Abstract
The major role and implication of bacterial biofilms in the case of bone and prosthesis infections have been highlighted and often linked to implant colonization. Management strategies of these difficult-to-treat infections consist in surgeries and antibiotic treatment, but the rate of relapse remains high, especially if Staphylococcus aureus, a high-virulent pathogen, is involved. Therapeutic approaches are not adapted to the specific features of biofilm in bone context whereas infectious environment is known to importantly influence biofilm structure. In the present study, we aim to characterize S. aureus SH1000 (methicillin-sensitive strain, MSSA) and USA300 (methicillin-resistant strain, MRSA) biofilm on different surfaces mimicking the periprosthetic environment. As expected, protein adsorption on titanium enhanced the number of adherent bacteria for both strains. On bone explant, USA300 adhered more than SH1000. The simultaneous presence of two different surfaces was also found to change the bacterial behaviour. Thus, proteins adsorption on titanium and bone samples (from bank or directly recovered after an arthroplasty) were found to be key parameters that influence S. aureus biofilm formation: adhesion, matrix production and biofilm-related gene regulation. These results highlighted the need for new biofilm models, more relevant with the infectious environment by using adapted culture medium and presence of surfaces that are representative of in situ conditions to better evaluate therapeutic strategies against biofilm.
Keywords: Periprosthetic environment, Biofilm, S. aureus, Bone explant, Titanium
1. Introduction
Bacterial colonization of medical implants and surrounding tissues are related to chronic and difficult-to-treat infections. More than a quarter of bone and joint infections occurs in presence of orthopedic material leading to severe clinical consequences, bone resorption and loss of mobility but also to high economic costs [[1], [2], [3]]. Bone and Prosthesis Infections (BPIs) are more difficult to treat if a bacterial biofilm is involved [4,5]. Eighty percent of BPIs are estimated to be biofilm-related, of which 30–40% are due to Staphylococcus aureus; including approximately 20%–50% S. aureus resistant to methicillin (MRSA) [6,7]. Biofilms are a very heterogeneous structure with a continuum of bacteria in different metabolic states, embedded in a complex matrix constituted of exopolysaccharides, exoproteins, extracellular DNA (eDNA) and lipids [8,9]. Thus, biofilm constitutes a super organization specifically adapted to surfaces colonization and to their environment besides being tolerant to antibiotics and immune system [[10], [11], [12]].
Moreover, biofilm formation is dependent of environmental factors, such as adhesion to biotic or abiotic surfaces, as well as availability of oxygen and nutrients, which strongly influence the structure and composition of biofilms [11,[13], [14], [15], [16], [17], [18]].
Thus, in BPI context, bone prostheses offer an ideal surface to biofilm formation. Indeed, the prosthesis surfaces are rapidly covered with host proteins, especially fibronectin which represents a key factor for S. aureus adhesion during infection [19]. S. aureus exhibits surface proteins named cell wall-anchored (CWA) proteins and for which the most prevalent group are microbial surface component recognizing adhesive matrix molecules (MSCRAMMS). The latter are adhesins that binds to host extracellular matrix and host proteins [20]. Fibronectin-binding proteins (FnBPs) and collagen-binding protein (Cna) are MSCRAMMS that ensure the first step of irreversible adhesion and biofilm formation [11,20].
Another important surface involved in BPI is the bone which complexity is hard to mimic using in vitro model. However, its presence could strongly affect bacterial behaviour. Indeed, bone is a permanently renewed tissue composed of a mineral part (calcium phosphate), an organic part containing mainly collagen (mostly type I collagen) and bone cells like osteoblasts, osteoclasts and osteocytes [21]. Then, a prosthesis as a foreign body may impair the bone homeostasis and induces immune responses that could influence bacterial adhesion [22,23].
Regarding biofilm formation, factors of the bone environment (e.g. hypoxia and a high concentration of magnesium) and the presence of highly active and communicating bone cells (e.g. osteoblasts and osteoclasts) have influences [15]. The physical and chemical properties of surfaces on which bacteria adhere can also affect biofilm formation [[24], [25], [26]].
In BPI context, a better comprehension of the periprosthetic environment influences on bacterial adhesion highlights the need for in vitro models that better represent infection conditions. This could facilitate the development of new strategies to prevent biofilm formation [27].
In this study, we investigated S. aureus adhesion on different surfaces found in bone context with the objective to decipher mechanisms involved (simple adhesion or stress response) but also to select parameters for the development of BPI in vitro model.
2. Materials and methods
2.1. Coating preparation
Titanium disks (diameter 12 mm × 5 mm) (ACNIS, France) were shotpeened by CRITT-MDTS (Charleville-Mézières, France), which corresponds to the processing of orthopedic femoral stem prosthesis surface. Titanium disk surfaces were then coated with different solutions: phosphate-buffer saline (PBS) corresponding to the control, pure fetal bovine serum (FBS) (Dutscher, Bernolsheim, France), and human fibronectin (Sigma, St Quentin-Fallavier, France) at a concentration of 100 μg/mL. To do so, a drop of 200 μL of solution was deposited on the top surface of titanium disks for 2 h at room temperature. After washing in PBS, the disks were dried overnight under the laminar flow.
2.2. Bone samples
Bones from bank were provided by O.S.T Laboratories (Saint-Beauzire, France). Femoral condyles and tibia samples from which trabecular bone explants were extracted were obtained by Orthopedic and Trauma Service of Reims University Hospital Center (Reims, France), from patients having undergone total knee arthroplasty, after written informed consent of patients (Conservation d’Élément du Corps Humain Authorization, DC-2014-2262 given by the French Ministry of Research). It should be noted that patients undergoing arthroplasty received a prophylactic cefazolin administration (intravenous, 2 g, with a renewal if the surgery lasts more than 4 h). After sampling with a 2 mm diameter trocar, bone explants were washed four times with Dulbecco's Phosphate Buffered Saline (Gibco Paisley, UK), then frozen at −80 °C and thawed three times in order to eliminate viable human cells. Fresh bone samples were extracted and washed in the same way but without freezing/thawing cycles. Scanning electron microscopy confirmed the presence of eukaryotic cells on the surface of fresh bone explants contrary to frozen bone explants (Supplementary data S1). Bone samples weighed between 10 and 80 mg.
2.3. Bacterial strains and culture media
Two different strains of S. aureus have been used: a methicillin-resistant strain USA300 discovered in the 1990s in the United States, and a methicillin-susceptible strains: SH1000 derived from strain 8325-4 with the rsbU gene reconstituted [28,29]. To prepare the inoculum, −20 °C glycerol stocks were streaked out on tryptic soy agar plates. Overnight cultures were performed by transferring one colony of a strain into 3 mL nutritive broth (Bio-Rad, Marnes-la-Coquette, France) for 18 h at 37 °C. Then the absorbance (at 600 nm) of the overnight cultures was adjusted to 1 and diluted to 1:100 in minimal medium (MM) [62 mM potassium phosphate buffer, pH 7.0, 7 mM (NH4)2SO4, 2 mM MgSO4, 10 μM FeSO4] containing 0.4% (w/v) glucose and 0.5% (w/v) casamino acids. Unless otherwise stated, for all experiments, 500 μL of MM inoculated with bacteria (corresponding to 106 bacteria) were distributed in 24-well plates containing titanium disk and/or bone samples (see below). Plates were incubated at 37 °C for 24 h in hypoxic atmosphere using GenBag system (Biomérieux, Marcy l’Étoile, France).
2.4. Enumeration of viable adherent bacteria
After incubation, the different surfaces (coated or uncoated titanium disks, bones samples) were washed with MM to remove non-adherent bacteria, then transferred to a 15-mL tube containing 2 mL of MM. The irreversibly adhered bacteria were then detached by placing the tube in ultrasonic bath (40 kHz) for 5 min. A volume of 100 μL from serial dilutions was plated on nutrient agar plates with an easySpiral Pro automatic plater (Interscience, St Nom, France) to determine the quantity of attached bacteria. After overnight incubation, CFU are enumerated with a Scan1200 automatic colony counter (Interscience, St Nom, France). To determine the number of CFU per mm2 on titanium disks, the number of CFU was correlated with the surface area of titanium disks diameter. Regarding bones samples, the number of CFU was correlated with the weight of the samples after determination of a linear correlation between weight and available bone surface using micro-computed tomography (Skyscan 1076, Bruker Optics, Germany) (Supplementary Fig. S2) [30]. For each experiment, at least four biological repeats were performed.
2.5. Scanning electron microscopy
Twenty-four hours after incubation, surfaces were washed twice in PBS for 5 min, then fixed with 2.5% (w/v) glutaraldehyde for 1 h (Sigma-Aldrich, St. Louis, MO, USA). Thereafter, surfaces were washed twice with distilled water for 10 min and dehydrated by successive treatment in graded ethanol solutions (50, 70, 90% and twice absolute) for 10 min. At this point, bank bones were treated differently than bone explants. Bank bones were desiccated in a drop of hexamethyldisilazane (Sigma-Aldrich, St. Louis, MO, USA) and air-dried at room temperature. To proceed with the desiccation of bone explants, critical-point drying was used with a critical point dryer CPD 030 (BAL-TEC, France). For this, after ethanol dehydration procedure, samples were placed in acetone which was replaced by liquid carbon dioxide with several cycles under stirring at 7 °C. All surfaces were then sputtered with a thin gold-palladium film using a JEOL ion sputter JFC 1100 instrument. Biofilms were observed using a Schottky Field Emission Scanning Electron Microscope (JEOL JSM-7900F, Tokyo, Japan). Images were obtained at a primary beam energy of 2 kV (SM-EXG65 electron emitter, Tokyo, Japan). At least two different biological cultures were used for acquisitions.
2.6. Confocal laser scanning microscopy (CLSM)
After 24 h of incubation, non-coated or coated titanium disks were washed twice in PBS and stained with SYTOTM 9 at 1 μM to label bacteria and (i) propidium iodide at 20 μM to label damaged or “dead” bacteria, (ii) SYPRO® Ruby (v/v) to label proteins, or (iii) wheat germ agglutinin (WGA) associated with the Alexa FluorTM 350 conjugate at 100 mg/mL to label polysaccharide intercellular adhesin (PIA) with TOTOTM-3 iodide (all from Thermo Fisher Scientific, USA) at 2 μM to label extracellular DNA. Each label was diluted in 0.9% NaCl. After 30 min of incubation in the dark at room temperature, each titanium disk was washed twice in PBS and placed in a 24-well Krystal plate with glass bottom (Porvair, Whiteley, UK) containing PBS, with the biofilm side down, before observation with confocal laser scanning microscopy (CLSM) (LSM 710 NLO, Zeiss, Germany). Fluorochromes-labeled compounds volume was quantified using IMARIS software (v 9.8.0). For each experiment, at least three areas per titanium disk were acquired (except for USA300 on titanium coated with fibronectin), and experiments were performed three times.
2.7. RT-qPCR (RNA Purification and reverse transcription)
Biofilm and stress-responses related-genes were studied: adhesion-related gene fnbpB, stress-response-related gene rsh, biofilm formation and environmental stress response-related gene sigB [[31], [32], [33]], the autoinducer peptide (AIP) and quorum sensing-related gene agrB [34], SOS-response-related genes recA and lexA [35], matrix formation-related gene cidA [36] and adhesion and early biofilm formation-related gene sarA [33].
During enumeration of viable adherent bacteria, remaining planktonic and sessile bacteria were independently centrifuge for 8 min at 900 g to remove the medium and freeze-stored at −80 °C. Total RNAs were extracted and cleaned up with MasterPureTM RNA Purification Kit (Lucigen, Middleton, WI, USA) in accordance with the manufacturer's protocol. Total RNAs were reverse transcribed into complementary DNA (cDNA) using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Sainte Genève des Bois, France) following the manufacturer's instructions. Transcription products were amplified by RT-qPCR using different primers (Eurogentec, Serraing, Belgium) (Table 1) on a StepOne PlusTM system (Applied Biosystems, Villebon-sur-Yvette, France). The TakyonTM Rox SYBR® MasterMix (Eurognetec, Belgium) was used for amplification. After a first denaturation step at 95 °C for 10 min, RT-qPCR reactions were performed according to a thermal profile that corresponds to 40 cycles of denaturation at 95 °C for 15 s, annealing and extension at 60 °C for 1 min. Data collection was performed at the end of each annealing/extension step. The third step that consists in a dissociation process was performed to ensure the specificity of the amplicons by measuring their melting temperature (Tm). Data analysis was performed with the StepOneTM Software v2.3 (Applied Biosystems, Villebon-sur-Yvette, France). Relative messenger RNA (mRNA) expression was calculated with the equation 2-ΔΔCt, where ΔΔCt is the subtraction of ΔCt of bacteria within biofilm after subtraction of ΔCt of bacteria in planktonic state, and ΔCt is the Ct value of the target gene after subtraction of Ct for the housekeeping gene (rho).
Table 1.
Nucleotide sequences of primers used for RT-qPCR and efficiency for each primer couple.
| Target gene | Sequences |
Efficiency | |
|---|---|---|---|
| Forward primer (5′→3′) | Reverse primer (5′→3′) | ||
| rho | AACGTGGGGATAAAGTAACTGG | TTCACTTCTTCTGCGTTATGGT | 1.9 |
| recA | ATAGGTCGCCGAGTTTCAAC | GCGCTACTGTTGTCTTACCA | 1.9 |
| lexA | TCAATATTTTCTACTGCGGTAATAGG | GAAACGATTCATGTGCCAGTT | 2 |
| sigB | TTGTCCCATTTCCATTGCTT | CAGTGAAATAGCTGATCGATTAGAAG | 2.1 |
| sarA | TTTCTCTTTGTTTTCGCTGATGT | TGTTATCAATGGTCACTTATGCTG | 2 |
| agrB | ACAGTGAGGAGAGTGGTGTAA | AGCTAAGACCTGCATCCCTA | 2 |
| rsh | CGAAACCTAATAACGTATCAAATGC | TGTATGTAGATCGAAAACCATCACT | 2 |
| cidA | GATTGTACCGCTAACTTGGGT | GCGTAATTTCGGAAGCAACAT | 1.8 |
| fnbpB | AATTAAATCAGAGCCGCCAGT | AATGGTACCTTCTGCATGACC | 2.0 |
2.8. Evaluation of minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC)
The MICs and MBCs of cefazolin against S. aureus strains were determined by the broth microdilution method. Briefly, cefazolin (Mylan, France) was reconstituted in distilled sterile water according to the manufacturer instructions and tested in final concentration range of 0.004–256 μg/mL prepared in 96-well cell culture plate by a series of two-fold dilutions in MM (100 μL at two-fold concentration). Absorbance at 600 nm of overnight cultures were adjusted to 1 and diluted 1:50 in MM. One hundred microliters of bacteria suspensions were added to cefazolin solutions. After 24 h incubation at 37 °C, the MICs were read as the lowest concentration of cefazolin at which visible growth was inhibited. A drop of 20 μL was harvested from wells equal or superior to MIC and then spotted on agar plates to determine the MBCs (minimal concentration able to kill 99.99% of bacteria after 24 h at 37 °C).
2.9. Graphical representation of data and statistical analysis
Data are presented as histograms (representing the average; error bars represent standard deviation) or whisker plots (whiskers represent minimum and maximum values, the bottom and top of the box are the 15th and 85th percentiles, and the band inside the box stands for the median). The significance of the results was assessed with and exact non-parametric Mann Whitney U test for independent samples (GraphPad Prism, 8.0.2). For confocal quantifications, acquisitions of different areas for the same biological replicates were considered for graphical representations and stratified Mann Whitney U test for independent samples was used. Differences were considered significant at p < 0.05.
3. Results
3.1. S. aureus adhesion on surfaces mimicking bone and joint prosthesis
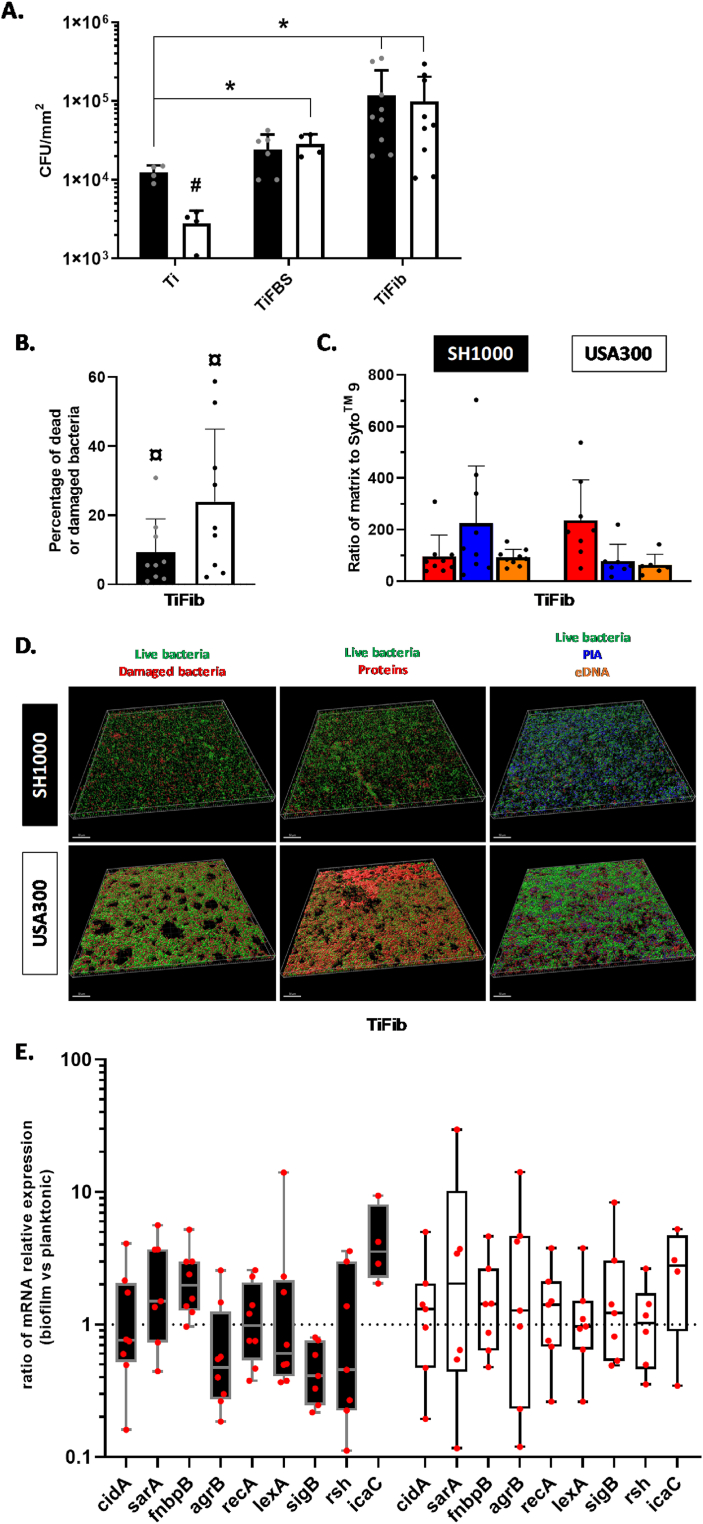
On titanium (Ti), SH1000 formed biofilms containing 1.2 × 104 CFU/mm2 and USA300 biofilms were composed with less bacteria 2.8 × 103 CFU/mm2 (p < 0.05) (Fig. 1A). On titanium coated with fetal bovine serum (TiFBS), SH1000 biofilms contained 2.5 × 104 CFU/mm2 and USA300 biofilms contained 2.9 × 104 CFU/mm2 which is significantly more than on Ti (p < 0.05). On titanium coated with fibronectin (TiFib), both strains presented significantly more adhered bacteria than on Ti: 1.2 × 105 CFU/mm2 for SH1000 and 1.0 × 105 CFU/mm2 for USA300 (p < 0.05).
Fig. 1.
S. aureus adhesion on uncoated or protein-coated titanium. (A) Number of viable adherent bacteria per mm2 on uncoated titanium (Ti), titanium coated with fetal bovine serum (TiFBS) or fibronectin (TiFib). *p < 0.05 versus Ti; #p < 0.05 versus the same condition between strains. n = 4 to 9. Black histograms = SH1000, white histograms = USA300. (B) Ratio of dead or damaged bacteria to live bacteria. SYTOTM 9 and propidium iodide were acquired by confocal microscopy. ¤ p < 0.05 versus SYTOTM 9. Black histograms = SH1000, white histogram = USA300. (C) Ratio of SYTOTM 9 to SYPRO® Ruby (red, protein), WGA (blue, polysaccharide intercellular adhesin, PIA), and TOTOTM-3 (orange, extracellular DNA) acquired by confocal microscopy. Results consist of three biological replicates with three different acquisitions for each. (D) Representative reconstructions of biofilm developed on TiFib are shown. Above each column is indicated the labeled-component and the corresponding colour. The scale bars indicate 50 μm. (E) Ratio of relative mRNA expressions of genes by bacteria within biofilms to those in planktonic state (TiFib condition). n = 4 to 8. Black histograms = SH1000, white histograms = USA300. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To quantify dead and damaged bacteria as well as matrix components, fluorescent staining and confocal microscopy were used. Regarding biofilms formed on TiFib, SH1000 and USA300 presented 9% and 26% of dead or damaged bacteria within their biofilms, respectively (Fig. 1B). To take into consideration the different amounts of bacteria per acquisition, results were standardized towards SYTOTM 9 volume (Fig. 1C). SH1000 biofilms proportionally contained more polysaccharide intercellular adhesin (PIA) than proteins and eDNA. USA300 biofilm matrix contained mainly proteins. Representative images of fluorescence reconstructions are shown in Fig. 1D underlining the difference in protein proportion in matrix between the two tested strain biofilms.
Several genes related to biofilm formation on TiFib were investigated through RT-qPCR (Fig. 1E). For SH1000, we observed an upward trend of fnbpB (1.98-fold) and icaC expression (3.55-fold) and a downward trend of sigB expression (2.45-fold).
3.2. S. aureus adhesion on bone samples
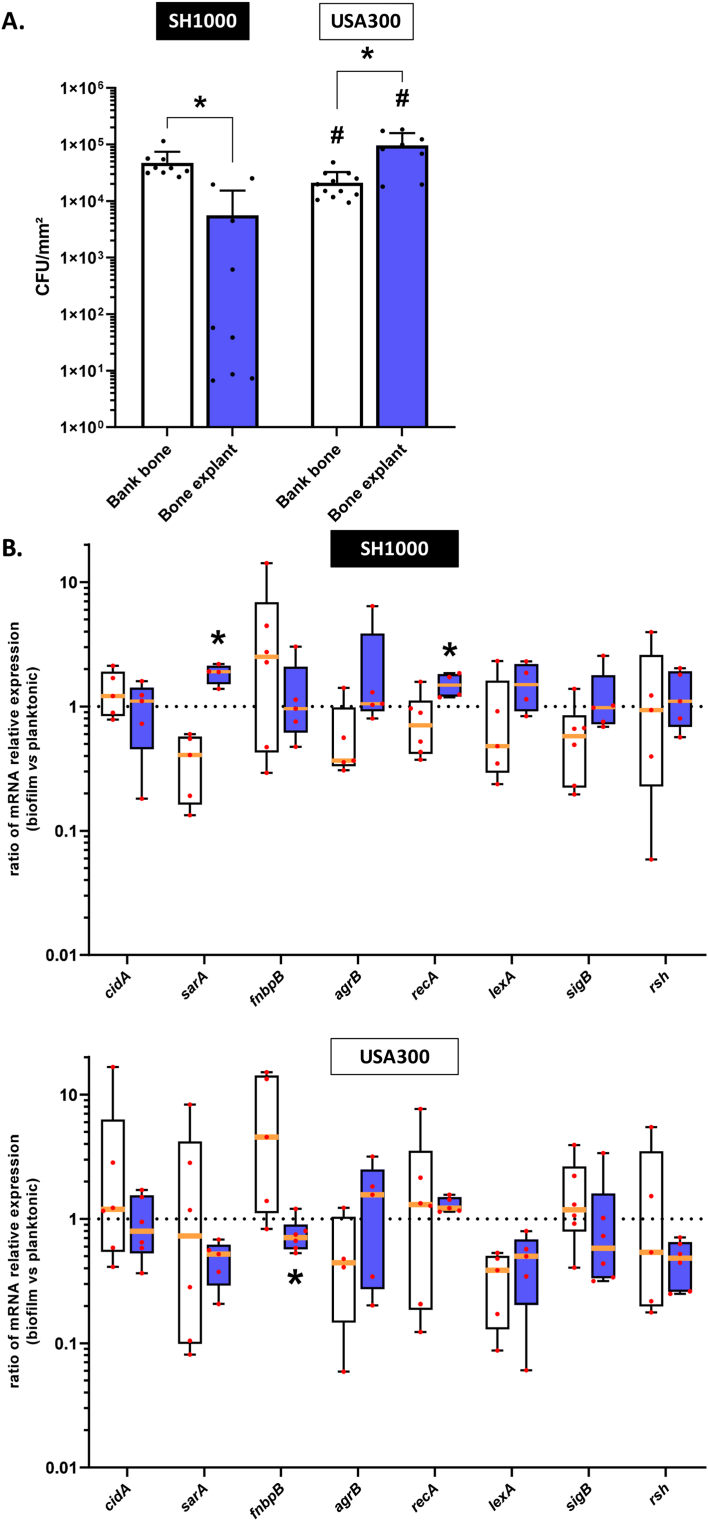
We next investigated biofilm formation of S. aureus on bank bones and bone explants frozen for more than one month and less than six months. On bank bones, SH1000 displayed 4.7 × 104 CFU/mm2 whereas 5.6 × 103 CFU/mm2 were counted for bone explants (significantly lower, p < 0.05) (Fig. 2A). USA300 displayed a lower number of adherent bacteria on bank bones than SH1000 (2.1 × 104 CFU/mm2, p < 0.05) but a higher number of adherent bacteria on bone explants (9.6 × 104 CFU/mm2, p < 0.05). For bank bones, SH1000 bacteria within biofilms demonstrated lower expression of sarA (2.46-fold decrease) and recA (1.41-fold decrease) contrary to bone explants (1.90- and 1.49-fold increase, respectively) (p < 0.05) (Fig. 2B). Regarding USA300 on bank bones, bacteria within biofilms expressed more fnbpB (4.54-fold increase) contrary to those on bone explants (1.41-fold decrease) (p < 0.05).
Fig. 2.
S. aureus adhesion on bank bone and bone explants. Bone explants were frozen for more than one month and less than six months. (A) Number of viable adherent bacteria per mm2. *p < 0.05 versus bank bone; #p < 0.05 versus the same condition between strains. n = 4 to 12. (B). Ratio of relative mRNA expressions of genes by bacteria within biofilm to those in planktonic states. *p < 0.05 versus bone bank. n = 4 to 5. White histograms = bank bone, blue histograms = bone explant. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To investigate the effect of the time of preservation of the bone explants, three other conditions were chosen and compared to the previous one: fresh bone explants (unfrozen) and bone explants frozen for less than one month or more than six months (Fig. 3). Bone explants frozen for more than one month significantly increased the number of viable adherent bacteria for SH1000 (99- and 1117-fold, respectively, p < 0.05) compared to bone frozen for less than a month. Only the preservation for more than six months significantly increased the bacterial burden on bone explant compared to fresh bones (248-fold, p < 0.05). Regarding USA300, the number of viable adherent bacteria was significantly reduced if the bone explants were frozen for more than six months compared to fresh bone (3.20-fold, p < 0.05). The other conditions did not present significant differences for USA300. Interestingly, comparing to USA300, SH1000 adhered less on fresh bone explants or bone explants frozen for less than six months (p < 0.05).
Fig. 3.
S. aureus adhesion on bone explant with different time of preservation. Number of viable adherent bacteria per mm2 on fresh bone explant (white histograms), frozen for less than one month (light blue histograms), for more than one month and less than six months (blue histograms) or for more than six months (dark blue histograms). *p < 0.05 versus fresh bone; & p < 0.05 versus bone frozen for less than one month, #p < 0.05 versus the same condition between strains. n = 4 to 9. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
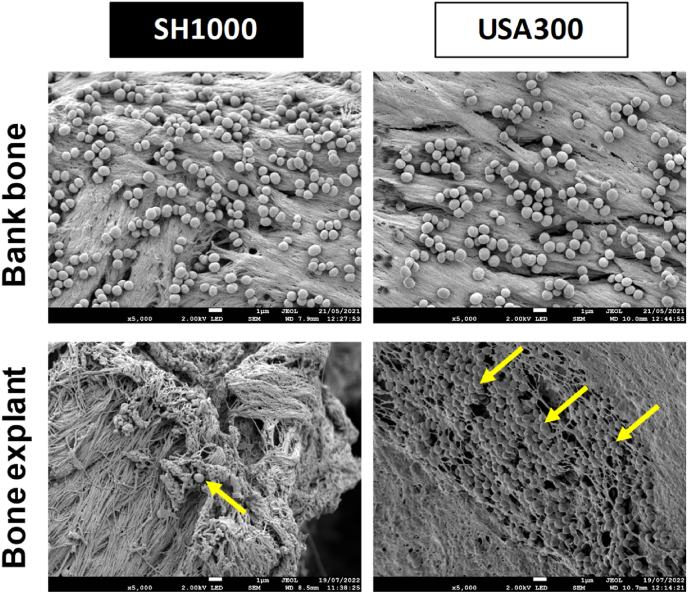
According to the previous results, we continued the experiments with bone explants preserved for less than one month. It allowed reproducing in a better way the bone environment during surgery time frame and these explants were easier to manage with the aim to create an easy-to-made in vitro model. We investigated the aspect of biofilms through scanning electron microscopy. Bank bones showed a different surface than bone explants. Indeed, the bone explants presented more reliefs and cavities than bank bones. No matrix structures were noticed on biofilms for both kinds of bones and almost no bacteria were observable on the surface of bone explants for SH1000 (Fig. 4).
Fig. 4.
S. aureus adhesion on bank bones and bone explants frozen for less than one month. Representative images of scanning electron microscopy acquisitions (magnification ×5,000). Scale bar = 1 μm. Yellow arrows indicate bacteria. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Influence of simultaneous presence of two different surfaces on S. aureus adhesion
In order to mimic the presence of different surfaces in periprosthetic bone and joint environment, bacterial adhesion was evaluated in presence of bone explant frozen for less than one month and TiFib deposited together into the same well.
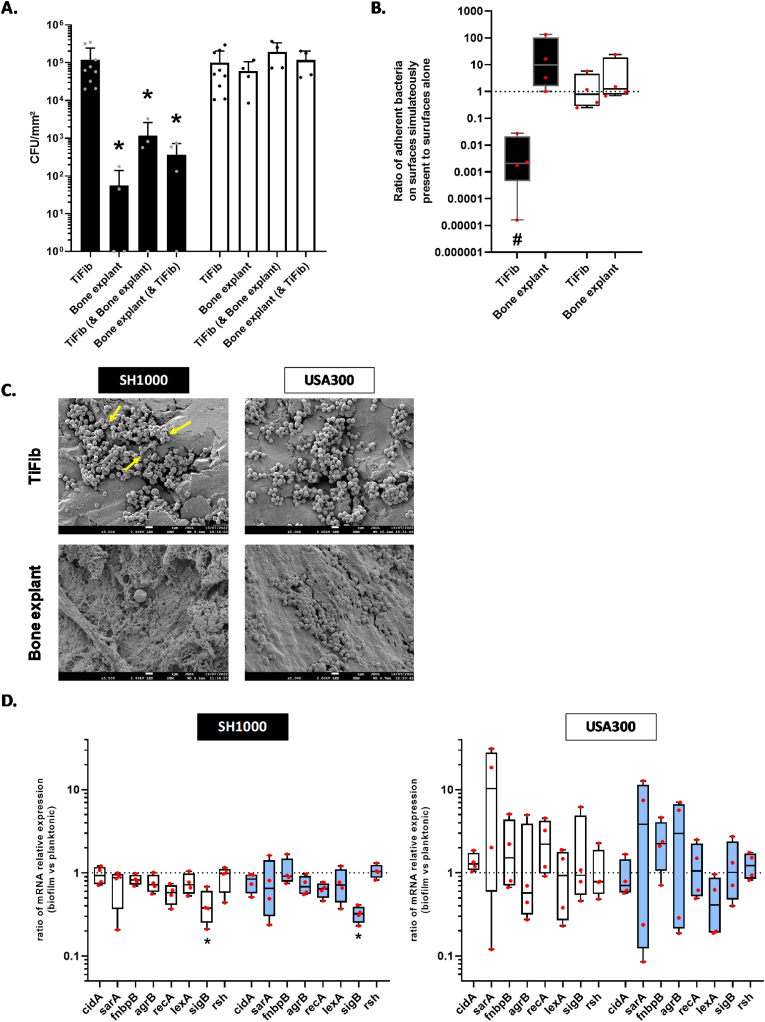
SH1000 displayed a significant decrease in adhesion on TiFib in presence of bone explant (101-fold decrease, p < 0.05) (Fig. 5A). USA300 strains kept the same behaviour if TiFib was alone or in presence of a bone explant. We observed a non-significant increase in bacterial adhesion on bone explants deposited next to TiFib for SH1000 (Fig. 5A and B). The analysis by scanning electron microscopy did not reveal difference between the tested conditions for USA300 (Fig. 5C). SH1000 displayed a production of matrix on TiFib. mRNA relative expression revealed a significant downregulation of SH1000 sigB gene expression on TiFib (2.65-fold, p < 0.05) and bone explant (3.10-fold, p < 0.05) (Fig. 5D). No significant changes of gene expression were notified for USA300 strain, this strain displaying a high heterogeneity of the relative mRNA expression compared to SH1000. Interestingly, USA300 showed important dysregulation (i.e. both important up- and downregulation) of sarA and agrB.
Fig. 5.
Comparison of S. aureus adhesion on titanium coated with fibronectin (TiFib) alone, bone explants (frozen for less than one month) alone or on each surface simultaneously present. (A) Number of live adherent bacteria per mm2. *p < 0.05 versus TiFib (alone). n = 4 to 11. (B) Ratio of adherent bacteria on surfaces simultaneously present to adherent bacteria on surfaces alone. Dark histograms = SH1000, White histograms = USA300. #p < 0.05 versus adherent bacteria on surface alone. (C) Representative images of scanning electron microscopy acquisitions (magnification ×5,000). Scale bar = 1 μm. Yellow arrows indicate bacterial matrix. (D) Results represent ratio of relative mRNA expressions by bacteria within biofilm to planktonic state, for SH1000 and USA300 grown on titanium coated with fibronectin (white histograms) or bone explants (light blue histograms) both present simultaneously. *p < 0.05 versus TiFib alone. n = 4. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
We evaluated in vitro bacterial adhesion of two S. aureus strains on different supports mimicking surfaces encountered by bacteria in the periprosthetic environment. The objective of this study was to determine the appropriate supports to create an in vitro BPI model, focusing on infection located outside the movable part of the joint. The development of such in vitro model for joint infection will include other parameters than those discussed here, like the presence of artificial synovial liquid or a rich medium and implementation of shear forces.
Implanted materials are rapidly covered by host proteins [11]. We therefore used protein-coated titanium to mimic this phenomenon. The medium FBS is a complex fluid, composed of non-proteinaceous components, which could also impact bacterial adhesion. Fibronectin-coated implants are known to promote bacterial adhesion [11]. However, S. aureus possess many adhesins allowing its adhesion to other polymers (fibrinogen, hyaluronic acid etc.) present in bone environment [11]. We confirmed that fibronectin-coating increases bacterial adhesion for SH1000 and USA300 in our model. Thanks to CLSM, we analyzed the composition of the biofilm matrix of both strains. SH1000 produced mainly PIA whereas USA300 produced mainly proteins. This confirmed the matrix production of MSSA and MRSA already described in literature [37].
To find out whether the observed modifications of biofilm formation were due to the regulation of biofilm-related genes, their expressions were assessed by RT-qPCR. Again, it seemed that the behaviours of SH1000 and USA300 were different. An over-expression of fnbpB and icaC was observed for SH1000. The first is involved in fibronectin binding and biofilm accumulation phase, which is consistent with the presence of fibronectin [38,39]. The second is involved in the operon ABCD, responsible of PIA production, which is consistent with the composition of the matrix quantify thanks to CLSM for SH1000 [40]. However, the two strains displayed different expression of sigB and agrB. SH1000 downregulated the expression of sigB, which is involved with biofilm maturation [32]. The gene agrB, involved in quorum sensing, was overexpressed by USA300 on TiFib. Agr operon is responsible for AIP production, which is involved in quorum sensing. This system allows bacteria to orchestrate biofilm formation: its structuration and detachment [34]. In conclusion, these strains seemed to form biofilm on TiFib through different pathways involving different genes.
Bone being an attractive surface for bacteria [41], the next step for the development of a BPI model was to compare bacterial adhesion on bones from bank and bone explants. As commercial products, bones from bank are easier to obtain than bone explants, but the latter gives a better representation of the composition of human trabecular bone. As patients undergoing arthroplasty were treated with cefazolin, bone explants may release unknown concentrations of this antibiotic, which could explain the obtained results (i.e. more adherent bacteria for USA300 MRSA). We performed MIC assay to investigate our hypothesis and tested one more MSSA: CIP 53.154 (Supplementary Table S1). This confirmed that these MSSA strains were more sensitive to this antibiotic than USA300. According to literature, the median cefazolin concentration for bone would be at the maximum of 13.5 μg/g of bone [42]. Our bone explants weighted around 40 mg and were placed in 1 mL of medium. According to theoretical calculations, the maximum dose of released cefazolin could be 0.54 μg/mL. This concentration is just over the MIC of both MSSA and under MIC for MRSA USA300. Furthermore, CIP 53.154 showed similar results to SH1000 regarding the number of adherent bacteria to TiFib and bone explants (Supplementary Fig. S3). More strains are required to conclude about the reason of such differences (difference between strains or methicillin sensitivity). Scanning electron microscopy acquisitions revealed no visible matrix structures on the observable surface of biofilm and bacterial matrix quantification was not technically possible.
The regulation of several genes involved in biofilm formation for SH1000 and USA300 bacteria adhered to bones from bank or bone explants was next investigated. SH1000 showed an opposite expression of sarA on the two surfaces. This gene involved in attachment and early biofilm formation could reveal an activation of the biofilm formation on bone explants. Furthermore, recA over-expression was observed regarding bone explants, suggesting an involvement of SOS response. This phenomenon was not observed for the second strain. However, USA300 adhered on bone explant and bank bone displayed a downregulation of lexA. Surprisingly, fnbpB was upregulated for USA300 on bank bone, suggesting a global enhancement of adhesion protein expression. It appeared that these two strains formed biofilm on the two surfaces by involving different metabolic pathways.
We noticed difference in bacteria adhered to bone explants according to the time of freezing. Thus, the consequence of different freezing times was explored. SH1000 showed an increase in the number of adherent bacteria according to the time of preservation. Cefazolin contained in bone explant could be degraded over time [43,44]. Further investigations are needed to determine if the phenomenon is due to the degradation of cefazolin, but other explanations are conceivable such as degradation of bone factors or alteration of the bone structure. Indeed, bone cells produce antimicrobial peptides as defensins efficient against MSSA but not MRSA [45,46]. Furthermore, bone explants released lipid droplets within the wells despite several washing step after sampling. The effect of these lipids on bacteria growth and biofilm formation remains to be elucidate. Finally, bone explant presents a more representative environment as bone composition is better preserved and structures such as cavities were observed thanks to SEM, contrary to bank bones. This highlighted the importance to use bone explant frozen for less than one month to investigate biofilm formation on bone in vitro.
Finally, the bacterial adhesion in the presence of both TiFib and bone explant surfaces (frozen for less than one month) was investigated to better mimic the presence of bone and coated prosthesis in the patient. The presence of bone explant decreased bacterial adhesion on TiFib. However, we noticed a non-significant increase in adhesion on bone explant for the three strains (see Fig. 5B and S3C). Interestingly, we observed a change in matrix production of SH1000 on TiFib in presence of bone explant and the under-expression of sigB gene. SigB is a sigma factor that ensure many environmental and stress response and among them biofilm formation [[31], [32], [33]]. This underlined a change of SH1000 behaviour between the conditions (surfaces alone or together). USA300 strains did not show any modification of biofilm under scanning electron microscopy observations but sarA and argB genes were overexpressed on bone explant. The non-significant increase in bacteria adhesion on bone explant simultaneously present with TiFib remains to be confirmed and explained. Nevertheless, it confirms the need to investigate on the simultaneous presence of these two surfaces in BPI model.
In perspective, the BPI in vitro biofilm model could also be completed with the presence of host factors. Indeed, the internalization of S. aureus in osteoblast and the secretion of bone cells also impact biofilm formation [2,47]. The study of a clinical strain panel will be interesting to characterize biofilm formed under BPI environment.
In conclusion, the type of surfaces and their structures strongly influence bacterial adhesion in bone context. Bank bone is not representative enough of bone, which mimics less BPIs. Thus, the use of a bone explants with the presence of coated titanium is necessary to get closer to in vivo conditions. Our results will help to build the foundations of an in vitro model of bone prosthesis infections.
Funding
The Ph.D. fellowship (COSIBIOOS program) of FL was co-funded by Région Grand Est, Strasbourg, France and Fondation URCA, Reims, France. ML was supported by the Société Française de Rhumatologie, France.
CRediT authorship contribution statement
Fabien Lamret : Formal analysis, Investigation, Methodology, Writing - original draft. Anaïs Lemaire : Investigation, Methodology, Writing - review & editing. Manon Lagoutte : Investigation, Methodology, Writing - review & editing. Jennifer Varin-Simon : Investigation, Methodology, Writing - review & editing. Laura Abrahams : Investigation, Methodology, Writing - review & editing. Marius Colin : Validation, Methodology, Writing - review & editing. Julien Braux : Formal analysis, Writing - review & editing. Frédéric Velard : Validation, Visualization, Writing - review & editing. Sophie C. Gangloff : Conceptualization, Data curation, Funding acquisition, Resources, Supervision, Validation, Writing - review & editing. Fany Reffuveille : Writing – original draft, Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing - review & editing.
Declaration of competing interest
Authors declare no competing interests.
Acknowledgments
The authors thank PICT-URCA platform (University of Reims Champagne-Ardenne) for imaging core facilities and Christine Terryn who provided methodologies to perform and analyze CLSM acquisitions. The authors also thank Yasser Dghoughi for having performed some of RT-qPCR experiments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2023.100120.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Tande A.J., Patel R. Prosthetic joint infection. Clin Microbiol Rev. 2014;27:302–345. doi: 10.1128/CMR.00111-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Josse J., Valour F., Maali Y., Diot A., Batailler C., Ferry T., Laurent F. Interaction between staphylococcal biofilm and bone: how does the presence of biofilm promote prosthesis loosening? Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murillo O., Grau I., Lora-Tamayo J., Gomez-Junyent J., Ribera A., Tubau F., Ariza J., Pallares R. The changing epidemiology of bacteraemic osteoarticular infections in the early 21st century. Clin Microbiol Infect. 2015;21:254.e1–254.e8. doi: 10.1016/j.cmi.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Potera C. Forging a link between biofilms and disease. Science. 1999;283:1837–1839. doi: 10.1126/science.283.5409.1837. [DOI] [PubMed] [Google Scholar]
- 5.Shoji M.M., Chen A.F. Biofilms in periprosthetic joint infections: a review of diagnostic modalities, current treatments, and future directions. J Knee Surg. 2020;33:119–131. doi: 10.1055/s-0040-1701214. [DOI] [PubMed] [Google Scholar]
- 6.Kaplan S.L. Recent lessons for the management of bone and joint infections. J Infect. 2014;68(Suppl 1):S51–S56. doi: 10.1016/j.jinf.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Parry M.C., Duncan C.P. The challenge of methicillin resistant staphylococcal infection after total hip replacement: overlooked or overstated? Bone Joint Lett J. 2014;96-B:60–65. doi: 10.1302/0301-620X.96B11.34333. [DOI] [PubMed] [Google Scholar]
- 8.Stewart P.S., Franklin M.J. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 9.Karygianni L., Ren Z., Koo H., Thurnheer T. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 2020;28:668–681. doi: 10.1016/j.tim.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Zapotoczna M., O’Neill E., O’Gara J.P. Untangling the diverse and redundant mechanisms of Staphylococcus aureus biofilm formation. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arciola C.R., Campoccia D., Montanaro L. Implant infections: adhesion, biofilm formation and immune evasion. Nat Rev Microbiol. 2018;16:397–409. doi: 10.1038/s41579-018-0019-y. [DOI] [PubMed] [Google Scholar]
- 12.Hall-Stoodley L., Costerton J.W., Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 13.Mashruwala A.A., Guchte A. van de, Boyd J.M. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. Elife. 2017;6 doi: 10.7554/eLife.23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliveira W.F., Silva P.M.S., Silva R.C.S., Silva G.M.M., Machado G., Coelho L.C.B.B., Correia M.T.S. Staphylococcus aureus and Staphylococcus epidermidis infections on implants. J Hosp Infect. 2018;98:111–117. doi: 10.1016/j.jhin.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Reffuveille F., Josse J., Velard F., Lamret F., Varin-Simon J., Dubus M., Haney E.F., Hancock R.E.W., Mongaret C., Gangloff S.C. Bone environment influences irreversible adhesion of a methicillin-susceptible Staphylococcus aureus strain. Front Microbiol. 2018;9:2865. doi: 10.3389/fmicb.2018.02865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crabbé A., Jensen P.Ø., Bjarnsholt T., Coenye T. Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol. 2019;27:850–863. doi: 10.1016/j.tim.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Bidossi A., Bottagisio M., De Grandi R., De Vecchi E. Ability of adhesion and biofilm formation of pathogens of periprosthetic joint infections on titanium-niobium nitride (TiNbN) ceramic coatings. J Orthop Surg Res. 2020;15:90. doi: 10.1186/s13018-020-01613-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamret F., Varin-Simon J., Velard F., Terryn C., Mongaret C., Colin M., Gangloff S.C., Reffuveille F. Staphylococcus aureus strain-dependent biofilm formation in bone-like environment. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.714994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.François P., Vaudaux P., Lew P.D. Role of plasma and extracellular matrix proteins in the physiopathology of foreign body infections. Ann Vasc Surg. 1998;12:34–40. doi: 10.1007/s100169900112. [DOI] [PubMed] [Google Scholar]
- 20.Foster T.J., Geoghegan J.A., Ganesh V.K., Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robey P.G., Boskey A.L. Primer on the metabolic bone diseases and disorders of mineral metabolism. John Wiley & Sons, Ltd; 2008. Chapter 6. The composition of bone; pp. 32–38. [Google Scholar]
- 22.Gibon E., Amanatullah D.F., Loi F., Pajarinen J., Nabeshima A., Yao Z., Hamadouche M., Goodman S.B. The biological response to orthopaedic implants for joint replacement: Part I: metals. J Biomed Mater Res B Appl Biomater. 2017;105:2162–2173. doi: 10.1002/jbm.b.33734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibon E., Córdova L.A., Lu L., Lin T.-H., Yao Z., Hamadouche M., Goodman S.B. The biological response to orthopedic implants for joint replacement. II: polyethylene, ceramics, PMMA, and the foreign body reaction. J Biomed Mater Res B Appl Biomater. 2017;105:1685–1691. doi: 10.1002/jbm.b.33676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nandakumar V., Chittaranjan S., Kurian V.M., Doble M. Characteristics of bacterial biofilm associated with implant material in clinical practice. Polym J. 2013;45:137–152. doi: 10.1038/pj.2012.130. [DOI] [Google Scholar]
- 25.Reffuveille F., Josse J., Vallé Q., Gangloff C.M., S.C. Staphylococcus aureus biofilms and their impact on the medical Field. Rise Virulence Antibiot Resist Staphylococcus aureus. 2017 doi: 10.5772/66380. [DOI] [Google Scholar]
- 26.Liu Y., Zhang J., Ji Y. Environmental factors modulate biofilm formation by Staphylococcus aureus. Sci Prog. 2020;103 doi: 10.1177/0036850419898659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjarnsholt T., Whiteley M., Rumbaugh K.P., Stewart P.S., Jensen P.Ø., Frimodt-Møller N. The importance of understanding the infectious microenvironment. Lancet Infect Dis. 2021;21(5):S1473–S3099. doi: 10.1016/S1473-3099(21)00122-5. 00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover F.C., Goering R.V. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J Antimicrob Chemother. 2009;64:441–446. doi: 10.1093/jac/dkp241. [DOI] [PubMed] [Google Scholar]
- 29.Horsburgh M.J., Aish J.L., White I.J., Shaw L., Lithgow J.K., Foster S.J. SigmaB modulates virulence determinant expression and stress resistance: characterization of a functional RsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol. 2002;184:5457–5467. doi: 10.1128/jb.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braux J., Jourdain M.-L., Guillaume C., Untereiner V., Piot O., Baehr A., Klymiuk N., Winter N., Berri M., Buzoni-Gatel D., et al. CFTR-deficient pigs display alterations of bone microarchitecture and composition at birth. J Cyst Fibros. 2020;19:466–475. doi: 10.1016/j.jcf.2019.10.023. [DOI] [PubMed] [Google Scholar]
- 31.Chan P., Foster S., Ingham E., Clements M. The Staphylococcus aureus alternative sigma factor SigmaB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol. 1998 doi: 10.1128/.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauderdale K.J., Boles B.R., Cheung A.L., Horswill A.R. Interconnections between sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Archer N.K., Mazaitis M.J., Costerton J.W., Leid J.G., Powers M.E., Shirtliff M.E. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan L., Li S.R., Jiang B., Hu X.M., Li S. Therapeutic targeting of the Staphylococcus aureus accessory gene regulator (agr) system. Front Microbiol. 2018;9:55. doi: 10.3389/fmicb.2018.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Podlesek Z., Žgur Bertok D. The DNA damage inducible SOS response is a key player in the generation of bacterial persister cells and population wide tolerance. Front Microbiol. 2020:11. doi: 10.3389/fmicb.2020.01785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice K.C., Mann E.E., Endres J.L., Weiss E.C., Cassat J.E., Smeltzer M.S., Bayles K.W. The CidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarthy H., Rudkin J.K., Black N.S., Gallagher L., O’Neill E., O’Gara J.P. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front Cell Infect Microbiol. 2015;5:1. doi: 10.3389/fcimb.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pietrocola G., Nobile G., Gianotti V., Zapotoczna M., Foster T.J., Geoghegan J.A., Speziale P. Molecular interactions of human plasminogen with fibronectin-binding protein B (FnBPB), a fibrinogen/fibronectin-binding protein from Staphylococcus aureus. J Biol Chem. 2016;291:18148–18162. doi: 10.1074/jbc.M116.731125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCourt J., O’Halloran D.P., McCarthy H., O’Gara J.P., Geoghegan J.A. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS (Fed Eur Microbiol Soc) Microbiol Lett. 2014;353:157–164. doi: 10.1111/1574-6968.12424. [DOI] [PubMed] [Google Scholar]
- 40.Nourbakhsh F., Namvar A.E. Detection of genes involved in biofilm formation in Staphylococcus aureus isolates. GMS Hyg Infect Control. 2016;11:Doc07. doi: 10.3205/dgkh000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masters E.A., Trombetta R.P., de Mesy Bentley K.L., Boyce B.F., Gill A.L., Gill S.R., Nishitani K., Ishikawa M., Morita Y., Ito H., et al. Evolving concepts in bone infection: redefining “biofilm”, “Acute vs. Chronic Osteomyelitis”, “the Immune Proteome” and “Local Antibiotic Therapy.”. Bone Res. 2019;7:20. doi: 10.1038/s41413-019-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeller V., Durand F., Kitzis M.-D., Lhotellier L., Ziza J.-M., Mamoudy P., Desplaces N. Continuous cefazolin infusion to treat bone and joint infections: clinical efficacy, feasibility, safety, and serum and bone concentrations. Antimicrob Agents Chemother. 2009;53:883–887. doi: 10.1128/AAC.00389-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mg B., Jl B., B M., W L. Bactericidal activity and pharmacology of cefazolin. Antimicrob Agents Chemother. 1973;4 doi: 10.1128/AAC.4.4.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schurman D.J., Hirshman H.P., Kajiyama G., Moser K., Burton D.S. Cefazolin concentrations in bone and synovial fluid. J Bone Joint Surg Am. 1978;60:359–362. [PubMed] [Google Scholar]
- 45.Dapunt U., Giese T., Stegmaier S., Moghaddam A., Hänsch G.M. The osteoblast as an inflammatory cell: production of cytokines in response to bacteria and components of bacterial biofilms. BMC Muscoskel Disord. 2016;17:243. doi: 10.1186/s12891-016-1091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Midorikawa K., Ouhara K., Komatsuzawa H., Kawai T., Yamada S., Fujiwara T., Yamazaki K., Sayama K., Taubman M.A., Kurihara H., et al. Staphylococcus aureus susceptibility to innate antimicrobial peptides, β-defensins and CAP18, expressed by human keratinocytes. Infect Immun. 2003;71:3730. doi: 10.1128/IAI.71.7.3730-3739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamret F., Varin-Simon J., Six M., Thoraval L., Chevrier J., Adam C., Guillaume C., Velard F., Gangloff S.C., Reffuveille F. Human osteoblast-conditioned media can influence Staphylococcus aureus biofilm formation. Int J Mol Sci. 2022;23 doi: 10.3390/ijms232214393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.