ABSTRACT
Macrophages are crucial components of the host’s defense against pathogens. Recent studies indicate that macrophage functions are influenced by lipid metabolism. However, knowledge of how bacterial pathogens exploit macrophage lipid metabolism for their benefit remains rudimentary. We have shown that the Pseudomonas aeruginosa MvfR-regulated quorum-sensing (QS) signaling molecule 2-aminoacetophenone (2-AA) mediates epigenetic and metabolic changes associated with this pathogen’s persistence in vivo. We provide evidence that 2-AA counteracts the ability of macrophages to clear the intracellular P. aeruginosa, leading to persistence. The intracellular action of 2-AA in macrophages is linked to reduced autophagic functions and the impaired expression of a central lipogenic gene, stearoyl-CoA desaturase 1 (Scd1), which catalyzes the biosynthesis of monounsaturated fatty acids. 2-AA also reduces the expression of the autophagic genes Unc-51-like autophagy activating kinase 1 (ULK1) and Beclin1 and the levels of the autophagosomal membrane protein microtubule-associated protein 1, light chain 3 isoform B (LC3B) and p62. Reduced autophagy is accompanied by the reduced expression of the lipogenic gene Scd1, preventing bacterial clearance. Adding the SCD1 substrates palmitoyl-CoA and stearoyl-CoA increases P. aeruginosa clearance by macrophages. The impact of 2-AA on lipogenic gene expression and autophagic machinery is histone deacetylase 1 (HDAC1) mediated, implicating the HDAC1 epigenetic marks at the promoter sites of Scd1 and Beclin1 genes. This work provides novel insights into the complex metabolic alterations and epigenetic regulation promoted by QS and uncovers additional 2-AA actions supporting P. aeruginosa sustainment in macrophages. These findings may aid in designing host-directed therapeutics and protective interventions against P. aeruginosa persistence.
KEYWORDS: Pseudomonas aeruginosa, persistence, quorum sensing, 2-aminoacetophenone, macrophages, immunometabolism, histone deacetylation, epigenetic reprogramming, MvfR, PqsR, autophagy, Scd1, fatty acids
INTRODUCTION
Therapeutic interventions to counter infections caused by ESKAPE pathogens are increasingly diminishing due to the continued rise in antimicrobial resistance (1, 2). Pseudomonas aeruginosa is a recalcitrant ESKAPE pathogen that causes persistent difficult-to-treat community-acquired and nosocomial infections (3–5). Elucidating mechanisms important for the survival and clearance of this pathogen are crucial in developing novel approaches to combat P. aeruginosa infections.
During infection, P. aeruginosa synthesizes and secretes several low molecular weight molecules regulated by cell density-dependent signaling systems, referred to as quorum sensing (QS), to synchronize and effectively modulate bacterial virulence functions (6–9). The P. aeruginosa multiple virulence transcription factor MvfR, also called PqsR, is one of the central QS systems in this pathogen. MvfR directly regulates the transcription of the pqsABCDE operon genes (10, 11) that control the synthesis of many small molecules produced and secreted in infected human tissues, including the aromatic volatile compound 2-aminoacetophenone (2-AA) (9, 12). 2-AA is an interkingdom effector molecule that has been shown to promote chronic infection phenotypes via its effects on both the pathogen (13, 14) and the host, leading to the long-term presence of P. aeruginosa in infected tissues (15, 16). Moreover, in previous studies, we have shown the systemic effects of 2-AA in skeletal muscle (17–19). It induces oxidative stress and apoptosis and promotes mitochondrial dysfunction (17, 18, 20). The 2-AA-mediated actions are not only limited to skeletal muscle but are also observed in immune cells. In macrophages, 2-AA dampens cytokine responses through epigenetic reprogramming implicating the histone deacetylase 1 (HDAC1) (21, 22). Epigenetic reprogramming is associated with the long-term presence of P. aeruginosa in infected mice (22).
While P. aeruginosa is considered an extracellular pathogen, an increasing number of studies indicate that this organism can also reside and survive intracellularly, utilizing specific functions (23–25). Macrophages are the first line of defense against pathogens, and they respond efficiently to bacterial invasion by engulfing and killing the bacteria. Many macrophage functions depend on membrane integrity and dynamics. Lipids are shown to be critical in cellular stabilization and signaling (26), and alterations in their composition and distribution can severely affect autophagy (27–29). Substantial contributions to macrophage functions come from the plasma membrane containing many glycoprotein receptors interacting with the membrane’s dynamic microdomains enriched with cholesterol and sphingolipids known as lipid rafts to facilitate cellular signaling (30, 31). These lipid rafts initiate antibacterial responses and are used by intracellular pathogens to enter the cell, propagate, and egress (32). Lipid biosynthesis is essential for membrane remodeling and synthesis of inflammatory mediators in M1 macrophages (33, 34). Although it is clear that macrophages reprogram their lipid metabolism in response to activation signals, knowledge of how different proinflammatory stimuli reshape the macrophage lipidome is limited. It has been shown that Toll-like receptor signals promote divergent fatty acid synthetic programs, including saturated fatty acids (SFAs) and monounsaturated fatty acids (MUFAs) (35). Among these, stearoyl-CoA desaturase 1 (SCD1) is an enzyme responsible for the desaturation of SFA into MUFAs (36).
Studies of P. aeruginosa internalization and clearance have implicated membrane functions (37, 38). While bacterial transport to lysosomal compartments is a well-orchestrated event carried out by various anchor proteins and lipids (39), less is known about the transport of P. aeruginosa and its mechanism of escape from the lysosomal compartments. Indirect evidence points to lipid homeostatic imbalances resulting from P. aeruginosa infection, including the decrease of host unsaturated immunomodulating lipids that result in the sustenance of bacteria inside the host neutrophils (40) and the triggering of the arachidonic acid-dependent inflammatory cascade by ExoU (41). While evidence points toward a role for lipids alterations during P. aeruginosa infection, however, the mechanistic basis of these membrane changes is unclear.
The primary purpose of this study was to interrogate further the 2-AA action on macrophages and their impact on P. aeruginosa persistence. Our findings point to specific actions of this small molecule on these professional phagocytes, involving autophagy and lipid metabolism components. They reveal that the dysregulation of autophagy and lipid metabolism functions implicate chromatin modifications. The results from the present study provide a new understanding of the P. aeruginosa-macrophage interactions and highlight the critical actions of this pathogen’s QS molecule 2-AA in promoting bacterial persistence.
RESULTS
2-AA sustains P. aeruginosa burden in macrophages.
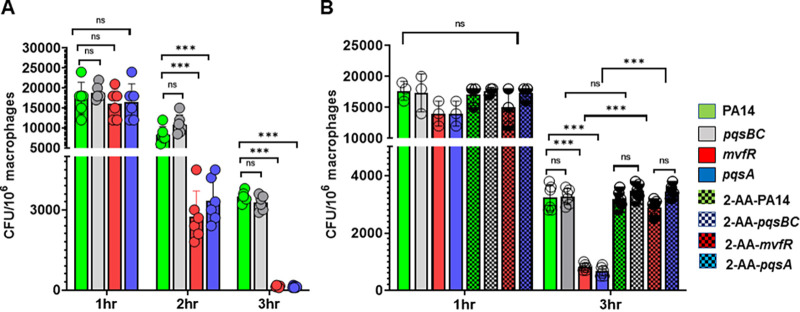
The P. aeruginosa MvfR-regulated QS signaling molecule 2-AA contributes to the persistence of this pathogen in host tissues (20, 21). Since macrophages are the main phagocytes of the innate immune system and are considered key players in bacterial clearance, we sought to determine the role of these immune cells in permitting the 2-AA-promoted persistence in infected host tissues. Using the P. aeruginosa clinical isolate PA14 (wild type [WT]) and the isogenic QS mutants mvfR and pqsA that do not produce 2-AA and mutant pqsBC that produces 2-AA but not hydroxyquinolines (14), we infected murine bone marrow-derived macrophages (BMDM). Similarly to mvfR and pqsA mutants, the pqsBC mutant does not produce hydroxyquinolines (14). We performed gentamicin protection assay studies to determine the intracellular presence of these bacterial strains over the course of 3 h. We found that the intracellular burden in the BMDM cells infected with mvfR and pqsA mutants significantly decreased by 2 h and almost cleared by 3 h, relative to cells infected with PA14 (Fig. 1A). In contrast, macrophages infected with PA14 or isogenic pqsBC mutant sustained similar and significant bacterial burdens throughout the course of infection. Exogenous addition of 2-AA (200 μM) to RAW264.7 cells infected with the 2-AA-deficient strains mvfR or pqsA restored the intracellular bacterial burden to levels similar to those observed in cells infected with PA14 and pqsBC (Fig. 1B). The supplementation of 2-AA in PA14- or pqsBC-infected macrophages did not have any additive effect on the intracellular bacterial load. These findings strongly support 2-AA’s role in interfering with the ability of these professional phagocytes to clear the presence of P. aeruginosa.
FIG 1.
The MvfR-regulated quorum sensing signaling molecule 2-AA sustains P. aeruginosa burden in macrophages. (A) Enumeration of the intracellular P. aeruginosa, as CFU/106 RAW 264.7 macrophages (mΦ) at 1, 2, and 3 h postinfection with WT PA14 (green) or the isogenic mutants nonproducing 2-AA mvfR (red) and pqsA (blue) and the 2-AA producing isogenic mutant pqsBC (gray). Bacterial loads were significantly reduced in mvfR and pqsA mutants relative to PA14 and pqsBC. (B) Enumeration of the P. aeruginosa intracellular CFU/106 RAW 264.7 mΦ in the absence or presence of 200 μM 2-AA (dotted) at 1 h and 3 h postinfection with PA14 or mutants mvfR, pqsA, or pqsBC. The addition of 2-AA significantly increases the intracellular bacterial load in mvfR and pqsA-infected macrophages relative to those infected with these nonproducing 2-AA mutants at 3 h postinfection. The error bars denote ± SD. One-way ANOVA followed by Tukey posttest was applied. ***, P < 0.001; ns indicates no significant difference. Data represent n ≥ 3 independent replicates. Each circle represents data from one replicate.
Impairment of autophagy contributes to the P. aeruginosa intracellular bacterial load in infected macrophages.
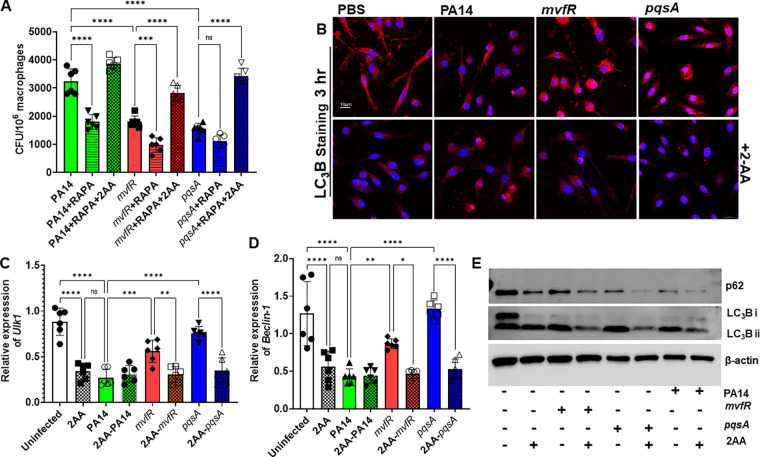
The 2-AA-mediated sustainment of the intracellular bacterial load prompted us to investigate whether 2-AA impacts autophagy. First, we used the autophagy inducer rapamycin to examine the contribution of autophagy in the 2-AA-mediated reduced intracellular bacterial clearance. Pretreatment of RAW264.7 cells with rapamycin starting 3 h before infection significantly decreased the intracellular bacterial load in cells infected with the strain PA14 and the isogenic mutants lacking 2-AA production (Fig. 2A). The exogenous addition of 2-AA to these cells increased the bacterial intercellular load, annulling the observed rapamycin effect. Subsequently, we performed punctate and diffuse staining of BMDM cells using the key autophagosomal membrane protein microtubule-associated protein 1, light chain 3 isoform B (LC3B) (Fig. 2B). Fluorescence confocal microscopy images of the infected BMDM (Fig. 2B) and quantification (Fig. S1 in the supplemental material) showed significantly lower (~3-fold) LC3B punctation in PA14 versus mvfR or pqsA infected cells. However, the exogenous addition of 2-AA in BMDM cells infected with mvfR or pqsA reduced the LC3B punctation to the levels observed in the cells infected with PA14 (Fig. 2B, bottom; Fig. S1). Consistent with these findings, the expression of the autophagic genes ULK1 (Unc-like kinase-1) and Beclin1 were significantly lower (~2.8-fold for ULK1 and ~2.7-fold for beclin1) in macrophages infected with PA14 compared to the levels observed in uninfected cells or those infected with mutants pqsA or mvfR. In corroboration, the expression of ULK1 and Beclin1 decreased following the addition of 2-AA in pqsA- or mvfR-infected macrophages, mimicking those observed in cells infected with PA14 (Fig. 2C and D). Similarly, Western Blot analyses of the LC3B autophagic protein and autophagy-specific substrate p62 from whole-cell lysates 3 h postinfection showed that in the presence of 2-AA, the proteins levels of LC3BII and p62 were hampered, further supporting that this small molecule promotes autophagic dysfunction (Fig. 2E).
FIG 2.
Impairment of autophagy contributes to the P. aeruginosa intracellular bacterial load in infected macrophages. (A) Enumeration of the intracellular P. aeruginosa CFU/106 RAW264.7 mΦ at 3 h postinfection with PA14 (green) strain or the isogenic mutants nonproducing 2-AA mvfR (red) and pqsA (blue) in the absence or in the presence of exogenously added 200 μM 2-AA (dotted), or 10 mM rapamycin (striped). The addition of the autophagic inducer rapamycin significantly decreases the intracellular CFU in PA14-infected macrophages (green striped) and isogenic mutants mvfR (red striped) compared to infected macrophages that did not receive rapamycin (solid color). This decrease is abolished in all macrophage-infected groups treated with 2-AA (dotted colors), despite the presence of rapamycin (striped colors). (B) Representative confocal images depict LC3B puncta (red) in BMDM cells infected with PA14, mvfR, or pqsA (top). PBS was used as a control. PA14-infected macrophages show fewer LC3B puncta than macrophages infected with mvfR and pqsA. However, supplementation with 2-AA (200 μM) in macrophages infected with mvfR and pqsA (bottom) also showed lower LC3B punctation than their nonsupplemented counterparts. The experiment was repeated independently three times with similar results. (C and D) RT-qPCR analysis of PA14 (green)-infected RAW264.7 macrophages reveal significantly lower ULK1 and Beclin1 expression levels relative to mvfR (red)- or pqsA (blue)-infected macrophages, while 2-AA (200 μM) addition in both mvfR (red dotted)- and pqsA (blue dotted)-infected cells decreases the expression of these genes. The error bars denote ± SD. One-way ANOVA followed by Tukey posttest was applied. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns indicates no significant difference. Data represent n = 6 independent replicates. Each dot represents data from one replicate. (E) Representative immunoblot of p62, LC3B protein levels in PA14-, mvfR-, or pqsA-infected macrophages ± 2-AA (200 μM). β-Actin was used as a control. The blots are representative of three independent experiments.
Graphical intensity plots of Fig. 2B showing arbitrary fluorescent units depicting the intensity of LC3B versus the intensity of DAPI in 2-AA treated or infected BMDM cells. A significant reduction in LC3B staining relative to the PBS-treated cells is observed within 3 h post-2-AA treatment. PA14-infected macrophages (green) have less LC3B staining than macrophages infected with mvfR (red) or pqsA (blue). However, supplementation with 2-AA (200 μM) in cells infected with mvfR and pqsA (checkered pattern) showed lower LC3B punctation than their nonsupplemented counterparts. Data represent n = 3 independent replicates. Each circle represents data from at least 2 replicates. The error bars denote ± SD. One-way ANOVA followed by Tukey posttest was applied. **, P < 0.01; ***, P < 0.001. Download FIG S1, TIF file, 0.1 MB (88.3KB, tif) .
Copyright © 2023 Chakraborty et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
2-AA impacts membrane lipids and decreases lipogenic gene expression.
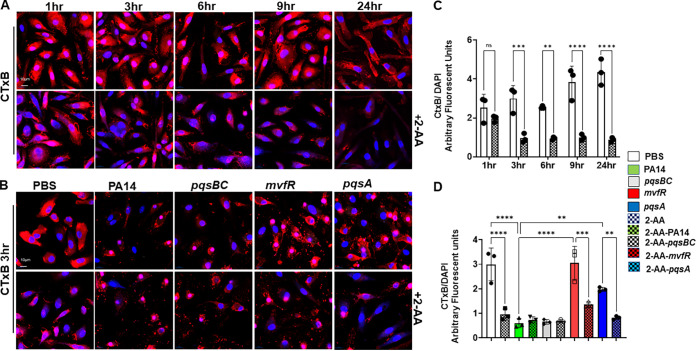
Autophagy is an active cellular process requiring interaction between membrane lipids and the autophagic machinery proteins (42, 43). Therefore, we assessed the effect of 2-AA on the BMDM membrane using fluorescent-labeled cholera toxin B (CTx-B) (31), which binds tightly to its receptor, the GM1 glycosphingolipids and is often used as a probe of membrane biology and marker of lipid rafts (43, 44). Since membrane alteration may be a dynamic event, we performed confocal microscopy to evaluate the potential effect of 2-AA on the membrane of fixed BMDM cells at different time points (3 h, 6 h, 9 h, and 24 h). The fluorescence confocal microscopy images (Fig. 3A) and quantification (Fig. 3B) show that the infected BMDM cells receiving 2-AA exhibit a significant reduction (~3-fold) in membrane staining starting as early as 3 h post-2-AA addition compared to the uninfected cells. A similar significant reduction (~5-fold) in staining was observed in cells infected with the 2-AA-producing strain PA14 and mutant strain pqsBC but appeared not as prominent with the non-2-AA-producing mutant strains mvfR and pqsA (Fig. 3C and D). RAW 264.7 macrophages infected with PA14 or the mvfR with or without exogenous addition of 2-AA gave similar CTx-B staining results to BMDM cells (Fig. S2C).
FIG 3.
2-AA affects membrane lipids. Representative confocal images of BMDM cells depicting membrane ganglioside staining with fluorescent-labeled cholera toxin B (red) (A) untreated (top) or treated (bottom) with 2-AA (200 μM) for 1 h, 3 h, 6 h, 9 h, and 24 h and infected (B) with PA14, mvfR, pqsA, and pqsBC (top), or treated with 2-AA (bottom) for 3 h. (C and D) Corresponding graphical intensity plots of panels A and B, respectively. Arbitrary fluorescent units depict the intensity of membrane staining (red) versus the intensity of DAPI (blue). Significant reduction in membrane staining relative to the PBS-treated cells is observed within 3 h post-2-AA treatment. The error bars denote ± SD. One-way ANOVA followed by Tukey posttest was applied. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns indicates no significant difference. The images are representative of three independent experiments.
Representative graphical intensity plots corresponding to Fig. 4A, respectively, depicting intensity over time of SCD1 (red) versus intensity of DAPI (blue) in BMDM cells treated (A) with 2-AA (white checkered/untreated white) and infected (B) with PA14 (green), mvfR (red), pqsA (blue), and pqsBC (gray) or infected with these strains and supplemented with 2-AA (200 μM) (checkered). Data represent n = 3 independent replicates. Each dot represents data from at least 2 replicates. The error bars denote ± SD. One-way ANOVA followed by Tukey’s HSD post hoc test was applied. **, P < 0.01; ns indicates no significant difference. Representative confocal microscopy images depicting staining with CTxB (red) (C) and SCD1 (red) (D) in RAW264.7 cells uninfected (control) and infected with PA14 or mvfR in the absence (top) and the presence of exogenously added 2-AA (400 μM) (bottom). Macrophage nuclei were counterstained with DAPI (blue). The images are representative of three independent experiments. Download FIG S2, TIF file, 0.6 MB (634.4KB, tif) .
Copyright © 2023 Chakraborty et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
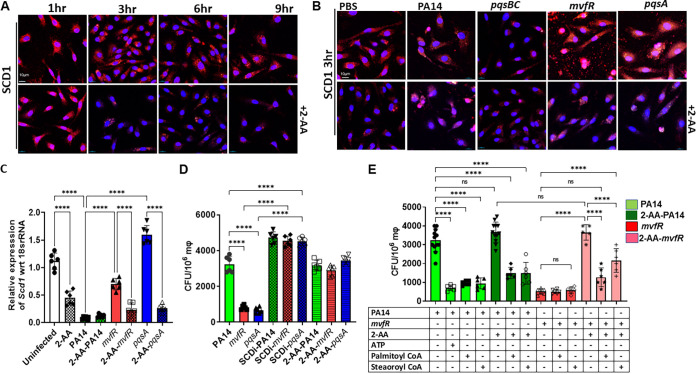
Membrane alterations in response to various stimuli depend heavily on the structural and functional aspects of their lipids and homeostatic levels of unsaturated fatty acids (45). Therefore, the observed alteration in the BMDM cell membranes suggested that fatty acid metabolism is likely affected (46). Accordingly, we interrogated the involvement of SCD1, a key enzyme in fatty acid metabolism catalyzing the biosynthesis of MUFAs also shown to be constituents of the cell membrane (47). SCD1 uses the preferred substrates palmitoyl coenzyme A (palmitoyl-CoA) and stearoyl coenzyme A (stearoyl-CoA) to form the MUFAs, palmitoleoyl-CoA and oleyl-CoA, two major constituents of membrane phospholipids. BMDM cells treated with 2-AA or infected with the 2-AA-producing strains PA14 or mutant pqsBC showed a significant reduction (~4-fold) in SCD1 staining starting at 3 h compared to the cells not receiving 2-AA (Fig. 4A; Fig. S2A) or infected with the mutant strains mvfR or pqsA (Fig. 4B; Fig. S2B). Further confirmation of these findings is provided by the exogenous addition of 2-AA in mvfR- or pqsA-infected macrophages where SCD1 staining is almost abolished (Fig. 4B, bottom). Similar results were obtained using RAW 264.7 macrophages (Fig. S2D). Consistent with these findings, Scd1 transcript levels in RAW 264.7 macrophages were significantly reduced in the presence of 2-AA (Fig. 4C). Notably, such reduction was accompanied by increased P. aeruginosa intracellular CFU in macrophages (Fig. 4D). Using an SCD1 inhibitor (ab142089) reverted the increased bacterial intracellular burden observed with non-2-AA-producing strains mvfR and pqsA, further strengthening the above findings and implicating the lipogenic enzyme SCD1 in the 2-AA-impeded bacterial clearance.
FIG 4.
2-AA affects lipogenic gene expression in macrophages. (A) Representative confocal images of SCD1 immunocytochemical staining (red) in BMDM treated (bottom)/untreated (top) with 2-AA. The macrophages were counterstained with DAPI (Blue). (B) Representative confocal images showing SCD1 expression (red) in BMDM infected with PA14, mvfR, pqsA, and pqsBC (top). Supplementation of 2-AA (200 μM) in macrophages infected with mvfR or pqsA (bottom) showed lower SCD1 staining than their nonsupplemented counterparts. (C) RT-qPCR analysis of Scd1 gene expression in RAW264.7 mΦ infected with PA14 (green), mvfR (red), or pqsA (blue). Supplementation of 2-AA in infected cells with mvfR (red dotted), pqsA (blue dotted), or uninfected (white dotted) cells decreases Scd1 expression levels. (D) Bacterial load represented as CFU/106 mΦ in the absence and presence of 10 μM SCD1 inhibitor (SCD1i) or 200 μM 2-AA. The bacterial load increases when infected RAW 264.7 cells are treated with either SCD1i or 2-AA. (E) Enumeration of PA14 and mvfR intracellular CFU/106 RAW 264.7 mΦ cells in the absence and presence of palmitoyl-CoA (50 μM) and stearoyl-CoA (50 μM). The addition of these compounds decreases the intracellular bacterial load in the presence of 2-AA. Addition of ATP (10 μM) was used as a control in PA14 cells. Data represent n ≥ 5 independent replicates. Each point represents data from one replicate. The error bars denote ± SD. One-way ANOVA followed by Tukey’s honestly significant difference (HSD) post hoc test was applied; ****, P < 0.0001; ns indicates no significant difference.
Given that derangement of Scd1 gene expression is accompanied by increased levels of intracellular bacterial burden in macrophages and that Scd1 utilizes the coenzymes palmitoyl-CoA and stearoyl-CoA to catalyze the biosynthesis of MUFAs (46), we hypothesized that addition of these substrates might impede the 2-AA effect in increasing the intracellular bacterial burden. We added 50 μM either stearoyl-CoA or palmitoyl-CoA 3 h prior to infection with PA14 or mvfR of RAW 264.7 macrophages with and without the addition of exogenous 2-AA at the time of infection and assessed the bacterial intracellular load (Fig. 4E). ATP, which has been known to induce bacterial killing in macrophages, was used as a control of the PA14 infected cells. Indeed, the PA14-infected macrophages that received stearoyl-CoA, palmitoyl-CoA, and ATP had a significantly lower number of PA14 compared to the infected-only macrophages even when 2-AA is added exogenously in these groups (Fig. 4E). More importantly, however, the addition of stearoyl-CoA, palmitoyl-CoA in mvfR-infected macrophages that received 2-AA exogenously decreased significantly the number of intracellular bacteria compared to the mvfR-infected macrophages in the presence of 2-AA (Fig. 4E). These results reaffirm the importance of these lipids in the reduction of the 2-AA-mediated intracellular P. aeruginosa burden in macrophages.
Impairment of lipid homeostasis and autophagy by 2-AA is HDAC1 dependent.
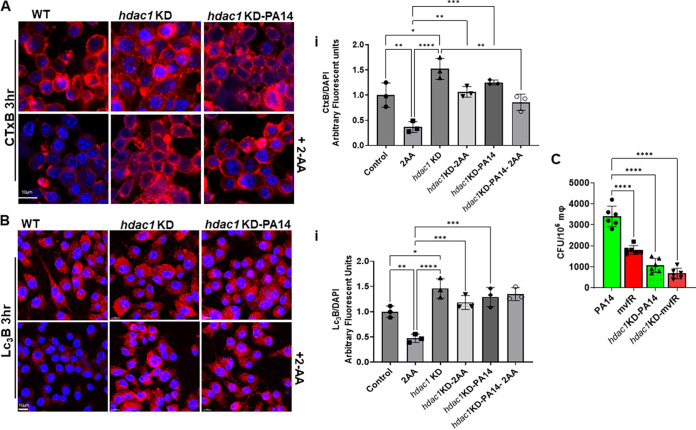
Given that 2-AA-mediated immune modulation and associated P. aeruginosa persistence in infected tissues implicates histone acetylation via HDAC1 (20), we examined the relevance of HDAC1 in the observed membrane effects observed, autophagy, and bacterial clearance. To verify whether 2-AA influences these processes in an HDAC1-dependent manner, we first compared CTx-B-stained membranes of RAW264.7 and isogenic hdac1-knockdown (KD) cells infected with PA14 or following 2-AA addition. Compared to the control cells receiving 2-AA, HDAC1 KD cells showed robust CTx-B staining, which was not affected by 2-AA addition or PA14 infection (Fig. 5Ai). Similarly, fluorescence confocal microscopy (Fig. 5B) and quantification (Fig. 5Bi) of the LC3B punctation showed that punctation was not altered in HDAC1 KD cells following infection or 2-AA exogenous addition compared to wild-type cells (2-fold reduction in LC3B). The effects observed in HDAC1 KD infected cells were accompanied by a significantly reduced (~3-fold decrease) bacterial load than that found in control isogenic RAW264.7-infected macrophages (Fig. 5C), indicating that 2-AA alters the clearance potential of macrophages in an HDAC1-dependent manner.
FIG 5.
Impairment of lipid homeostasis and autophagy by 2-AA is HDAC1-dependent. (A and B) Representative confocal images and graphical intensity plots (Ai and Bi) depicting enhanced membrane lipid staining with fluorescent-labeled cholera toxin B (red) (A) or LC3B puncta (Red) (B) in RAW 264.7 and hdac1 KD isogenic cells uninfected and infected with PA14 (top) and/or treated with 2-AA (bottom). (C) Enumeration of PA14 intracellular CFU/106 in RAW 264.7 or hdac1 KD mΦ cells. Loss of HDAC1 clears PA14 more efficiently than the wild-type macrophages. Data represent n = 6 independent replicates. Each dot represents data from one replicate. The error bars denote ± SD. One-way ANOVA followed by Tukey posttest was applied. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; and ns indicates no significant difference.
HDAC1-mediated deacetylation of H3K18 at the promoter sites of Beclin1 and Scd1 genes decreases their expression.
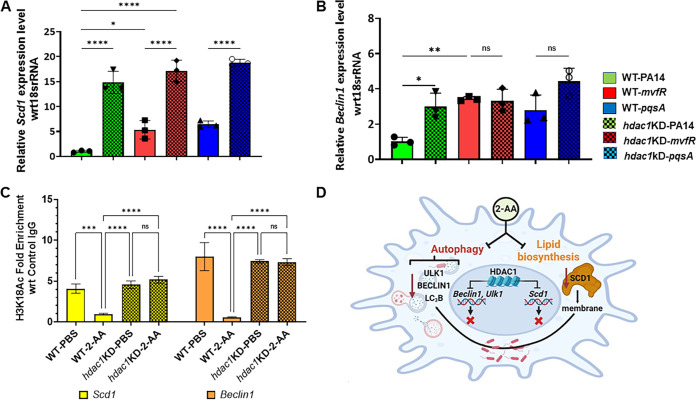
Low intracellular bacterial load, robust LC3B punctation, and CTx-B staining in RAW 264.7 isogenic HDAC1 KD-infected cells prompted us to assess the expression of Scd1 and Beclin1 genes, whose products are known to play central roles in autophagy. As shown in Fig. 6A, following infection with PA14, mvfR, or pqsA, the transcript levels of Scd1 were significantly higher in HDAC1 KD cells than in corresponding infected control macrophage cells. Moreover, the expression levels of Beclin1 in HDAC1 KD-infected cells with all strains used were higher than in control cells infected with the PA14 strain (Fig. 6B). To determine whether 2-AA alters gene expression by impacting histone marks at the promoters of Scd1 and Beclin1, we performed chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR). Enrichment of H3K18ac, specifically at the promoter region of Scd1 and Beclin1, was significantly attenuated in 2-AA treated control cells compared to the untreated control cells (Fig. 6C). In contrast, in HDAC1 KD cells, similar enrichment of H3K18 acetylation at the promoter region of both Scd1 and Beclin1 was observed regardless of the 2-AA addition.
FIG 6.
HDAC1-mediated deacetylation of H3K18 at the promoter of Beclin1 and Scd1 decreases their gene expression. Analysis of Scd1 (A) and Beclin1 (B) transcript levels by RT-qPCR. Shown is the relative gene expression in PA14 (green)-, mvfR (red)-, or pqsA (blue)-infected RAW 264.7 and hdac1 KD cells (dotted bars) compared to uninfected cells. The gene expression levels of Scd1 and Beclin1 in hdac1 KD cells (dotted) are significantly higher than in RAW 264.7-infected cells (plain). (C) Occurrence of H3K18ac marks on the Beclin1 and Scd1 promoters. ChIP-qPCR was used to show the enrichment of H3K18ac marks on the Beclin1 (orange) promoter and Scd1 promoter (yellow) in RAW 264.7 cells treated with 2-AA (400 mM) compared to untreated macrophages or HDAC1-knockdown treated macrophages. Each circle represents data from one replicate. The error bars denote ± SD. One-way ANOVA followed by Tukey’s HSD posttest was applied. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns indicates no significant difference. (D) Schematic representation of 2-AA effects on macrophages. Once P. aeruginosa is present in macrophages, 2-AA secures its intracellular sustenance through changes in the host autophagy by affecting autophagic nucleation and lipid biosynthesis. Alteration in macrophage lipid homeostasis results from changing the expression of the key lipogenic gene Scd1, which also affects the macrophage membrane, possibly through reduced monounsaturated fatty acid (MUFA) biosynthesis. The epigenetic eraser HDAC1 mediates inhibition of MUFAs biosynthesis and autophagy.
DISCUSSION
This study provides novel insights into the actions of P. aeruginosa in offsetting its clearance by macrophages. We show that the MvfR-regulated QS signaling molecule 2-AA impacts the autophagic machinery and lipids biosynthesis to sustain the presence of P. aeruginosa in macrophages after being engulfed by phagocytosis. A series of infection studies in BMDM and RAW 264.7 cells using the WT P. aeruginosa clinical isolate PA14 and isogenic mutants indicate that 2-AA is a contributing culprit of these dysfunctions. A 2-AA-mediated autophagic dysfunction is supported by the decreased expression of the autophagic genes ULK1 and Beclin1, along with the reduced levels of the LC3B protein and the autophagy-specific substrate p62. Our results show that in the presence of 2-AA, the rapamycin-induced autophagy effect is annulled. Rapamycin is an inducer of autophagy and an inhibitor of the mechanistic target of rapamycin complex 1 (mTORC1). Inhibition of mTORC1 is required to initiate the autophagy process. (48) P. aeruginosa has been reported to survive in epithelial cells by the ExoS-mediated simultaneous inhibition of both the mTOR pathway and autophagy through independent mechanisms (49) 2-AA’s ability to interfere in autophagy may involve mTOR signaling. Future studies aimed at exploring this aspect further are expected to be informative. Nevertheless, adding rapamycin further strengthens the 2-AA’s negative effect on autophagy initiation. At the same time, using the 2-AA-deficient mutant mvfR in this setting also suggests that additional 2-AA independent mechanism(s) impact the autophagic machinery.
The reduced expression of the lipogenic gene SCD1 and the membrane perturbations seen through the reduced SCD1 and CTx-B membrane staining of the BMDM cells infected with the PA14 strain pointed to dysfunctional fatty acid metabolism. The degree of unsaturation in membrane lipids is controlled partly by SCD1. Inhibition of SCD1 function in macrophages infected with PA14 isogenic non-2-AA-producing mutants increases the intracellular bacterial burden. In contrast, adding the SCD1 substrates palmitoyl-CoA and stearoyl-CoA reduces the bacterial burden in these macrophages, providing strong evidence of 2-AA’s impact on lipid biosynthesis. The 2-AA-dependent decrease of CTx-B in infected macrophages signifies the reduced ganglioside GM1 in the plasma membrane. However, since CTx-B binds to the sugar component of gangliosides, glycosylation may also be suppressed by 2-AA. Future studies are planned to address this aspect. Overall, the findings presented here offer novel insights into the actions of P. aeruginosa and provide strong evidence of 2-AA’s impact on fatty acid metabolism and its importance in the clearance of this pathogen.
The importance of fatty acids in P. aeruginosa infection has been previously reported. For example, oleate was shown to protect Caenorhabditis elegans from this pathogen’s infection (50). In addition, P. aeruginosa enzymes, including phospholipase C, lipoxygenase A, and ExoU, have been reported to impact host lipid pathways in the lung epithelial cells (51). The P. aeruginosa QS autoinducer N-3-oxo-dodecanoyl homoserine lactone has been reported to change the membrane dipole potential in lymphocytes by accumulating in the cholesterol-containing microdomains, leading to membrane conformation changes that impact immune signaling (52). Macrophage functions affecting P. aeruginosa clearance have also been reported. Loss of the protease-activated receptor 2 (PAR2) impairs the clearance of this pathogen in alveolar macrophages. PAR2 promotes P. aeruginosa clearance by inducing cAMP levels, showing that the cAMP-Rac1 signaling cascade activates the phagocytosis of this pathogen in alveolar macrophages (53). The nuclear hormone receptor peroxisome proliferator-activated receptor gamma (PPARg) also comes into play in P. aeruginosa clearance in mice lungs as it induces paraoxonase, an enzyme involved in hydrolysis and degradation of the QS homoserine lactones (54). Our results add to the limited repertoire of P. aeruginosa virulence functions and actions identified to counteract its clearance.
Another key finding in our study is the HDAC1 dependency of 2-AA-mediated effect on membrane lipids and autophagy. We show that 2-AA alters the killing potential of macrophages in an HDAC1-dependent manner by demonstrating that HDAC1 KD restores P. aeruginosa clearance in macrophages. HDACs catalyze the removal of acetyl groups from lysines on histones, increasing chromosome condensation and inhibiting transcription. Indeed, we find HDAC1 enrichment at Beclin1 and Scd1 promoters in support of the observed decreased expression of these genes. Our results are consistent with our previous findings on the HDAC1-mediated reprogramming of immune cells found to be associated with the sustained bacterial burden in host tissues (20, 22). The present findings provide new insights into 2-AA-mediated epigenetic reprogramming outcomes and reveal mechanistic actions and players contributing to P. aeruginosa persistence. The reported lipid and autophagic reprogramming in macrophages by a bacterial QS signaling molecule opens new avenues for designing and developing new therapeutics and interventions that may protect patients from recalcitrant persistent infections.
MATERIALS AND METHODS
Generation of bone marrow-derived macrophages.
BMDMs were generated by collecting femur and tibia from the hind legs of 6- to 8-week-old CD1 male mice (Charles River). Muscles attached to bones were removed using sterile scissors and forceps. Bone marrow was isolated by flushing the marrow into a sterile 50-mL tube with a syringe filled with RPMI 1640 medium. Upon centrifugation at 2,000 rpm for 5 min, cells were resuspended in RPMI 1640 medium supplemented with 10% FBS, macrophage colony-stimulating factor (M-CSF; 20 ng/mL; StemCell Technologies), 2% penicillin/streptomycin, and 2 mM l-glutamine (Gibco). Subsequently, the cells were plated on 90-mm nontissue culture-treated plates at a density of 5 × 106 cells/mL and incubated at 37°C in a 5% CO2 atmosphere. Three days after cell seeding, an extra 10 mL of fresh RPMI 1640 (supplemented with 10% FBS, M-CSF [20 ng/mL], 2% penicillin/streptomycin, and 2 mM l-glutamine) was added to each plate, and incubation continued for an additional 3 days. On the sixth day, supernatants were discarded. The attached cells were resuspended in RPMI 1640 after being dislodged by trypsin-EDTA. Finally, the cells were counted, and approximately 1.5 × 106 cells were seeded in tissue culture plates for 24 h before the experimental procedure. The culture medium was replaced by RPMI 1640 growth medium with 10% FBS.
Cell cultures.
The mouse macrophages cell line RAW264.7 (ATCC) was grown in DMEM medium, supplemented with 10% heat-inactivated FBS (endotoxin-free Certified FBS; Invitrogen), 2% penicillin/streptomycin, 2 mM l-glutamine, and 10 mM HEPES (all from Gibco). The cells were seeded in T-75 tissue culture flasks (Falcon, USA) and used between passages 2 and 3. For quality control, we tested the cells for mycoplasma with the PlasmoTest kit (Invivogen). HDAC1 knockdown cells were generated by shRNA and validated by Western blotting analyses as described in our previous publication (20) and grown in a complete DMEM medium in the presence of puromycin (10 μg/mL). The primary antibody to detect HDAC1 was specific to HDAC1 (cat no. 5356; Cell Signaling Technology).
P. aeruginosa strains and cell infection.
The P. aeruginosa clinical isolate PA14 (Rahme Lab) (55) and isogenic mutants mvfR, pqsA, or pqsBC were used (Rahme lab) (10, 14, 56, 57). The bacteria were grown at 37°C in Luria-Bertani (LB) broth under shaking and aeration overnight, diluted to 1:50,000,000 in fresh LB medium, and grown to an optical density at 600 nm (OD600nm) of 2.0. Bacterial cells were centrifuged at 5000 rpm for 5 min, resuspended in PBS, and used to infect BMDM, RAW264.7, or isogenic HDAC1 KD cells at a multiplicity of infection (MOI) of 5 or treated with 2-AA (200 μM).
Gentamicin protection assay.
To assess bacterial clearance of P. aeruginosa in macrophages, the same clinical isolate PA14 (Rahme Lab) (55) and isogenic mutants mvfR, pqsA, or pqsBC were used (Rahme lab) (10, 56, 57). Bacterial strains were grown to OD600nm 2.0 and used for the infection of RAW264.7 or HDCA1 KD cell lines. Macrophage cells were plated overnight on 100-mm cell culture-treated dishes. Subsequently, cells were washed with DMEM and infected with bacteria at 5 MOI for 30 min at 37°C in 5% CO2. Unbound bacteria were removed by one wash with cold DMEM medium, and to remove any excess extracellular bacteria, 100 μg of gentamicin was added for 30 min; cells were washed with DMEM and transferred to free medium without gentamicin and kept at 37°C in 5% CO2. Infected RAW cells were scraped at the indicated time points (1, 2, and 3 h), centrifuged at 500 × g, and lysed in distilled water. The lysed cells were immediately diluted in PBS and plated on LB agar plates to assess bacterial presence. Bacterial CFU were counted after incubating the plates overnight at 37°C.
Compounds supplementation and pharmacological inhibitors.
For experiments with 2-AA supplementation, 200 μM this compound was added. RAW264.7 cells were supplemented with 50 μM palmitoyl-CoA, stearoyl-CoA, or ATP 3 h prior to the infection. All three compounds were added after washing and immediately after infecting the cells. Rapamycin (10 μM; Sigma-Aldrich) or SCD1 inhibitor A939572 (10 μM; Calbiochem) was added to the RAW 264.7 cells 3 h prior to infection, washed, and subsequently infected.
Cholera toxin B staining for confocal microscopy.
BMDM, RAW264.7, or isogenic HDAC1 KD cells were grown on three-chamber glass slides (IBidi cat no. 80381) overnight. Macrophages were either subjected to infection or 2-AA treatment, as described earlier. At the end of the experiment, the glass slides were placed on ice and fixed with ice-cold 4% paraformaldehyde. Immediately after, the cells were incubated with ice-cold (1 μg/mL) Alexa594-cholera toxin B subunit conjugate (Alexa594-CTB) for 30 min. Cells were washed three times with PBS, counterstained with DAPI, and examined using a confocal microscope (Nikon ECLIPSE Ti2; Nikon Instruments Inc., Tokyo, Japan) at ×400 optical magnification. The assay was conducted in triplicate and repeated at least twice.
Immunocytochemistry.
BMDM or RAW264.7 cells were seeded at 1 × 105 cells per well on three-well chambered cover glass slides the day prior to an experiment. Cells were treated with PBS (control condition) or 200 μM 2-AA or infected with PA14 or isogenic mutants mvfR, pqsA, and pqsBC at an MOI of 5 for 3 h as described in the previous sections. At the end of the experiment, cells were fixed in 4% paraformaldehyde in 1× PBS (pH 7.4) for 15 min at room temperature. Slides were washed in 1× PBS (pH 7.4) and incubated with blocking solution (2.5% BSA, 0.05% Triton X-100, and 1× PBS [pH 7.4]) for an additional 1 h at room temperature. LC3A/B (catalog no. 13082S; Cell Signaling Technology; 1:100) or SCD1 primary antibody was added to designated wells overnight at 4°C protected from light. After being washed three times with PBS-T buffer, the cells were incubated in fluorescent-labeled respective secondary antibodies. Cells were stained with DAPI (1:10,000) at room temperature and washed in 1× PBS (pH 7.4) three times for 5 min each. The cells were examined using a confocal microscope (Nikon ECLIPSE Ti2; Nikon Instruments Inc., Tokyo) at ×400 optical magnification. The assay was conducted in triplicate and repeated twice.
Quantification of images.
Confocal images from each condition were collected using a confocal microscope (Nikon ECLIPSE Ti2; NIS-Elements 5.21; Nikon Instruments Inc., Tokyo, Japan). Fluorescence intensity from the red channel and the blue channel was measured using the ImageJ software. Fluorescence intensity arbitrary units were calculated by normalizing the red fluorescence of SCD1, LC3B, or CtxB values with the corresponding blue fluorescence values representing DAPI stain. Values from at least three experimental replicates were plotted using GraphPad Prism.
RNA isolation and quantitative RT-PCR.
Total RNA was isolated from approximately 1.2 × 106 cells with the RNeasy minikit (Qiagen, USA), and cDNA was prepared with the iScript Reverse transcription kit (Bio-Rad, USA), as per the manufacturer’s instruction. Real-time PCR was conducted using the PowerUP SYBR green Master mix (Applied Biosystem, USA) and primer sets for mouse Scd1 (forward: AAGATATTCACGACCCCACC; reverse: CAGCCGTGCCTTGTAAGTTC) and mouse Beclin1 (forward: CAGCCTCTGAAACTGGACACGA; reverse: CTCTCCTGAGTTAGCCTCTTCC) and ULK1 (forward: GCAGCAAAGACTCCTGTGACAC; reverse: CCACTACACAGCAGGCTATCAG) and mouse 18S rRNA (forward: GTTCCGACCATAAACGATGCC; reverse: TGGTGGTGCCCTTCCGTCAAT). The transcript levels of all the genes were normalized to 18S rRNA with the ΔCT method (58). The relative expression was calculated by dividing normalized transcript levels of infected cells by those in uninfected cells. The assay was conducted in triplicate; means and standard deviations were calculated for each group.
Western blot.
Whole-cell lysates were prepared in RIPA lysis buffer (Cell Signaling Technology, USA) supplemented with a protease inhibitor cocktail (Sigma). The concentration of the whole-cell total proteins was determined from each sample using the Bradford protein assays kit (Bio-Rad Laboratories, Hercules, CA, USA). Fifteen to twenty-five micrograms of the whole-cell lysates (total proteins) were added to 1× Laemmli buffer, boiled for 10 min, separated by SDS 7.5 to 15% PAGE in 25 mM Tris/250 mM glycine/0.1% SDS buffer (Bio-Rad), and transferred to PVDF membranes (Bio-Rad). After blocking with 5% BSA in TBS containing 0.1% Tween 20 for 1 h at room temperature, the membranes were incubated overnight with the primary antibodies specific for p62 or LC3B (Cell Signaling Technology), at a dilution of 1:1,000, and mouse anti-β-actin (Santa Cruz Biotechnology) at a dilution of 1:2,000. After being washed, the membranes were incubated with an anti-rabbit secondary antibody and mouse antibody for β-actin. The bands were detected by SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) reaction, according to the manufacturer’s instructions. The gels were visualized in the ChemiDOC Imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The bands were analyzed densitometrically using QuantityOne software (Bio-Rad).
Chromatin immunoprecipitation-qPCR.
Cells were cross-linked in 1% methanol-free formaldehyde for 10 min and then placed in 0.125 M glycine for 5 min at RT. Using the truChIP High Cell Chromatin Shearing kit (Covaris, United States), cells were prepared for sonication according to the Covaris protocol. Approximately 1 × 107 cells were plated in a 12 mm × 12 mm tube and subjected to shearing with the Covaris S220 sonicator for 8 min (140 peak power, 5 duty factor, 200 cycles/burst). The Magna ChIP A/G kit (Millipore, United States) was used for the subsequent immunoprecipitations according to the manufacturer’s protocol. Briefly, chromatin from approximately 106 cells was incubated overnight at 4°C with 4 μg of anti-acetyl H3K18 (Abcam, USA) ChIP-grade antibody and 20 μL of A/G magnetic beads. The beads were washed serially (5 min each) with low-salt wash buffer, high-salt wash buffer, LiCl wash buffer, and TE buffer from the kit at 4°C. Chromatin was eluted with elution buffer containing Proteinase K at 62°C for 4 h, then incubated at 95°C for 10 min. DNA was isolated by column purification (QIAquick PCR purification kit). Real-time PCR was performed with the Brilliant II SYBR green super mix (Agilent, USA) and primer sets to amplify various regions of Scd1 (forward: GGGTCATGTGTGTCCTGTGT, reverse: GTGAGCCCAACCCTATGTCC) and Beclin1 (forward: GACGTCACTTCTGGTCCTGG, reverse: ATCGGCTCCTTTGAACCTCG) loci. Normalized values were calculated by the percent-input method relative to the IgG. The assay was conducted in triplicate; means are reported with standard deviations.
Statistical analysis.
Whenever applicable, at least three independent experiments were performed. The error bars denote ± SD. Statistical analysis was carried out using GraphPad Prism software. One-way ANOVA followed by Tukey’s post hoc test was applied. Significant differences are indicated as *, P < 0.05; **, P < 0.01; and ***, P < 0.001, respectively; “ns” represents no significant difference.
ACKNOWLEDGMENTS
We thank Amy Tsurumi and the Rahme Lab members, Kelsey Wheeler, and Vijay Singh for the critical reading of the manuscript.
This work was supported by the NIH award R01AI134857 and The Massachusetts General Hospital Research Scholar Award to L.G.R. The funders had no role in the study design, data collection, analysis, decision to publish, or manuscript preparation.
L.G.R. has a financial interest in Spero Therapeutics, a company developing therapies to treat bacterial infections. L.G.R.’s financial interests are reviewed and managed by Massachusetts General Hospital and Partners Health Care in accordance with their conflict-of-interest policies. No funding was received from Spero Therapeutics, and it had no role in study design, data collection, analysis, interpretation, or the decision to submit the work for publication. The remaining authors declare no competing interests.
Footnotes
This article is a direct contribution from Laurence G. Rahme, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Stephen Lory, Harvard Medical School, and Koushik Roy, University of Utah School of Medicine.
Contributor Information
Laurence G. Rahme, Email: rahme@molbio.mgh.harvard.edu.
Shu-Sin Chng, National University of Singapore.
REFERENCES
- 1.el Zowalaty ME, Al Thani AA, Webster TJ, el Zowalaty AE, Schweizer HP, Nasrallah GK, Marei HE, Ashour HM. 2015. Pseudomonas aeruginosa: arsenal of resistance mechanisms, decades of changing resistance profiles, and future antimicrobial therapies. Future Microbiol 10:1683–1706. doi: 10.2217/fmb.15.48. [DOI] [PubMed] [Google Scholar]
- 2.Bassetti M, Vena A, Croxatto A, Righi E, Guery B. 2018. How to manage Pseudomonas aeruginosa infections. Drugs Context 7:212527. doi: 10.7573/dic.212527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee D, Stableforth D. 2000. The treatment of respiratory Pseudomonas infection in cystic fibrosis what drug and which way? Drugs 60: 1053–164. doi: 10.2165/00003495-200060050-00006. [DOI] [PubMed] [Google Scholar]
- 4.Öncül O, Öksüz S, Acar A, Ülkür E, Turhan V, Uygur F, Ulçay A, Erdem H, Özyurt M, Görenek L. 2014. Nosocomial infection characteristics in a burn intensive care unit: analysis of an eleven-year active surveillance. Burns 40:835–841. doi: 10.1016/j.burns.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Fujii A, Seki M, Higashiguchi M, Tachibana I, Kumanogoh A, Tomono K. 2014. Community-acquired, hospital-acquired, and healthcare-associated pneumonia caused by Pseudomonas aeruginosa. Respir Med Case Rep 12:30–33. doi: 10.1016/j.rmcr.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Déziel E, Gopalan S, Tampakaki AP, Lépine F, Padfield KE, Saucier M, Xiao G, Rahme LG. 2005. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting IasRI, rhIRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol 55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 7.Venturi V. 2006. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev 30:274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 8.Schuster M, Greenberg EP. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 9.Déziel E, Lépine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci USA 101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao G, Déziel E, He J, Lépine F, Lesic B, Castonguay MH, Milot S, Tampakaki AP, Stachel SE, Rahme LG. 2006. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol 62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 11.Maura D, Hazan R, Kitao T, Ballok AE, Rahme LG. 2016. Evidence for direct control of virulence and defense gene circuits by the Pseudomonas aeruginosa quorum sensing regulator, MvfR. Sci Rep 6:34083. doi: 10.1038/srep34083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Que Y-A, Hazan R, Ryan CM, Milot S, Lépine F, Lydon M, Rahme LG. 2011. Production of Pseudomonas aeruginosa intercellular small signaling molecules in human burn wounds. J Pathog 2011:549302. doi: 10.4061/2011/549302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Que YA, Hazan R, Strobel B, Maura D, He J, Kesarwani M, Panopoulos P, Tsurumi A, Giddey M, Wilhelmy J, Mindrinos MN, Rahme LG. 2013. A quorum sensing small volatile molecule promotes antibiotic tolerance in bacteria. PLoS One 8:e80140. doi: 10.1371/journal.pone.0080140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kesarwani M, Hazan R, He J, Que YA, Apidianakis Y, Lesic B, Xiao G, Dekimpe V, Milot S, Deziel E, Lépine F, Rahme LG. 2011. A quorum sensing regulated small volatile molecule reduces acute virulence and promotes chronic infection phenotypes. PLoS Pathog 7:e1002192. doi: 10.1371/journal.ppat.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapsetaki SE, Tzelepis I, Avgousti K, Livadaras I, Garantonakis N, Varikou K, Apidianakis Y. 2014. The bacterial metabolite 2-aminoacetophenone promotes association of pathogenic bacteria with flies. Nat Commun 5:4401. doi: 10.1038/ncomms5401. [DOI] [PubMed] [Google Scholar]
- 16.Bandyopadhaya A, Singh VK, Chakraborty A, Tzika AA, Rahme LG. 2021. Immunometabolic hijacking of immune cells by a Pseudomonas aeruginosa quorum-sensing signal. bioRxiv. doi: 10.1101/2021.10.03.462785. [DOI]
- 17.Tzika AA, Constantinou C, Bandyopadhaya A, Psychogios N, Lee S, Mindrinos M, Martyn JAJ, Tompkins RG, Rahme LG. 2013. A small volatile bacterial molecule triggers mitochondrial dysfunction in murine skeletal muscle. PLoS One 8:e74528. doi: 10.1371/journal.pone.0074528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bandyopadhaya A, Tzika AA, Rahme LG. 2019. Pseudomonas aeruginosa quorum sensing molecule alters skeletal muscle protein homeostasis by perturbing the antioxidant defense system. mBio 10:e02211-19. doi: 10.1128/mBio.02211-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bandyopadhaya A, Constantinou C, Psychogios N, Ueki R, Yasuhara S, Martyn JAJ, Wilhelmy J, Mindrinos M, Rahme LG, Tzika AA. 2016. Bacterial-excreted small volatile molecule 2-aminoacetophenone induces oxidative stress and apoptosis in murine skeletal muscle. Int J Mol Med 37:867–878. doi: 10.3892/ijmm.2016.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandyopadhaya A, Tsurumi A, Maura D, Jeffrey KL, Rahme LG. 2016. A quorum sensing signal promotes host tolerance training through hdac1-mediated epigenetic reprogramming. Nat Microbiol 1:16174. doi: 10.1038/nmicrobiol.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandyopadhaya A, Kesarwani M, Que YA, He J, Padfield K, Tompkins R, Rahme LG. 2012. The quorum sensing volatile molecule 2-amino acetophenon modulates host immune responses in a manner that promotes life with unwanted guests. PLoS Pathog 8:e1003024. doi: 10.1371/journal.ppat.1003024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandyopadhaya A, Tsurumi A, Rahme LG. 2017. NF-κBp50 and HDAC1 interaction is implicated in the host tolerance to infection mediated by the bacterial quorum sensing signal 2-aminoacetophenone. Front Microbiol 8:1211. doi: 10.3389/fmicb.2017.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heimer SR, Evans DJ, Stern ME, Barbieri JT, Yahr T, Fleiszig SMJ, Kaufmann GF. 2013. Pseudomonas aeruginosa utilizes the type III secreted toxin ExoS to avoid acidified compartments within epithelial cells editor. PLoS One 8:e73111. doi: 10.1371/journal.pone.0073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angus AA, Evans DJ, Barbieri JT, Fleiszig SMJ. 2010. The ADP-ribosylation domain of Pseudomonas aeruginosa ExoS is required for membrane bleb niche formation and bacterial survival within epithelial cells. Infect Immun 78:4500–4510. doi: 10.1128/IAI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroken AR, Chen CK, Evans DJ, Yahr TL, Fleiszig SMJ, Whiteley M, Wargo M. 2018. Impact of ExoS on Pseudomonas aeruginosa internalization by epithelial cells is independent of fleQ and correlates with bistability of type three secretion system gene expression. mBio 9:e00668-18. doi: 10.1128/mbio.00668-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson CL, Walch L, Verbavatz JM. 2016. Lipids and their trafficking: an integral part of cellular organization. Dev Cell 39:139–153. doi: 10.1016/j.devcel.2016.09.030. [DOI] [PubMed] [Google Scholar]
- 27.Bernard A, Klionsky DJ. 2014. Defining the membrane precursor supporting the nucleation of the phagophore. Autophagy 10:1–2. doi: 10.4161/auto.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai Y, Meng L, Han L, Jia Y, Zhao Y, Gao H, Kang R, Wang X, Tang D, Dai E. 2019. Lipid storage and lipophagy regulates ferroptosis. Biochem Biophys Res Commun 508:997–1003. doi: 10.1016/j.bbrc.2018.12.039. [DOI] [PubMed] [Google Scholar]
- 29.Xie Y, Li J, Kang R, Tang D. 2020. Interplay between lipid metabolism and autophagy. Front Cell Dev Biol 8:431. doi: 10.3389/fcell.2020.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varshney P, Yadav V, Saini N. 2016. Lipid rafts in immune signalling: current progress and future perspective. Immunology 149:13–24. doi: 10.1111/imm.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kannan S, Audet A, Huang H, Chen L-J, Wu M. 2008. Cholesterol-rich membrane rafts and Lyn are involved in phagocytosis during Pseudomonas aeruginosa infection. J Immunol 180:2396–2408. doi: 10.4049/jimmunol.180.4.2396. [DOI] [PubMed] [Google Scholar]
- 32.Kulkarni R, Wiemer EAC, Chang W. 2021. Role of lipid rafts in pathogen-host interaction–a mini review. Front Immunol 12:815020. doi: 10.3389/fimmu.2021.815020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogie JF, Grajchen E, Wouters E, Garcia Corrales A, Dierckx T, Vanherle S, Mailleux J, Gervois P, Wolfs E, Dehairs J, van Broeckhoven J, Bowman AP, Lambrichts I, Gustafsson A, Remaley AT, Mulder M, Swinnen JV, Haidar M, Ellis SR, Ntambi JM, Zelcer N, Hendriks JJ. 2020. Stearoyl-CoA desaturase-1 impairs the reparative properties of macrophages and microglia in the brain. J Exp Med 217:e20191660. doi: 10.1084/jem.20191660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei X, Song H, Yin L, Rizzo MG, Sidhu R, Covey DF, Ory DS, Semenkovich CF. 2016. Fatty acid synthesis configures the plasma membrane for inflammation in diabetes. Nature 539:294–298. doi: 10.1038/nature20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh W-Y, Zhou QD, York AG, Williams KJ, Scumpia PO, Kronenberger EB, Hoi XP, Su B, Chi X, Bui VL, Khialeeva E, Kaplan A, Son YM, Divakaruni AS, Sun J, Smale ST, Flavell RA, Bensinger SJ. 2020. Toll-like receptors induce signal-specific reprogramming of the macrophage lipidome. Cell Metab 32:128–143.e5. doi: 10.1016/j.cmet.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravaut G, Légiot A, Bergeron KF, Mounier C. 2020. Monounsaturated fatty acids in obesity-related inflammation. Int J Mol Sci 22:330. doi: 10.3390/ijms22010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grassmé H, Jendrossek V, Riehle A, von Kürthy G, Berger J, Schwarz H, Weller M, Kolesnick R, Gulbins E. 2003. Host defense against Pseudomonas aeruginosa requires ceramide-rich membrane rafts. Nat Med 9:322–330. doi: 10.1038/nm823. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto N, Yamamoto N, Petroll MW, Cavanagh HD, Jester JV. 2005. Internalization of Pseudomonas aeruginosa is mediated by lipid rafts in contact lens–wearing rabbit and cultured human corneal epithelial cells. Invest Ophthalmol Vis Sci 46:1348–1355. doi: 10.1167/iovs.04-0542. [DOI] [PubMed] [Google Scholar]
- 39.Stein MP, Müller MP, Wandinger-Ness A. 2012. Bacterial pathogens commandeer Rab GTPases to establish intracellular niches. Traffic 13:1565–1588. doi: 10.1111/tra.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flitter BA, Hvorecny KL, Ono E, Eddens T, Yang J, Kwak DH, Bahl CD, Hampton TH, Morisseau C, Hammock BD, Liu X, Lee JS, Kolls JK, Levy BD, Madden DR, Bomberger JM. 2017. Pseudomonas aeruginosa sabotages the generation of host proresolving lipid mediators. Proc Natl Acad Sci USA 114:136–141. doi: 10.1073/pnas.1610242114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saliba AM, Nascimento DO, Silva MCA, Assis MC, Gayer CRM, Raymond B, Coelho MGP, Marques EA, Touqui L, Albano RM, Lopes UG, Paiva DD, Bozza PT, Plotkowski MC. 2005. Eicosanoid-mediated proinflammatory activity of Pseudomonas aeruginosa ExoU. Cell Microbiol 7:1811–1822. doi: 10.1111/j.1462-5822.2005.00635.x. [DOI] [PubMed] [Google Scholar]
- 42.Vogel A, Brunner JS, Hajto A, Sharif O, Schabbauer G. 2022. Lipid scavenging macrophages and inflammation. Biochim Biophys Acta Mol Cell Biol Lipids 1867:159066. doi: 10.1016/j.bbalip.2021.159066. [DOI] [PubMed] [Google Scholar]
- 43.de la Ballina LR, Munson MJ, Simonsen A. 2020. Lipids and lipid-binding proteins in selective autophagy. J Mol Biol 432:135–159. doi: 10.1016/j.jmb.2019.05.051. [DOI] [PubMed] [Google Scholar]
- 44.Kenworthy AK, Schmieder SS, Raghunathan K, Tiwari A, Wang T, Kelly CV, Lencer WI. 2021. Cholera toxin as a probe for membrane biology. Toxins 13:543. doi: 10.3390/toxins13080543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adolph S, Fuhrmann H, Schumann J. 2012. unsaturated fatty acids promote the phagocytosis of P. aeruginosa and R. equi by RAW264.7 macrophages. Curr Microbiol 65:649–655. doi: 10.1007/s00284-012-0207-3. [DOI] [PubMed] [Google Scholar]
- 46.Weiss-Hersh K, Garcia AL, Marosvölgyi T, Szklenár M, Decsi T, Rühl R. 2020. Saturated and monounsaturated fatty acids in membranes are determined by the gene expression of their metabolizing enzymes SCD1 and ELOVL6 regulated by the intake of dietary fat. Eur J Nutr 59:2759–2769. doi: 10.1007/s00394-019-02121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paton CM, Ntambi JM. 2009. Biochemical and physiological function of stearoyl-CoA desaturase. Am J Physiol Endocrinol Metab 297:E28–E37. doi: 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim YC, Guan KL. 2015. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest 125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao L, de La Rosa I, Xu Y, Sha Y, Bhattacharya A, Holtzman MJ, Gilbert BE, Eissa NT. 2021. Pseudomonas aeruginosa survives in epithelia by ExoS-mediated inhibition of autophagy and mTOR. EMBO Rep 22:e50613. doi: 10.15252/embr.202050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson SM, Cheesman HK, Peterson ND, Salisbury JE, Soukas AA, Pukkila-Worley R. 2019. The fatty acid oleate is required for innate immune activation and pathogen defense in Caenorhabditis elegans. PLoS Pathog 15:e1007893. doi: 10.1371/journal.ppat.1007893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Constantino-Teles P, Jouault A, Touqui L, Saliba AM. 2022. Role of host and bacterial lipids in Pseudomonas aeruginosa respiratory infections. Front Immunol 13:931027. doi: 10.3389/fimmu.2022.931027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis BM, Jensen R, Williams P, O'Shea P. 2010. The interaction of N-acylhomoserine lactone quorum sensing signaling molecules with biological membranes: implications for inter-kingdom signaling. PLoS One 5:e13522. doi: 10.1371/journal.pone.0013522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rayees S, Joshi JC, Joshi B, Vellingiri V, Banerjee S, Mehta D. 2022. Protease-activated receptor 2 promotes clearance of Pseudomonas aeruginosa infection by inducing cAMP-Rac1 signaling in alveolar macrophages. Front Pharmacol 13:874197. doi: 10.3389/fphar.2022.874197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bedi B, Yuan Z, Joo M, Zughaier SM, Goldberg JB, Arbiser JL, Michael Hart C, Sadikot RT. 2016. Enhanced clearance of Pseudomonas aeruginosa by peroxisome proliferator-activated receptor gamma. Infect Immun 84:1975–1985. doi: 10.1128/IAI.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 56.Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, Rahme LG. 2001. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci USA 98:14613–14618. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dulcey CE, Dekimpe V, Fauvelle DA, Milot S, Groleau MC, Doucet N, Rahme LG, Lépine F, Déziel E. 2013. The end of a long-standing hypothesis: the Pseudomonas signalling molecules 4-hydroxy-2-alkylquinolines are derived from fatty acids, not 3-ketofatty acids. Chem Biol 20:1481–1491. doi: 10.1016/j.chembiol.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rao X, Huang X, Zhou Z, Lin X. 2013. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 3:71–85. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Graphical intensity plots of Fig. 2B showing arbitrary fluorescent units depicting the intensity of LC3B versus the intensity of DAPI in 2-AA treated or infected BMDM cells. A significant reduction in LC3B staining relative to the PBS-treated cells is observed within 3 h post-2-AA treatment. PA14-infected macrophages (green) have less LC3B staining than macrophages infected with mvfR (red) or pqsA (blue). However, supplementation with 2-AA (200 μM) in cells infected with mvfR and pqsA (checkered pattern) showed lower LC3B punctation than their nonsupplemented counterparts. Data represent n = 3 independent replicates. Each circle represents data from at least 2 replicates. The error bars denote ± SD. One-way ANOVA followed by Tukey posttest was applied. **, P < 0.01; ***, P < 0.001. Download FIG S1, TIF file, 0.1 MB (88.3KB, tif) .
Copyright © 2023 Chakraborty et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Representative graphical intensity plots corresponding to Fig. 4A, respectively, depicting intensity over time of SCD1 (red) versus intensity of DAPI (blue) in BMDM cells treated (A) with 2-AA (white checkered/untreated white) and infected (B) with PA14 (green), mvfR (red), pqsA (blue), and pqsBC (gray) or infected with these strains and supplemented with 2-AA (200 μM) (checkered). Data represent n = 3 independent replicates. Each dot represents data from at least 2 replicates. The error bars denote ± SD. One-way ANOVA followed by Tukey’s HSD post hoc test was applied. **, P < 0.01; ns indicates no significant difference. Representative confocal microscopy images depicting staining with CTxB (red) (C) and SCD1 (red) (D) in RAW264.7 cells uninfected (control) and infected with PA14 or mvfR in the absence (top) and the presence of exogenously added 2-AA (400 μM) (bottom). Macrophage nuclei were counterstained with DAPI (blue). The images are representative of three independent experiments. Download FIG S2, TIF file, 0.6 MB (634.4KB, tif) .
Copyright © 2023 Chakraborty et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.