Abstract
Background:
With functionally heterogeneous cells, tumors comprise a complex ecosystem to promote tumor adaptability and evolution under strong selective pressure from the given microenvironment. Diversifying tumor cells or intra-tumor heterogeneity is essential for tumor growth, invasion, and immune evasion. However, no reliable method to classify tumor cell subtypes is yet available. In this study, we introduced the single-cell sequencing combined with copy number characteristics to identify the types of tumor cells in microsatellite stable (MSS) colorectal cancer (CRC).
Methods:
To characterize the somatic copy number alteration (SCNA) of MSS CRC in a single cell profile, we analyzed 26 tissue samples from 19 Korean patients (GSE132465, the Samsung Medical Center [SMC] dataset) and then verified our findings with 15 tissue samples from five Belgian patients (GSE144735, the Katholieke Universiteit Leuven 3 [KUL3] dataset). The Cancer Genome Atlas (TCGA) cohort, GSE39582 cohort, and National Cancer Center (NCC) cohort (24 MSS CRC patients were enrolled in this study between March 2017 and October 2017) were used to validate the clinical features of prognostic signatures.
Results:
We employed single cell RNA-sequencing data to identify three types of tumor cells in MSS CRC by their SCNA characteristics. Among these three types of tumor cells, C1 and C3 had a higher SCNA burden; C1 had significant chromosome 13 and 20 amplification, whereas C3 was the polar opposite of C1, which exhibited deletion in chromosome 13 and 20. The three types of tumor cells exhibited various functions in the tumor microenvironment and harbored different mutations. C1 and C2 were linked to the immune response and hypoxia, respectively, while C3 was critical for cell adhesion activity and tumor angiogenesis. Additionally, one gene (OLFM4) was identified as epithelium-specific biomarker of better prognosis of CRC (TCGA cohort: P = 0.0110; GSE39582 cohort: P = 0.0098; NCC cohort: P = 0.0360).
Conclusions:
On the basis of copy number characteristics, we illustrated tumor heterogeneity in MSS CRC and identified three types of tumor cells with distinct roles in tumor microenvironment. By understanding heterogeneity in the intricate tumor microenvironment, we gained an insight into the mechanisms of tumor evolution, which may support the development of therapeutic strategies.
Keywords: Somatic copy number alteration, Microsatellite stable, Colorectal cancer, Single cell RNA-sequencing, Intra-tumor genetic heterogeneity
Introduction
Tumors are complex assemblages of diversiform tumor cells with remarkable variability in their genome and phenotype.[1] Genomic instability is a prominent source of genetic intra-tumor heterogeneity within tumors, generating a heterogeneous cell population that can be subjected to Darwinian selection to foster tumor adaptability and evolution in a given microenvironmental or therapeutic context.[2] Therefore, intra-tumor heterogeneity is crucial for tumor cell proliferation, invasion, immune surveillance evasion, and therapeutic resistance.[3,4]
Chromosomal instability exacerbates intercellular genetic heterogeneity and generates somatic copy number alterations (SCNAs) in >90% of solid tumors and many hematological cancers.[5] SCNAs affect tumor development and the composition of the tumor immune microenvironment.[6] SCNAs are also linked to the immune checkpoint therapy response or resistance by influencing the interferon-γ pathway and/or amplifying and deleting common tumor oncogenes and suppressors.[7] The circumstances by which SCNAs drive tumorigenesis are determined by the tumor stage, cell type, genetic makeup, tumor microenvironment, and immune system interactions.[8] In colorectal cancer (CRC), SCNAs are pertinent to adenoma–carcinoma progression (gains of 8q, 13q, and 20q, and losses of 8p, 15q, 17p, and 18q), clinical histological features, and clinical outcome.[9] This kind of chromosomal instability pathway is strongly associated with microsatellite stable (MSS) CRC, which causes 70% to 90% of CRC.[10]
CRC is the second most frequently diagnosed cancer in female and the third most prevalent cancer in male.[11] Although immunotherapy has revolutionized the treatment for various cancers, it has been less effective for CRC, particularly MSS CRC.[12] Consequently, there is a pressing need to investigate intra-tumor heterogeneity of MSS CRC and further stratify MSS CRC patients into subgroups with distinct genetic and clinical characteristics.
In the past few decades, bulk genomic and transcriptomic analyses have provided valuable insights into tumor research. However, the averaging of signals from large numbers of cells rendered by these methods often obscures specific subpopulations and/or cellular states.[13] Recently, single cell RNA-sequencing (scRNA-seq), which is a powerful sequencing technique based on single cells, has been used to characterize tumor heterogeneity and decipher the interactions between cancer cells and their microenvironmental components.[14,15] Dissecting the complexity of solid tumors is essential to better understand cancer and devise efficient early detection and therapeutic techniques.
This study aimed to investigate intra-tumor heterogeneity in MSS CRC by their SCNA characteristics. By analyzing both scRNA-seq data and bulk RNA sequencing data, we identified distinct SCNA patterns of tumor cells and further stratified MSS CRC patients into different subgroups with SCNA diversity.
Methods
Ethical approval
This study was approved by the Institutional Ethics Review Board of the Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (No. NCC2016JZ-06). All patients provided informed consent.
Public data availability and preprocessing
Processed scRNA-seq count and transcripts per million (TPM) data of CRC samples were obtained from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession codes GSE132465 (the Samsung Medical Center [SMC] dataset contains 63,689 cells from 23 primary CRC tissues and 10 matched normal mucosa tissues of 23 Korean CRC patients) and GSE144735 (the Katholieke Universiteit Leuven 3 [KUL3] dataset contains 27,414 cells from the core and border area of tumor regions as well as matched normal mucosa tissue of the six Belgian CRC patients).[16] These datasets had been filtered by the following criteria: unique molecular identifier counts >1000; 200 < genes < 6000; mitochondrial gene expression < 20% in unique molecular identifier counts; and the filter process took quality control with multiple canonical correlation analysis (CCA) and reference component analysis (RCA).[16] Considering that our MSS CRC analysis was substantial, we excluded microsatellite instability (MSI)-high patients and eventually included 19 patients from the SMC dataset and five patients from the KUL3 dataset [Supplementary Tables 1 and 2]. The Seurat V4 (version 4.0.3, https://satijalab.org/seurat/index.html)[17] of R package (version 4.1.3, https://cran.r-project.org/) was used to analyze the processed scRNA-seq data. For visualization, we reduced the dimensionality of each dataset using uniform manifold approximation and projection (UMAP) with the Seurat function RunUMAP. Cluster-specific differentially expressed genes (DEGs) were identified using the FindAllMarkers function of Seurat.
The TCGA biolinks (version 2.18.0, http://www.bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html) R package[18–20] was used to download bulk RNA-seq gene expression count and fragments per kilobase million data, microsatellite status data, copy number data, and mutation data of The Cancer Genome Atlas (TCGA) colon adenocarcinoma (TCGA-COAD) and rectum adenocarcinoma (TCGA-READ) cohorts. University of Cingifornia Sisha Cruz (UCSC) Xena (http://xena.ucsc.edu/) was used to download TCGA clinical characteristics data.[21] We excluded samples with incomplete copy number data and ultimately included 396 MSS CRC tumor samples from the TCGA cohort. The Maftools (version 2.6.05) R package[22] was applied to analyze and visualize mutation data. Moreover, the GEOquery (version 2.62.2) R package[23] was implemented to download the GSE39582 dataset with 444 MSS colon cancer samples.
Clinical sample collection and RNA sequencing
Colorectal tumor tissue samples were collected from 24 MSS CRC patients who were not treated with chemotherapy or radiotherapy prior to therapeutic resection [Supplementary Table 3]. All patients had undergone surgical treatment at the National Cancer Center (NCC) between March 2017 and October 2017. Twenty-two patients were followed up until death or until April 1, 2022 and the loss rate of follow-up was 8%. As all of the 24 MSS CRC patients had RNA sequencing data and clinical stage data, we used whole patient data for the following analysis. Total RNA was extracted from fresh frozen tissues using TRIzol (Invitrogen, Carlsbad, CA, USA) and then its integrity and concentration were measured by an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA) and ND-1000 (NanoDrop, Wilmington, DE, USA), respectively. Only samples with RNA integrity number > 7.0 were used for transcriptome sequencing with the Illumina NovaSeq6000 platform (Illumina, San Diego, CA, USA). Subsequently, 150 bp paired-end reads were aligned to the human reference genome (GRCh38) by Salmon (https://combine-lab.github.io/salmon/). TPM was used to normalize transcript abundances.
Copy number alteration prediction of scRNA-seq data and subgroup division of tumor cells
To predict SCNA in scRNA-seq data, we used CopyKAT (https://github.com/navinlabcode/copykat) that is designed for high-throughput scRNA-seq platform data and is capable of properly estimating genomic copy number profiles of single cells and characterization of the clonal substructure.[24] A total of 42,141 cells in the SMC dataset and 16,835 cells in the KUL3 dataset were retained after quality filtering in CopyKAT [Supplementary Table 4 and 5]. We excluded SMC05 from the subsequent analysis because the sample had < 10 filtered tumor cells. We used CopyKAT-filtered tumor cells to explore SCNA features, whereas total cells were used for tumor immune microenvironment analysis [Supplementary Table 6].
To ensure reliability of the copy number alteration analysis, we removed the sex chromosomes and performed SCNA analysis of tumor cells using the other 22 pairs of chromosomes. On the basis of copy number discrepancies, we used hierarchical clustering with Ward linkage and Euclidean distance to divide the tumor cells into subclusters. Considering the vital roles of chromosomes 8, 13, 15, 17, 18, and 20 in the adenoma–carcinoma progression, we took into account the pathological mechanism of MSS CRC and the hierarchical clustering results in our data, and then divided the hierarchical clustering tree into three subgroups: C1–C3.
The copy number variation (CNV) burden was estimated by summing the copy number absolute value. Cells with a high CNV burden had numerous copy number changes in a chromosome. Moreover, the CNV score was calculated by summation of the copy number values provided by the CopyKAT result. Consequently, a high CNV score indicated more chromosomal amplification, whereas a low CNV score indicated more chromosomal loss.
Enrichment analysis of DEGs
We filtered DEGs with Bonferroni-adjusted P-value of <0.05 and an absolute log2 fold change of >0.7. To further illustrate the function of DEGs, we applied the clusterProfiler (version 3.18.1, http://mirrors.nju.edu.cn/bioconductor/3.15/bioc/html/clusterProfiler.html) R package[25] for Gene Ontology (GO) analysis. Benjamini–Hochberg-adjusted P values of <0.05 were considered significantly enriched.
Gene set enrichment analysis
We used two types of gene sets for gene set enrichment analysis: the hallmark gene sets from the Molecular Signature Database[26,27] and the ferroptosis-related gene set from the FerrDb database.[28] We chose genes with confidence levels of “validated” or “screened” from the FerrDb database which is more reliable, and then eliminated duplicate genes from gene sets. We finally obtained a 34-gene ferroptosis marker gene set and a 75-gene ferroptosis driver gene set.
We used log_TPM_matrix data provided by GSE132465 and GSE144735 with the gene set variation analysis (GSVA) (version 1.38.2, http://www.bioconductor.org/packages/release/bioc/html/GSVA.html) R package[29] to perform the gene set enrichment analysis. Differences in pathway activity scores among the three SCNA patterns were calculated using the limma (version 3.46.0) R package. A heatmap was employed to display the results from limma with the P values < 0.05.
Gene regulatory network analysis
The single cell regulatory network inference and clustering (SCENIC) (version 1.2.4, https://scenic.aertslab.org/) R package[30] was applied to identify gene regulatory networks specificity in SCNA clusters. Species-specific databases for RcisTarget (motif rankings) were downloaded for human datasets: hg19-500bp-upstream-7species.mc9nr.feather and hg19-tss-centered-10kb-7species.mc9nr.featherare. We used both the regulon specificity score and regulon activity to calculate cluster-specific regulons. T-Distributed Stochastic Neighbor Embedding was used for visualization.
Cellular communications analysis
For cell–cell communication analysis, we used the CellChat (version 1.0.0, https://github.com/sqjin/CellChat) R package[31] to compare the differential cell interaction and signaling between SC1 and SC2 as well as SC1 and SC3 (SC1, SC2 and SC3 indicated that the dominate cells cluster was C1, C2, C3, respectively). Considering the lower number of cells and complex combinations with other cells, we deleted mucosal-associated invariant T cells, γδT cells, and neutrophils. When identifying upregulated signaling ligand–receptor pairs, we set ligand.logFC = 1 in the comparison between SC2 and SC1, and ligand.logFC = 0.5 in the comparison between SC3 and SC1.
Copy number analysis of the TCGA dataset
Masked copy number segment data of TCGA were downloaded by TCGAbiolinks, and the marker file was obtained from https://gdc.cancer.gov/about-data/gdc-data-processing/gdc-reference-files. Then, we applied GISTIC2.0[32] (ftp://ftp.broadinstitute.org/pub/GISTIC2.0/) to analyze gene- and arm-level copy number alterations. The following analysis was conducted using arm-level copy number alterations with “0” indicating a normal copy number, whereas positive and negative values indicated amplification and deletion, respectively. We first stratified MSS CRC samples into a low or high SCNA burden with the criterion of all arm-level copy number absolute values < 1 as the low SCNA burden. SC2 was assigned to samples with a low SCNA burden. For high SCNA burden samples, we further stratified them into SC1 and SC3 by the chromosome 13 copy number status. Just like the grouping of the scRNA-seq data, we grouped chromosome 13 amplification samples into SC1 and the others into SC3. For chromosomes, we employed the following copy number criteria: amplification (copy number > 0.2), diploid (copy number –0.2–0.2), and deletion (copy number < –0.2).
Chemotherapeutic or targeted drug response prediction
On the basis of the Cancer Therapeutics Response Portal (CTRP), we employed the R package OncoPredict (version 0.2, https://mirrors.sjtug.sjtu.edu.cn/cran/web/packages/oncoPredict/index.html)[33] to predict chemotherapeutic or targeted drug responses for patients in the TCGA dataset. The training dataset for OncoPredict was downloaded from https://osf.io/c6tfx/. For the training data, we used the CTRP2_Expr (TPM, log2[x+1] Transformed) data and CTRP2_Res data. The test TCGA data were also transformed into log2 (TPM+1) after removing 20% of the low varying genes. The training and test data matrices were homogenized with ComBat. Ridge regression was used to calculate the estimated half maximal inhibitory concentration (IC50) of each patient for a specific chemotherapeutic or targeted medication.
Identification of the prognostic signature of chromosomes 13 and 20
We eventually included 377 MSS CRC tumor samples from the TCGA cohort after excluding samples with overall survival (OS) time = 0. We applied log2 (TPM+1)-normalized values for gene expression evaluation in RNA-sequencing data. With the criteria of adjusted P values < 0.05 and log2 fold change > 1, we identified 156 upregulated genes in epithelial cells using the SMC scRNA-seq dataset. Then, we chose 15 of the upregulated genes on chromosomes 13 and 20, and divided patients into two groups with the best gene expression cutoff value of OS. On the basis of log-rank testing, Kaplan–Meier curves were used to evaluate overall survival differences between different gene expression level groups. And then, we validated the prognosis function of these genes in two independent external datasets (one from the NCC cohort with 24 MSS CRC patients and the other from the GSE39582 cohort with 444 MSS colon cancer patients).
Statistical analysis
All statistical analyses were conducted using R (version 4.1.3). We used the Wilcoxon rank-sum test to evaluate significant differences between two groups and the Kruskal–Wallis H test for three groups. A P-value of <0.05 was considered statistically significant.
Results
Single cell SCNA landscape in MSS CRC
To characterize the copy number alteration of MSS CRC in a single cell profile, we analyzed 26 tissue samples from 19 Korean patients (GSE132465, the SMC dataset). We next verified the results with 15 tissue samples from five Belgian patients (GSE144735, the KUL3 dataset). After excluding the patients with high MSI, 51,482 cells in the SMC dataset were classified into six major cell types (epithelial, stromal, myeloid, mast, T/natural killer [NK], and B cells) by specific gene markers [Supplementary Figures 1A–1C]. Similarly, we divided clusters into six major cell types for 21,351 cells in the KUL3 dataset [Supplementary Figures 2A–2C]. Notably, we found that myeloid cells were specifically enriched in tumor tissues, whereas B cells were predominantly present in normal tissue, indicating reprogramming of the tumor-dominated immune microenvironment [Supplementary Figures 1D and 2D].
Next, we used CopyKAT to identify copy number alterations. Cells were annotated into diploid and aneuploid. The aneuploid cells were primarily tumor epithelial cells [Supplementary Figures 1E and 2E]. Cells were clustered by cell types in diploid, but by samples in aneuploid, which suggested individual heterogeneity of the aneuploid cells in the SCNA profile [Supplementary Figures 1F and 2F]. Except for epithelial cells in tumors, SCNAs were also prevalent in other cell types, especially stromal cells, which exhibited remarkably higher SCNA levels than immune cells [Supplementary Figures 1F and 2F].
Identification of three distinct SCNA patterns in MSS CRC
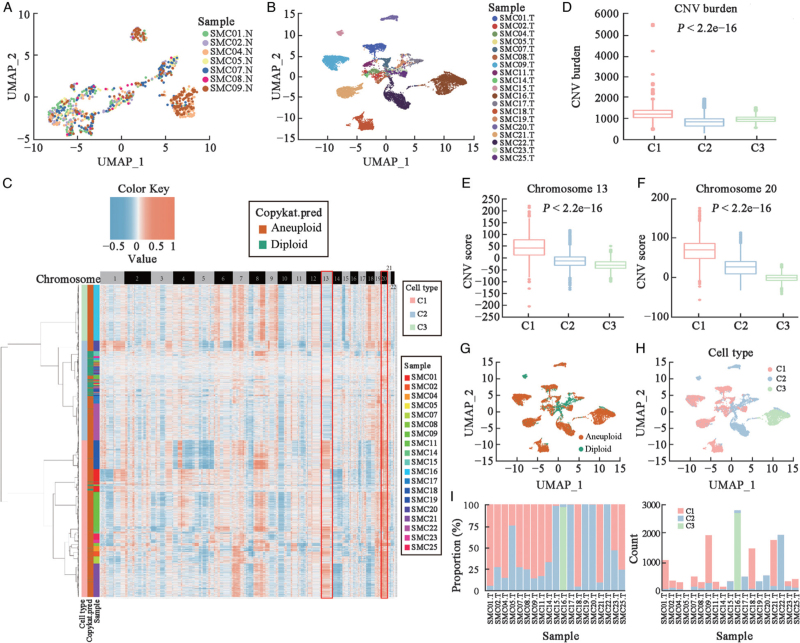
Unlike in normal tissue, we observed patient-specific clustering of tumor cells in tumors [Figures 1A, 1B and Supplementary Figures 3A, 3B]. To further illustrate the variety of tumor cells, we used SCNA features to classify tumor cells and identified three distinct SCNA patterns (C1–C3) in MSS CRC. C1 had considerable amplification of chromosomes 13 and 20 among these three patterns, whereas C3 exhibited deletion in above chromosomes [Figure 1C] (CNV burden: P < 2.2e–16 [Figure 1D], Chromosome 13 CNV score: P < 2.2e–16 [Figure 1E], Chromosome 20 CNV score: P < 2.2e–16 [Figure 1F]). C2 included mostly diploid cells and had the lowest SCNA burden compared with C1 and C3. Additionally, diploid and aneuploid cells were separated from each other in the UMAP plot, which was almost the same as the three types of tumor cells [Figures 1G, 1H and Supplementary Figures 3C, 3H]. The proportion of the three tumor cell clusters in each sample was then used to classify patients into SC1, SC2, or SC3, in which SC1 indicated that the C1 cluster was dominate in the patient [Figure 1I]. For the tumor cells were <10 in SMC05, we could not infer the real fraction of tumor cells in this sample. Therefore, we eliminated SMC05 from subsequent analysis. Finally, SC1 included 12 samples (SMC01, SMC02, SMC04, SMC07, SMC08, SMC09, SMC11, SMC14, SMC18, SMC21, SMC23, and SMC25). SC2 had five samples (SMC15, SMC17, SMC19, SMC20, and SMC22) and SC3 included only SMC16.
Figure 1.
Epithelial cells of normal and tumor tissues in the SMC dataset. (A) UMAP plot depicts the spatial distribution of epithelial cells in normal tissue for various samples. (B) UMAP plot depicts the spatial distribution of epithelial cells in tumor tissue for different samples. (C) SCNA heatmap of tumor cells in the SMC dataset. (D) Total CNV burden in three tumor cell clusters. (E) CNV scores of chromosome 13 in three tumor cell clusters. (F) CNV scores of chromosome 20 in three tumor cell clusters. (G) UMAP plot shows the spatial distribution of aneuploid and diploid cells in tumor tissue. (H) UMAP plot shows the spatial distribution of clusters C1–C3. (I) Comparison of the cell proportions of the three tumor cell clusters in each sample (left); comparison of the cell numbers of the three tumor cell clusters in each sample (right). CNV: Copy number variation; “N” indicates normal tissue; SCNA: Somatic copy number alteration; SMC: The Samsung Medical Center; “T” indicates tumor tissue; UMAP: Uniform manifold approximation and projection.
We used the same strategy to identify these three tumor cell types in the KUL3 dataset by SCNA features and found similar chromosome copy number alterations [Supplementary Figures 3D–G and 3I]. Furthermore, cells from the tumor core had a similar SCNA model as cells from the tumor border in the same patient [Supplementary Figure 3D]. Thus, we chose samples with more detectable tumor cells in core or border tumor samples to cluster those patients into SC1, SC2, or SC3. Finally, SC1 included KUL28, KUL30, and KUL31, and SC2 and SC3 included KUL19 and KUL21, respectively. Overall, we identified three types of tumor cells in MSS CRC by their SCNA characteristics.
Biological features of tumor cells with diverse SCNA patterns
To explore the biological features of C1–C3, we performed differentially expressed gene analysis of the three SCNA patterns. Gene Ontology enrichment analysis revealed that upregulated genes in C1 were enriched in immune-related pathways, especially antigen processing and presentation pathways [Supplementary Figure 4A]. Compared with the other clusters, C2 expressed high levels of tissue homeostasis-related genes, while cell adhesion activity genes were enriched in C3 [Supplementary Figures 4B and 4C].
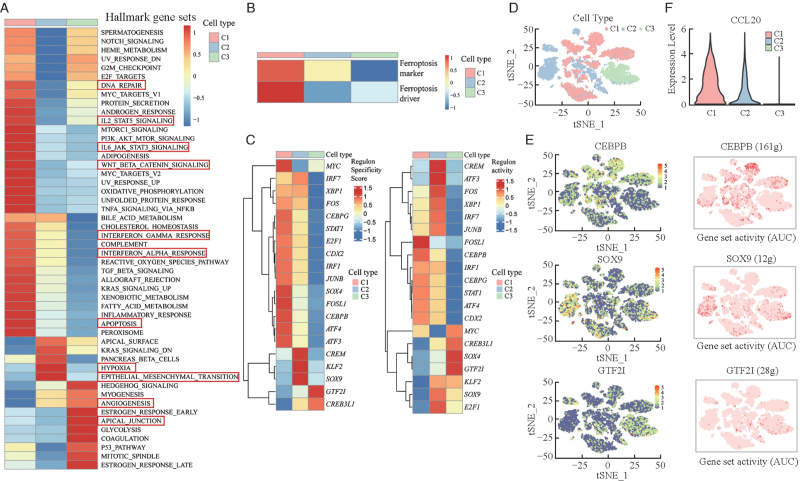
Pathway analysis by GSVA with hallmarker gene sets revealed that apoptosis, DNA repair, interferon-γ response, interleukin 2-signal transducer and activator of transcription 5 (IL2-STAT5) signaling, interleukin 6-Janus kinase-signal transducer and activator of transcription 3 (IL6-JAK-STAT3) signaling and Wntβ-catenin signaling pathways were enriched in C1, while hypoxia and epithelial–mesenchymal transition were upregulated in C2 [Figure 2A]. Furthermore, angiogenesis and the apical junction pathway were enriched in C3. In addition to the apoptosis pathway, another form of cell death pathway was enriched in C1, namely the ferroptosis pathway [Figure 2B]. In the KUL3 dataset, we found similar pathway enrichment, suggesting the active immune environment and ferroptosis reaction, which may restrain tumorigenesis in C1, and the upregulated angiogenesis pathway may prompt the formation of a tumor angiogenesis environment in C3 [Supplementary Figures 5A and 5B]. To decipher the gene regulatory network of the three clusters, we employed SCENIC. Considering both cell type-specific and average regulon activities, we found three specific regulons (CCAAT/enhancer binding protein beta [CEBPB], sex-determining region Y-box 9 [SOX9] and GTF2I) among the three clusters [Figure 2C]. Compared with the other two clusters, the CEBPB motif was highly active in the C1 cluster [Figures 2D and 2E]. We also found high expression of CCL20 in C1 compared with the other clusters [Figure 2F and Supplementary Figure 5C]. The specific regulons in C2 and C3 were SOX9 and GTF2I motifs, separately. Diverse SCNA patterns showed disparate biological features and formed different gene regulatory networks.
Figure 2.
GSVA and SCENIC analysis of three SCNA patterns in the SMC dataset. (A) Heatmap shows the different hallmark gene sets and pathway expression of the three SCNA clusters obtained by GSVA and is colored based on GSVA scores. (B) Heatmap shows different ferroptosis gene set expression of the three SCNA clusters obtained by GSVA and is colored based on GSVA scores. (C) Different regulon specificity scores of the three SCNA clusters in SCENIC (left), and different regulon activities of the three SCNA clusters in SCENIC (right). (D) tSNE plot of the spatial distribution of the three SCNA clusters. (E) tSNE plots of the expression levels of three distinctly different regulons (left) and AUC scores (right, the redder color represents the higher activity of the regulon). (F) Gene expression levels of CCL20 in the three SCNA clusters. AKT: Protein kinase B; AUC: Area under curve; DNA: Deoxyribonucleic acid; GSVA: Gene set variation analysis; IL-2: Interleukin 2; IL-6: Interleukin 6; JAK: Janus-activated kinase; KRAS: Kirsten rat sarcoma viral oncogene; MTOR: Mammalian target of rapamycin; MYC: Myelocytomatosis viral oncogene; NF-κB: Nuclear factor kappa-B; PI3K: Phosphoinositide 3-kinase; SCNA: Somatic copy number alteration; SCENIC: The single cell regulatory network inference and clustering; SMC: The Samsung Medical Center; STAT: Signal transducer and activator of transcription; tSNE: T-distributed stochastic neighbor embedding; TGF: Transforming growth factor; TNFA: Tumor necrosis factor α.
SCNA-specific cellular interaction network
To determine cell–cell communication differences in various SCNA patterns, we further classified cell clusters in accordance with specific gene markers [Supplementary Table 6] and performed CellChat analysis to compare SC2 with SC1 [Supplementary Figures 5D–5F and Supplementary Table 7] and SC3 with SC1 [Supplementary Figure 6 and Supplementary Table 8]. When we compared SC2 with SC1, we found stronger interaction strengths of cells in SC2 than in SC1 [Supplementary Figure 5D]. Anti-inflammatory macrophages, cancer-associated fibroblasts (CAFs), and endothelial cells had significantly enhanced communication with other cells in SC2 [Supplementary Figure 5E]. Considering the vital role of macrophages in the tumor immune milieu and the enhanced cell communication of macrophages in SC2, we further explored the macrophage signaling differences between SC2 and SC1. We noticed stronger upregulated signaling of secreted phosphoprotein 1 (SPP1)-related pathways in SC2 than in SC1, especially SPP1-CD44, which is pivotal to form an immunosuppressive microenvironment [Supplementary Figure 5F].
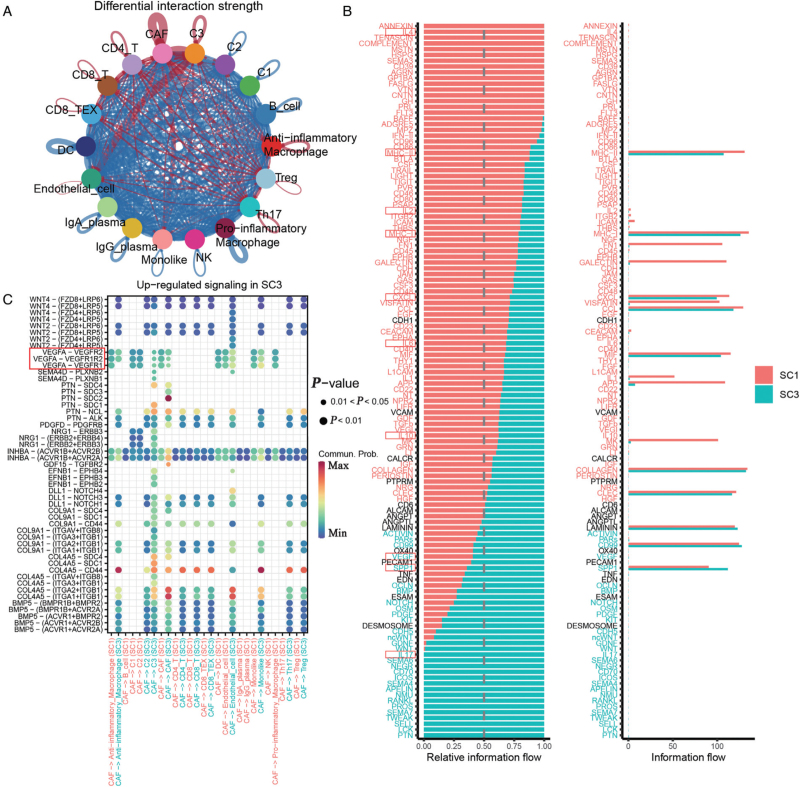
Next, we compared cell communication in SC3 and SC1, and found that SC3 had less cell interaction strength than SC1, but stronger cell interaction strength in CAFs and endothelial cells with other cells [Figure 3A]. When processing the signal analysis data, we noticed enhancement of vascular endothelial growth factor (VEGF) and interleukin-17 (IL-17) signals in SC3, indicating an angiogenic effect in SC3, and found a series of immune-related pathways, suggesting a better active immune microenvironment in SC1 [Figures 3B and 3C]. Combined with our GSVA analysis, these results indicated that CAFs might promote angiogenesis via the VEGF pathway together with IL-17 in SC3.
Figure 3.
Cell-cell communication differences between SC3 and SC1. (A) Circle plot shows the differential interaction strengths between SC3 and SC1. (B) Comparison of the overall signaling pathways between SC3 and SC1. (C) Dot plot shows the upregulated signaling of CAFs in SC3 compared with SC1. CAFs: Cancer-associated fibroblasts; DC: Dendritic cell; Ig: Immunoglobulin; NK: Natural killer cell; SC1: The dominate cells cluster was C1; SC3: The dominate cells cluster was C3; TEX: Exhausted T cell; Th17: T helper 17 cell; Treg: Regulatory T cell.
Different patient subgroups have variable mutation features, immune checkpoint molecule (ICM) expression, and chemotherapeutic or targeted drug responses
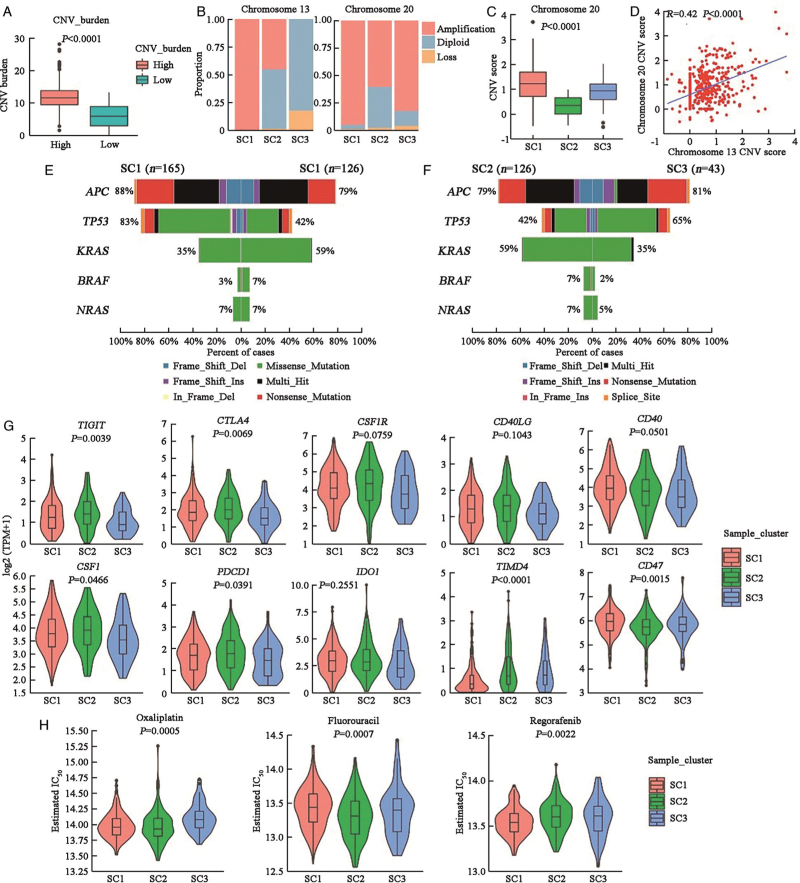
To explore the different molecular features of patients dominated by the three SCNA patterns, we stratified TCGA dataset patients into SC1, SC2, or SC3 by their SCNA characteristics. We first stratified MSS CRC samples into low and high SCNA burden groups, and classified samples with a low SCNA burden into SC2 [Figure 4A]. For high SCNA burden samples, we stratified them into SC1 and SC3 by the chromosome 13 copy number status [Figure 4B]. Similar to the scRNA-seq dataset, SC1 had a higher CNV score than SC3 in chromosome 20 (P < 0.0001) [Figure 4C]. Moreover, the chromosome 13 CNV score correlated positively with chromosome 20 (R = 0.42, P < 0.0001) [Figure 4D].
Figure 4.
Characteristics of the three SCNA clusters in the TCGA dataset. (A) Total CNV burden in high and low SCNA samples. The Wilcoxon rank-sum test was used for statistical analysis. (B) Proportion of patients with chromosome SCNA in SC1–SC3. (C) CNV scores of chromosome 20 in SC1–SC3. The Kruskal–Wallis test was used for statistical analysis. (D) Scatterplot shows the positive correlation between the CNV scores of chromosomes 13 and 20. The Pearson correlation coefficient and P-value are indicated. (E) Co-bar plot displays five pervasive gene mutations in SC1and SC2. (F) Co-bar plot displays five pervasive gene mutations in SC2 and SC3; (G) The expression levels of ten common immune checkpoint molecule in SC1–SC3. The Kruskal–Wallis test was used for statistical analysis. (H) Predicted IC50 values of three chemotherapy drugs (oxaliplatin, fluorouracil, and regorafenib) in SC1–SC3. The Kruskal–Wallis test was used for statistical analysis. CNV: Copy number variation; IC50: Half maximal inhibitory concentration; SCNA: Somatic copy number alteration; TCGA: The Cancer Genome Atlas.
We also noticed different mutation models with high frequencies of mutated TP53 in SC1 (83% in SC1, 42% in SC2, and 65% in SC3), and mutated KRAS (35% in SC1, 59% in SC2, and 35% in SC3) and BRAF (3% in SC1, 7% in SC2, and 2% in SC3) in SC2 [Figures 4E and 4F].
As immune checkpoint molecule (ICM) has been linked to the responsiveness of checkpoint inhibitor treatment,we compared the expression of certain common ICMs in various SCNA subgroups. The majority of ICMs were highly expressed in SC1 and SC2, whereas T cell immunoglobulin and mucin domain containing 4 (TIMD4) and CD47 were more expressed in SC3, implying that different individuals may benefit from various immune checkpoint inhibitor immunotherapies [Figure 4G]. OncoPredict was used to predict the response to three chemotherapeutic or targeted drugs and we found that various patients responded differently to the three medications [Figure 4H]. These findings imply that patients with distinct SCNA pattern-dominant malignancies will benefit from different immunotherapies, chemotherapies or targeted therapy. As a result, it will be critical to identify SCNA patterns and treat patients with a specific therapy.
Identification and verification of the prognosis-related signature
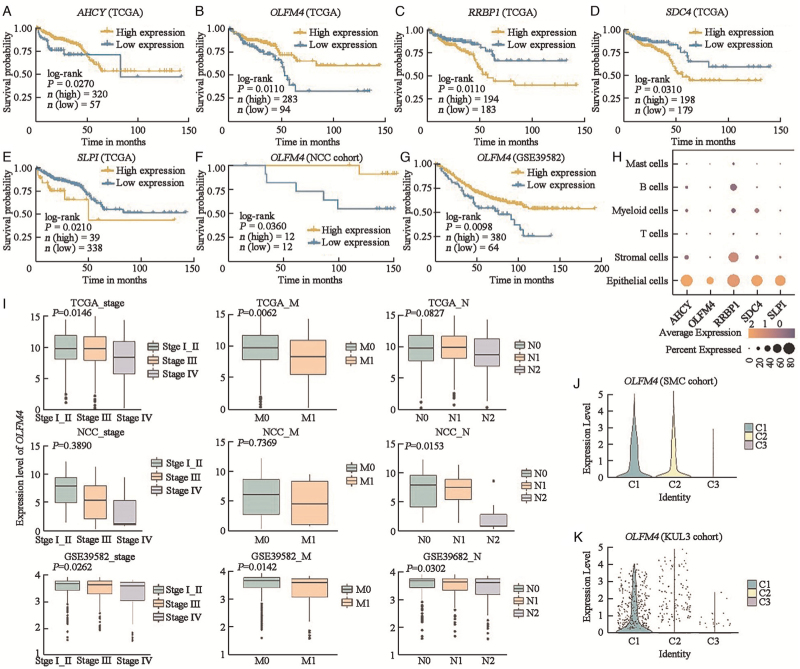
After exploring the biological function and molecular characteristics of the three types of tumor cells with chromosome 13 and 20 abnormalities, we further examined the prognosis contributions of distinct tumor cells to patient survival. To rule out the influence of gene expression from other cells, we used the SMC scRNA-seq dataset to identify 156 upregulated genes in epithelial cells with adjusted P value of <0.05 and a log2 fold change of >1 [Supplementary Table 9]. Next, we chose 15 of the upregulated genes on chromosomes 13 and 20 [Supplementary Table 10], and found that five of them were linked to prognosis in the TCGA CRC dataset (AHCY: P = 0.0270; OLFM4: P = 0.0110; RRBP1: P = 0.0110; SDC4: P = 0.0310; SLPI: P = 0.0210) [Figures 5A–5E]. Next, two independent external datasets (one from the NCC cohort with 24 MSS CRC patients and the other from the GSE39582 cohort with 444 MSS colon cancer patients) were used to validate the prognostic value of these five genes. Finally, we identified OLFM4, a gene expressed primarily in epithelial cells, which was associated with a better prognosis in the three datasets (TCGA cohort: P = 0.0110; NCC cohort: P = 0.0360; GSE39582 cohort: P = 0.0098) [Figures 5B, 5F–5H]. Furthermore, we found that patients with an earlier TNM stage and M stage showed enhanced expression of OLFM4, which had significant statistical differences in both TCGA (TNM stage: P = 0.0146, M stage: P = 0.0062) and GSE39582 (TNM stage: P = 0.0262, M stage: P = 0.0142) cohorts [Figure 5I]. In our NCC cohort, we found a similar tendency of OLFM4 expression without a statistical difference between patients at different TNM stages and M stages (TNM stage: P = 0.3890; M stage: P = 0.7369), and a significant dfference between patients at different N stages (P = 0.0153), which might be due to the small sample size. Additionally, in the scRNA-seq dataset (SMC and KUL3 cohorts), OLFM4 expression was the lowest in the C3 tumor cell cluster, which was related to angiogenesis and cell adhesion activity, and the highest in the C1 tumor cell, which was associated with immune activity and the classical pathway in cancer [Figures 5J and 5K].
Figure 5.
Identification of a prognosis-related signature. (A–E) Overall survival analysis of five genes (AHCY, OLFM4, RRBP1, SDC4, and SLPI) in primary CRC of the TCGA dataset. The log-rank test was used for statistical analysis. (F) Overall survival analysis of OLFM4 in primary CRC of NCC cohort. The log-rank test was used for statistical analysis. (G) Overall survival analysis of OLFM4 in primary CRC of GSE39582 cohorts. The log-rank test was used for statistical analysis. (H) Dot plot depicts expression of the five genes (AHCY, OLFM4, RRBP1, SDC4, and SLPI) in different cell types of the SMC dataset. (I) Expression level of OLFM4 in different TNM stages, N stage, and M stage. (J) Expression level of OLFM4 in three tumor clusters of the SMC cohort. (K) Expression level of OLFM4 in three tumor clusters of the KUL3 cohort. CRC: Colorectal cancer; KUL3: The Katholieke Universiteit Leuven 3; NCC: National Cancer Center; SMC: The Samsung Medical Center; TCGA: The Cancer Genome Atlas.
Discussion
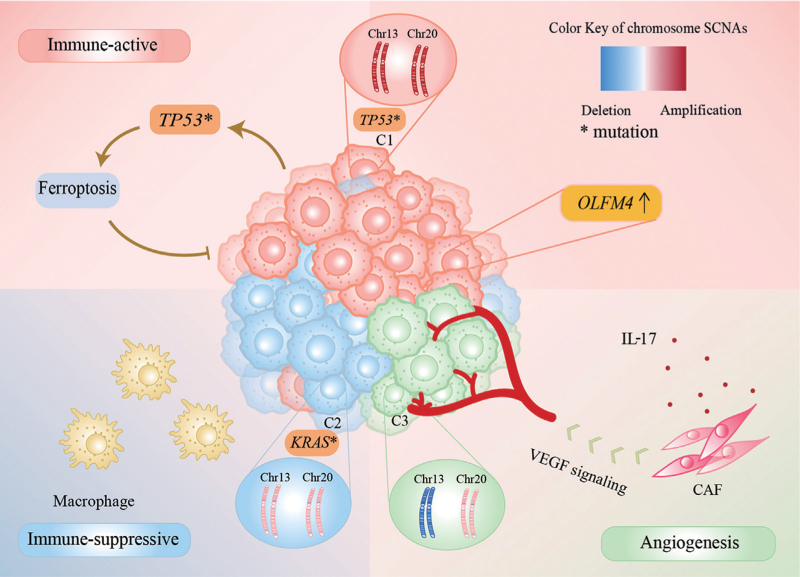
Intra-tumor genetic heterogeneity influences major pathways in cancer and drives phenotypic diversity, causing a huge hindrance to cancer medicine.[2] In this study, we clustered tumor cells in MSS CRC into three distinct SCNA patterns by their SCNA characteristics [Figure 6]. The major chromosome SCNA differences in these three tumor subcluster were on chromosomes 13 and 20. Chromosome 13, as the largest acrocentric human chromosome, is a chromosomal feature to distinguish mutant TP53.[34,35] Amplification of chromosome 13 occurs almost exclusively in CRC.[36] In terms of chromosome 20, 20q amplification has been linked to left-sided tumors, microsatellite stability, wild type KRAS and BRAF, and mutant TP53 in CRC patients.[37] Furthermore, a recent study has indicated that patients with chromosome 20q amplification had better recurrence-free survival, a higher frequency of mutant TP53, and a higher level of chromosomal instability than MSS patients without chromosome 20q amplification, who had more mutated KRAS and BRAF.[38]
Figure 6.
Tumor microenvironment of the three SCNA patterns in MSS CRC. CAF: Cancer associated fibroblasts; Chr: Chromosome; IL 17: Interleukin 17; MSS CRC: Microsatellite stable colorectal cancer; SCNA: Somatic copy number alteration; VEGF: Vascular endothelial growth factor.
Among the three SCNA patterns, the dominant SCNA feature of the C1 cluster was significant amplification of chromosomes 13 and 20. This subcluster of tumor cells expressed high levels of immune-related genes, especially those in antigen processing and presentation-related pathways. Furthermore, DNA repair, the interferon-γ response, IL-2-STAT5 signaling, IL-6-JAK-STAT3 signaling pathway, apoptosis, and the ferroptosis pathway were upregulated in C1. Unlike autophagy and apoptosis, ferroptosis is iron-dependent and reactive oxygen species-reliant cell death that plays a pivotal role in suppression of tumorigenesis.[39] Besides, we noticed the highly activation of CEBPB motif in the C1 cluster compared with the other two clusters. CRC cell-secreted CCL20 recruits regulatory T cells (Tregs) to promote chemoresistance via forkhead box O1 (FOXO1)/CEBPB/nuclear factor kappa B (NF-κB) signaling.[40] In the C1 dominant tumor, there was a lower interaction of CAFs and endothelial cells with other cells. SC1 had a high prevalence of mutated TP53 in the mutation profile. On the one hand, approximately 85% of the TP53 mutations in CRC are missense defects, which may positively contribute to the cancer phenotype via effects on tumor cell proliferation and increased tumor angiogenesis.[41] On the other hand, TP53 mutation may facilitate ferroptosis activation and inhibit tumor proliferation.[42] The higher TP53 mutation burden in C1 further confirmed that C1 activated the ferroptosis process, resulting in a favorable prognosis.
With a lower SCNA burden, the C2 cluster expressed high levels of genes in hypoxia and epithelial–mesenchymal transition pathways. Additionally, the highly active transcription factor SOX9 promotes tumor migration, invasion, and metastasis in CRC.[43] Notably, we found stronger SPP1-related signaling expression in SC2 than in SC1, especially the SPP1–CD44 interaction, which controls CD8+ T cell activation and promotes tumor immune evasion.[44] Interestingly, even with a lower SCNA burden, SC2 contained the most KRAS and BRAF mutations among the three clusters. Another study has also reported this aberrant phenomenon that SCNA levels positively correlate with the total number of mutations in most tumors, whereas tumors with activating oncogenic mutations in the receptor tyrosine kinase-RAS-phosphatidylinositol 3-kinase pathway have fewer SCNAs.[6] Oncogenic KRAS signaling is essential for tumor progression to invasive and metastatic growth.[45] Moreover, KRAS mutation promotes an immune-suppressive profile in CRC and leads to anti-PD1 immune therapy resistance by promoting myeloid-derived suppressor cell (MDSC) migration into the tumor microenvironment.[46] Taken together, we speculated that there was a suppressive immune microenvironment in SC2 with KRAS or BRAF mutations.
For C3, the combination of chromosome 13 deletion and chromosome 20 slight amplification or diploidy was more common in this cluster. Angiogenesis and cell adhesion activity were enriched in C3. Moreover, in C3, the GTF2I motif, which is related to angiogenesis and operates as a signal-induced transcription factor in response to various extracellular signaling pathways, was highly active.[47] In the C3-leading tumor microenvironment, CAFs were considered critical for tumorigenesis, forming an extracellular matrix structure, and immune reprogramming of the tumor microenvironment, impacting adaptive resistance to chemotherapy.[48] IL-17, mainly from Th17 cells, promotes tumor angiogenesis by stimulating fibroblasts to upregulate VEGF.[49,50] IL-17 also induces expression of granulocyte colony-stimulating factor through NF-κB and extracellular-related kinase signaling, leading to immature myeloid cell mobilization and recruitment into the tumor microenvironment.[51,52] In SC3, we also found dominant enhancement of VEGF and IL-17 signals compared with SC1 when processing the signal analysis data. Overall, in the presence of CAFs and IL-17, SC3 forms an angiogenic tumor microenvironment via the VEGF pathway.
The tumor microenvironment includes T, NK, B, myeloid, and stromal cells as well as tumor cells. The cellular interactions among them determine the development, progression, and clinical outcome of the tumor. In the different SCNA patterns, there were various constituents of immune and stromal cells. For example, macrophages, especially SPP1+ macrophages, were enriched in SC2, which promotes tumor immune evasion. Additionally, CAFs were vital for tumor angiogenesis in the C3-dominated tumor.
We also identified OLFM4, which is a prognosis-related signature with increased expression at an early stage. In CRC, OLFM4 initiates organoid culture growth and a differentiation capacity serving as a stem cell marker,[53,54] which demonstrated the major role of OLFM4+ tumor cells in the early stage, as we found enrichment of various important pathways in C1, such as Wnt, transforming growth factor beta (TGF-β), and Notch pathways. OLFM4 also inhibits cell adhesion and migration,[55] which was consistent with the cell adhesion activity of C3 with the lowest OLFM4 expression compared with the other two tumor clusters.
By understanding heterogeneity in the intricate tumor microenvironment, we gained an insight into the mechanisms of tumor evolution, which may support the development of therapeutic strategies. However, we only identified one C3-dominant patient in the SMC dataset because of the limited data sources. Thus, a greater scale of scRNA-seq data is needed to further validate the function of these three SCNA clusters in the tumor microenvironment.
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Song S, Feng L, Xi K, Sun Z, Kong D, Luo Z, Pei W, Zhang H. Single-cell profiling of the copy-number heterogeneity in colorectal cancer. Chin Med J 2023;136:707–718. doi: 10.1097/CM9.0000000000002469
Supplemental digital content is available for this article.
References
- 1.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer. Nat Rev Cancer 2012; 12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 2.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013; 501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 3.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017; 168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 4.Vitale I, Shema E, Loi S, Galluzzi L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat Med 2021; 27:212–224. doi: 10.1038/s41591-021-01233-9. [DOI] [PubMed] [Google Scholar]
- 5.McClelland SE. Role of chromosomal instability in cancer progression. Endocr Relat Cancer 2017; 24:T23–T31. doi: 10.1530/ERC-17-0187. [DOI] [PubMed] [Google Scholar]
- 6.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017; 355:eaaf8399.doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miao D, Margolis CA, Vokes NI, Liu D, Taylor-Weiner A, Wankowicz SM, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet 2018; 50:1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-David U, Amon A. Context is everything: aneuploidy in cancer. Nat Rev Genet 2020; 21:44–62. doi: 10.1038/s41576-019-0171-x. [DOI] [PubMed] [Google Scholar]
- 9.Ried T, Meijer GA, Harrison DJ, Grech G, Franch-Expósito S, Briffa R, et al. The landscape of genomic copy number alterations in colorectal cancer and their consequences on gene expression levels and disease outcome. Mol Aspects Med 2019; 69:48–61. doi: 10.1016/j.mam.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Dekker E, Tanis PJ, Vleugels J, Kasi PM, Wallace MB. Colorectal cancer. Lancet 2019; 394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 11.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021; 71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 12.Ciardiello D, Vitiello PP, Cardone C, Martini G, Troiani T, Martinelli E, et al. Immunotherapy of colorectal cancer: challenges for therapeutic efficacy. Cancer Treat Rev 2019; 76:22–32. doi: 10.1016/j.ctrv.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Gohil SH, Iorgulescu JB, Braun DA, Keskin DB, Livak KJ. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat Rev Clin Oncol 2021; 18:244–256. doi: 10.1038/s41571-020-00449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc 2018; 13:599–604. doi: 10.1038/nprot.2017.149. [DOI] [PubMed] [Google Scholar]
- 15.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 2018; 18:35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 16.Lee HO, Hong Y, Etlioglu HE, Cho YB, Pomella V, Van den Bosch B, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet 2020; 52:594–603. doi: 10.1038/s41588-020-0636-z. [DOI] [PubMed] [Google Scholar]
- 17.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021; 184:3573–3587. e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mounir M, Lucchetta M, Silva TC, Olsen C, Bontempi G, Chen X, et al. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput Biol 2019; 15:e1006701.doi: 10.1371/journal.pcbi.1006701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva TC, Colaprico A, Olsen C, D’Angelo F, Bontempi G, Ceccarelli M, et al. TCGA Workflow: analyze cancer genomics and epigenomics data using Bioconductor packages. F1000Res 2016; 5:1542.doi: 10.12688/f1000research.8923.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016; 44:e71.doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020; 38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018; 28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007; 23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 24.Gao R, Bai S, Henderson YC, Lin Y, Schalck A, Yan Y, et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol 2021; 39:599–608. doi: 10.1038/s41587-020-00795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G, Wang LG, Han Y. He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015; 1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford) 2020; 2020:baaa021.doi: 10.1093/database/baaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013; 14:7.doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017; 14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021; 12:1088.doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2. 0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011; 12:R41.doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform 2021; 22:bbab260.doi: 10. 1093/bib/bbab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunham A, Matthews LH, Burton J, Ashurst JL, Howe KL, Ashcroft KJ, et al. The DNA sequence and analysis of human chromosome 13. Nature 2004; 428:522–528. doi: 10.1038/nature02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A 2009; 106:7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wangsa D, Braun R, Stuelten CH, Brown M, Bauer KM, Emons G, et al. Induced chromosomal aneuploidy results in global and consistent deregulation of the transcriptome of cancer cells. Neoplasia 2019; 21:721–729. doi: 10.1016/j.neo.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ptashkin RN, Pagan C, Yaeger R, Middha S, Shia J, O’Rourke KP, et al. Chromosome 20q Amplification Defines a Subtype of Microsatellite Stable, Left-Sided Colon Cancers with Wild-type RAS/RAF and Better Overall Survival. Mol Cancer Res 2017; 15:708–713. doi: 10.1158/1541-7786.MCR-16-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B, Yao K, Zhou E, Zhang L, Cheng C. Chr20q amplification defines a distinct molecular subtype of microsatellite stable colorectal cancer. Cancer Res 2021; 81:1977–1987. doi: 10.1158/0008-5472.CAN-20-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mou Y, Wang J, Wu J, He D, Zhang C, Duan C, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol 2019; 12:34.doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Yang L, Yu W, Wu Q, Lian J, Li F, et al. Colorectal cancer cell-derived CCL20 recruits regulatory T cells to promote chemoresistance via FOXO1/CEBPB/NF-κB signaling. J Immunother Cancer 2019; 7:215.doi: 10.1186/s40425-019-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol 2011; 6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 42.Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J, et al. The Tumor Suppressor p53 Limits Ferroptosis by Blocking DPP4 Activity. Cell Rep 2017; 20:1692–1704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 43.Zhou T, Wu L, Ma N, Tang F, Yu Z, Jiang Z, et al. SOX9-activated FARSA-AS1 predetermines cell growth, stemness, and metastasis in colorectal cancer through upregulating FARSA and SOX9. Cell Death Dis 2020; 11:1071.doi: 10.1038/s41419-020-03273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klement JD, Paschall AV, Redd PS, Ibrahim ML, Lu C, Yang D, et al. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J Clin Invest 2018; 128:5549–5560. doi: 10.1172/JCI123360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boutin AT, Liao WT, Wang M, Hwang SS, Karpinets TV, Cheung H, et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev 2017; 31:370–382. doi: 10.1101/gad.293449.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, et al. KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell 2019; 35:559–572. e7. doi: 10.1016/j.ccell.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy AL. Pathophysiology of TFII-I: old guard wearing new hats. Trends Mol Med 2017; 23:501–511. doi: 10.1016/j.molmed.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016; 16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 49.Knochelmann HM, Dwyer CJ, Bailey SR, Amaya SM, Elston DM, Mazza-McCrann JM, et al. When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell Mol Immunol 2018; 15:458–469. doi: 10.1038/s41423-018-0004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murugaiyan G, Saha B. Protumor vs antitumor functions of IL-17. J Immunol 2009; 183:4169–4175. doi: 10.4049/jimmunol.0901017. [DOI] [PubMed] [Google Scholar]
- 51.Maniati E, Hagemann T. IL-17 mediates resistance to anti-VEGF therapy. Nat Med 2013; 19:1092–1094. doi: 10.1038/nm.3333. [DOI] [PubMed] [Google Scholar]
- 52.Chung AS, Wu X, Zhuang G, Ngu H, Kasman I, Zhang J, et al. An interleukin-17-mediated paracrine network promotes tumor resistance to anti-angiogenic therapy. Nat Med 2013; 19:1114–1123. doi: 10.1038/nm.3291. [DOI] [PubMed] [Google Scholar]
- 53.van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 2009; 137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 54.Okamoto T, duVerle D, Yaginuma K, Natsume Y, Yamanaka H, Kusama D, et al. Comparative analysis of patient-matched PDOs revealed a reduction in OLFM4-associated clusters in metastatic lesions in colorectal cancer. Stem Cell Reports 2021; 16:954–967. doi: 10.1016/j.stemcr.2021.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W, Liu Y, Zhu J, Wright E, Ding I, Rodgers GP. Reduced hGC-1 protein expression is associated with malignant progression of colon carcinoma. Clin Cancer Res 2008; 14:1041–1049. doi: 10.1158/1078-0432.CCR-07-4125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.