Abstract
A systematic understanding of the aetiology of neurodevelopmental disorders (NDDs) and their co-occurrence with other conditions during childhood and adolescence remains incomplete. In the current meta-analysis, we synthesized the literature on (1) the contribution of genetic and environmental factors to NDDs, (2) the genetic and environmental overlap between different NDDs, and (3) the co-occurrence between NDDs and disruptive, impulse control and conduct disorders (DICCs). Searches were conducted across three platforms: Web of Science, Ovid Medline and Ovid Embase. Studies were included only if 75% or more of the sample consisted of children and/or adolescents and the studies had measured the aetiology of NDDs and DICCs using single-generation family designs or genomic methods. Studies that had selected participants on the basis of unrelated diagnoses or injuries were excluded. We performed multilevel, random-effects meta-analyses on 296 independent studies, including over four million (partly overlapping) individuals. We further explored developmental trajectories and the moderating roles of gender, measurement, geography and ancestry. We found all NDDs to be substantially heritable (family-based heritability, 0.66 (s.e. = 0.03); SNP heritability, 0.19 (s.e. = 0.03)). Meta-analytic genetic correlations between NDDs were moderate (grand family-based genetic correlation, 0.36 (s.e. = 0.12); grand SNP-based genetic correlation, 0.39 (s.e. = 0.19)) but differed substantially between pairs of disorders. The genetic overlap between NDDs and DICCs was strong (grand family-based genetic correlation, 0.62 (s.e. = 0.20)). While our work provides evidence to inform and potentially guide clinical and educational diagnostic procedures and practice, it also highlights the imbalance in the research effort that has characterized developmental genetics research.
Subject terms: Behavioural genetics, Human behaviour
This meta-analysis examines how genetic variation is associated with neurodevelopmental disorders, their overlap and their co-occurrence with disruptive, impulse control and conduct disorders.
Main
Neurodevelopmental disorders (NDDs) are complex health concerns, starting from childhood1. NDDs affect around 15% of children and adolescents worldwide and lead to impaired cognition, communication, adaptive behaviour and psychomotor skills2. The fifth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-5) categorizes the following seven disorders under NDDs: intellectual disabilities, communication disorders, autism spectrum disorder (ASD), attention-deficit/hyperactivity disorder (ADHD), specific learning disorders, motor disorders and other NDDs3. NDDs often have lifelong trajectories: they can manifest before 12 months of age4 and can be diagnosed before children enter primary education3,5.
While some NDDs (for example, ASD and ADHD) may persist throughout adolescence and adulthood6,7, others are more likely to alleviate as children get older (for example, tic disorder8 and communication disorders9). Nevertheless, all NDDs can lead to social and behavioural difficulties and reduced independence over the lifespan6,7,10. For instance, ADHD in childhood has been associated with an increased risk of educational and occupational problems, risk-taking, and mood disorders in adulthood11; and an ASD diagnosis in childhood has been associated with increased occupational difficulties and a greater risk of psychopathologies in adulthood12,13. The difficulties are often more salient for children diagnosed with more than one NDD14.
A systematic understanding of the aetiology of NDDs remains incomplete. A disproportionate number of studies and systematic reviews have focused on ASD and ADHD, pointing to their substantial heritability—the extent to which observed individual differences are accounted for by underlying genetic differences. A meta-analysis of seven twin studies of clinically diagnosed ASD in child and adolescent samples yielded a grand heritability (h2) estimate of 0.74 (ref. 15). Similarly sizeable heritability estimates have also been obtained from twin studies of ADHD in childhood and adolescence16. Heritability estimates were found to differ across the two major components of ADHD, with genetic factors playing a more substantial role in the aetiology of hyperactivity (h2 = 0.71) than that of inattention (h2 = 0.56)17. However, other NDDs, despite showing similar prevalence rates and severity as ASD and ADHD, are less well understood and studied18.
In line with what has been observed for all complex traits, heritability estimates for ASD and ADHD obtained from DNA data are lower than those obtained from twin and family designs19. Single nucleotide polymorphism (SNP) heritability can be calculated using large samples of individual-level genotype data20 or summary statistics from genome-wide association studies21, hypothesis-free studies aimed at discovering associations between genetic variation across the genome and individual differences in traits and disorders. The two largest studies to date that have estimated the SNP heritability of ASD and ADHD report estimates of h2 = 0.12 for ASD22 and h2 = 0.22 for ADHD23.
It is now well established that NDDs often co-occur with one another (a phenomenon known as homotypic co-occurrence), and this points to a shared underlying liability between conditions24,25. Even in this instance, most studies have focused on examining the genetic correlations—the degree to which the same genetic variants contribute to the observed covariation between pairs of traits or disorders26—between ASD and ADHD, resulting in a meta-analytic genetic correlation of 0.59 (ref. 27) across twin and family studies, and a SNP-based genetic correlation of 0.35 (ref. 28). Aetiological sources of co-occurrence between all other NDDs have not been meta-analysed, but individual studies point to a moderate to strong shared liability between ASD/ADHD and other NDDs29–33.
Another category of disorders that begin in and progress through childhood and adolescence are disruptive, impulse control and conduct disorders (DICCs), which the DSM-5 describes as disorders that share the underlying features of impulsive behaviour, aggressiveness and pathological rule breaking3. The DSM-5 identifies eight main DICC categories: oppositional defiant disorder, intermittent explosive disorder, conduct disorder, antisocial personality disorder, pyromania, kleptomania, other specified DICC disorder and unspecified DICC disorders3 (Fig. 1). Similar to NDDs, DICCs have been linked to impaired social, emotional and educational outcomes34–37.
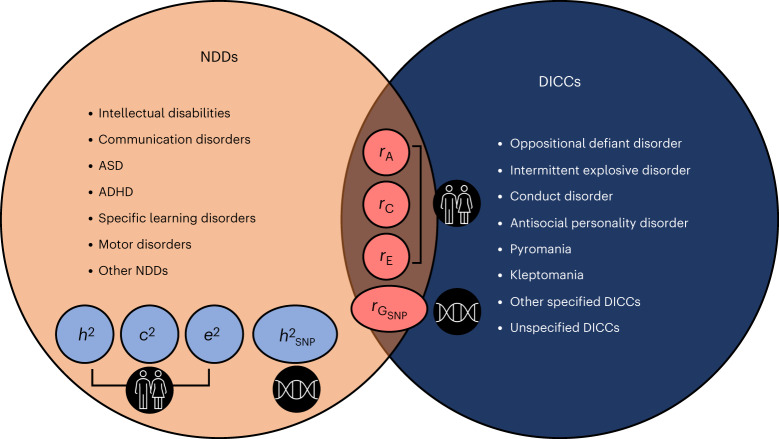
Fig. 1. Visual summary of the three core aims of the current meta-analysis.
Aim 1 (orange and light blue): estimate family-based genetic (h2), shared environmental (c2) and non-shared environmental (e2) influences as well as SNP heritability (h2SNP) for all NDDs identified by the DSM-5. Aim 2 (orange and red): provide grand estimates of family-based genetic (rA), shared environmental (rC) and non-shared environmental (rE) correlations and SNP-based genetic correlations () between different NDDs. Aim 3 (navy blue and red): provide grand estimates of rA, rC, rE and between NDDs and DICCs. The results for c2, e2, rC and rE are presented in Supplementary Note 1.
The developmental nature of DICCs makes them an ideal primary target for the investigation of how NDDs co-occur with other disorders (that is, heterotypic co-occurrence) during childhood and adolescence. However, the distinction between NDDs and DICCs in the published literature is often blurred, particularly for disorders that include clinical features that overlap across NDD and DICC categories, such as ADHD. The most investigated examples of symptom overlap between NDDs and DICCs involve ADHD and conduct disorder38,39, and ADHD and oppositional defiant disorder40. Studies highlight how these disorders are characterized by disturbances in emotion regulation, attention problems, cognitive inflexibility and impaired inhibition39,41,42. A shared symptomatology has also been observed between ASD and antisocial behaviour/personality disorder (which we refer to as conduct disorder in the current work since antisocial personality disorder describes adult diagnoses)3,43,44. However, studies on the association between NDDs and DICCs are characterized by a great deal of heterogeneity and inconsistencies across co-occurring conditions45,46.
With three core aims (Fig. 1), the current meta-analysis bridges gaps in our knowledge of the aetiology of NDDs and their co-occurrence with other developmental conditions in childhood and adolescence. First, we meta-analysed studies on the relative contributions of genetic and environmental influences to all NDD categories described in the DSM-5. Second, we meta-analysed estimates for the genetic and environmental overlap between different NDDs (homotypic co-occurrences). Third, given their developmental onset and progression and partly shared symptomatology, we examined the aetiology of the co-occurrence between NDDs and DICCs (heterotypic co-occurrences). In addition to addressing each disorder individually, we took a transdiagnostic approach by combining data across NDDs and including categorical (that is, the presence or absence of a disorder) and quantitative (that is, continuously measured symptoms) measures. Clarifying the genetic and environmental aetiology of all NDDs and their homotypic and heterotypic co-occurrences will advance our knowledge of how developmental disorders cluster together, which could inform educational and clinical practice47.
Results
This section presents meta-analytic findings on genetic influences on NDDs and on their genetic overlap with other NDDs and DICCs. Meta-analytic estimates for shared and non-shared environmental factors and their overlap are presented in Supplementary Note 1. The results for all sub-categories of NDDs and DICCs are reported in Supplementary Note 2, Supplementary Figs. 2 and 3, and Supplementary Tables 2, 4 and 6.
Searches and screening
Studies for this meta-analysis were selected during three screening stages including title and abstract screening, full text screening, and reference list screening (see Methods for a detailed description). This selection process resulted in a total of 296 studies (292 family-based and 34 SNP-based studies) included in the current meta-analysis (Fig. 2). The numbers of family-based and SNP-based studies do not add up because some studies provided both family-based and SNP-based estimates. These studies were counted only once towards the grand total but were included separately in family-based and SNP-based categories.
Fig. 2. Diagram of searches and screening.
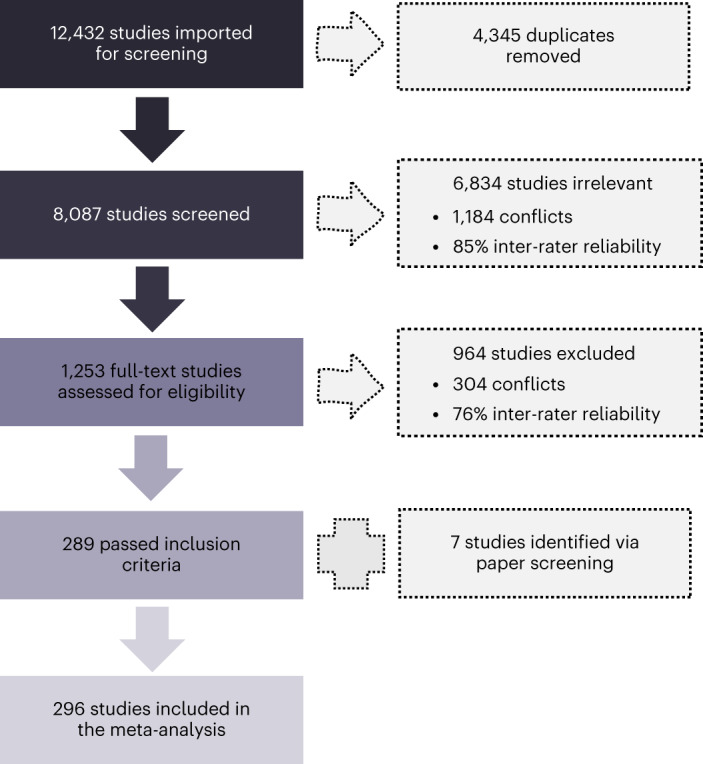
Overview of the screening and selection process across primary and secondary searches, along with statistics of inter-rater reliability.
Heritability of NDDs
Our first aim was to obtain reliable estimates of the contributions of genetic factors to individual differences in all NDDs. We considered two broad categories of methods that allow for the estimation of heritability: family-based designs including related individuals (such as sibling comparisons and twin studies) and SNP heritability48 (Methods). Given the substantial differences in methodology and outcomes, the findings across these two broad categories were meta-analysed separately.
Family-based heritability
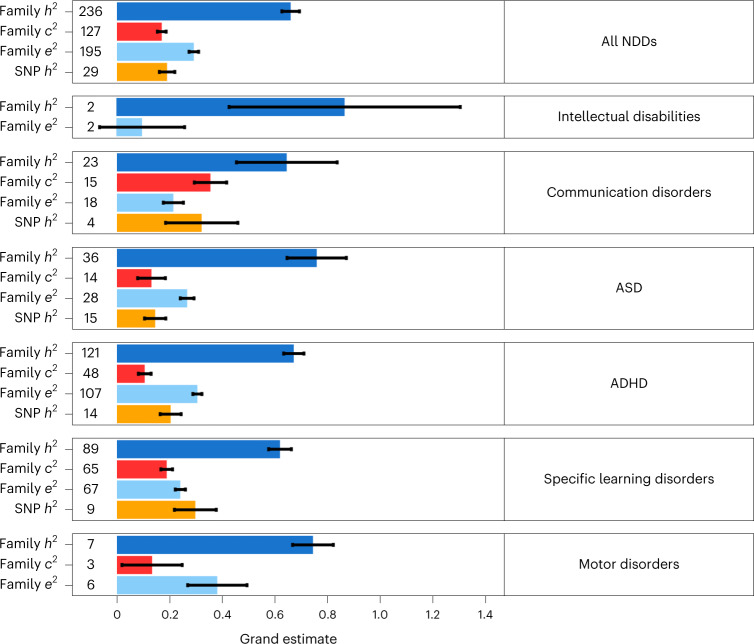
We identified a total of 236 family-based studies, comprising 2,792,511 partly overlapping individuals, that investigated the proportion of variance in NDDs that is accounted for by genetic factors. Out of the total, 121 studies (N = 682,340) investigated ADHD, 89 studies (N = 360,920) investigated specific learning disorders, 36 studies (N = 1,821,970) investigated ASD, 23 studies (N = 130,757) investigated communication disorders, 6 studies (N = 52,278) investigated motor disorders and 2 studies (N = 9,036) investigated intellectual disabilities. Across all NDDs and 236 studies, the grand h2 estimate was 0.66 (s.e. = 0.03). Grand h2 estimates differed, albeit not significantly, across NDD categories, ranging from 0.86 (s.e. = 0.44) for intellectual disabilities to 0.62 (s.e. = 0.04) for specific learning disorders (Fig. 3 and Supplementary Table 1). The distributions of genetic influences across studies and NDDs are presented in Supplementary Fig. 1.
Fig. 3. Genetic and environmental sources of variation in NDDs.
Meta-analytic family and SNP-based heritability (h2) of NDDs and shared environmental influences (c2) and non-shared environmental influences (e2) on variation in NDDs. The number preceding each bar on the y axis denotes the number of studies identified that provided estimates for specific NDDs. The error bars signify standard errors of the grand estimates of heritability and environmental influences. The results for c2 and e2 are discussed in Supplementary Note 1.
SNP heritability
Out of the total of 29 SNP-based studies, involving 893,896 partly overlapping individuals, the only disorders that were addressed by at least two independent studies49 included ASD (15 studies; N = 637,240), ADHD (14 studies; N = 725,168), specific learning disorders (9 studies; N = 40,637) and communication disorders (4 studies; N = 14,894). SNP heritability across all NDDs was moderate (0.19, s.e. = 0.03) and ranged from 0.15 (s.e. = 0.04) for ASD to 0.30 (s.e. = 0.14) for communication disorders (Fig. 3 and Supplementary Table 1). SNP heritability estimates were not found to differ significantly across disorders, although the degree of precision in the estimates varied substantially depending on the sample size and the number of individual studies included per disorder.
Genetic overlap between NDDs
Compared with the vast number of studies that examined the aetiology of individual differences in each NDD, only a limited body of research (37 studies; N = 212,569) investigated the co-occurrence between NDDs in childhood and adolescence. In fact, for some of the disorders, we were unable to find two independent statistics49 and therefore could not provide a meta-analytic estimate.
Family-based genetic correlations (rA)
When considering family-based designs (Methods and Supplementary Note 3), we obtained a sufficient number of studies to allow for meta-analysis for the following NDD pairs: ADHD and specific learning disorders (15 studies; N = 67,039), ASD and ADHD (6 studies; N = 58,518), ADHD and motor disorders (2 studies; N = 8,748), communication disorders and motor disorders (2 studies; N = 3,950), and communication disorders and specific learning disorders (2 studies; N = 42,098). Only one study was identified for the following pairs of NDDs: ASD and communication disorders (N = 12,174), ASD and specific learning disorders (N = 6,858), ASD and motor disorders (N = 6,858), and specific learning disorders and motor disorders (N = 6,858). These studies could therefore be included only in the transdiagnostic meta-analysis, capturing the degree of genetic and environmental co-occurrence across all NDD pairs. In addition, 9 studies (N = 46,000) examined the co-occurrence between subtypes of specific learning disorders, such as dyslexia and dyscalculia; these studies have been included in the transdiagnostic meta-analysis, and the results of these finer-grained analyses are reported in Supplementary Note 2.
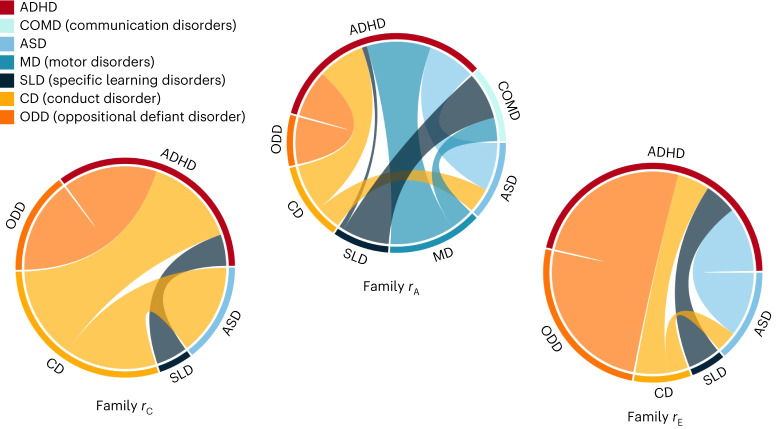
We first meta-analysed genetic correlations across all NDD categories (transdiagnostic genetic co-occurrence); this yielded a moderate grand estimate of rA = 0.36 (s.e. = 0.12). When considering NDD categories separately, we found the strongest genetic overlaps between ADHD and motor disorders (rA = 0.90, s.e. = 0.82) and between ASD and ADHD (rA = 0.67, s.e. = 0.30), while the weakest genetic correlation was found for the association between ADHD and specific learning disorders (rA = 0.07, s.e. = 0.12; Fig. 4 and Supplementary Table 3). However, given the considerable differences in sample sizes used to derive genetic correlations between pairs of disorders (for example, between ASD and ADHD or between communication disorders and motor disorders), the strength of these correlations may be difficult to compare. Low correlations could also reflect low power to detect the true overlap.
Fig. 4. Genetic and environmental correlations between NDDs and DICCs.
Strength of the meta-analytic genetic (rA), shared environmental (rC) and non-shared environmental (rE) correlations between NDDs and their homotypic (other NDDs) and heterotypic (DICCs) co-occurrences. The outer layer of each circle shows all the different NDDs and DICCs for which meta-analytic correlation estimates could be computed. Each coloured connector path indicates the strength of association between disorders; the thicker the connector path, the stronger the correlation between the two disorders. The results for family rC and rE are presented in Supplementary Note 1.
SNP-based genetic correlations ()
SNP-based designs in child and adolescent samples exclusively focused on the association between ASD and ADHD (five studies; N = 242,543) and between subtypes of specific learning disorders (one study; N = 4,500). The transdiagnostic genetic correlation obtained via meta-analysing SNP-based designs was 0.39 (s.e. = 0.19) (Supplementary Table 8), in line with the estimate obtained from family-based designs. A grand genetic correlation of 0.20 (s.e. = 0.14) was found for the co-occurrence between ADHD and ASD. The one remaining study examined the co-occurrence between dyslexia- and dyscalculia-related traits, specifically reading and mathematics abilities, which were strongly correlated ( = 0.74, s.e. = 0.17)50.
Genetic overlap between NDDs and DICCs
Our third aim was to obtain meta-analytic estimates of the genetic associations between NDDs and DICCs. Our search yielded only 15 eligible family-based studies (N = 42,718) and no SNP-based studies. Meta-analytic genetic correlations could be calculated for only a few NDD and DICC pairs—namely, ADHD and conduct disorder (6 studies; N = 11,308), ADHD and oppositional defiant disorder (6 studies; N = 10,748) and ASD and conduct disorder (3 studies; N = 24,564). In addition, we identified one study (N = 360) that examined the co-occurrence between specific learning disorders and disruptive behaviour, finding a weak negative genetic correlation (rA = −0.14, s.e. = 0.06)51.
Family-based genetic correlations (rA)
Across all co-occurrences between NDDs and DICCs (15 studies), the grand genetic correlation was 0.62 (s.e. = 0.20). Similarly strong genetic correlations were observed between ADHD and conduct disorder (6 studies) and between ADHD and oppositional defiant disorder (6 studies): rA = 0.66 (s.e. = 0.36) and rA = 0.66 (s.e. = 0.18), respectively—a similar level of aetiological overlap to that observed between strongly genetically correlated NDDs such as ADHD and ASD (Supplementary Table 5). In contrast, the genetic overlap between ASD and conduct disorder (3 studies) was much weaker, with a meta-analytic genetic correlation of 0.35 (s.e. = 0.10; Fig. 4). The similar extent of genetic overlap between ADHD and conduct disorder or oppositional defiant disorder and between ADHD and ASD may not be free from biases introduced by an unbalanced sample size used to derive these meta-analytic estimates. In addition, large meta-analytic standard errors make assessing the significance of differences between the estimates difficult.
Sex differences
Some NDDs do not affect males and females equally—for instance, males are four times more likely to be diagnosed with ASD52,53 and twice as likely to be diagnosed with ADHD54. Studies have suggested that these differences in prevalence may be caused by quantitative genetic sex differences, differences in the degree to which genes influence variation in NDDs in males versus females55. To provide an overview of sex differences in NDDs, we conducted separate meta-analyses including all studies that had reported sex-specific estimates.
Family-based heritability
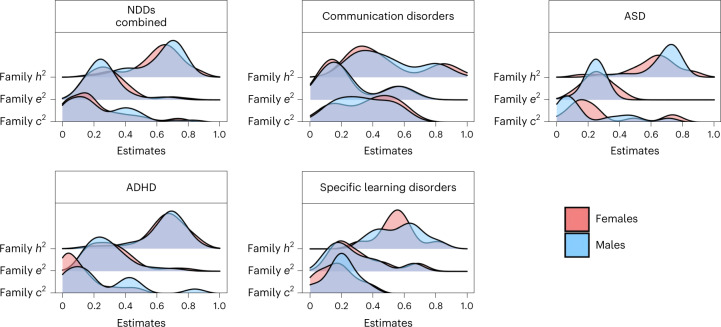
We identified 68 family-based studies that investigated the genetic aetiology of individual differences in NDDs in male samples and 67 studies that reported estimates for female samples. Of all the studies involving sex-stratified samples, 38 studies focused on ADHD, 21 focused on ASD, 8 focused on specific learning disorders, 4 focused on communication disorders and 2 focused on motor disorders. Across all NDDs, family-based heritability was not significantly different between males and females (h2 = 0.65, s.e. = 0.06 in males; h2 = 0.67, s.e. = 0.06 in females). The distributions of sex-specific family-based variance components for all NDDs (except motor disorders, for which a sufficient number of studies (>1) was not identified) are presented in Fig. 5 and Supplementary Table 16.
Fig. 5. Sex differences.
Distributions of the sex-specific meta-analytic estimates for the heritability (h2) of NDDs and environmental contributions to NDDs. The top left panel shows the distributions of sex-specific estimates for the transdiagnostic meta-analysis; the remaining panels show the same estimates for specific NDDs for which a sufficient number of studies (>2) reporting sex-specific estimates was identified. The results for sex-specific c2, e2, rC and rE estimates are presented in Supplementary Note 1.
SNP heritability
Marked differences in SNP heritability were observed between males and females across all NDDs (0.19, s.e. = 0.07 for males; 0.09, s.e. = 0.10 for females). However, these estimates were based on the only two studies to date that had calculated the SNP heritability of ASD and ADHD separately by sex (Supplementary Table 16).
Sex differences in genetic overlap between NDDs
We identified only four family-based studies that examined homotypic co-occurrences of NDDs in males and only two studies in females. Half of these studies considered the overlap between ASD and ADHD. The other half considered the co-occurrence between ASD and communication disorders (one study in both males and females) and between developmental coordination disorder and tic disorder, two subtypes of motor disorder (one study in males only). The grand family-based genetic correlation across all NDDs was estimated at 0.86 (s.e. = 0.58) for males and 0.25 (s.e. = 0.36) for females (Supplementary Table 17).
Sex-specific grand estimates of family-based genetic correlations between specific disorders could not be calculated due to the limited number of available studies. The only exception was the co-occurrence between ASD and ADHD in males, where two studies were identified (rA = 0.79, s.e. = 0.42) (Supplementary Table 17). SNP-based genetic correlations between NDDs could not be calculated for males and females separately due to a lack of studies that examined these associations separately by sex in samples of children and adolescents.
Sex differences in genetic overlap between NDDs and DICCs
Sources of co-occurrence between NDDs and DICCs could be estimated only between ADHD and conduct disorder and only in females. Of the only two studies that examined the sex-specific co-occurrence between ADHD and conduct disorders, one used a female-only sample. Hence, we could only meta-analyse the co-occurrence between ADHD and conduct disorder in females. We found a meta-analytic genetic correlation of 0.75 (s.e. = 0.58) (Supplementary Table 18).
Developmental trajectories
We investigated developmental change and continuity in the relative contributions of genetic factors to NDDs by examining age-related differences in their aetiology and sources of their homotypic and heterotypic co-occurrences. We distinguished among the three following developmental stages: childhood (4–7 years), middle childhood (8–10 years) and adolescence (11–24 years). We grouped estimates in any of those three categories or across multiple categories—that is, childhood and middle childhood (4–10 years), middle childhood and adolescence (8–24 years), and childhood and adolescence (4–24 years).
Family-based heritability
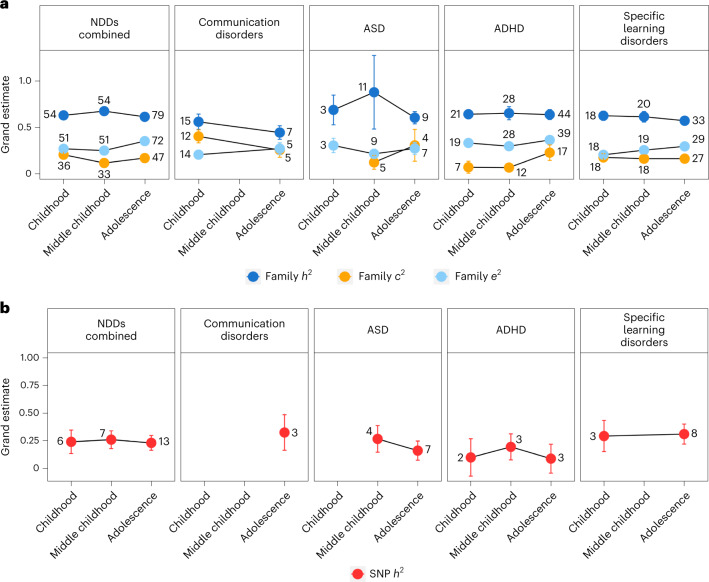
Across all NDDs, 54 family-based studies reported estimates in childhood (4–7 years), 54 studies reported estimates in middle childhood (8–10 years) and 79 studies reported estimates in adolescence (11–24 years). The remaining studies involved populations whose age range spanned across categories—that is, childhood and middle childhood (4–10 years; 14 studies), middle childhood and adolescence (8–24 years; 50 studies), and childhood and adolescence (4–24 years; 40 studies). We investigated age-related differences in heritability including all NDD categories (Fig. 6a), except motor disorders, for which we did not identify enough studies (>1) per age category. All estimates with standard errors, including those for age cross-categories, are presented in Supplementary Table 19.
Fig. 6. Developmental trajectories.
a,b, Age-related differences in family-based heritability (h2) and shared (c2) and non-shared (e2) environmental influences on NDDs (a) and SNP heritability (b). Developmental stages include childhood (4–7 years), middle childhood (8–10 years) and adolescence (11–24 years). The error bars represent standard errors of grand estimates of heritability and environmental influences. The number located near each point estimate denotes the number of studies identified that provided estimates for specific developmental stages. For intellectual disabilities and motor disorders, we could not identify a sufficient number of studies (>1) reporting age-dependent estimates, and we were consequently unable to derive meta-analytic estimates. The results for age-stratified c2, e2, rC and rE are reported in Supplementary Note 1.
Across all NDDs, grand heritability remained relatively stable developmentally, with the estimate of 0.63 (s.e. = 0.03) in childhood, a slight increase in middle childhood (0.68, s.e. = 0.04) and a subsequent drop back to 0.62 (s.e. = 0.08) in adolescence. This trend was consistent for some specific disorders (for example, ASD and ADHD) but not for others (for example, communication disorders and specific learning disorders), for which genetic influences decreased developmentally (Fig. 6a and Supplementary Table 19).
SNP heritability
Of a total of 29 SNP-based studies that were identified, 13 included adolescent samples, 7 included samples in middle childhood and 6 included samples in childhood, while 11 studies reported estimates across childhood and adolescence. SNP heritability was stable developmentally across NDDs, and the developmental trajectory mirrored that of family-based heritability (SNP h2 = 0.24, s.e. = 0.11 in childhood; SNP h2 = 0.26, s.e. = 0.08 in middle childhood; SNP h2 = 0.23, s.e. = 0.07 in adolescence) (Fig. 6b and Supplementary Table 19). For ASD, ADHD and specific learning disorders (the specific NDDs for which grand estimates could be calculated), the developmental trends were consistent with those observed for family-based heritability (Fig. 6b and Supplementary Table 19).
Developmental trajectories in genetic overlap between NDDs
Overall, we could not explore developmental trends in genetic correlations using either method due to a lack of available studies; the only exceptions were grand estimates for adolescence and across age categories (Supplementary Tables 20 and 21). Genetic correlations obtained for adolescent samples only were in line with those obtained for the total sample (for example, when considering the co-occurrence between ASD and ADHD, the genetic correlation was 0.66 (0.49) in adolescent samples and 0.67 (0.30) across all age categories).
Categorical versus continuous measurement
Although we meta-analysed categorical (binary phenotypes, such as clinical diagnoses and cut-offs) and quantitative (sub-threshold symptom counts or test/questionnaire scores) measures together, we also report separate grand estimates for both measurement types. Across all NDDs, categorical measures were observed to yield significantly higher family-based heritability estimates than continuous phenotypes (0.77 (s.e. = 0.07) versus 0.64 (s.e. = 0.03)). However, the opposite was found for SNP-based heritability (0.17 (s.e. = 0.03) for categorical measures versus 0.25 (s.e. = 0.06) for quantitative assessments). Differences in sources of variation in specific NDDs as well as specific homotypic and heterotypic co-occurrences are presented in Supplementary Note 4, Supplementary Fig. 26 and Supplementary Tables 28–30.
Geography and ancestry
Research into the genetic aetiology of NDDs and of their homotypic and heterotypic co-occurrences is largely limited to Western countries, even though, according to the Global Burden of Disease study56, the prevalence of diagnosed NDDs is not uniform across the globe. Furthermore, individuals of European ancestry represent 16% of the global population but 80% of participants in genomic (that is, DNA-based) research57. This Eurocentric bias58 has created a major gap in our knowledge of the genetic aetiology of NDDs and their co-occurrences in non-White populations. In the following section, we provide an overview of how behaviour genetics research into NDDs is distributed across countries and continents and how the estimates differ as a function of geographical location. Supplementary Note 5, Supplementary Fig. 27 and Supplementary Tables 25–27 contain meta-analytic results of how heritability and genetic correlations differ at different levels of sample ancestral diversity. We created a moderator with four levels of percentage of European-ancestry participants in samples: less than 50%, more than 50% but less than 75%, more than 75% but less than 100% and 100%.
Family-based heritability
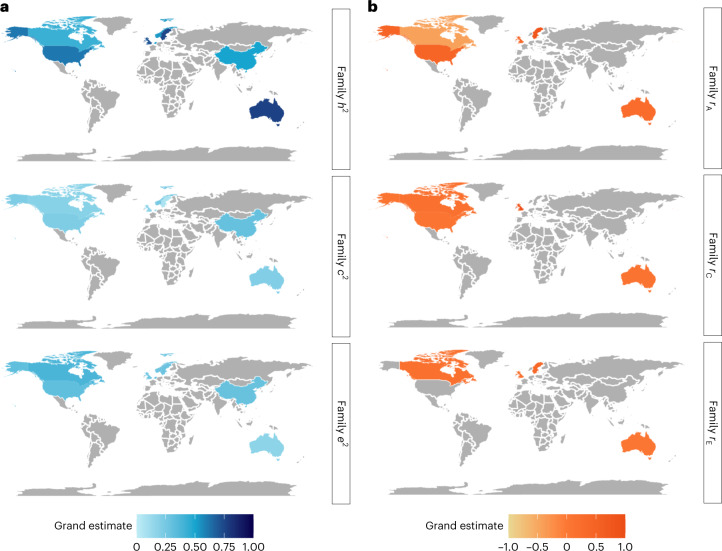
Of the 236 studies investigating sources of individual differences in NDDs, 41% (96 studies) involved samples and cohorts based in the United Kingdom, 77 studies involved samples based in the United States, 24 studies involved Swedish samples, 19 studies involved Dutch samples, 11 studies involved Australian samples, 7 studies involved Canadian samples, 4 studies involved samples from China and 2 studies involved samples from Norway. Other countries that contributed to the total grand estimate but did not have enough estimates for separate meta-analysis (that is, only one study was found from each country) included Finland, Japan, South Korea and Italy. Estimates differed significantly across countries. Considering all NDDs, the highest meta-analytic family-based heritability was estimated for Australian and Swedish samples (0.76 (s.e. = 0.17) and 0.74 (s.e. = 0.05), respectively), while the lowest was obtained for Canadian cohorts (0.43, s.e. = 0.09) (Fig. 7a and Supplementary Table 22).
Fig. 7. Geographical differences.
a, Differences in family-based heritability (h2) and shared environmental (c2) and non-shared environmental (e2) influences across all NDDs. b, Geographical differences in the genetic (rA), shared environmental (rC) and non-shared environmental (rE) overlap between NDDs. The areas shaded in grey are regions for which not enough relevant studies were identified (<2 studies). Geographical differences in rA, rC and rE between NDDs and DICCs are presented in Supplementary Fig. 28. The results for c2 and e2 as well as rC and rE are discussed in Supplementary Note 1.
Specific NDDs were investigated with different frequencies across countries: the aetiology of intellectual disabilities was exclusively investigated in Swedish cohorts (2 of 2 studies), and most studies addressing sources of variance in motor disorders also came from Sweden (4 of a total of 7 studies). Communication disorders were mostly researched in the United Kingdom (17 of a total of 23 studies), as were ASD (20 of 36 studies) and ADHD (42 of 121 studies). In contrast, 47 of a total of 89 studies investigating specific learning disorders were carried out in the United States.
SNP heritability
Studies exploring the SNP heritability of NDDs focused entirely on European cohorts and were primarily conducted in the United Kingdom and the Netherlands (14 and 3 of 29 SNP-based studies in total) (Supplementary Table 22).
Geography- and ancestry-related differences in the genetic overlap between NDDs
Sources of homotypic co-occurrence with NDDs were investigated in 37 independent family-based studies, of which the majority were conducted in the United Kingdom (49%) or the United States (30%). The highest genetic correlation across all co-occurrences was estimated in Swedish cohorts (0.80, s.e. = 0.26 across three studies), while the lowest grand genetic overlap was estimated in Canadian samples (−0.44, s.e. = 0.24 across only two studies that investigated the association between ADHD and specific learning disorders; Fig. 7b and Supplementary Table 23).
The genetic aetiology of the co-occurrence between ASD and ADHD during childhood and adolescence was exclusively researched in the United Kingdom and Sweden (three of a total of six studies each). The co-occurrence between ADHD and motor disorders was explored by only two studies, one conducted in Sweden and the other in Australia. Most studies examining the genetic overlap between ADHD and specific learning disorders came from the United States (8 of a total of 18 studies), whereas the overlap between communication disorders and motor disorders was addressed by only two studies conducted in the United Kingdom and Japan.
SNP-based studies (six in total) addressing the co-occurrence between NDDs were exclusively conducted in combined samples from the United Kingdom and Denmark (Supplementary Table 23).
Geography- and ancestry-related differences in the genetic overlap between NDDs and DICCs
A total of 15 family-based studies addressing the co-occurrence between NDDs and DICCs were identified, 40% of which were conducted in the United Kingdom, 20% in the United States and 20% in Sweden. The studies yielded consistently strong estimates of genetic correlations across the three regions: genetic correlations of 0.60 (s.e. = 0.29), 0.42 (s.e. = 0.15) and 0.68 (s.e. = 0.41), respectively (Supplementary Fig. 28 and Supplementary Table 24). The remaining 20% of the studies were conducted in Australia, Finland and South Korea but could not be meta-analysed separately as only one estimate was available for each country.
In terms of specific co-occurrences between NDDs and DICCs, half of the studies that explored genetic overlap between ADHD and conduct disorder and between ADHD and oppositional defiant disorder were conducted in the United States (three studies each). Three out of four studies examining the association between ASD and conduct disorder were conducted in the United Kingdom; the fourth was conducted in Sweden.
Bias and heterogeneity assessment
We applied I2 statistics to assess heterogeneity in the estimates, followed by outlier and influential case identification analyses. The results of these analyses are reported in Supplementary Note 6, Supplementary Tables 7–12 and Supplementary Figs. 4–7. We applied Egger’s regression and inspected funnel plots to examine the impact of publication bias on our results; the outcomes of these analyses are reported in Supplementary Note 7, Supplementary Tables 13–15 and Supplementary Figs. 8–24. The results of the risk-of-bias assessment are presented in Supplementary Fig. 25, where 93.8% of studies showed low risk of bias across the nine quality checklist items, and the remaining 6.2% showed moderate risk.
Discussion
The findings of the present meta-analysis synthesize the current state of knowledge on NDDs and have implications that can guide future research strategies as well as clinical and educational practice. First, by providing estimates of the relative contributions of genetic factors to all NDDs, our work responds to the need of moving beyond the nearly exclusive research focus on ASD and ADHD. Second, by providing an account of the genetic overlap between NDDs, we highlight how genetic influences are implicated in the co-occurrence between multiple NDDs, identifying patterns of shared aetiological liability. Third, by synthesizing the literature on the co-occurrence between NDDs and DICCs, we highlight how disorders from these two separate groups identified by the DSM-5 share as much of their genetic aetiology as do disorders all classified as NDDs.
Our work provides meta-analytic evidence for the substantial heritability of all NDDs (our first aim), particularly when considering family-based studies, which indicated that around two-thirds of the variation in NDDs is accounted for by genetic differences between individuals (in children and adolescents). Although males are up to four times more likely to be diagnosed with ASD and ADHD than females52–54, we showed that, when meta-analysed, the genetic effects associated with NDDs do not differ by sex. We also showed that genetic sources of variation in NDDs are remarkably stable across developmental stages, and this developmental stability was observed across all NDDs. Genetic effects were also mostly consistent when we separated studies that had considered diagnoses and clinical cut-offs from studies that had quantified NDDs as continuous traits.
Interestingly, we found that the genetic contributions to NDDs differed substantially as a function of geography. This highlights how estimates of genetic effects associated with disorders are sensitive to different environmental contexts59,60. Our work on geographical differences also highlights the major gap in our knowledge of the aetiology of NDDs in non-Western countries, a gap that is exceeded by the lack of ancestral diversity observed across all studies of NDDs. Importantly, the current study points to how genetic influences on NDDs are substantially reduced in more ancestrally diverse samples, again highlighting how heritability estimates are inextricably linked to our social context61,62, in the sense that increased ancestral homogeneity in the sample probably entails increased environmental homogeneity, reducing environmental variability and inflating heritability in these populations.
The lack of diversity in genetic research remains its most striking limitation to date, particularly when considering DNA-based methods, hampering the extension of genetic findings to the entire population63,64. Limited research resources in under-represented populations are likely to have profound cascading effects for future advances in clinical practice, including pharmacological and behavioural treatment. Fortunately, there are major initiatives underway to re-balance these biases65–67.
Our second aim was to provide a clear account of how close NDDs are to one another aetiologically. We found that, while meta-analytic estimates indicated moderate genetic overlap, the degree of heterogeneity in these associations across disorders was large. We found substantial genetic correlations between ASD and ADHD, between ADHD and motor disorders, and between communication disorders and specific learning disorders. In contrast, genetic overlap was only moderate between communication disorders and motor disorders and very weak between ADHD and specific learning disorders, which is consistent with the degree of symptom resemblance across these disorders.
Although we were able to explore general patterns of variation and co-occurrence, the aetiology of specific NDDs and of their associations could not be comprehensively characterized. The research gaps that we identified highlight an imbalance in focus across NDDs in developmental behaviour genetics research. When considering our first aim, we could identify only 2 family-based studies that investigated the genetic contributions to intellectual disabilities, compared with 121 family-based and 14 SNP-based studies for ADHD, and 36 family-based and 15 SNP-based studies for ASD. This lack of research on intellectual disabilities, an NDD affecting 2.5% of children in the United Kingdom68 (more than double the prevalence rate of ASD69), is reflected in and probably partly due to the lack of funding bodies devoted to researching NDDs other than ASD and ADHD, as well as a lack of publicly available data repositories and resources (for example, refs. 70–72).
We also identified very few studies that examined the aetiology of motor disorders, another neurodevelopmental condition showing significant prevalence rates of 5–6% in school-aged children73. This unbalanced research focus, which extends far beyond genetically informative research to touch developmental and therapeutic research74–77, has led to an uneven distribution of knowledge, which could lead to limited access to interventions for children with NDDs other than ASD, ADHD and dyslexia78.
The lack of equity in focus across NDDs was pronounced in analyses addressing our third aim. Sources of co-occurrence between NDDs and DICCs could only be investigated between ADHD and conduct disorder, between ADHD and oppositional defiant disorder, and between ASD and conduct disorder. Considering that in the DSM-5 the DICCs category comprises eight distinct disruptive disorders, this highlights a major gap in our knowledge.
This meta-analysis provides a holistic view of genetic and environmental contributions to all NDDs and commonly co-occurring developmental disorders, revealing that NDDs are just as strongly genetically correlated with other NDDs as most of them are with DICCs. Our work identifies a lack of balance in research across different NDDs, which calls for future genetic research to focus on less-investigated disorders. We provide knowledge about patterns of aetiological co-occurrence between NDDs as well as between NDDs and DICCs, which we hope will inform clinical and educational diagnostics and practice, resulting, for example, in expanded diagnostic screening.
Methods
The protocol for the current meta-analysis was registered with the international prospective register of systematic reviews (PROSPERO) and can be accessed at the following link: https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=230158. This meta-analysis was conducted in line with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines79. The PRISMA 2020 Checklist and PRISMA 2020 for Abstracts Checklist79 are included in Supplementary Notes 9 and 10. The code and master extraction tables are available at https://github.com/CoDEresearchlab/Meta_analysis_NDDs_DICCs.
Identification of relevant studies
A total of 296 studies were included in the meta-analysis (Fig. 2). Studies were identified during three searches: the primary search (Supplementary Fig. 29a), conducted on 20 January 2021; the secondary (confirmatory) search (Supplementary Fig. 29b), conducted on 15 April 2021; and the additional search of other relevant meta-analyses and reviews, finalized on 4 May 2021. The searches were conducted across three platforms: Web of Science, Ovid Medline and Ovid Embase. The outputs were managed with the aid of Covidence (Veritas Health Innovation), which is a web-based collaboration software platform that streamlines the production of systematic and other literature reviews (https://www.covidence.org/). An in-depth description of indexes, timespans, search strategy and key words is included in Supplementary Note 11. All studies included in the meta-analysis are listed in Supplementary Tables 31–36.
Screening and inclusion criteria
After the initial searches were conducted and duplicate studies were removed, 8,087 studies met the criteria for the first stage of screening, which involved title and abstract scanning. All titles and abstracts were screened by two independent, blinded reviewers to ensure inter-rater agreement. Conflicts were resolved by a third independent reviewer, and inter-rater reliability was calculated as the number of agreements divided by the total number of studies screened, multiplied by 100% (Fig. 2). After this initial screening phase, 6,834 studies were excluded as they were deemed not relevant to the purpose of the current meta-analysis.
The title and abstract screening process resulted in a total of 1,253 potentially eligible studies. The full text of each study was screened by two independent, blinded reviewers. Reviewer discrepancies were identified and resolved by a third independent reviewer. The inter-rater reliability statistic was calculated (Fig. 2). This resulted in 289 eligible articles. In addition, during full-text screening, relevant review articles, meta-analyses, editorials and conference abstracts were flagged to aid the potential discovery of further relevant studies by either screening the reference lists or contacting the authors of conference abstracts. Through this process, 7 additional studies were identified, which resulted in a total of 296 studies included in the current meta-analysis (Fig. 2). Studies were considered relevant and selected to be included at the next screening stage on the basis of the following criteria.
First, studies were included only if 75% or more of the sample consisted of children and/or adolescents. On the basis of guidelines from the World Health Organization (https://www.who.int/health-topics/adolescent-health#tab=tab_1), we defined the period from childhood to the end of adolescence as ranging from age 4, the earliest age for compulsory schooling, to age 24, the end of adolescence. Second, we included studies that had measured NDDs and DICCs considering formal clinical diagnoses, clinical cut-offs and/or quantitative measures of symptoms. Third, studies were selected only if they featured data on at least one NDD (Aim 1), at least two NDDs (Aim 2) or at least one NDD and one DICC (Aim 3).
Fourth, studies using family-based designs had to have reported at least one estimate of heritability (h2), shared environmental (c2) or non-shared environmental influence (e2), or genetic or environmental correlations. We included only single-generation family designs—that is, studies that used twin designs80, sibling comparisons81 or extended twin designs82. We excluded multiple-generation family designs (for example, children of twins83 and in vitro fertilization84) due to the potential confounding in the genetic and environmental estimates that could have resulted from including parental traits in the models decomposing the covariance between family members85.
Fifth, studies using genomic designs were included only if they reported at least one SNP-based heritability estimate and/or a genetic correlation (rA). Eligible SNP-based methods to quantify the proportion of phenotypic variance accounted for by common SNPs included genome-based restricted maximum likelihood86, linkage-disequilibrium score regression21 and SbayesS, which is a Bayesian approach to the analysis of genome-wide association summary data87. Each method is described in greater detail in Supplementary Note 12. Sixth, studies that selected participants on the basis of other diagnoses not related to NDD or DICC categories or on the basis of extreme vulnerability or environmental insult unrelated to NDDs or DICCs, such as alcohol abuse, were not included. Lastly, only studies published in English were included. Studies deemed eligible on the basis of full-text scanning were also scored in terms of their scientific quality and risk of bias by two reviewers (see the details on the quality-scoring checklist in Supplementary Note 13).
Data extraction
Data extraction was conducted by the primary reviewer. Issues and uncertainties were resolved through discussion with co-authors. Missing data were requested from study authors via email or ResearchGate (for details, see Supplementary Note 14). The extracted data were compiled in a table, including information on study reference, the project/cohort name, the study design (for example, classical twin study), the model reported (for example, the full ACE model; when multiple models were reported, the best-fitting model was selected for data synthesis), the overall number of participants and numbers of participants in subgroups (for example, the number of monozygotic versus dizygotic twins), the average age and age range of the sample, the cohort country or countries of origin, the participants’ ancestry (defined in terms of the percentage of participants of European ancestry in the samples), the broad types of NDD and DICC included (for example, specific learning disorder), the subtypes of NDD and/or DICC included (for example, dyslexia), the specific phenotypes measured (for example, reading fluency), the measure statistics (for example, binary (diagnosis) or continuous (symptom continua)), the measure (for example, Conners rating scale for ADHD) and rater (for example, parent reports), the covariates included in the analyses (for example, age and sex), statistics (for example, family-based heritability and SNP-based genetic correlation), and the estimated statistics and the provided index of measurement variance (for example, standard error). The master extraction tables, ‘Extraction_heritability’ and ‘Extraction_correlations’, are available at https://github.com/CoDEresearchlab/Meta_analysis_NDDs_DICCs.
Estimates of heritability and shared and non-shared environmental influences were extracted as reported by individual studies. When studies only reported twin correlations, the variance components were calculated using Falconer’s formula88, as follows:
where rMZ is the monozygotic twin correlation and rDZ is the dizygotic twin correlation.
Genetic, shared and non-shared environmental correlations were extracted only if reported by individual studies. For studies where neither standard deviations, standard errors nor 95% confidence intervals were reported, the 95% confidence intervals were calculated using the Cir function implemented in the R package psychometric89,90, on the basis of the sample size of the study, and subsequently converted to standard errors via dividing the difference between the upper and lower bounds by 3.92 (ref. 91).
Data synthesis
Heritability and environmental influences reported by the selected studies were synthesized using a multilevel random-effects meta-analysis in metafor for R55. We used heritability/environmental influences and genetic/environmental correlation coefficients, along with standard errors, as the measures of effect size27. However, to avoid the risk of type I error introduced by the distribution characteristics of the correlation coefficient92, we transformed all estimates using Fisher’s z. The effect sizes were then weighted by their inverse variance weights so that larger samples were given more weighting, and the standard error for the common effect size resulted as a function of the allocated weights. To present the results, Fisher’s z was transformed back to variance components and correlation coefficients93. Multilevel random-effects models enabled varying true effect sizes across studies. We introduced a two-level structure to account for nested effects underlying heterogeneity and clustering across studies (Level 1, individual clustering; Level 2, cohort clustering). Given that some NDDs have different prevalence rates in males and females52–54, we meta-analysed studies that provided sex-specific estimates in separate models to minimize sample heterogeneity across studies, and we report separate grand estimates for combined, male-only and female-only samples.
Data reporting
We report transdiagnostic grand estimates across all disorders and for broad NDD categories, comprising all studies that investigated the aetiology of a disorder using diagnoses, categorical measures or quantitative measures. For example, the broad ADHD phenotype includes studies that have measured ADHD using diagnoses, clinical cut-offs and continuous measures of ADHD traits, such as checklists and questionnaires. The only exception is intellectual disability. We did not consider quantitative measures of general intelligence as indexing a continuum of intellectual disability given that intellectual disability, as described in the DSM-5, is a complex disorder, characterized by impairments not only in intellectual performance but also in adaptive functioning and communication3,44. Finally, we considered specific manifestations of NDDs—for example, beyond ADHD, we also consider the hyperactive/impulsive and inattentive subtypes separately. The results for all sub-categories of NDDs and for their co-occurrence with other disorders are reported in Supplementary Note 2, Supplementary Figs. 2 and 3, and Supplementary Tables 2, 4 and 6.
Aggregation of non-independent effects
Multilevel meta-analytic models allow us to account for non-independence of estimates derived from partly or completely overlapping samples (that is, estimates obtained from multiple studies that have used the same cohort of participants). To further account for the non-independence of sampling variance (that is, when sampling errors correlate because data from partly the same individuals are used to estimate multiple effect sizes), we also aggregated multiple estimates within each individual study (for example, estimates at multiple time points derived from the same study). Dependent effect sizes were aggregated at the level of each study using the R package Meta-Analysis with Mean Differences90,94, applying a default correlation between estimates of 0.5. We conducted several sensitivity analyses comparing different aggregation methods—that is, aggregating at the level of the study, cohort and country, and varying the assumed correlation between dependent effect sizes (0.5, 0.3 and 0.9). The results of these additional checks are presented in Supplementary Fig. 30 and discussed in Supplementary Note 15. Since differences in aggregation strategy did not result in significant differences in meta-analytic effects, we report the results obtained when the correlation between dependent effect sizes was set to 0.5.
Bias and heterogeneity assessment
The potential for publication bias was explored using funnel plots and Egger’s linear regression95. The proportion of heterogeneity across estimates was estimated using the I2 statistic, which calculates the fraction of variance across studies that can be attributed to heterogeneity rather than chance96–98. The I2 statistic was computed as the proportion of the true variance of true effects to the variance of the observed effects, in line with the following formula:
where VTRUE is the variation of true effects and VOBS is the variation due to sampling error. In other words, I2 can be interpreted as the dispersion of observed effects compared with the dispersion that would be predicted just from sampling error. The I2 statistic also provides insight into the degree to which confidence intervals from individual studies are independent. We also conducted outlier case identification analysis, followed by re-calculation of the I2 estimates after removing studies considered to be outliers99. Studies having a substantial impact on the grand estimates and heterogeneity were identified using influential case identification analysis99. Heterogeneity assessment analyses were conducted using the metafor49, meta100 and dmetar101 packages in R90.
Moderation analyses
We tested for the effects of several moderators. The moderator terms were selected on the basis of the available data, considering the completeness of the reported moderator variables. We implemented a >50% rule of thumb—that is, if 50% or more studies reported data on the moderating variable, we included this moderator in our analyses. For example, less than 50% of studies reported the percentage of participants of Asian ancestry in the sample; hence, we did not include the percentage of Asian participants in the moderation analyses. We considered the following ten moderators: age group, design, type of model, rater, measurement, percentage of individuals who identified as White, number of covariates included in the analysis, measure adopted, country and specific phenotype measured. Each moderator is described in greater detail in Supplementary Note 3. The moderation analyses were conducted using a two-step procedure. First, only studies that reported data on the level of the moderator were selected (for example, only studies reporting estimates for adolescents). Second, analyses stratified by levels of the moderator were run using a multilevel random-effects meta-analysis in metafor for R—for example, a grand estimate was derived for adolescents and subsequently compared with estimates for other developmental stages (that is, childhood and middle childhood) using the same procedure. We report unstratified estimates (Supplementary Tables 1, 3 and 5) and estimates stratified by the specific phenotype measured (Supplementary Tables 2, 4 and 6), age category (Supplementary Tables 19–21), country (Supplementary Tables 22–24) and ancestry (Supplementary Tables 25–27) in the main text, whereas estimates stratified by all other moderators are reported in Supplementary Tables 37–50.
Deviations from the PROSPERO pre-registered protocol
Although we followed the preregistered plan step by step, we made some deviations from the plan on the basis of the availability of software and evidence. We describe our deviations from the preregistered protocol below.
As opposed to the first (primary) literature search, which followed the procedure described in the protocol, in the second (confirmatory) literature search we included an additional set of terms to identify studies that measured specific learning disorders and communication disorders on a quantitative scale. For the details, see Supplementary Note 11.
In the protocol, we indicated that study screening would be documented on an Excel spreadsheet. Instead, we used Covidence (https://www.covidence.org/), a software that automatically enables the double-blinded screening of title and abstract, as well as full-text screening and study selection, without the need for external recording of decisions.
Finally, while all 296 papers were assessed for publication reporting bias (Supplementary Note 7, Supplementary Tables 13–15 and Supplementary Figs. 8–24), the first 82 papers that were extracted (27.7% of the total) were also assessed for study quality using the checklist provided by Kmet et al.102 (Supplementary Note 13 and Supplementary Fig. 25).
Certainty assessment
We evaluated our confidence in the body of research included in the present meta-analysis on the basis of a number of key factors: (1) the sample size of each study, (2) the consistency of findings across studies, and (3) study quality and risk of publication bias.
Because differences in sample size can introduce an imbalance in the power to estimate effects reliably across studies, in our meta-analysis we weighted each estimate by the standard errors. Estimates reported by studies conducted in larger samples had smaller standard errors and were therefore given more weight than those reported by studies conducted in smaller samples.
The consistency of findings across studies was assessed by visually examining forest plots. Overall, we did not find significant differences between estimates.
Study quality and risk of bias were assessed in line with the framework proposed by Kmet et al.102 (Supplementary Note 13 and Supplementary Fig. 25). We applied Egger’s regression and inspected funnel plots to examine the impact of publication bias on our results; the outcomes of these analyses are reported in Supplementary Note 7, Supplementary Tables 13–15 and Supplementary Figs. 8–24.
On the basis of these criteria, we place confidence in the results of the current meta-analysis showing that (1) NDDs in childhood and adolescence are highly heritable; (2) the pattern of co-occurrence between NDDs is complex, and while some NDDs are closely related, others show little genetic overlap; and (3) NDDs show a moderate-to-strong genetic overlap with DICCs.
Limitations of the review process
The review process of the current meta-analysis does not come without limitations. One limitation is our sole focus on childhood and adolescence. A second limitation relates to our choice of focusing on specific co-occurring conditions, DICCs, without considering other neurological disorders that have been found to co-occur with NDDs, such as epilepsy, cerebral palsy, or sleep or psychiatric disorders. The inclusion of a wider range of co-occurring conditions could have resulted in a more detailed characterization of aetiological overlaps between NDDs and other conditions.
A third limitation is that the current meta-analysis focused only on single-generation studies—that is, twin and sibling studies—and excluded multi-generational family designs, such as children-of-twins and in-vitro-fertilization studies. Future studies focusing on multi-generational designs could provide valuable insights into the roles that parental genotypes and correlated environmental influences play in offspring’s NDDs and their co-occurring conditions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Notes 1–15, Tables 1–50, Figs. 1–30 and References.
Acknowledgements
This meta-analysis was funded by a starting grant awarded to M.M. by the School of Biological and Behavioural Sciences at Queen Mary University of London (grant no. QMULMAL18). A.G. is supported by a Queen Mary School of Biological and Behavioural Sciences PhD Fellowship awarded to M.M. G.M. was in receipt of a Klingenstein Third Generation Foundation fellowship (grant no. 20212999). F.P. is supported by a National Institutes of Health Subaward via the Regents of the University of California, Riverside (grant no. AG046938). K.R. is supported by a Sir Henry Wellcome Postdoctoral Fellowship. This research was funded in whole or in part by the Wellcome Trust (grant no. 213514/Z/18/Z). For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author contributions
A.G., M.M. and Y.I.A. conceived and designed the study. A.G., L.Y.L., M.D., F.P. and E.D. conducted the literature search and study screening. A.G. extracted and analysed the data. A.G. and M.M. wrote the paper with helpful contributions from Y.I.A., G.M., A.G.A., J.A.-B., F.P., A.R. and K.R. All authors contributed to the interpretation of the data, provided critical feedback on paper drafts and approved the final draft.
Peer review
Peer review information
Nature Human Behaviour thanks Elena Grigorenko, Hayley Mountford, Marietta Papadatou-Pastou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
The data that support the findings, including the master extraction tables, are available at https://github.com/CoDEresearchlab/Meta_analysis_NDDs_DICCs.
Code availability
The code for all analyses is available at https://github.com/CoDEresearchlab/Meta_analysis_NDDs_DICCs.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Agnieszka Gidziela, Email: a.gidziela@qmul.ac.uk.
Margherita Malanchini, Email: m.malanchini@qmul.ac.uk.
Supplementary information
The online version contains supplementary material available at 10.1038/s41562-023-01530-y.
References
- 1.Thapar, A. & Rutter, M. in Rutter’s Child and Adolescent Psychiatry (eds Thapar, A. et al.) 31–40 (Wiley, 2015).
- 2.Dietrich KN, et al. Principles and practices of neurodevelopmental assessment in children: lessons learned from the Centers for Children’s Environmental Health and Disease Prevention Research. Environ. Health Perspect. 2005;113:1437–1446. doi: 10.1289/ehp.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neurodevelopmental Disorders: DSM-5 Selections (American Psychiatric Association, 2015).
- 4.Hyman SL, Levy SE, Myers SM. Identification, evaluation, and management of children with autism spectrum disorder. Pediatrics. 2020;145:e20193447. doi: 10.1542/peds.2019-3447. [DOI] [PubMed] [Google Scholar]
- 5.Gillberg C. The ESSENCE in child psychiatry: early symptomatic syndromes eliciting neurodevelopmental clinical examinations. Res. Dev. Disabil. 2010;31:1543–1551. doi: 10.1016/j.ridd.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 6.McGovern CW, Sigman M. Continuity and change from early childhood to adolescence in autism. J. Child Psychol. Psychiatry. 2005;46:401–408. doi: 10.1111/j.1469-7610.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 7.Faraone SV, Biederman J, Mick E. The age-dependent decline of attention deficit hyperactivity disorder: a meta-analysis of follow-up studies. Psychol. Med. 2006;36:159–165. doi: 10.1017/S003329170500471X. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, et al. Provisional tic disorder is not so transient. Sci. Rep. 2019;9:3951. doi: 10.1038/s41598-019-40133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis EM, Thal DJ. Early language delay and risk for language impairment. Perspect. Lang. Learn. Educ. 2008;15:93–100. doi: 10.1044/lle15.3.93. [DOI] [Google Scholar]
- 10.McDowell MJ, Lesslie JM. Long-term outcomes for children with neurodevelopmental disorders: are they core business for paediatricians? J. Paediatr. Child Health. 2018;54:469–473. doi: 10.1111/jpc.13871. [DOI] [PubMed] [Google Scholar]
- 11.Hechtman L, et al. Functional adult outcomes 16 years after childhood diagnosis of attention-deficit/hyperactivity disorder: MTA results. J. Am. Acad. Child Adolesc. Psychiatry. 2016;55:945–952. doi: 10.1016/j.jaac.2016.07.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roux AM, et al. Postsecondary employment experiences among young adults with an autism spectrum disorder. J. Am. Acad. Child Adolesc. Psychiatry. 2013;52:931–939. doi: 10.1016/j.jaac.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCauley JB, Elias R, Lord C. Trajectories of co-occurring psychopathology symptoms in autism from late childhood to adulthood. Dev. Psychopathol. 2020;32:1287–1302. doi: 10.1017/S0954579420000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen P, Gillberg C. Natural outcome of ADHD with developmental coordination disorder at age 22 years: a controlled, longitudinal, community-based study. J. Am. Acad. Child Adolesc. Psychiatry. 2000;39:1424–1431. doi: 10.1097/00004583-200011000-00017. [DOI] [PubMed] [Google Scholar]
- 15.Tick B, Bolton P, Happé F, Rutter M, Rijsdijk F. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J. Child Psychol. Psychiatry. 2016;57:585–595. doi: 10.1111/jcpp.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faraone SV, Larsson H. Genetics of attention deficit hyperactivity disorder. Mol. Psychiatry. 2019;24:562–575. doi: 10.1038/s41380-018-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolas MA, Burt SA. Genetic and environmental influences on ADHD symptom dimensions of inattention and hyperactivity: a meta-analysis. J. Abnorm. Psychol. 2010;119:1–17. doi: 10.1037/a0018010. [DOI] [PubMed] [Google Scholar]
- 18.Bishop DV. Which neurodevelopmental disorders get researched and why? PLoS ONE. 2010;5:e15112. doi: 10.1371/journal.pone.0015112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheesman R, et al. Childhood behaviour problems show the greatest gap between DNA-based and twin heritability. Transl. Psychiatry. 2017;7:1284. doi: 10.1038/s41398-017-0046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulik-Sullivan BK, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grove J, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demontis D, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersson E, Anckarsäter H, Gillberg C, Lichtenstein P. Different neurodevelopmental symptoms have a common genetic etiology. J. Child Psychol. Psychiatry. 2013;54:1356–1365. doi: 10.1111/jcpp.12113. [DOI] [PubMed] [Google Scholar]
- 25.Brimo K, et al. The co-occurrence of neurodevelopmental problems in dyslexia. Dyslexia. 2021;27:277–293. doi: 10.1002/dys.1681. [DOI] [PubMed] [Google Scholar]
- 26.Knopik, V. S., Neiderhiser, J. M., DeFries, J. C. & Plomin, R. Behavioral Genetics (Worth, 2017).
- 27.Andersson A, et al. The strength of the genetic overlap between ADHD and other psychiatric symptoms—a systematic review and meta-analysis. J. Child Psychol. Psychiatry. 2020;61:1173–1183. doi: 10.1111/jcpp.13233. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, et al. Investigating shared genetic basis across tourette syndrome and comorbid neurodevelopmental disorders along the impulsivity-compulsivity spectrum. Biol. Psychiatry. 2021;90:317–327. doi: 10.1016/j.biopsych.2020.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichtenstein P, Carlström E, Råstam M, Gillberg C, Anckarsäter H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am. J. Psychiatry. 2010;167:1357–1363. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- 30.Paloyelis Y, Rijsdijk F, Wood AC, Asherson P, Kuntsi J. The genetic association between ADHD symptoms and reading difficulties: the role of inattentiveness and IQ. J. Abnorm. Child Psychol. 2010;38:1083–1095. doi: 10.1007/s10802-010-9429-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin NC, Piek JP, Hay D. DCD and ADHD: a genetic study of their shared aetiology. Hum. Mov. Sci. 2006;25:110–124. doi: 10.1016/j.humov.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Taylor MJ, et al. Language and traits of autism spectrum conditions: evidence of limited phenotypic and etiological overlap. Am. J. Med. Genet. B. 2014;165:587–595. doi: 10.1002/ajmg.b.32262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dworzynski K, et al. Developmental path between language and autistic-like impairments: a twin study. Infant Child Dev. Int. J. Res. Pract. 2008;17:121–136. doi: 10.1002/icd.536. [DOI] [Google Scholar]
- 34.Offord DR, et al. Outcome, prognosis, and risk in a longitudinal follow-up study. J. Am. Acad. Child Adolesc. Psychiatry. 1992;31:916–923. doi: 10.1097/00004583-199209000-00021. [DOI] [PubMed] [Google Scholar]
- 35.Frick, P. J. & Loney, B. R. in Handbook of Disruptive Behavior Disorders (eds Quay, H. C. & Hogan, A. E.) 507–524 (Springer, 1999).
- 36.Frick, P. J. in Handbook of Child Psychopathology (eds Ollendick, T. H. & Hersen, M.) 213–237 (Springer, 1998).
- 37.Lahey, B. B. & Loeber, R. in Disruptive Behavior Disorders in Childhood (ed. Routh, D. K.) 139–180 (Springer, 1994).
- 38.Thorell LB, Wåhlstedt C. Executive functioning deficits in relation to symptoms of ADHD and/or ODD in preschool children. Infant Child Dev. 2006;15:503–518. doi: 10.1002/icd.475. [DOI] [Google Scholar]
- 39.Bayard F, et al. Distinct brain structure and behavior related to ADHD and conduct disorder traits. Mol. Psychiatry. 2020;25:3020–3033. doi: 10.1038/s41380-018-0202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin NC, Levy F, Pieka J, Hay DA. A genetic study of attention deficit hyperactivity disorder, conduct disorder, oppositional defiant disorder and reading disability: aetiological overlaps and implications. Int. J. Disabil. Dev. Educ. 2006;53:21–34. doi: 10.1080/10349120500509992. [DOI] [Google Scholar]
- 41.Moffitt TE. The neuropsychology of conduct disorder. Dev. Psychopathol. 1993;5:135–151. doi: 10.1017/S0954579400004302. [DOI] [Google Scholar]
- 42.Rubia K. “Cool” inferior frontostriatal dysfunction in attention-deficit/hyperactivity disorder versus “hot” ventromedial orbitofrontal–limbic dysfunction in conduct disorder: a review. Biol. Psychiatry. 2011;69:e69–e87. doi: 10.1016/j.biopsych.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 43.Bronsard G, Botbol M, Tordjman S. Aggression in low functioning children and adolescents with autistic disorder. PLoS ONE. 2010;5:e14358. doi: 10.1371/journal.pone.0014358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moffitt, T. E., Caspi, A., Rutter, M. & Silva, P. A. Sex Differences in Antisocial Behaviour: Conduct Disorder, Delinquency, and Violence in the Dunedin Longitudinal Study (Cambridge Univ. Press, 2001).
- 45.Jones AP, et al. Phenotypic and aetiological associations between psychopathic tendencies, autistic traits, and emotion attribution. Crim. Justice Behav. 2009;36:1198–1212. doi: 10.1177/0093854809342949. [DOI] [Google Scholar]
- 46.O’Nions E, et al. Examining the genetic and environmental associations between autistic social and communication deficits and psychopathic callous-unemotional traits. PLoS ONE. 2015;10:e0134331. doi: 10.1371/journal.pone.0134331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skuse DH. Rethinking the nature of genetic vulnerability to autistic spectrum disorders. Trends Genet. 2007;23:387–395. doi: 10.1016/j.tig.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 48.Baselmans BM, Yengo L, van Rheenen W, Wray NR. Risk in relatives, heritability, SNP-based heritability, and genetic correlations in psychiatric disorders: a review. Biol. Psychiatry. 2021;89:11–19. doi: 10.1016/j.biopsych.2020.05.034. [DOI] [PubMed] [Google Scholar]
- 49.Viechtbauer, W. metafor. R package version 3.0.2 (2015).
- 50.Davis OS, et al. The correlation between reading and mathematics ability at age twelve has a substantial genetic component. Nat. Commun. 2014;5:4204. doi: 10.1038/ncomms5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newsome J, Boisvert D, Wright JP. Genetic and environmental influences on the co-occurrence of early academic achievement and externalizing behavior. J. Crim. Justice. 2014;42:45–53. doi: 10.1016/j.jcrimjus.2013.12.002. [DOI] [Google Scholar]
- 52.Christensen DL, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR Surveill. Summ. 2018;65:1–23. doi: 10.15585/mmwr.ss6513a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.May T, Sciberras E, Brignell A, Williams K. Autism spectrum disorder: updated prevalence and comparison of two birth cohorts in a nationally representative Australian sample. BMJ Open. 2017;7:e015549. doi: 10.1136/bmjopen-2016-015549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polanczyk G, De Lima MS, Horta BL, Biederman J, Rohde LA. The worldwide prevalence of ADHD: a systematic review and metaregression analysis. Am. J. Psychiatry. 2007;164:942–948. doi: 10.1176/ajp.2007.164.6.942. [DOI] [PubMed] [Google Scholar]
- 55.Martin J, et al. A genetic investigation of sex bias in the prevalence of attention-deficit/hyperactivity disorder. Biol. Psychiatry. 2018;83:1044–1053. doi: 10.1016/j.biopsych.2017.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Global Burden of Disease Collaborative Network Global Burden of Disease Study 2016 (GBD 2016) Results (Institute for Health Metrics and Evaluation, 2017).
- 57.Genetics for all. Nat. Genet. 51, 579 (2019). [DOI] [PubMed]
- 58.Whose genomics? Nat. Hum. Behav. 3, 409–410 (2019). [DOI] [PubMed]
- 59.Silventoinen K, et al. Genetic and environmental variation in educational attainment: an individual-based analysis of 28 twin cohorts. Sci. Rep. 2020;10:12681. doi: 10.1038/s41598-020-69526-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rimfeld K, et al. Genetic influence on social outcomes during and after the Soviet era in Estonia. Nat. Hum. Behav. 2018;2:269–275. doi: 10.1038/s41562-018-0332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abdellaoui A, et al. Genetic correlates of social stratification in Great Britain. Nat. Hum. Behav. 2019;3:1332–1342. doi: 10.1038/s41562-019-0757-5. [DOI] [PubMed] [Google Scholar]
- 62.Belsky DW, et al. Genetics and the geography of health, behaviour and attainment. Nat. Hum. Behav. 2019;3:576–586. doi: 10.1038/s41562-019-0562-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin AR, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finer S, et al. Cohort profile: East London Genes & Health (ELGH), a community-based population genomics and health study in British Bangladeshi and British Pakistani people. Int. J. Epidemiol. 2020;49:20–21i. doi: 10.1093/ije/dyz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wright J, et al. Cohort profile: the Born in Bradford multi-ethnic family cohort study. Int. J. Epidemiol. 2013;42:978–991. doi: 10.1093/ije/dys112. [DOI] [PubMed] [Google Scholar]
- 67.Ramsay M, Sankoh O, as members of the AWI-Gen study and the H3Africa Consortium. African partnerships through the H3Africa Consortium bring a genomic dimension to longitudinal population studies on the continent. Int. J. Epidemiol. 2016;45:305–308. doi: 10.1093/ije/dyv187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Estimates of the Population for the UK, England and Wales, Scotland and Northern Ireland (Office for National Statistics, 2020); https://www.ons.gov.uk/peoplepopulationandcommunity/populationandmigration/populationestimates/datasets/populationestimatesforukenglandandwalesscotlandandnorthernireland
- 69.Mehlmann-Wicks, J. Autism Spectrum Disorder (British Medical Association, 2020); https://www.bma.org.uk/what-we-do/population-health/improving-the-health-of-specific-groups/autism-spectrum-disorder
- 70.AGRE—Autism Genetic Resource Exchange (Autism Speaks, 2022); https://www.autismspeaks.org/agre
- 71.About iPSYCH (iPSYCH, 2022); https://ipsych.dk/en/about-ipsych/
- 72.Psychiatric Genomics Consortium (Psychiatric Genomics Consortium, 2022); https://www.med.unc.edu/pgc/
- 73.Zwicker JG, Missiuna C, Harris SR, Boyd LA. Developmental coordination disorder: a review and update. Eur. J. Paediatr. Neurol. 2012;16:573–581. doi: 10.1016/j.ejpn.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 74.Valentine AZ, et al. A systematic review evaluating the implementation of technologies to assess, monitor and treat neurodevelopmental disorders: a map of the current evidence. Clin. Psychol. Rev. 2020;80:101870. doi: 10.1016/j.cpr.2020.101870. [DOI] [PubMed] [Google Scholar]
- 75.Valentine AZ, et al. Implementation of telehealth services to assess, monitor, and treat neurodevelopmental disorders: systematic review. J. Med. Internet Res. 2021;23:e22619. doi: 10.2196/22619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McGregor KK. How we fail children with developmental language disorder. Lang. Speech Hear. Serv. Sch. 2020;51:981–992. doi: 10.1044/2020_LSHSS-20-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khan K, Hall CL, Davies EB, Hollis C, Glazebrook C. The effectiveness of web-based interventions delivered to children and young people with neurodevelopmental disorders: systematic review and meta-analysis. J. Med. Internet Res. 2019;21:e13478. doi: 10.2196/13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bigby C. Social inclusion and people with intellectual disability and challenging behaviour: a systematic review. J. Intellect. Dev. Disabil. 2012;37:360–374. doi: 10.3109/13668250.2012.721878. [DOI] [PubMed] [Google Scholar]
- 79.Page MJ, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Syst. Rev. 2021;10:89. doi: 10.1186/s13643-021-01626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rijsdijk FV, Sham PC. Analytic approaches to twin data using structural equation models. Brief. Bioinform. 2002;3:119–133. doi: 10.1093/bib/3.2.119. [DOI] [PubMed] [Google Scholar]
- 81.Kendler KS, et al. A novel sibling-based design to quantify genetic and shared environmental effects: application to drug abuse, alcohol use disorder and criminal behavior. Psychol. Med. 2016;46:1639–1650. doi: 10.1017/S003329171500224X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Posthuma D, Boomsma DI. A note on the statistical power in extended twin designs. Behav. Genet. 2000;30:147–158. doi: 10.1023/A:1001959306025. [DOI] [PubMed] [Google Scholar]
- 83.Eley TC, et al. The intergenerational transmission of anxiety: a children-of-twins study. Am. J. Psychiatry. 2015;172:630–637. doi: 10.1176/appi.ajp.2015.14070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rice F, et al. Disentangling prenatal and inherited influences in humans with an experimental design. Proc. Natl Acad. Sci. USA. 2009;106:2464–2467. doi: 10.1073/pnas.0808798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McAdams TA, et al. Revisiting the children-of-twins design: improving existing models for the exploration of intergenerational associations. Behav. Genet. 2018;48:397–412. doi: 10.1007/s10519-018-9912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang J, Zeng J, Goddard ME, Wray NR, Visscher PM. Concepts, estimation and interpretation of SNP-based heritability. Nat. Genet. 2017;49:1304–1310. doi: 10.1038/ng.3941. [DOI] [PubMed] [Google Scholar]
- 87.Zeng J, et al. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet. 2018;50:746–753. doi: 10.1038/s41588-018-0101-4. [DOI] [PubMed] [Google Scholar]
- 88.Falconer, D. S. Introduction to Quantitative Genetics (Pearson Education, 1996).
- 89.Fletcher, T. D. psychometric. R package version 2.2 (2013).
- 90.R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2020).
- 91.Cohen, J., Cohen, P., West, S. G. & Aiken, L. S. Applied Multiple Regression/Correlation Analysis for the Behavioral Sciences 3rd edn (Routledge, 2013).
- 92.Alexander RA, Scozzaro MJ, Borodkin LJ. Statistical and empirical examination of the chi-square test for homogeneity of correlations in meta-analysis. Psychol. Bull. 1989;106:329–331. doi: 10.1037/0033-2909.106.2.329. [DOI] [Google Scholar]
- 93.Malanchini M, et al. Aggressive behaviour in childhood and adolescence: the role of smoking during pregnancy, evidence from four twin cohorts in the EU-ACTION consortium. Psychol. Med. 2019;49:646–654. doi: 10.1017/S0033291718001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Del Re, A. C., Hoyt, W. T. & Del Re, M. A. MAd. R package version 0.8.2.1 (2014).
- 95.Sterne, J. A. & Egger, M. in Publication Bias in Meta-Analysis: Prevention, Assessment and Adjustments (eds Rothstein, H. R. et al.) 99–110 (Wiley, 2005).
- 96.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 97.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Brit. Med. J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Borenstein M, Higgins JP, Hedges LV, Rothstein HR. Basics of meta-analysis: I2 is not an absolute measure of heterogeneity. Res. Synth. Methods. 2017;8:5–18. doi: 10.1002/jrsm.1230. [DOI] [PubMed] [Google Scholar]
- 99.Viechtbauer W, Cheung MW-L. Outlier and influence diagnostics for meta-analysis. Res. Synth. Methods. 2010;1:112–125. doi: 10.1002/jrsm.11. [DOI] [PubMed] [Google Scholar]
- 100.Balduzzi S, Rücker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid. Based Ment. Health. 2019;22:153–160. doi: 10.1136/ebmental-2019-300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harrer, M., Cuijpers, P., Furukawa, T. & Ebert, D. D. dmetar: Companion R package for the guide ‘Doing meta-analysis in R’. R package version 0.0.9000 (2019).
- 102.Kmet, L. M., Cook, L. S. & Lee, R. C. Standard quality assessment criteria for evaluating primary research papers from a variety of fields. ERAhttps://era.library.ualberta.ca/items/48b9b989-c221-4df6-9e35-af782082280e (2004).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Notes 1–15, Tables 1–50, Figs. 1–30 and References.
Data Availability Statement
The data that support the findings, including the master extraction tables, are available at https://github.com/CoDEresearchlab/Meta_analysis_NDDs_DICCs.
The code for all analyses is available at https://github.com/CoDEresearchlab/Meta_analysis_NDDs_DICCs.