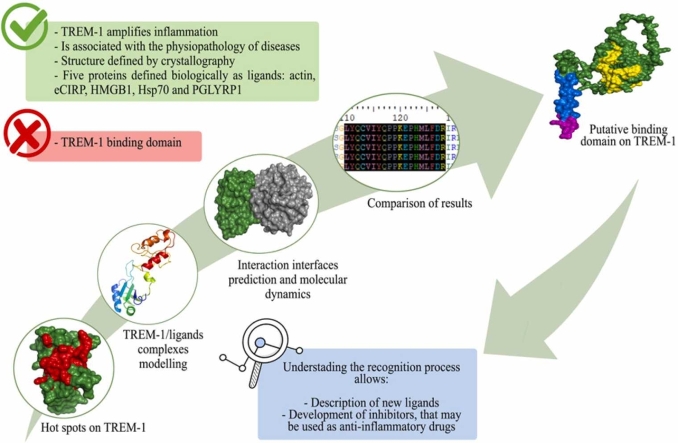
Abstract
The triggering receptor expressed on myeloid cells-1 (TREM-1) is a pattern recognition receptor heavily investigated in infectious and non-infectious diseases. Because of its role in amplifying inflammation, TREM-1 has been explored as a diagnostic/prognostic biomarker. Further, as the receptor has been implicated in the pathophysiology of several diseases, therapies aiming at modulating its activity represent a promising strategy to constrain uncontrolled inflammatory or infectious diseases. Despite this, several aspects concerning its interaction with ligands and activation process, remain unclear. Although many molecules have been suggested as TREM-1 ligands, only five have been confirmed to interact with the receptor: actin, eCIRP, HMGB1, Hsp70 and PGLYRP1. However, the domains involved in the interaction between the receptor and these proteins are not clarified yet. Therefore, here we used in silico approaches to investigate the putative binding domains in the receptor, using hot spots analysis, molecular docking and molecular dynamics simulations between TREM-1 and its five known ligands. Our results indicated the complementarity-determining regions (CDRs) of the receptor as the main mediators of antigen recognition, especially the CDR3 loop. We believe that our study could be used as structural basis for the elucidation of TREM-1′s recognition process, and may be useful for prospective in silico and biological investigations exploring the receptor in different contexts.
Keywords: TREM-1, Binding domain, CDR3, TREM-1 ligands, Pattern recognition receptor
Graphical Abstract
Highlights
-
•
TREM-1 is associated with the amplification of inflammation. To date, five proteins have been confirmed as TREM-1 ligands, but the binding domain in TREM-1 has not been described yet.
-
•
The interaction between TREM-1 and its ligands (actin, eCIRP, HMGB1, Hsp70 and PGLYRP1) predicted hot spots residues in the TREM-1 IgV-like domain.
-
•
The CDR1, and especially CDR3, are likely the main mediators of antigen recognition on TREM-1.
1. Introduction
The triggering receptor expressed on myeloid cells-1 (TREM-1) is a pattern recognition receptor (PRR), expressed mostly by immune cells from the myeloid lineage (e.g. neutrophils, monocytes, macrophages and dendritic cells) [1], [2], [3]. In general, TREM-1 seems to be associated with exacerbated inflammatory responses and, therefore, with the pathogenesis of parasitic, viral, bacterial and fungal infections (such as HIV, influenza, tuberculosis, aspergillosis, malaria and sepsis) [4], [5]. The receptor has also been involved in sterile inflammatory disorders, including inflammatory bowel disease, gastritis, arthritis, cardiovascular diseases and cancer [6], [7], [8], [9], [10]. Due to its essential role in inflammation, the receptor has been increasingly explored in the aforementioned scenarios, both in its soluble (sTREM-1) and membrane associated (mTREM-1) forms. The former, results from alternative splicing and/or cleavage of mTREM-1 by metalloproteinases [3], [11], [12], [13] and its levels have been explored as a disease and/or severity biomarker for several inflammatory and infectious diseases, including COVID-19 [14], [15]. On the other hand, mTREM-1 is responsible for triggering/amplifying the immune response, working in mutual cooperation with other PRRs, such as toll-like receptors (TLR), nucleotide oligomerization binding domain-like receptors (NLR), the retinoic acid-inducible gene-I-like-receptors (RLR), among others [16], [17], [18], [19]. Despite the elucidation of several mechanisms associated with TREM-1 activation [20], [21], [22], [23], [24], different features regarding the activity of the receptor remains unexplored, including the domains involved in antigen recognition.
TREM-1 is a glycoprotein composed by a 194 amino acids (aa) extracellular domain, a 29 aa transmembrane domain and a short cytoplasmic tail, with 5 aa [25]. So far, only the IgV-like portion of TREM-1 (aa 26–134), which is the portion most likely to interact with antigens/ligands, has been characterized by crystallography (18QM - [26]; 1SMO - [25]). This structure is composed by 10 β-strands and two small α-helixes, organized into two antiparallel sheets, formed by disulfide bonds between amino acids C41 and C113 [25], [26]. However, the antigen-binding domains within the receptor are still elusive, hindering the understanding of structural and molecular aspects regarding the activation of the receptor, the determination of new ligands and the development of therapeutic strategies targeting TREM-1. As a member of the immunoglobulin superfamily, TREM-1 shares some structural similarities with other members, including the presence of protruding loops that resemble the complementarity-determining regions (CDR) of antibodies, which are the domains associated with antigen recognition [25], [26]. These loops are important for cognate ligand recognition in other proteins from the same family, such as the T cell receptor (TCR), the cluster of differentiation 8 (CD8) and the cytotoxic T-lymphocyte associated protein 4 (CTLA-4) [26]. Therefore, for many years, it has been speculated that the three CDR-like regions of TREM-1 (the BC [CDR1], C′C’’ [CDR2] and FG [CDR3] loops) may mediate ligand/antigen recognition [25], [26], but this aspect has not been clarified yet.
Several molecules have been suggested to interact with TREM-1. Among them, it is possible to highlight some well-known microorganism associated molecular patterns (MAMPs) and danger/damage associated molecular patterns (DAMPs), such as: lipopolysaccharide (LPS), lipoteichoic acid (LTA), flagellin, the heat shock 70 kDa protein (Hsp70) and the high mobility group box 1 protein (HMGB1) [17], [27], [28], [29], [30], [31]. In general, such molecules are able to induce the expression of the receptor, the release of sTREM-1 and pro-inflammatory mediators, which are hallmarks of mTREM-1 activation. However, only five proteins had the physical interaction with TREM-1 confirmed by experimental assays and, therefore, can be considered as TREM-1 ligands: actin, the extracellular cold-inducible RNA-binding protein (eCIRP), HMGB1, Hsp70 and the peptidoglycan recognition protein 1 (PGLYRP1) [32].
Although the recognition of these five proteins by TREM-1 was observed, none of the aforementioned studies characterized the domains involved with this process. Therefore, the binding site in the structure of TREM-1 still needs to be determined. Considering the paramount importance of understanding the interactions between TREM-1 and its ligands and the regions of the receptor involved in this process, we have integrated several in silico approaches to determine the structural basis for the interaction of TREM-1 with its known ligands (i.e., actin, eCIRP, HMGB1, Hsp70 and PGLYRP1). Despite limitations in the simulations, due to structural characteristics of the ligands, our results suggest the CDR regions, in particular CDR3, as key regions for the interactions between the receptor and its ligands, most likely, for the activation of the receptor. With this approach, we hope to broaden the knowledge regarding the recognition process by the receptor, providing data and tools that might be useful for the discovery of new TREM-1 ligands and prospective computer-aided drug discovery studies.
2. Material and methods
2.1. Prediction of hotspots on TREM-1
The crystallographic structure of human TREM-1 was retrieved from the Protein Data Bank (PDB) website (https://www.rcsb.org). The FTMap [33], [34], [35], [36] and the SeRenDIP [37], [38], [39] web servers were used to predict hot spots (i.e. residues that are the major contributors to the interaction with ligands) in the TREM-1 extracellular domain. FTMap simulates the interaction between the input structure and 16 small organic probes, generating consensus sites in which probes clusters bind to, while SeRenDIP uses random-forest based methods to predict protein-protein interaction positions in an amino acid sequences. On the FTMap website, the assay was conducted with the advanced option “PPI mode” (specific for binding hot spots on protein-protein interfaces). For SeRenDIP, the sequence of TREM-1 IgV-like portion was submitted and the analysis was conducted using the combined dataset for random forest model training.
2.2. Generation of TREM-1/ligands complexes
The sequences of TREM-1, actin, CIRP, HMGB1, Hsp70 and PGLYRP1 were retrieved from the UniProt database (https://www.uniprot.org). We also selected Apolipoprotein A-I (APOA1) to act as a negative control, as a previous study showed, by biological assays, that this molecule was unable to bind to TREM-1 [40]. For the generation of the protein-protein complexes, we used the Alphafold2-multimer tool at ColabFold v1.5.2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb#scrollTo=G4yBrceuFbf3) [41], an artificial intelligence-based server for the prediction of protein structures. The default parameters were maintained for the modeling process, except for the selection of the pdb70 template mode (which searches for homologous proteins at the pdb70 database), and the relaxation of the five generated poses with the Amber force-field (to correct eventual stereochemical violations). ColabFold generates five models, which are ranked according to three scores: interface predicted template modeling score (ipTM score), the predicted template modeling score (pTM) score and the predicted local distance difference test (pLDDT) score. For each complex, the best ranked model was selected.
The selected TREM-1/ligands complexes were analyzed regarding their structural quality on Molprobity [42]b and PROCHECK [43], [44], the latter used for the generation of Ramachandran plots. The binding free energy of each pose was calculated with the molecular mechanics with generalized born and surface area solvation (MM/GBSA) method, with the HawkDock webserver [45], [46], [47]. We also predicted the binding affinity and the dissociation constants (Kd), at 36.5 ℃, with the Prodigy server, which uses a predictive model based on number of interfacial contacts and the properties of the residues [48], [49], [50]. The Computational Characterization of Protein-Protein Interactions (CCharPPI) webserver [51] was used to further investigate the intermolecular interactions, with the following descriptors: PYDOCK_TOT (total energy) [52], [53], HBOND (hydrogen bond potential) [54], VDW (van der Waals energy) [52], [53], ELE (total electrostatic energy) [52], [53], FA_ATR (attractive van der Waals forces) [55] and DESOLV (desolvation energy) [52], [53].
2.3. Characterization of surface interaction between TREM-1 and its ligands
The interaction between TREM-1 and its ligands was assessed with the combined results from the PPCheck [56] web server and the DIMPLOT software (LigPlot+ suite v. 2.2) [57]. The latter was used for the identification of hydrogen bonds and salt bridges, and water molecules were included in the analysis with DIMPLOT. The PyMol software was used for the visualization of poses and the generation of images.
2.4. Molecular dynamics
We performed molecular dynamics (MD) simulations to investigate the reliability of the models proposed by AlphaFold2-Multimer for two protein complexes consisting of TREM-1 complexed with either PGLYRP1 or actin.
To prepare the systems for MD simulations, we first used the propka3 software [58] to evaluate the protonation states of all residues, and we visually inspected the histidine tautomer. We then parametrized the protein atoms with the ff19SB force field [59] using the tLeap module [60]of AmberTools21. The complexes were solvated in an octahedral box, with a minimum dimension of 12 Å from each solute atom, using the OPC water model. To ensure neutralization of the simulation system and ionic strength to 150 mM, we added Na+ and Cl- ions. Hydrogen mass repartitioning (HMR) [61], [62] was performed using the parmed module [63], which allowed us to use a 4 ps timestep in the simulation.
Simulations were conducted using the pmemd.cuda engine of Amber20 [64]. Water and ions underwent double-precision mode minimization, followed by new water/ion minimization and unrestrained minimization with steps switching from steepest descent to conjugate gradient after 3500, 3500, and 10,000 cycles, respectively. The system was gradually heated to 310 K over 200 ps in the NVT ensemble using a Langevin thermostat with a weak force of 10 kcal.mol-1 applied to the protein atoms. The restraint force was maintained during density equilibration for 500 ps in the NPT ensemble, using a Langevin thermostat, Monte Carlo barostat, and NPT ensemble. An unrestrained simulation was then conducted in the NPT ensemble for 5 ns, using a Langevin thermostat and Monte Carlo barostat. Bonds involving hydrogen atoms were constrained using the SHAKE protocol [65], and long-range interactions were computed with Particle Mesh Ewald [66], applying a nonbonded cutoff of 10 Å. The simulation timestep was set to 4 ps.
To explore the conformational space of our systems, we utilized a stepwise heated molecular dynamics approach [67]. This involved consecutive simulations where the temperature was gradually increased: 30 ns at 310 K, 12.5 ns at 330 K, 12.5 ns at 360 K, and a final 15 ns at 390 K, while keeping all other parameters constant from the equilibration step. To validate our results, we conducted five new conventional simulations, each consisting of 200 ns, starting from the last frame of the stepwise heated molecular dynamics replicates but with a different random seed.
We employed the cpptraj module [68] within the AmberTools suite to perform trajectory analysis, specifically for the identification of hydrogen bonds and calculation of root-mean-square deviation (RMSD) values for the protein-protein interface. For the latter, we restricted our analysis to residues of the folded domain from one protein that were within a 5 Å distance of any atom from the folded domain of the corresponding complex partner.
3. Results
3.1. Hot spots residues on TREM-1
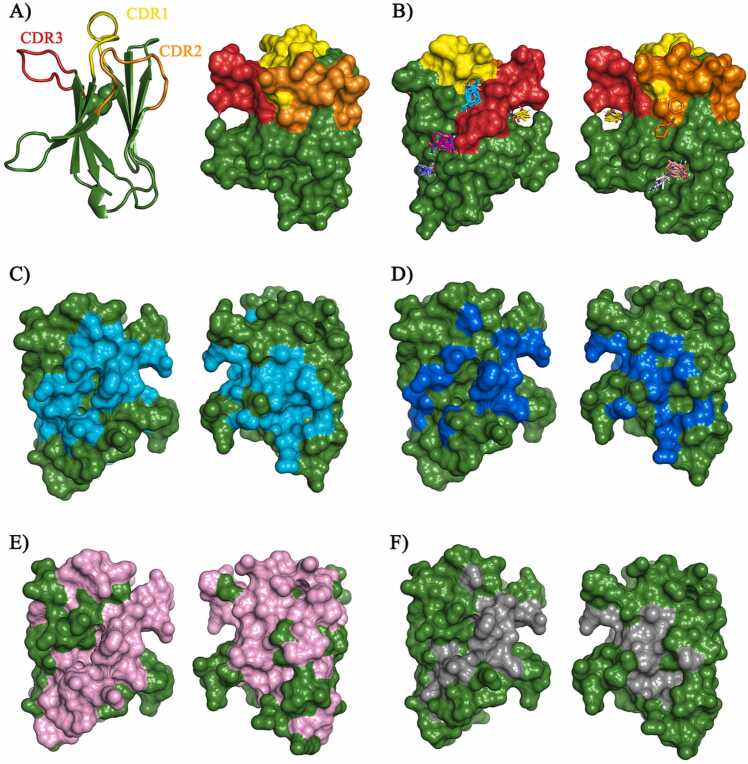
Initially, the 3D structure of human TREM-1 was retrieved from PDB (ID: 1SMO, resolution: 1.47 Å) [25]. This structure comprises the IgV-like portion of the receptor (aa 26–134), including its CDR loops. These loops present certain plasticity but, in the B chain of 1SMO (1SMO_B), the CDR regions are: Y43–G52 (CDR1), C70–P79 (CDR2) and Y116–R128 (CDR3) (yellow, orange and red, respectively, in Fig. 1A) [25]. Two web servers were used in the identification of hot spots residues: FTMap (Fig. 1B–D) and SeRenDIP (Fig. 1E). The results from FTMap indicated that the largest probe cluster (cyan, Fig. 1B) was located between CDR1 and CDR3, and two other clusters interacted with CDR3 (magenta and yellow, Fig. 1B), which implies the importance of such region for antigen recognition. Probes clusters also bonded to regions comprised by the B β-strand, CDR1, the small C”D loop and, especially, the EF loop and F strand portions (Fig. 1B). The FTMap server also assesses which residues are most likely involved in nonbonded (cyan, Fig. 1C) and H-bond (blue, Fig. 1D) interactions (full list on Supplementary Table S1) and the regions that concentrate the most interactions are: 26–31, 36–44, 56–59, 64–69, 82–85, 106–114, 121–129.
Fig. 1.
Hot spots analysis in the IgV-like domain of the triggering receptor expressed on myeloid cells (TREM-1). (A) The Complementarity-determining regions (CDRs) of TREM-1. CDR1 in yellow, CDR2 in orange and CDR3 in red. (B) The interaction of FTMap probe clusters with TREM-1, highlighting the interaction with CDR1 (yellow), CDR2 (orange) and CDR3 (red). TREM-1 amino acids mediating nonbonded, in cyan (C), and H-bonded, in blue (D) interactions, from the FTMap analysis. (E) Amino acids predicted as possibly “interactive” by SeRenDIP, in pink. (F) Amino acids predicted as hot spots by both FTMap and SeRenDIP, in gray.
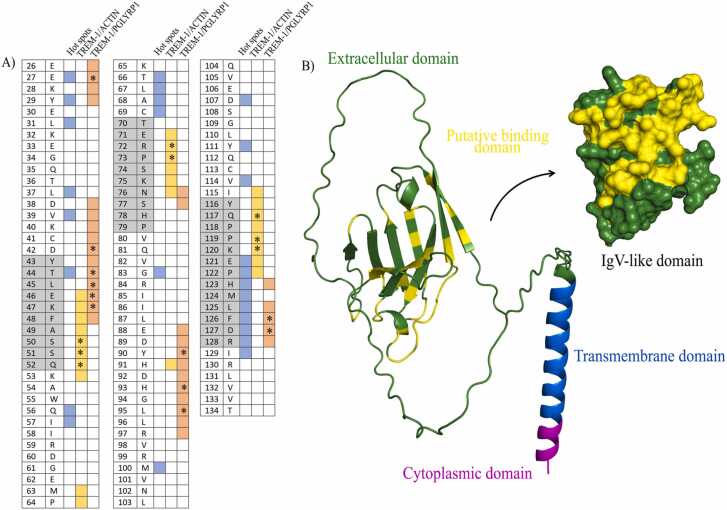
The TREM-1 IgV-like sequence (aa 26–134, UniProt: Q9NP99) was submitted to SeRenDIP and, as observed in the Supplementary Table S1, the output data from the server indicates the likelihood of each amino acid to be part of the interface, considering “interactive”, residues with a prediction score > 0.5. The results from SeRenDIP indicated that 74 of the residues within the Ig-like V-type portion of TREM-1 might be in the protein-protein interface (pink, Fig. 1E). Comparing the results obtained from FTMap and SeRenDIP, we found 26 residues that were indicated by both web servers as hot spots (Table 1). Therefore, these amino acids are crucial for the protein-protein interactions involving TREM-1 (gray, Fig. 1F), especially part of the CDR3 portion (aa 121 – 128).
Table 1.
Combined hot spots analysis in the TREM-1 IgV-like domain.
| FTMap |
SeRenDIP |
||||
|---|---|---|---|---|---|
| Position | Amino acid | NB interactions % | HB interactions % | Prediction score | Classification |
| 27 | E | 1.84 | 0.666 | I | |
| 29 | Y | 8.12 | 11.83 | 0.628 | I |
| 31 | L | 1.16 | 0.584 | I | |
| 37 | L | 1.66 | 0.504 | I | |
| 39 | V | 1.73 | 0.53 | I | |
| 44 | T | 1.21 | 5.48 | 0.514 | I |
| 56 | Q | 1.2 | 0.47 | 0.63 | I |
| 57 | I | 2.7 | 0.554 | I | |
| 66 | T | 1.73 | 0.67 | 0.564 | I |
| 67 | L | 11.55 | 2.1 | 0.606 | I |
| 68 | A | 0.9 | 0.642 | I | |
| 69 | C | 0.52 | 5.27 | 0.616 | I |
| 83 | G | 2.9 | 0.514 | I | |
| 100 | M | 1.57 | 0.528 | I | |
| 107 | D | 6.78 | 3.49 | 0.518 | I |
| 111 | Y | 2.11 | 5.25 | 0.506 | I |
| 114 | V | 0.35 | 0.544 | I | |
| 121 | E | 0.36 | 0.24 | 0.574 | I |
| 122 | P | 1.52 | 1.92 | 0.534 | I |
| 123 | H | 1.52 | 2.47 | 0.524 | I |
| 124 | M | 0.93 | 2.22 | 0.566 | I |
| 125 | L | 2.58 | 0.616 | I | |
| 126 | F | 2.6 | 10.14 | 0.528 | I |
| 127 | D | 2.23 | 5.77 | 0.538 | I |
| 128 | R | 0.6 | 0.514 | I | |
| 129 | I | 0.43 | 0.538 | I | |
NB – non bonded; HB – H-bond; I – interactive.
3.2. Simulation of TREM-1/ligands complexes
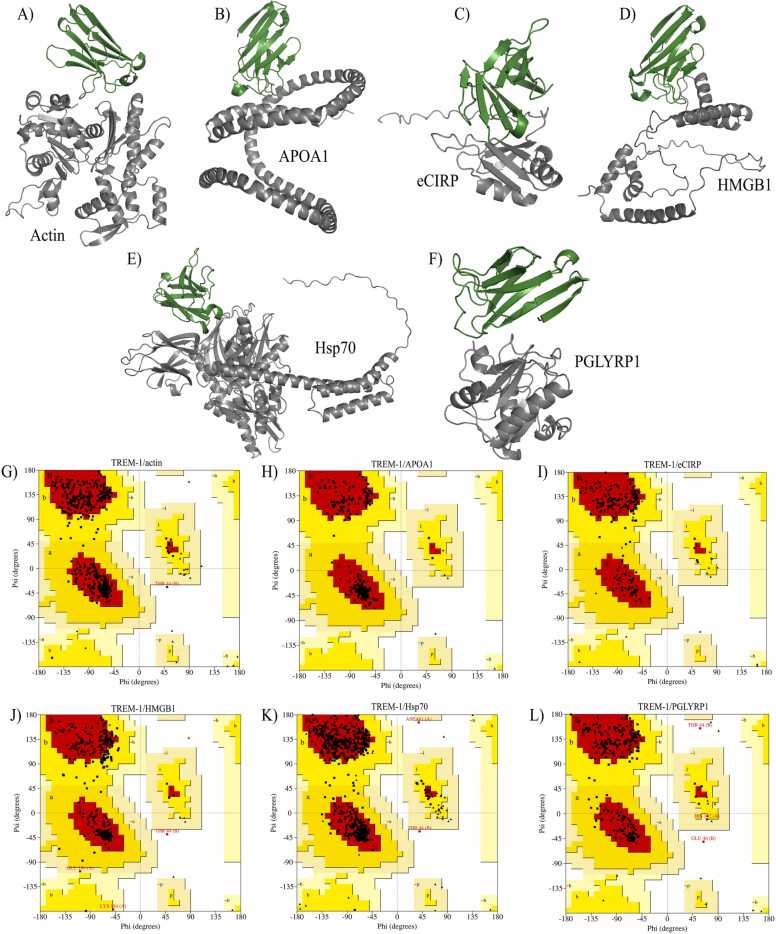
The prediction of the TREM-1/ligands complexes were conducted with Alphafold2-Multimer, with the amino acids sequences obtained from the following entries on Uniprot: P60709 (actin), Q14011 (eCIRP), P09429 (HMGB1), P0DMV8 (Hsp70), O75594 (PGLYRP1), P02647 (APOA1) and Q9NP99 (TREM-1) (Fig. 2A–F). We only included the sequence of the IgV-like domain of TREM-1 (aa 26–134), and excluded the peptide signal for the sequences of PGLYRP1 (aa 1–21) and APOA1 (aa 1–18). For eCIRP, we initially modeled its entire structure, however, due to an extensive disordered region (aa 81–175), we decided to predict a complex containing only the amino acids 1–110, which includes the region on eCIRP used for the development of a TREM-1 inhibitory peptide (M3, aa 101–107) [69].
Fig. 2.
Predicted TREM-1/ligands complexes. (A) TREM-1 and actin, (B) TREM-1 and APOA1, the negative control, (C) TREM-1 and eCIRP, (D) TREM-1 and HMGB1, (E) TREM-1 and Hsp70, (F) TREM-1 and PGLYRP1. Ramachandran plots for the complexes: TREM-1/actin (G), TREM-1/APOA1 (H), TREM-1/eCIRP (I), TREM-1/HMGB1 (J), TREM-1/Hsp70 (K), TREM-1/PGLYRP1 (L).
Regarding the confidence of the modeling process by Alphafold2-Multimer, predicted local distance difference test (pLLDT) values greater than 90 indicate high confidence, while values lower than 50, low confidence. In our prediction, all models had a pLLDT score higher than 50, but only the TREM-1/PGLYRP1 complex had a score indicating high confidence (pLLDT=94.3) (Table 2). Although the Molprobity analysis and Ramachandran plots (Fig. 2G–L) of the five TREM-1/ligands complexes indicated a relative quality of the modeled structures (Table 2), we decided to exclude the TREM-1/eCIRP and TREM-1/HMGB1 complexes from further analysis. This decision was based on uncertainties regarding the structures of these ligands, which could compromise the confidence of the predicted interactions: eCIRP still presented a large disordered region (aa 81–110), while HMGB1 has a long flexible domain (the acidic tail) and structural changes based on the its oxidative state [70], which we could not be simulated in silico.
Table 2.
Structural parameters of the selected TREM-1/ligands complexes.
| Protein | TREM-1 / Actin | TREM-1 / eCIRP1–110 | TREM-1 / HMGB1 | TREM-1 / Hsp70 | TREM-1 / PGRLYP1 | TREM-1 / APOA1 |
|---|---|---|---|---|---|---|
| pLDDT | 59.2 | 75.9 | 74 | 83.4 | 94.3 | 59.2 |
| pTM score | 0.337 | 0.547 | 0.393 | 0.766 | 0.908 | 0.337 |
| ipTM | 0.0939 | 0.231 | 0.206 | 0.387 | 0.872 | 0.0939 |
| MolProbity score | 1.33 | 1.99 | 1.82 | 1.62 | 1.48 | 2.06 |
| Clashscore | 3.29 | 9.59 | 4.4 | 6.53 | 3.6 | 7.68 |
| Poor rotamers | 1.91% | 0.53% | 0.70% | 1.11% | 1.22% | 6.98% |
| Favored rotamers | 94.99% | 97.33% | 94.41% | 96.66% | 95.93% | 84.13% |
| Bad bonds | 0% | 0% | 0% | 0% | 0% | 0% |
| Bad angles | 0.36% | 0.51% | 0.67% | 0.45% | 0.32% | 0.79% |
| Ramachandran’s most favored¹ | 93.6% | 88.2% | 82.1% | 93.4% | 89.3% | 98.1% |
| Ramachandran’s disallowed¹ | 0.2% | 0% | 0.4% | 0.3% | 0.8% | 0% |
APOA1 - Apolipoprotein A-I; eCIRP - extracellular cold-inducible RNA-binding protein; Hsp70–70 kDa heat shock protein; HMGB1 - high mobility group box 1 protein; dHMGB1 – disulfide HMGB1; rHMGB1 – reduced HMGB1; PGLYRP1 - peptidoglycan recognition protein 1; ¹Residues found in the most favored and disallowed regions of the Ramachandran plot, in analysis with PROCHECK.
The prediction of the intermolecular parameters between TREM-1 and actin, Hsp70 and PGLYRP1 was conducted with the HawkDock, Prodigy and CCharPPI webservers (Table 3). The TREM-1/Hsp70 pose presented a HawkDock’s free binding energy value of 14.49 kcal/mol, and results from CCharPPI’s PYDOCK_TOT and VDW outside the range expected for protein-protein complexes, therefore indicating that this pose was not stable and most likely did not represent the true conformation of this complex.
Table 3.
Results from the molecular docking assays.
| TREM-1 / actin | TREM-1 / PGLYRP1 | TREM-1 / Hsp70 | TREM-1 / APOA1 | |
|---|---|---|---|---|
| HawkDock's ∆G | -37.38 kcal/mol | -68.39 kcal/mol | 14.49 kcal/mol | -21.09 kcal/mol |
| Prodigy's ∆G | -7.4 | -9.3 | -7.2 | -6.2 |
| Prodigy's Kd (36.5 °C) | 6.4e-06 | 2.9e-07 | 7.8e-06 | 4.3e-05 |
| PYDOCK_TOT (−60 a −5) | -17.643 | -28.518 | -0.254 | 13.142 |
| VDW (−200 a −50) | -48.749 | -63.885 | -24.858 | -8.202 |
| HBOND (−15 a −1) | -3.72 | -5.46 | -6.21 | -8.09 |
| FA_ATR (−100 a −20) | -24.716 | -38.314 | -34.739 | -37.849 |
| ELE (−60 a 0) | -13.78 | -25.822 | -7.756 | -8.836 |
| DESOLV (−30 a 20) | 1.012 | 3.692 | 9.988 | 22.799 |
Regarding TREM-1/actin and TREM-1/PGLYRP1 complexes, we obtained negative values for the HawkDock’s free binding energy (−37.38 kcal/mol and −68.39 kcal/mol, respectively), and results for the CCharPPI descriptors within the expected ranges. These values were considerably better than those observed for the TREM-1/APOA1 complex, which is our negative control. The simulated TREM-1 and APOA1 complex had a positive value for the PYDOCK_TOT descriptor. Further, it also showed the worst values for the binding free energies and the dissociation constant (−21.09 kcal/mol, −6.2 kcal/mol, 4.3e-05), in comparison to TREM-1/actin and TREM-1/PGLYRP1. Furthermore, the TREM-1/PGLYRP1 showed the most intense predicted interaction, with the most negative values for both HawkDock’s and Prodigy’s binding free energies (−68.39 kcal/mol, −9.3 kcal/mol), besides the best overall dissociation constant (2.9e-07) (Table 3).
3.3. TREM-1/ligands interaction interfaces
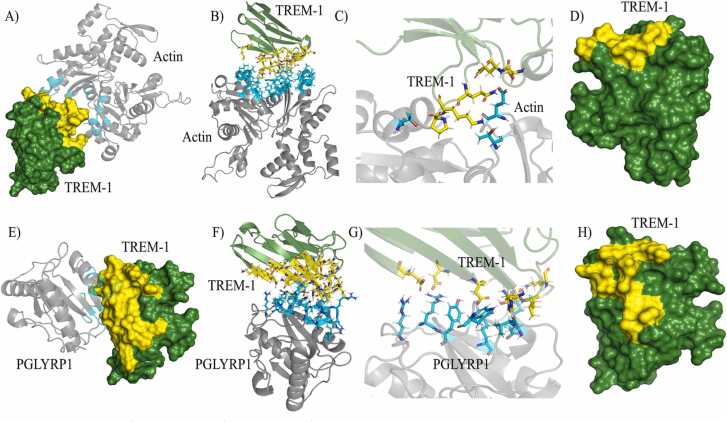
The regions comprising the interaction between TREM-1, actin and PGLYRP1, are depicted on Fig. 3. Regarding TREM-1/actin (Fig. 3A–D), from the 60 residues predicted by PPCheck, TREM-1 interacts with 25 residues as follows: I115, Y116, Q117, P118, P119, K120, E121, P122, E46, K47, F48, A49, S50, S51, Q52, K53, M63, P64, E71, R72, P73, S74, K75, N76 and H91 (Fig. 3A–B). Furthermore, the DIMPLOT software indicated five hydrogen bonds: E167/Q52, E167/Q117, E167/S51, T351/P119 and T148/K120 (actin/TREM-1) (Fig. 3C). Finally, the amino acids predicted as the main mediators of interactions by HawkDock (ie. had binding free energies <−1 kcal/mol) were: S51, P119, P73, Q117, R72, Q52 and S50.
Fig. 3.
The simulated TREM-1/actin and TREM-1/PGLYRP1 complexes. (A – D) TREM-1 (green) and actin (gray), with interacting residues in yellow and cyan, respectively. (C) Hydrogen bonds in TREM-1/actin predicted by DIMPLOT. (D) Combination of residues with binding free energies < −1 kcal/mol and/or involved in hydrogen bonds or salt bridges from TREM-1/actin. Yellow: TREM-1 residues, cyan: actin residues. (E – H) TREM-1 (green) and PGLYRP1 (gray), with interacting residues in yellow and cyan, respectively. (G) Hydrogen bonds and salt bridges in TREM-1/PGLYRP1 predicted by DIMPLOT. (H) Combination of residues with binding free energies < −1 kcal/mol and/or involved in hydrogen bonds or salt bridges from TREM-1/PGLYRP1. Yellow: TREM-1 residues, cyan: PGLYRP1 residues.
For TREM-1/PGLYRP1, the full interface predicted by PPCheck included 59 amino acids, from which, 32 were from TREM-1: H123, L125, F126, D127, R128, E26, E27, K28, Y29, D38, V39, K40, C41, D42, Y43, T44, L45, E46, K47, F48, N76, S77, E88, D89, Y90, H91, D92, H93, G94, L95, L96 and R97 (Fig. 3E–F). DIMPLOT detected 11 hydrogen bonds (two between R55/D127, R55/E27, R162/E27, Y56/D42, K168/D42, K168/T44, P196/K47, D172/L45, V173/H93 and W122/H93 [PGLYRP1/TREM-1]) and one salt bridge (R194/D127 [PGLYRP1/TREM-1]) (Fig. 3G). Furthermore, the amino acids predicted as the main mediators of interactions by HawkDock, were: L45, D127, T44, L95, H93, K47, E27, D42, F126 and Y90.
To highlight the residues that are likely the main mediators of antigen recognition in the TREM-1/actin and TREM-1/PGLYRP1 complexes, the residues with HawkDock’s binding free energies < −1 kcal/mol, and/or amino acids involved in hydrogen bonds or salt bridges, are depicted in yellow on Fig. 3D and H.
3.4. Molecular dynamics
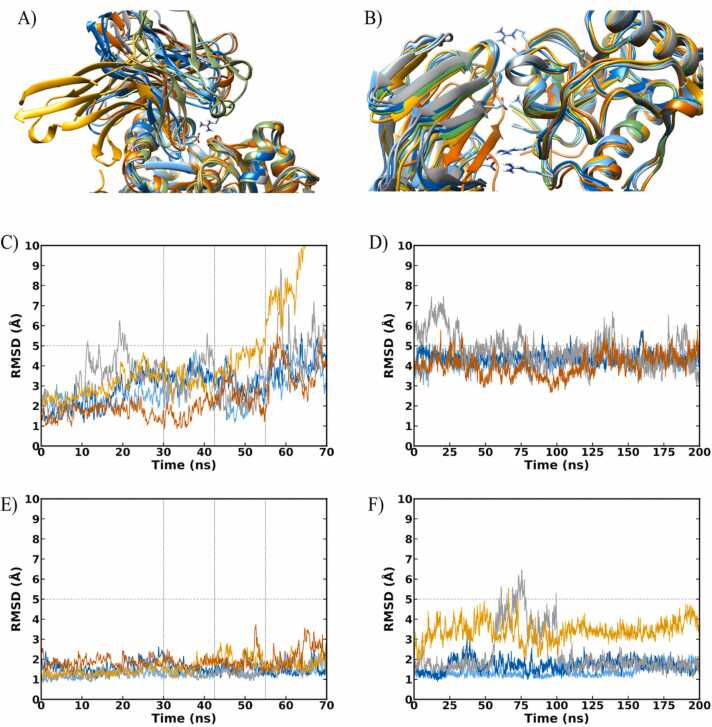
The selected docked poses of TREM-1/actin and TREM-1/PGLYRP1 were submitted to molecular dynamics simulations, to further evaluate the stability and intermolecular interactions of these poses (Fig. 4).
Fig. 4.
Molecular dynamics simulations for TREM-1/actin and TREM-1/PGLYRP1. (A) Last frames of each replicate of the TREM1 complex with (A) actin and (B) PGLYRP1, represented in ribbons. The interface RMSD during the five replicate stepwise heated simulations for the (C) TREM-1/actin and (D) TREM-1/PGLYRP1 complexes are also shown. The interface RMSD for each simulation during the extended conventional MD is illustrated in (E) and (F) for TREM1-Actin and TREM1-PGLYRP1, respectively. Each color of the lines represents one replicate and corresponds to the color of the ribbons. The green ribbons represent the reference structure obtained from AlphaFold2-Multimer. Sticks are used to depict the residues performing the most relevant interactions. Dashed lines represents a cutoff of acceptable interface RMSD during the simulations.
Our results demonstrated a stable interaction in the TREM-1/actin proposed complex, as four out of five simulations maintained an interface RMSD below 5 Å throughout the simulation (Fig. 4A–C). Further analysis showed that the final structures of each simulation converged to a distinctive pose, with a maximum RMSD interface below 4 Å when comparing the last frames of each simulation (Table S2). Notably, the loop comprising residues Q117 to P122 of TREM-1 consistently interacted with a cavity formed by actin residues Y133, Y143, L346, F352, M355 and C374 in all five replicates (Fig. S1). Another conserved interaction observed across the replicates was a salt bridge between TREM-1 R72 and the natural carboxylate termination of actin. This interaction was absent in the reference structure and was not observed in replicate 1, suggesting a possible reason for the divergent binding mode observed in our converged pose (Fig. S2). These findings suggest that the loop-cavity and R72-Cterm interactions may play crucial roles in establishing the TREM-1/actin complex.
The PGLYRP1 protein is suggested to interact with the outer sheet of TREM-1, which is distinct from its interaction with the loops observed in the actin complex. This finding is supported by the highly reliable Alphafold2-Multimer prediction, which was reinforced by interface RMSD values as low as 2 Å in four out of five replicates, even after performing a stepwise heated simulation followed by a 200 ns long MD (Fig. 4D–F, Table S3). The interaction between PGLYRP1 and TREM1 was found to be predominantly mediated by salt bridges, as evidenced by the occurrence of four specific pairs (E27/R162, D42/K168, D127/R55 and E46/R175 [TREM-1/PGLYRP1]) for over 50% of the last 10 ns of simulations in four out of the five replicates (Fig. 4D, Table S4). Notably, a hydrogen bond interaction between TREM-1 H93 and PGLYRP1 W122 was prevalent even in the simulation with the highest RMSD.
3.5. Putative binding domains on TREM-1
The residues predicted by the hot spot analysis, and molecular docking between TREM-1/actin and TREM-1/PGLYRP1, are displayed on Fig. 5A. There were some differences regarding the TREM-1 residues observed in each case. Thus, to predict the putative binding domains within the receptor, we combined the amino acids predicted as hot spots by both FTMap and SeRenDIP, and as the main mediators of antigen recognition in the TREM-1/actin and TREM-1/PGLYRP1 complexes (i.e. binding free energies <−1 kcal/mol and/or involved in hydrogen bonds or salt bridges, marked with * on Fig. 5A). As a result, the following residues were defined as the most likely involved in antigen recognition by TREM-1: E27, L31, V39, D42, T44, L45, K47, S50-Q52, Q56, I57, T66-C69, R72, P73, G83, Y90, H93, L95, M100, D107, Y111, V114, Q117, P119-I129 (yellow, Fig. 5B). Although a clear domain could not be predicted by our results, is possible to observe that CDR3 (aa 116–128) is by far the most involved region, followed by CDR1. The full monomeric TREM-1 structure, predicted by Alphafold (ID: AF-Q9NP99-F1), is depicted in Fig. 5B with the different domains, including the putative binding site, highlighted in different colors.
Fig. 5.
The putative binding domains on TREM-1. (A) Residues from the IgV-like domain of TREM-1 observed in the interaction interfaces (*residues with binding free energies <−1 kcal/mol and/or involved in hydrogen bonds or salt bridges) and in the hot spots analysis. (B) Full structure of TREM-1 with its different domains and the predicted putative binding domain (in yellow). PDB file downloaded from the Alphafold database (https://alphafold.ebi.ac.uk/entry/Q9NP99).
4. Discussion
In our study, we used different in silico strategies to predict the still elusive binding domain of TREM-1. Combining hot spot analysis by two different approaches, and the simulation of the TREM-1 complexes with its stablished protein ligands, we can suggest that the CDR1 and, specially, CDR3, seem to be the main regions of TREM-1 that mediate protein-protein interactions. The importance of the CDRs to antigen recognition by TREM-1 has been previously suggested, as these loop sites are described in receptors from the immunoglobulin superfamily as the antigen recognition domains [25].
The characterization of TREM-1 binding sites was previously attempted by Radaev et al. [26] and Kelker et al. [25], in their crystallographic studies with TREM-1′s IgV-like domain. However, they were unable to define such regions but suggested the CDR loops, especially CDR3 (the F-G loop, that appeared as a flexible area), as the domain most likely involved with ligand recognition. Another topic that remained poorly explored for many years was the nature of the TREM-1 ligands. The receptor was first described as a pro-inflammatory structure activated in neutrophils and monocytes stimulated by LPS [2]. Accordingly, subsequent studies showed that TREM-1 could be activated in the presence of several MAMPs and DAMPs [4], [5]. However, as the receptor was shown to work in cooperation with different PRRs, whether these molecules directly bound to TREM-1, or promoted its activation indirectly, was unknown. In the last decade, studies using techniques such as immunoblotting, affinity chromatography and surface plasmon resonance, identified five molecules that directly bonded to TREM-1 and, therefore, were considered ligands of the receptor [32].
Here, we started our search for the TREM-1 binding domain by conducting hot spots analysis with different approaches, resulting in the observation of 26 predicted hot spots residues, and the CDR3 as the most likely main domain of antigen recognition. Next, we tried to simulate the interactions between TREM-1 and its five known ligands with molecular docking and dynamics. In this step, we found some limitations regarding the confidence in the predicted structures of certain ligands, and in the selection of the correct docked pose. Therefore, from the five TREM-1/ligands complexes predicted, only two were further explored, regarding its stability and regions of interaction.
The peptidoglycan recognition protein 1, PGLYRP1, was described as a TREM-1 ligand in 2015, by Read and cols [71]. PGLYRP1 is a secreted protein with antimicrobial and antitumor activities in mammals, commonly detected in neutrophils tertiary granules [72], [73]. Interestingly, the interaction between TREM-1 and PGLYRP1 seems to require a co-factor. Carrasco et al. [17] showed that, in LPS-primed monocytes, the addition of PGLYRP1 was not enough to promote TREM-1 dimerization and, therefore, its activation. Peptidoglycan may be one co-factor, as Read et al. [71] showed that this molecule was essential for soluble PGLYRP1 to activate TREM-1. However, the existence of others co-factors able to bind to PGLYRP1, cannot be ruled out, as this protein was able to activate the receptor in monocytes in the absence of PGN [30], [74]. Despite this fact, we did not consider peptidoglycan or any other co-factors in our simulations. In this regard, it is important to mention that some scientific data suggest that, in fact, PGLYRP1 is the molecule that physically interacts with TREM-1, while the co-factor may mediates the cross-linking of TREM-1 on the cell surface [17], [71].
It was initially proposed that platelets express a TREM-1 ligand on its surface, however, a previous study was unable to identify such ligand [75]. Later, Fu et al. [76] showed the co-localization of actin and TREM-1 during the activation of macrophages by platelets, and demonstrated that extracellular actin could activate TREM-1 [76]. Actin is a ubiquitous protein, biologically observed as a monomer (G-actin) or polymer (F-actin), and in different isoforms, including α-, β- and γ-actin [77]. The isoform that was implicated as a TREM-1 ligand was β-actin [76], thus, this was the isoform used in our study. Despite this, a putative interaction between the other actin isoforms (α and γ) and the receptor has never been addressed. Although best known as a cytoskeleton protein, actin can be found on the cell surface and as an extracellular protein [75], [77], [78], [79]. Soluble actin is released by dying cells, and has been considered as a biomarker of tissue damage and a DAMP, with interactions described with DNGR-1 (also known as CLEC9A), C1q (a complement system protein), and TREM-1 [77], [80], [81], [82], [83].
Regarding the ligands that did not result in reliable complexes, we believe that the main limitation was the correct definition of the protein structures. Both eCIRP and HMGB1 still do not have full crystallographic structures available. Limiting factors also included the oxidative state-dependent conformations of HMGB1, besides the presence of the flexible acidic tail on this molecule and a long disordered region on eCIRP, which hinders the confidence of molecular docking and molecular dynamics simulations [70], [84], [85], [86]. Finally, we were able to predict a TREM-1/Hsp70 complex, but the energetic parameters of the selected pose did not indicated the stability of this interaction.
HMGB1 is a DNA-binding protein that can be passively or actively released by different cells, thus acting as a DAMP [87]. The different roles of this molecule depend on its location and, specially, its redox state [87]. Two DNA-binding boxes are present in the structure of HMGB1 and, within these portions, three cysteines (C23 and C45 in Box A, C106 in Box B) are critical for the conformation and function of HMGB1. Connected to Box A and B there is a flexible acidic tail, that is able to interact with specific region of the boxes [86]. The HMGB1 conformation with the three cysteines fully reduced (rHMGB1) promotes monocyte recruitment through the formation of a complex with CXCL12, thus resulting in the activation of the CXCR4 receptor [88]. The mildly oxidized form of HMGB1, with a disulfide bond between residues C23 and C45 (dHMGB1), is able to activate TLR4 after interacting with the co-receptor MD-2, leading to cytokine secretion [89]. The third form, originated from further oxidation, has sulfonyl groups on the cysteine residues and has no known function [87]. It is important to mention that Wu et al. [31] did not define which HMGB1 conformation was interacting with TREM-1, which represented a first obstacle for the in silico simulation of the TREM-1/HMGB1 complex. Furthermore, there are no crystallographic structures of the full rHMGB1 and dHMGB1 proteins, which also presented a major drawback in our study, as we could not predict such structural changes with acceptable accuracy. For the future, the use of different modeling strategies, and the use of the separated HMGB1 domains for the molecular docking simulations, may help the study of the TREM-1/HMGB1 interaction.
The interaction between TREM-1 and Hsp70, was initially ruled out by Wu et al. [31], however, it was confirmed by a more recent study [30]. The Hsp70 family of proteins is observed in physiological conditions, promoting proper protein folding, however, they are increased during stress, when they act to promote cell survival [90]. Extracellular Hsp70, which can be found free or associated with exomes and antigens, is able to induce the secretion of pro-inflammatory cytokines [90], [91], [92]. Many receptors have been identified to interact with Hsp70, such as CD14, CD35, CD40, CD91, TLR2, TLR4, some scavenger receptors and TREM-1 [90]. Among the Hsp70 family members, the HSPA1A isoform was shown to interact with TREM-1 [30], thus, this was the isoform selected by our group. Although the Hsp70 family is one of the most conserved protein families [90], it is possible that the discrepancies between studies regarding the recognition of Hsp70 by TREM-1, such as those observed by Wu et al. [31] and Sharapova et al. [30], rely on the use of distinct Hsp70 isoforms. However, we cannot confirm or deny such assumption as Wu and cols have not clarified the Hsp70 isoform used in their study.
CIRP, or CIRBP, was first describe in the late 1990s as an RNA chaperone controlling the cell cycle of hibernating animals [93]. This protein is composed by a RNA binding region (aa 6–84), a disordered domain (aa 70–172) and a region with polar residues (aa 143–172) [85]. Aside its role as regulator of the cellular response to stress, the presence of an extracellular form of this protein, eCIRP, was described and has been investigated in different cell types and disease contexts, especially for therapeutic purposes [94]. Initially shown as a ligand for TLR4/MD-2 [95], eCIRP was proposed to be a TREM-1 ligand by Denning and collaborators [96]. The authors also explored this interaction to develop a new inhibitor of TREM-1, M3, consisting of 7 amino acids from murine CIRP [96]. They compared the structure of murine PGLYRP1 and CIRP, and identified a portion in CIRP (RGFFRGG, aa 101–107) with similar sequence and conformation to that observed in PGLYRP1 (RGFLR, aa 145–149), which was also conserved in human CIRP. The M3 peptide inhibited exacerbated inflammation in sepsis and a murine model of intestinal ischemia-reperfusion injury [69], [96], [97], highlighting the potential of exploring the interfaces between TREM-1 and its ligands for drug development.
Despite the limitations of our study, we believe that our results might shed a light on the subject of ligand recognition by TREM-1, showing the prominent role of CDR3 in this process. These results may assist future experiments to confirm how TREM-1 binds to its ligands and can be useful for further in silico investigations with the receptor, besides the development of new therapeutic approaches targeting TREM-1 in inflammatory and infectious diseases.
5. Limitations
Our study had some limitations regarding the ability of our selected tools to predict the structure of two TREM-1 ligands, HMGB1 and eCIRP, and the right conformation of the TREM-1/Hsp70 complex. In addition, some ligands, more specifically HMGB1, presents a great flexibility, which impaired the molecular dynamics. Therefore, future biological assays may prove whether the binding domain that we observed here is biologically involved in interactions between TREM-1 and its ligands.
6. Conclusions
Using different in silico approaches, we simulated interactions between TREM-1 and its known ligands: actin, eCIRP, HMGB1, Hsp70 and PGLYRP1. Comparing the TREM-1 residues observed in the interfaces of the stable poses (TREM-1/actin and TREM-1/PGLYRP1) in addition to those predicted in the hot spots analysis, we proposed that the binding domain within the IgV-like portion of TREM-1 is mainly driven by the CDR3 region, which seems to play a pivotal role in the antigen recognition process. Finally, we believe our results can be used in future experimental studies involving the discovery of new TREM-1 ligands and may foster the discovery of new therapeutic approaches aiming at modulating the activity of the receptor.
CRediT authorship contribution statement
Amanda de Oliveira Matos: Conceptualization, Methodology, Formal analysis, Writing – original draft, Visualization, Project administration. Pedro Henrique dos Santos Dantas: Conceptualization, Methodology, Writing – review & editing. Mike Telemaco Contreras Colmenares: Methodology, Formal analysis, Writing – final draft. Geraldo Rodrigues Sartori: Methodology, Formal analysis, Writing – final draft. Marcelle Silva-Sales: Writing – review & editing. João Herminio Martins Da Silva: Methodology, Resources. Bruno Junior Neves: Methodology, Resources, Writing – review & editing, Funding acquisition. Carolina Horta Andrade: Methodology, Writing – review & editing, Funding acquisition. Helioswilton Sales-Campos: Conceptualization, Methodology, Writing – review & editing, Resources, Supervision, Funding acquisition. All authors have read and approved the final version of this manuscript.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Helioswilton Sales-Campos reports financial support was provided by Fundação de Apoio a Pesquisa (FUNAPE). Carolina Horta Andrade reports a relationship with Fundação de Amparo a Pesquisa do Estado de Goiás (FAPEG) that includes: funding grants. Bruno Junior Neves reports a relationship with Fundação de Amparo a Pesquisa do Estado de Goiás (FAPEG) that includes: funding grants. Carolina Horta Andrade reports a relationship with Conselho Nacional de Desenvolvimento Científico (CNPq) that includes: funding grants. Bruno Junior Neves reports a relationship with Conselho Nacional de Desenvolvimento Científico (CNPq) that includes: funding grants. Helioswilton Sales-Campos reports a relationship with Fundação de Amparo a Pesquisa (FUNAPE) that includes: funding grants. Bruno Junior Neves reports a relationship with Fundação de Amparo a Pesquisa (FUNAPE) that includes: funding grants.
Acknowledgments
The authors would like to thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for A.O.M and P.H.S.D scholarships, Conselho Nacional de Desenvolvimento Científico (CNPq – 308732/2022-7) for HSC fellowship, and Fundação de Amparo a Pesquisa (FUNAPE). CHA and BJN also thanks FAPEG (#202010267000272 CHA) and CNPq (# 441038/2020-4 CHA) for financial support.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.04.008.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Bosco M.C., Pierobon D., Blengio F., Raggi F., Vanni C., Gattorno M., et al. Hypoxia modulates the gene expression profile of immunoregulatory receptors in human mature dendritic cells: identification of TREM-1 as a novel hypoxic marker in vitro and in vivo. Blood. 2011;117:2625–2639. doi: 10.1182/blood-2010-06-292136. [DOI] [PubMed] [Google Scholar]
- 2.Bouchon A., Dietrich J., Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 1950;164(2000):4991–4995. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 3.Gingras M.C., Lapillonne H., Margolin J.F. TREM-1, MDL-1, and DAP12 expression is associated with a mature stage of myeloid development. Mol Immunol. 2002;38:817–824. doi: 10.1016/s0161-5890(02)00004-4. [DOI] [PubMed] [Google Scholar]
- 4.de Oliveira Matos A., Dos Santos Dantas P.H., Figueira Marques Silva-Sales M., Sales-Campos H. The role of the triggering receptor expressed on myeloid cells-1 (TREM-1) in non-bacterial infections. Crit Rev Microbiol. 2020;46:237–252. doi: 10.1080/1040841X.2020.1751060. [DOI] [PubMed] [Google Scholar]
- 5.Matos A.O., Dantas P., Silva-Sales M., Sales-Campos H. TREM-1 isoforms in bacterial infections: to immune modulation and beyond. Crit Rev Microbiol. 2021;47:290–306. doi: 10.1080/1040841X.2021.1878106. [DOI] [PubMed] [Google Scholar]
- 6.Bosco M.C., Raggi F., Varesio L. Therapeutic potential of targeting TREM-1 in inflammatory diseases and cancer. Curr Pharm Des. 2016;22:6209–6233. doi: 10.2174/1381612822666160826110539. [DOI] [PubMed] [Google Scholar]
- 7.Gao S., Yuan L., Wang Y., Hua C. Enhanced expression of TREM-1 in splenic cDCs in lupus prone mice and it was modulated by miRNA-150. Mol Immunol. 2017;81:127–134. doi: 10.1016/j.molimm.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Natale G., Biagioni F., Busceti C.L., Gambardella S., Limanaqi F., Fornai F. TREM receptors connecting bowel inflammation to neurodegenerative disorders. Cells. 2019;8 doi: 10.3390/cells8101124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palazzo S.J., Simpson T.A., Simmons J.M., Schnapp L.M. Soluble triggering receptor expressed on myeloid cells-1 (sTREM-1) as a diagnostic marker of ventilator-associated pneumonia. Respir Care. 2012;57:2052–2058. doi: 10.4187/respcare.01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tammaro A., Derive M., Gibot S., Leemans J.C., Florquin S., Dessing M.C. TREM-1 and its potential ligands in non-infectious diseases: from biology to clinical perspectives. Pharmacol Ther. 2017;177:81–95. doi: 10.1016/j.pharmthera.2017.02.043. [DOI] [PubMed] [Google Scholar]
- 11.Begum N.A., Ishii K., Kurita-Taniguchi M., Tanabe M., Kobayashi M., Moriwaki Y., et al. Mycobacterium bovis BCG cell wall-specific differentially expressed genes identified by differential display and cDNA subtraction in human macrophages. Infect Immun. 2004;72:937–948. doi: 10.1128/IAI.72.2.937-948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gómez-Piña V., Soares-Schanoski A., Rodríguez-Rojas A., Del Fresno C., García F., Vallejo-Cremades M.T., et al. Metalloproteinases shed TREM-1 ectodomain from lipopolysaccharide-stimulated human monocytes. J Immunol. 1950;179(2007):4065–4073. doi: 10.4049/jimmunol.179.6.4065. [DOI] [PubMed] [Google Scholar]
- 13.Mahdy A.M., Lowes D.A., Galley H.F., Bruce J.E., Webster N.R. Production of soluble triggering receptor expressed on myeloid cells by lipopolysaccharide-stimulated human neutrophils involves de novo protein synthesis. Clin Vaccin Immunol. 2006;13:492–495. doi: 10.1128/CVI.13.4.492-495.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao C., Gu J., Zhang J. Soluble triggering receptor expressed on myeloid cell-1 (sTREM-1): a potential biomarker for the diagnosis of infectious diseases. Front Med. 2017;11:169–177. doi: 10.1007/s11684-017-0505-z. [DOI] [PubMed] [Google Scholar]
- 15.Van Singer M., Brahier T., Ngai M., Wright J., Weckman A.M., Erice C., et al. COVID-19 risk stratification algorithms based on sTREM-1 and IL-6 in emergency department. J Allergy Clin Immunol. 2021;147:99–106. doi: 10.1016/j.jaci.2020.10.001. e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arts R.J., Joosten L.A., van der Meer J.W., Netea M.G. TREM-1: intracellular signaling pathways and interaction with pattern recognition receptors. J Leukoc Biol. 2013;93:209–215. doi: 10.1189/jlb.0312145. [DOI] [PubMed] [Google Scholar]
- 17.Carrasco K., Boufenzer A., Jolly L., Le Cordier H., Wang G., Heck A.J., et al. TREM-1 multimerization is essential for its activation on monocytes and neutrophils. Cell Mol Immunol. 2019;16:460–472. doi: 10.1038/s41423-018-0003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colonna M., Facchetti F. TREM-1 (triggering receptor expressed on myeloid cells): a new player in acute inflammatory responses. J Infect Dis. 2003;187(Suppl. 2):S397–S401. doi: 10.1086/374754. [DOI] [PubMed] [Google Scholar]
- 19.Zhu J., Duan G., Wang H., Cao M., Liu Y. TREM-1 activation modulates dsRNA induced antiviral immunity with specific enhancement of MAPK signaling and the RLRs and TLRs on macrophages. Exp Cell Res. 2016;345:70–81. doi: 10.1016/j.yexcr.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 20.Dantas P., Matos A.O., da Silva Filho E., Silva-Sales M., Sales-Campos H. Triggering receptor expressed on myeloid cells-1 (TREM-1) as a therapeutic target in infectious and noninfectious disease: a critical review. Int Rev Immunol. 2020;39:188–202. doi: 10.1080/08830185.2020.1762597. [DOI] [PubMed] [Google Scholar]
- 21.Gibot S., Massin F., Marcou M., Taylor V., Stidwill R., Wilson P., et al. TREM-1 promotes survival during septic shock in mice. Eur J Immunol. 2007;37:456–466. doi: 10.1002/eji.200636387. [DOI] [PubMed] [Google Scholar]
- 22.Klesney-Tait J., Keck K., Li X., Gilfillan S., Otero K., Baruah S., et al. Transepithelial migration of neutrophils into the lung requires TREM-1. J Clin Investig. 2013;123:138–149. doi: 10.1172/JCI64181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber B., Schuster S., Zysset D., Rihs S., Dickgreber N., Schürch C., et al. TREM-1 deficiency can attenuate disease severity without affecting pathogen clearance. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C., Zhao J., Lin L., Pan S., Fu L., Han L., et al. Targeting TREM-1 signaling in the presence of antibiotics is effective against streptococcal toxic-shock-like syndrome (STSLS) caused by Streptococcus suis. Front Cell Infect Microbiol. 2015;5:79. doi: 10.3389/fcimb.2015.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelker M.S., Foss T.R., Peti W., Teyton L., Kelly J.W., Wüthrich K., et al. Crystal structure of human triggering receptor expressed on myeloid cells 1 (TREM-1) at 1.47 A. J Mol Biol. 2004;342:1237–1248. doi: 10.1016/j.jmb.2004.07.089. [DOI] [PubMed] [Google Scholar]
- 26.Radaev S., Kattah M., Rostro B., Colonna M., Sun P.D. Crystal structure of the human myeloid cell activating receptor TREM-1. Structure. 2003:1527–1535. doi: 10.1016/j.str.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Bouchon A., Facchetti F., Weigand M.A., Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 28.El Mezayen R., El Gazzar M., Seeds M.C., McCall C.E., Dreskin S.C., Nicolls M.R. Endogenous signals released from necrotic cells augment inflammatory responses to bacterial endotoxin. Immunol Lett. 2007;111:36–44. doi: 10.1016/j.imlet.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rolli J., Loukili N., Levrand S., Rosenblatt-Velin N., Rignault-Clerc S., Waeber B., et al. Bacterial flagellin elicits widespread innate immune defense mechanisms, apoptotic signaling, and a sepsis-like systemic inflammatory response in mice. Crit Care. 2010;14:R160. doi: 10.1186/cc9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharapova T.N., Romanova E.A., Ivanova O.K., Yashin D.V., Sashchenko L.P. Hsp70 interacts with the TREM-1 receptor expressed on monocytes and thereby stimulates generation of cytotoxic lymphocytes active against MHC-negative tumor cells. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22136889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J., Li J., Salcedo R., Mivechi N.F., Trinchieri G., Horuzsko A. The proinflammatory myeloid cell receptor TREM-1 controls Kupffer cell activation and development of hepatocellular carcinoma. Cancer Res. 2012;72:3977–3986. doi: 10.1158/0008-5472.CAN-12-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh H., Rai V., Nooti S.K., Agrawal D.K. Novel ligands and modulators of triggering receptor expressed on myeloid cells receptor family: 2015-2020 updates. Expert Opin Ther Pat. 2021;31:549–561. doi: 10.1080/13543776.2021.1883587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozakov D., Grove L.E., Hall D.R., Bohnuud T., Mottarella S.E., Luo L., et al. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc. 2015;10:733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenke R., Kozakov D., Chuang G.Y., Beglov D., Hall D., Landon M.R., et al. Fragment-based identification of druggable 'hot spots' of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25:621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozakov D., Hall D.R., Chuang G.Y., Cencic R., Brenke R., Grove L.E., et al. Structural conservation of druggable hot spots in protein-protein interfaces. Proc Natl Acad Sci USA. 2011;108:13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohnuud T., Beglov D., Ngan C.H., Zerbe B., Hall D.R., Brenke R., et al. Computational mapping reveals dramatic effect of Hoogsteen breathing on duplex DNA reactivity with formaldehyde. Nucleic Acids Res. 2012;40:7644–7652. doi: 10.1093/nar/gks519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou Q., Stringer B., Waury K., Capel H., Haydarlou R., Xue F., et al. SeRenDIP-CE: sequence-based interface prediction for conformational epitopes. Bioinformatics. 2021;37:3421–3427. doi: 10.1093/bioinformatics/btab321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou Q., De Geest P.F.G., Griffioen C.J., Abeln S., Heringa J., Feenstra K.A. SeRenDIP: SEquential REmasteriNg to DerIve profiles for fast and accurate predictions of PPI interface positions. Bioinformatics. 2019;35:4794–4796. doi: 10.1093/bioinformatics/btz428. [DOI] [PubMed] [Google Scholar]
- 39.Hou Q., De Geest P.F.G., Vranken W.F., Heringa J., Feenstra K.A. Seeing the trees through the forest: sequence-based homo- and heteromeric protein-protein interaction sites prediction using random forest. Bioinformatics. 2017;33:1479–1487. doi: 10.1093/bioinformatics/btx005. [DOI] [PubMed] [Google Scholar]
- 40.Yeh F.L., Wang Y., Tom I., Gonzalez L.C., Sheng M. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron. 2016;91:328–340. doi: 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 41.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci Publ Protein Soc. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskowski R.A., Macarthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 44.Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 45.Chen F., Liu H., Sun H., Pan P., Li Y., Li D., et al. Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein-protein binding free energies and re-rank binding poses generated by protein-protein docking, Physical chemistry chemical physics. PCCP. 2016;18:22129–22139. doi: 10.1039/c6cp03670h. [DOI] [PubMed] [Google Scholar]
- 46.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun H., Li Y., Tian S., Xu L., Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys Chem Chem Phys. 2014;16:16719–16729. doi: 10.1039/c4cp01388c. [DOI] [PubMed] [Google Scholar]
- 48.Honorato R.V., Koukos P.I., Jiménez-García B., Tsaregorodtsev A., Verlato M., Giachetti A., et al. Structural biology in the clouds: the WeNMR-EOSC ecosystem. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.729513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vangone A., Bonvin A. PRODIGY: a contact-based predictor of binding affinity in protein-protein complexes. Bio-Protocol. 2017;7 doi: 10.21769/BioProtoc.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 51.Moal I.H., Jiménez-García B., Fernández-Recio J. CCharPPI web server: computational characterization of protein–protein interactions from structure. Bioinformatics. 2014;31:123–125. doi: 10.1093/bioinformatics/btu594. [DOI] [PubMed] [Google Scholar]
- 52.Cheng T.M., Blundell T.L., Fernandez-Recio J. pyDock: electrostatics and desolvation for effective scoring of rigid-body protein-protein docking. Proteins. 2007;68:503–515. doi: 10.1002/prot.21419. [DOI] [PubMed] [Google Scholar]
- 53.Grosdidier S., Pons C., Solernou A., Fernández-Recio J. Prediction and scoring of docking poses with pyDock. Proteins. 2007;69:852–858. doi: 10.1002/prot.21796. [DOI] [PubMed] [Google Scholar]
- 54.Andrusier N., Nussinov R., Wolfson H.J. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 55.Chaudhury S., Lyskov S., Gray J.J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics. 2010;26:689–691. doi: 10.1093/bioinformatics/btq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sukhwal A., Sowdhamini R. Oligomerisation status and evolutionary conservation of interfaces of protein structural domain superfamilies. Mol Biosyst. 2013;9:1652–1661. doi: 10.1039/c3mb25484d. [DOI] [PubMed] [Google Scholar]
- 57.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 58.Olsson M.H., Søndergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 59.Tian C., Kasavajhala K., Belfon K.A.A., Raguette L., Huang H., Migues A.N., et al. ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J Chem Theory Comput. 2020;16:528–552. doi: 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- 60.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput-Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 61.Feenstra K.A., Hess B., Berendsen H.J.C. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J Comput Chem. 1999;20:786–798. doi: 10.1002/(SICI)1096-987X(199906)20:8<786::AID-JCC5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 62.Hopkins C.W., Le Grand S., Walker R.C., Roitberg A.E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J Chem Theory Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 63.Shirts M.R., Klein C., Swails J.M., Yin J., Gilson M.K., Mobley D.L., et al. Lessons learned from comparing molecular dynamics engines on the SAMPL5 dataset. J Comput-Aided Mol Des. 2017;31:147–161. doi: 10.1007/s10822-016-9977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Case D.A., Cheatham III T.E., Darden T., Gohlke H., Luo R., Merz Jr K.M., et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 66.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 67.Radom F., Plückthun A., Paci E. Assessment of ab initio models of protein complexes by molecular dynamics. PLoS Comput Biol. 2018;14 doi: 10.1371/journal.pcbi.1006182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roe D.R., Cheatham T.E., 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 69.Denning N.L., Aziz M., Ochani M., Prince J.M., Wang P. Inhibition of a triggering receptor expressed on myeloid cells-1 (TREM-1) with an extracellular cold-inducible RNA-binding protein (eCIRP)-derived peptide protects mice from intestinal ischemia-reperfusion injury. Surgery. 2020;168:478–485. doi: 10.1016/j.surg.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang D., Kang R., Zeh H.J., 3rd, Lotze M.T. High-mobility group box 1, oxidative stress, and disease. Antioxid Redox Signal. 2011;14:1315–1335. doi: 10.1089/ars.2010.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Read C.B., Kuijper J.L., Hjorth S.A., Heipel M.D., Tang X., Fleetwood A.J., et al. Cutting edge: identification of neutrophil PGLYRP1 as a ligand for TREM-1. J Immunol. 1950;194(2015):1417–1421. doi: 10.4049/jimmunol.1402303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dziarski R., Kashyap D.R., Gupta D. Mammalian peptidoglycan recognition proteins kill bacteria by activating two-component systems and modulate microbiome and inflammation. Microb Drug Resist. 2012;18:280–285. doi: 10.1089/mdr.2012.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yashin D.V., Sashchenko L.P., Georgiev G.P. Mechanisms of action of the PGLYRP1/Tag7 protein in innate and acquired immunity. Acta Nat. 2021;13:91–101. doi: 10.32607/actanaturae.11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharapova T.N., Ivanova O.K., Soshnikova N.V., Romanova E.A., Sashchenko L.P., Yashin D.V. Innate immunity protein Tag7 induces 3 distinct populations of cytotoxic cells that use different mechanisms to exhibit their antitumor activity on human leukocyte antigen-deficient cancer cells. J Innate Immun. 2017;9:598–608. doi: 10.1159/000479382. [DOI] [PubMed] [Google Scholar]
- 75.Haselmayer P., Grosse-Hovest L., von Landenberg P., Schild H., Radsak M.P. TREM-1 ligand expression on platelets enhances neutrophil activation. Blood. 2007;110:1029–1035. doi: 10.1182/blood-2007-01-069195. [DOI] [PubMed] [Google Scholar]
- 76.Fu L., Han L., Xie C., Li W., Lin L., Pan S., et al. Identification of extracellular actin as a ligand for triggering receptor expressed on myeloid cells-1 signaling. Front Immunol. 2017;8:917. doi: 10.3389/fimmu.2017.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sudakov N.P., Klimenkov I.V., Byvaltsev V.A., Nikiforov S.B., Konstantinov Y.M. Extracellular actin in health and disease, biochemistry. Biokhimiia. 2017;82:1–12. doi: 10.1134/S0006297917010011. [DOI] [PubMed] [Google Scholar]
- 78.Miles L.A., Andronicos N.M., Baik N., Parmer R.J. Cell-surface actin binds plasminogen and modulates neurotransmitter release from catecholaminergic cells. J Neurosci J Soc Neurosci. 2006;26:13017–13024. doi: 10.1523/JNEUROSCI.2070-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moroianu J., Fett J.W., Riordan J.F., Vallee B.L. Actin is a surface component of calf pulmonary artery endothelial cells in culture. Proc Natl Acad Sci USA. 1993;90:3815–3819. doi: 10.1073/pnas.90.9.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahrens S., Zelenay S., Sancho D., Hanč P., Kjær S., Feest C., et al. F-actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity. 2012;36:635–645. doi: 10.1016/j.immuni.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 81.Lee W.M., Galbraith R.M. The extracellular actin-scavenger system and actin toxicity. N Engl J Med. 1992;326:1335–1341. doi: 10.1056/NEJM199205143262006. [DOI] [PubMed] [Google Scholar]
- 82.Nishioka M., Kobayashi K., Uchida M., Nakamura T. A binding activity of actin with human C1q. Biochem Biophys Res Commun. 1982;108:1307–1312. doi: 10.1016/0006-291x(82)92142-8. [DOI] [PubMed] [Google Scholar]
- 83.Srinivasan N., Gordon O., Ahrens S., Franz A., Deddouche S., Chakravarty P., et al. Actin is an evolutionarily-conserved damage-associated molecular pattern that signals tissue injury in Drosophila melanogaster. eLife. 2016;5 doi: 10.7554/eLife.19662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vakser I.A. Challenges in protein docking. Curr Opin Struct Biol. 2020;64:160–165. doi: 10.1016/j.sbi.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murao A., Tan C., Jha A., Wang P., Aziz M. Exosome-mediated eCIRP release from macrophages to induce inflammation in sepsis. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.791648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stott K., Watson M., Howe F.S., Grossmann J.G., Thomas J.O. Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J Mol Biol. 2010;403:706–722. doi: 10.1016/j.jmb.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 87.Andersson U., Tracey K.J., Yang H. Post-translational modification of HMGB1 disulfide bonds in stimulating and inhibiting inflammation. Cells. 2021;10 doi: 10.3390/cells10123323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schiraldi M., Raucci A., Muñoz L.M., Livoti E., Celona B., Venereau E., et al. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. J Exp Med. 2012;209:551–563. doi: 10.1084/jem.20111739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang H., Wang H., Ju Z., Ragab A.A., Lundbäck P., Long W., et al. MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J Exp Med. 2015;212:5–14. doi: 10.1084/jem.20141318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones. 2016;21:379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Asea A., Kraeft S.K., Kurt-Jones E.A., Stevenson M.A., Chen L.B., Finberg R.W., et al. HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat Med. 2000;6:435–442. doi: 10.1038/74697. [DOI] [PubMed] [Google Scholar]
- 92.Hulina A., Grdić Rajković M., Jakšić Despot D., Jelić D., Dojder A., Čepelak I., et al. Extracellular Hsp70 induces inflammation and modulates LPS/LTA-stimulated inflammatory response in THP-1 cells. Cell Stress Chaperones. 2018;23:373–384. doi: 10.1007/s12192-017-0847-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nishiyama H., Itoh K., Kaneko Y., Kishishita M., Yoshida O., Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137:899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aziz M., Brenner M., Wang P. Extracellular CIRP (eCIRP) and inflammation. J Leukoc Biol. 2019;106:133–146. doi: 10.1002/JLB.3MIR1118-443R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qiang X., Yang W.L., Wu R., Zhou M., Jacob A., Dong W., et al. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat Med. 2013;19:1489–1495. doi: 10.1038/nm.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Denning N.L., Aziz M., Murao A., Gurien S.D., Ochani M., Prince J.M., et al. Extracellular CIRP as an endogenous TREM-1 ligand to fuel inflammation in sepsis. JCI Insight. 2020;5 doi: 10.1172/jci.insight.134172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Denning N.L., Aziz M., Diao L., Prince J.M., Wang P. Targeting the eCIRP/TREM-1 interaction with a small molecule inhibitor improves cardiac dysfunction in neonatal sepsis. Mol Med. 2020;26:121. doi: 10.1186/s10020-020-00243-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material