S. stercoralis hierarchical tree was divided into 6 partitions using the Rand Index and adjusted Rand Index showed that the cluster memberships obtained using Plucinski’s method more closely resembled those obtained for Barratt’s heuristic compared to all other methods (Table 4).
Fig. 4.
Cluster dendrograms showing the population structure predicted for the S. stercoralis dataset using each of seven distance statistics. Distance matrices were computed from the 704 S. stercoralis genotypes using each of seven distance statistics. These matrices were clustered using Ward’s method to generate the dendrograms shown. Each dendrogram was dissected into 6 partitions to compute the Rand indices as shown in Table 4. The largest dendrogram was generated using Barratt’s heuristic, where the outer circle of colored bars shows the boundary between each of the 6 partitions. The inner circle of colored bars on the larger dendrogram is color coded to indicate genotypes obtained from one of four possible hosts (humans, dogs, cats, and chimpanzees). The color coding on the smaller dendrograms also reflects the host species from which the genotyped S. stercoralis isolates were derived. The orange circle shown on four of the smaller dendrograms is adjacent to or on a node that includes six specimens belonging to lineage B that were incorrectly assigned to lineage A using four of the distance statistics. The partition representing lineage B of S. stercoralis is labelled on each dendrogram. Isolates that were assigned incorrectly to lineage A are shown in Supplementary File S1 (colored in blue). Heuristic: Barratt’s heuristic, Bay: Plucinski’s Bayesian distances, Euc: Euclidean distances, Jaccard: Jaccard distances, Man: Manhattan distances, BCD: Bray-Curtis Dissimilarity, JSD: Jensen-Shannon Divergence.
3.3. The Strongyloides fuelleborni MLST dataset
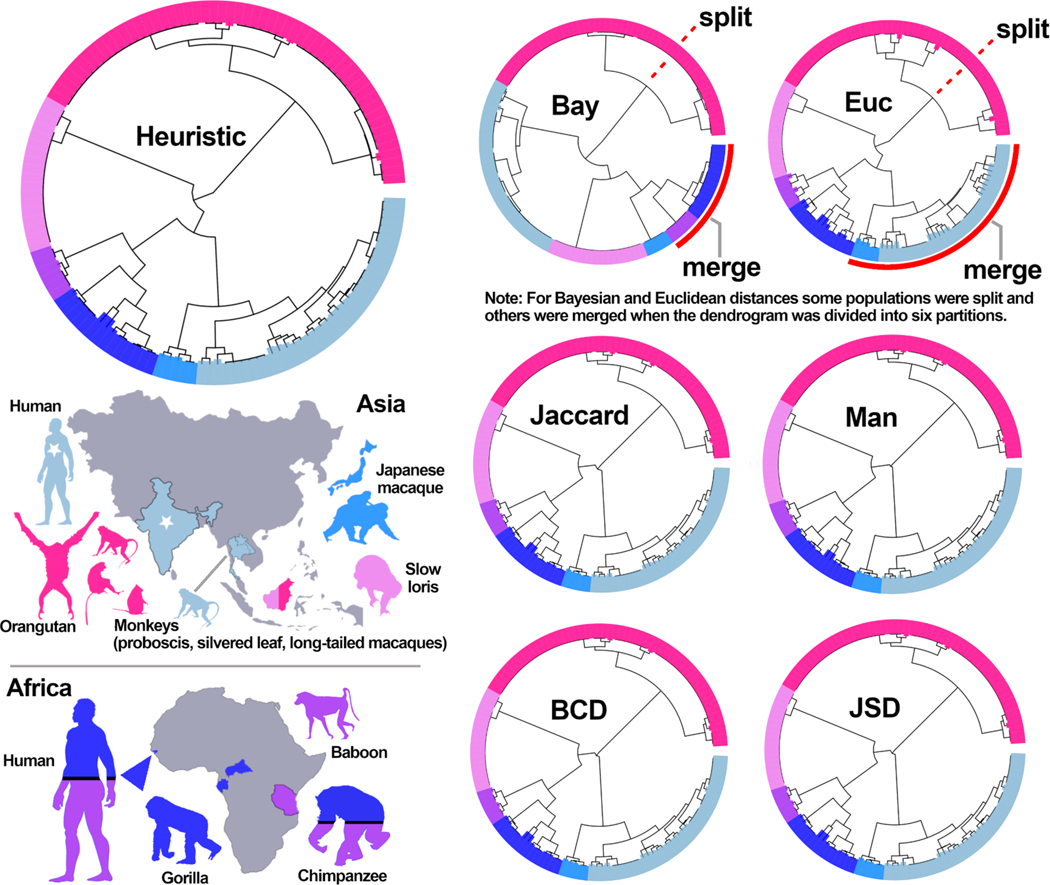
Information on the geographic origin and host origin of S. fuelleborni isolates facilitated a qualitative evaluation of clustering performance. All seven distance statistics applied to the S. fuelleborni dataset produced similar results that were supported by host and/or geographic trends. Using alignment-based phylogenetic methods, the Strongyloides isolate collected from Bornean slow lorises was shown previously to be distinct from S. fuelleborni found in monkeys and apes (Frias et al., 2018). In agreement with this observation, each of the seven distance statistics evaluated here supported the distinctness of isolates from Bornean slow lorises. Isolates from Central/West African primates formed a single cluster, and East African (Tanzanian) S. fuelleborni isolates formed another distinct cluster using each of the seven distance statistics (Fig. 5). Using all statistics, isolates from Japanese macaques were separated from all other isolates, as were those collected in Malaysian Borneo from various monkeys and orangutans. Isolates collected from long-tailed macaques in Thailand and Laos plus a single Indian human, formed a sixth distinct group using all distance statistics evaluated here. Subtle differences were observed for some of the distance statistics when the relationship between the six partitions was investigated via manual examination of the hierarchical trees (Fig. 5), where Barratt’s heuristic, Jaccard distances, Bray-Curtis dissimilarity, Jensen-Shannon divergence, and Manhattan distances shared a strong consensus in terms of overall tree structure. This tree structure was reflected in values obtained using the Rand Index and adjusted rand index, where the five distance statistics listed above gave rise to identical partition memberships while Plucinski’s Bayesian distances and Euclidean distances differed only slightly (Table 4). Despite these minor differences, all distance statistics supported the same general relationship between geographic origin and/or host origin for isolates assigned to each of the six partitions (Fig. 5).
Fig. 5.
Cluster dendrograms showing the population structure predicted for the S. fuelleborni dataset using each of seven distance statistics. Distances were computed from the 133 S. fuelleborni genotypes (including 18 from a distinct Strongyloides species) and were clustered using Ward’s method to generate the dendrograms shown. These dendrograms were divided into 6 partitions to compute the Rand indices as shown in Table 4. The largest dendrogram shows the result obtained using Barratt’s heuristic, where the bars are color coded to indicate genotypes obtained from various primates (i.e., monkeys, apes, humans, and lorises) from different locations, which match the boundary between the 6 partitions. Color coding on the smaller dendrograms also reflects the host species from which the Strongyloides isolates were derived. On the map of Asia, the star indicates a single isolate from an Indian human which was assigned to the partition colored in gray on each dendrogram. Long tailed macaques from Southeast Asia (gray without a star – indicating Laos and Thailand) were assigned to the same genetic partition as the Indian isolate. On the map of Africa, dark blue indicates S. fuelleborni isolates from chimpanzees, humans, and/or gorillas from Gabon, Guinea-Bissau (indicated with a triangle) and/or the Central African Republic. Purple indicates isolates from humans, chimpanzees, and baboons from Tanzania. Heuristic: Barratt’s heuristic, Bay: Plucinski’s Bayesian distances, Euc: Euclidean distances, Jaccard: Jaccard distances, Man: Manhattan distances, BCD: Bray-Curtis Dissimilarity, JSD: Jensen-Shannon Divergence.
4. Discussion
Seven distance statistics were evaluated for their ability to facilitate a robust haplotype-based phylogenetic reconstruction. Of these, Barratt’s heuristic was the most robust based on a qualitative and quantitative assessment against expected clustering outcomes. Haplotype-based methods require computation of distances using a chosen distance statistic. These distances are then clustered to construct hierarchical trees that reflect evolutionary relationships inferred from the genetic data. A key distinction between haplotype-based and alignment-based phylogenetic methods is that for haplotype-based methods, distances are computed based the number of intersecting haplotypes observed between pairs of isolates. This intersect is subsequently used as statistical evidence to numerically characterize the genetic relationship between each possible pair of isolates.
An advantage of haplotype-based analysis workflows is that genotypes are represented by a list of identifiers (i.e., haplotype names) which may include multiple alleles detected at the same locus (i.e., heterozygosity); a feature that would confound alignment-based approaches. Additionally, isolates with a partial genotype may be retained for analysis – within reason, and depending on the level of incompleteness – where loci without data would simply be absent from some isolates’ lists. Furthermore, because haplotype-based methods compare identifiers (not sequences), indels and/or repeat-based polymorphisms do not impact the comparison. The C. cayetanensis dataset includes a repeat-based locus called the mitochondrial junction, and haplotypes of this locus differ by varying numbers and combinations of multiple 15-mer repeat motifs (Nascimento et al., 2019). Because of its repetitive nature, haplotypes of this locus align poorly, with many large gaps, making it a poor candidate for alignment-based phylogeny (Nascimento et al., 2019). Similarly, the Strongyloides datasets includes different haplotypes of the HVR-I and HVR-IV loci, which possess various indels that culminate in poor alignments (Barratt and Sapp, 2020).
While they afford some clear advantages, haplotype-based methods also possess limitations. For instance, haplotype-based tree structures may lack granularity compared to alignment-based trees because they do not consider each variant base in the distance computation. Instead, haplotypes are defined across a span of multiple bases as defined by the investigator. A pair of haplotypes differing by one nucleotide and those differing by five nucleotides are considered equally different during distance computation because nucleotide composition is not considered.
Investigators should recognize this limitation when defining target loci by understanding that the main source of granularity for haplotype-based methods comes from heterozygosity and recombination of unlinked loci. Consequently, investigation of genetic kinship among sexually reproducing species will benefit most from haplotype-based methods, which will be of greatest value when applied to marker combinations that include unlinked loci (i.e., those subject to recombination), heterozygous loci, and loci encoded on different organellar genomes (i.e., nuclear, mitochondrial, and plastid genomes). Selecting combinations of loci possessing these characteristics considers sources of diversity that are poorly captured by alignment-based methods, and will likely improve granularity. Additionally, rather than defining haplotypes over a span of several bases, investigators could define every base that possesses a known variant as a distinct marker (i.e., where each nucleotide variant receives its own haplotype identifier) to improve granularity in the resultant tree structure. However, this could reduce the speed of distance computations (particularly for larger datasets with many variants), and would only work for SNP-based polymorphisms; indel- and repeat-based polymorphisms would still need to be defined over a span of multiple bases.
While tolerance for missing data is described as a clear advantage of haplotype-based methods, investigators must recognize that this comes with limitations: distances become increasingly tenuous as the amount of missing data increases. Investigators should therefore establish minimum data requirements that exclude isolates with too few loci sequenced. Implementation of such thresholds and suggestions on how thresholds could be defined are discussed elsewhere (Barratt and Sapp, 2020; Nascimento et al., 2020). Another consideration is that the distance statistics evaluated here do not treat missing data in the same way, so tolerance for missing loci varies between some of them. For example, Barratt’s heuristic attempts to impute distance values for missing loci rather than simply ignoring them as most of the other statistics do (Barratt and Sapp, 2020; Nascimento et al., 2020). This behavior may partially account for the robustness of Barratt’s heuristic relative to other statistics.
Of the statistics evaluated, only Barratt’s heuristic and Plucinski’s Bayesian methods were specifically designed for analysis of eukaryote-derived MLST datasets. Manhattan distances, Euclidean distances, Bray-Curtis dissimilarity, and Jensen-Shannon divergence have been applied to a diverse range of fields including genetics, quantum theory, ranking words in text documents, ecology, and clustering of spoken languages (Kartal et al., 2020; Majtey et al., 2005; Mehri et al., 2015; Ricotta and Podani, 2017; Strauss et al., 2017). The simplest of the statistics evaluated here is the Jaccard distance, which is computed by taking the value of one and subtracting the value obtained after dividing the number of intersecting haplotypes by the union (Ricotta and Podani, 2017). The robustness of Barratt’s heuristic relative to other statistics may be partly attributed to its consideration of several aspects of genetic data that are largely ignored by some other statistics. For instance, Barratt’s heuristic considers that sexual reproduction may account for the presence of two haplotypes in one parasite isolate but only one haplotype for the same locus in another (i.e., a heterozygote versus homozygote). In the context of genetic data, the absence of one haplotype in the homozygote could still indicate close kinship to the heterozygote (e.g., they may be siblings). Consequently, a single matching haplotype between a heterozygote and a homozygote is penalized less by Barratt’s heuristic compared to when two heterozygotes share only one allele (Nascimento et al., 2020). This ‘genetic rationale’ is not fundamental to the logic underpinning some of the other statistics which simply consider the homozygote to be lacking a haplotype present in the heterozygote.
Barratt’s heuristic also accounts for differences in the amount of information provided by each locus by weighting the contribution of loci by their Shannon entropy (Nascimento et al., 2020). Consequently, loci providing the most information contribute most to the final set of distances. Barratt’s heuristic also scales the distance contributed by each locus separately, using a frequentist probability to account for differences in haplotype frequency. The rationale for this is that a match observed between two isolates for a rare haplotype (e.g., found in 1 % of isolates) provides better evidence that a pair shares close genetic kinship compared to a match observed for a haplotype that occurs in 99 % of the population. In this example, the probability that a randomly selected pair of isolates would possess the more common haplotype would be 0.99^2 or 0.9801. In contrast, the probability of randomly selecting two isolates possessing the rarer haplotype would be 0.01^2 or 0.0001. Barratt’s heuristic also accounts for the fact that nuclear and mitochondrial genes possess different mechanisms of inheritance, so matches (and mismatches) at nuclear versus mitochondrial loci are scored differently (Barratt et al., 2019a). Most other statistics evaluated do not consider these aspects of genetic data.
The observation that each statistic performed similarly on the small S. fuelleborni dataset (133 isolates, 3 markers, no heterozygosity), while Barratt’s heuristic outperformed others when applied to the large and complex C. cayetanensis dataset (1137 isolates, 8 markers, high heterozygosity), are a likely consequence of the genetic considerations underpinning this heuristic algorithms’ design. Notably, while differences in performance were observed for Barratt’s heuristic and Plucinski’s method, the Rand Index and adjusted Rand Index supported a high degree of clustering similarity between these two statistics for the S. stercoralis and C. cayetanensis datasets. This is likely related to the fact that important genetic considerations informed the logic underpinning both methods (Nascimento et al., 2020), which is not the case for the five other distance statistics evaluated.
Importantly, our choice of the MLST datasets utilized here was driven by the availability of the MLST data itself, in addition to the availability of high-quality metadata accompanying the MLST data; these metadata were required for assessment of clustering performance against a set of expected outcomes. The organisms from which these data are derived are widely disparate from one another taxonomically (worms and protozoa), yet the heuristic algorithm remained the strongest performer when applied to the S. stercoralis and C. cayetanensis datasets. This supports that the conclusions of this analysis are likely generalizable to a range of taxa. Despite this, evaluation of these methods on MLST datasets from other taxa would be of great value, to better understand how generalizable the haplotype-based methods are.
While haplotype-based methods possess advantages over traditional alignment-based approaches, we must emphasize that haplotype-based methods should not be used as a replacement for alignment-based methods under all circumstances. Alignment-based methods are robust and widely used, and possess certain advantages, including the potential for increased granularity given that all nucleotide bases are considered in the distance computation. Another advantage of alignment-based methods is the ability to predict divergence times via molecular clock analysis; this type of analysis is not applicable to the haplotype-based approaches described here. Alignment-based and haplotype-based approaches are useful for evaluating both interspecific and intraspecific relationships; we demonstrate here that haplotype-based methods can be used to explore intraspecific phylogenetic relationships among C. cayetanensis isolates. We also explored relationships between a distinct Strongyloides sp. from slow lorises and S. fuelleborni isolates from different geographic locations (i.e., both interspecific and intraspecific relationships were examined in this case). While alignment-based phylogenetic approaches should remain the phylogenetic methods of choice, we do wish to emphasize that in the context of certain datasets that are not amenable to (or even preclude) the use of alignment-based phylogeny (i.e., due the presence of high heterozygosity, or missing data), certain haplotype-based methods represent a robust and viable alternative.
To conclude, Barratt’s heuristic definition of genetic distance performed more robustly than the other statistics evaluated based on a qualitative and quantitative assessment against expected clustering outcomes. Barratt’s heuristic detected genetic relationships that more closely reflected the epidemiologic linkage of 552 C. cayetanensis isolates, compared to all other distance statistics examined. Barratt’s heuristic was also among three of seven distance statistics that fully supported previously described relationships between two distinct lineages of S. stercoralis (Barratt et al., 2019b; Beknazarova et al., 2019; Jaleta et al., 2017; Ko et al., 2020; Nagayasu et al., 2017). For the S. fuelleborni dataset, all seven statistics produced a similar result that generally supported host and/or geographic trends. Ultimately, while each statistic detected many plausible genetic links, for the larger, more complex datasets, Barratt’s heuristic consistently produced a robust phylogenetic reconstruction based on expected clustering outcomes. It is therefore proposed that Barratt’s heuristic represents a viable phylogenetic approach for use in the context of challenging MLST datasets possessing features (i.e., high heterozygosity, partial genotypes, and indel-based and/or repeat-based polymorphisms) that preclude the use of alignment-based methods.
Supplementary Material
Acknowledgements
The authors acknowledge the US Centers for Disease Control and Prevention (CDC), Division of Parasitic Diseases and Malaria in support of this work. MMP was funded by the U.S. President’s Malaria Initiative. The authors acknowledge the support of the Parasitic Diseases Branch and ORAU in support of JB and DJ.
Abbreviations:
- MLST
multi-locus sequence typing
- JD
Jaccard Distances
- ED
Euclidean distances
- MD
Manhattan distances
- BC
Bray-Curtis
- JSD
Jensen-Shannon divergence
- PB
Plucinski’s Bayesian definition of genetic distance
- BH
Barratt’s heuristic definition of genetic distance
- TP
True positive
- TN
True negative
- FP
False positive
- FN
False negative
- PPV
Positive predictive value
- NPV
Negative predictive value
Footnotes
Disclaimer
The findings and conclusions in this paper are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ympev.2022.107608.
CRediT authorship contribution statement
David Jacobson: Data curation, Software, Writing – review & editing. Yueli Zheng: Data curation, Software, Writing – review & editing. Mateusz M. Plucinski: Conceptualization, Methodology, Writing – review & editing. Yvonne Qvarnstrom: Supervision, Project administration, Funding acquisition, Writing – review & editing. Joel L. N. Barratt: Conceptualization, Visualization, Supervision, Project administration, Writing – original draft, Data curation, Investigation, Formal analysis, Investigation, Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- Anonymous. 2018. Domestically Acquired Cases of Cyclosporiasis — United States, May–August 2018. Centers for Disease Control and Prevention; 2018 [cited 2020]; Available from: https://www.cdc.gov/parasites/cyclosporiasis/outbreaks/2018/c-082318/index.html. [Google Scholar]
- Anonymous. 2019a. Domestically Acquired Cases of Cyclosporiasis — United States, May–August 2019. Centers for Disease Control and Prevention; 2019 [cited 2020]; Available from: https://www.cdc.gov/parasites/cyclosporiasis/outbreaks/2019/a-050119/index.html. [Google Scholar]
- Anonymous. 2019b. Outbreak of Cyclospora Infections Linked to Fresh Basil from Siga Logistics de RL de CV of Morelos, Mexico. Centers for Disease Control and Prevention; 2019 [cited 2020]; Available from: https://www.cdc.gov/parasites/cyclosporiasis/outbreaks/2019/weekly/index.html. [Google Scholar]
- Anonymous. 2020. Domestically Acquired Cases of Cyclosporiasis — United States, May–August 2020. Centers for Disease Control and Prevention; 2020 [cited 2021]; Available from: https://www.cdc.gov/parasites/cyclosporiasis/outbreaks/2020/seasonal/index.html. [Google Scholar]
- Ashkenazy H, Cohen O, Pupko T, Huchon D, 2014. Nov 18. Indel reliability in indel-based phylogenetic inference. Genome Biol Evol. 6 (12), 3199–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt JLN, Lane M, Talundzic E, Richins T, Robertson G, Formenti F, Pritt B, Verocai G, Nascimento de Souza J, Mato Soares N, Traub R, Buonfrate D, Bradbury RS, Periago MV, 2019b. A global genotyping survey of Strongyloides stercoralis and Strongyloides fuelleborni using deep amplicon sequencing. PLoS Negl Trop Dis. 13 (9), e0007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt JLN, Park S, Nascimento FS, Hofstetter J, Plucinski M, Casillas S, Bradbury RS, Arrowood MJ, Qvarnstrom Y, Talundzic E, 2019a. Genotyping genetically heterogeneous Cyclospora cayetanensis infections to complement epidemiological case linkage. Parasitology 146 (10), 1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt JLN, Sapp SGH, 2020. Machine learning-based analyses support the existence of species complexes for Strongyloides fuelleborni and Strongyloides stercoralis. Parasitology 147 (11), 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M, 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13, 1–46. [DOI] [PubMed] [Google Scholar]
- Barratt J, Ahart L, Rice M, Houghton K, Richins T, Cama V, et al. , 2021. Genotyping Cyclospora cayetanensis from multiple outbreak clusters with an emphasis on a cluster linked to bagged salad mix - United States, 2020. J Infect Dis 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt J, Kaufer A, Peters B, Craig D, Lawrence A, Roberts T, Lee R, McAuliffe G, Stark D, Ellis J, Schönian G, 2017. Isolation of Novel Trypanosomatid, Zelonia australiensis sp. nov. (Kinetoplastida: Trypanosomatidae) Provides Support for a Gondwanan Origin of Dixenous Parasitism in the Leishmaniinae. PLoS Negl Trop Dis. 11 (1), e0005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt J, Houghton K, Richins T, Straily A, Threlkel R, Bera B, Kenneally J, Clemons B, Madison-Antenucci S, Cebelinski E, Whitney BM, Kreil KR, Cama V, Arrowood MJ, Qvarnstrom Y, 2021. Investigation of US Cyclospora cayetanensis outbreaks in 2019 and evaluation of an improved Cyclospora genotyping system against 2019 cyclosporiasis outbreak clusters. Epidemiol. Infect. 149 (e214), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beknazarova M, Barratt JLN, Bradbury RS, Lane M, Whiley H, Ross K, Rinaldi G, 2019. Detection of classic and cryptic Strongyloides genotypes by deep amplicon sequencing: A preliminary survey of dog and human specimens collected from remote Australian communities. PLoS Negl Trop Dis. 13 (8), e0007241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozidis P, Sakkas H, Pertsalis A, Christodoulou A, Kalogeropoulos CD, Papadopoulou C, 2021. Mar. Molecular Analysis of Dirofilaria repens Isolates from Eye-Care Patients in Greece. Acta Parasitol. 66 (1), 271–276. [DOI] [PubMed] [Google Scholar]
- Casillas SM, Bennett C, Straily A, 2018. Notes from the Field: Multiple Cyclosporiasis Outbreaks - United States, 2018. MMWR Morb Mortal Wkly Rep. 67 (39), 1101–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J, 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17 (4), 540–552. [DOI] [PubMed] [Google Scholar]
- Ellis J, Barratt J, Kaufer A, Pearn L, Armstrong B, Johnson M, Park Y, Downey L, Cao M, Neill L, Lee R, Ellis B, Tyler K, Lun Z-R, Stark D, 2021. A new subspecies of Trypanosoma cyclops found in the Australian terrestrial leech Chtonobdella bilineata. Parasitology 148 (10), 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias L, Stark DJ, Lynn MS, Nathan SK, Goossens B, Okamoto M, MacIntosh AJJ, 2018. Lurking in the dark: Cryptic Strongyloides in a Bornean slow loris. Int J Parasitol Parasites Wildl. 7 (2), 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodswen SJ, Barratt JLN, Kennedy PJ, Kaufer A, Calarco L, Ellis JT, 2021. Machine learning and applications in microbiology. FEMS Microbiol Rev 45 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavsa MC, Roellig DM, Seabolt MH, Kahler AM, Murphy JL, McKitt TK, Geeter EF, Dawsey R, Davidson SL, Kim TN, Tucker TH, Iverson SA, Garrett B, Fowle N, Collins J, Epperson G, Zusy S, Weiss JR, Komatsu K, Rodriguez E, Patterson JG, Sunenshine R, Taylor B, Cibulskas K, Denny L, Omura K, Tsorin B, Fullerton KE, Xiao L, 2017. Using Molecular Characterization to Support Investigations of Aquatic Facility-Associated Outbreaks of Cryptosporidiosis - Alabama, Arizona, and Ohio, 2016. MMWR Morb Mortal Wkly Rep. 66 (19), 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton KA, Lomsadze A, Park S, Nascimento FS, Barratt J, Arrowood MJ, VanRoey E, Talundzic E, Borodovsky M, Qvarnstrom Y, 2020. Development of a workflow for identification of nuclear genotyping markers for Cyclospora cayetanensis. Parasite. 27, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleta TG, Zhou S, Bemm FM, Schär F, Khieu V, Muth S, Odermatt P, Lok JB, Streit A, Fuehrer H-P, 2017. Different but overlapping populations of Strongyloides stercoralis in dogs and humans-Dogs as a possible source for zoonotic strongyloidiasis. PLoS Negl Trop Dis. 11 (8), e0005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janwan P, Rodpai R, Intapan PM, Sanpool O, Tourtip S, Maleewong W, Thanchomnang T, 2020. Possible transmission of Strongyloides fuelleborni between working Southern pig-tailed macaques (Macaca nemestrina) and their owners in Southern Thailand: Molecular identification and diversity. Infect Genet Evol. 85, 104516. [DOI] [PubMed] [Google Scholar]
- Johnson AD, 2010. An extended IUPAC nomenclature code for polymorphic nucleic acids. Bioinformatics 26 (10), 1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan G, Goldman N, 2012. The effects of alignment error and alignment filtering on the sitewise detection of positive selection. Mol Biol Evol. 29 (4), 1125–1139. [DOI] [PubMed] [Google Scholar]
- Kamvar ZN, Brooks JC, Grunwald NJ, 2015. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front Genet. 6, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan L, Wheeler WC, 2012. Maximum Parsimony on Phylogenetic networks. Algorithms Mol Biol. 7 (1), 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartal O, Schmid MW, Grossniklaus U, 2020. Cell type-specific genome scans of DNA methylation divergence indicate an important role for transposable elements. Genome Biol. 21 (1), 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufer A, Ellis J, Stark D, Barratt J, 2017. The evolution of trypanosomatid taxonomy. Parasit Vectors. 10 (1), 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T, Hino A, Tanaka T, Aung MPPTHH, Afrin T, Nagayasu E, Tanaka R, Higashiarakawa M, Win KK, Hirata T, Htike WW, Fujita J, Maruyama H, Cantacessi C, 2016. Genome-Wide Analyses of Individual Strongyloides stercoralis (Nematoda: Rhabditoidea) Provide Insights into Population Structure and Reproductive Life Cycles. PLoS Negl Trop Dis. 10 (12), e0005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko PP, Suzuki K, Canales-Ramos M, Aung MPPTHH, Htike WW, Yoshida A, Montes M, Morishita K, Gotuzzo E, Maruyama H, Nagayasu E, 2020. Phylogenetic relationships of Strongyloides species in carnivore hosts. Parasitol Int. 78, 102151. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG, 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23 (21), 2947–2948. [DOI] [PubMed] [Google Scholar]
- Leigh JW, Bryant D, Nakagawa S, 2015. popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 6 (9), 1110–1116. [Google Scholar]
- Lin CS, 1982. Grouping genotypes by a cluster method directly related to genotype-environment interaction mean square. Theor Appl Genet. 62 (3), 277–280. [DOI] [PubMed] [Google Scholar]
- Lischer HE, Excoffier L, Heckel G, 2014. Ignoring heterozygous sites biases phylogenomic estimates of divergence times: implications for the evolutionary history of microtus voles. Mol Biol Evol. 31 (4), 817–831. [DOI] [PubMed] [Google Scholar]
- Majtey AP, Lamberti PW, Prato DP, 2005. Jensen-Shannon divergence as a measure of distinguishability between mixed quantum states. Phys. Rev. A 72 (5). [Google Scholar]
- Martins JF, Marques C, Nieto-Andrade B, Kelley J, Patel D, Nace D, Herman C, Barratt J, Ponce de León G, Talundzic E, Rogier E, Halsey ES, Plucinski MM, 2020. Malaria Risk and Prevention in Asian Migrants to Angola. Am J Trop Med Hyg. 103 (5), 1918–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehri A, Jamaati M, Mehri H, 2015. Word ranking in a single document by Jensen-Shannon divergence. Phys. Lett. A 379 (28–29), 1627–1632. [Google Scholar]
- Mount DW, 2008b. Choosing a method for phylogenetic prediction. CSH Protoc. 2008 (4), pdb.ip49. [DOI] [PubMed] [Google Scholar]
- Mount DW, 2008a. Distance methods for phylogenetic prediction. CSH Protoc. 2008 (4), pdb.top33. [DOI] [PubMed] [Google Scholar]
- Munjal G, Hanmandlu M, Srivastava S, 2019. Phylogenetics Algorithms and Applications. Ambient Communications and Computer Systems. 904, 187–194. [Google Scholar]
- Nagayasu E, Aung MPPTHH, Hortiwakul T, Hino A, Tanaka T, Higashiarakawa M, Olia A, Taniguchi T, Win SMT, Ohashi I, Odongo-Aginya EI, Aye KM, Mon M, Win KK, Ota K, Torisu Y, Panthuwong S, Kimura E, Palacpac NMQ, Kikuchi T, Hirata T, Torisu S, Hisaeda H, Horii T, Fujita J, Htike WW, Maruyama H, 2017. A possible origin population of pathogenic intestinal nematodes, Strongyloides stercoralis, unveiled by molecular phylogeny. Sci Rep 7 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento FS, Barratt J, Houghton K, Plucinski M, Kelley J, Casillas S, Bennett C(., Snider C, Tuladhar R, Zhang J, Clemons B, Madison-Antenucci S, Russell A, Cebelinski E, Haan J, Robinson T, Arrowood MJ, Talundzic E, Bradbury RS, Qvarnstrom Y, 2020. Evaluation of an ensemble-based distance statistic for clustering MLST datasets using epidemiologically defined clusters of cyclosporiasis. Epidemiol Infect. 148, e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento FS, Barta JR, Whale J, Hofstetter JN, Casillas S, Barratt J, Talundzic E, Arrowood MJ, Qvarnstrom Y, 2019. Mitochondrial Junction Region as Genotyping Marker for Cyclospora cayetanensis. Emerg. Infect. Dis. 25 (7), 1314–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Nzigou Mombo B, Lalremruata A, Koehne E, Zoleko Manego R, Dimessa Mbadinga LB, et al. , 2019. DNA recovery from archived RDTs for genetic characterization of Plasmodium falciparum in a routine setting in Lambarene, Gabon. Malar J. 18 (1), 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettengill JB, Pightling AW, Baugher JD, Rand H, Strain E, Tang H, 2016. Real-Time Pathogen Detection in the Era of Whole-Genome Sequencing and Big Data: Comparison of k-mer and Site-Based Methods for Inferring the Genetic Distances among Tens of Thousands of Salmonella Samples. PLoS ONE 11 (11), e0166162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plucinski MM, Barratt JLN, 2021. Nonparametric Binary Classification to Distinguish Closely Related versus Unrelated P. falciparum Parasites. Am J Trop Med Hyg. Apr 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand WM, 1971. Objective Criteria for the Evaluation of Clustering Methods. J. Am. Stat. Assoc. 66 (336), 846–850. [Google Scholar]
- Redelings BD, Suchard MA, 2007. Incorporating indel information into phylogeny estimation for rapidly emerging pathogens. BMC Evol Biol. 14 (7), 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricotta C, Podani J, 2017. On some properties of the Bray-Curtis dissimilarity and their ecological meaning. Ecol. Complexity 31, 201–205. [Google Scholar]
- Rubio-Largo A, Vanneschi L, Castelli M, Vega-Rodriguez MA, 2018. A Characteristic-Based Framework for Multiple Sequence Aligners. IEEE Trans Cybern. 48 (1), 41–51. [DOI] [PubMed] [Google Scholar]
- Rzhetsky A, Nei M, 1995. Tests of applicability of several substitution models for DNA sequence data. Mol Biol Evol. 12 (1), 131–151. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4 (4), 406–425. [DOI] [PubMed] [Google Scholar]
- Sanpool O, Intapan PM, Rodpai R, Laoraksawong P, Sadaow L, Tourtip S, Piratae S, Maleewong W, Thanchomnang T, 2019. Dogs are reservoir hosts for possible transmission of human strongyloidiasis in Thailand: molecular identification and genetic diversity of causative parasite species. J Helminthol. 94, e110. [DOI] [PubMed] [Google Scholar]
- Schär F, Guo L.i., Streit A, Khieu V, Muth S, Marti H, Odermatt P, 2014. Strongyloides stercoralis genotypes in humans in Cambodia. Parasitol Int. 63 (3), 533–536. [DOI] [PubMed] [Google Scholar]
- Som A, 2006. Theoretical foundation to estimate the relative efficiencies of the Jukes-Cantor+gamma model and the Jukes-Cantor model in obtaining the correct phylogenetic tree. Gene 30 (385), 103–110. [DOI] [PubMed] [Google Scholar]
- Strauss T, von Maltitz MJ, Chen K, 2017. Generalising Ward’s Method for Use with Manhattan Distances. PLoS ONE 12 (1), e0168288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J, Kjer K, Page R, Sullivan J, 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56 (4), 564–577. [DOI] [PubMed] [Google Scholar]
- Tan G.e., Muffato M, Ledergerber C, Herrero J, Goldman N, Gil M, Dessimoz C, 2015. Current Methods for Automated Filtering of Multiple Sequence Alignments Frequently Worsen Single-Gene Phylogenetic Inference. Syst Biol. 64 (5), 778–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanchomnang T, Intapan PM, Sanpool O, Rodpai R, Sadaow L, Phosuk I, Somboonpatarakun C, Laymanivong S, Tourtip S, Maleewong W, 2019. First molecular identification of Strongyloides fuelleborni in long-tailed macaques in Thailand and Lao People’s Democratic Republic reveals considerable genetic diversity. J Helminthol. 93 (05), 608–615. [DOI] [PubMed] [Google Scholar]
- Zhou S, Fu X, Pei P, Kucka M, Liu J, Tang L, Zhan T, He S, Chan YF, Rödelsperger C, Liu D, Streit A, Taylan Ozkan A, 2019. Characterization of a non-sexual population of Strongyloides stercoralis with hybrid 18S rDNA haplotypes in Guangxi, Southern China. PLoS Negl. Trop. Dis. 13 (5), e0007396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.