Wider application of next generation genetic sequencing (NGS) has significantly improved diagnosis for patients with inborn errors of immunity (IEI) and is increasingly a routine part of clinical practice [1, 2]. However, functional validation of genetic variants of unknown significance (VUS) remains a significant challenge for confirming definitive diagnoses, both due to inconsistent access to specialized testing and poor standardization of currently available assays [3]. The challenges are greater for conditions where clinical phenotype is not confirmatory due to heterogeneity or incomplete penetrance.
STAT1 gain-of-function (GOF) immunodeficiency typically presents with chronic mucocutaneous candidiasis (CMC), with or without a combination of bacterial, viral or mycobacterial infections, along with other complications including bronchiectasis, autoimmunity and vascular abnormalities [4, 5]. Over 75 different individual genetic mutations have been described to confer gain of function in the STAT1 gene [6] and classically result in increased phosphorylation of STAT1 (pSTAT1) in immune cells, either at baseline or following cytokine stimulation [7]. Therefore, most immunology laboratories that offer validation assays for STAT1 VUS rely on comparing pSTAT1 levels between a potential index case and one or a group of healthy controls. However, levels of pSTAT1 at baseline and upregulation after cytokine stimulation vary considerably in the healthy population and overlap with levels observed in STAT1 GOF patients.
In this study, we performed technically standardized flow cytometry assays using peripheral blood obtained from six patients with five different STAT1 VUS, identified through whole genome or targeted chip panel sequencing for suspected IEI (see Supplementary Table 1 and Methods). We compared these against 4 patients with known pathogenic GOF mutations in STAT1 and 16 healthy controls. A group of six patients with Common Variable Immunodeficiency (CVID) who had no pathogenic mutations identified in currently known IEI genes by NGS and no rare VUS in STAT1 were used as disease controls. Healthy control samples were taken on the same day as patient or disease control samples and assays performed in parallel. STAT1 VUS tested here were rare (allele frequency <10−4 in reference databases) with variable prediction of deleterious impact using in silico prediction tools (Table 1 and Supplementary Fig. 1a and b).
Table 1.
Predicted functional impact of STAT1 variants evaluated by in silico analysis and functional assays
| Group | SIFT raw (<0.05) | PolyPhen2 Raw | CADD PHRED | Patient/mutation | pSTAT1 | pSTAT1 | IL-17 |
|---|---|---|---|---|---|---|---|
| <0.05 | >0.85 | >20 | Fold change > 2* (normalized for HC) | >Mean + SD of HC | <HC of the day | ||
| VUS | Yes | Yes | Yes | 1/G416R | Yes | No | Yes |
| Yes | Yes | Yes | 2/F404V | No | Yes | Yes | |
| No | No | No | 3/E284K | No | Yes | Yes | |
| No | No | No | 4/E284K | Yes | Yes | Yes | |
| Yes | Yes | Yes | 5/K344Q | Yes | Yes | Yes | |
| No | No | No | 6/T419K | Yes | Yes | Yes | |
| GOF | Yes | Yes | Yes | 7/K388E | Yes | Yes | Yes |
| No | No | No | 8/T720I | Yes | Yes | Yes | |
| Yes | Yes | Yes | 9/T385M | Yes | Yes | Yes | |
| Yes | Yes | No | 10/P293S | Yes | Yes | Yes |
*Normalized for the healthy control of the day; VUS = variant of unknown significance; GOF = gain of function; HC= healthy control; SD = standard deviation.
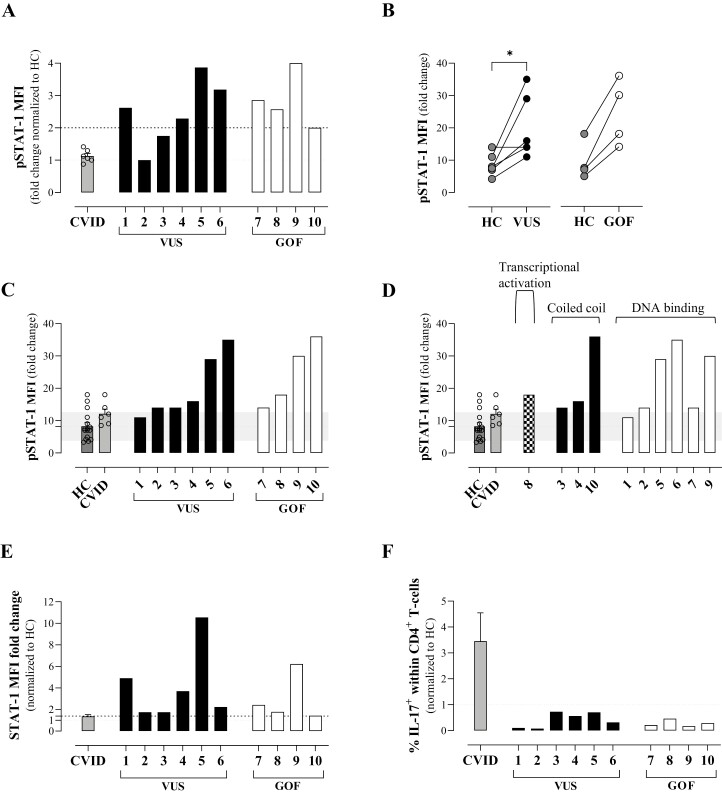
Significant variability was seen in STAT1 levels and in pSTAT1 upregulation in response to interferon-alpha (IFN-α) stimulation for all groups (Supplementary Figs 1c-e and 2a). When considered by group, both STAT1 GOF mutations and STAT1 VUS showed a significant increase in upregulation of pSTAT1 compared with healthy control (Supplementary Fig. 1d), although there was an overlap in the range observed for each group. We then considered each genetic variant individually to determine whether or not VUS could be assigned as GOF. When normalized against their matched healthy control sample, all patients with GOF mutations and 4/6 with VUS demonstrated at least 2-fold higher upregulation in pSTAT1 levels after stimulation (Fig. 1a). Two patients with VUS did not achieve this arbitrary cut off and one of these had a matched healthy control at the upper limit of the healthy control range (Fig. 1b). To reduce the impact of individual healthy control variation, we compared each VUS and GOF mutation against the whole healthy control range and identified that all patients with GOF and 5/6 with VUS upregulated pSTAT1 above a cut off of mean+1SD of the healthy control range (Fig. 1c). Variant location did not appear to impact degree of upregulation (Fig. 1d). Upregulation of total STAT1 was more variable; although all GOF and VUS showed upregulation to at least an arbitrary cut-off of 1.4x healthy control (Fig. 1e), this was considered to be not sufficiently robust to assist diagnosis (Supplementary Fig. 1e). CVID disease controls as a group behaved like healthy controls, with comparable upregulation of pSTAT1 and STAT1 (Fig. 1a, c–e, and Supplementary Fig. 1c–e). To test inclusion of other parameters that could improve diagnosis of GOF mutations, we quantified the frequency of CD4+ T cells producing IL-17 (Fig. 1f, Supplementary Figs 1f and 2b) and upregulation of the expression of STAT1-inducible CXCL-10 in monocytes (Supplementary Figs 3 and 4). In keeping with the known suppression of STAT3 signalling in STAT1 GOF [8], all known GOF mutations supported reduced frequency of Th17 cells (Fig. 1f; Supplementary Figs 1f and 2b). All patients with STAT1 VUS also had lower levels of Th17+ cells than the matched healthy control samples analysed on the same day. In contrast, CXCL-10 expression by monocytes at baseline and fold-change in levels following stimulation were variable between known GOF mutations (Supplementary Fig. 3), reducing the utility of this assay for functional validation of STAT1 VUS.
Figure 1.
STAT1 phosphorylation and Th17 cells. Levels of expression of pSTAT1 (a–d) or total STAT1 (e) on gated CD3+ T cells after stimulation with IFN-α. (f) Frequency of IL-17+ cells within CD4+ lymphocytes. Fold change was calculated as the MFI ratio between IFN-α-stimulated and non-stimulated cells. Each dot or number represents one individual. For (b and c), the mean of the HC group is shown as a dotted line and the standard deviation of the HC group as grey shading.
To support diagnostic reporting, we considered a combination of pSTAT1 and IL-17 production results for each patient (Table 1). All GOF mutations demonstrated elevated levels of pSTAT1 in response to IFN-α stimulation (at least 2-fold upregulation compared with the matched healthy control and above the mean+1SD of the healthy control group range) plus had a frequency of Th17 cells below the matched healthy control. All six patients with VUS also demonstrated increased upregulation of pSTAT1, either when considered against the matched healthy control of the day or against the healthy control group range (Fig. 1a–d) and all 6 had low levels of Th17 compared with the control (Fig. 1f). Thus, using this combination of pSTAT1 and IL-17 measurements, we concluded that all 5 VUS in 6 patients were GOF.
This study highlights the difficulty of definitively assigning GOF status to STAT1 VUS using the single assessment of pSTAT1 and the benefit of including Th17 quantification as an additional parameter. For laboratories performing pSTAT1 assays, highly standardized phosflow protocols and flow cytometer settings together with a local healthy control range are important to reduce variation and improve assessment of index cases. In our hands, pSTAT1 assays only perform reproducibly on fresh blood (taken same day) and older samples are not reliably interpretable. To mitigate against healthy control variation, we suggest that assessment of pSTAT1 is made both against a same-day matched control and the local healthy control range. Future refinement of functional assessment for STAT1 VUS requires development of additional robust assays that can be translated from research to diagnostic laboratories.
Supplementary Material
Acknowledgements
We thank NIHR BioResource volunteers for their participation, and gratefully acknowledge NIHR BioResource centres, NHS Trusts and staff for their contribution. We thank the National Institute for Health and Care Research, NHS Blood and Transplant, and Health Data Research UK as part of the Digital Innovation Hub Programme. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Glossary
Abbreviations
- CMC
chronic mucocutaneous candidiasis
- CVID
common variable immunodeficiency
- CXCL-10
C-X-C motif chemokine ligand 10
- GOF
gain of function
- HC
healthy control
- IEI
inborn errors of immunity
- IFN-α
interferon-alpha
- MFI
mean fluorescence intensity
- NGS
next generation genetic sequencing
- PBMC
peripheral blood mononuclear cell
- PID
primary Immunodeficiency
- PMA
phorbol 12-myristate 13-acetate
- SD
standard deviation
- pSTAT1
phosphorylation of signal transducer and activator of transcription 1
- STAT1
signal transducer and activator of transcription 1
- VUS
variant of unknown significance.
Contributor Information
Adriana S Albuquerque, Institute of Immunity and Transplantation, University College London, London, UK.
Jesmeen Maimaris, Institute of Immunity and Transplantation, University College London, London, UK; Department of Immunology, Royal Free London NHS Foundation Trust, London, UK.
Alexander J McKenna, Institute of Immunity and Transplantation, University College London, London, UK.
Jonathan Lambourne, Department of Infectious Diseases, Royal London Hospital, Barts Health NHS Trust, London, UK.
Fernando Moreira, Department of Immunology, Royal Free London NHS Foundation Trust, London, UK.
Sarita Workman, Department of Immunology, Royal Free London NHS Foundation Trust, London, UK.
Karyn Megy, NIHR BioResource-Rare Disease Consortium, Cambridge University Hospitals, Cambridge Biomedical Campus, Cambridge, UK.
Ilenia Simeoni, NIHR BioResource-Rare Disease Consortium, Cambridge University Hospitals, Cambridge Biomedical Campus, Cambridge, UK.
Hana Lango Allen, NIHR BioResource-Rare Disease Consortium, Cambridge University Hospitals, Cambridge Biomedical Campus, Cambridge, UK; Cambridge Genomics Laboratory, Cambridge University Hospitals NHS Foundation Trust, Cambridge, UK.
Emma C Morris, Institute of Immunity and Transplantation, University College London, London, UK; Department of Immunology, Royal Free London NHS Foundation Trust, London, UK.
Siobhan O Burns, Institute of Immunity and Transplantation, University College London, London, UK; Department of Immunology, Royal Free London NHS Foundation Trust, London, UK.
NIHR BioResource-Rare Disease Consortium:
Zoe Adhya, Hana Alachkar, Ariharan Anantharachagan, Richard Antrobus, Gururaj Arumugakani, Chiara Bacchelli, Helen Baxendale, Claire Bethune, Shahnaz Bibi, Barbara Boardman, Claire Booth, Michael Browning, Mary Brownlie, Siobhan Burns, Anita Chandra, Hayley Clifford, Nichola Cooper, Sophie Davies, John Dempster, Lisa Devlin, Rainer Doffinger, Elizabeth Drewe, David Edgar, William Egner, Tariq El-Shanawany, Bobby Gaspar, Rohit Ghurye, Kimberley Gilmour, Sarah Goddard, Pavel Gordins, Sofia Grigoriadou, Scott Hackett, Rosie Hague, Lorraine Harper, Grant Hayman, Archana Herwadkar, Stephen Hughes, Aarnoud Huissoon, Stephen Jolles, Julie Jones, Peter Kelleher, Nigel Klein, Taco Kuijpers, Dinakantha Kumararatne, James Laffan, Hana Lango Allen, Sara Lear, Hilary Longhurst, Lorena Lorenzo, Jesmeen Maimaris, Ania Manson, Elizabeth McDermott, Hazel Millar, Anoop Mistry, Valerie Morrisson, Sai Murng, Iman Nasir, Sergey Nejentsev, Sadia Noorani, Eric Oksenhendler, Mark Ponsford, Waseem Qasim, Ellen Quinn, Isabella Quinti, Alex Richter, Crina Samarghitean, Ravishankar Sargur, Sinisa Savic, Suranjith Seneviratne, Carrock Sewall, Fiona Shackley, Ilenia Simeoni, Kenneth G C Smith, Emily Staples, Hans Stauss, Cathal Steele, James Thaventhiran, Moira Thomas, Adrian Thrasher, Steve Welch, Lisa Willcocks, Sarita Workman, Austen Worth, Nigel Yeatman, Patrick Yong, Sofie Ashford, John Bradley, Debra Fletcher, Tracey Hammerton, Roger James, Nathalie Kingston, Willem Ouwehand, Christopher Penkett, F Lucy Raymond, Kathleen Stirrups, Marijke Veltman, Tim Young, Sofie Ashford, Matthew Brown, Naomi Clements-Brod, John Davis, Eleanor Dewhurst, Marie Erwood, Amy Frary, Rachel Linger, Jennifer Martin, Sofia Papadia, Karola Rehnstrom, William Astle, Antony Attwood, Marta Bleda, Keren Carss, Louise Daugherty, Sri Deevi, Stefan Graf, Daniel Greene, Csaba Halmagyi, Matthias Haimel, Fengyuan Hu, Roger James, Hana Lango Allen, Vera Matser, Stuart Meacham, Karyn Megy, Christopher Penkett, Olga Shamardina, Kathleen Stirrups, Catherine Titterton, Salih Tuna, Ernest Turro, Ping Yu, Julie von Ziegenweldt, Abigail Furnell, Rutendo Mapeta, Ilenia Simeoni, Simon Staines, Jonathan Stephens, Kathleen Stirrups, Deborah Whitehorn, Paula Rayner-Matthews, and Christopher Watt
Consortia
NIHR BioResource – Rare Diseases Consortium:
The members of the NIHR BioResource – Rare Diseases PID Consortium are Zoe Adhya, Hana Alachkar, Ariharan Anantharachagan, Richard Antrobus, Gururaj Arumugakani, Chiara Bacchelli, Helen Baxendale, Claire Bethune, Shahnaz Bibi, Barbara Boardman, Claire Booth, Michael Browning, Mary Brownlie, Siobhan Burns, Anita Chandra, Hayley Clifford, Nichola Cooper, Sophie Davies, John Dempster, Lisa Devlin, Rainer Doffinger, Elizabeth Drewe, David Edgar, William Egner, Tariq El-Shanawany, Bobby Gaspar, Rohit Ghurye, Kimberley Gilmour, Sarah Goddard, Pavel Gordins, Sofia Grigoriadou, Scott Hackett, Rosie Hague, Lorraine Harper, Grant Hayman, Archana Herwadkar, Stephen Hughes, Aarnoud Huissoon, Stephen Jolles, Julie Jones, Peter Kelleher, Nigel Klein, Taco Kuijpers (principal investigator), Dinakantha Kumararatne, James Laffan, Hana Lango Allen, Sara Lear, Hilary Longhurst, Lorena Lorenzo, Jesmeen Maimaris, Ania Manson, Elizabeth McDermott, Hazel Millar, Anoop Mistry, Valerie Morrisson, Sai Murng, Iman Nasir, Sergey Nejentsev, Sadia Noorani, Eric Oksenhendler, Mark Ponsford, Waseem Qasim, Ellen Quinn, Isabella Quinti, Alex Richter, Crina Samarghitean, Ravishankar Sargur, Sinisa Savic, Suranjith Seneviratne, Carrock Sewall, Fiona Shackley, Ilenia Simeoni, Kenneth G. C. Smith (principal investigator), Emily Staples, Hans Stauss, Cathal Steele, James Thaventhiran, Moira Thomas, Adrian Thrasher (principal investigator), Steve Welch, Lisa Willcocks, Sarita Workman, Austen Worth, Nigel Yeatman, and Patrick Yong.
The members of the NIHRBR-RD Management Team are Sofie Ashford, John Bradley, Debra Fletcher, Tracey Hammerton, Roger James, Nathalie Kingston, Willem Ouwehand, Christopher Penkett, F Lucy Raymond, Kathleen Stirrups, Marijke Veltman, and Tim Young.
The members of the NIHRBR-RD Enrolment and Ethics Team are Sofie Ashford, Matthew Brown, Naomi Clements-Brod, John Davis, Eleanor Dewhurst, Marie Erwood, Amy Frary, Rachel Linger, Jennifer Martin, Sofia Papadia, and Karola Rehnstrom.
The members of the NIHRBR-RD Bioinformatics Team are William Astle, Antony Attwood, Marta Bleda, Keren Carss, Louise Daugherty, Sri Deevi, Stefan Graf, Daniel Greene, Csaba Halmagyi, Matthias Haimel, Fengyuan Hu, Roger James, Hana Lango Allen, Vera Matser, Stuart Meacham, Karyn Megy, Christopher Penkett, Olga Shamardina, Kathleen Stirrups, Catherine Titterton, Salih Tuna, Ernest Turro, Ping Yu, and Julie von Ziegenweldt.
The members of the Cambridge Translational Genomics Laboratory are Abigail Furnell, Rutendo Mapeta, Ilenia Simeoni, Simon Staines, Jonathan Stephens, Kathleen Stirrups, Deborah Whitehorn, Paula Rayner-Matthews, and Christopher Watt.
Ethics approval and consent to participate
The local ethics committees have approved our study (reference numbers 04/Q0501/119 and 06/Q0508/16) and patients and healthy controls gave written informed consent.
Conflict of interests
S.O.B. has received grant support from CSL Behring and personal fees or travel expenses from Immunodeficiency Canada/IAACI, CSL Behring, Baxalta US Inc. and Biotest. E.C.M. has received honoraria from GlaxoSmithKline, AstraZenica, and Orchard Therapeutics. The rest of the authors declare that they have no conflicts of interest.
Funding
This work was supported by the National Institute for Health Research UCLH Biomedical Research Centre (ASA, ECM), the Medical Research Council (AJMcK), and the National Institute for Health Research Integrated Academic Training Scheme (J.M.). The NIHR BioResource Rare Disease Consortium was funded by the National Institute for Health Research (I.S., K.M., and H.L.A.; grant numbers RG65966 and RG94028).
Data availability
The datasets used and analysed in the current study are available from the corresponding author on reasonable request.
Author contributions
A.S.A. performed experiments and analysed data. J.M. and A.J.M. analysed data. E.C.M. and S.O.B. designed the project and supervised the work. A.S.A., J.M., A.J.M., E.C.M., and S.O.B. wrote and edited the manuscript. J.M., J.L., E.C.M., and S.O.B. collected clinical data. F.M. and S.W. consented the patients and healthy controls and collected blood. J.M., K.M., I.S., H.L.A., and NIHR BioResource-Rare Disease Consortium facilitated and identified the genetic variants. J.M. did the bioinformatics analysis. All authors contributed to the article and approved the submitted version.
References
- 1. Convers KD, Slack M, Kanarek HJ.. Take a leap of faith: implement routine genetic testing in your office. J Allergy Clin Immunol Pract 2022, 10, 1676–87. [DOI] [PubMed] [Google Scholar]
- 2. Thaventhiran JED, Lango Allen H, Burren OS, Rae W, Greene D, Staples E, et al. Whole-genome sequencing of a sporadic primary immunodeficiency cohort. Nature 2020, 583, 90–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014, 508, 469–76. doi: 10.1038/nature13127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Toubiana J, Okada S, Hiller J, Oleastro M, Gomez ML, Becerra JCA, et al. Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood 2016, 127, 3154–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Depner M, Fuchs S, Raabe J, Frede N, Glocker C, Doffinger R, et al. The extended clinical phenotype of 26 patients with chronic mucocutaneous candidiasis due to gain-of-function mutations in STAT1. J Clin Immunol 2016, 36, 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Okada S, Puel A, Casanova JL, Kobayashi M.. Chronic mucocutaneous candidiasis disease associated with inborn errors of IL-17 immunity. Clin Translational Immunol 2016, 5, e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med 2011, 208, 1635–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng J, van de Veerdonk FL, Crossland KL, Smeekens SP, Chan CM, al Shehri T, et al. Gain-of-function STAT1 mutations impair STAT3 activity in patients with chronic mucocutaneous candidiasis (CMC). Eur J Immunol 2015, 45, 2834–46. doi: 10.1002/eji.201445344 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analysed in the current study are available from the corresponding author on reasonable request.