Abstract
CRISPR-mediated genome editing of primary human lymphocytes is typically carried out via electroporation, which can be cytotoxic, cumbersome and costly. Here we show that the yields of edited primary human lymphocytes can be increased substantially by delivering a CRISPR ribonucleoprotein mixed with an amphiphilic peptide identified through screening. We evaluated the performance of this simple delivery method by knocking out genes in T cells, B cells and natural killer cells via the delivery of Cas9 or Cas12a ribonucleoproteins or an adenine base editor. We also show that peptide-mediated ribonucleoprotein delivery paired with an adeno-associated-virus-mediated homology-directed repair template can introduce a chimaeric antigen receptor gene at the T-cell receptor α constant locus, and that the engineered cells display antitumour potency in mice. The method is minimally perturbative, does not require dedicated hardware, and is compatible with multiplexed editing via sequential delivery, which minimizes the risk of genotoxicity. The peptide-mediated intracellular delivery of ribonucleoproteins may facilitate the manufacturing of engineered T cells.
Subject terms: Protein delivery, Gene therapy, Cancer immunotherapy
The yields of edited primary human lymphocytes can be increased substantially, with respect to those obtained via electroporation, by delivering a CRISPR ribonucleoprotein alongside an amphiphilic peptide identified via screening.
Main
Efficient, scalable and minimally perturbative manufacturing of autologous and allogeneic engineered T cells remains a central challenge in the development of effective and accessible cell therapies. Engineered immune cells have entered clinical trials to treat cancers, infections, genetic diseases and autoimmune disorders1, but manufacturing barriers in cell therapy pipelines persist, hindering cost-effective development of these products. Precise T-cell engineering offers advantages over pseudo-random integration with lentiviral or gammaretroviral (gRV) vectors2,3, but the prevailing delivery strategies for ex vivo clustered regularly interspaced short palindromic repeats (CRISPR)-mediated genome editing of human lymphocytes rely on electroporation, which is a substantial source of cytotoxicity, cost and manufacturing burden in cell therapy pipelines4. Peptide-enabled delivery to cells is an attractive alternative involving a low-cost reagent that is simply mixed with genome-editing cargo before application to cells in culture. With suitably low toxicity, this approach could facilitate serial rounds of precise genome engineering to manufacture next-generation cell therapies featuring combinations of precise knock-in and knockout edits, but peptide-based delivery has so far shown limited utility in human primary immune cells.
The use of peptides for delivering macromolecular cargo into cells has focused primarily on highly cationic cell-penetrating peptides5 (CPPs) such as the TAT sequence from HIV-1. CPPs, although often effective at facilitating membrane translocation of fused or associated cargo, are limited in use due to toxicity in many therapeutically relevant cell types6. Endosomolytic peptides, such as HA2 from influenza, are typically amphiphilic and undergo a pH-dependent conformational shift7, leading to disruption of the endosomal membrane and escape of associated cargo to the cytosol. On their own, sequences like HA2 are too hydrophobic and insoluble8 to be easily applied and require further engineering to have improved biochemical properties. Chimaeric peptides containing a fusion of an HA2-derived endosomolytic peptide with a cationic CPP such as TAT can retain the endosomolytic functionality and improve macromolecular delivery9. For instance, sequences derived from the chimaeric HA2-TAT fusion sequence (including INF7-TAT and E5-TAT) have been used for non-toxic delivery of short interfering RNAs10. HA2-TAT peptides and their derivatives can associate with other macromolecules through non-covalent associations and—with simple co-incubation and application to cells—facilitate transduction of associated cargo9.
Given the current limitations of non-viral methods for delivering CRISPR reagents, we sought to screen peptides that previously have been described to deliver short interfering RNAs, investigate peptide compatibility with CRISPR ribonucleoproteins (RNPs) and optimize efficacy in engineering primary human immune cells. In this Article, we report a convenient and effective strategy for delivering genome-editing reagents into primary human lymphocytes wherein a CRISPR RNP is simply mixed with a peptide reagent and applied to cells in ex vivo culture (Fig. 1a). This approach—here named peptide-enabled RNP delivery for CRISPR engineering (PERC)—mediates highly efficient, precise, multiplex T-cell engineering with multiple advantages over other delivery technologies. PERC facilitates non-toxic, minimally perturbative genome editing in primary T cells without the need for dedicated hardware, and it may enable point-of-care cell therapy manufacturing in settings where more cumbersome delivery technologies are not feasible.
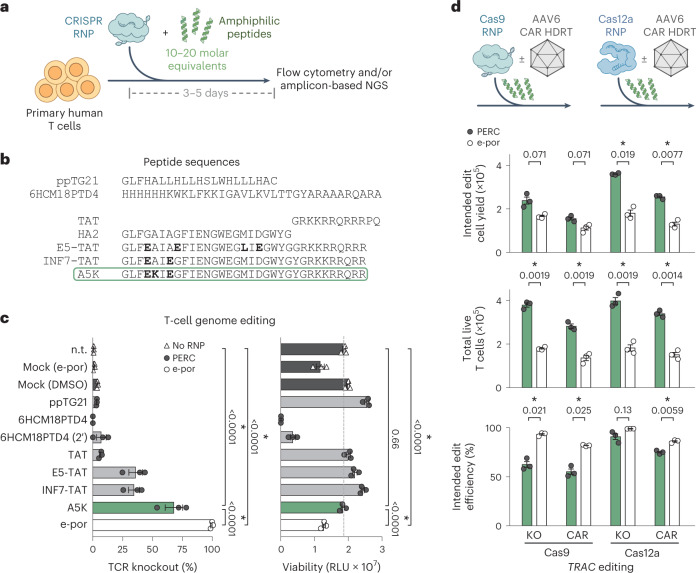
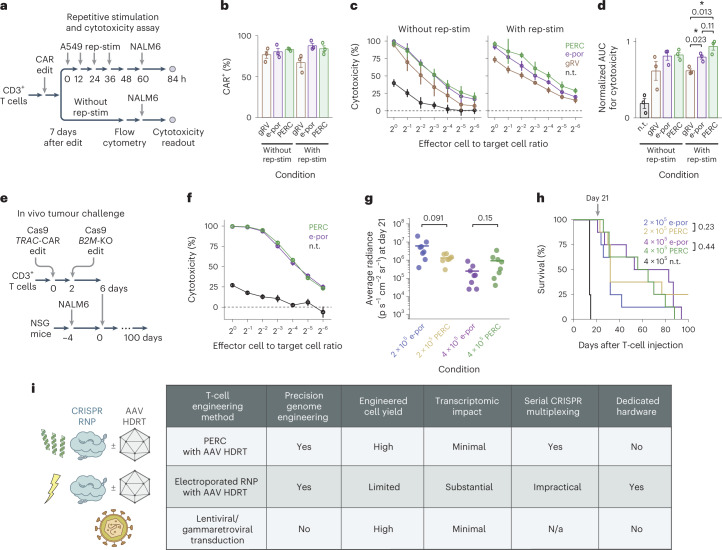
Fig. 1. PERC facilitates genome editing in primary human T cells.
a, Strategy for peptide-enabled RNP delivery. Peptides are mixed with pre-formed RNPs, allowing association, and the mixture is applied to primary human T cells. b, Tested peptide sequences. The influenza HA2 sequence is compared with TAT-fusion derivatives INF7-TAT and E5-TAT, as well as the A5K peptide. Differences are in bold. c, Cas9 TRAC-RNP delivery to CD4+ T cells, facilitated by peptides or electroporation (e-por). Knockout efficiency was assayed 4 days after delivery by flow cytometry for TCR surface expression. n.t. is non-treated, and mock is DMSO only or electroporated (no RNP). Total cell viability was assessed at day 2 and is reported in relative light units (RLU). P values are from an analysis of variance and Holm–Šidák multiple comparisons test. d, Editing of CD3+ T cells using Cas9 or Cas12a TRAC-RNP delivered by PERC or electroporation, paired with or without CAR AAV HDRT. n = 3 biological replicates from distinct human donors. Bars represent the mean. Error bars represent s.e.m. P values are from two-tailed Welch’s unpaired t-tests and Holm–Šidák multiple comparisons test.
Results
Identification of A5K peptide for CRISPR RNP delivery into T cells
We used a nuclear localization signal (NLS)-rich Streptococcus pyogenes Cas9 construct11 to assess a panel of 37 amphiphilic peptides derived from an HA2-TAT fusion scaffold (Fig. 1b and Supplementary Table 1) for the capacity to facilitate delivery of Cas9 RNP for editing the B2M locus in primary human CD4+ T cells (Supplementary Fig. 1). We mixed each peptide with pre-formed RNP in a 20:1 molar ratio (peptide:RNP; determined using initial, empirical screening), applied the mixture to cells in culture and evaluated genome editing at the B2M locus 3 days later using amplicon-based next-generation sequencing (NGS) to detect insertions and deletions (indels) (Supplementary Fig. 1a) and flow cytometry to detect loss of B2M surface expression following biallelic knockout (Supplementary Fig. 1b). This screen identified several promising peptides, with INF7-A5K-TAT (‘A5K’, depicted in green throughout) being the most potent for facilitating genome editing and the most consistent in producing high edited cell yields.
Using a Cas9 RNP targeting the T-cell receptor (TCR) α constant (TRAC) locus (subsequently ‘TRAC-RNP’), we used flow cytometry to further compare the activity of A5K with that of other amphiphilic peptides known to facilitate delivery of macromolecular cargo, for example, the HA2-TAT derivatives E5-TAT9,12 and INF7-TAT13–15 (Fig. 1c and Supplementary Fig. 2). Among the peptides screened, A5K again exhibited the highest activity: 68% knockout as compared with 99% knockout when using an electroporation protocol optimized for RNP delivery in T cells16. We also tested 6HCM18PTD4, a peptide previously reported to facilitate Cas9 RNP delivery to natural killer (NK) cells17, but observed low editing efficiency and substantial toxicity even with abbreviated treatment duration (Fig. 1c and Supplementary Fig. 3). The peptide ppTG21 was previously reported to aid ligand-mediated delivery of Cas9 RNP to hepatocarcinoma cells18, but we observed very low efficiency in T cells. Control conditions containing dimethyl sulfoxide (DMSO; the solvent for peptide stocks) and RNP exhibited nominal genome editing frequencies, which could be due to the slight capacity for NLS-rich Cas9 to promote its own intracellular delivery11. Among the peptides tested, A5K conferred efficient T-cell editing without substantial impact on viability (Fig. 1c and Supplementary Fig. 3), resulting in significantly higher viability compared with electroporation when assayed 2 days after treatment. Particle size analysis of our PERC formulations revealed ~60-nm-diameter particles, larger than that of Cas9 RNP alone (~20 nm diameter observed, resembling the ~15 nm diameter of the crystallized enzyme19) (Supplementary Fig. 4).
We evaluated the compatibility of A5K-driven PERC with different commercially available Cas9 reagents. In addition to our non-commercial NLS-rich Cas9 (ref. 11) and another NLS-rich construct20 used in this study, Cas9 protein obtained from Invitrogen, Integrated DNA Technologies (IDT) and Synthego were all compatible with PERC (Supplementary Fig. 5). Overall, these results establish PERC as a convenient, efficient and minimally toxic method for T-cell genome editing that involves just three readily available reagents (recombinant protein, synthetic RNA and synthetic peptide) and does not require dedicated hardware for preparation or application of the three-component formulation.
PERC represents a versatile delivery method
We next tested PERC in additional human cell types. PERC facilitated genome editing of primary human B cells (Supplementary Figs. 6 and 7) and NK cells (Supplementary Figs. 6 and 8). Following PERC with Cas9 CD45-RNP in B cells, we observed 67% knockout of CD45 surface expression as compared with 92% with electroporation. B-cell viability following PERC was similar to that of non-treated or DMSO-treated cells (Supplementary Fig. 6a). PERC in NK cells was also effective under these conditions, resulting in 17% CD45 knockout as compared with 52% with electroporation. Because PERC was substantially less toxic to NK cells than electroporation, edited cell yields were similar for both delivery methods (Supplementary Fig. 6b).
Base editing is an attractive strategy to knock out genes or correct pathogenic single nucleotide polymorphisms without the need for genomic double-strand breaks or co-delivering a homology-directed repair template (HDRT). We investigated the capability of PERC to deliver an adenine base editor (ABE) RNP to primary human T cells. As was recently reported21, expression of CCR5, the HIV co-receptor that is a clinical target for HIV therapeutics22, can be abolished via an A-to-G edit abrogating the start codon. Using a construct incorporating an evolved TadA-8e deaminase23 and S. pyogenes Cas9 engineered to recognize the protospacer adjacent motif (PAM) 5′-NG-3′24 (Supplementary Fig. 9), we delivered ABE CCR5-RNP to CD4+ T cells using PERC or electroporation and assessed editing frequencies after 9 days by NGS (Supplementary Figs. 10–12). A single dose of ABE RNP with PERC achieved 23% editing as compared with 73% with electroporation. To test whether editing could be improved, we performed two or three sequential PERC treatments and ultimately reached 35% editing efficiency. These findings establish PERC as a straightforward means of base editing with efficiencies that may have clinical relevance, in particular for inborn errors of immunity25,26.
We next tested if PERC could facilitate concomitant delivery of a non-viral DNA HDRT for making a knock-in in primary human T cells (Supplementary Figs. 13 and 14). As previously described16,27, a hybrid single-/double-stranded DNA HDRT containing truncated Cas9 target sequences to promote nuclear localization can be used to introduce a FLAG-tag to the CD5 N-terminus. Five days after PERC or electroporation-mediated delivery of CD5-Cas9 RNP and FLAG-tag HDRT into 200 × 103 T cells, we observed that PERC led to a FLAG-tag signal in 3% of T cells (yielding 2.7 × 103 CD5-FLAG-tag+ T cells), as compared with 39% with electroporation (yielding 22 × 103 FLAG-tag+ cells). We suspect that HDRT DNA might disrupt productive peptide–RNP particle formation and delivery, as the peptide-enabled CD5 knockout efficiency decreased from 76% to 14% in the presence of HDRT DNA. Despite the relatively low efficiency, peptide-enabled delivery of RNP and DNA in tandem facilitates precise knock-in without viral vectors or electroporation.
PERC with AAV HDRT facilitates high yields of precisely edited CAR-T cells
To achieve clinically relevant knock-in efficiency for T-cell engineering, we paired PERC using either Cas9 or Cas12a (a commercially available engineered Acidaminococcus sp. construct28) with adeno-associated virus (AAV)-mediated delivery of a 1928z chimaeric antigen receptor (CAR) HDRT for knock-in targeted to the TRAC locus in T cells (Fig. 1d). Compared with non-targeting retroviral vectors, precise knock-in at the TRAC locus has been demonstrated to improve CAR-T cell anti-tumour potency2,29. At 6 days after treatment, we observed that the frequency of edited cells was modestly higher with electroporation than with PERC (Fig. 1d, bottom), but the toxicity of electroporation dramatically decreased the total number of live cells (Fig. 1d, middle) and the edited cell yield (Fig. 1d, top). We thus observed that PERC using Cas9 or Cas12a resulted in respective 1.4-fold or 2-fold higher CAR-T cell yields as compared with electroporation. Notably, the same PERC protocol developed using Cas9 also enabled Cas12a delivery without requiring enzyme-specific optimization. These results demonstrate that pairing PERC with AAV HDRT is an efficient, straightforward and hardware-independent strategy for making precisely engineered CAR-T cells.
PERC minimally perturbs the T-cell transcriptome
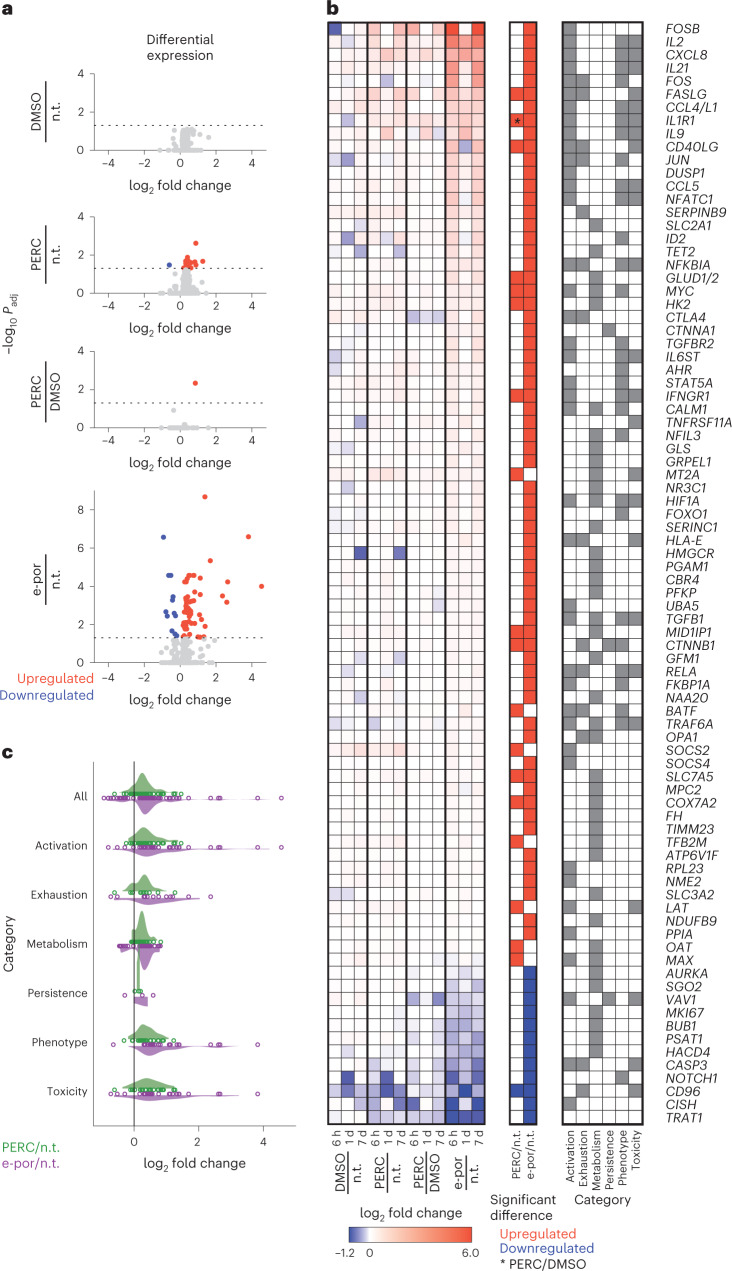
Because T-cell viability was higher after PERC than after electroporation, we sought to characterize the impact of these two methods on the T-cell transcriptome (Fig. 2 and Supplementary Fig. 15). We measured messenger RNA (NanoString nCounter CAR-T characterization panel; 770 genes) from T cells at 6 h, 1 day and 7 days after delivery of a Cas9 RNP targeting the AAVS1 safe harbour locus. The AAVS1 site was targeted to minimize impacts on cellular pathways in edited cells30. Outcomes were analysed for the following pairs of conditions: electroporation of RNP versus no treatment; PERC versus no treatment; PERC versus DMSO (the solvent in the peptide stock) with RNP; and DMSO with RNP versus no treatment. We found that, compared with electroporation of RNP, PERC induced far fewer changes in gene expression. Using a threshold of ≥1.5-fold change in expression as compared with non-treated, 28 genes were significantly affected by electroporation, compared with 7 by PERC. The impact of PERC was similar to that of cells treated with RNP plus the equivalent amount of DMSO used in peptide-mediated editing (0.1% v/v). Therefore, the modest effect of peptide with RNP appears to be primarily due to the DMSO solvent (and/or the presence of principally extracellular RNP) as opposed to peptide itself (Fig. 2a and Supplementary Fig. 15). Additionally, analysis of NanoString gene category annotations indicated that electroporation largely altered the expression of genes with roles spanning activation, exhaustion, cell phenotype and toxicity, whereas these effects were diminished or absent with PERC (Fig. 2b,c and Supplementary Fig. 15c). Because A5K is derived from the influenza peptide haemagglutinin, we tested whether T cells in PBMC culture might recognize this peptide sequence. In an assay for peptide-induced T-cell activation, we did not detect an A5K-specific response. In contrast, a library of H1N1-derived peptides elicited a mild albeit non-statistically significant response (Supplementary Fig. 16). In summary, PERC is minimally perturbative to T cells.
Fig. 2. Compared with electroporation, PERC minimally perturbs T-cell state.
a, Volcano plots depict gene expression fold changes and adjusted P values (Padj) incorporating data from 6 h, 1 day and 7 days after editing. Red denotes significant upregulation, and blue denotes significant downregulation. This plot combines all timepoints, and timepoint-specific outcomes are in Supplementary Fig. 15. n.t., non-treated. b, Set of 84 genes that were significantly differentially expressed in one or more conditions in a. Left: fold changes at each timepoint. Middle: outcomes in each condition; the asterisk indicates the one gene that was significantly affected in PERC/DMSO. Right: NanoString gene category annotations. c, Fold changes across gene categories using the genes and annotations in b. The dots represent individual genes, and the shaded curves represent distributions of fold changes.
PERC with AAV results in high yields of double knock-in CAR-T cells
Ideal manufacturing for T-cell-based immunotherapies should enable editing at multiple genomic sites, but simultaneous multiplex editing using a genome editing nuclease increases the frequency of translocations31,32, which are a cause for concern in therapeutic applications33. This issue has been addressed using multiplex base editing34,35, a strategy with some practical limitations due to the scarcity of commercially available base editor proteins as well as the inability to facilitate knock-ins. Performing sequential delivery of nucleases to T cells decreases the frequency of translocations32, but this practice has not been widely adopted due to the impracticality and cell death associated with repeated electroporation for CRISPR RNP delivery. On the basis of the limited phenotypic impact and high viability associated with PERC, we investigated its capacity for repeat dosing.
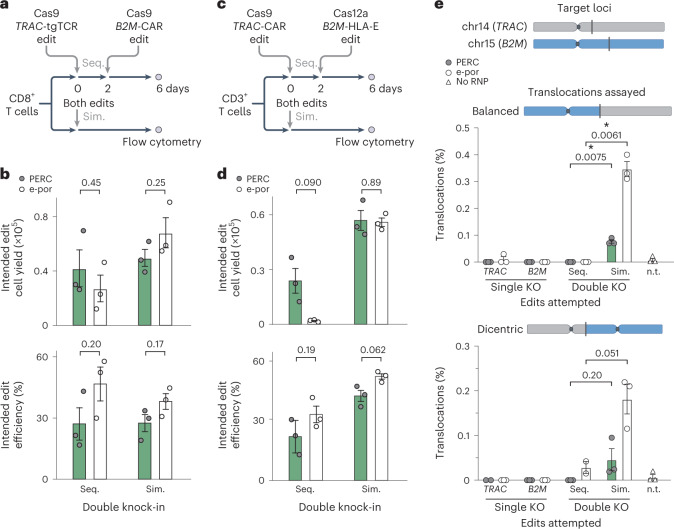
To test whether PERC could facilitate multiplex editing of T cells, we conducted sequential or simultaneous double knock-ins using Cas9 RNPs (Fig. 3a,b and Supplementary Fig. 17) or a combination of Cas9 RNP and Cas12a RNP (Fig. 3c,d and Supplementary Fig. 18). In the first case, the NY-ESO-1 TCR was targeted to TRAC and a 1928z CAR was targeted to B2M as an example of co-expressing two transgenic antigen receptors. The intent of knocking out TRAC and B2M is to ameliorate graft-versus-host and host-versus-graft responses, respectively36. In the second case, the CAR was targeted to TRAC and a human HLA-E antigen was targeted to B2M (as an N-terminal fusion to B2M). The goal of forced HLA-E expression is to prevent the host NK cell ‘missing self’ response37. We observed that PERC generated similar yields of double knock-in T cells when used with Cas9, either sequentially or simultaneously, which were also similar to electroporation-mediated editing (Fig. 3a,b). When pairing Cas9 and Cas12a enzymes (another approach to decrease translocations32), PERC resulted in edited cell yields comparable to electroporation when simultaneous double knock-in was performed (Fig. 3c,d) and substantially higher yields of double knock-in cells than electroporation after sequential rounds of editing using two different nucleases (Fig. 3d).
Fig. 3. Sequential editing with PERC minimizes translocations and maintains cell yield.
a–d, Schematic (a) and data for sequential (Seq.) and simultaneous (Sim.) double knock-in editing (b) in CD8+ T cells using Cas9. Schematic (c) and data for sequential and simultaneous double knock-in editing (d) in CD3+ T cells using Cas9 and Cas12a. Plots depict cells that underwent both knock-ins. Flow cytometry was conducted 6 days after the first edit. e, Analysis of translocation frequencies by ddPCR for treatment with one or two RNPs, sequentially or simultaneously, corresponding to the experiment in a and b. In each experiment, n = 2–3 biological replicates from distinct human donors. Bars represent the mean. Error bars represent s.e.m. P values are from two-tailed Welch’s unpaired t-tests. n.t., non-treated.
We quantified the frequency of balanced or dicentric translocations following multiplex editing of the TRAC and B2M loci using droplet digital PCR (ddPCR). For both types of translocation, simultaneous editing resulted in more translocations than did sequential editing, with no detectable translocations following two sequential applications of PERC (Fig. 3e). Because sequential delivery via electroporation causes substantial decreases in cell counts (Supplementary Figs. 17c and 18c), it is not a practical strategy for minimizing translocations when using a pair of targeted nucleases. In contrast, sequential PERC is less detrimental to cell viability and thus represents a strategy for minimizing translocation frequencies when using multiple targeted nucleases. In a separate experiment, we evaluated RNPs targeting two loci, EMX1 and HEKsite4, for off-target edits using amplicon-based NGS. These sites were used because other guide RNAs were selected for their lack of off-target sites, rendering them impractical for this analysis. Editing frequencies were low at both off-target sites (≤1% at EMX1; ≤4% at HEKsite4), and off-target versus on-target editing ratios were similar between PERC and electroporation (Supplementary Fig. 19). In all, PERC facilitates reliable and high-yield sequential editing that could be adopted to manufacture multiplex-edited cells with less risk of genotoxicity.
PERC in T cells for multiplex editing with improved preservation of naïve phenotype
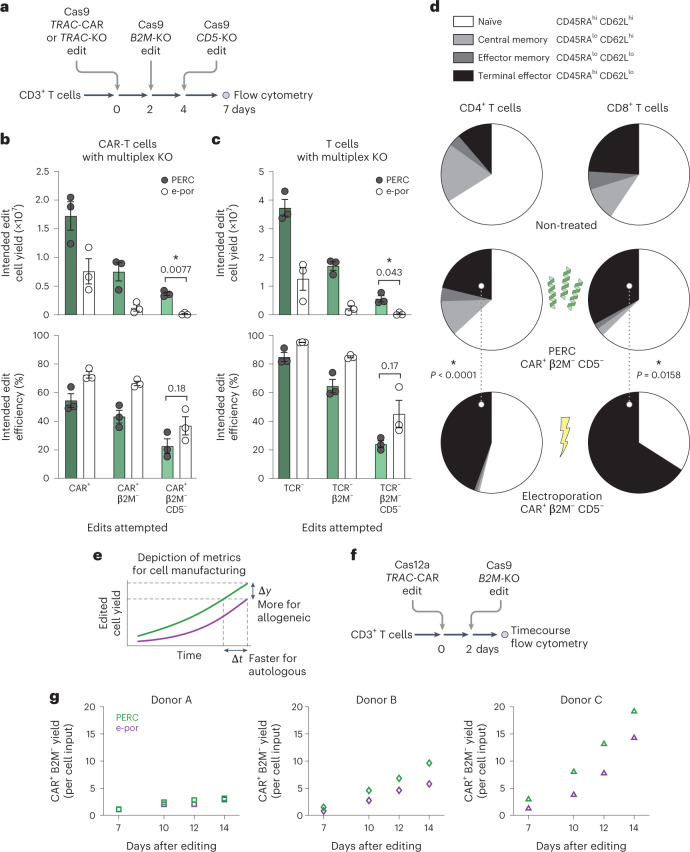
To test whether sequential RNP delivery could facilitate more complex editing, we used PERC to make single, double or triple knockouts of the clinically relevant TRAC, B2M and CD5 (ref. 38) genes in primary human T cells (Fig. 4a–c and Supplementary Figs. 20–22). We performed PERC-mediated TRAC knockout either with (Fig. 4b) or without (Fig. 4c) addition of CAR AAV HDRT and conducted analogous electroporation editing. PERC improved edited cell yields compared with electroporation: 3-fold higher for single knockout (TCR−), 8.5-fold higher for double knockout (TCR− B2M−) and 14-fold higher for triple knockout (TCR− B2M− CD5−) T cells (Fig. 4c). This trend was also apparent with AAV-treated cells (Fig. 4b), again suggesting that serial PERC could support improved manufacturing of complex and translocation-free cell therapy products, which has otherwise been impractical using electroporation.
Fig. 4. PERC supports cell viability and maintenance of phenotype in sequential editing.
a, Schematic of sequential editing of three loci in CD3+ T cells. b, Comparison of editing using PERC versus electroporation as measured by flow cytometry for TCR, CAR, B2M and CD5 surface expression. Reported CAR+ cells are also TCR−. c, Editing without CAR AAV. Cell counts for each condition are scaled to an initial input of 4 × 106 T cells. n = 3 biological replicates from distinct human donors. Bars represent the mean. Error bars represent s.e.m. P values are from two-tailed Welch’s unpaired t-tests. d, Comparison of CD4+ and CD8+ cell phenotypes between delivery methods (independent of editing outcome), as measured by flow cytometry for CD62L and CD45RA surface expression. Pie segments represent the mean proportion of each phenotype. The dotted lines denote comparisons, with P values from a two-way analysis of variance and Holm–Šidák multiple comparisons test (Supplementary Figs. 24 and 25 indicate the comparisons). e, Depiction of metrics for evaluating improvement in cell manufacturing. f, Schematic of sequential editing. g, Expansion of edited cells over time, relative to the number of cells used for editing; n = 3 biological replicates from distinct human donors, plotted separately.
Clinical outcomes with adoptive cell therapies can be affected by not only cellular doses but also cellular phenotypes of engineered T-cell products39. After three rounds of PERC, the proportion of T cells with a terminal effector phenotype (CD62Llo CD45RAhi) was similar to that of non-treated cells. In contrast, the few surviving thrice-electroporated cells exhibited an increase in this phenotype (Fig. 4d and Supplementary Figs. 23–26), which is associated with less robust anti-tumour activity40. Comparison of single versus triple delivery indicated that electroporation led to a decrease in the naïve-like phenotype (CD62Lhi CD45RAhi), whereas PERC maintained a stable proportion, which is important for the therapeutic application of CAR-T cells2,41 (Supplementary Fig. 24).
Considering the importance of edited cell yield through the duration of the manufacturing process to clinical transplantation (Fig. 4e), we tracked sequentially edited cells in extended culture (Fig. 4f). We delivered a Cas12a RNP and AAV for CAR knock-in at the TRAC locus, followed 2 days later by a Cas9 RNP for knocking out B2M. Cells were cultured for 14 days, and CAR+ B2M− cell yields resulting from PERC or electroporation were measured over time (Fig. 4g and Supplementary Fig. 27). Overall, peptide-treated cells survived and expanded better than did electroporated cells, resulting in similar or higher edited cell yields for the three donors tested.
PERC produces potent CAR-T cells
Having established the improved cell yields and reduced phenotypic impact associated with PERC (as compared with electroporation), we next tested the cytotoxicity of PERC CAR-T cells. We engineered bulk T cells to express a 1928z CAR using either gRV transduction, electroporated Cas12a TRAC-RNP plus AAV HDRT, or PERC with Cas12a TRAC-RNP plus AAV HDRT (Fig. 5a and Supplementary Fig. 28). The CAR+ T-cell percentages were similar across the three engineering strategies (Fig. 5b). We compared conditions with or without repetitive stimulation via co-culture with CD19+ A549 target cells. Repetitive stimulation diminished the naïve phenotype and increased the terminal effector phenotype for cells generated using each strategy (Supplementary Fig. 28b). Cells from each condition were then co-cultured for 1 day with luciferase-expressing NALM6 acute lymphoblastic leukaemia cells, and cytotoxicity was quantified. After repetitive stimulation, TRAC-CAR-T cells generated by either method outperformed the gRV condition (Fig. 5c,d), demonstrating that PERC CAR-T cells recapitulate the advantage of targeting a CAR to the TRAC locus2.
Fig. 5. Functional evaluation of CAR-T cells generated using PERC.
a, Schematic of the repetitive stimulation (rep-stim) and cytotoxicity assay. CD3+ T cells were edited to express a CAR using either gRV, Cas12a RNP electroporation and AAV, or Cas12a RNP PERC and AAV. b, Percentages of CAR+ cells in each condition with or without repetitive stimulation using CD19+ A549 cells. c,d, NALM6 cytotoxicity assay for T cells in each condition (c), and area under the curve analysis (d). n = 3 biological replicates from distinct human donors. Bars represent the mean. Error bars represent s.e.m. (3 biological replicates × 3 technical replicates). P values are from two-tailed Welch’s unpaired t-tests. e, Schematic of the in vivo tumour challenge experiment, using n = 8 mice per group. f, NALM6 cytotoxicity assay. Error bars represent s.e.m. from three technical replicates. g, BLI values on the last measurement day on which all CAR-T-cell-injected mice were alive. P values are from two-tailed Welch’s unpaired t-tests. h, Kaplan–Meier survival analysis. P values are from a log-rank test. P < 0.001 for each comparison of an edited condition versus non-treated (n.t.) cells. i, Table summarizing different delivery methods evaluated.
To evaluate the in vivo anti-tumour potency of multiplex edited cells, we conducted sequential TRAC-CAR B2M-KO editing using either PERC or electroporation (Fig. 5e and Supplementary Fig. 29). To generate a sufficient number of cells, the PERC protocol used in earlier experiments was scaled up from 100 μl culture to 9 ml with similar editing efficiency, viability and edited cell yield. In an in vitro cytotoxicity assay, T cells performed similarly between the two delivery methods (Fig. 5f). We performed an in vivo ‘stress test’, in which the CAR-T cell dose is lowered to reveal the functional limits of different CAR-T cell populations. A total of 4 × 105 or 2 × 105 CAR+ T cells (irrespective of the efficiency of B2M knockout, which is not expected to impact potency in this model system) generated using each method were injected intravenously into NSG mice engrafted with 5 × 105 NALM6 cells 4 days prior. Tumour burden was evaluated over time by bioluminescence imaging (BLI) (Supplementary Fig. 30). An assessment on day 21 (the last timepoint when all CAR-T-cell-injected mice were alive) indicated a similar tumour burden for each method within each CAR-T cell dose (Fig. 5g). PERC and electroporation also conferred similar survival within each dose over the course of the experiment (Fig. 5h). Taken together, these results demonstrate that PERC CAR-T cells have similar therapeutic potency in this preclinical model to their electroporation counterpart.
Discussion
Our findings establish PERC as an appealing alternative to electroporation for inexpensive, facile and efficient production of sophisticated cell therapy products generated from primary human T cells. PERC works well for Cas9 and Cas12a nucleases as well as an ABE, suggesting broad compatibility with CRISPR effectors. PERC opens up new possibilities for ex vivo manufacturing of both autologous and allogeneic (‘off the shelf’) products featuring sequential combinations of knockouts, knock-ins and/or therapeutic base editing of clinically relevant genes. As compared with electroporation, PERC offers several advantages: minimal perturbation of cell transcriptome and phenotype, compatibility with sequential delivery to avoid the genotoxicity of simultaneous edits, and independence from dedicated hardware. PERC is as easy to implement as viral transduction while enabling precision genome editing of multiple loci (Fig. 5i). Additionally, PERC produces edited cell yields surpassing those attained using electroporation. Edited cell yield is a more important metric compared with editing efficiency when generating a cell therapy product; even so, with careful guide RNA selection, it is indeed possible to attain editing efficiencies via PERC that approach those attained via electroporation of CRISPR enzymes.
PERC minimizes cell handling steps (as compared with electroporation) and could be incorporated into fully automated cell processing pipelines. Although middle-income regions may have access to therapeutic cell manufacturing hardware compatible with viral transduction, there is a critical lack of infrastructure such as clean rooms mandated by electroporation-based editing protocols42. PERC minimizes cell handling steps, is compatible with broadly available automated cell manufacturing hardware and could be incorporated into point-of-care manufacturing, thus enabling clinical applications that otherwise would be impractical.
Beyond accelerating broader adoption of CRISPR-mediated engineering for next-generation immunotherapy applications, PERC could be applied with base editing enzymes to treat inborn errors of immunity by correcting pathogenic mutations43. Because PERC requires only three chemically defined reagents that are straightforward to obtain (plus AAV for efficient knock-in), it can be a practical tool for laboratory research, industrial manufacturing and clinical pipelines. The adoption of PERC could reduce costs, minimize genotoxicity and expand access to precisely engineered therapeutic lymphocytes (including in a point-of-care production context), and with further development could facilitate in vivo genome editing.
Methods
Cell culture
Primary adult blood cells from anonymous healthy human donors were purchased as leukapheresis packs from STEMCELL Technologies. Specific lymphocytes were isolated by magnetic negative selection using EasySep isolation kits for human CD4+ T cells, CD8+ T cells, CD3+ T cells, B cells or NK cells (STEMCELL) per the manufacturer-provided instructions. T cells were stored in liquid nitrogen until use. B and NK cells were used fresh.
Isolated T cells were thawed (on day −3 relative to delivery) and cultured overnight in X-VIVO 15 medium (Lonza) supplemented with 5% foetal bovine serum (FBS), 50 μM 2-mercaptoethanol and 10 mM N-acetyl-l-cysteine. Cells were activated (on day −2 relative to delivery) and cultured at 1 × 106 cells ml−1 for 2 days in supplemented X-VIVO 15 medium with anti-human CD3/CD28 magnetic Dynabeads (Gibco 40203D) at a bead-to-cell ratio of 1:1, 200 U ml−1 IL-2 (Proleukin), 5 ng ml−1 IL-7 (R&D Systems) and 5 ng ml−1 IL-15 (R&D Systems). After 2 days of activation, Dynabeads were removed from the cell culture using an EasySep cell separation magnet (STEMCELL). For genome editing, 200 × 103 T cells per well were suspended in 100 μl Opti-MEM (Gibco) (treatments detailed below). After 1 h of treatment, FBS-supplemented X-VIVO-15 recovery medium (or in experiments involving AAV treatment, medium without FBS) was added to cells. Unless specified otherwise, after genome editing (as described below), cells were cultured in supplemented X-VIVO 15 at 0.5 × 106 cells ml−1 with 300 U ml−1 IL-2 (Proleukin) and split every 2–3 days.
Isolated B cells were activated and cultured at 1 × 106 cells ml−1 for 2 days in Iscove’s modified Dulbecco’s medium (Thermo Fisher) supplemented with 10% FBS, 50 µM 2-mercaptoethanol, 100 ng ml−1 MEGACD40L (Enzo), 200 ng ml−1 anti-human RP105 (BioLegend), 500 U ml−1 IL-2 (Proleukin), 50 ng mL−1 IL-10 (Thermo Fisher) and 10 ng ml−1 IL-15 (R&D Systems). For genome editing, 200 × 103 B cells per well were suspended in 100 μl consisting of a mixture of Opti-MEM (Gibco) and up to 20% peptide:RNP formulation (treatments detailed below). After 1 h of treatment, recovery medium was added to return cells to the original culture conditions. After treatment, B cells were cultured at 0.5 × 106 cells ml−1 and split every 2–3 days.
Isolated NK cells were cultured at 1 × 106 cells ml−1 for 6 days in X-VIVO 15 medium (Lonza) supplemented with 5% FBS, 50 μM 2-mercaptoethanol and 10 mM N-acetyl-l-cysteine and 1,000 U ml−1 IL-2 (Proleukin), and activated with MACSiBead particles pre-loaded with anti-human CD335 (NKp46) and anti-human CD2 antibodies (Miltenyi Biotec). After 6 days of activation, MACSiBead particles were removed from the cell culture using an EasySep cell separation magnet (STEMCELL). For genome editing, 200 × 103 NK cells per well were suspended in 100 μl consisting of a mixture of Opti-MEM (Gibco) and up to 20% peptide:RNP formulation (treatments detailed below). After 1 h of treatment, FBS-supplemented X-VIVO-15 recovery medium was added to cells. After genome editing, NK cells were cultured at 0.5 × 106 cells ml−1 with 1,000 U ml−1 IL-2 and split at 5 days.
Cell viability assays
Cell viability was assessed using a CellTiter-Glo assay (Promega G7570) according to the manufacturer-provided instructions. Luminescence was measured using a Spark plate reader.
Peptides
TAT peptide was purchased from GenScript (GSCRPT-RP20256), and other peptides were procured via custom solid phase synthesis (CPC Scientific; 95% purity). All peptides were stored lyophilized or as 10 mM stocks in DMSO at −20 °C in a desiccator. Peptide sequences are indicated in Fig. 1 and Supplementary Table 1.
Cas9 proteins
The S. pyogenes Cas9 protein used in all experiments (except those shown in Supplementary Fig. 5) has six SV40 nuclear localization sequences (Cas9-6 × NLS); four are N-terminal, and two are C-terminal11 (Addgene ID# 88917; referred to as ‘4 × NLS-Cas9-2 × NLS’ in Staahl et al., 2017). A cysteine-free Cas9-6 × NLS construct (Addgene ID# 194246; C80S / C574S) was also used without distinction, since the two constructs are functionally equivalent. Cas9-6 × NLS was expressed in Escherichia coli and purified via nickel affinity chromatography (using a detergent wash to remove lipopolysaccharides), heparin affinity chromatography and size exclusion chromatography as previously described18. Purified proteins were concentrated to ~50 µM in a buffer of 25 mM sodium phosphate pH 7.25, 300 mM NaCl and 200 mM trehalose and stored at −80 °C. Protein expression and purification were performed by UC Berkeley MacroLab. Cas9-triNLS (Addgene ID# 196245; referred to as 3 × NLS-Cas9 when originally reported20) was expressed and purified by the R.C.W. lab using the same protocol. Alt-R A.s. Cas12a Ultra (10001273), Alt-R S.p. Cas9 Nuclease V3 (1081058) and Alt-R S.p. HiFi Cas9 Nuclease V3 (1081061) were purchased from IDT. SpCas9 2NLS Nuclease was purchased from Synthego. Invitrogen TrueCut Cas9 Protein v2 (A36498) was purchased from Thermo Fisher.
Cas9 sgRNAs
S. pyogenes Cas9 single guide RNAs (sgRNAs) were purchased with manufacturer-recommended standard chemical modifications from Synthego and resuspended in water, or from IDT(Alt-R) and resuspended in IDT duplex buffer or diethyl pyrocarbonate (DEPC)-treated water. Before use, sgRNAs were diluted to 15 µM in 20 mM HEPES pH 7.5 and 150 mM NaCl, then refolded by warming to 95 °C for 5 min and slow cooling to room temperature for 25 min. Spacer sequences are listed in Supplementary Table 2.
RNP formation
For peptide-mediated delivery experiments, Cas9 protein was diluted to 10 µM in ‘RNP buffer’ (20 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol and 2 mM MgCl2). The sgRNA was diluted to 15 µM in 20 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol and 2 mM MgCl2. The molar ratio of Cas9:sgRNA was 1:1.5 unless otherwise specified. Cas9 was mixed with sgRNA in equal volumes yielding 5 µM RNP complex, with the RNP concentration defined by the amount of Cas9 protein. For non-viral knock-in experiments, Cas9 was diluted to 20 µM in RNP buffer and mixed with sgRNA at 30 µM in equal volumes, yielding a final concentration of 10 µM RNP. For electroporation, Cas9 was diluted in RNP buffer to 40 µM, and sgRNA was diluted to 60 µM in IDT duplex buffer (30 mM HEPES pH 7.5 and 100 mM potassium acetate) or DEPC-treated water. Cas9 was mixed with sgRNA in equal volumes, yielding 20 µM RNP complex.
Dynamic light scattering
Representative samples of Cas9 RNP and RNP:peptide formulation were assessed using a Zetasizer Nano ZS (Malvern Panalytical) instrument with a quartz low-volume cuvette (ZEN2112). Approximating a 3-fold scale-up of the typical conditions used in Cas9-based editing experiments, 27 μl of B2M-Cas9 RNP was prepared at 5 μM (135 pmol) in RNP buffer, and particle size was measured at room temperature. The 27 μl RNP sample was then supplemented with 3 μl A5K peptide at 1 mM in a 10% DMSO stock, a mixing step analogous to that used in editing experiments (following RNP formation and preceding addition of RNP:peptide to cells). The resulting 30 μl RNP:peptide formulation contained 4.5 μM Cas9 RNP (135 pmol) and 1 mM A5K peptide (3 nmol). This formulation was incubated at room temperature for 1 min, and particle size was determined using the same settings as used for the RNP-only solution. Data were analysed ‘by number’ using Zetasizer analysis software.
Peptide and RNP delivery formulations (T, B and NK cells)
Peptides (10 mM in 100% DMSO) were diluted in DEPC-treated water to 1 mM (resulting in a solution of 90% water and 10% DMSO) and added to RNP, resulting in a volume no greater than 20% of the eventual final volume (for example, ≤20 μl formulation for a well with 100 μl cells in Opti-MEM). The RNP/peptide mixture was added to a 96-well round-bottom plate, and 200 × 103 cells in 100 µl Opti-MEM (Gibco) per well was added to the RNP/peptide mixture. The final dose of RNP during cell treatment was 50 pmol per well with a final peptide concentration of 10 µM, unless stated otherwise. In all cases of peptide-enabled delivery, the final concentration of DMSO was proportional to the peptide concentration: 0.1% DMSO per 10 μM peptide; this concentration of DMSO was used for the ‘mock’ negative control conditions. For experiments not involving AAV, after a 1 h incubation at 37 °C, 100 μl of treated volume was split in half into two plates, then 150 µl culture medium was added per well, thus diluting but not removing the treatment. For experiments involving AAV, after editing, 200 × 103 cells in 100 µl Opti-MEM were centrifuged, supernatant was discarded, cells were recovered with 100 µl serum-free culture medium and AAV was added subsequently; after an overnight incubation, the medium was exchanged for fresh serum-containing medium, and cells were split starting 2 days after editing. The concentration of additives and stimulation cocktail in the recovery medium was such that the final concentrations matched the description above for each cell type.
RNP electroporation (T, B and NK cells)
In a 4D nucleofector (Lonza), 50 pmol of Cas9, Cas12a or ABE RNP was electroporated into 200 × 103 T, B or NK cells resuspended in 20 μl of P3 buffer and supplement (Lonza V4XP-3032) using the EH-115 pulse code. Cells were incubated for 10 min at 37 °C, then rescued with 80 µl of pre-warmed culture medium before diluting for further cell culture as above. For AAV knock-in experiments, 60 pmol of RNP was electroporated into 0.5 × 106 or 1 × 106 T cells per well.
Recombinant AAV6 vectors were used to deliver HDRT for making knock-ins. (1) The SpCas9-compatible TRAC-targeted CAR AAV contains homology arms for targeting near the beginning of TRAC exon 1. The cargo is P2A, 1928z CAR, P2A. In this editing strategy, after knock-in, the CAR is transcribed cocistronically with the mRNA encoding endogenous TCRα but translated as a separate protein, and the TCR is not surface expressed2. The CAR contains a CD8A signal sequence, SJ25C1 scFv, CD28 hinge, CD28 transmembrane domain, CD28 costimulatory domain and CD3ζ signalling domain. (2) The SpCas9-compatible TRAC-targeted NY-ESO-1 TCR AAV contains homology arms for targeting near the beginning of TRAC exon 1. The cargo is P2A, TCRβ chain, P2A and a portion of the TCRα chain; the C-terminal portion of NY-ESO-1 TCRα is encoded by endogenous TRAC exons. (3) The SpCas9-compatible B2M-targeted CAR AAV contains homology arms for targeting near the beginning of B2M exon 1. The cargo is 1928z CAR, stop codon and bGH polyA tail. (4) The Cas12a-compatible TRAC-targeted CAR AAV contains homology arms for targeting TRAC exon 1. This site is distinct from where the SpCas9-compatible CAR is targeted, but the cargo is the same. (5) The Cas12a-compatible B2M-targeted peptide-B2M-HLA-E AAV contains homology arms for targeting B2M exon 2. The cargo is P2A, signal sequence, HLA-G peptide (which is presented on HLA-E), linker, B2M rescue, linker, HLA-E, stop codon and bGH polyA tail. After knock-in, the B2M that is expressed is the fusion with HLA-E37.
Peptide immunogenicity assay
Primary human PBMCs were isolated from leukapheresis packs from STEMCELL Technologies and stored in liquid nitrogen until use. Cells were thawed and cultured in RPMI-1640 supplemented with non-essential amino acids, l-glutamine and 5% human AB serum at a concentration of 1 × 106 cells in 100 μl medium per well in 96-well TC plates. A5K peptide, influenza A (H1N1) HA protein PepTivator pool (Miltenyi Biotec) or SARS-CoV-2 spike protein PepTivator pool (Miltenyi Biotec) were each diluted in DMSO and added to PBMC culture at a final concentration of 1 μg peptide ml−1 or as otherwise indicated; phorbol 12-myristate 13-acetate (PMA, STEMCELL) at final concentration of 50 ng ml−1 plus ionomycin (STEMCELL) at final concentration of 1 μM were added to PBMCs as a positive control. After 2 h of incubation, brefeldin A (BD Biosciences) was added at 1 μg ml−1. After 8 h of incubation, cells were stained for T-cell surface markers (CD3, CD4 and CD8), fixed and permeabilized using a Cytofix/Cytoperm kit (BD Biosciences) according to manufacturer-provided protocol, stained for intracellular TNF and IFN-γ, and analysed by flow cytometry.
TCR knockout, AAV-mediated CAR knock-in and sequential B2M and CD5 knockouts
CD3+ T cells were prepared as described above, with activation on day −2 relative to delivery. On day 0, RNP formed with TRAC sgRNA (Cas9:sgRNA ratio of 1:1.2) was delivered using A5K peptide or electroporation. For electroporation, 60 pmol RNP was delivered for 1 × 106 cells per well, and cells were rescued in serum-free X-VIVO 15 medium with 300 U ml−1 IL-2 (Proleukin). For A5K peptide-mediated delivery, 50 pmol RNP was delivered using 1,500 pmol A5K for 2 × 105 cells per 100 μl of medium (15 μM final peptide concentration). After 1 h incubation at 37 °C, cells were pelleted by centrifugation (300g for 5 min) then resuspended in serum-free X-VIVO 15 medium with 300 U ml−1 IL-2 to 2 × 106 cells ml−1. Cells were transduced with AAV at a multiplicity of infection of 5 × 104. After overnight incubation, the serum-free AAV-containing medium was replaced with fresh X-VIVO 15 medium with 300 U ml−1 IL-2, and cells were resuspended to 1 × 106 cells ml−1. In parallel with the CAR knock-in condition, a similar condition was conducted with TRAC-targeting RNP without virus.
On day 2, cells were counted with trypan blue stain to estimate the number of live cells. A subset of TRAC knockout or CAR-knock-in treated cells were separated and pelleted by centrifugation (300g for 5 min), and cell density was normalized to 1 × 106 cells ml−1 in fresh medium. The remaining cells were pelleted and treated using the same protocols for electroporation and A5K peptide-mediated delivery as described above with RNP targeting B2M. After treatment, cells were rescued with X-VIVO 15 medium supplemented with 300 U ml−1 IL-2, and cell density was adjusted to 1 × 106 cells ml−1. On day 4, cells were counted with trypan blue stain, and cell density of each treatment condition was adjusted to 1 × 106 cells ml−1 in fresh medium. Another subset of double knockout/CAR-knock-in treated cells was separated for a third round of sequential editing as above with RNP targeting CD5. Cells were rescued with supplemented X-VIVO 15 medium containing 300 U ml−1 IL-2 and cell density adjusted to 1 × 106 cells ml−1. On day 6, each editing condition was counted with trypan blue stain, and cell density of each treatment was adjusted to 1 × 106 cells ml−1 in fresh medium. Because each treatment’s branch began with different starting cell numbers, the total live cell count for each condition (for example, for 1×, 2× or 3× editing) was normalized to a theoretical starting condition of 4 × 106 cells, and adjusted cell counts were extrapolated on the basis of the proliferation measured every 2 days. On day 7, flow cytometry was performed to assay editing efficiencies and cell numbers of TCR, B2M and CD5 knockout and CAR knock-in. On the basis of these values, a normalized cell yield for day 6 was calculated for A5K peptide-mediated delivery (or electroporation) for each condition. Separately, T-cell phenotypes were examined by staining for surface expression of CD45RA and CD62L.
CD8+ T-cell sequential editing experiment in Fig. 3
CD8+ T cells were activated from n = 3 donors and underwent editing on day 0 and/or day 2 after Dynabead removal. TRAC editing involved Cas9 RNP and NY-ESO-1 TCR AAV, and B2M editing involved Cas9 RNP and 1928z CAR AAV. Cells were passaged every 2 days. Flow cytometry was conducted 6 days after the first edit. Cells were pelleted and resuspended in QuickExtract DNA extraction solution (Biosearch Technologies #SS000035-D2), incubated at 65 °C for 10 min and then 96 °C for 5 min, and stored at −20 °C until genomic DNA extraction.
CD3+ T-cell sequential editing experiment in Fig. 3
CD3+ T cells were activated from n = 3 donors and underwent editing on day 0 and/or day 2 after Dynabead removal. TRAC editing involved Cas9 RNP and 1928z CAR AAV, and B2M editing involved Cas12a RNP and HLA-E AAV. Cells were passaged every 2 days. Flow cytometry was conducted 6 days after the first edit.
Base editing
ABE8e-SpCas9-NG base editor protein (Supplementary Fig. 14) was expressed and purified by Aldevron and stored in a buffer of 20 mM HEPES-KOH pH 7.5, 150 mM NaCl and 10% (v/v) glycerol at −80 °C. The construct comprises (from N-terminus to C-terminus) a bipartite SV40 NLS, a catalytically dead ‘8e’ TadA deaminase domain23, another ‘8e’ TadA that is catalytically active, a D10A nickase version of the ‘NG’ PAM Cas9 (ref. 24), a bipartite SV40 NLS and a nucleoplasmin NLS. ABE8e-NG Cas9 RNP was formed with ‘CCR5off-1’ sgRNA (Supplementary Table 2) following the RNP formation protocol above. Then 200 pmol RNP was applied to primary human T cells (protocol described above) with 20 µM A5K peptide (final concentration in 100 μL Opti-MEM). Cells were treated with either a single dose, followed up with a second dose 4 days later or followed up again with a third dose another 3 days later. Genomic DNA was extracted and flow cytometry analysis was done 9 days after the first dose (2 days after the third dose). Editing efficiency was analysed by deep sequencing as described below. Flow cytometry analysis was used for measuring knockout of CCR5 surface expression and edited cell yields.
Non-viral FLAG-tag knock-in
An HDRT was designed to append an N-terminal fusion of FLAG-tag to the extracellular portion of CD5 downstream of the signal peptide. The template was synthesized as a single-stranded 160-mer DNA oligonucleotide with a 40 nt left homology arm and a 40 nt right homology arm, as well as with truncated Cas9 binding sites as previously described16,27. Per treatment well, 200 pmol HDRT oligo (1:1 primer-to-template ratio) was mixed with 200 pmol RNP, to which 3,000 pmol A5K was added (for 30 µM final concentration of A5K in 100 μl Opti-MEM). At 5 days after editing, cells were stained for CD5 and FLAG-tag and analysed on an Attune NXT flow cytometer.
Flow cytometry
Flow cytometry was performed on an Attune NxT flow cytometer with a 96-well autosampler (Thermo Fisher Scientific) or on an LSRFortessa X-50 flow cytometer (BD Biosciences). Cells were resuspended in FACS buffer (phosphate-buffered saline (PBS), 2% FBS and 1 mM EDTA) and stained with live–dead stain and surface marker-targeting antibodies (Supplementary Table 3) according to manufacturer-provided instructions. Sampling was at defined volumes (60 µl per well) to quantify cell counts. Cytometry data were processed and analysed using FlowJo software (BD Biosciences).
Amplicon-based deep sequencing
Genome editing was quantified by NGS of PCR amplicons. Cells were pelleted by centrifugation at 300g for 5 min, washed once with PBS, resuspended in 50 µl of QuickExtract solution (Lucigen) per well and incubated at 65 °C for 10 min, then 95 °C for 5 min. Genomic DNA was stored at −20 °C. PCR amplification was performed using PrimeSTAR GXL DNA polymerase (Takara Bio) or Kapa HiFi (Roche) according to the manufacturer-provided protocol. Amplicons were purified using SPRI beads (UC Berkeley sequencing core), and concentrations were quantified via NanoDrop. Amplicon libraries were pooled and sequenced on an Illumina MiSeq at 300 bp paired-end reads to a depth of at least 10,000 reads per sample. Editing outcomes were analysed using Cortado (https://github.com/staciawyman/cortado). Briefly, reads were adapter trimmed and then merged into single reads. These joined reads were aligned to the target reference sequence to identify editing events at the cut site. Indel frequencies were calculated by counting any reads with an insertion or deletion overlapping the cut site or occurring within a 3 bp window on either side of the cut site. Single nucleotide polymorphisms occurring within the window around the cut site were not counted towards this value. The total number of indel reads was divided by the total number of aligned reads to arrive at % indels. For base editing experiments, the joined reads were aligned to the target reference sequence to identify base editing events at the targeted locus. The frequency of base editing was calculated as the number of reads with an edit divided by the total aligned reads, calculated individually for each site.
Translocation analysis
Cells were pelleted by centrifugation at 300g for 5 min, resuspended in 40 µl of QuickExtract solution (Lucigen) per well and incubated at 65 °C for 10 min, then 95 °C for 5 min. Genomic DNA was stored at −20 °C. Primers (Supplementary Table 4) and 5′ 6-FAM-labelled probes were synthesized by IDT (ZEN-3′ Iowa Black FQ quencher). ddPCR reactions were prepared using ddPCR Supermix for probes (no dUTP) (Bio-Rad #1863023), and droplets were generated and read using a QX200 droplet generator and reader. Data were analysed using the Bio-Rad QuantaSoft Analysis Pro software. Experimental samples were compared to control samples, scaled accordingly if amounts of template DNA differed.
Extended culture experiment
CD3+ T cells were edited with Cas12a TRAC-RNP and CAR AAV on day 0 and with Cas9 B2M-RNP on day 2 using either electroporation or A5K delivery. Edited cells and non-treated cells were maintained in culture for 2 weeks and passaged every 2 days.
gRV transduction
The gRV vector contains a 1928z CAR and a truncated LNGFR separated by a P2A tag. Virus was concentrated using Retro-X concentrator (Takara Bio). Tissue culture plates were pre-treated with RetroNectin (Takara Bio; 15 µg ml−1 in PBS, overnight at 4 °C). CD3+ T cells were mixed with polybrene (10 µg ml−1) and virus, plated, centrifuged (2,000g, 1 h, 30 °C), incubated overnight in a TC incubator and resuspended in fresh T-cell medium. Subsequently, cells underwent a second round of gRV transduction.
Target cells
mKate2-NLS+ CD19+ A549 cells were cultured in Dulbecco’s modified Eagle medium (Gibco) supplemented with FBS (Gibco, 10%), sodium pyruvate (Gibco, 1%), HEPES buffer (Corning, 1%) and penicillin–streptomycin (Gibco, 1%). Firefly luciferase+ CD19+ NALM6 cells were cultured in RPMI (Gibco) supplemented with FBS (10%), sodium pyruvate (1%), HEPES buffer (1%), penicillin–streptomycin (1%), non-essential amino acids (Gibco, 1%) and 2-mercaptoethanol (0.1%).
Repetitive stimulation
After 2 days of activation, CD3+ T cells from three donors were edited with Cas12a TRAC-RNP using electroporation or A5K peptide and transduced with CAR AAV. Other T cells were transduced with gRV or not edited. Flow cytometry was conducted 6 days after editing to determine CAR+ percentages and the numbers of T cells to co-culture with A549 cells (~1:1 effector cell to target cell ratio). At 12 h intervals, T cells were removed from the previous co-culture and seeded onto new pre-plated target cells, for four rounds in total. Other T cells for each editing condition were cultured without target cells. Afterwards, flow cytometry was conducted to determine CAR+, CD4+, and CD8+ percentages and CD62L/CD45RA phenotypes, and T cells were evaluated in a cytotoxicity assay with NALM6 target cells.
Cytotoxicity assays
The assay for the repetitive stimulation experiment used T cells that were edited by three different CAR delivery methods with or without having undergone repetitive stimulation, or that were non-treated. The assay corresponding to the in vivo experiment used T cells that were sequentially edited for TRAC-CAR knock-in and B2M knockout using either A5K or electroporation, or that were non-treated.
Seven 2-fold serial dilutions of T cells were plated in a 96-well flat clear-bottom plate (Thermo Fisher Scientific #165306), with 5 × 104 CAR+ (or non-treated) T cells in the first set of wells. A total of 5 × 104 NALM6 cells were added to each well. The medium was X-VIVO 15, human serum (5%), penicillin–streptomycin (0.5%), IL-7 (5 ng ml−1) and IL-15 (5 ng ml−1), and the total volume per well was 100 μl. The control for maximum signal was NALM6 cells, and the control for minimum signal was NALM6 cells and Tween-20 (0.2%). Co-cultures were plated in triplicate and incubated for approximately 24 h. Then, 100 μl d-luciferin (GoldBio, 0.75 mg ml−1) was added to each well, and luminescent signal was measured using a GloMAX Explorer microplate reader (Promega). Cytotoxicity was calculated as: 100% × (1 − (sample − minimum)/(maximum − minimum)). Error was propagated in calculating the standard error of the mean (s.e.m.).
In vivo tumour challenge experiment
After 2 days of Dynabead activation, CD3+ T cells were edited with Cas9 TRAC-RNP using electroporation or A5K peptide and transduced with CAR AAV. Two days later, Cas9 B2M-RNP editing was performed using electroporation or A5K peptide. Flow cytometry was conducted 6 days after the first edit.
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were handled ethically and in accordance with the protocol AN194345-01E approved by the University of California, San Francisco (UCSF) Institutional Animal Care and Use Committee. Before and during the experiment, mice were maintained on Clavamox antibiotic. A total of 5 × 105 NALM6 cells were injected into the tail vein of mice that were between 8 and 12 weeks old. After the first BLI measurement (after NALM6 injection and before T-cell injection), mice were assigned to each T-cell condition so as to maintain a similar average mass and tumour burden across conditions. Four days after NALM6 injection, 2 × 105 or 4 × 105 CAR+ T cells that had undergone sequential editing (cell numbers were based on CAR knock-in, independent of B2M knockout percentage) or 4 × 105 non-treated T cells were injected into the tail vein. Mouse health and survival were monitored over time. BLI was performed one or two times per week using a Xenogen in vivo imaging system. At each imaging session, mice were injected intraperitoneally with luciferin (3 mg luciferin per 0.2 ml DPBS) and anaesthetized with isoflurane (Medline Industries). The default imaging exposure was 1 min, and shorter exposures were used for images that had a saturating signal at 1 min. Luminescence was quantified using Living Image software (PerkinElmer). Reported BLI values are an average from imaging each mouse on its front and on its back. Mice were euthanized per the approved protocol in the event that they reached end points such as loss of mobility or other signs of morbidity.
Transcriptome analysis
CD4+ T cells were edited with a Cas9 RNP targeting AAVS1. The decision to target this locus enables effects on RNA levels to be attributed to the delivery methods under comparison, without the effects of disrupting a cellular component such as the TCR. RNA was extracted from treated cells (non-treated, DMSO, A5K, electroporation) from four donors at three timepoints (6 h, 1 day and 7 days after editing) using the RNeasy Micro kit (Qiagen #74004), quantified using a NanoDrop and stored at −80 °C. RNA samples were submitted to the UCSF-HDFCCC Laboratory for Cell Analysis shared resource facility for nCounter analysis using the NanoString CAR-T characterization panel. Gene expression fold changes and Benjamini–Yekutieli-adjusted P values for volcano plots were determined using nSolver analysis software. For each treatment comparison of interest, analysis was run for each timepoint separately and for all timepoints incorporated. Because the latter approach made use of more data as input, it was more sensitive at identifying significantly affected genes; for this reason, and because it was less biased to timepoint-specific outcomes, we focused on this analysis. Plots were generated in MATLAB.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary figures and tables.
Acknowledgements
S. Pyle and the study authors adapted CRISPR enzyme illustrations from the Innovative Genomics Institute Glossary Icon Collection, originally by C. Liu of Two Photon Art. We thank C. Jeans of the QB3 Berkeley MacroLab for assistance. We thank M. L. Hochstrasser for comments on the manuscript. D.V.F. is supported by a Canadian Institute of Health Research fellowship. J.J.M. is supported by a Cancer Research Institute Irvington postdoctoral fellowship. D.N.N. is supported by NIH grants L40AI140341 and K08AI153767, a UCSF LGR Innovation Award, and previously by a CIRM Alpha Stem Cell Clinic fellowship. V.S.V. was supported by the UCSF Multiple Myeloma Translational Initiative. B.R.S. is supported by a UCSF Herbert Perkins Cellular Therapy and Transfusion Medicine fellowship, a CIRM Alpha Stem Cell Clinic fellowship and an NIH LRP grant from the NCATS. A.M., S.U.S. and R.C.W. are supported by NIH award UG3AI150552 as part of the Somatic Cell Genome Editing (SCGE) consortium. V.A. is supported by the Fondation ARC pour la recherche sur le cancer. A.M. is supported by the Simons Foundation, a Lloyd J. Old STAR grant from the Cancer Research Institute, Parker Institute for Cancer Immunotherapy, and Innovative Genomics Institute. The UCSF Parnassus Flow Core RRID:SCR_018206 is supported by the DRC Center Grant NIH P30 DK063720. NanoString nCounter data were obtained in collaboration with the HDFCCC Laboratory for Cell Analysis Shared Resource Facility, which is supported by NIH grant P30CA082103. A549 cells were provided by the Kole Roybal laboratory.
Source data
Source data.
Author contributions
D.V.F., J.J.M., D.N.N., A.M., J.E. and R.C.W. conceived the study. D.V.F., J.J.M., D.N.N., D.C. and S.U.S. designed and performed experiments and analysed data. J.M.H. and R.M. conceived and prepared reagents. S.K.W. analysed data. N.K., E.V.S. and Z.L. performed experiments. B.R.S., V.S.V. and V.A. contributed to experimental design. D.V.F., J.J.M., D.N.N., D.C., S.U.S., A.M., J.E. and R.C.W. wrote and edited the manuscript. All authors read the manuscript and agree with its contents.
Peer review
Peer review information
Nature Biomedical Engineering thanks Meisam Kararoudi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
The data supporting the results in this study are available within the paper and its Supplementary Information. Sequencing data are available from the NCBI SRA with BioProject ID PRJNA923785. Cas9 plasmids are available on Addgene (ID# 196245 and 196246). All data, including the pre-processed nCounter data, are available from the corresponding authors on reasonable request. Source data are provided with this paper.
Competing interests
D.V.F., D.N.N., D.C., S.U.S., J.M.H., A.M. and R.C.W. are named inventors on a patent application related to this work. D.N.N. is a holder of patents pertaining to but not resulting from this work. D.N.N. is a consultant for and owns stock in Navan Technologies. B.R.S. and A.M. are holders of patents pertaining to but not resulting from this work. J.E. is a compensated co-founder at Mnemo Therapeutics, a compensated scientific advisor to Cytovia Therapeutics, owns stocks in Mnemo Therapeutics and Cytovia Therapeutics, is a compensated member of the scientific advisory board at Treefrog Therapeutics, and has received a consulting fee from Casdin Capital. The J.E. laboratory has received research support from Cytovia Therapeutics, Mnemo Therapeutics and Takeda. J.E. is a holder of patents pertaining to but not resulting from this work. A.M. is a co-founder of Arsenal Biosciences, Spotlight Therapeutics and Survey Genomics, serves on the boards of directors at Spotlight Therapeutics and Survey Genomics, is a board observer (and former member of the board of directors) at Arsenal Biosciences, is a member of the scientific advisory boards of Arsenal Biosciences, Spotlight Therapeutics, Survey Genomics, NewLimit and Amgen, owns stock in Arsenal Biosciences, Spotlight Therapeutics, NewLimit, Survey Genomics and PACT Pharma, and has received fees from Arsenal Biosciences, Spotlight Therapeutics, NewLimit, 23andMe, PACT Pharma, Juno Therapeutics, Trizell, Vertex, Merck, Amgen, Genentech, AlphaSights, Rupert Case Management, Bernstein and ALDA. A.M. is an investor in and informal advisor to Offline Ventures and a client of EPIQ. The A.M. laboratory has received research support from Juno Therapeutics, Epinomics, Sanofi, GlaxoSmithKline, Gilead and Anthem. The R.C.W. laboratory has received research support from Genentech, Roche and Pfizer; the associated research is distinct from what is reported here.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Dana V. Foss, Joseph J. Muldoon, David N. Nguyen.
Contributor Information
Alexander Marson, Email: alexander.marson@ucsf.edu.
Justin Eyquem, Email: justin.eyquem@ucsf.edu.
Ross C. Wilson, Email: rosswilson@berkeley.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41551-023-01032-2.
References
- 1.Weber EW, Maus MV, Mackall CL. The emerging landscape of immune cell therapies. Cell. 2020;181:46–62. doi: 10.1016/j.cell.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eyquem J, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rezalotfi A, Fritz L, Förster R, Bošnjak B. Challenges of CRISPR-based gene editing in primary T cells. Int. J. Mol. Sci. 2022;23:1689. doi: 10.3390/ijms23031689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aijaz A, et al. Biomanufacturing for clinically advanced cell therapies. Nat. Biomed. Eng. 2018;2:362–376. doi: 10.1038/s41551-018-0246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruseska I, Zimmer A. Internalization mechanisms of cell-penetrating peptides. Beilstein J. Nanotechnol. 2020;11:101–123. doi: 10.3762/bjnano.11.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br. J. Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han X, Bushweller JH, Cafiso DS, Tamm LK. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Mol. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 8.Dastpeyman M, Karas JA, Amin A, Turner BJ, Shabanpoor F. Modular synthesis of trifunctional peptide-oligonucleotide conjugates via native chemical ligation. Front. Chem. 2021;9:627329. doi: 10.3389/fchem.2021.627329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y-J, Erazo-Oliveras A, Pellois J-P. Delivery of macromolecules into live cells by simple co-incubation with a peptide. ChemBioChem. 2010;11:325–330. doi: 10.1002/cbic.200900527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tai W, Gao X. Functional peptides for siRNA delivery. Adv. Drug. Deliv. Rev. 2017;110–111:157–168. doi: 10.1016/j.addr.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staahl BT, et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 2017;35:431–434. doi: 10.1038/nbt.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danielson DC, Sachrajda N, Wang W, Filip R, Pezacki JP. A novel p19 fusion protein as a delivery agent for short-interfering RNAs. Mol. Ther. Nucleic Acids. 2016;5:e303. doi: 10.1038/mtna.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung MS, Mun J-Y, Kwon O, Kwon K-S, Oh D-B. Efficient myogenic differentiation of human adipose-derived stem cells by the transduction of engineered MyoD protein. Biochem. Biophys. Res. Commun. 2013;437:156–161. doi: 10.1016/j.bbrc.2013.06.058. [DOI] [PubMed] [Google Scholar]
- 14.Lin N, et al. A novel system enhancing the endosomal escapes of peptides promotes Bak BH3 peptide inducing apoptosis in lung cancer A549 cells. Target. Oncol. 2014;9:163–170. doi: 10.1007/s11523-013-0282-9. [DOI] [PubMed] [Google Scholar]
- 15.Algayer B, et al. Novel pH selective, highly lytic peptides based on a chimeric influenza hemagglutinin peptide/cell penetrating peptide motif. Molecules. 2019;24:2079. doi: 10.3390/molecules24112079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen DN, et al. Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol. 2020;38:44–49. doi: 10.1038/s41587-019-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del’Guidice T, et al. Membrane permeabilizing amphiphilic peptide delivers recombinant transcription factor and CRISPR-Cas9/Cpf1 ribonucleoproteins in hard-to-modify cells. PLoS ONE. 2018;13:e0195558. doi: 10.1371/journal.pone.0195558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouet R, et al. Receptor-mediated delivery of CRISPR–Cas9 endonuclease for cell-type-specific gene editing. J. Am. Chem. Soc. 2018;140:6596–6603. doi: 10.1021/jacs.8b01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang F, et al. Structures of a CRISPR–Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med. 2019;25:776–783. doi: 10.1038/s41591-019-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knipping F, et al. Disruption of HIV-1 co-receptors CCR5 and CXCR4 in primary human T cells and hematopoietic stem and progenitor cells using base editing. Mol. Ther. 2022;30:130–144. doi: 10.1016/j.ymthe.2021.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen AG, et al. Gene editing of HIV-1 co-receptors to prevent and/or cure virus infection. Front. Microbiol. 2018;9:2940. doi: 10.3389/fmicb.2018.02940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter MF, et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishimasu H, et al. Engineered CRISPR–Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vavassori V, et al. Modeling, optimization, and comparable efficacy of T cell and hematopoietic stem cell gene editing for treating hyper-IgM syndrome. EMBO Mol. Med. 2021;13:e13545. doi: 10.15252/emmm.202013545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox TA, Houghton BC, Booth C. Gene edited T cell therapies for inborn errors of immunity. Front. Genome Ed. 2022;4:899294. doi: 10.3389/fgeed.2022.899294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shy, B. R. et al. High-yield genome engineering in primary cells using a hybrid ssDNA repair template and small-molecule cocktails. Nat. Biotechnol. 10.1038/s41587-022-01418-8 (2022). [DOI] [PMC free article] [PubMed]
- 28.Zhang L, et al. AsCas12a ultra nuclease facilitates the rapid generation of therapeutic cell medicines. Nat. Commun. 2021;12:3908. doi: 10.1038/s41467-021-24017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyberg WA, et al. An evolved AAV variant enables efficient genetic engineering of murine T cells. Cell. 2023;186:446–460.e19. doi: 10.1016/j.cell.2022.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samulski RJ, et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stadtmauer EA, et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367:eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bothmer A, et al. Detection and modulation of DNA translocations during multi-gene genome editing in T cells. CRISPR J. 2020;3:177–187. doi: 10.1089/crispr.2019.0074. [DOI] [PubMed] [Google Scholar]
- 33.Sheridan C. Off-the-shelf, gene-edited CAR-T cells forge ahead, despite safety scare. Nat. Biotechnol. 2021 doi: 10.1038/d41587-021-00027-1. [DOI] [PubMed] [Google Scholar]
- 34.Webber BR, et al. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 2019;10:5222. doi: 10.1038/s41467-019-13007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgiadis C, et al. Base-edited CAR T cells for combinational therapy against T cell malignancies. Leukemia. 2021;35:3466–3481. doi: 10.1038/s41375-021-01282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian Y, Li Y, Shao Y, Zhang Y. Gene modification strategies for next-generation CAR T cells against solid cancers. J. Hematol. Oncol. 2020;13:54. doi: 10.1186/s13045-020-00890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gornalusse GG, et al. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat. Biotechnol. 2017;35:765–772. doi: 10.1038/nbt.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun I, et al. CRISPR–Cas9 knock out of CD5 enhances the anti-tumor activity of chimeric antigen receptor T cells. Blood. 2020;136:51–52. doi: 10.1182/blood-2020-136860. [DOI] [Google Scholar]
- 39.Spiegel JY, Komanduri KV. Treating CAR-T relapses: check not checkmate. Blood. 2022;139:955–957. doi: 10.1182/blood.2021013896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat. Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangare C, et al. Robust identification of suitable T-cell subsets for personalized CMV-specific T-cell immunotherapy using CD45RA and CD62L microbeads. Int. J. Mol. Sci. 2019;20:1415. doi: 10.3390/ijms20061415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timmins LM, et al. Selecting a cell engineering methodology during cell therapy product development. Cell Transplant. 2021;30:9636897211003022. doi: 10.1177/09636897211003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng Q, Sun H, Liu J. Precise somatic genome editing for treatment of inborn errors of immunity. Front. Immunol. 2022;13:960348. doi: 10.3389/fimmu.2022.960348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.
Data Availability Statement
The data supporting the results in this study are available within the paper and its Supplementary Information. Sequencing data are available from the NCBI SRA with BioProject ID PRJNA923785. Cas9 plasmids are available on Addgene (ID# 196245 and 196246). All data, including the pre-processed nCounter data, are available from the corresponding authors on reasonable request. Source data are provided with this paper.