Abstract
Background
Approximately 95% of Colorectal cancers (CRC) consist of adenocarcinomas originating from colonic Adenomatous polyps (AP). Increasing importance in CRC occurrence and progression has been attributed to the gut microbiota; however, a huge proportion of microorganisms inhabit the human digestive system. So, to comprehensively study the microbial spatial variations and their role in CRC progression, from AP to the different CRC phases, a holistic vision is imperative, including the simultaneous evaluation of multiple niches from the gastrointestinal system. Through an integrated approach, we identified potential microbial and metabolic biomarkers, able to discriminate human CRC from AP and/or also the different Tumor node metastasis (TNM) staging. In addition, as the microbiota contributes to the production of essential metabolic products detectable in fecal samples, we analysed and compared metabolites obtained from CRC and AP patients by using a Nuclear magnetic resonance (NMR) approach.
Methods
In this observational study, saliva, tissue and stool samples from 61 patients, have been collected, including 46 CRC and 15 AP patients, age and sex-matched, undergoing surgery in 2018 at the Careggi University Hospital (Florence, Italy). First, the microbiota in the three-district between CRC and AP patients has been characterized, as well as in different CRC TNM stages. Subsequently, proton NMR spectroscopy has been used in combination with multivariate and univariate statistical approaches, to define the fecal metabolic profile of a restricted group of CRC and AP patients.
Results
CRC patients display a different profile of tissue and fecal microbiota with respect to AP patients. Significant differences have been observed in CRC tissue microbial clades, with a rise of the Fusobacterium genus. In addition, significant taxa increase at the genus level has been observed in stool samples of CRC patients. Furthermore, Fusobacterium found in intestinal tissue has been positively correlated with fecal Parvimonas, for the first time. Moreover, as predicted by metagenomics pathway analysis, a significant increase of lactate (p=0.037) has been observed in the CRC fecal metabolic profiles, and positively correlated with Bifidobacterium (p=0.036). Finally, minor bacterial differences in CRC patients at stage T2 (TNM classification) have been detected, with a raise of the Spirochaetota phylum in CRC samples, with a slight increase of the Alphaproteobacteria class in fecal samples.
Conclusion
Our results suggest the importance of microbiota communities and oncometabolites in CRC development. Further studies on CRC/AP management with a focus on CRC assessment are needed to investigate novel microbial-related diagnostic tools aimed to improve therapeutic interventions.
Keywords: Microbiota, colorectal cancer, colonic adenomatous polyps, microbiota, metabolomics, biomarkers, cancer staging
Introduction
Colorectal adenocarcinoma, also known as Colorectal cancer (CRC), is the third most common type of cancer and the fourth leading cause of cancer mortalities with two million new cases reported per year [1]. The management of CRC patients, especially those with metastatic disease, is complex and expensive, and offers poor life quality. Thus, CRC prevention and screening programmes are crucial.
In addition to this, CRC aetiology is still unknown, only 10 to 15% of CRC cases are hereditary. The evolution of CRC is heterogeneous and follows genetic and epigenetic variations influenced by dietary patterns, environmental conditions, host immunity, and microbial adhesion [2]. Regarding microbial components, a large portion of microorganisms can survive and thrive in the human digestive system, establishing vast and complex communities from the oral cavity to the gastrointestinal tract [3]. Therefore, it is imperative to adopt a holistic approach, including a simultaneous study of multiple niches starting from the mouth to the gastrointestinal system, in order to comprehensively evaluate the spatial variations of microbial populations,
Recently, increasing importance in CRC progression has been attributed to the gut microbiota (GM) [4], [5], [6], [7]. In detail, the CRC “tumour microenvironment” (TME) sees a complex interaction occurring among the GM, the cancer-associated microbiome (oncobiome) and the immune system [7], [8]. It is well established that approximately 95% of CRCs consist of adenocarcinomas that originate as colonic Adenomatous polyps (AP) [10], [11] or adenomas. Several studies showed that adherent bacteria to colorectal adenomas or carcinomas were different from adherent bacteria to healthy gut mucosa [12], due to the altered tumour environment (i.e. decreased pH and modified metabolic conditions determined by hypoxia and necrosis’ onset) [13].
Indeed, specific microbiota strains members could potentially detect CRC and predict clinical outcomes, and could be used in acting screening tests (detecting high-risk adenomas or CRC in asymptomatic people [4]), as prognostic and/or predictive biomarkers (suggesting the clinical outcome, response to the treatment and its potential adverse effects [4]), or as modifiable factors influencing CRC prevention as well as CRC systemic treatment effectiveness [6], [7], [14]. In addition, other screening markers, such as metabolic and genotoxic metabolites of specific strains, may be used to recognize and screen CRC in its early stages [4], [15]. Other potential non-invasive biomarkers may also be represented by metabolites in fecal samples. In this previous study [16], for the first time, fecal metabolic alterations were identified between CRC and AP patients through Nuclear magnetic resonance (NMR) spectroscopy.
In this scenario, the main aim of this study is the exploration of potential microbial and metabolic biomarkers able to discriminate CRC from AP and/or also different TNM staging phases [17]. To this purpose, a holistic approach has been adopted to deeply explore the alterations in the oral-gut microbiome axis, assessing the microbial composition of three gastrointestinal human districts (saliva, gut tissue and stool), in CRC vs AP patients. Moreover, by a previously used NMR approach [16], the fecal metabolites profile of CRC and AP patients has been compared. Finally, the specific microbial profile of CRC patients at various TNM clinical stages, has been investigated for the first time in order to characterize the microbiota alterations during CRC evolution.
Our results suggest the importance of a simultaneous evaluation of microbiota communities and oncometabolites in the CRC progression and that could be a forerunner for future studies on CRC/AP management, focusing on the CRC assessment through novel non-invasive microbial-related diagnostic tools and on the development of therapeutic interventions.
Materials and Methods
Patients
In this observational study, a total of 61 patients including 46 CRC patients (affected by non-metastatic colorectal adenocarcinoma), and 15 AP patients, have been enrolled between January 2018 and February 2019 at the Careggi University Hospital, Florence, Italy (see the Table 1 for a summary of clinical features). The study was reviewed and approved by AOUC Careggi Institutional Review Board (Prot 2010/0012462). All study participants, or their legal guardians, have provided an informed written consent prior to the study enrollment in compliance with the national legislation and the Code of Ethical Principles for Medical Research Involving Human Subjects of the World Medical Association (Declaration of Helsinki).
Table 1.
Clinical features of the enrolled patients Clinical characteristics of colorectal cancer patients and adenomatous patients
| Code | CRC | AP |
|---|---|---|
|
Gender ratio M/F |
33 M-13 F; M/F = 2, 75 | 7M-8F; M/F = 0.85 |
|
Median age, yr |
70,8 | 62,2 |
|
Range of age, yr |
36-85 | 40-80 |
|
Tumor stage T0/T1/T2/T3/T4 (n of patients) |
1/13/21/10/1 | |
| Operative technique VL-Conv/VL/Open/Robot | 3/37/2/4 | 0/8/3/3 |
| Proximal location | 14 | 10 |
| Distal location | 32 | 5 |
|
Diet |
Mediterranean | Mediterranean |
|
Race |
Caucasian | Caucasian |
AP: Adenomatous patients; CRC: Colorectal cancer;
VL: Videolaparoscopic
VL-Conv: Videolaparoscopic-Conversion
Exclusion criteria were extended to extraperitoneal rectum localization of the tumor; previous surgery for cancer; previous chemo-radiotherapy treatment; immunodeficiency; travel to exotic countries in the last five years; treatment with immunosuppressive drugs, antibiotics, or regular probiotics in the previous two months; acute gastrointestinal infections in the month prior to enrolment; associated presence of established malignancies or chronic intestinal inflammatory diseases (Crohn's disease and ulcerative recto colitis).
In addition, all CRC patients were divided according to the pathological TNM staging system 7th edition and Dukes classification [18]. Moreover, all adenomas analysed in the study are tubulovillous adenomas with dysplasia, which can not be endoscopically removed.
Data collected included nutritional data, clinical history and status (Table 1).
Sample collection
All samples have been collected at surgery time, before treatments (e.g., chemotherapy, probiotic intake). CRC patients and AP patients, have been age and sex-matched.
Unstimulated saliva, tissue and stool have been collected from CRC and AP patients. On surgery day, in the morning, saliva samples have been collected from patients who were asked to hold the saliva for 1 minute and then spit it into a sterile tube; stool samples were collected in a sterile container. Fresh tissue samples of tumor/adenoma were collected in sterile conditions during surgery from each patient and stored in physiological solution (NaCl 0.9%). After collection, saliva, tissue and stool samples were immediately frozen and stored at −80°C until DNA extraction.
Microbiota Characterization
DNA Extraction
Genomic DNA was extracted using the DNeasy PowerLyzer PowerSoil Kit (Qiagen, Hilden, Germany) from frozen (-80°C) saliva, tissue and fecal samples according to the manufacturer's instructions with modifications as reported in our previous study [19]. Total genomic DNA was captured on a silica membrane in a spin column format and subsequently washed and eluted. The quality and quantity of extracted DNA were assessed using the NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, US) and the Qubit Fluorometer (Thermo Fisher Scientific), respectively. Then, genomic DNA was frozen at -20°C. Subsequently, total DNA samples were sent to IGA Technology Services (Udine, Italy) where amplicons of the variable V3–V4 region of the bacterial 16S rRNA gene, delimited through the primers 341F and 805R, were sequenced in paired-end (2 × 300 cycles) on the Illumina MiSeq platform, according to the Illumina 16S Metagenomic Sequencing Library Preparation protocol.
Bioinformatic Analysis of V3-VA 16S rDNA sequences
Demultiplexed sequence reads were processed using QIIME2 2021.4. The sequencing primers and the reads without primers were removed using Cutadapt tool. DADA2 was used to perform paired-end reads filtering, merging and chimeras removal steps after trimming low quality nucleotides from both forward and reverse reads (–p-trunclen-f 261 and –p-trunclen-r 184). Hence, ASVs (amplicon sequence variants) were generated and the taxonomic assignments were performed through global alignment in SILVA 138 using V-Search algorithm with a 99% identity threshold.
Statistical analyses on V3-VA 16S rDNA sequences data
The statistical analyses on bacterial communities were performed in R 4.1 with the help of the packages phyloseq 1.38.0, vegan 2.6.2, DESeq2 1.32.0 and other packages satisfying their dependencies. The packages ggplot2 3.3.6, dendextend 1.15.1 and ggpubr 0.40 were used to plot data and results. ASVs taxonomically unassigned to the prokaryotic domain in tissue sequencing data have been discarded prior to the statistical analysis being probable host DNA contaminants due to the low biomass of this specific environment [20]. After the decontamination, a rarefaction analysis on ASV was performed on every sample using the function rarecurve (step 100 reads), further processed to highlight saturated samples (arbitrarily defined as saturated samples with a final slope in the rarefaction curve with an increment in ASV number per reads < 1e-5). The observed richness, Shannon and Pielou's evenness indices were used to estimate the bacterial alpha-diversity in each sample using the function estimate_richness from phyloseq. The evenness index was calculated using the formula E = S/log(R), where S is the Shannon diversity index and R is the observed ASV richness in the sample. Differences in alpha-diversity indices were tested using the Kruskal-Wallis test or the Kruskal-Wallis test. Due to the non-normal distribution of those indexes, the related statistical tests performed are non-parametric. PCoAs was performed using Bray-Curtis index on proportional count data of each sample adjusted with square root transformation. A PERMANOVA was used to test the statistical significance of the beta-diversity distances. At different taxonomic ranks, the differential analysis of the abundances has been computed through DESeq2 on raw count data.
Moreover, potentially expressed Metacyc pathways in each group have been predicted through PICRUST2 v2.4.2 with EPA-ng algorithm and then significant differences of those between the CRC vs. AP in tissue and fecal samples, have been explored using LEFSE (LDA Effect Size) analysis.
NMR sample preparation and analyses
NMR analysis has been performed following [16]. In detail, fecal water was extracted to ratios of 1:2 (g/mL, weight of unthawed feces-to-buffer) in 0.75 M phosphate-buffered saline (PBS, pH 7.4) [21]. The buffered samples were homogenized by whirl mixing for 30 s and sonicated for 15 min. Each sample was then centrifuged at 10000 g for 10 min at 4°C, and 700 μL supernatant was transferred to 1.5 mL Eppendorf tubes and centrifuged again at 14000 rpm for 5 min at 4°C. The clear supernatant was used for NMR analyses.
A total of 70 μL buffer solution (1.5 M KH2PO4/D2O, pH 7.4; 2 mmol/L NaN3; 0.1% TMSP) was added to 630 μL of each fresh fecal water sample, and a total of 600 μL of this mixture was transferred to a 5 mm NMR tube. One-dimensional proton NMR (1H-NMR) spectra for all samples were acquired using the Bruker 600 MHz spectrometer (Bruker BioSpin srl; Rheinstetten, Germany) operating at 600.13 MHz proton Larmor frequency and equipped with a 5 mm PATXI 1 H-13C-15N and 2H-decoupling probe including a z axis gradient coil, an automatic tuning-matching, and an automatic and refrigerated sample changer (SampleJet, Bruker BioSpin srl; Rheinstetten). The BTO 2000 thermocouple served for temperature stabilization at the level of approximately 0.1 K at the sample. Before measurement, samples were kept for at least 3 min inside the NMR probe head for temperature equilibration. Two one-dimensional 1H-NMR spectra, namely one-dimensional (1D) NOESY and Carr-Purcell-Meiboom-Gill (CPMG), were acquired at 310 K with different pulse sequences: a standard nuclear Overhauser effect spectroscopy pulse sequence 1D NOESY PRESAT (noesygppr1d.comp; Bruker BioSpin) pulse sequence, using 64 scans, 98304 data points, a spectral width of 18028 Hz, an acquisition time of 2.7 s, a relaxation delay of 4 s, and a mixing time of 0.1 s; and a standard spin echo CPMG [16] (cpmgpr1d.comp; Bruker BioSpin) pulse sequence applied to a standard 1D sequence, with 64 scans, 73728 data points, a spectral width of 12019 Hz, and a relaxation delay of 4 s.
Spectral processing was performed following these [16] methods. In detail, free induction decays were multiplied by an exponential function equivalent to 0.3 Hz line-broadening factor before applying Fourier transform. Transformed spectra were automatically corrected for phase and baseline distortions and fecal spectra were calibrated to TMSP singlet at 0 ppm using the TopSpin version 4.1.0 (Bruker BioSpin GmbH). Before analysis of the generated data matrix, probabilistic quotient normalization [22] normalization and mean centring of the variables were performed.
Statistical analysis on metabolites
Statistical analyses on microbiota composition were performed using R [23]. The software GraphPad Prism (v. 5) was used for statistical data analysis while MetaboAnalyst 4.0 free online software was used for pathway analysis [24].
Metabolite identification was performed manually based on previous literature [21], the human metabolome database public database, and a library of pure organic compounds (BBIOREFCODE; Bruker BioSpin). The relative metabolite concentrations (expressed in arbitrary units) were calculated by integrating and calculating the area of the peaks [25]. To determine the discriminating molecules among all classes under study, the Wilcoxon test was chosen to infer differences between two groups of subjects. False discovery rate (FDR) correction was applied using the Benjamini & Hochberg method, and FDR < 0.05 was considered statistically significant.
Correlations between metabolites and taxonomies that have varied significantly between CRC and AP were evaluated using Spearman's rank-correlation analysis. Correlations with a p-value < 0.05 were considered significant.
Data availability statement
The microbial-related data (raw reads, ASV tables and taxonomic assignments) is freely available. To review GEO accession GSE217490: Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE217490.
The processing and analysis scripts are available at https://github.com/LeandroD94.
RESULTS
Microbiota comparison between CRC and AP patients
Overall Comparison of the Oral, Tissue and Fecal Microbiota
In order to investigate similarities/differences of bacterial abundance on patients in different samples (i.e. intestinal tissue vs. stool vs. saliva), a cluster analysis has been performed on normalized ASV counts of all enrolled patients (Table 1). A number of 19,071,701 reads has been obtained, and after all the steps of pre-processing (pair merging, trimming, quality filtering, and chimera detection), a total of 13,269,334 (69%) reads were available for further analysis.
In the first part of the study, saliva, tissue and fecal microbial communities have been characterised in both CRC and AP patients. The alpha diversity of saliva and tissue samples between CRC and AP patients did not show any significant difference in Observed richness, Shannon index and Evenness index (data not shown) (Figure 1 Supplementary material).
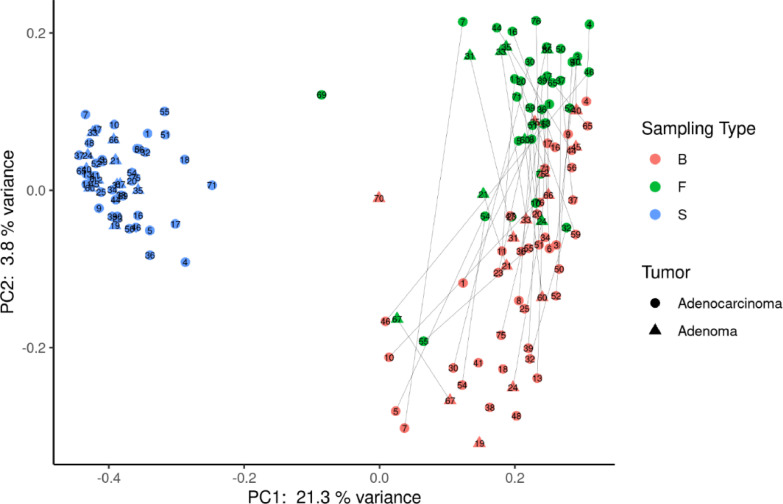
Figure 1.
Principal coordinate analysis using Bray–Curtis dissimilarity as a distance metric on square root–transformed percent abundance of identified ASVs in salivary, tissue and fecal samples in CRC and AP patients. B= biopsy; F =fecal samples; S= Saliva samples. The lines connect the samples from the same patient for tissue and fecal samples.
As expected, the PCoA analysis evidenced three distinct clusters, populated by saliva, tissue and stool samples, respectively (Figure 1). However, the microbiota composition of fecal and tissue samples appears more similar to each other than the bacterial composition of saliva samples, for both CRC and AP conditions.
CRC vs APC patients display a different profile of tissue and fecal microbiota
The microbiota composition of saliva, tissue and fecal samples was compared between CRC and AP patients (Figure 2A, B, C).
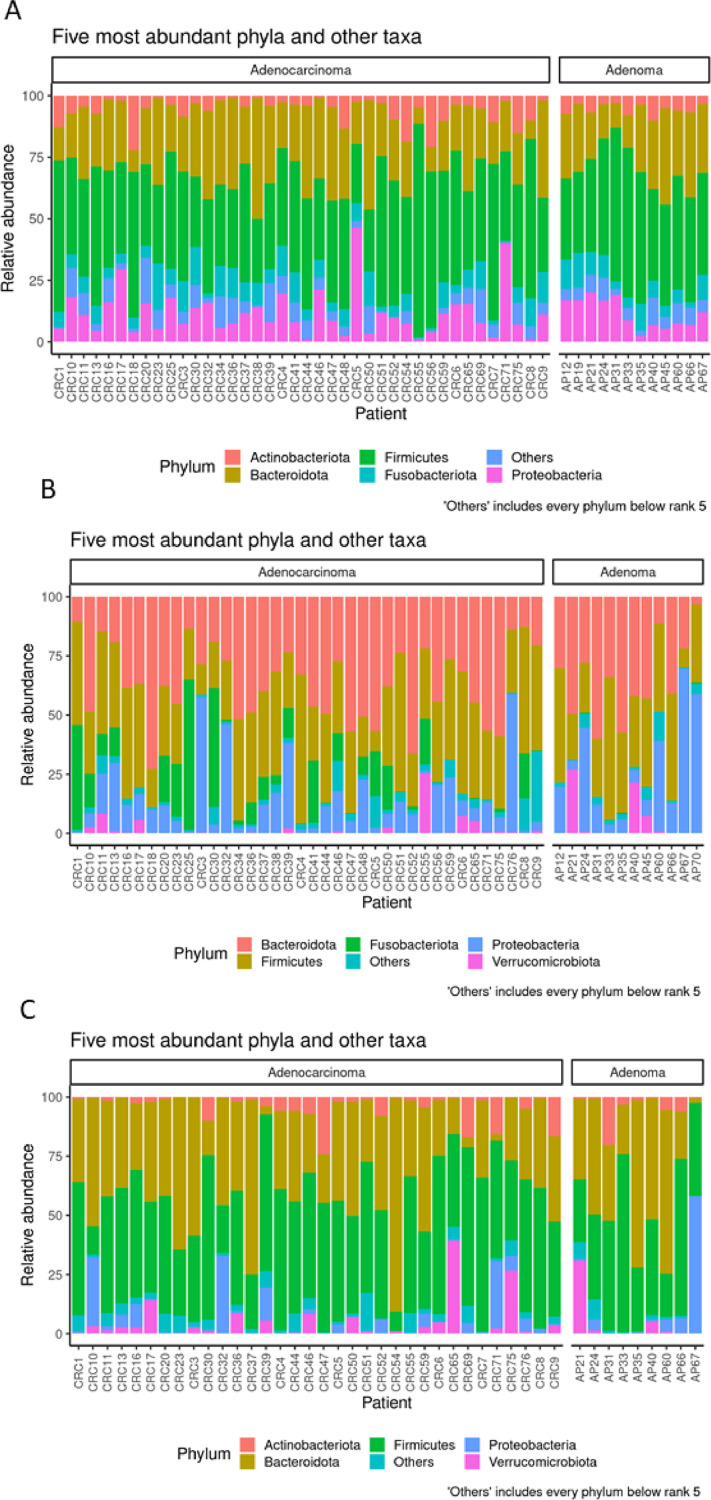
Figure 2.
Taxonomic composition of the five most abundant microbial phyla in saliva (A), intestinal tissue (B) and fecal (C) samples from CRC and AP samples. Bar plot shows the relative abundance of bacterial phyla in each sample.
Concerning the microbiota of saliva, the relative abundance of the five most represented microbial phyla revealed that the sequences collected were classified into: Firmicutes (43.79%), Bacteroidota (25.05%), Proteobacteria (11.64%), Fusobacteriota (7.43%), Actinobacteriota (6.11%), (Figure 2A). However, we did not observe any significant difference in alpha and beta diversity comparing the saliva of CRC to that of AP patients. Moreover, these results were also confirmed by PCA, PCoA and DeSeq2 analysis (data not shown).
Regarding the pathological tissue, the relative abundance of the most represented microbial phyla revealed that the sequences collected were classified into Bacteroidota (36.17%), Firmicutes (33.64%), Proteobacteria (15.77%) and Fusobacteriota (8.94%) (Figure 2B). In addition, at the genus level, we detected Bacteroides (19.16%), Escherichia –Shigella (10.55%), Fusobacterium (9.25%), Prevotella (7.12%) and Faecalibacterium (4.03%), (Figure 2, Supplementary material).
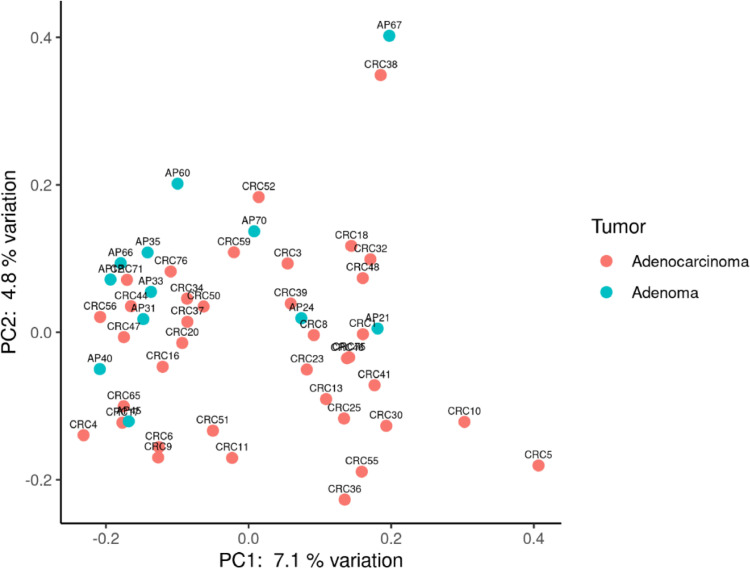
Moreover, PCoA analysis showed a slight differentiation between CRC and AP microbiota composition (Figure 3).
Figure 3.
Principal coordinate analysis using Bray–Curtis dissimilarity as a distance metric on square root–transformed percent abundance of identified ASVs in salivary, tissue and fecal samples in CRC and AP patients.
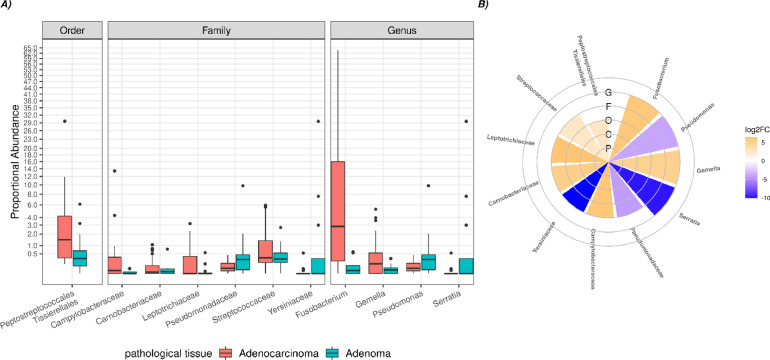
Significant differences were observed in microbial clades from intestinal tissue when comparing CRC vs AP patients. It should be noted that, a significant increase of the orders Peptostreptoccales Tissierellales was detected, as well as a significant increase of the families Campylobacteraceae, Carnobacteriacerae, Gemellaceae, Leptotrichiaceae, Streptococcaceae, and a decrease of families Pseudomonadaceae, Yersiniaceae in CRC tissue. Finally, at the genus level, a significant rise of Fusobacterium and Gemella has been recorded, as well as a significant reduction of Pseudomonas and Serratia in CRC compared to AP patients (Figure 4).
Figure 4.
Boxplot (A) and circoplot (B) respectively showing the results of differential abundances analysis and log2foldchange between taxa of the pathological tissue from CRC and AP patients. Letters indicate the taxonomic depth, in detail, G=genus, F= family, O=order, C= class, P= phylum. All results have an FDR < 0.05.
Concerning the fecal samples, the relative abundance of the most represented microbial phyla revealed that the sequences collected were classified into: Firmicutes (45.59%), Bacteroidota (36.80%), Proteobacteria (5.60%), Verrucomicrobia (5.29%) and Actinobacteriota (4.29%) (Figure 2C).
The alpha diversity displayed a significant difference in richness (observed number of species richness, p=0.054), while no significant differences were found for the Evenness and Shannon indices (Figure 3 supplementary material).
However, no differences were found between PCA and PCoA (data not shown).
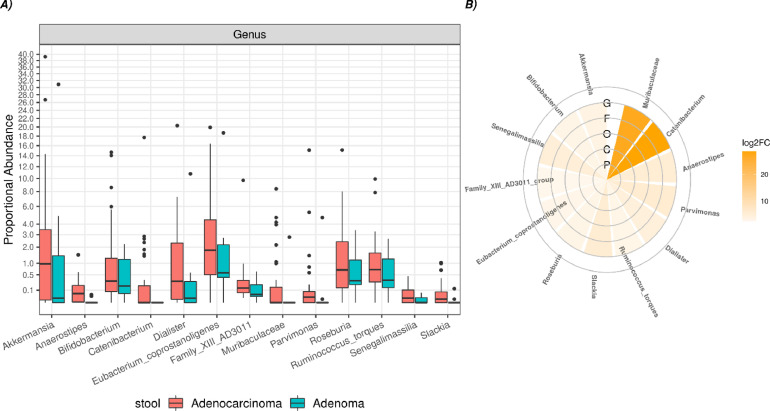
Moreover, several significant differences at the genus level were detected when comparing CRC vs AP patients (Figure 5). In particular, a significant increase of Akkermansia, Anaerostipes, Bifidobacterium, Catenibacterium, Dialister, Eubacterium_coprostanoligenes, Family_XIII_AD3011, Muribaculaceae, Parvimonas, Roseburia, Ruminococcus_torques, Senegalimassilia and Slakia was observed in stool of CRC patients.
Figure 5.
Boxplot (A) and circoplot (B) respectively showing the results of differential abundances analysis and log2foldchange between taxa of the fecal samples from CRC and AP patients. Letters indicate the taxonomic depth, in detail, G=genus, F= family, O=order, C= class, P= phylum. All results have a FDR < 0.05.
Correlation between intestinal tissue and fecal microbiota
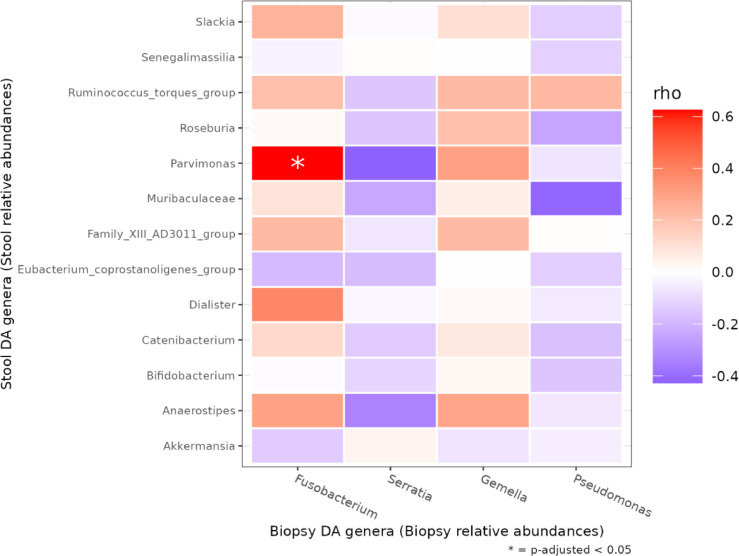
Despite the convenience and non-invasiveness of fecal sampling, the fecal microbiota does not fully represent that of the intestinal tract, and the efficacy of fecal sampling to accurately represent the gut microbiota is poorly understood. Since we observed remarkable alteration of intestinal tissue and fecal microbiota architecture between AP and CRC, we wondered if these changes might be interrelated, in order to explore the potential stool use as a gut proxy in the CRC development. Among the genera changes between CRC and AP tissue and fecal samples, we observed a significant positive correlation only between intestinal tissue Fusobacterium and fecal Parvimonas (p adj=0.0008) (Figure 6).
Figure 6.
Heatmap of Spearman correlation value between intestinal tissue (rows) and fecal (columns) taxa resulted differently abundant in DESeq2 analyses in CRC and AP fecal samples. FDR < 0.05 are marked with an asterisk.
Functional profiles of tissue and fecal microbiota in CRC vs AP
Since significant alterations in tissue and fecal microbiota were observed, the functional metagenomics inferred using PICRUST2 analysis has been evaluated to estimate how the bacterial functional profiles differed between the two groups. Pathways involved in microbial gene functions belonging to metabolism, genetic information processing, environmental information processing and cellular processes categories were included. Particular functional profiles associated with potentially expressed microbial genes in CRC vs AP have been recorded.
The CRC tissue was positively associated with the superpathways of homolactic fermentation, reductive acetyl coenzyme A pathway, dTDP-L-rhamnose biosynthesis I, glycolysis I (from glucose 6-phosphate), O-antigen building blocks biosynthesis (E. coli), L-glutamate degradation V (via hydroxyglutarate), L-lysine fermentation to acetate and butanoate, L-lysine biosynthesis II, glycolysis II (from fructose 6-phosphate), acetyl-CoA fermentation to butanoate II, succinate fermentation to butanoate, GDP-D-glycero-α-D-manno-heptose biosynthesis, chondroitin sulfate degradation I (bacterial), pyruvate fermentation to acetone, superpathway of thiamin diphosphate biosynthesis II, cob(II)yrinate a,c-diamide biosynthesis I (early cobalt insertion).
On the other hand, the AP tissue was positively associated with the superpathway of fatty acid biosynthesis initiation (E. coli), hexuronide and hexuronate degradation, glucose and glucose-1-phosphate degradation, superpathway of β-D-glucuronide and D-glucuronate degradation, myo-inositol degradation I, stearate biosynthesis II (bacteria and plants), palmitoleate biosynthesis I (from (5Z)-dodec-5-enoate), oleate biosynthesis IV (anaerobic), superpathway of L-alanine biosynthesis, (5Z)-dodec-5-enoate biosynthesis, mycolate biosynthesis, sulfate reduction I (assimilatory). (Figure 4A Supplementary material)
Regarding the fecal samples, the CRC was positively associated with the superpathay Bifidobacterium shunt and polyamine biosynthesis II. On the other hand, the AP stool was positively associated with the superpathway of hexuronide and hexuronate degradation, D-galacturonate degradation I, superpathway of β-D-glucuronide and D-glucuronate degradation, 4-deoxy-L-threo-hex-4-enopyranuronate degradation (Figure 4B Supplementary material)
Comparative (CRC vs APC) fecal metabolomics’ profile
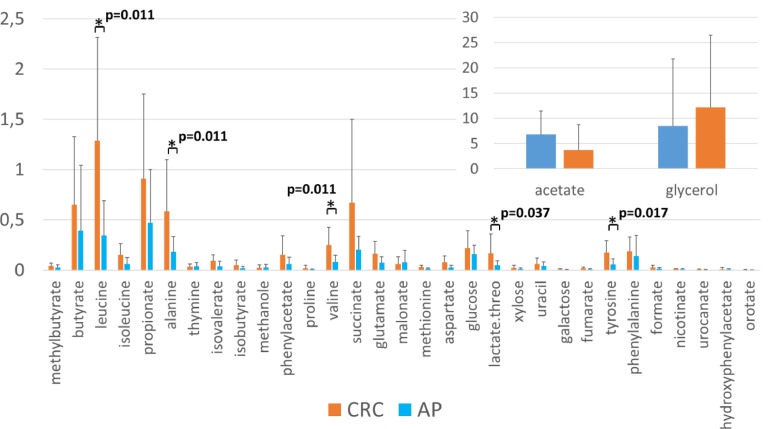
Due to the potential differences in pathways involved in microbial gene functions belonging to the metabolism, the NMR spectra of 29 fecal extract samples (20 CRC, 9 AP) have been acquired to assess the effective fecal metabolomics profile of CRC vs AP patients,. With the aim of distinguishing metabolite-level variations characteristic for the two groups, the Kruskal-Wallis test was applied to the identified fecal metabolites. We observed a significant increase of leucine, alanine, valine (respectively, p=0.011), lactate (p=0.037) and tyrosine (p=0.017) in the fecal metabolic of CRC compared to AP patients (Figure 7).
Figure 7.
Fecal metabolites levels in 29 fecal extract samples (20 CRC, 9 AP). Histogram reports the levels of metabolites (concentrations expressed in arbitrary units) evaluated respectively in CRC (orange) and AP (blue). Kruskal-Wallis test was performed to test the differences between CRC and AP samples. A p-value < 0,05 after multiple test correction is considered statistically significant. The asterisks (*) mark represent the FDR < 0.05.
Correlation between fecal microbiota and metabolites
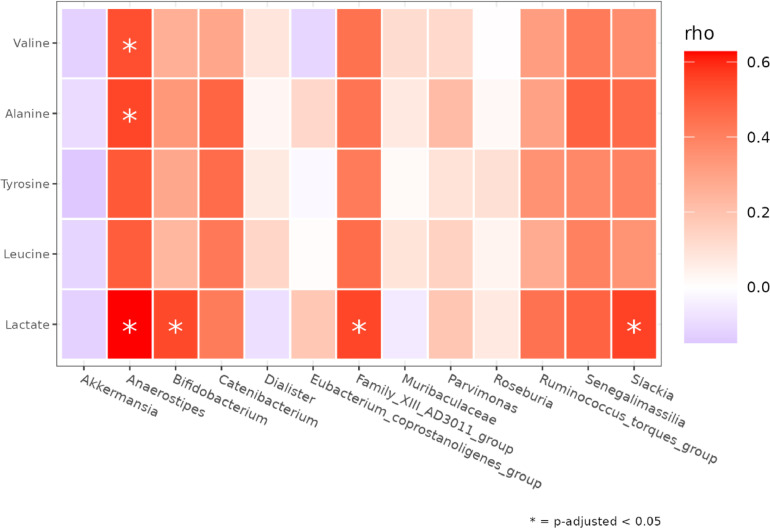
To explore the relationships between the stool microbiota and the fecal metabolites, the potential associations between leucine, alanine, valine, lactate and tyrosine (p=0.017), have been investigated since a) they could have represented significantly altered metabolites in CRC compared to AP condition, and b) the tissue and fecal microbiota at the genus level was differentially represented (Figure 8). The correlation between metabolites and tissue taxa did not show any statistical differences (data not shown) but regarding the fecal genera we registered different and significant associations. In detail we observed positive correlations between Anaerostipes with lactate (p=0.022), alanine (p=0.036) and valine (0.033); Bifidobacterium with lactate (p=0.036), Family_XIII_AD3011_group with lactate (0.036), as well as Slakia with lactate (p=0.036).
Figure 8.
Heatmap of Spearman correlation value between metabolites (rows) and differently abundant taxa in CRC vs AP fecal samples according toDESeq2 (columns) . P-values < 0.05 are marked with an asterisk.
Alterations of the oral-gut microbiota axis in the CRC staging
CRC staging displays a different profile of saliva, tissue and stool microbiota
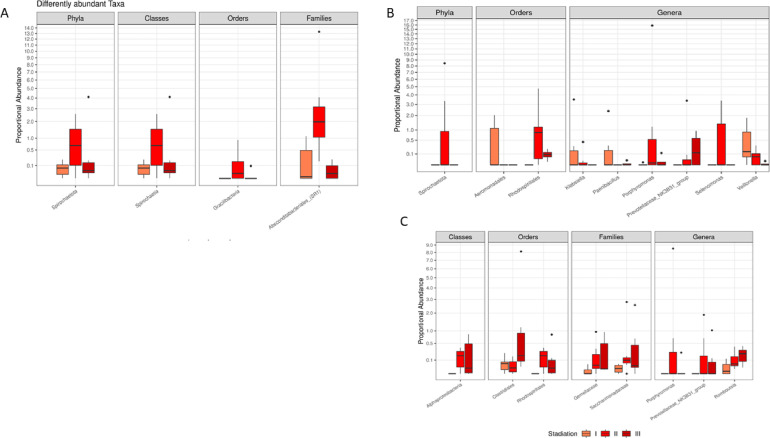
Since significant bacterial count variations between CRC and AP intestinal tissue and fecal samples was detected, a deep investigation has been performed to look for alteration of CRC microbiota architecture in salivary, tissue and faecal samples, at different stages, according to TNM classification and focusing on T parameter (size of the primary tumor and invasion of surrounding tissues). The CRC patients have been divided into three groups (I, II, II) and respectively compared for their microbial patterns in saliva, tissue and faecal samples. More specifically, patients with T0 and T1 staging were classified as Group I, while, patients with T2 staging were classified as Group II and patients T3 and T4 as Group III. Starting from the different comparisons, some significant variations have been detected for the first time. Notably, regarding saliva samples, a significant increase of Spirochaetota phylum, Spirochaetia classes, Gracilibacteria order and Absconditabacteriales in Group III, has emerged compared to Groups I and II (Figure 9A). Concerning tissue samples, an increase of the Aeromonadales order, Klebsiella, Paenibacillus Veilonella genera was detected in Group I. Moreover, in Group II an increase of Spirochaetota phylum, Rhodospirillaless order, as well as Porphyromonas and Selenomonas genera was observed while an increase of Prevotellaceae_NK3B31_group was documented in Group III (Figure 9B). Finally, in the fecal samples from Group II, a significant rise of Alphaproteobacteria class, Rhodospirillales order, as well as Gemellaceae, Saccarimonadaceae families, Porphyromonas, Prevotellaceae_NK3B31_group genera was evident. An increase in Clostridiales order and Romboutsia genus has been reported in Group III (Figure 9 C). Moreover, regarding alpha diversity, fecal samples showed a significant difference in richness (observed number of species) (p=0.043) and in the Shannon index (p=0.053) (Figure 5 supplementary material)
Figure 9.
Boxplot showing the results of differential abundance analysis between taxa in saliva (A), intestinal tissue (B) and fecal (C) samples from different TNM staging groups of CRC patients (I, II, III). All results have an FDR p-value < 0.05.
The comparative analysis of fecal metabolic profile at different CRC stages
Finally, a metabolic profile of fecal samples was performed, but no statistical difference could be detected amongst the different clinical staging groups (I, II, II)
Discussion
In this study, an integrated approach has been adopted to explore the microbial patterns of the oral-gut microbiota axis, fecal metabolites and their mutual involvement in the discrimination between CRC vs AP and in different TNM stages, in order to fully understand microbial variations during the malignancy progression.
As reported, overwhelming evidence suggests that gut microbiota play a critical role in the development of colorectal malignancies as well as in the AP progression into CRC [12]. There are several study that used an integrative approach to analyse both microbial and metabolic patterns in AP/CRC samples [26], [27], [28], however, to the best of our knowledge, this is the first study that aims to simultaneously delineate the compositional and functional changes in the oral-gut microbiota axis, related to different steps of neoplastic progression, ranging from the initial non-cancerous AP stage to the CRC and its T staging.
To investigate the microbiota profile residing in the whole oral-gut axis, we used an approach previously applied in our pilot study [19], where, for the first time, we simultaneously compared the microbial compositions in the three mentioned body sites (oral, tissue and stools), in CRC vs healthy controls [19]. In the first part of this present study, we expanded our previous results [19], exploring the saliva, intestinal tissue and fecal microbial communities in CRC and AP, to appreciate similarities and alterations of microbial architecture, potentially involved in AP progression to CRC.
Concerning the salivary microbiota, the relative abundance of the five most represented microbial phyla revealed that the sequences collected were classified into Firmicutes, Bacteroidota, Fusobacteriota, Actinobacteriota and Proteobacteria (Figure 2A). Variations of the salivary microbiota composition in oral health and disease conditions have been previously described [29], suggesting that the oral microbial compositions may theoretically mirror the oral and general health status [30]. According to previous data [31], we observed that sequences affiliated to the phylum Firmicutes dominated the bacterial communities in saliva samples, even though, similarly to our previous results [19], we did not observe any significant difference between CRC vs AP patients.
Moreover, the microbiota composition in neoplastic tissue significantly diverges from the intestinal lumen (stool), in agreement with Flemer et al. [32]. However, the microbiota composition of fecal and tissue samples appeared more similar to each other than the bacterial composition of saliva samples, for both CRC and AP conditions. Furthermore, we showed significant alterations in the tissue and fecal microbiota of CRC and AP patients.
Indeed, regarding the tissue-associated microbiota, in accordance with our previous results [9], [19], the relative abundance of the most represented microbial phyla revealed that sequences collected were classified into Firmicutes, Bacteroidota, Fusobacteriota and Proteobacteria (Figure 2B). In specific, we found a significant abundance of Proteobacteria in CRC, according to previous studies where an imbalanced microbiota is often associated with a sustained increase in Proteobacteria phylum members [33]. Moreover, we observed significant differences of microbial clades from intestinal tissue when comparing CRC vs AP patients: at order level, we detected a significant increase of Peptostreptoccales, Tissierellales, at family level, a significant increase of Campylobacteraceae, Carnobacteriacerae, Gemellaceae, Leptotrichiaceae, Streptococcaceae, while a decrease of Pseudomonadaceae, Yersiniaceae were observed in CRC. Finally, at genera level, we observed a significant rise of Fusobacterium and Gemella, as well as a significant reduction of Pseudomonas and Serratia in CRC patients compared to that in AP patients (Figure 2 Supplementary material).
In particular, we also confirm our recent data [19], demonstrating that Fusobacteria and Fusobacterium are associated with CRC and are amplified during carcinogenesis. In detail, the Fusobacterium, a gram-negative and strictly anaerobic genus, is a common bacterium present in healthy microbiota, but its increased amount has been detected in various inflammatory disorders [34], [35], [36] and cancers [37], including CRC [38]. Furthermore, the high level of Fusobacterium in CRC usually predicts poor prognosis [39], [40]. Additionally, the Gemella morbillorum, a member of this genus, has been proposed as potential non-invasive biomarkers for CRC in a recent study, establishing a predictive model with good sensitivity and specificity [41].
Regarding the stool samples, we observed a characteristic distribution of Bacteroidetes and Firmicutes, (Figure 2 C). Moreover, a number of significant differences at genus level was detected when comparing CRC vs AP samples (Figure 5). In particular, a significant increase of Akkermansia, Anaerostipes, Bifidobacterium, Catenibacterium, Dialister, Eubacterium_coprostanoligenes, Family_XIII_AD3011, Muribaculaceae, Parvimonas, Roseburia, Ruminococcus_torques, Senegalimassilia and Slakia was observed in CRC samples, suggesting their potential role in CRC progression. Interestingly, Akkermansia, a mucin-degrading bacterium in the phylum of Verrucomicrobia, has been reported to correlate with CRC in human and mouse models [42], [43]. Concerning the anaerobic bacteria Parvimonas, our data is in line with the study of Clos-Garcia et al. [27]. Moreover, we observed for the first time a significant correlation with Fusobacterium in the intestinal tissue. Curiously, a member of Parvimonas genus has been proposed as a putative faecal biomarker for CRC [44]. Additionally, different studies have simultaneously evaluated Parvimonas and Fusobacterium genera respectively as members of a bacterial biomarkers’ CRC panel [45] and investigated their association in tumour colonisation and immune events in an immunologically well-characterised cohort of CRC patients [46]. However, for the first time, we suggest an interdependence that needs to be further explored by additional in vitro/in vivo models. As previously mentioned, paramount evidence indicates these two genera as biomarkers of CRC progression. In other words, the detection of Parvimonas in fecal samples may mirror Fusobacterium abundance in intestinal tissue, leading the way to new non-invasive test for monitoring CRC progression. Moreover, Bifidobacterium was augmented in CRC samples, however many Bifidobacterium species demonstrated anticancer action on CRC cells by decreasing and boosting anti-apoptotic and pro-apoptotic genes [47]. Some Bifidobacterium species generate bile acid hydrolase [48], which participate in bile acid metabolism, thereby specifically affecting the development of CRC [49].
Intriguingly, in accordance with the increase of Bifidobacteria in our fecal CRC samples, we found that the predicted metagenomic pathway analysis was positively associated with the superpathway “Bifidobacterium shunt”. This is a unique fermentation pathway for hexose catabolism, which produces SCFA, primarily acetate and lactate [50]. So, to corroborate these predictive results, we deeply investigated the effective interplay of the fecal microbiome and perturbed metabolites in CRC vs. AP through a metabolomics approach. We observed a significant increase of leucine, alanine, valine, lactate and tyrosine in the fecal samples of CRC compared to AP patients (Figure 7). In general, the amino acid metabolism has been documented as significantly altered in cancer pathogenesis [51]. The leucine rise is likely a result of malabsorption due to large epithelial inflammation and damage associated with CRC [52]. In agreement with our results, five previous studies reported the metabolic alteration of leucine in CRC, in detail leucine upregulation was observed in fecal [53], urine [54], plasma [55], and tissue samples [56], while leucine downregulation was detected in serum samples [57]. However, in disagreement with our data, multiple studies showed the most significant decrease of alanine and tyrosine, known as proteinogenic amino acids, among metabolites identified in CRC patients [58]. On the other hand, consistently with our results, valine, were found upregulated in CRC stool but downregulated in most blood-related specimens [58].
Moreover, the increase in lactate in CRC samples is consistent with two previous studies, in urine [59] and [53] in stool, respectively. Interestingly, lactate, one of the key factors involved in glycolysis, can play an immunosuppressive role in the TME and promote the tumor development by recruiting and inducing the molecular activity of immunosuppressive cells. Finally, high lactate concentrations are important for tumor cell metastasis, angiogenesis, and treatment resistance. Of note, the lactate score has been recently shown as an independent prognostic factor for cancer that can be used as a clinical guide for predicting CRC progression and as an evaluation factor for immunotherapy effectiveness [60]. Interestingly, we also observed a positive correlation of Bifidobacterium with lactate, which further corroborates the predicted metagenomic pathway analysis. In addition, lactate was also positively correlated to Anaerostipes (previously found involved in lactate metabolism [61]). Family_XIII_AD3011_group and Slakia. However, we are aware that these results may be affected by several biases, due to other non-microbial factors that can influence the metabolism, like drug interactions and diet alterations. So, the investigation of microbe-metabolite relationships in the gut is fundamental to understanding and potentially reducing CRC risk.
Furthermore, we also detected differences in CRC saliva, tissue and stool specimens at different stage, taking into account only the T parameter. Intriguingly, we observed differences in the Group II (T2), with a rise of Spirochaetota phylum in intestinal tissue samples, in agreement with a previous study [62], and a slight rise of Alphaproteobacteria class in fecal samples. Interestingly, Alphaproteobacteria is a class of the phylum Proteobacteria, a conserved ecological pattern associated with the increase of inflammation [63]. The modifications of microbiota composition at tissue and stool levels during TNM staging may be related to TME alterations, able to change according to malignancy type. Notably, in T2 stage, the tumor invades the muscularis propria by growing into the muscle layer of the bowel wall. We recently demonstrated a different grading of molecular mediators of inflammation across the diverse gut layers (mucosa, submucosa, muscularis/serosa) in Crohn's disease patients, in relation to mucosal microbial composition [64]. Indeed, a recent theory by Yamauchi et al. proposes that CRCs differ molecularly in various parts of the large intestine and that gut microorganisms, along with factors like biochemical substances, epithelial cells, and innate immunity, could have a direct or indirect effect on tumour development [65]. Probably, the invasion of muscularis propria might be a crucial step in the tumor evolution, leading to relevant changes in the inflammatory response and thus in bacterial TME component. In addition to the intestinal tissue, these microbial alterations are observed also in stool, which may act as a gut proxy. Given these results, a better understanding of these spatial microbiotas’ networks might help to identify important predictive and prognostic tools as well as new targets for therapy.
However, this study shows some limits: the restricted number of patients, gender imbalance among groups to be compared (which doesn't look like affecting microbiome composition based on our preliminary analysis–data not shown), the lack of proper validation of all the results in this phase of the study; hence, considering the mentioned limits, these results must be cautiously taken at the moment.
Additionally, it is well known that GM composition changes with dietary patterns and lifestyle, which could be country-based [66]. More similar sample size or larger multi centre studies from different geographical locations, are needed to derive robust and generalizable patterns.
Conclusion
The overall taxonomic and metabolomics findings of our study suggest the importance of a simultaneous evaluation of microbiota structure and function in the CRC progression and staging. Our study, rather than providing final answers, offers a new integrated vision to explore new microbial and metabolic CRC biomarkers along the oral-gut microbiome axis. In detail, the shift in microbial taxa at different taxonomic levels in fecal samples suggests that faecal microbiome-based strategies may be useful for early diagnosis, staging and treatment of CRC and needs further extensive validation. Moreover, a comprehensive understanding of the efficacy of using fecal samples as a proxy to study the intestinal microbiota would help improve the application of stool analysis in the CRC field.
Additionally, fecal metabolic products may be used as potential biomarkers for the increased risk of CRC development and progression from AP to CRC. Unravelling the role specific tumour-subtype microbial communities could lead to tailored strategies of gut microbiome management through lifestyle and diet recommendations including probiotic and antimicrobial interventions for CRC patients.
Funding
The research was founded with a grant from the regional contribution of “The Programma Attuativo Regionale (Toscana)” funded by FAS (now FSC), the Italian Ministry of University and Research (MIUR), and the Foundation “Ente Cassa di Risparmio di Firenze.” In addition, this work was partially supported by MICAfrica (European Union's Horizon 2020 - grant No 952583). This project received funding from MUR under the umbrella of the European Joint Program Initiative “A Healthy Diet for a Healthy Life” (JPI-HDHL) and of the ERA-NET Cofound ERA-HDHL, ID: 1523 (GA N 696295 of the EU HORIZON 2020 Research and Innovation Programme).
Ethics approval
The study has been approved by the Ethical Committee of Careggi University Hospital.
Informed consent
All the subjects enrolled in this study signed the informed consent.
Author contributions
ER, AA, designed the research; EN, SB, GN, RB, AT, MNR collected the samples; GM, GM, data acquisition and generation of NMR; LDG, MR analyzed the data; ER wrote the paper; ER, AA, supervised all the experimental work. ER, AA, LT, GM, RF revised the manuscript; all authors critically read and approved the manuscript.
Declaration of Competing Interest
No conflict of interest.
Acknowledgments
The authors would like to acknowledge the support of all the patients that participated to this study. The authors thanks Dr. Ingrid Lamminpaa and Dr Mascha Stroobant for the English revision.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2023.100901.
Contributor Information
Matteo Ramazzotti, Email: matteo.ramazzotti@unifi.it.
Amedeo Amedei, Email: amedeo.amedei@unifi.it.
Appendix. Supplementary materials
References
- 1.Xi Y., Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14(10) doi: 10.1016/j.tranon.2021.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russo E., et al. The interplay between the microbiome and the adaptive immune response in cancer development. Therap Adv Gastroenterol. 2016;9(4):594–605. doi: 10.1177/1756283X16635082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russo Edda, A.A., The Role of the Microbiota in the Genesis of Gastrointestinal Cancers, in Frontiers in Anti-Infective Drug Discovery. Pp. 1-44 (44).
- 4.Wong S.H., Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 5.Mizutani S., Yamada T., Yachida S. Significance of the gut microbiome in multistep colorectal carcinogenesis. Cancer Sci. 2020;111(3):766–773. doi: 10.1111/cas.14298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McQuade J.L., et al. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019;20(2):e77–e91. doi: 10.1016/S1470-2045(18)30952-5. [DOI] [PubMed] [Google Scholar]
- 7.Scott A.J., et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut. 2019;68(9):1624–1632. doi: 10.1136/gutjnl-2019-318556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo E., Boem F., Curini L., Amedei A. Interdisciplinary Cancer Research. Springer; Cham: 2022. Gastrointestinal Cancers: What Is the Real Board of Microenvironment and the Role of Microbiota–Immunity Axis? [Google Scholar]
- 9.Niccolai E., et al. Significant and Conflicting Correlation of IL-9 With Prevotella and Bacteroides in Human Colorectal Cancer. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.573158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siskova A., et al. Colorectal Adenomas-Genetics and Searching for New Molecular Screening Biomarkers. Int J Mol Sci. 2020;21(9) doi: 10.3390/ijms21093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manne U., et al. Development and progression of colorectal neoplasia. Cancer Biomark. 2010;9(1-6):235–265. doi: 10.3233/CBM-2011-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valciukiene J., Strupas K., Poskus T. Tissue vs. Fecal-Derived Bacterial Dysbiosis in Precancerous Colorectal Lesions: A Systematic Review. Cancers (Basel) 2023;15(5) doi: 10.3390/cancers15051602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaupel P., Harrison L. Tumor hypoxia: causative factors, compensatory mechanisms, and cellular response. Oncologist. 2004;9(Suppl 5):4–9. doi: 10.1634/theoncologist.9-90005-4. [DOI] [PubMed] [Google Scholar]
- 14.Montalban-Arques A., Scharl M. Intestinal microbiota and colorectal carcinoma: Implications for pathogenesis, diagnosis, and therapy. EBioMedicine. 2019;48:648–655. doi: 10.1016/j.ebiom.2019.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yachida S., et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 16.Nannini G., et al. Fecal metabolomic profiles: A comparative study of patients with colorectal cancer vs adenomatous polyps. World J Gastroenterol. 2021;27(38):6430–6441. doi: 10.3748/wjg.v27.i38.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 18.Williams S.T., Beart R.W., Jr Staging of colorectal cancer. Semin Surg Oncol. 1992;8(2):89–93. doi: 10.1002/ssu.2980080208. [DOI] [PubMed] [Google Scholar]
- 19.Russo E., et al. Preliminary Comparison of Oral and Intestinal Human Microbiota in Patients with Colorectal Cancer: A Pilot Study. Front Microbiol. 2017;8:2699. doi: 10.3389/fmicb.2017.02699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker S.P., et al. Non-specific amplification of human DNA is a major challenge for 16S rRNA gene sequence analysis. Sci Rep. 2020;10(1):16356. doi: 10.1038/s41598-020-73403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamichhane S., et al. Strategy for Nuclear-Magnetic-Resonance-Based Metabolomics of Human Feces. Anal Chem. 2015;87(12):5930–5937. doi: 10.1021/acs.analchem.5b00977. [DOI] [PubMed] [Google Scholar]
- 22.Dieterle F., et al. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78(13):4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 23.Ihaka R G.R. R: A Language for Data Analysis and Graphics. J Comput Graph Stat. 1996;5:299. [Google Scholar]
- 24.Chong J., Yamamoto M., Xia J. MetaboAnalystR 2.0: From Raw Spectra to Biological Insights. Metabolites. 2019;9(3) doi: 10.3390/metabo9030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onitilo A.A., Kio E., Doi S.A. Tumor-related hyponatremia. Clin Med Res. 2007;5(4):228–237. doi: 10.3121/cmr.2007.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen F., et al. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut. 2022;71(7):1315–1325. doi: 10.1136/gutjnl-2020-323476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clos-Garcia M., et al. Integrative Analysis of Fecal Metagenomics and Metabolomics in Colorectal Cancer. Cancers (Basel) 2020;12(5) doi: 10.3390/cancers12051142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y., et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. 2019;9(14):4101–4114. doi: 10.7150/thno.35186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas C., et al. Oral Microbiota: A Major Player in the Diagnosis of Systemic Diseases. Diagnostics (Basel) 2021;11(8) doi: 10.3390/diagnostics11081376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo S., et al. Oral-Intestinal Microbiota in Colorectal Cancer: Inflammation and Immunosuppression. J Inflamm Res. 2022;15:747–759. doi: 10.2147/JIR.S344321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruan X., et al. The salivary microbiome shows a high prevalence of core bacterial members yet variability across human populations. NPJ Biofilms Microbiomes. 2022;8(1):85. doi: 10.1038/s41522-022-00343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flemer B., et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66(4):633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori G., et al. Shifts of Faecal Microbiota During Sporadic Colorectal Carcinogenesis. Sci Rep. 2018;8(1):10329. doi: 10.1038/s41598-018-28671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekizuka T., et al. Characterization of Fusobacterium varium Fv113-g1 isolated from a patient with ulcerative colitis based on complete genome sequence and transcriptome analysis. PLoS One. 2017;12(12) doi: 10.1371/journal.pone.0189319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohkusa T., et al. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol. 2002;17(8):849–853. doi: 10.1046/j.1440-1746.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- 36.Ohkusa T., et al. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52(1):79–83. doi: 10.1136/gut.52.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tahara T., et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74(5):1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kostic A.D., et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray K. Colorectal cancer: Fusobacterium nucleatum found in colon cancer tissue–could an infection cause colorectal cancer? Nat Rev Gastroenterol Hepatol. 2011;8(12):662. doi: 10.1038/nrgastro.2011.208. [DOI] [PubMed] [Google Scholar]
- 40.Hussan H., et al. Fusobacterium's link to colorectal neoplasia sequenced: A systematic review and future insights. World J Gastroenterol. 2017;23(48):8626–8650. doi: 10.3748/wjg.v23.i48.8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao Y., et al. A New Biomarker of Fecal Bacteria for Non-Invasive Diagnosis of Colorectal Cancer. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.744049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baxter N.T., et al. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome. 2014;2:20. doi: 10.1186/2049-2618-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weir T.L., et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8(8):e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowenmark T., et al. Parvimonas micra as a putative non-invasive faecal biomarker for colorectal cancer. Sci Rep. 2020;10(1):15250. doi: 10.1038/s41598-020-72132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osman M.A., et al. Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer. Sci Rep. 2021;11(1):2925. doi: 10.1038/s41598-021-82465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowenmark T., et al. Parvimonas micra is associated with tumour immune profiles in molecular subtypes of colorectal cancer. Cancer Immunol Immunother. 2022;71(10):2565–2575. doi: 10.1007/s00262-022-03179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faghfoori Z., et al. Anticancer effects of bifidobacteria on colon cancer cell lines. Cancer Cell Int. 2021;21(1):258. doi: 10.1186/s12935-021-01971-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winston J.A., Theriot C.M. Diversification of host bile acids by members of the gut microbiota. Gut Microbes. 2020;11(2):158–171. doi: 10.1080/19490976.2019.1674124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia B., et al. Metagenomic analysis of the human microbiome reveals the association between the abundance of gut bile salt hydrolases and host health. Gut Microbes. 2020;11(5):1300–1313. doi: 10.1080/19490976.2020.1748261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolin M.J., et al. NMR detection of 13CH313COOH from 3-13C-glucose: a signature for Bifidobacterium fermentation in the intestinal tract. J Nutr. 1998;128(1):91–96. doi: 10.1093/jn/128.1.91. [DOI] [PubMed] [Google Scholar]
- 51.Wei Z., et al. Metabolism of Amino Acids in Cancer. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.603837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu Rev Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- 53.Sinha R., et al. Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0152126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ning W., et al. Metabolic profiling analysis for clinical urine of colorectal cancer. Asia Pac J Clin Oncol. 2021;17(4):403–413. doi: 10.1111/ajco.13591. [DOI] [PubMed] [Google Scholar]
- 55.Geijsen A., et al. Plasma metabolites associated with colorectal cancer stage: Findings from an international consortium. Int J Cancer. 2020;146(12):3256–3266. doi: 10.1002/ijc.32666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cottet V., et al. Fatty acid composition of adipose tissue and colorectal cancer: a case-control study. Am J Clin Nutr. 2015;101(1):192–201. doi: 10.3945/ajcn.114.088948. [DOI] [PubMed] [Google Scholar]
- 57.Martin-Blazquez A., et al. Untargeted LC-HRMS-based metabolomics to identify novel biomarkers of metastatic colorectal cancer. Sci Rep. 2019;9(1):20198. doi: 10.1038/s41598-019-55952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gold A., et al. The Application of Metabolomics in Recent Colorectal Cancer Studies: A State-of-the-Art Review. Cancers (Basel) 2022;14(3) doi: 10.3390/cancers14030725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Udo R., et al. Urinary charged metabolite profiling of colorectal cancer using capillary electrophoresis-mass spectrometry. Sci Rep. 2020;10(1):21057. doi: 10.1038/s41598-020-78038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu D., et al. Lactate: A regulator of immune microenvironment and a clinical prognosis indicator in colorectal cancer. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.876195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shetty S.A., et al. Unravelling lactate-acetate and sugar conversion into butyrate by intestinal Anaerobutyricum and Anaerostipes species by comparative proteogenomics. Environ Microbiol. 2020;22(11):4863–4875. doi: 10.1111/1462-2920.15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi S., et al. Analysis of changes in microbiome compositions related to the prognosis of colorectal cancer patients based on tissue-derived 16S rRNA sequences. J Transl Med. 2021;19(1):485. doi: 10.1186/s12967-021-03154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winter S.E., Baumler A.J. Why related bacterial species bloom simultaneously in the gut: principles underlying the 'Like will to like' concept. Cell Microbiol. 2014;16(2):179–184. doi: 10.1111/cmi.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russo E., et al. Diving into Inflammation: A Pilot Study Exploring the Dynamics of the Immune-Microbiota Axis in Ileal Tissue Layers of Patients with Crohn's Disease. J Crohns Colitis. 2021;15(9):1500–1516. doi: 10.1093/ecco-jcc/jjab034. [DOI] [PubMed] [Google Scholar]
- 65.Yamauchi M., et al. Colorectal cancer: a tale of two sides or a continuum? Gut. 2012;61(6):794–797. doi: 10.1136/gutjnl-2012-302014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu A.H., et al. Gut microbiome associations with breast cancer risk factors and tumor characteristics: a pilot study. Breast Cancer Res Treat. 2020;182(2):451–463. doi: 10.1007/s10549-020-05702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microbial-related data (raw reads, ASV tables and taxonomic assignments) is freely available. To review GEO accession GSE217490: Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE217490.
The processing and analysis scripts are available at https://github.com/LeandroD94.