Abstract
Chemotherapy is one of the most common therapeutic methods in advanced and metastatic tumors. Cisplatin (CDDP) is considered as one of the main first-line chemotherapy drugs in solid tumors. However, there is a high rate of CDDP resistance in cancer patients. Multi-drug resistance (MDR) as one of the main therapeutic challenges in cancer patients is associated with various cellular processes such as drug efflux, DNA repair, and autophagy. Autophagy is a cellular mechanism that protects the tumor cells toward the chemotherapeutic drugs. Therefore, autophagy regulatory factors can increase or decrease the chemotherapy response in tumor cells. MicroRNAs (miRNAs) have a pivotal role in regulation of autophagy in normal and tumor cells. Therefore, in the present review, we discussed the role of miRNAs in CDDP response through the regulation of autophagy. It has been reported that miRNAs mainly increased the CDDP sensitivity in tumor cells by inhibition of autophagy. PI3K/AKT signaling pathway and autophagy-related genes (ATGs) were the main targets of miRNAs in the regulation of autophagy-mediated CDDP response in tumor cells. This review can be an effective step to introduce the miRNAs as efficient therapeutic options to increase autophagy-mediated CDDP sensitivity in tumor cells.
Keywords: Autophagy, Cisplatin, Chemotherapy, Drug resistance, Cancer, microRNA
Background
Chemotherapy, radiotherapy, and surgery are the most prominent clinical techniques in cancer therapy [1]. Since, chemotherapeutic medications can reach every organ in the body through the circulation, it is recognized as the most beneficial treatment option for the majority of patients with late-stage and metastatic cancer [2]. Despite pharmacological advances in tumor therapy, the emergence of multidrug resistance (MDR) limits the efficiency of chemotherapy in tumor cells [3, 4]. MDR is associated with genetic and growth factors, increased DNA repair capability, drug efflux, and xenobiotic metabolism [5]. Platinum-based drugs inhibit DNA replication and transcription, resulting in cell cycle arrest and apoptosis, through covalently interacting with purine bases to construct interstrand and intrastrand DNA adducts [6]. Cisplatin (CDDP) is an inorganic chemotherapeutic agent frequently used to treat diverse malignancies that has the potential to significantly raise the overall survival rates of cancer patients [7, 8]. It is a highly effective anticancer drug that is frequently utilized in the first-line treatment of solid tumors [9]. However, Cisplatin treatment is frequently accompanied with various side effects, such as nausea, vomiting, alopecia, liver disorders, and bone marrow inhibition [10]. Although, the majority of cancer patients respond to platinum, cisplatin-resistance and tumor relapse can be observed among cancer patients [9]. It has been discovered that half of the patients treated with cisplatin acquire multidrug or intrinsic resistance [9, 11]. Therefore, clarifying the molecular mechanisms to overcome cisplatin resistance can greatly enhance prognosis. Drug efflux, drug uptake, DNA repair, autophagy, and apoptosis are the key regulators of cisplatin resistance [12]. Autophagy disruption increases CDDP-mediated apoptosis in various cancer cells [13–15].
Autophagy is a conserved catabolic mechanism of organelle recycling that has a cytoprotective role against adverse conditions including nutrient deprivation, reactive oxygen species, and cell stress [16, 17]. Autophagy is the process by which autophagosomes wrap damaged cell components in bilayer lipid vesicles and then destruct them via lysosomal fusion [18]. It is involved in regulation of a variety of biological processes, including cell survival, differentiation, cell death, and tumorigenesis [19, 20]. Autophagy deregulation has been linked to a variety of diseases, such as diabetes, cardiomyopathy, and cancer [21–23]. It is a cell survival strategy that confers tumor cell survival and poor prognosis through induction of dormancy during therapeutic phases [24]. Therefore, its activation allows drug-resistant tumors to maintain their viability [25]. Autophagy suppression paired with chemotherapy resulted in enhanced tumor cell death, suggesting its pro-survival function in the development of chemotherapy resistance [26].
MicroRNAs (miRNAs) are a category of small non-coding RNAs that function as the negative post-transcriptional regulators by mRNA degradation or translational inhibition [27, 28]. They are involved in regulation of cell proliferation, migration, and cell death [29, 30]. MiRNAs have also been implicated in the tumor progression and chemo resistance [31, 32]. Since, circulating miRNAs are highly stable in urine and blood, they can be used as effective and non-invasive tumor markers [33–35]. MiRNAs are involved in autophagy by regulation of retrieval stages, induction, vesicle elongation, and vesicle nucleation [36]. They also play a role in the cisplatin response of tumor cells by regulation of autophagy [34]. Regarding the side effects of cisplatin on healthy tissues, it is required to clarify the molecular mechanisms of cisplatin resistance to provide novel efficient therapeutic modalities to reduce the side effects of chemotherapy in cancer patients. Accordingly, microRNAs as the non-invasive and more stable factors compared with mRNAs, can be introduced as valuable prognostic markers of cisplatin response in cancer patients through autophagy regulation. Taken together, considering the importance of the autophagy in response to cisplatin treatment, in the present review we discussed the role of miRNAs in regulation of autophagy-mediated cisplatin response in tumor cells (Table 1).
Table 1.
Role of miRNAs in autophagy-mediated cisplatin response
| miRNA | Type | Target | Samples | Autophagy | CDDP response | Clinical application | Study |
|---|---|---|---|---|---|---|---|
| Signaling pathways | |||||||
| miR-181 | Non-small cell lung cancer | PI3K/Akt/mTOR pathway |
6T 6 N* A549/DDP cell line |
Induction | Increased CDDP sensitivity | Diagnosis | Liu [42] |
| miR-22 | Osteosarcoma | PI3K/Akt/mTOR pathway |
MG-63, MG-63/CDDP, U2OS, Saos2 and OS9901 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Meng [43] |
| miR-5047 | Cervical canecr | VEGFA |
25T 25 N HeLa and SiHa cell lines |
Inhibition | Increased CDDP sensitivity | Diagnosis | Guo [50] |
| miR-99a-5p | Gastric cancer | MTMR3 |
25R 75 S* BGC823, SGC7901, and SGC7901CDDP cell lines Xenograft model |
Induction | Increased CDDP resistance | Diagnosis | Sun [51] |
| miR-339-5p | Laryngeal carcinoma | TAK1 | Hep-2 cell line | Inhibition | Increased CDDP sensitivity | Diagnosis | Li [52] |
| miR-222 | Bladder cancer | PPP2R2A | T24 and 5637 cell lines | Inhibition | Increased CDDP resistance | Diagnosis | Zeng [60] |
| miR-26a | Glioblastoma | GSK3β | U87MG and U251MG cell lines | Induction | Increased CDDP resistance | Diagnosis | Ma [65] |
| miR-205 | Nasopharyngeal carcinoma | HER3 | CNE1, CNE2, SUNE1, and HK1 cell lines | Induction | Increased CDDP sensitivity | Diagnosis | Hao [67] |
| miR-144 | Anaplastic thyroid carcinoma | TGF-α |
5T 5 N ARO and TPC1 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Liu [69] |
| miR-144-3p | Anaplastic thyroid carcinoma | TGF-α |
5T 5 N TPC1 and BHT101 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Liu [70] |
| miR-145-5p | Laryngeal squamous cell carcinoma | PRKCI |
FD-LSC-1 and Tu 177 cell lines Xenograft model |
Induction | Increased CDDP sensitivity | Diagnosis | Gao [73] |
| miR-425-3p | Non-small cell lung cancer | AKT1 |
19R 19 S 15 after first trial 15 after last trial A549 and A549/DDP cell lines |
Induction | Increased CDDP resistance | Diagnosis and prognosis | Ma [76] |
| miR-30a | Ovarian/ breast/ liver cancer | Beclin-1 |
HeLa, MCF-7, HepG2, and HepS cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Zou [81] |
| miR-30a-5p | Small Cell Lung Cancer | Beclin-1 |
22R 32 S H446 and Letp cell lines |
Inhibition | Increased CDDP sensitivity | Diagnosis | Yang [82] |
| miR-216b | Non-small cell lung cancer | Beclin-1 |
40T 40 N A549 cell line Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Chen [83] |
| miR-148a-3p | Gastric cancer | AKAP1/RAB12 |
105T 105 N BGC823CDDP, SGC7901CDDP, BGC823, and SGC7901 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis and prognosis | Li [90] |
| miR-136-5p | Laryngeal squamous cell carcinoma | ROCK1 | FD-LSC-1 and FaDu cell lines | Induction | Increased CDDP sensitivity | Diagnosis | Yang [95] |
| miR-142-3p | Gastric cancer | ROCK2 |
100T 100 N 48 GC blood sample 48 healthy blood sample AGS, SGC-7901, MKN-45, and BGC-823 cell lines xenograft model |
Inhibition | Increased CDDP resistance | Diagnosis and prognosis | Peng [97] |
| Apoptosis and drug efflux | |||||||
| miR-15a-3p | Non-small cell lung cancer | BCL2 | Calu1 cell line | Induction | Increased CDDP sensitivity | Diagnosis | Bozok [105] |
| miR-143 | Cervical cancer | BCL2 | HeLa and CaSki cell lines | Induction | Increased CDDP sensitivity | Diagnosis | Esfandyari [106] |
| miR-7-5p | Cervical cancer | PARP-1/Bcl-2 |
15T 15 N HeLa and SiHa cell lines |
Induction | Increased CDDP resistance | Diagnosis | Yang [107] |
| miR-30a | Oral squamous carcinoma | Beclin1 |
13T 14 N SCC084 cell line |
Inhibition | Increased CDDP sensitivity | Diagnosis | Kulkarni [108] |
| miR-30 | Gastric cancer | LC3II/ LC3-I | SGC-7901 cell line | Inhibition | Increased CDDP sensitivity | Diagnosis | Du [111] |
| miR-22 | Osteosarcoma | caspase-3/ Bcl-2/ ATG5/ beclin1/ LC3B |
MG-63, MG-63/CDDP, U2OS, Saos2 and OS9901 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Meng [116] |
| miR-22 | Osteosarcoma | MTDH | MG-63 cell line | Inhibition | Increased CDDP sensitivity | Diagnosis | Wang [117] |
| Ubiquitin-like modifiers and autophagy receptors | |||||||
| miR-199a-5p | Small cell lung cancer | p62 |
30T 30 N NCI-H446 and H69PR cell lines Xenograft model |
Induction | Increased CDDP resistance | Diagnosis | Li [124] |
| miR-146a | Lung cancer | CHOP |
69T 69 N A549 and H446 cell lines Xenograft model |
Inhibition | Increased CDDP resistance | Diagnosis | Tan [131] |
| miR-133a | Ovarian cancer | YES1 |
24R 12 S SKOV3 and A2780 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Zhou [134] |
| Transcription factors and DNA binding proteins | |||||||
| miR-29c-3p | Ovarian cancer | FOXP1/ATG14 |
SKOV3 and A2780 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Hu [136] |
| miR-125b | Thyroid cancer | ATG7/ Foxp3 |
30T 30 N WRO, FRO, and KAT18 cell lines Xenograft model |
Induction | Increased CDDP sensitivity | Diagnosis | Wang [137] |
| miR-152 | Ovarian cancer | ATG14 |
35T A2780/CP70, SKOV3/DDP, A2780 and SKOV3 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | He [139] |
| miR-579-3p | Osteosarcoma | MSH6 |
SCSP-5030 and TCHu124 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Zhan [145] |
| miR-216b | Non-small cell lung cancer | Beclin-1 |
40T 40 N A549 and cisplatin resistance A549/DDP cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Chen [151] |
| miR-181a-5p | Breast cancer | VDR | HS578T, HCC70, MDA-MB-231, MDA-MB-468 and BT549 cell lines | Induction | Increased CDDP sensitivity | Diagnosis | Lin [154] |
| Autophagy-related genes | |||||||
| miR-1278 | Nasopharyngeal carcinoma | ATG2B |
90T 90 N CNE-1, CNE-2, C666-1, 5–8 F and HONE-1 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis and prognosis | Zhao [161] |
| miR-376a | laryngocarcinoma | ATG2A |
30T 30 N SNU46 and M4E cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Feng [163] |
| miR-1 | Non-small cell lung cancer | ATG3 |
30R 30 S A549 and H1299 cell lines |
Inhibition | Increased CDDP sensitivity | Diagnosis | Hua [164] |
| miR-651 | Cervical canecr | ATG3 |
30T 30 N 30T blood sample 30 N blood sample C33A, HT-3, HeLa/S, and HeLa/DDP cell lines |
Inhibition | Increased CDDP sensitivity | Diagnosis and prognosis | Zhu [165] |
| miR-16 | Osteosarcoma | ATG4B |
30T 30 N SAOS2 and U2OS cell lines |
Inhibition | Increased CDDP sensitivity | Diagnosis | Liu [168] |
| miR-101-3p | Non-small cell lung cancer | ATG4D |
A549, PC-9, NCI-H1299, and HCC827 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Cui [169] |
| miR-101 | Hepatocellular carcinoma | RAB5A/ STMN1/ ATG4D | HepG2 cell lines | Inhibition | Increased CDDP sensitivity | Diagnosis | Xu [170] |
| miR-30e | Gastric cancer | ATG5 |
SGC7901 cell line Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Zhang [173] |
| miR-181a | Gastric cancer | ATG5 |
SGC7901/CDDP cell line Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Zhao [174] |
| miR-1301 | Ovarian cancer | ATG5/ Beclin1/ EMT | SKOV3 cell line | Inhibition | Increased CDDP sensitivity | Diagnosis | Yu [175] |
| miR-30b | Gastric cancer | ATG5 | AGS, HGC-27 and the 293T cell lines | Inhibition | Increased CDDP sensitivity | Diagnosis | Xi [176] |
| miR-17 | Non-small cell lung cancer | ATG7 |
50T 50 N A549 and H1299 cell lines |
Inhibition | Increased CDDP sensitivity | Diagnosis | Sun [180] |
| miR-138-5p | Non-small-cell lung cancer | TRIM65 |
30R 30 S A549 and A549/DDP cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Pan [185] |
| miR-199a-5p | Hepatocellular carcinoma | ATG7 |
21T blood sample before and after treatment Huh7 cells, HepG2 cell lines |
Inhibition | Increased CDDP sensitivity | Diagnosis | Xu [186] |
| miR-654-5p | Non-small cell lung cancer | ATG7 |
108T 108 N H1975, H820, H1299, H358, and A549 cell lines |
Inhibition | Increased CDDP sensitivity | Diagnosis | Kong [187] |
| miR-17 | Nonsmall cell lung cancer | ATG7 |
A549 and H1299 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Huang [188] |
| miR-7 | Bladder cancer | ATG7 |
47T 47 N T24T and EJ cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Wang [189] |
| miR-646 | Oral squamous cell carcinoma | ATG13 |
SCC-15 and CAL-27 cell lines Xenograft model |
Inhibition | Increased CDDP resistance | Diagnosis | Gao [190] |
| miR-186 | Colorectal cancer | ATG14 |
50T 50 N SW620 and SW480 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Han [191] |
| miR-410 | Osteosarcoma | ATG16L1 |
40T 40 N U2OS and MG-63 cell lines |
Inhibition | Increased CDDP sensitivity | Diagnosis and prognosis | Chen [195] |
| Structural factors and enzymes | |||||||
| miR-205 | Prostate cancer | RAB27A/LAMP3 |
DU145 and PC-3 PCa cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Pennati [199] |
| miR-140-5p | Non-small cell lung cancer | WEE1 |
30R 30 S A549 and H1299 cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Fu [200] |
| miR-329-3p | Neuroblastoma | MYO10 |
26T 26 N HUVEC and NB cell lines |
Inhibition | Increased CDDP sensitivity | Diagnosis | Wang [203] |
| miR-124/ miR-142 | Non-small cell lung cancer | SIRT1 |
36T 36 N H1299 and H1299/CDDP cell lines Xenograft model |
Inhibition | Increased CDDP sensitivity | Diagnosis | Song [208] |
*Tumor (T) tissues and Normal (N) margins*Resistant (R) patients and Sensitive (S) patients to CDDP
Role of miRNAs in autophagy-mediated CDDP response by regulation of signaling pathways
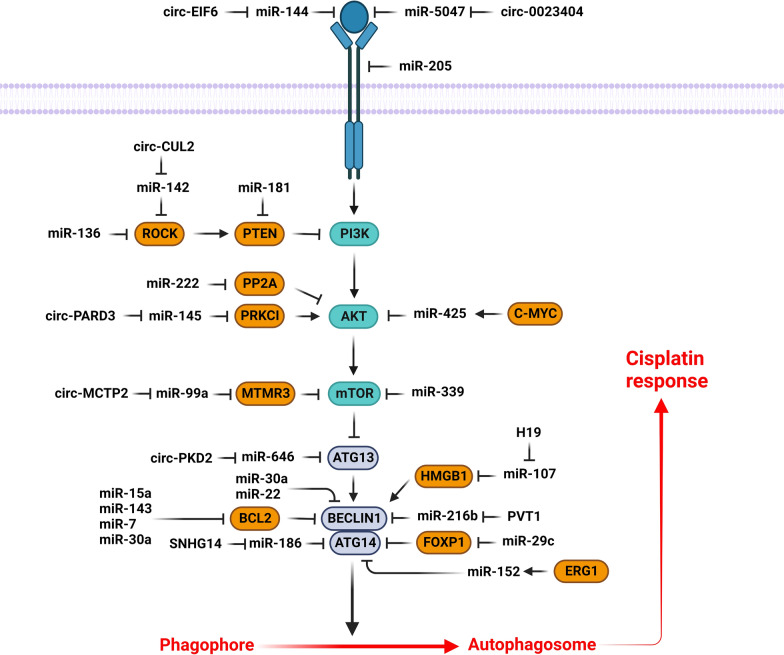
The PI3K/AKT/mTOR signaling pathway promotes the tumor progression by increased cell proliferation while reduced autophagy. MicroRNAs have a pivotal role in autophagy-mediated cisplatin response by regulation of PI3K/AKT signaling pathway (Fig. 1). PTEN is a crucial stimulator of autophagy by PI3K/PKB inhibition [37, 38]. Despite the recent improvements in Non-small-cell lung cancer (NSCLC) treatment, there is still a poor prognosis in advanced stage patients with an overall survival rate of 15% that can be associated with chemoresistance and tumor relapse [39, 40]. Cisplatin is one of the primary post-surgical adjuvant therapeutics for NSCLC [41]. It was shown that miR-181 was significantly down regulated in cisplatin-resistant NSCLC patients compared to healthy controls. MiR-181 inhibition was associated with Autophagy related 5 (ATG5) and Microtubule-associated protein light chain 3 (LC3) down regulations in A549/DDP cells. MiR-181 reduced cell proliferation while promoted autophagy via the PTEN/PI3K/AKT/mTOR pathway in A549/DDP cells [42]. MiR-22 repressed cisplatin resistance in osteosarcoma via autophagy suppression through the PI3K/AKT/mTOR axis [43]. MTMR3 is an inositol lipid 3-phosphatase from the myotubularin family that hydrolyzes the Phosphatidylinositol 3-phosphate (PI3P) autophagic effector [44, 45]. The autophagosome formation has been shown to be repressed by MTMR3-mediated inhibition of the PI3P [46]. It is also a negative regulator of mTORC1 [47]. Circular RNAs (circRNAs) are covalently closed loop non-coding RNAs lacking either polyadenylation or 5′ to 3′ polarity [48]. They are involved in regulation of cell proliferation, migration, apoptosis, autophagy, and chemoresistance [49, 50]. It has been shown that there was circMCTP2 down regulation in CDDP-resistant gastric cancer (GC) cells. CircMCTP2 enhanced CDDP sensitivity via sponging miR-99a-5p and upregulating MTMR3 in GC cells [51]. MiR-339-5p has been reported to reduce CDDP resistance of laryngeal carcinoma by hindering the autophagy process through TAK1 targeting. MiR-339-5p also down regulated the mTOR and AMPK in CDDP-resistant cells [52].
Fig. 1.
Role of miRNAs in autophagy-mediated cisplatin response via regulation of PI3K/AKT signaling pathway. (Created with BioRender.com)
The phosphatase 2 A (PP2A) is a pivotal regulator of cell cycle, metabolism, protein synthesis, and cell death [53]. PP2A downregulation has been associated with increased tumor progression via induction of various proliferative kinases in cancer cells [54, 55]. PPP2R2A is a member of regulatory B subunits of the PP2A family [56]. It was found to be downregulated by miR-222, which activated the AKT/mTOR axis [57–59]. MiR-222 enhanced bladder tumor cell proliferation and repressed CDDP-mediated apoptosis via regulation of PPP2R2A/AKT/mTOR axis. The AKT/mTOR pathway was markedly induced in bladder tumor cells with miR-222 upregulation. MiR-222 inhibited autophagy via activating the AKT/mTOR axis in bladder tumor cells [60]. GSK3 is one of the main substrates of AKT that inhibits GSK3 by phosphorylation [61]. Long non-coding RNAs (lncRNAs) interact with mRNAs, proteins, and miRNAs to regulate tumor progression [62]. They are also involved in regulation of chemoresistance and autophagy [63, 64]. The AC023115.3 upregulation has been observed in cisplatin-exposed glioma cells that stimulated the cisplatin-mediated apoptosis by autophagy inhibition. It also upregulated GSK3β and decreased autophagy through the miR-26a sponging [65].
HER3 is a receptor tyrosine kinase (RTK) that regulates cell growth and proliferation via the PI3K/AKT pathway [66]. It has been shown that there was significant miR-205 down regulation in nasopharyngeal cancer (NPC) cells. MiR-205 reduced the NPC cell proliferation while induced autophagy via LC3B II up regulation and p62 down regulation. MiR-205 increased CDDP sensitivity by HER3 targeting [67]. VEGFA is a growth factor that regulates cell proliferation via PI3K/AKT pathway. Circ_0023404 promoted the cervical cancer progression through miR-5047/VEGFA axis. It also induced CDDP resistance via inhibition of autophagy-mediated apoptosis in cervical tumor cells [50]. Transforming growth factor (TGF)-α is an EGF-like protein that functions as a ligand for the EGFR along with amphiregulin and EGF [68]. It was reported that miR-144 inhibited autophagy while enhanced CDDP sensitivity via TGF-α targeting in anaplastic thyroid cancer (ATC) cells [69]. CircEIF6 upregulated the TGF-α and increased cisplatin-resistance via autophagy enhancement through miR-144-3p sponging in thyroid tumor cells [70].
PRKCI belongs to the protein kinase C family that regulates tumor progression and chemosensitivity via regulation of the immune microenvironment and the WNT signaling pathway [71, 72]. CircPARD3 is an autophagy-suppressive circRNA abundantly expressed in LSCC tissues that was correlated with poor prognosis. It triggers proliferation, invasion, migration, and chemoresistance via suppression of autophagy in LSCC cells. CircPARD3 stimulated the AKT-mTOR axis and inhibited autophagy by upregulation of PRKCI via miR-145-5p sponging. There was a significant correlation between p62 upregulation and poor prognosis in LSCC patients. PRKCI was found to suppress autophagy while increased mTOR and AKT phosphorylation in LSCC cells [73]. Exosomes are currently being studied extensively due to their role in mediating miRNA transfer, which helps various malignancies resist chemotherapy [74, 75]. Additionally, exosomal miRNAs are regarded as intriguing biomarkers for determining treatment response or tracking disease advancement due to their accessibility from peripheral blood. The expression of miR-425-3p has been found to be triggered in exosomes and cells by c-Myc-mediated transactivation following cisplatin exposure. MiR-425-3p promoted drug resistance via enhancing autophagy through AKT1 targeting. Cisplatin treatment was associated with the upregulation of c-Myc together with β-catenin in NSCLC cells. C-Myc was observed to positively regulate miR-425-3p expression through direct binding to its promoter region [76].
Beclin-1, as the first recognized mammalian autophagic protein, is implicated in autophagy initiation and modulation of several tumor cell signaling pathways [77]. It is a component of the PI3K complex that is involved in regulation of vesicle-trafficking. Beclin-1 interacts with the Bcl-2 anti-apoptotic protein to induce cell death [78]. Its deregulation has been associated with the prognosis in many cancer types [79, 80]. Additionally, upregulation of Beclin-1 led to increased drug-resistance and autophagy upon CDDP treatment [81]. MiR-30a-5p was shown to downregulate Beclin-1 and hence promotes the efficacy of VP16/DDP chemotherapy in lung cancer [82]. PVT1 expression levels were found to be positively associated with tumor volume, TNM staging, lymph node involvement, poor survival, and cisplatin sensitivity in NSCLC. PVT1 promoted the autophagy-mediated cisplatin resistance via miR-216b/Beclin-1 axis [83]. MiR-30a inhibited the beclin1 mediated to induce CDDP mediated apoptosis in tumor cells [81].
Dynamin-Related Protein 1 (DRP1) is considered as an upstream regulator of mitochondrial fission, whose inhibition could be an efficient cancer treatment strategy [84, 85]. Mitochondrial fission 1 (FIS1) enhances CDDP sensitivity by functioning as a receptor to attract DRP1 into the mitochondria [86]. AKAP1 serves as a scaffold for the delivery of PKA to outer mitochondrial membrane in order to regulate the phosphorylation state of target proteins [87]. RAB12 belongs to the Ras family that can operate as a potential autophagy activator by blocking mTORC1 signaling or promoting autolysosome development [88, 89]. It has been reported that there was significant miR-148a-3p down regulation in CDDP-resistant GC cells. MiR-148a-3p induced CASP-3 and CDDP mediated cell death. AKAP1 inhibited the CDDP-induced mitochondrial fission by mediating the DRP1 phosphorylation and inactivation. MTORC1 prevented CDDP-induced death of GC cells via promoting early autophagosome production by RAB12 inhibiting. Additionally, miR-148a-3p concurrently targeted RAB12 and AKAP1 to make GC cells more susceptible to CDDP treatment [90].
Rho-associated protein kinase (ROCK) is a kinase that enhances the invasive capacity of tumor cells through interacting with the actin filaments to induce stress fibers and focal adhesions [91]. ROCK is a negative regulator of PI3K/AKT through PTEN phosphorylation and activation [92]. Its knockdown improves the CDDP effectiveness and prevents tumor progression and metastasis [93]. ROCK1 is a member of the ROCK family that functions as a downstream mediator of Rho A when activated by GTP binding [94]. MiR-136-5p suppressed cell migration while enhanced CDDP sensitivity by ROCK1 targeting in HPSCC and LSCC. MiR-136-5p upregulation coupled with cisplatin treatment was shown to downregulate P62 and repress the AKT/mTOR axis [95]. ROCK2 is a serine-threonine kinase that determines cell shape and migration by influencing the cytoskeleton [96]. It has been observed that there was circCUL2 down regulation in GC tissues, which was associated with the tumor differentiation, lymph node metastasis, and TNM stage. CircCUL2 suppressed GC cell proliferation and migration via regulating ROCK2 through miR-142-3p sponging. CircCUL2 also modulated the CDDP sensitivity via inducing autophagy through the miR-142-3p/ROCK2 axis [97].
Role of miRNAs in autophagy-mediated CDDP response by regulation of apoptosis and drug efflux
Apoptosis is the primary mechanism of cell death that is inhibited in tumor cells to resist against drug mediated DNA damage [98, 99]. Although, apoptosis is directly linked to cell death, autophagy has a dual effect on tumor cells [100]. Apoptosis can be modulated through the Bcl-2 protein family that involves multiple members such as Bcl-2, Bax, CASP-9 [101]. Bcl-2, which is a critical regulator of apoptosis, has been found to be upregulated in a wide range of human malignancies [102]. On the other hand, it suppresses autophagy via Beclin1 targeting [103]. Beclin1 uses its BH3 domain to interact with different homologs of Bcl-2, resulting in autophagy suppression [104]. It has been shown that miR-15a-3p increased cisplatin sensitivity by suppressing Bcl-2 that resulted in autophagy induction in NSCLC cells [105]. MiR-143 was shown to regulate the Bcl-2, Bax, and CASP-9 apoptotic genes, which in turn enhanced the cisplatin-mediated death in cervical tumor cells. Interestingly, the combined miR-143 and cisplatin treatment resulted in autophagy induction, cell cycle arrest, c-Myc downregulation, and cell migration suppression by vimentin down regulation. Therefore, application of miR-143 in conjunction with cisplatin may provide a potential therapeutic approach for cervical cancer patients [106]. MiR-7-5p was found to be upregulated in cisplatin-resistant cervical tumor cells, boosting energy production via targeting Bcl-2 and decreasing energy consumption through PARP-1 targeting. MiR-7-5p induced autophagy through Bcl-2 down regulation in cisplatin-resistant cells [107]. There was significant miR-30a down regulation in CDDP-resistant oral squamous cell carcinoma (OSCC) cells. Exosomal delivery of miR-30a was associated with enhanced CDDP response in OSCC cells by inhibiting autophagy and improving apoptosis through Beclin1 and Bcl-2 targeting, respectively [108].
P-glycoprotein (P-gp) is a transmembrane ABC transporter that acts as an efflux pump to discharge a variety of chemotherapy drugs from MDR tumor cells [109]. It also protects chemo-resistant tumor cells toward the caspase-dependent apoptosis [110]. It has been shown that miR-30a inhibited the GC cell proliferation and CDDP resistance by P-gp and MDR1 down regulations. CDDP treatment promoted autophagy while reduced apoptosis in the SGC7901/CDDP-resistant cells. MiR-30a was correlated with CDDP resistance-related autophagy in SGC7901 cells [111]. MTDH is involved in autophagy and chemoresistance by MDR1 up regulation [112]. MTDH was suggested to promote 5-FU resistance by inducing autophagy via AMPK/ATG5 axis [113]. Osteosarcoma (OS) is primarily managed with different approaches, including surgical resection, radiotherapy, neoadjuvant chemotherapy, and adjuvant chemotherapy [114]. High-dose cisplatin, doxorubicin, etoposide, and methotrexate are frequently used in chemotherapeutic regimens [115]. MiR-22 has been observed to improve the efficiency of CDDP treatment which in turn suppressed the proliferation of osteosarcoma cells via downregulation of the autophagy-related genes, including beclin1, ATG5, and LC3. It also diminished CDDP resistance via blocking autophagy. Moreover, miR-22 suppressed MTDH upregulation induced by CDDP [116]. Another study also reported that miR-22 suppressed autophagy and cell proliferation while triggered CDDP sensitivity by targeting MTDH in OS cells [117].
Role of miRNAs in autophagy-mediated CDDP response by regulation of ubiquitin-like modifiers and autophagy receptors
Autophagy is a cellular process that employs lysosomal machinery to recycle dysfunctional long half-life proteins and organelles. It begins with the creation of double membrane-bound vesicles known as autophagosomes and is regulated via conserved autophagy-related proteins [118]. It is vitally involved in tumor progression through autophagy-related (ATG) proteins [119]. Light chain 3 (LC3) is a critical autophagosome biomarker in the autophagy system which serves in substrate selection and autophagosome formation [120]. The cleavage of LC3 into the LC3-I variant with an exposed C-terminal glycine that permits association with phophatidylethanolamine to generate LC3-II, crucially involves in autophagosome formation. In addition, LC3 recycling occurs when LC3-II gets deconjugated from LC3-I through this proteolytic cleavage [121]. The p62 protein breaks down during autophagy and builds up as the autophagy declines [122]. The p62/LC3 interaction is one of the main ways to deliver the autophagic cargo [123]. It has been reported that miR-199a-5p has an important role in cisplatin resistance of SCLC via regulation of p62 mediated autophagy [124]. Cisplatin upregulates the UPR-related chaperones, including CALR, GRP78, and PDIA3, through inducing endoplasmic reticulum (ER) stress in tumor cells [125]. Conversely, ER stress tolerance (ERST) develops after repeated activation of the ER stress response by chemotherapeutic agents. CHOP is regarded as a crucial regulator of apoptosis induced by ER stress [126, 127]. Nutrient deficiency, UV rays, tunicamycin, and thapsigargin promote ER stress and CHOP expression [128–130]. CHOP was shown to be significantly downregulated in patients with recurrent lung cancer compared to those without recurrence that was associated with a worse overall survival rate. CHOP expression was also accompanied by an increase in the levels of DR5, LC3-II, and TRB3. Moreover, miR-146a induced chemoresistance by CHOP targeting in lung tumor cells [131]. YES1 has been characterized as an oncogene that could serve as a therapeutic target in several malignancies [132]. It was demonstrated that ovarian cancer patients with Yes1 up regulation had more sensitivity to platinum and a better prognosis compared with down regulated patients [133]. MiR-133a decreased CDDP resistance via targeting YES1 and autophagy regulation in ovarian tumor cells. YES1 induced CDDP resistance by upregulating LC3B in a xenograft tumor model [134].
Role of miRNAs in autophagy-mediated CDDP response by regulation of transcription factors and DNA binding proteins
Forkhead box gene P1 (FOXP1) is a transcription factor involved in embryogenesis and myocardial development [135]. FOXP1 was considered to be implicated in CDDP resistance by targeting ATG14 in ovarian cancer. MiR-29c-3p impeded autophagy and CDDP resistance via FOXP1/ATG14 axis in ovarian tumor cells [136]. It has been observed that miR-125b promoted autophagy in FTC and ATC cells by Foxp3 targeting. MiR-125b induced drug sensitivity to sorafenib and cisplatin in thyroid cancer cells. Autophagy was also induced by upregulation of ATG7 and LC3II and downregulation of Bcl-2 via Foxp3 suppression. Although, miR-125b or cisplatin could individually shrink the tumor size, the combination of miR-125b and cisplatin showed the most pronounced antitumor activity in a xenograft mouse model [137]. Early growth response factor 1 (EGR1) is a transcription factor that plays a pivotal role in transcriptional induction of the apoptotic pathway regulators such as TP53, TNF, TP53, BAX, and RB1 following chemotherapy or radiotherapy [138]. EGR1-MIR152 was shown to regulate cisplatin-mediated autophagy in ovarian tumor cells via ATG14 targeting. MiR-152 suppressed cisplatin-induced autophagy in tumor cells. EGR1 regulated miR-152 and ATG14, which then improved CDDP sensitivity in ovarian tumor cells [139].
CCCTC-binding factor (CTCF) is a zinc finger transcription factor that participates in several gene regulatory mechanisms by the chromatin structure modulation [140, 141]. CTCF as a key transcription factor influences tumor progression via regulation of lncRNAs [142, 143]. MSH6 is a MutS family member that is involved in the eukaryotic mismatch repair system by heterodimerizing with MSH2 to build a mismatch recognition complex [144]. CTCF upregulated IGF2-AS to promote autophagy by upregulating Beclin1, resulting in greater CDDP resistance in osteosarcoma cells. IGF2-AS increased CDDP resistance in OS cells by upregulating MSH6 via miR-579-3p sponging [145].
High mobility group box-1 (HMGB1) is a nucleoprotein that has key roles in regulation of DNA replication, damage repair, and apoptosis [146]. HMGB1 upregulation has been reported in numerous malignancies that were associated with poor prognosis and metastasis [147]. In addition, HMGB1 as an essential regulator of autophagy is also involved in chemo resistance of tumor cells [148]. A majority of tumor cells release HMGB1 during externally induced apoptosis and autophagy. Moreover, HMGB1 silencing was shown to sensitize tumor cells to chemo-radio therapeutic modalities [149]. HMGB1 also activate autophagy in response to stress by detaching Beclin-1 from Bcl-2 [150]. H19 knockdown significantly inhibited autophagy and limited CDDP resistance through regulation of miR-107/HMGB1 axis in LSCC cells [151]. Vitamin D receptor (VDR) is a nuclear receptor that regulates autophagy and a number of genes implicated in cell proliferation, differentiation, and calcium-phosphate homeostasis [152, 153]. MiR-181a-5p enhanced autophagy and CDDP sensitivity in breast tumor cells via VDR targeting [154].
Role of miRNAs in autophagy-mediated CDDP response by regulation of autophagy-related genes
Autophagy is involved in elimination of the endogenous substances through lysosomal degradation pathway, which is regulated by autophagy-related genes (ATGs) [155]. MicroRNAs have a pivotal role in autophagy-mediated cisplatin reponse by regulation of ATG proteins (Fig. 2). ATG2B is a member of the ATG2 family, which interacts with WIPI4 and GABARAP to generate phagophores and transport lipids [156–158]. ATG2B participates in the early stages of autophagosome assembly via associating with WDR45 and ATG2A [159]. It stimulates differentiation of the hematopoietic progenitor cells by making them more sensitive to thrombopoietin [160]. There was significant miR-1278 down regulation in NPC tissues that was correlated with poor chemotherapy outcomes and overall survival. MiR-1278 down regulated ATG2B to suppress autophagy and increase CDDP sensitivity in NPC cells [161]. It has been reported that there was significant miR-375 down regulation in CDDP resistant OS cells. It enhanced cell death while inhibited cell proliferation and autophagy through ATG2B targeting in CDDP-resistant OS cells [162]. MiR-376a was found to be involved in drug resistance conferred by circPGAM1 in laryngeal carcinoma. CircPGAM1 increased autophagy-mediated CDDP resistance by regulation of the miR-376a/ATG2A axis [163]. There was miR-1 down regulation in CDDP resistant NSCLC tissues. MiR-1 was inversely correlated with LC3B autophagy factor in NSCLC tissue samples. MiR-1 blocked autophagy-mediated by ATG3 and thus enhanced CDDP sensitivity in NSCLC cells [164]. It was shown that miR-651 reduced the CDDP resistance of cervical tumor cells by ATG3 targeting [165].
Fig. 2.
Role of miRNAs in autophagy-mediated cisplatin response via regulation of autophagy-related genes (ATGs). (Created with BioRender.com)
ATG4 is a cysteine protease that functions as an oncogene by promotion of autophagy. Meanwhile, its phosphorylation could inhibit tumor cell function and decrease autophagy [166, 167]. It has been shown that SNHG16 promoted the osteosarcoma cell survival, invasion, and autophagy-mediated chemo resistance via miR-16/ATG4B axis [168]. MiR-101-3p was reported to inhibit CDDP-mediated autophagy via ATG4D targeting in NSCLC cells. MiR-101a-3p and cisplatin therapy upregulated p62 while suppressed the LC3II/LC3I ratio and the formation of autolysosomes and autolysosomes in NSCLC cells [169]. Autophagy protects liver tumor cells from cell death mediated by anti tumor drugs. MiR-101 increased CDDP-mediated apoptosis and suppressed autophagy by ATG4D and mTOR targeting in HCC cells [170]. It has been reported that miR-24-3p inhibited autophagy and promoted the VP16-DDP response via targeting ATG4A in SCLC [171].
ATG5 is involved in autophagy via ATG12 and LC3 (ATG8) conjugation to produce autophagosomes [172]. Chemo-resistant GC cells had elevated levels of autophagy compared with chemo-sensitive GC cells. Propofol increased CDDP-mediated apoptosis via regulation of MALAT1/miR-30e/ATG5 axis in GC cells [173]. MiR-181a suppressed autophagy through ATG5 targeting in CDDP resistant GC cells [174]. It was observed that there was significant miR-1301 up regulation in ovarian tumor-resistant cells. MiR-1301 induced cell invasion and proliferation and up regulated the EMT-related genes such as Slug, Snail, and N-cadherin while down regulated E-cadherin, Beclin1, and ATG5 [175]. It has been observed that MALAT1 enhanced CDDP resistance via autophagy activation through regulation of the miR-30b/ATG5 pathway in CDDP-resistant GC cells [176].
Autophagy-related protein 7 (ATG7) is a critical positive regulator of autophagy by stimulating the ATG8 and ATG12 [177, 178]. The cell cycle alteration brought on by ATG7-mediated autophagy has been shown to promote the generation of neural crest cells [179]. ATG7 has been found to be up-regulated in various malignancies, correlating with tumor chemoresistance [180, 181]. TRIM65 E3 ubiquitin ligase is involved in regulation of autophagy, immunity, carcinogenesis, and chemoresistance [182–184]. TRIM65 down regulation suppressed autophagy while induced CDDP-mediated apoptosis via LC3-II and ATG7 down regulations in lung tumor cells. MiR-138-5p regulated the role of TRIM65 in CDDP resistance and autophagy [185]. A significant down regulation of miR-199a-5p has been shown in HCC patients undergoing cisplatin treatment, which improved autophagy via ATG7 targeting. Downregulated miR-199a-5p in cisplatin-treated HCC cells enhanced CDDP resistance through autophagy induction [186]. Tissue samples taken from cisplatin-resistant NSCLC patients showed circ_0085131 up regulation that was correlated with poor prognosis. There was a positive association between the expression levels of ATG7 and circ_0085131 in NSCLC cells. ATG7 played a pivotal oncogenic role in cisplatin-resistance of NSCLC patients. Circ_0085131 induced cisplatin resistance via miR-654-5p/ATG7 axis in NSCLC cells [187]. BLACAT1 up regulation has been observed in CDDP-resistant NSCLC cells. It increased the resistance of NSCLC cells to CDDP by enhancing autophagy through the miR-17/ATG7 axis [188]. MiR-7-5p inhibited the autophagy through ATG7 targeting in BCa. ATG7 was shown to diminish miR-7-5p-induced CDDP sensitivity in BCa cells [189]. It has been reported that there was lncRNA-XIST up regulation in NSCLC tissues and CDDP-resistance A549 cells that was correlated with TNM stage. LncRNA-XIST induced cisplatin resistance by activating autophagy through the miR-17/ATG7 axis in NSCLC cells [180]. Circ-PKD2 promoted autophagy and CDDP sensitivity by targeting miR-646 in OSCC cells. It also enhanced apoptosis by miR-646/ATG13 axis that up regulated CASP-8 in OSCC cells [190]. SNHG14 promoted CRC cisplatin resistance by inducing autophagy through regulation of the miR-186/ATG14 axis [191]. Autophagy impedes the effectiveness of cisplatin, doxorubicin, and methotrexate in osteosarcoma cells [192–194]. ATG16L1 is a member of a multimeric complex required for autophagy. It has been shown that miR-410 improved CDDP sensitivity by suppressing autophagy via ATG16L1 targeting in osteosarcoma cells [195].
Role of miRNAs in autophagy-mediated CDDP response by regulation of structural factors and enzymes
LAMP3 is a lysosomal membrane glycoprotein that has a key role in protein degradation and lysosome-autophagosome fusion [196, 197]. LAMP3 was found to be activated subsequent to the induction of the PERK/eIF2a/ATF4 axis in unfolded protein response (UPR) pathway [198]. MiR-205 was indicated to impede the autophagy by promoting the lysosomal disruption through down-regulating LAMP3 that enhaced cisplatin sensitivity due to interfering with the detoxification function of PCa cells [199]. WEE1 is a Ser/Thr kinase that inhibits the CDC2/cyclin B during cell cycle progression. FGD5-AS1 or WEE1 inhibition reduced CDDP-resistant cell viability and autophagy while promoted apoptosis in NSCLC. FGD5-AS1 induced NSCLC cell progression and CDDP resistance via miR-140-5p/WEE1 axis [200]. The motor protein myosin-X (MYO10) is significantly up regulated in metastatic tumors [201]. It connects integrins to microtubules in order to induce filopodia construction [202]. SNHG7 was found to increase cisplatin-mediated autophagy via regulating the miR-329-3p/MYO10 axis in NB cells [203]. SIRT1 belongs to the sirtuin family of histone deacetylases that is involved in a wide range of cellular mechanisms, including cell cycle, survival, metabolism, aging, chemoresistance, and apoptosis [204, 205]. SIRT1 has been observed to exhibit tumor-suppressive or oncogenic roles in various malignancies [206, 207]. It has been reported that miR-124 and miR-142 inhibited autophagy while induced apoptosis via SIRT1 targeting in CDDP-resistant NSCLC cells [208].
Conclusions
Cisplatin is considered as one of the most common first-line chemotherapy drugs in metastatic tumors. However, there is a noticeable rate of CDDP resistance in cancer patients. Autophagy is considered as one of the main causes of CDDP resistance in tumor cells. Therefore, autophagy regulatory factors can be involved in CDDP response. Considering the role of miRNAs in the regulation of autophagy, in the present review we discussed the role of miRNAs in CDDP response. It has been reported that miRNAs mainly increase the CDDP sensitivity through inhibition of autophagy. Therefore, this review can be an effective step to suggest miRNAs as the efficient therapeutic options to enhance the autophagy-mediated CDDP sensitivity in tumor cells.
Acknowledgements
None.
Abbreviations
- ATG5
Autophagy related 5
- ATC
Anaplastic thyroid cancer
- ATGs
Autophagy-related genes
- ATG7
Autophagy-related protein 7
- CTCF
CCCTC-binding factor
- circRNAs
Circular RNAs
- CDDP
Cisplatin
- DRP1
Dynamin-related protein 1
- EGR1
Early growth response factor 1
- ER
Endoplasmic reticulum
- ERST
ER stress tolerance
- FOXP1
Forkhead box gene P1
- GC
Gastric cancer
- HMGB1
High mobility group box-1
- LC3
Light chain 3
- lncRNAs
Long non-coding RNAs
- miRNAs
MicroRNAs
- LC3
Microtubule-associated protein light chain
- MYO10
Motor protein myosin-
- MDR
Multi-drug resistance
- NPC
Nasopharyngeal cancer
- NSCLC
Non-small-cell lung cancer
- OSCC
Oral squamous cell carcinoma
- OS
Osteosarcoma
- P-gp
P-glycoprotein
- PP2A
Phosphatase 2A
- PI3P
Phosphatidylinositol 3-phosphate
- RTK
Receptor tyrosine kinase
- ROCK
Rho-associated protein kinase
- TGF
Transforming growth factor
- UPR
Unfolded protein response
- VDR
Vitamin D receptor
Author contributions
FTG, IA, and AM were involved in search strategy and drafting. MM designed, revised, structured, and edited the manuscript. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kumari P, Ghosh B, Biswas S. Nanocarriers for cancer-targeted drug delivery. J Drug Target. 2016;24(3):179–91. doi: 10.3109/1061186X.2015.1051049. [DOI] [PubMed] [Google Scholar]
- 2.Chabner BA, Roberts TG., Jr Timeline: Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5(1):65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 3.Zhang JF, et al. Biological activities of novel pyrazolyl hydroxamic acid derivatives against human lung cancer cell line A549. Eur J Med Chem. 2014;83:516–25. doi: 10.1016/j.ejmech.2014.06.065. [DOI] [PubMed] [Google Scholar]
- 4.Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16(9):e447–59. doi: 10.1016/S1470-2045(15)00246-6. [DOI] [PubMed] [Google Scholar]
- 5.Bukowski K, Kciuk M, Kontek R. Mechanisms of multidrug resistance in cancer chemotherapy. Int J Mol Sci. 2020;21(9):3233. doi: 10.3390/ijms21093233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7(8):573–84. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 7.Farooq MA, et al. Recent progress in nanotechnology-based novel drug delivery systems in designing of cisplatin for cancer therapy: an overview. Artif Cells Nanomed Biotechnol. 2019;47(1):1674–92. doi: 10.1080/21691401.2019.1604535. [DOI] [PubMed] [Google Scholar]
- 8.Kim M, et al. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy. 2017;13(1):149–68. doi: 10.1080/15548627.2016.1239676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–78. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kentepozidis N, et al. Salvage treatment with irinotecan/cisplatin versus pemetrexed/cisplatin in patients with non-small cell lung cancer pre-treated with a non-platinum-based regimen in the first-line setting: a randomized phase II study of the Hellenic Oncology Research Group (HORG) Clin Transl Oncol. 2017;19(3):317–25. doi: 10.1007/s12094-016-1532-y. [DOI] [PubMed] [Google Scholar]
- 11.Velasco G, Sanchez C, Guzman M. Anticancer mechanisms of cannabinoids. Curr Oncol. 2016;23(2):S23–32. doi: 10.3747/co.23.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y, et al. Emerging role of long non-coding RNAs in cisplatin resistance. Onco Targets Ther. 2018;11:3185–94. doi: 10.2147/OTT.S158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem. 2014;289(24):17163–73. doi: 10.1074/jbc.M114.558288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu L, et al. Induction of autophagy counteracts the anticancer effect of cisplatin in human esophageal cancer cells with acquired drug resistance. Cancer Lett. 2014;355(1):34–45. doi: 10.1016/j.canlet.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, et al. MicroRNA-199a-5p inhibits cisplatin-induced drug resistance via inhibition of autophagy in osteosarcoma cells. Oncol Lett. 2016;12(5):4203–8. doi: 10.3892/ol.2016.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lapierre LR, et al. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy. 2015;11(6):867–80. doi: 10.1080/15548627.2015.1034410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson I, et al. The marine n-3 PUFA DHA evokes cytoprotection against oxidative stress and protein misfolding by inducing autophagy and NFE2L2 in human retinal pigment epithelial cells. Autophagy. 2015;11(9):1636–51. doi: 10.1080/15548627.2015.1061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, et al. Competitive endogenous RNA is an intrinsic component of EMT regulatory circuits and modulates EMT. Nat Commun. 2019;10(1):1637. doi: 10.1038/s41467-019-09649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang F, Wang B-R, Wang Y-G. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol. 2018;24(41):4643. doi: 10.3748/wjg.v24.i41.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrotta C, et al. Autophagy in the regulation of tissue differentiation and homeostasis. Front Cell Dev Biology. 2020;8:602901. doi: 10.3389/fcell.2020.602901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amaravadi RK, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17(4):654–66. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLendon PM, et al. Tubulin hyperacetylation is adaptive in cardiac proteotoxicity by promoting autophagy. Proc Natl Acad Sci U S A. 2014;111(48):E5178–86. doi: 10.1073/pnas.1415589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarparanta J, Garcia-Macia M, Singh R. Autophagy and mitochondria in obesity and type 2 diabetes. Curr Diabetes Rev. 2017;13(4):352–69. doi: 10.2174/1573399812666160217122530. [DOI] [PubMed] [Google Scholar]
- 24.Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. J Pathol. 2019;247(5):708–18. doi: 10.1002/path.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen HG, et al. Targeting autophagy overcomes enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene. 2014;33(36):4521–30. doi: 10.1038/onc.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho CJ, Gorski SM. Molecular mechanisms underlying autophagy-mediated treatment resistance in cancer. Cancers (Basel) 2019;11(11):1775. doi: 10.3390/cancers11111775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gandellini P, et al. MicroRNAs as new therapeutic targets and tools in cancer. Expert Opin Ther Targets. 2011;15(3):265–79. doi: 10.1517/14728222.2011.550878. [DOI] [PubMed] [Google Scholar]
- 28.Moghbeli M. Molecular interactions of miR-338 during tumor progression and metastasis. Cell Mol Biol Lett. 2021;26(1):13. doi: 10.1186/s11658-021-00257-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma X, et al. microRNA-501-5p promotes cell proliferation and migration in gastric cancer by downregulating LPAR1. J Cell Biochem. 2020;121(2):1911–22. doi: 10.1002/jcb.29426. [DOI] [PubMed] [Google Scholar]
- 30.Moghbeli M, et al. Molecular mechanisms of the microRNA-132 during tumor progressions. Cancer Cell Int. 2021;21(1):439. doi: 10.1186/s12935-021-02149-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moghbeli M. MicroRNAs as the critical regulators of cisplatin resistance in ovarian cancer cells. J Ovarian Res. 2021;14(1):127. doi: 10.1186/s13048-021-00882-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zangouei AS, Alimardani M, Moghbeli M. MicroRNAs as the critical regulators of doxorubicin resistance in breast tumor cells. Cancer Cell Int. 2021;21(1):213. doi: 10.1186/s12935-021-01873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorur A, et al. Determination of plasma microRNA for early detection of gastric cancer. Mol Biol Rep. 2013;40(3):2091–6. doi: 10.1007/s11033-012-2267-7. [DOI] [PubMed] [Google Scholar]
- 34.Zangouei AS, Moghbeli M. MicroRNAs as the critical regulators of cisplatin resistance in gastric tumor cells. Genes Environ. 2021;43(1):21. doi: 10.1186/s41021-021-00192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdollahi A, et al. A combined panel of circulating microRNA as a diagnostic tool for detection of the non-small cell lung cancer. QJM. 2019;112(10):779–85. doi: 10.1093/qjmed/hcz158. [DOI] [PubMed] [Google Scholar]
- 36.Shen M, et al. Crucial roles of microRNA-Mediated autophagy in urologic malignancies. Int J Biol Sci. 2021;17(13):3356–68. doi: 10.7150/ijbs.61175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang R, et al. Stellettin B induces G1 arrest, apoptosis and autophagy in human non-small cell lung cancer A549 cells via blocking PI3K/Akt/mTOR pathway. Sci Rep. 2016;6:27071. doi: 10.1038/srep27071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Y, et al. Thymosin alpha 1 suppresses proliferation and induces apoptosis in breast cancer cells through PTEN-mediated inhibition of PI3K/Akt/mTOR signaling pathway. Apoptosis. 2015;20(8):1109–21. doi: 10.1007/s10495-015-1138-9. [DOI] [PubMed] [Google Scholar]
- 39.Jamal-Hanjani M, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109–21. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 40.Sun JM, et al. Prognostic significance of PD-L1 in patients with non-small cell lung cancer: a large cohort study of surgically resected cases. J Thorac Oncol. 2016;11(7):1003–11. doi: 10.1016/j.jtho.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Scagliotti GV, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26(21):3543–51. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Xing Y, Rong L. miR-181 regulates cisplatin-resistant non-small cell lung cancer via downregulation of autophagy through the PTEN/PI3K/AKT pathway. Oncol Rep. 2018;39(4):1631–9. doi: 10.3892/or.2018.6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng CY, et al. MicroRNA–22 mediates the cisplatin resistance of osteosarcoma cells by inhibiting autophagy via the PI3K/Akt/mTOR pathway. Oncol Rep. 2020;43(4):1169–86. doi: 10.3892/or.2020.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker DM, et al. Characterization of MTMR3. an inositol lipid 3-phosphatase with novel substrate specificity. Curr Biol. 2001;11(20):1600–5. doi: 10.1016/S0960-9822(01)00501-2. [DOI] [PubMed] [Google Scholar]
- 45.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186(6):773–82. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taguchi-Atarashi N, et al. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010;11(4):468–78. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 47.Hao F, et al. The PtdIns3-phosphatase MTMR3 interacts with mTORC1 and suppresses its activity. FEBS Lett. 2016;590(1):161–73. doi: 10.1002/1873-3468.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. Embo j. 2013;32(7):923–5. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bell ES, Coelho PP, Park M. LC3C mediates selective autophagy of the MET RTK, inhibiting cancer cell invasion. Autophagy. 2020;16(5):959–61. doi: 10.1080/15548627.2020.1728099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo J, et al. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed Pharmacother. 2019;115:108957. doi: 10.1016/j.biopha.2019.108957. [DOI] [PubMed] [Google Scholar]
- 51.Sun G, et al. Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J Exp Clin Cancer Res. 2020;39(1):246. doi: 10.1186/s13046-020-01758-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li G, Cheng Z. miR-339-5p Inhibits autophagy to reduce the resistance of laryngeal carcinoma on cisplatin via targeting TAK1. Biomed Res Int. 2021;2021:9938515. doi: 10.1155/2021/9938515. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353(Pt 3):417–39. doi: 10.1042/bj3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang JY, et al. Silibinin suppresses the maintenance of colorectal cancer stem-like cells by inhibiting PP2A/AKT/mTOR pathways. J Cell Biochem. 2012;113(5):1733–43. doi: 10.1002/jcb.24043. [DOI] [PubMed] [Google Scholar]
- 55.Zimmerman R, et al. PP2A inactivation is a crucial step in triggering apoptin-induced tumor-selective cell killing. Cell Death Dis. 2012;3(4):e291. doi: 10.1038/cddis.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen W, et al. PP2A-mediated anticancer therapy. Gastroenterol Res Pract. 2013;2013:675429. doi: 10.1155/2013/675429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong R, et al. miR-222 overexpression may contribute to liver fibrosis in biliary atresia by targeting PPP2R2A. J Pediatr Gastroenterol Nutr. 2015;60(1):84–90. doi: 10.1097/MPG.0000000000000573. [DOI] [PubMed] [Google Scholar]
- 58.Shen WJ, et al. microRNA-222 modulates liver fibrosis in a murine model of biliary atresia. Biochem Biophys Res Commun. 2014;446(1):155–9. doi: 10.1016/j.bbrc.2014.02.065. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, et al. High-mobility group A1 proteins enhance the expression of the oncogenic miR-222 in lung cancer cells. Mol Cell Biochem. 2011;357(1–2):363–71. doi: 10.1007/s11010-011-0907-1. [DOI] [PubMed] [Google Scholar]
- 60.Zeng LP, et al. miR-222 attenuates cisplatin-induced cell death by targeting the PPP2R2A/Akt/mTOR axis in bladder cancer cells. J Cell Mol Med. 2016;20(3):559–67. doi: 10.1111/jcmm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hermida MA, Dinesh Kumar J, Leslie NR. GSK3 and its interactions with the PI3K/AKT/mTOR signalling network. Adv Biol Regul. 2017;65:5–15. doi: 10.1016/j.jbior.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 62.Rahmani Z, Mojarrad M, Moghbeli M. Long non-coding RNAs as the critical factors during tumor progressions among iranian population: an overview. Cell Biosci. 2020;10:6. doi: 10.1186/s13578-020-0373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol Res. 2018;129:151–5. doi: 10.1016/j.phrs.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 64.Khalili-Tanha G, Moghbeli M. Long non-coding RNAs as the critical regulators of doxorubicin resistance in tumor cells. Cell Mol Biol Lett. 2021;26(1):39. doi: 10.1186/s11658-021-00282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma B, et al. Long non-coding RNA AC023115.3 suppresses chemoresistance of glioblastoma by reducing autophagy. Biochim Biophys Acta Mol Cell Res. 2017;1864(8):1393–404. doi: 10.1016/j.bbamcr.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 66.Moghbeli M, et al. ErbB1 and ErbB3 co-over expression as a prognostic factor in gastric cancer. Biol Res. 2019;52(1):2. doi: 10.1186/s40659-018-0208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hao Y, et al. MicroRNA-205 targets HER3 and suppresses the growth, chemosensitivity and metastasis of human nasopharyngeal carcinoma cells. J Buon. 2020;25(1):350–6. [PubMed] [Google Scholar]
- 68.Madala SK, et al. Inhibition of the αvβ6 integrin leads to limited alteration of TGF-α-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2014;306(8):L726–35. doi: 10.1152/ajplung.00357.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu J, et al. Effects of miR-144 on the sensitivity of human anaplastic thyroid carcinoma cells to cisplatin by autophagy regulation. Cancer Biol Ther. 2018;19(6):484–96. doi: 10.1080/15384047.2018.1433502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu F, et al. Circular RNA EIF6 (Hsa_circ_0060060) sponges mir-144-3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging. 2018;10(12):3806–20. doi: 10.18632/aging.101674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin N, et al. Protein kinase Cι and Wnt/β-Catenin signaling: alternative pathways to Kras/Trp53-driven lung adenocarcinoma. Cancer Cell. 2019;36(2):156–67. doi: 10.1016/j.ccell.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kikuchi K, et al. Protein kinase C iota as a therapeutic target in alveolar rhabdomyosarcoma. Oncogene. 2013;32(3):286–95. doi: 10.1038/onc.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao W, et al. circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-mTOR pathway. Mol Cancer. 2020;19(1):166. doi: 10.1186/s12943-020-01279-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kong JN, et al. Guggulsterone and bexarotene induce secretion of exosome-associated breast cancer resistance protein and reduce doxorubicin resistance in MDA-MB-231 cells. Int J Cancer. 2015;137(7):1610–20. doi: 10.1002/ijc.29542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qin X, et al. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal mir-100-5p-dependent manner. Int J Nanomedicine. 2017;12:3721–33. doi: 10.2147/IJN.S131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma Y, et al. Exosomal transfer of cisplatin-induced mir-425-3p confers cisplatin resistance in NSCLC through activating autophagy. Int J Nanomedicine. 2019;14:8121–32. doi: 10.2147/IJN.S221383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maiuri MC, et al. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16(1):87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- 78.Kang R, et al. The beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin HX, et al. Decreased expression of beclin 1 correlates closely with Bcl-xL expression and poor prognosis of ovarian carcinoma. PLoS ONE. 2013;8(4):e60516. doi: 10.1371/journal.pone.0060516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu M, et al. Beclin 1 expression is an independent prognostic factor for gastric carcinomas. Tumour Biol. 2013;34(2):1071–83. doi: 10.1007/s13277-013-0648-8. [DOI] [PubMed] [Google Scholar]
- 81.Zou Z, et al. MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing beclin 1-mediated autophagy. J Biol Chem. 2012;287(6):4148–56. doi: 10.1074/jbc.M111.307405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang X, et al. Intensified beclin-1 mediated by low expression of Mir-30a-5p promotes chemoresistance in human small cell lung cancer. Cell Physiol Biochem. 2017;43(3):1126–39. doi: 10.1159/000481754. [DOI] [PubMed] [Google Scholar]
- 83.Chen L, et al. The PVT1/miR-216b/Beclin-1 regulates cisplatin sensitivity of NSCLC cells via modulating autophagy and apoptosis. Cancer Chemother Pharmacol. 2019;83(5):921–31. doi: 10.1007/s00280-019-03808-3. [DOI] [PubMed] [Google Scholar]
- 84.Cerveny KL, et al. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007;17(11):563–9. doi: 10.1016/j.tcb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 85.Li S, et al. The 1p36 tumor suppressor KIF 1Bβ is required for calcineurin activation, controlling mitochondrial fission and apoptosis. Dev Cell. 2016;36(2):164–78. doi: 10.1016/j.devcel.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 86.Fan S, et al. Mir-483-5p determines mitochondrial fission and cisplatin sensitivity in tongue squamous cell carcinoma by targeting FIS1. Cancer Lett. 2015;362(2):183–91. doi: 10.1016/j.canlet.2015.03.045. [DOI] [PubMed] [Google Scholar]
- 87.Merrill RA, Strack S. Mitochondria: a kinase anchoring protein 1, a signaling platform for mitochondrial form and function. Int J Biochem Cell Biol. 2014;48:92–6. doi: 10.1016/j.biocel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsui T, Fukuda M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep. 2013;14(5):450–7. doi: 10.1038/embor.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21(3):348–58. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li B, et al. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 2017;410:212–27. doi: 10.1016/j.canlet.2017.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28(1–2):65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 92.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27(41):5477–85. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 93.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 94.Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4(6):446–56. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 95.Yang B, et al. The miR-136-5p/ROCK1 axis suppresses invasion and migration, and enhances cisplatin sensitivity in head and neck cancer cells. Exp Ther Med. 2021;21(4):317. doi: 10.3892/etm.2021.9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pranatharthi A, Ross C, Srivastava S. Cancer stem cells and radioresistance: Rho/ROCK pathway plea attention. Stem Cells Int. 2016;2016:5785786. doi: 10.1155/2016/5785786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peng L, et al. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 2020;19(1):156. doi: 10.1186/s12943-020-01270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Z, Zhu H, Hu J. CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis. 2021;12(2):219. doi: 10.1038/s41419-021-03486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hashemi Sheikhshabani S, et al. Oleuropein reduces cisplatin resistance in ovarian cancer by targeting apoptotic pathway regulators. Life Sci. 2021;278:119525. doi: 10.1016/j.lfs.2021.119525. [DOI] [PubMed] [Google Scholar]
- 100.Ouyang L, et al. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45(6):487–98. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reed JC. Bcl-2 family proteins. Oncogene. 1998;17(25):3225–36. doi: 10.1038/sj.onc.1202591. [DOI] [PubMed] [Google Scholar]
- 102.Sánchez-Beato M, Sánchez-Aguilera A, Piris MA. Cell cycle deregulation in B-cell lymphomas. Blood. 2003;101(4):1220–35. doi: 10.1182/blood-2002-07-2009. [DOI] [PubMed] [Google Scholar]
- 103.Lindqvist LM, Vaux DL. BCL2 and related prosurvival proteins require BAK1 and BAX to affect autophagy. Autophagy. 2014;10(8):1474–5. doi: 10.4161/auto.29639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pattingre S, Levine B. Bcl-2 inhibition of autophagy: a new route to cancer? Cancer Res. 2006;66(6):2885–8. doi: 10.1158/0008-5472.CAN-05-4412. [DOI] [PubMed] [Google Scholar]
- 105.Bozok Çetintaş V, et al. miR-15a enhances the anticancer effects of cisplatin in the resistant non-small cell lung cancer cells. Tumour Biol. 2016;37(2):1739–51. doi: 10.1007/s13277-015-3950-9. [DOI] [PubMed] [Google Scholar]
- 106.Esfandyari YB, et al. MicroRNA-143 sensitizes cervical cancer cells to cisplatin: a promising Anticancer combination therapy. Reprod Sci. 2021;28(7):2036–49. doi: 10.1007/s43032-021-00479-5. [DOI] [PubMed] [Google Scholar]
- 107.Yang F, et al. MicroRNA-7-5p promotes cisplatin resistance of cervical cancer cells and modulation of cellular energy homeostasis by regulating the expression of the PARP-1 and BCL2 genes. Med Sci Monit. 2018;24:6506–16. doi: 10.12659/MSM.910969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kulkarni B, et al. Exosome-mediated delivery of miR-30a sensitize cisplatin-resistant variant of oral squamous carcinoma cells via modulating Beclin1 and Bcl2. Oncotarget. 2020;11(20):1832–45. doi: 10.18632/oncotarget.27557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Abraham I, et al. Marine sponge-derived sipholane triterpenoids reverse P-glycoprotein (ABCB1)-mediated multidrug resistance in cancer cells. Biochem Pharmacol. 2010;80(10):1497–506. doi: 10.1016/j.bcp.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smyth MJ, et al. The drug efflux protein, P-glycoprotein, additionally protects drug-resistant tumor cells from multiple forms of caspase-dependent apoptosis. Proc Natl Acad Sci U S A. 1998;95(12):7024–9. doi: 10.1073/pnas.95.12.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Du X, et al. miR-30 decreases multidrug resistance in human gastric cancer cells by modulating cell autophagy. Exp Ther Med. 2018;15(1):599–605. doi: 10.3892/etm.2017.5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wan L, et al. Genetic ablation of metadherin inhibits autochthonous prostate cancer progression and metastasis. Cancer Res. 2014;74(18):5336–47. doi: 10.1158/0008-5472.CAN-14-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pei G, et al. Autophagy facilitates metadherin-induced chemotherapy resistance through the AMPK/ATG5 pathway in gastric cancer. Cell Physiol Biochem. 2018;46(2):847–59. doi: 10.1159/000488742. [DOI] [PubMed] [Google Scholar]
- 114.Lindsey BA, Markel JE, Kleinerman ES. Osteosarcoma overview. Rheumatol Ther. 2017;4(1):25–43. doi: 10.1007/s40744-016-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ferrari S, Serra M. An update on chemotherapy for osteosarcoma. Expert Opin Pharmacother. 2015;16(18):2727–36. doi: 10.1517/14656566.2015.1102226. [DOI] [PubMed] [Google Scholar]
- 116.Meng CY, et al. MicroRNA–22 regulates autophagy and apoptosis in cisplatin resistance of osteosarcoma. Mol Med Rep. 2020;22(5):3911–21. doi: 10.3892/mmr.2020.11447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang P, et al. Roles of microRNA-22 in suppressing proliferation and promoting sensitivity of osteosarcoma cells via metadherin-mediated autophagy. Orthop Surg. 2019;11(2):285–93. doi: 10.1111/os.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12(9):814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19(1):12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 121.Tanida I, et al. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279(35):36268–76. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 122.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 123.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 124.Li T, et al. The regulation of autophagy by the miR-199a-5p/p62 axis was a potential mechanism of small cell lung cancer cisplatin resistance. Cancer Cell Int. 2022;22(1):120. doi: 10.1186/s12935-022-02505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581(19):3641–51. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chumakov AM, et al. Modulation of DNA binding properties of CCAAT/enhancer binding protein epsilon by heterodimer formation and interactions with NFkappaB pathway. Blood. 2007;109(10):4209–19. doi: 10.1182/blood-2005-09-031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kudo T. [Involvement of unfolded protein responses in neurodegeneration] Nihon Shinkei Seishin Yakurigaku Zasshi. 2003;23(3):105–9. [PubMed] [Google Scholar]
- 128.Chung H, et al. Ghrelin suppresses tunicamycin- or thapsigargin-triggered endoplasmic reticulum stress-mediated apoptosis in primary cultured rat cortical neuronal cells. Endocr J. 2011;58(5):409–20. doi: 10.1507/endocrj.K10E-396. [DOI] [PubMed] [Google Scholar]
- 129.Galehdar Z, et al. Neuronal apoptosis induced by endoplasmic reticulum stress is regulated by ATF4-CHOP-mediated induction of the Bcl-2 homology 3-only member PUMA. J Neurosci. 2010;30(50):16938–48. doi: 10.1523/JNEUROSCI.1598-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13(3):393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 131.Tan W, et al. miRNA 146a promotes chemotherapy resistance in lung cancer cells by targeting DNA damage inducible transcript 3 (CHOP) Cancer Lett. 2018;428:55–68. doi: 10.1016/j.canlet.2018.04.028. [DOI] [PubMed] [Google Scholar]
- 132.Hamanaka N, et al. YES1 is a targetable oncogene in cancers harboring YES1 gene amplification. Cancer Res. 2019;79(22):5734–45. doi: 10.1158/0008-5472.CAN-18-3376. [DOI] [PubMed] [Google Scholar]
- 133.Zhou Y, et al. Overexpression of YES1 is associated with favorable prognosis and increased platinum-sensitivity in patients with epithelial ovarian cancer. Histol Histopathol. 2020;35(7):721–8. doi: 10.14670/HH-18-203. [DOI] [PubMed] [Google Scholar]
- 134.Zhou Y, et al. miR-133a targets YES1 to reduce cisplatin resistance in ovarian cancer by regulating cell autophagy. Cancer Cell Int. 2022;22(1):15. doi: 10.1186/s12935-021-02412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Katoh M, et al. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328(2):198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 136.Hu Z, et al. miR-29c-3p inhibits autophagy and cisplatin resistance in ovarian cancer by regulating FOXP1/ATG14 pathway. Cell Cycle. 2020;19(2):193–206. doi: 10.1080/15384101.2019.1704537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang S, et al. MicroRNA-125b interacts with Foxp3 to induce autophagy in thyroid cancer. Mol Ther. 2018;26(9):2295–303. doi: 10.1016/j.ymthe.2018.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ahmed MM. Regulation of radiation-induced apoptosis by early growth response-1 gene in solid tumors. Curr Cancer Drug Targets. 2004;4(1):43–52. doi: 10.2174/1568009043481704. [DOI] [PubMed] [Google Scholar]
- 139.He J, et al. Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy. 2015;11(2):373–84. doi: 10.1080/15548627.2015.1009781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Choi Y, et al. Liver-specific deletion of mouse CTCF leads to hepatic steatosis via augmented PPARγ signaling. Cell Mol Gastroenterol Hepatol. 2021;12(5):1761–87. doi: 10.1016/j.jcmgh.2021.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kemp CJ, et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7(4):1020–9. doi: 10.1016/j.celrep.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu S, et al. Long noncoding RNAs control the modulation of immune checkpoint molecules in cancer. Cancer Immunol Res. 2020;8(7):937–51. doi: 10.1158/2326-6066.CIR-19-0696. [DOI] [PubMed] [Google Scholar]
- 143.Zhao X, et al. Risk-associated long noncoding RNA FOXD3-AS1 inhibits neuroblastoma progression by repressing PARP1-mediated activation of CTCF. Mol Ther. 2018;26(3):755–73. doi: 10.1016/j.ymthe.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 144.Hsu T, et al. Sublethal levels of cadmium down-regulate the gene expression of DNA mismatch recognition protein MutS homolog 6 (MSH6) in zebrafish (Danio rerio) embryos. Chemosphere. 2010;81(6):748–54. doi: 10.1016/j.chemosphere.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 145.Zhan H, et al. Exosomal CTCF confers cisplatin resistance in osteosarcoma by promoting Autophagy via the IGF2-AS/miR-579-3p/MSH6 axis. J Oncol. 2022;2022:9390611. doi: 10.1155/2022/9390611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Di X, et al. High-mobility group box 1 protein modulated proliferation and radioresistance in esophageal squamous cell carcinoma. J Gastroenterol Hepatol. 2019;34(4):728–35. doi: 10.1111/jgh.14371. [DOI] [PubMed] [Google Scholar]
- 147.Wu X, et al. High mobility group box protein 1 serves as a potential prognostic marker of lung cancer and promotes its invasion and metastasis by matrix metalloproteinase-2 in a nuclear factor-κB-dependent manner. Biomed Res Int. 2018;2018:3453706. doi: 10.1155/2018/3453706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen Y, et al. HMGB1 promotes HCC progression partly by downregulating p21 via ERK/c-Myc pathway and upregulating MMP-2. Tumour Biol. 2016;37(4):4399–408. doi: 10.1007/s13277-015-4049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Luo J, Chen J, He L. mir-129-5p attenuates irradiation-induced autophagy and decreases radioresistance of breast cancer cells by targeting HMGB1. Med Sci Monit. 2015;21:4122–9. doi: 10.12659/MSM.896661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kang R, et al. HMGB1: a novel beclin 1-binding protein active in autophagy. Autophagy. 2010;6(8):1209–11. doi: 10.4161/auto.6.8.13651. [DOI] [PubMed] [Google Scholar]
- 151.Chen L, et al. H19/miR-107/HMGB1 axis sensitizes laryngeal squamous cell carcinoma to cisplatin by suppressing autophagy in vitro and in vivo. Cell Biol Int. 2021;45(3):674–85. doi: 10.1002/cbin.11520. [DOI] [PubMed] [Google Scholar]
- 152.Sun J. VDR/vitamin D receptor regulates autophagic activity through ATG16L1. Autophagy. 2016;12(6):1057–8. doi: 10.1080/15548627.2015.1072670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang Y, Zhu J, DeLuca HF. Where is the vitamin D receptor? Arch Biochem Biophys. 2012;523(1):123–33. doi: 10.1016/j.abb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 154.Lin J, et al. Upregulation of microRNA-181a-5p increases the sensitivity of HS578T breast cancer cells to cisplatin by inducing vitamin D receptor-mediated cell autophagy. Oncol Lett. 2021;21(4):247. doi: 10.3892/ol.2021.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen Y, et al. Unravelling the multifaceted roles of atg proteins to improve cancer therapy. Cell Prolif. 2014;47(2):105–12. doi: 10.1111/cpr.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zheng JX, et al. Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy. 2017;13(11):1870–83. doi: 10.1080/15548627.2017.1359381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Valverde DP, et al. ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol. 2019;218(6):1787–98. doi: 10.1083/jcb.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bozic M, et al. A conserved ATG2-GABARAP family interaction is critical for phagophore formation. EMBO Rep. 2020;21(3):e48412. doi: 10.15252/embr.201948412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Behrends C, et al. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Saliba J, et al. Germline duplication of ATG2B and GSKIP predisposes to familial myeloid malignancies. Nat Genet. 2015;47(10):1131–40. doi: 10.1038/ng.3380. [DOI] [PubMed] [Google Scholar]
- 161.Zhao Y, Wang P, Wu Q. miR-1278 sensitizes nasopharyngeal carcinoma cells to cisplatin and suppresses autophagy via targeting ATG2B. Mol Cell Probes. 2020;53:101597. doi: 10.1016/j.mcp.2020.101597. [DOI] [PubMed] [Google Scholar]
- 162.Gao S, Wang K, Wang X. miR-375 targeting autophagy-related 2B (ATG2B) suppresses autophagy and tumorigenesis in cisplatin-resistant osteosarcoma cells. Neoplasma. 2020;67(4):724–34. doi: 10.4149/neo_2020_190423N366. [DOI] [PubMed] [Google Scholar]
- 163.Feng B, et al. circPGAM1 enhances autophagy signaling during laryngocarcinoma drug resistance by regulating miR-376a. Biochem Biophys Res Commun. 2021;534:966–72. doi: 10.1016/j.bbrc.2020.10.063. [DOI] [PubMed] [Google Scholar]
- 164.Hua L, Zhu G, Wei J. MicroRNA-1 overexpression increases chemosensitivity of non-small cell lung cancer cells by inhibiting autophagy related 3-mediated autophagy. Cell Biol Int. 2018;42(9):1240–9. doi: 10.1002/cbin.10995. [DOI] [PubMed] [Google Scholar]
- 165.Zhu X, et al. Cancer-derived exosomal miR-651 as a diagnostic marker restrains cisplatin resistance and directly targets ATG3 for cervical cancer. Dis Markers. 2021;2021:1544784. doi: 10.1155/2021/1544784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Huang T, et al. MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer Cell. 2017;32(6):840–855e8. doi: 10.1016/j.ccell.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ni Z, et al. AKT-mediated phosphorylation of ATG4B impairs mitochondrial activity and enhances the Warburg effect in hepatocellular carcinoma cells. Autophagy. 2018;14(4):685–701. doi: 10.1080/15548627.2017.1407887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu Y, et al. SNHG16 promotes osteosarcoma progression and enhances cisplatin resistance by sponging miR-16 to upregulate ATG4B expression. Biochem Biophys Res Commun. 2019;518(1):127–33. doi: 10.1016/j.bbrc.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 169.Cui D, Feng Y, Qian R. Up-regulation of microRNA mir-101-3p enhances sensitivity to cisplatin via regulation of small interfering RNA (siRNA) anti-human AGT4D and autophagy in non-small-cell lung carcinoma (NSCLC) Bioengineered. 2021;12(1):8435–46. doi: 10.1080/21655979.2021.1982274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xu Y, et al. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2013;29(5):2019–24. doi: 10.3892/or.2013.2338. [DOI] [PubMed] [Google Scholar]
- 171.Pan B, et al. Mir-24-3p downregulation contributes to VP16-DDP resistance in small-cell lung cancer by targeting ATG4A. Oncotarget. 2015;6(1):317–31. doi: 10.18632/oncotarget.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27(6):421–9. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 173.Zhang YF, et al. Propofol facilitates cisplatin sensitivity via lncRNA MALAT1/miR-30e/ATG5 axis through suppressing autophagy in gastric cancer. Life Sci. 2020;244:117280. doi: 10.1016/j.lfs.2020.117280. [DOI] [PubMed] [Google Scholar]
- 174.Zhao J, et al. MiR-181a suppresses autophagy and sensitizes gastric cancer cells to cisplatin. Gene. 2016;576(2 Pt 2):828–33. doi: 10.1016/j.gene.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 175.Yu JL, Gao X. MicroRNA 1301 inhibits cisplatin resistance in human ovarian cancer cells by regulating EMT and autophagy. Eur Rev Med Pharmacol Sci. 2020;24(4):1688–96. doi: 10.26355/eurrev_202002_20343. [DOI] [PubMed] [Google Scholar]
- 176.Xi Z, Si J, Nan J. LncRNA MALAT1 potentiates autophagy–associated cisplatin resistance by regulating the microRNA–30b/autophagy–related gene 5 axis in gastric cancer. Int J Oncol. 2019;54(1):239–48. doi: 10.3892/ijo.2018.4609. [DOI] [PubMed] [Google Scholar]
- 177.Noda NN, et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell. 2011;44(3):462–75. doi: 10.1016/j.molcel.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 178.Yamaguchi M, et al. Atg7 activates an autophagy-essential ubiquitin-like protein Atg8 through multi-step recognition. J Mol Biol. 2018;430(3):249–57. doi: 10.1016/j.jmb.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 179.Wang G, et al. Atg7-mediated autophagy is involved in the neural crest cell generation in chick embryo. Mol Neurobiol. 2018;55(4):3523–36. doi: 10.1007/s12035-017-0583-6. [DOI] [PubMed] [Google Scholar]
- 180.Sun W, et al. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep. 2017;38(6):3347–54. doi: 10.3892/or.2017.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wu J, et al. Long noncoding RNA UCA1 targets mir-582-5p and contributes to the progression and drug resistance of bladder cancer cells through ATG7-mediated autophagy inhibition. Onco Targets Ther. 2019;12:495–508. doi: 10.2147/OTT.S183940. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 182.Ozato K, et al. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8(11):849–60. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 184.Zhang LH, et al. TRIM24 promotes glioma progression and enhances chemoresistance through activation of the PI3K/Akt signaling pathway. Oncogene. 2015;34(5):600–10. doi: 10.1038/onc.2013.593. [DOI] [PubMed] [Google Scholar]
- 185.Pan X, et al. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 2019;10(6):429. doi: 10.1038/s41419-019-1660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xu N, et al. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Commun. 2012;423(4):826–31. doi: 10.1016/j.bbrc.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 187.Kong R. Circular RNA hsa_circ_0085131 is involved in cisplatin-resistance of non-small-cell lung cancer cells by regulating autophagy. Cell Biol Int. 2020;44(9):1945–56. doi: 10.1002/cbin.11401. [DOI] [PubMed] [Google Scholar]
- 188.Huang FX, et al. LncRNA BLACAT1 is involved in chemoresistance of non–small cell lung cancer cells by regulating autophagy. Int J Oncol. 2019;54(1):339–47. doi: 10.3892/ijo.2018.4614. [DOI] [PubMed] [Google Scholar]
- 189.Wang C, et al. MiR-7-5p suppresses invasion via downregulation of the autophagy-related gene ATG7 and increases chemoresistance to cisplatin in BCa. Bioengineered. 2022;13(3):7328–39. doi: 10.1080/21655979.2022.2037323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gao L, et al. Circ-PKD2 promotes Atg13-mediated autophagy by inhibiting miR-646 to increase the sensitivity of cisplatin in oral squamous cell carcinomas. Cell Death Dis. 2022;13(2):192. doi: 10.1038/s41419-021-04497-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Han Y, et al. SNHG14 stimulates cell autophagy to facilitate cisplatin resistance of colorectal cancer by regulating miR-186/ATG14 axis. Biomed Pharmacother. 2020;121:109580. doi: 10.1016/j.biopha.2019.109580. [DOI] [PubMed] [Google Scholar]
- 192.Zhang W, et al. Knockdown of autophagy-related protein 6, Beclin-1, decreases cell growth, invasion, and metastasis and has a positive effect on chemotherapy-induced cytotoxicity in osteosarcoma cells. Tumour Biol. 2015;36(4):2531–9. doi: 10.1007/s13277-014-2868-y. [DOI] [PubMed] [Google Scholar]
- 193.Xie Z, et al. Bafilomycin A1 inhibits autophagy and induces apoptosis in MG63 osteosarcoma cells. Mol Med Rep. 2014;10(2):1103–7. doi: 10.3892/mmr.2014.2281. [DOI] [PubMed] [Google Scholar]
- 194.Kim HJ, et al. Cytoprotective role of autophagy during paclitaxel-induced apoptosis in Saos-2 osteosarcoma cells. Int J Oncol. 2013;42(6):1985–92. doi: 10.3892/ijo.2013.1884. [DOI] [PubMed] [Google Scholar]
- 195.Chen R, et al. MicroRNA-410 regulates autophagy-related gene ATG16L1 expression and enhances chemosensitivity via autophagy inhibition in osteosarcoma. Mol Med Rep. 2017;15(3):1326–34. doi: 10.3892/mmr.2017.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dominguez-Bautista JA, et al. Loss of lysosome-associated membrane protein 3 (LAMP3) enhances cellular vulnerability against proteasomal inhibition. Eur J Cell Biol. 2015;94(3–4):148–61. doi: 10.1016/j.ejcb.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 197.Nagelkerke A, et al. LAMP3 is involved in tamoxifen resistance in breast cancer cells through the modulation of autophagy. Endocr Relat Cancer. 2014;21(1):101–12. doi: 10.1530/ERC-13-0183. [DOI] [PubMed] [Google Scholar]
- 198.Mujcic H, et al. Hypoxic activation of the unfolded protein response (UPR) induces expression of the metastasis-associated gene LAMP3. Radiother Oncol. 2009;92(3):450–9. doi: 10.1016/j.radonc.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 199.Pennati M, et al. miR-205 impairs the autophagic flux and enhances cisplatin cytotoxicity in castration-resistant prostate cancer cells. Biochem Pharmacol. 2014;87(4):579–97. doi: 10.1016/j.bcp.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 200.Fu J, et al. Elevation of FGD5-AS1 contributes to cell progression by improving cisplatin resistance against non-small cell lung cancer cells through regulating miR-140-5p/WEE1 axis. Gene. 2020;755:144886. doi: 10.1016/j.gene.2020.144886. [DOI] [PubMed] [Google Scholar]
- 201.Cao R, et al. Elevated expression of myosin X in tumours contributes to breast cancer aggressiveness and metastasis. Br J Cancer. 2014;111(3):539–50. doi: 10.1038/bjc.2014.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sousa AD, Cheney RE. Myosin-X: a molecular motor at the cell’s fingertips. Trends Cell Biol. 2005;15(10):533–9. doi: 10.1016/j.tcb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 203.Wang SY, Wang X, Zhang CY. LncRNA SNHG7 enhances chemoresistance in neuroblastoma through cisplatin-induced autophagy by regulating miR-329-3p/MYO10 axis. Eur Rev Med Pharmacol Sci. 2020;24(7):3805–17. doi: 10.26355/eurrev_202004_20847. [DOI] [PubMed] [Google Scholar]
- 204.Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69(5):1702–5. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 205.Lim CS. SIRT1: tumor promoter or tumor suppressor? Med Hypotheses. 2006;67(2):341–4. doi: 10.1016/j.mehy.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 206.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9(2):123–8. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Firestein R, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3(4):e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Song X, et al. miR-124 and miR-142 enhance cisplatin sensitivity of non-small cell lung cancer cells through repressing autophagy via directly targeting SIRT1. RSC Adv. 2019;9(9):5234–43. doi: 10.1039/C8RA09914F. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.