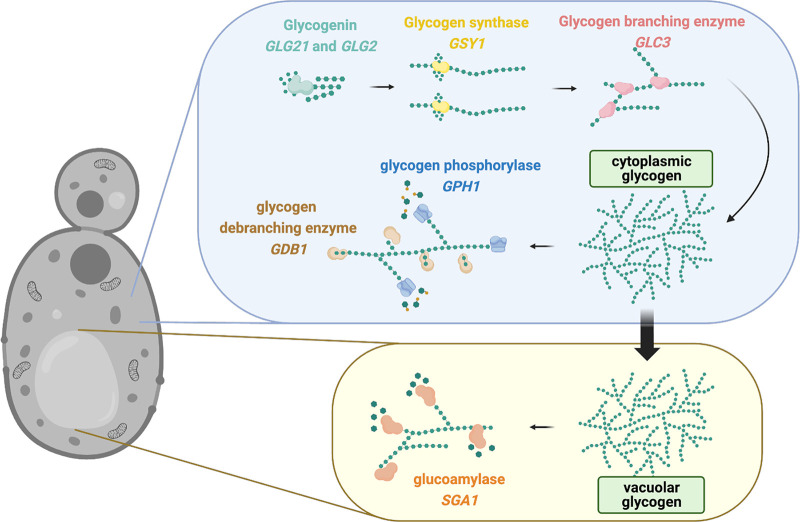
FIG 1.
Inferred glycogen synthesis and catabolism pathways in C. albicans, based on orthologous functions in S. cerevisiae. Glycogen synthesis is initiated by a glucosyltransferase (otherwise referred to as glycogenin and encoded by GLG2 or GLG21) that converts UDP-α-glucose into short α-1,4-linked glucosyl chains that are covalently attached to form a nucleate core. The glycogen synthase (encoded by GSY1) extends short α-1,4-linked glucosyl primers into elongated chains. The extensive branching of the mature glycogen molecule is achieved via the activity of the glycogen branching enzyme (encoded by GLC3) to introduce α-1,6-linkages. Glycogen is catabolized in a reverse fashion by the glycogen debranching enzyme (encoded by GDB1) to remove α-1,6-linkages, and this is followed by the activity of the glycogen phosphorylase (encoded by GPH1) to generate α-glucose-1-phosphate monomers from α-1,4-linked glucose via the addition of a phosphate. α-glucose-1-phosphate can be further metabolized to enter glycolysis. Yeast can also store glycogen in the vacuole, where glucoamylase (encoded by SGA1) can rapidly degrade glycogen to α-glucose-1-phosphate during sporulation. However, this pathway is less understood than that of conventional GDB1-GPH1-mediated glycogen catabolism. Orthologous sequences were identified by using the Candida Genome Database. The figure was created using BioRender.