Abstract
Numerous variations of the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), including D614G, B.1.1.7 (United Kingdom), B.1.1.28 (Brazil P1, P2), CAL.20C (Southern California), B.1.351 (South Africa), B.1.617 (B.1.617.1 Kappa & Delta B.1.617.2) and B.1.1.529, have been reported worldwide. The receptor-binding domain (RBD) of the spike (S) protein is involved in virus-cell binding, where virus-neutralizing antibodies (NAbs) react. Novel variants in the S-protein could maximize viral affinity for the human angiotensin-converting enzyme 2 (ACE2) receptor and increase virus transmission. Molecular detection with false-negative results may refer to mutations in the part of the virus's genome used for virus diagnosis. Furthermore, these changes in S-protein structure alter the neutralizing ability of NAbs, resulting in a reduction in vaccine efficiency. Further information is needed to evaluate how new mutations may affect vaccine efficacy.
Keywords: SARS-CoV-2, Mutations, Variants, Disease diagnosis, COVID-19
1. Introduction
The Severe Acute Respiratory Syndrome Corona 2 (Virus SARS-CoV-2, belonging to the family Coronaviridae, appeared in 2019 Wuhan-China [1,2]. By mid-September 2022, the pandemic had confirmed nearly 614 million cases worldwide and caused around six and a half million deaths (World Health Organization). The coronavirus, SARS-CoV-2, is an enveloped, positive-stranded virus that infects vertebrates [3]. The SARS-CoV-2 genome is coronavirus's largest single-stranded RNA genome, composed of 32 kb long nucleotides with polyadenylated 3′ end and cap structure at 5′-terminal [4]. The genome sequence includes a 265 base pair (bp) leader sequence that plays an essential role in coronavirus gene expression during its discontinuous sub-genomic replication [5]. In addition, genes responsible for the production of RNA replication and transcription proteins are part of open reading frames (ORFs) called ORF1ab. The first ORF signifies around 67% of the entire genome, encoding 16 non-structural proteins (NSPs), whereas the leftover ORFs encode auxiliary and structural proteins [6].
The SARS-CoV-2 genome also has genes for structural and non-structural proteins, such as papain-like proteinase and PL proteinase, such as NSP3 [7]. The main structural proteins are nucleocapsid protein (N), the spike surface glycoprotein (S), matrix protein (M), and a small envelope protein (E). In addition, Coronaviruses spike glycoprotein has two domains, the S1 domain manages host receptor ligation, and the S2 domain is used for cell fusion [8]. Angiotensin-converting enzyme 2 (ACE2) is a host receptor that binds spike protein S1 of the virus to start attaching. ACE2, the receptor protein SARS-CoV-2, is decidedly abundant in the human epithelia of the small intestine and the lungs [9].
SARS-CoV-2 has undergone mutations that have had a major influence on pathogenesis during the COVID-19 pandemic. The most important concern about different SARS-CoV-2 variants is the risky changes that could worsen the severity of the disease or reduce the effects of vaccines [10]. In the present review, we aim to summarize the major viral mutations that produce a new variant and their effect on the infectivity of SARS-CoV-2, transmissibility, disease diagnosis, and vaccination trials.
2. Genomic variations of SARS-cov-2
Coronaviruses are characterized by unusual genetic plasticity resulting from the accumulated genetic mutations that permit the virus to evolve rapidly. The analysis of the mutation of the SARS-CoV-2 genome may explain the behavior of a virus and alterations in the transmissibility and virulence of the coronavirus. RNA viruses, mainly SARS-CoV-2, have a high-frequency mutation rate, resulting in more genetic diversity [11]. Mutations, including S glycoprotein, ORF1ab, ORF3a, nucleocapsid (N) gene, membrane (M) gene, envelope (E) protein, ORF6, ORF7a, ORF7b, ORF8, and ORF10 can influence all segments of the SARS-CoV-2 genome. Analysis of mutations related to the S glycoprotein region is essential to understanding virus antigenicity and for vaccine development or therapeutic interventions [12]. [12] studied site-specific spontaneous mutations in the S glycoprotein of the SARS-CoV-2 genome from a different isolate. The S glycoprotein mutation analysis of the SARS-CoV-2 identifies, among 11 sequences tested, 80 mutated bases at 47 positions. The most common Single Nucleotide Polymorphism (SNP) is A > G at position 1,841, resulting in a change in codon from GAU - > GGU and, consequently, a change from aspartic acid to Glycine. Jia et al. (2020) found that the mutational rate of SARS-CoV-2 was less than that for other SARS. In addition, they identified that the SARS-CoV-2 has an amino acid mutation in the RBD of the S-protein and related it to the affinity to binding to the ACE2 receptor in humans. They confirmed that the other protein-encoding genes tend to be less conserved than the S protein-encoding gene of SARS-COV-2 [13]. compared the reference SARS-CoV-2 sequence with 12,343 SARS-CoV-2 genome sequences isolated from patients in different geographic areas. Two identified 1234 mutations, S-protein614G variants, and ORF1ab 4715L, were related to high fatality rates.
Zhang et al. (2020) examined the phylogenetic relationships of 169 genomes of SARS-CoV-2 by using whole-genome alignment. They classify the SARS-CoV-2 genome into Type I and Type II, which mainly differ at three nucleotide loci which are 8750, 28,112, and 29,063. Type I genotype is divided into type IA, related to ancestor SARS-CoV-2, and Type IB, based on nucleotide at the 29,063 sites. The researchers suggested that Type I might be more associated with the ancestral human-infecting strain, while Type II has derived from mutant Type IB. The analysis of the samples collected from the Wuhan Huanan market revealed that they are more related to Type II. There were no samples from type I directly linked with the Huanan market. The high transmissibility of Type II may be explained by the presence of two variants correlated to higher translation efficiency [[5], [14]]. genotype 558 SARS-CoV-2 isolated from different countries worldwide. It measures the frequency of gene mutations encoding the S protein, RNA primase, nucleoprotein, and RNA polymerase. Depending on the influence of mutation on protein, SNP mutations are divided into four groups: which are a single mutation in nsp 6 (11083G > T), ORF3a (26144G > T), RNA polymerase (NSP8) (8782C > T, 28144 T > C), and double mutations in RNA polymerase and S-protein: (23403 A > G, 14408C > T, 3037C > T, 241C > T) of the 558 genotypes (after the exclusion of the leader sequence and the synonymous mutations). In addition to that, the leader sequence mutation (241C > T) was reported to be common and coincides with three significant mutations, 14408C > T, 3037C > T, and 23403 A > G, which are synonymous mutations in the amino acid of nsp 3. The two SNP mutations (8782C > T, 28144 T- > C) in SARS-CoV-2 isolated were the most frequent in China, the USA, and Europe [15]. Mutations at other positions (28144 T > C, 8782C > T, 18060C > T) were identified later from the strain's early phase. In European countries, SARS-CoV-2 isolates have additional essential co-mutations (241C > T, 3037C > T, 23403 A > G) and extended mutations at (3037C > T, 241C > T, 23403 A > G, 14408C > T) positions were reported. SNP mutations are disseminated among the complete genomes of SARS-CoV-2 unevenly and concentrated in the RNA primase, RNA polymerase, S protein, and nucleoprotein.
RNA-dependent RNA polymerase (NSP12, RdRp) mutant 14,408C > T (P323L) and S proteins mutant 23,403 A > G (D614G) are the dominant non-synonymous mutations of SARS-CoV-2 in Europe and the USA. These two dominant mutants in SARS-CoV-2 correlate to accumulating non-synonymous and synonymous mutations in the S and ORF1a genes [15].
Sarder, R. et al. (2020) identified the genetic mutational spectrum of SARS-CoV-2 genomes from diverse geographical areas, primarily from Italy, India, the USA, China, and Nepal. The highest cases of mutations were recognized in the Indian sequence located in NSP2, ORF1ab, NSP3, ORF8 protein, helicase, and spike surface glycoprotein. Indian SARS-CoV-2 genome has a specific mutation in the spike surface glycoprotein (A930V (24351C > T), which is absent in other SARS-CoV-2 genomes of different locations. A sharp increase in the reported cases of deaths in Italy correlates to unique mutations reported in ORF1ab, ORF3a, ORF1a, and NSP6 [16].
D614G mutation is a non-synonymous dominant mutation in the SARS-CoV-2 spike protein. D614G is connected to conformational SARS-CoV-2 spike protein and may be combined with other mutations leading to increases in virus infectivity [[17], [18]]. Moreover, mutation Spike D614G may have first been identified in China or Europe and spread rapidly in Europe and other countries. In addition to the high frequency of the D614G mutation, it is accompanied by two different (C-to-T) mutations, one silent in the NSP3 gene and another in an RNA-dependent RNA polymerase amino acid (RdRp P323L). L8V is a signal peptide mutation with limited presence in a single lineage in Hong Kong and once in Canada. On the other hand, the L5F mutation is distributed in different geographic areas, and the G476S mutation frequently occurs in the binding surface of the RBD in the USA [19].
Ou, J. et al. (2020) analyzed 1609 SARS-CoV-2 complete genome sequences from various global locations, including China, Finland, the UK, France, Belgium, the USA, and India. W436R, V367F, and D364Y mutants have appeared in Shenzhen, Wuhan, and France, whereas RBD mutations V367F revealed high frequency in France and Hong Kong. Still, V483A and G476S were reported frequently in USA samples [20].
[21] characterized diverse mutations in 11 viruses isolated from some patients admitted to Hangzhou hospitals in China. Collectively 33 mutations were recognized. The newly discovered mutation was 19 in comparison to genomic sequences available at GISAID. A22301C and T22303G are new missense mutations that lead to the same effect at the protein level (S247R in the S-protein). Two mutations out of the defined four mutations in the ORF7b gene that affect protein structure are identified [21].
In December 2020, a new variant was reported in the UK as SARS-CoV-2 variants of concern (VOC) 202,012/01, also named B.1.1.7 (Alpha) [22]. The new variant, which was detected during regular sampling, shows several mutations in the RBD that were never reported before and started to replace other virus lineages in the area. There are 23 mutations in the B.1.1.7 variant, 14 of which are non-synonymous mutations: T1001I, I2230T, and A1708D in ORF1ab (open reading frame); Q27stop, Y73C, and R52I in ORF8; N501Y, P681H, A570D, T716I, D1118H, and S982A in spike protein; and S235F and D3L in nucleocapsid protein. Synonymous mutations are: Y144del and H69–V70del in spike protein; T26801C in membrane gene; C14676T, C5986T, C913T, T16176C, and C15279T in ORF1ab; and three are deletions: Y144del and H69/V70del in spike protein; and SGF 3675-3677del in ORF1ab. Apart from synonymous mutations, the gene encoding for the spike protein containing the RBD accounts for 47% of recorded variants in B.1.1.7 [23]. As such, the N501Y mutation of the B.1.1.7 variant that causes an amino acid transition from asparagine to tyrosine, in combination with spike deletion 69-70del and P681H, has the greatest possible biological impact [24]. At the same time, a different SARS-CoV-2 variant (20C/501Y·V2 or B.1.351 (Beta) lineage) developed independently of the B.1.1.7 lineage in South Africa. B.1.351 variant (also known as 20H/501Y·V2) is characterized by nine variations in spike protein related to the Wuhan-1 D614G spike mutant previously identified in South Africa. These mutations include N501Y, which represents the common mutation among the new UK and South African variants [25].
The virus transmission rates in Los Angeles (LA) and California have been among the highest rates worldwide, coinciding with the high positivity rate in this region. From the beginning of November 2020, an S-protein L452R mutation of the RBD region has been identified in Southern California as CAL.20C strain (B.1.427/429) that shares multiple mutations with UK and South African strains [26].
Brazil experienced high infection levels of SARS-CoV-2 in early 2021. The appearance of the new SARS-CoV-2, lineage P.1 (Gamma variant), had ten mutations in the spike protein (E484K, L18F, T20 N, K417T, P26S, D138Y, N501Y, R190S, H655Y, and T1027I V1176), were identified in Manaus virus's samples in addition to three RBD mutations (E484K, K417 N, and L18F) [27,28].
The B.1.617 lineage has been studied in India since the beginning of 2021; it was initially discovered in early 2021 in Maharashtra and spread all over India [[29], [30]]. B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.617.3. The L452R, D614G, and P681R spike protein mutations are three sub-lineages as common mutations [28,31]. L452R mutation has previously been linked to enhanced infectivity and slightly reduced susceptibility to neutralizing antibodies. Wuhan-Hu-1 D614G compared to B.1.617.2, has spike mutations T19R, E156G, R158del, L452R, D614G, and D950 N [30]. The E484Q, L452R, and P614R are critical mutations in the Delta variant that simplify the binding of the virus spike to ACE-2 receptors [32]. Delta Plus (B.1.617.2.1/(AY.1) is an Indian Delta variant with an additional K417 N mutation in Spike protein [32]. According to the Public Health England bulletin, the novel mutation was found in six genomes from India [33].
The CDC (USA) and WHO have categorized variants of a new SARS-CoV-2 as either variants of concern (VOC) or variants of interest (VOI) [34]. A new SARS-CoV-2 VOC reported on the November 25, 2021 was the Omicron B.1.1.529 variant, which emerged when vaccine immunity increased in different countries. The polymerase chain reaction (PCR) assay false negative result was associated with a 69–70del deletion mutation in the S-gene target. In addition to some deletion mutations, Omicron has many mutations, many of them (e.g., K417 N, T95I, G142D/143–145del, N655Y, T478K, N501Y, P681H, and N679K) cross-over with those in the beta, alpha, Delta or gamma VOC [35].
Among all SARS-CoV-2 variants studied Omicron possesses the most significant number of mutation sites. In addition to two deletions, amino acid 2083 and amino acid 3674–3676, six substitution mutations are identified within ORF1a (A2710T, T3255I, P3395H, K856R, L2084I, and I3758V). Moreover, the variant has two substitutions (I1566V and P314L) in ORF1b, and a P10S substitution accompanied by three residue deletions was reported in ORF9b. Structural protein mutations are distributed between the envelope with one substitution (T9I), three substitutions (D3G, Q19E, and A63T) in the membrane, and three residue omissions, respectively, in the nucleocapsid proteins. More than 5% of the total Omicron mutations described are found in the spike protein. Spike protein mutations consist of 30 substitutions of A67V, Y145D, S371L, L212I, S373P, K417 N, S375F, S477 N, G446S, N440K, E484A, T478K, G496S, Q498R, Y505H, Q493R, N501Y, H655Y, T95I, G339D N679K, P681H, D614G, T547K, N764K, Q954H, D796Y, N969K, L981F, N856K, and three deletions of V143/G142/Y144, H69/V70, and N211, besides one insertion of 3 amino acids (EPE) at position 214. The amino acid alteration D614G represents the shared spike protein mutation in all five VOC mutations [36].
Recently the Omicron variant has been divided into distinct sub-variants, BA.1, BA.2, BA.1.1, BA.3. BA.4, and BA.5 [37]. The twenty spike mutations (S375F, G142D, G339D, S373P, N440K, S477 N, K417 N, T478K, Q498R, D614G, E484A, N501Y, Y505H, N764K, H655Y, N679K, P681H, Q954H, D796Y, and N969K) are found in all five sub-varients [38]. Subvariant BA.1 (B.1.1.529.1) makes up the dominant circulating omicron variant, while subvariant BA.2 is predominant in India, Denmark, and the Philippines [39]. The BA.1 and BA.2 subvariants have 12 common substitution mutations out of 16. The BA.2 is distinguished from BA.1, which represents the original omicron strain, by the presence of 4 substitution mutations (D405 N, S371F, R408S, and T376A) in the RBD region of spike protein [39]. The BA.3 subvariant has more than 90% of the BA.1 spike protein mutations in addition to two similar mutations in BA.2. subvariant [40].
According to genome sequencing analysis, the nucleotide substitution rate of around 1 × 10−3 substitutions per year resulted in variants via point mutations, additions, recombination, and omissions [41]. An analysis study based on 48,635 samples confirmed that SARS-CoV-2 has an average number of 7.23 mutations for every viral sample compared to the reference SARS-CoV-2 genome sequences [42]. Compared with SARS-CoV-2, other RNA viruses have low sequence variation with mutation hot spots in the viral genome [43]. Compared to SARS-CoV or MERS-CoV, the SARS-CoV-2 genome is much more stable, with a comparatively high dynamic mutation rate compared to other RNA viruses [44]. SARS-COV-2 is well preserved, particularly “in the E, 7 b, six areas with hotspot mutations in ORFs 1a, 8, S, and the N region. According to Ref. [45] study, the primary locus of amino or acid nucleotide variation in SARS-CoV-2 isolate strains was in open-reading frames eight and 1a.
3. The impact of genetic variation on the SARS-cov-2 genome
Mutational analysis of SARS-CoV-2 genomes gives an excellent opportunity to understand how different genes undergo frequent mutations that may affect the viral features and disease manifestation. Transmissibility of the SARS-CoV-2 virus, one of the virus features, is affected by genetic variation in the virus genome. Zhang et al. (2020) stated that the dominance of the Type II SARS-CoV-2 virus is correlated to high translational efficiencies due to two synonymous mutations that facilitate the production of new virus particles. Moreover, these genetic variants in the Type II SARS-CoV-2 virus provide high transmissibility and lead to a virus outbreak. Leader sequence mutation (241C > T) with co-mutations (14408C > T, 3037C > T, 23403 A > G) is another synonymous mutation that increases virus rigorousness and transmissibility of infection in Europe [5].
The spike glycoprotein of SARS-CoV-2, which mediates the ligation of the virus to the host cell membrane, is accounted for by the high mutation frequency. D614G (23403 A > G) is a spike protein mutation targeting a region where S1/S2 junction area. D614G mutation was recorded in March 2020 and then spread rapidly by April and May in high frequency (70%). D614G is correlated to enhance transmission of the virus and increased virus infectivity due to low S1 domain shedding accompanied by high S-protein integration into the virion [46].
The spike protein receptor binding domain of SARS-CoV-2, mainly S1, is attached directly to human cell receptors ACE2. Mutations in the RBD region that alter protein conformation and enhance base rigidity affect the binding nature and stability of the ACE2 receptor [47]. Stabilization of the RBD beta-sheet scaffold was reported in three mutants, V367F, W436R, and D364Y, of SARS-CoV-2. The V367F mutant is primarily correlated to the high binding affinity of the virus to ACE2 receptors and the increases in virus infectivity and morbidity [20].
Mutation in the RdRp region, mainly 14408C > T mutation, causes a switch of proline amino acid to leucine, leading to an increase in the rigidity of the RdRp protein structure concurrent with increases in the mutation rate and evolution of SARS-CoV-2 genome [11]. RdRp mutant 14408C > T was significantly correlated to the strictly affected group compared to the mildly affected group [48].
Variability of the SARS-CoV-2 genome that resulted from different mutations in their sequence affects the pathogenicity of the disease differently. The functional impact of specific mutations and how they change the pathogenicity of SARS-CoV-2 was reported [21]. studied various mutations in 11 virus-isolated cases admitted to Zhejiang hospitals in China and the pathogenic effects of these mutations. After infecting Vero-E6 cells with 11 viral isolates, one isolate (ZJU-1), which clusters with the A23403G mutation (D614G in S protein), has a statistically significant higher viral load than other viral isolates at 48 h post-infection. Moreover, a greater viral load that may lead to a much higher cell death ratio correlates to low Cycle threshold (Ct) values in PCR results [21].
The new variant of SARS-CoV-2 in Britain acquires more than a dozen mutations, leading to changes in spike protein amino. The spike protein mutations, mainly N501Y mutant, alter the amino acid within the receptor-binding domain and increase protein tightness necessary to the ACE2 receptor. Furthermore, the CAL.20C strain of southern California with L452R mutation could enable easier binding of the SARS-CoV-2 virus to the cell receptor. The CAL.20C strain and disease severity remains unclear until now [26]. This alteration in virus protein structure influences the virus's transmissibility in humans and may lead to a higher viral load [25,49,50]. SARS-CoV-2 variant lineage P.1, which emerged in Brazil, correlated with enhanced virus binding to the human ACE2 receptor. Moreover, P.1 lineage increases virus transmissibility around 1.4–2.2 times compared to non-P.1 lineage accompanied by increases in immune invasion [27].
The dynamics, molecular interactions, and binding energy of the B.1.1.7 variant (spike N501Y mutant), B.1.617 variant (E484Q and L452R spike mutant), and wild-type (WT) were investigated recently in India using molecular dynamics simulations of three diverse spike-ACE2 complexes. The results revealed that mutation enhances binding energy amid the spike and human ACE2. The B.1.617 lineage, L452R, and E484Q mutations improved the intra-chain interactions and steadiness in the spike protein, which may affect the ability to interact with human antibodies with this spike variant. Moreover, variant B.1.1.7 has higher interaction of hydrogen with LYS353 of human ACE2 and a different mandatory affinity than the WT. The high transmissivity and infectivity of the virus mutants form may be explained by the amplified structural constancy of its spike protein and human ACE2 affinity [51].
Compared to B.1.617.1 (Kappa) variant, B.1.617.2 (Delta) variant showed greater replication and spike-mediated entry, which might explain its dominance [[30], [31]]. Delta variant blowouts are almost two times faster than the original SARS-CoV-2 virus. Moreover, this mutation was formerly responsible for amplified infection and an immune escape compared to the previous conditions. Compared to wild-type Wuhan-1 D614G, the Delta variant is around six folds less susceptible to neutralizing antibodies in sera from recuperated persons and nearly eight folds lower sensitivity to vaccine-produced antibodies [[31], [52]]. As such, the Delta variant has specific significant mutations in the spike protein of the virus, such as the E484Q, L452R, and P614R, which may enhance the ACE2 binding, whereas P681R mutation in the furin cleavage location might improve the rate of S1–S2 cleavage, leading to enhance virus transmissibility [53]. The Delta variant revealed greater reproduction efficacy in the human airway and airway organoid epithelial systems [54]. Remarkably, the escaping ability of the Delta variant from natural immunity was associated with increased transmissibility with greater viral load, high rates of re-infection, and longer time of infectiousness [55].
The genome of the Omicron variant has more than 30 mutations in the spike protein, which directly interacts with human cells before cell access. Omicron has some deletions and over 30 mutations associated with evolved viral binding affinity, enhanced antibody emission, and augmented transmissibility due to the mutations close to the furin cleavage site [55]. Omicron variant E484K in virus spike protein modified the conformation of the site that interacts with class 2 antibodies, making them less effective. According to preliminary findings, the Omicron variant has a greater risk of re-infection than the other variants. In South Africa, hospitalization rates are increased, and fewer people who needed oxygen support were accompanied by fewer severe symptoms than in the previous variants [56]. Due to the increased cases, Omicron is expected to infect 3–6 times as many individuals as Delta during the same time. However, recent information reveals that Omicron VOC is infectious, dispersing quicker than other variants. Additionally, Omicron infects people resistant to different variants and shows fewer symptoms than other coronavirus strains [57].
Among Omicron subvariants, BA.2 has higher transmissibility than other subvariants and becomes the dominant strain. The spike protein mutation of the Omicron BA.2 subvariant found in RBD and N-terminal domains is correlated to high binding strength to ACE2 receptors and high virus transmissibility [58]. The Omicron BA.2 subvariant mutations are associated with unsusceptibility to the neutralizing effect of monoclonal antibodies [59].
The ongoing evolution of SARS-CoV-2 variants from time to time has worsened the COVID-19 pandemic by nearly 2–5% mortality. Developing a new variant of SARS-CoV-2, such as VOC, that spreads worldwide causes different severity levels of the disease [60]. Islam et al. (2023) filtrate 16,954 distinct and widespread mutations out of 10,531 sequences for 12 strains to identify more deleterious mutations in the variants. Omicron variants reveal the highest mutation level pattern, followed by the Delta variant, which has more deleterious mutations than the Delta variant. L916S and V915S are spike protein mutations in omicron variants that decrease the likelihood of binding with T- and B- cells, reduce affinity, and improve molecular flexibility.
There are few studies examining the relationship between viral mutation and disease severity [61]. reviewed studies in PubMed, Google Scholar, and Web of Science that were published in English. They reported that the D614G mutation was significantly correlated with elevated fatality rates in a number of countries. Moreover, countries with an elevated fatality rate reported a significantly high frequency of the variant. As a result, the VOC appears to become more lethal and transmissible.
A cohort study linked the differences in infection mortality rates with the Alpha variant of SARS-CoV-2. A strong correlation was found between the participants who tested positive for SARS-CoV-2 and the PCR test results variant for Alpha for the period from Oct-01-2020 and Jan-29-2021, followed up until Feb-12- 2021. A high mortality hazard ratio reached 1.32 and 2.04, with 32% and 104% more susceptible to dying than comparable individuals infected with previous variants [62]. Another study used patients who tested positive for SARS-CoV-2 in England from Nov-16-2020 to Feb-05-2021 to estimate the disease severity of the VOC (B.1.1.7) compared to the non-VOC. A high risk of death with a hazard ratio of 1.67 (95% confidence interval (CI): 1.34–2.09; p < 0.0001) was recorded for patients with VOC with an estimated risk of dying within 28 days of a positive SARS-CoV-2 test, accompanied by a 35% mortality rate. Moreover, the Alpha variant can increase the mortality rate from 30 to 50% [63].
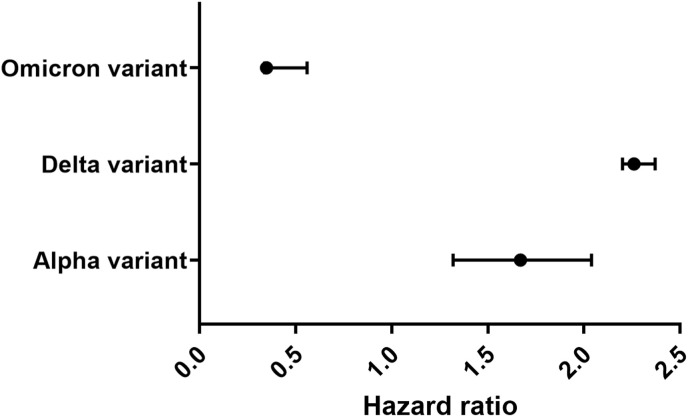
Compared to the preceding circulating variants, the mortality hazard ratio correlated with infection with the Delta variant was 2.26 (95% CI: 1.32–3.89) compared to Alpha variant infections in England [64]. Furthermore, the odd ratio for mortality of the Delta variant is 2.37 (95% CI: 1.50–3.30), with a 133% elevated mortality risk. Another study compared the Omicron variant with the Delta one, reducing the hazard ratio to 0.35 [65]. [66] compared the COVID-19 mortality risk to patients diagnosed with Omicron BA.1 or Delta variants. They found a 66% (95% CI: 54%–75%) reduction in the death risk of Omicron BA.1 compared to Delta variants. Moreover, Omicron BA.1 patients had a hazard ratio of 0.34 (95% CI: 0.25 to 0.46) for Omicron BA.1 infection compared to the delta infection model strongly associated with variation in mortality risk as a result of the age factor. These results are consistent with another study by Ref. [67] to estimate a hazard ratio for Omicron BA.1 patients in hospital attendance compared to Delta ones [67]. found that the hazard ratio of Omicron hospital attendance patients was 0·56 (95% CI 0·54–0·58) compared with delta [68]. After comparing different SARS-CoV-2 variants, the Omicron variant has a noticeable reduction in hazard ratio compared to other variants (Fig. 1 ).
Fig. 1.
Hazard ratio for SARS-CoV-2 death for Omicron BA.1 infection compared to Alpha and Delta infections.
3.1. The impact of genetic variation on SARS-CoV-2 diagnosis
The genomic diversity of SARS-CoV-2 could influence the performance of molecular tests used for virus diagnosis. The real-time reverse transcriptase-polymerase chain reaction (RT-PCR) test is a molecular tool that uses primer-probe designed to bind specifically to one of the highly conserved regions of structural genes (envelope, nucleocapsid, and RdRp) of SARS-CoV-2 [69]. Various diagnostic kits of RT-PCR have been developed and used as reliable tests to detect diverse nucleotide sequences of SARS-CoV-2 [70]. Viral RNA in SARS-CoV-2 patients measured by the cycle threshold (Ct), the number of replication cycles needed to generate a fluorescent signal, considered PCR positive with a Ct value lower than 40. The Ct value required to detect the virus's presence is proportional to the patient's viral load [71]. A particular assay kit with different primer-probe sets used in the University of Washington (in the Clinical Virology Lab) for SARS-CoV-2 revealed high reliability without cross-reactivity with other respiratory infections. The most heightened sensitivity of assay kits was achieved by those assays that used the set of E-gene primer-probe [72].
A study in South America used different probe/primer sequences to identify mutations in more than one gene frequently used to identify SARS-CoV-2. Several Single nucleotide polymorphisms (SNPs) in RdRp and N genes have been reported in the scrutinized genomes among several South American SARS-CoV-2 sequences able to alter the physicochemical features of the PCR assays. The sensitivities of PCR tests using different target genes are comparable with lower sensitivity for the RdRp-SARS-CoV-2 (Charité) primer probe [71]. Genetic variation in these two regions of the viral genome widely used for viral identification negatively affects diagnosis tests and leads to false-negative results. On the other hand, conserved envelope (E) gene sequences of more than 300 South American genomes were studied to provide a good choice for molecular diagnosis of SARS-CoV-2 in this region [73].
One control strategy for SARS-CoV-2 worldwide spreading is testing for the presence of the virus, which may result in false-negative outcomes with detection virus molecular tests. False-negative results may result from a mutation in the part of the virus genome used by the test. In the USA, the Food and Drug Administration revealed that false-negative results were detected in different molecular tests to detect SARS-CoV-2 if a mutation occurred in the viral genome part used by that test [74]. In addition, PCR tests that depend on identifying more than one part of the genome may be less affected by genetic variation in the SARS-CoV-2 genome than tests that rely on identifying a single genome part.
Molecular-based testing methods for SARS-CoV-2 recommend using multiple genetic targets to detect the presence of different genetic variants in the viral genome. For example, a new SARS-CoV-2 variant B.1.1.7 recognized in England possesses several mutations in different parts of the viral genome. Spike gene mutations have significantly reduced RT-PCR assay sensitivity and led to false-negative results [75]. In addition, the 69/70 del mutation, which occurs in the spike gene of the B.1.1.7 variant, has increased the percentage of diagnostic failure.
RT-PCR assays designed to identify multiple SARS-CoV-2 genetic targets, such as cobas (Roche) SARS-CoV-2 assay, are probably less affected by the augmented occurrence of genetic variants [76]. studied different quantitative qRT-PCR assays that were improved to investigate SARS-CoV-2 by identifying various positions in the viral genome. False-negative results of the cobas SARS-CoV-2 E gene were correlated to a C-to-U transition of the genome SARS-CoV-2. It should be emphasized that although the E gene qRT-PCR failed in the patients, the cobas analysis appropriately called these people progressive for SARS-CoV-2 because of ORF1ab qRT-PCR while the same mutation using other E gene qRT-PCR assay could not impair the assay's effectiveness [77]. [78] investigated some variants belonging to the binding sites of primers and probes that occur at a lower frequency and involve only one base. The exception includes a GGG to AAC mutation at genome positions 28,881–28883, which overlaps with the first three CDC N gene bases forward primer 5′ end [78]. This highlights the importance of targeting multiple positions of viral genomes in diagnostic assays.
The sequence variation at the primer binding sites may be one parameter leading to false negatives. The false negativity in RT-PCR tests may result from a high virus mutation rate. However, as recorded, the possibility of this probe strains and variation with mutations in two targets affecting an assay's sensitivity is low. Mutations, as such, at the 3′ ends of the primer areas are more expected to affect assay sensitivity [79].
The Omicron variant has about 50 mutations, most of which are in the spike proteins, which leads to an S-gene target failure [80]. This led to increased false-negative RT-PCR test results due to a 69–70del mutation, mainly in the PCR tests with S-gene target failure. The efficacy of several in vitro diagnostic tests is put at risk due to changes in the protein and viral nucleic acid sequences. If virus mutation occurs in a particular region critical for a primer in RT-PCR, a mismatch in primer binding areas will lead to negative results (Himanshu et al., 2021).
4. The impact of genetic variation on SARS-cov-2 vaccination
SARS-CoV-2 uses angiotensin-converting enzyme type 2 as a receptor to access human cells through its surface spike protein. The expression level of ACE2 at the lung alveolar epithelial cells surface is related to the susceptibility to the infection of SARS-CoV-2. Moreover, genetic polymorphism of the ACE2 gene and human ACE2 protein could affect the vulnerability to SARS-CoV-2 contagion and the consequences of the disease [81]. Significantly, the S-protein of the SARS-CoV-2 surface, mainly the S1 subunit, comprises a receptor-binding domain required for virus-cell binding and mediates the production of virus-neutralizing antibodies [82]. Vaccination trials induce virus-neutralizing antibodies to block the virus from binding to ACE2 receptors. Genetic variation in S-protein is a significant problem that impairs virus-neutralizing antibodies (NAbs) binding sites [83]. The virus developed various mutations under antibodies that patients produced to restrict virus replication as evolutionary escape mechanisms. Fortunately, genetic variants located in the S-protein region of either D614G or B.1.1.7 variants are detected by NAbs [83]. B.1.1.7 variants with or without N501Y change are equally neutralized by serum samples from Moderna mRNA and Pfizer-BioNTech vaccines [84].
The CAL.20C, southern California variant, with S-protein L452R mutation located within RBD, decreased sensitivity to definite counterbalancing monoclonal antibodies [26]. As such, the effect of the strain on the infection and disease severity needs to be determined. Moreover, the 501Y·V2 lineage that emerged during the second half of 2020 in South Africa is resistant to neutralizing antibodies were detected in nearly 50% of the patients with PCR-confirmed SARS-CoV-2 disease [85]. A laboratory study at Rockefeller University states that the proper sequence of N501Y·V2 fluctuates within the RBD, humbly limiting the effectiveness of mRNA vaccine-induced antibodies neutralizing test viruses. This coincident with neutralizing antibodies produced by strain. This reduction in vaccine effectiveness may be explained by SARS-CoV-2 vaccines being developed grounded on “immune responses to the original spike protein of the virus [45].
The Omicron variant's mutation is concentrated in the spike protein, representing the primary target for current vaccines [55]. The SARS-CoV-2 vaccines that Pfizer-BioNTech developed, Moderna, Janssen, and AstraZeneca, are created on a spike (S) glycoprotein version and vary in efficacy against SARS-CoV-2.
A study by Ref. [86] in England was designed to evaluate vaccine efficacy against symptomatic disease, which is triggered by the Delta (B.1.617.2) and Omicron variants, after being immunized with two dosages of BioNTech– Pfizer (BNT162b2), AstraZeneca (ChAdOx1 nCoV-19), or Moderna (mRNA-1273) vaccine after a supporter dosage of BNT162b2, or ChAdOx1 nCoV-1. The vaccine's efficacy counter to symptomatic disease tended to be lesser for the Omicron than for the Delta variant. After the two doses (ChAdOx1 nCoV-19), no response for the omicron variant was reported after 20 weeks. In contrast, vaccine efficacy after two doses of BNT162b2 was 65.5% at two to four weeks, declining to 8.8% (95% CI, 7.0 to 10.5) after 25 weeks or more [86]. stated that primary immunization with two BNT162b2 or ChAdOx1 nCoV-19 doses provided a restricted defense counter to symptomatic disease resulting from the omicron. Booster doses led to substantial enhancement of protection counter to minor contagion; nonetheless, a reduction in defense against the symptomatic disease was also noticed after booster doses. A study in the United Kingdom on the Delta variant explained that good protection against hospitalization was effectively maintained after two doses of the vaccine (Andrews et a., 2022).
Appropriate titers of neutralizing antibodies (NAb) levels are considered a good indication of resistance against the SARS-CoV-2 infection. Neutralizing activity in serum samples taken from persons who received two dosages of BNT162b2 is reduced by a factor of 20–40 when compared to the neutralizing counter to early pandemic virus and by a factor of 10 at least when matched with neutralization counter to the Delta variant, according to studies from South Africa, Germany, and the United Kingdom [87]. In addition, studies explained that vaccine boosters might be required to enhance neutralization titers countering SARS-CoV-2 variants [[88], [89]].
Mutations in Omicron BA.2 spike proteins produce different sequences of amino acids that lead to a high antibody immune response. Omicron BA.1 and BA.2 reveal solid immune responses to antibodies resulting from vaccination by ChAdOx1 and mRNA vaccines [37]. The vaccine's efficacy against severe coronavirus disease was low for Omicron variants compared to previous SARS-CoV-2 variants after one month of vaccination. For the Omicron variant, the vaccine's effectiveness declined rapidly from the first to the sixth months after the initial vaccine series was finished [90]. Moreover, the Omicron BA.2 subvariant is correlated with a high risk of infection for unvaccinated individuals, those who have received booster shots, and completely vaccinated individuals [59].
Evans et al. studied immune responses against Omicron sub-variants (BA.1, BA.2, and BA.1.1) in individuals who received the mRNA vaccine, boosted people, and cured COVID-19 [91]. The results revealed that all Omicron sub-variants, especially BA.1.1 and BA.1, show significant immune resistance, mainly prevented by vaccine boosters. Further research has been performed worldwide to understand better how different SARS-CoV-2 variants are affected by these other vaccines.
Ethics approval and consent to participate
N/A.
Human and animal rights
N/A.
Consent for publication
N/A.
Availability of Data and materials
Data will be available upon request via e-mail from the corresponding author.
Funding
None.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Declared none.
References
- 1.Hemida M.G., Abduallah M.M.B. One Health; 2020. The SARS-CoV-2 outbreak from a one health perspective. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W.…Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandes R.S., Freire M.C., Bueno R.V., Godoy A.S., Gil L.H., Oliva G. Reporter replicons for antiviral drug discovery against positive single-stranded RNA viruses. Viruses. 2020;12(6):598. doi: 10.3390/v12060598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E.…Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;112(5):3588–3596. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alanagreh L.A., Alzoughool F., Atoum M. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9(5):331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu A., Peng Y., Huang B., Ding X., Wang X.…Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host & Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Li X., Li T., Zhang S., Wang L., Wu X., Liu J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur J Clin Microbiol Infect Dis. 2020;39(9):1629–1635. doi: 10.1007/s10096-020-03899-4. https://link.springer.com/article/10.1007/s10096-020-03899-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J. Pathogenicity and transmissibility of 2019-nCoV—a quick overview and comparison with other emerging viruses. Microb Infect. 2020;22(2):69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokhansanj B.A., Rosen G.L. Predicting COVID-19 disease severity from SARS-CoV-2 spike protein sequence by mixed effects machine learning. Comput Biol Med. 2022;149 doi: 10.1016/j.compbiomed.2022.105969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eskier D., Karakülah G., Suner A., Oktay Y. RdRp mutations are associated with SARS-CoV-2 genome evolution. PeerJ. 2020;8 doi: 10.7717/peerj.9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee Sidharta. June 21, 2020. An Overview of Mutations Occurring within the Coronavirus-2 Genome: Mutations Data Reporting on SARS-CoV-2.https://ssrn.com/abstract=3632241 Available at SSRN: [DOI] [Google Scholar]
- 13.Toyoshima Y., Nemoto K., Matsumoto S., Nakamura Y., Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J Hum Genet. 2020:1–8. doi: 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S.…Izard T., et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 doi: 10.1101/2020.06.12.148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eskier D., Suner A., Karakülah G., Oktay Y. Mutation density changes in SARS-CoV-2 are related to the pandemic stage but to a lesser extent in the dominant strain with mutations in spike and RdRp. PeerJ. 2020;8 doi: 10.7717/peerj.9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sardar R., Satish D., Birla S., Gupta D. BioRxiv; 2020. Comparative analyses of SAR-CoV2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host-virus interaction and pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S.…Qin H. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J., He C.L., Gao Q., Zhang G.J., Cao X.X., Long Q.X., Tang N. 2020. The D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity. [BioRxiv] [Google Scholar]
- 19.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W.…Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ou J., Zhou Z., Dai R., Zhang J., Lan W., Zhao S.…Zhanga Q. 2020. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein 2. [DOI] [Google Scholar]
- 21.Yao H., Lu X., Chen Q., Xu K., Chen Y., Cheng L.…Li L. 2020. Patient-derived mutations impact pathogenicity of SARS-CoV-2.MedRxiv.https://www.medrxiv.org/content/10.1101/2020.04.14.20060160.abstract [Google Scholar]
- 22.Carrazco-Montalvo A., Bruno A., de Mora D., Olmedo M., Garces J., Paez M.…Patino L. First report of SARS-CoV-2 lineage B. 1.1. 7 (alpha variant) in Ecuador, january 2021. Infect Drug Resist. 2021;14:5183. doi: 10.2147/IDR.S319439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gómez C.E., Perdiguero B., Esteban M. Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccines. 2021;9(3):243. doi: 10.3390/vaccines9030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitz A., Weber A., Bayin M., Breuers S., Famulok M., Mayer G. A SARS-CoV-2 spike binding DNA aptamer that inhibits pseudovirus infection in vitro by an RBD independent mechanism. Angew Chem Weinheim Bergstr Ger. 2021;133(18):10367–10373. doi: 10.1002/ange.202100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahase E. Covid-19: what have we learnt about the new variant in the UK? 2020. BMJ. 2020;371:m4944. doi: 10.1136/bmj.m4944. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W., Davis B.D., Chen S.S., Martinez J.M.S., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA. 2021;325(13):1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S.…Sabino E.C. 2021. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil.medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi J.Y., Smith D.M. SARS-CoV-2 variants of concern. Yonsei Med J. 2021;62(11):961. doi: 10.3349/ymj.2021.62.11.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez J.C. 2021. The India mutations and B. 1.617.https://www.preprints.org/manuscript/202104.0689 Variant: Is There a Global" Strategy" for Mutations and Evolution of Variants of The SARS-CoV-2 Genome? [Google Scholar]
- 30.Mlcochova P., Kemp S., Dhar M.S., Papa G., Meng B., Mishra S.…Datir R. 2021. SARS-CoV-2 B. 1.617. 2 Delta variant emergence, replication and sensitivity to neutralising antibodies. bioRxiv. biorxiv. org/content/10.1101/2021.05, 8, vol. 3. [Google Scholar]
- 31.Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.…Gupta R.K. SARS-CoV-2 B. 1.617. 2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. https://www.nature.com/articles/s41586-021-03944-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexandar S., Ravisankar M., Kumar R.S., Jakkan K. A comprehensive review on Covid-19 Delta variant. Int J Pharmaceut Chem Res. 2021;5(83–85) [Google Scholar]
- 33.Richi F.T., Alam S., Ahmed F., Al-Hossain A.M. The outbreak of Delta Plus variant: the notorious and novel strain of SARS-CoV-2. Clinical Epidemiology and Global Health. 2022;14 doi: 10.1016/j.cegh.2022.100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborty C., Sharma A.R., Bhattacharya M., Agoramoorthy G., Lee S.S. Evolution, mode of transmission, and mutational landscape of newly emerging SARS-CoV-2 variants. mBio. 2021;12(4) doi: 10.1128/mBio.01140-21. e01140-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398(10317):2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He X., Hong W., Pan X., Lu G., Wei X. SARS‐CoV‐2 Omicron variant: characteristics and prevention. MedComm. 2021;2(4):838–845. doi: 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhawan M., Saied A.A., Mitra S., Alhumaydhi F.A., Emran T.B., Wilairatana P. Biomedicine & Pharmacotherapy; 2022. Omicron variant (B. 1.1. 529) and its sublineages: what do we know so far amid the emergence of recombinant variants of SARS-CoV-2? [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y., Zhi H., Teng Y. The outbreak of SARS‐CoV‐2 Omicron lineages, immune escape and vaccine effectivity. J Med Virol. 2023;95(1) doi: 10.1002/jmv.28138. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M.…Kawaoka Y. Efficacy of antiviral agents against the SARS-CoV-2 omicron subvariant BA. 2. N Engl J Med. 2022;386(15):1475–1477. doi: 10.1056/NEJMc2201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahase E. Covid-19: what do we know about omicron sublineages? BMJ. 2022:376. doi: 10.1136/bmj.o358. [DOI] [PubMed] [Google Scholar]
- 41.Ciotti M., Ciccozzi M., Pieri M., Bernardini S. The COVID-19 pandemic: viral variants and vaccine efficacy. Crit Rev Clin Lab Sci. 2022;59(1):66–75. doi: 10.1080/10408363.2021.1979462. [DOI] [PubMed] [Google Scholar]
- 42.Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeh T.Y., Contreras G.P. Full vaccination against COVID-19 suppresses SARS-CoV-2 delta variant and spike gene mutation frequencies and generates purifying selection pressure. medRxiv. 2021 doi: 10.1101/2021.08.08.21261768. [DOI] [Google Scholar]
- 44.Bakhshandeh B., Jahanafrooz Z., Abbasi A., Goli M.B., Sadeghi M., Mottaqi M.S., Zamani M. Mutations in SARS-CoV-2; Consequences in structure, function, and pathogenicity of the virus. Microb Pathog. 2021;154 doi: 10.1016/j.micpath.2021.104831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y.…Ho D.D. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. Nature. 2021:1–6. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L., Yang J.R., Zhang Z., Lin Z. Medrxiv; 2020. Genomic variations of SARS-CoV-2 suggest multiple outbreak sources of transmission. [Google Scholar]
- 47.Jia Y., Shen G., Nguyen S., Zhang Y., Huang K.S.…Ho H.Y., et al. Analysis of the mutation dynamics of SARS-CoV-2 reveals the spread history and emergence of RBD mutant with lower ACE2 binding affinity. BioRxiv. 2021;2020-04 doi: 10.1101/2020.04.09.034942. [DOI] [Google Scholar]
- 48.Biswas S.K., Mudi S.R. 2020. RNA-dependent RNA polymerase and spike protein mutant variants of SARS-CoV-2 predominate in severely affected COVID-19 patients. [Google Scholar]
- 49.Tang J.W., Tambyah P.A., Hui D.S. Emergence of a new SARS-CoV-2 variant in the UK. J Infect. 2021;82(4):e27–e28. doi: 10.1016/j.jinf.2020.12.024. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wise J. Vol. 371. 2020. p. m4857. (Covid-19: new coronavirus variant is identified in UK). [DOI] [PubMed] [Google Scholar]
- 51.Kumar V., Singh J., Hasnain S.E., Sundar D. Possible link between higher transmissibility of alpha, kappa and delta variants of SARS-CoV-2 and increased structural stability of its spike protein and hACE2 affinity. Int J Mol Sci. 2021;22(17):9131. doi: 10.3390/ijms22179131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sapoval N., Mahmoud M., Jochum M., Liu Y., Elworth R.L., Wang Q.…Treangen T.J. Genome Research; 2021. Hidden genomic diversity of SARS-CoV-2: implications for qRT-PCR diagnostics and transmission. gr-268961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., Rakshit P., Singh S., Abraham P., Panda S., Team N. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7):1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamble C., Kamble V., Kamble P. SARS-CoV-2 B. 1.617. 2 (Delta), B. 1.617. 2.1 (Delta Plus) Variant and vaccine strategies. Research & Reviews: a journal of medical science and technology. SARS-CoV-2 B. 2021;1(2):2. 10 (3), 63–75pp. [Google Scholar]
- 55.Torjesen I. 2021. Covid-19: omicron may be more transmissible than other variants and partly resistant to existing vaccines. [scientists fear] [DOI] [PubMed] [Google Scholar]
- 56.Ren S.Y., Wang W.B., Gao R.D., Zhou A.M. Omicron variant (B. 1.1. 529) of SARS-CoV-2: mutation, infectivity, transmission, and vaccine resistance. World J.Clin.Cases. 2022;10(1):1. doi: 10.12998/wjcc.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Araf Y., Akter F., Tang Y.D., Fatemi R., Parvez M.S.A., Zheng C., Hossain M.G. Omicron variant of SARS‐CoV‐2: genomics, transmissibility, and responses to current COVID‐19 vaccines. J Med Virol. 2022;94(5):1825–1832. doi: 10.1002/jmv.27588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mehta P., Ravi V., Devi P., Maurya R., Parveen S., Mishra P.…Pandey R. Mutational dynamics across VOCs in International travellers and Community transmission underscores importance of Spike-ACE2 interaction. Microbiol Res. 2022;262 doi: 10.1016/j.micres.2022.127099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lyngse F.P., Kirkeby C.T., Denwood M., Christiansen L.E., Mølbak K., Møller C.H.…Mortensen L.H. MedRxiv; 2022. Transmission of SARS-CoV-2 omicron VOC subvariants BA. 1 and BA. 2: evidence from Danish households. [Google Scholar]
- 60.Islam M.A., Shahi S., Al Marzan A., Amin M.R., Hasan M.N.…Hoque M.N., et al. Variant-specific deleterious mutations in the SARS-CoV-2 genome reveal immune responses and potentials for prophylactic vaccine development. Front Pharmacol. 2023;14:1090717. doi: 10.3389/fphar.2023.1090717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dao T.L., Hoang V.T., Colson P., Lagier J.C., Million M., Raoult D.…Gautret P. SARS-CoV-2 infectivity and severity of COVID-19 according to SARS-CoV-2 variants: current evidence. J Clin Med. 2021;10(12):2635. doi: 10.3390/jcm10122635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grint D.J., Wing K., Williamson E., McDonald H.I., Bhaskaran K., Evans D.…Eggo R.M. Case fatality risk of the SARS-CoV-2 variant of concern B. 1.1. 7 in England, 16 November to 5 February. Euro Surveill. 2021;26(11) doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Twohig K.A., Nyberg T., Zaidi A., Thelwall S., Sinnathamby M.A., Aliabadi S.…Bashton M. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B. 1.617. 2) compared with alpha (B. 1.1. 7) variants of concern: a cohort study. Lancet Infect Dis. 2022;22(1):35–42. doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ulloa A.C., Buchan S.A., Daneman N., Brown K.A. Canada.MedRxiv; Ontario: 2021. Early estimates of SARS-CoV-2 Omicron variant severity based on a matched cohort study. 2021-12. [Google Scholar]
- 66.Ward I.L., Bermingham C., Ayoubkhani D., Gethings O.J., Pouwels K.B.…Yates T., et al. Risk of covid-19 related deaths for SARS-CoV-2 omicron (B. 1.1. 529) compared with delta (B. 1.617. 2): retrospective cohort study. BMJ. 2022;378 doi: 10.1136/bmj-2022-070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nyberg T., Ferguson N.M., Nash S.G., Webster H.H., Flaxman S., Andrews N.…Thelwall S. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B. 1.1. 529) and delta (B. 1.617. 2) variants in England: a cohort study. Lancet. 2022;399(10332):1303–1312. doi: 10.1016/S0140-6736(22)00462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoteit R., Yassine H.M. Biological properties of SARS-CoV-2 variants: epidemiological impact and clinical consequences. Vaccines. 2022;10(6):919. doi: 10.3390/vaccines10060919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barreto H.G., de Pádua Milagres F.A., de Araújo G.C., Daúde M.M., Benedito V.A. Diagnosing the novel SARS-CoV-2 by quantitative RT-PCR: variations and opportunities. J Mol Med. 2020;98(12):1727–1736. doi: 10.1007/s00109-020-01992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jayamohan H., Lambert C.J., Sant H.J., Jafek A., Patel D., Feng H.…Gale B.K. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal Bioanal Chem. 2020:1–23. doi: 10.1007/s00216-020-02958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323(22):2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 72.Nalla A.K., Casto A.M., Huang M.L.W., Perchetti G.A., Sampoleo R.…Shrestha L., et al. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J Clin Microbiol. 2020;58(6):e00557–20. doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramírez J.D., Muñoz M., Hernández C., Flórez C., Gomez S., Rico A.…Paniz-Mondolfi A.E. Genetic diversity among SARS-CoV-2 strains in South America may impact performance of molecular detection. Pathogens. 2020;9(7):580. doi: 10.3390/pathogens9070580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jindal H., Jain S., Suvvari T.K., Kutikuppala L., Rackimuthu S., Rocha I.C.N., Goyal S. False-negative rt-pcr findings and double mutant variant as factors of an overwhelming second wave of covid-19 in India: an emerging global health disaster. SN Compr Clin Med. 2021;3(12):2383–2388. doi: 10.1007/s42399-021-01059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramírez J.D., Muñoz M., Patiño L.H., Ballesteros N., Paniz‐Mondolfi A. Will the emergent SARS‐CoV2 B. 1.1. 7 lineage affect molecular diagnosis of COVID‐19? J Med Virol. 2021;93(5):2566–2568. doi: 10.1002/jmv.26823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Artesi M., Bontems S., Göbbels P., Franckh M., Maes P., Boreux R.…Durkin K. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol. 2020;58(10) doi: 10.1128/JCM.01598-20. https://journals.asm.org/doi/abs/10.1128/jcm.01598-20 e01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.…Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vogels C.B., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C.…Grubaugh N.D. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reijns M.A., Thompson L., Acosta J.C., Black H.A., Sanchez-Luque F.J., Diamond A.…Jackson A.P. A sensitive and affordable multiplex RT-qPCR assay for SARS-CoV-2 detection. PLoS Biol. 2020;18(12) doi: 10.1371/journal.pbio.3001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balaji S.M. S-gene Dropout and False-negative reverse transcriptase-polymerase chain reaction tests. Ann Oral Maxillofac Surg. 2021;11(2):217–218. doi: 10.4103/ams.ams_2_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53(3):425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moore J.P., Offit P.A. SARS-CoV-2 vaccines and the growing threat of viral variants. JAMA. 2021;325(9):821–822. doi: 10.1001/jama.2021.1114. [DOI] [PubMed] [Google Scholar]
- 84.Wu K., Werner A.P., Moliva J.I., Koch M., Choi A.…Stewart-Jones G.B., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021:1–20. [Google Scholar]
- 85.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B.…Moore P.L. SARS-CoV-2 501Y. V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021:1–4. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 86.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T.…Gallagher E., et al. Covid-19 vaccine effectiveness against the Omicron (B. 1.1. 529) variant. N Engl J Med. 2022, 386:1532-1546 doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C.…Ciesek S. MedRxiv; 2021. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies.https://www.medrxiv.org/content/10.1101/2021.12.07.21267432.abstract [Google Scholar]
- 88.Ramesh S., Govindarajulu M., Parise R.S., Neel L., Shankar T., Patel S.…Moore T. Emerging SARS-CoV-2 variants: a review of its mutations, its implications and vaccine efficacy. Vaccines. 2021;9(10):1195. doi: 10.3390/vaccines9101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cromer D., Steain M., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A.…Davenport M.P. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. The Lancet Microbe. 2022;3(1):e52–e61. doi: 10.1016/S2666-5247(21)00267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Higdon M.M., Baidya A., Walter K.K., Patel M.K., Issa H., Espié E.…Knoll M.D. Duration of effectiveness of vaccination against COVID-19 caused by the omicron variant. Lancet Infect Dis. 2022;22(8):1114–1116. doi: 10.1016/S1473-3099(22)00409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Evans J.P., Zeng C., Qu P., Faraone J., Zheng Y.M., Carlin C., Liu S.L. 2022. Neutralization of SARS-CoV-2 Omicron sub-lineages BA. 1, BA. 1.1, and BA. 2.Cell host & microbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available upon request via e-mail from the corresponding author.