ABSTRACT
tRNA-derived fragments have been reported in many different organisms and have diverse cellular roles, such as regulating gene expression, inhibiting protein translation, silencing transposable elements, and modulating cell proliferation. In particular, tRNA halves, a class of tRNA fragments produced by the cleavage of tRNAs in the anti-codon loop, have been widely reported to accumulate under stress and regulate translation in cells. Here, we report the presence of tRNA-derived fragments in Entamoeba, with tRNA halves being the most abundant. We further established that tRNA halves accumulate in the parasites upon different stress stimuli such as oxidative stress, heat shock, and serum starvation. We also observed differential expression of tRNA halves during developmental changes of trophozoite-to-cyst conversion, with various tRNA halves accumulating during early encystation. In contrast to other systems, the stress response does not appear to be mediated by a few specific tRNA halves, as multiple tRNAs appear to be processed during the various stresses. Furthermore, we identified some tRNA-derived fragments associated with Entamoeba Argonaute proteins, EhAgo2-2 and EhAgo2-3, which have a preference for different tRNA-derived fragment species. Finally, we show that tRNA halves are packaged inside extracellular vesicles secreted by amoebas. The ubiquitous presence of tRNA-derived fragments, their association with the Argonaute proteins, and the accumulation of tRNA halves during multiple different stresses, including encystation, suggest a nuanced level of gene expression regulation mediated by different tRNA-derived fragments in Entamoeba.
KEYWORDS: Entamoeba, tRNA-derived fragments, tRNA halves, encystation
INTRODUCTION
Entamoeba histolytica is a protozoan parasite with a biphasic life cycle. A dormant cyst stage causes infection upon ingestion of contaminated food or water. The cysts transform into invasive trophozoites in the small intestine and proliferate upon reaching the colon (1). Entamoeba parasites can elicit disease symptoms if trophozoites invade the colonic wall, causing colitis. The life cycle is completed when trophozoites convert to cysts in the colon through a process known as encystation; these newly formed cysts can be excreted and spread to new hosts. Currently, the signals that cause the trophozoites to encyst in the colon are not completely understood (2). Our group has previously shown that extracellular vesicles (EVs) secreted by encysting parasites can promote encystation in Entamoeba parasites, though it was not ascertained what factors in the EV cargo were responsible for modulating encystation (3).
tRNA genes are extremely abundant in the amoeba genome, with approximately 4,500 copies. The tRNA genes are organized in unique tandem repeats clustered into arrays that make up over 10% of the genome. Though the function of the arrays is not yet completely clear, there is evidence to show that these have a structural role in amoebas in the absence of classical telomeres (4, 5). Only four tRNA genes are outside the arrays, and the rest are exclusively found in the arrays. The existence of these low-copy-number tRNA genes does not affect efficient translation, and no correlation has been found between codon usage and tRNA copy number (4).
tRNA-derived fragments (tRFs) are a class of small RNAs (sRNAs), approximately 16 to 40 bases in length, that have been identified in organisms from all domains of life (6). tRFs are produced through the cleavage of either precursor or mature tRNAs. Studies have shown that the cleavage can occur at any position on the tRNAs and is context dependent (7). The best-characterized tRFs are the tRNA halves, formed by the cleavage at the anticodon loop of mature tRNAs (8, 9). tRNA halves are 30 to 40 nucleotides (nt) in size and map to either the 5′ or 3′ end of the mature tRNA. In humans, angiogenin can cleave tRNAs at the anticodon loop during stress; the resultant tRNA halves have been shown to regulate translation (10). However, angiogenin-independent mechanisms of tRNA half production have also been reported in human cells, as well as in other organisms that lack this protein (11, 12). Other tRFs are tRF-3s, tRF-5s, tRF-1s, and misc-tRFs (6). misc-tRFs include the internal tRFs (itRFs), which span the internal regions of mature tRNAs but do not map to either the 5′ or 3′ end. tRFs generated from the 3′ end of mature tRNAs can sometimes contain a 3′ CCA tail that is added to the precursor tRNA upon maturation.
One of the first studies in parasites describing tRNA fragments was performed in Giardia lamblia (13). Li et al. described a class of small RNAs, approximately 46 nt in length, derived from the 3′ end of mature tRNAs. These small RNAs, referred to as stress-induced tRNAs (si-tRNAs), contained a 3′ CCA tail and accumulated in the cell during encystation. si-tRNAs are longer than tRNA halves and contain a cleavage site in the anticodon right arm. It was observed that si-tRNAs were generated indiscriminately from the entire tRNA family in the parasite. The authors further showed that si-tRNAs also accumulated during various stress stimuli, such as temperature shock and serum deprivation. More recently, tRNA halves have been shown to modulate translation during stress response in Trypanosoma brucei (12). Fricker et al. demonstrated that during serum starvation, 3′ tRNAThr accumulated in parasites and associated with ribosomes and polysomes to stimulate global protein translation (12). tRFs have also been reported in other parasites, such as Trypanosoma cruzi and Plasmodium falciparum (14, 15). Moreover, Leishmania parasites and T. cruzi are known to package tRFs in their secreted exosomes, suggesting that these parasites could use tRFs as a means of intercellular communication (16, 17).
tRFs have also been shown to associate with Argonaute (Ago) proteins in multiple organisms (18–21). This association suggests a possible role of tRFs in the RNA interference (RNAi) pathway. Indeed, there is increasing evidence demonstrating the loading of tRFs onto Argonaute proteins, resulting in transcript cleavage based on sequence complementarity (22, 23). In humans, the 3′ tRF from tRNAGlyGCC has been shown to bind with Ago and lead to degradation of RPA1 to arrest B cell lymphoma (24). In the E. histolytica genome, three Ago proteins have been identified (EHI_125650, EHI_186850, and EHI_177170); EhAgo2-2 (EHI_125650) is the most highly expressed Ago protein in E. histolytica. We previously demonstrated that EhAgo2-2 binds to populations of small RNA, 27 nt or 31 nt, and mediates transcriptional gene silencing (25, 26).
In our present work, we wished to determine whether tRFs are present in Entamoeba and whether there is a change in abundance in response to stress. We determined that tRFs can be identified in small-RNA sequencing data sets of Entamoeba, and this was confirmed experimentally by Northern blotting experiments. Additionally, we determined that tRNA halves accumulate in amoebas during stress and encystation. Furthermore, we observed short tRFs (24 to 30 nt) bound to E. histolytica Argonaute proteins, suggesting a possible role in the Argonaute-mediated gene silencing pathway. However, unlike in some other systems, such as Trypanosoma cruzi, where the stress response is mediated by elevation of only a few (or predominantly one) tRNA halves, stress conditions in amoebas lead to the elevation of a large subset of tRNA halves, suggesting that the tRNA fragment pool in this parasite is more diverse and varied and that it may work in a redundant manner to carry out any molecular regulatory role after stress induction.
RESULTS
tRNA fragments identified in E. histolytica small-RNA libraries and confirmed by Northern blotting analyses.
To check for the presence of tRNA fragments in Entamoeba, we interrogated previously published small-RNA sequencing data sets. These data sets consisted of small-RNA libraries generated from E. histolytica parasites under basal growth conditions and during two different stresses (oxidative stress or heat stress) (27, 28). For the purpose of our previous work, the libraries had been prepared by fractionating the total RNA into two size fractions: 15 to 30 nt and 30 to 45 nt. In the present study, the sequencing data sets from the two size-selected groups (for each sample) were combined for subsequent analysis. For mapping the sequence reads, an index of E. histolytica tRNA sequences was first created from genomic sequences using tRNAscan-SE (29). Mature tRNAs contain a nontemplated CCA tail at the 3′ end. Therefore, tRNA fragments derived from the 3′ ends of mature tRNAs contain a 3′ CCA tail, whereas those derived from immature pre-tRNAs do not. A CCA tail was added to the 3′ terminal end of these predicted tRNA sequences to create a final tRNA index. The tRNA sequence index was used for mapping the sequencing data sets mentioned above, using Bowtie version 1.0.0 with a previously established pipeline (27, 30).
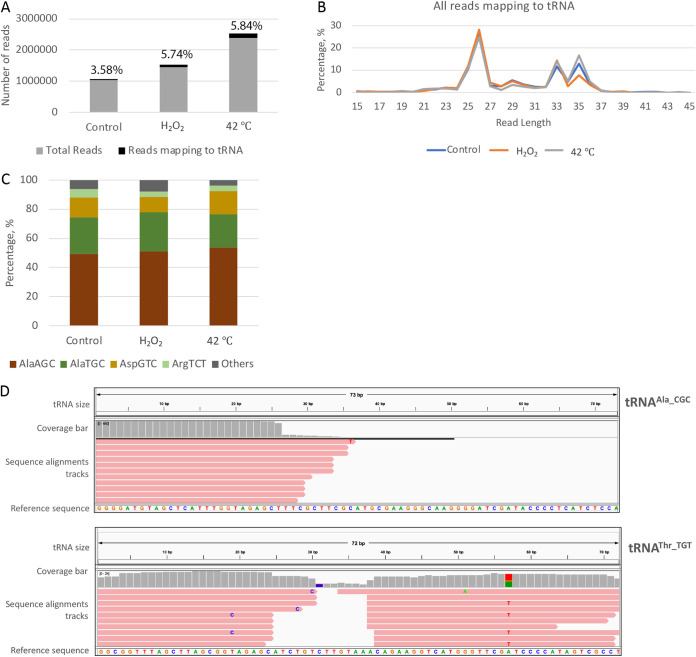
The total number of reads for each sample and the percentage of reads mapping to tRNAs are shown in Fig. 1A. Similar-sized libraries were obtained for the control and oxidative stress-treated samples (1,028,524 and 1,446,363 reads, respectively), and a much larger one was obtained for the heat shock samples (2,388,635 reads). As published earlier, almost 40% of the reads were unique reads, indicating that the sequencing was not saturated and was likely missing rare species (27). Overall, around 3 to 6% of the total sequence reads mapped to the tRNAs in each sample. The sequence reads that mapped to the tRNA sequences were selected and analyzed based on their lengths (Fig. 1B). A nonrandom distribution was observed, suggesting that the tRNA fragments were not generated by indiscriminate degradation. The peaks at ~35 nt corresponded to the expected size for tRNA halves. Other tRFs, besides tRNA halves, were also present in the sequence reads, as seen in peaks around 26 and 29 nt. A majority of these tRNA fragments mapped to the 5′ end of their respective tRNAs (>90% for each data set). This trend is not uncommon; for example, it was recently reported that tRFs originating from the 5′ end constituted around 86% of total tRFs in Plasmodium falciparum (14). However, there could have been a further bias generated during our library preparations, which involved a 5′ phosphate-dependent cloning step. As a result of this, tRFs containing 5′ phosphate groups would be preferentially cloned compared to those formed by cleavage of tRNA resulting in a 5′-OH group.
FIG 1.
Bioinformatics analyses of the small-RNA sequencing data set from E. histolytica demonstrates the presence of tRFs. (A) Small-RNA sequences generated from size-selected libraries from E. histolytica under either basal or stress conditions were mapped to tRNA sequences using Bowtie. Chart showing the percentage of sequence reads that map to tRNA in different samples. (B) Length distribution of the different reads that mapped to tRNA sequences. (C) Frequency of reads mapping to the four most abundant tRNAs seen. (D) Representative IGV mapping of sequence reads from E. histolytica (oxidative stress sample) mapping against the tRNA templates tRNAAla_CGC and tRNAThr_TGT. The top of each IGV mapping shows the size of the parent tRNA template (72 and 73 nt for tRNAAla_CGC and tRNAThr_TGT, respectively). The coverage bar in gray displays the depth of the reads at each locus as a bar chart. Below this, the sequence alignment tracks represent each sequence read aligned to the parent tRNA, whose sequence is shown as the reference sequence at the bottom. As seen for a majority of tRNAs, both 5′ and 3′ tRNA halves were observed for tRNAThr_TGT. However, only the 5′ tRNA half was observed for tRNAAla_CGC.
The frequency of the tRNAs to which the reads mapped (note that these data show the cumulative mapping for all sequence reads of different sizes) is shown in Fig. 1C. The tRNA frequency is identical for the control sample and the samples under different stresses. The tRFs derived from 4 different tRNAs (tRNAAla_AGC, tRNAAla_TGC, tRNAAsp_GTC, and tRNAArg_TCT) constituted around 80% of the total reads, and tRNAAla_AGC was the most abundant tRNA to which the sequence reads mapped. Closer inspection of the sequence reads showed that the data are skewed by multiple copies of a few reads, similar to what has been reported for other systems.
Approximately 10% of the sequence reads mapped to the 3′ end of tRNAs, within all the mapped tRNA reads. We found that approximately 80% of these reads that mapped to the 3′ end also contained a CCA tail, suggesting that the tRFs were formed predominantly by the cleavage of mature tRNAs.
The alignment of the sequence reads to representative tRNAs (tRNAAla_CGC and tRNAThr_TGT) in the Integrative Genomics Viewer (IGV) is shown in Fig. 1D (31). We did not compare the tRFs from different samples but used this alignment to display the different tRNA-derived fragments in our data set. Therefore, only the sequence reads from the oxidative-stress sample are used to show the mapping of different tRFs with respect to the parent tRNA. As seen in Fig. 1D, the mapping is not randomly distributed but instead is localized to specific regions on the parent tRNA template (predominantly around the 5′ end), which suggests that these reads are likely not degradation products. However, the diversity in the positions of 3′ termini suggests that some of the reads maybe the results of degradation/exonucleolytic process.
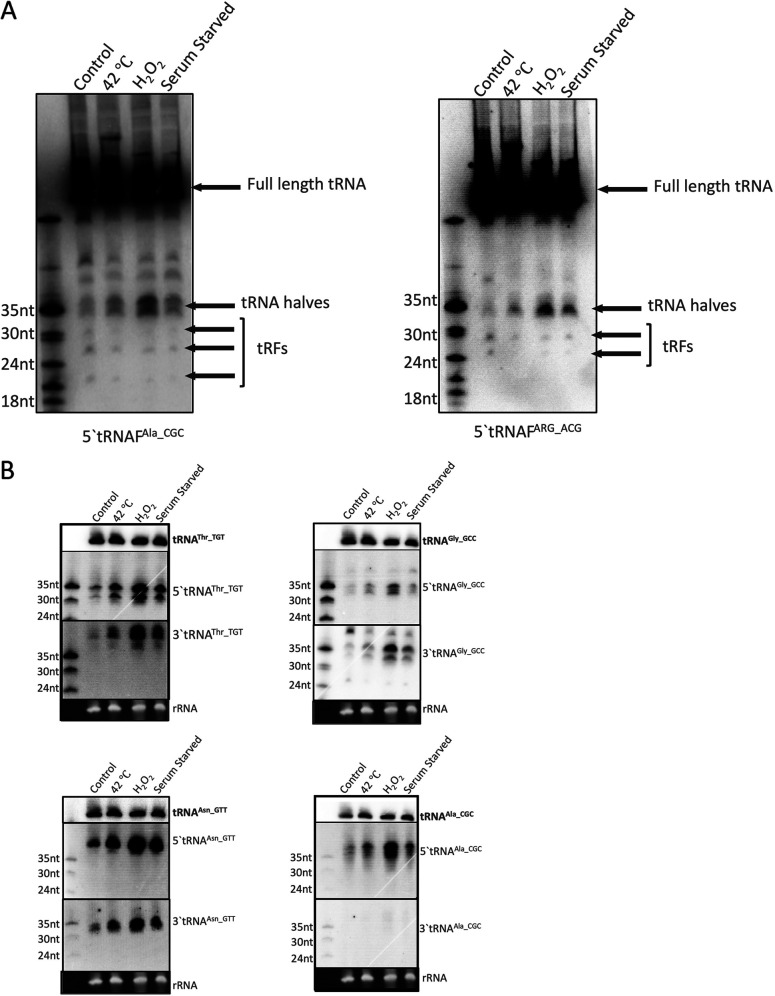
To confirm the presence of the tRFs in E. histolytica and to compare the tRF levels during different stress stimuli, Northern blot analysis of total RNA was performed. Results of Northern blot analysis for 5′ tRNAFAla_CGC and 5′ tRNAFARG_ACG, displaying full-length tRNA and tRFs of different sizes, is shown in Fig. 2A. tRNA halves (around 30 to 35 nt) were the most abundant tRFs and were found to accumulate under the different stress conditions tested. Distinct bands corresponding to smaller tRFs were also observed, but their relative amounts appeared to remain unaffected during the stress compared to the control. We checked for tRNA halves corresponding to other tRNAs and found that in each case there was a stress-induced accumulation of tRNA halves (Fig. 2B). Oxidative stress following H2O2 treatment had the most significant impact on the accumulation of different tRNA halves. tRNA halves were not observed for 3′ tRNAAla_CGC. The presence of the 5′ tRNAAla portion but the absence of its corresponding 3′ half has also been observed in other systems (32).
FIG 2.
Northern blot analysis of tRNA fragments from E. histolytica shows accumulation of tRNA halves during stress. (A) Total RNA was prepared from E. histolytica parasites under basal condition or stressed with oxidative stress (H2O2), with heat shock (42°C), or with serum starvation and analyzed by Northern blotting. Representative blots of 5′ tRNAFAla_CGC and 5′ tRNAFARG_ACG show the presence of abundant tRNA halves during stress as well as smaller tRFs of distinct sizes. (B) Northern blots showing tRNA halves for different tRNAs. Both 5′ and 3′ tRNA halves were seen to accumulate in response to the different stresses for tRNAThr_TGT, tRNAFGly_GCC, and tRNAFAsn_GTT, whereas only the 5′ tRNA half was observed for tRNAFAla_CGC.
tRNA halves accumulate in Entamoeba parasites during encystation.
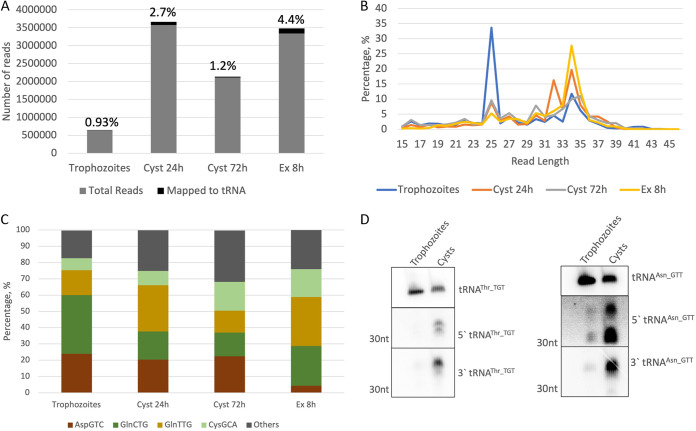
Since Entamoeba encystation also involves stress on the parasites in the form of nutritional depletion, we wanted to determine if tRNA halves are regulated during this developmental process. Efficient encystation in E. histolytica has not been achieved in vitro, and encystation of Entamoeba is studied using Entamoeba invadens (33, 34). We performed bioinformatics analyses of small-RNA sequencing data sets (size-selected RNA <45 nt in length) comparing developmental stages (trophozoites, early and late cysts, and excysting parasites) in E. invadens (27, 35). We observed that ~1 to 4% of sequence reads mapped to the tRNA sequences (Fig. 3A). Analysis of the length distribution showed that tRFs were present in all the different developmental stages, with peaks at around 25 nt and 34 nt (Fig. 3B). tRNA halves made up a larger fraction of the total reads mapping to tRNAs in early cysts and excysting parasites than in trophozoites. The 25-nt sequence reads were the most abundant tRNA fragment species in trophozoites. As observed with E. histolytica parasites, a majority of the reads mapped to the 5′ region of tRNAs (>80%). The frequency of reads mapping to different tRNAs was found to be similar across different stages, with a few tRNAs (tRNAAsp_GTC, tRNAGln_CTG, tRNAGln_TTG, and tRNACys_GCA) accounting for a majority of the sequence read mapping (Fig. 3C). However, there was some apparent variability in the tRFs belonging to different stages of development. For example, reads mapping to tRNAAsp_GTC constituted around 20% of total reads for trophozoites and early and late cysts but were only 5% of the reads in excysting parasites.
FIG 3.
tRNA halves accumulate during encystation of E. invadens. (A) Small-RNA sequences generated from size selected libraries from E. invadens parasites in different stages: trophozoites, 24-h cysts, 72-h cysts, or excysting parasites at 8 h. The chart shows percentage of reads mapping to tRNA. (B) Length distribution of reads mapping to tRNA sequences. (C) Frequency of reads mapping to most abundant tRNAs. (D) Northern blot analysis of total RNA from 24-h cysts and trophozoites probed for tRNA halves. Both 5′ and 3′ halves accumulated in the early cyst stage, in contrast to the trophozoites.
Northern blot analysis was carried out to compare the tRNA halves in E. invadens trophozoites and early cysts (24 h following the induction of encystation). Similar to the stress response in E. histolytica, various tRNA halves accumulated during encystation. Representative Northern blots for the 5′ and 3′ halves of tRNAThr_TGT and tRNAAsn_GTT are shown (Fig. 3D). Northern blots carried out for 5′ tRNA halves of tRNAArg_TCT, tRNAAla_CGC, tRNAGly_GCC, and tRNAAla_AGC are shown in Fig. S1 in the supplemental material. A densitometry analysis was performed to quantitate and compare the levels of full-length tRNA and tRNA halves in different samples (normalized to corresponding control tRNA levels) (Fig. S4).
Northern blot analysis of total RNA from 24 h cysts and trophozoites probed for tRNA halves. tRNA halves accumulated for the different tRNAs probed. Download FIG S1, PDF file, 0.08 MB (85.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Densitometry analysis of Northern blots shown in Fig. 2B. Total tRNA levels and tRNA halves for each tRNA were analyzed separately and compared to normalized control (unstressed) samples. Download FIG S4, PDF file, 0.07 MB (74.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
tRNA fragments in extracellular vesicles derived from E. histolytica.
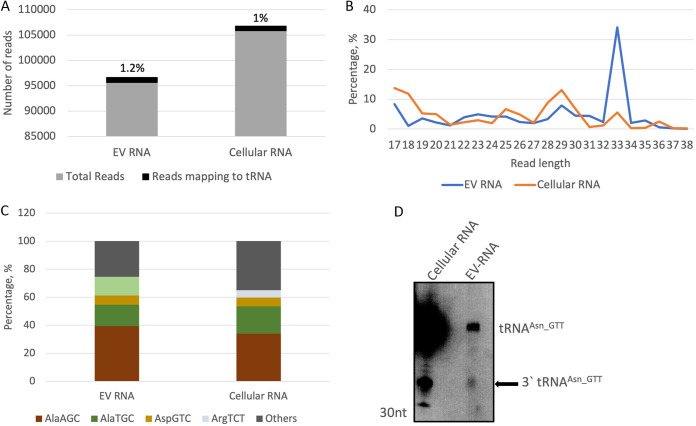
In our previous work, we characterized EVs prepared from amoebas and analyzed their RNA cargo by sequencing (3). In the present study, the RNA-seq data set from the size-selected EV RNA library was analyzed for tRFs as described above. Around 1% of the sequence reads mapped to tRNAs (Fig. 4A). Similar to what is shown in Fig. 1B, the length distribution of the cellular RNAs had peaks at around 26, 29, and 33 nt (Fig. 4B). The EV RNA had peaks at 29 nt and a more pronounced peak at 33 nt, suggesting that tRNA halves are the most abundant tRNA-derived fragments packaged in these EVs. Note that the protocol for EV preparation required the use of serum-free medium, and therefore, both the EV and cellular RNA samples had been serum starved (3). As seen in Fig. 1D, serum starvation is one of the stressors that lead to accumulation of tRNA halves, similar to what was reported in different systems (12, 36). The frequency of the reads mapping to different tRNAs is shown in Fig. 4C. As previously seen, a majority of the reads mapped to a few tRNAs (tRNAAla_AGC, tRNAAla_TGC, tRNAArg_TCT, and tRNAAsp_GTC). We did not find any major differences in the IGV profiles for EV and cellular reads for the different tRNAs (data not shown). Northern blot analysis was performed to check for tRNA halves in E. histolytica cellular and EV RNA. EV preparation was done under serum-free conditions to exclude contamination from exogenous exosomes in the serum (3). RNA isolated from the EVs was compared to cellular RNA isolated from parasites grown under similar serum-free conditions. As seen in Fig. 4D, the 3′ tRNAAsn_GTT tRNA half was observed in the EV RNA.
FIG 4.
tRNA fragments are identified in small-RNA sequence data sets obtained from size-selected libraries of EVs secreted by E. histolytica. Small-RNA sequence data sets reported previously were analyzed for tRNA fragments (3). Trophozoites were grown in serum-free medium for 24 h for EV preparation. Cellular RNA was prepared from these trophozoites grown in serum free medium. (A) Percentage of reads mapping to tRNA. (B) Length distribution of the reads that mapped to tRNA. (C) Frequency of reads mapping to the most abundant tRNAs. (D) Northern blot analysis of EV and cellular RNA prepared from E. histolytica parasites grown in serum-free medium. 3′ tRNAAsn_GTT was seen to be packaged in EVs.
tRNA-derived fragments are associated with EhAgo proteins.
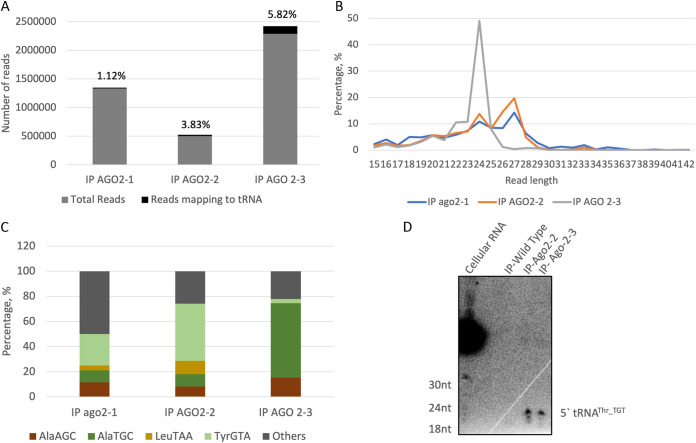
We previously demonstrated that two small-RNA populations, 27 and 31 nt, associate with E. histolytica Argonaute (EhAgo) proteins (25, 26, 28). Since Ago proteins have been reported to bind tRNA fragments in other systems (23, 37), we analyzed our sequencing data from size-fractionated total RNAs isolated from immunoprecipitations (IPs) of each of the 3 EhAgos: Ago2-1, Ago2-2 and Ago2-3. We found that the number of reads mapping to tRNA in IP RNA was comparable to what we observed in whole-cell sRNA samples (Fig. 5A). Length distribution of the mapped reads showed a nonrandom distribution with peaks at around 24 and 27 nt for these tRNA fragments (Fig. 5B). A majority of the sequence reads from the Ago2-3 samples were 24 nt in length, whereas Ago2-2 and Ago2-1 were predominantly 27 nt. Larger tRFs (e.g., tRNA halves around 33 nt) were not observed binding to Ago, suggesting that Ago proteins might have a preferred role in guiding shorter tRNA-derived fragments; this phenomenon has been observed in other systems (37). The mapped reads were viewed on IGV against tRNA templates, and two representative tRNAs, tRNAAla_TGC and tRNAArg_TCT, are shown in Fig. S2. The 5′ tRNA-derived fragments were the most abundant group of tRFs binding to the three Ago proteins. Interestingly, Ago2-2 seems to bind to itRF species for different tRNAs. Though we did not observe itRFs in the cellular small-RNA data set (analyzed in Fig. 1), it is possible that the itRFs were enriched and stabilized in the Ago overexpression cell lines that were used for the IP experiments.
FIG 5.
tRFs associate with the three EhAgo proteins in amoebas. (A) IPs of Myc-tagged overexpression cell lines for the three EhAgo proteins were performed, and RNA from beads was isolated. Small-RNA libraries were prepared by size selection and sequenced. Chart showing the percentage of sequence reads that map to tRNA in different samples. (B) Length distribution of the different reads that mapped to tRNA sequences. (C) Frequency of reads mapping to the most abundant tRNAs. (D) Norther blot analysis of IP RNA prepared from myc-EhAgo2-2 and myc-Eh-Ago2-3 overexpression cells lines. IP-Wild Type refers to IP performed with untransfected control cells. Cellular RNA was used as a positive control. tRFs were observed in both IP-Ago2-2 and IP-Ago2-3 samples for 5′ tRNAThr_TGT but not for 3′ tRNAThr_TGT.
IGV mapping of reads mapping against the tRNA templates tRNAAla_TGC and tRNAArg_TCT. A 24-nt 5′ tRF is associated with all Ago proteins for tRNAAla_TGC. An itRF species is associated with Ago2-2 for both tRNAs. Download FIG S2, PDF file, 0.2 MB (178.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The frequency of the reads mapping to different tRNAs is shown in Fig. 5C. We saw that a majority of the reads in trophozoites and in early or late cysts mapped to tRNAAsp_GTC.
Northern blot analysis was carried out for the Ago-IP samples. We focused on association of tRFs with EhAgo2-2 and EhAgo2-3, since those had the largest populations of mapping sequence reads. E. histolytica cell lines overexpressing Myc-tagged EhAgo2-2 or EhAgo2-3 were used for anti-Myc immunoprecipitation, and the total associated RNA was isolated as described elsewhere (28). Anti-Myc IP was carried out in parallel on untransfected E. histolytica trophozoites, and RNA isolation was performed to obtain a negative control. 5′ tRNAThr_TGT-derived fragments, around 27 nt in length, were observed in RNA isolated from both EhAgo2-2 and EhAgo2-3 IPs, but with a stronger association with the latter (Fig. 5D). Compared to 5′-tRNAThr_TGT-derived fragments, accumulation of other tRNA-derived fragments was seen at a much lower intensity not perceptible in our Northern blots. We could not identify tRNA-derived fragments from 3′ tRNAThr_TGT. This would make sense if binding of small RNAs to Ago proteins is dependent on the 5′ phosphate groups on RNAs.
In mammalian cells, generation of tRNA halves by angiogenin-dependent and angiogenin-independent mechanisms has been reported (11). However, we could not find homologs of angiogenin in the amoebic genome based on sequence similarity by reciprocal BLAST searches or the presence of a conserved domain by the use of Pfam (data not shown). This could suggest that an alternative tRNA cleavage mechanism is used to generate the tRNA halves in Entamoeba parasites. Mechanisms for tRNA generation are not uncommon and have been reported in various organisms. In yeasts, the RNase T2 family member Rny1 can cleave tRNAs at the anticodon loop, to generate tRNA halves (38). In T. brucei, no known homologs of angiogenin or Rny1 exist (12). Entamoeba species have RNases that belong to the RNase T2 family, and it is possible that these proteins could be involved in tRNA half biogenesis in response to stress. Further work would be required to investigate this.
tRNA half accumulation is concomitant with increased protein synthesis after heat shock-induced stress.
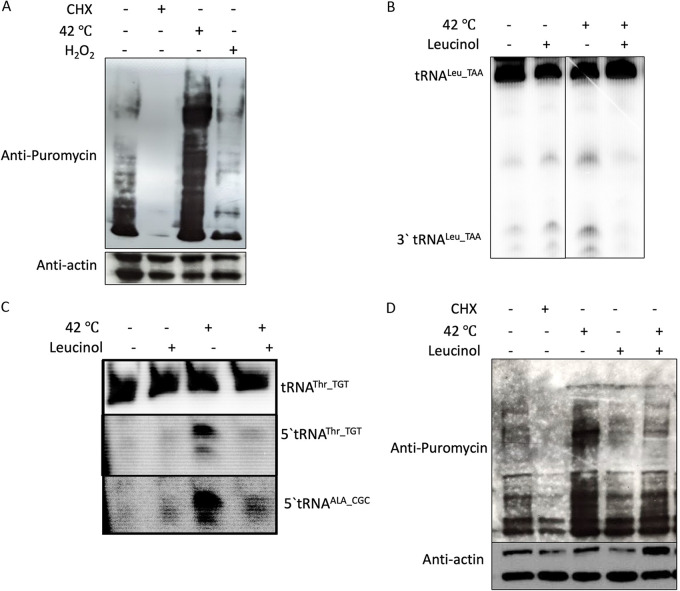
It has been shown before that protein synthesis is strongly inhibited in Entamoeba parasites exposed to oxidative stress by H2O2 (39, 40). Using the assay employed in these studies (surface sensing of translation assay [SUnSET], which compared the amount of puromycin incorporated in cells as a measure of translation levels), we also found that protein synthesis was inhibited during oxidative stress (Fig. 6A). However, the level of protein synthesis was increased after heat shock treatment (42°C for 1 h) (Fig. 6A). In agreement with this, we saw that treatment of parasites with 2.5 mM H2O2 resulted in around 15 to 20% cell cytotoxicity (measured by trypan blue staining), compared to less than 5% cytotoxicity in parasites after heat shock. We decided to further analyze heat shock samples.
FIG 6.
Heat shock results in increased protein synthesis and tRNA half accumulation. (A) Parasites were treated with 100 μg/mL cycloheximide (CHX), heat shock at 42°C, or 2.5 mM H2O2 for 1 h. 10 μg/mL Puromycin was added to all samples after 10 min of stress induction. After 1 h of puromycin (and stress) treatment, samples were collected and lysed for Western blot analysis. The membrane was first probed with an antipuromycin antibody and then stripped and reprobed for actin. (B) Total RNA was isolated from parasites under heat shock with or without the presence of leucinol or control and analyzed by Northern blotting. (Top) Full-length tRNALeu; (bottom) 3′ tRNALeu. (C) Northern blot showing levels of full-length tRNA and tRNA halves corresponding to tRNAThr and tRNAAla for the samples from panel B. (D) Western blot showing the decrease in protein synthesis upon leucinol treatment during heat shock. Parasites were treated with either CHX or heat shock at 42°C, and puromycin was added after 10 min, as before. After 1 h, cells were collected, lysed, and analyzed by Western blotting, as before.
Recently, Liu et al. showed that an analog of leucine called leucinol results in a reduction of the tRNALeu half in HeLa cells under normal growth conditions (41). We decided to check for the effect of leucinol on tRNA half synthesis in Entamoeba. Under normal growth conditions, leucinol did not have a perceptible effect on tRNALeu levels. However, in the presence of heat shock and leucinol, there was a marked reduction in tRNA half generation for tRNALeu (Fig. 6B). Since we had observed that various tRNA halves are accumulated in a similar fashion during stress, we also checked for two other tRNAs, 5′ tRNAThr_TGT and 5′ tRNAAla_CGC. Leucinol treatment also abrogated the accumulation of these tRNA halves during heat shock (Fig. 6C).
Liu et al. (41) had shown that amino acid charging was required for tRNA half generation, and in the presence of leucinol, only 50% of full-length tRNALeu was charged. We carried out the same experiment and checked for the charging of leucine in full-length tRNA and tRNA fragments (Fig. S3). We did not see any changes in the amino acid charging levels of full-length tRNA after leucinol treatment. As noted by Liu et al. (41), we found that 3′ tRNALeu was charged with an amino acid. We also checked for the charging of threonine in full-length tRNA and the 5′ tRNA half (5′ tRNAThr_TGT). Since there is no amino acid on the 5′ tRNAThr, there is no size reduction after exonuclease T treatment in the tRNA half, only in the full-length tRNA (Fig. S3).
Northern blot analysis of RNA samples from parasites treated with leucinol or control. RNA samples were either N-acetylated or deacylated. All samples were then treated with exonuclease T for 1 h. Amino acid charging is indicated by reduction in size of the deacetylated RNA samples. Download FIG S3, PDF file, 0.1 MB (147.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Next, we checked if protein translation level can be affected by leucinol treatment. Interestingly, we found that pretreatment of trophozoites with leucinol for 30 min before heat shock abrogated the puromycin incorporation seen in the control (Fig. 6D). Our results thus demonstrate that upon leucinol treatment, the tRNA half accumulation during heat shock is reduced and a concomitant decrease in protein synthesis is also observed.
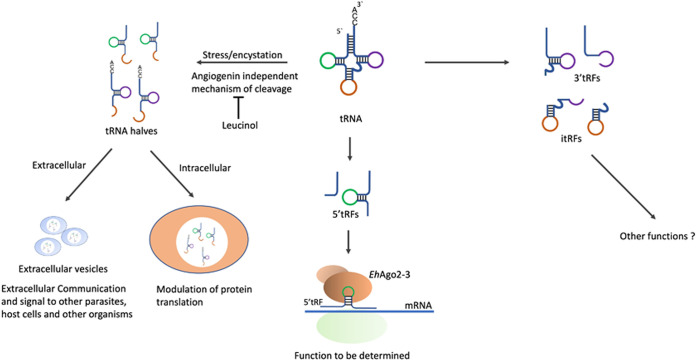
A schematic of our findings of tRFs in amoebas is shown in Fig. 7. Cleavage of tRNAs gives rise to tRFs of distinct sizes that appear to be precisely generated and are not degradation products. When the parasites are treated with different stress inducers, tRNA halves are the most abundant tRFs that accumulate. Both 5′ and 3′ tRNA halves were generated in response to stress, and these corresponded to various tRNA isoacceptors. Treatment with the inhibitor leucinol reduced the accumulation of tRNA halves during stress, and a concomitant reduction in protein synthesis was observed. Moreover, tRNA halves are packaged inside extracellular vesicles, where they could have a potential role in extracellular communication with other parasites. As reported for other systems, tRFs are found to bind to Ago proteins, and this could indicate a possible role in gene silencing as well. The role of these smaller tRFs in amebic biology is not clear at present, and our study opens the door for future functional studies of the various tRNA fragments in amoebas.
FIG 7.
Schematic outlining potential roles of tRFs in amoebas. Both 5′ and 3′ tRFs are produced in amoebas. tRNA halves accumulate during stress and during encystation, and these can be subsequently packed into EVs for extracellular communication. Smaller tRFs (3′ tRFs, 5′ tRFs, or itRFs) are also present in amoebas and are seen to bind to Ago proteins for possible roles in gene silencing.
DISCUSSION
tRNA genes are among the oldest genes that can be traced back to the putative RNA world that predated the separation of the three domains of life (42). It is no surprise, then, that tRNA-derived fragments have now been reported in Bacteria, Archaea, and Eukarya. Different classes of tRFs formed from the cleavage of either the mature or precursor tRNAs exist and have been shown to regulate gene expression in diverse contexts. Our present work provides evidence that tRF species exist in amoebas and may have the potential to regulate gene expression.
Bioinformatics analysis of preexisting sRNA libraries prepared from Entamoeba in our lab indicated the presence of various tRNA fragments. We found that around 80% of the tRFs originated from just four tRNAs: tRNAAla_AGC, tRNAAla_TGC, tRNAAsp_GTC, and tRNAArg_TCT. This has been reported in other systems as well (43–45). However, the tRF abundance did not correlate with either the codon usage or the tRNA copy numbers. Again, this is not surprising and other groups have reported a similar absence of correlation between codon usage and tRF abundance (46, 47). Ghosh et al. carried out a comprehensive analysis of available E. histolytica genome data sets to determine codon bias and found that the genome is AT rich and that, overall, A- or T-ending codons are strongly biased in the coding region. However, when only highly expressed genes were considered, there was a clear bias for C-ending codons, suggesting that these codons are translationally optimal for amoebas (48). Is there a correlation between the tRF fragment abundance (a majority originate from tRNAs with anticodons for G-ending codons) and translation efficiency in amoebas? The abundance of tRFs originating from different tRNAs varied during the development stages (analysis of E. invadens small-RNA data [Fig. 3C]); tRFs from tRNAGln_CTG were the most abundant tRFs in trophozoites, whereas tRNAAsp_GTC was the most abundant for late cysts. Further work would be required to understand if this tRF abundance is a cause or effect of the gene expression changes during stage conversion.
A majority of the tRFs identified through bioinformatic analysis mapped to the 5′ end of tRNAs. Similar results have been reported for other organisms, such as Plasmodium falciparum (14). One reason for the overrepresentation of 5′ tRFs could be the nature of our library preparation; the libraries used in this analysis involved a 5′ phosphate-dependent cloning step (these were not prepared for tRF analysis). If the cleavage of the tRNA yields fragments with a 5′-OH group, these fragments would not be efficiently cloned in the library. Additional issues in our bioinformatic analysis could arise, since we have included all reads that map to our selected tRNA sequences. A more thorough approach would omit sequence reads that also map elsewhere in the genome (49). However, in the present study, our aim was largely to check for the presence of tRNA fragments from existing sequencing data, rather than quantifying and comparing the tRNA fragments between samples.
tRNA halves were the most abundant species of tRFs that accumulated during stress in amoebas. Cleavage of tRNAs at the anticodon loop to generate tRNA halves in other organisms, including parasites, has been extensively reported. In G. lamblia, 46-nt si-tRNAs were reported to accumulate during encystation (13). Importantly, the si-tRNAs were representatives of the entire tRNA family and not restricted to a few members. We observed a similar accumulation of tRNA halves in response to both encystation and various stress inducers (oxidative stress, serum starvation, or heat shock). As in Giardia, almost the entire tRNA family seems to accumulate in response to stress.
Entamoeba is exposed to a variety of stresses during its life cycle inside the human host. The anaerobic parasite can survive extreme conditions, such as high oxygen content during tissue invasion, release of reactive oxygen and nitrogen species by the host immune response, and fluctuations in glucose availability (50, 51). The parasite employs a variety of strategies to cope with these stresses, including the regulation of antioxidant protein expression (such as thioredoxin) (52) or H2O2-responsive proteins (51, 53–55). Incorporation of puromycin is an elegant tool to measure protein translation levels. Like other groups, we observed that oxidative stress reduced puromycin incorporation in the parasites. However, we found that puromycin incorporation increased during heat shock treatment, indicating that the stress response by amoebas to 1 h of heat shock is different from that to 1 h of oxidative stress. Previously, a reduction in gene transcription upon heat shock in amoebas measured by gene expression microarrays was reported (56). That study probed for 1,131 unique genes with a microarray and found around 42% to be downregulated and around 5% to be upregulated during heat shock. That study used 4 h of heat shock treatment at 42°C. In contrast, in the present study, we used 1 h of heat shock treatment at 42°C. It is possible that the trophozoites regulate protein synthesis in a different way as they adapt to the heat shock for a prolonged period of time. However, upregulation of gene expression in response to stress (serum starvation) has also been reported in Entamoeba (57).
To check if the increased protein translation level was related to the accumulation of tRNA halves, we explored ways to block the tRNA half synthesis. Fricker et al. depleted tRNA halves in Trypanosoma brucei by transfecting the parasites with antisense oligonucleotides against tRNA halves using electroporation (12). However, this method was not possible in Entamoeba, since high transfection efficiency has not been accomplished. The finding that leucinol blocks different tRNA half accumulation was serendipitous. Liu et al. had reported that leucinol, an analog of leucine and inhibitor of leucyl-tRNA synthetase, reduced the levels of 3′ tRNALeu under normal growth conditions in mammalian cells (41). They found that amino acid charging was required for tRNA half generation, and therefore, in the presence of leucinol, only 50% of total tRNALeu was charged and generated the corresponding tRNA half. The effect of leucinol on any other tRNA, during resting or stress conditions, was not reported in that paper. We found that in the presence of leucinol, tRNA half accumulation was reduced not just for tRNALeu but for other tRNAs as well. However, we did not observe changes in the amino acid charging levels after leucinol treatment. Thus, it appears that the depletion of tRNA halves by leucinol is not because of an inhibition of tRNA charging but through a different mechanism. Although we do not have direct evidence that confirms that tRNA half accumulation is instrumental in regulating protein synthesis during stress in amoebas, our results indicate that these events occur concomitantly and are possibly related.
Amoebic parasites have a functional RNAi pathway mediated through a population of 27-nt antisense RNAs (25, 58–60), including 3 Ago proteins, which have been shown to bind sRNAs (26). The possibility of tRNA-derived fragments interacting with the RNAi pathways in amoebas offers an intriguing new role for the abundant tRNA genes in the amoebic genome. Furthermore, the packaging of tRNA halves in EVs could be a mechanism for affecting extracellular gene modulation, as seen elsewhere (16, 61).
Our data show that various tRNA fragments are present in amoebas and that, similar to other organisms, accumulation of tRNA halves under stress conditions is a mechanism that is conserved in amoebas. Further work is required to determine exactly how the tRNA halves that accumulate during stress modulate protein synthesis and what the physiological role of this modulation is during amoebic infection.
MATERIALS AND METHODS
Parasite culture and cell lines.
E. histolytica trophozoites (HM-1:IMSS) were grown axenically in TYI-S-33 (Trypticase, yeast extract, iron, and serum) medium under standard conditions described previously (62). E. invadens strain IP-1 was cultured in LYI-S-2 (Liver digest, yeast extract, iron, and serum) at 25°C (35). For encystation experiments, trophozoites at mid-log phase were iced, pooled, washed, seeded into tubes in encystation medium (47% liver digest, yeast extract, iron- Low Glucose [LYI-LG]), and incubated at 25°C. To measure encystation efficiency, total cells were counted using a hemocytometer before and after treatment with 0.1% Sarkosyl. Parasites were transfected using the transfection reagent Attractene (Qiagen) to generate stable E. histolytica cell lines. All parasite-transfected lines were maintained at 6 μg/mL G418.
Extracellular vesicle isolation.
Parasites, grown to confluence in T25 flasks, were washed with serum-free TYI medium and incubated with 10 mL serum-free TYI medium for 16 h in an anaerobic chamber (BD GasPak EZ gas-generating container systems with GasPak EZ Campy container sachets; catalog number 260680). The conditioned medium was collected and centrifuged at 2,000 rpm to remove cell debris, and EVs were pelleted using total exosome isolation reagent (Invitrogen, Carlsbad, CA, USA; catalog number 4478359) using the manufacturer’s protocol. The EV pellet was further purified using size exclusion chromatography with a SmartSEC Mini EV isolation system (Systems Biosciences, California) (3).
RNA isolation and analysis.
Cellular RNA was isolated using a standard TRIzol (Invitrogen)-based protocol according to the manufacturer’s protocol. For isolation of RNA from immunoprecipitation samples, 300 μL of TRIzol (Invitrogen) reagent was added to the final IP beads, and total RNA was isolated using the manufacturer’s protocol.
Immunoprecipitation.
Immunoprecipitation experiments were done as previously described (28). Briefly, cell lysate was diluted with lysis buffer for a final protein concentration of 1 μg/μL. Then, 20 to 30 μL packed anti-Myc beads (Thermo Scientific) was added to the IP mixture and rotated for 2 h at 4°C. The beads were washed 6 times at 4°C (5 min each) using a low-stringency wash buffer (the basic lysis buffer plus 0.1% (vol/vol) Tween 20, 0.1% (vol/vol) NP-40, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 0.5× Halt EDTA-free protease inhibitors). After the final wash step, the beads were pelleted and used for RNA preparation using the TRIzol reagent.
SUnSET assay to measure protein synthesis.
The protocol described by Schmidt et al. (63) and modified by Nagaraja et al. (40) for Entamoeba was used. Trophozoites were treated with 2.5 mM H2O2, with heat shock at 42°C, or with cycloheximide (100 μg/mL) (Sigma-Aldrich) after 30 min pretreatment with or without 2 mM leucinol (Santa Cruz Biotechnology, CA). After 10 min of pretreatment, 10 μg/mL of puromycin (Sigma-Aldrich) was added for 1 h. The parasites were lysed with cell lysis buffer (Promega), and protein concentration was measured using the Bradford assay (Bio-Rad). Samples were run on a Western blot and probed with antipuromycin antibody (clone 12D10, monoclonal puromycin antibody [Millipore]).
Amino acid charging.
Total RNA from cells treated with or without leucinol (Santa Cruz Biotechnology, California) was isolated using the TRIzol method, as described above. The RNA was divided into two aliquots; one aliquot was N-acetylated using acetic anhydride for 2 h, to stabilize the aminoacyl bond. The other aliquot was incubated at pH 9 (in 3% ammonium hydroxide) for 30 min for deacylation. After this, both aliquots were treated with exonuclease T, which can digest only a single-stranded RNA from an unprotected 3′ end. A second TRIzol preparation was done to remove the salt and enzymes from the RNA preparation, and the recovered RNA was analyzed by Northern blotting as described above.
Northern blot analysis.
Northern blot protocol was performed as previously described (25). Twenty-microgram RNA samples were separated on a denaturing 12% polyacrylamide gel and transferred to a membrane (Amersham Hybond N+ membrane; GE Healthcare). Probe DNA was 5′-end labeled by PNK reaction using γ-[32P]ATP and hybridized with the membrane in perfectHyb buffer (Sigma) overnight at 37°C. The membrane was washed under low-stringency conditions (2 × SSC [1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS at 37°C for 20 min) and medium-stringency conditions (1 × SSC, 0.1% SDS at 37°C for 20 min). Radioactive signal was detected using a Phosphor screen and imaged on a personal molecular imager (Bio-Rad). Densitometry analysis was performed on the raw images using the instrument’s software (Personal Molecular Imager).
The probes used for tRFs are listed in Table 1.
TABLE 1.
Probes used for tRFs
| Probe | tRNA | Sequence |
|---|---|---|
| 1 | 5′ tRNAAla-CGC | AGCTCTACCAAATGAGCTACATCCCC |
| 2 | 5′ tRNAGly-GCC | CAGGGATACGTCATACCATTGGACCACAAGTGC |
| 3 | 3′ tRNAGly-GCC | GTGCACTTGCCGGGAATCGAACCCGGGTCTCATCC |
| 4 | 3′ tRNAAsn-GTT | CGCTTCCGCTTGGGATCGAACCAAGGACCTAACGG |
| 5 | 5′ tRNAArg-TCT | TGCTCTTCCGACTGAGCTAACAAGGC |
| 6 | 5′ tRNAThr-TGT | ACAGATGCTCTACCGCTAAGCTAAACCGCC |
| 7 | 3′ tRNAThr-TGT | GGAGGCGACTATGGGGAACGAACCCATGACCTTCTGT |
| 8 | 5′ tRNAiMet-CAT | TGAGCCTGTCGCGCTACCACTGCGCCACCCCGCT |
| 9 | 5′ tRNATrp-CCA | AGTCAGCTGCTCTGCCACTGAGCTAAGCCCCC |
| 10 | 3′ tRNATrp-CCA | GGGTGAGGGCTAGAGGGATTGAACCACTGACCGGCTGA |
| 11 | 3′ tRNAGly-GCC | CGCUUCCGCUUGGGAUCGAACCAAGGACCUAACGG |
| 12 | 5′ tRNAAla-AGC | CGAATGCTCTACCATCTGAGCTACATCCCC |
| 13 | 5′ tRNAArg-ACG | TGATGCGTTTCCATTGCGCCACAAGCCC |
| 14 | 3′ tRNAArg-ACG | GGCGAGCTTGGCAGGGATCGAACCTGCAATCCCCAG |
| 15 | 3′ tRNAiMet-CAT | CAGCGGAGGGAAGTTTCGATCAACCGACCT |
| 16 | 5′ tRNAAsn-GTT | AGCCGTTCGCTCTGCCGACTGAGCCACGGAAGC |
| 17 | 3′ tRNALeu-TAA | GTTCGAACCCACGCATCTTGCGATAGCCGA |
Bioinformatics analyses.
RNA sequencing data were analyzed using a previously described pipeline (58). Raw reads were processed using Cutadapt (64). Sequences were then mapped to E. histolytica tRNA using Bowtie v1.2.2 (http://bowtie-bio.sourceforge.net) with the parameters –v1 and –best (30). tRNAscan-SE was used to predict tRNA genes from the genomes of E. histolytica and E. invadens (29). CCA was added to the genes at the 3′ end. The tRNAscan-SE command used in this prediction run was as follows: tRNAscan-SE -qQ –detail -o# -m# -f# -l# -c tRNAscan-SE.conf -b# -s# (FASTA file).
ACKNOWLEDGMENTS
The project was supported by funds from the National Institutes of Health (R01-AI121084 and R21-AI125764). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
We thank all members of the Singh lab for their help and contribution to this work.
Contributor Information
Upinder Singh, Email: usingh@stanford.edu.
Purnima Bhanot, Rutgers—New Jersey Medical School.
REFERENCES
- 1.Stanley SL. 2001. Pathophysiology of amoebiasis. Trends Parasitol 17:280–285. doi: 10.1016/s1471-4922(01)01903-1. [DOI] [PubMed] [Google Scholar]
- 2.Eichinger D. 2001. Encystation in parasitic protozoa. Curr Opin Microbiol 4:421–426. doi: 10.1016/s1369-5274(00)00229-0. [DOI] [PubMed] [Google Scholar]
- 3.Sharma M, Morgado P, Zhang H, Ehrenkaufer G, Manna D, Singh U. 2020. Characterization of extracellular vesicles from Entamoeba histolytica identifies roles in intercellular communication that regulates parasite growth and development. Infect Immun 88:e00349-20. doi: 10.1128/IAI.00349-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark CG, Ali IKM, Zaki M, Loftus BJ, Hall N. 2006. Unique organisation of tRNA genes in Entamoeba histolytica. Mol Biochem Parasitol 146:24–29. doi: 10.1016/j.molbiopara.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Tawari B, Ali IKM, Scott C, Quail MA, Berriman M, Hall N, Clark CG. 2008. Patterns of evolution in the unique tRNA gene arrays of the genus Entamoeba. Mol Biol Evol 25:187–198. doi: 10.1093/molbev/msm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YS, Shibata Y, Malhotra A, Dutta A. 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telonis AG, Loher P, Magee R, Pliatsika V, Londin E, Kirino Y, Rigoutsos I. 2019. tRNA fragments show intertwining with mRNAs of specific repeat content and have links to disparities. Cancer Res 79:3034–3049. doi: 10.1158/0008-5472.CAN-19-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson DM, Lu C, Green PJ, Parker R. 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. 1990. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J 9:1383–1389. doi: 10.1002/j.1460-2075.1990.tb08253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. 2011. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su Z, Kuscu C, Malik A, Shibata E, Dutta A. 2019. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J Biol Chem 294:16930–16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fricker R, Brogli R, Luidalepp H, Wyss L, Fasnacht M, Joss O, Zywicki M, Helm M, Schneider A, Cristodero M, Polacek N. 2019. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun 10:118. doi: 10.1038/s41467-018-07949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. 2008. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res 36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Wei C, Hao X, Deng W, Zhang L, Wang Z, Wang H. 2019. Genome-wide identification and characterization of transfer RNA-derived small RNAs in Plasmodium falciparum. Parasit Vectors 12:36. doi: 10.1186/s13071-019-3301-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, Rovira C, Robello C, Goldenberg S, Cayota A. 2010. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol 171:64–73. doi: 10.1016/j.molbiopara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Lambertz U, Oviedo Ovando ME, Vasconcelos EJR, Unrau PJ, Myler PJ, Reiner NE. 2015. Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both Old and New World Leishmania providing evidence for conserved exosomal RNA Packaging. BMC Genomics 16:151. doi: 10.1186/s12864-015-1260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayer-Santos E, Lima FM, Ruiz JC, Almeida IC, Da Silveira JF. 2014. Characterization of the small RNA content of Trypanosoma cruzi extracellular vesicles. Mol Biochem Parasitol 193:71–74. doi: 10.1016/j.molbiopara.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, Minis A, Sato F, Mimata H, Yoshimura A, Tamir I, Rainer J, Kofler R, Yaron A, Eggan KC, Woolf CJ, Glatzel M, Herbst R, Martinez J, Penninger JM. 2013. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. 2008. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 20.Nie Z, Zhou F, Li D, Lv Z, Chen J, Liu Y, Shu J, Sheng Q, Yu W, Zhang W, Jiang C, Yao Y, Yao J, Jin Y, Zhang Y. 2013. RIP-seq of BmAgo2-associated small RNAs reveal various types of small non-coding RNAs in the silkworm, Bombyx mori. BMC Genomics 14:661. doi: 10.1186/1471-2164-14-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JWS, Green PJ, Barton GJ, Hutvagner G. 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. 2018. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24:1093–1105. doi: 10.1261/rna.066126.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. 2013. TRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci USA 110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Ehrenkaufer GM, Pompey JM, Hackney JA, Singh U. 2008. Small RNAs with 5′-polyphosphate termini associate with a piwi-related protein and regulate gene expression in the single-celled eukaryote Entamoeba histolytica. PLoS Pathog 4:e1000219. doi: 10.1371/journal.ppat.1000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Tran V, Manna D, Ehrenkaufer G, Singh U. 2019. Functional Characterization of Entamoeba histolytica Argonaute Proteins Reveals a Repetitive DR-Rich Motif Region That Controls Nuclear Localization. mSphere 4:e00580-19. doi: 10.1128/mSphere.00580-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Ehrenkaufer GM, Manna D, Hall N, Singh U. 2015. High throughput sequencing of Entamoeba 27nt small RNA population reveals role in permanent gene silencing but no effect on regulating gene expression changes during stage conversion, oxidative, or heat shock stress. PLoS One 10:e0134481. doi: 10.1371/journal.pone.0134481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Ehrenkaufer GM, Hall N, Singh U. 2020. Identification of oligo-adenylated small RNAs in the parasite Entamoeba and a potential role for small RNA control. BMC Genomics 21:879. doi: 10.1186/s12864-020-07275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe TM, Eddy SR. 1997. TRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cognat V, Morelle G, Megel C, Lalande S, Molinier J, Vincent T, Small I, Duchene AM, Marechal-Drouard L. 2017. The nuclear and organellar tRNA-derived RNA fragment population in Arabidopsis thaliana is highly dynamic. Nucleic Acids Res 45:3460–3472. doi: 10.1093/nar/gkw1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitra BN, Pradel G, Frevert U, Eichinger D. 2010. Compounds of the upper gastrointestinal tract induce rapid and efficient excystation of Entamoeba invadens. Int J Parasitol 40:751–760. doi: 10.1016/j.ijpara.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vazquezdelara-Cisneros LG, Arroyo-Begovich A. 1984. Induction of encystation of Entamoeba invadens by removal of glucose from the culture medium. J Parasitol 70:629–633. doi: 10.2307/3281741. [DOI] [PubMed] [Google Scholar]
- 35.Ehrenkaufer GM, Weedall GD, Williams D, Lorenzi HA, Caler E, Hall N, Singh U. 2013. The genome and transcriptome of the enteric parasite Entamoeba invadens, a model for encystation. Genome Biol 14:R77. doi: 10.1186/gb-2013-14-7-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamasaki S, Ivanov P, Hu GF, Anderson P. 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar P, Anaya J, Mudunuri SB, Dutta A. 2014. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson DM, Parker R. 2009. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shahi P, Trebicz-Geffen M, Nagaraja S, Alterzon-Baumel S, Hertz R, Methling K, Lalk M, Ankri S. 2016. Proteomic identification of oxidized proteins in Entamoeba histolytica by resin-assisted capture: insights into the role of arginase in resistance to oxidative stress. PLoS Negl Trop Dis 10:e0004340. doi: 10.1371/journal.pntd.0004340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagaraja S, Cai MW, Sun J, Varet H, Sarid L, Trebicz-Geffen M, Shaulov Y, Mazumdar M, Legendre R, Coppée J-Y, Begley TJ, Dedon PC, Gourinath S, Guillen N, Saito-Nakano Y, Shimokawa C, Hisaeda H, Ankri S. 2021. Queuine is a nutritional regulator of Entamoeba histolytica response to oxidative stress and a virulence attenuator. mBio 12:e03549-20. doi: 10.1128/mBio.03549-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Kim HK, Xu J, Jing Y, Kay MA. 2021. The 3′ tsRNAs are aminoacylated: implications for their biogenesis. PLoS Genet 17:e1009675. doi: 10.1371/journal.pgen.1009675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szathmáry E. 1999. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends Genet 15:223–229. [DOI] [PubMed] [Google Scholar]
- 43.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, Qian J, Duan E, Zhai Q, Zhou Q. 2016. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 44.Krishna S, Yim DG, Lakshmanan V, Tirumalai V, Koh JL, Park JE, Cheong JK, Low JL, Lim MJ, Sze SK, Shivaprasad P, Gulyani A, Raghavan S, Palakodeti D, DasGupta R. 2019. Dynamic expression of tRNA‐derived small RNAs define cellular states. EMBO Rep 20:e47789. doi: 10.15252/embr.201947789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lakshmanan V, Sujith TN, Bansal D, Shivaprasad PV, Palakodeti D, Krishna S. 2021. Comprehensive annotation and characterization of planarian tRNA and tRNA-derived fragments (tRFs). RNA 27:477–495. doi: 10.1261/rna.077701.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gebetsberger J, Zywicki M, Künzi A, Polacek N. 2012. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guan L, Karaiskos S, Grigoriev A. 2020. Inferring targeting modes of Argonaute-loaded tRNA fragments. RNA Biol 17:1070–1080. doi: 10.1080/15476286.2019.1676633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghosh TC, Gupta SK, Majumdar S. 2000. Studies on codon usage in Entamoeba histolytica. Int J Parasitol 30:715–722. doi: 10.1016/s0020-7519(00)00042-4. [DOI] [PubMed] [Google Scholar]
- 49.Magee R, Rigoutsos I. 2020. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res 48:9433–9448. doi: 10.1093/nar/gkaa657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olivos-García A, Saavedra E, Nequiz M, Santos F, Luis-García ER, Gudiño M, Pérez-Tamayo R. 2016. The oxygen reduction pathway and heat shock stress response are both required for Entamoeba histolytica pathogenicity. Curr Genet 62:295–300. doi: 10.1007/s00294-015-0543-5. [DOI] [PubMed] [Google Scholar]
- 51.Nagaraja S, Ankri S. 2018. Utilization of different omic approaches to unravel stress response mechanisms in the parasite Entamoeba histolytica. Front Cell Infect Microbiol 8:19. doi: 10.3389/fcimb.2018.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeelani G, Nozaki T. 2016. Entamoeba thiol-based redox metabolism: a potential target for drug development. Mol Biochem Parasitol 206:39–45. doi: 10.1016/j.molbiopara.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 53.Pearson RJ, Morf L, Singh U. 2013. Regulation of H2O2 stress-responsive genes through a novel transcription factor in the protozoan pathogen Entamoeba histolytica. J Biol Chem 288:4462–4474. doi: 10.1074/jbc.M112.423467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Field J, Van Dellen K, Ghosh SK, Samuelson J. 2000. Responses of Entamoeba invadens to heat shock and encystation are related. J Eukaryot Microbiol 47:511–514. doi: 10.1111/j.1550-7408.2000.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 55.Weber C, Koutero M, Dillies MA, Varet H, Lopez-Camarillo C, Coppée JY, Hon CC, Guillén N. 2016. Extensive transcriptome analysis correlates the plasticity of Entamoeba histolytica pathogenesis to rapid phenotype changes depending on the environment. Sci Rep 6:35852. doi: 10.1038/srep35852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber C, Guigon G, Bouchier C, Frangeul L, Moreira S, Sismeiro O, Gouyette C, Mirelman D, Coppee JY, Guillén N. 2006. Stress by heat shock induces massive down regulation of genes and allows differential allelic expression of the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot Cell 5:871–875. doi: 10.1128/EC.5.5.871-875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naiyer S, Kaur D, Ahamad J, Singh SS, Singh YP, Thakur V, Bhattacharya A, Bhattacharya S. 2019. Transcriptomic analysis reveals novel downstream regulatory motifs and highly transcribed virulence factor genes of Entamoeba histolytica. BMC Genomics 20:206. doi: 10.1186/s12864-019-5570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H, Ehrenkaufer GM, Hall N, Singh U. 2013. Small RNA pyrosequencing in the protozoan parasite Entamoeba histolytica reveals strain-specific small RNAs that target virulence genes. BMC Genomics 14:53. doi: 10.1186/1471-2164-14-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H, Alramini H, Tran V, Singh U. 2011. Nucleus-localized antisense small RNAs with 5′-polyphosphate termini regulate long term transcriptional gene silencing in Entamoeba histolytica G3 strain. J Biol Chem 286:44467–44479. doi: 10.1074/jbc.M111.278184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Pompey JM, Singh U. 2011. RNA interference in Entamoeba histolytica: implications for parasite biology and gene silencing. Future Microbiol 6:103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szempruch AJ, Dennison L, Kieft R, Harrington JM, Hajduk SL. 2016. Sending a message: extracellular vesicles of pathogenic protozoan parasites. Nat Rev Microbiol 14:669–675. doi: 10.1038/nrmicro.2016.110. [DOI] [PubMed] [Google Scholar]
- 62.Diamond LS, Harlow DR, Cunnick CC. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other entamoeba. Trans R Soc Trop Med Hyg 72:431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 64.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Northern blot analysis of total RNA from 24 h cysts and trophozoites probed for tRNA halves. tRNA halves accumulated for the different tRNAs probed. Download FIG S1, PDF file, 0.08 MB (85.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Densitometry analysis of Northern blots shown in Fig. 2B. Total tRNA levels and tRNA halves for each tRNA were analyzed separately and compared to normalized control (unstressed) samples. Download FIG S4, PDF file, 0.07 MB (74.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
IGV mapping of reads mapping against the tRNA templates tRNAAla_TGC and tRNAArg_TCT. A 24-nt 5′ tRF is associated with all Ago proteins for tRNAAla_TGC. An itRF species is associated with Ago2-2 for both tRNAs. Download FIG S2, PDF file, 0.2 MB (178.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Northern blot analysis of RNA samples from parasites treated with leucinol or control. RNA samples were either N-acetylated or deacylated. All samples were then treated with exonuclease T for 1 h. Amino acid charging is indicated by reduction in size of the deacetylated RNA samples. Download FIG S3, PDF file, 0.1 MB (147.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.