ABSTRACT
Agrobacterium tumefaciens incites the formation of readily visible macroscopic structures known as crown galls on plant tissues that it infects. Records from biologists as early as the 17th century noted these unusual plant growths and began examining the basis for their formation. These studies eventually led to isolation of the infectious agent, A. tumefaciens, and decades of study revealed the remarkable mechanisms by which A. tumefaciens causes crown gall through stable horizontal genetic transfer to plants. This fundamental discovery generated a barrage of applications in the genetic manipulation of plants that is still under way. As a consequence of the intense study of A. tumefaciens and its role in plant disease, this pathogen was developed as a model for the study of critical processes that are shared by many bacteria, including host perception during pathogenesis, DNA transfer and toxin secretion, bacterial cell-cell communication, plasmid biology, and more recently, asymmetric cell biology and composite genome coordination and evolution. As such, studies of A. tumefaciens have had an outsized impact on diverse areas within microbiology and plant biology that extend far beyond its remarkable agricultural applications. In this review, we attempt to highlight the colorful history of A. tumefaciens as a study system, as well as current areas that are actively demonstrating its value and utility as a model microorganism.
KEYWORDS: cell polarity, crown gall, gene regulation, genetic models, horizontal gene transfer, host-pathogen interactions, plant pathogens, plasmids, quorum sensing, secretion systems
INTRODUCTION
Undoubtedly, one of the most fascinating and important areas of microbiology is the interaction of microorganisms with hosts. Indeed, the study of model bacterial pathogens has been an active component of microbial science from its earliest days, most notably, work on Bacillus anthracis and Streptococcus pneumoniae (1, 2). In studying these pathogenic bacteria and their host associations, many important biological phenomena have been discovered, including the elucidation of numerous processes that have broadly contributed to our fundamental understanding of biology. Among these, Agrobacterium tumefaciens, the causative agent of the plant neoplastic disease crown gall, is a prime example of how fundamental discoveries can result from comprehensively studying a pathogen. A. tumefaciens has a remarkable history of yielding important insights into microbial mechanisms of host interaction, as well as many core processes that are shared across different bacteria.
EARLY STUDIES OF AGROBACTERIUM
The earliest records reporting studies of plant crown galls are credited to Italian biologist Marcello Malpighi (of Malpighian tubule fame) (3). However, it was another Italian scientist, Fridiano Cavara who recognized that crown galls on grapevines could be correlated to the presence of a microbial isolate and that inoculation of this isolated “schizomycete” could induce tumors on young grapevines, and his observations were published in 1897 (4). It was not until 10 years later in 1907 that renowned U.S. plant pathologist Erwin F. Smith along with C. O. Townsend, applied bacteriological approaches to satisfy Koch’s postulates for the etiologic agent of crown gall (5). Smith is often credited as first isolating and characterizing the pathogen. The range of basic observations made in this initial characterization that remain relevant today is startling. There are several excellent sources that document the remarkable history of Agrobacterium research (6, 7) and an entire volume that compiles and comments on many of the landmark discoveries that led to our current knowledge of Agrobacterium biology (8). The history of A. tumefaciens research is highlighted by a striking series of discoveries during the 20th century and now into the 21st century (Fig. 1, Table 1), and this delightfully complex bacterium continues to yield unexpected and illuminating new findings.
FIG 1.
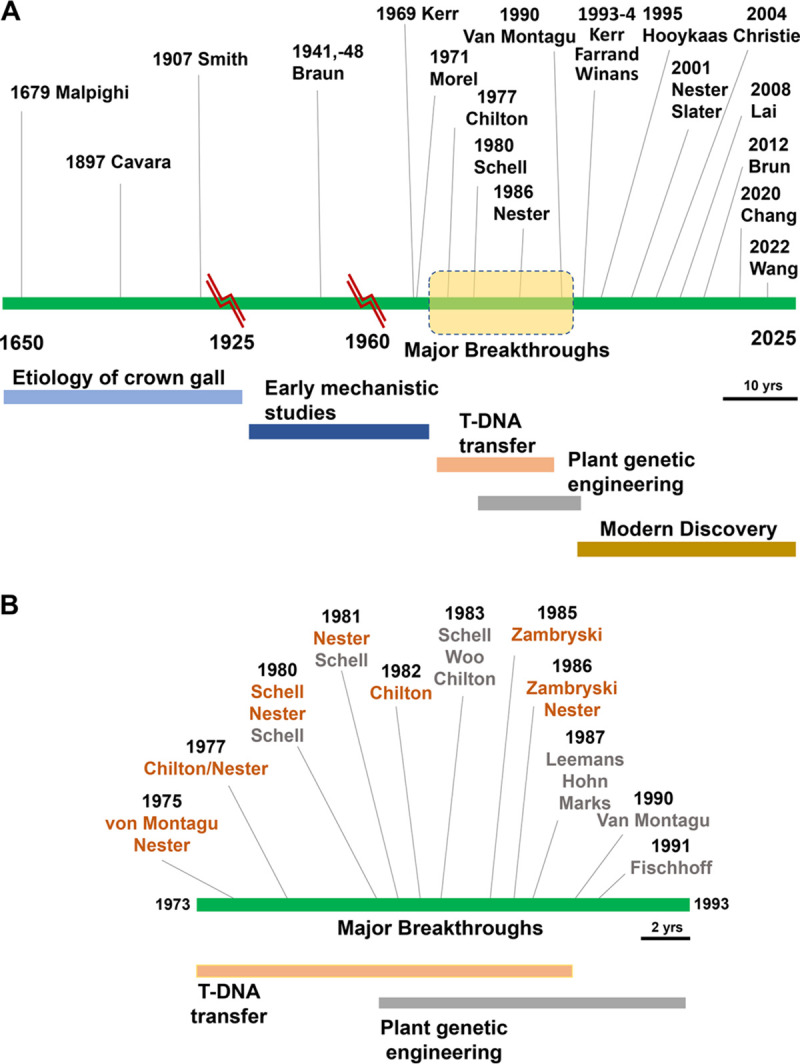
Timeline of publications reporting impactful Agrobacterium discoveries. (A) Timeline of selected discoveries arrayed linearly by year from 1650 to 2025. Lead scientists are indicated and usually match the corresponding author on the publications. Broad periods are indicated by the colored labeled bars below the timeline. “Major breakthroughs” refers to a very active period expanded in panel B. (B) Expanded view of discoveries that defined T-DNA transfer and harnessed it for use in plant transgenesis. Basic science discoveries are indicated by author names in orange text, and development of applications is indicated in silver text. All citations are referenced in Table 1.
TABLE 1.
Selected major discoveries and innovations in Agrobacterium research
| Yr | Discovery | Lead scientist(s) | Reference(s) |
|---|---|---|---|
| 1679 | Observation of crown galls on plants | Malpighi | 3 |
| 1897 | Isolation/reinoculation of pathogen | Cavara | 4 |
| 1907 | Etiological identification of pathogen | Smith | 5 |
| 1941 | Bacteria-free tumors | Braun | 16 |
| 1948 | Tumor-inducing principle | Braun | 14 |
| 1969 | Genetic transfer of virulence | Kerr | 17 |
| 1971 | Opine characterization | Morel | 18 |
| 1975 | Plasmid as the basis of virulence | von Montagu, Nester | 19, 20 |
| 1977 | Agrobacterial DNA in transformed plants | Chilton/Nester | 21 |
| 1980 | Tumor-inducing (Ti) plasmid genetic map | Schell | 137 |
| Organization of T-DNA genes in plant tumors | Nester | 138 | |
| Ti plasmid for gene introduction in plants | Schell | 139 | |
| 1981 | Fine structure genetic map of T-DNA in plants | Nester | 140 |
| Mendelian transmission of transferred genes | Schell | 141 | |
| 1982 | T-DNA border sequences and transfer | Chilton | 142 |
| 1983 | Expression of engineered genes in plants | Schell, Woo | 143, 144 |
| Engineered mini-Ti plasmids | Chilton | 145 | |
| 1985 | Plant phenolics induce virulence | Zambryski | 27 |
| 1986 | T-strand formation in early T-DNA transfer | Zambryski | 33 |
| VirA-VirG two-component systems | Nester | 28 | |
| 1987 | Engineered insect resistance | Leemans | 146 |
| Gene transfer to corn | Hohn | 147 | |
| Gene transfer to Arabidopsis | Marks | 148 | |
| 1990 | Nuclear targeting of the T-DNA | Van Montagu | 149 |
| 1991 | Transgene codon optimization | Fischhoff | 150 |
| 1993–1994 | Ti plasmid quorum sensing system | Kerr, Farrand, Winans | 57 – 59 |
| 1995 | T-DNA transfer to Saccharomyces | Hooykaas | 23 |
| 2001 | A. tumefaciens C58 genome sequence | Nester, Slater | 86, 87 |
| 2004 | Type IV secretion of T-DNA | Christie | 37 |
| 2008 | Type VI secretion in A. tumefaciens | Lai | 38 |
| 2012 | Polar budding in A. tumefaciens | Brun | 74 |
| 2020 | Ti plasmid diversity and evolution | Chang | 49 |
| 2022 | Multipartite replicon coordination | Wang | 92 |
TAXONOMIC CHALLENGES
The taxonomic classification of Agrobacterium has had a storied and contentious history that often confounds scientists who do not study this bacterium, and even those who do (9). Smith and Townsend originally named the causative agent of crown gall Bacterium tumefaciens (5). The bacterium subsequently went through several different taxonomic designations until 1942, when the genus/species name Agrobacterium tumefaciens was proposed (10). Three issues are at the heart of the confusion over the name of the pathogen and its close relatives. First is that early classification schemes attempted to capture differences in pathogenicity and host range. For example, Agrobacterium tumefaciens (biovar 1) and Agrobacterium rhizogenes (biovar 2) were classically used to infer those that cause crown gall and hairy root diseases, respectively. The Agrobacterium vitis (biovar 3) and Agrobacterium radiobacter species names were used to describe those that cause crown gall disease on grapevine and strains that are not pathogenic, respectively. However, pathogenicity is dependent upon a plasmid that can be lost but can also mobilize broadly across taxonomic groups, making pathogenicity an unstable and horizontally acquirable trait that does not accurately reflect relatedness among strains. For example, members of biovar 2, reflecting a species-level phylogenetic group, can cause crown gall, hairy root disease, infect grapevine, or be non-pathogenic. The second issue is the amalgamation of some lineages of Agrobacterium and rhizobia that drive nitrogen fixation in legumes (also a mobilizable trait) into a genus and renaming them all Rhizobium (11, 12). Adoption of this naming nomenclature has met limited acceptance among both agrobacterial and rhizobial research communities. The third issue is related to the second in that effectiveness in communicating relationships among strains is undermined by repeated revisions to taxonomic classifications. Consequently, multiple schemes persist and are variably adopted or abandoned by researchers who contribute to the long history of publication on these bacteria (13). We do not tackle this topic here but recommend looking toward an invited review that will attempt to relate some classification schemes to the inferred evolutionary relationships among strains of Agrobacterium (Alexandra J. Weisberg, Yu Wu, Jeff H. Chang, Erh-Min Lai, Chih-Horng Kuo, in preparation). The association of arguably ephemeral properties such as pathogenicity with bacterial nomenclature understandably has important legacy effects on Agrobacterium taxonomy, and throughout this review we use Agrobacterium tumefaciens for simplicity.
A BURST OF BASIC RESEARCH PROPELLED APPLIED STUDIES
The initial work and much of the history of A. tumefaciens research was rightfully focused on its interaction with plant hosts and eventually its development as a tool to manipulate plants in profound ways. Following the isolation of A. tumefaciens and the establishment of its etiological relationship with crown gall disease, progress on the microorganism was initially relatively sparse, with just a few key labs contributing. The laboratory of Armin Braun at the Rockefeller Institute in the 1940s provided several critical insights into the formation of crown gall (14, 15) and, importantly, developed the concept of the “tumor-inducing principle” to describe the factor(s) that stimulates gall formation (Fig. 1A, Table 1). Certainly, Braun and his lab members must have been influenced by the seminal studies of Avery, Macleod, and McCarty (also performed at the Rockefeller Institute) on the “transforming principle” in Streptococcus pneumoniae, which produced the first evidence that DNA is the genetic material (1). The Braun group made the critical insight that, once formed, the neoplastic growth in plants could continue to be propagated in the absence of the infecting bacterium, implicating a genetic or heritable change (14, 16). The true nature of the tumor-inducing principle would require many more years to be revealed. Nonetheless, it is striking how in the cases of both Avery and Braun, and their colleagues, who were studying very different biological phenomena, they were also constructing the conceptual framework for horizontal gene transmission that would contribute so importantly to our understanding of heritability and evolution, enabling the dawn of molecular biology.
The late 1960s and early 1970s brought pivotal discoveries from laboratories in multiple countries about the host-microbe interactions that underlie crown gall. Initially, Australian plant pathologist Allen Kerr recognized that the ability to cause crown gall tumors was encoded on an element transmissible between bacteria (17). Next, crown gall tumors were found to produce unique metabolites called opines that are semiexclusive nutrients for A. tumefaciens (18). Finally, studies by several groups including those of Allan Kerr, Jeff Schell and Marc van Montagu in Belgium, and Eugene Nester in the United States, among others, demonstrated the relationship between crown gall disease and the episomal element, named the tumor-inducing plasmid (or Ti plasmid), as the genetic prerequisite for crown gall disease (17, 19, 20). Together, these discoveries led to the then radical hypothesis that A. tumefaciens transfers its own genetic material to plants. The groundbreaking studies of Mary-Dell Chilton in the Nester laboratory provided definitive proof of the transfer of agrobacterial DNA to plant tissues (21). Chilton’s findings initiated a barrage of research within both academia and industry to understand the transferred DNA (T-DNA) and how it is generated, as well as to harness the power of interkingdom horizontal gene transmission (Fig. 1B). A barrage of brilliant work led to major advances in engineering the T-DNA and utilizing this natural process to transfer genes into plants. In several cases, the labs elucidating the basis for T-DNA transfer were the same ones propelling the applications for plant transgenesis, working in concert with applied scientists in the private sector. The burst of fundamental observations led to a wave of novel applications (Fig. 1B, Table 1). This research also yielded significant insights into plant cell biology and eventually provided transformation approaches for not only plants, but also fungal systems (22–24). It was a remarkable period of discovery and innovation and is an excellent example of basic research fueling applied science.
A MODEL FOR FUNDAMENTAL PROCESSES
The major breakthroughs on pathogenicity and interkingdom gene transfer propelled a sustained period of modern discoveries with A. tumefaciens that has continued to add crucial fundamental knowledge to inform molecular microbiology (Fig. 1A). Seminal work in A. tumefaciens has impacted our understanding of microbe-host and microbe-microbe interactions, particularly in the areas of secretion mechanisms and cell-cell communication. Furthermore, A. tumefaciens is emerging as a model system for bacterial cell biology with recent mechanistic insights related to polar growth, multipartite replicons, and plasmid biology. A. tumefaciens has been and remains a treasure trove for important, fundamental findings that have influenced the entire field of microbiology.
Host-microbe signaling and the two-component paradigm.
For pathogenic microorganisms, we now take for granted the concept of interkingdom communication in which pathogens perceive and respond to host-released signals. A. tumefaciens-plant interactions are among the first where such host-released signals were identified, as was the signal transduction cascade that leads to virulence gene expression in response to these signals (25, 26). Early studies used transposon mutagenesis to disrupt genes required for A. tumefaciens virulence (vir genes), also fusing lacZ encoding β-galactosidase with the disrupted loci (Fig. 1B, Table 1). The mutants were often avirulent, but also, several of the lacZ fusions were inducible in the presence of plant extracts (25). Chemical characterization of the plant exudates using the lacZ fusions as a bioassay identified phenolic precursors of lignin, acetosyringone (AS), and related molecules, which are produced during the wound healing response of plants (27). It was also demonstrated that many of the vir genes required for T-DNA transfer are only significantly expressed in the presence of the plant signals (25). Subsequent genetic studies demonstrated that two of the vir genes, virA and virG, were required for A. tumefaciens to respond to inducing conditions (26). Sequence analysis of these loci showed first that VirG is similar to several proteins from E. coli now called response regulators, including NtrC, OmpR, and PhoB, and that VirA is similar to proteins now known as sensor kinases, NtrB, EnvZ, and PhoR (28, 29). These and other studies of VirA and VirG established the two-component signal transduction mechanism in A. tumefaciens but also contributed greatly to the establishment of this general model of environmental response in bacteria (30). Demonstration of phosphotransfer between VirA and VirG in response to acetosyringone not only reinforced findings from the Escherichia coli two-component systems, but also broadened the scope of these regulatory systems to include host perception by pathogens (31). The two-component regulatory paradigm is now recognized as one of the most pervasive and influential systems in microbiology.
Type IV secretion systems.
The type IV secretion system (T4SS) is central to horizontal gene transfer, as this system is the apparatus used by conjugative plasmids and integrative conjugative elements to mediate transfer (32). The T4SS is also a protein secretion system used by pathogens important to human health. The T-DNA transfer process in agrobacteria is dependent on a T4SS and has long been a model for these systems. The virB operon on Ti plasmids encodes 11 proteins, all of which are involved in export of the T-DNA. It was reported by Stachel et al. in 1986 (Fig. 1B) that induction of virulence and T-DNA transfer created a single-stranded nick similar to that generated during the conjugation of bacterial plasmids (33). As DNA sequences from diverse plasmids emerged, and broad comparisons became possible, it was recognized that the virB gene products are similar to those of conjugative transfer systems (34, 35). Furthermore, secretion systems for bacterial toxins in unrelated systems, such as the Bordetella pertussis toxin liberation system (Ptl) were also found to be similar to the products of the virB genes (36). Several labs began focusing on the VirB proteins, and they rapidly became a model for the emergent type IV secretion systems. A large number of studies on the VirB T4SS from multiple research groups revealed many of the key mechanisms of T4SS, including an elegant study biochemically tracking the route of T-DNA through the VirB T4SS (Fig. 1A, Table 1) (37). This work propelled the A. tumefaciens VirB system to be the prototype for T4SS for many years, as more and more of these systems in mammalian pathogens were shown to export protein substrates (32). It is now clear that the A. tumefaciens system independently secretes protein effectors and T-DNA into plant cells during infection.
Type VI secretion systems.
The type VI secretion system (T6SS) was identified in A. tumefaciens strain C58 by Erh-Min Lai and colleagues during a screen for proteins secreted under conditions that induce virulence gene expression (Fig. 1A, Table 1) (38). T6SSs deploy toxic effector proteins to antagonize other bacteria (39). Studies of the T6SS of A. tumefaciens have yielded key discoveries about mechanisms of effector loading, such as the molecular features that determine specificity during engagement, and revealing a checkpoint for loading during T6SS activation (40–46). Studies have also revealed similarities and differences between T6SS and virulence gene regulation and provided a more complete view on the ecology of pathogenicity (46). Acidic pH, predicted to mimic the environment of the plant and rhizosphere, is crucial for inducing and activating the T6SS and virulence. Conversely, unlike virulence genes, sugars had no detectable effect, while phenolics attenuated T6SS activation. Overall, findings support a model predicting that the T6SS is activated when cells are near or associated with plants, but once virulence is induced, the T6SS is downregulated.
T6SS-associated effectors will cause self-intoxication, and cells must encode for immunity against each of their effectors (39, 47). Consequently, the activity and diversification of the T6SS is predicted to have a tremendous effect on the composition of bacterial communities (48). The Agrobacterium group has been deeply sequenced, and genomic data sets are valuable resources for understanding the natural variation of the T6SS (49). The presence of T6SS loci is variable across Agrobacterium taxa, but when present, genes encoding most of the structural and regulatory proteins are conserved in sequence and organization (50, 51). The presence/absence polymorphism was inferred to reflect recurrent loss, at various points in the history of these bacteria. Biovar 2, multiple genomospecies groups, and individual strains entirely lack a T6SS locus. Strains with T6SS-encoding loci are variable in the number and types of effector genes. The T6SSs in taxa that represent the diversity within the genus have different patterns of activation, suggesting that T6SSs of different strains or species-level groups are regulated under different environmental conditions (51). The T6SS of Agrobacterium is a model for understanding processes that have shaped its evolution and mechanistic diversification (52). Overall, the role of the T6SS in interbacterial competition among Agrobacterium cells is well supported, but its involvement directly or indirectly in virulence toward plants remains unresolved (43, 52).
Quorum sensing.
A. tumefaciens has been a critical model system for understanding cell-cell communication, particularly via quorum sensing. These findings have their origin in the studies of Kerr and colleagues in the late 1960s (Fig. 1A, Table 1) on the conjugative transfer of the Ti plasmid between bacteria during plant infection (17). Others later found that Ti plasmid conjugation was stimulated by specific opines, called conjugal opines (53, 54). Alan Kerr and Lian-Hui Zhang isolated a small molecule that they called conjugation factor that was produced by A. tumefaciens grown in the presence of specific opines (55). Chemical characterization of the conjugation factor identified it as N-3-oxo-octanoyl-homoserine lactone (3-oxo-C8-HSL), similar to the so-called autoinducer (N-3-oxo-hexanoyl- homoserine lactone) that regulates bioluminescence in Vibrio fischeri, the symbiont of certain fishes and bobtail squid (Fig. 1A, Table 1) (56, 57). In parallel with this chemical characterization was the genetic identification of the TraR transcription factor from two different Ti plasmids and the evidence that it regulated conjugal transfer gene expression in response to 3-oxo-C8-HSL (58, 59). This inducer molecule was synthesized by an enzyme called TraI, also encoded on the Ti plasmid (59, 60). TraR and TraI were among the founding members of the larger LuxR-LuxI family of regulators. As with bioluminescence regulation by LuxR and its autoinducer synthase LuxI, TraI and TraR imparted population density-dependent expression control on their target genes and Ti plasmid conjugation (61, 62), and the general population density response was subsequently described as quorum sensing (63). Several other LuxR-LuxI type systems were also discovered in diverse bacteria in this same period, most prominently LasR-LasI from Pseudomonas aeruginosa (64), and acylated homoserine lactone (AHL) autoinducers were detected for multiple bacteria (65), resulting in a wave of research on quorum sensing that continues today.
In addition to its foundational role in expanding quorum sensing beyond the bioluminescent marine vibrios, A. tumefaciens has provided several major contributions to the mechanistic understanding of quorum sensing. These include TraI being the first LuxI-type protein for which there was in vitro evidence that AHLs could be synthesized from acylated-acyl carrier protein (ACP) and S-adenosylmethionine (AdoMet) precursors (66), the identification of antagonistic regulators such as TraM that function with TraR and TraI as an integral part of the quorum sensing mechanism (67, 68), and the first three-dimensional structure of a LuxR-type protein (69). The structure reported for TraR was associated with the AHL ligand, and double-stranded DNA, providing a wealth of biochemical insights for other LuxR-type proteins. These and other fundamental insights shaped the field of bacterial cell-cell communication and established a quorum sensing paradigm.
Polar growth.
While most rod-shaped bacteria are assumed to elongate through growth in the lateral cell wall, it is becoming increasingly clear that asymmetric modes of peptidoglycan synthesis are prevalent among bacteria. It was in the early 1970s when Tamio Fujiwara and Sakuzo Fukui first proposed that A. tumefaciens grows unipolarly based upon striking morphologies of mutants and careful observations of microcolony formation (70, 71); however, it was not until the late 2000s that genome sequencing revealed that the entire clade of Rhizobiales, including Agrobacterium species, lacks the genes to encode the canonical elongation machinery (72). This opened up new research directions for using A. tumefaciens to answer the question, how do these bacteria maintain rod-like morphologies and elongate? Use of cell wall probes and timelapse microscopy to track A. tumefaciens cell growth clearly demonstrated that elongation is mediated by unipolar cell wall insertion (Fig. 1A, Table 1) (73, 74). As with more overtly asymmetric bacteria such as Caulobacter species, there are a number of extracellular polar structures such as a unipolar polysaccharide (UPP) adhesin and a tuft of polar flagella, that are integrated with the polar growth process (75, 76). These observations and processes have enabled A. tumefaciens to recently emerge as a model for mechanistic studies of unipolar growth in bacteria.
Systematic characterization of enzymes involved in cell wall biosynthesis and hydrolysis has resulted in several surprising findings that revealed diversification of these functional bacterial processes. First, unlike the proposed auxiliary function of penicillin binding protein 1a (PBP1a) in other rod-shaped bacteria, in A. tumefaciens, PBP1a is an essential enzyme required for polar growth (77). Second, while some ld-transpeptidases (LDTs) likely have broadly conserved roles in the modification of existing cell wall material, Rhizobiales-specific LDTs may function in peptidoglycan cross-linking during polar growth (77, 78). Third, cell wall hydrolytic enzymes have unique functions in establishing polarity to enable polar growth. While amidases and their regulators typically function in cell separation of many bacteria at the culmination of division, amidase AmiC and its regulator EnvC are required by A. tumefaciens to establish new poles following cell division (79). These observations suggested that AmiC-mediated modifications of the cell wall may serve as a signal for the recruitment of the polar growth machinery to the correct pole. Finally, scaffolding proteins such as PopZ and growth pole ring (GPR) protein reside at the growth pole and may function to facilitate the organization of specific growth pole proteins, including those important for cell wall and membrane biogenesis (80–85). The mechanism of polar growth in A. tumefaciens and other members of the Rhizobiales has evolved through the expansion, diversification, and altered regulation of the core cell wall synthesis machinery, and its further investigation has promise in providing a more comprehensive understanding of bacterial growth processes.
Multipartite replicons.
A. tumefaciens has emerged as a model organism for understanding the evolution and mechanisms of DNA segregation in bacteria with multiple replicons. The genome of the type strain A. tumefaciens C58 consists of four replicons: a circular chromosome, a linear chromosome, and the pAtC58 and pTiC58 plasmids (86, 87). The circular chromosome contains an oriC-type origin of replication found in many bacteria chromosomes, whereas the other replicons contain repABC origins, typical of plasmids in this group of bacteria. Consequently, the linear chromosome is thought to have been a plasmid into which gene transfer events from the circular chromosome occurred over evolution, enabling the transfer of some essential genes and an rRNA operon (86–88). These findings provided the foundation for a more generalizable model of genome evolution among bacteria with multiple replicons (88). It has been suggested that multipartite replicons may allow for acquisition of additional genetic material, enable faster genome duplication, and provide an advantage for microbes in changing environments, including host invasion (88–90). While the precise advantage for multipartite replicons remains unknown, the maintenance and management of multiple replicons pose some unique challenges.
In A. tumefaciens, the origin of replication of the circular chromosome is localized to the old pole of the cell, and during chromosomal replication the replicated origin migrates to the new pole (Fig. 1A, Table 1) (91–94). This polar localization is dependent on the ParABS chromosome segregation system, and docking of ParBI at the new poles is dependent on PopZ, while PodJ is indirectly responsible for docking of ParBI at the old poles (85, 91, 92). Like the circular chromosome, the origin of the linear chromosome is also dependent on the presence of PopZ and PodJ for efficient localization to cell poles (91, 93). The absence of GPR causes formation of spherical cells with origin location disrupted and a stochastic appearance (93). PodJ and GPR play indirect roles in localizing origins to the cell poles, while PopZ directly interacts with ParB and, to a lesser extent, RepB. Partition systems of bacterial replicons have sequences analogous to centromeres that are recognized by centromere-binding proteins. In A. tumefaciens, centromere clustering occurs independently of the polar organizing proteins (93). The circular and linear chromosomes have interarm interactions that depend on the SMC complex and require direct interaction of the centromere-binding ParB and RepB proteins (92, 93). These interactions result in a linear alignment of the chromosomes and may reduce chromosome entanglement, which if formed and not resolved can have severe fitness effects on cells. Indeed, disruption of this clustering pattern leads to loss of the linear chromosome and plasmids from progeny, indicating that coordination of centromere clustering is important for maintaining the integrity of multipartite genomes (93). These findings in A. tumefaciens suggest that centromeric clustering is a solution for ensuring that secondary replicons, such as virulence plasmids, are maintained in the absence of the selective forces that would otherwise promote their retention.
Plasmid biology.
Plasmids innovate Agrobacterium spp. with several important traits. Pathogenicity is the most renowned and the one that has elevated the prominence of A. tumefaciens as a model and an indispensable biotechnology tool. Multiple excellent and detailed reviews on the mechanism of Ti plasmid-dependent genetic transformation of plants have been published (95–97). The oncogenic Ti plasmid encodes five core functions: (i) the vir gene products that are required for pathogenicity and T-DNA transfer, (ii) the T-DNA that is delivered to and integrates with the plant genome to cause crown gall disease and opine production, (iii) opine catabolic functions, (iv) the interbacterial conjugative transfer system, and (v) the plasmid replication functions. Several oncogenic plasmids have been “disarmed” by removing genes within the T-DNA and separating the disarmed T-DNA from the vir genes to make “binary” and “helper” vectors, respectively (98). Engineering the Ti plasmid was not trivial, as these plasmids are large and maintained at low copy numbers. The binary system is a foundational technology for plant transformation and remains central to both agricultural applications and basic research.
The study of Agrobacterium plasmids has also been illustrative for plasmid and bacterial evolution. While plasmids carry cargo genes that give their bacterial hosts a fitness benefit, there are costs for bearing plasmids, and advantages are realized only in certain environments (99). Studies of the Ti plasmid demonstrated that costs are context dependent, with higher costs measured under conditions depleted of resources and those that induced expression of vir genes (100). These observations explain why copy numbers of Ti plasmids and expression of vir genes are tightly regulated (101–103). In addition, observations were consistent with a fundamental theory of cooperation, which is that pathogenic cells bear costs to direct benefits to related cells, which in this case are presumably those with related plasmids (104). This perspective of “taking one for the team” helps translate the understanding of molecular processes in single cells to understanding the evolution and ecology of populations.
Plasmids have traditionally been difficult to study because their sequences are challenging to resolve and analyze (105, 106). Conversely, because of the intensive work on oncogenic Ti and Ri plasmids (related to Ti plasmids, but they incite hairy root formation rather than galls, on plants), there is a depth of knowledge that positions them well as models for tackling the challenge of inferring plasmid evolution. Phylogenetic and network-based methods were used to analyze multiple scales of information and classify well over 100 oncogenic plasmids (Fig. 1A, Table 1) (49, 107). One of the major unexpected findings was that the oncogenic plasmids formed a limited number of types that could be related to each other and to a conceptional ancestral proto-oncogenic plasmid. Another major finding was that these plasmids are highly modular, with the most conserved regions being potential hot spots for homologous recombination, T-DNAs being extremely flexible, and virulence being conferred by vir genes mixed and matched from different plasmids or even distributed across plasmids (107). Nevertheless, despite the potential for tremendous diversity, structural organization of plasmids can constrain recombination such that certain events are more permissive, limiting most plasmids to the few extant types observed to date.
Agrobacterium spp. have another group of diverse nononcogenic plasmids often loosely referred to as “At” plasmids (108, 109). Although their existence has long been known, we still have little understanding of their roles in agrobacterial fitness. Catabolism may be one of the primary functions, as they can have homology to oncogenic plasmids, can catabolize opines, and can provide access to other nutrients (109–111). Regardless, nononcogenic and oncogenic plasmids can shape each other in multiple ways. Both classes can be simply viewed as members of a common pool of molecules that can recombine and diversify (107, 110). The members of this common pool can also influence signaling and stability across the two classes of plasmids (112, 113). It is also notable that these plasmids may have shaped the evolution of Agrobacterium genomes by capturing and shuffling genes across multiple large replicons. Plasmids have had a significant effect accessorizing Agrobacterium spp. with novel traits and shaping the structure and organization of their genomes.
FROM BASIC SCIENCE BACK TO APPLICATION: BIOCONTROL
Species of Agrobacterium are exemplary for demonstrating societal benefits of genetic engineering and biocontrol. Strain K84 of Agrobacterium was the first organism to be genetically engineered and commercialized for use as a live biocontrol product (114). K84 was discovered in 1970 from a plot with a history of crown gall (115). It is a nonpathogenic biovar 2 strain that lacks an oncogenic plasmid and, unlike others tested, consistently prevented crown gall disease following a 1:1 coinoculation with pathogenic strains, and was found to be a hyperparasite of A. tumefaciens. The value of K84 was recognized immediately and it emerged quickly as a highly successful preventative biocontrol strain (116). However, biocontrol is specific and reportedly effective against only strains that induce synthesis of the opine nopaline in crown galls (117). Furthermore, toxicity and immunity are mediated by pAgK84, a nononcogenic, nonconjugative but mobilizable plasmid, which raised concerns over potential for plasmid transmission causing breakdown of biocontrol (118–122). Concerns were mollified by deleting a small region to eliminate plasmid transfer (123). K1026, the strain carrying the engineered plasmid, is approved as a commercial biocontrol product in many countries and has no evidence for breakdown of plant protection.
Biocontrol is mediated by agrocin 84, a disubstituted adenine nucleotide and structural mimic of two molecules (124, 125). The protoxin is a mimic of a conjugative opine, agrocinopine A, and is consequently taken up by the cognate transporter (126, 127). The sugar moiety of agrocin 84, necessary for uptake but not toxicity, is subsequently released to yield a molecule that mimics leucyl-adenylate and interferes with translation by binding the catalytic domain of leucyl-tRNA synthase, an enzyme that catalyzes the transfer of leucine to its cognate tRNA (128, 129). Cells carrying pAgK84 are resistant to agrocin 84 because the plasmid also encodes a novel variant of leucyl-tRNA synthase that is less sensitive to inhibition (130). The success of agrocin 84 in limiting infection by A. tumefaciens is exceptional and has helped make these bacteria potential sources for other forms of control. These include agrocin variants, secondary metabolites, and other compounds that have yet to be detailed at the genetic and molecular levels (131–134). Lastly, it has long been known that avirulent Agrobacterium cells can prevent disease presumably through competition and limiting attachment of pathogens to plant surfaces (135, 136). While this trait by itself may not be as useful for biocontrol, it makes nonpathogenic strains competitive for plants and contributes in important ways toward the effectiveness of other control mechanisms.
FUTURE OUTLOOK FOR THE AGROBACTERIUM MODEL SYSTEM
The purpose of this review is to highlight some of the rich history of A. tumefaciens as a model pathogenic bacterial system, and even so, there is a great deal of important biology and applications that we have been unable to cover. A model system allows researchers to investigate phenomena in detail and manipulate processes in ways that take advantage of the system’s natural attributes and properties, to reveal new insights into general properties that are otherwise opaque. The interkingdom gene transfer from A. tumefaciens to plants certainly has warranted the intense level of attention it has received, and it is hard to argue with the scientific, societal, and economic impact its study and application have generated. Even with the tremendous and insightful research findings on Agrobacterium-plant interactions, there remains much to be learned. For example, how do molecular interactions with plant cells lead to nuclear import of T-DNA and integration of the T-DNA into the plant genome? These remain active areas of investigation and will yield new findings that have the potential to translate to innovations in genetic engineering. Beyond its utility as a genetic engineering tool, Agrobacterium research has arguably had as large an impact on our understanding of fundamental biological processes in bacteria, including but not limited to cell-cell communication, secretion, cell growth, and chromosome dynamics. As with all good model systems, there are many phenomena and insights that remain to be uncovered with the appropriate experiments, approaches, and perspective. Agrobacterium research continues to inform multiple areas of biology and expands in multiple new directions at the forefront of microbiology.
ACKNOWLEDGMENTS
We thank Alexandra Weisberg and Jennifer Greenwich for critical reading of the manuscript and engaging discussions and Patricia Zambryski for additional insights. We express our regrets for the many excellent researchers in the Agrobacterium community who due to space constraints, could not be included.
Agrobacterium research in the Brown lab is supported by the National Science Foundation (IOS1557806), in the Chang lab in part by the National Institutes of Food and Agriculture, U.S. Department of Agriculture (award 2022-67013-36883), and in the Fuqua lab by the National Institutes of General Medical Sciences (GM120337). The funders had no role in deciding the content or preparation of the manuscript.
Contributor Information
Clay Fuqua, Email: cfuqua@indiana.edu.
Anke Becker, Philipps University Marburg.
REFERENCES
- 1.Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–158. 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koch R. 2010. Die Ätiologie der Milzbrand-Krankheit, Begründet auf die Entwicklungsgeschichte des Bacillus anthracis. Robert Koch-Institut, Berlin, Germany. [Google Scholar]
- 3.Malpighi M. 1679. On galls. Anotomia Plantarum. [Google Scholar]
- 4.Cavara F. 1897. Tuberculosi della vite. Intorno alla eziologia di alcune malattie di piante cultivate. Le Stazioni Sperimentale Agraric Itliana 30:483–487. [Google Scholar]
- 5.Smith EF, Townsend CO. 1907. A plant-tumor of bacterial origin. Science 25:671–673. 10.1126/science.25.643.671. [DOI] [PubMed] [Google Scholar]
- 6.Zambryski P. 2013. Fundamental discoveries and simple recombination between circular plasmid DNAs led to widespread use of Agrobacterium tumefaciens as a generalized vector for plant genetic engineering. Int J Dev Biol 57:449–452. 10.1387/ijdb.130190pz. [DOI] [PubMed] [Google Scholar]
- 7.Kado CI. 2014. Historical account on gaining insights on the mechanism of crown gall tumorigenesis induced by Agrobacterium tumefaciens. Front Microbiol 5:340. 10.3389/fmicb.2014.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nester EW, Gordon MP, Kerr A (ed). 2005. Agrobacterium tumefaciens: from plant pathology to biotechnology. American Phytopathological Society Press, St. Paul, MN. [Google Scholar]
- 9.Flores-Félix JD, Menéndez E, Peix A, García-Fraile P, Velázquez E. 2020. History and current taxonomic status of genus Agrobacterium. System Appl Microbiol 43:126046. 10.1016/j.syapm.2019.126046. [DOI] [PubMed] [Google Scholar]
- 10.Conn HJ. 1942. Validity of the genus Alcaligenes. J Bacteriol 44:353–360. 10.1128/jb.44.3.353-360.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrand SK, van Berkum PB, Oger P. 2003. Agrobacterium is a definable genus of the family Rhizobiaceae. Int J Syst Evol Microbiol 53:1681–1687. 10.1099/ijs.0.02445-0. [DOI] [PubMed] [Google Scholar]
- 12.Young JM, Kuykendall LD, Martínez-Romero E, Kerr A, Sawada H. 2001. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int J Syst Bacteriol 51:89–103. 10.1099/00207713-51-1-89. [DOI] [PubMed] [Google Scholar]
- 13.Baltrus DA. 2016. Divorcing strain classification from species names. Trends Microbiol 24:431–439. 10.1016/j.tim.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Braun AC, Mandle RJ. 1948. Studies on the inactivation of the tumor-inducing principle in crown gall. Growth 12:255–269. [PubMed] [Google Scholar]
- 15.Braun AC, Elrod RP. 1946. Stages in the life history of Phytomonas tumefaciens. J Bacteriol 52:695–702. 10.1128/jb.52.6.695-702.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White PR, Braun AC. 1941. Crown gall production by bacteria-free tumor tissues. Science 94:239–241. 10.1126/science.94.2436.239. [DOI] [PubMed] [Google Scholar]
- 17.Kerr A. 1969. Transfer of virulence between isolates of Agrobacterium. Nature 223:1175–1176. 10.1038/2231175a0. [DOI] [Google Scholar]
- 18.Petit A, Delhaye S, Tempe J, Morel G. 1970. Recherches sur les guanidines des tissus de crown gall: mise en evidence d’une relation biochimique specifique entre les souches d’Agrobacterium tumefaciens es les tumeurs qu’elles induisent. Physiol Veg 8:205–213. [Google Scholar]
- 19.Van Larebeke N, Genetello C, Schell J, Schilperoort RA, Hermans AK, Hernalsteens JP, Van Montagu M. 1975. Acquisition of tumour-inducing ability by non-oncogenic agrobacteria as a result of plasmid transfer. Nature 255:742–743. 10.1038/255742a0. [DOI] [PubMed] [Google Scholar]
- 20.Watson B, Currier TC, Gordon MP, Chilton MD, Nester EW. 1975. Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol 123:255–264. 10.1128/jb.123.1.255-264.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chilton M-D, Drummond MH, Merlo DJ, Sciaky D, Montoya AL, Gordon MP, Nester EW. 1977. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11:263–271. 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Nam J, Humara JM, Mysore KS, Lee L-Y, Cao H, Valentine L, Li J, Kaiser AD, Kopecky AL, Hwang H-H, Bhattacharjee S, Rao PK, Tzfira T, Rajagopal J, Yi H, Yadav BS, Crane YM, Lin K, Larcher Y, Gelvin MJ, Knue M, Ramos C, Zhao X, Davis SJ, Kim S-I, Ranjith-Kumar CT, Choi Y-J, Hallan VK, Chattopadhyay S, Sui X, Ziemienowicz A, Matthysse AG, Citovsky V, Hohn B, Gelvin SB. 2003. Identification of Arabidopsis rat mutants. Plant Physiol 132:494–505. 10.1104/pp.103.020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas PJJ. 1995. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J 14:3206–3214. 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ballas N, Citovsky V. 1997. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc Natl Acad Sci USA 94:10723–10728. 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stachel SE, Nester EW, Zambryski PC. 1986. A plant cell factor induces Agrobacterium tumefaciens vir gene expression. Proc Natl Acad Sci USA 83:379–383. 10.1073/pnas.83.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stachel SE, Zambryski PC. 1986. virA and virG control the plant induced activation of the T-DNA transfer process in A. tumefaciens. Cell 46:325–333. 10.1016/0092-8674(86)90653-7. [DOI] [PubMed] [Google Scholar]
- 27.Stachel SE, Messens E, Van Montagu M, Zambryski P. 1985. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318:624–629. 10.1038/318624a0. [DOI] [Google Scholar]
- 28.Winans SC, Ebert PR, Stachel SE, Gordon MP, Nester EW. 1986. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci USA 83:8278–8282. 10.1073/pnas.83.21.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leroux B, Yanofsky MF, Winans SC, Ward JE, Ziegler SF, Nester EW. 1987. Characterization of the virA locus of Agrobacterium tumefaciens: a transcriptional regulator and host range determinant. EMBO J 6:849–856. 10.1002/j.1460-2075.1987.tb04830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nixon BT, Ronson CW, Ausubel FM. 1986. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci USA 83:7850–7854. 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin S, Roitsch T, Ankenbauer RG, Gordon MP, Nester EW. 1990. The VirA protein of Agrobacterium tumefaciens is autophosphorylated and is essential for vir gene regulation. J Bacteriol 172:525–530. 10.1128/jb.172.2.525-530.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59:451–485. 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stachel SE, Timmerman B, Zambryski P. 1986. Generation of single-stranded T-DNA molecules during the initial stages of T-DNA transfer from Agrobacterium tumefaciens to plant cells. Nature 322:706–712. 10.1038/322706a0. [DOI] [Google Scholar]
- 34.Lessl M, Balzer D, Pansegrau W, Lanka E. 1992. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem 267:20471–20480. 10.1016/S0021-9258(19)88726-4. [DOI] [PubMed] [Google Scholar]
- 35.Pohlman RF, Genetti HD, Winans SC. 1994. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol Microbiol 14:655–668. 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 36.Weiss AA, Johnson FD, Burns DL. 1993. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci USA 90:2970–2974. 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cascales E, Christie PJ. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170–1173. 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H-Y, Chung PC, Hsiao-Wei -S, Wen S-R, Lai E-M. 2008. Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J Bacteriol 190:2841–2850. 10.1128/JB.01775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347. 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bondage DD, Lin J-S, Ma L-S, Kuo C-H, Lai E-M. 2016. VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc Natl Acad Sci USA 113:e3931–e3940. 10.1073/pnas.1600428113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hachani A, Allsopp LP, Oduko Y, Filloux A. 2014. The VgrG proteins are “à la carte” delivery systems for bacterial type VI effectors. J Biol Chem 289:17872–17884. 10.1074/jbc.M114.563429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin J-S, Ma L-S, Lai E-M. 2013. Systematic dissection of the Agrobacterium type VI secretion system reveals machinery and secreted components for subcomplex formation. PLoS One 8:e67647. 10.1371/journal.pone.0067647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma L-S, Hachani A, Lin J-S, Filloux A, Lai E-M. 2014. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16:94–104. 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitney JC, Beck CM, Goo YA, Russell AB, Harding BN, De Leon JA, Cunningham DA, Tran BQ, Low DA, Goodlett DR, Hayes CS, Mougous JD. 2014. Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol 92:529–542. 10.1111/mmi.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu C-F, Lien Y-W, Bondage D, Lin J-S, Pilhofer M, Shih Y-L, Chang JH, Lai E-M. 2020. Effector loading onto the VgrG carrier activates type VI secretion system assembly. EMBO Rep 21:e47961. 10.15252/embr.201947961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu CF, Lin JS, Shaw GC, Lai E-M. 2012. Acid-induced type VI secretion system is regulated by ExoR-ChvG/ChvI signaling cascade in Agrobacterium tumefaciens. PLoS Pathog 8:e1002938. 10.1371/journal.ppat.1002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lien Y-W, Lai E-M. 2017. Type VI secretion effectors: methodologies and biology. Front Cell Infec Microbiol 7:254. 10.3389/fcimb.2017.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas J, Watve SS, Ratcliff WC, Hammer BK. 2017. Horizontal gene transfer of functional Type VI killing genes by natural transformation. mBio 8:e00654-17. 10.1128/mBio.00654-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weisberg AJ, Davis EW II, Tabima J, Belcher MS, Miller M, Kuo CH, Loper JE, Grunwald NJ, Putnam ML, Chang JH. 2020. Unexpected conservation and global transmission of agrobacterial virulence plasmids. Science 368:eaba5256. 10.1126/science.aba5256. [DOI] [PubMed] [Google Scholar]
- 50.Chou L, Lin YC, Haryono M, Santos MNM, Cho ST, Weisberg AJ, Wu CF, Chang JH, Lai EM, Kuo CH. 2022. Modular evolution of secretion systems and virulence plasmids in a bacterial species complex. BMC Biol 20:16. 10.1186/s12915-021-01221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu C-F, Weisberg AJ, Davis EW, Chou L, Khan S, Lai E-M, Kuo C-H, Chang JH. 2021. Diversification of the Type VI secretion system in agrobacteria. mBio 12:e01927-21. 10.1128/mBio.01927-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu C-F, Santos MNMC, Cho ST, Chang H-H, Tsai Y-M, Smith DA, Kuo C-H, Chang JH, Lai E-M. 2019. Plant-pathogenic Agrobacterium tumefaciens strains have diverse Type VI effector-immunity pairs and vary in in-planta competitiveness. MPMI 32:961–971. 10.1094/MPMI-01-19-0021-R. [DOI] [PubMed] [Google Scholar]
- 53.Ellis JG, Kerr A, Petit A, Tempé J. 1982. Conjugal transfer of nopaline and agropine Ti-plasmids - the role of agrocinopines. Mol Gen Genet 186:269–274. 10.1007/BF00331861. [DOI] [Google Scholar]
- 54.Klapwijk PM, Scheulderman T, Schilperoort R. 1978. Coordinated regulation of octopine degradation and conjugative transfer of Ti plasmids in Agrobacterium tumefaciens: evidence for a common regulatory gene and separate operons. J Bacteriol 136:775–785. 10.1128/jb.136.2.775-785.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L, Kerr A. 1991. A diffusible compound can enhance conjugal transfer of the Ti plasmid in Agrobacterium tumefaciens. J Bacteriol 173:1867–1872. 10.1128/jb.173.6.1867-1872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444–2449. 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 57.Zhang L, Murphy PJ, Kerr A, Tate ME. 1993. Agrobacterium conjugation and gene regulation by N-acyl-L-homoserine lactones. Nature 362:446–448. 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 58.Piper KR, Beck von Bodman S, Farrand SK. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448–450. 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 59.Fuqua WC, Winans SC. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol 176:2796–2806. 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hwang I, Li P-L, Zhang L, Piper KR, Cook DM, Tate ME, Farrand SK. 1994. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc Natl Acad Sci USA 91:4639–4643. 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Piper KR, Beck von Bodman S, Hwang I, Farrand SK. 1999. Hierarchical gene regulatory systems arising from fortuitous gene associations: controlling quorum sensing by the opine regulon in Agrobacterium. Mol Microbiol 32:1077–1089. 10.1046/j.1365-2958.1999.01422.x. [DOI] [PubMed] [Google Scholar]
- 62.Fuqua C, Winans SC. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol 178:435–440. 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR/LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275. 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gambello MJ, Kaye S, Iglewski BH. 1993. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxon A expression. Infect Immun 61:1180–1184. 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bainton NJ, Bycroft BW, Chhabra SR, Stead P, Gledhill L, Hill PJ, Rees CE, Winson MK, Salmond GP, Stewart GS, Williams P. 1992. A general role for the lux autoinducer in bacterial cell signalling: control of antibiotic biosynthesis in Erwinia. Gene 116:87–91. 10.1016/0378-1119(92)90633-Z. [DOI] [PubMed] [Google Scholar]
- 66.Moré MI, Finger LD, Stryker JL, Fuqua C, Eberhard A, Winans SC. 1996. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science 272:1655–1658. 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 67.Hwang I, Cook DM, Farrand SK. 1995. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J Bacteriol 177:449–458. 10.1128/jb.177.2.449-458.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fuqua C, Burbea M, Winans SC. 1995. Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the traM gene. J Bacteriol 177:1367–1373. 10.1128/jb.177.5.1367-1373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang RG, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneaux JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971–974. 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 70.Fujiwara T, Fukui S. 1974. Unidirectional growth and branch formation of a morphological mutant, Agrobacterium tumefaciens. J Bacteriol 120:583–589. 10.1128/jb.120.2.583-589.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fujiwara T, Fukui S. 1972. Isolation of morphological mutants of Agrobacterium tumefaciens. J Bacteriol 110:743–746. 10.1128/jb.110.2.743-746.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Margolin W. 2009. Sculpting the bacterial cell. Curr Biol 19:R812–R822. 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl 51:12519–12523. 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. 2012. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA 109:1697–1701. 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Onyeziri MC, Hardy GG, Natarajan R, Xu J, Reynolds IP, Kim J, Merritt PM, Danhorn T, Hibbing ME, Weisberg AJ, Chang JH, Fuqua C. 2022. Dual adhesive unipolar polysaccharides synthesized by overlapping biosynthetic pathways in Agrobacterium tumefaciens. Mol Microbiol 117:1023–1047. 10.1111/mmi.14887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mohari B, Thompson MA, Trinidad JC, Setayeshgar S, Fuqua C. 2018. Multiple flagellin proteins have distinct and synergistic roles in Agrobacterium tumefaciens motility. J Bacteriol 200:e00327-18. 10.1128/JB.00327-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams MA, Aliashkevich A, Krol E, Kuru E, Bouchier JM, Rittichier J, Brun YV, VanNieuwenhze MS, Becker A, Cava F, Brown PJB. 2021. Unipolar peptidoglycan synthesis in the Rhizobiales requires an essential class A penicillin-binding protein. mBio 12:e02346-21. 10.1128/mBio.02346-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cameron TA, Anderson-Furgeson J, Zupan JR, Zik JJ, Zambryski PC. 2014. Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. mBio 5:e01219-14. 10.1128/mBio.01219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Figueroa-Cuilan WM, Randich AM, Dunn CM, Santiago-Collazo G, Yowell A, Brown PJB. 2021. Diversification of LytM protein functions in polar elongation and cell division of Agrobacterium tumefaciens. Front Microbiol 12:729307. 10.3389/fmicb.2021.729307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zupan JR, Grangeon R, Robalino-Espinosa JS, Garnica N, Zambryski P. 2019. GROWTH POLE RING protein forms a 200-nm-diameter ring structure essential for polar growth and rod shape in Agrobacterium tumefaciens. Proc Natl Acad Sci USA 116:10962–10967. 10.1073/pnas.1905900116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zupan J, Guo Z, Biddle T, Zambryski P. 2021. Agrobacterium tumefaciens Growth Pole Ring Protein: C terminus and internal apolipoprotein homologous domains are essential for function and subcellular localization. mBio 12:e00764-21. 10.1128/mBio.00764-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Howell M, Aliashkevich A, Salisbury AK, Cava F, Bowman GR, Brown PJB. 2017. Absence of the polar organizing protein PopZ results in reduced and asymmetric cell division in Agrobacterium tumefaciens. J Bacteriol 199:e00101-17. 10.1128/JB.00101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grangeon R, Zupan JR, Anderson-Furgeson J, Zambryski PC. 2015. PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens. Proc Natl Acad Sci USA 112:11666–11671. 10.1073/pnas.1515544112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grangeon R, Zupan J, Jeon Y, Zambryski PC. 2017. Loss of PopZ (At) activity in Agrobacterium tumefaciens by deletion or depletion leads to multiple growth poles, minicells, and growth defects. mBio 8:e01881-17. 10.1128/mBio.01881-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ehrle HM, Guidry JT, Iacovetto R, Salisbury AK, Sandidge DJ, Bowman GR. 2017. Polar organizing protein PopZ Is required for chromosome segregation in Agrobacterium tumefaciens. J Bacteriol 199:e00111-17. 10.1128/JB.00111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, et al. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323. 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 87.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C, Mullin L, Houmiel K, Gordon J, Vaudin M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, Slater S. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323–2328. 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 88.Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW, Burr TJ, Banta L, Dickerman AW, Paulsen I, Otten L, Suen G, Welch R, Almeida NF, Arnold F, Burton OT, Du Z, Ewing A, Godsy E, Heisel S, Houmiel KL, Jhaveri J, Lu J, Miller NM, Norton S, Chen Q, Phoolcharoen W, Ohlin V, Ondrusek D, Pride N, Stricklin SL, Sun J, Wheeler C, Wilson L, Zhu H, Wood DW. 2009. Genome sequences of three agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J Bacteriol 191:2501–2511. 10.1128/JB.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Misra HS, Maurya GK, Kota S, Charaka VK. 2018. Maintenance of multipartite genome system and its functional significance in bacteria. J Genet 97:1013–1038. 10.1007/s12041-018-0969-x. [DOI] [PubMed] [Google Scholar]
- 90.diCenzo GC, Finan TM. 2017. The divided bacterial genome: structure, function, and evolution. Microbiol Mol Biol Rev 81:e00019-17. 10.1128/MMBR.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robalino-Espinosa JS, Zupan JR, Chavez-Arroyo A, Zambryski P. 2020. Segregation of four Agrobacterium tumefaciens replicons during polar growth: PopZ and PodJ control segregation of essential replicons. Proc Natl Acad Sci USA 117:26366–26373. 10.1073/pnas.2014371117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ren Z, Liao Q, Karaboja X, Barton IS, Schantz EG, Mejia-Santana A, Fuqua C, Wang X. 2022. Conformation and dynamic interactions of the multipartite genome in Agrobacterium tumefaciens. Proc Natl Acad Sci USA 119:e2115854119. 10.1073/pnas.2115854119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ren Z, Liao Q, Barton IS, Wiesler EE, Fuqua C, Wang X. 2022. Centromere interactions promote the maintenance of the multipartite genome in Agrobacterium tumefaciens. mBio 13:e0050822. 10.1128/mbio.00508-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kahng LS, Shapiro L. 2003. Polar localization of replicon origins in the multipartite genomes of Agrobacterium tumefaciens and Sinorhizobium meliloti. J Bacteriol 185:3384–3391. 10.1128/JB.185.11.3384-3391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nester EW. 2015. Agrobacterium: nature’s genetic engineer. Front Plant Sci 5:730. 10.3389/fpls.2014.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gordon JE, Christie PJ. 2015. The Agrobacterium Ti plasmids, p 295–313. In Tolmasky ME, Alonso JC (ed), Plasmids: biology and impact in biotechnology and discovery. ASM Press, Washington, DC. [Google Scholar]
- 97.Gelvin SB. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67:16–37. 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee L-Y, Gelvin SB. 2008. T-DNA binary vectors and systems. Plant Physiol 146:325–332. 10.1104/pp.107.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brockhurst MA, Harrison E. 2022. Ecological and evolutionary solutions to the plasmid paradox. Trends Microbiol 30:534–543. 10.1016/j.tim.2021.11.001. [DOI] [PubMed] [Google Scholar]
- 100.Platt TG, Bever JD, Fuqua C. 2012. A cooperative virulence plasmid imposes a high fitness cost under conditions that induce pathogenesis. Proc Biol Sci 279:1691–1699. 10.1098/rspb.2011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suzuki K, Iwata K, Yoshida K. 2001. Genome analysis of Agrobacterium tumefaciens: construction of physical maps for linear and circular chromosomal DNAs, determination of copy number ratio and mapping of chromosomal virulence genes. DNA Res 8:141–152. 10.1093/dnares/8.4.141. [DOI] [PubMed] [Google Scholar]
- 102.Pappas KM, Winans SC. 2003. A LuxR-type regulator from Agrobacterium tumefaciens elevates Ti plasmid copy number by activating transcription of plasmid replication genes. Mol Microbiol 48:1059–1073. 10.1046/j.1365-2958.2003.03488.x. [DOI] [PubMed] [Google Scholar]
- 103.Cho H, Winans SC. 2005. VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound-released chemical signals. Proc Natl Acad Sci USA 102:14843–14848. 10.1073/pnas.0503458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Platt TG, Fuqua C, Bever JD. 2012. Resource and competitive dynamics shape the benefits of public goods cooperation in a plant pathogen. Evolution 66:1953–1965. 10.1111/j.1558-5646.2011.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Orlek A, Stoesser N, Anjum MF, Doumith M, Ellington MJ, Peto T, Crook D, Woodford N, Walker AS, Phan H, Sheppard AE. 2017. Plasmid classification in an era of whole-genome sequencing: Application in studies of antibiotic resistance epidemiology. Front Microbiol 8:182. 10.3389/fmicb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen K, Otten L. 2017. Natural Agrobacterium transformants: recent results and some theoretical considerations. Front Plant Sci 8:1600. 10.3389/fpls.2017.01600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weisberg AJ, Miller M, Ream W, Grunwald NJ, Chang JH. 2022. Diversification of plasmids in a genus of pathogenic and nitrogen-fixing bacteria. Philos Trans R Soc Lond B Biol Sci 377:20200466. 10.1098/rstb.2020.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zaenen I, van Larebeke N, Teuchy H, van Montagu M, Schell J. 1974. Supercoiled circular DNA in crown-gall inducing Agrobacterium strains. J Mol Biol 86:109–127. 10.1016/S0022-2836(74)80011-2. [DOI] [PubMed] [Google Scholar]
- 109.Merlo DJ, Nester EW. 1977. Plasmids in avirulent strains of Agrobacterium. J Bacteriol 129:76–80. 10.1128/jb.129.1.76-80.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vaudequin-Dransart V, Petit A, Chilton WS, Dessaux Y. 1998. The cryptic plasmid of Agrobacterium tumefaciens cointegrates with the Ti plasmid and cooperates for opine degradation. MPMI 11:583–591. 10.1094/MPMI.1998.11.7.583. [DOI] [Google Scholar]
- 111.Burr TJ, Otten L. 1999. Crown gall of grape: biology and disease management. Annu Rev Phytopathol 37:53–80. 10.1146/annurev.phyto.37.1.53. [DOI] [PubMed] [Google Scholar]
- 112.Barton IS, Platt TG, Rusch DB, Fuqua C. 2019. Destabilization of the tumor-inducing plasmid from an octopine-type Agrobacterium tumefaciens lineage drives a large deletion in the co-resident At megaplasmid. G3 (Bethesda) 9:3489–3500. 10.1534/g3.119.400554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barton IS, Eagan JL, Nieves-Otero PA, Reynolds IP, Platt TG, Fuqua C. 2020. Co-dependent and interdigitated: Dual quorum sensing systems regulate conjugative transfer of the Ti plasmid and the At megaplasmid in Agrobacterium tumefaciens 15955. Front Microbiol 11:605896. 10.3389/fmicb.2020.605896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kerr A, Bullard G. 2020. Biocontrol of crown gall by Rhizobium rhizogenes: Challenges in biopesticide commercialisation. Agronomy 10:1126. 10.3390/agronomy10081126. [DOI] [Google Scholar]
- 115.New PB, Kerr A. 1972. Biological control of crown gall: field measurements and glasshouse experiments. J Appl Bacteriol 35:279–287. 10.1111/j.1365-2672.1972.tb03699.x. [DOI] [Google Scholar]
- 116.Moore LW, Warren G. 1979. Agrobacterium radiobacter Strain 84 and biological control of crown gall. Annu Rev Phytopathol 17:163–179. 10.1146/annurev.py.17.090179.001115. [DOI] [Google Scholar]
- 117.Kerr A, Roberts WP. 1976. Agrobacterium: correlations between and transfer of pathogenicity, octopine and nopaline metabolism and bacteriocin 84 sensitivity. Physiol Plant Pathol 9:205–211. 10.1016/0048-4059(76)90041-2. [DOI] [Google Scholar]
- 118.Slota JE, Farrand SK. 1982. Genetic isolation and physical characterization of pAgK84, the plasmid responsible for agrocin 84 production. Plasmid 8:175–186. 10.1016/0147-619X(82)90055-5. [DOI] [PubMed] [Google Scholar]
- 119.Ryder MH, Slota JE, Scarim A, Farrand SK. 1987. Genetic analysis of agrocin 84 production and immunity in Agrobacterium spp. J Bacteriol 169:4184–4189. 10.1128/jb.169.9.4184-4189.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim J-G, Park BK, Kim S-U, Choi D, Nahm B-H, Moon JS, Reader JS, Farrand SK, Hwang I. 2006. Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc Natl Acad Sci USA 103:8846–8851. 10.1073/pnas.0602965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Farrand SK, Slota JE, Shim JS, Kerr A. 1985. Tn5 insertions in the agrocin 84 plasmid: the conjugal nature of pAgK84 and the locations of determinants for transfer and agrocin 84 production. Plasmid 13:106–117. 10.1016/0147-619X(85)90063-0. [DOI] [PubMed] [Google Scholar]
- 122.Ellis JG, Kerr A, Van Montagu M, Schell J. 1979. Agrobacterium: genetic studies on agrocin 84 production and the biological control of crown gall. Physiol Plant Pathol 15:311–319. 10.1016/0048-4059(79)90082-1. [DOI] [Google Scholar]
- 123.Jones DA, Ryder MH, Clare BG, Farrand SK, Kerr A. 1988. Construction of a Tra– deletion mutant of pAgK84 to safeguard the biological control of crown gall. Mol Gen Genet 212:207–214. 10.1007/BF00334686. [DOI] [Google Scholar]
- 124.Tate ME, Murphy PJ, Roberts WP, Keer A. 1979. Adenine N6-substituent of agrocin 84 determines its bacteriocin-like specificity. Nature 280:697–699. 10.1038/280697a0. [DOI] [PubMed] [Google Scholar]
- 125.Roberts WPT, Tate ME, Kerr A. 1977. Agrocin 84 is a 6-N-phosphoramidate of an adenine nucleotide analogue. Nature 265:379–381. 10.1038/265379a0. [DOI] [PubMed] [Google Scholar]
- 126.Murphy PJ, Roberts WP. 1979. A basis for agrocin 84 sensitivity in Agrobacterium radiobacter. J Gen Microbiol 114:207–213. 10.1099/00221287-114-1-207. [DOI] [Google Scholar]
- 127.El Sahili A, Li SZ, Lang J, Virus C, Planamente S, Ahmar M, Guimaraes BG, Aumont-Nicaise M, Vigouroux A, Soulere L, Reader J, Queneau Y, Faure D, Morera S. 2015. A pyranose-2-phosphate motif Is responsible for both antibiotic import and quorum-sensing regulation in Agrobacterium tumefaciens. PLoS Pathog 11:e1005071. 10.1371/journal.ppat.1005071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Reader JS, Ordoukhanian PT, Kim JG, de Crecy-Lagard V, Hwang I, Farrand S, Schimmel P. 2005. Major biocontrol of plant tumors targets tRNA synthetase. Science 309:1533. 10.1126/science.1116841. [DOI] [PubMed] [Google Scholar]
- 129.Chopra S, Palencia A, Virus C, Tripathy A, Temple BR, Velazquez-Campoy A, Cusack S, Reader JS. 2013. Plant tumour biocontrol agent employs a tRNA-dependent mechanism to inhibit leucyl-tRNA synthetase. Nat Commun 4:1417. 10.1038/ncomms2421. [DOI] [PubMed] [Google Scholar]
- 130.Chopra S, Palencia A, Virus C, Schulwitz S, Temple BR, Cusack S, Reader J. 2016. Structural characterization of antibiotic self-immunity tRNA synthetase in plant tumour biocontrol agent. Nat Commun 7:12928. 10.1038/ncomms12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zheng DB, Burr TJ. 2016. Inhibition of grape crown gall by Agrobacterium vitis F2/5 requires two nonribosomal peptide synthetases and one polyketide synthase. MPMI 29:109–118. 10.1094/MPMI-07-15-0153-R. [DOI] [PubMed] [Google Scholar]
- 132.Noutoshi Y, Toyoda A, Ishii T, Saito K, Watanabe M, Kawaguchi A. 2020. Complete genome sequence data of nonpathogenic strain Rhizobium vitis VAR03-1, a biological control agent for grapevine crown gall disease. MPMI 33:1451–1453. 10.1094/MPMI-07-20-0181-A. [DOI] [PubMed] [Google Scholar]
- 133.Kawaguchi A, Nita M, Ishii T, Watanabe M, Noutoshi Y. 2019. Biological control agent Rhizobium (=Agrobacterium) vitis strain ARK-1 suppresses expression of the essential and non-essential vir genes of tumorigenic R. vitis. BMC Res Notes 12:1. 10.1186/s13104-018-4038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Donner SC, Jones DA, McClure NC, Rosewarne GM, Tate ME, Kerr A, Fajardo NN, Clare BG. 1993. Agrocin 434, a new plasmid encoded agrocin from the biocontrol Agrobacterium strains K84 and K1026, which inhibits biovar 2 agrobacteria. Physiol Plant Pathol 42:185–194. 10.1006/pmpp.1993.1017. [DOI] [Google Scholar]
- 135.Lippincott JA, Lippincott BB. 1969. Bacterial attachment to a specific wound site as an essential stage in tumor initiation by Agrobacterium tumefaciens. J Bacteriol 97:620–628. 10.1128/jb.97.2.620-628.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cooksey DA. 1982. Biological control of crown gall with an agrocin mutant of Agrobacterium radiobacter. Phytopathol 72:919. 10.1094/Phyto-72-919. [DOI] [Google Scholar]
- 137.Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inze D, Engler G, Villarroel R, Van Montagu M, Schell J. 1980. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid 3:212–230. 10.1016/0147-619X(80)90110-9. [DOI] [PubMed] [Google Scholar]
- 138.Thomashow MF, Nutter R, Postle K, Chilton MD, Blattner FR, Powell A, Gordon MP, Nester EW. 1980. Recombination between higher plant DNA and the Ti plasmid of Agrobacterium tumefaciens. Proc Natl Acad Sci USA 77:6448–6452. 10.1073/pnas.77.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hernalsteens JP, Van Vliet F, De Beuckeleer M, Depicker A, Engler G, Lemmers M, Holsters M, Van Montagu M, Schell J. 1980. The Agrobacterium tumefaciens Ti plasmid as a host vector system for introducing foreign DNA in plant cells. Nature 287:654–656. 10.1038/287654a0. [DOI] [PubMed] [Google Scholar]
- 140.Garfinkel DJ, Simpson RB, Ream LW, White FF, Gordon MP, Nester EW. 1981. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27:143–153. 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 141.Otten L, De Greve H, Hernalsteens JP, Van Montagu M, Schieder O, Straub J, Schell J. 1981. Mendelian transmission of genes introduced into plants by the Ti plasmids of Agrobacterium tumefaciens. Mol Gen Genet 183:209–213. 10.1007/BF00270619. [DOI] [PubMed] [Google Scholar]
- 142.Yadav NS, Vanderleyden J, Bennett DR, Barnes WM, Chilton MD. 1982. Short direct repeats flank the T-DNA on a nopaline Ti plasmid. Proc Natl Acad Sci USA 79:6322–6326. 10.1073/pnas.79.20.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zambryski P, Joos H, Genetello C, Leemans J, Montagu MV, Schell J. 1983. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J 2:2143–2150. 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fraley RT, Rogers SG, Horsch RB, Sanders PR, Flick JS, Adams SP, Bittner ML, Brand LA, Fink CL, Fry JS, Galluppi GR, Goldberg SB, Hoffmann NL, Woo SC. 1983. Expression of bacterial genes in plant cells. Proc Natl Acad Sci USA 80:4803–4807. 10.1073/pnas.80.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de Framond AJ, Barton KA, Chilton M-D. 1983. Mini-Ti: A new vector strategy for plant genetic engineering. Biotechnology (NY) 1:262–269. [Google Scholar]
- 146.Vaeck M, Reynaerts A, Hofte H, Jansens S, De Beuckeleer M, Dean C, Zabeau M, Van Montagu M, Leemans J. 1987. Transgenic plants protected from insect attack. Nature 327:33–37. [Google Scholar]
- 147.Grimsley N, Hohn T, Davies JW, Hohn B. 1987. Agrobacterium-mediated delivery of infectious maize streak virus into maize plants. Nature 325:177–179. 10.1038/325177a0. [DOI] [Google Scholar]
- 148.Feldman KA, Marks MD. 1987. Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: a non-tissue culture approach. Mol Gen Genet 208:1–7. 10.1007/BF00330414. [DOI] [Google Scholar]
- 149.Herrera-Estrella A, Van Montagu M, Wang K. 1990. A bacterial peptide acting as a plant nuclear targeting signal: the amino-terminal portion of Agrobacterium VirD2 protein directs a beta-galactosidase fusion protein into tobacco nuclei. Proc Natl Acad Sci USA 87:9534–9537. 10.1073/pnas.87.24.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Perlak FJ, Fuchs RL, Dean DA, McPherson SL, Fischhoff DA. 1991. Modification of the coding sequence enhances plant expression of insect control protein genes. Proc Natl Acad Sci USA 88:3324–3328. 10.1073/pnas.88.8.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]


