Highlights
-
•
C9orf72 mutation causes extensive disease-specific patterns of thalamo-cortico-striatal atrophy.
-
•
Cortical thinning is most pronounced in posterior areas and extends to frontal regions.
-
•
Reduced functional connectivity supports the central role of thalamus in C9orf72-mediated disease.
-
•
A mutation in C9orf72 affects ALS neurodegenerative/developmental clinical profiles.
Keywords: Amyotrophic Lateral Sclerosis, C9orf72 mutation, MRI, fMRI
Abstract
Objective
C9orf72 mutation carriers with different neurological phenotypes show cortical and subcortical atrophy in multiple different brain regions, even in pre-symptomatic phases. Despite there is a substantial amount of knowledge, small sample sizes, clinical heterogeneity, as well as different choices of image analysis may hide anatomical abnormalities that are unique to amyotrophic lateral sclerosis (ALS) patients with this genotype or that are indicative of the C9orf72-specific trait overlain in fronto-temporal dementia patients.
Methods
Brain structural and resting state functional magnetic imaging was obtained in 24 C9orf72 positive (ALSC9+) ALS patients paired for burden disease with 24 C9orf72 negative (ALSC9-) ALS patients. A comprehensive structural evaluation of cortical thickness and subcortical volumes between ALSC9+ and ALSC9- patients was performed while a region of interest (ROI)-ROI analysis of functional connectivity was implemented to assess functional alterations among abnormal cortical and subcortical regions. Results were corrected for multiple comparisons.
Results
Compared to ALSC9- patients, ALSC9+ patients exhibited extensive disease-specific patterns of thalamo-cortico-striatal atrophy, supported by functional alterations of the identified abnormal regions. Cortical thinning was most pronounced in posterior areas and extended to frontal regions. Bilateral atrophy of the mediodorsal and pulvinar nuclei was observed, emphasizing a focal rather than global thalamus atrophy. Volume loss in a large portion of bilateral caudate and left putamen was reported. The marked reduction of functional connectivity observed between the left posterior thalamus and almost all the atrophic cortical regions support the central role of the thalamus in the pathogenic mechanism of C9orf72-mediated disease.
Conclusions
These findings constitute a coherent and robust picture of ALS patients with C9orf72-mediated disease, unveiling a specific structural and functional characterization of thalamo-cortico-striatal circuit alteration. Our study introduces new evidence in the characterization of the pathogenic mechanisms of C9orf72 mutation.
1. Introduction
Amyotrophic lateral sclerosis (ALS) is a progressive and heterogeneous neurodegenerative disorder characterized by the involvement of motor neurons and some associated neuronal pathways (Chiò et al., 2014). C9orf72 repeat expansion is the most common genetic cause associated with ALS and frontotemporal dementia (FTD) (Renton et al., 2011). C9orf72 mutation carriers show some peculiar clinical and neuropsychological features in comparison with wild-type ALS patients (Iazzolino et al., 2021, Van Der Ende et al., 2021), such as earlier disease onset, more frequent bulbar impairment, a faster disease progression that also determines a lower survival (Glasmacher et al., 2020). Moreover, they are characterized by a higher frequency of cognitive impairment, such as language alterations, associated with extrapyramidal signs and psychiatric disorders (Esselin et al., 2020).
Magnetic resonance imaging (MRI) is a widely used neuroimaging technique capable of unraveling the pathological changes in-vivo during disease progression. By assessing both anatomical and functional consequences of the spread of the disease, MRI allows for distinguishing various ALS endophenotypes with possible different genetic causes and prognoses (Chiò et al., 2014). In previous structural MRI-based studies, C9orf72 mutation carriers reported different cortical alterations when compared to non-mutation carriers. In particular, C9orf72 mutation carriers showed diffuse symmetrical cortical volume loss involving the primary motor areas, but also extending beyond motor regions to include orbitofrontal, opercular, fusiform, lingual, cingulate, and temporal areas (Bede et al., 2013a, Byrne et al., 2012). Moreover, C9orf72-related ALS showed greater structural alterations in the cortical thickness of extra-motor regions, particularly in posterior areas, such as the occipital cortex and cuneus, when compared to ALS patients (Agosta et al., 2017, Bede et al., 2013a, Westeneng et al., 2016). Subcortical atrophy in C9orf72 mutation carriers is mainly localized in the thalamus (Agosta et al., 2017, Bede et al., 2013a, Bede et al., 2013b, Chipika et al., 2020b, Chipika et al., 2020c, Floeter et al., 2016, McKenna et al., 2022, Westeneng et al., 2016). Interestingly, pulvinar alterations have been proposed as a C9orf72-specific trait based on findings in FTD patients (Bocchetta et al., 2020, Lee et al., 2014, McKenna et al., 2022), but have not been confirmed by others in ALS patients (Chipika et al., 2020b, Chipika et al., 2020c).
From a functional perspective, only a few studies have investigated C9orf72-related functional alterations, identifying heterogeneous patterns of altered connectivity in ALS and FTD patients (Agosta et al., 2017, Lee et al., 2017, Smallwood Shoukry et al., 2020).
Despite this considerable amount of knowledge, there is still a lack of comprehensive information about the anatomical-structural peculiarities - and functional repercussions - that characterize C9orf72 genetically mediated ALS. Small sample sizes of C9orf72-related ALS patients, clinical heterogeneity, as well as the different choices of image analyses in previous studies could have hidden peculiar anatomical and functional features that can characterize these patients in comparison to sporadic ALS patients and possible structural imaging signature overlapping with FTD patients.
In this study, we aim at investigating the structural alterations in cortical and subcortical structures, and the functional impairments underpinning these structural changes, between C9orf72 ALS mutation carriers and wild-type ALS patients. Concerning all previous studies, participants were matched for disease burden, to characterize the genotype signature controlling for possible differences due to the clinical phenotype.
2. Materials and methods
2.1. Data collection
We collected the clinical data of all patients that underwent 1.5 T MRI assessment at diagnosis in the Turin ALS centre. We paired 24 ALS patients carrying C9orf72 expansion (ALSC9+) with 24 wild-type patients (ALSC9-) matched for age, gender, ALSFRS- total score, and disease duration, using a one-by-one pairing technique. Age was considered similar in a range of ± 5 years, ALSFRS-R at time of MRI performance in a range of ± 5 points, and disease duration from the onset in a range of ± 3 months.
2.2. C9orf72 repeat expansion detection
All participants were screened for the C9orf72 intronic expansion using a conventional repeat-primed Polymerase Chain Reaction (PCR). Repeat-primed PCR was performed as follows: 100 ng of genomic DNA were used as template in a final volume of 28 ml containing 14 ml of FastStart PCR Master Mix (Roche Applied Science, Indianapolis, IN, USA), and a final concentration of 0.18 mM 7-deaza-dGTP (New England Biolabs, Ipswich, MA, USA), 13 Q-Solution (QIAGEN, Valencia, CA, USA), 7% DMSO (Sigma-Aldrich), 0.9 mM MgCl2 (QIAGEN), 0.7 mM reverse primer consisting of ∼ four GGGGCC repeats with an anchor tail, 1.4 mM 6FAM-fluorescent labeled forward primer located 280 bp telomeric to the repeat sequence, and 1.4 mM anchor primer corresponding to the anchor tail of the reverse primer (sequences available in Supplemental Experimental Procedures). A touchdown PCR cycling program was used where the annealing temperature was gradually lowered from 70 °C to 56 °C in 2 °C increments with a 3 min extension time for each cycle. The repeat-primed PCR is designed so that the reverse primer binds at different points within the repeat expansion to produce multiple amplicons of incrementally larger size. The lower concentration of this primer in the reaction means that it is exhausted during the initial PCR cycles, after which the anchor primer is preferentially used as the reverse primer. Fragment length analysis was performed on an ABI 3730xl genetic analyzer (Applied Biosystems, Foster City, CA, USA), and data were analyzed using GeneScan software (version 4, ABI). Repeat expansions produce a characteristic sawtooth pattern with a 6 bp periodicity. Repeat lengths of ≥ 30 units with the characteristic sawtooth pattern were considered to be pathogenic (Renton et al, 2011). All patients included had no mutation in SOD1, TARDBP, and FUS genes.
2.3. MRI acquisition
Imaging data were acquired using a Signa HDxt EchoSpeed 1.5-MRI scanner (General Electric) equipped with a 8-channels head coil. A high-resolution 3D T1-weighted (T1w) structural sequence (TR = 11.82 ms, TE = 4.92 ms, FOV = 240 × 240 mm2, voxel size = 0.47 × 0.47 × 1 mm3, flip angle = 12°, 120 axial slices) was acquired for each participant in a MRI session including other sequences. Among these additional sequences, a eye-closed resting-state functional magnetic resonance imaging (rs-fMRI) acquisition (TR = 2250 ms, TE = 5 ms, FOV = 210 × 210 mm2, voxel size = 3.28 × 3.28 × 5 mm3, flip angle = 90°, number of volumes = 585, 25 axial slices) was obtained in a subset of patients (n = 19 ALSC9+, n = 19 ALSC9-).
2.4. Structural MRI data analysis
To identify alteration of cortical and subcortical regions, we performed two different analyses: vertex-wise analyses on cortical thickness and subcortical nuclei shape and volume analyses on subnuclei of thalamus, amygdala, and hypothalamus.
For each participant, N4 bias field correction was applied as a preprocessing step on T1w image to normalize variations in image intensity across the volume (Nicholas J. Tustison et al., 2011).
First, the vertex-wise analysis (i.e cortical thickness vertex-wise analysis and subcortical shape vertex-wise analysis) was conducted on cortical and subcortical nuclei surfaces.
For cortical thickness vertex-wise analysis we used FreeSurfer software (version 7; https://surfer.nmr.mgh.harvard.edu/). Preprocessed T1w images underwent the “recon-all” pipeline which included affine registration to the MNI305 atlas (Collins et al., 1994), skull stripping, segmentation of grey matter, white matter, and cerebrospinal fluid (CSF), based on maps of prior probabilities, and reconstruction of white matter and pial surfaces. Visual inspection and manual correction were conducted on the output by an expert operator (A.N.) blind to the genetic classification, to verify and remove the presence of inaccuracies of pial and white boundary surfaces. Cortical thickness was obtained as the distance between the vertices of the white and pial surface. “-qcache” flag was used to process and resample the individual cortical surfaces onto common (fsaverage) space. To compare the cortical thickness between pathological groups, a vertex-wise analysis on surfaces was performed using a General Linear Model (GLM) approach, as implemented in “qdec” module, including as covariates age, gender, disease progression, and number of body regions involved (NBRI) (Manera et al., 2019) at the date of MRI scans. Monte Carlo simulation with a cluster threshold of p < 0.05 was used to correct for multiple comparisons. In order to perform all analyses in the same space, coordinates of the peaks of the observed cortical abnormalities were transformed from MNI305 to MNI152 using the matrix provided in https://surfer.nmr.mgh.harvard.edu/fswiki/CoordinateSystems.
For subcortical shape vertex-wise analysis we used FIRST tool (Patenaude et al., 2011), part of FMRIB Software Library – FSL (Jenkinson et al., 2012). The T1w images were transformed to the MNI152 standard space and then segmented to obtain the subcortical surfaces (i.e. bilateral putamen, caudate, pallidum, thalamus, hippocampus). Amygdala was not included in this shape analysis due to the lower sensitivity in detecting differences between groups (Alexander et al., 2020). Then, the segmentation outputs were carefully checked by the same operator (A.N.) and a vertex-wise analysis was performed for each single subcortical structure using a GLM as implemented in “first_utils” and employing age, gender, disease progression, and NBRI as confounding variables. Results were corrected for multiple comparisons using p < 0.05 familywise cluster correction (FWE) (“randomize” module).
Second, volume analyses were conducted on subnuclei of the thalamus and amygdala and hypothalamic subunits. After the “recon-all” pipeline, thalamic nuclei (Iglesias et al., 2018), amygdala nuclei (Saygin et al., 2017), and hypothalamic subunits (Billot et al., 2020) segmentation modules were applied as implemented in FreeSurfer. Thalamic nuclei segmentation was performed with the updated version of the “quantifyThalamicNuclei.sh” script, as recommended by FreeSurfer’s guidelines (https://freesurfer.net/fswiki/ThalamicNuclei). After a visual check for errors, no manual interventions were performed on the data. Total intracranial volume (TIV) was obtained from the “aparc” file. Volumes of thalamic, amygdalar, and hypothalamic nuclei were obtained. To reduce the number of comparisons, the 25 thalamic nuclei for each hemisphere were merged in 14 subregions according to Bocchetta et al. (2020): anteroventral (AV), laterodorsal (LD), lateroposterior (LP), ventral anterior (VA), ventral lateral anterior (VLa), ventral lateral posterior (VLp), ventral posterolateral (VPL), ventromedial (VM), Intralaminar, Midline, mediodorsal (MD), lateral (LGN) and medial (MGN) geniculate, pulvinar [the association between Freesurfer parcellation and these subregions is reported in Supplementary Table 1 according to (Bocchetta et al., 2020)]. For the amygdala and hypothalamus, each subunit was considered separately. All extracted volumes were expressed as a percentage of TIV.
Statistical analyses were performed on volumes using R software 4.0.3. We tested the normal distribution using Shapiro-Wilk test, then we applied Mann-Whitney U-test to compare the extracted volumes data between ALSC9+ e ALSC9-. Results were corrected for multiple comparisons using Holm correction of p < 0.05.
2.5. Resting-state fMRI data analysis
To identify functional alterations among cortical and subcortical regions that were significantly atrophic between the two pathological groups, we performed a region of interest (ROI)-to-ROI functional connectivity analysis. Connectivity analyses were carried out using the CONN connectivity toolbox (v. 20.b) (Whitfield-Gabrieli & Nieto-Castanon, 2012). The “default_MNI” preprocessing pipeline was used. For each participant, functional images were realigned to the first volume, unwarped, and slice-timing corrected (slice order = interleaved, bottom-up), to reduce motion artifacts and geometric distortions. Outlier volume identification was performed with the Artifact Rejection Toolbox (ART) implemented in CONN. Structural images were segmented into gray matter, white matter (WM), and cerebrospinal fluid (CSF) tissue classes. Structural and functional images were registered and normalized into standard MNI space. Functional data resampling to 2 mm isotropic voxels was performed and images were smoothed with a full-width at half-maximum (FWHM) 6 mm Gaussian kernel. The anatomical component-based noise correction (aCompCor) was adopted to extract noise signals from WM and CSF components on a voxel-by-voxel level. Subsequently, a 0.008 – 0.1 Hz bandpass filter was applied to the time-series to remove low-frequency drifts and high-frequency noise.
At the first-level analysis, a ROI-to-ROI approach was adopted to investigate functional connectivity patterns among cortical and subcortical regions resulting significantly different in structural MRI analyses between groups. Spherical ROIs (radius = 6 mm) centered on the significant peak coordinates resulting from vertex-wise analyses were generated (coordinates are reported in Supplementary Tables 2 and 3). For each participant, realignment parameters, ART outlier volumes scrubbing parameters, physiological noise components obtained using aCompCor, and ‘‘effect of rest’’ (initial magnetization transient effects) were entered as covariates in the first-level analysis (Nieto-Castanon, 2020). At the second-level analysis, participants’ ROI-to-ROI connectivity matrices were compared between ALSC9+ and ALSC9- groups, with age, gender, disease progression, and NBRI as confounding variables. The Functional Network Connectivity (FNC) method implemented in CONN was adopted to analyze the connectivity patterns between the ROIs (Nieto-Castanon, 2020; (Jafri et al., 2008). FNC first identifies networks of related ROIs with a data-driven hierarchical clustering procedure. The entire set of connections between all pairs of ROIs is then analyzed using a multivariate parametric General Linear Model analysis. A False Discovery Rate (FDR) p < 0.05 cluster-level threshold is applied to adjust for multiple comparisons and control the false discovery rate. Specifically, among all network-to-network connectivity sets only those deemed significant were selected. Additionally, a connection-level post-hoc uncorrected p threshold was set at < 0.01 (CONN’s default = 0.05) to select only the most robust individual connections within each significant cluster.
Standard Protocol Approvals, Registrations, and Patient Consents. The study design was approved by the Ethical Committee of the Azienda Ospedaliero-Universitaria Città della Salute of Turin (Prot. N.0021674–24/02/2022).
3. Results
The main clinical features of included patients are summarized in Table 1. As a result of the matching, no significant differences were found between groups.
Table 1.
Demographic and clinical characteristics of patients. § Results for Chi-square test between ALSC9+ and ALSC9-; * results for two-sample independent t-test between ALSC9+ and ALSC9. Abbreviations: IQR = interquartile range; ALSbi = ALS patients with behavioural impairment; ALSci = ALS patients with cognitive impairment; ALScbi = ALS patients with cognitive and behavioural impairment; ALS-FTD = amyotrophic lateral sclerosis – frontotemporal dementia.
| ALSC9+ (n = 24) | ALSC9- (n = 24) | p§ | |
|---|---|---|---|
| Male/Female ratio | 14/10 | 13/11 | 0.771 |
| Bulbar/spinal onset | 6/18 | 8/16 | 0.525 |
| Cognitive classification | n (%) | n (%) | 0.665 |
| Normal cognition | 10 (41.7) | 14 (58.3) | |
| ALSbi | 2 (8.3) | 2 (8.3) | |
| ALSci | 2 (8.3) | 2 (8.3) | |
| ALScbi | 1 (4.2) | 1 (4.2) | |
| ALS-FTD | 4 (16.7) | 1 (4.2) | |
| Unknown | 5 (20.8) | 4 (16.7) | |
| median (IQR) | median (IQR) | p* | |
| Age (years) | 58.7 (50.9–68.3) | 60.6 (52.9–66.6) | 0.902 |
| ALSFRS-R score | 42.5 (37.3–45.0) | 43.0 (37.0–45.0) | 0.983 |
| Disease duration (months) | 11.0 (8.0–14.7) | 11.0 (9.0–19.0) | 0.717 |
| Overall survival (years) | 2.61 (2.17–3.55) | 2.70 (2.11–3.99) | 0.845 |
| Disease progression (ΔALSFRS-R/months) | 0.51 (0.32–0.91) | 0.47 (0.31–0.69) | 0.606 |
| Number of body regions involved | 2.00 (1.00–3.00) | 2.00 (1.00–2.75) | 0.979 |
3.1. Structural MRI
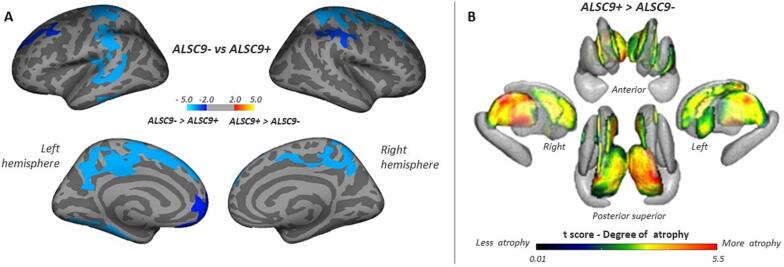
Vertex-wise analysis showed a decrease of cortical thickness mainly in posterior regions, but also extended to frontal regions, in ALSC9+ compared to ALSC9- patients. In particular, we showed a decreased cortical thickness in bilateral precentral and postcentral cortex, superior frontal gyrus, and precuneus, in left inferior temporal cortex, fusiform and parahippocampal gyri, lateral and medial orbital frontal cortex, posterior and rostral anterior cingulate, in right superior parietal and supramarginal cortex (Fig. 1A, Supplementary Table 2). Vertex-wise analysis also showed an extensive decrease in bilateral thalamic volume in ALSC9+ when compared to ALSC9- patients, more prominent in the superior-lateral-posterior portions of the thalamus, in the lateral and medial portions of bilateral caudate, and left putamen (Fig. 1B, Supplementary Table 3).
Fig. 1.
Cortical and subcortical alterations between ALSC9+ and ALSC9- patients. Panel A: vertex-wise analysis showing significant changes in cortical brain thickness (panel A) ALSC9+ patients compared to ALSC9- patients. All results were reported with p < 0.05 corrected for multiple comparisons using Monte Carlo cluster-based simulation. Clusters color-coded in blue indicate significantly decreased cortical thickness in ALSC9+ patient group compared to ALSC9- patient group. Panel B: vertex-wise analysis showing a significant decrease in the shape of subcortical regions in ALSC9+ compared to ALSC9- patients. Results were corrected using p < 0.05 family-wise error (FWE) -corrected threshold. Only significant results are shown. Abbreviations: ALSC9- = amyotrophic lateral sclerosis C9orf72 non mutation carriers; ALSC9+ = amyotrophic lateral sclerosis C9orf72 mutation carriers. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
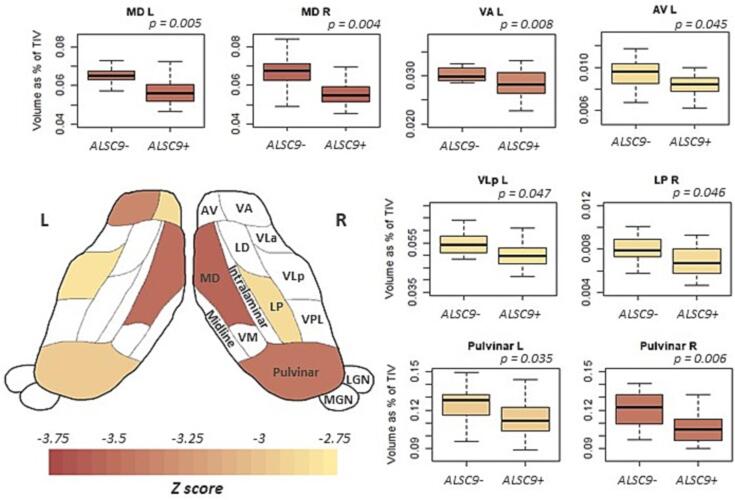
Regarding the volume measures in the subnuclei/subunits of the thalamus, amygdala, and hypothalamus, we did not observe any group differences in amygdala nuclei and hypothalamic subunits, while we identified significant differences in thalamic nuclei. In particular, we observed an important reduction of volume in ALSC9+ compared to ALSC9- patients in bilateral MD, followed by bilateral Pulvinar. Moreover, significantly decreased volumes were also observed in left AV, VA, VLp, and right LP (Fig. 2, Table 2).
Fig. 2.
Thalamic nuclei alterations between ALSC9+ and ALSC9- patients. Pattern of atrophy in the thalamic nuclei in the two pathological groups. Boxplot of significant statistical differences in volume of thalamic nuclei between ALSC9+ and ALSC9- patients. The cartoon, adapted from (Bocchetta et al., 2020), is a schematic representation of an axial view of the thalamic nuclei. Abbreviations: ALSC9- = amyotrophic lateral sclerosis C9orf72 non mutation carriers; ALSC9+ = amyotrophic lateral sclerosis C9orf72 mutation carriers; TIV = total intracranial volume; AV = anteroventral; LD = latero-dorsal; LP = latero posterior; VA = ventral anterior; VLa = ventral lateral anterior; VLp = ventral lateral posterior; VPL = ventral posterolateral; VM = ventromedial; MD = mediodorsal; LGN = lateral geniculate; MGN = medial geniculate.
Table 2.
Thalamic nuclei: significant statistical differences in volume of thalamic nuclei between ALSC9+ and ALSC9- patients. Abbreviations: ALSC9- = amyotrophic lateral sclerosis C9orf72 non mutation carriers; ALSC9+ = amyotrophic lateral sclerosis C9orf72 mutation carriers; TIV = total intracranial volume; AV = anteroventral; LD = laterodorsal; LP = latero posterior; VA = ventral anterior; VLa = ventral lateral anterior; VLp = ventral lateral posterior; VPL = ventral posterolateral; VM = ventromedial; MD = mediodorsal; LGN = lateral geniculate; MGN = medial geniculate.
|
Thalamus - Left side |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AV | LD | LP | VA | VLa | VLp | VPL | VM | Intralaminar | Midline | MD | LGN | MGN | Pulvinar | ||
| ALSC9+ | Mean | 0.008 | 0.002 | 0.007 | 0.028 | 0.038 | 0.050 | 0.056 | 0.001 | 0.026 | 0.001 | 0.057 | 0.016 | 0.008 | 0.113 |
| SD | 0.001 | 0.001 | 0.002 | 0.003 | 0.004 | 0.005 | 0.007 | 0.000 | 0.003 | 0.000 | 0.007 | 0.003 | 0.001 | 0.014 | |
| ALSC9- | Mean | 0.009 | 0.002 | 0.008 | 0.031 | 0.041 | 0.054 | 0.059 | 0.001 | 0.028 | 0.001 | 0.065 | 0.018 | 0.007 | 0.125 |
| SD | 0.001 | 0.001 | 0.001 | 0.003 | 0.004 | 0.006 | 0.006 | 0.000 | 0.003 | 0.000 | 0.007 | 0.003 | 0.001 | 0.013 | |
| z score | −2.89 | −0.95 | −2.27 | −3.38 | −2.75 | −2.84 | −2.51 | −1.95 | −2.25 | −2.25 | −3.54 | −2.19 | 1.34 | −2.97 | |
| p-value | 0.045 | 0.511 | 0.172 | 0.008 | 0.059 | 0.047 | 0.102 | 0.203 | 0.172 | 0.172 | 0.005 | 0.172 | 1 | 0.035 | |
|
Thalamus - Right side |
|||||||||||||||
| AV | LD | LP | VA | VLa | VLp | VPL | VM | Intralaminar | Midline | MD | LGN | MGN | Pulvinar | ||
| ALSC9+ | Mean | 0.009 | 0.001 | 0.007 | 0.029 | 0.039 | 0.051 | 0.054 | 0.001 | 0.026 | 0.001 | 0.057 | 0.016 | 0.008 | 0.108 |
| SD | 0.001 | 0.001 | 0.001 | 0.004 | 0.004 | 0.006 | 0.007 | 0.000 | 0.003 | 0.000 | 0.008 | 0.002 | 0.001 | 0.015 | |
| ALSC9- | Mean | 0.010 | 0.002 | 0.008 | 0.031 | 0.042 | 0.054 | 0.057 | 0.001 | 0.028 | 0.001 | 0.067 | 0.017 | 0.008 | 0.121 |
| SD | 0.001 | 0.000 | 0.001 | 0.003 | 0.004 | 0.005 | 0.005 | 0.000 | 0.003 | 0.000 | 0.009 | 0.002 | 0.001 | 0.013 | |
| z score | −2.66 | −1.83 | −2.86 | −2.71 | −2.19 | −2.08 | −1.26 | −1.70 | −1.64 | −2.06 | −3.64 | −2.45 | 0.15 | −3.50 | |
| p-value | 0.069 | 0.237 | 0.046 | 0.064 | 0.172 | 0.187 | 0.414 | 0.267 | 0.267 | 0.187 | 0.004 | 0.115 | 1 | 0.006 | |
3.2. Resting-state fMRI
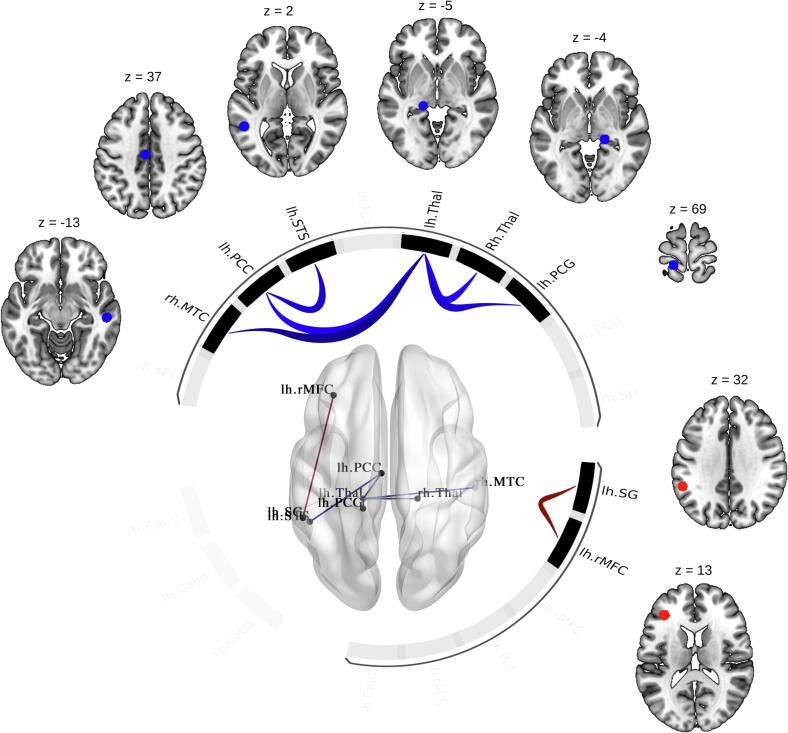
ROI-ROI functional connectivity analyses revealed that ALSC9+ patients, compared to patients with ALSC9- patients, showed a significant decrease in functional connectivity in one cluster of multiple connections (F(2,31) = 7.56; p-FDR-corrected for multiple comparisons = 0.012) and a significant increase in functional connectivity in one cluster including one connection (F(2,31) = 6.21; p-FDR-corrected for multiple comparisons = 0.018). Notably, the left thalamus exhibited a significant decrease in functional connectivity with the right thalamus, left postcentral cortex, left posterior cingulate cortex, and right middle temporal cortex, while the left banks of the superior temporal sulcus with the left posterior cingulate cortex. In contrast, the left rostral middle frontal cortex showed increased functional connectivity with the left supramarginal gyrus (Fig. 3, Supplementary Table 4).
Fig. 3.
Functional connectivity analysis between ALSC9+ and ALSC9- patients. ROI-to-ROI functional connectivity analysis between ALSC9+ and ALSC9- groups, with age, gender, disease progression, and NBRI as confounding variables. Cluster-forming significance threshold: p < 0.05 false-discovery-rate (FDR)-corrected for multiple comparisons; connection-level post-hoc: p < 0.01 uncorrected. Blue colour scale indicates a decrease in functional connectivity of ALSC9+ compared to ALSC9-; red colour scale indicates an increase in functional connectivity of ALSC9+ compared to ALSC9-. Abbreviations: rh = right hemisphere; lh = left hemisphere; MTC = middle temporal cortex; PCC = posterior cingulate cortex; STS = banks of the superior temporal sulcus; Thal = thalamus; PCG = postcentral gyrus; SG = supramarginal gyrus; rMFC = rostral middle frontal cortex. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Using two cohorts of ALS patients with similar disease burdens, our study reveals extensive disease-specific patterns of thalamo-cortico-striatal atrophy and underlying functional connectivity alterations in ALSC9+ compared to wild-type ALSC9- patients. In ALSC9+ patients, cortical thinning was most pronounced in posterior areas, but also extended into frontal regions. Moreover, genetically mediated patients showed bilateral thalamic volume decrease, as well as loss of volume in large portions of bilateral caudate nuclei and left putamen. Among thalamic nuclei, the atrophy was more prominent in bilateral MD nuclei, followed by bilateral pulvinar nuclei. Additionally, the marked reduction of functional connectivity between the left posterior thalamus and almost all the atrophic cortical regions identified indicates the thalamus' central role in the pathogenic mechanisms of C9orf72.
The underlying mechanisms through which C9orf72 repeat expansions exert neuronal degeneration are not completely understood (Xu et al., 2021). Indeed, although disease progression affects both motor, behavioral, and cognitive networks for ALSC9+ patients, there is still a need to measure disease-driven impairment. Recent studies conducted on ALSC9+ carriers explored lifelong behavioural and personality characteristics, pointing out that subtle personality traits like fixed behavioural patterns and limited empathy are present in early life (Gossink et al., 2021). Language, especially as assessed by verbal fluency performance, is also altered in ALSC9+ carriers before symptomatic disease (Lule et al., 2020). These observations, confirmed by neuroimaging studies (Bertrand et al., 2018), reinforce the hypothesis that considers C9orf72 mutation as a neurodevelopmental disease that could facilitate neurodegenerative processes (Bede et al., 2020) and the resulting increased neuropsychiatric vulnerability (Esselin et al., 2020). C9orf72-mediated disease in ALS exhibits a highly precise pattern of cortical and subcortical atrophy. Our results confirm the prevalence of extensive cortical thickness reductions in the posterior regions, encompassing parietal, occipital, and temporal areas, but also extended to frontal regions among ALSC9+ as compared to ALSC9- patients (Agosta et al., 2017, Bede et al., 2013a, Querin et al., 2022, Westeneng et al., 2016). The diffusion studies on the staging of pathology indicate similar white matter impairments in ALSC9+ and ALSC9- patients compared to healthy participants (Agosta et al., 2017, Müller et al., 2020, Müller et al., 2021, Kassubek et al., 2014). In this pattern, motor tracts, such as the corticospinal tract and motor mid-segment of the corpus callosum, were affected. These tracts are extensively connected to the regions where we observed reduced cortical thickenss. Besides extensive cortical atrophy, our study emphasizes that thalamic alterations play a crucial role in the C9orf72 genotype as previously reported in neuroimaging studies in both presymptomatic (Bertrand et al., 2018, Lee et al., 2017, Querin et al., 2022, Rohrer et al., 2015) and symptomatic patients (Bede et al., 2013a, Bede et al., 2013b, Querin et al., 2022, Westeneng et al., 2016). Unquestionably, vertex-wise and subnuclear analyses revealed specific atrophy of the thalamic profile in ALS patients.
Indeed, vertex-wise analysis showed bilateral thalamic atrophy extending to the superior-inferior-lateral portion, with a peak of atrophy around the pulvinar, very similar to the pattern previously reported on the right thalamus only (Bede et al., 2013b) and in bilateral (Chipika et al., 2020b). According to the fine-grained parcellation method of the thalamus in our study, ALSC9+ patients clearly showed bilateral atrophy of the MD and Pulvinar nuclei, high order nuclei, in comparison to non-mutated patients, emphasizing a focal rather than global thalamus atrophy. These two nuclei are critical to high-order associative and integrative functions of stimuli, such as behavioral modulation, visual-spatial, and executive functions (Benarroch, 2015, Halassa and Kastner, 2017, Ouhaz et al., 2018, Saalmann et al., 2012, Schmahmann, 2003). These findings are in line with observations reported in the C9orf72 ALS-FTD continuum in comparison to healthy participants (Bocchetta et al., 2022, Chipika et al., 2020b), where MD is affected across the whole spectrum (Bocchetta et al., 2022, Chipika et al., 2020b), while pulvinar alterations were reported as a unique feature of C9orf72 carriers in FTD (Bocchetta et al., 2020, 2022). By controlling for disease burden between the two pathological groups we were able to pinpoint these volume changes that were previously reported either as a trend towards significant or non-significant at all in ALSC9+ versus ALSC9- patients (Bocchetta et al., 2022, Chipika et al., 2020b, Chipika et al., 2020c). These results suggest that dysfunction of these high-order nuclei - which regulate synchrony and information flow at the cortico-cortical and cortico-subcortical levels (Mitchell, 2015) - may contribute to cognitive, but also neuropsychiatric symptoms that are seen in ALSC9+ compared to ALSC9- patients (Esselin et al., 2020). In support of this assumption, these thalamic nuclei are reported in post-mortem and in-vivo studies to be sites of neuropathological alteration in patients carrying the C9orf72 gene (Bocchetta et al., 2020, Vatsavayai et al., 2016), as well as in patients with psychiatric disorders such as schizophrenia (Byne et al., 2009, Kemether et al., 2003, Ouhaz et al., 2018). Furthermore, we report a unilateral involvement of functional limbic and motor thalamic nuclei (Schmahmann, 2003) such as the right LP, a higher-order region relevant for somatosensory and visuo-spatial integration (Schmahmann, 2003), as well as the anterior (i. e. AV, VA) and ventrolateral (VLp) first-order nuclei of the left thalamus primarily engaged in spatial learning, navigation, and motor programming (Jankowski et al., 2013, Nelson, 2021, Schmahmann, 2003).
It is not surprising that the left thalamus, whose seed coordinates are mainly located at the pulvinar (i.e., maximum atrophy on the vertex-wise analysis), showed decreased functional connectivity with the greatest cortical thickness changes in the posterior regions. Presymptomatic mutation carriers and patients with C9orf72-related FTD display similar functional connectivity changes in the thalamus (i.e. medial pulvinar) (Lee et al., 2014, Lee et al., 2017, Smallwood Shoukry et al., 2020). On the other hand, we observed higher functional connectivity between the left medial frontal and supramarginal gyri in ALSC9+ patients, which can be interpreted as a result of a compensatory effect limited by the degree of cognitive-behavioral deficits. This observation is in line with an earlier study that found an association between frontal-executive dysfunction and an increase in frontoparietal network functional connectivity of these two regions in ALS patients (Castelnovo et al., 2020).
Focusing on other subcortical regions, portions of the caudate and putamen resulted structurally altered, consistent with previous MRI studies that showed typical basal ganglia involvement in ALS and FTD associated with C9orf72 mutations (Bede et al., 2013b, Lee et al., 2014, Westeneng et al., 2016).
However, neither the amygdala nor the hippocampus, nor the hypothalamus showed a specific signature of atrophy in mutated versus wild-type patients. A similar result was reported by Chipika et al., 2020a, Chipika et al., 2020c, showing a selective involvement of specific amygdala nuclei in ALS patients compared to healthy participants, but not between mutated or non-mutated patients. Therefore, although the amygdala, hippocampus, and hypothalamus have been reported among the first brain regions which appear structurally altered in C9orf72 expansion carriers without evident clinical symptoms (Bocchetta et al., 2021), our observations suggest that these abnormalities may be unrelated to the C9orf72 genotype.
Our study is not without limitations. First, it was not possible to include the control group. Therefore, we could not compare the patterns obtained in patients with healthy participants. However, this does not influence the goodness of our inferences because the primary objective of the study was to characterize the pattern of C9orf72-mediated disease in ALS, controlling the main sources of clinical heterogeneity. Second, despite matching many clinical and demographic variables between subjects in the two groups, both samples included patients who did not undergo cognitive testing, and it was not possible to retrieve data regarding psychiatric symptoms for all patients.
Despite these shortcomings, this research has several strengths. The ALS sample of patients is relatively unique, both in terms of size and match for disease burden. This may have led to the identification of specific alterations that might have been underestimated or not sufficiently highlighted in previous studies due to the clinical heterogeneity of the patients. Finally, we used extensive multimodal analysis methods to uncover a complete C9orf72 brain signature when compared to ALS non-carriers.
5. Conclusion
Our study unveiled a specific structural and functional characterization of thalamo-cortico-striatal circuit alteration in ALS patients with C9orf72-mediated disease with robust data due to extreme care in sample match and comprehensive analyses. These findings highlight a unique contribution of C9orf72 expansions in the clinical and neurodegenerative/neurodevelopmental profiles of ALS patients with structural and functional imaging signatures overlapping with FTD. These observations introduce new evidence in the characterization of the C9orf72 patients in the continuum ALS-FTD.
Funding
This work was in part supported by the Italian Ministry of Health (Ministero della Salute, Ricerca Sanitaria Finalizzata and Giovani ricercatori, grant RF-2016-02362405, GR-2019-12371291, Ricerca corrente RRC), the European Commission’s Health Seventh Framework Programme (FP7/2007-2013 under grant agreement 259867), the Italian Ministry of Education, University and Research (Progetti di Ricerca di Rilevante Interesse Nazionale, PRIN, grant 2017SNW5MB), the Joint Programme - Neurodegenerative Disease Research (ALS-Care, Strength and Brain-Mend projects), granted by Italian Ministry of Education, University and Research. This study was performed under the Department of Excellence grant of the Italian Ministry of Education, University and Research to the ‘Rita Levi Montalcini’ Department of Neuroscience, University of Torino, Italy.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Our special thanks also to our colleagues. The authors are grateful to Martin Monti and Evan Lutkenhoff for support in subcortical shape vertex-wise analysis and visualization. Special acknowledgment should be expressed to the patients who participated in the study and to all the technicians (Francesco Primo, Roberto Agliata, Lucy Stefanelli, Rosalba Caruana, Nicola Mazzei, Manuela D’Ambrosio, Gianfranco Tagliente, Francesca Stancati) and radiologists (Giovanna Carrara, Maurizio Cogogni, Ivan Gomez Pavanello, Paola Sciortino, Fabrizio Venturi) at the Neuroradiology Unit of CTO Hospital (AOU Città della Salute e della Scienza di Torino) who supervised the MRI scans.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2023.103400.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Agosta F., Ferraro P.M., Riva N., Spinelli E.G., Domi T., Carrera P., Copetti M., Falzone Y., Ferrari M., Lunetta C., Comi G., Falini A., Quattrini A., Filippi M. Structural and functional brain signatures of C9orf72 in motor neuron disease. Neurobiol. Aging. 2017;57:206–219. doi: 10.1016/j.neurobiolaging.2017.05.024. [DOI] [PubMed] [Google Scholar]
- Alexander B., Georgiou-Karistianis N., Beare R., Ahveninen L.M., Lorenzetti V., Stout J.C., Glikmann-Johnston Y. Accuracy of automated amygdala MRI segmentation approaches in Huntington’s disease in the IMAGE-HD cohort. Hum. Brain Mapp. 2020;41(7):1875–1888. doi: 10.1002/hbm.24918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Bokde A.L., Byrne S., Marwa Elamin M., Russell McLaughlin M.L., Kenna K., Fagan A.J., Pender N., Bradley D.G., Hardiman O. Multiparametric MRI study of ALS stratified for the C9orf72 genotype. Neurology. 2013;81(4):361–369. doi: 10.1212/WNL.0b013e31829c5eee. www.neurology.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Elamin M., Byrne S., McLaughlin R.L., Kenna K., Vajda A., Pender N., Bradley D.G., Hardiman O. Basal ganglia involvement in amyotrophic lateral sclerosis. Neurology. 2013;81(24):2107–2115. doi: 10.1212/01.wnl.0000437313.80913.2c. [DOI] [PubMed] [Google Scholar]
- Bede P., Siah W.F., McKenna M.C., Li H., Shing S. Consideration of C9orf72 -associated ALS-FTD as a neurodevel opmental disorder: Insights from neuroimaging. J. Neurol. Neurosurg. Psychiatry. 2020;91(11):1138. doi: 10.1136/jnnp-2020-324416. [DOI] [PubMed] [Google Scholar]
- Benarroch E.E. Pulvinar: Associative role in cortical function and clinical correlations. Neurology. 2015;84(7):738–747. doi: 10.1212/WNL.0000000000001276. [DOI] [PubMed] [Google Scholar]
- Bertrand A., Wen J., Rinaldi D., Houot M., Sayah S., Camuzat A., Fournier C., Fontanella S., Routier A., Couratier P., Pasquier F., Habert M.-O., Hannequin D., Martinaud O., Caroppo P., Levy R., Dubois B., Brice A., Durrleman S., Colliot O., Le Ber I. Early cognitive, structural, and microstructural changes in presymptomatic C9orf72 carriers younger than 40 years. JAMA Neurol. 2018;75(2):236. doi: 10.1001/jamaneurol.2017.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billot B., Bocchetta M., Todd E., Dalca A.V., Rohrer J.D., Iglesias J.E. Automated segmentation of the hypothalamus and associated subunits in brain MRI. Neuroimage. 2020;223(July) doi: 10.1016/j.neuroimage.2020.117287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchetta M., Iglesias J.E., Neason M., Cash D.M., Warren J.D., Rohrer J.D. Thalamic nuclei in frontotemporal dementia: Mediodorsal nucleus involvement is universal but pulvinar atrophy is unique to C9orf72. Hum. Brain Mapp. 2020;41(4):1006–1016. doi: 10.1002/hbm.24856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchetta M., Todd E.G., Peakman G., Cash D.M., Convery R.S., Russell L.L., Thomas D.L., Iglesias J.E., van Swieten J.C., Jiskoot L.C., Seelaar H., Borroni B., Galimberti D., Sanchez-Valle R., Laforce R., Moreno F., Synofzik M., Graff C., Masellis M., Zulaica M. Differential early subcortical involvement in genetic FTD within the GENFI cohort. NeuroImage: Clinical. 2021;30(March):8–11. doi: 10.1016/j.nicl.2021.102646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchetta M., Todd E.G., Tse N.Y., Devenney E.M., Tu S., Caga J., Hodges J.R., Halliday G.M., Irish M., Piguet O., Kiernan M.C., Rohrer J.D., Ahmed R.M. Thalamic and Cerebellar Regional Involvement across the ALS–FTD Spectrum and the Effect of C9orf72. Brain Sci. 2022;12(3):1–16. doi: 10.3390/brainsci12030336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byne W., Hazlett E.A., Buchsbaum M.S., Kemether E. The thalamus and schizophrenia: current status of research. Acta Neuropathol. 2009;117(4):347–368. doi: 10.1007/s00401-008-0404-0. [DOI] [PubMed] [Google Scholar]
- Byrne S., Elamin M., Bede P., Shatunov A., Walsh C., Corr B., Heverin M., Jordan N., Kenna K., Lynch C., McLaughlin R.L., Iyer P.M., O’Brien C., Phukan J., Wynne B., Bokde A.L., Bradley D.G., Pender N., Al-Chalabi A., Hardiman O. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: A population-based cohort study. The Lancet Neurology. 2012;11(3):232–240. doi: 10.1016/S1474-4422(12)70014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelnovo V., Canu E., Calderaro D., Riva N., Poletti B., Basaia S., Solca F., Silani V., Filippi M., Agosta F. Progression of brain functional connectivity and frontal cognitive dysfunction in ALS. NeuroImage: Clinical. 2020;28(November) doi: 10.1016/j.nicl.2020.102509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A., Pagani M., Agosta F., Calvo A., Cistaro A., Filippi M. Neuroimaging in amyotrophic lateral sclerosis: Insights into structural and functional changes. The Lancet Neurology. 2014;13(12):1228–1240. doi: 10.1016/S1474-4422(14)70167-X. [DOI] [PubMed] [Google Scholar]
- Chipika, R. H., Christidi, F., Finegan, E., Li Hi Shing, S., McKenna, M. C., Chang, K. M., Karavasilis, E., Doherty, M. A., Hengeveld, J. C., Vajda, A., Pender, N., Hutchinson, S., Donaghy, C., McLaughlin, R. L., Hardiman, O., & Bede, P. (2020). Amygdala pathology in amyotrophic lateral sclerosis and primary lateral sclerosis. Journal of the Neurological Sciences, 417(May), 117039. 10.1016/j.jns.2020.117039. [DOI] [PubMed]
- Chipika, R. H., Finegan, E., Li Hi Shing, S., McKenna, M. C., Christidi, F., Chang, K. M., Doherty, M. A., Hengeveld, J. C., Vajda, A., Pender, N., Hutchinson, S., Donaghy, C., McLaughlin, R. L., Hardiman, O., & Bede, P. (2020). “Switchboard” malfunction in motor neuron diseases: Selective pathology of thalamic nuclei in amyotrophic lateral sclerosis and primary lateral sclerosis. NeuroImage: Clinical, 27(February), 102300. 10.1016/j.nicl.2020.102300. [DOI] [PMC free article] [PubMed]
- Chipika R.H., Siah W.F., Shing S.L.H., Finegan E., McKenna M.C., Christidi F., Chang K.M., Karavasilis E., Vajda A., Hengeveld J.C., Doherty M.A., Donaghy C., Hutchinson S., McLaughlin R.L., Hardiman O., Bede P. MRI data confirm the selective involvement of thalamic and amygdalar nuclei in amyotrophic lateral sclerosis and primary lateral sclerosis. Data Brief. 2020;32 doi: 10.1016/j.dib.2020.106246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins D., Neelin P., Peters T., Evans A. Automatic 3D Inter-Subject Registration of MR Volumetric Data in Standardized Talairach Space. J. Comput. Assist. Tomogr. 1994;18(2):192–205. [PubMed] [Google Scholar]
- Esselin F., Mouzat K., Polge A., Juntas-Morales R., Pageot N., De la Cruz E., Bernard E., Lagrange E., Danel V., Alphandery S., Labar L., Nogué E., Picot M.C., Lumbroso S., Camu W. Clinical Phenotype and Inheritance in Patients With C9ORF72 Hexanucleotide Repeat Expansion: Results From a Large French Cohort. Front. Neurosci. 2020;14(April):1–8. doi: 10.3389/fnins.2020.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floeter M.K., Bageac D., Danielian L.E., Braun L.E., Traynor B.J., Kwan J.Y. Longitudinal imaging in C9orf72 mutation carriers: Relationship to phenotype. NeuroImage: Clinical. 2016;12:1035–1043. doi: 10.1016/j.nicl.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasmacher S.A., Wong C., Pearson I.E., Pal S. Survival and Prognostic Factors in C9orf72 Repeat Expansion Carriers: A Systematic Review and Meta-analysis. JAMA Neurol. 2020;77(3):367–376. doi: 10.1001/jamaneurol.2019.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossink F., Dols A., Stek M.L., Scheltens P., Nijmeijer B., Cohn Hokke P., Dijkstra A., Van Ruisssen F., Aalfs C., Pijnenburg Y.A.L. Early life involvement in C9orf72 repeat expansion carriers. J. Neurol. Neurosurg. Psychiatry. 2021;93–100 doi: 10.1136/jnnp-2020-325994. [DOI] [PubMed] [Google Scholar]
- Halassa M.M., Kastner S. Thalamic functions in distributed cognitive control. Nat. Neurosci. 2017;20(12):1669–1679. doi: 10.1038/s41593-017-0020-1. [DOI] [PubMed] [Google Scholar]
- Iazzolino, B., Peotta, L., Zucchetti, J. P., Canosa, A., Manera, U., Vasta, R., Grassano, M., Palumbo, F., Brunetti, M., Barberis, M., Sbaiz, L., Moglia, C., Calvo, A., & Chiò, A. (2021). Differential Neuropsychological Profile of Patients With Amyotrophic Lateral Sclerosis With and Without C9orf72 Mutation. Neurology, 96(1), e141–e152. 10.1212/WNL.0000000000011093. [DOI] [PubMed]
- Iglesias J.E., Insausti R., Lerma-Usabiaga G., Bocchetta M., Van Leemput K., Greve D.N., van der Kouwe A., Fischl B., Caballero-Gaudes C., Paz-Alonso P.M. A probabilistic atlas of the human thalamic nuclei combining ex vivo MRI and histology. Neuroimage. 2018;183(June):314–326. doi: 10.1016/j.neuroimage.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri M.J., Pearlson G.D., Stevens M., Calhoun V.D. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. Neuroimage. 2008;39(4):1666–1681. doi: 10.1016/j.neuroimage.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski M.M., Ronnqvist K.C., Tsanov M., Vann S.D., Wright N.F., Erichsen J.T., Aggleton J.P., O’Mara S.M. The anterior thalamus provides a subcortical circuit supporting memory and spatial navigation. Front. Syst. Neurosci. 2013;7(August):1–12. doi: 10.3389/fnsys.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C.F., Behrens T.E.J., Woolrich M.W., Smith S.M. FSL. NeuroImage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Kassubek J., Müller H.P., Del Tredici K., Brettschneider J., Pinkhardt E.H., Lulé D., Böhm S., Braak H., Ludolph A.C. Diffusion tensor imaging analysis of sequential spreading of disease in amyotrophic lateral sclerosis confirms patterns of TDP-43 pathology. Brain. 2014;137(6):1733–1740. doi: 10.1093/brain/awu090. [DOI] [PubMed] [Google Scholar]
- Kemether E.M., Buchsbaum M.S., Byne W., Hazlett E.A., Haznedar M., Brickman A.M., Platholi J., Bloom R. Magnetic resonance imaging of mediodorsal, pulvinar, and centromedian nuclei of the thalamus in patients with schizophrenia. Arch. Gen. Psychiatry. 2003;60(10):983–991. doi: 10.1001/archpsyc.60.9.983. [DOI] [PubMed] [Google Scholar]
- Lee S.E., Khazenzon A.M., Trujillo A.J., Guo C.C., Yokoyama J.S., Sha S.J., Takada L.T., Karydas A.M., Block N.R., Coppola G., Pribadi M., Geschwind D.H., Rademakers R., Fong J.C., Weiner M.W., Boxer A.L., Kramer J.H., Rosen H.J., Miller B.L., Seeley W.W. Altered network connectivity in frontotemporal dementia with C9orf72 hexanucleotide repeat expansion. Brain. 2014;137(11):3047–3060. doi: 10.1093/brain/awu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Sias A.C., Mandelli M.L., Brown J.A., Brown A.B., Khazenzon A.M., Vidovszky A.A., Zanto T.P., Karydas A.M., Pribadi M., Dokuru D., Coppola G., Geschwind D.H., Rademakers R., Gorno-Tempini M.L., Rosen H.J., Miller B.L., Seeley W.W. Network degeneration and dysfunction in presymptomatic C9ORF72 expansion carriers. NeuroImage: Clinical. 2017;14:286–297. doi: 10.1016/j.nicl.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lule D.E., Müller H.P., Finsel J., Weydt P., Knehr A., Winroth I., Andersen P., Weishaupt J., Uttner I., Kassubek J., Ludolph A.C. Deficits in verbal fluency in presymptomatic C9orf72 mutation gene carriers - A developmental disorder. J. Neurol. Neurosurg. Psychiatry. 2020;91(11):1195–1200. doi: 10.1136/jnnp-2020-323671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manera U., Calvo A., Daviddi M., Canosa A., Vasta R., Torrieri M.C., Grassano M., Brunetti M., D’Alfonso S., Corrado L., De Marchi F., Moglia C., D’Ovidio F., Mora G., Mazzini L., Chiò A. Regional spreading of symptoms at diagnosis as a prognostic marker in amyotrophic lateral sclerosis: A population-based study. J. Neurol. Neurosurg. Psychiatry. 2019;291–297 doi: 10.1136/jnnp-2019-321153. [DOI] [PubMed] [Google Scholar]
- McKenna, M. C., Li Hi Shing, S., Murad, A., Lope, J., Hardiman, O., Hutchinson, S., & Bede, P. (2022). Focal thalamus pathology in frontotemporal dementia: Phenotype-associated thalamic profiles. Journal of the Neurological Sciences, 436(March), 120221. 10.1016/j.jns.2022.120221. [DOI] [PubMed]
- Mitchell A.S. The mediodorsal thalamus as a higher order thalamic relay nucleus important for learning and decision-making. Neurosci. Biobehav. Rev. 2015;54:76–88. doi: 10.1016/j.neubiorev.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Müller H.P., Del Tredici K., Lulé D., Müller K., Weishaupt J.H., Ludolph A.C., Kassubek J. In vivo histopathological staging in C9orf72-associated ALS: A tract of interest DTI study. Neuroimage Clin. 2020;27 doi: 10.1016/j.nicl.2020.102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.P., Lulé D., Roselli F., Behler A., Ludolph A.C., Kassubek J. Segmental involvement of the corpus callosum in C9orf72-associated ALS: a tract of interest-based DTI study. Ther Adv Chronic Dis. 2021;12:1–9. doi: 10.1177/20406223211002969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson A.J.D. The anterior thalamic nuclei and cognition: A role beyond space? Neurosci. Biobehav. Rev. 2021;126(February):1–11. doi: 10.1016/j.neubiorev.2021.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Castanon A., editor. Handbook of functional connectivity Magnetic Resonance Imaging methods in CONN. Hilbert Press; 2020. [Google Scholar]
- Ouhaz Z., Fleming H., Mitchell A.S. Cognitive functions and neurodevelopmental disorders involving the prefrontal cortex and mediodorsal thalamus. Front. Neurosci. 2018;12(FEB):1–18. doi: 10.3389/fnins.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude B., Smith S.M., Kennedy D.N., Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56(3):907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querin G., Biferi M.G., Pradat P.F. Biomarkers for C9orf7-ALS in Symptomatic and Pre-symptomatic Patients: State-of-the-art in the New Era of Clinical Trials. Journal of Neuromuscular Diseases. 2022;9(1):25–37. doi: 10.3233/JND-210754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton, A. E., Majounie, E., Waite, A., Simón-Sánchez, J., Rollinson, S., Gibbs, J. R., Schymick, J. C., Laaksovirta, H., van Swieten, J. C., Myllykangas, L., Kalimo, H., Paetau, A., Abramzon, Y., Remes, A. M., Kaganovich, A., Scholz, S. W., Duckworth, J., Ding, J., Harmer, D. W., … Traynor, B. J. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron, 72(2), 257–268. 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed]
- Rohrer J.D., Nicholas J.M., Cash D.M., van Swieten J., Dopper E., Jiskoot L., van Minkelen R., Rombouts S.A., Cardoso M.J., Clegg S., Espak M., Mead S., Thomas D.L., De Vita E., Masellis M., Black S.E., Freedman M., Keren R., MacIntosh B.J., Rogaeva E., Tang-Wai D., Tartaglia M.C., Laforce R., Tagliavini F., Tiraboschi P., Redaelli V., Prioni S., Grisoli M., Borroni B., Padovani A., Galimberti D., Scarpini E., Arighi A., Fumagalli G., Rowe J.B., Coyle-Gilchrist I., Graff C., Fallström M., Jelic V., Ståhlbom A.K., Andersson C., Thonberg H., Lilius L., Frisoni G.B., Binetti G., Pievani M., Bocchetta M., Benussi L., Ghidoni R., Finger E., Sorbi S., Nacmias B., Lombardi G., Polito C., Warren J.D., Ourselin S., Fox N.C., Rossor M.N. Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the Genetic Frontotemporal dementia Initiative (GENFI) study: A cross-sectional analysis. The Lancet Neurology. 2015;14(3):253–262. doi: 10.1016/S1474-4422(14)70324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalmann Y.B., Pinsk M.A., Wang L., Li X., Kastner S. The pulvinar regulates information transmission between cortical areas based on attention demands. Science. 2012;337(6095):753–756. doi: 10.1126/science.1223082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saygin, Z. M., Kliemann, D., Iglesias, J. E., van der Kouwe, A. J. W., Boyd, E., Reuter, M., Stevens, A., Van Leemput, K., McKee, A., Frosch, M. P., Fischl, B., & Augustinack, J. C. (2017). High-resolution magnetic resonance imaging reveals nuclei of the human amygdala: manual segmentation to automatic atlas. NeuroImage, 155(May 2016), 370–382. 10.1016/j.neuroimage.2017.04.046. [DOI] [PMC free article] [PubMed]
- Schmahmann J.D. Vascular syndromes of the thalamus. Stroke. 2003;34(9):2264–2278. doi: 10.1161/01.STR.0000087786.38997.9E. [DOI] [PubMed] [Google Scholar]
- Smallwood Shoukry R.F., Clark M.G., Floeter M.K. Resting State Functional Connectivity Is Decreased Globally Across the C9orf72 Mutation Spectrum. Front. Neurol. 2020;11(November):8–15. doi: 10.3389/fneur.2020.598474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison N.J., Avants B.B., Cook P.A., Zheng Y., Egan A., Yushkevich P.A., Gee J.C. N4ITK: Improved N3 Bias Correction. J. Rehabil. Res. Dev. 2011;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908.N4ITK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Ende E.L., Jackson J.L., White A., Seelaar H., Van Blitterswijk M., Van Swieten J.C. Unravelling the clinical spectrum and the role of repeat length in C9ORF72 repeat expansions. J. Neurol. Neurosurg. Psychiatry. 2021;92(5):502–509. doi: 10.1136/jnnp-2020-325377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsavayai S.C., Yoon S.J., Gardner R.C., Gendron T.F., Vargas J.N.S., Trujillo A., Pribadi M., Phillips J.J., Gaus S.E., Hixson J.D., Garcia P.A., Rabinovici G.D., Coppola G., Geschwind D.H., Petrucelli L., Miller B.L., Seeley W.W. Timing and significance of pathological features in C9orf72 expansion-associated frontotemporal dementia. Brain. 2016;139(12):3202–3216. doi: 10.1093/brain/aww250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westeneng H.J., Walhout R., Straathof M., Schmidt R., Hendrikse J., Veldink J.H., Van Den Heuvel M.P., Van Den Berg L.H. Widespread structural brain involvement in ALS is not limited to the C9orf72 repeat expansion. J. Neurol. Neurosurg. Psychiatry. 2016;87(12):1354–1360. doi: 10.1136/jnnp-2016-313959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S., Nieto-Castanon A. Conn: A Functional Connectivity Toolbox for Correlated and Anticorrelated Brain Networks. Brain Connect. 2012;2(3):125–141. doi: 10.1089/brain.2012.0073. [DOI] [PubMed] [Google Scholar]
- Xu X., Su Y., Zou Z., Zhou Y., Yan J. Correlation between c9orf72 mutation and neurodegenerative diseases: A comprehensive review of the literature. Int. J. Med. Sci. 2021;18(2):378–386. doi: 10.7150/ijms.53550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.