Abstract
Background:
Heterogeneity of tumor cells and the tumor microenvironment (TME) is significantly associated with clinical outcomes and treatment responses in patients with urothelial carcinoma (UC). Comprehensive profiling of the cellular diversity and interactions between malignant cells and TME may clarify the mechanisms underlying UC progression and guide the development of novel therapies. This study aimed to extend our understanding of intra-tumoral heterogeneity and the immunosuppressive TME in UC and provide basic support for the development of novel UC therapies.
Methods:
Seven patients with UC were included who underwent curative surgery at our hospital between July 2020 and October 2020. We performed single-cell RNA sequencing (scRNA-seq) analysis in seven tumors with six matched adjacent normal tissues and integrated the results with two public scRNA-seq datasets. The functional properties and intercellular interactions between single cells were characterized, and the results were validated using multiplex immunofluorescence staining, flow cytometry, and bulk transcriptomic datasets. All statistical analyses were performed using the R package with two-sided tests. Wilcoxon-rank test, log-rank test, one-way analysis of variance test, and Pearson correlation analysis were used properly.
Results:
Unsupervised t-distributed stochastic neighbor embedding clustering analysis identified ten main cellular subclusters in urothelial tissues. Of them, seven urothelial subtypes were noted, and malignant urothelial cells were characterized with enhanced cellular proliferation and reduced immunogenicity. CD8+ T cell subclusters exhibited enhanced cellular cytotoxicity activities along with increased exhaustion signature in UC tissues, and the recruitment of CD4+ T regulatory cells was also increased in tumor tissues. Regarding myeloid cells, coordinated reprogramming of infiltrated neutrophils, M2-type polarized macrophages, and LAMP3+ dendritic cells contribute to immunosuppressive TME in UC tissues. Tumor tissues demonstrated enhanced angiogenesis mediated by KDR+ endothelial cells and RGS5+/ACTA2+ pericytes. Through deconvolution analysis, we identified multiple cellular subtypes may influence the programmed death-ligand 1 (PD-L1) immunotherapy response in patients with UC.
Conclusion:
Our scRNA-seq analysis clarified intra-tumoral heterogeneity and delineated the pro-tumoral and immunosuppressive microenvironment in UC tissues, which may provide novel therapeutic targets.
Keywords: Transitional cell carcinoma, Single-cell RNA Seq, Tumor microenvironment, Prognosis
Introduction
Urothelial carcinoma (UC), also known as transitional cell carcinoma, includes bladder cancer (BC), urethra cancer, and upper tract urothelial carcinoma (UTUC). Approximately 90% to 95% of clinical UCs are BCs, which rank as the 12th most commonly diagnosed cancer worldwide.[1] Patients with advanced-stage disease (pT3-T4a or lymph node-positive disease) have a 5-year overall survival (OS) of only 10% to 40%, with limited clinical benefits of neoadjuvant cisplatin-based chemotherapy.[2] Several immune checkpoint-based therapies have been recommended for surgery ineligible patients or patients with metastatic UC, but only approximately 30% of patients respond to these immunotherapies.[3]
Single-cell RNA sequencing (scRNA-seq) has recently enabled sequencing of thousands of cells in a single sample, and gene expression profiling of all individual cells has been performed in multiple cancers.[4,5] Tumor cell heterogeneity and certain tumor microenvironment (TME) cell subtypes or characteristics are significantly associated with tumor progression and treatment responses.[4] Recent scRNA-seq analyses have dissected the functional characteristics of tumor-infiltrating T cells and the TME cellular profiles in BC tissues.[6] However, the diverse characteristics and intercellular interactions in normal and tumor urothelial tissues, which may account for the mechanisms underlying UC development and progression, are not yet fully understood.
In this study, we analyzed seven UC and six matched normal tissues using plate-based single-cell sequencing. Our in-house scRNA-seq data were integrated with public data from two previous studies that included a further eight primary UC and six normal urothelial tissues to provide a more comprehensive view of the inter- and intra-tumoral heterogeneity of urothelial cells.[6,7] The current study may extend our understanding of intra-tumoral heterogeneity and the immunosuppressive TME in UC and provide basic support for the development of novel UC therapies.
Methods
Ethical approval
This research was reviewed and approved by the Ethics Committee of the Shanghai Jiao Tong University Medical School Affiliated Ruijin Hospital (No. 202034) and was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all the participants.
Patient recruitment and sample collection
Seven patients with UC who underwent curative surgery at Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, between July 2020 and October 2020, were recruited for the current study. The patients were histopathologically diagnosed with primary UC (UTUC or BC) by two independent pathologists. None of the patients had received any prior anticancer treatments before surgery, and none of the patients had any other cancer history. Detailed patient characteristics and individual scRNA-seq quality are shown in [Supplementary Tables 1 and 2].
Single-cell sample preparation
After surgery, primary UC and adjacent normal samples were washed in ice-cold saline, cut into pieces (2–4 mm), and dissociated in a tumor-dissociation solution at 37°C for 60 min. Tumor-dissociation solution comprised RPMI-1640 medium supplemented with 60 U/mL type I collagenase (#A004194, Sangon Biotech, Shanghai, China), 100 U/mL type IV collagenase (#A004186, Sangon Biotech), 0.1 mg/mL neutral protease (#A002100, Sangon Biotech), and 44 U/mL DNase I (#B006331, Sangon Biotech). Digestion was stopped with excess RPMI-1640 medium supplemented with 10% fetal bovine serum (#16140071, Gibco, Life Technologies, Carlsbad, CA, USA), and the cell supernatants were passed through 70-μm nylon cell strainers. Red blood cells were lysed using a red blood cell lysis buffer (#420301; BioLegend, San Diego, CA, USA). Single-cell quality control was first assessed via trypan blue staining, and the cell viability and concentration was then assessed with the Countess® II FL Automated Cell Counter (Invitrogen, Carlsbad, CA, USA). Samples with cell viability >80%, concentration >300 cells/μL, and without obvious cell debris or clumps would be used for following experiment.
Plate-based single-cell library preparation and sequencing
Microbead-captured single-cell library construction was performed using a BD Rhapsody™ Single-Cell Analysis System (#633701, BD Biosciences, San Jose, CA, USA) following the manufacturer's instructions as described previously.[8] The constructed libraries were sequenced on a NovaSeq instrument (Illumina, San Diego, CA, USA).
Raw single-cell sequencing data processing and quality control
Raw sequencing data were analyzed using the standard BD Rhapsody™ Whole Transcriptome Analysis platform on Seven Bridges (https://www.sevenbridges.com) following the standard recommendations. Low-quality cells with <200 detected genes or >25% of mitochondrial genes were removed.
Public scRNA-seq dataset processing
We included two independent droplet-based single-cell sequencing datasets based on the 10× Genomics platform reported by Chen et al[6] and Yu et al[7] to increase the statistical power of the current study. Raw sequencing data reported by Chen et al[6] were downloaded from the SRA database under BioProject No. PRJNA662018 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA662018) and processed using the Cell Ranger (version 4.0.0; 10× Genomics, Pleasanton, CA, USA, http://10xgenomics.com) count platform. Regarding dataset GSE129845 reported by Yu et al[7], the processed gene expression matrix was downloaded from GEO datasets. Detailed information on the participants in these two datasets was provided in Supplementary Tables 1 and 2.
scRNA-seq data processing and multiple datasets integration
The cell-gene count matrix was processed using the Seurat (version 4.0.1; https://satijalab.org/seurat/) package of the R software (version 3.6.1; The R Foundation, Vienna, Austria, https://www.r-project.org/). The obtained Seurat objects from each tissue were integrated using the Harmony algorithm (Harmony package of R, version 1.0; https://github.com/immunogenomics/harmony/) to generate an integrated and batch-corrected Seurat object.[9] Top 50 principal components were used for graph-based clustering to identify distinct groups of cells. Significant principal components were used for graph-based clustering for each cell cluster to identify cell subgroups based on t-stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP) analysis.
Differentially expressed genes (DEGs) and gene set enrichment analysis (GSEA)
Specific markers were identified using the FindAllMarkers function in the Seurat package to normalize expression data. Genes with an adjusted P < 0.05 were performed with Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses. The ClusterProfiler package (version 3.14.3; https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) was used for the enrichment analysis of cluster-specific biomarker genes.[10] Competitive GSEA was performed using Molecular Signatures Database (MSigDB) gene sets to identify differentially activated pathways or gene signatures, as previously described.[11] Fifty hallmark gene sets were retrieved from the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb).
Calculation of functional module scores
We evaluated the functional activities of a specific cell sub-cluster by calculating the averaged functional gene signature score. Umbrella urothelial-specific genes (including KRT20, UPK2, UPK3A, UPK3B, UPK1A, and UPK1B) and basal urothelial marker genes (including KRT5, KRT10, KRT13, KRT15, KRT17, KRT19, and KRT23) were used to calculate the umbrella to basal score ratio of the single cells, respectively. The full T cell signature gene lists were extracted from the curated gene list reported by Chung et al.[5] The gene sets for dendritic cells (DCs) were extracted from a study reported by Liu et al.[12]
Correlation analysis with prognosis and clinical responses to immunotherapy treatments UC patients
Raw RNA-seq count data and related clinical information of The Cancer Genome Atlas-Bladder Endothelial Carcinoma (TCGA-BLCA) patients was downloaded from the National Cancer Institute Genomic Data Commons database. After removing patients without follow-up information or duplicated samples, 401 patients were included. Raw data were processed and normalized using the DESeq2 package (version 1.34.0; https://bioconductor.org/packages/release/bioc/html/DESeq2.html), and reference-based decomposition was performed using the BisqueRNA package (version 1.0.5; https://github.com/cran/BisqueRNA) algorithm based on the single-cell expression data in our in-house plate-based scRNA-seq study.[13] Patients were categorized into groups with higher or lower distinct cell proportion levels using the optimal cutoff point, which corresponded to the most significant association with OS, as determined by the R package survminer (version 0.4.9; www.sthda.com/english/wiki/survminer). Kaplan–Meier plots and the corresponding log-rank tests, as well as the univariate cox proportional hazards regression method, were used to compare the OS of UC patients between groups.
To evaluate the influence of subtype cells in response to anti-programmed death-ligand 1 (PD-L1)-based immunotherapies, we evaluated the cellular abundance in the RNA-seq data derived from a patient cohort consisting of 192 metastatic UCs treated with atezolizumab.[14] Raw RNA-seq count data and clinical information were retrieved from the Imvigor210CoreBiologies package (version 1.0.0; https://github.com/SiYangming/IMvigor210CoreBiologies) and processed in the similar manner as in TCGA-BLCA data. Clinical responses to atezolizumab treatment were categorized as complete response (CR, n = 25), partial response (PR, n = 40), stable disease (SD, n = 44), or progressive disease (PD, n = 128).
Inference of the developmental trajectory of immune cells
We characterized the potential lineage differentiation process of immune cells by trajectory analysis of monocytes to macrophages using the Monocle 2 (version 2.18.0; http://cole-trapnell-lab.github.io/monocle-release/docs/) algorithm.[15] The enrichment of GO terms in each gene cluster was determined using the clusterProfiler package (version 3.18.0; https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html).
Copy number variation (CNV) estimation and malignant cell identification
We separated non-malignant urothelial cells from definitive tumor cells by evaluating the genetic CNV score, inferred from the scRNA-seq data reported by Kim et al.[16] Urothelial cells (CNV signal perturbation > 0.02 mean square of CNV estimate, or > 0.2 for correlation with top 5% CNV score cells) were classified as malignant cells. Differences in CNV scores between groups were compared using two-tailed Wilcoxon rank-sum test.
Putative interactions between cell types
Intercellular ligand–receptor (L–R) interactions between single cells from normal and tumor tissues were mapped using the CellChat algorithm (version 0.0.2; https://github.com/sqjin/CellChat).[17] The overexpressed ligands or receptors in each cell group were determined and projected to the known L–R protein interaction network. Biologically significant cell–cell communication was assigned a probability value by integrating gene expression with prior knowledge of the interactions between signaling ligands, receptors, and their cofactors by using the law of mass action. The significance of the L–R interactions was defined by permutation test with a threshold of P < 0.05. Potential L–R communication networks between cell subclusters were visualized using a bubble plot. We applied CellChat to compare the number of interactions and interaction strengths between different cell populations.
Single-cell regulatory network inference and clustering (SCENIC) analysis
The regulatory networks were evaluated based on scRNA-seq data using the SCENIC algorithm, as reported, based on the normalized matrix generated from the Seurat object.[18] The top 15 regulons with significant fold-changes between cell subclusters were included in further analyses.
Statistical analysis
All statistical analyses were performed using the R language (version 4.0.3, R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/) with two-sided tests. Cellular proportions were compared using two-tailed Wilcoxon signed-rank test. Statistical significance was set at P < 0.05. Other detailed statistical tools, methods, and thresholds are described in the Methods section.
Methods of flow cytometry, immunofluorescence, and immunohistochemistry were shown in Supplementary Methods.
Results
Overview of single-cell subclusters in normal and tumor urothelial tissues
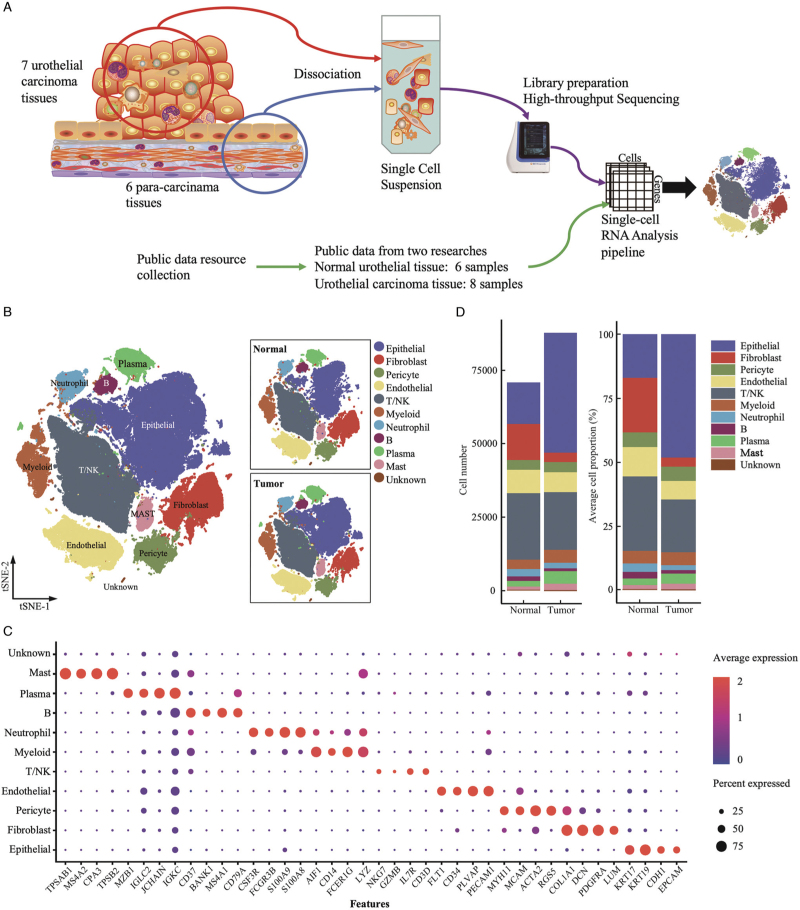
We examined intra-tumoral heterogeneity and the corresponding TME using scRNA-seq analysis of viable cells derived from tumors and matched adjacent normal biopsies from seven patients with UC (seven tumors and six normal tissues, Figure 1A). We increased the analytical power of the analysis by integrating our data with public published data from two previous scRNA-seq studies with a total of eight tumors and six normal tissues.[6,7] Detailed information of the included samples is provided in Supplementary Tables 1 and 2. A total of 87,898 cells were derived from 15 UC tumor tissues, and 70,969 cells were obtained from 12 normal tissues. Based on t-SNE and UMAP dimension reduction analyses, ten major cellular subclusters [Figure 1B and Supplementary Figure 1A–1C] were identified as urothelial epithelial cells (EPCAM, CDH1, KRT17, and KRT19), fibroblasts (COL1A1, DCN, PDGFRA, and LUM), pericytes (RGS5, and ACTA2), endothelial cells (ECs) (PECAM1, CD34, and FLT1), T/NK cells (CD3D, NKG7, and IL7R), myeloid cells (LYZ, CD14, and AIF1), neutrophils (S100A8, S100A9, FCGR3B, and CSF3R), B cells (CD79A and MS4A1), plasma cells (IGKC, JCHAIN, and MZB1), and mast cells (CPA3, MS4A2, and TPSAB1) according to the expression levels of canonical biomarkers [Figure 1C]. Distributions of cell subclusters in the three datasets matched well with each other under unsupervised cluster t-SNE and UMAP dimensional reduction plots [Supplementary Figure 1A and 1B], indicating consistently identified cell types by different platforms or batches. The proportions of cellular subclusters in each sample and dataset were shown in Supplementary Figure 1C and 1D. Total cell numbers and average cellular proportions were shown in Figure 1D, and urothelial epithelial cells largely contributed to the cellular composition in tumor tissues. Due to the bias that might be introduced during tissue dissociations by tumoral and normal epithelial cells and aimed to provide deep insights into the TME contexture of bladder cancer, we assessed the compositions of TME cell subsets after removing the epithelial and potential doublets cellular populations [Supplementary Figure 1E]. More myeloid cells and fewer fibroblasts were observed in the tumor tissues compared to the normal tissues (P < 0.05; Supplementary Figure 1E). We also compared the average cellular distribution between tumor tissues from bladder cancer and upper tract urothelial cancer; more T/NK cells and fewer myeloid, neutrophil, and mast cells were observed in bladder cancer tissues compared to UTUC [Supplementary Figure 2].
Figure 1.
Landscape profiling of single cells in tumor and normal urothelial tissues. (A) Overview of experimental design and data integration. (B) t-SNE projection identified ten main cell types (N = 158,867 cells). Dots represent single cells, and colors indicate cell types. (C) Dot plot indicating mean expression of canonical marker genes for the ten main identified cell types in tumor and normal urothelial tissues. (D) The total identified cell number (left) and the average cellular proportions (right) between the tumor (N = 15) and normal (N = 12) urothelial tissues colored by the cell types. t-SNE: t-Stochastic neighbor embedding.
Diversity of normal and tumor urothelial cells with seven subpopulations
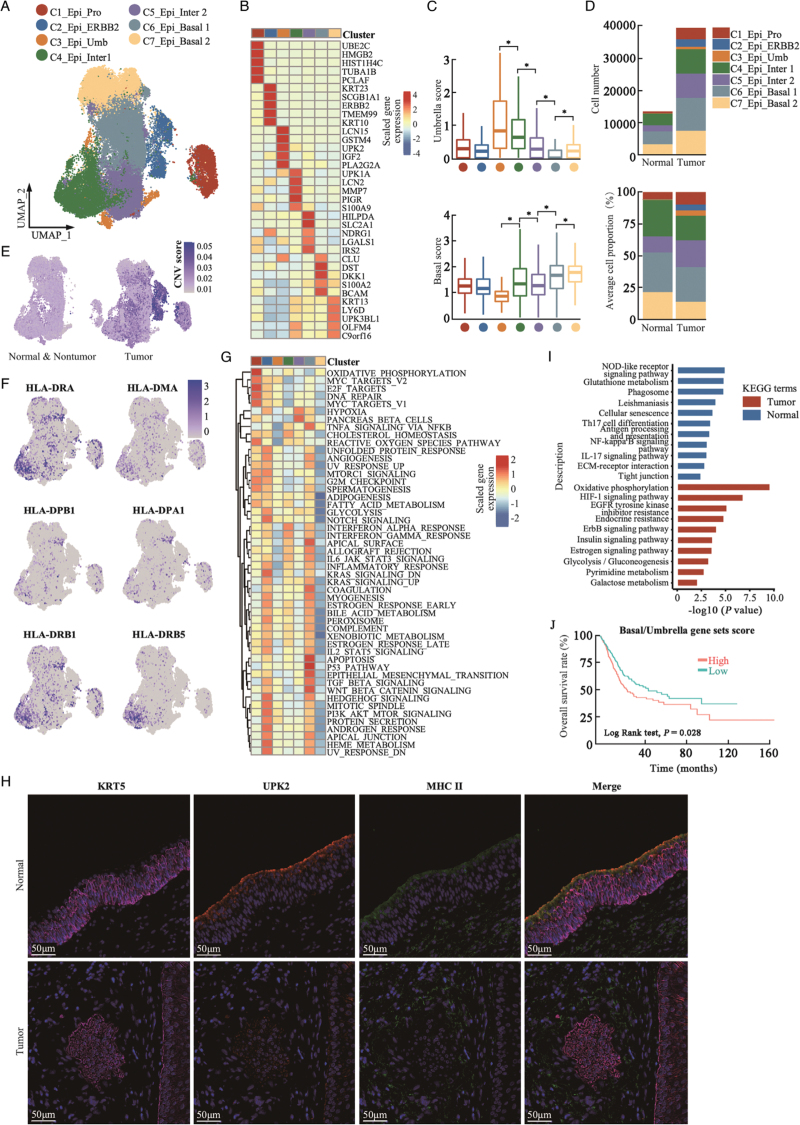
Seven subpopulations of urothelial epithelial cells were identified using unsupervised t-SNE clustering analysis [Figure 2A and 2B and Supplementary Figure 3A–3C]. Subcluster 1 consisted of proliferating urothelial cells that highly expressed UBE2C and TOP2A (C1_Epi_pro). Subcluster 2 highly expressed oncogene ERBB2 (C2_Epi_ERBB2). Subcluster 3 highly expressed the umbrella cell biomarker KRT20 (C3_Epi_Umb). Cells in subclusters 4 and 5 expressed uroplakins and keratin genes, except for the basal-specific keratin gene KRT5, indicating that these were intermediate urothelial cells (C4_Epi_Inter 1 and C5_Epi_Inter 2). Subclusters 6 and 7 showed relatively higher KRT5 expression but lower levels of uroplakin genes and were assigned as basal urothelial cells (C6_Epi_Basal 1 and C7_Epi_Basal 2). Based on the well-known canonical umbrella and basal biomarkers, we calculated the umbrella-like and basal-like scores of the single cells in each cell subcluster and found a stepwise increment of basal gene signature score and a reduction in umbrella gene signature score across the umbrella, intermediate, and basal cell subclusters (C3–C7; Figure 2C).
Figure 2.
Heterogeneity of urothelial cells in urothelial tissues. (A) UMAP plot of urothelial cell subgroups, colored by assigned cell type (N = 52,785 cells). (B) Heatmap of top five DEGs among urothelial cell subclusters. (C) Box plots showing umbrella (upper panel) and basal gene signature (lower panel) scores in the seven urothelial cell subgroups. ∗P < 0.001, two-sided unpaired Wilcoxon-rank test. Groups were color labeled in dots as in (A). (D) Total urothelial cell number (upper panel) and average cell proportion (lower panel) in tumor (N = 15) and normal (N = 12) urothelial tissues. (E) Stratified feature plots of CNV scores of single urothelial cells determined as normal and nonmalignant (left) or malignant (right) urothelial cells by the infer CNV algorithm. (F) Feature plots of selected MHC-II molecules in normal urothelial cells. (G) Heatmap showing enrichment of 50 hallmark gene sets (rows) in MSigDB database between malignant urothelial cell subgroups (columns). (H) Representative immunofluorescence staining image of basal marker KRT5, umbrella marker UPK2, and MHC-II molecules in normal (upper panel) and tumor (lower panel) urothelial tissues. (I) Bar plots showing comparison of KEGG pathways corresponding to DEGs between malignant and normal urothelial cell subgroups. (J) Kaplan-Meier curves of OS in TCGA-BLCA patients (n = 401) with higher or lower basal vs. umbrella gene set scores. P value was calculated using two-sided log-rank test. CNV: Copy number variation; DEGs: Differentially expressed genes; KEGG: Kyoto Encyclopedia of Genes and Genomes; OS: Overall survival; UMAP: Uniform manifold approximation and projection; MigDB: Molecular Signatures Database; MHC-II: Histocompatibility complex class II; TCGA-BLCA: The Cancer Genome Atlas-Bladder Endothelial Carcinoma.
As presented in Figure 2D and Supplementary Figure 3D and 3E, the average proportion of C1_Epi_pro was higher in tumor tissues than in normal tissues, indicating the high proliferation potential of tumor cells. We noted a reduction in C4_Epi_Inter 1 and an increase in C5_Epi_Inter 2 cell subcluster in tumors compared with normal urothelial tissues. The mean proportion of C6_Epi_Basal 1 was comparable between normal and tumor tissues; however, the C7_Epi_Basal 2 cell subcluster was significantly reduced in tumor tissues [Supplementary Figure 3E]. Non-malignant and malignant cells were identified in UC tumor tissues by inferring a larger scale CNV score, and C2_Epi_ERBB2 cells had the highest CNV score [Figure 2E and Supplementary Figure 4A]. According to scRNA-seq data, malignant cells could be derived from basal, intermediate, and umbrella cells but show significant heterogeneity between patients.[19] C3_Epi_Umb cells were predominantly identified in SC295 and SRR12603785 [Supplementary Figure 3D], which were consistent with the high protein expression level of the umbrella biomarker KRT20 in SC295 but rarely noticed in other patients [Supplementary Figure 4B]. C2_Epi_ERBB2 was exclusively identified in two tumor samples, SC340 and SC298 (C2_Epi_ERBB2), indicating that these cells exhibit ERBB2 genomic amplification [Supplementary Figure 3D]. The other patients were positive for KRT5/6, with a rarely positive umbrella gene, KRT20, indicating that they originated from basal or intermediate cells [Supplementary Figure 4B].
Basal signature enriched tumors were associated with poorer prognosis due to loss of immunogenicity activities
When comparing the DEGs between the cell subgroups, we noted that C4_Epi_Inter 1 cells expressed higher levels of histocompatibility complex class II (MHC-II) genes [Figure 2F]. GSEA revealed that C4_Epi_Inter 1 cells had relatively higher complement, interleukin (IL)-6/JAK/STAT3, and inflammatory response activities [Figure 2G], suggesting that C4_Epi_Inter 1 cells mediate immune surveillance in urothelial tissues. Multiplex immunofluorescence staining showed that normal umbrella (UPK2+/KRT5−) and intermediate (UPK2+/KRT5+) urothelial cells were positive for MHC-II molecules, and basal cells (UPK2−/KRT5+) showed minimal MHC-II expression [Figure 2H], indicating cell-specific expression of MHC-II molecules in normal urothelial tissues; however, minimal or weak MHC-II expression was observed in UPK2+/KRT5+ intermediate cells in the tumor tissues [Figure 2H and Supplementary Figure 4C]. When comparing the DEGs between all normal and tumor single epithelial cells, KEGG enrichment analysis suggested that oncogenic signaling pathways were significantly enhanced as well as the loss of inflammatory signaling pathways in malignant tumor cells [Figure 2I]. In the TCGA-BLCA cohort, patients with relatively higher level of basal vs. umbrella cell-specific gene set score showed poorer OS [Figure 2J]. Using the deconvolution methods, patients with higher proportions of C1_Epi_Pro, C2_Epi_ERBB2, and C5_Epi_inter 2 cells were associated with poorer OS, whereas patients with a higher proportion of C3_Epi_Umb, C4_Epi_Inter 1, and C7_Epi_Basal 2 cells were associated with better OS [Supplementary Figure 4D]. These results suggest that the loss of immunogenicity in urothelial cells is involved in UC development, which may influence the prognosis of patients with UC.
Characteristics of tumor-infiltrated lymphocytes in TME of UC tissues
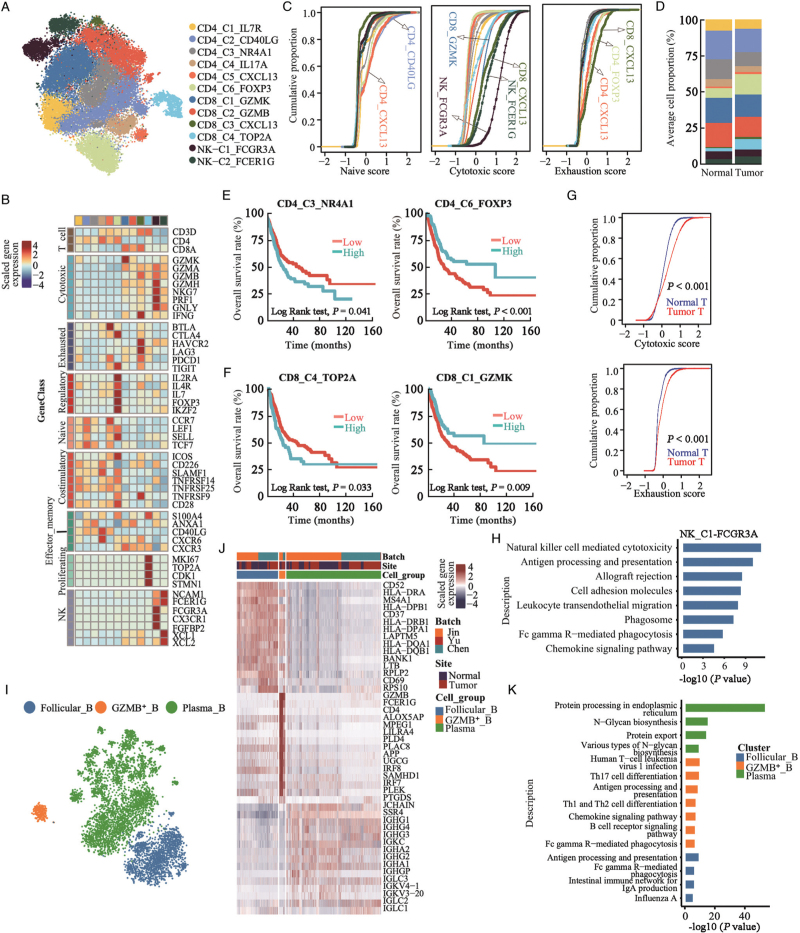
Graph-based unsupervised t-SNE analysis of 37,077 T/NK cells identified 12 subclusters, including 6 types of CD4+ T cells, 4 subtypes of CD8+ T cells, and 2 subtypes of NK cells [Figure 3A]. Regarding CD4+ T cells, there were naive (CD4_C1_IL7R and CD4_C2_CD40LG; CCR7+/SELL+/LEF1+) cells, memory cells (CD4_C3_NR4A1; ANXA1+), Th17 cells (CD4_C4_IL17A; IL17A+/CXCR6+/CD40LG+), exhausted T follicular helper (CD4_C5_CXCL13; CXCL13+/BTLA+/PDCD1+), and regulatory T cells (Tregs; CD4_C6_FOXP3; FOXP3+/IL2RA+/IL4R+) [Figure 3B and Supplementary Figure 5A]. CD4_C2_CD40LG and CD4_C5_CXCL13 cells showed the highest naive score compared with the other T cell subclusters [Figure 3C]. Interestingly, CD4_C6_FOXP3 Tregs showed relatively higher expression levels of immune checkpoint biomarkers including CTLA4 and TIGIT, while exhausted CD4_C5_CXCL13 follicular helper cells had higher expression of checkpoint inhibitors BTLA and PDCD1, suggesting that these two cell subclusters received immunosuppressive signals through distinct pathways [Figure 3B]. The proportion of CD4_C6_FOXP3 regulatory cells in CD4+ T cells was significantly higher in tumor tissues compared to normal tissues but not for the other cell types [Figure 3D and 3E and Supplementary Figure 5B]. UC patients in the TCGA-BLCA cohort with higher CD4_C3_NR4A1 cell proportion were significantly associated with poorer OS while with higher CD4_C6_FOXP3 proportion were associated with better OS [Figure 3E].
Figure 3.
T/NK and B cell-mediated tumor suppressive activities in urothelial tissues. (A) t-SNE projection of T and NK cells (N = 37,077 cells), colored according to cell clusters. (B) Heatmap of mean expression of functional gene sets in indicated cell clusters. (C) Cumulative distribution function showing distributions of naive, cytotoxic, and exhaustion scores for single T and NK cells. (D) Average proportions of T and NK cell subclusters in tumor (N = 15) and normal (N = 12) urothelial tissues, colored according to cell types. (E) Kaplan-Meier curves for OS among TCGA-BLCA patients (n = 401) with higher or lower CD4_C3_NR4A1 or CD4_C6_FOXP3 cellular proportion. P values from two-sided log-rank test were provided. (F) Kaplan-Meier curves for OS among TCGA-BLCA patients with higher or lower CD8_C4_TOP2A or CD8_C1_GZMK cellular proportion. P values from two-sided log-rank test were provided. (G) Cumulative distribution of cytotoxic and exhaustion scores for CD8+ T cells in normal and tumor urothelial tissues. P value was calculated using two-sided Wilcoxon rank-sum test. (H) Enriched KEGG pathways related to genes with increased expression levels in NK-C1-FCGR3A cells compared to NK-C2-FCER1G cells. (I) t-SNE projections of B cells, colored according to cell cluster. (J) Heatmap showing top 15 differentially expressed genes (DEGs, in row) among the three B cell clusters. (K) Bar plots showing enriched KEGG pathways related to DEGs among B cell types. DEGs: Differentially expressed genes; KEGG: Kyoto Encyclopedia of Genes and Genomes; OS: Overall survival; t-SNE: t-Stochastic neighbor embedding; TCGA-BLCA: The Cancer Genome Atlas-Bladder Endothelial Carcinoma.
Regarding CD8+ T cells, CD8_C1_GZMK represented pre-effector T cells. CD8_C2_ANXA1 represents effector memory T cells [Figure 3B]. CD8_C3_CXCL13 was assigned as terminally exhausted CD8+ cytotoxic T cells because they showed the highest cytotoxic and exhaustion activity [Figure 3C]. CD8_C4_TOP2A cells showed proliferating properties among CD8+ T cells, which showed the lowest cytotoxicity among CD8+ T cells. The proportion of CD8_C4_TOP2A cells was significantly higher in tumor tissues and associated with poorer OS [Supplementary Figure 5B and Figure 3F]. In total, CD8+ T cells in tumor tissues showed increased cytotoxicity and exhaustion activities compared to those in normal urothelial tissues [Figure 3G], indicating that CD8+ T cells became exhausted after the initial activation phase in UC tissues. In TCGA-BLCA cohort, UC patients with a higher CD8_C1_GZMK cellular proportion had better OS [Figure 3F], and no significant association with OS was noticed for the other CD8+ T cells.
We identified two NCAM1 (also known as CD56) positive NK cell subpopulations, including NK_C1_FCGR3A, with higher expression of FCGR3A (also known as CD16) and CX3CR1, and NK_C2_FCER1G with higher NCAM1, XCL1, and XCL2 [Figure 3A,B]. NK_C1_FCGR3A cells showed the highest cytotoxic score for lymphocytes [Figure 3C]. KEGG analysis revealed that NK-mediated tumor immunity signaling pathways were enriched in NK_C1_FCGR3A compared to NK_C2_FCER1G [Figure 3H]. The proportions of NK cell subclusters were comparable between normal and tumor urothelial tissues for all T/NK cells [Figure 3D and Supplementary Figure 5B]. However, higher proportions of NK_C1_FCGR3A in tumor tissues were associated with better OS [Supplementary Figure 5C]. The existence of two NK cell populations in urothelial tumor tissues was confirmed by flow cytometry [Supplementary Figure 6A]. Consistent with the scRNA-seq data, CD16 higher NK cells showed lower NCAM1 expression levels than CD16 lower NK cells [Supplementary Figure 6B].
Three types of B cells were identified, including follicular B cells (MHC-II genes+), GZMB-secreting B cells (MHC-II genes+/GZMB+), and plasma B cells (immunoglobulins+) [Figure 3I, J and Supplementary Figure 7A]. KEGG analysis [Figure 3K] confirmed that follicular B cells were enriched in antigen processing, presentation, and IgA production. In addition to antigen processing and presentation, GZMB+ B cells have been reported to inhibit CD4+ T lymphocyte proliferation in contact- and GZMB-dependent manner.[20] No difference in B cell subcluster infiltration was detected between normal and tumor tissues [Supplementary Figure 7B]. Plasma cells showed biological activities of immunoglobulin synthesis and secretion activities, and UC patients with higher plasma cells were associated with better OS according to decomposition analysis in TCGA-BLCA cohort, but not in follicular B and GZMB_B cells [Supplementary Figure 7C].
Immunosuppressive microenvironment modulated by myeloid cells
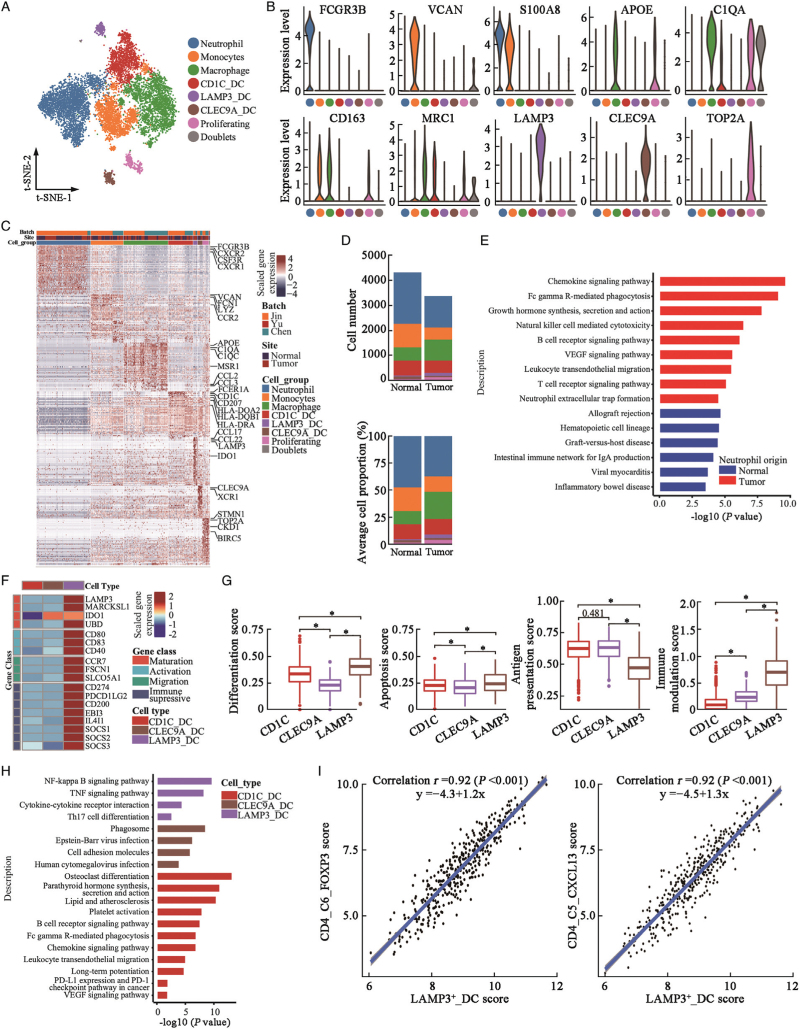
Seven myeloid cell subtypes were identified: neutrophils (FCGR3B+/CXCR2+/CSF3R+), monocytes (S100A8+/VCAN+/IL1B+), macrophages (C1QA+/C1QC+), proliferating myeloid cells (TOP2A+/C1QA), and three DC types (CD1c+ DC, LAMP3+ DC, and CLEC9A+ DC) [Figure 4A–4C and Supplementary Figure 8A]. Interestingly, neutrophils were predominantly detected in our BD Rhapsody™ platform, with few neutrophils detected in the 10 × Genomics platform reported by Chen et al[6] and Yu et al[7], further demonstrating that the 10 × Genomics platform rarely identifies neutrophils.[21] Tumor-infiltrated neutrophils were validated by flow cytometry according to the expression of CD45, CD11b, and CD66b on immune cells in UC tissues [Supplementary Figure 8B]. We noted a non-significant reduction in neutrophils in UC vs. normal urothelial tissues in the BD Rhapsody™ platform-based dataset [Figure 4D and Supplementary Figure 8C]; however, the neutrophils in tumor tissues showed enhanced activities of chemokines, B and T cell receptors, and vascular endothelial-derived growth factor (VEGF)-related signaling pathways, natural killer cell-mediated cytotoxicity activities, and neutrophil extracellular trap formation activities [Figure 4E], suggesting that neutrophils recruited in tumor tissues were reprogrammed with tumor-associated neutrophil-like properties.[22] Macrophages in tumors showed extensive expression of anti-inflammatory biomarkers (C1QA, C1QC, APOE, MRC1, and CD163; Supplementary Figure 8A] and relatively lower levels of proinflammatory genes (such as IL1B and CXCL8), suggesting that they were mainly M2-type tumor-associated macrophages (TAMs). Cellular proportion of macrophages was significantly increased in tumor tissues compared to normal tissues [Supplementary Figure 8C].
Figure 4.
Diversity of myeloid cells in urothelial tissues. (A) t-SNE projection of myeloid cells (N = 10,973 cells) identified in urothelial tissues, colored by cellular cluster. (B) Violin plots showing expression levels of canonical biomarker genes in each cell subcluster. (C) Heatmap showed the top 50 differentially expressed genes (DEGs, in row) among myeloid cell clusters (columns as indicated). (D) Cell number (upper panel) and average cellular proportion (lower panel) of myeloid cells in tumor (N = 15) and normal (N = 12) urothelial tissues. (E) Bar plot showing enrichment of KEGG pathways of over-expressed genes in neutrophils from tumor tissues compared with normal urothelial tissues. (F) Heatmap showed the normalized mean expression of genes related to DCs maturation, activation, migration and immune modulation activities. Filled color from blue to red indicated low to high expression level. (G) Boxplot showing differentiation, apoptosis, antigen presentation, and immune modulation scores of DC cell subclusters. ∗P < 0.001 from two-sided unpaired Wilcoxon rank-sum test. (H) Bar plot showing enrichment of KEGG pathways of over-expressed genes in each DC cluster. (I) Correlation of cell subcluster-specific score of LAMP3_DC cells and CD4_C6_FOXP3 T-regs or CD4_C5_CXCL13 T cells in TCGA-BLCA patients (N = 401). Coefficient calculated with Pearson correlation analysis. DC: Dendritic cell; DEGs: Differentially expressed genes; KEGG: Kyoto Encyclopedia of Genes and Genomes; t-SNE: t-Stochastic neighbor embedding; TCGA-BLCA: The Cancer Genome Atlas-Bladder Endothelial Carcinoma.
Three DC subclusters were identified [Figure 4A–4C and Supplementary Figure 8A]. CD1C_DCs were type 2 conventional DCs (CD1C+/CLEC10A+/FCER1A+; cDC2s), LAMP3_DCs were mature DCs (LAMP3+/CCR7+), and CLEC9A_DCs were type 1 conventional DCs (CD141+/CLEC9A+/XCR1+; cDC1s). The proportion of LAMP3_DCs was significantly higher in the tumor tissues than in the normal tissues [Figure 4D and Supplementary Figure 8C]. We found that LAMP3_DCs showed the highest maturation, activation, and migration properties among DC subclusters [Figure 4F]. LAMP3_DCs showed lower antigen-presentation activities; however, the apoptosis and differentiation gene signatures were higher [Figure 4G], and they also showed higher expression levels of immunosuppressive genes [Figure 4F]. KEGG analyses suggested that LAMP3_DCs were associated with increased cytokine–cytokine receptor interaction and TNF-α, NF-κB, and Th17 cell differentiation signaling pathways [Figure 4H]. LAMP3_DCs also expressed higher levels of CCL17, CCL19, and CCL22 [Supplementary Figure 9A], which were known to recruit Tregs and T helper cells.[23] LAMP3_DCs gene signatures were highly correlated with CD4_C6_FOXP3 and CD4_C5_CXCL13 cell signatures in the tumor tissues of the TCGA-BLCA cohort [Figure 4I]. The SCENIC algorithm suggested that activation of the POLR2A, RELB, NF-B, BCL3, and IRF4 motifs was correlated with upregulated immunosuppressive activity in LAMP3_DCs [Supplementary Figure 9B]. TCGA-BLCA patients with higher LAMP3_DCs showed poorer OS [Supplementary Figure 9C], and no significant association was observed for the other two cell subclusters. Overall, neutrophils, macrophages, and LAMP3+ DCs create an immunosuppressive microenvironment in UC tissues, which may serve as novel immunotherapy targets for UC in future.
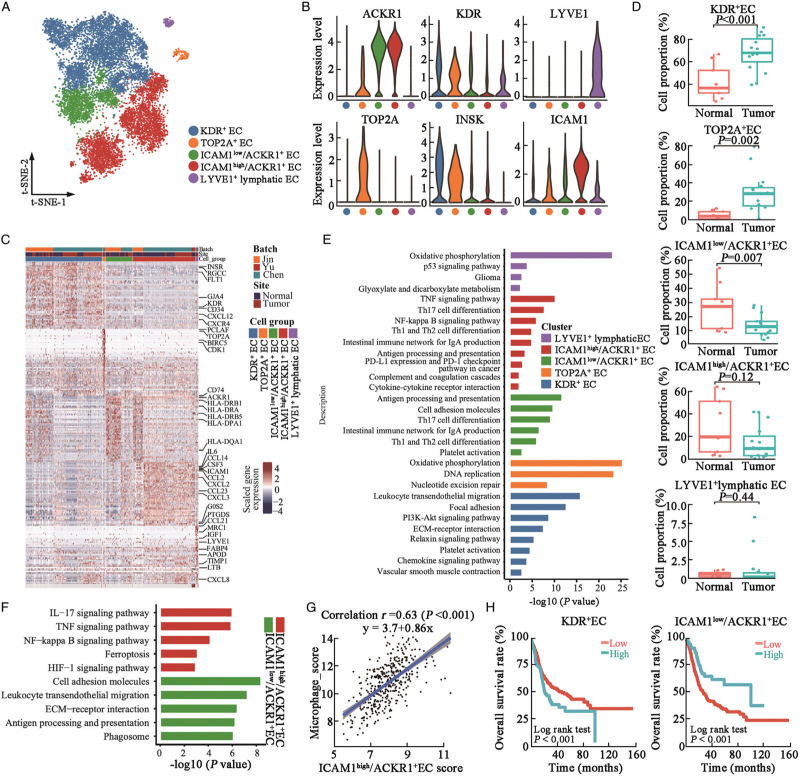
Enhanced angiogenesis and immunosurveillance deficiency caused by endothelial cells (ECs) in urothelial tumors
t-SNE analysis subclustered ECs into KDR+ (also known as VEGFR-2) ECs, proliferating ECs (TOP2A+ EC), ICAM1high/ACKR1+ ECs, ICAM1low/ACKR1+ ECs, and LYVE+ lymphatic ECs [Figure 5A–C]. The proportion of KDR+ and TOP2A+ ECs was significantly higher in the UC tissues [Figure 5D]. The KDR+ EC cluster highly expressed receptors of the VEGF signaling pathways including KDR and FLT1 [Figure 5B and 5C], and its proportion was significantly increased in UC tissues [Figure 5D], indicating the enhanced angiogenesis in UC tissues. KEGG enrichment analysis suggested that KDR+ EC cluster was enriched in leukocyte transendothelial migration, focal adhesion, platelet activation, and vascular smooth muscle contraction, demonstrating the neovascularization roles of the KDR+ ECs in UC tissues [Figure 5E]. ICAM1low/ACKR1+ ECs showed higher levels of MHC-II molecules [Figure 5C] and were enriched with antigen processing and presentation and Th17/Th1/Th2 cell differentiation activities [Figure 5E], indicating tumor immunosurveillance and CD4+ T cell activation activities of ICAM1low/ACKR1+ ECs.[24] KEGG analysis suggested that ICAM1low/ACKR1+ ECs were enriched in cell adhesion molecules, leukocyte transendothelial migration, and antigen processing and presentation activities, whereas ICAM1high/ACKR1+ ECs were enriched in IL-17/TNF/NF-κB inflammatory signaling pathways [Figure 5F]. Ferroptosis and HIF-1α signaling pathways were also increased in ICAM1high/ACKR1+ ECs, further confirming their tumor-stimulatory roles in urothelial tumors [Figure 5F]. The proportion of ICAM1low/ACKR1+ ECs was significantly reduced in the tumor tissues [Figure 5D]. ICAM1high/ACKR1+ cells highly express CCL2, CXCL2, CXCL3, and CCL23 [Figure 5C], which may be involved in the recruitment and activation of myeloid cells in UC tissues.[24] We noted a significant positive correlation between ICAM1high/ACKR1+ ECs and the macrophage gene signature in the TCGA-BLCA cohort, which further suggests the potential chemotaxis of ICAM1high/ACKR1+ ECs on macrophages [Figure 5G]. Decomposition analysis suggested that TCGA-BLCA patients with higher KDR+ ECs were associated with poorer OS, while patients with higher ICAM1low/ACKR1+ EC were associated with better OS [Figure 5H]. These results suggest that targeting KDR+ and ICAM1high/ACKR1+ ECs may inhibit angiogenesis and reverse the immunosuppressive microenvironment in UC tissues.
Figure 5.
ECs enhance angiogenesis and mediated immunosurveillance in UC tissues. (A) t-SNE plot of the identified ECs (N = 13,287 cells), colored by cell subset. (B) Violin plots showing expression levels of selected canonical biomarker genes in each EC subcluster. (C) Heatmap showing top 50 differentially expressed genes (DEGs, in row) among the five EC clusters (columns as indicated). (D) Box plot showing proportion of ECs in normal and tumor urothelial tissues. P value was derived from two-sided unpaired Wilcoxon rank-sum test. (E) Bar plot showing enrichment of KEGG pathways of over-expressed genes in each DC cluster, colored by cell subclusters. (F) Bar plot showing enriched KEGG pathways of over-expressed genes in ICAM1high/ACKR1+ EC or ICAM1low/ACKR1+ EC subclusters as color indication. (G) Correlation between ICAM1high/ACKR1+ EC specific gene signature score and macrophage cell signature score in TCGA-BLCA patients (N = 401). Coefficient calculated with Pearson's correlation analysis. (H) Kaplan-Meier OS curves of TCGA-BLCA patients (n = 401) with higher or lower KDR+ EC or ICAM1low/ACKR1+ EC cellular proportion. P value was calculated using two-sided log-rank test. DEGs: Differentially expressed genes; ECs: Endothelial cells; KEGG: Kyoto Encyclopedia of Genes and Genomes; OS: Overall survival; t-SNE: t-Stochastic neighbor embedding; IL-17: Interleukin 17; TNF: Tumor necrosis factor; HIF-1: Hypoxia-inducible factor-1; ECM: Extracellular matrix; UC: Urothelial carcinoma; TCGA-BLCA: The Cancer Genome Atlas-Bladder Endothelial Carcinoma.
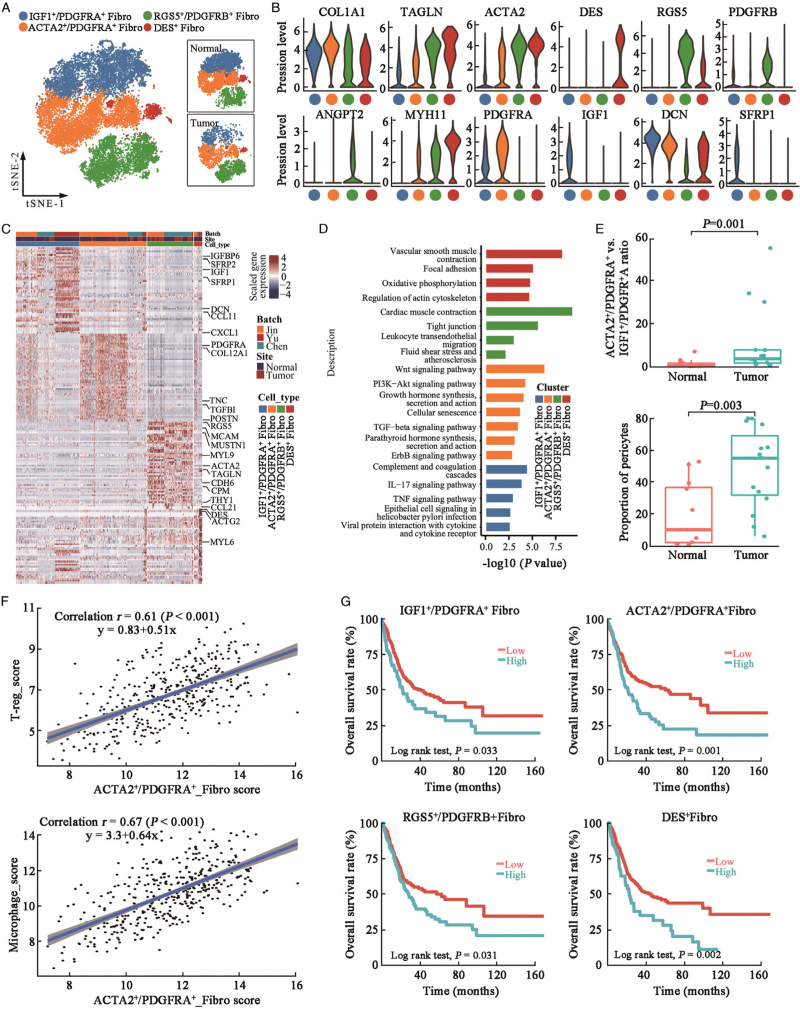
Distinct stromal cell subpopulations in UC and normal urothelial tissues
Fibroblasts and pericytes constitute the main COL1A1+ stromal cells in the urothelial tissues. A previous study proposed two different fibroblast subtypes: PDGFRA1+ and RGS5+ fibroblasts.[6] In our study, four stromal cell subtypes were identified, including IGF1+/PDGFRA+, ACTA2+/PDGFRA+, RGS5+/PDGFRB+, and DES+ cell clusters [Figure 6A–6C]. IGF1+/PDGFRA+ fibroblasts showed high levels of IGF1 and pro-inflammatory cytokines (IL-6 and CXCL1) [Supplementary Figure 10A], indicating that they were inflammatory fibroblasts. These cells also express SFRP1, which inhibits fibroblast proliferation and collagen synthesis, promotes fibroblast apoptosis, and inhibits myofibroblasts transformation.[25] These results suggested that IGF1+/PDGFRA+ fibroblasts were normal urothelial tissue fibroblasts. ACTA2+/PDGFRA+ fibroblasts showed higher ACTA2 and MYH11 expression levels than cluster 1 [Figure 6B and 6C and Supplementary Figure 10A], and they were enriched in Wnt, PI3K-Akt, growth hormone synthesis, TGF-β, and ErbB signaling pathways [Figure 6D], indicating that these fibroblasts were cancer-associated myofibroblasts. ACTA2+/PDGFRA+ myofibroblasts were one of the major sources of bone morphogenetic protein (BMP) and Wnt signaling pathway factors [Supplementary Figure 10A], which may induce the M2 polarization of TAMs and regulate T cell differentiation.[26] RGS5+/PDGFRB+ stromal cells expressed angiopoietin ANGPT2 [Figure 6B] and were enriched in leukocyte transendothelial migration, tight junction, and fluid shear stress signaling pathways [Figure 6D], indicating that they were pericytes in bladder tissues. Cluster 4 cells were positive for DES but negative for PDGFRA and PDGFRB, suggesting that these were smooth muscle cells [Figure 6B].
Figure 6.
Distinct cell types of stromal cells in urothelial tissues. (A) t-SNE projection of stromal cells in (n = 20,315) urothelial tissues, colored by cellular cluster. (B) Violin plots showing expression levels of selected canonical biomarker genes in each stromal cell subcluster. (C) Heatmap showing top differentially expressed genes (DEGs, in row) among the four stromal cell clusters (columns as indicated). (D) Bar plot showing enrichment of KEGG pathways of over-expressed genes in each DC cluster, colored by cell subcluster. (E) Box plot showing ratio of ACTA2+/PDGFRA+vs. IGF1+/PDGFRA+ fibroblasts number (upper panel) and total proportion of pericytes (RGS5+/PDGFRB+ cells) in normal (n = 9) and tumor (n = 15) urothelial tissues (lower panel). P value was derived from two-sided unpaired Wilcoxon rank-sum test. (F) Correlation between ACTA2+/PDGFRA+ specific gene signature score and CD4+ T-reg cell signature score or microphage cell signature score in TCGA-BLCA patients (N = 401). Coefficient calculated with Pearson's correlation analysis. (G). Kaplan-Meier survival curves of TCGA-BLCA patients (n = 401) with higher or lower of stromal cellular subcluster proportion as indicated. P values were calculated using two-sided log-rank tests. DEGs: Differentially expressed genes; KEGG: Kyoto Encyclopedia of Genes and Genomes; t-SNE: t-Stochastic neighbor embedding; TCGA-BLCA: The Cancer Genome Atlas-Bladder Endothelial Carcinoma.
We compared the relative stromal cellular proportions between normal and tumor urothelial tissues [Figure 6E and Supplementary Figure 10B], the ratio of ACTA2+/PDGFRA+vs. IGF1+/PDGFRA+ cells was significantly increased in tumor tissues. Besides, the proportion of RGS5+/PDGFRB+ pericytes was also significantly increased in the tumor tissues [Figure 6E]. In the TCGA-BLCA cohort, we identified a significant correlation between ACTA2+/PDGFRA+ myofibroblasts and macrophage or Treg-specific gene signature scores, suggesting that ACTA2+/PDGFRA+ myofibroblasts may stimulate UC progression by regulating immune cell activity [Figure 6F]. Using decomposition analysis, we found that UC patients with the higher proportion of any stromal cell subgroups were associated with poorer OS among TCGA-BLCA patients [Figure 6G]. These results demonstrate the tumor-supporting activities of stromal cells in the progression of UC.
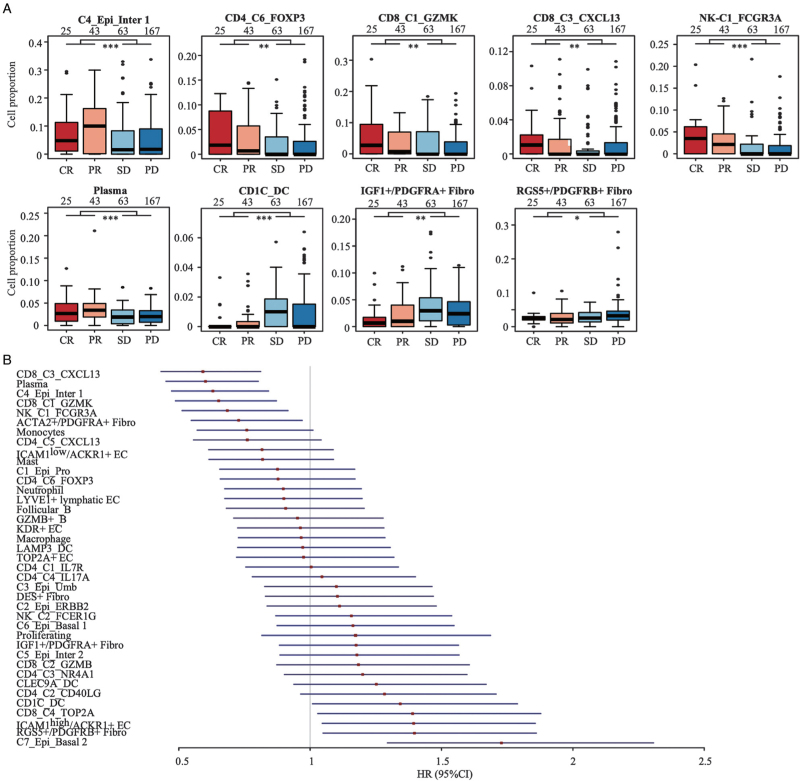
Decomposition analysis of the cellular subtypes that associated with immunotherapy responses
To evaluate whether the cellular subtypes may influence the response to immunotherapy in urothelial cancer patients, we applied decomposition analysis in the patient cohort of IMvigor210, which consisted of 298 metastatic urothelial cancer patients treated with atezolizumab (a PD-L1 inhibitor).[14] We found that patients sensitive to atezolizumab (CR or PR) had higher proportions of C4_Epi_Inter 1, CD4_C6_FOXP3, CD8_C1_GZMK, CD8_C3_CXCL13, NK_C1_FCGR3A, and plasma cells, whereas those resistant to atezolizumab (SD and PD) had higher proportions of CD1c_DC, RGS5+/PDGFRB+ pericytes, and IGF1+/PDGFRA+ fibroblasts [Figure 7A]. Patients with higher proportion of C6_Epi_Basal 1, C7_Epi_Basal 2, CD4_C2_CD40LG, CD4_C3_NR4A1, CD8_C2_GZMB, CD8_C4_TOP2A, CD1C_DC, CLEC9A_DC, ICAM1high/ACKR1+ EC, LYVE1+ lymphatic EC, IGF1+/PDGFRA+ fibroblasts, and RGS5+PDGFRB+ pericytes were associated with poorer OS in IMvigor210 cohort, whereas patients with higher proportion of C4_Epi_Inter 1, CD4_C5_CXCL13, CD4_C6_FOXP3, CD8_C1_GZMK, CD8_C3_CXCL13, NK_C1_FCGR3A, NK_C2_FCER1G, monocytes, plasma cells, mast cells, and ICAM1low/ACKR1+ EC were associated with better OS [Figure 7B and Supplementary Figure 11].
Figure 7.
Cellular subcluster proportions in bladder cancer tumors were associated with sensitivity of anti-PD-L1 treatment in mUC patients. (A). Boxplot showed the cellular proportion of subtype cells in patients sensitive (CR or PR) or resistant (SD or PD) to anti-PD-L1 treatment as indicated. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 for two-tailed Wilcoxon rank-sum test. Sample size in each group was given above the boxplot. (B). Forest plot of the estimates in univariate Cox regression analysis that assessed the association between relative cell abundance and mUC patients survival treated with atezolizumab in IMvigor210 (https://github.com/SiYangming/IMvigor210CoreBiologies) cohort (N = 298). CR: Complete response; PD: Progressive disease; PR: Partial response; SD: Stable disease; DC: Dendritic cell; mUC: metastatic urothelial carcinoma.
Intercellular interactions between cell subclusters in UC and normal urothelial tissues
We constructed tissue-specific cellular communication networks using the CellChat algorithm.[17] A total of 16,702 and 17,572 significant intercellular interactions were detected in the tumor and normal urothelial tissues, respectively, and the sum of intercellular strength was reduced in the tumor tissues [Supplementary Figure 12A]. The number of interactions and interaction strengths between CD4+ T cells, CD8+ T cells, and myeloid cells decreased, while interactions between RGS5+/PDGFRB+ pericytes and ECs were enhanced [Supplementary Figure 12B]. The total incoming and outcoming L–R interactions by cellular subgroups were shown in Supplementary Figures 13 and 14. We observed novel cell subgroup incoming L–R interactions mediated by the leukemia inhibitory factor receptor (LIFR), LIGHT, CD137, a proliferation-inducing ligand (APRIL), PERIOSTIN, CD80, T cell immunoreceptor with Ig and ITIM domains (TIGIT), and PD-L1 signaling pathways in tumor tissues compared to normal tissues [Supplementary Figure 13]. Outgoing signaling pathways, including the angiopoietin-like protein (ANGPTL), NECTIN, VEGF, and insulin-like growth factor (IGF) pathways mediated by urothelial cells, and the heparan sulfate proteoglycan (HSPG) and AGRN pathways mediated by ECs were also enhanced in tumor tissues [Supplementary Figure 14]. Network centrality analysis revealed that IGF and pro-epidermal growth factor (EGF) ligands were predominantly produced by urothelial cells and ECs and that their receivers were urothelial cells [Supplementary Figure 12C]. VEGF factors were predominantly derived from urothelial and stromal cells and act on ECs to stimulate angiogenesis in UC tissues. Platelet-derived growth factors (PDGFs) were produced by fibroblasts and ECs and act on stromal cells. C-C motif chemokine (CCL) and C-X-C motif chemokine (CXCL) chemokines were widely produced in urothelial, immune, and stromal cells and recruit and regulate myeloid cells and ECs by interacting with CCRs, C-X-C chemokine receptors (CXCRs), and atypical chemokine receptors (ACKRs) [Supplementary Figure 12C].
Discussion
Here, we delineated the transcriptomic landscape and heterogeneity of tumor, immune, and stromal cells, as well as the intercellular interactions among cell subclusters in UC and normal urothelial tissues. Compared with normal urothelial cells, tumor urothelial cells showed a significantly higher genomic CNV and enhanced activation of multiple oncogenic signaling pathways associated with a loss of antigen processing and presentation activities. M2-type TAMs, LAMP3+ DCs, and exhausted tumor-infiltrated lymphocytes create an immunosuppressive microenvironment in UC tissues. Infiltrated KDR+ ECs stimulated angiogenesis, whereas ICAM1high/ACKR1+ ECs modulated immune cell homing and activation in tumor tissues. In addition, remodeling of stromal cells in the urothelial TME, especially ACTA2+/PDGFRA+ myofibroblasts and pericytes, enhanced tumor cell growth and angiogenesis in UCs. These results reveal multiple hallmarks of UC development and progression, which may provide potential novel therapeutic targets.
The UC subtypes are associated with distinct prognoses and clinical therapeutic outcomes. Sjodahl et al[27] clustered five subtypes and found that higher expression of the umbrella cell biomarker KRT20 was associated with better OS. Choi et al[28] classified muscle-invasive UC into basal, luminal, and p53-like types, and the basal group was associated with increased aggressiveness and shorter disease-free survival and OS. According to the current study, each UC tissue sample included various types of urothelial epithelial cells, but with different cellular proportions, indicating intra-tumoral heterogeneity of UCs. TCGA-BLCA patients with higher umbrella cell-specific scores had better OS. These results were in accordance with genetic lineage studies showing that urothelial intermediate-origin tumors were usually low-grade, non-invasive papillary lesions, whereas a KRT5 basal cell origin was associated with muscle-invasive or squamous cell carcinoma.[19] According to scRNA-seq data, KRT5+ cells showed MHC-II molecules deficiency, while umbrella and intermediate cells showed MHC-II molecule expression and relatively higher immune surveillance activities. These cellular properties may underlie the different prognoses of basal and luminal tumors. Moreover, intra-tumoral heterogeneity of tumor cells may lead to drug resistance and tumor progression under mono-targeting drug treatments; thus, combination therapies may be warranted in patients with UC.
The biological activities of myeloid cells depend on their cellular state and the TME. Neutrophils have demonstrated multifactorial roles in tumorigenesis and progression and have been associated with detrimental outcomes in several solid tumors.[29] Previous scRNA studies based on the droplet 10× Genomics platform rarely detected neutrophils in UC and normal urothelial tissues.[6,7] However, using the BD Rhapsody platform, we noted significant neutrophil infiltration in the urothelial TME and increased tumor-supportive activities, suggesting that normal neutrophils had switched to tumor-associated neutrophil properties in UC.[29] In addition, macrophages in UC tissues were mostly M2-like TAMs, which have been shown to promote angiogenesis and distant metastasis of UCs.[30] The macrophages expressed high levels of immunosuppressive ligands, including CD274, LGALS9, and PDCD1LG2, suggesting that they may contribute to T-cell immunosuppressive activities in the TME of UC. CCL2 secreted by malignant urothelial cells and myofibroblasts may recruit monocytes and stimulate their reprogramming into macrophages by interacting with CCR2.[31] ANGPTL2 also binds to TLR4, which may stimulate an M1- to M2-type macrophage transition in the TME.[32] DC_C3_LAMP3 cells were also significantly enriched myeloid cell clusters, showing relatively higher expression levels of multiple immunosuppressive genes, and were considered as regulatory and tolerogenic DCs.[12,33] The DC_C3_LAMP3 subset also showed strong potential interactions with multiple T-cells, contributing to T cell dysfunction. Overall, these results indicated that myeloid cells created an immunosuppressive environment in UCs, and targeting these cells may help to reverse immunosurveillance in UC patients.
Clinical studies have revealed that some patients were sensitive to immune checkpoint inhibitor-based therapy.[34] We noted a subset of CD4+ and CD8+ T cells in the urothelial TME that highly expressed CXCL13, a key regulator of B cell migration and lymphoid tissue architecture, through interaction with CXCR5.[35,36] The recruited B cells may contribute to antitumor immunity by directly damaging cancer cells,[37] generating tumor-specific antibodies,[38] and promoting tumor antigen uptake by DCs.[39] Moreover,CXCL13 can improve immune cell tumor infiltration.[40] In bladder tissues, CD4_C5_CXCL13 cells showed higher BTLA and PDCD1 expression, and CD8_C3_CXCL13 cells showed higher levels of HAVCR2, LAG3, and PDCD1, which may serve as novel therapeutic targets. Compared to other T cells, CXCL13+/CD8+ T cells showed the highest cytotoxicity and exhaustion score. Deconvolution analysis identified that metastatic urothelial carcinoma (mUC) patients with higher CD8_C3_CXCL13 levels were more sensitive to atezolizumab treatment and associated with better prognosis, which was consistent with a study reporting that tissues with higher CXCL13 levels were more sensitive to anti-PD-1 immunotherapy and associated with better OS in patients with metastatic UC.[41] However, CXCL13 has dual effects on tumor promotion and immunity. CXCL13 induces IL-10 production by B cells and T cells, allowing the tumor to escape host immune surveillance.[42,43] CXCL13 also recruits immunosuppressive myeloid-derived suppressor cells and Tregs.[44,45] Further studies are needed to elucidate the roles of CXCL13 in UC progression and the immune response.
Normal fibroblasts can transform into cancer-associated fibroblasts in malignant tissues, which may, in turn, stimulate tumor proliferation, metastasis, and drug resistance.[46] In the current study, ACTA2+/PDGFRA+ fibroblasts were enriched in UC tissues as cancer-associated fibroblasts. Compared with IGF1+/PDGFRA+ cells, ACTA2+/PDGFRA+ cells showed enhanced activation of genes such as WNT5A and BMP4. WNT5A was associated with arsenic-associated malignant transformation and invasion[47] and was associated with advanced clinical stage in patients.[48] BMP4 induces M2-type TAM polarization and stimulates epithelial–mesenchymal transition of urinary epithelial carcinoma cells.[49] These results suggest that ACTA2+/PDGFRA+ stromal cells may promote UC progression by directly modulating malignant urothelial cells and TME in UC. Chen et al[6] identified an RGS5+/PDGFRA− fibroblast subcluster with myofibroblast properties; however, there was significant heterogeneity among PDGFRA− stromal cells. In the current study, a subgroup of RGS5+/PDGFRA− stromal cells showed high expression of PDGFRB, MYH11, MCAM, and angiopoietin ANGPT2, suggesting that these stromal cells were pericytes in urothelial tissues.[50] These results highlight the heterogeneous nature of stromal cells and their tumor-supportive roles in the urothelial TME, which may serve as potential therapeutic targets in UC.
The current study has several limitations. We integrated publicly available scRNA-seq datasets with our in-house dataset, which significantly increased the statistical power of the analysis and provided a more comprehensive profile of single cells in normal and tumor urothelial tissues. However, these datasets were generated by different research groups using different platforms. Although we integrated the datasets using the Harmony algorithm and normalized them, batch effects caused by differences in patient recruitment, sample preparation, library preparation, and bioinformatics analysis cannot be fully ruled out. Spatial information for single cells, which may influence patient prognosis and treatment outcomes, is largely lacking in current scRNA-seq data. Therefore, more studies with larger sample sizes are warranted to clarify the heterogeneous nature of urothelial cells and TME in UC tissues.
In conclusion, the current study provides a comprehensive transcriptomic atlas for single cells, which highlights the heterogeneity of the multicellular ecosystem and intercellular interactions occurring in normal and tumoral urothelial tissues. These results may pave the way for the development of novel methods for prognosis prediction and treatment of UC.
Funding
This study was supported by grants from the National Nature Science Foundation (No. 81970658) and Natural Science Foundation of Shanghai (No. 20ZR1434100).
Conflicts of interest
None.
Data available statement
The data reported in this study are available in the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number: CNP0003434). Specific code will be available upon reasonable request.
Supplementary Material
Footnotes
How to cite this article: Jin X, Wang Q, Luo F, Pan J, Lu T, Zhao Y, Zhang X, Xiang E, Zhou C, Huang B, Lu G, Chen P, Shao Y. Single-cell transcriptomic analysis of tumor heterogeneity and intercellular networks in human urothelial carcinoma. Chin Med J 2023;136:690–706. doi: 10.1097/CM9.0000000000002573
Xingwei Jin and Qizhang Wang contributed equally to this work.
Supplemental digital content is available for this article.
References
- 1.Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J 2021; 134:783–791. doi: 10.1097/CM9.0000000000001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drakaki A, Pantuck A, Mhatre SK, Dhillon PK, Davarpanah N, Degaonkar V, et al. “Real-world” outcomes and prognostic indicators among patients with high-risk muscle-invasive urothelial carcinoma. Urol Oncol 2021; 39:76.e15–76.e22. doi: 10.1016/j.urolonc.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Morales-Barrera R, Suárez C, de Castro AM, Racca F, Valverde C, Maldonado X, et al. Targeting fibroblast growth factor receptors and immune checkpoint inhibitors for the treatment of advanced bladder cancer: new direction and new hope. Cancer Treat Rev 2016; 50:208–216. doi: 10.1016/j.ctrv.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, Yang D, Yang Q, Lv X, Huang W, Zhou Z, et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat Commun 2020; 11:1–17. doi: 10.1038/s41467-020-20059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung W, Eum HH, Lee HO, Lee KM, Lee HB, Kim KT, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun 2017; 8:1–12. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Zhou L, Liu L, Hou Y, Xiong M, Yang Y, et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun 2020; 11:1–12. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Z, Liao J, Chen Y, Zou C, Zhang H, Cheng J, et al. Single-cell transcriptomic map of the human and mouse bladders. J Am Soc Nephrol 2019; 30:2159–2176. doi: 10.1681/ASN.2019040335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen P, Wang Y, Li J, Bo X, Wang J, Nan L, et al. Diversity and intratumoral heterogeneity in human gallbladder cancer progression revealed by single-cell RNA sequencing. Clin Transl Med 2021; 11:1–20. doi: 10.1002/ctm2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat Methods 2019; 16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu G, Wang LG, Han Y, He QY. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cillo AR, Kürten CHL, Tabib T, Qi Z, Onkar S, Wang T, et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity 2020; 52:183–199. doi: 10.1016/j.immuni.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, He S, Wang XL, Peng W, Chen QY, Chi DM, et al. Tumour heterogeneity and intercellular networks of nasopharyngeal carcinoma at single cell resolution. Nat Commun 2021; 12:1–18. doi: 10.1038/s41467-021-21043-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jew B, Alvarez M, Rahmani E, Miao Z, Ko A, Garske KM, et al. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat Commun 2020; 11:1–11. doi: 10.1038/s41467-020-15816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018; 554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017; 14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun 2020; 11:1–15. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021; 12:1–20. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017; 14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Batavia J, Yamany T, Molotkov A, Dan H, Mansukhani M, Batourina E, et al. Bladder cancers arise from distinct urothelial sub-populations. Nat Cell Biol 2014; 16:982–991. 1-5. doi: 10.1038/ncb3038. [DOI] [PubMed] [Google Scholar]
- 20.Chesneau M, Mai HL, Danger R, Le Bot S, Nguyen TV, Bernard J, et al. Efficient expansion of human granzyme B-expressing B cells with potent regulatory properties. J Immunol 2020; 205:2391–2401. doi: 10.4049/jimmunol.2000335. [DOI] [PubMed] [Google Scholar]
- 21.Qi JDSD, Dawson T, Geanon D, Stefanos H, Marvin R, Walker L, et al. Multimodal single-cell characterization of the human granulocyte lineage. bioRxiv 2021; doi: 10.1101/2021.06.12.448210. [Google Scholar]
- 22.Masucci MT, Minopoli M, Carriero MV. Tumor associated neutrophils. Their role in tumorigenesis, metastasis, prognosis and therapy. Front Oncol 2019; 9:1–16. doi: 10.3389/fonc.2019.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizukami Y, Kono K, Kawaguchi Y, Akaike H, Kamimura K, Sugai H, et al. CCL17 and CCL22 chemokines within tumor microenvironment are related to accumulation of Foxp3+ regulatory T cells in gastric cancer. Int J Cancer 2008; 122:2286–2293. doi: 10.1002/ijc.23392. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Zhu Z, Wang L, Lin Y, Fang H, Lei J, et al. Single-cell transcriptome profiling reveals vascular endothelial cell heterogeneity in human skin. Theranostics 2021; 11:6461–6476. doi: 10.7150/thno.54917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Liu Y, Liang X, Yang G, Liu Y, Li F, et al. Effects of secreted frizzled-related protein 1 on inhibiting cardiac remodeling. Eur Rev Med Pharmacol Sci 2020; 24:6270–6278. doi: 10.26355/eurrev_202006_21525. [DOI] [PubMed] [Google Scholar]
- 26.Quandt J, Arnovitz S, Haghi L, Woehlk J, Mohsin A, Okoreeh M, et al. Wnt-beta-catenin activation epigenetically reprograms treg cells in inflammatory bowel disease and dysplastic progression. Nat Immunol 2021; 22:471–484. doi: 10.1038/s41590-021-00889-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sjodahl G, Lauss M, Lovgren K, Chebil G, Gudjonsson S, Veerla S, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res 2012; 18:3377–3386. doi: 10.1158/1078-0432.CCR-12-0077-T. [DOI] [PubMed] [Google Scholar]
- 28.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014; 25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ocana A, Nieto-Jiménez C, Pandiella A, Templeton AJ. Neutrophils in cancer: prognostic role and therapeutic strategies. Mol Cancer 2017; 16:1–7. doi: 10.1186/s12943-017-0707-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharifi L, Nowroozi MR, Amini E, Arami MK, Ayati M, Mohsenzadegan M. A review on the role of M2 macrophages in bladder cancer; pathophysiology and targeting. Int Immunopharmacol 2019; 76:1–11. doi: 10.1016/j.intimp.2019.105880. [DOI] [PubMed] [Google Scholar]
- 31.Yang H, Zhang Q, Xu M, Wang L, Chen X, Feng Y, et al. CCL2-CCR2 axis recruits tumor associated macrophages to induce immune evasion through PD-1 signaling in esophageal carcinogenesis. Mol Cancer 2020; 19:1–14. doi: 10.1186/s12943-020-01165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei X, Nie S, Liu H, Sun J, Liu J, Li J, et al. Angiopoietin-like protein 2 facilitates non-small cell lung cancer progression by promoting the polarization of M2 tumor-associated macrophages. Am J Cancer Res 2017; 7:2220–2233. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 2019; 179:829–845.e20. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Apolo AB, Infante JR, Balmanoukian A, Patel MR, Wang D, Kelly K, et al. Avelumab, an anti-programmed death-ligand 1 antibody, in patients with refractory metastatic urothelial carcinoma: results from a multicenter, phase Ib study. J Clin Oncol 2017; 35:2117–2124. doi: 10.1200/JCO.2016.71.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansel KM, Ngo VN, Hyman PL, Luther SA, Förster R, Sedgwick JD, et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature 2000; 406:309–314. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 36.Cosgrove J, Novkovic M, Albrecht S, Pikor NB, Zhou Z, Onder L, et al. B cell zone reticular cell microenvironments shape CXCL13 gradient formation. Nat Commun 2020; 11:1–15. doi: 10.1038/s41467-020-17135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kemp TJ, Moore JM, Griffith TS. Human B cells express functional TRAIL/Apo-2 ligand after CpG-containing oligodeoxynucleotide stimulation. J Immunol 2004; 173:892–899. doi: 10.4049/jimmunol.173.2.892. [DOI] [PubMed] [Google Scholar]
- 38.Schlößer HA, Thelen M, Lechner A, Wennhold K, Garcia-Marquez MA, Rothschild SI, et al. B cells in esophago-gastric adenocarcinoma are highly differentiated, organize in tertiary lymphoid structures and produce tumor-specific antibodies. Oncoimmunology 2019; 8:1–12. doi: 10.1080/2162402X.2018.1512458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carmi Y, Spitzer MH, Linde IL, Burt BM, Prestwood TR, Perlman N, et al. Allogeneic IgG combined with dendritic cell stimuli induce antitumour T-cell immunity. Nature 2015; 521:99–104. doi: 10.1038/nature14424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013; 39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Goswami S, Chen Y, Anandhan S, Szabo PM, Basu S, Blando JM, et al. ARID1A mutation plus CXCL13 expression act as combinatorial biomarkers to predict responses to immune checkpoint therapy in mUCC. Sci Transl Med 2020; 12:1–11. doi: 10.1126/scitranslmed.abc4220. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Yuling H, Yanping J, Xinti T, Yaofang Y, Feng Y, et al. CCL19 and CXCL13 synergistically regulate interaction between B cell acute lymphocytic leukemia CD23+CD5+ B cells and CD8+ T cells. J Immunol 2007; 179:2880–2888. doi: 10.4049/jimmunol.179.5.2880. [DOI] [PubMed] [Google Scholar]
- 43.Cha Z, Qian G, Zang Y, Gu H, Huang Y, Zhu L, et al. Circulating CXCR5+CD4+ T cells assist in the survival and growth of primary diffuse large B cell lymphoma cells through interleukin 10 pathway. Exp Cell Res 2017; 350:154–160. doi: 10.1016/j.yexcr.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 44.Ding Y, Shen J, Zhang G, Chen X, Wu J, Chen W. CD40 controls CXCR5-induced recruitment of myeloid-derived suppressor cells to gastric cancer. Oncotarget 2015; 6:38901–38911. doi: 10.18632/oncotarget.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, Takemoto Y, Deng H, Middelhoff M, Friedman RA, Chu TH, et al. Histidine decarboxylase (HDC)-expressing granulocytic myeloid cells induce and recruit Foxp3+ regulatory T cells in murine colon cancer. Oncoimmunology 2017; 6:1–13. doi: 10.1080/2162402X.2017.1290034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu F, Yang J, Liu J, Wang Y, Mu J, Zeng Q, et al. Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer. Signal Transduct Target Ther 2021; 6:1–35. doi: 10.1038/s41392-021-00641-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jensen TJ, Wozniak RJ, Eblin KE, Wnek SM, Gandolfi AJ, Futscher BW. Epigenetic mediated transcriptional activation of WNT5A participates in arsenical-associated malignant transformation. Toxicol Appl Pharmacol 2009; 235:39–46. doi: 10.1016/j.taap.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malgor R, Crouser S, Greco D, Brockett C, Coschigano K, Nakazawa M, et al. Correlation of Wnt5a expression with histopathological grade/stage in urothelial carcinoma of the bladder. Diagn Pathol 2013; 8:1–9. doi: 10.1186/1746-1596-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martínez VG, Rubio C, Martínez-Fernández M, Segovia C, López-Calderón F, Garín MI, et al. BMP4 induces M2 macrophage polarization and favors tumor progression in bladder cancer. Clin Cancer Res 2017; 23:7388–7399. doi: 10.1158/1078-0432.CCR-17-1004. [DOI] [PubMed] [Google Scholar]
- 50.Chang L, Scott MA, Meyers CA, James AW. Pericytes in sarcomas and other mesenchymal tumors. Adv Exp Med Biol 2019; 1147:109–124. doi: 10.1007/978-3-030-16908-4_4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.