Abstract
Mesenchymal stem cell (MSC) exosomes have been found to attenuate cardiac systolic and diastolic dysfunction in animal models of ischemia. Exosomes carry a plethora of active and inactive proteins as their cargo, which are readily available to the recipient cell for use in intracellular signaling pathways-depending on the stresses, such as ischemia or hypoxia. Among the exosomal proteins are the often-overlooked cargo of transcriptional regulators. These transcriptional regulators influence the transcriptome and subsequently the proteome of recipient cell. Here, we report the transcriptional factors and regulators differentially modulated and their potential role in modulating cardiac function in MSC exosome treated ischemic mice hearts. Our analysis shows ischemic stress modulating transcriptional regulators and factors such as HSF1 and HIF1A in the infarct and peri-infarct areas of ischemic hearts which is mitigated by MSC exosomes. Similarly, STAT3 and SMAD3 are also modulated by MSC exosomes. Interestingly, NOTCH1 and β-catenin were detected in the ischemic hearts. The differential expression of these regulators and factors drives changes in various biological process governed in the ischemic cardiac cells.
We believe these studies will advance our understanding of cardiac dysfunction occurring in the ischemic hearts and lay the groundwork for further studies on the modulation of cardiac function during ischemia by MSC exosomes.
Keywords: Mesenchymal stem cell exosomes, Myocardial ischemia, Post-ischemic fibrosis, Proteomics, Transcription factors
Graphical abstract
Highlights
-
•
MSC exosomes alter myocardial proteomic profiles in ischemic mouse hearts, observed in infarct and peri-infarct regions.
-
•
Activation of dormant developmental signals (Notch-1 and beta-catenin) in ischemic myocardium is activated by MSC exosomes.
-
•
MSC exosomes influence differential regulation of transcription factors in infarct and peri-infarct areas, determining protein expression and ischemic event outcomes.
-
•
Our analysis illuminates the differentially regulated transcription factors in ischemic mouse hearts treated with MSC exosomes.
1. Introduction
Heart failure following myocardial infarction (MI) is the leading cause of morbidity and mortality in the US and worldwide. Advances have led to a decline in death rates following MI; however, there has been an increase in number of patients suffering post-ischemic heart failure [1,2]. The cardiomyocytes and fibroblasts in and around the infarct areas continue to experience hypoxia. Molecular changes in these areas result in death of cardiomyocytes (resulting in systolic dysfunction), and growth of fibroblasts resulting in fibrosis and diastolic dysfunction [3]. Adverse extracellular matrix (ECM) remodeling begins in the heart soon after the index ischemic event triggered by expression and activation of various transcriptional regulators and persists for weeks and months afterwards [4]. The resulting targets of these transcriptional activators in the myocardium govern apoptosis in damaged cardiomyocytes, recruit monocytes, and activate resident cardiac macrophages [5,6], and fibroblast proliferation and trans-differentiation [7,8]. These changes govern adverse ECM turnover leading to cardiac dysfunction [[9], [10], [11]].
Studies using various stem cell exosomes, in particular those derived from mesenchymal stem cells (MSCs), have shown promise in terms of preventing or limiting cardiac damage [[12], [13], [14], [15], [16]]. Our recent studies have shown that MSC exosome treatment reduces infarct size and preserves cardiac systolic function. These effects are mediated through attenuation of markers of inflammation (leukocytosis, activation of p-38MAPK, NF-κB and NLRP3 inflammasome, and expression of TLR9, TLR4 and LOX-1), release of inflammatory cytokines (IL-1β, Il-18), and occurrence of apoptosis and pyroptosis. Other unpublished preliminary studies from our laboratory have shown improvement in post-ischemic diastolic function via changes in collagen and fibronectin expression.
Exosomal proteins have been shown to regulate a variety of biological pathways in recipient cells. These proteins can downregulate apoptosis, oxidation, and caspases and toll-receptor cascade pathways. Exosomes themselves secrete a variety of proteins and transcription factors (TFs) that regulate cell survival, differentiation and metabolic pathways [17].
In a previous study, Gude et al. [18], demonstrated that NOTCH1 is expressed in the ischemic myocardium and is cardioprotective. Building on our previous work investigating the cardioprotective effects of MSC exosomes, we were interested in exploring the molecular mechanisms underlying these effects. We hypothesized that MSC exosomes might activate epigenetically silenced NOTCH1 signaling. Our proteomics data revealed that NOTCH1 signaling was activated in both the infarct and peri-infarct areas of ischemic mice hearts treated with MSC exosomes.
In this report, we present proteomic data on the biological processes in the infarct and peri-infarct areas and differentially regulated TFs in MSC exosome-treated mice hearts (vs. saline-treated mice hearts) subjected to ischemia. These changes in transcription activities and patterns are some of the earliest events in the cascade of changes that ensue after an ischemic event, and as such represent innovative and potentially clinically useful targets for mitigating cardiac damage and rescuing cardiac function.
2. Results
The data are based on the analysis of MS data in the infarct and peri-infarct areas of saline-treated mice hearts and MSC exosome-treated mice hearts subjected to ischemia. Details of the induction of ischemia by LCA ligation and methodology for MS analysis are described under Materials and Methods.
2.1. Proteomic analysis of the effect of MSC exosomes on the ischemic heart
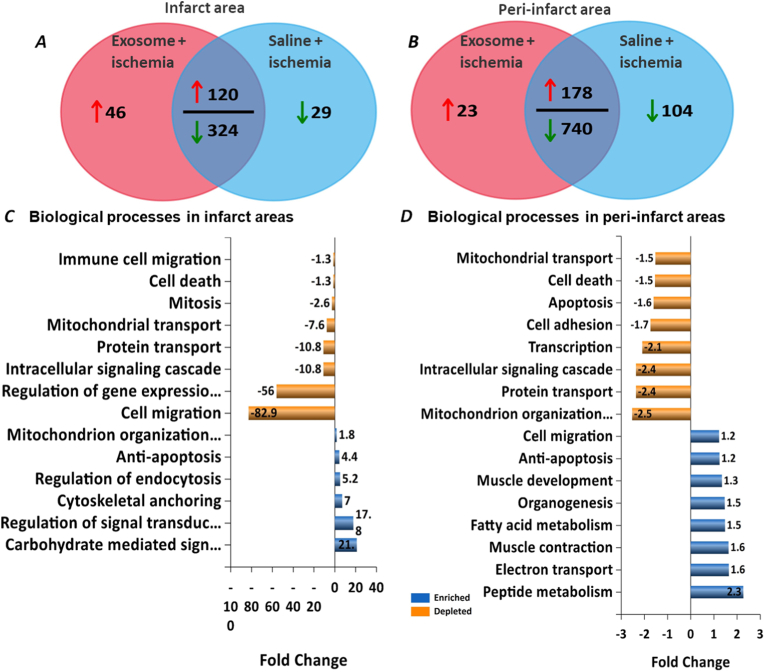
Compared to the saline treated ischemic mice hearts, FunRich™ analysis of proteins [19] detected in the infarct areas of MSC exosome treated ischemic hearts showed 166 proteins elevated, out of which 46 were detected only in infarct areas with MSC exosome treatment. Saline -treated ischemic hearts showed decrease in levels of 353 proteins of which 29 were exclusive to saline treatment (Fig. 1A). Similarly, FunRich™ analysis of the peri-infarct areas showed increase in the levels of 201 proteins with MSC exosome treatment with 23 being exclusive to MSC exosome treatment, and saline treatment downregulated 844 proteins with 104 being exclusive to peri-infarct areas of saline-treated ischemic hearts (Fig. 1B).
Fig. 1.
Analysis of ischemic heart proteome. FunRich analysis of the mice heart proteome by quantitative Venn diagrams show differential regulation of expressed proteins in infarct area (A) and peri-infarct area (B). Biological processes modulated y identified proteins in infarct areas (C) and peri-infarct areas (D).
Compared to the saline-treated hearts, biological processes with MSC exosome treatment such as those governing anti-apoptosis, mitochondrial organization, regulation of signal transduction were enhanced while mitochondrial and protein transport, cell migration, intracellular signaling cascade and cell death were depleted in the infarct areas (Fig. 1C). In the peri-infarct areas, MSC exosome treatment enriched processes governing cell migration, anti-apoptosis, muscle development, muscle contraction and organogenesis, while processes governing cell adhesion, apoptosis, mitochondrial transport, protein transport were depleted (Fig. 1D).
2.2. Proteomic analysis of the effect of MSC exosomes on transcriptional regulators in the ischemic heart
Next, we focused on differential regulation of TFs governing pathways leading to cardiac dysfunction and its rescue by MSC exosomes. We analyzed mice heart proteomes for regulators of post-ischemic pathways.
Integrated Pathway Analysis (IPA Qiagen Bioinformatics) [20], revealed the potential of MSC exosomes to affect differential expression of TFs in infarct (Supplementary Table 1) and peri-infarct areas (Supplementary Table 2) of ischemic mice hearts. Among the upstream regulators modified by MSC exosomes, HSF1, HIF1A, SIRT1, MYCN, NOTCH1 and β-CATENIN 1 were further explored.
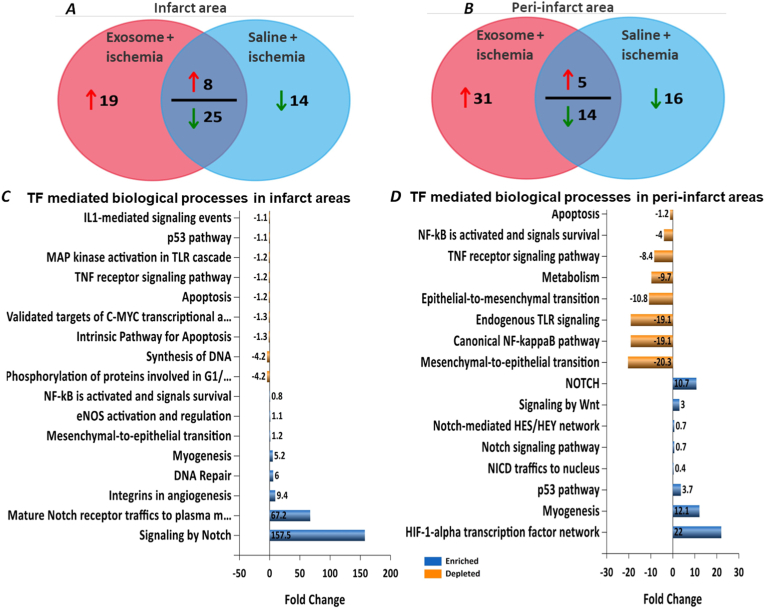
Compared to the saline-treated ischemic mice hearts, FunRich™ analysis of proteins [19] detected in the infarct areas of MSC exosome-treated ischemic hearts showed 27 TFs elevated, out of which 19 were detected only in infarct areas with MSC exosome treatment. Saline treatment only showed decrease in levels of 39 TFs of which 14 were exclusive to saline treatment (Fig. 2A). Similarly, FunRich™ analysis of the peri-infarct areas showed increase in the levels of 36 TFs with MSC exosome treatment with 31 being exclusive to MSC exosome treatment, and saline treatment downregulated 30 TFs with 16 being exclusive to peri-infarct areas of saline treated ischemic hearts (Fig. 2B).
Fig. 2.
FunRich analysis of TFs identified in ischemic heart proteome. FunRich analysis of the TFs identified in the mice heart proteome by quantitative Venn diagrams show differential regulation of expressed TFs in infarct area (A) and peri-infarct area (B). Biological processes governed by identified transcriptional factors in infarct areas (C) and peri-infarct areas (D).
Compared to saline treated heart areas, biological processes with MSC exosome treatment such as those governing NOTCH1 signaling, myogenesis, DNA repair, mesenchymal-to-endothelial transition, eNOS activation were enhanced while p53 pathway, TNF receptor signaling pathway, IL1-mediated signaling, MAP kinase activation and apoptosis were depleted in the infarct areas (Fig. 2C). In the peri-infarct areas, MSC exosome treatment enriched processes governing HIF1-alpha transcription network, myogenesis, and NOTCH signaling pathways while processes governing NF-kappaB pathway, endogenous TLR signaling, epithelial-to-mesenchymal transition apoptosis, TNF receptor signaling pathway were depleted (Fig. 2D).
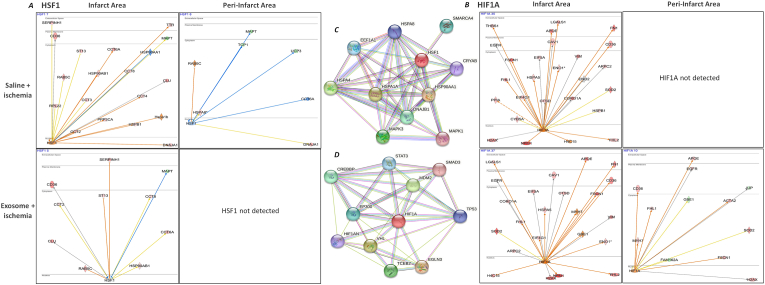
IPA analysis of upstream regulators and transcriptional factors showed an increase in HSF1 and HIF1A in the infarct areas of saline treated ischemic hearts. However, while HSF1 was decreased, HIF1A was not detected in the peri-infarct areas of saline treated ischemic hearts (Fig. 3A and B). MSC exosomes decreased levels of HSF1 but elevated HIF1A levels in the infarct areas. In the peri-infarct areas, IPA analysis failed to detect HSF1, while HIF1A levels were elevated with MSC exosome treatment (Fig. 3A and B). Both functional and physical network STRING [21,22] database analysis showed HSF1 interacting with HSPA1A, HSPA4, MAPK3, DNAJB1, CRYAB, SMARCA4, HSP90AA1 among other. Similarly, STRING analysis of HIF1A displayed its interactions with TP53, STAT3, SMAD3 among others (Fig. 3C and D).
Fig. 3.
Pathways of TFs HSF1 and HIF1A in the ischemic mice hearts
IPA network analysis showed that ischemia stimulates changes in the expression of HSF1 (A) and HIF1A (B) in saline-and MSC exosome-treated ischemic hearts, MSC exosome treatment revealed dramatic changes in these processes. STRING database analysis of HSF1 (C) and HIF1A (D) show their interacting proteins. Prediction legend in Supplementary Fig. 1 describes the network relationships between different molecules.
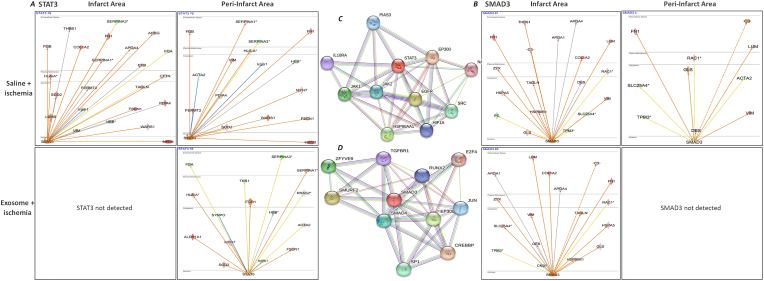
We next analyzed STAT3 and SMAD3. IPA analysis showed an increase in STAT3 and a decrease in SMAD3 in the infarct areas of saline-treated ischemic hearts. There was an increase in both STAT3 and SMAD3 in the peri-infarct areas (Fig. 4A and B). STAT3 was not detected in the infarct areas of MSC exosome-treated hearts while SMAD3 levels were elevated. In the peri-infarct areas, IPA showed decrease in STAT3 levels while SMAD3 was not detected with MSC exosome treatment (Fig. 4A and B). STRING database analysis showed JAK1&2, HIF1A, IL10RA, HSP90AA1 as proteins interacting with STAT3. Similarly, SMAD3 is shown interacting with SMAD4, TGFBR1, E2F4, JUN, CREBBP and SP1 (Fig. 4C and D).
Fig. 4.
Comparison of TFs STAT3 and SMAD3 in the ischemic mice hearts
IPA network analysis showed differences in the regulation of the processes governed by some STAT3 (A) and SMAD3 (B). STAT3 mediated signaling networks were upregulated in saline-treated ischemic hearts. MSC exosome treatment suppressed STAT3 and SMAD3 mediated signaling networks. Prediction legend in Supplementary Fig. 1 describes the network relationships between different molecules.
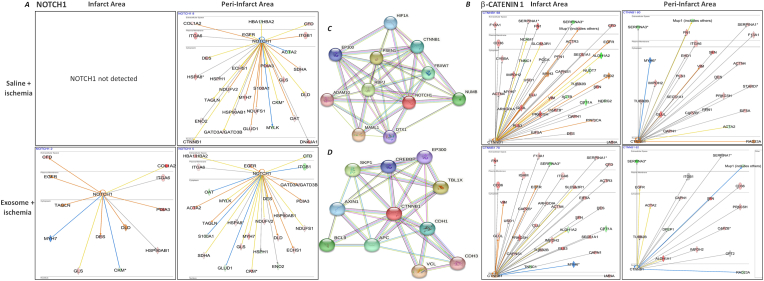
An interesting observation was the detection of NOTCH1 levels in the saline-treated ischemic hearts. Compared to the saline-treated hearts, IPA analysis showed a further increase of NOTCH1 in the MSC exosome treated infarct areas while NOTCH1 was also, but barely, detected in the peri-infarct areas of these hearts. In contrast, NOTCH1 was absent in the infarct areas and decreased in the peri-infarct areas of saline treated ischemic hearts (Fig. 5A and B). IPA analysis of the mice heart proteome also showed differential levels of β-catenin. In saline-treated infarct and peri-infarct areas, β-catenin levels were elevated while MSC exosome treatment decreased its levels in both infarct and peri-infarct areas (Fig. 5A and B). ADAM10, HIF1A, CTNNB1, PSEN1, MAML1 were the NOTCH1 interacting proteins identified through STRING database analysis. CREBBP, TBL1X, SKP1, BCL9 CDH3 were among the proteins shown interacting with β-catenin using the STRING database (Fig. 5C and D).
Fig. 5.
Comparison of TFs NOTCH1 and β-catenin in the ischemic mice hearts
IPA network analysis showed differences in the regulation of the processes governed by some NOTCH1 (A) and beta-catenin-1 (B) mediated signaling networks. Prediction legend in Supplementary Fig. 1 describes the network relationships between different molecules.
3. Discussion
MSC exosomes offer a promising novel way for recovery of various tissues from stress-induced injury [[23], [24], [25]]. These exosomes constitute a class of constitutively secretory vesicles of endosomal origin. They carry a cargo of proteins and genetic material encompassed in a spherical lipid bilayer. Unlike stem cells, MSC exosomes can alter and affect both the transcriptome and the proteome of a recipient cell, leading to a modified cardiac function but do not respond to the microenvironment they are introduced into. Exosomes, through their cargo, can modulate pathways governing apoptosis, cell growth, proliferation, and differentiation. Exosomal cargo depends on the status of the secreting cell as well as the microenvironment in which the secreting cell finds itself [26,27]. We surmise that MSCs could be engineered to produce optimally effective exosomes with added growth factors or other targeted stimuli.
To understand the basis of functional changes in the heart, we studied the proteome of the saline- and exosome-treated mice ischemic hearts. We identified profound changes in the signals leading to cardiac dysfunction and fibrogenesis following ischemia (Fig. 1A).
As expected, when we looked at transcriptomic variations in the tissue samples in the present study, several upstream regulators and TFs in mice hearts were differentially regulated (Supplementary Tables 1 and 2). Interesting among these, is the heat shock factor-1 (HSF1). HSF1 plays an important role in regulating transcription of genes which govern response to ischemic injury. Under stress conditions, HSF1 activates expression of heat shock proteins which, due to their chaperone properties, protect stressed cells against unfolded protein response due to accumulation of proteins due to suppression of proteosome function or over-expressed stress/ischemia related response proteins.
Hypoxia-inducible factor 1-alpha (HIF1A). HIF1A acts as the master transcriptional regulator of genes in response to hypoxic stress. Under ischemic conditions in the myocardium leading to hypoxia, sustained HIF1A activation triggers the transcription response genes which govern various processes such as resistance to oxidative stress, mitochondrial function, cell survival, and apoptosis, and other transcription targets, the protein products of which stimulate oxygen delivery and or stimulate adaptive metabolic mechanisms to hypoxia. (Fig. 3).
We also observed that ischemia in the mice hearts induced the expression of STAT3 and SMAD3 (Fig. 4). STAT3 and SMAD3 signaling pathways complement each other in that SMAD3 binds to non-phosphorylated STAT3. Phosphorylation of SMAD3 increases the phosphorylation of STAT3 leading to increased expression of pro-fibrotic proteins in the fibroblasts [28]. Expression of SMAD3 by fibroblasts is crucial for scar formation and alignment of myofibroblasts in the heart tissue [29]. Our MS and proteomic analysis showed that MSC exosome treatment markedly downregulated STAT3 and SMAD3 mediated pathways in the ischemic mice hearts.
NOTCH1 is an evolutionarily conserved signaling pathway that controls cell differentiation, fate and tissue pattern formation in the developing embryo and in adults [30]. Epigenetic modifications in the NOTCH1 responsive elements irreversibly repress NOTCH1 transcription making it ineffective in promoting cardiac regeneration in adults [31]. As an adaptive response to injury, several dormant pre-natal genes, including NOTCH1 are activated in the adult myocardium [18]. NOTCH1 plays an important role protecting heart tissue from ischemic injury [32,33]. NOTCH signaling is also involved in suppressing cardiac fibrosis, by inhibiting fibroblast to myofibroblast transformation, while inhibiting Notch signaling promotes myofibroblast formation [34]. It is likely that NOTCH1 expression inhibits myofibroblast proliferation while it stimulates cardiomyocyte precursor cells thereby promoting pro-regenerative repair. Activation of NOTCH1 signaling mitigates ischemic damage, reduces myocardial fibrosis and improves cardiac function [18,33,[35], [36], [37]]. We observed that NOTCH1 signaling pathway was stimulated by MSC exosomes (Fig. 5).
Wnt/β-catenin signaling which plays an important role in development of the heart [38]. Wnt signaling has been shown to be re-activated in the ischemic myocardium, both in the infarct and peri-infarct areas [39,40]. Renin-angiotensin system activation also leads to activation of Wnt signaling/β-catenin signaling [41]. Wnt regulates proliferation of cells in the epicardium and the endocardium, proliferation and differentiation of cardiac progenitor cells [39], leading to hypertrophy of the cardiac muscle and hypertension, increased fibrosis, impaired fractional shortening and ventricular tachycardia. Inhibition of wnt/β-catenin signaling mitigates impaired fractional shortening, decreases myocardial fibrosis and reduces fibroblast activation [[38], [39], [40], [41], [42]].
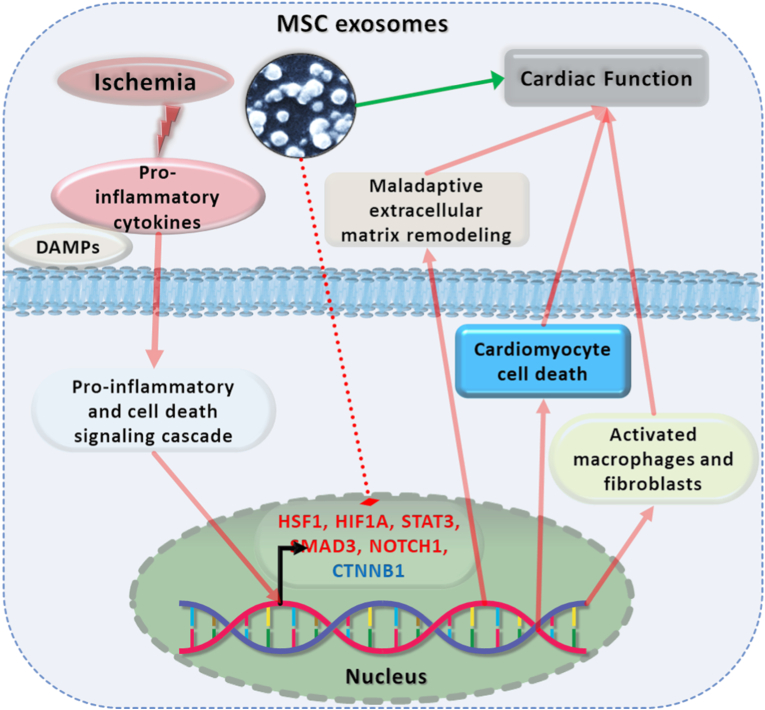
Multiple transcriptional regulators are necessary to maintain normal cardiac homeostasis and function. Ischemic insult disrupts this homeostasis resulting in cell death, loss of cardiac function and fibrotic scar formation. MSC exosomes reduce the ischemia-induced inflammation and related cardiac injury, in part through their modulation of expression of various TFs (Fig. 6).
Fig. 6.
Central illustration: Mitigation of cardiac dysfunction in the ischemic heart by MSC exosomes
MSC exosomes protect against ischemia mediated inflammation related fibrotic events and cardiac dysfunction in the ischemic mice hearts. The inflammatory response stimulates expression of various TFs which lead to fibrotic remodeling in ischemic hearts leading to compromised cardiac function. Administration of MSC exosomes differentially regulates levels of such TFs leading to suppression of fibrotic remodeling and promoting reparative remodeling of the cardiac extracellular matrix.
In summary, we have described for the first time a host of changes in the proteome of the ischemic heart. Many of these alterations in upstream regulators and TFs may relate to the structural and physiologic changes following ischemia. The identity and relevance of some of these upstream regulators and TFs need to be confirmed and subsequently whether they are a druggable target for prevention or treatment can be fully explored. At a minimum, we believe that proper identification and validation of these regulators may lead to improved understanding of the pathophysiologic alterations in the heart that take place following ischemia.
4. Materials and Methods
4.1. Cell culture and reagents; culture of MSCs
The MSCs were cultured in complete DMEM media (Thermo Fischer Scientific, Waltham, MA), (containing 10% FBS (Atlanta Biologicals Inc., Flowery Branch, GA) and penicillin/streptomycin (100 units/ml obtained from Thermo Fischer Scientific, Waltham, MA)) at 37 °C in a 5% CO2 incubator. The cells cultured overnight were washed with PBS and fresh serum-free DMEM media was added to further culture the cells for 16–18 h. This serum-free conditioned media was harvested for exosome isolation.
4.2. Exosome isolation and characterization
Isolation of exosomes from MSC serum-free culture media was carried out by sequential centrifugation as previously described [26]. In brief, pooled media from MSC cultures was processed through a series of centrifugation steps (3000 g for 10 min, 10,000 g for 30 min and finally ultracentrifugation at 100,000 g for 3–4 h). The exosome pellet collected at the end of final centrifugation step was washed with PBS and then resuspended in PBS for further use. JEOL JSM7000F scanning electron microscope was used to image MSC exosomes as previously reported [27]. Markers for exosomal vesicles (CD9 and CD63) were detected by Western blots. Please refer our previous report for detailed exosome characterization [17,43].
4.3. Mouse ischemia model
Adult male C57BL/6 mice were obtained from the Jackson Laboratory (Bar Harbor, ME). The animal study protocol was approved by the local institutional animal use care committee and conformed to the Guide for the Care and Use of Laboratory Animals published by the National Academies Press (Washington, DC). The animals were euthanatized by CO2 inhalation at the end of 7 days after LCA ligation.
Myocardial ischemia was induced by total LCA occlusion in the anaesthetized mice as described previously [43,44]. In brief, following ketamine hydrochloride (60 mg kg−1) and xylazine hydrochloride (8 mg kg−1) administered intraperitoneally, the anaesthetized animals were endotracheally intubated and mechanically ventilated (1.2 mL min−1 tidal volume and, 110/min respiration rate). An 8-0 silk suture was passed around the LCA at a point two-third of the way between its origins near the pulmonary conus. Another group of animals underwent the same procedure, but without LCA occlusion (sham ischemia). A third group of mice was pre-treated with a single dose of MSC exosomes (0.5 mg/kg body weight, administered intravenously through the lateral tail vein) 30 min prior to left coronary artery (LCA) ligation. The quantity of MSC exosomes administered to the animals was based on our previous reported study [43] and studies by Lai et al., 2010, and Arsalan et al., 2013 [45,46].
4.4. Mass spectrometry
Infarct and peri-infarct areas of the freshly isolated LCA ligated mice hearts were visually identified and carefully dissected from euthanized mice. The tissues were then homogenized, and total protein was extracted using a lysis buffer. The proteins from these lysates were subjected to mass spectrometric analysis using a previously described method for identification and analysis [17]. Scaffold 4 software was used to compile identified proteins and FunRich™ and Ingenuity Pathway Analysis [20] was used for detailed analysis of identified proteins, as previously described method [17,43].
Funding
Current grant support from Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development (Washington, DC, USA) (grant No BX-000282-05) (JLM), and Stebbins Chair Funds. AJT acknowledges funding from the National Institutes of Health (P20GM121293 and S10OD018445) and support from the UAMS Proteomics Core Facility.
Disclosures
Dr. Mehta has previously served as consultant to and received grant support from Bayer, Boehringer Ingelheim, AstraZeneca, MedImmmune and Pfizer. Dr. Griffin is serving as a consultant to Accuray, IGF Oncology, Rejuvenix Technologies. Other authors have nothing to declare.
Credit author statement
Rajshekhar A. Kore: Conceptualization, Investigation, Methodology, Software, Data Curation, Original Draft Preparation, Samir V. Jenkins: Writing- Reviewing and Editing. Azemat Jamshidi-Parsian: Sample Preparation, Alan J. Tackett, Robert J. Griffin, Srinivas Ayyadevara: Methodology, Writing- Reviewing and Editing: Jawahar L. Mehta: Supervision, Investigation, Conceptualization, Writing- Reviewing and Editing.
Declaration of competing interest
Dr. Mehta has previously served as consultant to and received grant support from Bayer, Boehringer Ingelheim, AstraZeneca, MedImmmune and Pfizer. Dr. Griffin is serving as a consultant to Accuray, IGF Oncology, Rejuvenix Technologies. Other authors have nothing to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2023.101463.
Contributor Information
Rajshekhar A. Kore, Email: rakore@uams.edu.
Jawahar L. Mehta, Email: mehtajl@uams.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Supplementary Table-1: List of TFs detected in the infarct areas of ischemic mice hearts.
Supplementary Table-2: List of TF detected in the peri-infarct areas of ischemic mice hearts.
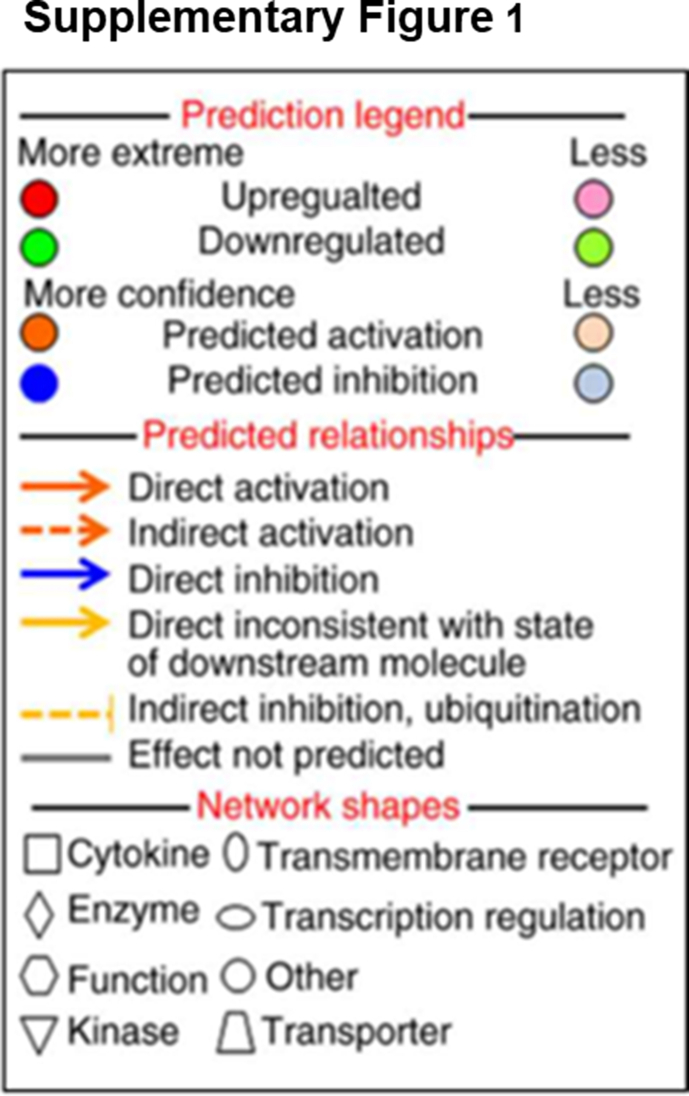
Supplementary Fig. 1.
Prediction legend describes the network relationships between different molecules.
Data availability
Data will be made available on request.
References
- 1.Mozaffarian D., Benjamin E.J., Go A.S., Arnett D.K., Blaha M.J., Cushman M., Das S.R., de Ferranti S., Despres J.P., Fullerton H.J., Howard V.J., Huffman M.D., Isasi C.R., Jimenez M.C., Judd S.E., Kissela B.M., Lichtman J.H., Lisabeth L.D., Liu S., Mackey R.H., Magid D.J., McGuire D.K., Mohler E.R., 3rd, Moy C.S., Muntner P., Mussolino M.E., Nasir K., Neumar R.W., Nichol G., Palaniappan L., Pandey D.K., Reeves M.J., Rodriguez C.J., Rosamond W., Sorlie P.D., Stein J., Towfighi A., Turan T.N., Virani S.S., Woo D., Yeh R.W., Turner M.B. Heart disease and stroke statistics-2016 update: a report from the American heart association. Circulation. 2016;133:e38–e360. doi: 10.1161/cir.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 2.Jackson S.L., Tong X., King R.J., Loustalot F., Hong Y., Ritchey M.D. National burden of heart failure events in the United States, 2006 to 2014, circulation. Heart Fail. 2018;11 doi: 10.1161/CIRCHEARTFAILURE.117.004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abbate A., Bussani R., Biondi-Zoccai G.G.L., Santini D., Petrolini A., Giorgio F.D., Vasaturo F., Scarpa S., Severino A., Liuzzo G., Leone A.M., Baldi F., Sinagra G., Silvestri F., Vetrovec G.W., Crea F., Biasucci L.M., Baldi A. Infarct-related artery occlusion, tissue markers of ischaemia, and increased apoptosis in the peri-infarct viable myocardium. Eur. Heart J. 2005;26:2039–2045. doi: 10.1093/eurheartj/ehi419. [DOI] [PubMed] [Google Scholar]
- 4.Konstam M.A., Kramer D.G., Patel A.R., Maron M.S., Udelson J.E. Left ventricular remodeling in heart failure. JACC (J. Am. Coll. Cardiol.): Cardiovascular Imaging. 2011;4:98. doi: 10.1016/j.jcmg.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Daseke M.J., 2nd, Tenkorang M.A.A., Chalise U., Konfrst S.R., Lindsey M.L. Cardiac fibroblast activation during myocardial infarction wound healing: fibroblast polarization after MI, Matrix biology. journal of the International Society for Matrix Biology. 2020;91–92:109–116. doi: 10.1016/j.matbio.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manso A.M., Kang S.M., Ross R.S. Integrins, focal adhesions, and cardiac fibroblasts. J. Invest. Med. : the official publication of the American Federation for Clinical Research. 2009;57:856–860. doi: 10.2310/JIM.0b013e3181c5e61f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wynn T.A. Cellular and molecular mechanisms of fibrosis. J. Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van den Borne S.W., Diez J., Blankesteijn W.M., Verjans J., Hofstra L., Narula J. Myocardial remodeling after infarction: the role of myofibroblasts. Nat. Rev. Cardiol. 2010;7:30–37. doi: 10.1038/nrcardio.2009.199. [DOI] [PubMed] [Google Scholar]
- 9.Burchfield J.S., Xie M., Hill J.A. Pathological ventricular remodeling: mechanisms: part 1 of 2. Circulation. 2013;128:388–400. doi: 10.1161/CIRCULATIONAHA.113.001878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christia P., Bujak M., Gonzalez-Quesada C., Chen W., Dobaczewski M., Reddy A., Frangogiannis N.G. Systematic characterization of myocardial inflammation, repair, and remodeling in a mouse model of reperfused myocardial infarction. J. Histochem. Cytochem. : official journal of the Histochemistry Society. 2013;61:555–570. doi: 10.1369/0022155413493912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westman P.C., Lipinski M.J., Luger D., Waksman R., Bonow R.O., Wu E., Epstein S.E. Inflammation as a driver of adverse left ventricular remodeling after acute myocardial infarction. J. Am. Coll. Cardiol. 2016;67:2050. doi: 10.1016/j.jacc.2016.01.073. [DOI] [PubMed] [Google Scholar]
- 12.Lazar E., Benedek T., Korodi S., Rat N., Lo J., Benedek I. Stem cell-derived exosomes - an emerging tool for myocardial regeneration. World J. Stem Cell. 2018;10:106–115. doi: 10.4252/wjsc.v10.i8.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang P.C. Induced pluripotent stem cell (iPSC)-Derived exosomes for precision medicine in heart failure. Circ. Res. 2018;122:661–663. doi: 10.1161/circresaha.118.312657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L., Jin X., Hu C.F., Li R., Zhou Z., Shen C.X. Exosomes derived from mesenchymal stem cells rescue myocardial ischaemia/reperfusion injury by inducing cardiomyocyte autophagy via AMPK and akt pathways, cellular physiology and biochemistry. international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;43:52–68. doi: 10.1159/000480317. [DOI] [PubMed] [Google Scholar]
- 15.Kishore R., Khan M. More than tiny sacks: stem cell exosomes as cell-free modality for cardiac repair. Circ. Res. 2016;118:330–343. doi: 10.1161/circresaha.115.307654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju C., Shen Y., Ma G., Liu Y., Cai J., Kim I.M., Weintraub N.L., Liu N., Tang Y. Transplantation of cardiac mesenchymal stem cell-derived exosomes promotes repair in ischemic. Myocardium. 2018;11:420–428. doi: 10.1007/s12265-018-9822-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kore R.A., Henson J.C., Hamzah R.N., Griffin R.J., Tackett A.J., Ding Z., Mehta J.L. Molecular events in MSC exosome mediated cytoprotection in cardiomyocytes. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-55694-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gude N.A., Emmanuel G., Wu W., Cottage C.T., Fischer K., Quijada P., Muraski J.A., Alvarez R., Rubio M., Schaefer E., Sussman M.A. Activation of Notch-mediated protective signaling in the myocardium. Circ. Res. 2008;102:1025–1035. doi: 10.1161/circresaha.107.164749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A., Bacic A., Hill A.F., Stroud D.A., Ryan M.T., Agbinya J.I., Mariadason J.M., Burgess A.W., Mathivanan S. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 20.Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Mering C.V. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., Jensen L.J., von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–d612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adamiak M., Sahoo S. Exosomes in myocardial repair: advances and challenges in the development of next-generation therapeutics. Mol. Ther. : the journal of the American Society of Gene Therapy. 2018;26:1635–1643. doi: 10.1016/j.ymthe.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagno L., Hatzistergos K.E., Balkan W., Hare J.M. Mesenchymal stem cell-based therapy for cardiovascular disease: progress and challenges. Mol. Ther. : the journal of the American Society of Gene Therapy. 2018;26:1610–1623. doi: 10.1016/j.ymthe.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan Y., Du W., Liu J., Ma W., Zhang L., Du Z., Cai B. Stem cell-derived exosome in cardiovascular diseases: macro roles of micro particles. Front. Pharmacol. 2018;9:547. doi: 10.3389/fphar.2018.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kore R.A., Abraham E.C. Inflammatory cytokines, interleukin-1 beta and tumor necrosis factor-alpha, upregulated in glioblastoma multiforme, raise the levels of CRYAB in exosomes secreted by U373 glioma cells. Biochem. Biophys. Res. Commun. 2014;453:326–331. doi: 10.1016/j.bbrc.2014.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kore R.A., Edmondson J.L., Jenkins S.V., Jamshidi-Parsian A., Dings R.P.M., Reyna N.S., Griffin R.J. Hypoxia-derived exosomes induce putative altered pathways in biosynthesis and ion regulatory channels in glioblastoma cells. Biochemistry and biophysics reports. 2018;14:104–113. doi: 10.1016/j.bbrep.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakraborty D., Sumova B., Mallano T., Chen C.W., Distler A., Bergmann C., Ludolph I., Horch R.E., Gelse K., Ramming A., Distler O., Schett G., Senolt L., Distler J.H.W. Activation of STAT3 integrates common profibrotic pathways to promote fibroblast activation and tissue fibrosis. Nat. Commun. 2017;8:1130. doi: 10.1038/s41467-017-01236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong P., Shinde A.V., Su Y., Russo I., Chen B., Saxena A., Conway S.J., Graff J.M., Frangogiannis N.G. Opposing actions of fibroblast and cardiomyocyte Smad3 signaling in the infarcted myocardium. Circulation. 2018;137:707–724. doi: 10.1161/CIRCULATIONAHA.117.029622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luxán G., D'Amato G., MacGrogan D., de la Pompa J.L. Endocardial notch signaling in cardiac development and disease. Circ. Res. 2016;118:e1–e18. doi: 10.1161/circresaha.115.305350. [DOI] [PubMed] [Google Scholar]
- 31.Felician G., Collesi C., Lusic M., Martinelli V., Ferro M.D., Zentilin L., Zacchigna S., Giacca M. Epigenetic modification at Notch responsive promoters blunts efficacy of inducing notch pathway reactivation after myocardial infarction. Circ. Res. 2014;115:636–649. doi: 10.1161/circresaha.115.304517. [DOI] [PubMed] [Google Scholar]
- 32.Fan Y.H., Dong H., Pan Q., Cao Y.J., Li H., Wang H.C. Notch signaling may negatively regulate neonatal rat cardiac fibroblast-myofibroblast transformation. Physiol. Res. 2011;60:739–748. doi: 10.33549/physiolres.932149. [DOI] [PubMed] [Google Scholar]
- 33.Nemir M., Metrich M., Plaisance I., Lepore M., Cruchet S., Berthonneche C., Sarre A., Radtke F., Pedrazzini T. The Notch pathway controls fibrotic and regenerative repair in the adult heart. Eur. Heart J. 2014;35:2174–2185. doi: 10.1093/eurheartj/ehs269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X., Chen X., Cai J.J., Chen L.Z., Gong Y.S., Wang L.X., Gao Z., Zhang H.Q., Huang W.J., Zhou H. Relaxin inhibits cardiac fibrosis and endothelial-mesenchymal transition via the Notch pathway. Drug Des. Dev. Ther. 2015;9:4599–4611. doi: 10.2147/dddt.s85399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kratsios P., Catela C., Salimova E., Huth M., Berno V., Rosenthal N., Mourkioti F. Distinct roles for cell-autonomous Notch signaling in cardiomyocytes of the embryonic and adult heart. Circ. Res. 2010;106:559–572. doi: 10.1161/circresaha.109.203034. [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Hiroi Y., Ngoy S., Okamoto R., Noma K., Wang C.Y., Wang H.W., Zhou Q., Radtke F., Liao R., Liao J.K. Notch1 in bone marrow-derived cells mediates cardiac repair after myocardial infarction. Circulation. 2011;123:866–876. doi: 10.1161/circulationaha.110.947531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrari R., Rizzo P. The Notch pathway: a novel target for myocardial remodelling therapy? Eur. Heart J. 2014;35:2140–2145. doi: 10.1093/eurheartj/ehu244. [DOI] [PubMed] [Google Scholar]
- 38.Ozhan G., Weidinger G. Wnt/β-catenin signaling in heart regeneration. Cell Regen. 2015;4:3. doi: 10.1186/s13619-015-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haybar H., Khodadi E., Shahrabi S. Wnt/β-catenin in ischemic myocardium: interactions and signaling pathways as a therapeutic target. Heart Fail. Rev. 2019;24:411–419. doi: 10.1007/s10741-018-9759-z. [DOI] [PubMed] [Google Scholar]
- 40.Wang J., Xia Y., Lu A., Wang H., Davis D.R., Liu P., Beanlands R.S., Liang W. Cardiomyocyte-specific deletion of β-catenin protects mouse hearts from ventricular arrhythmias after myocardial infarction. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-97176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y., Wang C., Wang C., Hong X., Miao J., Liao Y., Zhou L., Liu Y. An essential role for Wnt/β-catenin signaling in mediating hypertensive heart disease. Sci. Rep. 2018;8:8996. doi: 10.1038/s41598-018-27064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baurand A., Zelarayan L., Betney R., Gehrke C., Dunger S., Noack C., Busjahn A., Huelsken J., Taketo M.M., Birchmeier W., Dietz R., Bergmann M.W. β-Catenin downregulation is required for adaptive cardiac remodeling. Circ. Res. 2007;100:1353–1362. doi: 10.1161/01.RES.0000266605.63681.5a. [DOI] [PubMed] [Google Scholar]
- 43.Kore R.A., Wang X., Ding Z., Griffin R.J., Tackett A.J., Mehta J.L. MSC exosome-mediated cardioprotection in ischemic mouse heart comparative proteomics of infarct and peri-infarct areas. Mol. Cell. Biochem. 2021 doi: 10.1007/s11010-020-04029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding Z., Wang X., Liu S., Shahanawaz J., Theus S., Fan Y., Deng X., Zhou S., Mehta J.L. PCSK9 expression in the ischemic heart and its relationship to infarct size, cardiac function and development of autophagy. Cardiovasc. Res. 2018 doi: 10.1093/cvr/cvy128. [DOI] [PubMed] [Google Scholar]
- 45.Arslan F., Lai R.C., Smeets M.B., Akeroyd L., Choo A., Aguor E.N., Timmers L., van Rijen H.V., Doevendans P.A., Pasterkamp G., Lim S.K., de Kleijn D.P. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013;10:301–312. doi: 10.1016/j.scr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Lai R.C., Arslan F., Lee M.M., Sze N.S., Choo A., Chen T.S., Salto-Tellez M., Timmers L., Lee C.N., El Oakley R.M., Pasterkamp G., de Kleijn D.P., Lim S.K. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table-1: List of TFs detected in the infarct areas of ischemic mice hearts.
Supplementary Table-2: List of TF detected in the peri-infarct areas of ischemic mice hearts.
Data Availability Statement
Data will be made available on request.