ABSTRACT
The ferric uptake regulator (Fur) protein is the founding member of the FUR superfamily of metalloregulatory proteins that control metal homeostasis in bacteria. FUR proteins regulate metal homeostasis in response to the binding of iron (Fur), zinc (Zur), manganese (Mur), or nickel (Nur). FUR family proteins are generally dimers in solution, but the DNA-bound complex can involve a single dimer, a dimer-of-dimers, or an extended array of bound protein. Elevated FUR levels due to changes in cell physiology increase DNA occupancy and may also kinetically facilitate protein dissociation. Interactions between FUR proteins and other regulators are commonplace, often including cooperative and competitive DNA-binding interactions within the regulatory region. Further, there are many emerging examples of allosteric regulators that interact directly with FUR family proteins. Here, we focus on newly uncovered examples of allosteric regulation by diverse Fur antagonists (Escherichia coli YdiV/SlyD, Salmonella enterica EIIANtr, Vibrio parahaemolyticus FcrX, Acinetobacter baumannii BlsA, Bacillus subtilis YlaN, and Pseudomonas aeruginosa PacT) as well as one Zur antagonist (Mycobacterium bovis CmtR). Small molecules and metal complexes may also serve as regulatory ligands, with examples including heme binding to Bradyrhizobium japonicum Irr and 2-oxoglutarate binding to Anabaena FurA. How these protein-protein and protein-ligand interactions act in conjunction with regulatory metal ions to facilitate signal integration is an active area of investigation.
KEYWORDS: metal homeostasis, transcription, metalloregulation, allosteric regulation, repressor, activator
INTRODUCTION
Iron is an essential element in all domains of life and is used as a cofactor in numerous enzymes that are required for metabolism and growth (1). When iron levels are low, the enzymes that are required for central metabolism, respiration, and DNA synthesis may fail and thereby limit growth. Conversely, high concentrations of iron (Fe2+) accelerate reactions with hydrogen peroxide (H2O2) (via the Fenton reaction), which produces toxic hydroxyl radicals that damage DNA and proteins (2, 3). Thus, it is crucial that iron be maintained at optimal levels within the cell (4, 5). Fur (ferric uptake regulation protein) was first purified from Escherichia coli in 1987 and was found to be an iron-activated repressor (6). Unlike many DNA-binding regulators, Fur is abundant, with an estimate of 10,000 proteins per cell in E. coli (7), which suggests that Fur may additionally serve as an iron buffer. Fur is now recognized as the founding member of the FUR superfamily, a diverse set of metal-regulated transcription factors (8). Here, we briefly summarize the role of FUR proteins in the metal-dependent regulation of gene expression, a topic that has been covered in more detail in several recent reviews (9–13). Then, we focus on the ability of diverse proteins and small molecules to allosterically regulate FUR protein function, often in response to yet unknown signals.
MODEL FOR REGULATION BY FUR FAMILY PROTEINS
Fur is a homodimeric protein containing at least one iron-sensing site on each protomer (3, 10). Each Fur protomer may additionally have a structural zinc site that is important for protein folding (14, 15). In some cases, a secondary metal-sensing site may be present (10, 16). At low ambient levels of cytosolic ferrous iron (Fe2+), the regulatory metal-sensing site is unoccupied, and the dimeric Fur protein is in an “open” conformation that is not conducive to DNA-binding. The binding of Fe2+ to the regulatory site generates a “closed” protein conformation in which the two metal-binding domains are appropriately positioned for high-affinity DNA-binding. The precise conformation changes, and the molecular details of metal-binding and selectivity have been visualized for several Fur family members and are reviewed elsewhere (10). This simple model, in which Fur functions as an iron-dependent transcriptional repressor, accounts for most Fur regulation, as characterized using genome-wide methods (17–25). Because cytosolic pools of Fe2+ are affected by the oxygen tension in the cell, Fur can also function indirectly as a sensor of O2 (24).
FUR family proteins include the ferric uptake regulator (Fur) (6), zinc sensor Zur (12), nickel sensor Nur (26, 27), manganese sensor Mur (28–30), and their orthologs (sometimes with different names). Each of these metalloregulators senses changes in ambient metal levels to regulate metal ion homeostasis (see [9] for a recent review). In addition, members of the PerR (peroxide stress) subfamily of regulators sense H2O2 stress rather than metal status (reviewed in [11]). The prototype for this group, namely, B. subtilis PerR, senses H2O2 when the oxidation of bound Fe2+ generates a hydroxyl radical that covalently modifies a proximal histidine side chain, thereby leading to protein inactivation (31, 32) and, ultimately, degradation (33). Bound ferrous ions can also be nitrosylated, leading to the derepression of Fur and PerR-regulated genes in response to nitric oxide (34, 35). Orthologs of PerR are widespread in bacteria, including many pathogens, although there are variations in the precise signals that are sensed, the biochemical mechanisms of sensing, and the suite of genes under PerR regulation (11).
Although many FUR regulators are specific for a single cognate metal ion in vivo, biochemical studies reveal that DNA binding can also be activated by noncognate metal ions. Indeed, the ability of Mn2+ to mismetalate Fur proteins and thereby inappropriately repress iron acquisition likely accounts for the ability of mutations in fur to suppress Mn2+ intoxication in several systems (36–38). This type of regulatory crosstalk can be aggravated by mutations that perturb metal ion homeostasis, often with deleterious consequences, as noted for B. subtilis (39–41). For PerR subfamily proteins, the abilities of both Fe2+ and Mn2+ to activate DNA binding allows these proteins to tune their H2O2 sensitivity in response to the Fe/Mn ratio in the cell (41).
Although FUR proteins typically act as classic transcriptional repressors, other regulatory modes are possible, including transcriptional activation by the metalated regulator and regulation by the DNA-bound apo-protein (42). Direct transcriptional activation by DNA-bound FUR proteins was first documented for Neisseria meningitidis Fur (43) and Helicobacter pylori Fur (44–46). A direct activation mechanism has also been documented for E. coli Fur (25), B. subtilis Fur (47), Vibrio cholerae Fur (48), Campylobacter jejuni Fur (49), Streptomyces coelicolor Zur (50), and Xanthomonas campestris Zur (51). Many organisms contain multiple FUR proteins, and the DNA-binding sites are often similar in sequence (9, 52). Despite this, regulatory crosstalk in which one binding site is recognized by more than one FUR protein is rare, although some examples have been noted (47, 53). FUR binding may, at some sites, also be regulated by epigenetic DNA modifications, such as adenine methylation (54).
Fur proteins may also indirectly activate gene expression via the repression of negatively acting small RNAs (sRNA). In turn, these sRNAs reduce the translation of abundant iron-containing proteins and iron-storage proteins, thereby allowing Fur to indirectly activate their expression when iron is abundant. This type of regulation is generically referred to as an iron-sparing response (reviewed in references [55, 56]). Though rarer, apo-Fur can also act as a direct activator, as documented in E. coli (25), C. jejuni (22), and Staphylococcus aureus (57). Borrelia burgdorferi BosR may also be an example of a Fur homolog that functions as an activator in the absence of a regulator metal (although it requires a structural Zn ion for protein folding) (58).
Homotypic interactions: oligomerization and polymerization of FUR proteins.
Functioning as a traditional repressor, iron-bound Fur (holo-Fur) binds to DNA to prevent the transcription of genes involved in iron acquisition and in the iron-sparing response (17–25, 59). As originally defined, the Fur consensus (Fur box) is a 19 bp inverted nucleotide repeat in DNA (60). This sequence is best viewed as two overlapping 15 bp (7-1-7) inverted repeats (61). At such sites, two Fur dimers interact with overlapping repeat sequences on opposing faces of the DNA (Fig. 1), as visualized in structures of the Magnetospirillum gryphiswaldense Fur protein bound to DNA (62) and in the structure of E. coli Zur, which is a zinc-sensing Fur paralog (63). This general architecture is conserved across bacteria, and searches using a modified information theory model of overlapping Fur-binding sites was successful at the identification of Fur-regulated genes (64). A similar dimer-of-dimers model likely applies to other members of the extended FUR superfamily (65). FUR proteins can also regulate genes when bound as a single dimer, as when B. subtilis Fur represses the expression of the feuA promoter controlling ferri-siderophore uptake functions (66).
FIG 1.
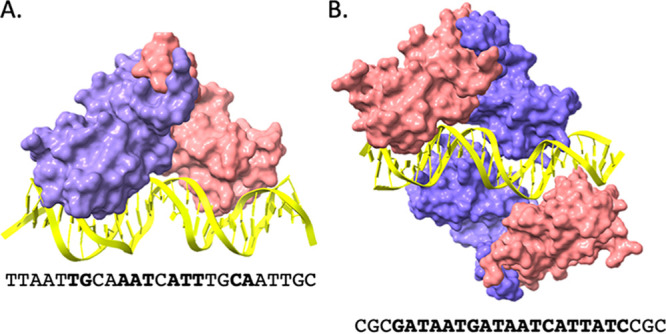
Space-filling representation of Fur protein (from Magnetospirillum gryphiswaldense MSR-1) (62) that was activated by Mn2+ and bound to operator DNA. (A) A dimer of Fur protein (with one protomer in purple and another in pink) bound to the P. aeruginosa feoAB1 operator site (PDB: 4RB3). Bases that match the 7-1-7 consensus (TGATAATnATTATCA) for Fur binding are in bold. (B) Two Fur dimers in complex with a consensus 19 bp Fur box (PDB: 4RB1), which can be represented as two overlapping 7-1-7 consensus sites (61).
Although FUR proteins are commonly observed as stable dimers in solution, other oligomeric states may also be relevant. For example, purified Fur proteins from Pseudomonas aeruginosa, Francisella tularensis, and Legionella pneumophila form stable tetramers, and tetramer dissociation is required to generate dimers that are competent to bind DNA (67, 68). Similarly, Anabaena FurC (a PerR ortholog) forms both noncovalent and disulfide-linked tetramers in solution (69).
At some regulatory regions, FUR dimers may oligomerize to form extended arrays or interact to form compacted nucleoprotein structures (70, 71). The extended complexes formed by Fur may result from contiguous DNA-binding sites or result from favorable protein-protein interactions that are nucleated by an initially bound Fur dimer. Other FUR family members can also form extended arrays on DNA. The precise arrangement of the oligomerized FUR proteins along or around the DNA helix has not been resolved. Streptomyces coelicolor Zur has been studied in detail, and repression was associated with Zur binding to target sites as dimers or tetramers, whereas activation of the zitB efflux gene was associated with the formation of an extended Zur protein array (50). FUR proteins may also bind cooperatively to spatially separated sites. For example, H. pylori Fur forms a repression complex at the arsRS acid acclimation operon with as many as three separated binding sites condensed together to form a compacted nucleoprotein complex. In sum, FUR proteins may bind as dimers, as a dimer-of-dimers (Fig. 1), or in more extensive complexes, including extended protein arrays or compact complexes that are mediated by DNA looping or wrapping.
In addition to increasing DNA occupancy, dimers from solution may kinetically facilitate protein dissociation from DNA (Fig. 2) (72). This type of protein-assisted DNA dissociation can be observed via the single-molecule tracking of fluorescently labeled proteins in vivo by monitoring the on and off rates of DNA-bound complexes as a function of protein concentration. In the first noted example, it was found that the increasing concentrations of the E. coli CueR metalloregulator (a member of the MerR family) led to an increase in the DNA dissociation rate (73). Similarly, E. coli Zur dissociates from its operator site in a concentration-dependent manner that is indicative of protein-assisted dissociation (74). The ability of metalloregulators to kinetically facilitate protein dissociation may be particularly important for those regulators that bind metal ions, such as Cu+ and Zn2+, which bind with high affinity to proteins. These ions can have low rates of dissociation from their cognate metalloregulators, and, when the dissociation of bound protein is also slow, this can create a kinetic barrier to gene induction in response to the falling levels of metals in the cell.
FIG 2.
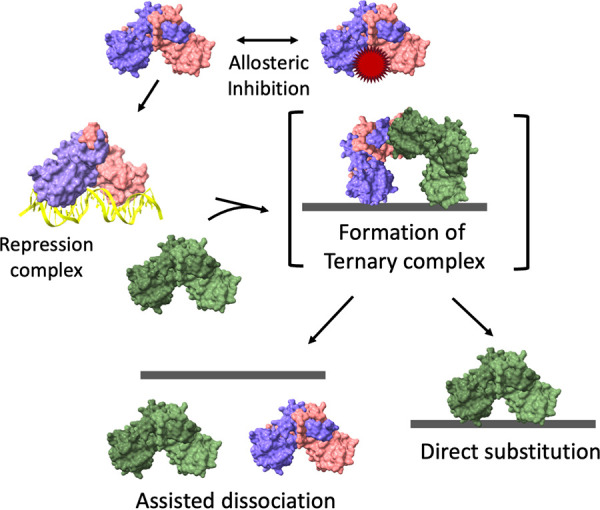
Schematic illustration of a representative Fur family protein binding to DNA as a dimer and the regulatory impact of allosteric regulators and protein-facilitated operator dissociation. Most antagonists likely act from solution to impede the binding of the metalloregulator to DNA (gray line). Once bound, protein still in solution is postulated to interact transiently with DNA-bound protein to form an unstable ternary complex that can then resolve to yield either a direct substitution (with no change in regulation) or the dissociation of both dimers (leading to either derepression or a loss of activation), as described in (72–74).
Heterotypic interactions: competition and cooperation in metalloregulation.
FUR proteins, like most transcription factors, can also be affected by interactions with other regulators that bind in the vicinity of their cognate operator sites or even compete for the same sites. The binding of multiple regulatory proteins may be independent, cooperative, or competitive. Transcription regulation is thereby dependent on the precise set of bound proteins, which allows the integration of multiple signals to control gene expression.
Competition often results when regulatory binding sites are positioned to allow one protein to sterically exclude another. This type of interaction is seen in the competitive binding of a Mn2+-responsive Fur homolog (Mur) and an iron-responsive regulator Irr in the regulatory region of the irr gene in Bradyrhizobium japonicum (75, 76). At this promoter, Mur represses transcription, and Irr acts as an antirepressor under iron limitation by binding to an overlapping operator site and occluding Mur binding. A similar type of antagonism has been suggested to affect DNA occupancy by B. subtilis Fur, depending on the activity of two other DNA-binding regulators that signal redox changes in the cell: ResD and NsrR (77). ResD activity is correlated with anaerobiosis, whereas NsrR signals the presence of nitric oxide, a reactive gas that antagonizes Fur function via the nitrosylation of bound iron (34, 35). Similarly, the DNA binding of the H. pylori Fur protein can be antagonized by NikR, a Ni-responsive repressor (78, 79). While direct competition for a single binding site is rare, in the regulatory region controlling the expression of the B. subtilis pfeT gene (encoding an iron efflux ATPase), Fur and PerR compete for binding at one of three DNA-binding sites (47). This type of overlapping DNA recognition reflects the fact that the B. subtilis Fur, PerR, and Zur consensus sites differ by only one or two bases per half-site. At the pfeT regulatory region, PerR binds cooperatively to two upstream sites with the promoter proximal site overlapping the promoter. The competitive binding of Fur to the promoter distal site allows for the replacement of the PerR repression complex with the Fur protein, which, at this promoter, functions as a transcription activator (47).
Cooperative binding between FUR proteins and other DNA-binding regulators has also been observed. The B. subtilis catDE operon, encoding a catechol dioxygenase, is repressed by Fur, and this repression is cooperative with CatR, which is a sensor of catechols (19). Similarly, NsrR and ResD bind cooperatively with Fur at some coregulated sites under anaerobic conditions (77). In S. aureus, the synthesis and uptake of the nicotianamine-like metallophore staphylopine is regulated by both Fur and Zur (53). In this case, the biosynthesis gene cluster (cntK operon) is regulated independently by Fur and Zur from separate binding sites. However, the uptake genes (cntA operon) are apparently regulated by the cooperative binding of both Fur and Zur. This uptake operon is most sensitively repressed by low levels of Zn, but this repression requires both the Fur and Zur proteins (53). Thus, FUR proteins can bind competitively or cooperatively with other regulators, a property shared with many other DNA-binding transcription factors.
PROTEIN-PROTEIN INTERACTIONS THAT ANTAGONIZE DNA BINDING
In addition to cooperative and competitive interactions between FUR proteins and other DNA-binding regulators, numerous examples have emerged of allosteric regulatory proteins that bind FUR proteins to prevent DNA binding. In the simplest case, a regulator binds a FUR protein to form an inactive complex, but more complex types of interactions are also likely involved.
Uropathogenic E. coli: modulation of Fur activity by YdiV and SlyD.
Uropathogenic E. coli (UPEC) infections require the successful navigation of environments with highly variable iron availability. During the early stages of infection, the bacteria require flagellar motility to colonize the host, whereas at later stages, flagellar motility is turned off to evade the host’s innate immune system, and high-affinity iron uptake is induced (80). Recently, the EAL (Glu-Ala-Leu) domain protein YdiV and SlyD, which is a peptidyl-prolyl cis-trans isomerase, were found to function together to reduce DNA binding by Fur in UPEC (81). The ydiV gene is itself induced in response to the low iron conditions, but this induction is independent of Fur. In a strain overexpressing YdiV, the DNA-binding activity of Fur decreases 300-fold, and this decrease depends on the presence of the SlyD prolyl isomerase. Fur protein purified from cells with YdiV and SlyD is in an altered conformation, and this conformational change depends on a Pro residue at position 18. A bacterial two-hybrid study revealed formation between both Fur-SlyD and Fur-YdiV, but not between SlyD-YdiV. Thus, a model emerges in which YdiV binds to Fur, and this may enhance the ability of the SlyD prolyl isomerase to convert Fur into an inactive conformer (Fig. 3). It is not yet clear whether this isomerization occurs only during the folding of nascent Fur protein or whether it might act on extant Fur molecules. The two conformers, presumably differing in the presence of a cis- versus trans-Pro bond, still bind iron with similar affinities, but they differ greatly in their ability to bind DNA. The two Fur conformers also differ in their propensity to form disulfide bonds, but it was not established whether a disulfide bond occurs in vivo; disulfide bonds in cytoplasmic proteins are generally rare and transient. Together, YidV and SlyD are important for the efficient expression of iron uptake functions, and, therefore, for establishing an infection within host bladder epithelial cells (81). In addition to its role in helping counter the Fur repression of iron uptake functions, YdiV also functions as a protein antagonist of the major activator of flagellar motility, namely, the FlhD4C2 protein complex. This regulator may thereby help coordinate the induction of iron uptake functions with the shutoff of flagellar motility during the invasion into epithelial cells.
FIG 3.
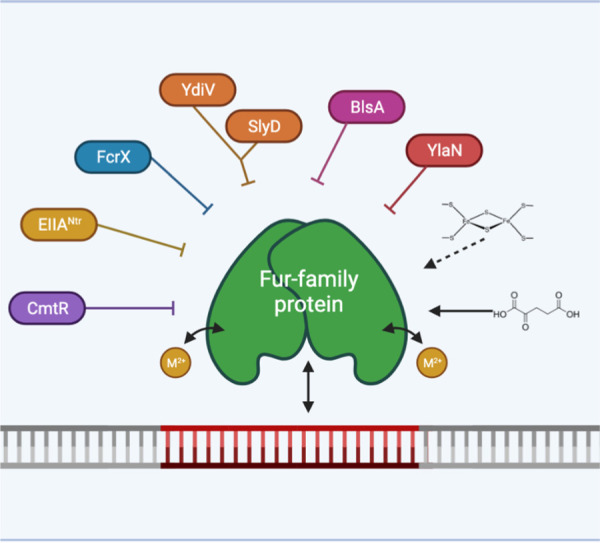
Summary of protein and small molecule antagonists that affect the function of FUR regulators. FUR regulators most commonly require a divalent metal ion (M2+) to activate DNA-binding. However, numerous other factors have now been revealed that can also modulate FUR function, often by acting as antagonists of DNA-binding. The signals that control the expression and activity of each of these effectors are only partially understood (see the text for details).
The details of how the YdiV-SlyD system is itself regulated remains to be clarified. Although ydiV is induced by low iron conditions, the nature of this regulation is unclear. Further, whereas many EAL domain proteins bind and hydrolyze cyclic-di-GMP, the YdiV family of standalone EAL domain proteins are catalytically inactive and do not bind c-di-GMP (82). The activity of SlyD is also puzzling, as it is not yet clear whether it only acts during protein folding to affect the activity of newly synthesized Fur protein, nor is it established whether the reported effects of protein conformation on disulfide bond formation are relevant in vivo.
Salmonella enterica: EIIANtr antagonizes Fur DNA binding.
Salmonella enterica, like many bacteria, employs phosphotransferase (PTS)-dependent sugar import systems that couple the phosphorylation of sugars to their import. This system is initiated via the phosphoenolpyruvate (PEP)-dependent phosphorylation of enzyme I (EI) followed by phosphotransfer to histidine protein (HPr), membrane tethered EIIA and EIIB proteins, and, ultimately, to sugars being imported into the cell (83). In addition, several proteobacteria encode a parallel nitrogen phosphotransferase system (PTSNtr) that comprises a homologous set of proteins: EINtr, Npr, and EIIANtr (84). The EINtr protein binds l-glutamine and 2-oxoglutarate (2-OG) to integrate information about cellular nitrogen and carbon fluxes. The output of the PTSNtr system is phosphorylated EIIANtr, which interacts with a variety of proteins to allosterically regulate their activity (85).
Iron homeostasis is of paramount importance during infection of the mammalian host, which is often an iron-restricted environment. Choi and Ryu have shown that EIIANtr regulates iron uptake genes through a protein-protein interaction with Fur (86). S. enterica encounters iron limitation within the macrophage phagosome. In response, S. enterica derepresses Fur-regulated iron uptake systems. However, the derepression of the Fur regulon is compromised in ptsN mutant strains that lack the EIIANtr protein (87). Interestingly, this regulatory effect of EIIANtr is dependent on the presence of Fur, suggesting that EIIANtr may physically interact with Fur to decrease iron uptake repression. Indeed, a bacterial two hybrid assay revealed a physical interaction between EIIANtr and Fur, and, in vitro, EIIANtr antagonizes Fur binding to DNA.
There are over 600 species of bacteria that contain both Fur and EIIANtr proteins, and, thus, EIIANtr may affect iron homeostasis in other related organisms (86). Because EIIANtr is involved in regulating other cellular processes, it is possible that EIIANtr regulation may allow Fur to integrate signals besides iron starvation. Intriguingly, phosphorylation was not necessary for the interaction of EIIANtr and Fur (86). Thus, it is unlikely that nitrogen or glutamine concentrations are the key signals for this EIIANtr-dependent regulation. Instead, EIIANtr may function by helping titrate free Fur protein to assist in the derepression of the Fur regulon under low iron conditions. A mechanism for the titration of free metalloregulator, and possibly for protein-assisted dissociation, may be particularly important for Fur, as this protein can be abundant, with up to 10,000 proteins per cell in E. coli (7).
Vibrio parahaemolyticus: FcrX antagonizes Fur DNA binding.
Iron can be a limiting nutrient for many marine species, including the marine pathogen Vibrio parahaemolyticus, which exists as both swimming cells in marine environments as well as swarming cells during the colonization of surfaces, such as the chitinous shells of crustaceans (88). V. parahaemolyticus differentiates from unipolarly to laterally flagellated cells on surfaces, thereby allowing for swarming motility and colonization. Differentiation into swarmer cells requires a temporal gene expression cascade that is mediated by the LafK activation of σ54-depedent promoters. This differentiation is inhibited by polar flagella and is induced by iron limitation.
Morabe and McCarter discovered that the fcrX gene is repressed by Fur in iron-replete conditions (89). In a fur mutant, or upon overexpression of fcrX, cells exhibit increased swarming motility and derepress iron uptake. Together, this suggests that FcrX plays an antagonistic role toward Fur. Indeed, FcrX binds directly to Fur, as judged by coimmunoprecipitation evidence. The authors propose a model in which Fur binds iron under iron-sufficient conditions and represses the transcription of genes containing an upstream Fur box (90). As iron concentrations fall, the expression of Fur-regulated genes, including fcrX, is increased. FcrX then binds to Fur to block iron loading or to inhibit repression by holoFur. Alternatively, FcrX might work to kinetically facilitate the dissociation of holoFur from DNA. Further studies are needed to discern how Fur regulation is integrated with the LafK-dependent activation of swarming-related genes (91–94). Since swarmer cell differentiation genes lack Fur boxes, iron regulation may be indirect (90). One possibility is that Fur regulates the expression of another regulator that is required for the expression of these swarmer cell genes.
Acinetobacter baumannii: photoreceptor BlsA allows for the light-sensitive modulation of the Fur regulon.
In the multidrug resistant human pathogen Acinetobacter baumannii, light intensity influences biofilm formation, metabolism, antibiotic resistance, virulence, and iron limitation responses (95). Blue light using flavin (BLUF) proteins sense blue light and transmit this signal to downstream effector proteins through protein-protein interactions. The A. baumannii BLUF photoreceptor BlsA antagonizes the Fur-dependent repression of gene expression in the dark and at 23°C but not at 30°C (96). Consistently, even under iron-limited conditions, the expression of the Fur-regulated acinetobactin siderophore was only induced in the dark and in a blsA-dependent manner. The BLUF protein BlsA allows both light and temperature signals to regulate A. baumannii Fur when the organism is outside the host. How these light and temperature signals may affect the sensing of iron status during infection is not yet clear.
B. subtilis: YlaN modulates Fur DNA binding.
B. subtilis Fur represses a large regulon of genes involved in siderophore biosynthesis and iron import pathways, and it indirectly activates numerous Fe-containing enzymes through the small RNA FsrA (97). The transcription of fur is repressed by another FUR protein, namely, the peroxide-sensor PerR. In a perR null mutant, Fur protein levels are elevated by approximately 2.2-fold, and this increase results in the repression of iron uptake functions due to the binding of intracellular Mn2+ to the Fe2+-sensing sites in Fur (40). This inappropriate repression of iron uptake contributes to a severe growth defect in perR mutant strains. Indeed, perR is essential in B. licheniformis, unless fur is also inactivated (98). These findings highlight the importance of tightly regulating Fur protein levels. This also illustrates the deleterious consequences of the mismetalation of metalloregulatory proteins. For example, Mn2+ can act as a toxic agonist of Fur proteins in many Gram-negative organisms.
Genome-wide efforts in B. subtilis led to the identification of approximately 260 essential genes (99). Among these, several have no functionally characterized homologs, and these were therefore considered to be attractive candidates for the development of new antimicrobials. One such gene, namely, ylaN, encodes a small, dimeric protein that is comprised of 3 alpha-helices (100). Using a CRISPRi inhibition approach to characterize the consequences of depletion, Peters et al. discovered that the severe growth defects due to ylaN depletion (and even deletion) could be rescued by iron supplementation (101). In a genome-wide screen of protein-protein interactions that were detected using a bifunctional cross-linker, YlaN was found to bind to Fur protein, with the cross-linked lysine residue in Fur (K74) being near the regulatory metal-binding site (102). Thus, YlaN is postulated to bind to and inhibit Fur, and, in the absence of YlaN, an unregulated increase in Fur-mediated repression and subsequent iron starvation may account for the apparently essential role of ylaN. YlaN proteins are largely restricted to Bacilli and relatives (COG4838), and the gene seems to be constitutively expressed. It is not yet clear which signals may regulate the amount or activity of YlaN in the cell to modulate iron homeostasis.
Mycobacterium bovis CmtR antagonizes Zur DNA binding.
Mycobacterial species encode up to five paralogous type VII secretion systems (T7SS), and they are known as ESX-1 through ESX-5 (103, 104). The ESX-3 system is intimately linked to metal homeostasis, as judged by the inability of the Mycobacterium tuberculosis mutants in this locus to grow in culture, unless supplemented with iron or, even better, iron and zinc (105). The ESX-3 system secretes effector proteins that are important for siderophore-mediated iron acquisition (106), and they may additionally be involved in other metal homeostasis pathways. Indeed, the expression of the ESX-3 locus is repressed by three different metalloregulators: the iron sensor IdeR (107), the zinc sensor Zur (108), and the manganese sensor MntR (109).
The M. bovis ESX-3 system is additionally regulated by CmtR, which is a metalloregulatory protein of the SmtB/ArsR family (110). CmtR likely functions physiologically as a thiol-dependent sensor of redox stress (111). In response to peroxide stress, CmtR dissociates from an autoregulatory binding site in its own promoter region, thereby leading to an increase in CmtR protein. Unexpectedly, CmtR enhanced the expression of the ESX-3 T7SS, despite having no binding activity to the promoter region of this operon. This effect results from binding between CmtR and Zur, which reduces Zur DNA binding. These results support a model in which oxidative stress derepresses cmtR expression and elevated CmtR antagonizes the Zur repression of the esx3 operon (111).
Products of the esx3 operon, including EsxG and EsxH, help combat oxidative stress, although the mechanism is still unclear. EsxGH may act as external zinc scavengers that chaperone zinc to importers. Such external zinc chaperones have been identified in B. subtilis (ZinT) and in P. dentrificans (AtzD). It has been suggested that Zn import may help alleviate oxidative stress, but the molecular basis for this effect is not clear. In fact, H2O2 can lead to the demetallation of Fe-dependent enzymes that can then be inactivated via metalation by Zn (112). Under these conditions Zn import might be disadvantageous. Further efforts are needed to clarify how these multiple regulators bind to the esx3 promoter region (Zur, IdeR, MntR) and, acting from solution (CmtR), work together to sense diverse signals to control ESX3 expression.
Pseudomonas aeruginosa PacT inhibits Fur DNA binding.
Toxin-antitoxin (TA) systems in bacteria consist of a stable toxin protein and a cognate unstable antitoxin. The bacterial pathogen Pseudomonas aeruginosa encodes multiple type II TA systems, which are those with a toxin that is regulated by a protein antitoxin (113). One recently characterized type II TA system, namely, PacTA, contains a toxin (PacT) that has a GCN5-related N-acetyltransferase (GNAT) domain and is thought to arrest translation via the acetylation of charged tRNAs. The cognate anti-toxin, namely, PacA, is a member of the AraC family of DNA-binding proteins (113). In addition to its effects on translation arrest, the PacTA system also affects iron homeostasis. A proteomics analysis of pacTA deletion mutants revealed that several Fur-regulated iron-uptake proteins had reduced expression, suggesting that PacTA modulates iron homeostasis in P. aeruginosa (114). PacT specifically bound to P. aeruginosa Fur in vitro to inhibit DNA binding. The inhibition of Fur DNA-binding activity by PacT derepresses genes for iron uptake and biofilm formation, two traits that are important for P. aeruginosa virulence. Interestingly, the coexpression of Fur and PacT reduced the toxic effect of PacT, suggesting that Fur binding to PacT also provides some antitoxin activity. In addition, pacT and pacA expression is enhanced upon the addition of the iron chelator dipyridine, indicating that iron starvation acts as a signal for the induction of the PacTA system via a mechanism that is still unknown. Together, these results support a model in which PacT inhibits bacterial translation, unless it is bound to Fur or attenuated by the antitoxin PacA.
SMALL MOLECULES THAT AFFECT FUR FAMILY PROTEIN FUNCTION
Bradyrhizobium japonicum Irr: regulation by the binding of heme.
In alphaproteobacteria, the Irr subfamily of FUR proteins sense iron in the form of the biologically important chelate heme. The best studied representative of this subfamily is Bradyrhizobium japonicum Irr (9, 115). B. japonicum, like many other alphaproteobacteria, also encodes Mur, which is a FUR protein that senses Mn2+. In turn, Mur regulates the expression of Irr, which responds physiologically to changes in iron status (76). Despite its role in regulating iron homeostasis, Irr (like Mur) binds DNA in complex with Mn2+ (115). Indeed, Irr binds Mn2+ with high affinity, suggesting that Mn2+ is the relevant metal cofactor for DNA binding under all but the most Mn2+-limited conditions. Mn2+ binds to Irr in a site resembling the metal activation site in B. subtilis PerR, which also binds to Mn2+ with a higher affinity (Kd of approximately 2.8 μM) (15) than does the dedicated Mn2+ sensor MntR (Kd of approximately 13 μM) (116). Under iron replete conditions, heme synthesized by ferrochelatase is delivered to Irr, where it competitively displaces Mn2+, thereby antagonizing DNA binding (117). An additional layer of regulatory control results from the proteolytic instability of the heme-bound Irr protein. The displaced Irr-heme complex can be oxidized and degraded, which further reinforces the derepression of Irr-regulated genes. The heme-dependent release of Irr from DNA leads to the expression of heme synthesis enzymes, iron-containing proteins, and proteins involved in iron storage and efflux. Irr is also an activator of transcription, and, when heme levels are low, Irr binds DNA to activate the expression of iron uptake genes (76). At sites where Irr functions as an activator, heme may antagonize activation, independent of DNA dissociation or degradation (118). The picture that emerges is that Irr senses heme rather than free Fe2+ to monitor iron status, and it does so by competition with Mn2+. Together, Mur and Irr coordinate gene expression and allow cells to integrate signals that are related to Mn2+, Fe2+, redox status, and heme availability.
Anabaena sp. PCC 7120: 2-oxoglutarate enhances DNA binding by Fur at some sites.
The extent to which FUR proteins regulate gene expression through interactions with other small molecules is not yet clear. In one example, Anabaena sp. PCC 7120 FurA senses both Fe2+ and 2-oxoglutarate (2-OG) (119), which is a central metabolite at the intersection of the Krebs cycle (carbon metabolism) and nitrogen metabolism. The accumulation of 2-OG serves as a signal of nitrogen limitation. FurA binds to 2-OG with a high affinity (Kd of approximately 2.8 μM) (119), and this allows FurA to repress ntcA. NtcA is a CRP (cAMP receptor protein) family protein that regulates nitrogen scavenging and assimilation genes, and it is important for the differentiation of nitrogen-fixing heterocysts.
The FurA regulation of ntcA appears to be part of a complex feedback loop. The binding of 2-OG to NtcA activates nitrogen assimilation genes and also upregulates the expression of FurA. FurA also senses 2-OG and represses ntcA, thereby completing a feedback loop to shut off further NtcA synthesis. The repression of ntcA by FurA requires a bound metal cofactor (Mn2+ is used for in vitro studies), and it is enhanced when 2-OG is present. In contrast, 2-OG did not affect FurA binding to a different target site, namely, isiB (119). Thus, nitrogen assimilation genes can be regulated by NtcA and FurA, and they are affected by iron and redox homeostasis as well as nitrogen limitation (120).
FurA may also sense other signals, in addition to Fe2+ and 2-OG. Some FUR regulators have a Cys-rich metal-binding site that coordinates a Zn2+ ion that is necessary for proper protein folding. Although FurA has two CxxC motifs, the protein does not copurify with Zn2+. The FurA Cys residues are redox active and can be found in either reduced or disulfide bond form in the cytosol (121). The C-terminal CxxC motif is maintained in a reduced state by thioredoxin A (TrxA) (122), and one of these Cys residues (C141) contributes to the ability of FurA to bind heme, which prevents DNA binding (123–125). How redox status may regulate FurA activity in vivo is not fully resolved, but only the fully reduced form of FurA is competent to sense 2-OG and to respond to heme.
E. coli Fur: iron-sulfur cluster assembly.
E. coli Fur is one of the best characterized metalloregulators, and it has both a structural Zn2+ site and an Fe2+-sensing regulator site. In addition, there is a third potential metal-binding site that is rich in Cys residues. Recently, it has been noted that E. coli Fur also has the potential to bind an iron-sulfur cluster [2Fe-2S] at this auxiliary sensing site (126, 127). The in vivo relevance of the [2Fe-2S] cluster assembly on Fur function is not yet clear. Only a small fraction (approximately 4%) of Fur protein purified from wild-type E. coli was found to contain a [2Fe-2S] cluster. However, upon the deletion of two iron-sulfur cluster assembly proteins, namely, IcsA and paralogous SufA, intracellular iron concentrations were significantly increased, and Fur was purified in a red form with approximately 31% of the protein containing a bound [2Fe-2S] cluster at site 3 (126). Site 3 is an auxiliary site of variable (and often uncertain) relevance that is found in a subset of Fur family members (10, 16). The third site contains three cysteine residues that, upon mutation, abolish the [2Fe-2S] cluster binding to Fur. Additionally, Fur homologs from H. influenzae and Vibrio cholerae can also bind substoichiometric levels of an [2Fe-2S] cluster when expressed in E. coli mutants with elevated intracellular iron levels (126, 128). To date, no evidence has been presented to demonstrate that the [2Fe-2S] cluster assembly impacts DNA binding or the expression of genes in the Fur regulon, nor is it clear whether the [2Fe-2S] cluster assembly is relevant in wild-type cells.
Conclusions.
FUR proteins are ubiquitous in bacteria and most commonly function to sense divalent metal ions (including zinc, manganese, iron, and nickel) to regulate metal ion homeostasis. Here, we have focused on emerging insights into the ability of FUR proteins to integrate signals via protein-protein or small molecule-protein interactions (Fig. 3). While the demonstration that a diverse set of proteins can function as antagonists of metalloregulator activity is an important first step, additional efforts are required to better define how these allosteric regulators are themselves controlled and to discern the molecular messages that are represented by their binding. Small molecules also have regulatory potential, as is most carefully defined for heme and the 2-OG regulation of FurA in Anabaena, but here, too, there are many complexities that still remain to be explored. Metal ions are central to cell physiology, and metalloregulatory proteins and riboswitches provide the most direct measurement of intracellular metal status. Therefore, it is fitting that metalloregulators serve as such central regulatory hubs, controlling not only metal homeostasis but also diverse metabolic pathways, developmental programs, and virulence gene expression.
ACKNOWLEDGMENTS
We thank Ankita Sachla, Mark O’Brian, and María Fillat for their helpful comments on the manuscript.
Research reported in this publication was supported by the National Institutes of Health under award number R35GM122461 to J.D.H. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
John D. Helmann, Email: jdh9@cornell.edu.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Chandrangsu P, Rensing C, Helmann JD. 2017. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol 15:338–350. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen A, Imlay JA. 2021. How microbes defend themselves from incoming hydrogen peroxide. Front Immunol 12:667343. doi: 10.3389/fimmu.2021.667343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley JM, Svistunenko DA, Wilson MT, Hemmings AM, Moore GR, Le Brun NE. 2020. Bacterial iron detoxification at the molecular level. J Biol Chem 295:17602–17623. doi: 10.1074/jbc.REV120.007746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelis P, Wei Q, Andrews SC, Vinckx T. 2011. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3:540–549. doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 5.Faulkner MJ, Helmann JD. 2011. Peroxide stress elicits adaptive changes in bacterial metal ion homeostasis. Antioxid Redox Signal 15:175–189. doi: 10.1089/ars.2010.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 7.Zheng M, Doan B, Schneider TD, Storz G. 1999. OxyR and SoxRS regulation of fur. J Bacteriol 181:4639–4643. doi: 10.1128/JB.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fillat MF. 2014. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 546:41–52. doi: 10.1016/j.abb.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 9.Sevilla E, Bes MT, Peleato ML, Fillat MF. 2021. Fur-like proteins: beyond the ferric uptake regulator (Fur) paralog. Arch Biochem Biophys 701:108770. doi: 10.1016/j.abb.2021.108770. [DOI] [PubMed] [Google Scholar]
- 10.Sarvan S, Butcher J, Stintzi A, Couture JF. 2018. Variation on a theme: investigating the structural repertoires used by ferric uptake regulators to control gene expression. Biometals 31:681–704. doi: 10.1007/s10534-018-0120-8. [DOI] [PubMed] [Google Scholar]
- 11.Pinochet-Barros A, Helmann JD. 2018. Redox sensing by Fe2+ in bacterial Fur family metalloregulators. Antioxid Redox Signal 29:1858–1871. doi: 10.1089/ars.2017.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kandari D, Joshi H, Bhatnagar R. 2021. Zur: zinc-sensing transcriptional regulator in a diverse set of bacterial species. Pathogens 10:344. doi: 10.3390/pathogens10030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikhaylina A, Ksibe AZ, Scanlan DJ, Blindauer CA. 2018. Bacterial zinc uptake regulator proteins and their regulons. Biochem Soc Trans 46:983–1001. doi: 10.1042/BST20170228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Althaus EW, Outten CE, Olson KE, Cao H, O'Halloran TV. 1999. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry 38:6559–6569. doi: 10.1021/bi982788s. [DOI] [PubMed] [Google Scholar]
- 15.Lee JW, Helmann JD. 2006. Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J Biol Chem 281:23567–23578. doi: 10.1074/jbc.M603968200. [DOI] [PubMed] [Google Scholar]
- 16.Ma Z, Faulkner MJ, Helmann JD. 2012. Origins of specificity and cross-talk in metal ion sensing by Bacillus subtilis Fur. Mol Microbiol 86:1144–1155. doi: 10.1111/mmi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee R, Weisenhorn E, Schwartz KJ, Myers KS, Glasner JD, Perna NT, Coon JJ, Welch RA, Kiley PJ. 2020. Tailoring a global iron regulon to a uropathogen. mBio 11. doi: 10.1128/mBio.00351-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pi H, Helmann JD. 2017. Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis. Proc Natl Acad Sci USA 114:12785–12790. doi: 10.1073/pnas.1713008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pi H, Helmann JD. 2018. Genome-wide characterization of the Fur regulatory network reveals a link between catechol degradation and bacillibactin metabolism in Bacillus subtilis. mBio 9. doi: 10.1128/mBio.01451-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riediger M, Hernandez-Prieto MA, Song K, Hess WR, Futschik ME. 2021. Genome-wide identification and characterization of Fur-binding sites in the cyanobacteria Synechocystis sp. PCC 6803 and PCC 6714. DNA Res 28. doi: 10.1093/dnares/dsab023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butcher J, Handley RA, van Vliet AH, Stintzi A. 2015. Refined analysis of the Campylobacter jejuni iron-dependent/independent Fur- and PerR-transcriptomes. BMC Genomics 16:498. doi: 10.1186/s12864-015-1661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butcher J, Sarvan S, Brunzelle JS, Couture JF, Stintzi A. 2012. Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc Natl Acad Sci USA 109:10047–10052. doi: 10.1073/pnas.1118321109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butcher BG, Bronstein PA, Myers CR, Stodghill PV, Bolton JJ, Markel EJ, Filiatrault MJ, Swingle B, Gaballa A, Helmann JD, Schneider DJ, Cartinhour SW. 2011. Characterization of the Fur regulon in Pseudomonas syringae pv. tomato DC3000. J Bacteriol 193:4598–4611. doi: 10.1128/JB.00340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beauchene NA, Mettert EL, Moore LJ, Keleş S, Willey ER, Kiley PJ. 2017. O2 availability impacts iron homeostasis in Escherichia coli. Proc Natl Acad Sci USA 114:12261–12266. doi: 10.1073/pnas.1707189114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo SW, Kim D, Latif H, O'Brien EJ, Szubin R, Palsson BO. 2014. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun 5:4910. doi: 10.1038/ncomms5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An YJ, Ahn BE, Han AR, Kim HM, Chung KM, Shin JH, Cho YB, Roe JH, Cha SS. 2009. Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucleic Acids Res 37:3442–3451. doi: 10.1093/nar/gkp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manley OM, Myers PD, Toney DJ, Bolling KF, Rhodes LC, Gasparik JL, Grossoehme NE. 2020. Evaluation of the regulatory model for Ni2+ sensing by Nur from Streptomyces coelicolor. J Inorg Biochem 203:110859. doi: 10.1016/j.jinorgbio.2019.110859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hohle TH, O'Brian MR. 2016. Metal-specific control of gene expression mediated by Bradyrhizobium japonicum Mur and Escherichia coli Fur is determined by the cellular context. Mol Microbiol 101:152–166. doi: 10.1111/mmi.13381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnston AW, Todd JD, Curson AR, Lei S, Nikolaidou-Katsaridou N, Gelfand MS, Rodionov DA. 2007. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals 20:501–511. doi: 10.1007/s10534-007-9085-8. [DOI] [PubMed] [Google Scholar]
- 30.Diaz-Mireles E, Wexler M, Sawers G, Bellini D, Todd JD, Johnston AWB. 2004. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology (Reading) 150:1447–1456. doi: 10.1099/mic.0.26961-0. [DOI] [PubMed] [Google Scholar]
- 31.Lee JW, Helmann JD. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 32.Traore DA, El Ghazouani A, Jacquamet L, Borel F, Ferrer JL, Lascoux D, Ravanat JL, Jaquinod M, Blondin G, Caux-Thang C, Duarte V, Latour JM. 2009. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat Chem Biol 5:53–59. doi: 10.1038/nchembio.133. [DOI] [PubMed] [Google Scholar]
- 33.Ahn BE, Baker TA. 2016. Oxidization without substrate unfolding triggers proteolysis of the peroxide-sensor, PerR. Proc Natl Acad Sci USA 113:E23–E31. doi: 10.1073/pnas.1522687112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Autreaux B, Touati D, Bersch B, Latour JM, Michaud-Soret I. 2002. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc Natl Acad Sci USA 99:16619–16624. doi: 10.1073/pnas.252591299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore CM, Nakano MM, Wang T, Ye RW, Helmann JD. 2004. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J Bacteriol 186:4655–4664. doi: 10.1128/JB.186.14.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hantke K. 1987. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol Gen Genet 210:135–139. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]
- 37.Lam MS, Litwin CM, Carroll PA, Calderwood SB. 1994. Vibrio cholerae fur mutations associated with loss of repressor activity: implications for the structural-functional relationships of fur. J Bacteriol 176:5108–5115. doi: 10.1128/jb.176.16.5108-5115.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas CE, Sparling PF. 1996. Isolation and analysis of a fur mutant of Neisseria gonorrhoeae. J Bacteriol 178:4224–4232. doi: 10.1128/jb.178.14.4224-4232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandrangsu P, Helmann JD. 2016. Intracellular Zn(II) intoxication leads to dysregulation of the PerR regulon resulting in heme toxicity in Bacillus subtilis. PLoS Genet 12:e1006515. doi: 10.1371/journal.pgen.1006515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faulkner MJ, Ma Z, Fuangthong M, Helmann JD. 2012. Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J Bacteriol 194:1226–1235. doi: 10.1128/JB.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helmann JD. 2014. Specificity of metal sensing: iron and manganese homeostasis in Bacillus subtilis. J Biol Chem 289:28112–28120. doi: 10.1074/jbc.R114.587071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 43.Delany I, Rappuoli R, Scarlato V. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol Microbiol 52:1081–1090. doi: 10.1111/j.1365-2958.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- 44.Delany I, Spohn G, Rappuoli R, Scarlato V. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol Microbiol 42:1297–1309. doi: 10.1046/j.1365-2958.2001.02696.x. [DOI] [PubMed] [Google Scholar]
- 45.Danielli A, Scarlato V. 2010. Regulatory circuits in Helicobacter pylori: network motifs and regulators involved in metal-dependent responses. FEMS Microbiol Rev 34:738–752. doi: 10.1111/j.1574-6976.2010.00233.x. [DOI] [PubMed] [Google Scholar]
- 46.Gilbreath JJ, West AL, Pich OQ, Carpenter BM, Michel S, Merrell DS. 2012. Fur activates expression of the 2-oxoglutarate oxidoreductase genes (oorDABC) in Helicobacter pylori. J Bacteriol 194:6490–6497. doi: 10.1128/JB.01226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinochet-Barros A, Helmann JD. 2020. Bacillus subtilis Fur is a transcriptional activator for the PerR-repressed pfeT gene, encoding an iron efflux pump. J Bacteriol 202. doi: 10.1128/JB.00697-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao H, Ma L, Qin Q, Qiu Y, Zhang J, Li J, Lou J, Diao B, Zhao H, Shi Q, Zhang Y, Kan B. 2020. Fur represses Vibrio cholerae biofilm formation via direct regulation of vieSAB, cdgD, vpsU, and vpsA-K transcription. Front Microbiol 11:587159. doi: 10.3389/fmicb.2020.587159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarvan S, Charih F, Askoura M, Butcher J, Brunzelle JS, Stintzi A, Couture JF. 2018. Functional insights into the interplay between DNA interaction and metal coordination in ferric uptake regulators. Sci Rep 8:7140. doi: 10.1038/s41598-018-25157-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi SH, Lee KL, Shin JH, Cho YB, Cha SS, Roe JH. 2017. Zinc-dependent regulation of zinc import and export genes by Zur. Nat Commun 8:15812. doi: 10.1038/ncomms15812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang DL, Tang DJ, Liao Q, Li HC, Chen Q, He YQ, Feng JX, Jiang BL, Lu GT, Chen B, Tang JL. 2008. The Zur of Xanthomonas campestris functions as a repressor and an activator of putative zinc homeostasis genes via recognizing two distinct sequences within its target promoters. Nucleic Acids Res 36:4295–4309. doi: 10.1093/nar/gkn328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuangthong M, Helmann JD. 2003. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J Bacteriol 185:6348–6357. doi: 10.1128/JB.185.21.6348-6357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fojcik C, Arnoux P, Ouerdane L, Aigle M, Alfonsi L, Borezee-Durant E. 2018. Independent and cooperative regulation of staphylopine biosynthesis and trafficking by Fur and Zur. Mol Microbiol 108:159–177. doi: 10.1111/mmi.13927. [DOI] [PubMed] [Google Scholar]
- 54.Brunet YR, Bernard CS, Cascales E. 2020. Fur-Dam regulatory interplay at an internal promoter of the enteroaggregative Escherichia coli type VI secretion sci1 gene cluster. J Bacteriol 202. doi: 10.1128/JB.00075-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oglesby-Sherrouse AG, Murphy ER. 2013. Iron-responsive bacterial small RNAs: variations on a theme. Metallomics 5:276–286. doi: 10.1039/c3mt20224k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chareyre S, Mandin P. 2018. Bacterial iron homeostasis regulation by sRNAs. Microbiol Spectr 6. doi: 10.1128/microbiolspec.RWR-0010-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng X, Sun F, Ji Q, Liang H, Missiakas D, Lan L, He C. 2012. Expression of multidrug resistance efflux pump gene norA is iron responsive in Staphylococcus aureus. J Bacteriol 194:1753–1762. doi: 10.1128/JB.06582-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samuels DS, Lybecker MC, Yang XF, Ouyang Z, Bourret TJ, Boyle WK, Stevenson B, Drecktrah D, Caimano MJ. 2021. Gene regulation and transcriptomics. Curr Issues Mol Biol 42:223–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies BW, Bogard RW, Mekalanos JJ. 2011. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc Natl Acad Sci USA 108:12467–12472. doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Escolar L, Pérez-Martín J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181:6223–6229. doi: 10.1128/JB.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baichoo N, Helmann JD. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol 184:5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng Z, Wang Q, Liu Z, Zhang M, Machado AC, Chiu TP, Feng C, Zhang Q, Yu L, Qi L, Zheng J, Wang X, Huo X, Qi X, Li X, Wu W, Rohs R, Li Y, Chen Z. 2015. Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat Commun 6:7642. doi: 10.1038/ncomms8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilston BA, Wang S, Marcus MD, Canalizo-Hernandez MA, Swindell EP, Xue Y, Mondragon A, O'Halloran TV. 2014. Structural and mechanistic basis of zinc regulation across the E. coli Zur regulon. PLoS Biol 12:e1001987. doi: 10.1371/journal.pbio.1001987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Z, Lewis KA, Shultzaberger RK, Lyakhov IG, Zheng M, Doan B, Storz G, Schneider TD. 2007. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res 35:6762–6777. doi: 10.1093/nar/gkm631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berg K, Pedersen HL, Leiros I. 2020. Biochemical characterization of ferric uptake regulator (Fur) from Aliivibrio salmonicida. Mapping the DNA sequence specificity through binding studies and structural modelling. Biometals 33:169–185. doi: 10.1007/s10534-020-00240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baichoo N, Wang T, Ye R, Helmann JD. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol 45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- 67.Nader S, Pérard J, Carpentier P, Arnaud L, Crouzy S, Michaud-Soret I. 2019. New insights into the tetrameric family of the Fur metalloregulators. Biometals 32:501–519. doi: 10.1007/s10534-019-00201-8. [DOI] [PubMed] [Google Scholar]
- 68.Pérard J, Covès J, Castellan M, Solard C, Savard M, Miras R, Galop S, Signor L, Crouzy S, Michaud-Soret I, de Rosny E. 2016. Quaternary structure of Fur proteins, a new subfamily of tetrameric proteins. Biochemistry 55:1503–1515. doi: 10.1021/acs.biochem.5b01061. [DOI] [PubMed] [Google Scholar]
- 69.Sarasa-Buisan C, Emonot E, Martínez-Júlvez M, Sevilla E, Velázquez-Campoy A, Crouzy S, Bes MT, Michaud-Soret I, Fillat MF. 2022. Metal binding and oligomerization properties of FurC (PerR) from Anabaena sp. PCC7120: an additional layer of regulation? Metallomics 14. doi: 10.1093/mtomcs/mfac077. [DOI] [PubMed] [Google Scholar]
- 70.de Lorenzo V, Giovannini F, Herrero M, Neilands JB. 1988. Metal ion regulation of gene expression. Fur repressor-operator interaction at the promoter region of the aerobactin system of pColV-K30. J Mol Biol 203:875–884. doi: 10.1016/0022-2836(88)90113-1. [DOI] [PubMed] [Google Scholar]
- 71.Le Cam E, Frechon D, Barray M, Fourcade A, Delain E. 1994. Observation of binding and polymerization of Fur repressor onto operator-containing DNA with electron and atomic force microscopes. Proc Natl Acad Sci USA 91:11816–11820. doi: 10.1073/pnas.91.25.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen TY, Cheng YS, Huang PS, Chen P. 2018. Facilitated unbinding via multivalency-enabled ternary complexes: new paradigm for protein-DNA interactions. Acc Chem Res 51:860–868. doi: 10.1021/acs.accounts.7b00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joshi CP, Panda D, Martell DJ, Andoy NM, Chen TY, Gaballa A, Helmann JD, Chen P. 2012. Direct substitution and assisted dissociation pathways for turning off transcription by a MerR-family metalloregulator. Proc Natl Acad Sci USA 109:15121–15126. doi: 10.1073/pnas.1208508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jung W, Sengupta K, Wendel BM, Helmann JD, Chen P. 2020. Biphasic unbinding of a metalloregulator from DNA for transcription (de)repression in live bacteria. Nucleic Acids Res 48:2199–2208. doi: 10.1093/nar/gkaa056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hohle TH, O'Brian MR. 2010. Transcriptional control of the Bradyrhizobium japonicum irr gene requires repression by fur and antirepression by Irr. J Biol Chem 285:26074–26080. doi: 10.1074/jbc.M110.145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O'Brian MR. 2015. Perception and homeostatic control of iron in the rhizobia and related bacteria. Annu Rev Microbiol 69:229–245. doi: 10.1146/annurev-micro-091014-104432. [DOI] [PubMed] [Google Scholar]
- 77.Chumsakul O, Anantsri DP, Quirke T, Oshima T, Nakamura K, Ishikawa S, Nakano MM. 2017. Genome-wide analysis of ResD, NsrR, and Fur binding in Bacillus subtilis during anaerobic fermentative growth by in vivo footprinting. J Bacteriol 199. doi: 10.1128/JB.00086-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Delany I, Ieva R, Soragni A, Hilleringmann M, Rappuoli R, Scarlato V. 2005. In vitro analysis of protein-operator interactions of the NikR and fur metal-responsive regulators of coregulated genes in Helicobacter pylori. J Bacteriol 187:7703–7715. doi: 10.1128/JB.187.22.7703-7715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roncarati D, Pelliciari S, Doniselli N, Maggi S, Vannini A, Valzania L, Mazzei L, Zambelli B, Rivetti C, Danielli A. 2016. Metal-responsive promoter DNA compaction by the ferric uptake regulator. Nat Commun 7:12593. doi: 10.1038/ncomms12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lüthje P, Brauner A. 2014. Chapter seven - virulence factors of uropathogenic E. coli and their interaction with the host, p 337–372. In Poole RK (ed), Advances in Microbial Physiology, vol 65. Academic Press. [DOI] [PubMed] [Google Scholar]
- 81.Zhang F, Li B, Dong H, Chen M, Yao S, Li J, Zhang H, Liu X, Wang H, Song N, Zhang K, Du N, Xu S, Gu L. 2020. YdiV regulates Escherichia coli ferric uptake by manipulating the DNA-binding ability of Fur in a SlyD-dependent manner. Nucleic Acids Res 48:9571–9588. doi: 10.1093/nar/gkaa696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mouali YE, Kim H, Ahmad I, Brauner A, Liu Y, Skurnik M, Galperin MY, Römling U. 2017. Stand-alone EAL domain proteins form a distinct subclass of EAL proteins involved in regulation of cell motility and biofilm formation in enterobacteria. J Bacteriol 199:e00179-17. doi: 10.1128/JB.00179-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pflüger-Grau K, Görke B. 2010. Regulatory roles of the bacterial nitrogen-related phosphotransferase system. Trends Microbiol 18:205–214. doi: 10.1016/j.tim.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 85.Gravina F, Degaut FL, Gerhardt ECM, Pedrosa FO, Souza EM, Antônio de Souza G, Huergo LF. 2021. The protein–protein interaction network of the Escherichia coli EIIANtr regulatory protein reveals a role in cell motility and metabolic control. Res Microbiol 172:103882. doi: 10.1016/j.resmic.2021.103882. [DOI] [PubMed] [Google Scholar]
- 86.Choi J, Ryu S. 2019. Regulation of iron uptake by fine-tuning the iron responsiveness of the iron sensor Fur. Appl Environ Microbiol 85. doi: 10.1128/AEM.03026-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gravina F, Sanchuki HS, Rodrigues TE, Gerhardt ECM, Pedrosa FO, Souza EM, Valdameri G, de Souza GA, Huergo LF. 2018. Proteome analysis of an Escherichia coli ptsN-null strain under different nitrogen regimes. J Proteomics 174:28–35. doi: 10.1016/j.jprot.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 88.Freitas C, Glatter T, Ringgaard S. 2020. The release of a distinct cell type from swarm colonies facilitates dissemination of Vibrio parahaemolyticus in the environment. ISME J 14:230–244. doi: 10.1038/s41396-019-0521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Muranishi K, Ishimori K, Uchida T. 2022. Regulation of the expression of the nickel uptake system in Vibrio cholerae by iron and heme via ferric uptake regulator (Fur). J Inorg Biochem 228:111713. doi: 10.1016/j.jinorgbio.2022.111713. [DOI] [PubMed] [Google Scholar]
- 90.Morabe ML, McCarter LL. 2020. Vibrio parahaemolyticus FcrX, a Fur-controlled regulator that inhibits repression by Fur. Mol Microbiol 114:77–92. doi: 10.1111/mmi.14497. [DOI] [PubMed] [Google Scholar]
- 91.Tsang J, Hoover TR. 2014. Themes and variations: regulation of RpoN-dependent flagellar genes across diverse bacterial species. Scientifica (Cairo) 2014:681754. doi: 10.1155/2014/681754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Merino S, Shaw JG, Tomás JM. 2006. Bacterial lateral flagella: an inducible flagella system. FEMS Microbiol Lett 263:127–135. doi: 10.1111/j.1574-6968.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 93.Jaques S, McCarter LL. 2006. Three new regulators of swarming in Vibrio parahaemolyticus. J Bacteriol 188:2625–2635. doi: 10.1128/JB.188.7.2625-2635.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stewart BJ, McCarter LL. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J Bacteriol 185:4508–4518. doi: 10.1128/JB.185.15.4508-4518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mussi MA, Gaddy JA, Cabruja M, Arivett BA, Viale AM, Rasia R, Actis LA. 2010. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J Bacteriol 192:6336–6345. doi: 10.1128/JB.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tuttobene MR, Cribb P, Mussi MA. 2018. BlsA integrates light and temperature signals into iron metabolism through Fur in the human pathogen Acinetobacter baumannii. Sci Rep 8:7728. doi: 10.1038/s41598-018-26127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, Helmann JD. 2008. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci USA 105:11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim JH, Yang YM, Ji CJ, Ryu SH, Won YB, Ju SY, Kwon Y, Lee YE, Youn H, Lee JW. 2017. The inability of Bacillus licheniformis perR mutant to grow is mainly due to the lack of PerR-mediated fur repression. J Microbiol 55:457–463. doi: 10.1007/s12275-017-7051-x. [DOI] [PubMed] [Google Scholar]
- 99.Commichau FM, Pietack N, Stulke J. 2013. Essential genes in Bacillus subtilis: a re-evaluation after ten years. Mol BioSyst 9:1068–1075. doi: 10.1039/c3mb25595f. [DOI] [PubMed] [Google Scholar]
- 100.Xu L, Sedelnikova SE, Baker PJ, Hunt A, Errington J, Rice DW. 2007. Crystal structure of S. aureus YlaN, an essential leucine rich protein involved in the control of cell shape. Proteins 68:438–445. doi: 10.1002/prot.21377. [DOI] [PubMed] [Google Scholar]
- 101.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo B-M, Marta E, Shiver AL, Whitehead EH, Weissman JS, Brown ED, Qi LS, Huang KC, Gross CA. 2016. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.de Jong L, Roseboom W, Kramer G. 2021. A composite filter for low FDR of protein-protein interactions detected by in vivo cross-linking. J Proteomics 230:103987. doi: 10.1016/j.jprot.2020.103987. [DOI] [PubMed] [Google Scholar]
- 103.Vaziri F, Brosch R, Sandkvist M, Cascales E, Christie PJ. 2019. ESX/Type VII secretion systems–an important way out for mycobacterial proteins. Microbiol Spectr 7:7.4.10. doi: 10.1128/microbiolspec.PSIB-0029-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gröschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. 2016. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol 14:677–691. doi: 10.1038/nrmicro.2016.131. [DOI] [PubMed] [Google Scholar]
- 105.Serafini A, Boldrin F, Palù G, Manganelli R. 2009. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol 191:6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng T-Y, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. 2009. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci USA 106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun 70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, Smith I, Palù G, Riccardi G, Manganelli R. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol 189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pandey R, Russo R, Ghanny S, Huang X, Helmann J, Rodriguez GM. 2015. MntR (R v2788): a transcriptional regulator that controls manganese homeostasis in Mycobacterium tuberculosis. Mol Microbiol 98:1168–1183. doi: 10.1111/mmi.13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y, Hemmingsen L, Giedroc DP. 2005. Structural and functional characterization of Mycobacterium tuberculosis CmtR, a PbII/CdII-sensing SmtB/ArsR metalloregulatory repressor. Biochemistry 44:8976–8988. doi: 10.1021/bi050094v. [DOI] [PubMed] [Google Scholar]
- 111.Li X, Chen L, Liao J, Hui J, Li W, He ZG. 2020. A novel stress-inducible CmtR-ESX3-Zn2+ regulatory pathway essential for survival of Mycobacterium bovis under oxidative stress. J Biol Chem 295:17083–17099. doi: 10.1074/jbc.RA120.013017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Imlay JA. 2014. The mismetallation of enzymes during oxidative stress. J Biol Chem 289:28121–28128. doi: 10.1074/jbc.R114.588814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fraikin N, Goormaghtigh F, Melderen LV. 2020. Type II toxin-antitoxin systems: evolution and revolutions. J Bacteriol 202:e00763-19. doi: 10.1128/JB.00763-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Song Y, Zhang S, Ye Z, Song Y, Chen L, Tong A, He Y, Bao R. 2022. The novel type II toxin-antitoxin PacTA modulates Pseudomonas aeruginosa iron homeostasis by obstructing the DNA-binding activity of Fur. Nucleic Acids Res 50:10586–10600. doi: 10.1093/nar/gkac867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nam D, Matsumoto Y, Uchida T, O'Brian MR, Ishimori K. 2020. Mechanistic insights into heme-mediated transcriptional regulation via a bacterial manganese-binding iron regulator, iron response regulator (Irr). J Biol Chem 295:11316–11325. doi: 10.1074/jbc.RA119.011855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Osman D, Martini MA, Foster AW, Chen J, Scott AJP, Morton RJ, Steed JW, Lurie-Luke E, Huggins TG, Lawrence AD, Deery E, Warren MJ, Chivers PT, Robinson NJ. 2019. Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat Chem Biol 15:241–249. doi: 10.1038/s41589-018-0211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Qi Z, O'Brian MR. 2002. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol Cell 9:155–162. doi: 10.1016/S1097-2765(01)00431-2. [DOI] [PubMed] [Google Scholar]
- 118.Jaggavarapu S, O'Brian MR. 2014. Differential control of Bradyrhizobium japonicum iron stimulon genes through variable affinity of the iron response regulator (Irr) for target gene promoters and selective loss of activator function. Mol Microbiol 92:609–624. doi: 10.1111/mmi.12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guio J, Sarasa-Buisan C, Velazquez-Campoy A, Bes MT, Fillat MF, Peleato ML, Sevilla E. 2020. 2-oxoglutarate modulates the affinity of FurA for the ntcA promoter in Anabaena sp. PCC 7120. FEBS Lett 594:278–289. doi: 10.1002/1873-3468.13610. [DOI] [PubMed] [Google Scholar]
- 120.González A, Bes MT, Valladares A, Peleato ML, Fillat MF. 2012. FurA is the master regulator of iron homeostasis and modulates the expression of tetrapyrrole biosynthesis genes in Anabaena sp. PCC 7120. Environ Microbiol 14:3175–3187. doi: 10.1111/j.1462-2920.2012.02897.x. [DOI] [PubMed] [Google Scholar]
- 121.Botello-Morte L, Bes MT, Heras B, Fernández-Otal Á, Peleato ML, Fillat MF. 2014. Unraveling the redox properties of the global regulator FurA from Anabaena sp. PCC 7120: disulfide reductase activity based on its CXXC motifs. Antioxid Redox Signal 20:1396–1406. doi: 10.1089/ars.2013.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Guio J, Bes MT, Balsera M, Calvo-Begueria L, Sevilla E, Peleato ML, Fillat MF. 2021. Thioredoxin dependent changes in the redox states of FurA from Anabaena sp. PCC 7120. Antioxidants 10:913. doi: 10.3390/antiox10060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hernández JA, Peleato ML, Fillat MF, Bes MT. 2004. Heme binds to and inhibits the DNA-binding activity of the global regulator FurA from Anabaena sp. PCC 7120. FEBS Lett 577:35–41. doi: 10.1016/j.febslet.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 124.Pellicer S, González A, Peleato ML, Martinez JI, Fillat MF, Bes MT. 2012. Site-directed mutagenesis and spectral studies suggest a putative role of FurA from Anabaena sp. PCC 7120 as a heme sensor protein. FEBS J 279:2231–2246. doi: 10.1111/j.1742-4658.2012.08606.x. [DOI] [PubMed] [Google Scholar]
- 125.Botello-Morte L, Pellicer S, Sein-Echaluce VC, Contreras LM, Neira JL, Abian O, Velazquez-Campoy A, Peleato ML, Fillat MF, Bes MT. 2016. Cysteine mutational studies provide insight into a thiol-based redox switch mechanism of metal and DNA binding in FurA from Anabaena sp. PCC 7120. Antioxid Redox Signal 24:173–185. doi: 10.1089/ars.2014.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fontenot CR, Tasnim H, Valdes KA, Popescu CV, Ding H. 2020. Ferric uptake regulator (Fur) reversibly binds a [2Fe-2S] cluster to sense intracellular iron homeostasis in Escherichia coli. J Biol Chem 295:15454–15463. doi: 10.1074/jbc.RA120.014814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lill R. 2020. Do FeS clusters rule bacterial iron regulation? J Biol Chem 295:15464–15465. doi: 10.1074/jbc.H120.016190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fontenot CR, Ding H. 2022. Ferric uptake regulators (Fur) from Vibrio cholerae and Helicobacter pylori bind a [2Fe-2S] cluster in response to elevation of intracellular free iron content. Biometals 35:591–600. doi: 10.1007/s10534-022-00390-9. [DOI] [PubMed] [Google Scholar]


