Abstract
Butyrate is a microbiota-produced metabolite, sensed by host short-chain fatty acid receptors FFAR2 (Gpr43), FFAR3 (Gpr41), HCAR2 (Gpr109A), and Histone deacetylase (HDAC) that promotes microbiota-host crosstalk. Butyrate influences energy uptake, developmental and immune response in mammals. This microbial metabolite is produced by around 79 anaerobic genera present in the mammalian gut, yet little is known about the role of butyrate in the host-microbiota interaction in salmonid fish. To further our knowledge of this interaction, we analyzed the intestinal microbiota and genome of Atlantic salmon (Salmo salar), searching for butyrate-producing genera and host butyrate receptors. We identified Firmicutes, Proteobacteria, and Actinobacteria as the main butyrate-producing bacteria in the salmon gut microbiota. In the Atlantic salmon genome, we identified an expansion of genes orthologous to FFAR2 and HCAR2 receptors, and class I and IIa HDACs that are sensitive to butyrate. In addition, we determined the expression levels of orthologous of HCAR2 in the gut, spleen, and head-kidney, and FFAR2 in RTgutGC cells. The effect of butyrate on the Atlantic salmon immune response was evaluated by analyzing the pro and anti-inflammatory cytokines response in vitro in SHK-1 cells by RT-qPCR. Butyrate decreased the expression of the pro-inflammatory cytokine IL-1β and increased anti-inflammatory IL-10 and TGF-β cytokines. Butyrate also reduced the expression of interferon-alpha, Mx, and PKR, and decreased the viral load at a higher concentration (4 mM) in cells treated with this molecule before the infection with Infectious Pancreatic Necrosis Virus (IPNV) by mechanisms independent of FFAR2, FFAR3 and HCAR2 expression that probably inhibit HDAC. Moreover, butyrate modified phosphorylation of cytoplasmic proteins in RTgutGC cells. Our data allow us to infer that Atlantic salmon have the ability to sense butyrate produced by their gut microbiota via different specific targets, through which butyrate modulates the immune response of pro and anti-inflammatory cytokines and the antiviral response.
Keywords: Microbiota, Butyrate, Atlantic salmon
Graphical Abstract
1. Introduction
Multicellular eukaryotes (plants and animals) have been traditionally classified as highly complex organisms independent of the microorganisms (prokaryotes, archaea, fungi and yeasts) that conform their commensal microbiota [1], [2]. This categorization has changed in recent years, as in mammals there is abundant evidence showing that microbiota participates directly in the protection against pathogens, behavior, energy balance, and stimulation and maturation of the immune system [3].
In Fish, the microbiota-host relationship has been less well-defined. However, studies in zebrafish (Danio rerio) have indicated that the microbiota also influences resistance against pathogens [4], behavior and social interactions [5], energy balance [6], and stimulation and maturation of the immune system [7], as well as modulating the maturation of the intestinal epithelial [8]. The microbiota of zebrafish is also able to produce the Short Chain Fatty Acids (SCFAs) acetate, propionate, and butyrate. Butyrate promotes an anti-inflammatory response, reducing the recruitment of neutrophils and macrophages, by mechanisms that depend on the SCFA hydroxycarboxylic receptor HCAR2 and the inhibition of Histone deacetylase, respectively [9].
The physiological role of SCFAs in non-model species of fish is less well understood; nevertheless, there are similarities to those roles described in mice, and zebrafish [10], [11], [12]. SCFAs are found in the intestinal tract of herbivorous and carnivorous fish [13], [14]. Although their origin is unclear, it is hypothesized that their production is via microbiota. Oral administration of butyrate increases intestinal microvilli and nutrient absorption in Sparus aurata [15], while in Cyprinus carpio, this SCFA increases the expression of heat shock protein-70 (HSP70), pro-inflammatory cytokines (IL-1β and TNF-α) and anti-inflammatory cytokines (transforming growth factor-β, TGF-β) [16]. The use of the butyrate-producing bacteria (BPB) Clostridium butyricum as an additive in the feed of the fish Cyprinus carpio, increases butyric and propionic acid, raises the expression of antioxidant enzymes, immune genes and epithelial integrity, improves histological parameters of the intestine, modifies gut microbiota abundance, resulting in enhanced fish growth [17]. Similar effects have been observed in Tilapia (Oreochromis niloticus) [18]. BPBs are phylogenetically and metabolically diverse, grouped in clusters XVIa, XVI, IV, comprising between 5% and 10% of the bacteria detected in human feces [19], [20].
The action of SCFAs on cell metabolism can occur by passive diffusion using transporters such as monocarboxylate transporter 1 and sodium-coupled monocarboxylate transporter 1 (SMCT1), as well as by directly activating G protein-coupled receptors (GPRs) [21]. The classical receptors reported in mammals for the detection of SCFAs are free fatty acid receptor FFAR3 (GPR41), FFAR2 (GPR43) and HCAR2 (GPR109A), which show different affinities for acetate, propionate and butyrate [22]. Studies carried out on FFAR4 in the fish Larimichthys crocea showed that this receptor is capable of regulating the inflammatory effects induced by bacterial Lipopolysaccharide (LPS) [23]. Studies of the HCAR1 receptor in zebrafish showed that embryos lacking this gene were unable to respond to the anti-inflammatory effect triggered by butyrate in controls [9]. Characterization of the FFAR1 receptor in carp identified 10 copies, which possess conserved regions and phylogenetic proximity to FFAR2 and FFAR3 of humans [24]. These studies demonstrate that receptors of the FFAR family are phylogenetically related, respond to microbiota metabolites triggering immunomodulatory effects, and are conserved among various teleost fish. Besides their interaction with FFAR2, FFAR3 and HCAR2, SCFA n-butyrate and n-propionate are strong histone-deacetylase (HDAC) inhibitors, inducing chromatin remodeling [25], [26]. In mammalians, HDAC inhibition with SCFA stimulates the expression of foxP3, which promotes the differentiation of T CD4 + lymphocytes in Treg lymphocytes, favoring the anti-inflammatory response [27]. In intestinal macrophages of mice, SCFA also reduces the production of pro-inflammatory mediators via HDAC inhibition [28]. In Salmonids, the effects of butyrate and other microbial SCFAs on fish physiology have not been extensively studied. The dietary administration of butyrate improves the expression of genes related with the complement in Atlantic salmon [29], and the expression of intestinal lysozyme, IL-1β, TGF-β, and IL-10 in Rainbow trout, suggesting that butyrate also has a non-pathological immunostimulant effect [30]. However, the elements that transduce microbiota-host communication mediated by SCFA, such as BPB and the host SCFA receptors, have not been fully characterized.
In this communication, we report the presence of genera associated with the production of butyrate within the gut microbiota of Atlantic salmon, together with the characterization of the genes encoding for putative butyrate receptors present in the genome of this species. Their characterization was performed by combining syntenic and expression studies of the genes, and phylogenetic and structural analysis of their encoded proteins. We complemented these studies by analyzing the effects of butyrate on the expression of cytokines and the antiviral response in SHK-1 cells, a macrophage-like cell from Atlantic salmon, to understand whether Atlantic salmon cells are sensitive to this fermentation product.
2. Materials and methods
2.1. Fish maintenance and sampling
The Atlantic salmon (Salmo salar) specimens used in this study were purchased from the El Copihue fish farm in the municipality of Puerto Octay, Región de Los Lagos, Chile. The pre-smolt fish arrived at the experimental station of Universidad de Los Lagos located in Rupanco lake, with an average weight of 44.6 g and a length of 14.6 cm. The fish were disposed in three 200-liter open-circulation tanks, with a water flow of 12 L / minute, at a density of 0.77 kg / m3, and a photoperiod of 24 h light. The water averaged 13.2 ºC, with an average oxygen saturation of 9.3 mg/mL. Feeding was carried out using the commercial diet Protec RC 60 (Skretting), with daily feeding ad libitum based on 1.6% of fish body weight. After 40 days of acclimatization, six specimens per tank (three for microbiota analysis, and three for further SCFA determination) were sampled every seven days for one month (Days 7, 14, 21, and 28). The fish were sacrificed by an overdose of Benzocaine (Veterquímica) at 20%, followed by the application of brain trauma. Prior to sampling, 70% alcohol was applied to the abdomen of the fish with a paper towel. The post pyloric intestine (section containing the mid and distal intestine) was aseptically removed, then the digesta of the mid and distal intestine were directly drained into a sterile polypropylene tube, whilst the intestinal tissue was washed aseptically with saline and subsequently stored in tubes with 1 mL of RNAlater (ThermoFisher). To analyze the gene expression of butyrate receptors, the spleen and head-kidney were also sampled from fish collected at day 28. Once the samples were taken, they were immediately kept at − 20 °C then stored long-term at − 80 ºC. The health status of the fish was monitored by evaluating swimming behavior, and the average weight and length of each one during the experiment. The weight and length were monitoring by sampling six fish every week, which were then returned to the tank. Fish with abnormal behavior or appearance (erratic swimming, fin wounds, or lack of appetite) or death were removed from the experiment. All the procedures used were designed to minimize fish suffering and authorized by the bioethics committee of the University of Santiago de Chile (nº471, 2020).
2.2. Metagenomic analysis of fecal samples
2.2.1. DNA Isolation
Microbial DNA from Atlantic salmon feces was isolated following the procedure reported by Han et al., 2018 [31], and optimized according to the guidelines described by Gaur et al., 2019 [32]. Briefly, the digesta samples were kept at − 80 ºC until extraction, and 200 mg were aliquoted in a 2 mL tube with 500 µL of homogenization buffer (100 mM Tris-HCl pH 8.0; 50 mM EDTA pH 8.0 and 1 M NaCl). The sample was homogenized with a HG-15A homogenizer (Witeg, 17500 rpm, 40 s). At the end of the homogenization, 300 µL of homogenization buffer were added and the mixture was centrifuged for 2 min at 2500 rpm. The supernatant was transferred to a new 2 mL tube and the pellet was discarded. The sample was centrifuged at 10000 rpm for one minute. The supernatant was discarded, the pellet was resuspended in 900 µL of 1X PBS, and centrifuged for two minutes at 3000 rpm. The supernatant was transferred to a new 2 mL tube and the pellet was discarded. The sample was subsequently centrifuged at 10000 rpm for one minute. The supernatant was discarded, and the pellet was resuspended in 700 µL of homogenization buffer. Lysozyme (100 µL of 10 mg/mL) was added and the sample was incubated for one hour at 60 ºC. Subsequently, 3 µL of RNAse (4 mg/mL) were added and the sample was incubated for thirty minutes at 37 ºC. SDS (100 µL of 10%) and 10 µL proteinase K (20 mg/mL) were added, and incubated for one hour at 60 ºC. An equal volume of Phenol: Chloroform: Isoamyl Alcohol (25: 24: 1) was added and centrifuged for 15 min at 9500 rpm. The supernatant was carefully transferred to a new 2 mL tube and a 1:1 vol of Chloroform: Isoamyl Alcohol (24: 1) was added. The sample was centrifuged for 15 min at 9500 rpm, the supernatant transferred to a new 2 mL tube and a 1:10 vol of 3 M sodium acetate (pH 5.4) and 1:1 of absolute isopropanol was added. Precipitation was carried out for 20 min at − 80 °C. The sample was then centrifuged for 15 min at 9500 rpm, and after carefully discarding the supernatant, the pellet was washed twice with ethanol 70%, centrifuged for 5 min at 10000 rpm and then at 14000 rpm for two minutes. The supernatant was discarded, and the pellet was allowed to dry at room temperature, before being resuspended in 20 µL TE buffer. The concentration and purity of the DNA were analyzed by absorbance to 260 and 280 nm. For the subsequent metagenomic analyses, a DNA sample (the one with the highest purity) was chosen for each tank on each sampling day. Thus, a total of 12 samples were analyzed, representing days 7, 14, 21 and 28 post-acclimatization.
2.2.2. 16S ribosomal sequencing
The 16 S rRNA amplification and high-throughput sequencing was performed by Molecular Research LP (MR DNA; Shallowater, TX, USA). Briefly, the DNA extracted from fecal samples collected at days 7, 14, 21, and 28 was used as template for amplification of the V4 variable region of the 16 S rRNA gene. The amplification was performed using the primers 515-F and 806-R [33] with a barcode on the forward primer and the HotStarTaq Plus Master Mix Kit (Qiagen, USA). The PCR conditions were 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 53 °C for 40 s and 72 °C for 1 min, followed by a final elongation step at 72 °C for 5 min. After amplification, PCR products were checked in a 2% agarose gel to determine the success of amplification and the relative intensity of bands. PCR products were purified using Ampure XP beads and used to prepare an Illumina DNA library with a TruSeq Nano kit. High-throughput sequencing of 16 S rRNA amplicons was performed with the MiSeq reagent kit v3 on the Illumina MiSeq platform (2 × 300-bp paired ends [PE]) following the manufacturer’s guidelines. Raw sequence reads were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under accession number PRJNA834799.
2.2.3. Bioinformatic analysis
The data derived from sequencing were processed using QIIME2 version 2020.2 for 16S-based microbiota analyses [34] and the construction of the pipeline was carried out based on the documents present on the QIIME2 webpage (https://docs.qiime2.org/2020.2/tutorials/). Initially, barcodes and adapters were removed from the demultiplexed paired-end sequences. For quality filtering and feature (Amplicon Sequence Variants (ASV)) prediction, we used DADA2 [35]. Forward and reverse reads were each truncated to 250 nts. Representative sequences were aligned using MAFFT (Multiple Alignment using Fast Fourier Transform) [36]. A phylogenetic tree of the aligned sequences was elaborated with FastTree 2 [37]. ASVs/features were taxonomically classified using a pre-trained Naive Bayes taxonomy classifier, Greengenes 13_8 99% ASVs [38]. Tables of taxonomic counts and percentages (relative frequency) were generated. We obtained a mean of 108898 (SD: 22768) individual sequencing reads per sample (min = 60791; max = 147222). After data processing, the average number of sequences for each sample passing through ASV classification was 60258 (SD: 12800). The average number of ASVs per sample was 37218 (SD: 7416). Rarefaction was used to sample the same number of random reads from each sample for the diversity analyses. The sampling depth was set at 26200 sequences per sample. Taxon-level abundance data were filtered to remove very low-abundance taxa (<0.05%) and taxa not represented in at least half the samples before further analysis.
2.3. Identification of classical HDAC and Butyrate receptor orthologues in the Atlantic salmon genome
To identify the presence and number of butyrate receptors FFAR2, FFAR3 and HCAR2, a bioinformatic search was carried out in the annotated genomes of Atlantic salmon (Ssal_3.1 (GCF_905237065.1)) and Rainbow trout (USDA_OmykA_1.1 (GCF_013265735.2)), deposited in the NCBI database (https://www.ncbi.nlm.nih.gov). The initial search was performed using the human FFAR2, FFAR3, and HCAR2 protein sequences (NP_001357016.1; AAI13696.1 and NP_808219.1, respectively), with the Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Subsequently, the sequences that had an identity percentage of 50% or higher and had the specific domains of the FFAR and HCAR2 receptors (7tmA_FFAR2_FFAR3 and 7tmA_HCAR1–3, respectively) were used for the other analyses. The presence of these domains was determined using the Conserved Domain Database tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The classical HDACs from Atlantic salmon were downloaded from the UniProt database (https://www.uniprot.org).
2.4. Phylogenetic reconstruction and divergence time estimation
The evolutionary history of the butyrate receptors FFAR2, FFAR3, and HCAR2 was inferred using the Maximum Likelihood method and the JTT matrix-based model [39], with a bootstrap of 1000 replicates [40]. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model and then selecting the topology with a superior log-likelihood value [41], [42]. Evolutionary analyses were conducted in MEGA X [41], [42].
For each receptor, molecular dating analysis was performed in BEAST 1.10.4 [43], using an uncorrelated relaxed clock model with lognormal rate heterogeneity [44], and a prior Yule speciation tree [45]. We used the JTT+I+G model of evolution for the three amino acid datasets, which was selected with BIC criteria in ModelTest-NG [46]. To calibrate the estimation of divergence time, we used the oldest known fossil of Salmonidae (Eosalmo driftwoodensis) [47] which has been dated to 51.8 ± 0.3 Mya [48]. We used 50 Mya (Ln offset: 50, mean: 1.0, SD: 1.0) as a conservative minimum boundary to calibrate the most recent common ancestor of Salmonidae, as employed previously in other studies [49], [50].
2.5. Modelling the 3D structure of butyrate receptors and prediction of ligand binding pockets and binding sites
The tri-dimensional structure of FFAR from Atlantic salmon was modelled by homology using the program Modeller 10 v2 [51] and the protocols for advanced modelling that include multiple templates and slow refining protocols. As templates for FFAR2/3 from Atlantic salmon, we used six structures that correspond to FFAR2 and FFAR3 from human (O15552, O14843), mouse (Q8VCK6, Q3UFD7) and rat (Q76EI6, B2GV46). For the modelling of HCAR2, we employed three structures that correspond to those predicted for the receptor in human (Q8TDS4), mouse (Q9EP66), and rat (Q80Z39). All these structures were predicted by Alphafold [52] and are available in Uniprot [53]. A total of 5 models were predicted for each structure, that were then analyzed by PROSA-Web [54] and GA341 [55]. The structure with the lowest z-value and a GA341 score closest to 1 was chosen as the model to be analyzed. The ligand-binding pocket was evaluated with the PrankWeb method [56], [57] and the binding sites of butyrate were studied with SwissDock [58], [59], [60], using the modelled structure as target and the butyrate structure (ZINC895132) as ligand.
2.6. Expression of butyrate receptors in Atlantic salmon tissues
The evaluation of the expression of butyrate receptors in the tissues of the intestine, head-kidney, and spleen of Atlantic salmon was carried out by RT-PCR using total RNA and specific primers for butyrate receptors in salmonids, FFAR2, FFAR3 and HCAR2, designed in this work (Table 1).
Table 1.
List of primers used.
| Gene product | Primer name | Sequence 5′− 3′ | Reference |
|---|---|---|---|
| 16 S rRNA | 515-F | GTGCCAGCMGCCGCGGTAA | [61] |
| 806-R | GGACTACVSGGGTATCTAAT | ||
| FFAR2 (XP_014021565.1) | FFAR2_1F | TCTCGGACCTCCTCTTCCTG | This work |
| FFAR2_1R | CTGGTGCAGAACAACACAGC | ||
| FFAR2 (XP_014021519.1) | FFAR2_2F | ATTGGCGGGCAAACTTCCTA | This work |
| FFAR2_2R | ACGCTGGTCAGCTATTATCG | ||
| FFAR2 (XP_014056041.1) | FFAR2_3F | TGACCGCAATCAGTGTCGAA | This work |
| FFAR2_3R | GCATAGTCCTCGGTCTTTTCCA | ||
| FFAR2 (XP_014056042.1) | FFAR2_4F | TACAACCCACCGCAAGACTC | This work |
| FFAR2_4R | TGAAAGTCTCTCCCGTCTCAC | ||
| FFAR2 (XP_014055668.1) | FFAR2_5F | TGGGTTATGCTTGACGTGCT | This work |
| FFAR2_5R | AGCACTGCGTTATTGAGCCT | ||
| FFAR2 (XP_014055669.1) | FFAR2_6F | AACATCCTGGCACTCTACGC | This work |
| FFAR2_6R | AGTTGTGGGTGGGTTACTGT | ||
| FFAR2 (XP_014019408.1) | FFAR2_7F | CGTTACCTGGCGGTTGCT | This work |
| FFAR2_7R | CGGGTGGTGCTGGATGATGA | ||
| FFAR3 (XP_014032532.1, XP_014032531.1) | FFAR36_F | CCTTCTCCCGGTACGCTTAG | This work |
| FFAR36_R | GGCTGATGTTGGGAAGCCTA | ||
| HCAR2 (XP_013993962.1, XP_013982987.1) | HCAR25_F | TGACGTTCTAGGAACAGTGGG | This work |
| HCAR25_R | CCCACATAGCTACAGAATCGGA | ||
| TGF-β | TGFb_Fw | AGCTCTCGGAAGAAACGACA | [62] |
| TGFb_Rv | AGTAGCCAGTGGGTTCATGG | ||
| IL-6 | IL-6_Fw | CCTTGCGGAACCAACAGTTTG | [63] |
| IL-6_Rv | CCTCAGCAACCTTCATCTGGTC | ||
| IL-1β | IL-1b_Fw | CCCCATTGAGACTAAAGCCA | [62] |
| IL-1b_Rv | GCAACCTCCTCTAGGTGCAG | ||
| IL-10 | IL-10_Fw | TAATGACGAGCTGGAGGCTT | This work |
| IL-10_Rv | AGATGTTTCCGATGGAGTCG | ||
| eF1α | eF1a_Fw | GGGTGAGTTTGAGGCTGGTA | [62] |
| eF1a_Rv | TTCTGGATCTCCTCAAACCG | ||
| Mx | MX_R | GACGTCAGGGGAGCCAATC | [64] |
| MX_FW | TGTAACACGATGCCCTCTCG | ||
| PKR | PKR_R | CCCTTATTTATGCTAATCCAG | [65] |
| PKR_FW | CAATGACCGATTCCAGCTCC | ||
| IFNα1 | IFNα1_Fw | ATCACAAAACAGAATGCCCC | [62] |
| IFNα1_Rv | GACTGACAGGGTCCCACAT | ||
| 18 S rRNA | 18SF | CCTTAGATGTCCGGGGCT | [63] |
| 18SR | CTCGGCGAAGGGTAGACA | ||
| VP2-IPNV | VP2_Fw | GACCAAGTTCGACTTCCAGC | This work |
| VP2_Rv | ATCGGCTTGGTGATGTTCTC |
2.6.1. RNA isolation and cDNA synthesis
RNA extraction was performed using 50–100 mg of Atlantic salmon tissue (intestine, spleen and head-kidney) from fish sampled on day 28. The tissues were mechanically homogenized in 400 µL TRIzol™ Reagent (Invitrogen) and completed to 1 mL. The manufacturer's protocol was then followed, until the RNA pellet was obtained. The pellet was air-dried and subsequently resuspended in RNAse-free water in a volume dependent on the size obtained. RNA integrity was assessed on a 1% agarose gel. cDNA synthesis was performed using the enzyme M-MLV (Promega). To do so, 2 μg of total RNA, 1 μg of Oligo dT 15-mer (IDT) and nuclease-free water was added up to 10 µL. Subsequently, it was incubated for 5 min at 70 ºC and immediately placed on ice. To the tube, 5 µL of M-MLV buffer, 10 mM of dNTPs, 200 U of M-MLV enzyme, and 25 U of Recombinant RNasin® Ribonuclease Inhibitor (Promega) were added and filled with water up to 25 µL. The tube was vortexed and incubated for 1 h at 42 ºC. Subsequently, the cDNA was stored at − 80 ºC until use.
2.6.2. Polymerase chain reaction (PCR)
PCR reactions were performed using 1 µL cDNA, 1X GoTaq® G2 Hot Start Green Master Mix (Promega), 10 mM primer forward and reverse, and adjusted with nuclease-free water up to a volume of 25 µL. The amplification programs consisted of an initial denaturation cycle of 2 min at 95 ºC, followed by 30 cycles of: denaturation for 30 s at 95 ºC, annealing for 30 s (58 ºC for FFAR2 and 3, and 55 ºC for HCAR2), extension for 60 s at 72 ºC; and a final extension cycle of 5 min at 72 °C. These programs were executed in the MyCycler™ Thermal Cycler System (Bio-Rad). The sequence, name and targets of the primers used to amplify the FFAR2, FFAR3 and HCAR2 sequences are listed in Table 1.
2.7. Effect of butyrate on the Poly I:C dependent induction of Mx, PKR and IFNα in SHK-1 cells
The effect of butyrate on the antiviral response induced by Poly I:C transfection of SHK-1 cells was evaluated by quantifying by RT-qPCR the changes in the expression levels of Mx, PKR and IFNα in cells pre-treated with butyrate (2 mM) and transfected with Poly I:C, with respect to untreated SHK-1 cells. Expression was evaluated for 168 h post-stimulation.
2.7.1. Culture of SHK-1 cells
SHK-1 is a macrophage cell line isolated from Atlantic salmon head-kidney (Salmo salar, ECACC Number 97111106, European Collection of Cell Culture, UK). These cells were grown in Leibovitz’s 15 media (L-15, Gibco®, Invitrogen®, Carlsbad, CA, USA) supplemented with 4 mM L-glutamine (Corning cellgro®, Mediatech), Fetal Serum Bovine at 10% (Hyclone®, Thermo Fischer Scientific), gentamycin at 50 µg/mL (US Biological) and 40 μM β-mercaptoethanol (ChemCruz®). Cells were grown at 16 ºC and sub-cultured on reaching a confluence between 80% and 90%.
2.7.2. Antiviral response in SHK-1 cells treated with Poly I:C and butyrate
To evaluate the effect of butyrate on the antiviral response induced by Poly I:C, we evaluated the kinetics of Mx, PKR and IFNα expression in four groups of SHK-1 cells: a) pre-treated with butyrate (2 mM) and transfected with Poly I:C, b) treated with Poly I:C or c) with butyrate, and d) without treatment. We evaluated these four conditions in triplicate for each time point analyzed. SHK-1 cells were sub-cultured on six-well plates and grown at 16 ºC. On reaching a confluence of 70% (1.0 ×106 cells/mL), groups a and c were stimulated with butyrate at a final concentration of 2 mM in each well for 24 h. After the stimulation, the cells from groups a and c were transfected with Poly I:C (Sigma Aldrich) at 1 μg/mL using Fugene X-Treme Gene HP DNA (Roche Diagnostic) according to the instructions of the manufacturer. The medium in each group was replaced with fresh media, that contained 2 mM butyrate in the case of groups a and c. The butyrate stock solution (Sigma Aldrich) was prepared in distilled water to a final concentration of 0.5 M and sterilized by filtration using a microfilter of 0.25 µm (Jet Biofil) at 0, 12-, 24-, 48-, 72-, and 96-hours post-treatment, cells were collected.
2.7.3. Isolation of total RNA and cDNA synthesis
Total RNA from cell cultures was isolated using the E.Z.N.A Total RNA Kit I (Omega Bio-Tek). Briefly, the culture media was removed, lysed from the culture plates and the cells attached were suspended in 350 µL TRK buffer. The nucleic acids were precipitated by adding one volume of 70% ethanol. This suspension was passed through a HiBind RNA mini-column by centrifugation at 10000 rpm for 1 min at room temperature. The RNA bound to the mini-column was washed with RNA wash buffer according to the kit’s instruction and eluted in 50 µL of water pre-treated with DEPC and heated at 70 ºC. To obtain the viral RNA, the culture media was added in 700 µL of TRK buffer and passed through a HiBind RNA mini-column and processed as above. Total RNA was quantified by spectrophotometry using a TEKAN® Infinite M200PRO. The purity of total RNA was evaluated by the 260/280 nm ratio and its integrity was analyzed by electrophoresis in agarose gels. cDNA synthesis was performed using 0.5 pmol of oligo-dT (15 mer), 100 U of M-MLV Reverse Transcriptase (200 U/µL) (Invitrogen ®), 10 pmoles of dNTPs (Kappa Biosystems®) and 1 µg of total RNA that had been previously treated with DNAse I (RQ1 RNAse free DNAse, Promega®) during 30 min at 37 ºC in a total volume of 10 µL. Next, the DNAse I was inactivated with stop solution (Promega®) by heating to 65 ºC for 10 min. The cDNA synthesis reaction mix was incubated at 42 ºC for 1 h in a total volume of 20 µL, then inactivated by heat at 70 ºC for 10 min. After cDNA synthesis, reactions were stored at − 80 ºC until use.
2.7.4. Quantification of gene expression of Mx, PKR and IFNα
The effects of butyrate on the gene expression of cytokines and antiviral genes were quantified by RT-qPCR in an Aria MX® (Agilent Technologies®) thermocycler. Each reaction was performed using 5 µL of Kapa SYBR® FAST Master Mix (2x) (KR0389, KAPA BIOSYSTEMS®), 4 pmol of each primer, and cDNA from 400 ng of total RNA. To quantify the antiviral response, the primers MX_R and MX_FW were used to amplify Mx, PKR_R and PKR_FW for PKR, and IFNα1_Fw and IFNα1_Rv for Interferon-alpha. The reference gene used in this analysis was the expression of 18 S rRNA using primers 18SF and 18SR. All primers are shown in Table 1. Each reaction was performed in a volume of 20 µL, using the following thermal program: 3 min at 95 ºC, followed by 40 cycles of denaturation at 90 ºC for 30 s, annealing at 60 ºC for 15 s and extension at 72 ºC for 20 s. The amplification was concluded with a final cycle of 30 s at 95 ºC, 30 s at 65 ºC and 30 s at 95 ºC. The fold of induction was calculated based on the method described by Pfaffl et al., 2001 [66]. The efficiency of each pair of primers was calculated by serial dilution of the template.
2.8. Effect of Butyrate on the viral replication of IPNV in SHK-1 cells
The effect of butyrate on viral replication of IPNV was assessed by evaluating the changes in IPNV load present in the supernatant of SHK-1 cell cultures infected with IPNV and pre-treated with 1, 2 and 4 mM of butyrate. As control, SHK-1 cells previously transfected with Poly I:C and without pre-treatment, were infected with IPNV. Each condition was assessed in triplicate.
2.8.1. SHK-1 culture and viral infection
The SHK-1 cells were cultured as above. Briefly, SHK-1 cells were sub-cultured on six-well plates and grown at 16 ºC. On reaching 70% confluence, cells were exposed to 1, 2, and 4 mM of butyrate for 24 h, or transfected with Poly I:C as mentioned before. Later, treated and untreated cells were infected with IPNV Sp (ATCC VR-1318) with a m.o.i of 0.1 and cultured at 16 ºC. The culture media was collected at 0, 12-, 24-, 48-, 72-, 96-, and 168-hours post-infection, and viral RNA was extracted. The butyrate stock solution (Sigma Aldrich) used to achieve the final concentration of 1, 2 and 4 mM was prepared in distilled water as indicated before. For each time of sampling, a different set of wells were used.
2.8.2. Viral load
The viral load of IPNV was determined by RT-qPCR. Total RNA was isolated from the media of cell cultures previously infected with IPNV, using the E.Z.N.A Total RNA Kit I. cDNA was prepared using the SensiFAST ™ SYBR Hi-ROX One-Step Kit (Bioline). Briefly, one-step RT-qPCR was performed using 5 µL of total isolated RNA, and 10 pmol of primer VP2_Fw and VP2_Rv, in a total volume of 20 µL. To quantify the viral load in the culture media, a titration curve was set graphing Ct values against molar concentration of a plasmid containing the VP2 gene. The Ct obtained from the RNA samples was interpolated into the titration curve. Primers are listed in Table 1.
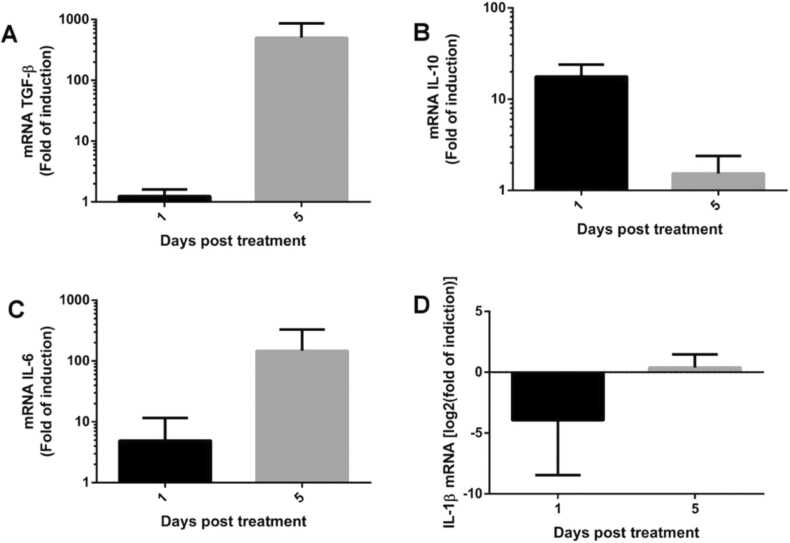
2.9. Effect of butyrate on the expression of IL-1β, IL-6, IL-10 and TGF-β in SHK-1 cells
To evaluate the effect of butyrate on the expression of IL-1β, IL-6, IL-10 and TGF-β in SHK-1, cells were cultured at 16 ºC on six-well plates, grown until 70% confluency, and then treated with 2 mM butyrate. SHK-1 cell cultures without treatment with butyrate were used as control. The cells were sampled at 1 and 5 days after treatment with butyrate and each condition was evaluated in triplicate. Gene expression was assessed by RT-qPCR. Total RNA extraction, cDNA synthesis, and qPCR procedures were performed as described above for the quantification of Mx, PKR and IFNα, using primers TGFb_Fw and TGFb_Rv for TGF-β, IL-6_Fw and IL-6_Rv for IL-6, IL-1b_Fw and IL-1b_Rv for IL-1β, and IL-10_Fw and IL-10_Rv for IL-10. Expression was normalized by that of eF1α using primers eF1a_Fw and eF1a_Rv (Table 1).
2.10. Detection of phosphoproteins in RTgutGC cell line
The RTgutGC cell line was maintained in Leibovitz (L-15) medium (HyClone) containing 10% Fetal calf serum (FCS), plus 100 U/mL penicillin and 100 μg/mL streptomycin (P/S, Invitrogen) at 18 °C. For stimulation, 1 mL of cell suspension (at 1 × 106 cells/mL) was seeded into each well of a six-well plate (Falcon) and left overnight in the incubator to create a monolayer of ∼70% confluence. The cells were divided into 5 wells per condition, including different times from 0, 5, 15, 30, and 45 s. Before stimulation, cells were incubated with L-15 medium without supplementation for 2 h. After that time, the medium was removed, and the cells were stimulated with butyrate at 1 mM diluted in L-15 medium. As a control, the cells were incubated with L-15 medium without supplementation, or with L-15 medium containing 10% FCS. After each time point, the medium was removed, cells were washed with PBS, and RIPA buffer was added.
The obtained samples were separated by 15% SDS-PAGE under reducing conditions (80 V for 20 min, 120 V for 90 min). Protein bands in gels were transferred to nitrocellulose membranes at 350 mA for 90 min. The membrane was stained with Ponceau S stain (Sigma-Aldrich) to confirm equal loading. Next, the membranes were blocked with 5% (w/v) Bovine Serum Albumin in PBS containing 0.1% (v/v) Tween 20 (Sigma-Aldrich), and then incubated with 1:2000 phospho-Tyrosine Mouse mAb (cell signaling) or 1:1000 β-actin monoclonal antibody (Sigma), followed by HRP-conjugated goat anti-mouse IgG (Thermos), diluted at 1:8000. These membranes were then visualized with a SuperSignal West Pico Chemiluminescent Substrate kit (Thermos).
2.11. Statistical analyses
Statistical analyses were performed using the GraphPad Prism 6.0 software. The ANOVA test was used to analyze the significance of the fold induction. The significance was evaluated with a non-parametric Student T-test (p < 0.05).
3. Results
3.1. Identification of butyrate-producing taxa in the gut microbiota from Atlantic salmon
To identify the presence of microorganisms related to those producing butyrate in mammals, the intestinal microbiome of twelve pre-smolt specimens of Atlantic salmon was determined using total DNA extracted from feces present in the fish intestine as template. The fish were sampled at four different times from three tanks to avoid interference produced by the differences in the environment where the fish were grown. According to Vital et al. [67], about 76 genera associated with BPB exist, distributed in 38 families and 11 phyla, with most belonging to the Firmicutes phylum. The microbiota analysis of the feces using Illumina next-generation sequencing of the V4 amplicon from the 16 S rRNA allowed us to identify ten genera in Atlantic salmon that have been associated with butyrate production in mammals. These genera belong mainly to the Firmicutes and Proteobacteria phyla. Additionally, we identified nine taxa (ASVs) belonging to families associated with BPB which could not be assigned to any previously reported genus (Table 2). Since these nine taxa could represent new unculturable BPB, they were assigned as putative butyrate producers. The quantification of the bacterial population potentially involved in butyrate production in the gut of Atlantic salmon indicated that this group represents between 1.2% and 2.7% of the total population of the taxa identified (Figs. 1a and 1b). This population shows only slight variation between fish collected at the same time of culture; however statistical differences (p < 0.05) were observed in the older fish, where the total population almost duplicated the percentage of BPB observed in younger fish (Fig. 1b). When the prevalence of each taxon was compared between fish, the most prevalent ones associated with BPB were an unknown genus from the Comamonaceae and Lachnospiraceae families, and the genus Clostridium from the Clostridiaceae family which were present in 100% (12/12) of the fish sampled. The other highly prevalent taxa (>75%) were the unknown genus from the Ruminococcaceae family (11/12; 91.6%) and the genus Thermoanaerobacterium (9/12; 75%), followed by Clostridium (9/12; 75%) from Clostridiaceae and Peptostreptococcaceae families respectively (Fig. 1c). Consistent with these results, we were also able to identify butyrate in the feces of the sampled fish, at around 1.2 mM (Vargas, unpublished data). Overall, these results indicate the presence of BPB in the gut of Atlantic salmon and that they are constituents of the commensal microbiota.
Table 2.
Taxa associated with butyrate-producing bacteria identified in the feces of Atlantic salmon.
| Phylum | Family | Genus | Genus identified in Atlantic salmon feces |
|---|---|---|---|
| Actinobacteria | Glycomycetaceae | Stackebrandtia | Non-Identified |
| Intrasporangiaceae | Intrasporangium | Non-Identified | |
| Micromonosporaceaea | Micromonospora, Salinispora, Verrucosispora | Unknown taxa | |
| Nocardioidaceae | Kribbella, Nocardioides | Nocardioides, Unknown taxa | |
| Thermomonosporaceae | Thermomonospora | Non-Identified | |
| Bacteroidetes | Porphyromonadaceae | Bacteroidetes, Odoribacter, Porphyromonas, Propiomibacterium | Non-Identified |
| Rikenellaceae | Alistipes | Alistipes | |
| Chrysiogenetes | Desulfurispirillum | Non-Identified | |
| Deferribacteres | Deferribacter | Non-Identified | |
| Firmicutes | C. Incertae Sedis III | Thermoanaerobacterium | Thermoanaerobacterium |
| C. Incertae Sedis XI | Anaerococcus, Peptoniphilus | Non-Identified | |
| C. Incertae Sedis XVIII | Symbiobacterium | Non-Identified | |
| Carnobacteriaceae | Carnobacterium | Non-Identified | |
| Clostridiaceae | Clostridium | Clostridium | |
| Erysipelotrichaceae | Clostridium, Eubacterium | Clostridium | |
| Eubacteriaceae | Anaerofustis, Eubacterium, Pseudoramibacter | Eubacterium | |
| Halanaerobiaceae | Halanaerobium, Heliobacterium | Non-Identified | |
| Lachnospiraceae | Anaerotipes, Bytyrivibrio, Clostridiales, Coprococcus, Eubacterium, Roseburia, Shuttleworthia | Unknown, Coprococcus, Roseburia, Shuttleworthia | |
| Natranaerobiaceae | Dethiobacter | Non-Identified | |
| Natranaerobiaceae | Nathanaerobius | Non-Identified | |
| Peptococcaceae I | Desulfitobacterium | Non-Identified | |
| Peptococcaceae II | Desulfotomaculum | Desulfotomaculum | |
| Peptostreptococcaceae | Alkaliphilus, Clostridium, Eubacterium | Unknown | |
| Ruminococcaceae | Anaerotruncus, Faecalibacterium, Subdoligranulum | Unknown, Faecalibacterium | |
| Syntrophomonadaceae | Syntrophomonas | Non-Identified | |
| Syntrophothermus | Non-Identified | ||
| Thermoactinomycetaceae | Desmospora, Carboxydibrachium, Carboxydothermus, Thermoanaerobacter | Non-Identified | |
| Veillonellaceae | Acetonema, Acidaminococcus, Megasphaera, Thermosinus | Unknown | |
| Fusobacteria | Fusobacteriaceae | Fusobacterium, Ilyobacter | Unknown |
| Proteobacteria | Comamonadaceae | Rhodoferax | Unknown |
| Cystobacteraceae | Stigmatella | Non-Identified | |
| Desulfarculaceae | Desulfarculus | Non-Identified | |
| Desulfobulbaceae | Desulfobulbus | Non-Identified | |
| Geobacteraceae | Geobacter | Non-Identified | |
| Haliangiaceae | Haliangium | Non-Identified | |
| Myxococcaceae | Anaeromyxobacter, Myxococcus | Non-Identified | |
| Polyangiaceae | Sorangium | Unknown | |
| Syntrophobacteraceae | Syntrophobacter | Non-Identified | |
| Unclassified | Delta | Non-Identified | |
| Spirochaetes | Brachyspiraceae | Brachyspira | Non-Identified |
| candidate division SHA-4 | Candidatus Cloacamonas | Non-Identified | |
| Treponemaceae | Treponema | Non-Identified | |
| Teniricutes | Haloplasma | Non-Identified | |
| Thrermotogae | Thermotogaceae | Fervidobacterium, Kosmotoga, Petroga, Themosipho, Thermotoga | Non-Identified |
Fig. 1.
Abundance and prevalence of butyrate-producing bacteria in the intestinal microbiota of Atlantic salmon. The figure shows the abundance of taxa associated with butyrate-producing bacteria in each fish analyzed at a) the highest level of taxonomic classification (phylum), b) the lower level of taxonomical classification, genus or family if the taxa are not associated to any known genus described. The figure also shows the fish sampled on day 7 (fish 1, 2, and 3), day 14 (fish 4, 5, and 6), day 21 (fish 7, 8, and 9) and day 28 (fish 10, 11 and 12) after acclimatization. Panel c) shows the prevalence of taxa in the fish analyzed, where 100% of prevalence implies that the taxa were identified in each fish analyzed. The superscript index on the Clostridium genus indicates the family, aClostridiaceae, bPeptostreptococcaceae, cLachnospiraceae, and dErysipelotrichaceae.
3.2. Phylogenetic analysis of the classic HDAC and butyrate receptors from Atlantic salmon
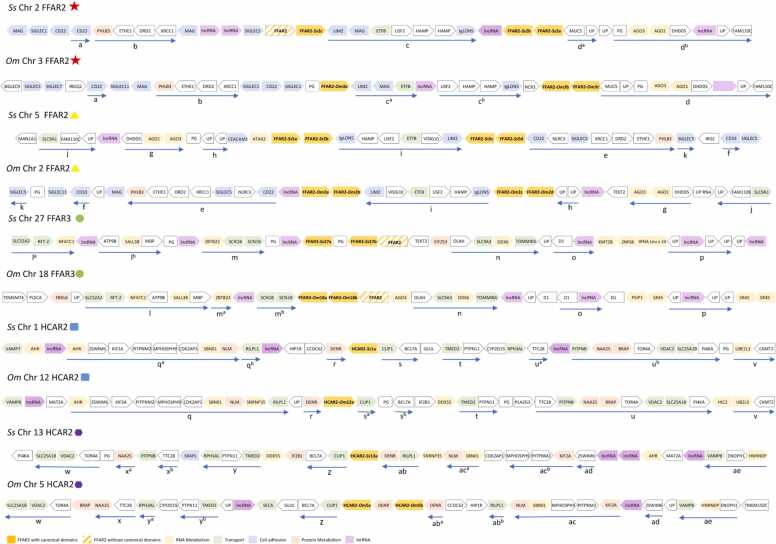
Once we had identified in the gut microbiota of Atlantic salmon the presence of microorganisms known to produce butyrate in mammals, we focused on identifying in the Atlantic salmon genome the presence of genes encoding proteins that are homologous to those have been reported to sense butyrate in mammals, specifically the Class I and IIa HDAC, and the butyrate receptors. Histone Deacetylases are classified into four classes, of which Classes I, II (a,b) and IV are denominated as classical HDACs. The analysis of the Atlantic salmon proteins homologous to the classical Human HDACs allowed the identification of 69 HDACs, most of them clustered together with the HDACs inhibited by butyrate (12 Class I HDACs and 48 Class IIa HDACs), indicating that in Atlantic salmon a specific expansion of genes encoding for the HDACs that respond to this microbial SCFA has occurred (Supplementary Figure 1). With respect to FFAR2, FFAR3 and HCAR2 in Atlantic salmon and Rainbow trout, we identified the presence of seven copies of FFAR2 and two copies of FFAR3 in both species, while HCAR2 is in four copies in Rainbow trout and two copies in Atlantic salmon. In Atlantic salmon, the 7 FFAR2s were distributed in Chromosome 2 (3 copies) and 5 (4 copies), while both FFAR3s were located in Chromosome 27 (2 copies), and HCAR2 was distributed in Chromosome 1 and 13, with one copy in each one. Rainbow trout shows a similar organization of FFAR2s, located in Chromosome 2 (4 copies) and 3 (3 copies), whilst FFAR3 is located in Chromosome 18 (2 copies) and HCAR2 is located in Chromosome 5 (two copies) and 12 (one copy) (Fig. 2, supplementary Table 1).
Fig. 2.
Ideogram representing the chromosomal location of genes encoding for butyrate receptors FFAR2, FFAR3, and HCAR2, in the chromosomes of Atlantic salmon and Rainbow trout.
Phylogenetic reconstruction using Maximum Likelihood and Bayesian probability assumptions, together with the estimation of the divergence times of the butyrate receptors FFAR2, FFAR3 and HCAR2, showed that all the receptors are present in paraphyletic groups between Atlantic salmon and Rainbow trout in their final clades (Fig. 2). The exception was the monophyletic clades for the HCAR2 receptors of Rainbow trout (Fig. 2b). When we analyzed the divergence times, the three receptors showed a basal divergence around 50 Myr. However, the internal clades of each receptor showed variable divergence times, between 20 and 2 Myr, depending on the receptor and clade.
The phylogenetic reconstruction of the FFAR2 receptor showed basal divergence of two main branches, which would have separated approximately 50.68 Myr ago, while the two main sub-branches would have diverged between 37 and 30 Myr. The upper main branch (Fig. 3 A) possessed three clades, all of which are composed of taxa belonging to Atlantic salmon and Rainbow trout. The terminal clades of the upper clade showed divergence times ranging from 7 to 2 Myr. The lower main clade comprises two subclades that show separation times of 4 and 13 Myr, respectively. These clades are also composed of taxa from both species, with divergence times ranging between 9 and 3 Myr.
Fig. 3.
Phylogenetic reconstruction and estimation of butyrate receptor divergence times in Atlantic salmon and Rainbow trout. The figure shows the phylogenetic reconstruction of butyrate receptor FFAR2 (a), FFAR3 (b) and HCAR2 (c) encoded in the Atlantic salmon and Rainbow trout genomes. The phylogenetic trees were constructed under the assumptions of Maximum Likelihood (ML) and Bayesian Posterior Probabilities Before each clade, the Bayesian Posterior Probability is shown followed by the bootstrap value of ML; below these estimates is the divergence time of each clade. A black arrow indicates some divergence time to facilitate their location in the tree. Each taxon is shown with its species name and NCBI accession number. Divergence times for salmonid species are in bold. The timeline is shown on the X-axis.
Phylogenetic reconstruction of the FFAR3 receptor from Atlantic salmon and Rainbow trout showed divergence into two main clades at 51.51 Myr. The upper main clade is composed of copies of both species, which would have separated 5.75 Myr ago. In turn, the lower main clade is composed of two copies from Rainbow trout and Atlantic salmon, which would have diverged 2.91 Myr ago (Fig. 3B). This implies that the second round of duplication observed in FFAR2 (30–40 Myr) did not affect FFAR3.
Phylogenetic reconstruction of the HCAR2 receptor showed basal divergence at 50.67 Myr, giving rise to two main clades. The upper main clade diverged at 20.27 Myr, with a gene copy from Atlantic salmon outside an internal clade composed exclusively of HCAR2 from Rainbow trout, which would have diverged 9.85 Myr ago. On the other hand, the lower clade diverged at 19.71 Myr, and has the same topology as the upper clade, as it is composed of one HCAR2 from Atlantic salmon outside of an internal clade consisting of two HCAR2s from Rainbow trout. When we analyzed the chromosomal location of HCAR2 in Rainbow trout, the copies located in the same chromosome formed a monophyletic clade, except for the gene encoding for CDQ79106.1 whose location could not be identified in the genome (Fig. 3 C).
To identify which genes, correspond to events that happened before or after the diversification of the Salmonidae family or were present in its common ancestor, we performed phylogenetic analysis. Under the assumption of Maximum Likelihood, the phylogenetic reconstruction of the FFAR2 genes, using sequences from the main vertebrate groups, resulted in a tree with a general topology of four main clades (Supplementary Figure 2). One of the main clades groups humans, monotremes, marsupials and birds. A second clade clusters amphibians and the fish orders Coelacanthiformes, Dipnoi and Petromyzontiformes. The third clade clusters reptiles and in a subclade, the fish of the orders Acipenseriformes and Lepisosteiformes. Representatives of the order Salmoniformes (Atlantic salmon and Rainbow trout) were found in three subclades (Supplementary Figure 2). FFAR2 from Atlantic salmon and Rainbow trout were clustered in clades I and II, respectively. These results suggest that the ancestor of the Salmoniform group harbored these copies prior to diversification and genomic duplication, and later diverged separately in each species.
The phylogenetic reconstruction of FFAR3s, highlighted a tree with a general topology of three main clades (Supplementary Figure 3). The first main clade groups is humans, marsupials, monotremes, birds, amphibians, and fish of the orders Dipnoi and Coelacanthiformes. Amphibians and fish of the order Dipnoi are grouped in a subclade, and fish of the order Coelacanthiformes are found outside. In the second main clade, fish of the order Acipenseriformes and the marine lampreys of the order Petromyzontiformes are gathered into a subclade; outside this clade are the reptiles, while fish of the Order Salmoniformes are grouped into two subclades (Supplementary Figure 3). Like FFAR2, these data demonstrate that a gene encoding for FFAR3 was present in the common ancestor of the Salmoniformes group, which later diversified in each species.
The phylogenetic tree of the HCAR2 genes (Supplementary Figure 4) was constructed using protein sequences from the main vertebrate groups, except for the group of birds, in which a homologous protein could not be identified. The topology of this tree showed that the protein belonging to the species Protopterus annectens is outside the two main clades, probably due to the retention of an ancestral variant of the gene prior to the diversification of the other vertebrate groups. One of the main clades contains humans, monotremes, marsupials, amphibians, and the fish orders Lepisosteiformes and Acipenseriformes. The other major clade contains exclusively the fish orders Coelacanthiformes and Salmoniformes. At the level of the order Salmoniformes, the topology of the subclades of this group is consistent with the phylogenetic relationships shown in (Fig. 3B). Therefore, this subclade is divided into two, where we find a distribution of two copies of the HCAR2 gene belonging to Rainbow trout together with a copy of Atlantic salmon, in both subclades. These results are consistent with phylogenetic reconstruction using the butyrate receptor present in other members of the Salmonidae family and teleosts (Supplementary Figures 5 and 6).
Therefore, according to our analysis, we infer that the paraphyletic clades of the FFAR2, FFAR3 and HCAR2 receptors are probably the result of the divergence of paralogs in the common ancestor before the diversification of the salmonids. The divergence data showed that for all the receptors analyzed, the main branches arose around 50 Myr, the approximate time in which the genomic duplication has been dated prior to the diversification of the salmonid group [68]. On the other hand, the monophyletic clades of Rainbow trout for the HCAR2 receptor could be a consequence of a species-specific duplication, after the divergence of the genus Oncorhynchus.
3.3. Syntenic analyses of butyrate receptors from Atlantic salmon
Considering that all butyrate receptor copies identified are distributed in five chromosomes within Atlantic salmon and Rainbow trout, and that most of the copies are located within the same chromosome, we characterized the genetic context in which each copy was found. The comparison between the genetic contexts of butyrate receptors in Atlantic salmon and Rainbow trout showed that they have a similar structure in terms of gene identity and orientation. In these chromosomes, we identified a total of 30 conserved regions (named with letters in Fig. 4). e.g. chromosome three of Rainbow trout and chromosome two of Atlantic salmon share 18 genes distributed in four conserved regions (from a to d) (Fig. 4).
Fig. 4.
Genetic context and collinearity of FFAR2, FFAR3 and HCAR2 receptors in the Atlantic salmon and Rainbow trout genomes. The analyzed genes are ordered based on the species, chromosome number, receptor and a comparative symbol (star, circle, square, hexagon) that links it to the most similar sequence within the analysis. Genes are abbreviated using human nomenclature and gene orientation is in order 5′ to 3′ (arrow to the right) and 3′ to 5′ (arrow to the left). Blue arrows indicate contiguous collinear genes in similar gene contexts (e.g. stars, circles, etc.), reverse arrows mean reversed orientation of collinear genes, and dashed arrows mean interruption of collinearity in one of the sequences. Letters under blue arrows identify the regions in different chromosomes.
The total number of gene elements present in the ten analyzed chromosomes (five from each fish species), excluding butyrate receptors, was 331, where a putative function could be assigned to 102 elements, generating a redundancy of 69.18%. Of the total elements, 8.5% corresponds to long noncoding RNA (lncRNA) and 3.6% to pseudogenes, 20.4% correspond to RNA metabolism, 16.2% to transport, 12.12% to cell adhesion and 8.08% to protein metabolism.
When the distribution of genes encoding for FFAR2 or FFAR3 was analyzed, we observed that several copies are arranged so as to form continuous pairs or are separated by a single gene (e.g. Ss Chr 2 FFAR2-Ss2b - FFAR2-Ss2a). We also identified the presence of three copies of FFAR2 that did not meet our assignment parameters for FFAR2 or FFAR3 (presence of the FFAR2/3 domain). These copies are found upstream of FFAR3-Ss27b (XM_014177057.2, XP_014032532.1), FFAR3-Om18b, and FFAR2-Ss2c (Fig. 4). In the case of HCAR2, all copies were found uniquely on their respective chromosomes, except for chromosome five of Rainbow trout where we find that two copies are interrupted by a gene.
We also noted that the genetic context of FFAR2, independent of the chromosome in which it is located, contains genes related mainly to the mucosal and cellular immune response, and regulation by RNAi. On the other hand, FFAR3 is associated mainly with genes related to epigenetic regulation, translation, and neuronal function. Finally, the chromosome regions where HCAR2 are encoded contain genes related with cilia formation, cytoskeleton, cell division, intracellular traffic, and secondary messengers, suggesting that these receptors could have different functions associated with the genes with which they are collocated. When we analyzed the presence of genes encoding proteins related with the microbiota-host interaction, we identified genes encoding for cell adhesion molecules (CAM), mucosa integrity, receptors and enzymes for microbial metabolites, cellular immune response, iron and riboflavin uptake, epigenetic regulation and neuronal function. In particular we identified homologous receptors for Dopamine (D2R) [69], Aryl hydrocarbons (AHR) [70], and for one enzyme related to the metabolism of H2S (ETHE1) [71], Hepcidine for iron uptake [72], and riboflavin transporters [73], in addition to the butyrate receptors (Supplementary File 2).
Taken together, these results show that genes encoding for butyrate receptors are in chromosomal regions conserved between Atlantic salmon and Rainbow trout. Such genes are surrounded by those related with the microbiota-host interaction. The syntenic analysis also shows that the diversification process of these receptors has resulted mainly from intrachromosomal tandem duplication, but in the case of FFAR2 and HCAR2, a genome duplication event has also been a source of diversity since the function of the neighboring genes is conserved independently of the chromosome where the genes encoding for FFAR2 or HCAR2 are located.
3.4. Expression of butyrate receptors in Atlantic salmon tissues
Since the genetic context of each kind of butyrate receptor is different and could be related to a specific function, we analyzed the expression of HCAR2, FFAR3 and FFAR2 in the gut, head-kidney and spleen from pre-smolt fish (day 21), by RT-PCR. We designed primers able to identify all gene copies of HCAR2 and FFAR3. However, this approach was unsuccessful in the case of Atlantic salmon FFAR2; thus, we generated primers specific for each of the seven copies of FFAR2. We also analyzed their expression in SHK-1 and RTgutGC cultures. Our results show that HCAR2 is expressed in the gut, spleen, and head-kidney, but not in SHK-1 and RTgutGC. Furthermore, we were unable to detect the expression of FFAR3 in any of the tissues and cell cultures analyzed, while for FFAR2, the only gene whose expression was detected was XP_014056042.1 in RTgutGC. Given this latter result, the functionality of this FFAR2 was evaluated by exposing cell cultures of RTgutGC to butyrate and evaluating changes in the pattern of tyrosine-phosphorylated proteins. We detected several bands (proteins) between 30 and 41 kDa whose intensity changed, increasing after 5 min and decreasing after 15 min of exposure to butyrate, supporting the presence of a protein able to detect butyrate in RTgutGC (Supplementary Figure 7). Our results show that HCAR2 and FFAR2 are expressed differentially depending on the tissue, cell culture and the gene copy analyzed, with HCAR2 being the more widely-expressed receptor in the samples of Atlantic salmon analyzed. Our results also suggest that FFAR2 (XP_014056042.1) can detect the presence of butyrate, thus stimulating the phosphorylation/dephosphorylation of cytoplasmatic proteins.
3.5. Structural characterization of butyrate receptors from Atlantic salmon
3.5.1. Sequence analysis of FFAR2/3 from Atlantic salmon
The putative FFARs identified in Atlantic salmon contain the conserved domain cd15170 that is characteristic for FFAR2 and FFAR3 [74]; however, their functionality and specificity for SCFA cannot be defined only by the presence of this domain. To characterize more deeply the potential functionality and specificity of these receptors, a structure-sequence analysis was performed focusing on identifying amino acids previously described as important for the binding and specificity of SCFAs in other FFAR2s and FFAR3s. Since there are no reported empirical 3D structures of these receptors, to model by homology the structures of Atlantic salmon FFAR2/3, we used the structural information of human, mouse and rat FFAR2 and FFAR3, recently predicted by Alphafold [52]. The amino acid residues important for SCFA interactions were obtained from the works of Tikhonova [75], which combine homology modelling of human and mouse receptors with in silico docking and mutagenesis for their identification. These residues were located in the 3D structure of Atlantic salmon FFAR2/3 models in order to identify whether their spatial distribution is consistent with the biological function predicted from human or mice.
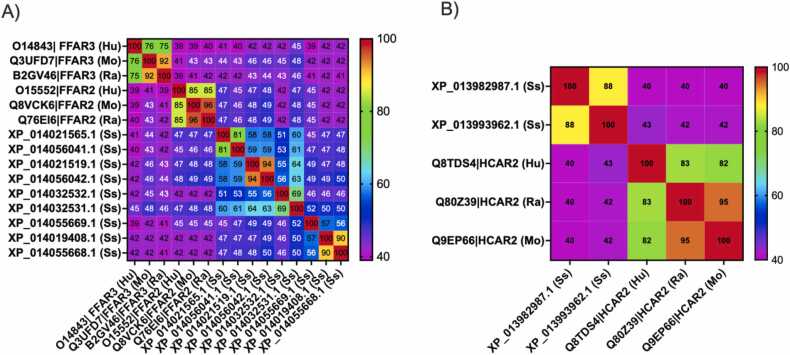
Multiple alignments between FFAR2/3 identified in Atlantic salmon, human, rat and mouse showed that FFAR3 from mammalian species share at least 75% identity, rising to 85% in the case of FFAR2. However, the average identity in mammals between FFAR2 and FFAR3 was just 40%. When Atlantic salmon FFAR2/3 were analyzed, they showed identities ranging from 39% to 94% between them, falling to 42%− 45% identity when compared with mammalian FFAR3 and FFAR2, respectively (Fig. 5). This result reinforces the idea that FFAR2/3 from Atlantic salmon are more related to mammalian FFAR2. The prediction of transmembrane regions and protein topology using Predic Protein [76] indicated that all the putative FFAR2/3 present in Atlantic salmon contain seven transmembrane helices, with the NH2-terminal located outside of the cell membrane and a cytoplasmic carboxy-terminus (Table 3).
Fig. 5.
Identity between butyrate receptors from Atlantic salmon, human, rat and mouse. The figure shows the percentage of identity between FFAR2 and FFAR3 (A) and HCAR2 (B) encoded in the genome of Atlantic salmon (Ss), human (Hu), rat (Ra) and mouse (Mo). The percentage of identity is indicated using a color scale and by the number indicated in each cell. The percentage of identity was calculated using ClustalW.
Table 3.
Prediction of Transmembrane helices and topology [76].
| ID | Species | Receptor | NH2 | TM1 | TM2 | TM3 | TM4 | TM5 | TM6 | TM7 | COOH |
|---|---|---|---|---|---|---|---|---|---|---|---|
| XP014019408 | S. salar | FFAR2/3 | out | 8–32 | 44–64 | 82–105 | 123–145 | 171–193 | 218–239 | 252–271 | In |
| XP014021519 | S. salar | FFAR2/3 | out | 8–31 | 44–64 | 82–104 | 124–145 | 203–223 | 247–268 | 281–300 | In |
| XP014021565 | S. salar | FFAR2/3 | out | 15–39 | 51–71 | 89–110 | 130–152 | 180–202 | 224–247 | 259–279 | In |
| XP014032531 | S. salar | FFAR2/3 | out | 6–30 | 42–62 | 80–104 | 121–143 | 174–196 | 222–243 | 256–275 | In |
| XP014032532 | S. salar | FFAR2/3 | out | 31–54 | 67–87 | 107–131 | 147–168 | 194–215 | 235–259 | 272–291 | In |
| XP014055668 | S. salar | FFAR2/3 | out | 8–32 | 44–64 | 82–105 | 123–145 | 170–192 | 216–239 | 251–271 | In |
| XP014055669 | S. salar | FFAR2/3 | out | 7–31 | 43–63 | 81–105 | 123–144 | 172–194 | 214–235 | 247–267 | In |
| XP014056041 | S. salar | FFAR2/3 | out | 16–40 | 52–72 | 90–114 | 132–153 | 183–205 | 228–249 | 265–284 | In |
| XP014056042 | S. salar | FFAR2/3 | out | 8–32 | 44–64 | 82–106 | 124–145 | 200–221 | 244–265 | 278–297 | In |
| Q8VCK6 | FFAR2 | out | 9–33 | 46–66 | 83–107 | 126–149 | 176–198 | 223–244 | 257–276 | In | |
| Q76EI6 | FFAR2 | out | 9–32 | 46–66 | 84–110 | 125–147 | 175–197 | 223–244 | 257–276 | In | |
| O15552 | Human | FFAR2 | out | 9–33 | 44–66 | 83–107 | 125–147 | 175–197 | 223–246 | 257–276 | In |
| O14843 | Human | FFAR3 | out | 16–38 | 52–72 | 90–112 | 131–153 | 180–202 | 226–247 | 259–279 | In |
| Q3UFD7 | FFAR3 | out | 12–36 | 48–68 | 85–110 | 127–149 | 183–204 | 222–243 | 256–275 | In | |
| B2GV46 | FFAR3 | out | 12–36 | 48–68 | 85–109 | 127–149 | 176–197 | 222–243 | 256–275 | In |
In order to identify whether FFAR2/3 from Atlantic salmon present the residues related with their interaction with SCFAs, we used the information previously obtained from the structures of human FFAR2 and FFAR3 modelled using as template the β2-adrenergic receptor, FFAR1, and the empirical data obtained from mutagenic experiments. Altogether, these studies show that K65, W75, Y90, H140, I145, Q148, E166, R180, Y238, H242, and R255 from human FFAR2 are related with SCFA interaction and selectivity [77], [78]. The multiple alignments showed that W75, R180, Y238, H242, and R255 were the only residues conserved between Atlantic salmon and mammalian FFAR2/3. According to the homology models of FFAR2 and FFAR3 from humans, and the docking simulation with agonist and orthostatic ligands, the arginine and histidine are involved in the interaction with the carboxylic region of SCFA or the agonist, while the aromatic residues are related with the hydrophobic core of the binding site [77]. To evaluate whether the FFAR2/3 identified in Atlantic salmon show different specificities for SCFAs (butyrate, propionate or acetate), as is the case for FFAR2 and FFAR3 in mammalians [77], [78], we analyzed the presence of residues associated with the specificity of FFAR2 (Y90, I145, and E166) and FFAR3 (F96, Y151 and L171) in the FFAR2/3 from Atlantic salmon. We observed that the sequence XP014056042.1 was the only FFAR2/3 containing all three residues previously described in mammal, specifically the equivalents to Y90, I145 and E166 from FFAR2. The remaining FFAR2/3 s from Atlantic salmon were classified into six types according to the amino acids present in the alignments. The first position, which is Y90 in FFAR2 and F96 in FFAR3, also contains the same amino acids in FFAR2/3 present in Atlantic salmon. In the second position (I145 or Y151) the amino acids present in FFAR2/3 s from Atlantic salmon were V, Y, and F, while the amino acids present in the third position (E166 or L171) corresponded to E, L, T, N and G. Not a single FFAR2/3 from Atlantic salmon showed exactly the same amino acid disposition as found in mammalian FFAR3 (Table 4).
Table 4.
Amino acids involved in the interaction of Free Fatty Acid Receptor 2 and 3 with Short-Chain Fatty Acids.
| Amino acid | Classification | ID sequence | Organism |
|---|---|---|---|
| Y90, I145, E166 | FFAR2 | Q76EI6 | Rattus norvegicus |
| Y90, I145, E166 | FFAR2 | Q8VCK6 | Mus musculus |
| Y90, I145, E166 | FFAR2 | O15552 | Homo sapiens |
| Y90, I145, E166 | FFAR2 | XP014056042.1 | Salmo salar |
| F96, Y151, L171 | FFAR3 | B2GV46 | Rattus norvegicus |
| F96, Y151, L171 | FFAR3 | Q3UFD7 | Mus musculus |
| F96, Y151, L171 | FFAR3 | O14843 | Homo sapiens |
| Y90, V145, E166 | FFAR2/3 | XP_014021519.1 | Salmo salar |
| Y90, Y145, E166 | FFAR2/3 | XP_014056041.1 | Salmo salar |
| Y90, Y145, L166 | FFAR2/3 | XP_014032531.1 | Salmo salar |
| Y90, Y145, T166 | FFAR2/3 | XP_014021565.1 | Salmo salar |
| F96, Y151, N171 | FFAR2/3 | XP_014055669.1 | Salmo salar |
| F96, F151, G171 | FFAR2/3 | XP_014032532.1 | Salmo salar |
| F96, F151, E171 | FFAR2/3 | XP_014019408.1 | Salmo salar |
| F96, F151, E171 | FFAR2/3 | XP_014055668.1 | Salmo salar |
Altogether, these results show that FFAR2 and FFAR3 from Atlantic salmon are more related to mammalian FFAR2 than to mammalian FFAR3, and are more diverse in the amino acids that are putatively related with the interaction and specificity for SCFA than in mammalian FFAR2 and FFAR3.
3.5.2. Structural analysis of the FFAR2/3 from Atlantic salmon
To identify whether the protein sequences of FFAR2/3 from Atlantic salmon are able to form protein structures that are potentially functional and explain the conserved residues identified, we modeled by homology their structures using as template the ab initio models obtained for Alphafold [52] onto the FFAR2 and FFAR3 structures from human, mouse and rat. We also predicted the potential binding sites and docked the butyrate molecule in these structures.
In the case of FFAR2 (O1552) from human, we identified four potential binding sites. Pockets 1 and 4 are located in the extracytoplasmic side of the transporter, of which ligand-binding pocket (LBP) 1 is the largest, spanning 26 residues, and pocket 4 only nine residues (Supplementary File 3). These pockets contain the amino acids previously described as important in the binding site of SCFAs, and share residues R255, R180 and V176. The docking of butyrate in the modeled structure shows that butyrate has a potential binding site in pockets 2 and 3, both of which are located on the cytoplasmic side and which may represent allosteric sites. Butyrate also docks in a site shared by pockets 1 and 2, interacting with E166, and R255 and R180 networks (Fig. 6 A). This interaction is more superficial than that previously reported when human FFAR2 was modelled using the β2-adrenergic receptor as template [77]. This suggests that both models could represent different conformations of the same receptor, with one being more open allowing the interaction with residues located inside pocket 1, as reported previously, whilst a second tighter conformation could represent the initial interaction of FFAR2 with the SCFA.
Fig. 6.
The predicted structure of FFAR2 and FFAR3 from human. The figure shows the structure of FFAR2 O15552 (A) and FFAR3 O14843 (B) predicted by Alphafold. The amino acid that conforms the putative Ligand Binding Pocket (LBP) predicted by P2RANK is shown in surface representation. The putative binding sites for butyrate predicted by docking using the SwissProt platform are shown as the clusters of possible conformations that the ligand (butyrate) could adopt on the surface of the target. The blue dashed line represents the putative position of the cytoplasmatic membrane. The arrows between the figures show the rotation of the view and the axes of this rotation.
In the case of human FFAR3 (O14843), the prediction of LBPs indicates seven possible regions, of which four are located in the extracellular side (LBP1, 2, 4, 7), one on the cytoplasmatic side (LBP5) and two in the transmembrane region (LBP3 and 6). LBP1 comprises four residues that have been experimentally associated with the function of FFAR3 [77], [78], while LBP4 contains three residues (R185, R258 and F96) which have been implicated in the SCFA selectivity of FFAR3 [77], [78]. LBP4 shares F96 with LBP1, which has been suggested to participate in the specificity of FFAR3 [77], [78]. LBP4, LBP2 and LBP1 from FFAR3 are equivalent to LBP1 from FFAR2, while LBP3-FFAR3 is equivalent to LBP3-FFAR2, sharing a similar topology and location. FFAR3 and FFAR2 also possess a pocket on the cytoplasmic side identified as LBP5-FFAR3 and LBP2-FFAR2, respectively. LBP6-FFAR3 has no equivalent in FFAR2 but is also located in the transmembrane region. LBP7-FFAR3 is close to LBP1-FFAR3 that also has no equivalent in FFAR2, and is formed by the interaction of the NH2 terminal of FFAR3 and the loops between transmembrane helices TMH5 and TMH6. When the localization of the potential binding site to butyrate was explored using molecular docking, we identified that butyrate binds to a surface formed by the interaction of LBP1-FFAR3 and LBP4-FFAR3 via Y70 and R258. LBP2-FFAR3 and LBP7-FFAR3 also show interactions with butyrate in a similar way as observed in its equivalent LBP in FFAR2, which suggests that it could also be a site for allosteric regulation (Fig. 6B).
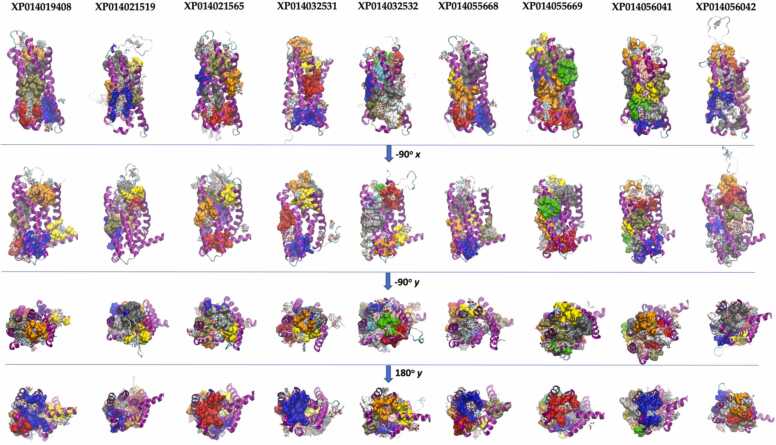
Given that FFAR2 and FFAR3 modeled by Alphafold harbor structural information that is consistent with that previously obtained by homology modeling, we used these structures and those of FFAR2 and FFAR3 from rat and mouse as template for modeling FFAR2/3 s identified in Atlantic salmon. In the seven models obtained, an overall analysis of their quality demonstrates GA341 values equal to 1, indicating that the alignment between templates and targets has enough accuracy to produce feasible models of the Atlantic salmon FFAR2/3 receptors. Nevertheless, the analysis of the energy based on contacts of amino acids in the predicted structures using PROSA, shows that in all models, the amino acids from TMH2 and 3 have high energy, suggesting that the side chains of amino acids positioned in these helices could have a different packing or the helices may have a different conformational arrangement, as is observed in the models for FFAR2 and FFAR3 in mammals [77], [78]. The main structural differences identified in FFAR2/3 from Atlantic salmon were in the extracellular loop between TMH4 and TMH5. In this loop, XP014056042 and XP014021519 have an insertion of approximately 25 amino acids which probably acquires a secondary structure of an alpha-helix according to secondary structure prediction using the Phyre 2.0 server (Supplementary Figure 8). The analysis of the LBPs indicated that the presence of the pockets in the cytoplasmic and extracellular sides of the FFAR are conserved, together with the pocket located in the membrane formed by TMH3 and TMH4. Substantial variability in the number of pockets and the amino acids that are part of these pockets was identified on the extracellular sides of the Atlantic salmon FFAR studied. These differences are related to the variable size of the loop region between TMH4 and TMH5, which depending on the sequences, can block the access to the pocket that contains the amino acids involved in the interaction with the carboxylic acids, equivalent to Y90, R180 and R255 of human FFAR2. The prediction of potential binding sites for butyrate using molecular docking simulation shows similar results; butyrate can bind to some of these pockets in several different conformations, suggesting that at least in the conformation proposed by the modelling of 3D structures, these binding sites are not specific and could represent a point of interaction involved in allosteric regulation of the receptors. Like in FFAR3, the FFAR2/3 from Atlantic salmon harbor binding sites for butyrate in several pockets located in the extracellular side of the receptor (Fig. 7).
Fig. 7.
Molecular modeling of FFAR2/3 identified in the genome of Atlantic salmon. The figure shows the structure predicted by homology modeling of FFAR2/3 found in the genome of Atlantic salmon. The figure also shows the binding pocket predicted by P2RANK (atoms in the surface display) and the docking site for butyrate predicted using SwissDock. The structure was modelled using as templates the structures predicted by Alphafold for FFAR2 and FFAR3 of human, rat and mouse. The color of the atom surface represents the ranking on the binding pocket predicted by P2RANK, with blue (1st), red (2nd), gray (3rd), orange (4th), yellow (5th), tan (6th), silver (7th), green (8th), white (9th), pink cyan (10th), and purple (11th). The figure was generated using VMD software.
Altogether, these results show that FFAR2/3 from Atlantic salmon share the same structure as FFAR2 and FFAR3 from mammalian species, but show a great diversity in the predicted binding pockets and binding sites for butyrate in the loops located in the extracellular sides of the receptors, suggesting that each receptor presents a different affinity for butyrate, possibly responding at different concentrations.
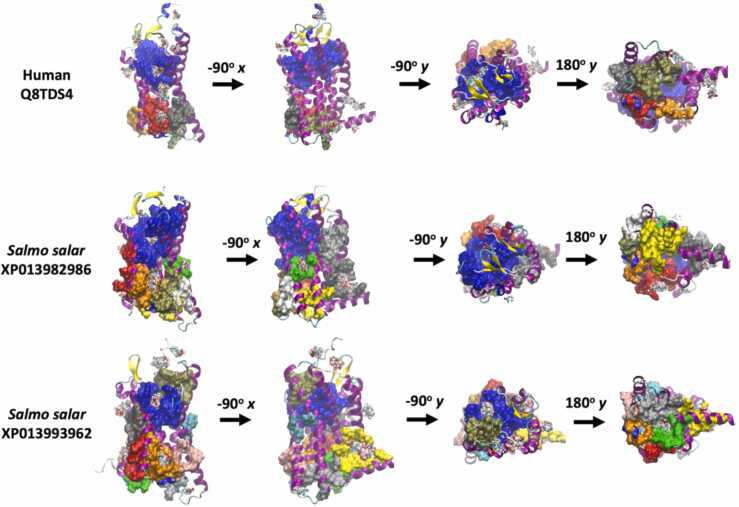
3.5.3. Sequence and structural analysis of HCAR2 from Atlantic salmon
The analysis of the HCAR2 receptor shows similar results. The HCARs identified in Atlantic salmon, XP013982987 and XP013993962, share 88% identity and an average identity of 41% with mammalian HCAR2. The alignment shows several conserved regions across the sequence, but since structural studies have not been reported, the function of these regions is currently unknown. Nevertheless, molecular docking predictions based on structures predicted by homology modelling have shown that R111 and R251 interact with the carboxylate, while S178 interacts with amide carbonyl of nicotinic acid [79], [80]. I254, F255, F276, N171, S179 and H259 are also predicted to be involved in the interactions with the ligands [79], [80]. The comparison between HCAR2 from Atlantic salmon and mammalian species shows that the equivalent to S178, R111 and R251 are conserved, suggesting that both receptors share this property to bind molecules with carboxylate groups. Regarding the other residues, F276 was the only one conserved among mammalian and Atlantic salmon HCAR2, suggesting that both kinds of receptors have different affinities for nicotinic acids (Supplementary Figure 9). The identification of LBP showed that human HCAR2 possesses a pocket composed of residues that are involved in the interaction with the carboxylate group. This pocket also interacts with butyrate according to the molecular docking predictions. When HCAR2 from Atlantic salmon was analyzed by searching for LBPs and sites of interaction with butyrate, both receptors showed pockets equivalent to the LBP identified in human HCAR2, where butyrate is also predicted to interact (Fig. 8).
Fig. 8.
Molecular modeling of HCAR2 identified in the genome of Atlantic salmon. The figure shows the structure predicted by homology modeling of HCAR2 found in the genome of Atlantic salmon. The figure also shows the binding pocket predicted by P2RANK (atoms in surface display) and the docking site for butyrate predicted using SwissDock. The structure was modelled using as templates the structures predicted by Alphafold for HCAR2 of human, rat and mouse. The color of the atom surface represents the ranking on the binding pocket predicted by P2RANK, with blue (1st), red (2nd), gray (3rd), orange (4th), yellow (5th), tan (6th), silver (7th), green (8th), white (9th), pink cyan (10th), and purple (11th). The figure was generated using VMD software.
Altogether these predictions indicate that putative FFAR2/3 and HCAR2 from Atlantic salmon could bind butyrate, but with different affinities, as also observed for mammalian receptors.
3.6. In vitro effect of butyrate on immune genes and antiviral response
In order to determine whether butyrate can modify the innate immune response in Atlantic salmon cells, such in mammalian cells [81], we evaluated the effects of butyrate on the antiviral response in SHK-1. This cell line was isolated from head-kidney of Atlantic salmon and has a macrophage-like phenotype [82]. Since in mammalian butyrate inhibit HDAC from both blood peripherical and intestinal macrophages, we hypothesized that immune function of SHK-1 should be affected by butyrate if the microbiota-host communication are conserved. Cultures of SHK-1 cells were pre-treated for 24 h with butyrate (2 mM), and then transfected with Poly I:C to stimulate the innate immunity against viral infections. Poly I:C induced the expression of antiviral genes, Interferon-alpha (INFα), Mx and PKR. INFα and PKR rose to a maximum of induction at 24 h (>30 fold and>15 fold, respectively) (Fig. 9c, Fig. 9b), while Mx peaked at 36 h post-transfection (>20 fold) (Fig. 9a). In cells treated with butyrate, Mx and PKR reached maximum expression at 96 h post-transfection (>20 fold and>10 fold, respectively) (Fig. 9a, Fig. 9b), with peak induction values similar to those observed in the untreated cells. The expression of INFα shows a similar pattern; treated cells peaked at 36 h (>20 folds) (Fig. 9c). However, after this time the fold-induction was 15 times lower than the maximum seen in untreated cells, and remained constant until the end of the experiment. Our results show that butyrate could delay the transcription of genes related with the antiviral response in SHK-1 cells.
Fig. 9.
Effects of butyrate on the antiviral response in SHK-1 cells. The figure shows the effects of butyrate on the expression of the genes encoding Mx (Panel A), PKR (panel B), INFα (Panel C), and on the replication of IPNV (panel D). Cultures of SHK-1 cells were exposed to 2 mM butyrate for 24 h and then transfected with Poly I:C to stimulate the antiviral response (blue inverted triangle). Cells only treated with IPNV are in red circles, while cells treated only with butyrate are in black squares. The effect on the replication of IPNV (Panel D) was achieved by exposing SHK-1 cultures for 24 h to 1 mM (blue circles), 2 mM (white circles) and 4 mM (green circles) butyrate. Cells without treatment and cells pretreated with Poly I:C are represented in yellow and red circles, respectively. After treatment with butyrate, IPNV infection was performed with a m.o.i of 0.1. Gene expression and IPNV load were quantified by RT-qPCR. Gene expression was quantified using the ΔΔCt method and normalized by the expression of the 18 S rRNA in cultures without treatment. Bars represent means± SD n = 3. Data were analyzed with non-parametric Student t-test (* p < 0.05, ** p < 0.01, *** p < 0.001).
In order to assess whether butyrate modifies viral replication, SHK-1 cells were pre-incubated for 24 h with different concentrations of butyrate and infected with IPNV (Fig. 9d). As a positive control of antiviral response, some culture wells were transfected with Poly I:C after butyrate treatment, prior to the infection with IPNV. As expected, cells pretreated with Poly I:C show lower viral load in the culture media supernatant with respect to the untreated cells (from 1 ×104 to 1 ×106 copies/mL). However, at the assayed concentrations of butyrate, viral load did not show statistical differences between the butyrate-treated and untreated cells, but there was a tendency to reduce the viral load in cell cultures concomitant with the increased concentration of butyrate.
To determine whether butyrate is able to modify the basal expression of cytokines, we assessed the gene expression of several cytokines in SHK-1 cells kept at normal growth conditions and in the presence of 2 mM butyrate for 24 h. IL-10 reached a maximum of induction after one-day post-treatment with butyrate (>10 fold). This effect decayed over time, as judged by the lower values of induction on day 5 (>2 fold, p < 0.01) (Fig. 10b). In contrast, TGF-β showed the opposite behavior, peaking at 5 days after treatment (>500 fold, p < 0.001) (Fig. 10a). Although a non-statistical difference was observed in the expression of IL-1β, a decreasing tendency was observed in IL-1β one day after treatment (Fig. 10d). With IL-6, a tendency to increase expression was observed on the fifth-day post-treatment (>100 fold, p < 0.05) (Fig. 10c). These results suggested that butyrate modifies the expression of cytokines in order to promote an anti-inflammatory state.
Fig. 10.
Effects of butyrate on the expression of anti- and pro-inflammatory cytokines. The figure shows the effect on anti-inflammatory (Panel A) and pro-inflammatory (Panel B) cytokines. Cultures of SHK-1 cells were treated with 2 mM butyrate for 24 h. Samples were taken after 1- and 5-days post-treatment. Expression was quantified using RT-qPCR, by the ΔΔCt method using as housekeeping the expression of the gene encoding for Elongation factor 1α. Changes in expression are shown as the logarithm in base 2. As a control, the data were also normalized with respect to the expression of the genes in cell cultures without butyrate treatment. Bars represent means± SD n = 3.
4. Discussion
In this work, we combine the use of metagenomic, bioinformatic, phylogenomics, structural, and biochemical approaches to enlighten our knowledge concerning microbiota-host interactions in Atlantic salmon. We focus on identifying the elements that allow the interplay between microbiota and host by using the SCFA butyrate as a model, as it is the best characterized microbial metabolite in crosstalk in mammalian microbiota-host systems. Our first goal was to identify in the microbiota of Atlantic salmon, microorganisms belonging to the genus associated with BPB. In mammals, most BPB correspond to anaerobic microorganisms, which metabolize acetyl-CoA from cellulose metabolism to butyrate. Besides this pathway, microbiota can produce butyrate by metabolizing amino acids through the glutaric, 4-aminobutyrate or lysine pathways [83], or processing lactate through the Acetyl CoA pathway [84]. Our results show that intestinal microbiota from Atlantic salmon have lower diversity (nine families and ten genera) of butyrate-producing genus than humans [83], with taxon associated with butyrate production from glutaric, 4 aminobutyric and lysine pathways, in agreement with the carnivorous diet of Atlantic salmon and a low amount of plant fiber such as cellulose. Moreover, Atlantic salmon microbiota is rich in lactic acid bacteria [85], [86], in which lactate production could be used by bacterial cross feeding [87] to produce butyrate for some bacteria belonging to the Eubacterium genus [84]. The presence of taxa associated with BPB, together with the presence of butyrate (1.2 mM) in the gut of Atlantic salmon, strongly suggest a microbial origin of this SCFA. Nimalan et al. reported a similar concentration of butyrate in Atlantic salmon digesta from post- smolts specimens, varying depending on the diet and the presence of probiotics suggesting a microbiota origin of this SCFA [88]. Our findings regarding the taxonomic composition of BPBs show that the dominant phyla are Proteobacteria and Firmicutes (Fig. 1a). This result differs from that established for BPBs in humans, where the dominant ranks are Firmicutes and Bactereoides [89]. BPB analysis at lower taxonomic levels (Fig. 1b) show a predominance of members of the family Comamonadaceae and the genus Clostridium. The predominance of the family Comamonadaceae, accounting for almost 50% in fecal samples of all fish evaluated is interesting, since members of this family are not abundant BPB in studies in other vertebrates [90]. Probably many BPB of the Comamonadaceae family are not identified in the databases because these collections are built based on the human microbiota and model vertebrates [91]. This bias, could also explain the low taxonomic diversity of BPB identified in the intestine of Atlantic salmon. Similar results were reported by Gupta et al., which by 16 S rRNA metagenomic identified in the intestinal-microbiota of Atlantic salmon genera that encodes for butyrate producing pathways (Brevinema, Achromobacter, Aquabacterium and Phyllobacterium) different from those described in the mammalian microbiota [92]. Further characterization of microbiota metabolism either by whole metagenome sequencing or isolation and sequencing of butyrate-producing microorganisms from the Atlantic salmon intestinal microbiota will enlighten the main microorganisms and butyrate-producing pathways present in this species. In agreement with this hypothesis, our group has recently isolated a strain belonging to the genus Paeniclostridium from the gut of Rainbow trout. This strain is able to produce butyric acid when cultured in Yeast Brain Heart Infusion Broth (unpublished data), suggesting that similar bacteria could be present in the intestinal microbiota of Atlantic salmon.
In the second stage, we analyzed whether the host targets for butyrate that have been previously described in mammalian systems were present in Atlantic salmon. We identified an expansion of genes encoding for the type IIa and I HDACs which are inhibited by butyrate and for orthologues of the butyrate receptors FFAR2, FFAR3 and HCAR2 described in mammals.
These expansions suggests an increase in the relevance of butyrate or BPB in Atlantic salmon physiology, as has been observed in vertebrates olfactory receptors where their number of genes is related with the importance of smell sense in the proper interaction with the environment [93]. Since FFAR2, FFAR3 and HCAR2 receptors conserve the residues involved in detecting butyrate, we hypothesized that neofunctionalization involving the recognition of other molecules different from SCFA has not been the evolutive force that prompted the duplications and conservation of FFAR2 and HDAC, in Atlantic salmon or Rainbow trout. The expansion in the number of genes encoding for HDAC inhibited by butyrate, also suggest an important role of BPB in the control of the process regulated by chromatin condensation in Atlantic salmon. To our knowledge, in Atlantic salmon, the role of HDAC in cell fate or physiological processes has not been studied. However, in zebrafish, HDAC has been implicated in thermal plasticity [94] while in mammals, butyrate-dependent inhibition of HDACs modifies the cell fate of T-cells and macrophages, promoting the differentiation of T-cells into a phenotype involved in the anti-inflammatory process [95] and macrophages into an improved antimicrobial phenotype [96]. Our data suggest that these expansions (or at least the expansions of Atlantic salmon FFAR2, FFAR3 and HCAR2) happened after or concomitant with the last autotetraploidization of salmonids, predicted to have occurred approximately 80 Myr ago [68]. We dated this event to approximately 50 Mya, a similar time as the date of duplication proposed for MHC-I in Rainbow trout (60 Mya) [97]. Our data also shows that the receptors suffered several expansion events in an ancestor prior to the divergence between Atlantic salmon and Rainbow trout (before 10 Mya). However, this data is not consistent with the previously-reported date of speciation between Atlantic salmon and Rainbow trout that is predicted around 21 Mya [68]. HCAR2 is the only gene whose date of diversification is coherent with the previously-reported data. This inconsistency in the speciation date could be explained by an increment in the selection pressure on the genes encoding for FFAR2 and FFAR3, or by recent interchromosomal duplication of genes encoding for FFAR2 and duplication in tandem for FFAR3 that occurred independently in Atlantic salmon and Rainbow trout. This model also suggests that FFAR2 diversification started prior to the separation between Atlantic salmon and Rainbow trout.
The syntenic analysis also highlights some clues about the putative physiological role of the FFAR2/3 receptors in Atlantic salmon. The genes encoding FFAR2 were located in regions that code for proteins related to the immune function, particularly the cellular immune response mediated by the interaction of microbial sialylated proteins with SIGLEC receptors. Among them we identified SIGLEC-like CD33 and CD22 which inhibit the microbial stimulation of leukocytes and macrophages, and SIGLECs orthologous to those involved in the activation of phagocytosis [98]. Moreover, this region also contains receptors involved in detecting dopamine and aril-aliphatic molecules such as tryptophan, which is involved in the crosstalk between the microbiota and the neuroimmune system [99], and genes encoding for enzymes required for the metabolism of H2S, a molecule that has been implicated in the interaction between microbiota and host [71]. Similar results were found by Petit et al., 2022, in a syntenic analysis of the butyrate receptor from Common carp, human and teleost [24]. Whether butyrate controls the expression of these genes and then modulates the microbiota-host interaction in Atlantic salmon is currently unknown. However, in mice butyrate induce the expression of the gene encoding for the hepcidin antimicrobial peptide (HAMP), which we identified in several copies adjacent to the butyrate receptor FFAR2 in Atlantic salmon and Rainbow trout [100]. The phenomenon of co-expression of physically close elements has been documented across multiple taxa such as Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, among others [101], [102], [103]. Further studies are necessary to identify if/how microbiota or SCFA modify the expression of the genes located proximal to FFAR2, 3 and HCAR2.
A higher number of genes encoding for butyrate receptors could imply a more complex regulatory network if each copy or group of copies are expressed differentially according to the tissue, growth stage and/or environmental condition. When we evaluated the expression of FFAR2, FFAR3 and HCAR2, the latter was the only one detected in all three tissues analyzed (the gut, spleen and head-kidney), suggesting that this receptor could possess an immune function, as reported for its orthologues in zebrafish [9]. However, it still remains to be seen whether in the regions where HCAR2 is encoded in zebrafish, nearby genes are related with immune functions; nevertheless, in Atlantic salmon we also found genes related with cell-traffic, the receptor for aromatic molecules (AHR) and signal transduction, among others. Interestingly, the genetic context of HCAR2 seems to be conserved between mammalian and fish species, since the genes ccdc62, denr and hip1R are present in human, mice and zebrafish [9] as well as Rainbow trout or Atlantic salmon, although it is unknown whether these genes are also related with immune functions.
In relation to FFAR, despite the higher number of copies of FFAR2, we were unable to detect their expression in spleen, head-kidney and the gut, suggesting that they are expressed at a very low level, or in a particular type of cell, or their main expression is in organs different to those evaluated, or in a stage of growth different from the pre-smolt. This idea is consistent with the expression analyzed in the cell culture line RTgutGC, where we could only detect the expression of the orthologues to XP_014056042.1. The RTgutGC cell line is an intestinal epithelial cell type from Rainbow trout [104]. In this cell line, the expression of this receptor, together with changes in the phosphorylation pattern of cytoplasmatic proteins, suggest that at least a subtype of intestinal epithelial cells are able to sense butyrate produced by the intestinal microbiota of Rainbow trout. The changes in the phosphorylation pattern of RTgutGC after treatment with butyrate are similar to those observed in mammals after the stimulation of FFAR2 with butyrate. Mammalian FFAR2 both induces and represses the phosphorylation of proteins whose phosphotypes have an estimate mass of approximately 41 kDa (p38) and 30 kDa (p27), depending on the cell type [105]. Regarding FFAR3, we were unable to detect its expression in the Atlantic salmon tissues analyzed and in cell cultures of RTgutGC and SHK-1, the latter representing a macrophage-like cell type isolated from the head-kidney of Atlantic salmon [82]. In mammals, FFAR3 is expressed mainly in cell types related with the nervous system or in neuromodulated immune cells [106], corresponding to tissues that were not evaluated in this present study. Thus, if FFAR3 from Atlantic salmon plays the same role, expression should be detected in brain tissue or in the terminal nerves present in the gut. The stage of fish growth evaluated may also explain why we could not detect the expression of some of these genes. In the genome data viewer from NCBI, which contains information from RNAseq data, we could identify expression of Atlantic salmon FFAR2, FFAR3 and HCAR2, mainly in adult specimens [107], and not in the pre-smolt specimens with the size used in this work. By mining this information, bioinformatic analysis should help to unveil the tissues/cells and timing of the expression of these receptors.
Once the diversity and the expression of FFAR2 and FFAR3 in Atlantic salmon had been analyzed, we then evaluated whether these putative receptors are able to recognize butyrate or other SCFAs. To achieve this goal, we combined three different approaches: identification of amino acids important for the recognition of SCFAs and sequence comparison, modelling the 3D structure of these receptors, and identification of putative binding sites for butyrate. We found that the residues predicted to interact with butyrate and SCFAs were conserved, but the residues related with the specificity of mammalian FFAR2 and FFAR3 for butyrate and another SCFA were different; in fact, the analysis of these residues showed that XP014056042 was the only FFAR2 from Atlantic salmon that contains the same residues involved in interacting with butyrate observed in mammalian FFAR2, suggesting that this fish receptor also interacts with butyrate. We did not observe the residues described for mammalian FFAR3 in Atlantic salmon FFAR2 or FFAR3, although XP014055669 possessed a similar distribution as found in mammalian FFAR3, harboring just a single mutation (L171N). The variation I145V in FFAR2 was also functional and responsive to butyrate, as only in RTgutGC expressing the closest orthologue to XP014056042 in Rainbow trout (XP021416718), were increments in the pattern of cytoplasmatic phosphotyrosine observed after exposure to butyrate. Considering that XP014021519 from Atlantic salmon shows this distribution of key amino acids (Y90, V145, E166), we extrapolate that it is also able to recognize butyrate.
The other FFAR members identified may recognize other SCFAs, since they have differences in the amino acids related to ligand binding, or respond at different concentrations of butyrate, increasing the complexity of the microbiota-host interaction mediated by SCFAs. This hypothesis is supported by the 3D models which show that pockets involved in the interactions with butyrate are more diverse in the FFAR2/3 of Atlantic salmon than in the reported structures of mammals. Further characterization based on molecular docking using refined homology models of FFAR that consider the membrane, together with the use of molecular dynamics tools should help to predict the affinity of each FFAR with the different SCFAs. This diversification in function is common in the receptor coupled to G-proteins, as found in the olfactory receptors in sensory and non-chemosensory organs [108].
After the bioinformatic analysis of the butyrate targets in the Atlantic salmon genome, we evaluated whether this SCFA could modify the immune response. We used SHK-1 cells as a model, a macrophage-like cell line isolated from head-kidney of Atlantic salmon. Butyrate increased the expression of anti-inflammatory cytokines IL-10, and TGF-β, however the maximum effect on TGF-β expression was achieved after 5 days of exposure to this SCFA. Although butyrate is reported to induce TGF-β expression in mammalian cells [109], this effect is not observed at high concentrations of butyrate [110]. Thus, the basal levels of expression after 24 h and the high relative expression level of TGF-β mRNA, could be a consequence of the reduction in butyrate concentration due to its use as an energy source in SHK-1 cells, or by a major cell rearrangement (probably involving epigenetic changes and expression of several proteins) that take more than 24 h to increase the expression of TGF-β. The former explanation agrees with the reduction of IL-10 expression and with the loss of the effects over IL-1β after five days of stimuli. The effect on IL-6 is similar to that observed in TGF-β, although in mammalian cells, butyrate is a strong inhibitor of IL-6 expression [111]. One possible explanation is that the regulation of IL-6 in Atlantic salmon or at least in SHK-1 cells is different from that observed in mammals. Interestingly, in Rainbow trout, IL-6 shows effects not observed in mammals, promoting macrophage proliferation and controlling the inflammatory response [112]. Thus, in Salmonids, the induction of IL-6 expression in response to butyrate agrees with the general anti-inflammatory role of this SCFA. Since SHK-1 does not express FFAR2, FFAR3 and HCAR2, we conclude that the effects observed on SHK-1 (Fig. 9, Fig. 10) could be the result of HDAC inhibition, as has been observed in intestinal and blood peripherical mammalian macrophages [96], [111]. Although SHK-1 cells represent an immune tissues distant from the intestinal microbiota, in mammals butyrate is able to be absorbed at intestinal level reaching the blood and thus non intestinal immune cells related with the allergic asthma such as eosinophils, mast cells, Treg, and dendritic cells among others, on which produce effects on their functions [113].
Our results also show that butyrate modifies the antiviral response in SHK-1 cells. We observed a delay of 48–60 h in the maximal expression of the Interferon Stimulated Genes (ISG) Mx and PKR in response to Poly I:C, while butyrate reduces the expression of INFα, reaching half of its expression in response to Poly I:C. The butyrate consumption could explain the delay in the response of Mx and PKR after several days of treatment. The effects on the expression of INFα could be the consequence of an epigenetic mark, as suggested by Chen et al. [114]. The inhibition of the antiviral response by butyrate has also been described in mammalian cells, increasing the replication of IAV and other viruses by reduction in the expression of Interferon Stimulated Genes (60%), in a mechanisms that dependent on HDAC inhibition [81], [115].
When we analyzed the effects of butyrate over the replication of IPNV, we observed a tendency where a higher concentration of butyrate results in a lower viral load. This result is different from that observed in Influenza A, HIV and others [81], where the exposure to butyrate promotes viral replication. The mechanisms by which butyrate could reduce the replication of IPNV in SHK-1 remain unknown, but could be explained at least in part by the increment in the basal expression of INFα, Mx and PKR observed in SHK-1 cells treated with butyrate.
5. Conclusions
The results obtained in our research represent the first identification and characterization of the elements required for intercommunication between Atlantic salmon and their intestinal microbiota through butyrate. Both Atlantic salmon and Rainbow trout possess several copies of highly conserved canonical butyrate receptors (FFAR2, FFAR3, HCAR2) with structural capabilities to detect butyrate and other short-chain fatty acids. In turn, Atlantic salmon harbor an intestinal microbiota composed of genera and bacterial species with the capacity to produce butyrate.
Furthermore, Atlantic salmon have a wider diversity of butyrate targets/receptors than mammals, and their expansion began probably after or concomitant with genome re-diploidization. These receptors have a differential pattern of expression, of which HCAR2 is the most widely expressed in the tissues analyzed. The effects of butyrate promoting an anti-inflammatory stage and on the innate antiviral response on SHK-1, a macrophage-like cell line of Atlantic salmon, that is, the impairment of the expression of INFα, Mx and PKR after exposure to Poly I:C, are probably consequences of HDAC inhibition, since FFAR2, FFAR3, and HCAR2 expression were not detected.
Further experiments that combine predictions of microbiota metabolism based on whole metagenome sequencing, data mining of RNAseq data from Atlantic salmon organs, measurements of SCFA in the gut of Atlantic salmon, and epigenetics studies based on the effects of butyrate on cell lines or tissues from Atlantic salmon, will help to decipher the role of butyrate in the interaction between microbiota and host in salmonids.
Funding
This research was funded by ECOS-ANID, grant number 180024, to MT and LBH, CORFO-INNOVA 19CVID-118939 to MT, VRIDEI-USACH to APC and CONICYT-PFCHA/Doctorado Nacional/2018-21181108 to RV.
CRediT authorship contribution statement
Rodrigo A. Vargas: Conceptualization, Methodology, Writing – original draft, Visualization. Sarita Soto-Aguilera: Methodology, Investigation. Mick Parra: Investigation, Methodology, Supervision. Sebastian Herrera: Validation, Investigation. Alvaro Santibañez: Validation, Investigation. Camila Kossack: Validation, Investigation. Claudia P. Saavedra: Writing – review & editing. Oscar Mora: Resources. Mauricio Pineda: Methodology, Investigation, Supervision. Oscar Gonzalez: Supervision. Alex Gonzalez: Conceptualization, Resources, Writing – review & editing, Funding acquisition. Kevin Maisey: Writing — review & editing, Validation. Edgar Torres-Maravilla: Writing – review & editing. Luis G. Bermúdez-Humarán: Resources, Writing – review & editing, Funding acquisition. Elkin Y. Suárez-Villota: Software. Mario Tello: Conceptualization, Methodology, Resources, Writing – original draft, Writing – review & editing, Visualization, Funding acquisition.
Conflicts of interest
“The authors declare no conflict of interest.” “The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results”.
Acknowledgments
This research was supported by the high-performance computing system PIDi-UTEM (SCC-PIDi-UTEM - CONICYT - FONDEQUIP - EQM180180). MT and AG thank the “Convenio de cooperación USACH-ULagos”.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.03.050.
Appendix A. Supplementary material
Supplementary material
.
Supplementary File 2: Excel file “Supplementary File 2.xls” containing information about the genes present in the chromosomic region where the butyrate receptors are encoded.
.
Supplementary File 3: Excel file “Supplementary File 3.xls” containing information about the residues that form potential ligand binding sites predicted using PrankWeb and the modelled 3D structure of the butyrate receptors encoded in the genome of Atlantic salmon.
.
References
- 1.Methé B.A., Nelson K.E., Pop M., Creasy H.H., Giglio M.G., Huttenhower C., et al. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huttenhower C., Gevers D., Knight R., Abubucker S., Badger J.H., Chinwalla A.T., et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomaa E.Z. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek. 2020;113:2019–2040. doi: 10.1007/S10482-020-01474-7. [DOI] [PubMed] [Google Scholar]
- 4.Stressmann F.A., Bernal-Bayard J., Perez-Pascual D., Audrain B., Rendueles O., Briolat V., et al. Mining zebrafish microbiota reveals key community-level resistance against fish pathogen infection. ISME J. 2021;15:702. doi: 10.1038/S41396-020-00807-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis D.J., Bryda E.C., Gillespie C.H., Ericsson A.C. Microbial modulation of behavior and stress responses in zebrafish larvae. Behav Brain Res. 2016;311:219–227. doi: 10.1016/J.BBR.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semova I., Carten J.D., Stombaugh J., Mackey L.C., Knight R., Farber S.A., et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rawls J.F., Samuel B.S., Gordon J.I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li P., Zhang J., Liu X., Gan L., Xie Y., Zhang H., et al. The function and the affecting factors of the zebrafish gut microbiota. Front Microbiol. 2022;13 doi: 10.3389/FMICB.2022.903471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cholan P.M., Han A., Woodie B.R., Watchon M., Kurz A.R.M., Laird A.S., et al. Conserved anti-inflammatory effects and sensing of butyrate in zebrafish. Gut Microbes. 2020;12:1–11. doi: 10.1080/19490976.2020.1824563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egerton S., Culloty S., Whooley J., Stanton C., Ross R.P. The gut microbiota of marine fish. Front Microbiol. 2018;9:873. doi: 10.3389/fmicb.2018.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim M.H., Kang S.G., Park J.H., Yanagisawa M., Kim C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013:145. doi: 10.1053/J.GASTRO.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 12.Safari R., Hoseinifar S.H., Kavandi M. Modulation of antioxidant defense and immune response in zebra fish (Danio rerio) using dietary sodium propionate. Fish Physiol Biochem. 2016;42:1733–1739. doi: 10.1007/S10695-016-0253-Z. [DOI] [PubMed] [Google Scholar]
- 13.Holben W.E., Williams P., Gilbert M.A., Saarinen M., Särkilahti L.K., Apajalahti J.H.A. Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon. Micro Ecol. 2002;44:175–185. doi: 10.1007/s00248-002-1011-6. [DOI] [PubMed] [Google Scholar]
- 14.Mountfort D.O., Campbell J., Clements K.D. Hindgut fermentation in three species of marine herbivorous fish. Appl Environ Microbiol. 2002;68:1374–1380. doi: 10.1128/AEM.68.3.1374-1380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robles R., Lozano A.B., Sevilla A., Márquez L., Nuez-Ortín W., Moyano F.J. Effect of partially protected butyrate used as feed additive on growth and intestinal metabolism in sea bream (Sparus aurata. Fish Physiol Biochem. 2013;39:1567–1580. doi: 10.1007/s10695-013-9809-3. [DOI] [PubMed] [Google Scholar]
- 16.Liu W., Yang Y., Zhang J., Gatlin D.M., Ringø E., Zhou Z. Effects of dietary microencapsulated sodium butyrate on growth, intestinal mucosal morphology, immune response and adhesive bacteria in juvenile common carp (Cyprinus carpio) pre-fed with or without oxidised oil. Br J Nutr. 2014;112:15–29. doi: 10.1017/S0007114514000610. [DOI] [PubMed] [Google Scholar]
- 17.Meng X., Wu S., Hu W., Zhu Z., Yang G., Zhang Y., et al. Clostridium butyricum improves immune responses and remodels the intestinal microbiota of common carp (Cyprinus carpio L.) Aquaculture. 2021;530 doi: 10.1016/J.AQUACULTURE.2020.735753. [DOI] [Google Scholar]
- 18.Li H., Zhou Y., Ling H., Luo L., Qi D., Feng L. The effect of dietary supplementation with Clostridium butyricum on the growth performance, immunity, intestinal microbiota and disease resistance of tilapia (Oreochromis niloticus) PLoS One. 2019;14 doi: 10.1371/JOURNAL.PONE.0223428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis P., Flint H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/J.1574-6968.2009.01514.X. [DOI] [PubMed] [Google Scholar]
- 20.Louis P., Hold G.L., Flint H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/NRMICRO3344. [DOI] [PubMed] [Google Scholar]
- 21.Venegas D.P., De La Fuente M.K., Landskron G., González M.J., Quera R., Dijkstra G., et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019:10. doi: 10.3389/FIMMU.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown A.J., Goldsworthy S.M., Barnes A.A., Eilert M.M., Tcheang L., Daniels D., et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/JBC.M211609200. [DOI] [PubMed] [Google Scholar]
- 23.Wu M., Li Q., Mai K., Ai Q. Regulation of free fatty acid receptor 4 on inflammatory gene induced by LPS in large yellow croaker ( Larimichthys crocea) Front Immunol. 2021:12. doi: 10.3389/FIMMU.2021.703914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petit J., Wiegertjes G.F. Conservation of members of the free fatty acid receptor gene family in common carp. Dev Comp Immunol. 2022:126. doi: 10.1016/J.DCI.2021.104240. [DOI] [PubMed] [Google Scholar]
- 25.Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132:1012–1017. doi: 10.1093/jn/132.5.1012. [DOI] [PubMed] [Google Scholar]
- 26.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang P V, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jalili M., Gerdol M., Greco S., Pallavicini A., Buonocore F., Scapigliati G., et al. Differential effects of dietary supplementation of krill meal, soybean meal, butyrate, and bactocell® on the gene expression of atlantic salmon head kidney. Int J Mol Sci. 2020;21:886. doi: 10.3390/IJMS21030886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirghaed A.T., Yarahmadi P., Soltani M., Paknejad H., Hoseini S.M. Dietary sodium butyrate (Butirex® C4) supplementation modulates intestinal transcriptomic responses and augments disease resistance of rainbow trout (Oncorhynchus mykiss. Fish Shellfish Immunol. 2019;92:621–628. doi: 10.1016/J.FSI.2019.06.046. [DOI] [PubMed] [Google Scholar]
- 31.Han Z., Sun J., Lv A., Sung Y.Y., Sun X., Shi H., et al. A modified method for genomic DNA extraction from the fish intestinal microflora. AMB Express. 2018;8:52. doi: 10.1186/S13568-018-0578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaur M., Vasudeva A., Singh A., Sharma V., Khurana H., Negi R.K., et al. Comparison of DNA extraction methods for optimal recovery of metagenomic DNA from human and environmental samples. Indian J Microbiol. 2019;59:482–489. doi: 10.1007/S12088-019-00832-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852. doi: 10.1038/S41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High resolution sample inference from Illumina amplicon data Benjamin. Nat Methods. 2016;13:4–5. doi: 10.1038/nmeth.3869.DADA2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/MOLBEV/MST010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price M.N., Dehal P.S., Arkin A.P. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010:5. doi: 10.1371/JOURNAL.PONE.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 40.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evol (N Y) 1985;39:783. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 41.Stecher G., Tamura K., Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol. 2020;37:1237–1239. doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suchard M.A., Lemey P., Baele G., Ayres D.L., Drummond A.J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018:4. doi: 10.1093/VE/VEY016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drummond A.J., Ho S.Y.W., Phillips M.J., Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:699–710. doi: 10.1371/JOURNAL.PBIO.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gernhard T. The conditioned reconstructed process. J Theor Biol. 2008;253:769–778. doi: 10.1016/J.JTBI.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Darriba Di, Posada D., Kozlov A.M., Stamatakis A., Morel B., Flouri T. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 2020;37:291–294. doi: 10.1093/MOLBEV/MSZ189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson M. Osteology and systematic position of the Eocene salmonid †Eosalmo driftwoodensisWilson from western North America. Zool J Linn Soc. 1999;125:279–311. doi: 10.1006/ZJLS.1997.0166. [DOI] [Google Scholar]
- 48.Greenwood D.R., Archibald S.B., Mathewes R.W., Moss P.T. Fossil biotas from the Okanagan Highlands, southern British Columbia and northeastern Washington State: climates and ecosystems across an Eocene landscape. Https://DoiOrg/101139/E04–100 2005;42:167–185. https://doi.org/10.1139/E04–100.
- 49.Crête-Lafrenière A., Weir L.K., Bernatchez L. Framing the salmonidae family phylogenetic portrait: a more complete picture from increased taxon sampling. PLoS One. 2012:7. doi: 10.1371/JOURNAL.PONE.0046662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lecaudey L.A., Schliewen U.K., Osinov A.G., Taylor E.B., Bernatchez L., Weiss S.J. Inferring phylogenetic structure, hybridization and divergence times within Salmoninae (Teleostei: Salmonidae) using RAD-sequencing. Mol Phylogenet Evol. 2018;124:82–99. doi: 10.1016/J.YMPEV.2018.02.022. [DOI] [PubMed] [Google Scholar]
- 51.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinforma. 2016;54:5.6.1–5.6.37. doi: 10.1002/CPBI.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nat. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. 2021 5967873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.UniProt n.d. 〈https://www.uniprot.org/〉 (accessed January 30, 2019).
- 54.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007:35. doi: 10.1093/NAR/GKM290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melo F., Sánchez R., Sali A. Statistical potentials for fold assessment. Protein Sci. 2002;11:430. doi: 10.1002/PRO.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jendele L., Krivak R., Skoda P., Novotny M., Hoksza D. PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res. 2019;47:W345–W349. doi: 10.1093/NAR/GKZ424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krivák R., Hoksza D. P2Rank: machine learning based tool for rapid and accurate prediction of ligand binding sites from protein structure. J Chemin- 2018;10:1–12. doi: 10.1186/S13321-018-0285-8/TABLES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grosdidier A., Zoete V., Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011:39. doi: 10.1093/NAR/GKR366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.SwissDock - The online docking web server of the Swiss Institute of Bioinformatics - Docking n.d. 〈http://www.swissdock.ch/docking〉 (accessed January 30, 2019).
- 60.Grosdidier A., Zoete V., Michielin O. Fast docking using the CHARMM force field with EADock DSS. J Comput Chem. 2011;32:2149–2159. doi: 10.1002/JCC.21797. [DOI] [PubMed] [Google Scholar]
- 61.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/PNAS.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reyes-Cerpa S., Reyes-López F., Toro-Ascuy D., Montero R., Maisey K., Acuña-Castillo C., et al. Induction of anti-inflammatory cytokine expression by IPNV in persistent infection. Fish Shellfish Immunol. 2014;41:172–182. doi: 10.1016/j.fsi.2014.08.029. [DOI] [PubMed] [Google Scholar]
- 63.Maisey K., Montero R., Corripio-Miyar Y., Toro-Ascuy D., Valenzuela B., Reyes-Cerpa S., et al. Isolation and characterization of salmonid CD4+ T cells. J Immunol. 2016;196:4150–4163. doi: 10.4049/jimmunol.1500439. [DOI] [PubMed] [Google Scholar]
- 64.Parra M., Espinoza D., Valdes N., Vargas R., Gonzalez A., Modak B., et al. Microbiota modulates the immunomodulatory effects of filifolinone on atlantic salmon. Microorganisms. 2020;8:1–21. doi: 10.3390/microorganisms8091320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muñoz C., González-Lorca J., Parra M., Soto S., Valdes N., Sandino A.M., et al. Lactococcus lactis expressing type I interferon from atlantic salmon enhances the innate antiviral immune response in vivo and in vitro. Front Immunol. 2021:12. doi: 10.3389/FIMMU.2021.696781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vital M., Karch A., Pieper D.H. Colonic butyrate-producing communities in humans: an overview using omics data. MSystems. 2017:2. doi: 10.1128/msystems.00130-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lien S., Koop B.F., Sandve S.R., Miller J.R., Kent M.P., Nome T., et al. The Atlantic salmon genome provides insights into rediploidization. Nature. 2016;533:200–205. doi: 10.1038/nature17164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018;1693:128–133. doi: 10.1016/j.brainres.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong F., Perdew G.H. The aryl hydrocarbon receptor as a mediator of host-microbiota interplay. Gut Microbes. 2020:12. doi: 10.1080/19490976.2020.1859812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mimoun S., Andriamihaja M., Chaumontet C., Atanasiu C., Benamouzig R., Blouin J.M., et al. Detoxification of H(2)S by differentiated colonic epithelial cells: implication of the sulfide oxidizing unit and of the cell respiratory capacity. Antioxid Redox Signal. 2012;17:1–10. doi: 10.1089/ARS.2011.4186. [DOI] [PubMed] [Google Scholar]
- 72.Bessman N.J., Mathieu J.R.R., Renassia C., Zhou L., Fung T.C., Fernandez K.C., et al. Dendritic cell-derived hepcidin sequesters iron from the microbiota to promote mucosal healing. Science. 2020;368:186–189. doi: 10.1126/SCIENCE.AAU6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.LeBlanc J.G., Milani C., de Giori G.S., Sesma F., van Sinderen D., Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160–168. doi: 10.1016/J.COPBIO.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 74.CDD Conserved Protein Domain Family: 7tmA_FFAR2_FFAR3 n.d. 〈https://www.ncbi.nlm.nih.gov/Structure/cdd/cd15170〉 (accessed November 5, 2019).
- 75.Tikhonova I.G. Application of GPCR structures for modelling of free fatty acid receptors. Handb Exp Pharm. 2017;236:57–77. doi: 10.1007/164_2016_52. [DOI] [PubMed] [Google Scholar]
- 76.Bernhofer M., Kloppmann E., Reeb J., Rost B. TMSEG: novel prediction of transmembrane helices. Proteins. 2016;84:1706–1716. doi: 10.1002/PROT.25155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmidt J., Smith N.J., Christiansen E., Tikhonova I.G., Grundmann M., Hudson B.D., et al. Selective orthosteric free fatty acid receptor 2 (FFA2) agonists: identification of the structural and chemical requirements for selective activation of FFA2 versus FFA3. J Biol Chem. 2011;286:10628–10640. doi: 10.1074/JBC.M110.210872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tikhonova I.G., Poerio E. Free fatty acid receptors: structural models and elucidation of ligand binding interactions Computational analysis. BMC Struct Biol. 2015;15:1–13. doi: 10.1186/S12900-015-0044-2/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tunaru S., Lättig J., Kero J., Krause G., Offermanns S. Characterization of determinants of ligand binding to the nicotinic acid receptor GPR109A (HM74A/PUMA-G) Mol Pharm. 2005;68:1271–1280. doi: 10.1124/MOL.105.015750. [DOI] [PubMed] [Google Scholar]
- 80.Deng Q., Frie J.L., Marley D.M., Beresis R.T., Ren N., Cai T.Q., et al. Molecular modeling aided design of nicotinic acid receptor GPR109A agonists. Bioorg Med Chem Lett. 2008;18:4963–4967. doi: 10.1016/J.BMCL.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 81.Chemudupati M., Kenney A.D., Smith A.C., Fillinger R.J., Zhang L., Zani A., et al. Butyrate reprograms expression of specific interferon-stimulated genes. J Virol. 2020:94. doi: 10.1128/JVI.00326-20/SUPPL_FILE/JVI.00326-20-SD001.XLSX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dannevig B.H., Falk K., Namork E. Isolation of the causal virus of infectious salmon anaemia (ISA) in a long-term cell line from Atlantic salmon head kidney. J Gen Virol. 1995;76(Pt 6):1353–1359. doi: 10.1099/0022-1317-76-6-1353. [DOI] [PubMed] [Google Scholar]
- 83.Vital M., Howe A.C., Tiedje J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014:5. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duncan S.H., Louis P., Flint H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gajardo K., Rodiles A., Kortner T.M., Krogdahl Å., Bakke A.M., Merrifield D.L., et al. A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): a basis for comparative gut microbial research. Sci Rep. 2016;6:30893. doi: 10.1038/srep30893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robertson P.A.W., O’Dowd C., Burrells C., Williams P., Austin B. Use of Carnobacterium sp. as a probiotic for Atlantic salmon (Salmo salar L.) and rainbow trout (Oncorhynchus mykiss, Walbaum) Aquaculture. 2000;185:235–243. doi: 10.1016/S0044-8486(99)00349-X. [DOI] [Google Scholar]
- 87.Smith N.W., Shorten P.R., Altermann E., Roy N.C., McNabb W.C. The classification and evolution of bacterial cross-feeding. Front Ecol Evol. 2019:7. doi: 10.3389/FEVO.2019.00153/FULL. [DOI] [Google Scholar]
- 88.Nimalan N., Sørensen S.L., Fečkaninová A., Koščová J., Mudroňová D., Gancarčíková S., et al. Mucosal barrier status in Atlantic salmon fed marine or plant-based diets supplemented with probiotics. Aquaculture. 2022:547. doi: 10.1016/j.aquaculture.2021.737516. [DOI] [Google Scholar]
- 89.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/SCIENCE.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rivière A., Selak M., Lantin D., Leroy F., De Vuyst L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front Microbiol. 2016:7. doi: 10.3389/FMICB.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dueholm M.S., Andersen K.S., McIlroy S.J., Kristensen J.M., Yashiro E., Karst S.M., et al. Generation of comprehensive ecosystem-specific reference databases with species-level resolution by high-throughput full-length 16s rrna gene sequencing and automated taxonomy assignment (Autotax) MBio. 2020;11:1–14. doi: 10.1128/mBio.01557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gupta S., Lokesh J., Abdelhafiz Y., Siriyappagouder P., Pierre R., Sørensen M., et al. Macroalga-Derived Alginate Oligosaccharide Alters Intestinal Bacteria of Atlantic Salmon. Front Microbiol. 2019:10. doi: 10.3389/FMICB.2019.02037/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Niimura Y. Olfactory Receptor Multigene Family in Vertebrates: From the Viewpoint of Evolutionary Genomics. Curr Genom. 2012;13:103–114. doi: 10.2174/138920212799860706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seebacher F., Simmonds A.I.M. Histone deacetylase activity mediates thermal plasticity in zebrafish (Danio rerio) Sci Rep 2019 91. 2019;9:1–13. doi: 10.1038/s41598-019-44726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 96.Schulthess J., Pandey S., Capitani M., Rue-Albrecht K.C., Arnold I., Franchini F., et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. 2019;50:432. doi: 10.1016/J.IMMUNI.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shiina T., Dijkstra J.M., Shimizu S., Watanabe A., Yanagiya K., Kiryu I., et al. Interchromosomal duplication of major histocompatibility complex class I regions in rainbow trout (Oncorhynchus mykiss), a species with a presumably recent tetraploid ancestry. Immunogenetics. 2005;56:878–893. doi: 10.1007/S00251-004-0755-1. [DOI] [PubMed] [Google Scholar]
- 98.Chang Y.C., Nizet V. The interplay between Siglecs and sialylated pathogens. Glycobiology. 2014;24:818–825. doi: 10.1093/GLYCOB/CWU067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bosi A., Banfi D., Bistoletti M., Giaroni C., Baj A. Tryptophan metabolites along the microbiota-gut-brain axis: an interkingdom communication system influencing the gut in health and disease. Int J Tryptophan Res. 2020:13. doi: 10.1177/1178646920928984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Das N.K., Schwartz A.J., Barthel G., Inohara N., Liu Q., Sankar A., et al. Microbial metabolite signaling is required for systemic iron homeostasis. Cell Metab. 2020;31:115–130. doi: 10.1016/j.cmet.2019.10.005. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blumenthal T., Evans D., Link C.D., Guffanti A., Lawson D., Thierry-Mieg J., et al. A global analysis of Caenorhabditis elegans operons. Nat. 2002;417:851–854. doi: 10.1038/nature00831. 2002 4176891. [DOI] [PubMed] [Google Scholar]
- 102.Boutanaev A.M., Kalmykova A.I., Shevelyov Y.Y., Nurminsky D.I. Large clusters of co-expressed genes in the Drosophila genome. Nat. 2002;420:666–669. doi: 10.1038/nature01216. 2002 4206916. [DOI] [PubMed] [Google Scholar]
- 103.Williams E.J.B., Bowles D.J. Coexpression of neighboring genes in the genome of arabidopsis thaliana. Genome Res. 2004;14:1060–1067. doi: 10.1101/GR.2131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kawano A., Haiduk C., Schirmer K., Hanner R., Lee L.E.J., Dixon B., et al. Development of a rainbow trout intestinal epithelial cell line and its response to lipopolysaccharide. Aquac Nutr. 2011;17:e241–e252. doi: 10.1111/J.1365-2095.2010.00757.X. [DOI] [Google Scholar]
- 105.Priyadarshini M., Kotlo K.U., Dudeja P.K., Layden B.T. Role of short chain fatty acid receptors in intestinal physiology and pathophysiology. Compr Physiol. 2018;8:1091. doi: 10.1002/CPHY.C170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nøhr M.K., Pedersen M.H., Gille A., Egerod K.L., Engelstoft M.S., Husted A.S., et al. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013;154:3552–3564. doi: 10.1210/EN.2013-1142. [DOI] [PubMed] [Google Scholar]
- 107.Salmo salar Annotation Report n.d. 〈https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Salmo_salar/100/〉 (accessed May 24, 2019).
- 108.Ferrer I., Garcia-Esparcia P., Carmona M., Carro E., Aronica E., Kovacs G.G., et al. Olfactory receptors in non-chemosensory organs: the nervous system in health and disease. Front Aging Neurosci. 2016:8. doi: 10.3389/FNAGI.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martin-Gallausiaux C., Béguet-Crespel F., Marinelli L., Jamet A., Ledue F., Blottière H.M., et al. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-28048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matsumoto N., Riley S., Fraser D., Al-Assaf S., Ishimura E., Wolever T., et al. Butyrate modulates TGF-β1 generation and function: potential renal benefit for Acacia(sen) SUPERGUM™ (gum arabic)? Kidney Int. 2006;69:257–265. doi: 10.1038/SJ.KI.5000028. [DOI] [PubMed] [Google Scholar]
- 111.Chang P.V., Hao L., Offermanns S., Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Costa M.M., Maehr T., Diaz-Rosales P., Secombes C.J., Wang T. Bioactivity studies of rainbow trout (Oncorhynchus mykiss) interleukin-6: Effects on macrophage growth and antimicrobial peptide gene expression. Mol Immunol. 2011;48:1903–1916. doi: 10.1016/j.molimm.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 113.Yip W., Hughes M.R., Li Y., Cait A., Hirst M., Mohn W.W., et al. Butyrate shapes immune cell fate and function in allergic asthma. Front Immunol. 2021:12. doi: 10.3389/FIMMU.2021.628453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen X., Barozzi I., Termanini A., Prosperini E., Recchiuti A., Dalli J., et al. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci. 2012;109:E2865–E2874. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nusinzon I., Horvath C.M. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci USA. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary File 2: Excel file “Supplementary File 2.xls” containing information about the genes present in the chromosomic region where the butyrate receptors are encoded.
Supplementary File 3: Excel file “Supplementary File 3.xls” containing information about the residues that form potential ligand binding sites predicted using PrankWeb and the modelled 3D structure of the butyrate receptors encoded in the genome of Atlantic salmon.